Abstract
Background & Aims
The enteroendocrine cell (EEC) lineage is important for intestinal homeostasis. It was recently shown that EEC progenitors contribute to intestinal epithelial growth and renewal, but the underlying mechanisms remain poorly understood. MicroRNAs are under-explored along the entire EEC lineage trajectory, and comparatively little is known about their contributions to intestinal homeostasis.
Methods
We leverage unbiased sequencing and eight different mouse models and sorting methods to identify microRNAs enriched along the EEC lineage trajectory. We further characterize the functional role of EEC progenitor-enriched miRNA, miR-7, by in vivo dietary study as well as ex vivo enteroid in mice.
Results
First, we demonstrate that miR-7 is highly enriched across the entire EEC lineage trajectory and is the most enriched miRNA in EEC progenitors relative to Lgr5+ intestinal stem cells. Next, we show in vivo that in EEC progenitors miR-7 is dramatically suppressed under dietary conditions that favor crypt division and suppress EEC abundance. We then demonstrate by functional assays in mouse enteroids that miR-7 exerts robust control of growth, as determined by budding (proxy for crypt division), EdU and PH3 staining, and likely regulates EEC abundance also. Finally, we show by single-cell RNA sequencing analysis that miR-7 regulates Xiap in progenitor/stem cells and we demonstrate in enteroids that the effects of miR-7 on mouse enteroid growth depend in part on Xiap and Egfr signaling.
Conclusions
This study demonstrates for the first time that EEC progenitor cell-enriched miR-7 is altered by dietary perturbations and that it regulates growth in enteroids via intact Xiap and Egfr signaling.
Keywords: miR-7, Enteroendocrine Lineage, Small Intestine, Enteroid, Proliferation
Abbreviations used in this paper: aISC, actively cycling intestinal stem cell; BSA, bovine serum albumin; EdU, 5-ethynyl-2′deoxyuridine; EEC, enteroendocrine cell; FACS, fluorescence-activated cell sorting; HFD, high-fat diet; IAP, inhibitor of apoptosis protein; ISC, intestinal stem cell; LFD, low-fat diet; LNA, locked nucleic acid; LNA-7, locked nucleic acid inhibitors against microRNA 7; LNA-scr, locked nucleic acid scramble control; LSP, lower side population; miR-7, microRNA 7; miRNA, microRNA; rISC, reserve/slowly cycling intestinal stem cell; PBS, phosphate-buffered saline; RNA-seq, RNA sequencing; RT-qPCR, reverse transcriptase quantitative polymerase chain reaction; USP, upper side population
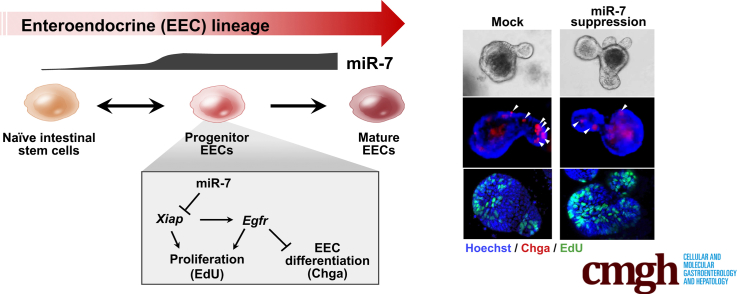
Graphical abstract
Summary.
It was recently shown that progenitors of enteroendocrine cells contribute to intestinal epithelial growth, but the underlying mechanisms remain poorly understood. We uncover the role of enteroendocrine cell–progenitor enriched microRNA 7 in regulating intestinal epithelial growth via Xiap and Egfr signaling.
The intestinal epithelium is the most rapidly renewing tissue in the body. This feature is driven by crypt-based intestinal stem cells (ISCs), which exhibit self-renewal properties and are responsible for giving rise to all of the differentiated cell types in the absorptive (enterocyte) and secretory lineages (Paneth cell, tuft cell, goblet cell and enteroendocrine cells [EECs]).1 So far, 2 distinct populations of ISCs have been defined: actively cycling ISCs (aISCs) at the base of the crypt and reserve/slowly cycling ISCs (rISCs) at the +4 position from the crypt base.2 More recently, though, several other intermediate cell populations, notably progenitors of EECs, have been shown to participate in the control of crypt behavior under certain conditions.3,4
EEC progenitors, which were thought to be fully committed to EEC differentiation, have recently been recognized to have proliferative potential and thereby contribute to the control of cell proliferation, crypt growth, and related behaviors.3,4 A recent study identified Prospero homeobox protein 1 (Prox1) as a novel marker labeling intermediates in the EEC lineage and demonstrated that sorted Prox1+ cells are sufficient for establishing enteroids ex vivo. Despite this advance, much remains unknown about the mechanisms that control EEC lineage behavior. It is of substantial interest to map the molecular landscape of the cells in the entire EEC lineage trajectory to define the mechanisms that control intestinal epithelial cell proliferation, crypt division or growth, or EEC differentiation.
MicroRNAs (miRNAs) are prominent posttranscriptional regulators of growth and cell fate decisions in many organ systems and disease models5,6; however, very little is known about their role in the regulation of intestinal crypt behavior. In fact, it is not even known which miRNAs are expressed along the entire EEC lineage trajectory, particularly the EEC progenitors or whether they are sensitive to perturbations that influence crypt division or EEC differentiation.7 In this study, using 8 different reporter mice and several sorting methods, we profile miRNAs in several lineages of the small intestinal epithelium, identify microRNA 7 (miR-7) as the most highly enriched miRNA in EEC progenitors (Prox1+) relative to Lgr5+ stem cells, show that miR-7 in EEC progenitors is among the most sensitive miRNAs to dietary conditions that favor crypt growth and reduced EEC abundance, and demonstrate through ex vivo functional studies and single cell analyses that miR-7 controls enteroid growth in part by regulation of Xiap.
Results
MiR-7 Is the Most Enriched miRNA in EEC Progenitors Relative to Lgr5+ Stem Cells
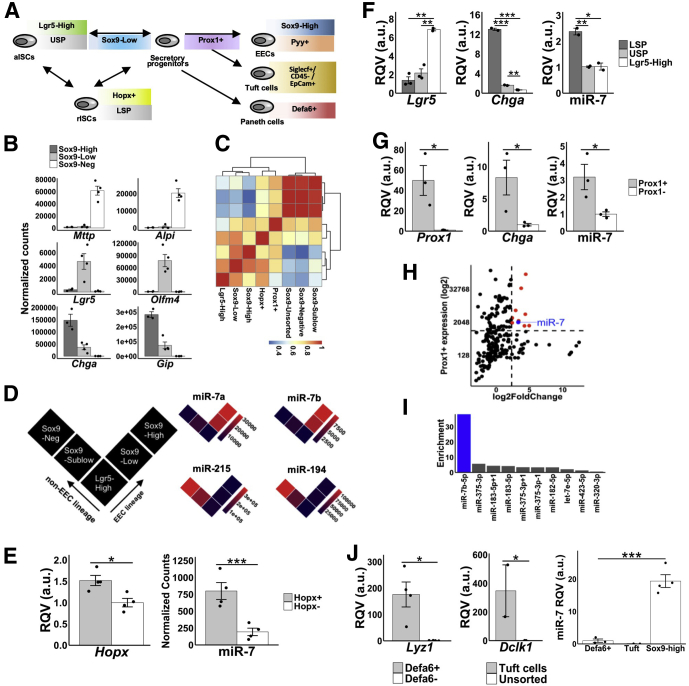
In this study, we defined the EEC lineage trajectory as the following: (1) Lgr5+ aISCs, (2) Sox9-Low cells and Hopx+ cells that exhibit features of the EEC lineage, (3) Prox1+ EEC progenitors, (4) Sox9-High and lower side population (LSP) cells that represent a mixed population of rISCs and mature EECs, and (5) Pyy+ cells that represent mature EECs. To define the miRNA landscape across the EEC lineage trajectory, we first investigated Sox9-EGFP reporter mice (Figure 1A). From the jejunal crypts of the Sox9-EGFP mice, we sorted and performed small RNA sequencing (RNA-seq) analysis on 4 different epithelial cell populations enriched in enterocytes (Sox9-Negative), stem cells or EEC progenitors (Sox9-Low) (hereafter referred to as EEC progenitors), transit amplifying cells (Sox9-Sublow), and mature EECs (Sox9-High), and demonstrated that each fraction is enriched for the expected markers (Figure 1B). We then focused our analysis on the cell populations in the EEC lineage trajectory, Sox9-Low and Sox9-High. The small RNA-seq analysis identified a total of 187 miRNAs in these 2 populations. Of these, we found that only 8 miRNAs are enriched (>5-fold) in mature EECs (class A), 2 in stem or EEC progenitors (class B), and 14 in both (class C) relative to unsorted intestinal epithelial cells (Table 1). Class A miRNAs represent candidate regulators of mature EEC function, class B miRNAs represent candidate regulators of EEC progenitor cell behavior, and class C miRNAs represent candidate regulators of both mature EEC function and EEC progenitor cell behavior. Notably, class C miRNAs include miR-7b, which has been previously extensively studied in the context of endocrine pancreatic development and function.8, 9, 10, 11, 12, 13, 14, 15 MiR-7 was also shown to be enriched in a specific subtype of mature EECs and cholecystokinin-producing EECs,16 and also in enterochromaffin cell-derived tumors17; however, importantly, the expression pattern of miR-7 (or any other miRNA) across the entire EEC lineage trajectory has never before been reported.
Figure 1.
MicroRNA-7 is highly enriched in the enteroendocrine (EEC) lineage trajectory. (A) Schematic diagram of different sorted cell populations representing specific cell lineages in the small intestine. (B) Level of expression (RNA-seq) of specific marker genes in each of the Sox9-Low (n = 4), Sox9-High (n = 3), and Sox9-Negative (n = 4) populations of cells. (C) Hierarchical clustering analysis based on the expression profiles of the top 50 most variable miRNAs across the different sorted cell populations shown in the heat map (Sox9-Low, n = 3; Sox9-High, n = 3; Sox9-Negative, n = 3; Sox9-Sublow, n = 3; Sox9-Unsorted, n = 2; Lgr5-High, n = 2; Hopx+, n = 4; Prox1+, n = 3). (D) MiR-7a/b expression in the EEC lineage vs non-EEC absorptive lineage. Similar data for miR-194 and miR-215 provided for sake of comparison. (E) RT-qPCR data showing enrichment of Hopx and miR-7 in Hopx+ cells (n = 4) relative to Hopx– cells (n = 4). (F) RT-qPCR data showing enrichment of miR-7, Lgr5, and Chga in LSP (n = 2) relative to USP (n = 2) and Lgr5+ cells (n = 2). (G) RT-qPCR data showing enrichment of Prox1, miR-7, and Chga in Prox1+ cells (n = 3) compared with Prox1– cells (n = 3). (H) Scatter plot showing abundance (y-axis) and enrichment (x-axis) of all detected miRNAs in Prox1+ cells (n = 3) relative to Prox1– cells (n = 3). MiRNAs above expression of 1000 reads per million mapped to miRNAs and 5-fold enrichment are shown in red (n = 10). Among these, miR-7b is highlighted in blue. (I) Fold-difference in expression of the 10 miRNAs highlighted in panel F in Prox1+ cells (n = 3) relative to Lgr5+ cells (n = 2) highlights miR-7 (blue) as a robust EEC progenitor cell enriched miRNA. (J) The left panel shows RT-qPCR data showing enrichment of Lyz1 (marker of Paneth cells) in Defa6+ (n = 4) relative to Defa6– cells (n = 4). The middle panel shows RT-qPCR data showing enrichment of Dclk1 (marker of tuft cells) in Siglecf+/CD45-/EpCam+ cells (n = 2) relative to unsorted cells (n = 2). The right panel shows RT-qPCR data showing miR-7 enrichment in EECs (Sox9-High; n = 3) compared with Paneth and tuft cells. * P < .05, ** P < .01, *** P < .001 by 2-tailed Student t test. RQV, relative quantitative value.
