Abstract

Cancer cell–matrix interactions have been shown to enhance cancer cell survival via the activation of pro-survival signaling pathways. These pathways are initiated at the site of interaction, i.e., integrins, and thus, their inhibition has been the target of therapeutic strategies. Individual roles for fibronectin-binding integrin subtypes αvβ3 and α5β1 have been shown for various cellular processes; however, a systematic comparison of their function in adhesion-dependent chemoresistance is lacking. Here, we utilize integrin subtype-specific peptidomimetics for αvβ3 and α5β1, both as blocking agents on fibronectin-coated surfaces and as surface-immobilized adhesion sites, in order to parse out their role in breast cancer cell survival. Block copolymer micelle nanolithography is utilized to immobilize peptidomimetics onto highly ordered gold nanoparticle arrays with biologically relevant interparticle spacings (35, 50, or 70 nm), thereby providing a platform for ascertaining the dependence of ligand spacing in chemoprotection. We show that several cellular properties—morphology, focal adhesion formation, and migration—are intricately linked to both the integrin subtype and their nanospacing. Importantly, we show that chemotherapeutic drug sensitivity is highly dependent on both parameters, with smaller ligand spacing generally hindering survival. Furthermore, we identify ligand type-specific patterns of drug sensitivity, with enhanced chemosurvival when cells engage αvβ3 vs α5β1 on fibronectin; however, this is heavily reliant on nanoscale spacing, as the opposite is observed when ligands are spaced at 70 nm. These data imply that even nanoscale alterations in extracellular matrix properties have profound effects on cancer cell survival and can thus inform future therapies and drug testing platforms.
Keywords: Integrin, extracellular matrix, nanoparticle, peptidomimetic, chemoresistance
Cells adhere to the extracellular matrix (ECM) via specific receptors, mainly integrins, which subsequently activate signaling pathways that regulate a variety of cellular functions, e.g., motility, proliferation, polarity, differentiation, and survival.1,2 In the tumor microenvironment, healthy ECM becomes deregulated, causing changes in both composition and architecture, which cancer cells have been shown to utilize to their advantage.3−5 Interactions with the ECM can promote tumor cell extravasation, metastasis, invasion, and most importantly, survival post-chemotherapeutic intervention via the activation of various pro-survival signaling pathways.3,6−10 Consequently, researchers have taken two main approaches to mitigating such adhesion-conferred chemoprotection—either via the perturbation of activated pro-survival signaling pathways11−13 or via blocking specific integrin-mediated adhesions in order to prevent the onset of signaling pathway initiation.14−16 A seminal example of the latter approach, which has also been extensively studied in the clinic, is the blocking of the RGD cell binding sequence via an engineered cyclic RGD pentapeptide, cilengitide (Merck KGaA). Cilengitide has been shown to greatly hinder angiogenesis, thereby starving the tumor and improving subsequent treatment efficacy and patient outcome in specific cases.16−18
Despite the exploration of numerous other integrin-targeting strategies (reviewed by Holle et al.)19 with varied outcomes, none have advanced into clinic practice. The primary obstacle in interpreting the limited data available on adhesion-dependent chemoresistance arises from the enormous molecular complexity of the extracellular environment, thereby making it difficult to identify the specific epitopes that are responsible for protective effects. Moreover, recent studies have shown that adhesion-mediated signaling is affected not only by the chemical nature of the adhesive environment but also by multiple physical features of the matrix, including the spatial patterning of adhesive epitopes available for cell binding, the topography of the substrate, and the rigidity of the matrix.20
Numerous previous reports have identified specific integrin subtypes that contribute to tumor progression (reviewed by Seguin et al.),21 including αvβ3’s role in breast tumor metastasis to the bone22 and α5β1’s role in breast cancer cell invasiveness.23 While various integrins shown to affect chemosurvival have been the target of therapeutic interventions, a direct comparison of different integrin subtypes in a highly controlled system has not yet been made. Fibronectin-binding integrin subtypes αvβ3 and α5β1 exhibit individual roles for various cellular processes including force sensing, mediating structural adaptation to forces, focal adhesion formation, and migration.24−27 Most importantly, they have also been shown to regulate chemoresistance in a variety of cancer cells,9 which leads us to believe that they could be sensitive to alterations in matrix properties.
When studying cell–ligand interactions, it would be prudent to consider the nanoscale spacing between ligands that are presented to cells. Previous work has shown that cells can indeed sense and differentially respond to nanometer-scale alterations in ligand spacing, affecting their migration, morphology, focal adhesion assembly, cell adhesion, and traction force generation.28−32 The spatial organization of available ligand binding partners has been shown to mediate ligand clustering, thereby affecting force-mediated contractility of the cell via “molecular clutches” between integrins and the ECM.31,33,34 Most important to this study, nanoscale ligand spacing has been shown to affect melanoma cancer cell behavior and plasticity,35,36 yet the degree to which nanoscale ligand spacing affects chemosurvival in cancer cells is still unknown.
While the exact distribution of ligands in physiological tissues is difficult to discern due to limitations in the resolution and accessibility of current imaging techniques, researchers have been able to shed some light on the range of physiologically relevant ligand spacing of proteins and focal complexes within cells using approaches such as immunogold labeling,37 crystallography,38,39 and advanced imaging techniques.40,41 For instance, collagen bundles exhibit a periodic structure occurring around 70 nm which is believed to be the length scale at which cells attach to the protein,42 while periodic binding domains in fibronectin have been identified to be as close as 42 nm.37 Furthermore, heterotypic protein assemblies have been shown to alter binding site spacing, decreasing the distance between sites by up to ∼20% from that of homotypic fibrils.37 While in vivo measurements are still lacking, these investigations are complicated by the fact that cancer cells can actively reorganize the ECM, likely altering the density of ligand binding sites dynamically.43,44 Taken together, we aim to better understand the interplay of integrin subtypes and nanoscale spacing of ligands in the chemosurvival of breast cancer cells by utilizing a highly defined platform.
