Abstract
Increased activity of amino acid transporters has been observed in a wide variety of cancers. However, whether amino acid metabolism is related to estrogen receptor-positive (ER+) breast cancer has been less well studied. We identified the rate-limiting enzyme involved in amino acid metabolism associated with ER+ breast cancer by integrating numerous bioinformatics tools and laboratory studies. The bioinformatics analysis revealed that highly expressed genes in ER+ breast cancer patients were correlated with breast cancer-related pathways, including ESR1 and PI3K signaling. The metabolic signaling and the amino acid metabolism were significantly regulated in breast neoplasms. We used the ER+ breast cancer cell line MCF-7 and breast cancer tissue from National Cheng Kung University Hospital to validate our findings in bioinformatics. In estradiol-treated MCF-7 cells, genes associated with anabolic metabolism of serine and methionine and genes associated with catabolic metabolism of tyrosine, phenylalanine and arginine were upregulated. Furthermore, the expression levels of ARG2, PSAT1, PSPH, TH, PAH, and MAT1A mRNA were increased in breast cancer patients relative to controls. The aforementioned genes were also found to be highly correlated with distant metastasis-free survival in breast cancer patients. High expression levels of ARG2, CBS, PHGDH, AHCY, HAL, TDO2, SHMT2, MAT1A, MAT2A, GLDC, GLS2, BCAT2, GLUD1, PAH and MTR contributed to poor prognoses, whereas high mRNA expression levels of HECA, CTH, PRODH, TAT, and MAT2B were correlated with good prognoses. FDA-approved drugs, including piperlongumine, ellipticine, etidronic acid, harmine, and meclozine, may have novel therapeutic effects in ER+ patients based on connectivity map (CMap) analyses. Collectively, our present study demonstrated that amino acid metabolism genes play crucial roles in tumor development and may serve as prospective drug targets or biomarkers for ER+ breast cancer.
Keywords: Breast cancer, estrogen receptor, amino acid metabolism
Introduction
Cancer cells have an unlimited potential for division and sustained growth. This process depends on the acquisition of essential nutrients from the impoverished and hypoxic microenvironment [1-3]. The metabolic flexibility of cancer cells is determined by their ability to reprogram anabolic and catabolic pathways by intracellular alteration of gene expression and intercellular interactions within the tumor microenvironment. The process of oncogenesis depends on amino acids as sources for protein synthesis, energy and metabolites [4-6]. The overexpression of amino acid-degrading enzymes has been detected in many cancers. The degrading amino acids provide metabolites for cellular energy and anabolic processes, also serve as modulators of immune evasion in cancer cells. The overexpression of indoleamine-2,3-dioxygenase and arginase depletes tryptophan and arginine respectively within the tumor microenvironment. Decreased level of tryptophan and arginine suppress proliferation of cytotoxic T-cells in tumor [7-10]. Breast cancer cells promote their ability to survive by using amino acid-degrading enzymes as immunosuppressive factors [11-13]. Several of these amino acid-related genes encode rate-limiting en-zymes that are associated with metabolic pathways.
According to cancer statistics reported in 2019, about 30% of new cancer cases in American women are diagnosed as breast cancer [14]. Immunohistochemical markers for subtyping of breast cancer are the estrogen receptor (ER), the progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER-2). Up to 80% of breast cancer is classified as ER-positive, which implies a crucial role for estrogen in the development of breast cancer [15,16]. Consequently, the ER signaling pathway serves as a potential target to develop new therapies for the majority of breast cancer patients. Estradiol is the predominant form of estrogen, which can be converted from testosterone by the enzyme aromatase [17,18]. Regulation of metabolism is one of the essential routes to tumorigenesis and progression of cancer [19-24]. The expression of lipid metabolism genes is different between ER+ and ER- breast cancer [25]. However, the functions of estradiol and amino acid metabolism in ER+ breast cancer patients remain largely unknown. Therefore, it is important to study the metabolism of estrogen and amino acids during breast cancer development.
Material and methods
Bioinformatics and functional enrichment analysis
Genomic data from ER+ breast cancer patient were collected from The Cancer Genome Atlas (TCGA; n = 1,105) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC; n = 2,509). Clinically, ER signaling was activated in ER+ patients. The MCF-7 cell line expresses estrogen receptor and ER signaling was stimulated after treatment with estradiol. We compared ER+ patients from TCGA and METABRIC and the estradiol-treated MCF-7 cell line from the GSE11352 dataset to identify novel ER-related genes. A list of genes with statistically significant differences (P < 0.05) was obtained and functional enrichment analyses were performed. Molecular functions and disease pathway of Gene Ontology (GO) term in MetaCore (GeneGo, Inc., St. Joseph, MI, USA) was used to screen and analyze the signaling networks upregulated in ER+ patients. The networks were built from the input gene list. Comparison with the ER- tissue, the genes overexpressed in ER+ patients were selected and the top 10% genes were uploaded to the MetaCore database. The REVIGO web-based tool was used to summarize and remove redundant GO terms [26]. In addition, the METABRIC database was applied to explore the pathway maps for each breast cancer subtype by computing the fold changes in gene expression. The p-value was set at 0.05 to represent statistical significance.
Patients
A total of 16 unlinked fresh specimens from ER+ breast cancer patients was collected from the Human Biobank within the Research Center of Clinical Medicine in National Cheng Kung University Hospital (NCKUH). The collection period was from August 2005 to August 2010, with random patient selection. The pathological stage was complied with the guidelines defined by the American Joint Committee on Cancer (AJCC) Cancer Staging Manual, 7th edition. The study was followed the formal guidelines and approved by Institutional Review Board of NCKUH with the IRB number ER-97-175.
Cell culture
Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) was used for cell growth. For estradiol (Sigma, St Louis, MO, USA) treatments, MCF-7 cells were washed twice with phosphate-buffered saline (PBS) and then grown in phenol red-free medium containing charcoal-stripped FBS (10%) overnight. Cells were washed and then treated with 10 nM estradiol for 0, 16 and 24 hours.
