Overall, our studies demonstrate that proteolytic cleavage is the primary barrier to infection for a subset of zoonotic coronaviruses. Moving forward, the results argue that both receptor binding and proteolytic cleavage of the spike are critical factors that must be considered for evaluating the emergence potential and risk posed by zoonotic coronaviruses. In addition, the findings also offer a novel means to recover previously uncultivable zoonotic coronavirus strains and argue that other tissues, including the digestive tract, could be a site for future coronavirus emergence events in humans.
KEYWORDS: MERS-CoV, PDF2180, coronavirus, emergence, spike, zoonotic
ABSTRACT
Traditionally, the emergence of coronaviruses (CoVs) has been attributed to a gain in receptor binding in a new host. Our previous work with severe acute respiratory syndrome (SARS)-like viruses argued that bats already harbor CoVs with the ability to infect humans without adaptation. These results suggested that additional barriers limit the emergence of zoonotic CoV. In this work, we describe overcoming host restriction of two Middle East respiratory syndrome (MERS)-like bat CoVs using exogenous protease treatment. We found that the spike protein of PDF2180-CoV, a MERS-like virus found in a Ugandan bat, could mediate infection of Vero and human cells in the presence of exogenous trypsin. We subsequently show that the bat virus spike can mediate the infection of human gut cells but is unable to infect human lung cells. Using receptor-blocking antibodies, we show that infection with the PDF2180 spike does not require MERS-CoV receptor DPP4 and antibodies developed against the MERS spike receptor-binding domain and S2 portion are ineffective in neutralizing the PDF2180 chimera. Finally, we found that the addition of exogenous trypsin also rescues HKU5-CoV, a second bat group 2c CoV. Together, these results indicate that proteolytic cleavage of the spike, not receptor binding, is the primary infection barrier for these two group 2c CoVs. Coupled with receptor binding, proteolytic activation offers a new parameter to evaluate the emergence potential of bat CoVs and offers a means to recover previously unrecoverable zoonotic CoV strains.
IMPORTANCE Overall, our studies demonstrate that proteolytic cleavage is the primary barrier to infection for a subset of zoonotic coronaviruses. Moving forward, the results argue that both receptor binding and proteolytic cleavage of the spike are critical factors that must be considered for evaluating the emergence potential and risk posed by zoonotic coronaviruses. In addition, the findings also offer a novel means to recover previously uncultivable zoonotic coronavirus strains and argue that other tissues, including the digestive tract, could be a site for future coronavirus emergence events in humans.
INTRODUCTION
Since the beginning of the 21st century, public health infrastructures have been required to periodically respond to new and reemerging zoonotic viral diseases, including influenza, Ebola, and Zika virus outbreaks (1). Severe acute respiratory syndrome coronavirus (SARS-CoV), the first major outbreak of the century, highlighted the global impact of a newly emerging virus in the context of expanding development, increased globalization, and poor public health infrastructures (2–4). A decade later, the emergence and continued outbreaks of the Middle East respiratory syndrome coronavirus (MERS-CoV) further illustrate the ongoing threat posed by circulating zoonotic viruses (5). Together, the outbreaks of the early part of this century argue that continued preparations and vigilance are needed to maintain global public health.
Despite their spontaneous emergence, several research approaches to rapidly respond and even predict outbreak strains already exist. During the MERS-CoV outbreak, our group and others were able to leverage reagents generated against related group 2C coronaviruses, namely, HKU4- and HKU5-CoV (6, 7). These reagents, created independent of viable virus replication, provided valuable insights and models for testing serologic responses during the early stages of the MERS-CoV outbreak. Similarly, reverse genetics systems permitted the exploration of zoonotic coronaviruses (8); using the known SARS spike/ACE2 receptor interaction, chimeric viruses containing the backbones of bat CoVs were generated to evaluate the efficacy of both vaccines and therapeutics (9–12). The inverse approach placed the zoonotic spike proteins in the context of the epidemic SARS-CoV backbone (13, 14). These studies provided insight into potential threats circulating in bats as well as the efficacy of current therapeutic treatments (15). While far from comprehensive, the results indicated that these approaches, reagents, and predictions may prove useful in preparations for future CoV outbreaks.