Table 1.
Small RNA-seq Profiling Followed by Enrichment Analysis of miRNAs in Stem/EEC Progenitors (Sox9-Low, n = 3) and in Mature EECs (Sox9-High, n = 3) Relative to Unsorted Intestinal Epithelial Cells (n = 2)
| Class A miRNAs enriched in mature EEC (Sox9-High) |
Class B miRNAs enriched in progenitor EEC (Sox9-Low) |
Class C miRNAs enriched in both mature and progenitor EECs |
|---|---|---|
| miR-139-3p | miR-181c-3p | let-7e-5p |
| miR-182-5p | miR-181d-5p | miR-1224-5p |
| miR-182-5p_+_1 | miR-125a-5p | |
| miR-183-5p | miR-132-3p | |
| miR-183-5p_+_1 | miR-184-3p | |
| miR-328-3p | miR-375-3p | |
| miR-672-5p | miR-375-3p_-_1 | |
| miR-744-5p | miR-375-3p_+_1 | |
| miR-375-3p_+_2 | ||
| miR-7a-2-3p | ||
| miR-7b-5p | ||
| miR-7b-5p_+_1 | ||
| miR-92b-3p | ||
| miR-99b-5p |
Only miRNAs that have an average reads per million mapped to miRNAs (reads per million mapped to miRNAs) >100 in either Sox9-Low or Sox9-High and have <2-fold enrichment in Sox9-Sublow (transit-amplifying cells) and Sox9-Negative (enterocytes) are shown.
EEC, enteroendocrine cell; miRNA, microRNA; RNA-seq, RNA sequencing.
Next, from the jejunal crypts of Lgr5-EGFP, Prox1-EGFP, and Hopx-CreERT2;Rosa26-tdTomato reporter mice (Figure 1A), we sorted Lgr5+, Prox1+, and Hopx+ cells, respectively, and performed small RNA-seq to define miRNA profiles in each population (Figure 1C). We found that the level of expression of miR-7a and miR-7b increases steadily along the EEC trajectory from Lgr5+ aISCs to Sox9-Low EEC progenitors to Sox9-High mature EECs, in contrast to other miRNAs such as miR-194 and miR-215, which are depleted in the EEC lineage and enriched in the non-EEC, absorptive lineage (Sox9-Sublow and Sox9-Negative) (Figure 1D). We also found by quantitative polymerase chain reaction (qPCR) that miR-7 is significantly enriched in Hopx+ cells (Figure 1E), which have been shown previously to exhibit molecular features of EEC progenitors.18 Moreover, through small RNA-seq analysis, we found that miR-7b is one of the top 3 miRNAs to be significantly enriched (>5-fold enrichment based on expression as measured by reads per million mapped to miRNAs and P < .05 by 2-tailed Student t test) in Hopx+ cells relative to Hopx– cells, further underscoring the potential importance of miR-7b in EEC progenitors.
To validate that the miR-7 family is enriched in EEC progenitors, we next performed side population sorting of the intestinal epithelium and isolated the LSP and upper side population (USP) of cells, which correspond to rISCs and aISCs, respectively (Figure 1A). Consistent with the notion of overlapping identity between rISCs and cell populations in the EEC lineage,18 we found that LSP cells exhibit molecular features of mature EECs and EEC progenitors, including expression of Chga (Figure 1F), and are depleted for markers of aISCs, including Lgr5 (Figure 1F). RT-qPCR analysis showed that miR-7 is significantly enriched in LSP relative to both USP and Lgr5+ cells (Figure 1F), confirming miR-7 enrichment in cells with EEC progenitor features.
To cement the finding of miR-7 enrichment in EEC progenitors, we next turned our attention to the Prox1+ cells sorted from the intestinal epithelium of Prox1-EGFP reporter mice (Figure 1A, C). Prox1 was recently shown to mark intestinal secretory progenitors with the capacity to either differentiate to mature EECs or exhibit proliferative stem cell–like activity,4 and our small RNA-seq analysis showed that the miRNA profile of intestinal epithelial Prox1+ cells most closely resembles that of Hopx+ cells (rISCs/EEC progenitors) and Sox9-Low cells (EEC progenitors) (Figure 1C). We first demonstrated by qPCR that the traditional EEC lineage marker, Chga, and miR-7 are significantly enriched in Prox1+ cells (Figure 1G). Then, we analyzed the small RNA-seq data and found that only 10 miRNAs are >5-fold enriched in Prox1+ cells relative to Prox1– cells (Figure 1H). Several of these, including miR-7b, overlap with the class C miRNAs defined in Table 1. Notably, among these 10 miRNAs, we found that miR-7b is by far the most dramatically enriched (∼41-fold) in Prox1+ EEC progenitors relative to Lgr5+ aISCs (Figure 1I).
Prox1+ progenitor cells are thought to give rise not only to mature EECs but also to differentiated tuft cells. To determine whether miR-7 is truly enriched along the EEC lineage trajectory, or also highly abundant in tuft cells, we next measured miR-7 in mouse jejunal tuft cells (Epcam+/Siglecf+/Cd45– cells sorted from wild-type C57BL/6J mice), which are highly enriched as expected for the tuft cell marker Dclk1 (Figure 1J). This analysis revealed that miR-7 is >350-fold enriched in Sox-9 High EECs relative to Dclk1+ tuft cells (Figure 1J). As a control, we also included Lyz1+ Paneth cells sorted from the Defa6-Cre;tdTomato line (Figure 1J), and demonstrated that miR-7 is indeed significantly depleted in these cells relative to Sox9-High EECs (Figure 1J). As additional validation, we sorted Pyy+ EECs from the jejunum of Pyy-EGFP reporter mice and found that miR-7 is >600-fold more highly expressed in Pyy+ cells than in tuft cells (data not shown). These findings provide strong support for the enrichment of miR-7 along the entire EEC lineage trajectory.
Taken together, these data define a clear EEC lineage trajectory (from Lgr5+ aISCs to mature EECs) miRNA signature for the first time and reveal that miR-7 is the most enriched miRNA in EEC progenitors compared with Lgr5+ aISCs.
MiR-7 Expression in EEC Progenitors, Not Mature EECs, Is Suppressed Under Conditions of Increased Intestinal Crypt Division and Reduced EEC Abundance
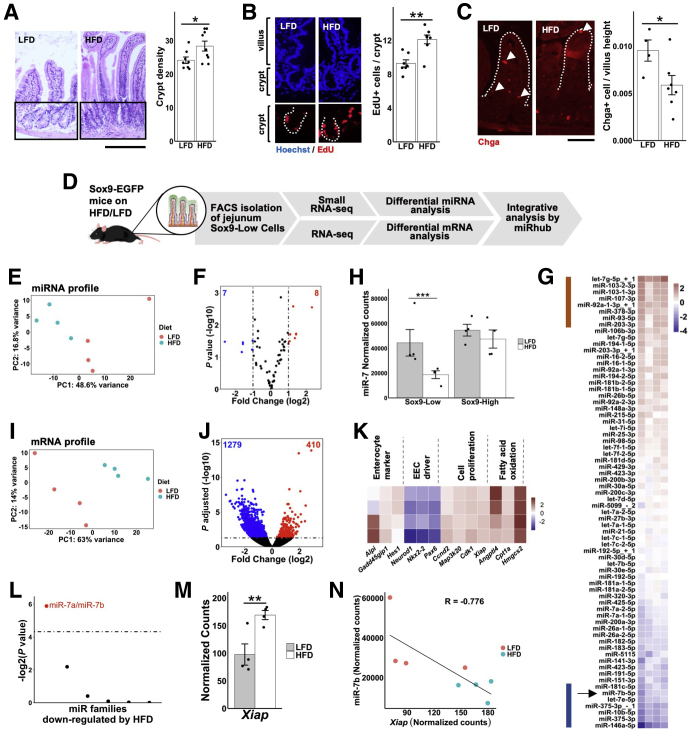
Several different types of environmental perturbations, including dietary modifications, have been reported to alter both crypt growth and EEC differentiation.19,20 For example, a high-fat diet (HFD) robustly increases intestinal crypt division (indicative of enhanced proliferation) and decreases EEC abundance in mice compared with a low-fat diet (LFD).20 Our previously described expression analysis demonstrated that miR-7 is highly expressed in Sox9-Low EEC progenitor cells (Figure 1D), which exhibit both proliferative and differentiative capacity, but relatively low expressed in Lgr5+ aISCs (Figure 1D). If miR-7 is critical to the control of crypt division or EEC abundance, we reasoned that it may be modulated by HFD. To determine whether miR-7 is significantly altered by HFD, we performed a HFD study for 16 weeks, identical to the one previously described in Mah et al.20 First, we confirmed that HFD-fed mice exhibit a significant increase relative to LFD-fed mice in body weight as expected (58.8% increase; P = 1.271e-9; LFD, n = 8; HFD, n = 9). Next, we showed by histomorphometry that crypt density, a marker of the rate of crypt division, is significantly elevated in the jejunum of HFD-fed mice (Figure 2A). We also observed a significant increase in the number of 5-ethynyl-2′deoxyuridine–positive (EdU+) cells/crypt (proliferating cells) in jejunal crypts from HFD-fed mice (Figure 2B). Finally, we demonstrated by immunofluorescence that HFD-fed animals exhibit significantly lower Chga+ cell/villus count, indicative of reduced EEC number (Figure 2C).
Figure 2.
MiR-7 expression is suppressed and predicted miR-7 targets are elevated in EEC progenitors under conditions of increased intestinal crypt division and reduced EEC abundance. (A) Representative hematoxylin and eosin images and crypt density quantification of mid-jejunum from HFD-fed (n = 9) and LFD-fed (n = 8) mix of Lgr5-EGFP reporter mice and naïve C57BL/6 wild-type (WT) mice. Bar = 100 μm. (B) Representative images of tissue sections stained with Hoechst (blue) and EdU (red) and quantification of EdU+ cells per crypt from HFD-fed (n = 8) and LFD-fed (n = 7) Lgr5-EGFP reporter mice and naïve C57BL/6 WT mice. (C) Representative images of tissue sections stained with Chga and quantification of Chga+ cells per villi (normalized by villi height) from HFD-fed (n = 7) and LFD-fed (n = 4) mix of Lgr5-EGFP reporter mice and C57BL/6 naïve WT mice. (D) Schematic of experimental workflow using Sox9-EGFP reporter mice. (E) Principal component analysis plot of small RNA-seq data of Sox9-Low cells from HFD-fed and LFD-fed Sox9-EGFP reporter mice. (F) Volcano plot of differentially expressed miRNAs in HFD-fed relative to LFD-fed Sox9-EGFP reporter mice (dashed lines represent fold-change > 2). (G) miRNAs that are significantly (P < .05) upregulated or downregulated in jejunal Sox9-Low sorted cells from HFD-fed relative to LFD-fed mice. Red and blue bars highlight miRNAs with >2-fold change up or down in HFD-fed relative to LFD-fed mice, respectively. (H) Expression level of miR-7b (small RNA-seq) in Sox9-Low and Sox9-High cells from HFD-fed and LFD-fed Sox9-EGFP mice. (I) Principal component analysis plot of RNA-seq data of Sox9-Low cells from HFD-fed and LFD-fed mice. (J) Volcano plot of differentially expressed genes in HFD-fed relative to LFD-fed Sox9-EGFP reporter mice (dashed line represents adjusted P < .05). (K) Heatmap showing changes in genes involved in proliferation, EEC differentiation, and enterocyte differentiation of jejunal Sox9-Low cells of HFD-fed relative to LFD-fed mice. (L) Enrichment analysis of miRNA target sites (using the Monte Carlo simulation tool miRhub) in genes upregulated in response to HFD (dashed line represents P < .05). Only miRNAs downregulated greater than 2-fold in Sox9-Low cells from HFD-fed relative to LFD-fed mice are included in the analysis. (M) Sequencing data showing upregulation of miR-7 target gene Xiap in Sox9-Low cells from HFD-fed relative to LFD-fed mice. (N) Inverse correlation between expression level of miR-7b and Xiap among the HFD-fed and LFD-fed Sox9-EGFP reporter mice. All figure panels from panel E onward correspond to data from n = 4 HFD-fed and n = 4 LFD-fed Sox9-EGFP reporter mice. * P < .05, ** P < .01 by 2-tailed Student t test.
