Significance
Glycogen is the primary form of glucose storage in mammals and plays a vital role in cellular energy homeostasis. Mapping glycogen in vivo is useful in diagnosis and assessment of diseases where glucose metabolism is altered, such as diabetes, tumors, and liver diseases. Currently, imaging glycogen in the clinic is not done due to the lack of a practical approach. We report a noninvasive MRI method for imaging glycogen with enhanced detection sensitivity and high specificity. We performed mouse liver glycogen mapping, showing a heterogenous distribution of glycogen and regions of metabolism. This work opens up opportunities for studying glycogen metabolism in vivo at high temporal and spatial resolution.
Keywords: saturation transfer, Z-spectrum, liver, fasting, glucagon
Abstract
Glycogen plays a central role in glucose homeostasis and is abundant in several types of tissue. We report an MRI method for imaging glycogen noninvasively with enhanced detection sensitivity and high specificity, using the magnetic coupling between glycogen and water protons through the nuclear Overhauser enhancement (NOE). We show in vitro that the glycogen NOE (glycoNOE) signal is correlated linearly with glycogen concentration, while pH and temperature have little effect on its intensity. For validation, we imaged glycoNOE signal changes in mouse liver, both before and after fasting and during glucagon infusion. The glycoNOE signal was reduced by 88 ± 16% (n = 5) after 24 h of fasting and by 76 ± 22% (n = 5) at 1 h after intraperitoneal (i.p.) injection of glucagon, which is known to rapidly deplete hepatic glycogen. The ability to noninvasively image glycogen should allow assessment of diseases in which glucose metabolism or storage is altered, for instance, diabetes, cardiac disease, muscular disorders, cancer, and glycogen storage diseases.
Glycogen, a branched polymeric form of glucose, plays a central role in maintaining glucose (short-term energy) homeostasis in different tissues. For instance, brain glycogen [with a concentration of ∼5.1 mM (1)] is almost exclusively localized in astrocytes (2) and acts as an energy reserve for neuronal activity. Liver glycogen [100–800 mM (3)] maintains appropriate levels of blood glucose (4), while glycogen in heart muscle is a crucial contributor to myocardial energy production (5). Skeletal muscle glycogen [40–100 mM (6)] storage is closely correlated to fatigue resistance during prolonged or high-intensity exercise (7). Considering its role in cellular energy homeostasis and its wide abundance in vivo, changes in glycogen concentration are often an endogenous marker for a variety of diseases such as cancers (8, 9), diabetes (10, 11), glycogen storage diseases (12), and liver diseases (13). A method allowing noninvasive in vivo measurement of glycogen levels would thus be of importance for the assessment of these diseases and disorders.
Several techniques are currently available for quantifying glycogen noninvasively, including ultrasound (14), 18F-N-(methyl-(2-fluoroethyl)-1H-[1,2,3]triazole-4-yl)glucosamine Positron Emission Tomography (18F-NFTG-PET) (15), and 13C and 1H magnetic resonance spectroscopy (MRS) (1, 3, 6, 16–18). The specificity of the MRS methods is superior to ultrasound. While 1H MRS has been shown to detect the total glycogen pool when dissolved in D2O in vitro (17), it may underestimate the amount of hepatic glycogen in vivo (19, 20). In vivo 13C MRS, one of the most popular methods in the past several decades, has been performed at natural abundance (1, 3, 18) (1.1% of all carbons) as well as with infusion of substrate (1), but its impact in the clinic has been limited due to its much lower detection sensitivity compared to 1H MR and its requirement for specialized equipment that often is unavailable on clinical MRI scanners. 18F-NFTG-PET study of exogenous isotope-labeled agents detects only the synthesis of labeled glycogen instead of total glycogen pool.
The glycoCEST (chemical exchange saturation transfer imaging of glycogen) MRI method (21–23) was proposed more than a decade ago as a promising method that doesn’t require specialized hardware or exogenous contrast agents and provides maps with relatively high spatial resolution. It detects glycogen indirectly through the exchange interaction between the glycogen hydroxyl protons and water protons, a principle that has been widely utilized to image several other metabolite molecules (24). However, the fast exchange properties of glycogen hydroxyl protons in vivo and the presence of magnetic resonance signals from several other molecular sources at the hydroxyl proton frequencies hinder the quantification of the glycoCEST signal. Consequently, the glycoCEST MRI method has only had limited success for in vivo studies (25, 26).