In this study, we extend the examination of zoonotic viruses to a novel MERS-like CoV strain isolated from a Ugandan bat, namely, PDF-2180 CoV (MERS-Uganda). Our initial attempt to cultivate a chimeric MERS-CoV containing the Ugandan MERS-like spike produced viral subgenomic transcripts but failed to result in infectious virus after electroporation (16). However, in the current study, we demonstrate that exogenous trypsin treatment produced high-titer virus capable of plaque formation and continued replication. These results are consistent with the recovery of enteric CoVs like porcine epidemic diarrhea virus (17) but have not previously been described as a major barrier for bat-derived CoVs. The chimeric Ugandan MERS-like spike virus could replicate efficiently in both Vero and Huh7 cells in the context of trypsin-containing media but failed to produce infection of either continuous or primary human respiratory cell cultures. Importantly, the MERS-Uganda chimeric virus successfully infected cells of the human digestive tract, potentially identifying another route for cross-species transmission and emergence. Notably, blockade of human DPP4, the receptor for MERS-CoV, had no significant impact on replication of the chimeric MERS-Uganda virus, suggesting the use of an alternative receptor. Similarly, the addition of trypsin also rescued replication of full-length HKU5-CoV, a related group 2C bat CoV, and showed no replication defect during DPP4 blockade. Together, the results indicate that proteolytic activation of the spike protein is a potent constraint to infection for zoonotic CoVs and expand the correlates for CoV emergence beyond receptor binding alone.
(This article was submitted to an online preprint archive [18].)
RESULTS
Utilizing the MERS-CoV infectious clone (19), we previously attempted to evaluate the potential of PDF-2180 CoV to emerge from zoonotic populations. Replacing the wild-type MERS-CoV spike with the PDF-2180 spike produced a virus capable of generating viral transcripts in vitro but not sustained replication (16). These results suggested that the significant amino acid differences observed within the receptor-binding domain precluded infection of Vero cells. However, amino acid changes were not confined only to the receptor-binding domain (RBD); highlighting changes between the Uganda spike on the MERS-CoV trimer revealed significant differences throughout the S1 region of spike (Fig. 1A and B). While the S2 remained highly conserved (Fig. 1C), changes in the C- and N-terminal domains of S1, in addition to the RBD, may also influence entry and infection compatibility. Notably, recent reports had also indicated differential protease cleavage of wild-type MERS-CoV based on cell types, suggesting that spike processing influences docking and entry of pseudotyped virus (20). To explore if spike cleavage impaired infectivity, we evaluated MERS-Uganda virus replication in the presence of trypsin-containing media. The addition of trypsin to the chimeric virus resulted in cytopathic effect, fusion of the Vero monolayer, formation of plaques under a trypsin-containing overlay, and collection of high-titer infectious virus stock (Fig. 1D). Utilizing the trypsin-treated stock, we subsequently examined the MERS-Uganda spike by Western blotting, finding that increasing amounts of trypsin produced more robust spike expression and cleavage (Fig. 1E). Together, these data indicate that the PDF-2180 spike can mediate infection of Vero cells in a trypsin-dependent manner.
FIG 1.
Exogenous trypsin rescues MERS-Uganda spike replication. (A and B) Structure of the MERS-CoV spike trimer in complex with the receptor human DPP4 (red) from the side (A) and top (B). Consensus amino acids are outlined for the S1 (gray) and S2 (black) domains, with PDF-2180 differences noted in magenta. (C) Spike protein sequences of the indicated viruses were aligned according to the bounds of total spike, S1, S2, and receptor-binding domain (RBD). Sequence identities were extracted from the alignments, and a heatmap of sequence identity was constructed using EvolView (www.evolgenius.info/evolview) with MERS-CoV as the reference sequence. (D) MERS-Uganda chimera stocks were grown in the presence or absence of trypsin and were quantitated by plaque assay with a trypsin-containing overlay (n = 2). (E) Protein expression of MERS-Uganda spike (S) and actin 24 and 48 hours postinfection of Vero cells in the presence of increasing amounts of trypsin (none, 0.25 μg/ml, and 0.5 μg/ml) in the media.