To determine whether the EEC progenitor enriched miR-7 is associated with this hyperproliferative phenotype, we next performed small RNA-seq in sorted Sox9-Low cells (EEC progenitors) from the jejunal epithelial tissue of both LFD- and HFD-fed mice (Figure 2D). Principal component analysis on miRNA profiles showed clear separation of LFD- and HFD-fed mice (Figure 2E) and differential miRNA expression analysis identified 15 miRNAs (7 up, 8 down) in Sox9-Low cells altered by more than 2-fold (Figure 2F). Notably, the downregulated group of miRNAs includes the EEC lineage-enriched miR-7b (Figure 2G). In addition, the downregulation of miR-7b by the dietary perturbation appears specific to EEC progenitors, as it is downregulated by HFD only in Sox9-Low (EEC progenitors) but not in Sox9-High (mature EECs) (Figure 2H). This finding indicates that miR-7 is suppressed in EEC progenitors under physiologic conditions that promote crypt division and reduce EEC abundance in the intestinal epithelium.
Genes Upregulated in EEC Progenitors Under Conditions of Increased Crypt Division Are Enriched for Predicted Targets of miR-7
Given that HFD alters miR-7 in Sox9-Low (EEC progenitors) but not Sox9-High cells (mature EECs) (Figure 2G), we next performed RNA-seq in sorted Sox9-Low cells from the jejunal epithelial tissue of both LFD- and HFD-fed mice (Figure 2D). Principal component analysis on gene expression profiles showed clear separation of LFD- and HFD-fed mice (Figure 2I) and differential expression analysis identified nearly 1700 significantly (adjusted P < .05) altered genes (410 up, 1279 down) (Figure 2J). After multiple testing correction, the genes upregulated by HFD are most significantly enriched in the Pparδ transcriptional network (Enrichr KEGG, P = 9.89e-6), including genes involved in fatty acid oxidation (Figure 2K), which is consistent with the finding from a recent study in the colon after a much longer HFD regimen.21 In the HFD condition, we also found that several genes involved in crypt division and enterocyte differentiation are significantly upregulated whereas genes that encode transcription factors that drive EEC maturation are significantly downregulated (Figure 2K), which is in line with the observed cellular phenotypes (Figure 2A–2C). Using the bioinformatic tool miRhub,22 we showed that upregulated genes are significantly enriched for predicted target sites of only 1 downregulated miRNA, miR-7 (Figure 2L). This shows that genes upregulated in EEC progenitors under conditions of increased intestinal epithelial proliferation are overrepresented for predicted targets of miR-7. Notably, among the upregulated genes is X-linked inhibitor of apoptosis (Xiap) (Figure 2M), which encodes a protein that promotes crypt survival and growth. While Xiap has been reported as a direct target of miR-7 in the context of cervical cancer,23 it has never been reported previously in the small intestine. Indeed, the levels of Xiap and miR-7b are strongly inversely correlated in EEC progenitors across LFD- and HFD-fed mice (Figure 2N).
MiR-7 Controls Intestinal Epithelial Growth
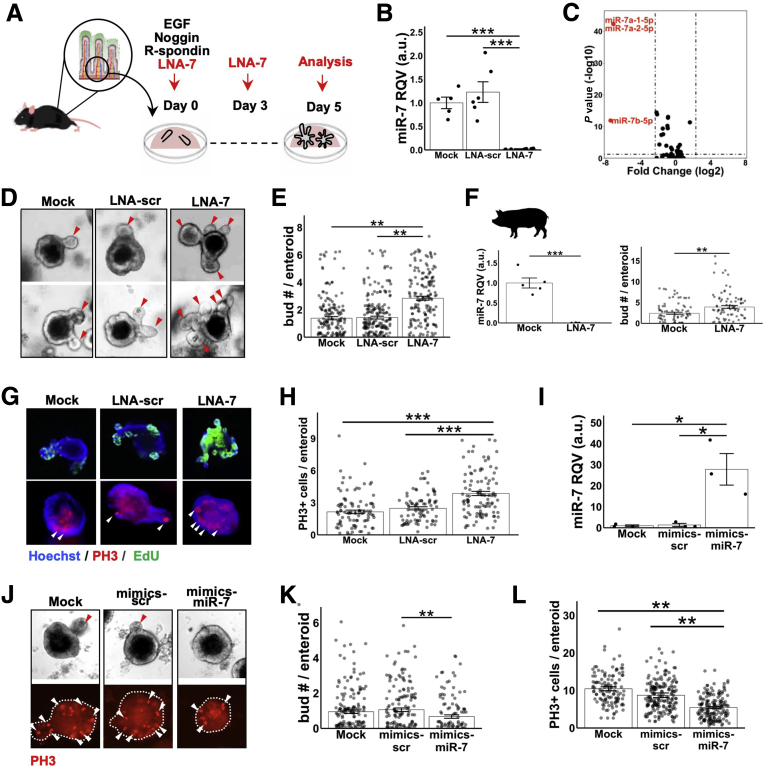
The previous findings, most notably that miR-7 in EEC progenitors is strongly suppressed under conditions of increased crypt division and that the genes upregulated during increased crypt division are enriched for miR-7 predicted targets, led us to hypothesize that miR-7 may regulate crypt division. To test this hypothesis, we established 3-dimensional enteroids from jejunal crypts of C57BL6/J mice and treated them with locked nucleic acid inhibitors against miR-7 (LNA-7) and compared with treatment with LNA-scramble control (LNA-scr) (Figure 3A). RT-qPCR analysis showed that treatment with LNA-7 led to a robust suppression (∼55-fold) of the miR-7 family in enteroids (Figure 3B). Also, small RNA-seq analysis of LNA-7–treated enteroids revealed that miR-7 is suppressed by many orders of magnitude more than any other miRNA (Figure 3C, Table 2), demonstrating the specificity of the effects of LNA-7 on miR-7 expression.
Figure 3.
MiR-7 controls intestinal epithelial growth ex vivo. (A) Experimental design for testing the effect of miR-7 suppression in enteroid culture established from C57BL/6 WT mouse jejunum. (B) RT-qPCR data showing effective suppression of miR-7 expression by LNA-7 treatments in mouse enteroids compared with mock (no treatment) and LNA-scr control (n = 5–6 wells per condition pooled from 2 independent experiments). (C) Small RNA-seq followed by differential expression analysis showing highly robust and specific suppression of all the miR-7 family members in the enteroids treated with LNA-7 compared with LNA-scr (LNA-7, n = 6; LNA-scr, n = 5). (D) Representative bright field images of enteroids treated with mock, LNA-scr, and LNA-7. (E) Bar graph depicting average number of buds per enteroid in the indicated treatment groups. Data were pooled from 3 independent experiments (mock, n = 174 enteroids; LNA-scr, n = 214 enteroids; LNA-7, n = 163 enteroids). (F) The left panel shows RT-qPCR data showing effective suppression of miR-7 by LNA-7 (n = 5 wells/condition) in porcine enteroids. The right panel shows bar graph depicting average number of buds per porcine enteroid in the indicated treatment groups. Enteroids were pooled from 5 independent experiments (mock, n = 83; LNA-7, n = 86 enteroids). (G) Representative images of mouse enteroids showing whole-mount staining signal for EdU (green; top) and PH3 (red; bottom). (H) Bar graph depicting average number of PH3+ cells per enteroid. All of the enteroids across multiple wells were examined (mock, n = 100 enteroids; LNA-scr, n = 83 enteroids; LNA-7, n = 106 enteroids). (I) RT-qPCR data showing effective increase in miR-7 levels in mouse enteroids treated with mimics of miR-7 (mimics-miR-7) compared with mock (no treatment) and mimics-scramble (mimics-scr) (n = 3 wells per group). (J) Representative brightfield images of wild-type mouse enteroids (top) and whole-mount staining signal for PH3 (red; bottom) in the indicated treatments. (K) Quantification of the average number of buds per enteroid in the indicated treatments. Enteroids per treatment group were pooled from multiple wells for quantification (mock, n = 175; mimics-scr, n = 168; mimics-miR-7, n = 116). (L) Quantification of the average number of PH3+ cells per enteroid in the indicated treatments. All of the enteroids across multiple wells were examined (mock, n = 123 enteroids; mimics-scr, n = 190 enteroids; mimics-miR-7, n = 165 enteroids). * P < .05, ** P < .01 and *** P < .001 by 2-tailed Student t test. RQV, relative quantitative value.
Table 2.
Small RNA-seq Analysis Showing Differentially Expressed miRNAs (Adjusted P < .05) in Mouse Enteroids Treated With LNA-7 (n = 6) Relative to LNA-scr (n = 5)
| miRNA | log2FoldChange | Adjusted P |
|---|---|---|
| mmu-mir-7a-1-5p | –7.109435038 | 4.64E-43 |
| mmu-mir-7a-2-5p | –7.103216777 | 5.31E-43 |
| mmu-let-7f-1-5p | –0.854266518 | 8.57E-14 |
| mmu-let-7f-2-5p | –0.862650621 | 1.58E-13 |
| mmu-mir-215-5p | –1.460451901 | 3.96E-10 |
| mmu-mir-148a-3p | 0.495197848 | 5.41E-05 |
| mmu-mir-215-5p_+_1 | –1.050094118 | 1.02E-04 |
| mmu-mir-192-5p_+_2 | 0.309404578 | 2.82E-04 |
| mmu-mir-98-5p | –0.509809453 | 4.18E-04 |
| mmu-let-7i-5p | –0.402567428 | .001535664 |
| mmu-mir-30d-5p | 0.317770352 | .004397959 |
| mmu-mir-26a-2-5p | 0.149129242 | .004685784 |
| mmu-mir-26a-1-5p | 0.149100629 | .004685784 |
| mmu-mir-203-3p_+_1 | 0.592648674 | .009726977 |
| mmu-mir-30e-3p | 0.276607739 | .010577384 |
| mmu-mir-192-5p_+_1 | 0.204704048 | .010887894 |
| mmu-mir-192-5p | 0.206058351 | .01488005 |
| mmu-let-7g-5p | –0.48204363 | .035030773 |
miRNA, microRNA; RNA-seq, RNA sequencing.
Brightfield imaging showed that treatment with LNA-7 (miR-7 loss-of-function) significantly increases enteroid budding, which is an ex vivo proxy for crypt division,24 relative to LNA-scr and mock (no treatment) controls (Figure 3D and 3E). We also confirmed this result in porcine enteroids (Figure 3F). Furthermore, we demonstrated that LNA-7 treatment dramatically increases EdU+ cells/enteroid (Figure 3G) and PH3+ cells/enteroid (Figure 3G and 3H). Next, we performed complementary gain-of-function studies in mouse enteroids using miR-7 mimics. Our results showed that overexpression of miR-7 (Figure 3I) suppresses budding (Figure 3J and 3K) and leads to a reduction in PH3+ cells/enteroid (Figure 3J–3L), which is the opposite of what occurs upon inhibition of miR-7. These studies together demonstrate the functional role of miR-7 in the control of enteroid budding.