Determination of mRNA expression using RT-qPCR
Total RNA was extracted from fresh specimens of ER+ breast cancer or MCF-7 cells after estradiol treatment by TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. The integrity of the 28S and 18S rRNA was examined to determine total RNA quality. Primers and fluorescent probes used for RT-qPCR reactions were synthesized and designed as previous described [27,28], and RT-qPCR experiments were performed using an ABI-prism 7700 sequence detector (PE Applied Biosystems, Foster City, CA, USA). Samples were incubated at 50°C for 2 min and 95°C for 10 min, with an amplification step comprising 40 cycles of 95°C for 15 s and 60°C for 1 min. Sequence Detector™ software (PE Applied Biosystems, Foster City, CA, USA) was used to detect the fluorescent signal according to threshold cycle number (CT). The delta CT (ΔCT) value was calculated as the difference between the CT value of the target mRNA (Target_CT) and that of GAPDH (GAPDH_CT). Then, the relative gene expression level of the target was presented as 2-ΔΔCT [29]. The results represent the averages of 3 independent experiments. Gene expression was calculated using the formula ΔCT = Target_CT - GAPDH_CT. The primers and TaqMan probes used in this study are listed below. #41 AHCY-F ggttcatttcctgcccaag, #41 AHCY-R tggtcaacttcacattcagctt; #87 ARG2-F ggcttgatgaaaaggctctc, #87 ARG2-R tggggactggagtaaaactca; #72 CBS-F acatgctggtggcttcagt, #72 CBS-R gggatccaccccaatgat; #34 CTH-F tttcttttgttgattgttccaaaa, #34 CTH-R ttctgggtggggtttgtg; #46 GLS2-F tgccatcggctattatctcaa, #46 GLS2-R tccacagaacacagctggaa; #31 GLUL-F ggcaaccctaacgatacgc, #31 GLUL-R agtgggaacttgctgaggtg; #31 GLUD1-F gggattctaactaccacttgctca, #31 GLUD1-R aactctgccgtgggtacaat; #66 HAL-F ctctgggttcatgatagctcact, #66 HAL-R atggcacagagccttgttct; #33 HDC-F tgcctgagagtgctcctga, #33 HDC-R ggctctgccaatgtaccac; #26 MAT1A-F ggacgaagacaccgtctacc, #26 MAT1A-R tcttacggccagtgacacc; #18 MAT2A-F ggcacattccttttcacctc, #18 MAT2A-R tcacaagctactttggcatca; #22 MAT2B-F gaaagagctctctatacactttgttcc, #22 MAT2B-R ccagtaaccagaaccctcctatt; #34 ODC1-F aaaacatgggcgcttacact, #34 ODC1-R tggaattgctgcatgagttg; #11 PAH-F gtggcttccatgaagataacatt, #11 PAH-R gcggaaaccagtgcaagt; #23 PHGDH-F agaagctctggggacactga, #23 PHGDH-R gcattcttcagggatgttcc; #62 PRODH-F agaccctgggagtgtctgg, #62 PRODH-R cttggacagcttggtcctg; #20 PYCR2-F acacatcgtggtctcctgtg, #20 PYCR2-R atgcagcgaatcactttgg; #31 SHMT1-F acgtccagccctactcagg, #31 SHMT1-R aaggtccaggcccatgat; #67 TDO2-F cgatgacagccttggacttc, #67 TDO2-R cggaattgcaaactctgga; #4 TH-F ctgacctggacttggaccac, #4 TH-R tgtactccacacggggaatc; #65 TPH2-F aaatttcaaaccactattgtgacg, #65 TPH2-R gggcacatcctctagctcttc; #37 TAT-F ccatgatttccctgtccatt, #37 TAT-R ggatggggcatagccattat; #66 ASL-F gtggcactgacccgagac, #66 ASL-R cagctctcggtccacacc; #37-MTR-F cagagtgcttaacggcacag, #37-MTR-R taacagtggccagcacgat; #46-DDC-F tcttagaagtcggtcctatctgc, #46-DDC-R ctgcgtaggctgcatcaa; #23-MTHFR-F aactcacagcccaacatcaa, #23-MTHFR-R cgcgggaagtgaaaaactc; #60-GAPDH-F agccacatcgctcagacac, #60-GAPDH-R gcccaatacgaccaaatcc; #89-PR-F ggcatggtccttggaggt, #89-PR-R cactggctgtgggagagc [26].
Connections between small molecules and genes via connectivity map
A signature of amino acid metabolism-related genes from the Connectivity Map (CMap) database was used to compare ER+ breast cancer and normal breast [30,31]. CMap applies a systematic approach to reveal interactions among drugs, compounds, and diseases based on alterations in the genetic backgrounds of ER+ breast cancer patients.
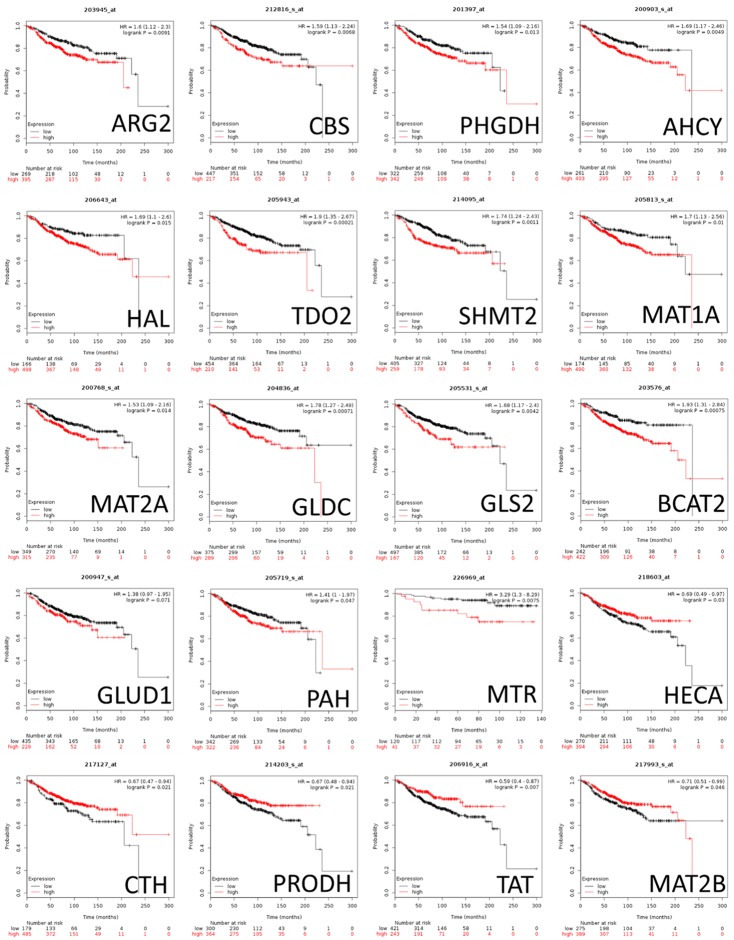
Kaplan-meier plot database analysis for survival probability
To correlate the mRNA expression levels of amino acid metabolism-related genes with the distant metastasis-free survival (DMFS) status of ER+ breast cancer patients, Kaplan-Meier plotter database (KM plot) was applied [32,33]. The KM plot analyzes the effects of 54,675 genes on survival of breast cancer patients by using the gene transcript data from 18,674 cancer samples in the NCBI GEO (Gene Expression Omnibus), EGA (European Genome-phenome Atlas), METABRIC, and TCGA datasets. To perform the analyses in KM plot, we selected “use multigene classifier” to upload the amino acid metabolism-related genes and explored their relationships with the DMFS in ER+ patients. In the present study, the following probe set IDs were evaluated: 203945_at (ARG2), 212816_s_at (CBS), 201397_at (PHGDH), 200903_s_at (AHCY), 206643_at (HAL), 205943_at (TDO2), 214095_at (SHMT2), 205813_s_at (MAT1A), 200768_s_at (MAT2A), 204836_at (GLDC), 205531_s_at (GLS2), 203576_at (BCAT2), 200947_s_at (GLUD1), 205719_s_at (PAH), 226969_at (MTR), 217127_at (CTH), 201397_at (PHGDH), 206916_x_at (TAT), 217993_s_at (MAT2B). Gene expression levels and survival statuses were used. Patients were divided into high and low gene expression groups based on the autoselected best cutoff values. The two patient cohorts were then compared using Kaplan-Meier survival plots, and the hazard ratios, 95% confidence intervals, and log-rank P values were calculated using the default algorithm.
Colony formation assay
MCF-7 cells were trypsinized and seeded in six-well plates at low density (500 cells/well) and grown in the presence of FDA-approved drugs that were predicted based on our bioinformatics analysis. After two weeks, the media was aspirated from the six-well plate, and the MCF-7 cells were fixed with methanol. Then, 0.1% crystal violet in distilled water was added to each well for ten minutes to stain colonies. After ten minutes, the crystal violet solution was removed, and the plates were then washed with distilled water five times and air dried. Colonies in each well were enumerated under low magnification light microscopy.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (version 4.00 for Windows; GraphPad Software, San Diego, CA, USA). Data are presented as the mean ± standard deviation (S.D). To identify statistically significant differences between two groups, the Student’s t-test was applied. One-way ANOVA adjusted by the Bonferroni test was used for multiple group comparisons. The p-value was set as 0.05.
