Abstract
The activity of antimicrobial peptides (AMPs) has been investigated extensively using model membranes composed of phospholipids or lipopolysaccharides in aqueous environments. However, from a biophysical perspective, there is a large scientific interest regarding the direct interaction of membrane-active peptides with whole bacteria. Working with living bacteria limits the usability of experimental setups and the interpretation of the resulting data because of safety risks and the overlap of active and passive effects induced by AMPs. We killed or inactivated metabolic-active bacteria using γ-irradiation or sodium azide, respectively. Microscopy, flow cytometry, and SYTOX green assays showed that the cell envelope remained intact to a high degree at the minimal bactericidal dose. Furthermore, the tumor-necrosis-factor-α-inducing activity of the lipopolysaccharides and the chemical lipid composition was unchanged. Determining the binding capacity of AMPs to the bacterial cell envelope by calorimetry is difficult because of an overlapping of the binding heat and metabolic activities of the bacteria-induced by the AMPs. The inactivation of all active processes helps to decipher the complex thermodynamic information. From the isothermal titration calorimetry (ITC) results, we propose that the bacterial membrane potential (Δψ) is possibly an underestimated modulator of the AMP activity. The negative surface charge of the outer leaflet of the outer membrane of Gram-negative bacteria is already neutralized by peptide concentrations below the minimal inhibitory concentration. This proves that peptide aggregation on the bacterial membrane surface plays a decisive role in the degree of antimicrobial activity. This will not only enable many biophysical approaches for the investigation between bacteria and membrane-active peptides in the future but will also make it possible to compare biophysical parameters of active and inactive bacteria. This opens up new possibilities to better understand the active and passive interaction processes between AMPs and bacteria.
Significance
Bacterial resistance to drugs makes a reliable treatment of infectious diseases difficult. Because we are running out of treatment options, it is extremely important to have a method portfolio that enables the investigation of molecular mechanisms in microbial systems, in particular, bacterial membrane systems. Here, we inactivated bacteria to switch off metabolic and cell division processes. Thus, the biophysical interaction of AMPs can be deciphered directly on the bacterial outer membrane by ITC. We present a way to bridge the gap between molecular and cellular investigation methods. Related disciplines can benefit from our strategy as the bacteria are inactivated, but modification of the bacterial cell envelope is reduced. At the same time, biosafety problems are minimized.
Introduction
The misuse and overuse of antibiotics for the treatment of prevalent diseases leads to the increasing proliferation of bacteria that are no longer susceptible to these drugs (1). Therefore, the development of bioactive molecules with a novel mechanism of action has critical priority (2,3). This need is particularly urgent in the case of drugs targeting Gram-negative bacteria (4). In this context, antimicrobial peptides (AMPs) are among the most promising candidates (5,6) because of their broad spectrum of antimicrobial activity and low propensity to induce resistance. AMPs are present in virtually every life form in which they play an active role in the clearance of infections (7). Compared to other antimicrobials (i.e., cephalosporins), bacteria are less likely to become resistant to AMPs because they interact with multiple targets on the bacterial envelope and in other cell compartments (8, 9, 10, 11). Most AMPs feature high affinity to lipopolysaccharides (LPSs) and lipoproteins (LPs) in particular. This facilitates peptide binding and insertion into the bacterial outer membrane (12). After reaching a critical peptide concentration, the bacterial membrane starts to disintegrate and permeabilizes. The latter event is almost instantaneously lethal for the pathogen. At subinhibitory levels, many AMPs can still permeabilize membranes and enhance the activity of a coadministered antibiotic (13). Interestingly, the ability of AMPs to bind to LPSs and LPs explains their therapeutic efficiency against endotoxin-mediated pathologies such as sepsis and septic shock (6,7,14,15). In addition, there are also AMPs with immunomodulatory activities. Those peptides form a class of their own and are known as host defense peptides because they comprise a first defense barrier of the native immune system and are involved in the neutralization of the damage-associated molecular patterns (16).
The initial interaction of AMPs with the membrane and the subsequent lipid displacement is a process governed by electrostatic forces and topological parameters (5,17,18). The elucidation of these mechanisms was based on physicochemical investigations performed with model membranes. However, this methodology only offers a simplified view of the peptide-membrane interaction concerning the whole bacteria situation because it neglects the high structural complexity of lipids present in the membrane and the contribution of membrane-associated proteins (19,20). Therefore, the degree of applicability of these studies to the real cell situation is controversial (21). On the other hand, experiments with viable bacterial cells are confronted with two major problems: 1) working with live pathogenic bacteria entails safety risks, and 2) the metabolic activity and cell division process can hinder or even prevent an accurate assessment of the AMP-membrane interaction. There are certain bacteria that express AMP-specific efflux pump systems to which the peptides bind and are expelled from the cell (22). Moreover, in thermodynamic investigations, binding events are overlaid by active processes of bacterial metabolism. Therefore, killing or metabolic inactivation of bacteria before peptide exposure can be a way to decipher passive and active effects of the interaction between AMPs and bacterial components. Killing of bacteria by classical antibiotics or heat has strong effects on the bacteria and is therefore not suitable. Here, we present results demonstrating that γ-irradiation and sodium azide (NaN3) are helpful tools to inactivate bacteria for biophysical experiments by killing or inhibition of the metabolic activity, respectively.
For this study, two peptides were selected: 1) LL-32, a fragment of the human endogenous peptide LL-37, comprising amino acid residues 1–32 but lacking the unstructured C-terminal part of its parental peptide. LL-37 is expressed in numerous immune cells and tissues and is used as a reference for antimicrobial, immunomodulatory and anti-endotoxin activities (6,23,24). Compared to its parental peptide, LL-32 has enhanced antimicrobial activity against Gram-positive and Gram-negative bacteria. This was correlated with its ability to form a complete α-helix upon interaction on the membrane interface by peptide intercalation (25, 26, 27). 2) Polymyxin B (PMB), the best-characterized AMP from Paenibacillus polymyxa, is endowed with potent bactericidal activity against Gram-negative bacteria (28, 29, 30, 31, 32). The mechanism by which PMB binds to LPS and neutralizes the proinflammatory activity of this molecule has been analyzed in great detail. However, the clinical use of PMB for treatment of bacterial infections and sepsis is restricted because of its nephrologic and neurologic side effects (29).
Here, we present a method to kill and metabolically inactivate bacteria while minimizing cell alteration. Moreover, we performed a detailed characterization of the interaction of PMB and LL32 with bacterial cells previously inactivated according to this method. Specifically, this procedure relies on the exposure of viable bacteria to γ-irradiation or sodium azide (NaN3). The radionuclide 137Cs emits γ-photons (0.662 MeV) with sufficient energy to initiate double-strand breaks (DSBs) and to induce the production of reactive oxygen species (ROS) that are lethal to bacteria (33). In contrast to ultraviolet rays, γ-photons are not shielded by water and, therefore, can kill bacteria in suspension efficiently. NaN3 inhibits ATP production by ATP synthase and its reverse operation by lowering the catalytic cooperativity of the ATP synthase machinery (34). We demonstrated that this approach preserves cell ultrastructure. Hence, we expect that our findings will allow conducting biochemical and biophysical studies on the interaction of antimicrobial agents with bacteria under conditions closer to the real cell situation. In addition to stopping bacterial metabolic activity, the γ-radiation kills the bacteria when using sufficient doses, and therefore bacteria of high biological safety level can then be used in conventional biophysical experiments.
Materials and Methods
Bacteria, reagents, and peptide synthesis
Bacterial strains were obtained from our internal reference strain collection. A panel of five Gram-negative strains were used in this study: 1) three Gram-negative LPS deep-rough mutant strains (Salmonella enterica sv Minnesota R595, Escherichia coli Re-mutant WBB01, and Proteus mirabilis R45), 2) three Gram-negative strains with complete LPS core oligosaccharides (S. enterica sv Minnesota R60 (S. enterica R60), E. coli ATCC 23716, and Klebsiella pneumoniae extended spectrum β-lactamase-resistant (ESBL)), and 3) the clinical isolate Staphylococcus aureus wild-type (WT) SA113 as a Gram-positive representative strain.
Fluorescent dyes, 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionic acid, succinimidyl ester (BODIPY FL, SE) and succinimidyl 6-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoate (NBD-X, SE) were purchased from Molecular Probes (Eugene, OR) and were used for conjugation to PMB and LL-32 peptides at the N-terminal end, respectively. SYTOX green nucleic acid stain was bought from Thermo Fisher Scientific, ref. S7020 (Waltham, MA). All other chemicals were analytical grade reagents and acquired from Merck (Darmstadt, Germany).
The amidated C-terminus peptide LL-32 was synthesized by solid-phase peptide synthesis in an automatic peptide synthesizer (model 433 A; Applied Biosystems, Foster City, CA) according to the FastMoc synthesis protocol of the manufacturer (35), including the removal of the N-terminal Fmoc group. In the case of fluorescent labeled LL-32 peptide (NBDLL-32), NBD-X, SE was attached at the N-terminal position in the last step of the condensation reaction. The amide at the C-terminus was synthesized on a Fmoc-amide resin. The peptides were deprotected and cleaved from the resin with 90% trifluoroacetic acid, 5% anisole, 2% thioanisole and 3% dithiothreitol for 3 h at room temperature (RT). After cleavage, the suspension was filtered, and the soluble peptides were precipitated with ice-cold diethyl ether, followed by centrifugation and extensive washing with cold ether. PMB was purchased in its sulfate salt form from Sigma (Deisenhofen, Germany) and was used for synthesizing BODIPYPMB. Briefly, a concentrated solution of 10 mg/mL of PMB was prepared in 0.1 M of sodium hydrogen carbonate (NaHCO3) buffer (pH 10.0). In parallel, BODIPY FL, SE (5 mg) was dissolved in 0.5 mL of dimethylformamide (DMF). The aqueous PMB solution was mixed with the organic BODIPY ester solution (5:1 v/v). An additional 0.5 mL of DMF was introduced, and the reaction was incubated for 1.5 h at RT with constant shaking at 150 rpm/min. This solution was diluted with ultrapure water in a volumetric ratio of 1:2.
Peptides were purified by RP-HPLC at 214 nm, using an Aqua-C18 column (Phenomenex, Aschaffenburg, Germany). The elution was performed with a gradient of 0–70% acetonitrile in 0.1% trifluoroacetic acid. Purity levels of up to 98% were checked by matrix-assisted laser desorption ionization-time of flight mass spectrometry.