MiR-7 Controls Intestinal Epithelial Proliferation in Drosophila Midgut In Vivo
To determine whether this function of miR-7 is conserved beyond mammals, we turned to the fruit fly model (Figure 4A). We first demonstrated that miR-7 is the most significantly reduced miRNA in the Drosophila melanogaster midgut during the well-documented hyperproliferative response to infection by Erwinia carotovora carotovora 15 (Ecc15) (Figure 4B and 4C).25 To study miR-7 function specifically in proliferating intestinal epithelial cells, we overexpressed miR-7 in esg+ cells of the midgut, which represent the stem or progenitor populations. We found that the number of PH3+ (proliferating) cells is dramatically reduced upon miR-7 overexpression in the background of Ecc15 infection compared with control (Figure 4D, E), which demonstrates that miR-7 downregulation is required for the hyperproliferative response to bacterial insult and that miR-7 is a key regulator of epithelial proliferative capacity in the midgut. While our focus is on mammalian miR-7, and while there are notable differences between fruit fly and mammalian gut tissue (eg, lack of well-established EEC progenitors in the fruit fly gut), the results from the Drosophila experiments convey a remarkable evolutionary conservation of miR-7 function in the intestinal epithelium.
Figure 4.
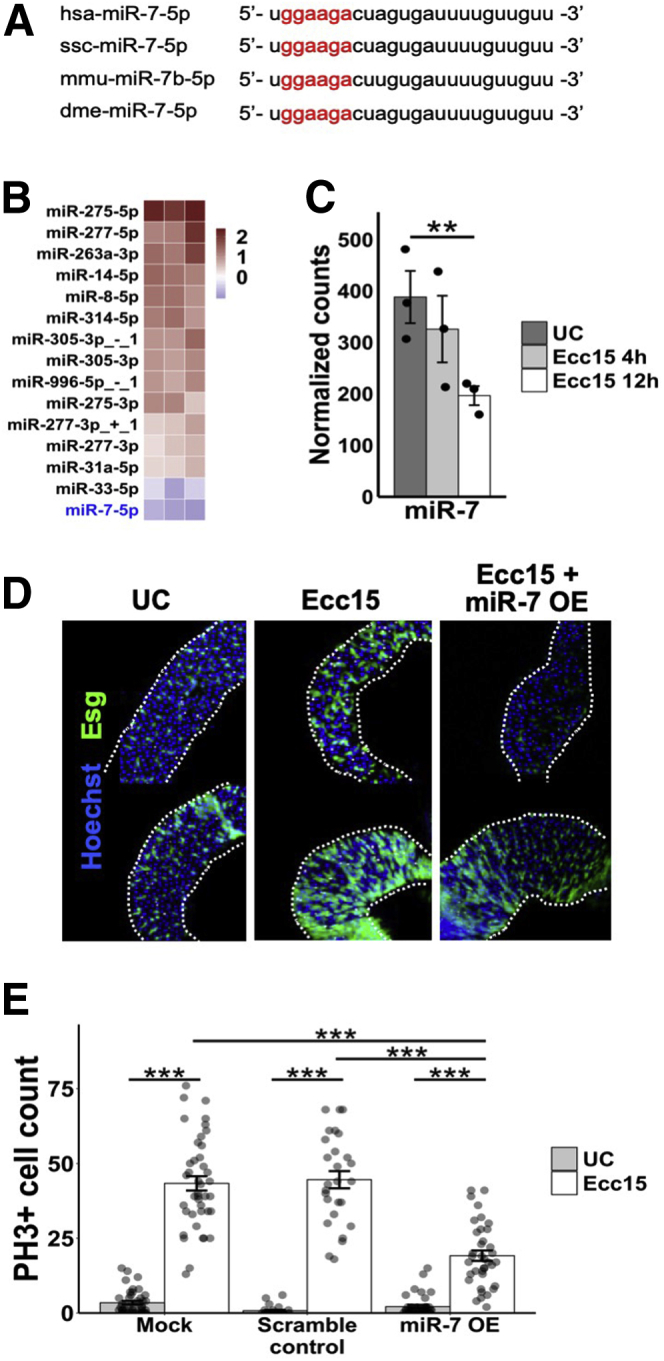
MiR-7 controls intestinal epithelial proliferation in Drosophila midgut in vivo. (A) Mature miR-7 sequence across different species, including human, pig, mouse, and fruit fly. The seed sequence is denoted in red. (B) Heatmap results of small RNA-seq analysis of Drosophila melanogaster midgut showing miRNAs that are significantly (P < .05) upregulated or downregulated upon exposure to the pro-proliferative Ecc15 pathogen (n = 3) relative to unchallenged (UC) condition (n = 3). Only showing those miRNAs with average reads per million mapped to miRNAs >100 in either Ecc15 challenge or UC. (C) Downregulation of miR-7 in the D. melanogaster midgut at 4 hours (minimal proliferative response, n = 3) and 12 hours (peak proliferative response, n = 3) after Ecc15 challenge compared with the UC control group. (D) Immunofluorescent staining of DAPI (blue) and Esg (green) in D. melanogaster midgut with mock (left), Ecc15 alone (middle), and Ecc15 with miR-7 overexpression driven by the Esg promoter (overexpressed miR-7 [miR-7 OE]) (right). (E) Bar graph showing counts of PH3+ replicating cells in the D. melanogaster midgut of each. * P < .05, ** P < .01, and *** P < .001 by 2-tailed Student t test.
MiR-7 Control of Enteroid Budding Is Dependent on Intact Xiap
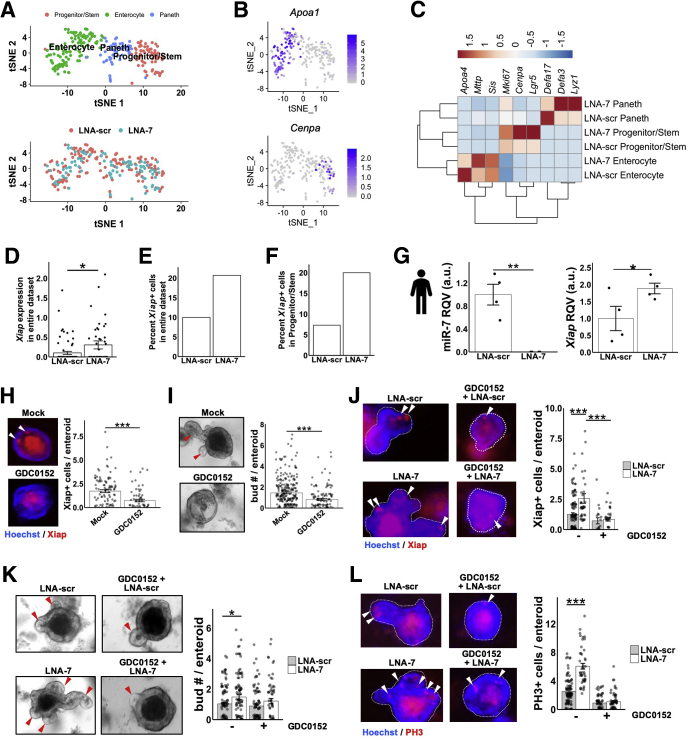
As mentioned previously, Xiap encodes a protein that promotes cell survival and growth, and it is an established direct target of miR-7 by 3′ UTR reporter gene assays in a cancer cell context.23 However, the functional relevance of the miR-7:Xiap regulatory relationship has not been examined in a noncancer context and definitely not in intestinal epithelial growth. To determine whether miR-7 regulates Xiap in enteroids, and to determine whether this regulation takes place in proliferating cells (stem or progenitor cells), we performed single-cell RNA-seq in mouse enteroids treated with either LNA-scr or LNA-7. We sequenced just enough cells to discern 3 clusters of different cell types, progenitor/stem, Paneth, and enterocyte (Figure 5A). These cell clusters are enriched for markers that are known to be associated with each cell type (Figure 5B and 5C). Notably, the signal for markers of proliferation increased in the progenitor/stem population after treatment with LNA-7 (Figure 5C). Next, we focused on Xiap and found that its average level across all cells is significantly elevated by treatment with LNA-7 relative to LNA-scr (Figure 5D). Moreover, the percentage of Xiap+ progenitor cells is ∼2-fold increased by LNA-7 (∼21%) relative to LNA-scramble (∼10%) (Figure 5E). In progenitor/stem cells, the fold-increase of Xiap+ cells after LNA-7 treatment is even greater (∼3-fold) (Figure 5F). These results suggest that more progenitor or stem cells express Xiap, and at higher levels, after LNA-7 treatment. Although mouse enteroids comprise many differentiated cell types, most of which do not robustly express miR-7, human enteroids tend to retain a more stem or progenitor state in culture.26 Therefore, to test miR-7 regulation of XIAP, we treated human enteroids with LNA-7, which led to dramatic suppression of miR-7 and a significant increase in XIAP (Figure 5G).
Figure 5.
MiR-7 control of enteroid budding is dependent on intact Xiap. (A) t-SNE plot of single-cell RNA-seq profiles reveals 3 major cell populations: enterocytes, Paneth cells, and progenitor or stem cells, from mouse enteroids treated with LNA-7 or LNA-scr (enterocytes, n = 107 cells; Paneth cells, n = 54; progenitor/stem, n = 71). The clusters were assigned based on overlap with marker genes for every major intestinal epithelial cell type defined in Haber et al., 2017. (B) t-SNE plot of single-cell RNA-seq data showing expression level for Apoa1 (top, enterocyte marker) and Cenpa (bottom, progenitor cell marker). (C) Heatmap displaying average expression of cell type-specific markers for cells in each cluster from LNA-scr and LNA-7–treated mouse enteroids. Expression is scaled by row. (D) Average Xiap expression across all cells from LNA-scr– or LNA-7–treated mouse enteroids (Wilcox). (E) Percentage of Xiap+ cells present in the single-cell RNA-seq data from mouse enteroids treated with LNA-scr or LNA-7. (F) Percentage of Xiap+ progenitor/stem cells present in the single-cell RNA-seq data from mouse enteroids treated with LNA-scr or LNA-7. (G) RT-qPCR data showing suppression of miR-7 (left panel) and the upregulation of XIAP (right panel) in human intestinal organoids treated with LNA-7 compared with LNA-scr (n = 3 wells per condition). (H) Representative images and quantification of whole-mount staining signal for Xiap (red) and Hoechst (blue) in mouse enteroids with the indicated treatment groups (mock, n = 99 enteroids; GDC0152, n = 69 enteroids). (I) Representative bright field images and quantification of average number of buds per enteroid with mock treatment (no treatment) or Xiap inhibitor treatment (GDC0152) (mock, n = 274 enteroids; GDC0152, n = 120 enteroids). (J) Representative images and quantification of whole-mount staining signal for Xiap (red) and Hoechst (blue) in the indicated treatment groups. Data from enteroids across multiple wells were examined (LNA-scr, n = 112 enteroids; LNA-7, n = 41 enteroids; GDC0152+LNA-scr, n = 18 enteroids; GDC0152+LNA-7 = 32 enteroids). (K) Representative bright field images and quantification of the average number of buds per enteroid after LNA-scr treatment or LNA-7 treatment with and without GDC0152 (LNA-scr, n = 192 enteroids; LNA-7, n = 118 enteroids; GDC0152+LNA-scr, n = 96 enteroids; GDC0152+LNA-7, n = 44 enteroids). (L) Representative images and quantification of whole-mount staining signal for PH3 (red) and Hoechst (blue) in the indicated treatment groups. Data from enteroids across multiple wells were examined (LNA-scr, n = 92 enteroids; LNA-7, n = 45 enteroids; GDC0152+LNA-scr, n = 58 enteroids; GDC0152+LNA-7 = 62 enteroids). * P < .05, ** P < .01 and *** P < .001 by 2-tailed Student t test. RQV, relative quantitative value.