Results
Breast neoplasm metabolism regulation was correlated with ER+ patients
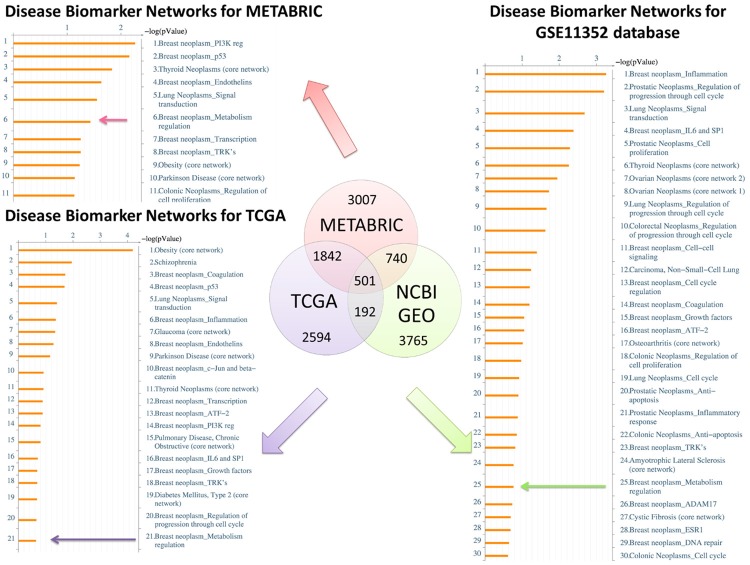
We used bioinformatics approach to identify the potential pathways associated with highly expressed genes in ER+ patients and estradiol-exposed MCF-7 cells. We calculated the fold changes of gene expression in ER+ patients comparing with ER- patients in the METABRIC (n = 2,509) and TCGA (n = 1,105). Highly expressed genes were also identified in estradiol-exposed MCF-7 database from GSE11352 dataset in NCBI GEO (National Center for Biotechnology Information Gene Expression Omnibus) database (MCF-7-estradiol vs. MCF-7-control). The upper quartile of overexpressed genes was selected for Venn analysis to reveal intersection and union among these three databases. The highly expressed ER+-related genes were analyzed for disease or biomarker in the MetaCore system (Figure 1). The highly expressed genes in ER+ breast cancer patients were correlated with breast-related diseases and pathways, such as ESR1, PI3K, TRK, IL-6, and SP1 signaling (Figure S1A and S1C). Of note, “Breast neoplasm_Metabolism regulation” was significantly enriched in the TCGA (the 6th space), METABRIC (the 21th space) ER+ patients, and in the MCF-7-estradiol group (the 25th space, Figures 1 and S1B).
Figure 1.
Venn diagram of overexpressed pathways and network in public databases. The red circle represents the upper quartile of genes that are overexpressed in ER+ patients compared with ER- patients according to the METABRIC database (n = 2,509), the blue circle represents the upper quartile of genes that are overexpressed in ER+ patients compared with ER- patients according to the TCGA database (n = 1,105), and the green circle represents the upper quartile of genes that are overexpressed in estradiol-treated groups compared with control groups according to the GSE11352 database (n = 18). For each database, the number of genes in each fraction is indicated. Arrows from each fraction point to the lists of disease and biomarker networks that were significantly enriched according to the MetaCore database. Downstream pathway analyses based on these top quartile genes revealed that “breast neoplasm metabolism regulation” was significantly correlated with ER+ patients and estradiol-treated groups.
Gene ontology enrichment analysis from ER+ breast cancer
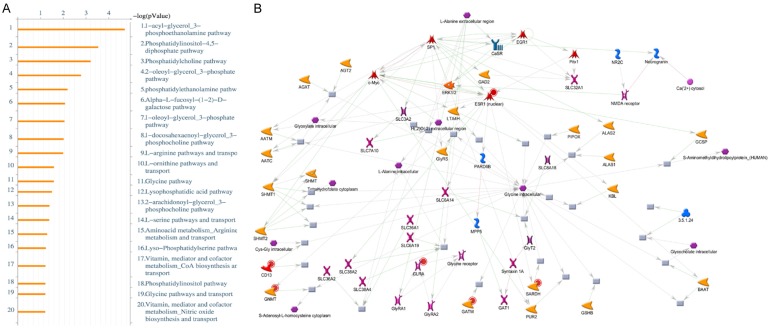
To investigate the function of ER in breast cancer, gene ontology (GO) analysis was performed in public database. The gene list in the first paragraph containing the top 501 highly-expressed ER-related genes with p-values < 0.001 was used for this functional analysis [34]. Several GO terms were identified, including amino acid transport, regulation of secretion, cell communication, lactose biosynthesis, and regulation of epithelial cell differentiation from bioinformatics analyzed. We calculated fold changes of these genes using the TCGA, METABRIC (ER+ patients vs. ER- patients), and GSE11352 (MCF-7-estradiol vs. MCF-7-control) databases. These upregulated genes were intersected with highly-expressed ER-related genes in Figure 1 and uploaded for the GO analysis. REVIGO was used for GO enrichment analysis. Amino acid metabolism and other related pathways were significantly upregulated in the ER+ patient group and the MCF-7-estradiol group (Figure 2A). MCF-7 cells treated with estradiol for different exposure time (12/24/48 hours, 12E/24E/48E) in GSE11352 database were compared with MCF-7-control groups. The amino acid transmembrane transporter activity-related pathway was present in the estradiol treatment group (p-value = 0.005) (Figure 2B). We identified networks of disease biomarker in the MCF-7-estradiol group via the MetaCore database (Figure S2A). The “breast neoplasm ESR1” and other related pathways were significantly expressed in the MCF-7-estradiol group compared with the MCF-7-control (Figure S2B).
Figure 2.
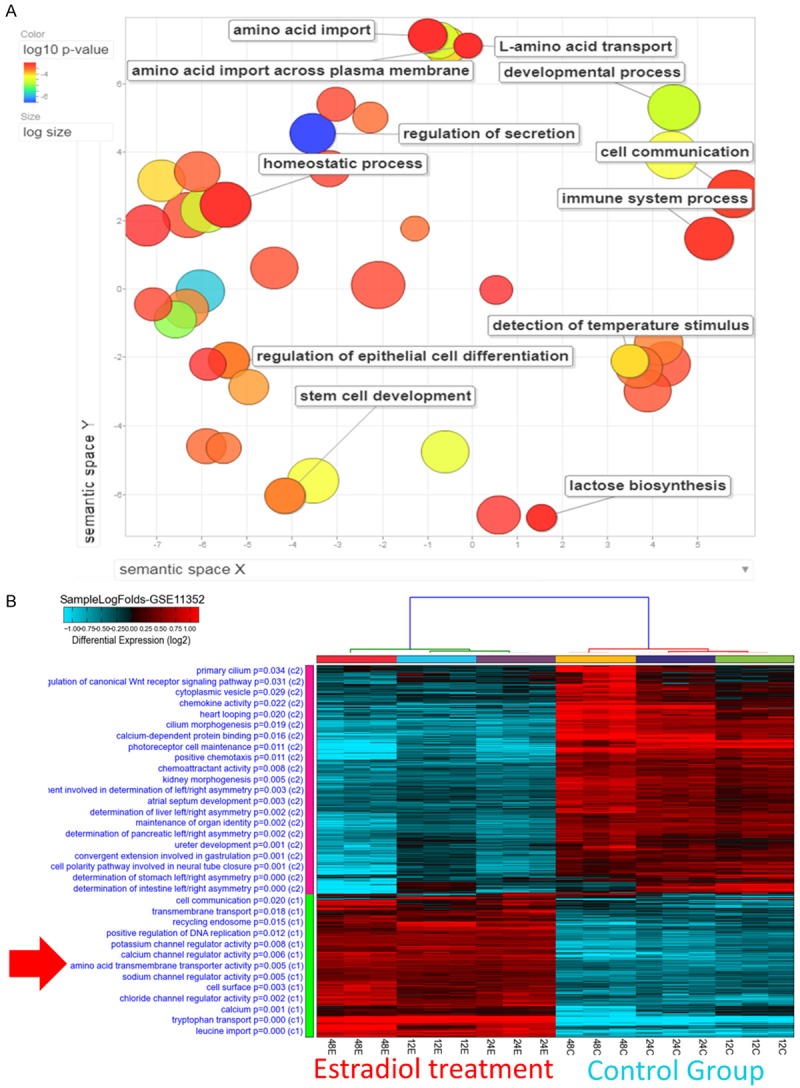
Gene ontology enrichment analysis using REVIGO and heatmap visualization of GSE11352. A. Significantly enriched GO terms related to biological processes in ER+ vs. ER- patients and in estradiol-treated MCF-7 cells. The scatterplot represents functional clusters, the bubble color indicates the p-values of the GO analysis, and the bubble size indicates the frequency of the GO term in the underlying gene ontology database. B. Analysis of the GSE11352 database revealed that the amino acid transmembrane transporter activity-related pathway appeared significantly in estradiol-treated 12 hours (12E), 24 hours (24E), 48 hours (48E) groups compared with control groups.