The requirement for trypsin complicated our studies due to cell toxicity; to overcome this issue, we utilized both trypsin-adapted Vero cells and a MERS-Uganda chimera encoding RFP in place of open reading frame 5 (ORF5), similar to a previously generated MERS-CoV reporter virus (19). Following MERS-Uganda infection, cultures with trypsin-containing medium showed evidence for replication of viral genomic RNA (Fig. 2A). Similarly, the nucleocapsid protein was observed only in the presence of exogenous trypsin following infection with the MERS-Uganda chimera (Fig. 2B). Notably, wild-type MERS-CoV expressing RFP was also augmented in the presence of trypsin with increased genomic RNA and nucleocapsid protein relative to no trypsin control (Fig. 2A and B). Examination of RFP signal confirmed these RNA and protein results (Fig. 2C), as RFP was only observed in MERS-Uganda chimeric infection in the presence of trypsin. Similarly, viral RNA, RFP expression, and N protein expression were more robust in trypsin-treated cells following MERS-CoV infection. Together, these data indicate that both the PDF-2180 and MERS-CoV spike have augmented infection of Vero cells in the presence of trypsin.
FIG 2.
Trypsin treatment augments MERS and Uganda spike-mediated infection. (A) Trypsin-resistant Vero cells were infected with MERS-CoV (black) or MERS-Uganda chimera (magenta) and were monitored for expression of genomic RNA in the presence or absence of trypsin (n = 3 for each time point). (B) Protein expression of MERS-CoV nucleocapsid (N) and actin 18 hours postinfection of Vero cells in the presence or absence of trypsin in the media. (C) RFP expression microscopy in Vero cells infected with MERS-CoV, MERS-Uganda spike chimera, or mock in the presence or absence of trypsin.
MERS-Uganda spike replicates in human cells.
Having demonstrated infection and replication, we next sought to determine the capacity of MERS-Uganda chimeric virus to grow in human cells. Previously, MERS-CoV had been shown to replicate efficiently in Huh7 cells (21). In the Huh7 liver cell line, infection with MERS-Uganda RFP chimeric virus resulted in RFP-positive cells and cell fusion (Fig. 3A). In contrast, while a few RFP-positive cells were observed in the non-trypsin-treated group, neither expanding RFP expression nor cytopathic effect were seen in the absence of trypsin. Our observation may have been the result of residual trypsin activity from the undiluted virus stock, resulting in low-level infection. Exploring further, N protein analysis by Western blotting indicated that the PDF-2180 spike chimera could produce significant viral proteins in the presence of trypsin (Fig. 3B); only low levels of protein were observed in the control-treated infection. While replication of the MERS-Uganda chimera was not equivalent to that of wild-type MERS-CoV, the results clearly demonstrate the capacity of the PDF-2180 spike to mediate infection of human cells in the presence of trypsin.
FIG 3.
MERS-Uganda spike chimera replicates in human cells. (A and B) Huh7 cells were infected with MERS-CoV or MERS-Uganda chimeric viruses, showing microscopy images of cell monolayer and RFP expression with and without trypsin treatment (A) and N protein expression following infection of Huh7 cells in the presence or absence of trypsin (B). (C and D) Primary HAE cultures were infected with MERS-CoV or MERS-Uganda chimera, showing RFP expression (C) and genomic viral RNA following infection (D) (n = 3 for 8 and 24 hours postinfection [hpi]). (E and F) Caco-2 cells were infected with MERS-CoV or MERS-Uganda chimeric viruses expressing RFP, showing microscopy images of cell monolayer and RFP expression with and without trypsin treatment (E) and N protein expression following infection of Caco-2 cells in the presence or absence of trypsin (F).
We next examined the capacity of the MERS-Uganda spike to infect human respiratory cells, the primary targets of SARS-CoV, MERS-CoV, and other common cold-causing human CoVs. Using Calu3 cells, a human lung epithelial cell line, we observed robust replication of wild-type MERS-CoV based on RFP expression, consistent with previous studies (19). However, no evidence of infection was noted in MERS-Uganda-infected Calu3 cells in the presence or absence of trypsin. We subsequently explored infection of primary human airway epithelial (HAE) cultures. Grown on an air-liquid interface, HAE cultures have a propensity to facilitate improved infections of several human CoVs and may be more permissive for infection with the PDF-2180 spike chimera (22). To infect, PDF-2180 chimeric virus grown in the presence of trypsin was inoculated onto the apical surface of the HAE culture; cultures were subsequently washed with phosphate-buffered saline (PBS) containing 5 μg/ml trypsin at 0, 18, 24, and 48 hours postinfection for 10 minutes and then removed. Following infection, no evidence of RFP expression was observed even after trypsin washes of the apical surface (Fig. 3C). Similarly, RNA expression analysis found no evidence for accumulation of viral genomic RNA, indicating no evidence for replication in HAE cultures (Fig. 3D). In contrast, wild-type MERS-CoV efficiently infects these HAE cultures, as demonstrated by both RFP expression and viral genomic RNA accumulation. Together, the Calu3 and HAE results suggest that the PDF-2180 spike is unable to infect respiratory cells in humans, even in the presence of exogenous trypsin.