To determine whether miR-7 control of enteroid growth is mediated in part by regulation of Xiap, we examined the effect of LNA-7 in mouse enteroids in the context of Xiap suppression. Treatment of enteroids with GDC0152, an Xiap inhibitor, effectively reduced Xiap+ cells per enteroid (Figure 5H) and suppresses the budding phenotype (Figure 5I). We then showed that treatment with GDC0152 significantly blunts the effects of LNA-7 on Xiap+ cells/enteroid (Figure 5J), budding (Figure 5K), and PH3+ cells/enteroid (Figure 5L). These results convey that the effects of miR-7 on enteroid growth are mediated at least in part through regulation of Xiap.
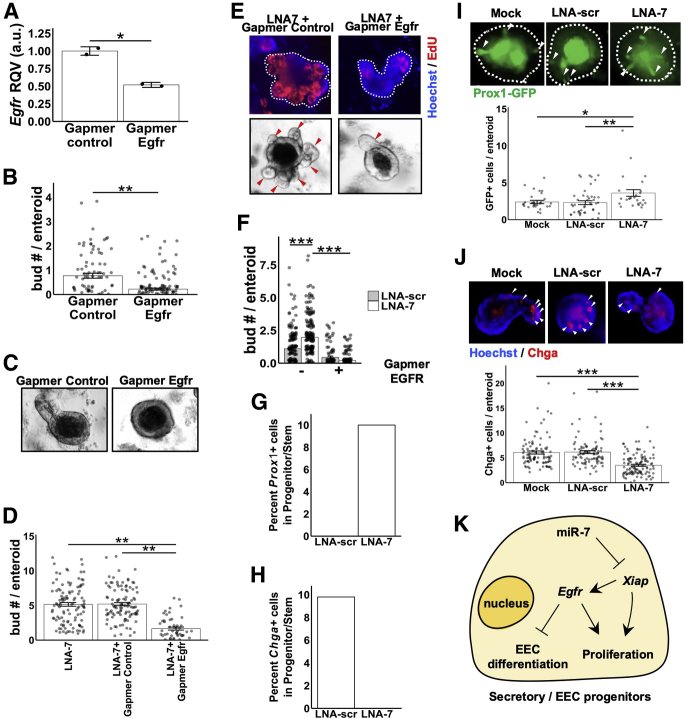
Xiap is known to promote Egfr.27 To evaluate further the dependency on Egfr, we next tested the effect of LNA-7 in the context of direct Egfr suppression. We first demonstrated in mouse enteroids that treatment with an oligonucleotide inhibitor of Egfr (Gapmer Egfr) downregulates Egfr messenger RNA levels by ∼60% (Figure 6A) and significantly reduces enteroid budding as expected (Figure 6B and 6C). Then we showed that treatment of enteroids with Gapmer Egfr significantly blunts the pro-proliferative phenotype observed with LNA-7 alone or with Gapmer Control, as determined by budding (Figure 6D and 6E) and EdU staining (Figure 6E). Also, we demonstrated that Egfr suppression greatly blunts the hyperbudding phenotype in LNA-7–treated enteroids compared with LNA-scr–treated enteroids (Figure 6F).
Figure 6.
MiR-7 control of enteroid budding depends on Egfr signaling. (A) RT-qPCR data showing Egfr expression in mouse enteroids treated with control Gapmer inhibitor (n = 2) or Egfr Gapmer inhibitor (n = 2) on day 5. (B) Quantification of the average number of buds per enteroid treated with mock (no treatment), Gapmer Control, or Gapmer Egfr (Mock, n = 194 enteroids; Gapmer Control, n = 88 enteroids; Gapmer Egfr, n = 149 enteroids). (C) Representative bright field images of mouse enteroids with treatment of Gapmer Control and Gapmer Egfr. (D) Quantification of the average number of buds per mouse enteroid in the indicated treatments (LNA-7, n = 55 enteroids; LNA-7 + Gapmer control, n = 53 enteroids; LNA-7 + Egfr Gapmer, n = 27 enteroids). (E) Representative images showing whole-mount staining of EdU (top; red), Hoechst (top; blue), and bright field (bottom) in mouse enteroids with the indicated treatments. (F) Quantification of the average number of buds per mouse enteroid in the indicated treatments (LNA-scr, n = 165 enteroids; LNA-7, n = 182 enteroids; LNA-scr + Gapmer Egfr, n = 91 enteroids; LNA-7 + Gapmer Egfr, n = 148 enteroids). (G) Percentage of Prox1+ progenitor or stem cells present in the single-cell RNA-seq data from mouse enteroids treated with LNA-scr or LNA-7. (H) Percentage of Chga+ progenitor/stem cells present in the single-cell RNA-seq data from mouse enteroids treated with LNA-scr or LNA-7. (I) Representative images of Prox1+ cells (green) and quantification of Prox1+ cells per mouse enteroid established from Prox1-EGFP reporter mice in the indicated treatments (mock, n = 30 enteroids; LNA-scr, n = 46 enteroids; LNA-7, n = 26 enteroids). (J) Representative images of Chga+ cells (red) and quantification of Chga+ cells per mouse enteroid in the indicated treatments (mock, n = 96 enteroids; LNA-scr, n = 92 enteroids; LNA-7, n = 113 enteroids pooled from 2 independent experiments). (K) The proposed partial working model of miR-7 control of intestinal epithelial proliferation. * P < .05, ** P < .01 and *** P < .001 by 2-tailed Student t test.
A previous study established that Egfr signaling can suppress EEC differentiation.28 Given the finding that miR-7 function is dependent in part on Egfr signaling, we hypothesized that loss of miR-7 may shift the balance of secretory progenitor cells toward proliferation and away from EEC maturation. Indeed, in our single-cell RNA-seq data from mouse enteroids, we found that LNA-7 treatment increases the proportion of Prox1+ cells (proliferative EEC progenitors) and decreases the proportion of Chga+ cells (mix of EEC progenitors primed to differentiation and mature EECs) in the progenitor population (Figure 6G and 6H). To validate this observation, we showed that treatment of enteroids with LNA-7 leads to a significant increase in Prox1-EGFP+ cells/enteroid (Figure 6I) and a significant decrease in Chga+ cells/enteroid (Figure 6J). Taken together, these data strongly suggest that EEC progenitor enriched miR-7, in part through Xiap and Egfr, controls enteroid budding (proxy for crypt division) and may also be involved in the regulation of EEC abundance (Figure 6K).
Discussion
Although miRNAs are well appreciated as regulators of cell division, growth, and differentiation in many organ systems, their contributions in the intestinal epithelium remain largely unknown. Recently, several studies have reported that EEC progenitors represent a plastic cell population that can either drive the formation of mature EECs or exhibit stem cell-like proliferative capacity.3,4 Here, we have (1) identified miR-7 as a highly enriched miRNA across the entire EEC lineage trajectory, using 8 different reporter mice, FACS, and small RNA-seq; (2) defined miR-7 as the most enriched miRNA in EEC progenitors relative to Lgr5+ stem cells; (3) demonstrated that miR-7 in the EEC progenitor population is dramatically suppressed under physiological conditions of increased crypt division; (4) showed that miR-7 controls enteroid budding and EEC progenitor cell behavior; and (5) showed that miR-7 function depends at least in part on intact Xiap and downstream Egfr signaling. To our knowledge, this is the first report of a miRNA enriched along the EEC lineage trajectory that exerts substantive control of enteroid growth and likely EEC abundance. Although previously miR-7 has been reported to have roles in specific cancer contexts,17,29,30 our study is the first to our knowledge to demonstrate that miR-7 regulates intestinal growth under normal physiological conditions. It is noteworthy that modest but significant changes in miRNA expression, similar to what we show for miR-7 in this study, have been linked functionally to important phenotypes in numerous prior studies.31, 32, 33
Although our data suggests that miR-7 has important roles in EEC progenitor cells, we also cannot rule out functions in other stem or progenitor cell types. Also, though we have demonstrated that manipulating miR-7 expression regulates intestinal epithelial proliferative capacity, whether miR-7 controls the abundance of specific subtypes of mature EECs requires future detailed investigation. In addition, we have observed downregulation of miR-7 only in EEC progenitors but not in mature EECs upon HFD (Figure 2), motivating the question of what upstream mechanisms regulate miR-7 in this cell-type specific manner. A study in Drosophila showed that Ato, the mammalian ortholog of which is Atoh1 (a known EEC lineage driver in mouse), is upstream of miR-7 during development.34 Whether Atoh1 regulates miR-7 in the mammalian intestinal epithelium and whether Atoh1 is responsible for the alteration of miR-7 expression upon HFD remain to be determined in the future.
Though mature EECs constitute only ∼1% of cells in the intestinal epithelium, they are critical for orchestrating proper responses to nutritional and microbial input to maintain energy homeostasis and immune function.35, 36, 37 They are known to secrete many different kinds of hormones that function through endocrine and paracrine signaling, and they are thought to be major contributors to the immediate, weight loss–independent positive metabolic effects of bariatric surgery.38 Also, recent studies have implicated EECs in the gut-brain axis and the immune-endocrine axis through direct connection to enteric nervous system and surrounding immune cells, respectively.39 While changes in EEC abundance or function have been linked to a wide array of diseases including diabetes, inflammatory bowel disease, and psychiatric or neurologic disorders,40,41 the key regulators of the EEC lineage remain incompletely characterized due in part to the rareness, high molecular heterogeneity, and plasticity of this cell population. In the present study, we uncovered the functional role of EEC lineage-enriched miR-7 particularly in regulating intestinal epithelial growth and also provided observations suggesting its role in regulating EEC abundance. The potential role of miR-7 in regulating EEC maturation and function warrants future studies.
In vivo loss-of-function studies of miR-7 are made extremely challenging by the fact that there are 3 different miR-7 paralogs in mice: miR-7a-1, miR-7a-2, and miR-7b. We believe that the current study provides strong motivation to invest in the generation of an EEC progenitor cell–specific miR-7 triple-knockout mouse colony. These mice will be useful for future in vivo investigation of the role of miR-7 in intestinal epithelial proliferation and renewal under baseline conditions and in response to different perturbations that affect crypt behavior, including HFD, pathogenic gastrointestinal infection, irradiation, or even bariatric surgery. Finally, we note that this study identifies several other miRNAs in addition to miR-7 that are enriched in the EEC lineage trajectory and altered under conditions of increased crypt budding. It will also be of great interest to investigate the role of these miRNAs too, in isolation and in conjunction with miR-7, in controlling the biology and pathophysiology of the crypt in the intestinal epithelium.
Materials and Methods
Animal Models and Diet Study
The following transgenic reporter mice were utilized: female and male Sox9-EGFP,42 female Lgr5-EGFP,24 female Prox1-EGFP,43 female Pyy-EGFP mice,44 male and female Defa6-CreERT;Rosa26-tdTomato mice,45 and male and female Hopx-CreERT;Rosa26-tdTomato mice (JAX strains 017606, 007914; Jackson Laboratory, Bar Harbor, ME). For the diet study, 8- to 10-week-old C57BL/6 wild-type mice and, either 8- to 10-week-old Lgr5-EGFP female mice (for histomorphometry and metabolic phenotyping) or adult 8- to 10-week-old Sox9-EGFP female mice (for cell sorting and RNA-seq), were fed ad libitum with either an LFD (14% kcal from fat; Prolab RMH3000 [LabDiet, St. Louis, MO]) or an HFD (45% kcal from fat; Research Diets D12451) for 16 weeks. Body weight, body composition and blood parameters were measured to confirm obesity phenotype. To mark cells in S-phase, EdU was administered by intraperitoneal injection (100 μg/25 g body weight) 90 minutes before euthanizing. The harvested small intestine was divided into 3 equal segments. The middle region was considered the jejunum. All animal procedures were performed with the approval and authorization of the Institutional Animal Care and Use Committee at each participating institution.
