Abstract
MicroRNAs (miRNAs) are short and non-coding RNAs binding to 3’UTR of target mRNAs to downregulate their expression. Recent studies have shown that miRNAs indirectly regulated alternative splicing (AS) by targeting splicing factors and caused shifts in splicing patterns of target genes. However, the roles of miRNA-regulating splicing factors in pancreatic cancer progression remain unknown. Herein, we reported that miR-193a-5p was markedly upregulated in pancreatic cancer tissues and cells and correlated with clinical outcomes of pancreatic cancer patients. Overexpression of miR-193a-5p contributed to the metastasis of pancreatic cancer cells both in vitro and in vivo. The mechanistic investigation suggested that miR-193a-5p modulated oxoglutarate dehydrogenase-like (OGDHL) and extracellular matrix protein 1 (ECM1) AS by targeting serine/arginine-rich splicing factor 6 (SRSF6), leading to the activation of the epithelial-to-mesenchymal transition (EMT) process. Together, our findings highlighted the role of miR-193a-5p-targeting SRSF6 in pancreatic cancer metastasis, which may serve as a novel target for pancreatic cancer diagnosis and therapy.
Keywords: Pancreatic cancer, miR-193a-5p, SRSF6, alternative splicing, invasion, migration, OGDHL, ECM1
Introduction
Pancreatic cancer, mainly pancreatic adenocarcinoma (PDAC), is the seventh leading cause of cancer death worldwide because of its poor prognosis [1]. pancreatic cancer is a highly invasive and metastatic disease: 60% of patients have distant metastasis within the first 24 months after surgery [2], and it is considered as one of the primary causes of mortality in these patients [3]. In contrast to the steady increase in survival observed for most cancer types with improved surgical techniques and development of new chemotherapy drugs, advances have been slow for pancreatic cancer [4]. Although previous research revealed multiple pathophysiological mechanisms underlying pancreatic cancer pathogenesis [5-7], the biology of pancreatic cancer is still largely unknown.
MicroRNAs (miRNAs) are a class of single-strand, non-encoding endogenous RNAs, which have approximately 19-24 nucleotides [8]. miRNAs exert their biological functions by directly binding to the 3’-untranslated region (3’UTR) of target mRNAs to inhibit translation or promote mRNA degradation. Aberrant expression of miRNA is frequently observed in cancers including pancreatic cancer and their role as promoters or suppressors of the oncogenic process is well established [9-11].
Alternative splicing (AS) of pre-mRNA produces a wide variety of differentially spliced mRNA transcripts, which determines the proteomic complexity of mammals and contributes to temporal and spatial diversification of biological functions. Recent studies have shown that AS was involved in multiple oncological processes, including proliferation [12], metastasis [13], angiogenesis [14], apoptosis [15] and drug resistance [16]. The “cancerous” splice variants of specific genes have turned into novel molecular biomarkers as well as therapeutic targets to outwit the cancer treatment. Serine/arginine-rich (SR) RNA binding protein family is one of the major classes of splicing factors composed of RNA recognition motifs (RRMs) and a C-terminal SR domain, which are concentrated in nuclear speckles and play critical roles in constitutive and alternative splicing [17]. SR proteins could recruit spliceosome assembly via binding to a specific RNA sequence, regulating the AS of exons or introns. Aberrant expression of serine/arginine-rich splicing factors (SRSFs) has been reported in multiple types of cancers such as leukemia [18], kidney [19], breast [20], colon [21], glioma [22] and lung [23]. Remarkably, several recent studies have demonstrated that miRNAs as indirect regulators of AS could target the expression of splicing factors and cause shifts in splicing patterns of target genes [24-26]. However, few studies have reported the regulation of SRSFs by miRNAs [19,27], or the functional roles of SRSFs in pancreatic cancer.
Our previous study has shown that miR-193a-5p was upregulated in Panc-1 spheroid cells that enriched in pancreatic cancer stem cells (pancreatic cancerSCs) [28]. However, the biological role of miR-193a-5p in pancreatic cancer need to be further elucidated. In the present study, we showed for the first time that the expression of SRSF6 is regulated by miR-193a-5p, which was upregulated in pancreatic cancer cells, pancreatic cancerSCs and tissues. We also indicated that miR-193a-5p was involved in pancreatic cancer metastasis by targeting SRSF6. Further mechanistic investigations suggested that SRSF6 participated in pancreatic cancer cell migration and invasion by modulating the expression pattern of oxoglutarate dehydrogenase-like (OGDHL) and extracellular matrix protein 1 (ECM1) splice variants, then activating the epithelial-to-mesenchymal transition (EMT) process. Therefore, our results provided the first evidence that miR-193a-5p-targeting SRSF6 plays a crucial role in pancreatic cancer metastasis by modulating AS, which may serve as a novel target for pancreatic cancer diagnosis and therapy.
Materials and methods
Patients and clinical specimens
Forty pairs of human pancreatic cancer tissues and normal tissue samples (located > 5 cm away from the tumor) were collected from patients who underwent primary surgical treatment at Zhongda Hospital of Southeast University (Nanjing, China) with written informed consent. All the clinical specimens were snapped-frozen and stored in liquid nitrogen. The ethical approval was granted from Committees for Ethical Review in China Pharmaceutical University (Nanjing, China). Pathological diagnosis was made according to the histology of tumor specimens or biopsy and examined by experienced pathologists, and the clinical features of the patients are listed in Table S1. The study is compliant with all relevant ethical regulations for human research participants, and all participants provided written informed consent.
Pancreatic cancer cell lines and cell culture
The human pancreatic cancer cell lines (Panc-1, MIApaca-2, SW1990, and BXpc-3), and human pancreatic duct epithelial (HPDE) cells used in the study were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The HPDE, Panc-1 and BXpc-3 cells were cultured in RPMI-1640 (Gibco, USA); the MIApaca-2 cells were cultured in DMEM medium (Gibco, USA), and SW1990 cells were cultured in L-15 medium (Gibco, USA). All these mediums were supplemented with 10% FBS Gibco, USA), 100 U/mL penicillin and 100 ng/mL streptomycin (Beyotime, China). The cells were maintained at 37°C in a humidified chamber supplemented with 5% CO2.
RT-pancreatic cancerR
Total RNAs from patient tissues or cultured cells were isolated using the Trizol (Invitrogen, USA). Then reverse transcription-polymerase chain reaction (RT-pancreatic cancerR) was performed using Takara reagent (RR047A; Dalian, China) according to the manufacturer’s manual.
RNA extraction and quantitative real-time pancreatic cancerR (qRT-pancreatic cancerR)
For the detection of miR-193a-5p expression level, total RNA was reverse-transcribed with a miR-193a-5p specific RT primer (GenePharma, China) and amplified with pancreatic cancerR primers (GenePharma, China) by the LightCycler480 Real-time pancreatic cancerR system (Roche Molecular Systems, Indianapolis, USA). The relative expression level of miR-193a-5p was normalized with U6. SYBR Green pancreatic cancerR kit (Takara) was used to quantify the mRNA levels by normalizing to GAPDH according to the manufacturer’s protocols. The primers used were listed in Table S2. Results were normalized with the expression of GAPDH or U6. The relative quantification was performed using the 2-ΔΔCt method.
Immunohistochemistry (IHC)
Paraffin-embedded tissues were sliced into sections with four μm thickness, followed by dewaxing with dimethylbenzene and hydration in graded ethanol. Antigens were retrieved by microwave heating. Endogenous peroxidase was blocked using 3% hydrogen peroxide, and tissues were kept in darkness at room temperature for 10 min. Then the primary antibody was added dropwise into sections, followed by incubation at 4°C overnight. Horseradish peroxidase-conjugated secondary antibody (Dako North America Inc., Carpinteria, CA, USA) was added dropwise into sections, and were then incubated at room temperature for 30 min. The incubated sections were stained by diaminobenzidine (DAB), and then counterstained by hematoxylin, before being mounted on slides with neutral balsam. Primary antibody was replaced with phosphate-buffered saline (PBS) as negative control, and human GC biopsies were selected as the positive control. Five high-power fields (400×) were randomly selected for each section (100 cells were counted in each visual field), and positive cells were counted. Antibodies used are provided in Table S3.
Cell transfection and virus infection
For transient transfection, miRNA mimics, inhibitors and siRNAs (Genepharma) were transfected by Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. The coding domain sequences of human SRSF6 and ECM1 mRNA variants were amplified by pancreatic cancerR using human cDNA as a template, and inserted into the pcDNA 3.0 vector (Invitrogen, Grand Island, NY, USA). Plasmids were also transfected by Lipofectamine 2000. Lentivirus encoding miR-193a-5p or miR-193a-5p sponge (Genepharma) or SRSF6 were imported into MIApaca-2 cells. The clones with the stable miR-193a-5p or SRSF6 expression were selected by green fluorescence protein (GFP) expression. The related sequences are listed in Table S2.
Wound healing assay
Cells were seeded into 6-well plates and incubated for 24 h after transfection. The wounds were made with a sterile pipette tip in the confluent cell layer, and were then photographed under a phase-contrast microscope immediately after scratching (0 h) and 24 h. The ability of the cells to wound healing was determined by comparing the results of 0 h and 24 h.
Transwell migration and invasion assay
After transfection for 48 h, cells were harvested and suspended in serum-free culture medium. After diluted (1:7) in serum-free medium, the Matrigel (Corning, Corning, NY) was added into the Transwell upper chamber, and maintained in an incubator for 30 min. Then 1 × 105 cells were seeded into the Transwell upper chamber (6.5 mm, Costar, Cambridge, MA). The lower chamber was filled with 1640 medium supplemented with 10% FBS. After incubation for 24 h to 48 h, the cells on the upper membrane surface were removed, and cells that had entered the lower surface of the filter membrane were fixed with 4% paraformaldehyde for 25 min and stained for 15 min with 1% crystal violet dye. Then, invasion cells were counted in five or six randomly selected fields per chamber by a photomicroscope (BX51, Olympus, Japan). For cell migration assay, similar procedures were performed compared to those in the invasion assay except for the Matrigel coating.