Kinetic of killing bacteria by γ-radiation and mononuclear cell stimulation
Bacterial suspensions (1.0 × 108 CFU/mL) in phosphate-buffered saline (PBS) were used for this assay. 20 μL of bacterial suspension was plated (control) on Luria-Bertani (LB) agar, and the rest was subjected to γ-irradiation on ice (0°C). The samples were positioned into the irradiation chamber of a BIOBEAM 8000 (Gamma-Service Medical GmbH, Leipzig, Deutschland). The instrument is equipped with 137Cs γ-source (E γ = 0.662 MeV) with a half-life time of 30.17 years and an activity of 81.4 TBq. The γ-irradiation was performed with an energy dose rate of 5.5 Gy/min. Every 15 min, 20 μL of the bacterial suspension was plated on LB agar. Three independent experiments were performed to determine the killing efficacy of bacteria (minimal bactericidal dose) by counting colonies after incubation.
To analyze the effect of the bacterial membrane on the tumor necrosis factor (TNF)-α release in vitro, peripheral blood mononuclear cells (PBMCs) isolated from heparinized blood of healthy donors were resuspended in medium (RPMI 1640 with 2% PS/L-glutamine and 4% AB-serum), and their number was equilibrated at 5.0 × 106 PBMCs/mL. For stimulation, 200 μL PBMCs (1.0 × 106 cells) were transferred into each well of a 96-well culture plate and incubated with untreated (control), heat-killed, and γ-irradiated (1000 and 8000 Gy) S. enterica R60 (from 1.0 × 102 to 1.0 × 107 bacteria/well) for 5 h at 37°C with 5% of CO2. Cell-free supernatants were collected after centrifugation of the culture plates for 10 min at 400 × g and stored at −20°C until immunological determination. TNF-α was monitored with a sandwich enzyme-linked immunosorbent assay using a monoclonal antibody against TNF-α (BD Biosciences, Heidelberg, Germany) as described previously in detail (36).
Ultrastructural alterations on the bacterial membrane
Aliquots of 2 mL of the stock bacterial suspension (1.0 × 109 CFU/mL) of S. enterica R60, ESBL K. pneumoniae, and S. aureus were heat-killed (submerged in 100 mL of boiling water for 10 min) or γ-irradiated (1000 and 8000 Gy). 500 μL of the bacterial suspensions were incubated alone (control) or with LL-32 (final peptide concentration: 128 and 64 μg/mL) or PMB (final peptide concentration: 32 and 16 μg/mL) at 37°C for 30 min at 150 rpm/min.
Morphological alterations of the bacterial cell surface caused by γ-irradiation or by peptide interactions were imaged by atomic force microscopy (AFM) or transmission electron microscopy (TEM). For AFM, bacteria were placed on mica, and the excess of liquid was removed with filter paper and then air-dried at RT for 24 h. After rinsing three times with 1 mL of water and air-drying overnight, images were recorded with an MFP-3D (Asylum Research, Santa Barbara, CA) in contact mode with a CSG 11 cantilever (κ = 0.1 N/m; NT-MDT; Llandderfel, Bala, Gwynedd, UK). Frequencies of 1.0 Hz and 512 points and lines and the set point were always adjusted to guarantee that minimal forces were applied to the sample. Further image processing (flattening and plane fitting) was done with the MFP-3D software under IGOR Pro (Lake Oswego, OR). Images shown here are representative of the respective samples.
For TEM, samples were fixed using 2% osmium tetroxide during 1.5 h. After fixation, the samples were rinsed three times with distilled water. For the positive stain, a new fixation was carried out with a 2% aqueous uranyl-acetate solution (1 h) and rinsed again. Then, the material was dehydrated in a series of ethanol washes (30, 50, 70, 90, and 100%), each for 15 min. The material was transferred two times to propylene oxide for 15 min. All procedures were carried out at RT. Infiltration of the samples was done overnight in a 1:1 propylene oxide-Epon-resin-mix, at 4°C. Then, the samples were embedded in Epon-resin, and polymerization was carried out at 60°C overnight. Ultrathin-sectioning (80–100 nm) was performed with a diamond knife, using a pyramytom. The slices were placed on a copper grid (200 squares) and counterstained with lead citrate. Pictures were taken with the EM Zeiss 910. Images shown are representative for the respective sample.
Analysis of bacterial membrane integrity and potential by flow cytometry
To confirm the integrity of the bacterial membranes and the adsorption of PMB and LL-32, we used flow cytometry. S. enterica R60 bacterial suspension (1.0 × 107 CFU/mL in 20 mM HEPES (pH 7.4) buffer + 150 mM NaCl) was incubated in ice-cold PBS in the absence or presence of the indicated concentrations of PMB, LL-32, or dye-conjugated PMB (BODIPYPMB) or LL-32 (NBDLL-32) for 1 h. Subsequently, bacteria were centrifuged (800 × g, 10 min), and pellets were resuspended in ice-cold PBS and washed twice. Flow cytometry analysis was performed on a FACSCantoTMII, (BD Bioscience). Data were analyzed by FCS Express 4 Flow Cytometry software (DeNovo Software).
Flow cytometric measurement of membrane potential (ΔΨ) in bacteria can be performed using diethyloxacarbocyanine (DiOC2 (3)) in PBS buffer, which is described within the BacLight Bacterial Membrane Potential Kit (B34950; Molecular Probes) and in the literature (37, 38, 39). Briefly, the method is based on the fluorescence shift by DiOC2 (3) toward red emission (from green fluorescence in all bacterial cell walls) due to the self-association of the dye molecules at higher cytosolic concentrations caused by larger membrane potential. Green fluorescence was collected using a 530/30 bandpass filter, and the red emission was measured using a 600 nm long-pass filter (both filters: AHF Analysentechnik AG, Tübingen, Germany). The proton ionophore carbonylcyanide 3-chlorophenylhydrazone (CCCP) increases the proton permeability of the membrane, thereby dissipating both the transmembrane proton gradient (ΔpH) and ΔΨ. Therefore, it is used as a negative control in bacterial suspensions. 1 mL S. enterica R60 bacterial suspension (1.0 × 106 CFU/mL of viable, NaN3–treated, and γ-irradiated bacteria; 1000 Gy) were exposed with and without CCCP (5 μM) for 60 min, then DiOC2 (3) (10 μM) was added. After incubation for 4 min at RT in the dark, the green and red emissions of the bacteria were measured for 70–100 s. To optimize the DiOC2 (3) permeability by the outer membrane, we used EDTA (1 mM). Analysis of at least 20,000 measured bacteria (forward scatter channel (FSC)/side scatter channel (SSC) dot blot and doublets excluded) were used to calculate the membrane potential by using the red/green ratio of the fluorescence intensities on a BD LSRII flow cytometer (Becton Dickinson, Franklin Lakes, NJ). The mean fluorescence intensity (MFI) values of the red population were divided by the green population MFI after the calculation of the difference of the ratio (red/green) between samples with and without CCCP. Data analysis was performed utilizing the FCSExpress5 program (DeNovo Software).
Influence of γ-irradiation on the phospholipid profile of S. enterica sv. Minnesota R60
A bacterial S. enterica R60 suspension in the middle-exponential growth phase was washed three times and adjusted to a concentration of 1.0 × 108 CFU/mL in buffer (20 mM HEPES (pH 7.4) + 150 mM NaCl). Aliquots of 1 mL were γ-irradiated with energy doses of 500, 1000, 1500, 2000, and 8000 Gy and compared with the non-γ-irradiated samples. After the γ-irradiation, the bacterial suspension was centrifuged at 13,200 rpm. A total pellet of 25 μL was mixed with 270 μL of CH3OH (mass spectrometry-liquid chromatography (MS-LC) grade solvents from Fluka chemicals; Buchs, Switzerland) with a glass tip. 35 μL of water was added to all samples, and subsequently, the samples were rigorously mixed. Then, 1 mL methyl tert-butyl ether was added (40,41). The mixture was incubated for 1 h at RT with rigorous agitation. Afterward, 250 μL water was added to induce phase separation, and the mixture was centrifuged at maximum speed for 2 min at 13,400 × g using a tabletop centrifuge. Then, 800 μL of the upper organic phase was collected into a new vial (2 mL size, Eppendorf). The organic phase was dried and taken up in 100 μL storage solution.
For lipidomics measurements, the organic phase was diluted by a factor of 20 in the MS-Mix consisting of chloroform/methanol/isopropanol (v/v/v; 1/2/4) with 2.9 mM ammonium acetate addition for measurements in both ion modes. All mass spectrometric measurements were performed on a Q Exactive Plus (Thermo, Bremen, Germany) equipped with a Triversa Nanomate (Advion, Ithaca, NY) as reported earlier (42,43). Lipids were identified using the accurate mass of precursor ions as well as tandem mass spectrometry (MS/MS) information of fatty acids in the negative mode using LipidXplorer (44). Quantities were determined using 1,2-di-O-phytanyl-sn-glycero-3-phosphoethanolamine (999985P; Avanti Polar Lipids, Alabaster, AL) as internal standard using MS1 intensities for normalization (41,45).
Analysis of the bacterial membrane-peptide interaction by ITC
S. enterica R60 bacteria were grown in LB broth at 37°C until the middle-exponential phase. 8 mL of culture was centrifuged at 4600 rpm and resuspended in the same volume of 20 mM HEPES (pH 7.4) buffer. The bacterial suspension was γ-irradiated with an energy dose of 1000 Gy. The suspension was pelleted and washed carefully three times more with previously degasified sterile HEPES buffer to remove growth medium components and to reduce air bubbles. After this procedure, the concentration of bacteria was carefully adjusted to a density of 1.0 × 109 CFU/mL with the same buffer.
Microcalorimetry measurements of PMB and LL-32 binding to the bacteria were performed on an MCS isothermal titration calorimeter (MicroCal, Northampton, MA) at 37°C. Bacterial samples were filled into the microcalorimetry cell (1500 μL). After thorough degassing of the peptide samples (1 mM peptides in sterile filter 20 mM HEPES (pH 7.4)), these were placed into the syringe (140 μL). After thermal equilibration, aliquots of 6 μL of peptide solution were injected every 4 min into the bacteria-containing cell. During each injection, the enthalpy change was measured by the instrument, and the area underneath each injection peak was integrated (Origin; MicroCal) and plotted versus peptide molecules/bacteria ratio. Because the instrument works in temperature equilibrium at a constant “current feedback,” an exothermic reaction leads to a lowering of this current and an endothermic reaction to an increase. As a control, PMB or LL-32 were titrated into HEPES buffer, and the minor endothermic reaction due to dilution was subtracted from the plotted curves. Three independent experiments were done.