Flow Cytometry
Six distinct reporter mouse strains that facilitate the sorting of a variety of different intestinal epithelial cell types were used to isolate aISCs (Lgr5-EGFP), EEC progenitors (Sox9-EGFP and Prox1-EGFP), mature EECs (Sox9-EGFP and Pyy-EGFP), Paneth cells (Defa6-CreERT2;Rosa26-tdTomato), and rISCs (Hopx-CreERT2;Rosa26-tdTomato). Mouse intestinal epithelial cells from the jejunum were dissociated and prepared for fluorescence-activated cell sorting (FACS) as described previously.20 CD31-APC (BioLegend, San Diego, CA), CD45-APC (BioLegend), and Annexin-V-APC (Life Technologies, Carlsbad, CA), and Sytox-Blue (Life Technologies) staining were used to exclude endothelial cells, immune cells, and apoptotic cells, respectively. The gating parameters of FACS sorting were described previously.20 The Lgr5, Sox9, and Defa6 sorts were performed using a Mo-Flo XDP cell sorter (Thermo Fisher Scientific, Fullerton, CA) at the University of North Carolina Flow Cytometry Core Facility. The Prox1 sorts were performed using BD FACS Aria Fusion Fluorescence Activated Cell Sorter (BD Biosciences, San Jose, CA) at Cornell University Flow Cytometry Core Facility at the Biotechnology Resource Center. The Pyy sorts were performed using BD FACSAria II Cell Sorter (BD Biosciences) at Kings BRC flow cytometry Core facility. Sorting of Hopx+ cells was conducted at North Carolina State University, College of Veterinary Medicine using a Mo-Flo XDP cell sorter (Beckman Coulter). The cells were sorted directly into cold Dulbecco’s modified Eagle medium or lysis buffer.
Tuft cells were sorted by use of the cell surface marker Siglecf (clone E50-2440; Thermo Fisher Scientific, Waltham, MA). Mouse jejnual cells were prepared as described previously.46 Aqua Live Dead fixable viability stain Live dye (Thermo Fisher Scientific) and anti-mouse EpCam-APC (clone G8.8; Thermo Fisher Scientific) staining was used to include viable, epithelial cells, respectively, while anti-mouse CD45 BV605 staining (clone 30-F11; BD Biosciences) was used to exclude immune cells. Anti-mouse Siglecf-PE antibody (clone E50-2440; Thermo Fisher Scientific) was employed to enrich for tuft cells. Sorting for tuft cells was performed using BD FACS Aria Fusion II Fluorescence Activated Cell Sorter (BD Biosciences) at Cornell University Flow Cytometry Core Facility, Biotechnology Resource Center. Cells were sorted directly into cold lysis buffer (Norgen Biotek, Thorold, Canada).
Side population sorting was used to separate the subfraction of slowly cycling from active cycling intestinal stem cells, as described previously.18 Mouse intestinal epithelial cells from the jejunum of female C57BL/6 mice were prepared and sorted into either upper side population (consisting of actively cycling stem cells) or lower side population (consisting of slowly cycling stem cells) by the previously described gating methods.18 The side population sorting46 was performed using a Mo-Flo XDP cell sorter (Beckman Coulter) at the University of North Carolina Flow Cytometry Core Facility. Cells were sorted directly into cold lysis buffer (Norgen).
Histological Analysis
Mouse jejunal tissue was fixed in 4% (v/v) neutral-buffered paraformaldehyde, embedded in paraffin, and cut into 5-μm sections for various staining experiments. Hematoxylin and eosin staining was performed for morphometric analyses (villi height and crypt density). Crypt density was calculated by dividing the number of well-oriented crypts per millimeter of submucosal circumference. EdU staining was performed to visualize cells in S-phase using the Click-iT EdU AlexaFluor 594 Kit (Invitrogen, Carlsbad, CA). Immunofluorescent staining of α-chromogranin (Chga) was performed for assessing mature EEC number. Briefly, sections were incubated with primary antibody (rabbit Chga, 1:100 dilution in immunofluorescence buffer; ab15160; Abcam, Cambridge, United Kingdom) overnight at 4°C followed by Alexa Fluor 594 (Abcam) secondary antibody incubation for 2 hours at room temperature. DAPI (1:1000) was used to visualize nuclei. Images were captured using ZEISS (Jena, Germany) Axiovert 200M inverted microscope. The numbers of Chga+ cells per villus were normalized by average villus height (millimeters) in each of the diet groups. For the villus height measurements, 1 data point from LFD group and 1 data point from HFD were removed as outliers after applying the criterion of value beyond average ± 1.5*standard deviation.
RNA Extraction and Real-Time qPCR
Total RNA was isolated using the Total or Single-cell RNA Purification kit (Norgen Biotek). High Capacity RNA to cDNA kit (Life Technologies, Grand Island, NY) was used for reverse transcription of RNA. TaqMan microRNA Reverse Transcription kit (Life Technologies) was used for reverse transcription of miRNA. Both miRNA and gene expression qPCR were performed using TaqMan assays (Life Technologies) with either TaqMan Universal PCR Master Mix (miRNA qPCR) or TaqMan Gene Expression Master Mix (messenger RNA qPCR) per the manufacturer’s protocol, on a Bio-Rad CFX96 Touch Real Time PCR Detection System (Bio-Rad Laboratories, Richmond, CA). Reactions were performed in triplicate using either U6 (miRNA qPCR), Rps9 (mouse messenger RNA qPCR) or RNU48 (human messenger RNA qPCR) as the normalizer.
Small RNA Library Preparation and Sequencing
The small RNA-seq of cells from the various cell sorts and D. melanogaster midgut was conducted at Genome Sequencing Facility of Greehey Children's Cancer Research Institute at University of Texas Health Science Center at San Antonio. Libraries were prepared using the TriLink CleanTag Small RNA Ligation kit (TriLink Biotechnologies, San Diego, CA). Seven to 8 libraries were sequenced per lane, with single-end 50× on the HiSeq2500 platform. Raw sequencing data and miRNA quantification tables can be accessed through GEO record GSE118814.
RNA Library Preparation and Sequencing
RNA-seq libraries from the Sox9 sorts of LFD-fed and HFD-fed C57BL/6J mice were prepared using the Clonetech SMARTer Ultra Low Input library preparation kit combined with Nextera XT DNA sample preparation kit (Illumina, San Diego, CA) and sequenced with single-end 100 bp on a HiSeq2000 platform at the UNC High Throughput Sequencing Core Facility, as previously described.47 Raw sequencing data as well as normalized counts are available through GEO accession GSE118814.
Single Cell Suspension Preparation, RNA Library Preparation, and Sequencing
Enteroids treated with either LNA-7 or LNA-scr were harvested at day 5. Briefly, media was removed from enteroid cultures and the enteroids were washed twice with ice-cold phosphate-buffered saline (PBS). Enteroids were collected in cold PBS, spun down at 300 g for 5 minutes, and resuspended as a pellet in 10 mL of ice-cold D-PBS with 0.04% (w/v) bovine serum albumin (BSA). Centrifugation at 1000 g at 4°C was carried out for 5 minutes. Supernatant was removed and the pellet was resuspended in 10 mL of room temperature Hank’s Balanced Salt Solution with 0.3-U/mL dispase. Incubation in a water bath was performed at 37°C for 12–16 minutes. The tubes were gently shaken every 2 minutes. An aliquot was examined under the microscope to determine whether >50% of the cells were single cells. Dispase activity was stopped by adding fetal bovine serum to 10% of the total volume of samples and DNaseI to a final concentration of 50 μg/mL (50 μL). Samples were filtered with 40-μm strainer and centrifuged at 500 g at 4°C for 5 minutes. The supernatant was removed and the pellet was resuspended in 10 mL of ice-cold Hank’s Balanced Salt Solution, and the samples were filtered again with 40 μm strainer and centrifuged at 500 g at 4°C for 5 minutes. The supernatant was removed and the pellet was resuspended in 100 μl of ice-cold D-PBS with 0.04% (w/v) BSA. The samples were gently mixed several times with wide-bore pipet tip (1000-μL pipet tip), and then checked under the microscope for any clumping. If clumps were present, the filtration, centrifugation, and resuspension steps were repeated. Finally, cell viability and cell number were determined using Trypan blue and Bio-Rad cell counter to proceed with single cell library preparation and sequencing. For single cell sequencing, we used the 10X Genomics Chromium instrument at the Cornell Genomics Facility.
Bioinformatics Analysis
Bulk Small RNA and RNA-Seq Analysis
Small RNA-seq reads were aligned to the mouse genome (mm9) and quantified using miRquant 2.0 as previously described,48 with the exception that raw miRNA counts were normalized using DESeq2,49 to determine significance. Fruit fly sequences were depleted for a dominant rRNA species before alignment to the full genome (dm3). miRNA annotation was performed using miRbase (r18 for mouse and r21 for fruit fly). RNA-seq reads were mapped to mouse genome release mm10 using STAR (v2.5.3a)50 and transcript quantification was performed using Salmon (v0.6.0).51 Differential gene expression analysis was accomplished using DESeq2.49
Single Cell RNA-Seq Analysis
Single-cell RNA-seq reads for 107 LNA-7 and 158 LNA-scr cells were aligned to the mouse genome (mm10) and quantified using 10X Genomics CellRanger count. Cells with low gene complexity (<200 genes/cell) or with a strong mitochondrial signature (>25% mitochondrial genes) were filtered, resulting in 92 LNA-7 and 140 LNA-scr cells. Cluster identification, differential expression analysis, and t-SNE plot generation were performed with Seurat_3.0.1.
Other
MiRNA binding site enrichment among differentially expressed genes was determined using miRhub.22 Transcription factor enrichment was determined by inputting the top 20 significant upregulated genes into Enrichr52,53 and using the ENCODE and ChEA Consensus TFs from the ChIP-X dataset.
Enteroid Culture
Mouse Enteroids
Jejunal crypts were isolated from 3- to 5-month-old female C57BL/6 mice as previously described.47 The isolated crypts (day 0) were grown into Reduced Growth Factor Matrigel (cat. 356231; Corning, Corning, NY). Advanced DMEM/F12 (cat. 12634-028; Gibco, Gaithersburg, MD) supplemented with GlutaMAX (cat. 35050-061; Gibco), Pen/Strep (cat. 15140; Gibco), HEPES (cat. 15630-080; Gibco), N2 supplement (cat. 17502-048; Gibco), 50-ng/mL EGF (cat. 2028-EG; R&D Systems, Minneapolis, MN), 100-μg/mL Noggin (cat. 250-38; PeproTech, Rocky Hill, NJ), 250-ng/μL murine R-spondin (cat. 3474-RS-050, R&D Systems), and 10-mM Y27632 (cat. ALX270-333-M025; Enzo Life Sciences, East Farmingdale, NY) was added. For miRNA loss-of-function studies, miRCURY LNA Power Inhibitor against mouse miR-7 (mmu-miR-7a-5p) (cat. YI04100818-DDA; Qiagen, Hilden, Germany) or Power Negative Control A (cat. YI00199006-DDA; Qiagen) was added at 500 nM on day 0 and supplemented at 250 nM on day 3. For miRNA gain-of-function studies, miRCURY LNA mimic of miR-7 (cat. YM00472714-AGA; Qiagen) or Negative Control (cat. YM00479902-AGA; Qiagen) was added at 500 nM on day 0 and supplemented at 250 nM on day 3. Enteroids at day 5 were harvested for RNA isolation or fixed in 2% (v/v) paraformaldehyde for whole-mount staining.