Amino acid metabolism-related pathways were associated with ER+ cancer
The “breast neoplasm metabolism regulation” under estradiol treatment conditions were also identified (Figure S2C). The integration of upregulated genes from ER+ patients in the TCGA and METABRIC, and from MCF-7-estradiol group in the GSE11352 database was uploaded to the MetaCore database for analysis of endogenous metabolic networks (Figure 3A). The “L-arginine pathways and transport” was recognized in both the ER+ patient group and the MCF-7-estradiol groups (Figure 3B).
Figure 3.
MetaCore pathway analysis of the upregulated genes in ER+ patients from different public databases. The 501 significantly expressed genes in ER+ patients and in the MCF-7-estradiol group that were identified by the Venn diagram analysis were exported to the MetaCore pathway analysis tool to explore potential gene networks and signaling pathways impacted by the selected genes. (A) MetaCore pathway analysis indicated that the amino acid metabolism-related pathway and (B) the L-arginine pathway were significantly associated with ER+ patients and the MCF-7-estradiol group.
Amino acid metabolism genes profile in ER+ breast cancer model and clinical tissue
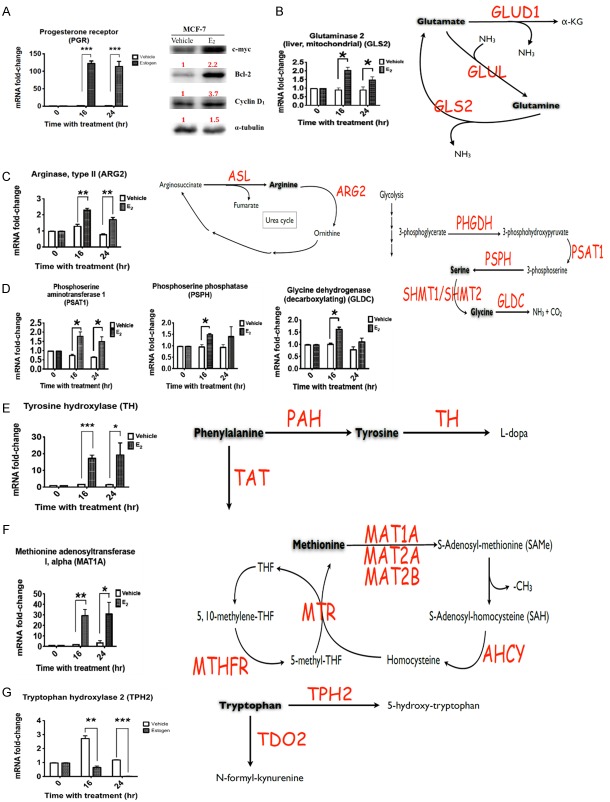
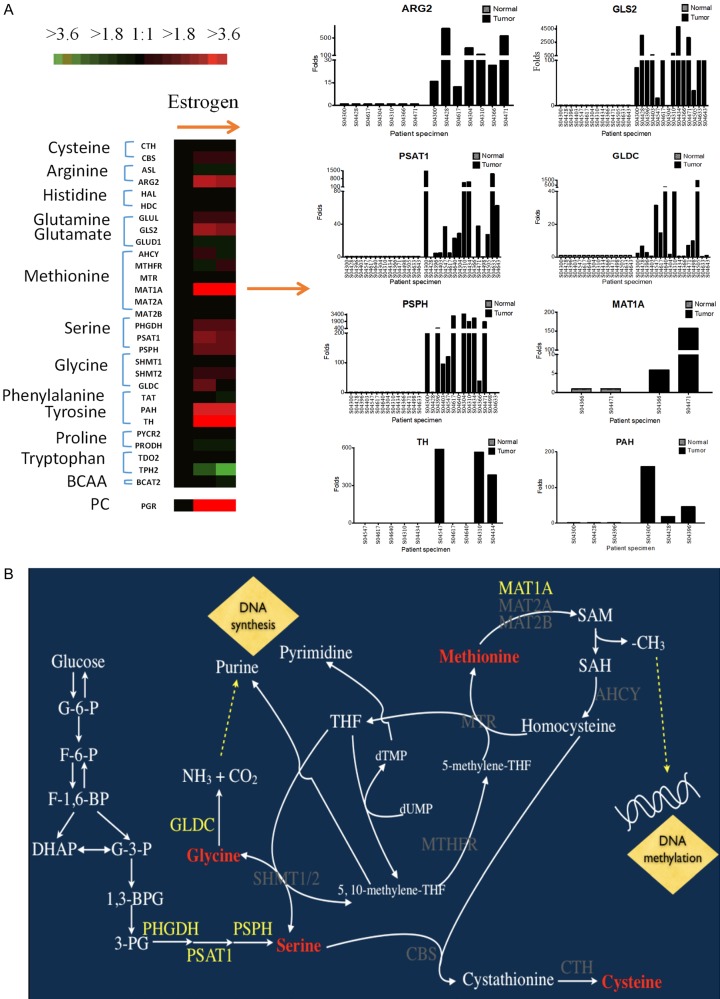
To evaluate the effects of estradiol on breast cancer cells in vitro, MCF-7 cells were cultured in six-well dishes and treated with 10 nM of estradiol for 16-18 hours. Total RNA was collected and analyzed by RT-qPCR (reverse transcription-quantitative polymerase chain reaction). The progesterone receptor (PR) and other oncogenic molecules, such as c-myc, Bcl-2 (B-cell lymphoma 2), and Cyclin D1, were used as positive controls during the evaluation of the effects of estradiol (Figure 4A). An increase in gene expression was observed for glutamine decomposing enzyme (GLS2), arginine catabolic enzyme (ARG2), glycine-degrading enzyme (GLDC) (Figure 4B and 4C). The synthetic enzymes for phosphoserine and serine, PSAT1 (phosphoserine aminotransferase 1) and PSPH (phosphoserine phosphatase), was increased after 16- and 24-hour treatment with estradiol (Figure 4D and 4E). We further analyzed six enzymes involving in the one-carbon cycle of cancer cells, including MTR (5-methyltetrahydrofolate-homocysteine methyltransferase) and MTHFR (methylenetetrahydrofolate reductase). The methionine decomposing enzyme has three isoforms: MAT1A (methionine adenosyltransferase 1A), MAT2A (methionine adenosyltransferase 2A), and MAT2B (methionine adenosyltransferase 2B). After estradiol treatment, only MAT1A increased by approximately 30-fold while comparing with the control group (Figure 4F). However, the tryptophan decomposing enzyme TPH2 (tryptophan hydroxylase 2) was significantly reduced after estradiol treatment for 16 and 24 hours (Figure 4G). TPH2 can metabolize tryptophan to 5-hydroxytryptophan. The expression levels of other enzymes were slightly altered, including the CBS (cystathionine-beta-synthase), CTH (cystathionine gamma-lyase), PYCR2 (pyrroline-5-carboxylate reductase family member 2), PRODH (proline dehydrogenase 1), and BCAA (branched-chain amino acid) decomposing enzymes (Figure S3). Meanwhile, we further analyzed the expression levels of amino acid metabolism genes in breast cancer and corresponding normal breast from 16 patients. The expression levels of PSAT1, PSPH, GLDC, and GLS2, were increased in cancer than normal breast (Figure 5). However, we were unable to detect ARG2, TH (tyrosine hydroxylase), and MAT1A expression due to the low expression levels of these genes.