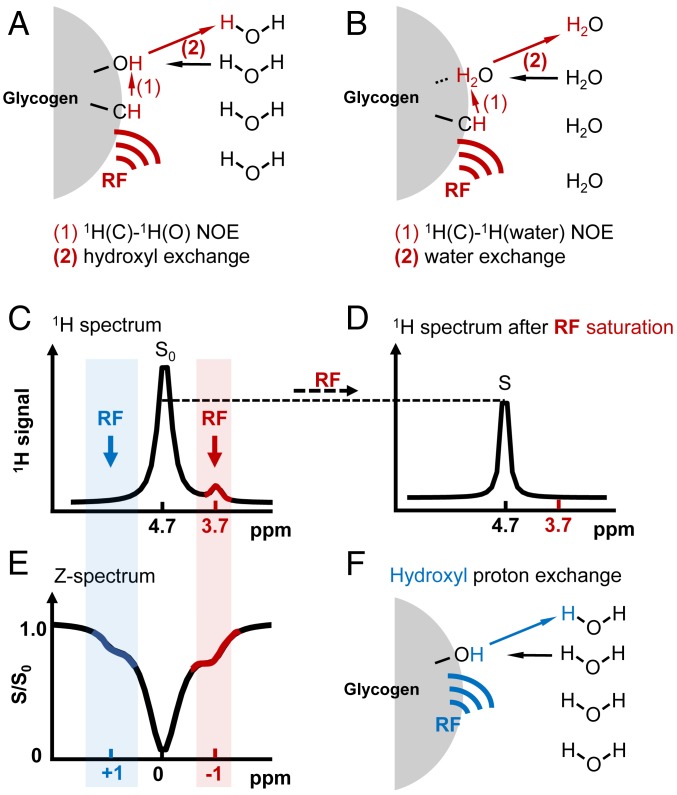
Here, we report an MRI method for imaging glycogen based on the nuclear Overhauser enhancement (NOE) phenomenon (also called nuclear Overhauser effect, when multiple enhancements are measured together) (27). NOE, a fundamental magnetization transfer mechanism, can be detected using saturation transfer (ST) experiments (24, 28), in which the relative saturation of water signal is measured as a function of saturation frequency, generating a so-called Z-spectrum (Fig. 1). Like a 1H MR spectrum (Fig. 1 C and D), the Z-spectrum (Fig. 1E) displays signals from different molecular or chemical origin, but here with enhanced sensitivity and from protons undergoing saturation transfer via either chemical exchange (Fig. 1F), relayed or direct NOEs (Fig. 1 A and B), or transferred magnetization due to transient molecular binding (29). Based on this rationale, we hypothesized that NOEs between water protons and glycogen aliphatic protons (27) should be detectable at one or more chemical shift positions in the Z-spectrum. In order to test our hypothesis, we first characterized glycogen NOE signals in vitro by varying glycogen concentration, pH, and temperature and then used the in vivo NOE signals to image glycogen in mouse liver. Both types of experiments showed a strong signal around −1 ppm (parts per million) relative to the water resonance in the aliphatic region of the Z-spectrum. To validate that the in vivo signal source was from glycogen, we examined the signal changes in the liver of fed mice before and after 24 h fasting as well as the effect of intraperitoneal (i.p.) injection of glucagon, which is known to rapidly deplete hepatic glycogen.
Fig. 1.
Basics of the glycoNOE saturation transfer experiment. (A and B) Two possible NOE-based saturation transfer pathways from glycogen aliphatic protons to water: (A) saturation on glycogen aliphatic protons by a radio frequency (RF) pulse is transferred to a neighboring hydroxyl proton and subsequently to water via proton chemical exchange between glycogen hydroxyl protons and water; (B) saturation on aliphatic protons is transferred directly to a nearby bound water and then to the free water pool via water molecular exchange. (C and D) Schematic 1H MR spectrum for glycogen in H2O before (C) and after (D) the RF saturation of the aliphatic protons. S0 and S represent water intensities in the two spectra. (E) Z-spectrum, showing S/S0 as a function of RF irradiation frequency. Signal shown at around −1 ppm in the Z-spectrum is from glycogen aliphatic protons undergoing the possible NOE-based saturation transfer mechanisms (A and B), while signal at around +1 ppm is from hydroxyl protons undergoing chemical exchange with water (F). Because of multiple saturation transfer events in a single MR sequence, the glycogen signal in the Z-spectrum is enhanced compared to that in the 1H MR spectrum. Note that the Z-spectrum is referenced to the water frequency.
Results
The occurrence of a glycogen NOE with water was first confirmed by saturation transfer experiments in vitro. Fig. 2B displays a 1H NMR spectrum showing the spectral positions of glycogen aliphatic protons (assigned in Fig. 2A) relative to water. Corresponding signals are observed in the Z-spectra acquired from rabbit liver glycogen and bovine liver glycogen (Fig. 2C). At the positive frequency offset in Fig. 2C, a broad saturation curve is visible from +0.3 ppm to about +1.5 ppm, a characteristic of intermediate to fast chemical exchange between water and glycogen hydroxyl protons (glycoCEST), which resonate at +0.7 and +1.2 ppm (22). The H1-4 proton at +0.6 ppm is also expected to generate an NOE (27), but at pH 7.4 (Fig. 2C) this NOE is hidden under the stronger glycoCEST effect and water direct saturation. The H1-4 NOE peak starts to become visible when glycoCEST effects are small at low or high pH (SI Appendix, Fig. S4). In the negative frequency range of Z-spectra, both rabbit and bovine liver glycogen show NOE peaks between −0.6 and −1.5 ppm, which are attributed to a combination of glycogen aliphatic protons H3, H5, and H2+H4-1 (Fig. 2 A–C) based on previously published assignments in MR spectra (27, 30). At a magnetic field of 11.7 T, these combined signals appear as a single broad peak at approximately −1 ppm, while two composite peaks (at −0.7 and −1.0 ppm) are visible at 17.6 T (SI Appendix, Fig. S1), corresponding to H3 (−0.7 ppm) and the overlapping resonances of H5 (−0.9 ppm) and H2+H4-1 (−1.1 ppm). Interestingly, the rabbit liver glycogen sample (with an average particle size of ∼52 nm, SI Appendix, Fig. S2) generates a much stronger NOE signal than the bovine liver glycogen (∼7 nm) (Fig. 2C), suggesting glycoNOE signal increases with glycogen particle size, in line with the expectation of stronger NOE with slower motion.