Western blotting
Total proteins were extracted from cells or tissues with RIPA buffer (10 mM Tris-HCl, pH 7.4, 1% Triton X-100, 0.1% SDS, 1% NP-40, 1 mM MgCl2) containing protease inhibitors. The total protein concentration was determined by BCA Protein Assay Kit (Beyotime, China). Proteins were separated on a 10% SDS-polyacrylamide gel and then transferred onto polyvinylidene fluoride (PDVF) membranes (Millipore, Billerica, MA, USA). Membranes were then blocked with 5% skimmed milk and probed with primary antibodies against E-cadherin, ZEB1, MMP9, MMP2, vimentin, snail, SRSF6, OGDHL, ECM1, and GAPDH at 4°C overnight. On the following day, the blots were washed with PBST and incubated with horseradish-peroxidase-conjugated secondary antibodies (Abcam, Cambridge, UK) for 2 h at room temperature. Protein bands were visualized using the ECL-kit (Millipore, MA, USA), according to the manufacturer’s instructions. GAPDH was used as a loading control and protein bands were analyzed using Bandscan software (Image J). Antibodies used are listed in Table S3.
Luciferase reporter assay
SRSF6 was identified as a potential miR-193a-5p target in TargetScan 7.1 (http://www.targetscan.org/). The wild-type (WT) or mutant (MUT) 3’-UTR of SRSF6 containing the miR-193a-5p binding site was cloned and inserted into the pGL3 plasmid (Ambion, Austin, TX, USA), named as SRSF6-WT and SRSF6-MUT, respectively. For luciferase assays, cells were cultured in 24-well plates and co-transfected with SRSF6-WT or SRSF6-MUT together with miR-193a-5p or control by Lipofectamine 2000 reagent according to the manufacturer’s instructions. The relative luciferase activity was calculated after 48 h by normalizing the Firefly luminescence to the Renilla luminescence using the Dual-Luciferase Reporter Assay (Promega, Madison, WI, USA).
Minigene assay
The OGDHL exon 3 or ECM1 exon 4 (part) minigene was constructed by insertion of exons and its flanking intron region into pGint (Addgene, Cambridge, USA) Vector. pGint has an EGFP open reading frame (ORF) which was divided into two exons by a constitutively spliced intron. OGDHL exon 3 or ECM1 exon 4 (part) and its flanking intron region were amplified with OGDHL or ECM1 minigene-F and minigene-R from genome DNA and inserted into the intron. The EGFP ORF could be disrupted once the above exons have been inserted; thus, green fluorescence signal will be impaired or vanished. Exons skipping bring a functional EGFP ORF and leads to its expression. pRint was co-transfected with the minigene to exclude the interfering signals.
In vivo xenograft experiments
Male BALB/c nude mice (four-weeks-old) were purchased from Vital River Laboratory Animal Technology Company (Beijing) and maintained in SPF level animal room. The procedure of all animal experiments complied with Institutional Animal Care and Use Committee (IACUC) regulations. 2 × 106 different infected Panc-1 cells were injected intravenously into each mouse through the tail vein (n=5 per group). Mice were anesthetized with 3% isoflurane, and then imaged in an IVIS spectrum imaging system (Caliper, Newton, USA) every 10 days. After 30 days, the liver tissues were extracted and fixed in 10% buffered formalin, immersed in an ascending series of alcohol washes, and embedded with paraffin. The tissues were then sectioned and stained with hematoxylin and eosin (H&E) and immunohistochemical staining. For all animal experiments, the operators and investigators were blinded to the group allocation. All animal experiments were approved by the Ethics Committee of China Pharmaceutical University (Permit No. 2162326).
Statistical analysis
These above experiments were performed in triplicate, and each was repeated several times. The results are presented as the means ± SEM of at least three independent experiments. The differences were considered statistically significant at P<0.05 using the Student’s t-test.
Results
miR-193a-5p is upregulated in both pancreatic cancer tissues and cells
To assess the relevance of miR-193a-5p in human pancreatic cancer, we first compared the expression levels of miR-193a-5p in 40 paired PADC tissues and corresponding adjacent normal tissues. As shown in Figure 1A, miR-193a-5p expression was significantly upregulated in 92.5% (37 of 40 paired) of the PADC tissues. We next divided the samples into high (above the median, n=20) and low (below the median, n=20) miR-193a-5p expression groups according to the median value of miR-193a-5p levels and explored the correlation between miR-193a-5p expressions and the clinicopathological factors of PADC patients (Table S1). As shown in Figure 1B, miR-193a-5p level was positively associated with tumor-node-metastasis (TNM) stage and N classification in this study. We further evaluated the correlation between the expression level of miR-193a-5p and clinical outcomes of pancreatic cancer patients from the TCGA database using Kaplan-Meier Plotter (www.kmplot.com). As shown in Figure 1C, the overall survival time of patients with miR-193a-5p high expression is significantly shorter (n=178, P<0.05).
Figure 1.
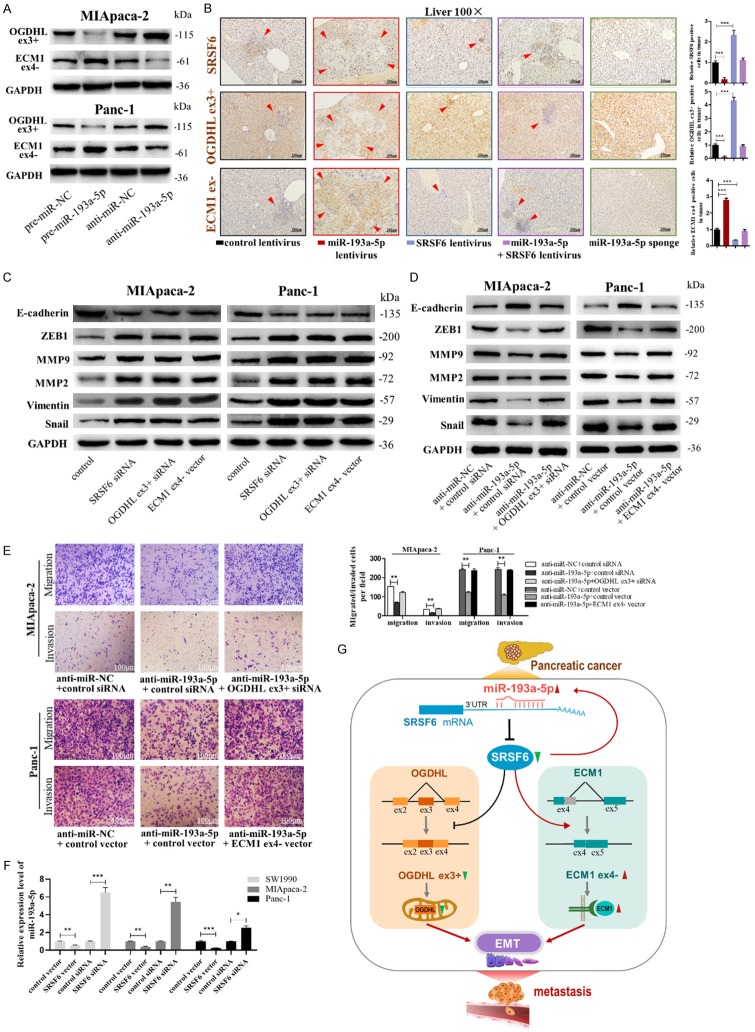
miR-193a-5p is upregulated in pancreatic cancer tissues and cells. A. miR-193a-5p levels were detected in 40 paired pancreatic ductal carcinoma tissues by qRT-pancreatic cancerR. B. miR-193a-5p levels in pancreatic ductal carcinoma (PADC) patients with different N classification and TNM stage. C. miR-193a-5p high expression was correlated with a poor survival rate of pancreatic cancer patients (n=188, P<0.05). Data was analyzed using Kaplan Meier Plotter (www.kmplot.com). D. miR-193a-5p levels were detected in pancreatic cancerSCs by qRT-pancreatic cancerR. E. miR-193a-5p levels in MIApaca-2 and Panc-1 cells under stimulation of bFGF and EGF. F. miR-193a-5p levels were detected in different pancreatic cancer cells by qRT-pancreatic cancerR. All data are shown as the mean ± SEM. *P<0.05, **P<0.01, and ***P<0.001 by two-tailed Student’s t-test.
Next, the upregulation of miR-193a-5p was validated in pancreatic cancerSCs and pancreatic cancer cell lines. A novel 3D semi-solid culture system [28] was used to culture MIApaca-2 and Panc-1 spheroid cells enriched in either CD133+ or CD24+CD44+ESA+ pancreatic cancerSCs. Analysis of MIApaca-2 and Panc-1 spheroid cells and pancreatic cancerSCs indicated that the expression of miR-193a-5p in spheroid cells and pancreatic cancerSCs were significantly higher than that in parental pancreatic cancer cells (Figure 1D). Epidermal growth factor (EGF) and b-fibroblast growth factors (b-FGF) were two major components added in our sphere-formation pelletizing system, which also exist in the tumor microenvironment [29]. We therefore exposed MIApaca-2 and Panc-1 cells to EGF and bFGF separately and found miR-193a-5p expression could be upregulated by EGF (Figure 1E). Also, we compared miR-193a-5p levels in different pancreatic cancer cell lines (Bxpc-3, SW1990, MIApaca-2 and Panc-1) and normal human pancreatic duct epithelial (HPDE) cells. We found miR-193a-5p levels were not only upregulated in pancreatic cancer cells compared with HPDE cells, but also showed a positive correlation with the malignancy degree [31] in four pancreatic cell lines (Bxpc-3, SW1990, MIApaca-2 and Panc-1; Figure 1F). Collectively, these results indicated that miR-193a-5p expression is upregulated in pancreatic cancer and that its high expression predicts a poor prognosis in pancreatic cancer patients.