To achieve this, we investigated two integrin subtypes that are known to be expressed in solid tumors, αvβ3 and α5β1.21,45 These integrin subtypes recognize the RGD-binding sequence found in proteins commonly present or overexpressed in the tumor microenvironment, including fibronectin, osteopontin, and/or vitronectin. In order to study their individual roles, highly specific integrin subtype peptidomimetics were synthesized.46 We first plated human metastatic MDA-MB-231 breast cancer cells on human fibronectin (Fn), which contains binding sites for both αvβ3 and α5β1 integrin subtypes, and examined cell morphology with or without the addition of subtype-specific peptidomimetics capable of blocking integrins αvβ3 or α5β1 (experimental design outlined in Figure 1A). When blocking αvβ3 (thereby engaging α5β1), cells maintain a similar spread area compared to Fn alone, and become slightly more rounded (Figure 1B, top row; Figure 1C). When blocking α5β1, thereby engaging αvβ3, cells become significantly smaller and rounder (Figure 1B, top row; Figure 1C,D), which is consistent with cells plated on vitronectin (Vn), where αvβ3 is the major binding integrin (Figure S1A–C).
Figure 1.
Integrin-specific engagement on fibronectin alters cellular and focal adhesion morphology. (A) MDA-MB-231s were plated on immobilized fibronectin and treated with blocking peptidomimetics as follows: no blocking peptidomimetics engages both integrins [left]; blocking of αvβ3 (blue ^) results in engagement of α5β1 (purple) [middle]; and blocking of α5β1 (purple ^) results in engagement of αvβ3 (blue) [right]. (B) Cells were stained for actin (red), paxillin (green), and nucleus (blue) for (1) both α5β1 and αvβ3 engagement (left column), (2) α5β1 engagement (middle column), and (3) αvβ3 engagement (right column), with or without drug treatment (no drug, top row; +5-FU, middle row; +paclitaxel, bottom row). Insets show zoomed in focal adhesions. Scale bar: 50 μm. All images without a scale bar have the same scaling as the bottom right image. Cell morphology in terms of (C) cell area (in μm2) and (D) form factor was quantified for all conditions, i.e., α5β1 and αvβ3 engagement (“Both”, green bars), α5β1 engagement (purple bars), and αvβ3 engagement (blue bars), with or without drug treatment as indicated (no drug, no outline; +5-FU, black outline; +paclitaxel, gray outline). Focal adhesion (FA) morphology in terms of (E) FA area (in μm2) and (F) major axis length (in μm) was quantified for all conditions and graphed as in parts C and D. Data are mean ±95% CI from ncells > 186 and nFAs > 771. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-way ANOVA. All significance comparisons are listed in Table S1.
In order to assess effects of chemotherapeutic drugs, we employed two commonly used compounds from which cells have been reported to have developed chemoresistance: 5-fluorouracil (5-FU), which blocks DNA replication, and paclitaxel (also commonly referred to as Taxol), which disrupts microtubule breakdown.47 When treated with 5-FU, cells on Fn become more elongated in all conditions (Figure 1B, middle row; Figure 1D), while paclitaxel treatment causes cells to become very large and rounded (Figure 1B, bottom row; Figure 1C,D), highlighting that the different mechanisms of action of the two drugs can affect subsequent cell morphology in surviving cells. The fact that these responses are also dependent upon specific ligand interactions underscores the synergy between these two biochemical pathways (Figure 1B).
Focal adhesion (FA) formation and the subsequent activation of downstream signaling pathways have been shown to be important indicators of cancer cell invasiveness.48 As FAs play a major role in substrate sensing and mechanotransduction,49 previous studies have linked alterations in FA formation to various substrate properties, e.g., stiffness, ligand type, topography, dimensionality, etc.50,51 Our results show that FA formation is affected not only by integrin subtype engagement but also by drug treatment. First, we observe that cells engaging only αvβ3 form small, short adhesions (Figure 1B, top row; Figure 1E,F; Figure S1A,D,E). While drug treatment does not greatly affect FA formation on Fn alone, engagement of α5β1 greatly hinders FA clustering in drug-treated cells (Figure 1B, middle and bottom rows; Figure 1E,F). Conversely, drug treatment in conjunction with αvβ3 engagement results in FA size enhancement, with paclitaxel inducing the largest FAs (Figure 1B, middle and bottom rows; Figure 1E,F). Overall, both cell and FA morphology were most notably affected by αvβ3 engagement, further underscoring the role this integrin subtype plays in breast cancer cell behavior.
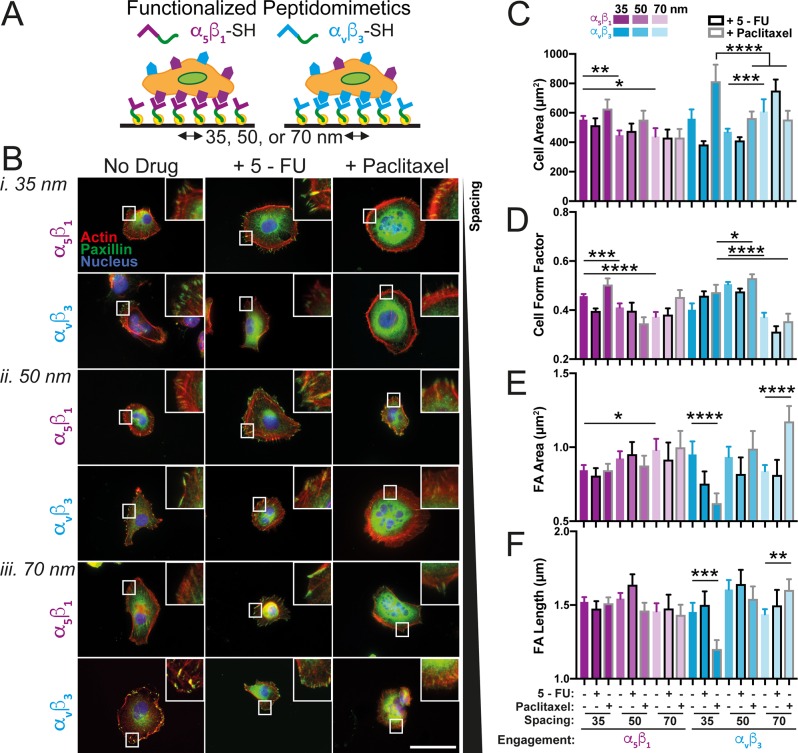
While native proteins are an important substrate for studying integrin subtype-specific effects, uncontrolled parameters are inherent to such systems (e.g., ligand spacing, the presence of other integrin subtype binding sites, etc.) that can obscure the role of primary mediators of cell-ECM behavior. To better isolate these variables, we synthesized highly ordered gold nanoparticle (AuNP) arrays via the previously described block copolymer micelle nanolithography (BCMN) technique52 in order to control both ligand spacing/density and type. AuNP arrays with interparticle spacings of 35, 50, and 70 nm were produced and characterized via SEM (Figure S3). Such biologically relevant spacings were chosen based on previous reports of protein binding site organization, conformation of proteins, and in vitro experiments showing restricted integrin clustering on substrates with ligand spacing greater than ∼73 nm.20 Thiolated versions of the aforementioned integrin-subtype-specific peptidomimetics were synthesized that could specifically attach to the AuNPs,46 thereby resulting in a highly controlled and highly defined nanoscale substrate (schematic of experimental outline in Figure 2A).