Fig. 2.
The glycoNOE signal in vitro for liver glycogen. (A) Chemical structure and proton assignment of glycogen. (B) One-dimensional 1H NMR spectrum of bovine liver glycogen [100 mM, pH 7.4; peak assignments are based on the work by L.-H. Zang et al. (30)]. (C) Z-spectra for bovine and rabbit glycogen solutions (300 mM glucose unit, pH 7.4, 20 °C.) The bottom row shows the residual signal after subtracting out the fitted direct water saturation signal from the Z-spectra. (D) Bovine liver glycogen and rabbit liver glycoNOE signal (peak value at −1.0 ppm) as a function of glucose-unit concentration in vitro (pH 7.4, 20 °C). (E) Rabbit liver glycogen (100 mM) NOE signal (peak value) as a function of pH in vitro at 20 °C and 37 °C. All data in C–E were acquired with an RF saturation power (B1) = 1.0 µT.
To characterize the glycoNOE signal, rabbit liver glycogen Z-spectra were acquired in vitro as a function of concentration and pH and at two temperatures (20 °C and 37 °C). The glycoNOE signal intensity was found to be linearly dependent on concentration (Fig. 2D and SI Appendix, Fig. S3) and showed negligible dependence on pH and temperature (Fig. 2E and SI Appendix, Fig. S4). Importantly, these dependencies are in strong contrast with glycoCEST signals (+0.7 and +1.2 ppm) which depend on concentration nonlinearly (21), and are sensitive to pH and temperature (SI Appendix, Figs. S3 and S4). Both glycoNOE and glycoCEST show large signal enhancement ratios: for the 100 mM (equivalent concentration of glucose units) rabbit liver glycogen sample, the glycoNOE peak reaches an intensity equivalent to about 5.3 M proton signal (Fig. 2E, calculated from water percentage), while the glycoCEST effect reaches up to 10 M (SI Appendix, Fig. S4) proton signal depending on pH and temperature.
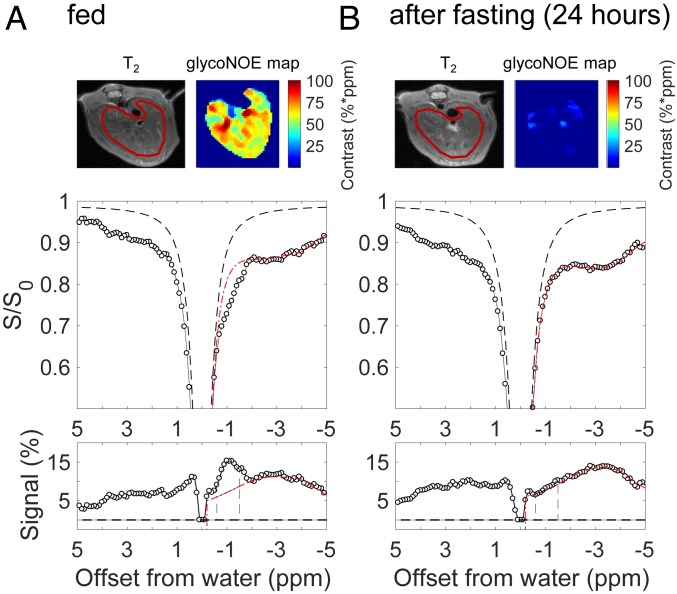
Following the in vitro experiments, glycoNOE experiments were performed in vivo in mouse liver. Fig. 3A shows the Z-spectrum from the liver of a fed healthy mouse. Noticeably, there is a peak at approximately −1 ppm, which is tentatively attributed to the overlapped NOE peaks from glycogen protons H3, H5, and H2+H4-1 and assigned to be the glycoNOE peak. To estimate the glycoNOE peak intensity in vivo, the smooth part of the negative range (−0.1 to −8 ppm) of the Z-spectrum was fitted using a multi-Lorentzian approach (31), and the magnetization transfer background was subtracted from the experimental data. The residual signal (Fig. 3 A, Bottom) shows overlapping peaks from different origins in the liver. The glycoNOE peak intensity was quantified by taking the integral from −0.6 to −1.5 ppm (shaded area in Fig. 3A). The glycoNOE maps (top row of Fig. 3A and SI Appendix, Figs. S5 and S7–S11) generated using this approach on a voxel-by-voxel basis show a heterogeneous distribution of glycogen over the liver.
Fig. 3.
Effect of fasting on liver Z-spectrum of a mouse. The same mouse was scanned before (A) and after (B) 24 h fasting. Data were acquired with an RF saturation power of B1 = 1.0 µT. The glycoNOE maps were estimated from the voxel-wise integral (unit: %*ppm, which is % times ppm range) of the glycoNOE peak over the shaded area (−0.6 to −1.5 ppm range). The bottom row shows residual Z-spectral signal after subtracting the fitted baseline. The T2 image and glycoNOE map have been magnified 1.3×.
To calibrate the glycoNOE signal with liver glycogen content, mouse liver glycogen was immediately extracted and quantified after glycoNOE MRI. The average glycoNOE signal was found to be linearly correlated with measured glycogen content (SI Appendix, Fig. S12). To further confirm the origin of the glycoNOE signal in liver, glycogen depletion experiments were performed using (1) a 24-h fasting protocol or (2) glucagon injections. The liver Z-spectra for mice in the fasted state show that the glycoNOE peak is greatly depleted (Fig. 3B and also SI Appendix, Figs. S7–S11). For five mice studied after fasting for 24 to 28 h, the integrated liver glycoNOE signal (−0.6 to −1.5 ppm) decreased on average from 49 ± 8%*ppm (equivalent to a glycogen concentration of about 47 mg/g wet liver; signal unit: % times ppm range) at fed state to 6 ± 8%*ppm (∼6 mg/g) in the fasted state (n = 5).
