Abstract
Objectives
To determine the association of Neisseria gonorrhoeae antimicrobial resistance and genotypes using N. gonorrhoeae sequence typing for antimicrobial resistance (NG-STAR).
Methods
We characterized 124 N. gonorrhoeae isolates for their antimicrobial susceptibility profiles and NG-STAR ST characteristics using the guidelines of CLSI and EUCAST. The NG-STAR STs of seven loci were analysed. N. gonorrhoeae multiantigen sequence typing (NG-MAST) and MLST analysis was conducted in isolates with specific NG-STAR STs.
Results
NG-STAR differentiated 124 N. gonorrhoeae isolates into 84 STs, of which 66 STs were novel to the NG-STAR database. NG-STAR ST-199, ST-348, ST-428, ST-497 and ST-1138 were the predominant STs. Three N. gonorrhoeae isolates with ceftriaxone and cefixime MICs ≥1.0 mg/L were grouped as NG-STAR ST-233. NG-STAR ST-202 isolates (n=4) were associated with high azithromycin MICs and had an identical NG-MAST ST. The NG-STAR ST-348 group (n=5) comprised more isolates with reduced susceptibility to cefixime (n=4) than cefixime-susceptible isolates (n=1).
Conclusions
NG-STAR analysis differentiated N. gonorrhoeae isolates in settings with a high prevalence of antimicrobial resistance. Specific NG-STAR STs are associated with reduced susceptibility to ceftriaxone or cefixime and resistance to azithromycin in N. gonorrhoeae.
Introduction
Neisseria gonorrhoeae has developed antimicrobial resistance to most antimicrobials in the past few decades.1–3 Molecular determinants of chromosomally mediated resistance have been identified.4 Quinolone resistance of N. gonorrhoeae is associated with the quinolone resistance determinants in the gyrA and parC genes.5 Penicillin resistance and reduced susceptibility to the third-generation cephalosporins (cefixime or ceftriaxone) have been associated with mutations in the penA, porB, mtrR and ponA genes.4 Determinants of high-level tetracycline resistance include the presence of tet(M) or mutations in rpsJ.6 Azithromycin resistance is associated with mutations in mtrR and 23S rRNA.7
Molecular typing methods have been used to differentiate genotypes of N. gonorrhoeae isolates and to study the association between N. gonorrhoeae genotypes and antimicrobial resistance phenotypes, such as porB sequence typing,8 MLST,8N. gonorrhoeae multiantigen sequence typing (NG-MAST)8,9 and N. gonorrhoeae sequence typing for antimicrobial resistance (NG-STAR).10 NG-STAR STs are determined using an allelic profile of the seven antimicrobial-resistance-associated loci: penA, mtrR, porB, ponA, gyrA, parC and 23S rRNA.10 NG-STAR analysis has been used in studies of N. gonorrhoeae isolates from Canada,10 Australia11,12 and Japan.13 Specific NG-STAR STs are associated with unique antimicrobial resistance phenotypes. Reduced susceptibility to ceftriaxone or cefixime is associated with NG-STAR ST-90; resistance to azithromycin is associated with NG-STAR ST-58, ST-61 and ST-64.10 This is the first study in China, to the best of our knowledge, that has employed the NG-STAR database to differentiate antimicrobial-resistant N. gonorrhoeae recovered from settings with a high prevalence of N. gonorrhoeae antimicrobial resistance.