Figure 2.
Integrin subtype-specific engagement at defined nanospacing alters cellular and focal adhesion morphology. (A) MDA-MB-231s were plated on integrin-specific peptidomimetics, i.e., α5β1 (purple) or αvβ3 (blue), immobilized on AuNPs with interspacings of 35, 50, or 70 nm. (B) Cells were stained for actin (red), paxillin (green), and nucleus (blue) on samples with AuNP spacing of (i) 35 nm, (ii) 50 nm, or (iii) 70 nm functionalized with either α5β1 (top of each set) or αvβ3 (bottom of each set), with or without drug treatment (no drug, left column; +5-FU, middle column; +paclitaxel, right column). Insets show zoomed in focal adhesions. Scale bar: 50 μm. (C) Cell morphology in terms of area (C, μm2) and form factor (D) was quantified for all conditions, i.e., α5β1 (purple) or αvβ3 (blue) engagement at 35, 50, or 70 nm (where decreasing opacity corresponds with increasing spacing), with or without drug treatment (no drug, no outlines; +5-FU, black outline; +paclitaxel, gray outline). Focal adhesion (FA) morphology in terms of area (E, μm2) and major axis length (F, μm) was quantified and graphed for all conditions as in parts C and D. Data are mean ±95% CI from ncells > 71 and nFAs > 376. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-way ANOVA. All significance comparisons are listed in Table S2.
In general, cells were smaller on AuNP surfaces compared to Fn-coated substrates (Figure 2C vs Figure 1C), likely due to differences in binding site availability and organization, which would be enhanced on Fn-coated substrates. Compared to cells on α5β1-coated surfaces, cells on αvβ3-coated surfaces were much larger and more fully spread on 70 nm spacing (Figure 2B–D). Overall, both integrin subtypes were affected by spacing, albeit at different length scales, as previously reported for various other cell types.29,53 On α5β1, cell spread area and form factor were significantly higher on 35 nm compared to 50 or 70 nm, while on αvβ3, cells on 50 nm spacing elicited a significantly different morphology compared to 70 nm (Figure 2C,D). Drug treatment was found to enhance this spacing-sensitive response. Paclitaxel-treated cells plated on αvβ3 exhibited significant differences in cell area between 35 and 50 or 70 nm (Figure 2C). When comparing the effects of specific spacings on the two integrin subtypes, larger nanoscale ligand spacing resulted in the greatest changes between the two, regardless of condition, while cells on 35 nm were more highly affected by drug treatment on αvβ3 vs α5β1 (Figure 2C,D). Additionally, cells plated on αvβ3-coated surfaces were more affected by drug treatment than those on α5β1, exhibiting a decrease in cell area with 5-FU treatment at 35 and 50 nm, and an increase at 70 nm. The exact opposite of this pattern was observed in response to paclitaxel treatment, suggesting that the nanoscale environment can impact the morphological response to chemo drugs in a pathway-specific fashion (Figure 2B,C).
Mirroring morphological observations, FA formation in cells on αvβ3-coated surfaces was more affected by drug treatment than FAs in cells on α5β1-coated surfaces (Figure 2B,E,F). FA area, but not necessarily length, on α5β1 is generally enhanced on larger spacing vs smaller spacing (Figure 2B,E,F), as has been previously reported in osteosarcoma cells.53 Interestingly, on αvβ3-coated surfaces, FA area and length were particularly sensitive to paclitaxel treatment. Compared to control groups, FA area and length in paclitaxel-treated cells decreased on 35 nm, remained unchanged on 50 nm, and increased on 70 nm (Figure 2E,F). In general, drug treatment significantly altered FA characteristics in almost all conditions. This shows that, while neither drug specifically targets focal contacts, global changes in cellular organization in response to chemo drugs are connected to dynamic alterations of focal adhesion presentation, and that these alterations are modulated by nanoscale ECM organization. Taken together, cellular and FA morphology data indicate that cancer cells are sensitive to both ligand type and nanoscale ligand spacing, and that this sensitivity may play a role in ECM-modulated chemoresistance.