Peptide susceptibility testing of the bacterial cell envelope
We used the SYTOX green nucleic acid stain to evaluate the integrity and peptide susceptibility of the bacterial cell envelope. Binding of SYTOX green stain to nucleic acids in the case of the permeable bacterial membrane results in an enhancement in green fluorescence emission (46). For this assay, we used 1 mL of 1) untreated, 2) metabolically inhibited (with 0.1% of sodium azide NaN3), or 3) γ-irradiated S. enterica R60 irradiated with an energy dose of 1000 Gy. Bacterial suspensions were prepared at a concentration of 1.0 × 108 CFU/mL in 20 mM HEPES buffer + 150 mM NaCl. As a positive control, a bacterial suspension was submerged in 100 mL of boiling water for 10 and 2 min sonification to induce 100% lysis.
Serial twofold dilutions of LL-32 (from 128 μg/mL) or PMB (from 32 μg/mL) were made in the same buffer in 96-well U-bottom polystyrene microtiter plates. 80 μL of each peptide was incubated during 60 min with 10 μL of 1) untreated, 2) metabolically inhibited, or 3) γ-irradiated bacterial suspensions and 10 μL of a stock SYTOX green solution (50 μM of SYTOX green stain). The fraction of permeabilized cells in the population was quantified by fluorometry. Kinetics of the fluorescence emission was measured with a Tecan Infinite PRO 200M fluorescence spectrophotometer (Tecan, Männedorf, Switzerland) every 5 min at 37°C. SYTOX green stain was excited at 480 nm, and the intensity of the emission light was detected at 530 nm. Three independent experiments were performed in duplicates.
Results and Discussion
Effect of γ-irradiation on the bacterial survival
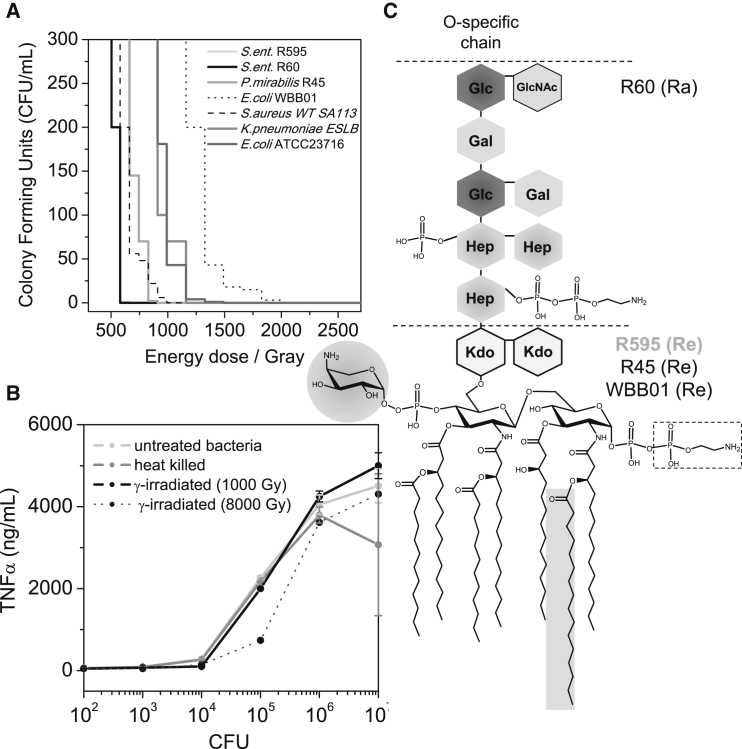
To test the susceptibility of viable bacteria to ionizing radiation (γ-irradiation), we selected bacteria expressing LPS chemotypes that are commonly used for peptide structure and activity studies (Fig. 1 C). The bacterial killing induced by γ-irradiation was apparent at minimal bactericidal doses ranging from 400 to 2000 Gy, although the lethality varies greatly depending on the individual strain (Fig. 1 A). Interestingly, for S. enterica R60 and R595, only low doses were required to achieve killing, and their responses to γ-irradiation were identical. This was in contrast to E. coli Re-mutant WBB01 (E. coli WBB01), which displayed the highest level of resistance to γ-irradiation (Fig. 1 A). The opposite is true for the Gram-positive strain S. aureus WT ESBL, which was significantly more sensitive than the Gram-negative strains P. mirabilis R45, K. pneumoniae ESBL, and E. coli ATCC 23716. From this, we conclude that the killing by γ-irradiation is independent of the respective cell envelope. Further, we could not draw any correlation between the sensitivity to ionizing radiation and the length of the LPS carbohydrate moiety of the Gram-negative strains tested (Fig. 1 C).
Figure 1.
Bacterial survival and mononuclear cell stimulation. (A) A total volume of 1 mL of 1.0 × 108 CFU/mL was γ-irradiated at different energy doses and then plated on LB agar. Survival curves obtained are representative for three independent experiments. (B) Stimulation of hMNCs (1.0 × 106/well) by untreated (control), heat-killed, and γ-irradiated (doses 1000 or 8000 Gy) S. enterica R60 (1.0 × 102 until 1.0 × 107 bacteria/well) cells for 1 h at 37°C. The γ-irradiation was performed at ∼5.5 Gy/min at 0°C. The mean value of the TNF-α secretion of three independent experiments is shown together with error bars representing the standard deviation (s.d.). (C) Schematic chemical structures of different LPS rough strains. The phosphate residue at the second heptose and the 2-aminoethyl diphosphate residues at the first heptose are only present in the strain R60. The LPS structures of S. enterica R595, P. mirabilis R45, and E. coli Re-mutant WBB01 differ mostly in the degree of amino-arabinose linked to the 4′-phosphate of glucosamine II (gray shaded circle), which leads to a reduction of the negative charge. 50% of the LPS R45 structure contains amino-arabinose at the carboxy group of the first 3-deoxy-D-manno-oct-2-ulosonic acid sugar. Furthermore, strain R595 has a nonstoichiometric acyloxyacyl chain C-16 in position 2 of the glucosamine I backbone. The bacterial chemotype is given in parentheses.
Gamma irradiation exert their bactericidal activity through two different mechanisms: 1) directly, by hitting DNA; or 2) indirectly, by interacting with water and generating ROS. In the indirect mechanism, ROS-induced protein oxidation is the main bactericidal process, which is counteracted by high ratios of [Mn2+]/[Fe2+] (33,47). Thus, we hypothesize that low intracellular levels of mangan (II) can be derived from the high sensitivity of the LPS rough mutant strain S. enterica R60. However, other factors, such as the efficiency of DNA repair mechanisms, affect bacterial resistance to radiation (48). We achieved 100% killing at 600 Gy (∼110 min of irradiation time) equivalent to six DSBs (Fig. 1 A; (33,47)), but just one DSB can be lethal for one bacterium. Therefore, our results show that the sensitivity to γ-irradiation is traced back to a multifaceted process including oxidative stress, the level of antioxidants, and bacterial DNA repair activity.
To indirectly assess the molecular integrity of bacterial pathogen-associated molecular patterns after γ-irradiation, we measured the release of TNF-α after the stimulation of human mononuclear cells (hMNCs) with γ-irradiated S. enterica R60 cells. In these experiments, the LPS Ra chemotype was used as model endotoxin (49). Upon the addition of untreated, heat-killed, and γ-irradiated bacteria, we observed a biomass-dependent increase of TNF-α production by hMNCs (Fig. 1 B). Identical TNF-α secretion levels were induced by untreated and γ-irradiated bacteria (1000 Gy), demonstrating that the molecular patterns recognized by hMNCs after γ-irradiation were retained. High γ-irradiation doses (8000 Gy) caused a slight reduction of TNF-α production, which is indicative of a γ-irradiation-induced chemical modification of those lipids’ A sections interacting with hMNC receptors (Fig. 1 B; (50,51)).
Ultrastructural alterations of the bacterial cell envelope caused by γ-irradiation
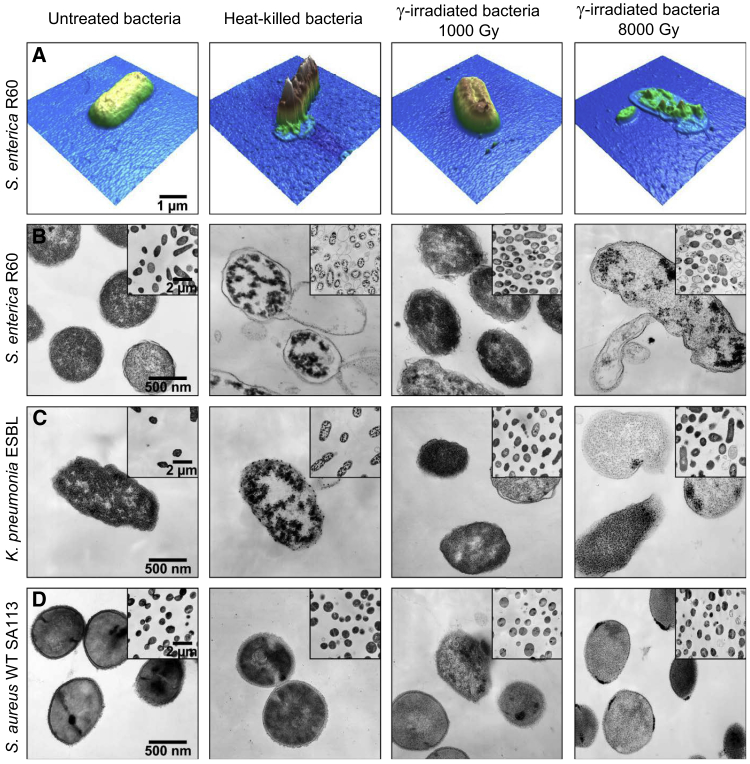
We hypothesize that oxidative stress could promote lipid or membrane protein modifications of the bacterial membrane, leading to significant alterations of its fluidity (52), or even its morphology. To evaluate potential membrane alterations, γ-irradiated cells were visualized by both AFM and TEM (Fig. 2). High-resolution images taken by AFM revealed only minor changes in the cell surface of γ-irradiated S. enterica R60. Specifically, a dose of 1000 Gy caused a slight increase in the surface roughness of γ-irradiated bacteria compared to untreated bacteria. In contrast, bacteria subjected to heat (95°C, 10 min) or γ-irradiated with 8000 Gy appeared severely damaged (Fig. 2 A). In fact, the surface of heat-killed bacteria presented large jagged-like protrusions associated with loss of the cytoplasm content, whereas cells receiving a γ-ray dose of 8000 Gy were totally distorted and destroyed. We confirmed these specific membrane alterations by TEM. Even a slight increase of the electron density was observed for γ-irradiated bacteria (energy dose 1000 Gy). These effects were dramatic for organisms exposed to either heat or a high dose (8000 Gy) of γ-rays as compared to the control (Fig. 2 B). The heat or high-gamma-dose treatments induced the formation of high electron-dense clusters in the cytoplasm and vesicles, blebs, and finger-like extensions. These severe morphological alterations are indicative for cell lysis and the release of intracellular content to the surrounding medium.