We next evaluated the capacity of the PDF-2180 chimera to infect cells of the digestive tract. While uncommon in humans, several animal CoVs have been shown to cause severe disease via the enteric pathway (23, 24). In addition, most bat CoV sequences, including PDF-2180-CoV, were isolated from bat guano samples, suggesting an enteric etiology. Importantly, the presence of trypsin and other soluble host proteases in the digestive tract may facilitate infection with the PDF-2180 spike in humans. To test this question, we infected Caco-2 cells, a human epithelial colorectal adenocarcinoma cell line, with wild-type MERS-CoV and MERS-Uganda spike chimera in the presence or absence of trypsin (Fig. 3E). For MERS-CoV, infection of Caco-2 cells resulted in robust infection and spread with or without trypsin in the media. For the MERS-Uganda chimera, the addition of trypsin facilitated infection with many RFP-positive Caco-2 cells; however, infection was not as robust as in the wild-type MERS-CoV infection. Examination of N protein by Western blotting indicated that the MERS-Uganda spike could produce infection in Caco-2 cells but confirmed replication at levels lower than that with wild-type MERS-CoV (Fig. 3F). Together, the results indicate that human cells, including gut cells, can support infection with the MERS-Uganda chimera in the presence of trypsin.
MERS-Uganda spike does not use DPP4 for entry.
The absence of infection of human respiratory cells coupled with significant changes in the RBD suggested that MERS-Uganda does not utilize the MERS-CoV receptor human DPP4 for entry (16). To explore this question, we utilized antibodies to block DPP4 in Vero cells to determine the effect on MERS-Uganda chimeric virus replication. As expected, the anti-DPP4 antibody successfully ablated replication of wild-type MERS-CoV in both the presence and the absence of trypsin treatment, as measured by both RFP and N protein expression (Fig. 4A and B). In contrast, the human DPP4-blocking antibody had no impact on infection with the MERS-Uganda chimera virus in the presence of trypsin, confirming that the MERS-CoV receptor is not required to mediate infection with the PDF-2180 spike. Together, these results indicate that while the MERS-Uganda spike infects human cells, it does not require human DPP4 to mediate infection.
FIG 4.
MERS-Uganda spike does not utilize DPP4 for infection. (A and B) Vero cells were infected with MERS-CoV or MERS-Uganda chimeric virus in the presence or absence of trypsin and a blocking antibody against human DPP4. (A) Fluorescent microscopy showing RFP expression 24 hours postinfection for each treatment group. (B) Western blot of N protein and actin 24 hours postinfection.
MERS-CoV therapeutics are ineffective against MERS-Uganda spike.
Having established replication capacity in human cells, we next sought to determine if therapeutics developed against the MERS-CoV spike could disrupt infection with the MERS-Uganda spike chimera. Several monoclonal antibodies have been identified as possible therapeutic options for the treatment of MERS-CoV, including LCA60 and G4. We first evaluated LCA60, a potent antibody that binds adjacent to the spike RBD of MERS-CoV (25). However, the major changes in the RBD region of the MERS-Uganda spike predicted a lack of efficacy (Fig. 5A). LCA60 potently neutralized wild-type MERS-CoV grown in both the presence and the absence of trypsin (Fig. 5B). However, consistent with expectations, the LCA60 antibody had no impact on infection with the MERS-Uganda chimera, failing to neutralize the bat spike-expressing virus (Fig. 5B). We subsequently examined a second monoclonal antibody, G4, which had previously mapped to a conserved portion of the S2 region of the MERS-spike (Fig. 5A) (26). With the epitope relatively conserved in the MERS-Uganda spike, we tested the efficacy against the zoonotic spike chimera. However, the results demonstrate no neutralization of MERS-Uganda spike virus by the S2-targeted antibody (Fig. 5C). Notably, G4 also failed to neutralize wild-type MERS-CoV grown in the presence of exogenous trypsin (Fig. 5C). Together, the results indicate that both group 2C CoV spikes could escape neutralization by the S2-targeted antibody in the presence of exogenous trypsin. Overall, these experiments suggest that antibodies targeted against MERS-CoV, even to regions in the highly conserved S2 domain, may not have utility against viruses expressing the PDF-2180 spike.