Figure 4.
The mRNA expression levels of amino acid metabolism genes in the MCF-7 cell line. MCF-7 cells were seeded and treated with 10 nM estradiol for 16-18 hours. Total RNA was extracted and the expression levels of amino acid metabolism genes were analyzed by RT-qPCR, using the mRNA levels of PR, which is activated by the estrogen receptor and served as a positive control. A. Progesterone receptor/PR. B. Glutaminase 2/GLS2. C. Arginase type II/ARG2. D. Phosphoserine amino transferase I/PSAT1, phosphoserine phosphatase/PSPH, and glycine dehydrogenase (decarboxylating)/GLDC. E. Tyrosine hydroxylase/TH. F. Methionine adenosyltransferase 1-MAT1A. G. Tryptophan hydroxylase 2/TPH2.
Figure 5.
Expression of amino acid metabolism genes in ER+ breast cancer. A. Total RNA from 16 ER+ patients at NCKUH was used to analyze the mRNA expression levels by RT-qPCR. B. Schematic summarizing how amino acid metabolism-related signaling pathways regulate breast cancer development. Estradiol activates the estrogen receptor and further upregulates PHGDH, PSAT1, PSPH, and GLDC transcriptional expression via the serine-glycine pathway and may facilitate purine and pyrimidine synthesis. Meanwhile, active estrogen receptor can promote the degradation of methionine by MAT1A and further elevate S-adenosylmethionine (SAM), leading to DNA methylation.
The survival status of amino acid metabolism genes
Next, we investigated the correlation between the expression of amino acid metabolism genes and distant metastasis-free survival (DMFS) in breast cancer patients. The gene expression levels and clinical outcomes of 1,764 breast cancer patient were acquired from the Kaplan-Meier plotter database (KM plot) database. High mRNA expression levels of ARG2, CBS, PHGDH (phosphoglycerate dehydrogenase), AHCY (adenosylhomocysteinase), HAL (histidine ammonia-lyase), TDO2 (tryptophan 2,3-dioxygenase), SHMT2 (serine hydroxymethyltransferase 2), MAT1A, MAT2A, GLDC (glycine decarboxylase), GLS2, BCAT2 (branched chain amino acid transaminase 2), GLUD1 (glutamate dehydrogenase 1), PAH (phenylalanine hydroxylase), and MTR were correlated with poor prognoses of breast cancer patients. In contrast with the aforementioned genes, high mRNA expression levels of HECA (headcase protein homolog), CTH, PRODH, TAT (tyrosine aminotransferase), and MAT2B were correlated with better prognoses (Figure 6).
Figure 6.
The relationship between distant metastasis-free survival and amino acid metabolism-related genes in breast cancer patients. The impacts of amino acid metabolism-related genes on distant metastasis-free survival (DMFS) of breast cancer patients were studied using the KMplot database (n = 1,764). The red colored lines indicate high transcriptional expression levels, whereas the black colored lines indicate low expression levels. The hazard ratios (HRs), with 95% confidence intervals and log-rank P-values, are illustrated. High expression levels of ARG2, CBS, PHGDH, BCAT1, MAT, TDO2, SHMT2, PSAT1, PSPH, MAT2A, GLDC, and ASL predicted poor prognoses. High expression levels of MTHFR, GLUL, MAT2B, TAT, PYCR2, SHMT1, GLS2, PIG6, CTH, BCAT2, GLUD1, and HECA predicted good prognoses.
The survival status of amino acid metabolism genes
Based on the Venn diagram analysis, we uploaded both up- and downregulated genes to the CMap (connectivity map) database to predict potential drug targets for the treatment of ER+ breast cancer (Figure 7). The top 15 drugs/molecules with positive correlations and the top 15 drugs/molecules with negative correlations were obtained from CMap. These drugs/molecules were ranked by p-values and were determined based on the gene signatures of ER+ cancer patients against the CMap database. Fulvestrant is recommended drug for metastatic ER+ breast cancer patients. Other drugs/molecules, including piperlongumine, ellipticine, etidronic acid, harmine, and levamisole, may serve as a potential drugs for ER+ breast cancer patients. We performed colony formation assays to validate our predictions. Fulvestrant, dirithromycin, meclizine, ellipticine, etidronic acid, piperlongumine, and harmine exhibited abilities to decrease the colonogenic propensity of MCF-7 breast cancer cells. These data confirm the usefulness of the CMap database (Figure 7C). These findings require further investigation to determine the therapeutic potential of these drugs/molecules in ER+ breast cancer patients.
Figure 7.
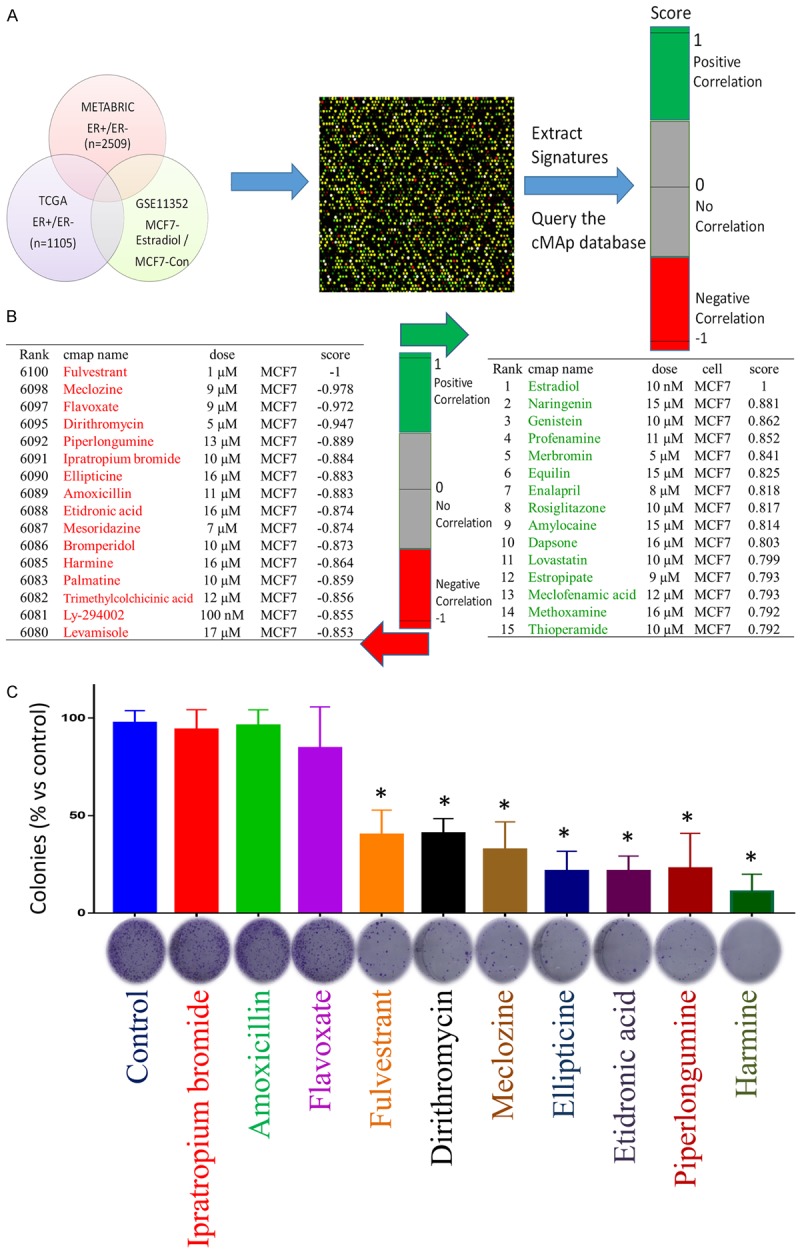
Connectivity map analysis of ER+ versus ER- patients. A. We uploaded the up- and downregulated genes identified in ER+ patients compared with ER- patients according to the METABRIC and TCGA databases and the estradiol-treated dataset into the CMap database to predict potential drug targets. The 15 top and bottom drugs/molecules represented positive and negative correlations, respectively, with ER+ patients. B. CMap analysis identified piperlongumine, ellipticine, etidronic acid, harmine and levamisole as potential therapeutic drugs for ER+ patients. C. Dirithromycin, meclozine, ellipticine, etidronic acid, piperlongumine, harmine treatments resulted in decreased clonogenic propensities during colony formation assays. MCF-7 cells were cultured in six-well plates to allow colony growth, cells were treated with FDA approved drugs for two weeks, and then colonies were stained with crystal violet and counted (n = 3). The graph represents the quantification of crystal violet stained MCF-7 colonies (*P < 0.05 is considered significant).