Glucagon injection experiments were also conducted to induce a change of glycogen level in mouse liver. Glucagon accelerates liver glycogen breakdown, thus allowing the monitoring of glycogen depletion within 2 h. Fig. 4 (also SI Appendix, Figs. S13–S17) shows the changes in the liver Z-spectrum as a function of time after i.p. injection of glucagon. The Z-spectral differences between pre- and postinjection show a great reduction of the integrated glycoNOE signal (−0.6 to −1.5 ppm) due to the effect of glucagon (Fig. 4 B–D). By fitting the Z-spectra voxel by voxel, we also constructed glycoNOE maps at different time points. Fig. 4E shows how the glycoNOE signal in the liver changed spatially with time after glucagon injection. At 65 min postinjection, the average glycoNOE signal (n = 5) in the liver was greatly depleted from 45 ± 15%*ppm (∼48 mg/g) to 11 ± 9%*ppm (∼12 mg/g). Fig. 4F shows the average (n = 5) integrated glycoNOE signal (and corresponding concentration) and “glycoCEST + H1-4 NOE” signal changes as a function of time. The glycoNOE signal reached a stationary phase ∼60 min after glucagon injection. The results clearly show that the glycoNOE signal is reduced with the breaking down of hepatic glycogen. There are also signal changes in the region of 0 ∼ +1.5 ppm with a peak of around +0.6 ppm, corresponding to the combined effect of hydroxyl protons (glycoCEST) and the glycogen H1-4 NOE. It is uncertain how much the H1-4 NOE and hydroxyl exchange each contribute to this signal.
Fig. 4.
Effects of glucagon on glycoNOE contrast in the liver of fed mice. (A) The experimental scheme: UTE-ST MRI experiments (B1 = 0.7 µT) were conducted before and after glucagon (100 µL, 1 mg/mL) injection. (B–D) Z-spectral changes after glucagon for the indicated region of interest. Shaded areas indicate signal changes over the glycoNOE region (−0.6 to −1.5 ppm). (E) glycoNOE maps as a function of time before and after injection. (F) Average (n = 5) glycoNOE and “glycoCEST + H1-4 NOE” signal changes as a function of time in mouse liver. The average glycogen concentration changes (right y axis) were calculated from glycoNOE signal. The T2 image and glycoNOE map have been magnified 1.3×.
Discussion
The Z-spectra (Fig. 2 and SI Appendix, Fig. S4) of glycogen in vitro show resonances around both +0.6 and −1 ppm. The results are consistent with previous MRS measurements on the glycogen NOE with water (27). Chen and coworkers (27) showed that saturation of water resulted in a reduction of H1-4 (+0.6 ppm) signal and that the inversion of H1-4 resulted in NOEs for H3 (−0.7 ppm), H5 (−0.9 ppm), and H2+H4-1 (−1.1 ppm), demonstrating the NOE network of glycogen protons (H1-4, H3, H5, H2+H4-1) with water. However, in the Z-spectrum, the extent of the H1-4 NOE is small compared with the composite glycoNOE peak around −1 ppm (see pH 2.0 in SI Appendix, Fig. S4), and it overlaps with the broad CEST signal from exchangeable OH protons. We therefore focused our analysis on the composite −1 ppm glycoNOE signal, where four aliphatic protons (H3, H5, H2+H4-1) contribute with a signal enhanced by about two orders of magnitude relative to proton MRS. This relatively large signal enhancement using glycoNOE MRI allowed fast acquisition (currently 9 s per frequency point) at acceptable resolution (∼0.3 mm) for in vivo imaging.
The results in vivo show that the distribution of the glycoNOE signal in the liver is heterogeneous. This agrees with previous observations that the liver glycogen content is highly dynamic and can vary dramatically depending on several factors such as health, the time after feeding, and the distances to portal tracts and central veins (32). Considering that glycoNOE depends linearly on glycogen concentration (Fig. 2D and SI Appendix, Fig. S12), the glycoNOE distribution should reflect local differences of glycogen levels. However, care has to be taken with such an interpretation, because particle size may in principle also contribute to the observed heterogeneity (Fig. 2 and SI Appendix, Fig. S2). Note that different forms of glycogen coexist in liver, namely particles (∼106−107 Da) that have various diameters between 10 and 30 nm and much larger particles (up to over 109 Da) that have diameters as high as 300 nm and likely have stronger NOE effects due to slower motion.