FIG 5.
Antibodies against MERS-CoV fail to neutralize MERS-Uganda chimera. (A) Structure of the MERS-CoV spike trimer with therapeutic antibody LCA60 bound adjacent to the receptor-binding domain and the antibody G4 bound to the S2 portion. Consensus amino acids are outlined for the S1 (gray) and S2 (black) domains, with PDF-2180 differences noted in magenta. (B and C) Plaque neutralization curves for LCA60 (B) and G4 (C) with (solid) and without (dotted) trypsin treatment for MERS-CoV (black) and MERS-Uganda chimera (magenta) (n = 3 per concentration).
Trypsin treatment rescues the replication of zoonotic HKU5-CoV.
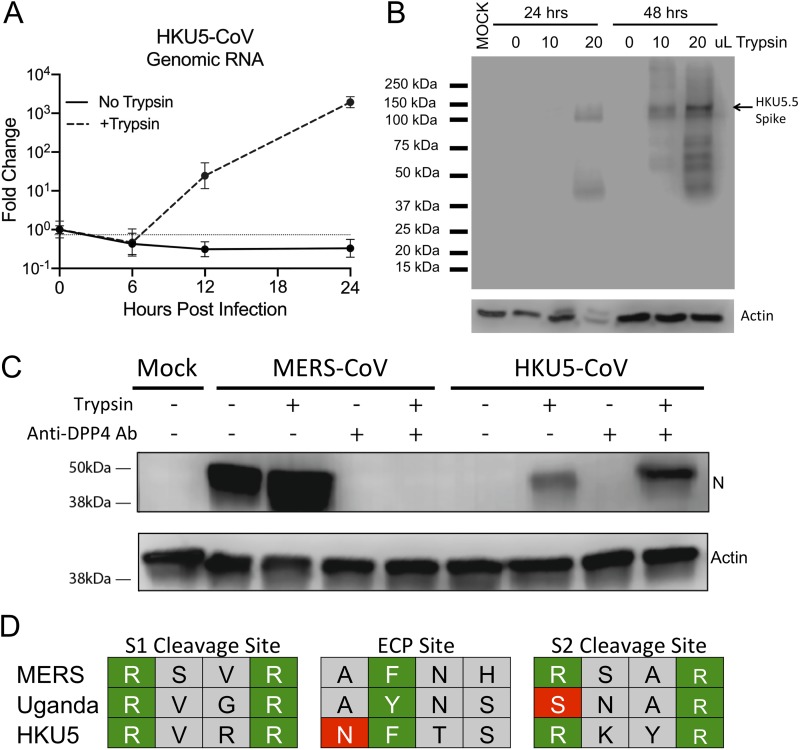
Based on the MERS-Uganda chimera virus, we wondered if a similar barrier prevented replication of other zoonotic CoVs. Previously, our group had generated a full-length infectious clone for HKU5-CoV, another group 2C coronavirus sequence isolated from bats. Similar to the MERS-Uganda chimera, the infectious clone of HKU5-CoV produced subgenomic transcripts but failed to achieve productive infection (6). Revisiting the full-length recombinant virus, we sought to determine if trypsin treatment could also rescue HKU5-CoV. Following HKU5-CoV infection, the addition of trypsin to the media resulted in cytopathic effect and cell fusion. In contrast, cultures lacking trypsin showed no signs of viral infection. Exploring viral genomic RNA, trypsin in the culture media permitted robust infection with HKU5-CoV that increased over time and was absent in cells not treated with trypsin (Fig. 6A). Similarly, trypsin in the media also permitted the accumulation and proteolytic cleavage of the HKU5 spike protein in a dose- and time-dependent manner (Fig. 6B). Importantly, the addition of the anti-DPP4 antibody had no impact on HKU5-CoV infection, suggesting the use of a different receptor than that used by wild-type MERS-CoV, similar to the findings with the MERS-Uganda spike (Fig. 6C). Together, these results demonstrate that protease cleavage is also the primary barrier to infection of Vero cells with HKU5-CoV. Examining further, we compared the predicted cleavage at S1/S2 border, S2’, and the endosomal cysteine protease site across MERS, PDF2180, and HKU5 spikes (Fig. 6D) (26). For the S1/S2 site, MERS, Uganda, and HKU5 maintain the RXXR cleavage motif, although the different interior amino acids may alter efficiency. For the S2’ sequence, MERS and HKU5 also retain the RXXR motif; however, the Uganda spike lacks the first arginine (SNAR), potentially impacting cleavage. Finally, all three spikes maintain an aromatic residue at position two in the endosomal cysteine protease (ECP) site (27). However, the HKU5 spike maintains N at position 1 which is similar to a MERS mutant previously shown to inactivate cathepsin L activation (28). Together, the results suggest that potential sequence changes in the protease cleavage sites may contribute to the trypsin dependency of MERS-Uganda and HKU5 spike-mediated infections.