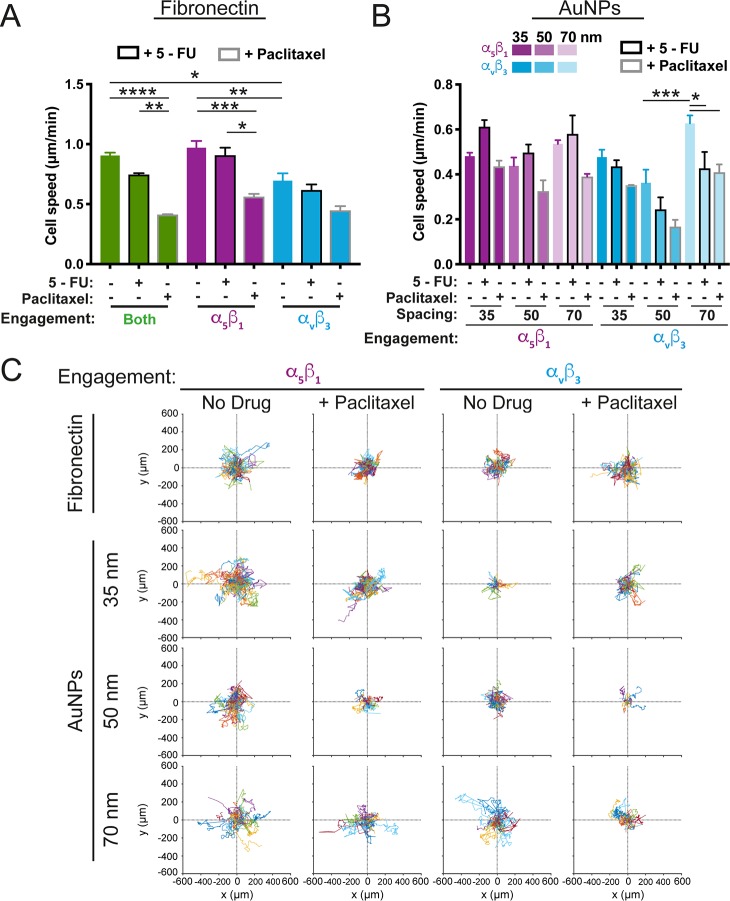
Altered cell motility usually accompanies the transformation of a cancer cell to a metastatic or chemoresistant phenotype.54,55 Additionally, subtype-specific integrin expression and abundance have been shown to be altered in cancer progression.56 Therefore, we analyzed cancer cell migration as a function of integrin engagement, dimensionality, and drug treatment (Figure 3). We found that cells plated on Fn slowed in response to either drug treatment, but most significantly with paclitaxel. Cells engaging only α5β1 on Fn behaved similarly to cells in which both integrins could be bound (i.e., on Fn alone), mirroring cell morphology patterns (Figure 1). When engaging only αvβ3 on Fn, cell speed decreased, a pattern which was enhanced with paclitaxel treatment (Figure 3A). On AuNP arrays, cells on αvβ3-coated surfaces were slower, in general, than cells on α5β1-coated surfaces (Figure 3B). Interestingly, cells of all conditions were slower on 50 nm spacing than on 35 or 70 nm spacing, where they exhibited similar speeds. While both drug treatments resulted in decreased cell speed on αvβ3, only paclitaxel reduced speed on α5β1. Lastly, cell trajectories on Fn or AuNPs treated with paclitaxel were examined compared to controls due to the fact that paclitaxel treatment hindered cell movement in nearly all conditions (Figure 3C). Cells on 70 nm spaced AuNPs were more exploratory than those on narrower spacings, which was most pronounced on αvβ3-coated surfaces. This was echoed on fibronectin-coated substrates, where paclitaxel-treated cells engaging αvβ3 were also more exploratory than those engaging α5β1 (Figure 3C). Previous studies have found differing roles for integrin subtypes in migration; e.g., β1 promotes a more random migration pattern compared to β3, which is ultimately regulated through Rho GTPases.57 These data suggest that this is not only a function of integrin subtype but also the nanoscale presentation of extracellular ligands in combination with drug treatment.
Figure 3.
Integrin subtype-specific and nanospacing-specific cell motility. (A) Cell motility (cell speed in μm/min) for integrin subtype-specific engagement on fibronectin using blocking peptidomimetics vs ligand type (both α5β1 and αvβ3 engagement, green; α5β1 engagement, purple; αvβ3 engagement, blue) and drug treatment (no drug, no outline; +5-FU, black outline; +paclitaxel, gray outline). (B) Cell motility for integrin subtype-specific engagement on peptidomimetics immobilized on AuNPs with interspacings of 35, 50, or 70 nm. Data plotted as in Figure 2, i.e., α5β1 (purple) or αvβ3 (blue) engagement at 35, 50, or 70 nm (where decreasing opacity corresponds with increasing spacing), with or without drug treatment (no drug, no outlines; +5-FU, black outline; +paclitaxel, gray outline). (C) Representative cell trajectories depicted as roseplots from the first 17 h of the experiment of cells engaging α5β1 (two left columns) or αvβ3 (two right columns) without drug treatment or with paclitaxel treatment (left and right column, respectively, of each set) for cells plated on fibronectin (top row) or on AuNPs with interspacing of 35 nm (second row), 50 nm (third row), and 70 nm (last row). nFn,tracks > 287; nAuNP,tracks > 93; r = 2. Data are mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-way ANOVA.
While cell metrics like morphology, focal adhesion formation, and motility show that both integrin subtype and ligand nanospacing are important mediators of cancer cell behavior, how these two parameters influence cell survival has important implications in therapeutic applications and drug screening. Therefore, cell survival was assessed after 48 h of drug treatment based on integrin subtype on both Fn and AuNP arrays, with either 5-FU or paclitaxel treatment (Figure 4). When treated with either drug, cells on Fn alone exhibited similar survival as when only α5β1 was engaged on Fn (Figure 4A). However, when only αvβ3 was engaged on Fn, survival was enhanced by up to ∼20% with 5-FU treatment and ∼10% with paclitaxel (Figure 4A).
Figure 4.
Chemosurvival is dependent on integrin subtypes and nanoscale dimensionality. (A) Percent survival of cells treated with 5-FU (black outline) or paclitaxel (gray outline) plated on fibronectin with integrin-specific engagement as indicated (both, i.e., α5β1 and αvβ3, green; α5β1, purple; αvβ3, blue). (B) Percent survival of cells treated with 5-FU (black outline) or paclitaxel (denoted as “P” on the x-axis, gray outline) on AuNPs functionalized with integrin-subtype-specific peptidomimetics (α5β1 engagement, purple; αvβ3 engagement, blue) at interspacing of 35, 50, or 70 nm (where decreasing opacity corresponds with increasing spacing). For samples in part A, n = 8; part B, n = 7. Data are mean ± SEM. **p < 0.01, ***p < 0.001 by one-way ANOVA for part A and *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by two-way ANOVA for part B.
The similarities found between the engagement of both integrin subtypes and the α5β1-only engagement can be explained by the fact that MDA-MB-231 cells express β1 more readily than αv,58 indicating that α5β1 is likely the dominant ligand partner for MDA-MB-231 cells on Fn. Although, expression alone does not necessarily indicate functional output, as distinct but complementary roles for these two integrin subtypes in regulating contractility and adhesion strength have been shown in fibroblasts.24,59 Furthermore, it is well-known that αvβ3 promotes breast cancer metastasis to the bone niche,22 which could explain the enhancement in chemosurvival we observe when engaging only this integrin subtype.