After fasting the mice for 24 h, glycoNOE signal (n = 5) decreases from 49 ± 8%*ppm (equivalent to a glycogen concentration of about 47 mg/g wet liver, SI Appendix, Fig. S12) to 6 ± 8%*ppm (∼6 mg/g). This is in agreement with a previous report that hepatic glycogen in mice is only 7% of its original concentration after 24 h of fasting (33). The fasting data strongly support our assignment that the −1 ppm NOE signal in liver Z-spectra is mainly from glycogen. This was further confirmed by the glucagon injection experiments showing a strong reduction in the glycoNOE signal upon glucagon injection. These results are in agreement with the previous study of glycoCEST on perfused mouse liver, which showed signal reduction at both −1.0 and +1.0 ppm under glucagon infusion (21). The reduction in glycoNOE (Fig. 4) is interpreted to be a result of glycogen breaking down into glucose due to the effect of glucagon, while the decrease in signal at +0.5 to +1.4 ppm cannot be interpreted as a direct consequence of this conversion process. The reason is that the liver glucosyl pool (the number of total glucosyl units in glucose and glycogen) is constant in the conversion reactions. Therefore, unless the glucose is removed by perfusion, the signal at around +1 ppm should not be reduced in this conversion and may even increase due to a higher number of OH groups in the +0.6 to +3 ppm range. The reasons for the observed signal reduction at +0.5 to +1.4 ppm could be a result of both glycogen H1-4 NOE signal loss and active export of glucose from the liver.
The decrease in glycoNOE signal in liver under the effects of fasting or glucagon due to metabolism may in principle be affected by a change in glycogen particle size. Glycogen average particle size has been suggested to fluctuate in the small range of about 15 to 30 nm (34) in mouse liver during this process. Similarly, possible spatial variations of glycogen particle size in liver may affect the glycoNOE signal distribution. Interestingly, however, we found a linear correspondence between chemical concentration measurement and the glycoNOE signal during fasting (SI Appendix, Fig. S12), which suggests that particle size change is not a major issue (a notable role would cause a nonlinear correlation). As a consequence, we have therefore now used this measurement in SI Appendix, Fig. S12 as a calibration and converted the glycoNOE into glycogen concentration.
Respiratory motion, blood flow, and arbitrary bodily movements may also increase the uncertainty in the glycoNOE signal. Although the glycoNOE maps can be generated with any type of MRI readout, we used an ultrashort echo time saturation transfer (UTE-ST) pulse sequence with a radial sampling scheme (35) to suppress respiratory motion artifacts in vivo. The observation that the glycoNOE signal for fasted liver is homogenously low (Fig. 3B) suggests artifacts from motion and small vessels are minimal. The consistency of the glycoNOE maps is further supported by measurements as a function of B1 (SI Appendix, Fig. S5).
The current study demonstrates a way of imaging glycogen noninvasively in vivo with enhanced sensitivity. It is based on the NOE between glycogen aliphatic protons and water protons, in contrast to the glycoCEST method which is based on the chemical exchange between glycogen hydroxyl protons and water protons. This glycoNOE approach is advantageous over glycoCEST for several reasons. First of all, the glycoNOE signal intensity was found to be minimally sensitive to pH and temperature (Fig. 2), while glycoCEST experiments are affected by both pH and concentration changes rendering data interpretation less straightforward. While this temperature insensitivity may seem counterintuitive at first due to the sensitivity of NOEs to molecular motion, this can be understood by the glycoNOE effect being proportional to the ratio of two relaxation rates (SI Appendix, Supplementary Discussion). Second, glycoNOE MRI can be readily translated to human scanners (see data at 3 T, SI Appendix, Fig. S18) because the NOE transfer is slow, allowing efficient saturation and high signal visibility to be achieved with reasonably low B1 powers (on the order of 1 µT or less). Third, the glycoNOE signal is easier to extract, while the glycoCEST signal is coalesced with the water peak. In addition, glycoCEST effects are often estimated based on an asymmetry analysis that quantifies glycoCEST as the saturation signal difference between −1 and +1 ppm (21). The current study suggests that such asymmetry analysis should not be used to assess glycogen changes as the glycoNOE and glycoCEST signals compensate each other in a proportion that will vary with concentration and the applied saturation power, thus complicating absolute quantification. Last but not least, glycoNOE has better signal specificity for glycogen. For example, glycoNOE MRI can differentiate glycogen from glucose, while glycoCEST MRI cannot. The glycoCEST method is based on the exchange between the hydroxyl protons of glycogen and water, but many other molecules in vivo, especially glucose and its analogs, have hydroxyl protons resonating at ∼ +1 ppm as well. The glycoCEST in reality measures the total contribution of any molecules that contain hydroxyl groups resonating at ∼ +1 ppm. In contrast, the signal at glycoNOE position (−1 ppm) in the liver could be mostly removed by fasting or glucagon injection, showing that its contamination by other metabolites is limited. When studying muscle, the interpretation of glycoCEST becomes even more difficult because signals from abundant metabolites such as creatine (guanidinium protons at +2 ppm) and phosphocreatine (36) may contaminate the glycoCEST effect at lower field strengths, where the exchange regime is faster than at 11.7 T. This field dependence of the proton exchange regime minimally affects the glycoNOE signals (SI Appendix, Fig. S18).
It is important to note that almost 100% of glycogen is visible in both in vivo and in vitro 13C MRS (16) and also in 1H MRS of glycogen in D2O (17), while glycogen in vivo is greatly underestimated by 1H MRS (20). This underestimation of glycogen by 1H MRS could not be explained by the existence of a significant “hidden” population of glycogen with extremely short T2 relaxation time, as 13C MRS (16, 37) results showed the opposite. The current study suggests the underestimation of glycogen in vivo by 1H MRS likely results from the water suppression techniques used in these experiments (20). The presaturation of water peak can reduce the glycogen proton signal by as high as ∼55% (27) due to the saturation transfer discussed above, causing glycogen to be underestimated by 1H MRS with water suppression in H2O but not in D2O.