Figure 2.
Ultrastructural alterations of the bacterial membrane. (A) AFM images of S. enterica R60. The images were taken in contact mode. TEM images of (B) S. enterica R60, (C) K. pneumonia ESBL, and (D) S. aureus WT SA113. Columns from left to right: untreated bacteria (untreated), heat-killed for 10 min at 100°C, and γ-inactivated with a dose of 1000 and 8000 Gy, respectively. The figures shown are representative of three independent experiments. To see this figure in color, go online.
To answer the question of whether these findings could be extrapolated to other bacteria or whether they are specific to S. enterica R60, we extended our characterization to two different bacterial strains (Fig. 2, C and D). Although K. pneumonia ESBL cells exposed to 1000 Gy of γ-irradiation had similar morphology to that of untreated controls, some of them exhibited a decrease in electron density. Like S. enterica R60, heat-treated Klebsiella cells presented electron-dense clusters in their cytoplasm, whereas cells receiving a dose of 8000 Gy appeared electron-lucent with poorly defined cell envelopes (Fig. 2 C). Finally, S. aureus WT SA 113 cells maintained their overall ultrastructure when subjected to low doses of γ rays (1000 Gy). Treatment with heat did not significantly alter the cell envelope of S. aureus. In contrast, high doses of γ rays (8000 Gy) induced the formation of electron-dense clusters of unknown nature at this bacterial cell membrane but kept the cell envelope intact.
S. enterica R60 bacterial cell envelope shows no structural impairment of the lipid membrane at the molecular level after γ-irradiation
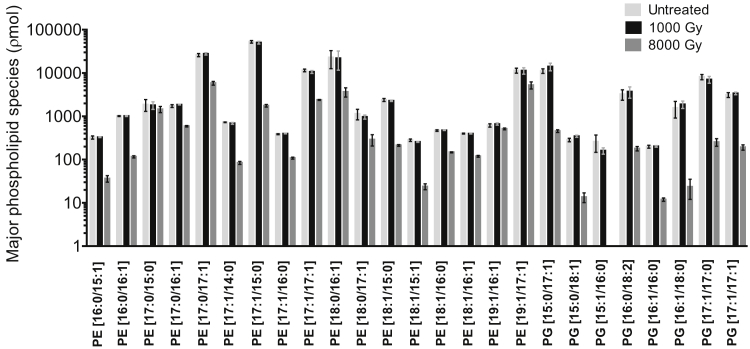
To study peptide interaction with γ-irradiated bacteria, ensuring the integrity of the cell envelope at the molecular level after inactivation is crucial. Analysis of the S. enterica R60 phospholipid profile after γ-irradiation showed no impact on membrane lipid profiles, and no peroxidation events were detectable at doses equal or lower than 1000 Gy, (Fig. 3; Fig. S1). The amounts of the most abundant phospholipids (i.e., phosphatidylethanolamine and phosphatidylglycerol) decreased in cells γ-irradiated with 8000 Gy but not in those treated with doses below 2000 Gy. These results were consistent with our previous observations using AFM and TEM (Fig. 2).
Figure 3.
Influence of γ-irradiation on the major phospholipid species in S. enterica R60. Phospholipid profiles of major abundant phosphatidylethanolamine and phosphatidylglycerol species of S. enterica R60 under two γ-irradiation doses (1000 and 8000 Gy). The quantitative determination was performed using 1,2-di-O-phytanyl-sn-glycero-3-phosphoethanolamine as internal standard and MS1 intensities for normalization. The major phospholipid species concentrations are shown with a log10 scale in ρmol. Mean ± s.d. of two independent experiments with two technical replicates shown.
In addition, when untreated, γ-irradiated (1000 Gy), and NaN3-treated bacteria were analyzed by Fourier-transform infrared spectroscopy, no significant differences in membrane fluidity were detected (Fig. S2). Two-phase transitions, deduced from the peak position of the symmetric stretching vibrational band of the methylene groups (-CH2-) in the lipid acyl chains in the range 2850–2853 cm−1, were observed and could be assigned to that of the inner membrane (∼22°C) and outer membrane (∼38°C), respectively. This strongly suggests that the low γ-irradiation dose used (1000 Gy) was not high enough to induce covalent bond breaks or to impair functionality and complexity within the phospholipid backbone. Lipid peroxidation has a strong effect on eukaryotic systems but seems to be less relevant in most bacteria because they lack polyunsaturated fatty acid moieties (53, 54, 55). These results are consistent with our observations on the preservation of the LPS Ra structure after γ-irradiation at 1000 Gy (Fig. 1, B and C).
To substantiate the finding that γ-irradiation has only minor effects on the overall structural integrity of the lipid membrane of the bacterial species tested, we analyzed untreated, γ-treated (1000 Gy), and heat-killed (95°C, 10 min) S. enterica R60 cells by flow cytometry (Fig. 4). Treatment of bacteria with heat leads to significantly enhanced signal intensities in the forward scatter (FSC-A) and strongly potentiated signals in the sideward scatter (SSC-A) when compared to untreated bacteria. In contrast, γ-irradiation did not change FSC-A or SSC-A values (Fig. 4, A and B). This suggests that the exposure to a relatively low dose of γ-radiation does not change surface properties of bacteria, including their granularity and structural complexity. These results agree with our AFM and TEM imaging experiments. Taken together, our findings indicate that the lipid part of the bacterial cell envelope does not undergo significant structural changes when exposed to a low dose (1000 Gy) of γ-irradiation. Therefore, the use of this technique to kill bacteria may allow studying the interaction of bacteria with antimicrobials under conditions closer to physiology.
Figure 4.
Effect of γ-irradiation on biophysical properties of S. enterica R60. (A) Representative density plots of untreated (1.0 × 108 CFU/mL), γ-irradiated (1000 Gy), or heat-killed (10 min; 95°C) bacteria. Bacteria were analyzed by flow cytometry on a BD FacsCantoII. Size and granularity of bacterial cultures are indicated by the forward scatter (FSC-A) and sideward scatter signal intensities (SSC-A), respectively. (B) The mean ± s.d. of FSC-A and SSC-A values of two (heat-killed) or three independent experiments are shown. (C–E) Flow cytometric measurement of ΔΨ using DiOC2 (3) analyzing differently treated bacteria (untreated, NaN3-treated, and γ-irradiated). (C) FSC-A versus SSC-A was used to gate on bacteria and exclude debris. Additionally, the red channel (RC) versus the width of SSC-W was used to exclude doublets. (D) To calculate the ΔΨ of different treated bacteria, the MFI of the RC and green channel (GC) was measured, and the RC/GC ratio was calculated as described in the Materials and Methods using CCCP treatment for depolarization as individual control for each condition. (E) The ΔΨ of viable bacteria was used as a reference to show its reduction within differently treated bacteria. Measurements were performed using PBS with or without EDTA-treated samples as recommended and described within the BacLight Bacterial Membrane Potential Kit. Calculations and figures regarding the ΔΨ are representative of two independent experiments.
Bacterial metabolism inhibition and γ-irradiation impact the bacterial membrane potential
Many cell processes such as signal transduction, bioenergetics, cell division, active transport, and membrane interactions with exogenous compounds are strongly dependent on the membrane potential (ΔΨ). Depolarization of the ΔΨ using CCCP versus nontreated samples was measured in untreated, NaN3-treated, and γ-irradiated bacteria (Fig. 4, C–E) with the membrane potential probe DiOC2 (3). NaN3 is known to produce a marked bacteriostatic action against Gram-negative bacteria (56). In flow cytometry assays, the MFI of the red and green channels (RC, GC) were measured in the absence and presence of CCCP. The difference between the RC/GC ratio in CCCP-treated and nontreated samples was positively correlated with ΔΨ (Fig. 4, C and D). Stronger GC intensity was correlated with lower RC intensity and lower ratio values, reflecting depolarization of ΔΨ. In these assays, untreated bacteria with and without CCCP treatment were used as the control and reference values, respectively. Data were calculated for NaN3-treated and γ-irradiated bacteria, and the percentage of depolarization of ΔΨ compared to untreated bacteria was calculated and plotted (Fig. 4 E). NaN3-treated bacteria showed a reduction of their ΔΨ down to 43%, the ΔΨ of γ-irradiated bacteria was reduced down to 28%, and both scenarios were compared to nontreated bacteria suspended in PBS. Repetition of the measurement in the presence of EDTA enables an easier ΔΨ depolarization with reductions in the same range: down to 36% for NaN3-treated and 43% for γ-irradiated compared to untreated bacteria. ΔΨ analyzed using DiOC2 (3) resulted in similar depolarization for bacteria treated with NaN3 or γ-irradiation.
The bacterial ΔΨ is dramatically depolarized by azide ions. Formation of ATP by the ATP synthase is undoubtedly driven by the proton motive force (Δp), which is associated with the ΔΨ and the transmembrane proton gradient (ΔpH) (57). ATP production by ATP synthase and its reverse operation (ATP-driven pumping of Na+ or H+ to the bacterial periplasm) is inhibited by NaN3 by lowering the catalytic cooperativity of the ATP synthase machinery (34). Such inhibition, however, interferes with the transport of H+ from the periplasmic space to the cytoplasm and hence with antiporter mechanisms in the bacterial inner membrane. As a result, this inhibitory effect leads to the perturbation of ΔpH and the intracellular pH (58,59), which contributes to the ΔΨ reduction detected in azide-treated bacteria (60) (Fig. 4 E).