Next, we looked at drug responsiveness as a function of ligand spacing and integrin subtype using the AuNP arrays. Importantly, we observed that cancer cell survival in response to chemotherapeutic treatment is highly dependent on nanoscale ligand spacing, an observation not yet reported in the literature. We demonstrated differences in survival not only between the two integrin subtypes, but also between drug type and ligand spacing. When cancer cells engage α5β1 integrins, higher spacing improves survival with both 5-FU and paclitaxel treatment, but the length scale at which this occurs is drug-dependent; i.e., 50 nm enhances survival with 5-FU, while larger spacing is required for paclitaxel (Figure 4B). On αvβ3, we observe the same trend with paclitaxel treatment, but a different trend in how spacing affects cell survival with 5-FU treatment, namely, that 50 nm spacing enhances chemosurvival (Figure 4B).
Optimal ligand spacing for cell spreading has previously been reported in various cell lines to be between ∼58 and 73 nm.28 Indeed, we observed that survival was low on 35 nm substrates vs larger spacing for all conditions, indicating that integrin-mediated pro-survival signaling could be hindered at such length scales. Supporting this, we found impaired focal adhesion formation at 35 nm, as has been previously demonstrated in melanoma cells. In that investigation, the authors linked this impaired FA formation to the restricted size of focal adhesion proteins, i.e., talin at ∼60 nm, and the restricted lateral motion of integrins during clustering.35
Most notably, we find that, by altering only nanoscale ligand spacing, we were able to increase cancer cell chemosurvival by over 40% (Figure 4B, α5β1/paclitaxel/50 vs 70 nm), a surprisingly large magnitude given the small change in ECM conditions. These data suggest that even nanoscale alterations in ligand presentation can significantly alter cancer cell survival in response to chemotherapeutic treatment, and that these alterations are affected by not only the ligand type but also the type of drug utilized in treatment. The observed differential responses in chemosurvival among the different conditions likely stem from integrin subtype-specific activation of pro-survival signaling pathways, e.g., FAK, PI3K, or Akt,60,61 which would be worthwhile to explore in future studies.
Although this study examined nanoscale ligand spacing on rigid glass substrates, ECM stiffness has been shown to be an important mediator in mechanotransduction. Moreover, it has been shown that matrix stiffness affects chemoresistance through such mechanisms as cellular stiffening, focal adhesion-driven signaling, or promoting the epithelial-to-mesenchymal transition.10,62,63 Intriguingly, the α5β1 and αvβ3 integrin subtypes have been shown to distinctly modulate force transmission at the cell–ECM interface64 via the activation of unique signaling pathways.65−67 Thus, future studies will focus on untangling the complex interplay of matrix stiffness and ligand presentation in regulating chemoresistance. This can be realized by utilizing the well-established technique of transfer nanolithography, in which AuNP arrays can be transferred onto soft polymeric substrates.68 Taken together, these data call not only for a better understanding of the intricate tumor microenvironment but also for more faithful ECM mimics for cancer drug screening.
Acknowledgments
This work was supported by the Max Planck Society, the German Federal Ministry of Education and Research, and the European Research Council (to J.L.Y. and J.P.S.). The authors would like to thank Dr. Andrew Holle, PD Dr. Dr. Elisabetta Ada Cavalcanti-Adam, Dr. Jacopo Di Russo, and Dr. Dimitris Missirlis for experimental discussions, as well as Janis Grigoridis for SEM support. The authors would like to acknowledge funding from the MaxSynBio Consortium, which is jointly funded by the Federal Ministry of Education and Research of Germany and the Max Planck Society. They further acknowledge support from the VolkswagenStiftung (priority call ‘Life?’). J.P.S. is the Weston Visiting Professor at the Weizmann Institute of Science and part of the excellence cluster CellNetworks at the University of Heidelberg.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.9b04607.
Materials and methods section, description of supporting experiments, tables of signficance, and additional data and figures including cells plated on vitronectin, integrin subtype immunostaining, SEM of samples produced by block copolymer micelle nanolithography, cellular proliferation, drug dose curves, and low magnification images of cells (PDF)
Author Contributions
J.L.Y. and J.P.S. designed the research. J.L.Y., X.H., and H.S. performed the research. J.L.Y. and X.H. analyzed the data. F.R. and H.K. synthesized the peptidomimetics. J.L.Y. and J.P.S. wrote the paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Hynes R. O. The Extracellular Matrix: Not Just Pretty Fibrils. Science 2009, 326, 1216–1219. 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak M. A.; Modzelewska K.; Kwong L.; Keely P. J. Focal Adhesion Regulation of Cell Behavior. Biochim. Biophys. Acta, Mol. Cell Res. 2004, 1692, 103–119. 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Oskarsson T. Extracellular Matrix Components in Breast Cancer Progression and Metastasis. Breast 2013, 22, S66–S72. 10.1016/j.breast.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Larsen M.; Artym V. V.; Green J. A.; Yamada K. M. The Matrix Reorganized: Extracellular Matrix Remodeling and Integrin Signaling. Curr. Opin. Cell Biol. 2006, 18, 463–471. 10.1016/j.ceb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Allinen M.; Beroukhim R.; Cai L.; Brennan C.; Lahti-Domenici J.; Huang H.; Porter D.; Hu M.; Chin L.; Richardson A. Molecular Characterization of the Tumor Microenvironment in Breast Cancer. Cancer Cell 2004, 6, 17–32. 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Glinsky G. V. Anti-Adhesion Cancer Therapy. Cancer Metastasis Rev. 1998, 17, 177–185. 10.1023/A:1006050302406. [DOI] [PubMed] [Google Scholar]