Conclusion
We demonstrated the specific mapping of hepatic glycogen with enhanced sensitivity using glycoNOE MRI. We further validated the hypothesis that glycogen protons have a −1 ppm composite NOE signal in Z-spectra both in vitro, and in mouse liver in vivo, using fasting and glucagon injection experiments on mice. As glycogen is present in the heart, liver, skeletal muscle, brain, and even tumor tissues, the proposed glycoNOE MRI method has the potential to be applied to assess function and diseases in these tissues.
Materials and Methods
UTE-ST MRI.
All MRI experiments were performed on an 11.7 T (500 MHz) Bruker Biospec preclinical scanner (Bruker) equipped with a 72-mm quadrature volume resonator for transmission and an 8-channel phased array RF coil for reception, unless specified otherwise. A UTE-ST MRI pulse sequence with a radial acquisition scheme, described in detail elsewhere (35), was used to acquire all data. In each repetition time (TR) of UTE-ST, a 20-ms Gaussian-shaped saturation pulse was used to label the proton pool at a certain frequency followed by the UTE readout. The interpulse delay mixing time was 10 ms, and the excitation pulse for the UTE readout was a 0.3-ms Gaussian pulse. The effective echo time (TE) was 0.38 ms, and total TR was 30 ms. The number of radial spokes was 302 for each frequency. The scan time for one saturation frequency image was 9 s (302 × 30 ms).
In Vitro Experiments on Glycogen.
Rabbit liver glycogen (Type III G8876, chemical abstract service (CAS) registry no. 9005-79-2) and bovine liver glycogen (Type 1X G0885, CAS no. 9005-79-2) from Sigma were dissolved in PBS (152 mM Na/9.6 mM Pi). (1) For both rabbit liver glycogen and bovine liver glycogen, solutions with glucose unit concentrations of 25, 50, 100, 150, 200, and 300 mM and the same pH of 7.4 were prepared. (2) To examine pH effects, rabbit liver glycogen solutions with pH of 2.0, 3.0, 4.0, 5.5, 6.0, 6.5, 7.0, 7.4, 8.0, and 9.0 and the same glucose unit concentration of 100 mM were made. Experiments were carried out at 20 °C or 37 °C. (3) To examine temperature effects on the measurements, scans on a rabbit liver sample set (see 2, above) were first conducted at 20 °C and subsequently at 37 °C (1 h of waiting for thermal equilibrium) using a 23-mm volume coil (for both transmit and receive) that was equipped with a heater and temperature sensor module.
Four different saturation B1 powers (0.5, 0.7, 1.0, and 1.5 µT) were used for UTE-ST scans on glycogen in vitro. The saturation frequency offsets were 200, 200, 200, 8, 7, 6, and 5 ppm, then every 0.1 to 0 ppm; and then 200, 200, 200, 200, −8, −7, −6, and −5 ppm, then every 0.1 to −0.1 ppm (a “converge sampling” scheme). The total time used for one Z-spectrum was 17 min 22 s.
In Vivo Liver glycoNOE MRI of Fed and Fasted Mice.
All experiments were performed with the approval of and in accordance with Johns Hopkins University Animal Care and Use Committee guidelines. Five healthy fed adult mice were scanned with MRI before fasting and after 24 to 28 h of fasting. Fed mice were studied at least 6 h after starting food consumption. A 2-mm slice thickness with in-plane resolution of 0.3 mm was used, and UTE-ST scans were acquired with the same parameters as the in vitro experiments.
In Vivo Liver glycoNOE MRI of Mouse Liver before and after i.p. Infusion of Glucagon.
Prior to the start of glucagon infusion, three baseline Z-spectra (B1 = 0.7 µT) were acquired. To reduce the hepatic glycogen, 100 µL of glucagon (porcine glucagon, CAS no. 16941-32-5; purity, 95.13%, MedChemExpress) solution with a concentration of 1.0 mg/mL was injected intraperitoneally within 10 s. Immediately after the injection, 12 repetitive UTE-ST scans with a B1 power of 0.7 µT were performed (10 min per scan). Additional scans with B1 of 0.5 and 1.0 µT were conducted before infusion and at 120 and 130 min postinfusion. The saturation frequencies were: 200, 200, 200, 8, 7, 6, 5, 4.5, 4, 3.5, 3, 2.5, 2, 1.9, 1.8, …, 0.1, 0, 200, 200, 200, −8, −7, −6, −5, −4.5, …, −2.5, −2, −1.9, −1.8, …, and −0.1. ppm (converge sampling). Mice were sacrificed after the experiments.
Chemical Assay of Liver Glycogen Content.
After glycoNOE MRI, liver tissues were immediately isolated from the mice, “flash frozen” on dry ice, and stored at −80 °C. The protocol for extraction of liver glycogen has been fully described elsewhere (16, 38). Briefly, each 200 mg minced liver tissue was mixed with 800 µL 30% KOH solution, heated in boiling water for 30 min, and centrifuged at 2,000 g for 10 min to remove insoluble components. The supernatant was mixed with 1.5 volume of 100% ethanal to precipitate glycogen and centrifuged at 4,000 g for 20 min. The glycogen pellet was lyophilized and measured using a fluorometric Glycogen Assay Kit (Cayman Chemical #700480). As water in liver is about 71% of liver weight (39), 1 mg glycogen per g wet liver is equivalent to ∼8.4 mM glucosyl units (168 g/mol) in solution.
Data Analysis.