Discussion
In the present study, we identified the rate-limiting enzyme for amino acid metabolism associated with ER+ breast cancer through the integration of data from public databases. The TCGA and METABRIC databases were utilized to analyze potential amino acid metabolism genes associated with ER+ breast cancer. A regulatory network of amino acid metabolism-related genes after estradiol exposure was also evaluated from the NCBI GEO database. We used fresh specimens of breast cancer and corresponding normal breast from NCKU and the ER+ breast cancer cell line MCF-7 to perform validating experiments. These genes involving amino acid metabolism have potential to predict survival of breast cancer patients and provide target for further therapy [35-37]. To the best of our knowledge, the present study is the first to provide comprehensive evidence of a novel association between ER+ breast cancer prognosis and rate-limiting genes of amino acid metabolism as well as their downstream networks.
Growing evidence has demonstrated a close relationship between oncogene activation and metabolic changes in cancer cells [38,39]. The alteration of tumor metabolism is considered to be a target for therapy, and tumor metabolic imaging provides a method for monitoring therapeutic effect [40-42]. Increases activity of amino acid transporter and metabolism of choline/ethanolamine phospholipid have been observed in a wide variety of cancers [43-47]. Therefore, regulation of metabolism is important in cancer. In the present study, we performed bioinformatics analysis from 5,378 breast cancer patients, including the METABRIC, TCGA, and NCBI GEO databases. The analysis revealed several amino acid-related pathways in ER+ breast cancer, including the taurine and hypotaurine, alanine, aspartate, and glutamate metabolic pathways. These pathways were dysregulated in ER+ breast cancer.
We used the ER+ MCF-7 breast cancer cell line and ER+ breast cancer tissue acquired from NCKUH to validate our predictions. The results demonstrated that the activation of the estrogen receptor by estradiol significantly increased the levels of serine metabolism genes. Meanwhile, the increased expression levels of the enzymes PSAT1 and PSPH, together with increases in serine levels, may increase the formation of downstream products, such as glycine and 5,10-Methylenetetrahydrofolate (5,10-Methylene-THF). Glycine is converted into downstream products through increased GLDC expression levels, and CO2, NH3, and 5,10-methylene-THF may contribute to cellular synthesis pathways. We speculate that the activation of the serine-glycine metabolic pathway increases purine and pyrimidine levels and promotes proliferation [48].
In addition, serine-glycine and methionine metabolism via the one-carbon metabolic pathway is also dysregulated in ER+ breast cancer. The expression levels of MTR and MTHFR, which are involved in the one-carbon metabolic pathway, were not affected by estradiol exposure. The expression level of the MAT1A was remarkably elevated by 30-fold after estradiol treatment compared with the control. The synthesis of serine is related to methionine decomposition and caused by the overactivation of MAT1A. The decomposition of methionine promotes increases in S-adenosylmethionine (SAM) levels, which is the most important methylation factor in cells and may alter the methylation of cellular DNA [49,50]. The regulation of tumor repressor genes is also associated with SAM [51]. This metabolism-related enzyme may function as either an oncogene or a tumor suppressor gene during cancer development.
The in vitro study of MCF-7 cells revealed the increased expression levels of arginine and decomposing glutamine enzymes, such as ARG2 and GLS2, after treatment with estradiol. These results are consistent with previous study. Increased ARG2 leads to the metabolism of arginine to ornithine, which is further metabolized to produce polyamine. Polyamine metabolism is a target for cancer therapy because its’ role in anti-apoptosis of cancer cells [52,53]. GLS2 can metabolize glutamine to glutamate, which contributes to ornithine level through the urea cycle and increases polyamine production. Finally, mRNA expression levels of TH increased by 15-fold relative to the control sample after treatment with estradiol. The TH enzyme can decompose the tyrosine into L-dopa [54]. The metabolism of L-dopa to dopamine is catalyzed by dopa decarboxylase (DDC); however, in our study, the expression of DDC in MFC-7 cells was quite low. However, TH levels were not consistent among breast cancer patients in this study. The GLS2 expression was able to be detected in most patients, and it may be a better target for the treatment of breast cancer patients.
Through CMap analysis, we revealed that certain drugs/molecules with highly negative correlations might serve as a potential treatments for ER+ breast cancer patients. Then, we performed colony formation assays to validate the CMap predictions. Fulvestrant was reported to confer a 19% reduction in risk of death in patients with advanced primary or metastatic ER+ breast cancer [55]. Fulvestrant had a moderate ability to decrease colony formation by breast cancer cells in present study. This is consistent with previous research that lapatinib and fulvestrant are more efficient in combination for the inhibition of tumor growth than either drug alone [56]. Piperlongumine was reported to downregulate the gene expression of HER family receptors in breast cancer cells [57], and ellipticine reduces the proliferation of breast cancer stem cells [58]. Treating the ER+ breast cancer cell line MCF-7 with etidronic acid was reported to interrupt cell cycle progression and attenuate cell viability [59]. Harmine inhibits cellular growth in the MDA-MB-231 breast cancer cell line. Levamisole suppresses tumor cell proliferation and apoptosis activation in animal models [60,61]. Collectively, our CMap data suggest that these FDA-approved drugs are potential therapeutic drugs for ER+ breast cancer. These drugs may be tested in combination with current therapies in ER+ breast cancer patients.
In conclusion, the present findings demonstrate the crucial roles played by amino acid metabolism in breast cancer patients. The one-carbon metabolic pathway, the urea cycle, and other metabolic pathways related to glycine are highly implied to be involved in the development and progression of breast cancer. These amino acid metabolic pathways could potentially be targeted for treatment and prevention of breast cancer. Because the current study focuses on the pathways of amino acid-related signatures, further studies will be required to explore these findings and to validate the usefulness of our findings for the diagnosis and treatment of breast cancer.
Acknowledgements
Bioinformatics analyses and data mining were conducted at the Bioinformatics Core at the National Cheng Kung University (Tainan, Taiwan). We thank American Journal Experts for providing professional editing assistance (https://www.aje.com/). The authors were grateful for the support from the Human Biobank, Research Center of Clinical Medicine, National Cheng Kung University Hospital. The study was supported by the Ministry of Science and Technology (MOST) of Taiwan (grants MOST105-2325-B-006-003 to M-D. L., MOST 108-2314-B-006-082 to H-P. H., and MOST109-2320-B-038-009-MY2 to C-Y. W.), the National Cheng Kung University Hospital (grant NCKUH-10601002 to M-D. L., and NCKUH-10802026 to H-P. H.), the Chi Mei Medical Center (grant CMNCKU-10708 to H-P. H.) and the Taipei Medical University (grant TMU-108-AE1-B16 and TMU 108-6602-003-112 to C-Y. W.).