miR-193a-5p contributes to pancreatic cancer cell migration and invasion in vitro
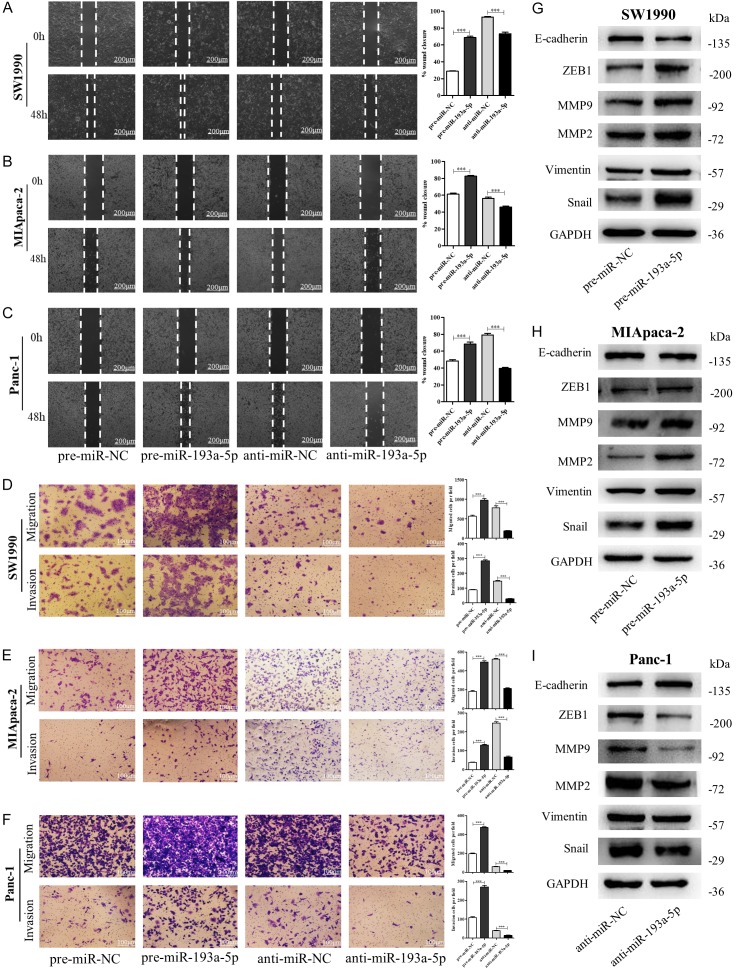
To explore the potential role of miR-193a-5p played in tumorigenesis and progression of pancreatic cancer, we overexpressed or knocked down miR-193a-5p levels by pre-miR-193a-5p or anti-miR-193a-5p (Figure S1A), and then evaluated the abilities of cell proliferation, migration, self-renewal. The wound healing assays (Figure 2A-C) and transwell assays (Figure 2D-F) showed that the migration and invasion capacities of SW1990, MIApaca-2 and Panc-1 cells were significantly enhanced by overexpression of miR-193a-5p whereas suppressed by depleting miR-193a-5p. However, alternation of miR-193a-5p in these cells did not have stable and significant effects on their proliferation, self-renewal and stemness (Figure S2). We further examined EMT-related proteins in miR-193a-5p overexpressed or knocked-down SW1990, MIApaca-2 and Panc-1 cells. As shown, miR-193a-5p overexpression activated the EMT process in pancreatic cancer cells, whereas miR-193a-5p knockdown inhibited the process (Figure 2G-I). Taken together, these data suggested that miR-193a-5p could contribute to pancreatic cancer metastasis.
Figure 2.
miR-193a-5p promotes pancreatic cancer cells migration and invasion in vitro. (A-C) Migration of SW1990 cells (A), MIApaca-2 cells (B) and Panc-1 cells (C) transfected with pre-miR-NC, pre-miR-193a-5p, anti-miR-NC, or anti-miR-193a-5p detected by wound healing assay. Scale bar, 100 μm. (D-F) Migration and invasion of SW1990 cells (D), MIAPACA-2 cells (E), and Panc-1 cells (F) transfected with pre-miR-NC, pre-miR-193a-5p, anti-miR-NC, or anti-miR-193a-5p detected by transwell migration and invasion assay. Scale bar, 100 μm. (G-I) The protein levels of EMT target genes in SW1990 cells (G), MIAPACA-2 cells (H) and Panc-1 cells (I) transfected with pre-miR-NC, pre-miR-193a-5p, anti-miR-NC or anti-miR-193a-5p detected by western blotting. All data are shown as the mean ± SEM. ***P<0.001.
SRSF6 is a direct target gene of miR-193a-5p in pancreatic cancer cells
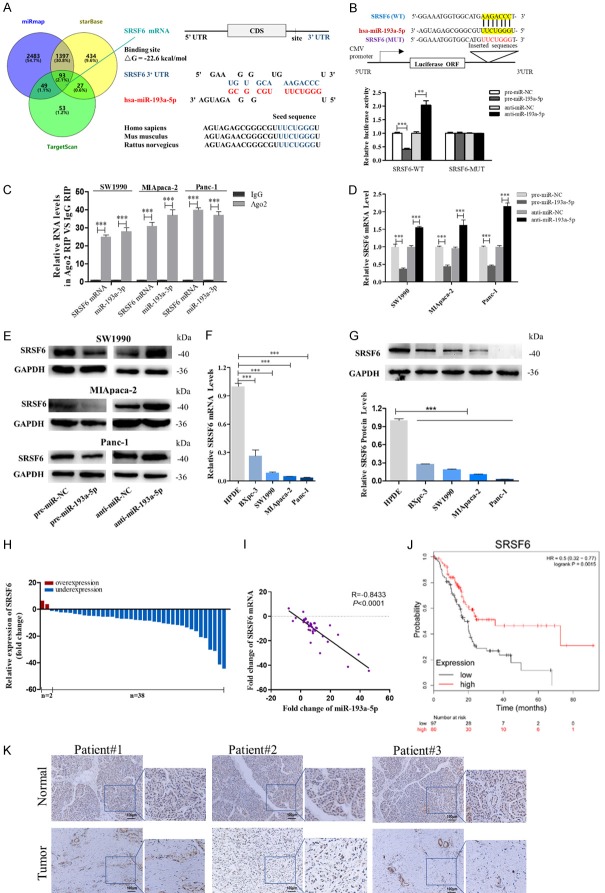
To elucidate the molecular mechanisms by which miR-193a-5p promotes pancreatic cancer cell metastasis, we used bioinformatic prediction software (TargetScan7.1, miRmap and starBase) to search for the potential targets of miR-193a-5p, and found 93 mRNAs in the intersection part (Figure 3A, left panel). SRSF6 was selected for further verification based on the following criteria (Table S4): its function had been reported in cancers but not in pancreatic cancer, low expressed in pancreatic cancer, positively correlated with survival of pancreatic cancer patients (Kaplan-Meier Plotter), related to cancer migration or invasion, and had not been reported as a miR-193a-5p target. The predicted interaction between miR-193a-5p and the target sites in the SRSF6 3’-UTR was illustrated in the right panel of Figure 3A. There was a perfect base-pairing between the seed region and the cognate target. The free energy value of the hybrid was well within the range of genuine miRNA-target pairs (-22.6 kcal/mol). Subsequently, luciferase reporter assay confirmed that miR-193a-5p directly targeted the predicted binding sites in the SRSF6 3’-UTR (Figure 3B), and RIP assays using antibodies against Ago2 demonstrated that miR-193a-5p and SRSF6 mRNA were all enriched in Ago2-immunoprecipitation (Ago2-IP; Figure 3C). Moreover, transfection of pre- or anti-miR-193a-5p significantly down or upregulated the expression of SRSF6 mRNA and protein in pancreatic cancer cells (Figure 3D and 3E).
Figure 3.
Identification of SRSF6 as a direct target gene of miR-193a-5p in pancreatic cancer cells. (A) Left: Venn diagram analysis of four independent databases reveals 93 possible targets of miR-193a-5p. Right: Schematic description of the hypothetical duplexes formed by the interactions between the binding sites in the SRSF6 3’-UTR and miR-193a-5p. The predicted free energy value of the hybrid is indicated. The seed recognition sites are denoted, and all nucleotides in these regions are highly conserved across species, including human, mouse, and rat. (B) Luciferase activity in Panc-1 cells co-transfected with a luciferase reporter containing either SRSF6-WT or SRSF6-MUT (miR-193a-5p-binding sequence mutated), and mimics NC, miR-193a-5p mimics, inhibitor NC or miR-193a-5p inhibitor. Data are presented as the relative ratio of renilla luciferase activity and firefly luciferase activity. (C) Relative enrichment of SRSF6 mRNA and miR-193a-5p associated with AGO2 in SW1990 cells, MIApaca-2 cells and Panc-1 cells detected by anti-AGO2 RIP (non-specific IgG as negative control). (D, E) SRSF6 mRNA (D) and protein (E) levels in SW1990 cells, MIApaca-2 cells and Panc-1 cells transfected with mimics NC, miR-193a-5p mimics, inhibitor NC, or miR-193a-5p inhibitor. (F, G) SRSF6 mRNA (F) and protein levels (G) in different pancreatic cancer cells. (H, I) SRSF6 mRNA levels (H) and Pearson’s correlation scatter plot of the fold change of miR-193a-5p and SRSF6 mRNA levels (I) in 40 pairs of human PADC tissues and corresponding adjacent normal tissues. (J) SRSF6 low expression was correlated with a poor survival rate of pancreatic cancer patients (n = 177, P<0.01). Data was analyzed using Kaplan Meier Plotter (www.kmplot.com). (K) In situ hybridization (ISH) analysis of miR-193a-5p expression levels in PADC tissues (Tumor) and their adjacent normal tissues (Normal). Representative LNA ISH images from patients #1, #2 and #3 are shown. Scale bar, 100 μm. All data are shown as the mean ± SEM. **P<0.01; ***P<0.001.