Static field (B0) inhomogeneities were corrected in each scan on a voxel-by-voxel basis by fitting for the drift of the direct water saturation chemical shift in each Z-spectrum (40). For each in vitro glycogen Z-spectrum, the negative half (−0.2 to −5 ppm) was assumed to consist of two resonances, one centered at 0 ppm (water peak) and another around −1.0 ppm (glycoNOE). These were fitted with Lorentzian shapes (31) on a voxel-by-voxel basis. The water line in the positive range of Z-spectrum was assumed to be the mirror of that in the negative half. The fitted glycoNOE peak height was used as the estimated glycoNOE signal intensity. For each in vivo Z-spectrum, the negative range was assumed to consist of a constant magnetization transfer contrast background plus four resonances, each centered at 0 ppm (water peak), −1.0 ppm (glycoNOE), around −3.0 ppm (broad NOE component), and −3.9 ppm. This range was fitted using a multi-Lorentzian shape analysis. The glycoNOE map was constructed based on the integral of the estimated −1 ppm glycoNOE peak (10 points from −0.6 to −1.5 ppm) for each voxel.
To calculate the glycoNOE change after glucagon injection, the scan right before the glucagon injection was used as reference. Z-spectra from each voxel were frequency corrected. To account for spectral baseline drift after the infusion of glucagon, the spectral intensities between −8 to −2 ppm and +2 to +8 ppm were used as a reference for spectral intensity alignment. Aligned Z-spectra were subtracted from the reference Z-spectra to obtain the differences spectra. The integral of the region from −0.6 to −1.5 ppm in the Z-spectral differences was used as the measure of glycoNOE changes.
Data Availability.
The MRI data and processing code used in this study are available on Open Science Framework at https://osf.io/xmbst/ (41).
Supplementary Material
Acknowledgments
This work was supported by NIH grants EB015032 (to P.C.M.v.Z) and EB025295 (to N.N.Y.).
Footnotes
Competing interest statement: The authors are filing a patent application for the proposed glycoNOE method.
This article is a PNAS Direct Submission. C.R.M. is a guest editor invited by the Editorial Board.
Data deposition: The MRI data and processing code used in this study are available on Open Science Framework at https://osf.io/xmbst/.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909921117/-/DCSupplemental.
References
- 1.van Heeswijk R. B., Morgenthaler F. D., Xin L., Gruetter R., Quantification of brain glycogen concentration and turnover through localized 13C NMR of both the C1 and C6 resonances. NMR Biomed. 23, 270–276 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Brown A. M., Ransom B. R., Astrocyte glycogen and brain energy metabolism. Glia 55, 1263–1271 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Gruetter R., et al. , Validation of 13C NMR measurements of liver glycogen in vivo. Magn. Reson. Med. 31, 583–588 (1994). [DOI] [PubMed] [Google Scholar]
- 4.König M., Bulik S., Holzhütter H.-G., Quantifying the contribution of the liver to glucose homeostasis: A detailed kinetic model of human hepatic glucose metabolism. PLOS Comput. Biol. 8, e1002577 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henning S. L., Wambolt R. B., Schönekess B. O., Lopaschuk G. D., Allard M. F., Contribution of glycogen to aerobic myocardial glucose utilization. Circulation 93, 1549–1555 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Price T. B., Rothman D. L., Avison M. J., Buonamico P., Shulman R. G., 13C-NMR measurements of muscle glycogen during low-intensity exercise. J. Appl. Physiol. 70, 1836–1844 (1991). [DOI] [PubMed] [Google Scholar]
- 7.Ørtenblad N., Westerblad H., Nielsen J., Muscle glycogen stores and fatigue. J. Physiol. 591, 4405–4413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rousset M., Zweibaum A., Fogh J., Presence of glycogen and growth-related variations in 58 cultured human tumor cell lines of various tissue origins. Cancer Res. 41, 1165–1170 (1981). [PubMed] [Google Scholar]
- 9.Favaro E., et al. , Glucose utilization via glycogen phosphorylase sustains proliferation and prevents premature senescence in cancer cells. Cell Metab. 16, 751–764 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Magnusson I., Rothman D. L., Katz L. D., Shulman R. G., Shulman G. I., Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. J. Clin. Invest. 90, 1323–1327 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krssak M., et al. , Alterations in postprandial hepatic glycogen metabolism in type 2 diabetes. Diabetes 53, 3048–3056 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Adeva-Andany M. M., González-Lucán M., Donapetry-García C., Fernández-Fernández C., Ameneiros-Rodríguez E., Glycogen metabolism in humans. BBA Clin. 5, 85–100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krähenbühl L., et al. , Reduced hepatic glycogen stores in patients with liver cirrhosis. Liver Int. 23, 101–109 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Nieman D. C., Shanely R. A., Zwetsloot K. A., Meaney M. P., Farris G. E., Ultrasonic assessment of exercise-induced change in skeletal muscle glycogen content. BMC Sports Sci. Med. Rehabil. 7, 9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witney T. H., et al. , A novel radiotracer to image glycogen metabolism in tumors by positron emission tomography. Cancer Res. 74, 1319–1328 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sillerud L. O., Shulman R. G., Structure and metabolism of mammalian liver glycogen monitored by carbon-13 nuclear magnetic resonance. Biochemistry 22, 1087–1094 (1983). [DOI] [PubMed] [Google Scholar]