Interestingly, a similar reduction in ΔΨ was also observed in γ-irradiated bacteria, a phenomenon that could be due to protein oxidation damages produced by γ-photons (33,47,61,62). Oxidation of membrane proteins greatly restricts protein flexibility within the lipid bilayer and often results in aggregate formation at the bacterial membrane. Moreover, proteins can undergo restrictions in mobility, probably by cross-linking of amino acids on the primary structure. We cannot rule out the damage of the membrane-associated protein structures at 1000 Gy and their unique functions. This aspect is very complex because of the high number of proteins and possible γ-irradiation-induced modifications. Although the reduction in ΔΨ detected in γ-irradiated bacteria could also be due to an increase in the state of order of the lipid chains, this explanation is not compatible with the unmodified membrane fluidity observed in those cells (Fig. S2). In addition, a possible damage of membrane-associated proteins did not impair the membrane fluidity. Finally, alteration of intracellular pH in γ-irradiated bacteria can lead to misfolded proteins, which in turn contributes to further reduction in ΔΨ because damaged proteins cannot maintain ion gradients properly (63).
Depolarization of the bacterial membrane potential increases peptide binding
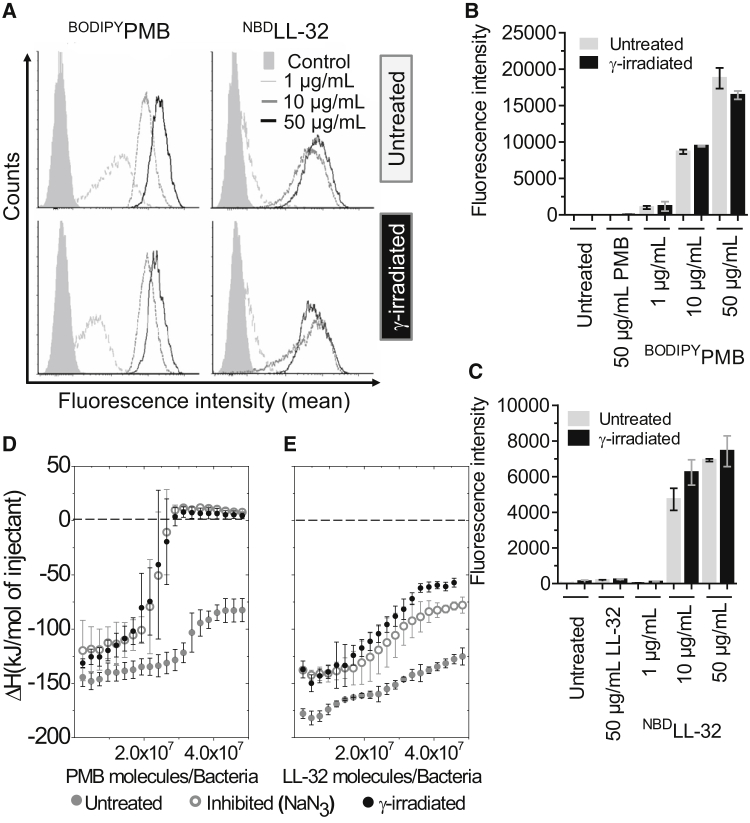
Most AMPs are polycationic, a feature that facilitates binding to negatively charged lipids and LPSs of the bacterial cell envelope (5). A second property, the amphipathic character of AMPs, is necessary for insertion in and perturbation of the bacterial membrane (18). AMP insertion can permeabilize the lipid bilayer, and as a consequence, the bacterial ΔΨ is depolarized (64, 65, 66, 67). However, our approach intends to depolarize the bacterial ΔΨ first and then measure the binding capacity of the peptides to the bacterial outer membrane. To study whether the inactivation method modifies the overall peptide adsorption on the bacterial membrane, we compared the fluorescence intensity of labeled AMPs for untreated and γ-irradiated cells. In this setup, we used dye-conjugated PMB (BODIPYPMB) or LL-32 (NBDLL-32) that were added to γ-irradiated and non-γ-irradiated bacteria. The changes in fluorescence intensity were quantified upon adsorption in flow cytometry experiments. As depicted in the representative histogram (Fig. 5 A), a dose-dependent rise of the fluorescence intensity was observed after incubating S. enterica R60 bacteria with increasing doses of BODIPYPMB or NBDLL-32. The fluorescence intensity measured in the case of γ-irradiated (1000 Gy) bacteria was of a similar magnitude to that recorded in the case of untreated cells (Fig. 5 B), suggesting the proper preservation of both the bacterial membrane and the molecules targeted by the AMP (Fig. 5, B and C).
Figure 5.
Adsorption and direct thermodynamic characterization of peptide binding. (A) Representative histograms of S. enterica R60 (1.0 × 107 CFU/mL) incubated for 1 h at 4°C with various concentrations of dye-conjugated PMB (BODIPYPMB) and LL-32 (NBDLL-32) that were previously either left untreated (upper panel) or that were exposed to γ-irradiation (lower panel); Control = unlabeled cells. The calculated PMB and LL-32 concentrations necessary for the bacterial neutralization were ∼0.6 and 1.5 μg/mL, respectively; values obtained from Table 1. (B and C) The mean ± s.d. of three independent experiments of the same samples as shown in (A) is shown. Unlabeled PMB or LL-32 were used as an additional control. (D and E) Binding curves for the interaction of PMB and LL-32 with untreated, metabolically inhibited (0.1% NaN3) and γ-irradiated S. enterica R60 (1000 Gy). The heat of the reaction (ΔH) is presented as a function of the number of peptide molecules/bacteria. The mean ± s.d. of three independent experiments are shown in the error bars. We did not expand the range of peptide concentration used to obtain the binding curves. The reason was to prevent the self-association ability of peptide in aqueous media.
To test whether the inactivation methods alter bacterial interaction with AMPs, we characterized the binding affinity of PMB and LL-32 to suspensions of S. enterica R60 that were treated with either γ-irradiation or NaN3 by ITC. In case of untreated cells, the thermodynamic response of the S. enterica R60 membrane upon the addition of PMB was highly exothermic, but no complete saturation was achieved at the used peptide concentrations (Fig. 5, D and E). Higher peptide concentrations could not be used because of peptide aggregation. The binding isotherms observed when bacteria were metabolically inhibited by NaN3 or killed with γ-irradiation were comparable. In these two settings, binding isotherms with comparable saturation values were obtained (Fig. 5 D). A strong exothermic reaction could be observed without saturation when using untreated bacteria and LL-32. However, after NaN3-inhibition or γ-inactivation, two binding isotherms were evident and comparable (Fig. 5 E). Probably, the binding affinity of PMB and LL-32 to the bacterial cell envelope was influenced by bacterial metabolism (energized cells with full ΔΨ) (Fig. 5, D and E).
The binding reaction of both peptides to the bacterial membrane was highly driven by enthalpy (ΔH) rather than by entropy (ΔS), with an overall free-energy change (ΔG) favorable for the binding reaction (Table 1). Regardless of whether bacteria were untreated, NaN3-inhibited, or γ-inactivated, the high negative ΔH indicated a strong electrostatic interaction between PMB or LL-32 and the bacterial outer membrane. In all likelihood, the conformational transition of peptides to more organized structures, when inserted into the bacterial membrane, maximizes the hydrogen bonding between the residues at the secondary structure level, leading to a significant reduction in entropy. This effect is reflected by the unfavorable entropy term (−TΔS) from the Gibbs equation (Table 1). We also performed experiments by using only isolated LPS Ra aggregates. Interestingly, PMB had a lower affinity for LPS Ra than LL-32, and the driving force of peptide binding to the LPS aggregates was both a gain in enthalpy and entropy (Fig. S3; Table S1). These results demonstrate that AMPs interact in a very different way with their targets depending on whether those targets are part of the bacterial membrane or isolated as aggregates. In addition, the differences in ΔH found when analyzing the interaction between the peptides and LPS either as aggregate or as the constituent of the bacterial outer membrane were very significant. This demonstrates that AMPs indeed bind to LPS but also to other biomolecules like lipopeptides and proteins.
Table 1.
Thermodynamic Parameters of PMB and LL-32 Binding to the Bacterial Outer Membrane of S. enterica R60.
| Thermodynamic parameters | Polymyxin B |
LL-32 |
||||
|---|---|---|---|---|---|---|
| Viable bacteriaf | Viable bacteria + NaN3 0.1 % |
γ-irradiated bacteria 3h | Viable bacteria | Viable bacteria + NaN3 0.1 %f |
γ-irradiated bacteria 3h f | |
| n (ƒg/bacteria)a | 75 ± 6 | 65 ± 14 | 55 ± 8 | n.d | 172 ± 42 | 152 ± 12 |
| Peptide s.a/Bacteria s.ab | 3.7 ± 0.3 | 3.2 ± 0.7 | 2.7 ± 0.4 | n.d | 9.6 ± 2.3 | 8.5 ± 0.7 |
| Peptide/LPS Ra mol ratiob | 5.2 ± 0.4 | 4.5 ± 1.0 | 3.8 ± 0.6 | n.d | 4.2 ± 1.0 | 3.7 ± 0.3 |
| KD (nM)c | 397± 326 | 80 ± 14 | 84 ± 59 | n.d | 958 ± 458 | 855 ± 401 |
| ΔH (kJ/mol) | - 59 ± 4 | - 114 ± 17 | - 133 ± 17 | n.d | - 64 ± 4 | - 81 ± 1 |
| -TΔS (kJ/mol)d | 20 ± 7 | 72 ± 17 | 91 ± 16 | n.d | 28 ± 5 | 45 ± 1 |
| ΔG (kJ/mol)e | - 39 ± 2 | - 42 ± 1 | - 42 ± 2 | n.d | - 36 ± 1 | - 36 ± 1 |
The values represent the mean of three independent experiments with the s.d.