In most cases, miRNAs generally have the expression patterns that are opposite to that of their targets. Therefore, we detected the expression level of SRSF6 in pancreatic cancer cells and clinical specimens. In contrast with miR-193a-5p, we found SRSF6 levels were downregulated in pancreatic cancer cells compared with HPDE cells, and showed a negative correlation with the malignancy degree in four pancreatic cell lines (Figure 3F and 3G). Consistently, SRSF6 mRNA levels were found downregulated in 95% (38 of 40 paired) PADC tissues and SRSF6 protein levels were shown to have an inverse correlation with miR-193a-5p levels using Spearman’s correlation scatter plots (Figure 3H and 3I). In addition, SRSF6 low expression predicts a poor prognosis in pancreatic cancer patients (n=177, P<0.005, www.kmplot.com; Figure 3J). IHC analysis of SRSF6 also showed reduced expression of SRSF6 in the PADC tissues compared to the corresponding adjacent normal tissues (Figure 3K). The above results demonstrated that SRSF6 can be a direct target of miR-193a-5p in pancreatic cancer and might be correlated with clinical outcomes of pancreatic cancer patients.
miR-193a-5p facilitates pancreatic cancer cell migration and invasion via targeting SRSF6
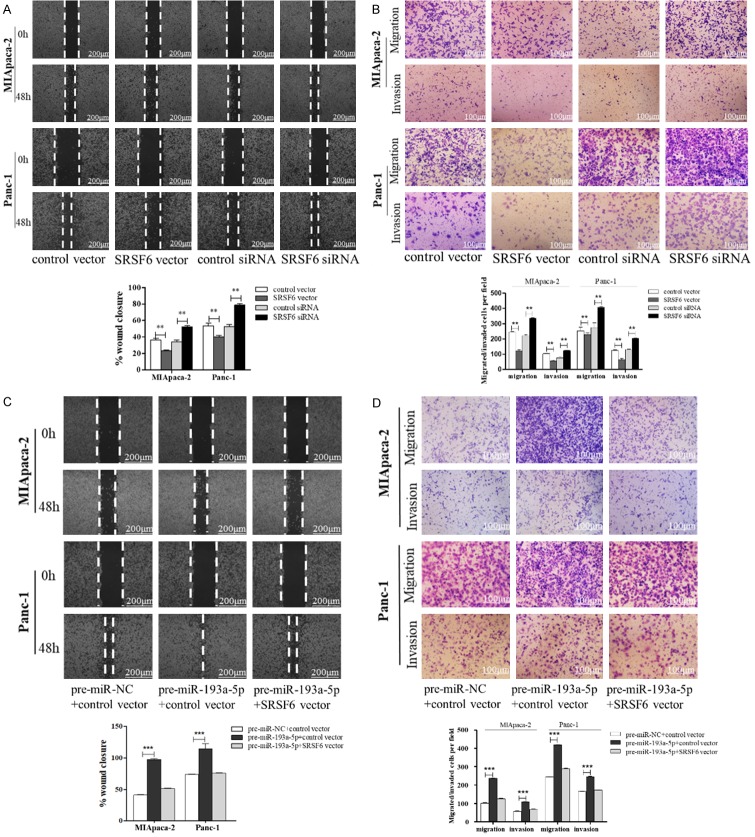
To prove that the pro-migration and invasion effects of miR-193a-5p were mediated by downregulating SRSF6, we conducted the rescue experiments. Firstly, we observed that transfection of SRSF6 vector repressed SW1990, MIApaca-2 and Panc-1 cell migration and invasion, while transfection SRSF6 siRNAs markedly promoted the behavior (Figures S1B, S3A, S3B, 4A and 4B). Secondly, transfection of SRSF6 vector not only successfully mitigated miR-193a-5p overexpression-induced restriction of SRSF6 protein expression (Figure S1C), but also attenuated the enhanced migration and invasion abilities induced by miR-193a-5p (Figures 4C, 4D, S3C and S3D). These results suggested that miR-193a-5p promotes the migration and invasion of pancreatic cancer cells by directly targeting SRSF6.
Figure 4.
miR-193a-5p promotes pancreatic cancer cell migration and invasion via targeting SRSF6. (A, B) Migration and invasion of MIApaca-2 cells (upper) and Panc-1 cells (lower) transfected with control vector, SRSF6 vector, control siRNA or SRSF6 siRNA detected by wound healing assay (A) or migration and invasion assay (B). Scale bar, 100 μm. (C, D) Migration and invasion of MIApaca-2 cells and Panc-1 cells transfected with pre-miR-NC plus control vector, pre-miR-193a-5p plus control vector, or pre-miR-193a-5p plus SRSF6 vector detected by wound healing assay (C) or migration and invasion assay (D). Scale bar, 100 μm. All data are shown as the mean ± SEM. **P<0.01; ***P<0.001.
miR-193a-5p promoted pancreatic cancer cell metastasis in vivo by targeting SRSF6
Furthermore, we explored the function of SRSF6-targeted miR-193a-5p in regulating pancreatic cancer cell metastasis in a mouse model. First, the Panc-1 cells were transfected with control lentivirus, miR-193a-5p lentivirus, SRSF6 lentivirus, miR-193a-5p lentivirus plus SRSF6 lentivirus, or miR-193a-5p sponge. Then, the five groups of cells were injected into nude mice via the tail vein. The metastasis was assessed by bioluminescent imaging (BLI) on days 10, 20 and 30 after implantation (Figure 5A-C). The image assay indicated that the GFP-labeled migrating cells were mainly distributed in the livers or lungs of the mice. The fluorescent intensities were significantly stronger in miR-193a-5p lentivirus group and weaker in SRSF6 lentivirus group and miR-193a-5p sponge group compared to their control group. Moreover, SRSF6 overexpression can attenuate the pro-metastasis effect caused by miR-193a-5p-overexpression (Figure 5D and 5E). After 30 days, mice were euthanized and the whole liver and lung tissues were extracted and subjected to H&E staining for evaluating tumor metastasis. We observed that the number of metastatic lesions (red arrows) at the of liver surface was increased in the miR-193a-5p lentivirus group and reduced in the SRSF6 lentivirus group and the miR-193a-5p sponge group compared with the control group, while SRSF6 overexpression recovered the levels to those of the control cells (Figure 5F). In addition, the H&E staining results showed significant differences in tumor number and growth in liver tissues (Figure 5F). Although we did not observe significant differences in the surface of lungs between different groups (data not shown), the results of H&E staining in lung tissues displayed the similar trend with liver tissues (Figure 5G). These results confirmed that miR-193a-5p promotes pancreatic cancer cell metastasis in the mouse model by suppressing SRSF6 expression.
Figure 5.
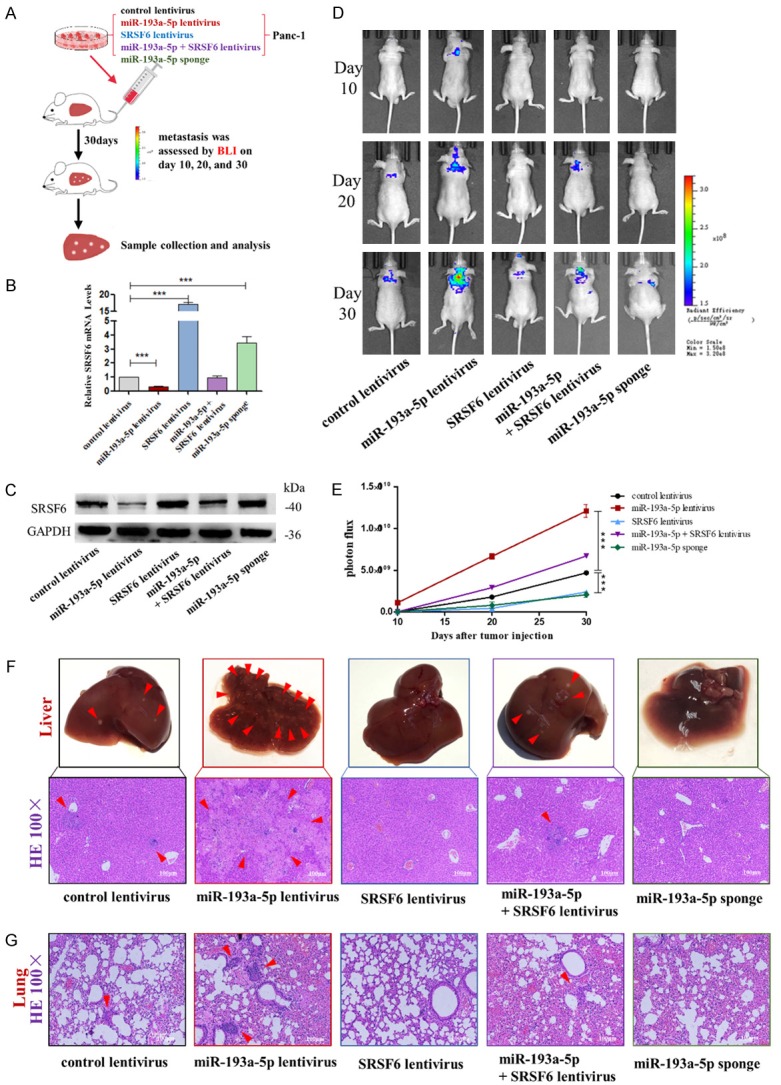
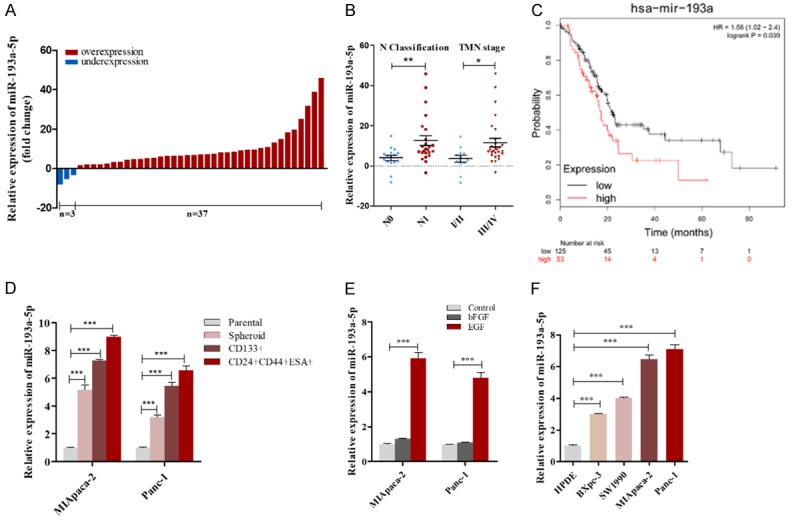
Effects of SRSF6-targeted miR-193a-5p on the liver colonization of Panc-1 cells xenografts in mice. (A) Experimental design: immunocompromised mice were injected through tail vein with Panc-1 cells transfected with either the control lentivirus, miR-193a-5p lentivirus, SRSF6 lentivirus, miR-193a-5p lentivirus plus SRSF6 lentivirus, or miR-193a-5p sponge. (B, C) miR-193a-5p levels (B) and SRSF6 protein levels (C) in Panc-1 cells transfected with different lentiviruses. (D, E) Representative BLI images (D) and quantitative analysis of the fluorescence intensities (E) of mice of five groups. The BLI was performed on days 10, 20, and 30 after injection. The intensity of BLI is represented by the color. (F) Upper: Representative liver tissues isolated from the intravenously injected mice. Red arrows indicate metastatic nodules. Lower: Representative H&E staining of liver tissues. (G) Representative H&E staining in lung tissues. Scale bar, 100 μm. All data are shown as the mean ± SEM. ***P<0.001.