Materials and methods
N. gonorrhoeae isolates were collected from male patients at Shanghai Skin Disease Hospital from January to December 2017 through the national Gonococcal Antimicrobial Susceptibility Surveillance Program. The first 10 N. gonorrhoeae isolates of each month were used in this study (n=120). Additionally, a ciprofloxacin-susceptible N. gonorrhoeae isolate and three N. gonorrhoeae isolates with ceftriaxone or cefixime MICs ≥1.0 mg/L, collected in 2017, were also included in this study. These 124 N. gonorrhoeae isolates were isolated and identified as previously described.5N. gonorrhoeae isolates were cultivated on Thayer–Martin (T–M) medium supplemented with 1% IsoVitaleX and identified using the oxidase test, Gram staining and glucose utilization tests. MICs for six antimicrobials were determined using the agar dilution method according to CLSI.14N. gonorrhoeae ATCC 49226 was used as a reference strain. The criteria for resistance phenotypes were as follows: MICs ≥2 mg/L for penicillin or tetracycline; MICs ≥1 mg/L for ciprofloxacin;14 MICs ≥1 mg/L for azithromycin; and MICs ≥0.25 mg/L for ceftriaxone or cefixime, using the breakpoints described previously.15
For genotyping, specific primers were used to amplify and to sequence various loci (Table S1, available as Supplementary data at JAC Online). Seven NG-STAR loci (penA, mtrR, porB, ponA, gyrA, parC and 23S rRNA) were PCR-amplified as previously described.5 DNA sequencing was performed at Sangon Biotech Co. DNA sequences were analysed and edited using Geneious and Vector NTI.
NG-STAR analysis was conducted using the NG-STAR database (https://ngstar.canada.ca).10 NG-MAST was performed using the porB and tbpB loci (Table S1)9 and NG-MAST STs were assigned using the NG-MAST database (http://www.ng-mast.net). MLST analysis was performed using seven loci (abcZ, adk, aroE, fumC, phdC, gdh and pgm) (Table S1)8 and MLST STs were assigned using the MLST database (https://pubmlst.org/neisseria).
Phylogenetic analysis based on seven NG-STAR loci was performed using Molecular Evolutionary Genetics Analysis (MEGA) and neighbour-joining trees and bootstrapping were analysed (https://www.megasoftware.net). Simpson’s diversity index was used to evaluate the discriminatory levels of NG-STAR.16 Statistical analysis was performed using Fisher’s exact test (SPSS Statistics version 22.0). A P value of <0.05 was considered statistically significant. DNA sequences were submitted to GenBank of the NCBI. The GenBank accession numbers are listed in Table S2.
Results
A high level of antimicrobial resistance was observed among the 124 N. gonorrhoeae isolates (Table S3). The proportions of N. gonorrhoeae isolates with resistance to penicillin, tetracycline, ciprofloxacin and azithromycin were 81.5% (n=101), 59.7% (n=74), 99.2% (n=123) and 8.1% (n=10), respectively. The proportions of N. gonorrhoeae isolates with resistance to ceftriaxone or cefixime (MICs ≥0.25 mg/L) were 6.5% (n=8) and 16.9% (n=21), respectively.
NG-STAR analysis differentiated the 124 N. gonorrhoeae isolates into 84 STs. There were 66 new NG-STAR STs, comprising 79 N. gonorrhoeae isolates (Table 1). ST-199 (n=7) was the predominant ST. The next most common STs were ST-348, ST-428, ST-497 and ST-1138; each of these STs was represented by five N. gonorrhoeae isolates. Four isolates were ST-202. Four NG-STAR STs were each associated with three isolates. Seven NG-STAR STs were each represented by two isolates. Sixty-seven NG-STAR STs were each associated with a single isolate. Three N. gonorrhoeae isolates with ceftriaxone or cefixime MICs ≥1.0 mg/L exhibited NG-STAR ST-233, having PenA type 60. Four N. gonorrhoeae isolates with NG-STAR ST-202 showed an identical NG-MAST ST (ST1866) and two MLST STs (ST10899 and ST12039). Five N. gonorrhoeae isolates with NG-STAR ST-348 had four different NG-MAST STs and two MLST STs (ST7363 and ST14283). The discrimination index of NG-STAR was 0.989.
Table 1.