FIG 6.
Exogenous trypsin rescues replication of HKU5-CoV. Vero cells were infected with full-length HKU5-CoV in the presence or absence of trypsin. (A) Expression (reverse transcription-quantitative PCR [qRT-PCR]) of HKU5-CoV viral genome in the presence or absence of trypsin (n = 3). (B) Immunoblotting of HKU5 spike protein and cellular actin 24 and 48 hours postinfection with various concentrations of trypsin in the media. (C) Immunoblotting for MERS N protein and cellular actin following infection in the presence or absence of trypsin and human DPP4 antibody. (D) Alignment of amino acid sequence from the S1/S2, endosomal cysteine protease (ECP), and the S2 cleavage sites. Green boxes represent key residues conserved, and red boxes outline amino acid changes that potentially impact cleavage.
DISCUSSION
In the manuscript, we expanded our examination of circulating zoonotic viruses and identified protease cleavage as an important barrier to emergence of some group 2C zoonotic CoVs. The chimeric virus containing the spike protein from PDF-2180 was capable of replication in Vero cells and human cells (Huh7 and Caco-2) if treated with exogenous trypsin. However, neither continuous nor primary human airway cultures were susceptible to infection, in contrast to wild-type MERS-CoV. The MERS-Uganda chimera also maintained replication despite treatment with antibodies blocking human DPP4, suggesting use of either an alternative receptor or a different entry mechanism for infection. Importantly, current therapeutics targeting the MERS spike protein showed no efficacy against the MERS-Uganda chimera, highlighting a potential public health vulnerability to this and related group 2C CoVs. Finally, the trypsin-mediated rescue of a second zoonotic group 2C CoV, HKU5-CoV, validates findings that suggested that protease cleavage may represent a critical barrier to zoonotic CoV infection in new hosts (29, 30). Together, the results highlight the importance of spike processing in CoV infection, expand the correlates associated with emergence beyond receptor binding alone, and provide a platform strategy to recover previously noncultivatable zoonotic CoVs.
With the ongoing threat posed by circulating zoonotic viruses, understanding the barriers for viral emergence represents a critical area of research. For CoVs, receptor binding has been believed to be the primary constraint to infection in new host populations. Following the SARS-CoV outbreak, emergence in humans was attributed to mutations within the receptor-binding domain that distinguished the epidemic strain from progenitor viruses harbored in bats and civets (31). Yet, work by our group and others has indicated that zoonotic SARS-like viruses circulating in Southeast Asian bats are capable of infecting human cells by binding to the known human ACE2 receptor without adaptation (13, 14, 32). Similarly, pseudotyped virus studies have identified zoonotic strains HKU4-CoV and NL140422-CoV as capable of binding to human DPP4 without mutations to the spike (30, 33). In this study, we demonstrate that both PDF-2180 and HKU5-CoV spikes are capable of binding to and infecting human cells if primed by trypsin cleavage. Together, the results argue that several circulating zoonotic CoV strains have the capacity to bind to human cells without adaption and that receptor binding may not be the only barrier to CoV emergence.