SRSF6 inhibits pancreatic cancer cell migration and invasion by regulating OGDHL AS
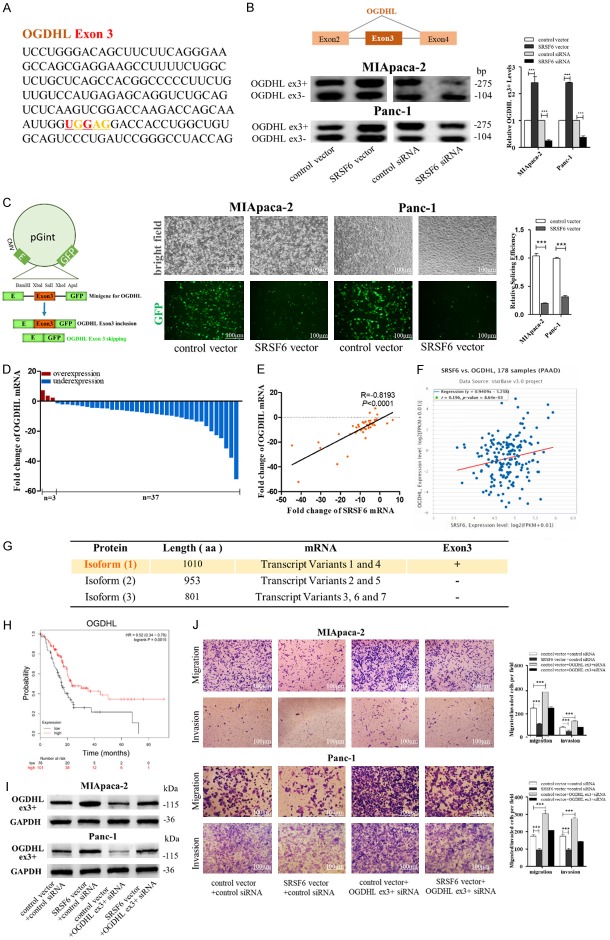
To clarify the underlying mechanisms of pancreatic cancer cell migration and invasion promoted by SRSF6, we attempted to find the AS targets of SRSF6 in these cells. By searching the potential AS targets of SRSF6 in colorectal cancer [30], we noticed the expression of OGDHL exon3 inclusion (OGDHL ex3+) splice variants was significantly decreased and part of ECM1 exon4 exclusion splice variants was significantly increased by SRSF6 shRNA. Since OGDHL and ECM1 were both involved cancer metastasis [31-34], which might be related to the molecular function of SRSF6, we searched the SRSF6 binding motif in OGDHL and the ECM1 mRNA sequence. Strikingly, we found a consensus motif sequence (UGGAG) with a predicted SRSF6 binding motif in exon3 of OGDHL (Figure 6A), and a consensus motif sequence (UGGAA) with a predicted SRSF6 binding motif in exon4 of ECM1 (Figure 7A). We first determined whether OGDHL is the AS target of SRSF6. By RT-pancreatic cancerR, we confirmed that the expression of OGDHL exon3 inclusion splice variants was significantly increased by SRSF6 overexpression, whereas decreased by SRSF6 knockdown (Figure 6B). To address whether SRSF6 could activate the splicing of OGDHL exon3, we constructed a OGDHL exon3 minigene reporter system [35], in which the entire ORF was retained after EGFP exon3 skipping (OGDHL ex3-), but the exon23 inclusion (OGDHL ex3+) disrupted the ORF and reduced the expression of EGFP (Figure 6C, left). As shown, SRSF6 overexpression caused a significant reduction in EGFP signal (Figure 6C, middle and right), indicating that SRSF6 overexpression promoted OGDHL exon3 inclusion.
Figure 6.
SRSF6 promotes pancreatic cancer cell migration and invasion through regulating OGDHL alternative splicing. (A) A consensus motif sequence (UGGAG) with predicted SRSF6 binding motif in exon3 of OGDHL. (B) RT-pancreatic cancerR for detecting OGDHL exon3 splicing isoforms expression in MIApaca-2 and Panc-1 cells transfected with control vector, SRSF6 vector, control siRNA or SRSF6 siRNA. (C) Minigene reporter system for detecting OGDHL exon3 splicing (left). The fluorescence signal in the GFP channel represents exon3 splicing efficiency in control and SRSF6 overexpression MIApaca-2 and Panc-1 cells (middle). Quantification of splicing efficiency by measuring the relative expression of intact EGFP transcript (right). (D, E) OGDHL mRNA levels (D) and Pearson’s correlation scatter plot of the fold change of OGDHL and SRSF6 mRNA levels (E) in 40 pairs of human PADC tissues and corresponding adjacent normal tissues. (F) Pearson’s correlation analysis of the fold change of OGDHL mRNA and SRSF6 mRNA in 178 human pancreatic adenocarcinoma (PAAD) tissues by ENCORI database from the TCGA project. (G) Different isoforms of OGDHL from NCBI database. Isoform 1 (1100aa) that contains exon3 is the canonical protein isoform. (H) OGDHL low expression was correlated with a poor survival rate of pancreatic cancer patients (n=177, P<0.01). Data was analyzed using Kaplan Meier Plotter (www.kmplot.com). (I) OGDHL exon3 included (ex3+) protein levels in MIApaca-2 and Panc-1 cells transfected with or without SRSF6 vector and with or without OGDHL (ex3+) siRNA detected by western blotting. (J) Migration and invasion of MIApaca-2 cells (upper) and Panc-1 cells (lower) transfected with or without SRSF6 vector and with or without OGDHL (ex3+) siRNA detected by transwell migration and invasion assay. Scale bar, 100 μm. All data are shown as the mean ± SEM. ***P<0.001.
Figure 7.
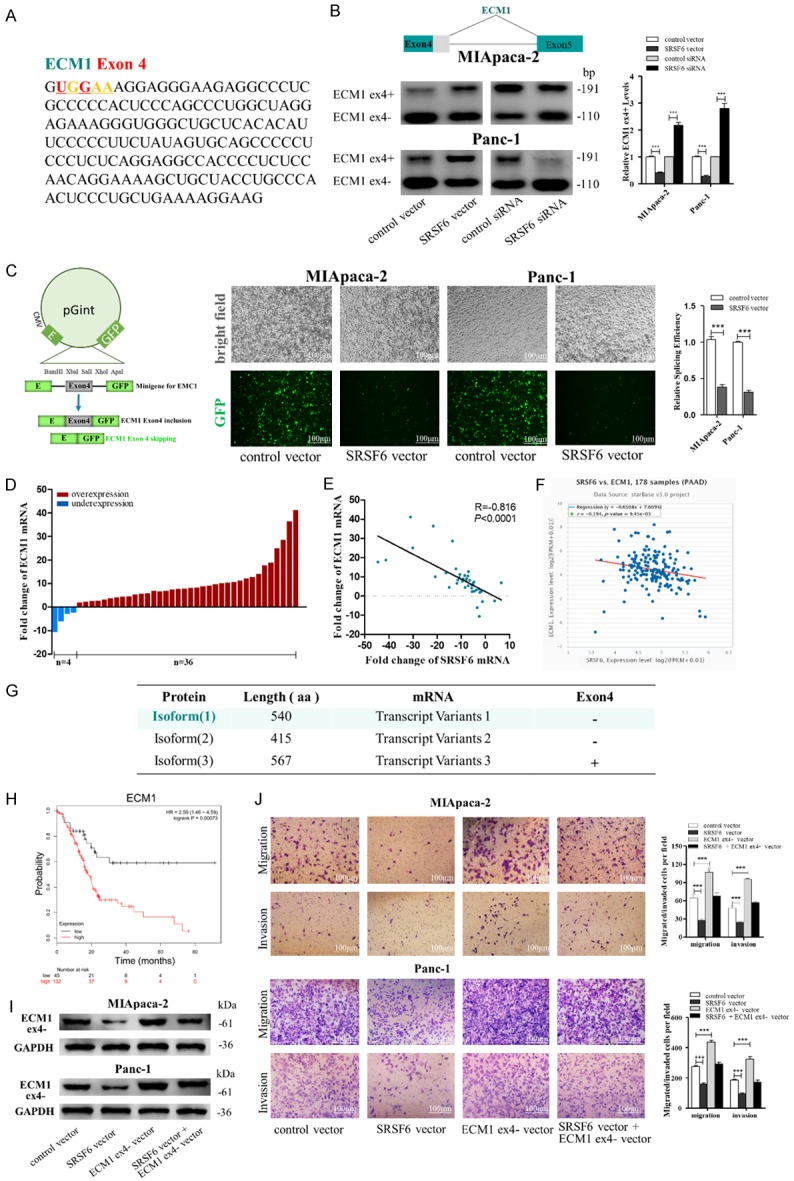
SRSF6 promotes pancreatic cancer cell migration and invasion through regulating ECM1 alternative splicing. (A) A consensus motif sequence (UGGAA) with predicted SRSF6 binding motif in exon4 of ECM1. (B) RT-pancreatic cancerR for detecting ECM1 exon4 splicing isoforms expression in MIApaca-2 and Panc-1 cells transfected with control vector, SRSF6 vector, control siRNA or SRSF6 siRNA. (C) Minigene reporter system for detecting ECM1 exon4 splicing (left). The fluorescence signal in the GFP channel represents exon4 splicing efficiency in control and SRSF6 overexpression MIApaca-2 and Panc-1 cells (middle). Quantification of splicing efficiency by measuring the relative expression of intact EGFP transcript (right). (D, E) ECM1 mRNA levels (D) and Pearson’s correlation scatter plot of the fold change of ECM1 and SRSF6 mRNA levels (E) in 40 pairs of human PADC tissues and corresponding adjacent normal tissues. (F) Pearson’s correlation analysis of the fold change of ECM1 mRNA and SRSF6 mRNA in 178 human pancreatic adenocarcinoma (PAAD) tissues by ENCORI database from the TCGA project. (G) Different isoforms of ECM1 from NCBI database. Isoform 1 (540aa) that excludes part of exon4 is the canonical protein isoform. (H) ECM1 high expression was correlated with a poor survival rate of pancreatic cancer patients (n=177, P<0.001). Data was analyzed using Kaplan Meier Plotter (www.kmplot.com). (I) ECM1 exon4 partially excluded (ex4-) protein levels in MIApaca-2 and Panc-1 cells transfected with or without SRSF6 or without ECM1 (ex4-) vector detected by western blotting. (J) Migration and invasion of MIApaca-2 cells (upper) and Panc-1 cells (lower) transfected with or without SRSF6 or ECM1 (ex4-) vector detected by transwell migration and invasion assay. Scale bar, 100 μm. All data are shown as the mean ± SEM. ***P<0.001.