- Juliano R. L.; Varner J. A. Adhesion Molecules in Cancer: the Role of Integrins. Curr. Opin. Cell Biol. 1993, 5, 812–818. 10.1016/0955-0674(93)90030-T. [DOI] [PubMed] [Google Scholar]
- Aoudjit F.; Vuori K. Integrin Signaling Inhibits Paclitaxel-Induced Apoptosis in Breast Cancer Cells. Oncogene 2001, 20, 4995–5004. 10.1038/sj.onc.1204554. [DOI] [PubMed] [Google Scholar]
- Aoudjit F.; Vuori K. Integrin Signaling in Cancer Cell Survival and Chemoresistance. Chemother. Res. Pract. 2012, 2012, 1–16. 10.1155/2012/283181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce M. H.; Lu C.; James E. R.; Hegab R.; Allen S. C.; Suggs L. J.; Brock A. Phenotypic Basis for Matrix Stiffness-Dependent Chemoresistance of Breast Cancer Cells to Doxorubicin. Front. Oncol. 2018, 8, 57. 10.3389/fonc.2018.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranen T.; Selfors L. M.; Worster D. T.; Iwanicki M. P.; Song L.; Morales F. C.; Gao S.; Mills G. B.; Brugge J. S. Inhibition of PI3K/mTOR Leads to Adaptive Resistance in Matrix-Attached Cancer Cells. Cancer Cell 2012, 21, 227–239. 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J. Targeting the Phosphoinositide-3 (PI3) Kinase Pathway in Breast Cancer. Oncologist 2011, 16 (S1), 12–19. 10.1634/theoncologist.2011-S1-12. [DOI] [PubMed] [Google Scholar]
- Courtney K. D.; Corcoran R. B.; Engelman J. A. The PI3K Pathway as Drug Target in Human Cancer. J. Clin. Oncol. 2010, 28, 1075–1083. 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Wang F.; Chen X. Integrin αVβ3-Targeted Cancer Therapy. Drug Dev. Res. 2008, 69, 329–339. 10.1002/ddr.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersey P.; Sosman J.; O’Day S.; Richards J.; Bedikian A.; Gonzalez R.; Sharfman W.; Weber R.; Logan T.; Buzoianu M.; et al. A Randomized Phase 2 Study of Etaracizumab, a Monoclonal Antibody Against Integrin αVβ3, ± Dacarbazine in Patients with Stage IV Metastatic Melanoma. Cancer 2010, 116, 1526–1534. 10.1002/cncr.24821. [DOI] [PubMed] [Google Scholar]
- Stupp R.; Hegi M. E.; Gorlia T.; Erridge S. C.; Perry J.; Hong Y.-K.; Aldape K. D.; Lhermitte B.; Pietsch T.; Grujicic D.; et al. Cilengitide Combined with Standard Treatment for Patients with Newly Diagnosed Glioblastoma with Methylated MGMT Promoter (CENTRIC EORTC 26071–22072 Study): a Multicentre, Randomised, Open-Label, Phase 3 Trial. Lancet Oncol. 2014, 15, 1100–1108. 10.1016/S1470-2045(14)70379-1. [DOI] [PubMed] [Google Scholar]
- Dechantsreiter M. A.; Planker E.; Mathä B.; Lohof E.; Hölzemann G.; Jonczyk A.; Goodman S. L.; Kessler H. N-Methylated Cyclic RGD Peptides as Highly Active and Selective αVβ3 Integrin Antagonists. J. Med. Chem. 1999, 42, 3033–3040. 10.1021/jm970832g. [DOI] [PubMed] [Google Scholar]
- Mas-Moruno C.; Rechenmacher F.; Kessler H. Cilengitide: the First Anti-Angiogenic Small Molecule Drug Candidate. Design, Synthesis and Clinical Evaluation. Anti-Cancer Agents Med. Chem. 2010, 10, 753–768. 10.2174/187152010794728639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holle A. W.; Young J. L.; Spatz J. P. In Vitro Cancer Cell–ECM Interactions Inform In Vivo Cancer Treatment. Adv. Drug Delivery Rev. 2016, 97, 270–279. 10.1016/j.addr.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Young J. L.; Holle A. W.; Spatz J. P. Nanoscale and Mechanical Properties of the Physiological Cell–ECM Microenvironment. Exp. Cell Res. 2016, 343, 3–6. 10.1016/j.yexcr.2015.10.037. [DOI] [PubMed] [Google Scholar]
- Seguin L.; Desgrosellier J. S.; Weis S. M.; Cheresh D. A. Integrins and Cancer: Regulators of Cancer Stemness, Metastasis, and Drug Resistance. Trends Cell Biol. 2015, 25, 234–240. 10.1016/j.tcb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan E. K.; Pouliot N.; Stanley K. L.; Chia J.; Moseley J. M.; Hards D. K.; Anderson R. L. Tumor-Specific Expression of αvβ3 Integrin Promotes Spontaneous Metastasis of Breast Cancer to Bone. Breast Cancer Res. 2006, 8, R20. 10.1186/bcr1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke C. T.; Frey B.; Fellner M.; Herrmann M.; Fabry B. Integrin α5β1 Facilitates Cancer Cell Invasion Through Enhanced Contractile Forces. J. Cell Sci. 2011, 124, 369–383. 10.1242/jcs.071985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H. B.; Hermann M.-R.; Polleux J.; Vignaud T.; Zanivan S.; Friedel C. C.; Sun Z.; Raducanu A.; Gottschalk K.-E.; Théry M.; et al. β1- and αV-Class Integrins Cooperate to Regulate Myosin II During Rigidity Sensing of Fibronectin-Based Microenvironments. Nat. Cell Biol. 2013, 15, 625–636. 10.1038/ncb2747. [DOI] [PubMed] [Google Scholar]