Association of NG-STAR STs with MICs for 124 N. gonorrhoeae isolates collected in 2017 in Shanghai
| NG-STAR STa | No. of isolates | Ceftriaxone MIC range (mg/L) | Cefixime MIC range (mg/L) | Azithromycin MIC range (mg/L) | Ciprofloxacin MIC range (mg/L) | Penicillin MIC range (mg/L) | Tetracycline MIC range (mg/L) |
|---|---|---|---|---|---|---|---|
| 38 | 3 | 0.008–0.06 | 0.015–0.125 | 0.03–0.25 | 2.0–16.0 | 0.25–16.0 | 0.5–2.0 |
| 199 | 7 | 0.03–0.06 | 0.03–0.06 | 0.06–0.5 | 4.0–16.0 | 1.0–16.0 | 0.5–32.0 |
| 202 | 4 | 0.015–0.06 | 0.03–0.06 | 8.0 | 8.0 | 2.0–16.0 | 32.0 |
| 233 | 3 | ≥1.0 | ≥1.0 | 0.125–0.25 | 16.0 | 2.0–4.0 | 1.0–2.0 |
| 346 | 2 | 0.06 | 0.06–0.125 | 0.25 | 16.0 | 2.0–16.0 | 2.0–4.0 |
| 348 | 5 | 0.03–0.125 | 0.015–1.0 | 0.06–0.25 | 16.0 | 1.0–4.0 | 1.0–2.0 |
| 428 | 5 | 0.03–0.25 | 0.06–0.25 | 0.06–0.5 | 8.0–16.0 | 1.0–16.0 | 1.0–16.0 |
| 497 | 5 | 0.03–0.25 | 0.06–0.25 | 0.125–1 | 8.0–16.0 | 1.0–32.0 | 1.0–4.0 |
| 501 | 2 | 0.06–0.125 | 0.06–0.125 | 0.06–0.25 | 16.0 | 2.0–4.0 | 1.0 |
| 1138 | 5 | 0.015–0.06 | 0.03–0.25 | 0.25–2 | 16.0 | 4.0–16.0 | 2.0–4.0 |
| 1144 | 2 | 0.03 | 0.06 | 0.125–0.25 | 8.0–16.0 | 16.0 | 32.0 |
| 1146 | 3 | 0.06 | 0.06–0.125 | 0.125–0.25 | 2.0–8.0 | 2.0–16.0 | 1.0 |
| 1150 | 2 | 0.03 | 0.06–0.125 | 0.125 | 8.0 | 1.0–16.0 | 16.0 |
| 1151 | 2 | 0.03–0.06 | 0.06–0.125 | 0.25 | 8.0–16.0 | 2.0 | 2.0–16.0 |
| 1155 | 2 | 0.03–0.06 | 0.06–0.125 | 0.25–0.5 | 4.0–8.0 | 1.0–2.0 | 16.0 |
| 1187 | 3 | 0.06–0.125 | 0.125 | 0.125–0.5 | 16.0 | 4.0–16.0 | 2.0–4.0 |
| 1190 | 2 | 0.06 | 0.06 | 0.25 | 16.0 | 16.0 | 1.0 |
| Other STs (67)b | 1/ST | 0.008–0.25 | 0.008–1.0 | 0.003–16.0 | 0.004–16.0 | 0.5–16.0 | 0.125–32.0 |
| Total no. of STs = 84 | total = 124 | 0.008 to ≥1.0 | 0.008 to ≥1.0 | 0.003–16.0 | 0.004–16.0 | 0.25–16.0 | 0.125–32.0 |
Eight STs in bold are novel to the NG-STAR database.
STs with single isolates are STs 51, 90, 138, 493, 496, 506, 515, 909, 1109, 1136, 1137, 1140–1143, 1145, 1147–1149, 1152–1154, 1156–1186, 1188, 1189, 1191–1198, 1217, 1436, 1463 and 1464, among which the 58 STs in bold are new to the NG-STAR database.