For studies knocking down Egfr, LNA-7–treated enteroids were treated with a custom LNA GapmeR against Egfr (Egfr GapmeR-A Design ID: LG00204888-DDA, Egfr GapmeR-B Design ID: LG00204889-DDA, Egfr GapmeR-C Design ID: LG00204890-DDA; Qiagen) or Negative Control A Gapmer (Design ID: LG00000001-DDA; Qiagen) at 500 nM at day 0 and supplemented at 250 nM at day 3. Enteroids at day 5 were harvested for RNA isolation or fixed in 2% (v/v) paraformaldehyde for whole-mount staining. For EdU staining assays, enteroids were treated with 10-mM EdU 6 hours before the harvest time point follow by the manufacturer’s protocol of Click-iT EdU Alexa Fluor 488 Flow Cytometry Assay Kit (C10425; Thermo Fisher Scientific).
For studies inhibiting Xiap function, enteroids were treated with GDC0152 (Cat. # S7010; SelleckChem, Houston, TX), which inhibits the activity of inhibitors of apoptosis proteins (IAPs) including X chromosome-linked IAP (XIAP) and IAPs 1 and 2 and promote apoptosis. The enteroid culture was treated alone with GDC0152 at 0.25 μm or co-treated with LNA at day 0. At day 5, enteroids were fixed in 2% (v/v) paraformaldehyde for brightfield imaging and whole-mount staining.
Porcine Enteroids
Jejunal crypts were isolated from 6- to 10-week-old, mixed gender, wild type Yorkshire cross pigs as previously described.54 The isolated crypts (day 0) were cultured in Reduced Growth Factor Matrigel (cat. 356231; Corning) and maintained in DMEM/F12 medium (cat. 12634-010; Life Technologies) supplemented with 50-ng/mL EGF (cat. 236-EG; R&D Systems), 100-ng/mL Noggin (cat. 120-10C; PeproTech), 1 μg/mL R-Spondin (cat. 4645RS/CF, R&D Systems), 500-nM A83-01 (cat. 2939; Tocris Bioscience, Bristol, United Kingdom), 10-μM SB202190 (cat. S7067; Sigma-Aldrich, St. Louis, MO), 1-mM Nicotinamide (cat. N0636, Sigma), 10-nM Gastrin (cat. G9145; Sigma), 10-μM Y-27632 (cat. Y0503; Sigma), and 100-ng/mL Wnt3a (cat. 5036-WN/CF; R&D Systems). For miRNA studies, miRCURY LNA Power Inhibitor against miR-7 (mmu-miR-7a-5p miRCURY LNA miRNA Power Inhibitor, cat. YI04100818-DDA; Qiagen) or Power Negative Control A (cat. YI00199006-DDA; Qiagen) were added on day 0 and enteroids were harvested on day 3.
Human Enteroids
Human duodenal enteroids were previously established from tissue collected from deceased donors through the Gift of Life, Michigan (University of Michigan IRB REP00000105; not regulated designation). For this study, specimen Duo-87 from a 21-year-old male were used (Translational Tissue Modeling Laboratory). Enteroids were cultured in medium containing 25% (v/v) L-WRN conditioned medium. The complete medium contained Advanced DMEM/F-12 (cat. 12634028; Gibco), 2-mM GlutaMax (cat. 35050-061; Gibco), 10-mM HEPES (cat. 15630080; Gibco), N-2 (cat. 17502048; Gibco), B-27 supplement minus vitamin A (cat. 12587010; Gibco), 50 units/mL of penicillin, 0.05-mg/mL streptomycin (cat. 15070063; Gibco), 50-μg/mL Primocin (##ant-pm-1; InvivoGen, San Diego, CA), 1-mM N-Acetyl-L-cysteine (A9165; Sigma-Aldrich), 50-ng EGF/mL (236-EG; R&D Systems), 10-μM SB202190 (S7067; Sigma-Aldrich), 500-nM A83-01 (#2939; R&D Tocris), and 10-μM Y27632 (125410; Tocris Bioscience). Cultures were grown in Matrigel (diluted to 8 mg/mL with growth media; #354234; Corning). Cultures were passaged by triturating and dissociating the Matrigel in cold DPBS, centrifuging at 300 g, and plating the first day with 2.5-μM CHIR99021 (4423; Tocris Bioscience). For miRNA studies, miRCURY LNA Power Inhibitor against miR-7 (mmu-miR-7a-5p miRCURY LNA miRNA Power Inhibitor; cat YI04100818-DDA; Qiagen) or Power Negative Control A (cat. YI00199006-DDA; Qiagen) were added on day 0 and enteroids were harvested on day 5.
Whole-Mount Enteroids Immunostaining and Imaging
The fixed mouse enteroids were permeabilized with 0.5% (v/v) Triton X-100/PBS, washed by PBS containing 0.1% (w/v) BSA/0.02% (v/v) Triton-X/0.05% (v/v) Tween 20 and blocked with 10% (v/v) normal goat serum. Primary antibodies were used to stain Chga (rabbit anti-Chga, 1:100; ab15160; Abcam) and PH3 (rabbit anti-Phospho-Histone H3 (Ser10), 1:100, 9701S; Cell Signaling) and Xiap (1:100; rabbit, NBP2-20918; Novus Biologicals, Centennial, CO). The staining was visualized by fluorescence microscopy with fluorescent-conjugated secondary antibodies (goat anti-rabbit; Alexa Fluor 488, 1:400; Cat. #A-11034; Thermo Fisher Scientific). Nuclei were counterstained with Hoechst 33258 dye (1:1000). For EdU staining assays, enteroids were treated with 10-mM EdU 6 hours before the harvest time point follow by the manufacturer’s protocol of Click-iT Plus EdU Alexa Fluor 488/594 imaging Kit (C10637, C10639; Thermo Fisher Scientific). The immunofluorescent staining was visualized by ZEISS Axiovert 200M inverted microscope. The z-stack bright field images were taken by ZEISS Axiovert 200M inverted microscope for bud count analysis.
Ecc15 Infection in D. melanogaster
Wide type line Canton-S (BDSC: 64349) was maintained at room temperature (∼23°C) on standard fly medium (50-g baker yeast, 30-g cornmeal, 20-g sucrose,15-g agar, 5-mL 99% [v/v] propionic acid mix, 0.5-mL 85% [v/v] phosphoric acid, 26.5-mL methyl paraben in ethanol per 1 L) in a 12-hour light/dark cycle. Oral infection of pathogen Erwinia carotovora ssp. carotovora 15 (Ecc15) was performed as previously described.25 Orally treated flies were incubated at 29°C until dissection for analyses.
Generation of Genetically Modified Lines of Drosophila
Esg-Gal4; UAS-GFP, tub-Gal80TS (EsgTS, progenitor specific)55 and Uas lines (BDSC 41137) were used for creating flies with miR-7 overexpression in Esg stem or progenitor cells. Genetic crosses for flies containing Gal4-UAS-Gal80 system were crossed using ∼15 female flies and 5 males, and transferred during development in a 12-hour light/dark 18°C incubator. Parental generation was removed after 5 days in the 18°C incubator to control for fly density of the F1 progeny.
Immunostaining of Drosophila Midgut
The excised Drosophila midguts were fixed in 4% paraformaldehyde and washed with 0.1% (w/v) Triton X-100 in PBS. The samples then incubated for an hour in blocking solution (1% (w/v) BSA, 1% (v/v) normal donkey serum, and 0.1% (w/v) Triton X-100 in PBS) followed by overnight primary antibody incubation and 2 hours secondary antibody staining. Primary antibodies used in this study were rabbit anti-PH3 (1:000; EMD Millipore, Burlington, MA). Secondary antibodies used in this study were donkey anti-rabbit-555 (1:2000; Thermo Fisher Scientific). DAPI (1:50,000) was used to visualize nuclei. Imaging was performed on a Zeiss LSM 700 fluorescent/confocal inverted microscope. PH3-positive cells were manually counted along the surface of the midgut.
Statistics
In most figure panels, quantitative data are reported as an average of biological replicates ± SEM. In figure panels pertaining to bright field bud count analysis and whole-mount immunofluorescent staining in enteroids, quantitative data are reported as an average of values from all the enteroids pooled from multiple independent experiments ± SEM (n = 2–7 wells per condition per experiment). In all analyses, statistical differences were assessed by 2-tailed Student t test with threshold P value <.05, unless otherwise specifically noted. All authors had access to the study data and had reviewed and approved the final manuscript.
Acknowledgments
The authors would like to thank members of the Sethupathy laboratory and Susan Henning for helpful comments and feedback at various stages of the study, Dr Richard Blumberg for permission to use the Defa6-Cre-tdTomato mice in this study, Dr Zhao Li and the Greehey Children's Cancer Research Institute at University of Texas Health Science Center at San Antonio for small RNA library preparation and sequencing, Dr Jen Grenier and the Cornell RNA sequencing core facility for RNA sequencing, the Cornell University and UNC Chapel Hill Flow Cytometry Core Facilities, and the Cornell University East Campus Mouse Research Facility.
Footnotes
Author contributions A.P.S. and Y.-H.H. contributed to study design and were involved in all of the assays and data collection for the experiments done in the mouse system. A.B. and N.B. carried out the assays for all fruit fly experiments. M.K. performed all bioinformatic analyses. M.T.S. and S.D. contributed to experimental design and histology. M.T.S., B.C.P., and E.G.C. contributed to cell sorting and mouse enteroid experiments. M.K.D. and J.R.S. carried out experiments in human intestinal organoids. M.B. and C.M.D. performed cell sorting from the Defa6-Cre-tdTomato line. J.M.F. and L.M.G. carried out experiments in porcine enteroids. G.A.B. contributed to cell sorting from the Pyy-GFP line. O.O.O. and E.D.T. helped sort and analyze tuft cells. R.L.C. and N.A.K. helped established the Prox1-GFP colony. P.S. initiated the project, secured funding, led the experimental designs, and supervised the study. A.P.S., Y.-H.H., and P.S. wrote the manuscript. All of the authors made important intellectual contributions to the experiments and discussion.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by a Pathway to Stop Diabetes Research Accelerator (1-16-ACE-47 ADA) (to Praveen Sethupathy), the Empire State Stem Cell Fund (C30293GG) (to Yu-Han Hung), the National Science Foundation (IOS-1656118 and IOS-1653021) (to Nicolas Buchon), the National Institute of Diabetes and Digestive and Kidney Diseases/National Institutes of Health (U01DK103141 [to Jason R. Spence] and R01DK100508 [to Christopher M. Dekaney]), the National Institute of Allergy and Infectious Diseases/National Institutes of Health (U19AI116482) (to Jason R. Spence), project grants from the European Association for the Study of Diabetes and JDRF (to Gavin A. Bewick), a Special Emphasis Research Career Award from the National Institutes of Health (K01OD019911-01A1) (to Liara M. Gonzalez), and the National Institute on Aging/National Institutes of Health (R01AG041198) (to Shengli Ding).