The amount of bonded peptide was calculated from the inflection point of the isotherm curves. Instead of a stoichiometry value, it represents the overall peptide needed to neutralize all the constitutive negative charges on the outer leaflet of the outer membrane. The molecular weights of LPS Ra, PMB, and LL-32 are 4300, 1385.6, and 3921.7 g/mol, respectively.
Peptide surface area (s.a.) was calculated assuming that a single bound peptide molecule covers an area of ∼1.0×10−6 and ∼3.5×10−6 μm2 for PMB and LL-32, respectively. The bacteria s.a. was estimated to be 9.5 μm2, which corresponds to a cylinder of 1 μm diameter and 3.5 μm height. The Peptide/LPS Ra mol ratio was calculated assuming an LPS Ra area of 1.5×10−6 μm2 (80). We assumed a bacterial surface completely covered by LPS Ra. Based on the LPS Ra dry mass yield, we calculated 52 μg of LPS Ra in 1.5×109 titrated bacteria, a value that was close to the 66 μg obtained by theoretical calculation.
KD represents an apparent dissociation constant due to the high chemical diversity on the bacterial membrane.
Entropy changes of binding were calculated with
Free energy changes were calculated according to
Enthalpy changes were corrected for incomplete bacterial membrane binding. As a consequence, the energy released upon the addition of PMB and LL-32 only represents the total energy minus the average residual heat of the last three titration points from the isotherms. The reason for this procedure was the possibility to use the one-side model function to get the thermodynamic values. The peptide binding to the bacterial membrane was strongly exothermic for both peptides, and the maximal values were observed for viable bacteria (Fig. 5, D and E). The binding reaction was entropically unfavorable with an overall Gibbs energy change favorable for both peptides under all conditions tested. n.d., not detected.
In principle, these data support that water molecules are not released upon AMP-membrane interaction to favor the peptide penetration or intercalation into the outer leaflet of the outer membrane. However, a consideration only regarding enthalpy and entropy can be misleading. The hydrophobic effect and its contribution are the main driving forces for partitioning, but the response of the bilayer to partitioning is responsible for modifying the overall thermodynamics of the process (bilayer effect) (68,69).
Our data on the heat released during the binding of the two AMPs were comparable to results from the few previously published ITC studies (70,71). However, the isothermal curves were not described in those previous studies, limiting their informative value. It is well-known that transitions from unordered peptide structures in suspension to α-helix or β-sheet at the membrane surface environment reduce the free energy and modulate the balance of electrostatic interactions of amphipathic peptides (64, 65, 66).
We show that the required amount in femtograms to saturate S. enterica R60 bacterial membrane was lower for PMB than for LL-32 (Fig. 5; Table 1). These calculated values (see the legend of Table 1) represent an extremely high cover and exceed the outer membrane surface. This observation might indicate a multilayer arrangement of the peptides due to self-association or peptide association with polysaccharide and proteins (Table 1). Moreover, the apparent dissociation constant (KD) of both peptides decreased in metabolically inhibited or γ-irradiated bacteria in comparison to untreated bacteria (increased affinity). Furthermore, the interaction of PMB with the bacterial membrane was stronger than that of LL-32, as judged by the magnitude of reduction of the apparent KD. This could explain why higher concentrations of LL-32 were required to inhibit the growth of bacteria (Table S2). Notably, the apparent KD reduction occurred only in metabolically inhibited and γ-irradiated bacteria but not in untreated bacteria. Two possible reasons can be argued to explain these effects: first, peptides can self-associate to form aggregates upon the interaction with the bacterial outer membrane, which can, in turn, reduce the effective peptide concentration for neutralization (21). Here, the overall result is the binding isotherms with nonsaturation characteristics mostly observed for LL-32. Second, the structural requirements (peptide conformation) for bacterial outer membrane neutralization by the peptides can be directly influenced by the bacterial physiological state. In this case, we need to refer to one useful parameter in bacterial physiology for a plausible explanation, the bacterial membrane potential (ΔΨ).
Untreated bacteria are fully energized cells (60). In these cells, the development and maintenance of Δp are accompanied by rapid Na+ extrusion and the maintenance of a substantial inwardly directed Na+ gradient (60). In addition, the periplasm can increase the level of free Ca2+, resulting in the outer leaflet of the inner membrane being exposed to free Ca2+ severalfold higher than the bulk of external fluid (72). In contrast, bacterial exposure to NaN3 and γ-irradiation drastically depolarizes ΔΨ as a natural consequence of disturbing the Δp (Fig. 4 E). This inactivation of active processes leaves Na+ and H+ free to flow down the membrane’s concentration gradient. This reduces the concentration of the cations in the periplasmic space (60). The depolarization of the ΔΨ leads to a Donnan potential being more negative than in untreated bacteria, which is explained by the reduction of cations, including H+, in the periplasmic space. This increase in Donnan potential can also increase or modify the interaction between the polycationic peptides with the outer membrane. Several mechanisms are discussed in literature describing the influence of the Donnan potential on the peptide conformation, partitioning, dipole reorientation, and insertion (73,74). These effects probably can facilitate a more stable association of the AMPs into the bacterial membrane and might explain the differences we observed in the ITC experiments (Fig. 5, D and E).
These experiments demonstrate that diminishing of the metabolic activity reduces the complexity of the ITC data, leading to results allowing a more precise interpretation of the passive binding properties. Furthermore, the difference between data obtained for active and nonactive bacteria can be attributed to active metabolic activities. Therefore, in the future, ITC experiments will be performed at various growth conditions to decipher the active processes in bacteria upon interaction with antimicrobials.
Permeabilization activities of PMB and LL-32 were altered by γ-irradiation
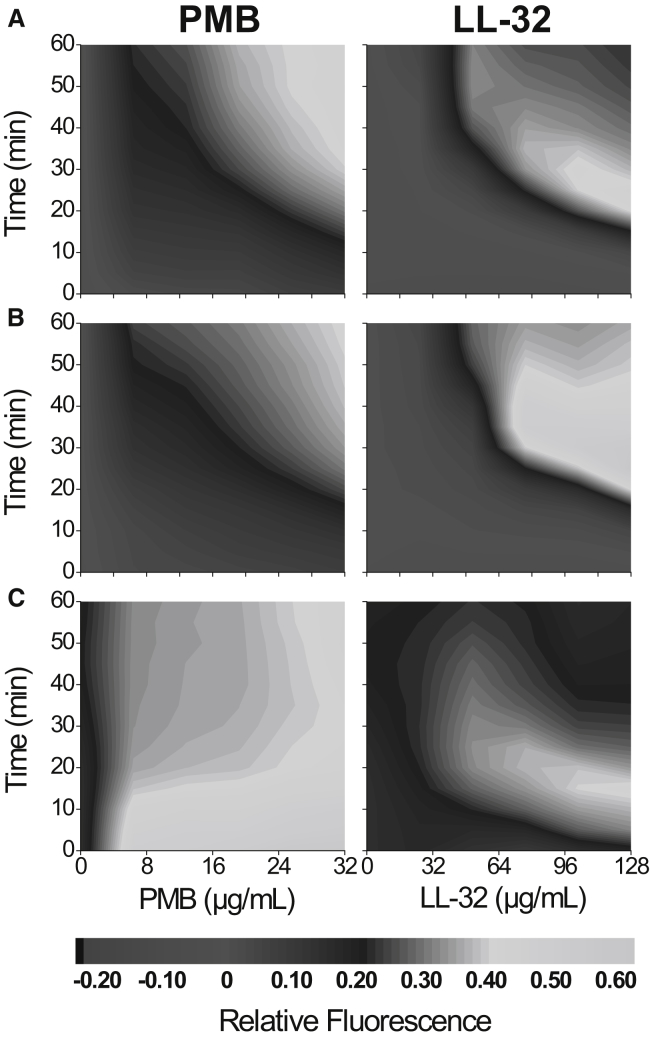
At subinhibitory concentrations, peptides could bind (i.e., adsorb to the lipid-water interface) and insert into the hydrophobic core of the bilayer without causing complete disruption of the membrane but increasing cell permeability by lesions or pores. To quantify the permeabilization of the bacterial cell envelope during exposure to antimicrobial agents, the SYTOX green assay was used (46,75,76). The integrity of the cell envelope of untreated, NaN3 metabolically inhibited (34), and γ-irradiated bacteria at 1000 Gy was analyzed after exposure to PMB and LL-32 by comparing the kinetics of SYTOX green cell uptake (Fig. 6). The permeabilizing activity of PMB on untreated bacteria was time- and concentration-dependent, reaching a relative fluorescence maximum of ∼0.40 after 40 min, corresponding to a PMB concentration of 28 μg/mL. In general, kinetics of PMB permeabilization was faster than that of LL-32, and a lower concentration of PMB was required to induce permeabilization (Fig. 6 A). This observation is in good correlation with the lower amount of PMB needed for bacterial membrane charge neutralization (Table 1). The decrease in fluorescence intensity observed for LL-32 after reaching the maximum permeabilization value is due to the competitive binding of the peptide and the SYTOX green to the nucleic acids (Fig. 6, A–C).
Figure 6.
Fluorometric measurement of SYTOX green stain labeling of peptide-treated S. enterica R60. (A) Untreated, (B) metabolically inhibited (0.01% of sodium azide (NaN3)), and (C) γ-irradiated (1000 Gy) bacteria. Contour plots show the enhancement of SYTOX green stain fluorescence due to the permeabilization of the bacterial membrane at a different concentration of PMB and LL-32. Fluorescence emission was measured in a kinetic experiment for 60 min. SYTOX green was exited at 480 nm. Light and dark gray represent high and no bacterial permeability, respectively. The figures shown are representative of three independent experiments in duplicates.
Metabolically inhibited bacteria were used to evaluate the effect of bacterial metabolism in the permeabilization activity of the peptides. Only a minor decrease in fluorescence intensity was detected for PMB compared to LL-32, suggesting that bacterial metabolism influences the permeabilizing activity of the peptides (Fig. 6, A and B). As we showed above, γ-irradiation (1000 Gy) preserved to a great extent the bacterial membrane ultrastructure. However, the SYTOX green stain clearly revealed a loss in membrane integrity of γ-irradiated bacteria compared with untreated and NaN3-treated bacteria (Fig. 6 C; Fig. S4) even before peptide addition. This phenomenon results in a dramatic increase in the kinetics of membrane permeabilization induced by either PMB or LL-32. In addition, the fluorescence intensity decreased after 15 min, suggesting that PMB also binds to the nucleic acids, thereby displacing the fluorescent stain (Fig. 6 C).