Disclosure of conflict of interest
None.
Abbreviations
- AHCY
adenosylhomocysteinase
- ARG2
arginase 2
- ASL
argininosuccinate lyase
- BCAT1
branched chain amino acid transaminase 1
- BCAT2
branched chain amino acid transaminase 2
- CBS
cystathionine-beta-synthase
- COPD
Pulmonary disease, chronic obstructive, severe early-onset
- CTH
cystathionine gamma-lyase
- DDC
dopa decarboxylase
- ErbB2
erb-b2 receptor tyrosine kinase 2
- SR1
estrogen receptor 1
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GLDC
glycine decarboxylase
- GLS2
glutaminase 2
- GLUD1
glutamate dehydrogenase 1
- GLUL
glutamate-ammonia ligase
- HAL
histidine ammonia-lyase
- HDC
histidine decarboxylase
- HECA
Headcase Protein Homolog
- MAT1A
methionine adenosyltransferase 1A
- MAT2A
methionine adenosyltransferase 2A
- MAT2B
methionine adenosyltransferase 2B
- Met
MET proto-oncogene, receptor tyrosine kinase
- MTHFR
methylenetetrahydrofolate reductase
- MTR
Methionine Synthase
- ODC1
ornithine decarboxylase 1
- PAH
phenylalanine hydroxylase
- PR
progesterone receptor
- PHGDH
phosphoglycerate dehydrogenase
- PRODH
proline dehydrogenase 1
- PSAT1
phosphoserine aminotransferase 1
- PSPH
phosphoserine phosphatase
- PYCR2
pyrroline-5-carboxylate reductase family member 2
- SHMT1
serine hydroxymethyltransferase 1
- SHMT2
serine hydroxymethyltransferase 2
- SP1
Sp1 transcription factor
- TAT
tyrosine aminotransferase
- TDO2
tryptophan 2,3-dioxygenase
- TH
tyrosine hydroxylase
- TPH2
tryptophan hydroxylase 2
Supporting Information
References
- 1.Schworer S, Vardhana SA, Thompson CB. Cancer metabolism drives a stromal regenerative response. Cell Metab. 2019;29:576–591. doi: 10.1016/j.cmet.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 3.Liu W, Shen SM, Zhao XY, Chen GQ. Targeted genes and interacting proteins of hypoxia inducible factor-1. Int J Biochem Mol Biol. 2012;3:165–178. [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. 2017;14:113. doi: 10.1038/nrclinonc.2017.1. [DOI] [PubMed] [Google Scholar]
- 5.Wang P, Wu SH, Zeng XF, Zhang YQ, Zhou Y, Su LL, Lin W. BCAT1 promotes proliferation of endometrial cancer cells through reprogrammed BCAA metabolism. Int J Clin Exp Pathol. 2018;11:5536. [PMC free article] [PubMed] [Google Scholar]
- 6.Yaw HP, Ton SH, Amanda S, Kong IG, Cheng HS, Fernando HA, Chin HF, Kadir KA. Irregularities in glucose metabolism induced by stress and high-calorie diet can be attenuated by glycyrrhizic acid. Int J Physiol Pathophysiol Pharmacol. 2014;6:172–184. [PMC free article] [PubMed] [Google Scholar]
- 7.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sikalidis AK. Amino acids and immune response: a role for cysteine, glutamine, phenylalanine, tryptophan and arginine in T-cell function and cancer? Pathol Oncol Res. 2015;21:9–17. doi: 10.1007/s12253-014-9860-0. [DOI] [PubMed] [Google Scholar]
- 9.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 10.Omar HA, Tolba MF, Saber-Ayad MM. Potential targets of energy restriction mimetic agents in cancer cells. Future Oncol. 2014;10:2547–2550. doi: 10.2217/fon.14.191. [DOI] [PubMed] [Google Scholar]
- 11.Lanz TV, Becker S, Mohapatra SR, Opitz CA, Wick W, Platten M. Suppression of Th1 differentiation by tryptophan supplementation in vivo. Amino Acids. 2017;49:1169–1175. doi: 10.1007/s00726-017-2415-4. [DOI] [PubMed] [Google Scholar]
- 12.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 15.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L, Gursky A, Siddiqui J, Tomlins SA, Roychowdhury S, Pienta KJ, Kim SY, Roberts JS, Rae JM, Van Poznak CH, Hayes DF, Chugh R, Kunju LP, Talpaz M, Schott AF, Chinnaiyan AM. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li AQ, Zhou SL, Li M, Xu Y, Shui RH, Yu BH, Yang WT. Clinicopathologic characteristics of oestrogen receptor-positive/progesterone receptor-negative/Her2-negative breast cancer according to a novel definition of negative progesterone receptor status: a large populationbased study from China. PLoS One. 2015;10:e0125067. doi: 10.1371/journal.pone.0125067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulun S, Lin Z, Zhao H, Lu M, Amin S, Reierstad S, Chen D. Regulation of aromatase expression in breast cancer tissue. Ann N Y Acad Sci. 2009;1155:121–131. doi: 10.1111/j.1749-6632.2009.03705.x. [DOI] [PubMed] [Google Scholar]
- 18.Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- 19.Murray PJ. Amino acid auxotrophy as a system of immunological control nodes. Nat Immunol. 2016;17:132–139. doi: 10.1038/ni.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sousa CM, Biancur DE, Wang XX, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying HQ, Asara JM, Evans RM, Cantley LC, Lyssiotis CA, Kimmelman AC. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–83. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loftus RM, Assmann N, Kedia-Mehta N, O’Brien KL, Garcia A, Gillespie C, Hukelmann JL, Oefner PJ, Lamond AI, Gardiner CM, Dettmer K, Cantrell DA, Sinclair LV, Finlay DK. Amino acid-dependent cMyc expression is essential for NK cell metabolic and functional responses in mice. Nat Commun. 2018;9:2341. doi: 10.1038/s41467-018-04719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menghini R, Fiorentino L, Casagrande V, Lauro R, Federici M. The role of ADAM17 in metabolic inflammation. Atherosclerosis. 2013;228:12–17. doi: 10.1016/j.atherosclerosis.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 23.Iurlaro R, Leon-Annicchiarico CL, Munoz-Pinedo C. Regulation of cancer metabolism by oncogenes and tumor suppressors. Methods Enzymol. 2014;542:59–80. doi: 10.1016/B978-0-12-416618-9.00003-0. [DOI] [PubMed] [Google Scholar]
- 24.Abbasi S, Rasouli M, Nouri M, Kalbasi S. Association of estrogen receptor-alpha A908G (K303R) mutation with breast cancer risk. Int J Clin Exp Med. 2013;6:39–49. [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Scholtens D, Holko M, Ivancic D, Lee O, Hu H, Chatterton RT Jr, Sullivan ME, Hansen N, Bethke K, Zalles CM, Khan SA. Lipid metabolism genes in contralateral unaffected breast and estrogen receptor status of breast cancer. Cancer Prev Res (Phila) 2013;6:321–330. doi: 10.1158/1940-6207.CAPR-12-0304. [DOI] [PubMed] [Google Scholar]
- 26.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang YS, Tsai CT, Huangfu CA, Huang WY, Lei HY, Lin CF, Su IJ, Chang WT, Wu PH, Chen YT, Hung JH, Young KC, Lai MD. ACSL3 and GSK-3beta are essential for lipid upregulation induced by endoplasmic reticulum stress in liver cells. J Cell Biochem. 2011;112:881–893. doi: 10.1002/jcb.22996. [DOI] [PubMed] [Google Scholar]
- 28.Wang CY, Li CY, Hsu HP, Cho CY, Yen MC, Weng TY, Chen WC, Hung YH, Lee KT, Hung JH, Chen YL, Lai MD. PSMB5 plays a dual role in cancer development and immunosuppression. Am J Cancer Res. 2017;7:2103–2120. [PMC free article] [PubMed] [Google Scholar]