- Ballestrem C.; Hinz B.; Imhof B. A.; Wehrle-Haller B. Marching at the Front and Dragging Behind: Differential αVβ3-Integrin Turnover Regulates Focal Adhesion Behavior. J. Cell Biol. 2001, 155, 1319–1332. 10.1083/jcb.200107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danen E. H. J.; Sonneveld P.; Brakebusch C.; Fässler R.; Sonnenberg A. The Fibronectin-Binding Integrins α5β1 and αvβ3 Differentially Modulate RhoA–GTP Loading, Organization of Cell Matrix Adhesions, and Fibronectin Fibrillogenesis. J. Cell Biol. 2002, 159, 1071–1086. 10.1083/jcb.200205014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D. P.; Caswell P. T.; Norman J. C. αvβ3 and α5β1 Integrin Recycling Pathways Dictate Downstream Rho Kinase Signaling to Regulate Persistent Cell Migration. J. Cell Biol. 2007, 177, 515–525. 10.1083/jcb.200609004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M.; Cavalcanti-Adam E. A.; Glass R.; Blümmel J.; Eck W.; Kantlehner M.; Kessler H.; Spatz J. P. Activation of Integrin Function by Nanopatterned Adhesive Interfaces. ChemPhysChem 2004, 5, 383–388. 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- Cavalcanti-Adam E.; Micoulet A.; Blummel J.; Auernheimer J.; Kessler H.; Spatz J. Lateral Spacing of Integrin Ligands Influences Cell Spreading and Focal Adhesion Assembly. Eur. J. Cell Biol. 2006, 85, 219–224. 10.1016/j.ejcb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Cavalcanti-Adam E. A.; Volberg T.; Micoulet A.; Kessler H.; Geiger B.; Spatz J. P. Cell Spreading and Focal Adhesion Dynamics Are Regulated by Spacing of Integrin Ligands. Biophys. J. 2007, 92, 2964–2974. 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oria R.; Wiegand T.; Escribano J.; Elosegui-Artola A.; Uriarte J. J.; Moreno-Pulido C.; Platzman I.; Delcanale P.; Albertazzi L.; Navajas D.; et al. Force Loading Explains Spatial Sensing of Ligands by Cells. Nature 2017, 552, 219. 10.1038/nature24662. [DOI] [PubMed] [Google Scholar]
- Selhuber-Unkel C.; Erdmann T.; López-García M.; Kessler H.; Schwarz U. S.; Spatz J. P. Cell Adhesion Strength Is Controlled by Intermolecular Spacing of Adhesion Receptors. Biophys. J. 2010, 98, 543–551. 10.1016/j.bpj.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine D. J.; Hue K.-A.; Mayes A. M.; Griffith L. G. Simulations of Cell-Surface Integrin Binding to Nanoscale-Clustered Adhesion Ligands. Biophys. J. 2002, 82, 120–132. 10.1016/S0006-3495(02)75379-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Medda R.; Liu Z.; Galior K.; Yehl K.; Spatz J. P.; Cavalcanti-Adam E. A.; Salaita K. Nanoparticle Tension Probes Patterned at the Nanoscale: Impact of Integrin Clustering on Force Transmission. Nano Lett. 2014, 14, 5539–5546. 10.1021/nl501912g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amschler K.; Erpenbeck L.; Kruss S.; Schön M. P. Nanoscale Integrin Ligand Patterns Determine Melanoma Cell Behavior. ACS Nano 2014, 8, 9113–9125. 10.1021/nn502690b. [DOI] [PubMed] [Google Scholar]
- Amschler K.; Kossmann E.; Erpenbeck L.; Kruss S.; Schill T.; Schön M.; Möckel S. M. C.; Spatz J. P.; Schön M. P. Nanoscale Tuning of VCAM-1 Determines VLA-4–Dependent Melanoma Cell Plasticity on RGD Motifs. Mol. Cancer Res. 2018, 16, 528–542. 10.1158/1541-7786.MCR-17-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzamba B. J.; Peters D. M. Arrangement of Cellular Fibronectin in Noncollagenous Fibrils in Human Fibroblast Cultures. Journal of Cell Science 1991, 100 (3), 605–612. [DOI] [PubMed] [Google Scholar]
- Leahy D. J.; Aukhil I.; Erickson H. P. 2.0 Å Crystal Structure of a Four-Domain Segment of Human Fibronectin Encompassing the RGD Loop and Synergy Region. Cell 1996, 84, 155–164. 10.1016/S0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- Schiefner A.; Gebauer M.; Skerra A. Extra-Domain B in Oncofetal Fibronectin Structurally Promotes Fibrillar Head-to-Tail Dimerization of Extracellular Matrix Protein. J. Biol. Chem. 2012, 287, 17578–17588. 10.1074/jbc.M111.303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Früh S. M.; Schoen I.; Ries J.; Vogel V. Molecular Architecture of Native Fibronectin Fibrils. Nat. Commun. 2015, 6, 7275. 10.1038/ncomms8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patla I.; Volberg T.; Elad N.; Hirschfeld-Warneken V.; Grashoff C.; Fässler R.; Spatz J. P.; Geiger B.; Medalia O. Dissecting the Molecular Architecture of Integrin Adhesion Sites by Cryo-Electron Tomography. Nat. Cell Biol. 2010, 12, 909–915. 10.1038/ncb2095. [DOI] [PubMed] [Google Scholar]
- Wallace J. M.; Chen Q.; Fang M.; Erickson B.; Orr B. G.; Holl M. M. B. Type I Collagen Exists as a Distribution of Nanoscale Morphologies in Teeth, Bones, and Tendons. Langmuir 2010, 26, 7349–7354. 10.1021/la100006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y.; Feng X.; Zhan Y.; Wang R.; Zheng S.; Liu W.; Zeng X. Matrix Metalloproteinase-2 Promotes αvβ3 Integrin-Mediated Adhesion and Migration of Human Melanoma Cells by Cleaving Fibronectin. PLoS One 2012, 7, e41591 10.1371/journal.pone.0041591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.; Wu F.; Seo B. R.; Fischbach C.; Chen W.; Hsu L.; Gourdon D. Breast Cancer Cells Alter the Dynamics of Stromal Fibronectin-Collagen Interactions. Matrix Biol. 2017, 60–61, 86–95. 10.1016/j.matbio.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner F.; Ray A.; Dontenwill M. Integrin α5β1, the Fibronectin Receptor, as a Pertinent Therapeutic Target in Solid Tumors. Cancers 2013, 5, 27–47. 10.3390/cancers5010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechenmacher F.; Neubauer S.; Polleux J.; Mas-Moruno C.; De Simone M.; Cavalcanti-Adam E. A.; Spatz J. P.; Fässler R.; Kessler H. Functionalizing αvβ3- or α5β1-Selective Integrin Antagonists for Surface Coating: A Method to Discriminate Integrin Subtypes In Vitro. Angew. Chem., Int. Ed. 2013, 52, 1572–1575. 10.1002/anie.201206370. [DOI] [PubMed] [Google Scholar]