Data from this study implicate the processing of the spike protein as a critical factor for CoV infection. In the absence of trypsin, the MERS-Uganda and HKU5-CoV spikes were unable to mediate infection and initially suggested a lack of receptor compatibility (6, 16). However, exogenous trypsin treatment produced robust infection, indicating that despite binding to human cells, CoVs cannot overcome incomplete spike processing. As such, evaluating zoonotic virus populations for emergence threats must also consider the capacity for CoV spike activation in addition to receptor binding. While exogenous processing of the spike has been well described as necessary for enteric CoVs (17), spike processing has not been considered a primary barrier for bat CoVs despite their enteric origins. In this new paradigm for bat CoVs, the combination of receptor binding and proteolytic activation by endogenous proteases permits zoonotic CoV infection, as with MERS-CoV and SARS-CoV (Fig. 7). The absence of receptor binding (Fig. 7A) or compatible host protease activity (Fig. 7B) restricts infection with certain zoonotic strains like PDF-2180 or HKU5-CoV. These barriers can be overcome with the addition of exogenous proteases, disrupting the need for host proteases and permitting receptor-dependent or receptor-independent entry (Fig. 7C). Overall, the new paradigm argues that both receptor binding and protease activation barriers must be overcome for successful zoonotic CoV infection of a new host.
FIG 7.
Barriers to zoonotic coronavirus emergence. Both receptor binding and protease activitation are key correlates that govern zoonotic coronavirus emergence. (A) A lack of receptor binding with zoonotic CoVs precludes the infection of new host cells. (B) Despite receptor binding, the absence of compatible host proteases for spike cleavage restricts infection in new hosts. (C) The addition of exogenous protease overcomes the host protease barriers and may or may not require receptor binding.
The requirement for exogenous trypsin treatment is not unique to MERS-Uganda or HKU5-CoV. Influenza strains are well known to require trypsin treatment to facilitate their release in cell culture (34). In addition, highly pathogenic avian influenza strains have been linked to mutations that improve cleavage by ubiquitous host protease, augmenting their tissue tropism and virulence (35). Similarly, a wealth of enteric viruses, including polio, cowpox, and rotaviruses, depend on trypsin to prime, modulate, and/or expand infection (36, 37). Even within the CoV family, enteric viruses, including porcine epidemic diarrhea virus (PEDV), porcine delta CoV, and swine acute diarrhea syndrome (SADS) CoV require trypsin for replication in vitro (38–40). Together, these prior studies illustrate the importance of protease activation in virus infections. However, the protease barrier to PDF-2180 and HKU5-CoV spike-mediated infection may also reflect on the emergence of SARS-CoV and MERS-CoV. While initial studies argued that receptor binding was the primary barrier, the existence of zoonotic strains capable of efficiently using the same human entry receptors contradicts that suggestion (13, 14). It is possible that emergence of epidemic CoV strains also requires modifying protease cleavage in either humans or an intermediate host, such as camels or civets, in addition to increased receptor-binding affinity. Consistent with this idea, reports have detailed differential infection with MERS-CoV based on host protease expression (20). Similarly, mouse adaptation of MERS-CoV resulted in spike modifications that alter protease activation and entry in vivo (41). Coupled with the augmented replication and protein production of wild-type MERS-CoV in the presence of trypsin, the results suggest that the proteolytic cleavage epidemic MER-CoV could still be enhanced, potentially by augmenting the cell surface entry mechanism, as previously described for SARS-CoV (42). While group 2B bat CoV strains (WIV1-CoV, WIV16-CoV, and SHC014-CoV) do not require trypsin for infection (9, 13, 14, 43), differences in protease activation may contribute to infection changes relative to the epidemic SARS-CoV. In this context, our findings expand the importance of protease cleavage as a criterion to consider for zoonotic virus emergence in a new host population.
In evaluating the threat to humans posed by PDF-2180 and HKU5-CoV, the results demonstrate a pathway to emergence. Neither CoV spike uses human DPP4 for entry, and the PDF-2180 chimera failed to replicate in human respiratory models, even in the presence of trypsin. As many different proteases can promote CoV entry, future studies must determine if other protease treatments or other components can promote virus replication in HAE cultures (29, 44, 45). However, replication in Huh7 and Caco-2 cells indicates human infection compatibility and may portend differential tropism, possibly in the alimentary or biliary tracts, as has been described for several mammalian CoVs (38–40). MERS-Uganda or HKU5-CoV could utilize this same trypsin-rich environment in the gut to emerge as an enteric pathogen in humans, although its pathology and virulence would be hard to predict. Evidence from both SARS-CoV and MERS-CoV outbreaks suggests the involvement of enteric pathways during infection (46, 47). Replication in the gut might select for mutations that expand spike processing/tropism and allow replication in other tissues, including the lung, and lead to virulent disease in the new host population, as seen with porcine respiratory coronavirus (48). In examining the threat posed by PDF-2180 and HKU5-CoV, we must consider the emergence of these CoVs in tissues other than the lung and that they harbor distinct pathologies compared with epidemic SARS and MERS-CoV.