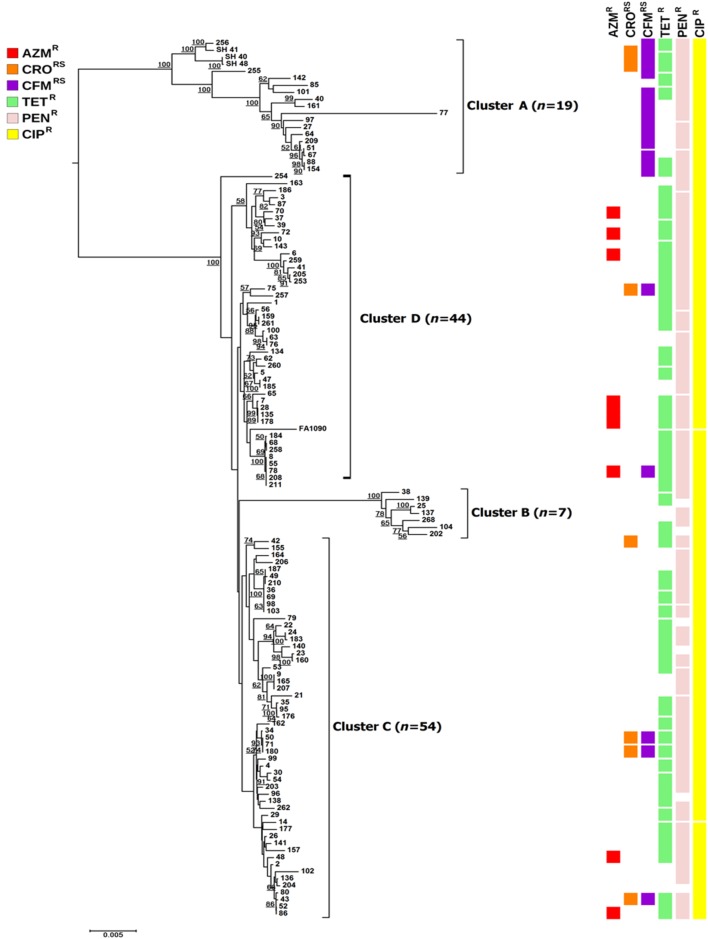
Phylogenetic analysis based on the seven NG-STAR loci differentiated the N. gonorrhoeae isolates into four clusters (Figure 1). Cluster A had 15.3% (19/124) of the isolates, including 37.5% (3/8) of the isolates with reduced susceptibility to ceftriaxone and 76.2% (16/21) of the isolates with reduced susceptibility to cefixime. The three isolates with ceftriaxone and cefixime MICs ≥1.0 mg/L were present in cluster A. Cluster B contained seven isolates, which had ciprofloxacin resistance phenotypes (resistance to both ciprofloxacin and penicillin, or resistance to both ciprofloxacin and tetracycline). The N. gonorrhoeae isolates in clusters C (n=54) and D (n=44) had diverse resistance phenotypes. N. gonorrhoeae isolates with azithromycin resistance were scattered across the tree.
Figure 1.
Phylogenetic analysis based on the seven loci for NG-STAR. The phylogenetic tree was constructed using MEGA 6 software. Horizontal lines are proportional to phylogenetic distance. Resistance phenotypes are shown as coloured squares for each isolate and each square represents one isolate. Numbers on the tips are N. gonorrhoeae isolate numbers. Bootstrap values of ≥50 are shown at the nodes of the tree branches. Four clusters (clusters A–D) are defined arbitrarily. AZMR, azithromycin resistance; CRORS, reduced susceptibility to ceftriaxone; CFMRS, reduced susceptibility to cefixime; TETR, tetracycline resistance; PENR, penicillin resistance; CIPR, ciprofloxacin resistance.
Discussion
This is the first study, to the best of our knowledge, to investigate the association of NG-STAR STs and antimicrobial resistance in N. gonorrhoeae clinical isolates from settings in which MDR is prevalent. We found that N. gonorrhoeae isolates with reduced susceptibility to ceftriaxone or cefixime could be grouped into a distinguishable cluster. Three isolates with ceftriaxone or cefixime MICs ≥1.0 mg/L exhibited a single NG-STAR type (ST-233) and appeared in the same cluster. Our results suggest that NG-STAR STs are associated with antimicrobial resistance phenotypes and can be potentially used in determining clonal expansion of antimicrobial resistance in settings in which MDR is prevalent.