- Desoize B.; Jardillier J.-C. Multicellular Resistance: a Paradigm for Clinical Resistance?. Critical Reviews in Oncology/Hematology 2000, 36, 193–207. 10.1016/S1040-8428(00)00086-X. [DOI] [PubMed] [Google Scholar]
- Carragher N. O.; Frame M. C. Focal Adhesion and Actin Dynamics: A Place Where Kinases and Proteases Meet to Promote Invasion. Trends Cell Biol. 2004, 14, 241–249. 10.1016/j.tcb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Holle A. W.; Engler A. J. More Than a Feeling: Discovering, Understanding, and Influencing Mechanosensing Pathways. Curr. Opin. Biotechnol. 2011, 22, 648–654. 10.1016/j.copbio.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham R. J.; Wang Y. L. Cell Locomotion and Focal Adhesions Are Regulated by the Mechanical Properties of the Substrate. Biol. Bull. 1998, 194, 348–349. and discussion 349–350 10.2307/1543109. [DOI] [PubMed] [Google Scholar]
- Geiger B.; Spatz J. P.; Bershadsky A. D. Environmental Sensing Through Focal Adhesions. Nat. Rev. Mol. Cell Biol. 2009, 10, 21–33. 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- Glass R.; Möller M.; Spatz J. P. Block Copolymer Micelle Nanolithography. Nanotechnology 2003, 14, 1153–1160. 10.1088/0957-4484/14/10/314. [DOI] [Google Scholar]
- Schaufler V.; Czichos-Medda H.; Hirschfeld-Warnecken V.; Neubauer S.; Rechenmacher F.; Medda R.; Kessler H.; Geiger B.; Spatz J. P.; Cavalcanti-Adam E. A. Selective Binding and Lateral Clustering of α5β1 and αvβ3 Integrins: Unraveling the Spatial Requirements for Cell Spreading and Focal Adhesion Assembly. Cell Adhesion & Migration 2016, 10, 505–515. 10.1080/19336918.2016.1163453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P.; Wolf K. Tumour-Cell Invasion and Migration: Diversity and Escape Mechanisms. Nat. Rev. Cancer 2003, 3, 362–374. 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Jung J.-W.; Hwang S.-Y.; Hwang J.-S.; Oh E.-S.; Park S.; Han I.-O. Ionising Radiation Induces Changes Associated with Epithelial-Mesenchymal Transdifferentiation and Increased Cell Motility of A549 Lung Epithelial Cells. Eur. J. Cancer 2007, 43, 1214–1224. 10.1016/j.ejca.2007.01.034. [DOI] [PubMed] [Google Scholar]
- Desgrosellier J. S.; Cheresh D. A. Integrins in Cancer: Biological Implications and Therapeutic Opportunities. Nat. Rev. Cancer 2010, 10, 9–22. 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danen E. H. J.; van Rheenen J.; Franken W.; Huveneers S.; Sonneveld P.; Jalink K.; Sonnenberg A. Integrins Control Motile Strategy Through a Rho–Cofilin Pathway. J. Cell Biol. 2005, 169, 515–526. 10.1083/jcb.200412081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taherian A.; Li X.; Liu Y.; Haas T. A. Differences in Integrin Expression and Signaling Within Human Breast Cancer Cells. BMC Cancer 2011, 11, 293. 10.1186/1471-2407-11-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj M.; Strohmeyer N.; Colo G. P.; Helenius J.; Beerenwinkel N.; Schiller H. B.; Fässler R.; Müller D. J. αV-Class Integrins Exert Dual Roles on α5β1 Integrins to Strengthen Adhesion to Fibronectin. Nat. Commun. 2017, 8, 14348. 10.1038/ncomms14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelló-Cros R.; Khan D. R.; Simons J.; Valianou M.; Cukierman E. Staged Stromal Extracellular 3D Matrices Differentially Regulate Breast Cancer Cell Responses Through PI3K and Beta1-Integrins. BMC Cancer 2009, 9, 94. 10.1186/1471-2407-9-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F.; Li L.; Guan L.; Yang H.; Wu C.; Liu Y. Roles for GP IIb/IIIa and αvβ3 Integrins in MDA-MB-231 Cell Invasion and Shear Flow-Induced Cancer Cell Mechanotransduction. Cancer Lett. 2014, 344, 62–73. 10.1016/j.canlet.2013.10.019. [DOI] [PubMed] [Google Scholar]
- Zustiak S.; Nossal R.; Sackett D. L. Multiwell Stiffness Assay for the Study of Cell Responsiveness to Cytotoxic Drugs. Biotechnol. Bioeng. 2014, 111, 396–403. 10.1002/bit.25097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda E. Y.; Leight J. L.; Anseth K. S. Modulation of Matrix Elasticity with PEG Hydrogels to Study Melanoma Drug Responsiveness. Biomaterials 2014, 35, 4310–4318. 10.1016/j.biomaterials.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcioglu H. E.; van Hoorn H.; Donato D. M.; Schmidt T.; Danen E. H. J. The Integrin Expression Profile Modulates Orientation and Dynamics of Force Transmission at Cell–Matrix Adhesions. J. Cell Sci. 2015, 128, 1316–1326. 10.1242/jcs.156950. [DOI] [PubMed] [Google Scholar]
- Miao H.; Li S.; Hu Y.-L.; Yuan S.; Zhao Y.; Chen B. P. C.; Puzon-McLaughlin W.; Tarui T.; Shyy J. Y. J.; Takada Y.; et al. Differential Regulation of Rho GTPases by β1 and β3 Integrins: the Role of an Extracellular Domain of Integrin in Intracellular Signaling. J. Cell Sci. 2002, 115, 2199–2206. [DOI] [PubMed] [Google Scholar]
- Roca-Cusachs P.; Gauthier N. C.; del Rio A.; Sheetz M. P. Clustering of α5β1 Integrins Determines Adhesion Strength Whereas αvβ3 and Talin Enable Mechanotransduction. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 16245–16250. 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P.; Iskratsch T.; Sheetz M. P. Finding the Weakest Link – Exploring Integrin-Mediated Mechanical Molecular Pathways. J. Cell Sci. 2012, 125, 3025–3038. 10.1242/jcs.095794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeter S. V.; Huang J.; Perschmann N.; López-García M.; Kessler H.; Ding J.; Spatz J. P. Mimicking Cellular Environments by Nanostructured Soft Interfaces. Nano Lett. 2007, 7, 1413–1418. 10.1021/nl070098g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.