- 17.Zang L.-H., Rothman D. L., Shulman R. G., 1H NMR visibility of mammalian glycogen in solution. Proc. Natl. Acad. Sci. U.S.A. 87, 1678–1680 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinicke K., et al. , Reproducibility and absolute quantification of muscle glycogen in patients with glycogen storage disease by 13C NMR spectroscopy at 7 Tesla. PLoS One 9, e108706 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roser W., Beckmann N., Wiesmann U., Seelig J., Absolute quantification of the hepatic glycogen content in a patient with glycogen storage disease by 13C magnetic resonance spectroscopy. Magn. Reson. Imaging 14, 1217–1220 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Ouwerkerk R., Pettigrew R. I., Gharib A. M., Liver metabolite concentrations measured with 1H MR spectroscopy. Radiology 265, 565–575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Zijl P. C., Jones C. K., Ren J., Malloy C. R., Sherry A. D., MRI detection of glycogen in vivo by using chemical exchange saturation transfer imaging (glycoCEST). Proc. Natl. Acad. Sci. U.S.A. 104, 4359–4364 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller C. O., et al. , Noninvasive measurements of glycogen in perfused mouse livers using chemical exchange saturation transfer NMR and comparison to 13C NMR spectroscopy. Anal. Chem. 87, 5824–5830 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simegn G. L., Van der Kouwe A. J. W., Robertson F. C., Meintjes E. M., Alhamud A., Real-time simultaneous shim and motion measurement and correction in glycoCEST MRI using double volumetric navigators (DvNavs). Magn. Reson. Med. 81, 2600–2613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Zijl P. C., Yadav N. N., Chemical exchange saturation transfer (CEST): What is in a name and what isn’t? Magn. Reson. Med. 65, 927–948 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagiyama K., Zhang S., Dimitrov I., Sherry A., Takahashi M., “In vivo monitoring of liver glycogen by chemical exchange saturation transfer imaging (GlycoCEST) in live mice” in Proceedings of the International Society for Magnetic Resonance in Medicine (International Society for Magnetic Resonance in Medicine, 2014), (abstract) p. 0762.
- 26.Deng M., et al. , Chemical exchange saturation transfer (CEST) MR technique for liver imaging at 3.0 Tesla: An evaluation of different offset number and an after-meal and over-night-fast comparison. Mol. Imaging Biol. 18, 274–282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen W., Avison M. J., Zhu X. H., Shulman R. G., NMR studies of 1H NOEs in glycogen. Biochemistry 32, 11483–11487 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Ling W., Regatte R. R., Navon G., Jerschow A., Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST). Proc. Natl. Acad. Sci. U.S.A. 105, 2266–2270 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yadav N. N., et al. , Detection of dynamic substrate binding using MRI. Sci. Rep. 7, 10138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zang L.-H., Howseman A. M., Shulman R. G., Assignment of the 1H chemical shifts of glycogen. Carbohydr. Res. 220, 1–9 (1991). [DOI] [PubMed] [Google Scholar]
- 31.Desmond K. L., Moosvi F., Stanisz G. J., Mapping of amide, amine, and aliphatic peaks in the CEST spectra of murine xenografts at 7 T. Magn. Reson. Med. 71, 1841–1853 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Giffin B. F., Drake R. L., Morris R. E., Cardell R. R., Hepatic lobular patterns of phosphoenolpyruvate carboxykinase, glycogen synthase, and glycogen phosphorylase in fasted and fed rats. J. Histochem. Cytochem. 41, 1849–1862 (1993). [DOI] [PubMed] [Google Scholar]
- 33.Jensen T. L., Kiersgaard M. K., Sørensen D. B., Mikkelsen L. F., Fasting of mice: A review. Lab. Anim. 47, 225–240 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Sullivan M. A., et al. , Changes in glycogen structure over feeding cycle sheds new light on blood-glucose control. Biomacromolecules 15, 660–665 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Chen L., et al. , Protein aggregation linked to Alzheimer’s disease revealed by saturation transfer MRI. Neuroimage 188, 380–390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L., Barker P. B., Weiss R. G., van Zijl P. C. M., Xu J., Creatine and phosphocreatine mapping of mouse skeletal muscle by a polynomial and Lorentzian line-shape fitting CEST method. Magn. Reson. Med. 81, 69–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zang L. H., Laughlin M. R., Rothman D. L., Shulman R. G., 13C NMR relaxation times of hepatic glycogen in vitro and in vivo. Biochemistry 29, 6815–6820 (1990). [DOI] [PubMed] [Google Scholar]
- 38.Shokri-Afra H., Ostovar-Ravari A., Rasouli M., Improvement of the classical assay method for liver glycogen fractions: ASG is the main and metabolic active fraction. Eur. Rev. Med. Pharmacol. Sci. 20, 4328–4336 (2016). [PubMed] [Google Scholar]
- 39.Strubelt O., et al. , The influence of fasting on the susceptibility of mice to hepatotoxic injury. Toxicol. Appl. Pharmacol. 60, 66–77 (1981). [DOI] [PubMed] [Google Scholar]
- 40.Kim M., Gillen J., Landman B. A., Zhou J., van Zijl P. C., Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn. Reson. Med. 61, 1441–1450 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y., et al. , Data from “Magnetic resonance imaging of glycogen using its magnetic coupling with water.” Open Science Framework. https://osf.io/xmbst/. Deposited 13 October 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MRI data and processing code used in this study are available on Open Science Framework at https://osf.io/xmbst/ (41).