In our study, the majority of NG-STAR STs were novel to the NG-STAR database (66/84), similar to that observed in a previous report.11 The most prevalent NG-STAR STs in our study were ST-199, ST-348, ST-428, ST-497 and ST-1138; whereas NG-STAR ST-90, ST-42, ST-91, ST-64 and ST-139 were most common in Canada10 and the frequencies of NG-STAR ST-755 and ST-NV9 were high in Australia.11 This regional distribution of NG-STAR STs suggests a correlation with geographic locations.
As previously reported,10,12 specific NG-STAR STs tended to be associated with characteristic antimicrobial resistance phenotypes such as ST-233 isolates with high ceftriaxone and cefixime MICs (≥1.0 mg/L). The first isolate reported as NG-STAR ST-233 was from Japan in 2015 (FC428).12 NG-STAR ST-233 was also reported in 2017 from Australia,12 Canada12 and France,17 followed by a report from the UK in 2018.18 The NG-STAR ST-233 isolates reported in all of these countries were associated with travel to Southeast Asia. The NG-STAR ST-233 N. gonorrhoeae isolates would, to the best of our knowledge, be the first report in China. However, further investigation of the social or epidemiological connections of the patients is warranted. In-depth genomic analysis would provide detailed information on the association of NG-STAR ST-233 and clone FC428. The NG-STAR STs associated with reduced susceptibility to ceftriaxone (ST-91 or ST-97) reported by another group10 were not observed in this study, which might be due to the criteria for reduced susceptibility to ceftriaxone used in the Canadian report (≥0.06 mg/L).10
The percentage of isolates identified as NG-STAR ST-202 was significantly higher among azithromycin-resistant N. gonorrhoeae isolates than azithromycin-susceptible isolates (P<0.001), indicating that ST-202 is associated with resistance to azithromycin, which is consistent with another report.10N. gonorrhoeae isolates identified as NG-STAR ST-202 (n=4) exhibited an identical NG-MAST ST (ST1866) and two MLST STs (ST10899 and ST12039). It has been reported that NG-MAST ST1866 and MLST ST10899/ST12039 were the predominant STs among high-level azithromycin-resistant N. gonorrhoeae isolates.19 The percentage of isolates identified as NG-STAR ST-348 was significantly higher among N. gonorrhoeae isolates with reduced susceptibility to cefixime (4/21) than that for isolates susceptible to cefixime (1/103) (P<0.05), indicating that NG-STAR ST-348 could be associated with reduced susceptibility to cefixime. However, the five N. gonorrhoeae isolates with NG-STAR ST-348 had various cefixime MICs and had four different NG-MAST STs and two MLST STs (Table 1).
This study only investigated a low number of isolates (<2%) from all of the 5711 N. gonorrhoeae cases reported in Shanghai in 2017 (data not shown). However, it is representative of the institution where the isolates were collected. A study with a larger sample size is required to extrapolate a broader strain distribution.
In conclusion, we preliminarily characterized NG-STAR STs in N. gonorrhoeae in Shanghai. The majority of NG-STAR STs were novel to the database. The most prevalent NG-STAR STs differed from those in the reports from Canada and Australia. NG-STAR ST-233 is associated with high ceftriaxone or cefixime MICs (≥1.0 mg/L). NG-STAR ST-348 is associated with reduced susceptibility to cefixime. NG-STAR ST-202 is associated with azithromycin resistance in N. gonorrhoeae.
Supplementary Material
Acknowledgements
We thank Dr Mingmin Liao (University of Saskatchewan, Canada) for assistance in data analysis and in the preparation of the manuscript.
Funding
The Wu Jieping Medical Foundation of China (grant number 320.6750.17339, to W. Gu) and the National Natural Science Foundation of China (grant number 81602905, to Y. Dong) supported this study.
Transparency declarations
None to declare.