Although the loss of membrane integrity was evident, in all experiments, a background intensity could be observed that did not change over time (cells without peptide addition). This indicates that the chromophores no longer accumulate inside the γ-irradiated cells, indicating that the bacterial membrane was not fully permeabilized or not all bacteria were affected (46). The accumulation of the stain, and therefore the fluorescence enhancement, was clearly detected after the addition of PMB and LL-32 in untreated, NaN3-treated, and γ-inactivated bacteria. Kinetics of membrane permeabilization after peptide binding seemed to be independent of the physiological state of the bacteria because once the ΔΨ is depolarized after NaN3 treatment, kinetics of SYTOX green uptake remains unaltered (Fig. 6, A and B). Finally, the physical γ-irradiation treatment also leads to the depolarization of the bacterial membrane, but, in contrast to the NaN3 induced inhibition, the kinetics of permeabilization increased dramatically (Fig. 6 C). The increase in the kinetics of membrane permeabilization detected in γ-inactivated bacteria is possibly due to 1) membrane protein oxidation induced by ROS and 2) by predamage of the bilayer integrity. Our results clearly show that such damages are not sufficient to alter the ultrastructure of the bacterial membrane but lead to a certain loss of the permeability barrier of the cell.
Conclusions
The use of γ-irradiated or metabolically inactivated bacteria allows the direct and precise quantification of the peptide required to neutralize the bacterial membrane, as well as disclosing the role of bacterial inner-membrane potential (ΔΨ) in this process. Rather than quantifying the ΔΨ depolarization upon the peptide addition, we used metabolically inactive or killed bacteria in which the ΔΨ was depolarized. Bacterial inactivation opens up the possibility of obtaining defined isothermal curves, which become valuable to characterize the bacterial cell envelope interactions with AMPs and to distinguish active and passive processes. These thermodynamic models cannot be extended to untreated bacteria. However, the membrane potential (ΔΨ) depolarization suggests that the peptide affinity to the cell envelope tends to increase. This behavior can be explained by the inability of bacteria to maintain transmembrane proton gradients and the proton motive force. We propose that, in addition to the electrostatic interactions, the bacterial membrane potential is a modulating parameter of the antimicrobial peptide activity. The use of SYTOX green revealed that the concentration of PMB and LL-32 required to cause a cytoplasmic membrane perturbation or to inhibit the bacterial growth (minimum inhibitory concentration (MIC) values) are much higher than the concentrations needed to neutralize the charges of the bacterial LPSs. This strongly suggests that the outer membrane neutralization (bound peptide) does not imply bacterial killing for these two peptides. Therefore, the number of molecules required to kill should increase drastically on the membrane surface by accumulation until reaching a threshold value. This value is crucial to promote the formation of defects that, for instance, cause leakage and cell death. We calculated that ∼3.0 × 107 PMB molecules and ∼2.6 × 107 LL-32 molecules are required to neutralize each bacteria. Similar values were reported recently for E. coli and the DNS-PMAP23 and ARVA-D peptides (77,78) and synthetic anti-LPS peptides (79). The study with the short, broad-spectrum, and cationic peptide ARVA-D strongly supports our finding, showing that for killing bacteria, the peptide needed to reach values >1.0 × 108 peptides per bacteria to achieve sterilization (78).
Finally, the robustness of the NaN3-treated or γ-irradiated bacterial membranes as a suitable model for testing bacterial susceptibility against peptides allows the conclusion that the permeabilization activities of PMB and LL-32 were independent of the membrane potential (ΔΨ) depolarization. The proposed method reveals, to our knowledge, new insights into the mechanism of action of the AMPs with the bacterial membrane under physiological conditions and is a further step to close the gap between biological systems and biochemical and biophysical models. Our approach opens new perspectives for the use of other biophysical techniques reducing biosafety problems or techniques that predamage or strongly modify the bacterial envelope. This knowledge will be valuable for our understanding of the mechanism of membrane-disruptive peptide antibiotics.
Author Contributions
W.C. designed and performed the γ-irradiation, membrane potential, lipidomics, ITC, and Fourier-transform infrared spectroscopy experiments and wrote the manuscript and the scientific protocols. J. Brandenburg. and N.R. evaluated the peptide adsorption on the bacterial membrane by flow cytometry and analyzed data. J. Behrends. contributed to the evaluation of membrane potential by flow cytometry. L.H. designed mononuclear cell stimulation experiments. L.P. designed the BODIPYPMB synthesis. D.S. designed the lipidomics measurements. K.S. performed bacterial survival, AFM, and TEM measurements and evaluated the bacterial membrane permeabilization by the SYTOX green assay. G.M.-d.-T. and K.B. critically reviewed the manuscript and contributed to its scientific foundation, and T.G. performed the AFM experiments and conceived the project. All the authors reviewed the manuscript before submission.
Acknowledgments
The authors thank Rainer Bartels for synthesizing LL-32 and the fluorescently labeled peptides and Carlotta Ober-Blöbaum for critical reading of the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft ExC306 Excellence Cluster Inflammation at Interfaces (EXC 2167–390884018, to T.G.), and G.M.-d.-T. was supported by the Proyectos de Investigación Universidad de Navarra, Spain (PIUNA-P2011-17).
Editor: Julie Biteen.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.10.012.
Supporting Materials
References
- 1.Brown E.D., Wright G.D. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 2.Berendonk T.U., Manaia C.M., Martinez J.L. Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 2015;13:310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 3.Spellberg B., Shlaes D. Prioritized current unmet needs for antibacterial therapies. Clin. Pharmacol. Ther. 2014;96:151–153. doi: 10.1038/clpt.2014.106. [DOI] [PubMed] [Google Scholar]
- 4.Tacconelli E., Carrara E., Magrini N., WHO Pathogens Priority List Working Group Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 5.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 6.Hancock R.E., Sahl H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 7.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 8.Hurdle J.G., O’Neill A.J., Lee R.E. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 2011;9:62–75. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epand R.M., Epand R.F. Bacterial membrane lipids in the action of antimicrobial agents. J. Pept. Sci. 2011;17:298–305. doi: 10.1002/psc.1319. [DOI] [PubMed] [Google Scholar]
- 10.Payne D.J., Gwynn M.N., Pompliano D.L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 11.Richter M.F., Drown B.S., Hergenrother P.J. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature. 2017;545:299–304. doi: 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez de Tejada G., Heinbockel L., Brandenburg K. Lipoproteins/peptides are sepsis-inducing toxins from bacteria that can be neutralized by synthetic anti-endotoxin peptides. Sci. Rep. 2015;5:14292. doi: 10.1038/srep14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bárcena-Varela S., Martínez-de-Tejada G., Heinbockel L. Coupling killing to neutralization: combined therapy with ceftriaxone/Pep19-2.5 counteracts sepsis in rabbits. Exp. Mol. Med. 2017;49:e345. doi: 10.1038/emm.2017.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutsmann T., Razquin-Olazarán I., Brandenburg K. New antiseptic peptides to protect against endotoxin-mediated shock. Antimicrob. Agents Chemother. 2010;54:3817–3824. doi: 10.1128/AAC.00534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinbockel L., Weindl G., Brandenburg K. Inhibition of lipopolysaccharide- and lipoprotein-induced inflammation by antitoxin peptide Pep19-2.5. Front. Immunol. 2018;9:1704. doi: 10.3389/fimmu.2018.01704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin L., van Meegern A., Schuerholz T. Antimicrobial peptides in human sepsis. Front. Immunol. 2015;6:404. doi: 10.3389/fimmu.2015.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 18.Paterson D.J., Tassieri M., Cooper J.M. Lipid topology and electrostatic interactions underpin lytic activity of linear cationic antimicrobial peptides in membranes. Proc. Natl. Acad. Sci. USA. 2017;114:E8324–E8332. doi: 10.1073/pnas.1704489114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silhavy T.J., Kahne D., Walker S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savini F., Bobone S., Stella L. From liposomes to cells: filling the gap between physicochemical and microbiological studies of the activity and selectivity of host-defense peptides. Pept. Sci. 2018;110:e24041. [Google Scholar]
- 22.Bengoechea J.A., Skurnik M. Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol. Microbiol. 2000;37:67–80. doi: 10.1046/j.1365-2958.2000.01956.x. [DOI] [PubMed] [Google Scholar]
- 23.Chromek M., Slamová Z., Brauner A. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med. 2006;12:636–641. doi: 10.1038/nm1407. [DOI] [PubMed] [Google Scholar]
- 24.Scott M.G., Davidson D.J., Hancock R.E. The human antimicrobial peptide LL-37 is a multifunctional modulator of innate immune responses. J. Immunol. 2002;169:3883–3891. doi: 10.4049/jimmunol.169.7.3883. [DOI] [PubMed] [Google Scholar]
- 25.Gutsmann T., Hagge S.O., Wiese A. Interaction of CAP18-derived peptides with membranes made from endotoxins or phospholipids. Biophys. J. 2001;80:2935–2945. doi: 10.1016/S0006-3495(01)76259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dannehl C., Gutsmann T., Brezesinski G. Surface activity and structures of two fragments of the human antimicrobial LL-37. Colloids Surf. B Biointerfaces. 2013;109:129–135. doi: 10.1016/j.colsurfb.2013.03.030. [DOI] [PubMed] [Google Scholar]
- 27.Dannehl C., Brezesinski G., Möhwald H. Interactions of two fragments of the human antimicrobial peptide LL-37 with zwitterionic and anionic lipid monolayers. Z. Phys. Chem. 2015;229:1141–1159. [Google Scholar]