We next investigated the relationship between SRSF6 and OGDHL expression in pancreatic cancer tissues. As shown in Figure 6D and 6E, OGDHL expression was significantly down regulated in 92.5% (37 of 40 paired) of the PADC tissues, and had a positive correlation with SRSF6 expression using Spearman’s correlation scatter plots. The positive correlation was also supported by using 178 pancreatic tissues from the TCGA database (Figure 6F).
Furthermore, we attempted to clarify that OGDHL ex3+ variant has the similar inhibitory effect on pancreatic cancer cell migration and invasion with SRSF6. By searching the NCBI Gene database, we found the isoform 1 (1010 aa) of OGDHL that contained exon3 was the canonical protein isoform of OGDHL (Figure 6G). Also, we observed that OGDHL low expression predicts a poor prognosis in pancreatic cancer patients (n=177, P<0.005, www.kmplot.com; Figure 6H). Moreover, we constructed specific siRNAs to specifically knockdown OGDHL ex3+ variant. As shown in Figure 6I and 6J, SRSF6 overexpression increased the protein levels of OGDHL ex3+ and decreased the migrated/invaded MIApaca-2 and Panc-1 cells while OGDHL ex3+ knockdown had opposite effects, and OGDHL ex3+ knockdown attenuated the increased OGDHL ex3+ protein levels and decreased migrated/invaded cells induced by SRSF6 overexpression. Overall, we inferred that SRSF6 could regulate OGDHL AS and thus inhibit pancreatic cancer cell migration and invasion.
SRSF6 downregulation promotes pancreatic cancer cell migration and invasion by regulating ECM1 AS
We next determined whether ECM1 is an AS target of SRSF6. We confirmed that the expression of ECM1 exon4 (part) exclusion (ECM1 ex4-) splice variants was significantly decreased by SRSF6 overexpression, whereas increased by SRSF6 knockdown using RT-pancreatic cancerR (Figure 7B). The results of ECM1 ex4- minigene reporter system showed that SRSF6 overexpression weakened the EGFP signal (Figure 7C), indicating that SRSF6 overexpression inhibited ECM1 exon4 (part) skipping. We also investigated the relationship between SRSF6 and OGDHL expression in pancreatic cancer tissues. ECM1 expression was significantly upregulated in 90% (36 of 40 paired) of the PADC tissues (Figure 7D), and had a negative relationship with SRSF6 expression (Figure 7E). The inverse correlation was also supported by using 178 pancreatic tissues from the TCGA database (Figure 7F). Moreover, we attempted to clarify that ECM1 ex4- variant has the opposite inhibitory effect on pancreatic cancer cell migration and invasion with SRSF6. We found the isoform 1 (540aa) of ECM1 that excluded part of exon4 that could be AS by SRSF6 was the canonical protein isoform of ECM1 by searching the NCBI Gene database (Figure 7G). Moreover, ECM1 high expression was corrected with poor outcomes of pancreatic cancer patients (n=177, P<0.005, www.kmplot.com; Figure 7H). Also, we observed that the SRSF6 overexpression could decrease the protein levels of ECM1 ex4- and the migrated/invaded MIApaca-2 and Panc-1 cells while ECM1 ex4- overexpression had opposite effects, attenuated the decreased ECM1 ex4- protein levels and migrated/invaded cells induced by SRSF6 overexpression (Figure 7I and 7J). The above results suggested that SRSF6 downregulation promotes pancreatic cancer cell migration and invasion via upregulating ECM1 ex4-.
miR-193a-5p activates EMT process by downregulating OGDHL (ex3+) and upregulating ECM1 (ex4-)
Finally, we evaluated whether miR-193a-5p modulated pancreatic cancer cell migration and EMT were mediated by the two AS targets of SRSF6, OGDHL ex3+ and ECM1 ex4-. Firstly, we detected that miR-193a-5p overexpression caused the downregulation of OGDHL ex3+ and upregulation of ECM1 ex4- protein, whereas miR-193a-5p knockdown had reverse effects in MIApaca-2 and Panc-1 cells (Figure 8A). Next, we measured the protein levels of SRSF6, OGDHL ex3+ and ECM1 ex4- in the hepatic metastasis focused by IHC staining. As shown in Figure 8B, tumors with miR-193a-5p overexpression had lower levels of SRSF6 and OGDHL ex3+ and higher levels of ECM1 ex4- than the control group, while tumors with SRSF6 overexpression or miR-193a-5p knockdown displayed the reverse results. Tumors with both miR-193a-5p and SRSF6 overexpression exhibited similar levels of SRSF6, OGDHL ex3+ and ECM1 ex4- compared with the control group.
Figure 8.
miR-193a-5p activates EMT process through down-regulating OGDHL (ex3+) and upregulating ECM1 (ex4-). (A) Protein levels of OGDHL (ex3+) and ECM1 (ex4-) in MIApaca-2 cells (upper) and Panc-1 cells (lower) transfected with pre-miR-NC, pre-miR-193a-5p, anti-miR-NC, or anti-miR-193a-5p. (B) IHC staining for SRSF6, OGDHL (ex3+) and ECM1 (ex4-) in the mouse livers. (C) The protein levels of EMT target genes in MIApaca-2 cells (left) and Panc-1 cells (right) transfected with control siRNA, SRSF6 siRNA, OGDHL (ex3+) siRNA, or ECM1 (ex4-). (D, E) The protein levels of EMT target genes (D), migrated and invaded cells (E) in MIApaca-2 cells transfected with anti-miR-NC plus control siRNA, anti-miR-193a-5p plus control siRNA, or anti-miR-193a-5p plus OGDHL (ex3+) siRNA, and Panc-1 cells transfected with anti-miR-NC plus control vector, anti-miR-193a-5p plus control vector, or anti-miR-193a-5p plus ECM1 (ex4-) vector. (F) miR-193a-5p levels in SW1990, MIApaca-2 and Panc-1 cells transfected with control vector, SRSF6 vector, control siRNA or SRSF6 siRNA. (G) A working model for the role of SRSF6-targeted miR-193a-5p in pancreatic cancer metastasis. During pancreatic tumorigenesis, upregulation of miR-133a-3p facilitated migration and invasion of pancreatic cancer cells through downregulating the expression of SRSF6, which regulated OGDHL and EMC1 alternative splicing thereby activated EMT process. All data are shown as the mean ± SEM. ***P<0.001.
As OGDHL [31,32] and ECM1 [33,34] were reported to inhibit and activate EMT process, separately, we verified that miR-193a-5p promotes pancreatic cancer cell migration, invasion and EMT by OGDHL ex3+ and ECM1 ex4- by using rescue experiments. Firstly, we demonstrated that SRSF6 or OGDHL ex3+ knocking down, or ECM1 ex4- overexpression could all activate EMT process (Figure 8C). Next, we observed that OGDHL ex3+ knocking down, or ECM1 ex4- overexpression attenuated the inhibitory effects of miR-193a-5p knocking down on EMT activation, migration and invasion in MIApaca-2 and Panc-1 cells (Figure 8D and 8E). Overall, these results indicated that miR-193a-5p, by targeting SRSF6, inhibited OGDHL ex3+ expression and promoted ECM1 ex4- expression, promoting EMT process of pancreatic cancer cells which leads to the cell migration and invasion.
Feedback regulation of miR-193a-5p and SRSF6
It was previously found that splicing factors can regulate the expression of its own regulatory miRNAs [19,36,37]. Therefore, we checked the effect of silencing of SRSF6 on the expression of miR-193a-5p. As shown in Figure 8F, the expression of miR-193a-5p was increased after depletion of SRSF6 in SW1990, MIApaca-2 and Panc-1 cells, whereas was downregulated after overexpression of SRSF6 in these cell lines. This result indicated SRSF6 might influence the processing of miR-193a-5p.
Discussion
In this study, we elucidated the critical roles of miR-193a-5p and SRSF6 in pancreatic cancer and the underlying mechanism for the first time. The upregulation of miR-193a-5p promotes the metastasis and EMT activation of pancreatic cancer by targeting SRSF6, which mediated the AS of OGDHL exon3 inclusion and ECM1 exon4 (part) exclusion splice variants (Figure 8G).
An increasing number of evidences have suggested that miR-193a-5p participated in cancer development and progression. miR-193a-5p was found upregulated in prostate cancer tissues [38] and hepatocellular carcinoma tissues [39], in which it significantly suppressed prostate cancer cell apoptosis induced by oxidative stress, and accelerated the proliferation and suppressed the apoptosis of hepatocellular carcinoma cells. Xu et al. found that miR-193a-5p expression is negatively correlated with the survival of invasive bladder cancer patients [40]. However, miR-193a-5p acts as a tumor suppressor miRNA in breast cancer [41], colon cancer [42], glioblastoma [43], non-small-cell lung cancer [44], and osteosarcoma cells [45]. Specific miRNA has various expression profiles in different tissues and presents biological activity by targeting different proteins, resulting in the inconsistent activity observed in different cell types. However, the expression and the role of miR-193a-5p, instead of miR-193a-3p [46,47], in pancreatic cancer remain largely unknown. In this study, we found the upregulation of miR-193a-5p in pancreatic cancer cells and tissues, and high expression levels of miR-193a-5p were correlated with TNM stage and poor prognosis in pancreatic cancer patients. Overexpression of miR-193a-5p promotes the migration and invasion of pancreatic cancer cell both in intro and in vivo. These findings indicated miR-193a-5p acts as an oncogene in pancreatic cancer.