References
- 1. Gu W, Chen Y, Yang Y. et al. Twenty-five-year changing pattern of gonococcal antimicrobial susceptibility in Shanghai: surveillance and its impact on treatment guidelines. BMC Infect Dis 2014; 14: 731.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wi T, Lahra MM, Ndowa F. et al. Antimicrobial resistance in Neisseria gonorrhoeae: global surveillance and a call for international collaborative action. PLoS Med 2017; 14: e1002344.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen SC, Yin YP, Dai XQ. et al. First nationwide study regarding ceftriaxone resistance and molecular epidemiology of Neisseria gonorrhoeae in China. J Antimicrob Chemother 2016; 71: 92–9. [DOI] [PubMed] [Google Scholar]
- 4. Unemo M, Shafer WM.. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 2014; 27: 587–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang Y, Liao M, Gu WM. et al. Antimicrobial susceptibility and molecular determinants of quinolone resistance in Neisseria gonorrhoeae isolates from Shanghai. J Antimicrob Chemother 2006; 58: 868–72. [DOI] [PubMed] [Google Scholar]
- 6. Hu M, Nandi S, Davies C. et al. High-level chromosomally mediated tetracycline resistance in Neisseria gonorrhoeae results from a point mutation in the rpsJ gene encoding ribosomal protein S10 in combination with the mtrR and penB resistance determinants. Antimicrob Agents Chemother 2005; 49: 4327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chisholm SA, Dave J, Ison CA.. High-level azithromycin resistance occurs in Neisseria gonorrhoeae as a result of a single point mutation in the 23S rRNA genes. Antimicrob Agents Chemother 2010; 54: 3812–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Unemo M, Dillon JA.. Review and international recommendation of methods for typing Neisseria gonorrhoeae isolates and their implications for improved knowledge of gonococcal epidemiology, treatment, and biology. Clin Microbiol Rev 2011; 24: 447–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liao M, Helgeson S, Gu WM. et al. Comparison of Neisseria gonorrhoeae multi antigen sequence typing and porB sequence analysis for identification of clusters of N. gonorrhoeae isolates. J Clin Microbiol 2009; 47: 489–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Demczuk W, Sidhu S, Unemo M. et al. Neisseria gonorrhoeae sequence typing for antimicrobial resistance, a novel antimicrobial resistance multilocus typing scheme for tracking global dissemination of N. gonorrhoeae strains. J Clin Microbiol 2017; 55: 1454–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Al Suwayyid BA, Coombs GW, Speers DJ. et al. Genomic epidemiology and population structure of Neisseria gonorrhoeae from remote highly endemic Western Australian populations. BMC Genomics 2018; 19: 165.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lahra MM, Martin I, Demczuk W. et al. Cooperative recognition of internationally disseminated ceftriaxone-resistant Neisseria gonorrhoeae strain. Emerg Infect Dis 2018; 24: doi:10.3201/eid2404.171873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hiyama Y, Takahashi S, Sato T. et al. Evaluation of susceptibilities to carbapenems and faropenem against cephalosporin-resistant Neisseria gonorrhoeae clinical isolates with penA mosaic alleles. Microb Drug Resist 2019; 25: 427–33. [DOI] [PubMed] [Google Scholar]
- 14.CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Twenty-Ninth Edition: M100 2019.
- 15.EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 9.0, 2019. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf.
- 16. Dillon JA, Rahman M, Yeung KH.. Discriminatory power of typing schemes based on Simpson’s index of diversity for Neisseria gonorrhoeae. J Clin Microbiol 1993; 31: 2831–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poncin T, Fouere S, Braille A. et al. Multidrug-resistant Neisseria gonorrhoeae failing treatment with ceftriaxone and doxycycline in France, November 2017. Euro Surveill 2018; 23: pii=1800264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eyre DW, Town K, Street T. et al. Detection in the United Kingdom of the Neisseria gonorrhoeae FC428 clone, with ceftriaxone resistance and intermediate resistance to azithromycin, October to December 2018. Euro Surveill 2019; 24: pii=1900147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu YH, Wang YH, Liao CH. et al. Emergence and spread of Neisseria gonorrhoeae strains with high-level resistance to azithromycin in Taiwan from 2001 to 2018. Antimicrob Agents Chemother 2019; 63: e00773–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.