- 28.Srimal S., Surolia N., Surolia A. Titration calorimetric studies to elucidate the specificity of the interactions of polymyxin B with lipopolysaccharides and lipid A. Biochem. J. 1996;315:679–686. doi: 10.1042/bj3150679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garidel P., Brandenburg K. Current understanding of polymyxin B applications in bacteraemia/sepsis therapy prevention: clinical, pharmaceutical, structural and mechanistic aspects. Anti-Infect. Agents Med. Chem. 2009;8:367–385. [Google Scholar]
- 30.Vanaja S.K., Russo A.J., Rathinam V.A.K. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell. 2016;165:1106–1119. doi: 10.1016/j.cell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandenburg K., Moriyon I., Seydel U. Biophysical investigations into the interaction of lipopolysaccharide with polymyxins. Thermochim. Acta. 2002;382:189–198. [Google Scholar]
- 32.Brandenburg K., Arraiza M.D., Zahringer U. The interaction of rough and smooth form lipopolysaccharides with polymyxins as studied by titration calorimetry. Thermochim. Acta. 2002;394:53–61. [Google Scholar]
- 33.Ghosal D., Omelchenko M.V., Daly M.J. How radiation kills cells: survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiol. Rev. 2005;29:361–375. doi: 10.1016/j.femsre.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 34.Noumi T., Maeda M., Futai M. Mode of inhibition of sodium azide on H+-ATPase of Escherichia coli. FEBS Lett. 1987;213:381–384. doi: 10.1016/0014-5793(87)81526-0. [DOI] [PubMed] [Google Scholar]
- 35.Miranda L.P., Alewood P.F. Accelerated chemical synthesis of peptides and small proteins. Proc. Natl. Acad. Sci. USA. 1999;96:1181–1186. doi: 10.1073/pnas.96.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinbockel L., Sánchez-Gómez S., Brandenburg K. Preclinical investigations reveal the broad-spectrum neutralizing activity of peptide Pep19-2.5 on bacterial pathogenicity factors. Antimicrob. Agents Chemother. 2013;57:1480–1487. doi: 10.1128/AAC.02066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hargittai P.T., Youmans S.J., Lieberman E.M. Determination of the membrane potential of cultured mammalian Schwann cells and its sensitivity to potassium using a thiocarbocyanine fluorescent dye. Glia. 1991;4:611–616. doi: 10.1002/glia.440040608. [DOI] [PubMed] [Google Scholar]
- 38.Novo D., Perlmutter N.G., Shapiro H.M. Accurate flow cytometric membrane potential measurement in bacteria using diethyloxacarbocyanine and a ratiometric technique. Cytometry. 1999;35:55–63. doi: 10.1002/(sici)1097-0320(19990101)35:1<55::aid-cyto8>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Novo D.J., Perlmutter N.G., Shapiro H.M. Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob. Agents Chemother. 2000;44:827–834. doi: 10.1128/aac.44.4.827-834.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matyash V., Liebisch G., Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwudke D., Hannich J.T., Shevchenko A. Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal. Chem. 2007;79:4083–4093. doi: 10.1021/ac062455y. [DOI] [PubMed] [Google Scholar]
- 42.Schuhmann K., Herzog R., Shevchenko A. Bottom-up shotgun lipidomics by higher energy collisional dissociation on LTQ Orbitrap mass spectrometers. Anal. Chem. 2011;83:5480–5487. doi: 10.1021/ac102505f. [DOI] [PubMed] [Google Scholar]
- 43.Schwudke D., Schuhmann K., Shevchenko A. Shotgun lipidomics on high resolution mass spectrometers. Cold Spring Harb. Perspect. Biol. 2011;3:a004614. doi: 10.1101/cshperspect.a004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herzog R., Schwudke D., Shevchenko A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;12:R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walser P.J., Ariotti N., Parton R.G. Constitutive formation of caveolae in a bacterium. Cell. 2012;150:752–763. doi: 10.1016/j.cell.2012.06.042. [DOI] [PubMed] [Google Scholar]
- 46.Roth B.L., Poot M., Millard P.J. Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl. Environ. Microbiol. 1997;63:2421–2431. doi: 10.1128/aem.63.6.2421-2431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daly M.J., Gaidamakova E.K., Fredrickson J.K. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 2007;5:e92. doi: 10.1371/journal.pbio.0050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slade D., Radman M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 2011;75:133–191. doi: 10.1128/MMBR.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiao B.H., Freudenberg M., Galanos C. Characterization of the lipid A component of genuine smooth-form lipopolysaccharide. Eur. J. Biochem. 1989;180:515–518. doi: 10.1111/j.1432-1033.1989.tb14676.x. [DOI] [PubMed] [Google Scholar]
- 50.Garidel P., Kaconis Y., Brandenburg K. Self-organisation, thermotropic and lyotropic properties of glycolipids related to their biological implications. Open Biochem. J. 2015;9:49–72. doi: 10.2174/1874091X01509010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rietschel E.T., Kirikae T., Di Padova F. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 52.Spector A.A., Yorek M.A. Membrane lipid composition and cellular function. J. Lipid Res. 1985;26:1015–1035. [PubMed] [Google Scholar]
- 53.Nichols D.S., McMeekin T.A. Biomarker techniques to screen for bacteria that produce polyunsaturated fatty acids. J. Microbiol. Methods. 2002;48:161–170. doi: 10.1016/s0167-7012(01)00320-7. [DOI] [PubMed] [Google Scholar]
- 54.Bielski B.H., Arudi R.L., Sutherland M.W. A study of the reactivity of HO2/O2- with unsaturated fatty acids. J. Biol. Chem. 1983;258:4759–4761. [PubMed] [Google Scholar]
- 55.Ayala A., Muñoz M.F., Argüelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014;2014:360438. doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lichstein H.C., Soule M.H. Studies of the effect of sodium azide on microbic growth and respiration: I. The action of sodium azide on microbic growth. J. Bacteriol. 1944;47:221–230. doi: 10.1128/jb.47.3.221-230.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dimroth P., Kaim G., Matthey U. Crucial role of the membrane potential for ATP synthesis by F(1)F(o) ATP synthases. J. Exp. Biol. 2000;203:51–59. doi: 10.1242/jeb.203.1.51. [DOI] [PubMed] [Google Scholar]
- 58.Krulwich T.A., Sachs G., Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andrés M.T., Fierro J.F. Antimicrobial mechanism of action of transferrins: selective inhibition of H+-ATPase. Antimicrob. Agents Chemother. 2010;54:4335–4342. doi: 10.1128/AAC.01620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castle A.M., Macnab R.M., Shulman R.G. Measurement of intracellular sodium concentration and sodium transport in Escherichia coli by 23Na nuclear magnetic resonance. J. Biol. Chem. 1986;261:3288–3294. [PubMed] [Google Scholar]
- 61.Imlay J.A. Pathways of oxidative damage. Annu. Rev. Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 62.Cabiscol E., Tamarit J., Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 2000;3:3–8. [PubMed] [Google Scholar]
- 63.Lund P., Tramonti A., De Biase D. Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol. Rev. 2014;38:1091–1125. doi: 10.1111/1574-6976.12076. [DOI] [PubMed] [Google Scholar]
- 64.Seelig J. Titration calorimetry of lipid-peptide interactions. Biochim. Biophys. Acta. 1997;1331:103–116. doi: 10.1016/s0304-4157(97)00002-6. [DOI] [PubMed] [Google Scholar]
- 65.Seelig J. Thermodynamics of lipid-peptide interactions. Biochim. Biophys. Acta. 2004;1666:40–50. doi: 10.1016/j.bbamem.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 66.Klocek G., Schulthess T., Seelig J. Thermodynamics of melittin binding to lipid bilayers. Aggregation and pore formation. Biochemistry. 2009;48:2586–2596. doi: 10.1021/bi802127h. [DOI] [PubMed] [Google Scholar]
- 67.Strahl H., Hamoen L.W. Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. USA. 2010;107:12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wimley W.C., White S.H. Membrane partitioning: distinguishing bilayer effects from the hydrophobic effect. Biochemistry. 1993;32:6307–6312. doi: 10.1021/bi00076a001. [DOI] [PubMed] [Google Scholar]
- 69.Fernández-Vidal M., White S.H., Ladokhin A.S. Membrane partitioning: “classical” and “nonclassical” hydrophobic effects. J. Membr. Biol. 2011;239:5–14. doi: 10.1007/s00232-010-9321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Howe J., Hammer M.U., Brandenburg K. Calorimetric investigations of the effect of polymyxin B on different Gram-negative bacteria. Thermochim. Acta. 2007;458:34–37. [Google Scholar]
- 71.Alves I.D., Bechara C., Sagan S. Relationships between membrane binding, affinity and cell internalization efficacy of a cell-penetrating peptide: penetratin as a case study. PLoS One. 2011;6:e24096. doi: 10.1371/journal.pone.0024096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones H.E., Holland I.B., Campbell A.K. Direct measurement of free Ca(2+) shows different regulation of Ca(2+) between the periplasm and the cytosol of Escherichia coli. Cell Calcium. 2002;32:183–192. doi: 10.1016/s0143416002001537. [DOI] [PubMed] [Google Scholar]
- 73.Duclohier H., Wróblewski H. Voltage-dependent pore formation and antimicrobial activity by alamethicin and analogues. J. Membr. Biol. 2001;184:1–12. doi: 10.1007/s00232-001-0077-2. [DOI] [PubMed] [Google Scholar]
- 74.Latorre R., Alvarez O. Voltage-dependent channels in planar lipid bilayer membranes. Physiol. Rev. 1981;61:77–150. doi: 10.1152/physrev.1981.61.1.77. [DOI] [PubMed] [Google Scholar]
- 75.Lebaron P., Catala P., Parthuisot N. Effectiveness of SYTOX green stain for bacterial viability assessment. Appl. Environ. Microbiol. 1998;64:2697–2700. doi: 10.1128/aem.64.7.2697-2700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Makovitzki A., Avrahami D., Shai Y. Ultrashort antibacterial and antifungal lipopeptides. Proc. Natl. Acad. Sci. USA. 2006;103:15997–16002. doi: 10.1073/pnas.0606129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roversi D., Luca V., Stella L. How many antimicrobial peptide molecules kill a bacterium? The case of PMAP-23. ACS Chem. Biol. 2014;9:2003–2007. doi: 10.1021/cb500426r. [DOI] [PubMed] [Google Scholar]
- 78.Starr C.G., He J., Wimley W.C. Host cell interactions are a significant barrier to the clinical utility of peptide antibiotics. ACS Chem. Biol. 2016;11:3391–3399. doi: 10.1021/acschembio.6b00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Correa W., Heinbockel L., Brandenburg K. Antibacterial action of synthetic antilipopolysaccharide peptides (SALP) involves neutralization of both membrane-bound and free toxins. FEBS J. 2019;286:1576–1593. doi: 10.1111/febs.14805. [DOI] [PubMed] [Google Scholar]
- 80.Qiao S., Luo Q., Huang Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature. 2014;511:108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.