SRSF6 was then identified as the target of miR-193a-5p in pancreatic cancer. As an main member of SRSFs, SRSF6 shuttles between the nucleus and the cytoplasm [48] and is involved in many RNA-associated processes including not only pre-mRNA splicing but also mRNA stability, export, and translation [49]. Recent studies have considered SRSF6 as a proto-oncogene that is frequently overexpressed in human skin cancer [50], lung cancer [51] and colon cancer [30] that promotes aberrant AS. Intriguingly, we found SRSF6 high expression predicts good survival rate in pancreatic cancer patients, which implied its tumor suppressor role in pancreatic cancer. We also observed the downregulation of SRSF6 in pancreatic cancer tissues and cells, which was negatively correlated with the expression of miR-193a-5p. Our biological assays in vitro and in vivo also confirmed that SRSF6 inhibits pancreatic cancer metastasis, and all of the prometastatic effects caused by miR-193a-5p could be rescued by SRSF6 restoration. These findings suggested the potential value of miR-193a-5p combining with SRSF6 as prognostic biomarkers in pancreatic cancer patients.
Partly due to the context-dependent nature of SRSFs target selection, the key SRSFs-governed AS networks responsible for tumorigenesis usually differ greatly among different tumor types [52], and unfortunately, little is known about SRSF6-affected AS in pancreatic cancer up to now. Previously, SRSF6-regulated AS targets and its binding motif was identified by next-generation RNA-sequencing and RNA immunoprecipitation sequencing, and was preliminarily validated by gel shift in colon cancer cells [30]. Among these AS events, OGDHL ex3+ and ECM1 ex4- splicing changes were identified. As both OGDHL and ECM1 participated in regulating metastasis of pancreatic cancer and the EMT process in other cancers [31-34], which might be related to SRSF6’s molecular function, we confirmed that OGDHL ex3+ splice variants were increased, while ECM1 ex4- splice variants were decreased by SRSF6 in pancreatic cancer cells using the gel shift and minigene reporter assay. We also noticed that their related protein isoforms are both the canonical ones and played a similar trend with the mRNA variants in both cells and mouse tissues. The downregulation of OGDHL and upregulation of ECM1 were both correlated with SRSF6, and predicted poorer outcome in pancreatic cancer patients. Consistently, we observed that downregulation of OGDHL ex3+ isoform and upregulation of ECM1 ex4- isoform contributed to the effects of miR-193a-5p targeting SRSF6 on pancreatic cancer cell metastasis and EMT activation. These results are adequate to demonstrate that OGDHL and ECM1 are crucial “bridge” molecules that mediate the oncogenic effects of miR-193a-5p targeting SRSF6 in pancreatic cancer.
Previous studies have demonstrated the important roles of miRNA-dependent regulation of AS in the cellular development and disease [53-55]. The examples of miRNAs affecting splicing patterns in tumorigenesis including miR-10a/b targeting SRSF1 contributing to retinoic acid-induced differentiation of neuroblastoma cells [27], miR-30a/181 forming regulatory feedback loop with SRSF7 promoting renal cancer cell proliferation [19], miR-200c/375 controlling epithelial plasticity-associated AS by repressing Quaking-5 in breast cancer [56], and miR-133b promoting hepatocellular carcinoma cell proliferation and metastasis by targeting splicing factor 3b subunit 4 [57]. It was also found that splicing factors can actively influence the processing of pre-miRNAs [19,36,37]. For instance, SRSF1 promotes maturation of miR-7 that in turn downregulates the expression of SRSF1 [37], hnRNP A1 binds to the loop of pri-miR-18a and promotes its cleavage. Together with the results of the present study may broaden the network of post-transcriptional changes orchestrated by miRNAs in human cancer.
In conclusion, we highlighted the functional importance of miR-193a-5p targeting SRSF6 in pancreatic cancer metastasis. Along with further research, miR-193a-5p-SRSF6-OGDHL/ECM1 axis may become useful prognostic biomarkers and provide effective targets for anti-metastasis therapies for pancreatic cancer.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81702941 and No. 81570696), Priority Academic Program Development of Jiangsu Higher Education Institutions, Science and Technology Development Fund Project of Nanjing Medical University (NMUB2018099), and Qing Lan Project.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Konstantinidis IT, Warshaw AL, Allen JN, Blaszkowsky LS, Castillo CF, Deshpande V, Hong TS, Kwak EL, Lauwers GY, Ryan DP, Wargo JA, Lillemoe KD, Ferrone CR. Pancreatic ductal adenocarcinoma: is there a survival difference for R1 resections versus locally advanced unresectable tumors? What is a “true” R0 resection? Ann Surg. 2013;257:731–6. doi: 10.1097/SLA.0b013e318263da2f. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida BA, Sokoloff MM, Welch DR, Rinker-Schaeffer CW. Metastasis-suppressor genes: a review and perspective on an emerging field. J Natl Cancer Inst. 2000;92:1717–30. doi: 10.1093/jnci/92.21.1717. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 5.Lee JJ, Perera RM, Wang H, Wu DC, Liu XS, Han S, Fitamant J, Jones PD, Ghanta KS, Kawano S, Nagle JM, Deshpande V, Boucher Y, Kato T, Chen JK, Willmann JK, Bardeesy N, Beachy PA. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A. 2014;111:E3091–100. doi: 10.1073/pnas.1411679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang CM, Guo YQ, Guo YB, Zhang ZQ, Lian GD, Chen YT, Li J, Su YH, Li JJ, Yang K, Chen SJ, Su H, Huang KH, Zeng LJ. MMP1/PAR1/SP/NK1R paracrine loop modulates early perineural invasion of pancreatic cancer cells. Theranostics. 2018;8:3074–86. doi: 10.7150/thno.24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu C, Chen SY, Guo YT, Sun CY. Oncogenic TRIM31 confers gemcitabine resistance in pancreatic cancer via activating the NF-kappa B signaling pathway. Theranostics. 2018;8:3224–36. doi: 10.7150/thno.23259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 9.Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38. doi: 10.1186/s12935-015-0185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G, Gormley M, Qiao J, Zhao Q, Wang M, Di Sante G, Deng S, Dong L, Pestell T, Ju X, Casimiro MC, Addya S, Ertel A, Tozeren A, Li Q, Yu Z, Pestell RG. Cyclin D1-mediated microRNA expression signature predicts breast cancer outcome. Theranostics. 2018;8:2251–63. doi: 10.7150/thno.23877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang T, Yang Z, Zhu Q, Wu Y, Sun K, Alahdal M, Zhang Y, Xing Y, Shen Y, Xia T, Xi T, Pan Y, Jin L. Up-regulation of miR-210 induced by a hypoxic microenvironment promotes breast cancer stem cells metastasis, proliferation, and self-renewal by targeting E-cadherin. FASEB J. 2018 doi: 10.1096/fj.201801013R. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Qi Y, Yu J, Han W, Fan X, Qian H, Wei H, Tsai YH, Zhao J, Zhang W, Liu Q, Meng S, Wang Y, Wang Z. A splicing isoform of TEAD4 attenuates the Hippo-YAP signalling to inhibit tumour proliferation. Nat Commun. 2016;7:ncomms11840. doi: 10.1038/ncomms11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braeutigam C, Rago L, Rolke A, Waldmeier L, Christofori G, Winter J. The RNA-binding protein Rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene. 2014;33:1082–92. doi: 10.1038/onc.2013.50. [DOI] [PubMed] [Google Scholar]
- 14.Hamdollah Zadeh MA, Amin EM, Hoareau-Aveilla C, Domingo E, Symonds KE, Ye X, Heesom KJ, Salmon A, D’Silva O, Betteridge KB, Williams AC, Kerr DJ, Salmon AH, Oltean S, Midgley RS, Ladomery MR, Harper SJ, Varey AH, Bates DO. Alternative splicing of TIA-1 in human colon cancer regulates VEGF isoform expression, angiogenesis, tumour growth and bevacizumab resistance. Mol Oncol. 2015;9:167–78. doi: 10.1016/j.molonc.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara H, Takeda T, Yamamoto N, Furuya K, Hirose K, Kamiya T, Adachi T. Zinc-induced modulation of SRSF6 activity alters Bim splicing to promote generation of the most potent apoptotic isoform BimS. FEBS J. 2013;280:3313–27. doi: 10.1111/febs.12318. [DOI] [PubMed] [Google Scholar]