References
- 1.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Smith N.R., Gallagher A.C., Wong M.H. Defining a stem cell hierarchy in the intestine: markers, caveats and controversies. J Physiol. 2016;594:4781–4790. doi: 10.1113/JP271651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jadhav U., Saxena M., O'Neill N.K., Saadatpour A., Yuan G.C., Herbert Z., Murata K., Shivdasani R.A. Dynamic reorganization of chromatin accessibility signatures during dedifferentiation of secretory precursors into Lgr5+ intestinal stem cells. Cell Stem Cell. 2017;21:65–77.e5. doi: 10.1016/j.stem.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan K.S., Gevaert O., Zheng G.X.Y., Anchang B., Probert C.S., Larkin K.A., Davies P.S., Cheng Z.F., Kaddis J.S., Han A., Roelf K., Calderon R.I., Cynn E., Hu X., Mandleywala K., Wilhelmy J., Grimes S.M., Corney D.C., Boutet S.C., Terry J.M., Belgader P., Ziraldo S.B., Mikelsen T.S., Wang F., von Furstenberg R.J., Smith N.R., CHandrakesan P., May R., Chrissy M.A.S., Jain R., Cartwright C.A., Niland J.C., Hong Y.K., Carrington J., Breault D.T., Epstein J., Houchen C.W., Lynch J.P., Martin M.G., Plevritis S.K., Curtis C., Ji H.P., Li L., Henning S.J., Wong M.H., Kuo C.J. Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell. 2017;21:78–90.e6. doi: 10.1016/j.stem.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivey K.N., Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Peng Y., Croce C.M. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung Y.H., Sethupathy P. MicroRNAs in the mammalian gut endocrine lineage. Endocrinology. 2018;159:866–868. doi: 10.1210/en.2017-03117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Latreille M., Hausser J., Stutzer I., Zhang Q., Hastoy B., Gargani S., Kerr-Conte J., Pattou F., Zavolan M., Esguerra J.L., Eliasson L., Stoffel M. MicroRNA-7a regulates pancreatic beta cell function. J Clin Invest. 2014;124:2722–2735. doi: 10.1172/JCI73066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poy M.N. MicroRNAs: An adaptive mechanism in the pancreatic beta-cell...and beyond? Best Pract Res Clin Endocrinol Metab. 2016;30:621–628. doi: 10.1016/j.beem.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Beas J., Capilla-Gonzalez V., Aguilera Y., Mellado N., Lachaud C.C., Martin F., Smani T., Soria B., Hmadcha A. miR-7 modulates hESC differentiation into insulin-producing beta-like cells and contributes to cell maturation. Mol Ther Nucleic Acids. 2018;12:463–477. doi: 10.1016/j.omtn.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu H., Guo S., Li W., Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Liu J., Liu C., Naji A., Stoffers D.A. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic beta-cells. Diabetes. 2013;62:887–895. doi: 10.2337/db12-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bravo-Egana V., Rosero S., Molano R.D., Pileggi A., Ricordi C., Dominguez-Bendala J., Pastori R.L. Quantitative differential expression analysis reveals miR-7 as major islet microRNA. Biochem Biophys Res Commun. 2008;366:922–926. doi: 10.1016/j.bbrc.2007.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kredo-Russo S., Mandelbaum A.D., Ness A., Alon I., Lennox K.A., Behlke M.A., Hornstein E. Pancreas-enriched miRNA refines endocrine cell differentiation. Development. 2012;139:3021–3031. doi: 10.1242/dev.080127. [DOI] [PubMed] [Google Scholar]
- 15.Downing S., Zhang F., Chen Z., Tzanakakis E.S. MicroRNA-7 directly targets Reg1 in pancreatic cells. Am J Physiol Cell Physiol. 2019;317:C366–C374. doi: 10.1152/ajpcell.00013.2019. [DOI] [PubMed] [Google Scholar]
- 16.Knudsen L.A., Petersen N., Schwartz T.W., Egerod K.L. The MicroRNA Repertoire in enteroendocrine cells: identification of miR-375 as a potential regulator of the enteroendocrine lineage. Endocrinology. 2015;156:3971–3983. doi: 10.1210/en.2015-1088. [DOI] [PubMed] [Google Scholar]
- 17.Heverhagen A.E., Legrand N., Wagner V., Fendrich V., Bartsch D.K., Slater E.P. Overexpression of MicroRNA miR-7-5p is a potential biomarker in neuroendocrine neoplasms of the small intestine. Neuroendocrinology. 2018;106:312–317. doi: 10.1159/000480121. [DOI] [PubMed] [Google Scholar]
- 18.von Furstenberg R.J., Buczacki S.J., Smith B.J., Seiler K.M., Winton D.J., Henning S.J. Side population sorting separates subfractions of cycling and non-cycling intestinal stem cells. Stem Cell Res. 2014;12:364–375. doi: 10.1016/j.scr.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards P., Pais R., Habib A.M., Brighton C.A., Yeo G.S., Reimann F., Gribble F.M. High fat diet impairs the function of glucagon-like peptide-1 producing L-cells. Peptides. 2016;77:21–27. doi: 10.1016/j.peptides.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mah A.T., Van Landeghem L., Gavin H.E., Magness S.T., Lund P.K. Impact of diet-induced obesity on intestinal stem cells: hyperproliferation but impaired intrinsic function that requires insulin/IGF1. Endocrinology. 2014;155:3302–3314. doi: 10.1210/en.2014-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beyaz S., Mana M.D., Roper J., Kedrin D., Saadatpour A., Hong S.J., Bauer-Rowe K.E., XIfaras M.E., Akkad A., Arias E., Pinello L., Katz Y., Shinagare S., Abu-Remaileh M., Mihaylova M.M., Lamming D.W., Dogum R., Guo G., Bell G.W., Selig M., Nielsen G.P., Gupta N., Ferrone C.R., Deshpande V., Yuan G.C., Orkin S.H., Sabatini D.M., Yilmaz O.H. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531:53–58. doi: 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baran-Gale J., Fannin E.E., Kurtz C.L., Sethupathy P. Beta cell 5'-shifted isomiRs are candidate regulatory hubs in type 2 diabetes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S., Zhang P., Chen Z., Liu M., Li X., Tang H. MicroRNA-7 downregulates XIAP expression to suppress cell growth and promote apoptosis in cervical cancer cells. FEBS Lett. 2013;587:2247–2253. doi: 10.1016/j.febslet.2013.05.054. [DOI] [PubMed] [Google Scholar]
- 24.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., Van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 25.Buchon N., Broderick N.A., Poidevin M., Pradervand S., Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Tsai Y.H., Czerwinski M., Wu A., Dame M.K., Attili D., Hill E., Colacino J.A., Nowacki L.M., Shroyer N.F., Higgins P.D.R., Kao J.Y., Spence J.R. A method for cryogenic preservation of human biopsy specimens and subsequent organoid culture. Cell Mol Gastroenterol Hepatol. 2018;6:218–222.e7. doi: 10.1016/j.jcmgh.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C., Zeng X., Jiang G., Liao X., Liu C., Li J., Jin H., Zhu J., Sun H., Wu X.R., Huang C. XIAP BIR domain suppresses miR-200a expression and subsequently promotes EGFR protein translation and anchorage-independent growth of bladder cancer cell. J Hematol Oncol. 2017;10:6. doi: 10.1186/s13045-016-0376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basak O., Beumer J., Wiebrands K., Seno H., van Oudenaarden A., Clevers H. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell. 2017;20:177–190.e4. doi: 10.1016/j.stem.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Liu X., Li G., Su Z., Jiang Z., Chen L., Wang J., Yu S., Liu Z. Poly(amido amine) is an ideal carrier of miR-7 for enhancing gene silencing effects on the EGFR pathway in U251 glioma cells. Oncol Rep. 2013;29:1387–1394. doi: 10.3892/or.2013.2283. [DOI] [PubMed] [Google Scholar]
- 30.Sun X., Li J., Sun Y., Zhang Y., Dong L., Shen C., Yang L., Yang M., Li Y., Shen G., Tu Y., Tao J. miR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget. 2016;7:53558–53570. doi: 10.18632/oncotarget.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., Heim M.H., Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 32.Poy M.N., Hausser J., Trajkovski M., Braun M., Collins S., Rorsman P., Zavolan M., Stoffel M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soh J., Iqbal J., Queiroz J., Fernandez-Hernando C., Hussain M.M. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19:892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X., Cassidy J.J., Reinke C.A., Fischboeck S., Carthew R.W. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunawardene A.R., Corfe B.M., Staton C.A. Classification and functions of enteroendocrine cells of the lower gastrointestinal tract. Int J Exp Pathol. 2011;92:219–231. doi: 10.1111/j.1365-2613.2011.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beumer J., Artegiani B., Post Y., Reimann F., Gribble F., Nguyen T.N., Zeng H., Van den Morn M., Van Es J.H., Clevers H. Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nat Cell Biol. 2018;20:909–916. doi: 10.1038/s41556-018-0143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shivdasani R.A. Limited gut cell repertoire for multiple hormones. Nat Cell Biol. 2018;20:865–867. doi: 10.1038/s41556-018-0153-9. [DOI] [PubMed] [Google Scholar]
- 38.Hutch C.R., Sandoval D. The role of GLP-1 in the metabolic success of bariatric surgery. Endocrinology. 2017;158:4139–4151. doi: 10.1210/en.2017-00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worthington J.J. The intestinal immunoendocrine axis: novel cross-talk between enteroendocrine cells and the immune system during infection and inflammatory disease. Biochem Soc Trans. 2015;43:727–733. doi: 10.1042/BST20150090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moran G.W., Leslie F.C., Levison S.E., Worthington J., McLaughlin J.T. Enteroendocrine cells: neglected players in gastrointestinal disorders? Therap Adv Gastroenterol. 2008;1:51–60. doi: 10.1177/1756283X08093943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zietek T., Rath E. Inflammation meets metabolic disease: gut feeling mediated by GLP-1. Front Immunol. 2016;7:154. doi: 10.3389/fimmu.2016.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Formeister E.J., Sionas A.L., Lorance D.K., Barkley C.L., Lee G.H., Magness S.T. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1108–G1118. doi: 10.1152/ajpgi.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi I., Chung H.K., Ramu S., Lee H.N., Kim K.E., Lee S., Yoo J., Choi D., Lee Y.S., Aguilar B., Hong Y.K. Visualization of lymphatic vessels by Prox1-promoter directed GFP reporter in a bacterial artificial chromosome-based transgenic mouse. Blood. 2011;117:362–365. doi: 10.1182/blood-2010-07-298562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bohorquez D.V., Chandra R., Samsa L.A., Vigna S.R., Liddle R.A. Characterization of basal pseudopod-like processes in ileal and colonic PYY cells. J Mol Histol. 2011;42:3–13. doi: 10.1007/s10735-010-9302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.King S.L., Mohiuddin J.J., Dekaney C.M. Paneth cells expand from newly created and preexisting cells during repair after doxorubicin-induced damage. Am J Physiol Gastrointest Liver Physiol. 2013;305:G151–G162. doi: 10.1152/ajpgi.00441.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerbe F., Sidot E., Smyth D.J., Ohmoto M., Matsumoto I., Dardalhon V., Cesses P., Garnier L., PouzollesM, Brulin B., Bruschi M., Harcus Y., Zimmermann V.S., Taylor N., Maizels R.M., Jay P. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature. 2016;529:226–230. doi: 10.1038/nature16527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peck B.C., Mah A.T., Pitman W.A., Ding S., Lund P.K., Sethupathy P. Functional transcriptomics in diverse intestinal epithelial cell types reveals robust MicroRNA sensitivity in intestinal stem cells to microbial status. J Biol Chem. 2017;292:2586–2600. doi: 10.1074/jbc.M116.770099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanke M., Baran-Gale J., Villanueva J., Sethupathy P. miRquant 2.0: an expanded tool for accurate annotation and quantification of MicroRNAs and their isomiRs from small RNA-sequencing data. J Integr Bioinform. 2016;13:307. doi: 10.2390/biecoll-jib-2016-307. [DOI] [PubMed] [Google Scholar]
- 49.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gonzalez L.M., Williamson I., Piedrahita J.A., Blikslager A.T., Magness S.T. Cell lineage identification and stem cell culture in a porcine model for the study of intestinal epithelial regeneration. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Micchelli C.A., Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]