- 29.Xu D, Lu Q, Hu X. Down-regulation of P-glycoprotein expression in MDR breast cancer cell MCF-7/ADR by honokiol. Cancer Lett. 2006;243:274–280. doi: 10.1016/j.canlet.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 30.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 31.Lamb J. The connectivity map: a new tool for biomedical research. Nat Rev Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 32.Wang CY, Lai MD, Phan NN, Sun Z, Lin YC. Meta-analysis of public microarray datasets reveals voltage-gated calcium gene signatures in clinical cancer patients. PLoS One. 2015;10:e0125766. doi: 10.1371/journal.pone.0125766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 34.Sun Z, Wang CY, Lawson DA, Kwek S, Velozo HG, Owyong M, Lai MD, Fong L, Wilson M, Su H, Werb Z, Cooke DL. Single-cell RNA sequencing reveals gene expression signatures of breast cancer-associated endothelial cells. Oncotarget. 2018;9:10945–10961. doi: 10.18632/oncotarget.23760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S METABRIC Group. Langerød A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Børresen-Dale AL, Brenton JD, Tavaré S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plathow C, Weber WA. Tumor cell metabolism imaging. J Nucl Med. 2008;49:43S. doi: 10.2967/jnumed.107.045930. [DOI] [PubMed] [Google Scholar]
- 41.Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PE, Harzstark AL, Ferrone M, van Criekinge M, Chang JW, Bok R, Park I. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-13C] pyruvate. Sci Transl Med. 2013;5:198ra108. doi: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campanella M, Casswell E, Chong S, Farah Z, Wieckowski MR, Abramov AY, Tinker A, Duchen MR. Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab. 2008;8:13–25. doi: 10.1016/j.cmet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Fuchs BC, Bode BP. Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? Semin Cancer Biol. 2005;15:254–266. doi: 10.1016/j.semcancer.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 44.Wang Q, Holst J. L-type amino acid transport and cancer: targeting the mTORC1 pathway to inhibit neoplasia. Am J Cancer Res. 2015;5:1281–1294. [PMC free article] [PubMed] [Google Scholar]
- 45.Imai H, Kaira K, Oriuchi N, Shimizu K, Tominaga H, Yanagitani N, Sunaga N, Ishizuka T, Nagamori S, Promchan K. Inhibition of Ltype amino acid transporter 1 has antitumor activity in non-small cell lung cancer. Anticancer Res. 2010;30:4819–28. [PubMed] [Google Scholar]
- 46.Podo F. Tumour phospholipid metabolism. NMR Biomed. 1999;12:413–439. doi: 10.1002/(sici)1099-1492(199911)12:7<413::aid-nbm587>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 47.Dueck DA, Chan M, Tran K, Wong JT, Jay FT, Littman C, Stimpson R, Choy PC. The modulation of choline phosphoglyceride metabolism in human colon cancer. Mol Cell Biochem. 1996;162:97–103. doi: 10.1007/BF00227535. [DOI] [PubMed] [Google Scholar]
- 48.Marino M, Acconcia F, Bresciani F, Weisz A, Trentalance A. Distinct nongenomic signal transduction pathways controlled by 17β-estradiol regulate DNA synthesis and cyclin D1 gene transcription in HepG2 cells. Mol Biol Cell. 2002;13:3720–3729. doi: 10.1091/mbc.E02-03-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strous RD, Ritsner MS, Adler S, Ratner Y, Maayan R, Kotler M, Lachman H, Weizman A. Improvement of aggressive behavior and quality of life impairment following S-adenosylmethionine (SAM-e) augmentation in schizophrenia. Eur Neuropsychopharmacol. 2009;19:14–22. doi: 10.1016/j.euroneuro.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Kim KC, Friso S, Choi SW. DNA methylation, an epigenetic mechanism connecting folate to healthy embryonic development and aging. J Nutr Biochem. 2009;20:917–926. doi: 10.1016/j.jnutbio.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo J, Li YN, Wang F, Zhang WM, Geng X. S-adenosylmethionine inhibits the growth of cancer cells by reversing the hypomethylation status of c-myc and H-ras in human gastric cancer and colon cancer. Int J Biol Sci. 2010;6:784–95. doi: 10.7150/ijbs.6.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerner EW, Meyskens FL Jr. Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004;4:781–92. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 53.Zaytouni T, Tsai PY, Hitchcock DS, DuBois CD, Freinkman E, Lin L, Morales-Oyarvide V, Lenehan PJ, Wolpin BM, Mino-Kenudson M, Torres EM, Stylopoulos N, Clish CB, Kalaany NY. Critical role for arginase 2 in obesity-associated pancreatic cancer. Nat Commun. 2017;8:242. doi: 10.1038/s41467-017-00331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alvarsson A, Zhang X, Stan TL, Schintu N, Kadkhodaei B, Millan MJ, Perlmann T, Svenningsson P. Modulation by trace amine-associated receptor 1 of experimental Parkinsonism, L-DOPA responsivity, and glutamatergic neurotransmission. J Neurosci. 2015;35:14057–14069. doi: 10.1523/JNEUROSCI.1312-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Leo A, Jerusalem G, Petruzelka L, Torres R, Bondarenko IN, Khasanov R, Verhoeven D, Pedrini JL, Smirnova I, Lichinitser MR, Pendergrass K, Garnett S, Lindemann JP, Sapunar F, Martin M. Results of the CONFIRM phase III trial comparing fulvestrant 250 mg with fulvestrant 500 mg in postmenopausal women with estrogen receptor-positive advanced breast cancer. J. Clin. Oncol. 2010;28:4594–4600. doi: 10.1200/JCO.2010.28.8415. [DOI] [PubMed] [Google Scholar]
- 56.Li Z, Yang SS, Yin PH, Chang T, Shi LX, Fang L, Fang GE. Activated estrogen receptor-mitogen-activated protein kinases cross talk confer acquired resistance to lapatinib. Thorac Cancer. 2015;6:695–703. doi: 10.1111/1759-7714.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin HO, Park JA, Kim HA, Chang YH, Hong YJ, Park IC, Lee JK. Piperlongumine downregulates the expression of HER family in breast cancer cells. Biochem Biophys Res Commun. 2017;486:1083–1089. doi: 10.1016/j.bbrc.2017.03.166. [DOI] [PubMed] [Google Scholar]
- 58.Pandrangi SL, Chikati R, Chauhan PS, Kumar CS, Banarji A, Saxena S. Effects of ellipticine on ALDH1A1-expressing breast cancer stem cells--an in vitro and in silico study. Tumour Biol. 2014;35:723–737. doi: 10.1007/s13277-013-1099-y. [DOI] [PubMed] [Google Scholar]
- 59.Zhou Y, Beyene D, Zhang R, Kassa A, Ashayeri E, Sridhar R. Cytotoxicity of etidronic acid to human breast cancer cells. Ethn Dis. 2008;18 S2-87-92. [PubMed] [Google Scholar]
- 60.Hegde M, Karki SS, Thomas E, Kumar S, Panjamurthy K, Ranganatha SR, Rangappa KS, Choudhary B, Raghavan SC. Novel levamisole derivative induces extrinsic pathway of apoptosis in cancer cells and inhibits tumor progression in mice. PLoS One. 2012;7:e43632. doi: 10.1371/journal.pone.0043632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Friis T, Engel AM, Klein BM, Rygaard J, Houen G. Levamisole inhibits angiogenesis in vitro and tumor growth in vivo. Angiogenesis. 2005;8:25–34. doi: 10.1007/s10456-005-3588-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.