- 16.Wang BD, Ceniccola K, Hwang S, Andrawis R, Horvath A, Freedman JA, Olender J, Knapp S, Ching T, Garmire L, Patel V, Garcia-Blanco MA, Patierno SR, Lee NH. Alternative splicing promotes tumour aggressiveness and drug resistance in African American prostate cancer. Nat Commun. 2017;8:15921. doi: 10.1038/ncomms15921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong S. SR proteins: binders, regulators, and connectors of RNA. Mol Cells. 2017;40:1–9. doi: 10.14348/molcells.2017.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou L, Zhang H, Du C, Liu X, Zhu S, Zhang W, Li Z, Gao C, Zhao X, Mei M, Bao S, Zheng H. Correlation of SRSF1 and PRMT1 expression with clinical status of pediatric acute lymphoblastic leukemia. J Hematol Oncol. 2012;5:42. doi: 10.1186/1756-8722-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boguslawska J, Sokol E, Rybicka B, Czubaty A, Rodzik K, Piekielko-Witkowska A. microRNAs target SRSF7 splicing factor to modulate the expression of osteopontin splice variants in renal cancer cells. Gene. 2016;595:142–9. doi: 10.1016/j.gene.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 20.Anczukow O, Akerman M, Clery A, Wu J, Shen C, Shirole NH, Raimer A, Sun S, Jensen MA, Hua Y, Allain FH, Krainer AR. SRSF1-regulated alternative splicing in breast cancer. Mol Cell. 2015;60:105–17. doi: 10.1016/j.molcel.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong J, Sun W, Li C, Wan L, Wang S, Wu Y, Xu E, Zhang H, Lai M. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016;380:476–84. doi: 10.1016/j.canlet.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X, Wang R, Li X, Yu L, Hua D, Sun C, Shi C, Luo W, Rao C, Jiang Z, Feng Y, Wang Q, Yu S. Splicing factor SRSF1 promotes gliomagenesis via oncogenic splice-switching of MYO1B. J Clin Invest. 2019;129:676–93. doi: 10.1172/JCI120279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng J, Zhao Q, Zhao J, Zhang W, Sun Y, Qin P, Lv Y, Bai L, Yang Q, Chen L, Qi Y, Zhang G, Zhang L, Gu C, Deng X, Liu H, Meng S, Gu H, Liu Q, Coulson JM, Li X, Sun B, Wang Y. SRSF1 modulates PTPMT1 alternative splicing to regulate lung cancer cell radioresistance. EBioMedicine. 2018;38:113–26. doi: 10.1016/j.ebiom.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardinali B, Cappella M, Provenzano C, Garcia-Manteiga JM, Lazarevic D, Cittaro D, Martelli F, Falcone G. MicroRNA-222 regulates muscle alternative splicing through Rbm24 during differentiation of skeletal muscle cells. Cell Death Dis. 2016;7:e2086. doi: 10.1038/cddis.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith PY, Delay C, Girard J, Papon MA, Planel E, Sergeant N, Buee L, Hebert SS. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum Mol Genet. 2011;20:4016–24. doi: 10.1093/hmg/ddr330. [DOI] [PubMed] [Google Scholar]
- 26.Zhang BW, Cai HF, Wei XF, Sun JJ, Lan XY, Lei CZ, Lin FP, Qi XL, Plath M, Chen H. miR-30-5p regulates muscle differentiation and alternative splicing of muscle-related genes by targeting MBNL. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meseguer S, Mudduluru G, Escamilla JM, Allgayer H, Barettino D. MicroRNAs-10a and -10b contribute to retinoic acid-induced differentiation of neuroblastoma cells and target the alternative splicing regulatory factor SFRS1 (SF2/ASF) J Biol Chem. 2011;286:4150–64. doi: 10.1074/jbc.M110.167817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z, Zhang Y, Tang T, Zhu Q, Shi W, Yin X, Xing Y, Shen Y, Pan Y, Jin L. Transcriptome profiling of panc-1 spheroid cells with pancreatic cancer stem cells properties cultured by a novel 3D semi-solid system. Cell Physiol Biochem. 2018;47:2109–25. doi: 10.1159/000491479. [DOI] [PubMed] [Google Scholar]
- 29.De Luca A, Carotenuto A, Rachiglio A, Gallo M, Maiello MR, Aldinucci D, Pinto A, Normanno N. The role of the EGFR signaling in tumor microenvironment. J Cell Physiol. 2008;214:559–67. doi: 10.1002/jcp.21260. [DOI] [PubMed] [Google Scholar]
- 30.Wan L, Yu W, Shen E, Sun W, Liu Y, Kong J, Wu Y, Han F, Zhang L, Yu T, Zhou Y, Xie S, Xu E, Zhang H, Lai M. SRSF6-regulated alternative splicing that promotes tumour progression offers a therapy target for colorectal cancer. Gut. 2019;68:118–29. doi: 10.1136/gutjnl-2017-314983. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Meng F, Wang J, Liu M, Yang G, Song R, Zheng T, Liang Y, Zhang S, Yin D, Wang J, Yang H, Pan S, Sun B, Han J, Sun J, Lan Y, Wang Y, Liu X, Zhu M, Cui Y, Zhang B, Wu D, Liang S, Liu Y, Song X, Lu Z, Yang J, Li M, Liu L. A novel oxoglutarate dehydrogenase-like mediated miR-214/TWIST1 negative feedback loop inhibits pancreatic cancer growth and metastasis. Clin Cancer Res. 2019;25:5407–5421. doi: 10.1158/1078-0432.CCR-18-4113. [DOI] [PubMed] [Google Scholar]
- 32.Sen T, Sen N, Noordhuis MG, Ravi R, Wu TC, Ha PK, Sidransky D, Hoque MO. OGDHL is a modifier of AKT-dependent signaling and NF-kappaB function. PLoS One. 2012;7:e48770. doi: 10.1371/journal.pone.0048770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang W, Huang Y, Gu J, Zhang J, Yang J, Liu S, Xie C, Fan Y, Wang H. miR-23a-5p inhibits cell proliferation and invasion in pancreatic ductal adenocarcinoma by suppressing ECM1 expression. Am J Transl Res. 2019;11:2983–94. [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Jia W, Li J. ECM1 promotes migration and invasion of hepatocellular carcinoma by inducing epithelial-mesenchymal transition. World J Surg Oncol. 2016;14:195. doi: 10.1186/s12957-016-0952-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonano VI, Oltean S, Garcia-Blanco MA. A protocol for imaging alternative splicing regulation in vivo using fluorescence reporters in transgenic mice. Nat Protoc. 2007;2:2166–81. doi: 10.1038/nprot.2007.292. [DOI] [PubMed] [Google Scholar]
- 36.Michlewski G, Guil S, Semple CA, Caceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32:383–93. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z, Chen JS, Wen JK, Gao HT, Zheng B, Qu CB, Liu KL, Zhang ML, Gu JF, Li JD, Zhang YP, Li W, Wang XL, Zhang Y. Silencing of miR-193a-5p increases the chemosensitivity of prostate cancer cells to docetaxel. J Exp Clin Cancer Res. 2017;36:178. doi: 10.1186/s13046-017-0649-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang JT, Wang ZH. Role of miR-193a-5p in the proliferation and apoptosis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:7233–9. doi: 10.26355/eurrev_201811_16257. [DOI] [PubMed] [Google Scholar]
- 40.Xu Z, Yu YQ, Ge YZ, Zhu JG, Zhu M, Zhao YC, Xu LW, Yang XB, Geng LG, Dou QL, Jia RP. MicroRNA expression profiles in muscle-invasive bladder cancer: identification of a four-microRNA signature associated with patient survival. Tumour Biol. 2015;36:8159–66. doi: 10.1007/s13277-015-3559-z. [DOI] [PubMed] [Google Scholar]
- 41.Xie F, Hosany S, Zhong S, Jiang Y, Zhang F, Lin L, Wang X, Gao S, Hu X. MicroRNA-193a inhibits breast cancer proliferation and metastasis by downregulating WT1. PLoS One. 2017;12:e0185565. doi: 10.1371/journal.pone.0185565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang P, Ji DB, Han HB, Shi YF, Du CZ, Gu J. Downregulation of miR-193a-5p correlates with lymph node metastasis and poor prognosis in colorectal cancer. World J Gastroenterol. 2014;20:12241–8. doi: 10.3748/wjg.v20.i34.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin L, Li H, Wang J, Lin D, Yin K, Lin L, Lin Z, Lin G, Wang H, Ying X, Wang L, Zhang Y, Teng L. MicroRNA-193a-5p exerts a tumor suppressor role in glioblastoma via modulating NOVA1. J Cell Biochem. 2019;120:6188–97. doi: 10.1002/jcb.27906. [DOI] [PubMed] [Google Scholar]
- 44.Yu T, Li J, Yan M, Liu L, Lin H, Zhao F, Sun L, Zhang Y, Cui Y, Zhang F, Li J, He X, Yao M. MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34:413–23. doi: 10.1038/onc.2013.574. [DOI] [PubMed] [Google Scholar]
- 45.Pu Y, Zhao F, Cai W, Meng X, Li Y, Cai S. MiR-193a-3p and miR-193a-5p suppress the metastasis of human osteosarcoma cells by down-regulating Rab27B and SRR, respectively. Clin Exp Metastasis. 2016;33:359–72. doi: 10.1007/s10585-016-9783-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang C, Dai CY, Mei Z, Jiang MJ, Gu DN, Huang Q, Tian L. microRNA-193a stimulates pancreatic cancer cell repopulation and metastasis through modulating TGF-beta2/TGF-betaRIII signalings. J Exp Clin Cancer Res. 2018;37:25. doi: 10.1186/s13046-018-0697-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen ZM, Yu Q, Chen G, Tang RX, Luo DZ, Dang YW, Wei DM. MiR-193a-3p inhibits pancreatic ductal adenocarcinoma cell proliferation by targeting CCND1. Cancer Manag Res. 2019;11:4825–37. doi: 10.2147/CMAR.S199257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anko ML. Regulation of gene expression programmes by serine-arginine rich splicing factors. Semin Cell Dev Biol. 2014;32:11–21. doi: 10.1016/j.semcdb.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 50.Jensen MA, Wilkinson JE, Krainer AR. Splicing factor SRSF6 promotes hyperplasia of sensitized skin. Nat Struct Mol Biol. 2014;21:189–97. doi: 10.1038/nsmb.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen-Eliav M, Golan-Gerstl R, Siegfried Z, Andersen CL, Thorsen K, Orntoft TF, Mu D, Karni R. The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers. J Pathol. 2013;229:630–9. doi: 10.1002/path.4129. [DOI] [PubMed] [Google Scholar]
- 52.Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M Jr, Fu XD. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell. 2013;50:223–35. doi: 10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–48. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24:653–8. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pillman KA, Phillips CA, Roslan S, Toubia J, Dredge BK, Bert AG, Lumb R, Neumann DP, Li X, Conn SJ, Liu D, Bracken CP, Lawrence DM, Stylianou N, Schreiber AW, Tilley WD, Hollier BG, Khew-Goodall Y, Selth LA, Goodall GJ, Gregory PA. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. EMBO J. 2018;37 doi: 10.15252/embj.201899016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Z, Li W, Pang Y, Zhou Z, Liu S, Cheng K, Qin Q, Jia Y, Liu S. SF3B4 is regulated by microRNA-133b and promotes cell proliferation and metastasis in hepatocellular carcinoma. EBioMedicine. 2018;38:57–68. doi: 10.1016/j.ebiom.2018.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.