The receptor dynamics of MERS-Uganda and HKU5-CoV also remain unclear in the context of this study. In the presence of trypsin, neither spike protein requires the MERS-CoV receptor DPP4 for entry, which is consistent with the differences between the receptor-binding domains of the bat and epidemic strains. Therefore, it was not surprising that antibodies that target the RBD of the MERS-CoV spike were ineffective in blocking infection of the PDF-2180 chimera. However, the S2-targeted antibody G4 also had no efficacy against MERS-Uganda, despite a relatively conserved binding epitope. This result is possibly explained by differing amino acid sequences between MERS-CoV and PDF-2180 at the G4 epitope, specifically residue 1175, which is associated with G4 escape mutants in MERS-CoV (49). Alternatively, the G4 antibody also failed to neutralize wild-type MERS-CoV grown in the presence of trypsin, indicating that entry is still possible, despite treatment with antibody binding the S2 domain. These results suggest that trypsin treatment may permit a conformational change either masking the G4 epitope or facilitating fusion/entry prior to antibody binding. Conversely, the presence of trypsin may prime a receptor-independent entry for the MERS-Uganda chimera, similar to the JHVM strain of MHV (50). Yet, this result would contrast with that of PEDV, which requires receptor binding prior to trypsin activation to facilitate infection (39). Importantly, the lack of infection in respiratory cells suggests that some receptor or attachment factor is necessary to mediate entry with the PDF-2180 spike. Recent work with MERS-CoV binding sialic acid supports this idea (51) and indicates that the PDF-2180 spike may not have a similar binding motif. Overall, further experimental studies are required to fully understand the receptor dynamics of the PDF-2180 spike.
While providing a new strategy to recover zoonotic CoVs, the manuscript highlights proteolytic cleavage of the spike as a major barrier to group 2C zoonotic CoV infection. For both MERS-Uganda and HKU5-CoV, the addition of exogenous trypsin rescues infection, indicating that spike cleavage, not receptor binding, limits these strains in new hosts and tissues. The adaptation of the protease cleavage sites or infection of tissues with robust host protease expression could permit these two zoonotic CoV strains to emerge and may pose a threat to public health due to the absence of effective spike-based therapeutics. In considering cross-species transmission, our results using reconstructed bat group 2C CoVs confirm spike processing as a correlate associated with emergence. Adding spike processing to receptor binding as primary barriers offers a new framework to evaluate the threat of emergence for zoonotic CoV strains.
MATERIALS AND METHODS
Cells, viruses, in vitro infection, and plaque assays.
Vero cells were grown in Dulbecco’s modified Eagle medium (DMEM; Gibco, CA) supplemented with 5% FetalClone II (HyClone, UT) and antibiotic/antimycotic (anti/anti) (Gibco). Huh7 cells were grown in DMEM supplemented with 10% FetalClone II and anti/anti. Caco-2 cells were grown in MEM (Gibco) supplemented with 20% fetal bovine serum (HyClone) and anti/anti. Human airway epithelial cell (HAE) cultures were obtained from the University of North Carolina (UNC) Cystic Fibrosis (CF) Center Tissue Procurement and Cell Culture Core from human lungs procured under University of North Carolina at Chapel Hill Institutional Review Board-approved protocols. Wild-type MERS-CoV, chimeric MERS-Uganda, and HKU5-CoV were cultured on Vero cells in Opti-MEM (Gibco) supplemented with anti/anti. For indicated experiments, trypsin (Gibco) was added at 0.5 μg/ml unless otherwise indicated.
Generation of wild-type MERS-CoV, MERS-Uganda, and HKU5-CoV viruses utilized reverse genetics and have been previously described (6, 16, 19). For MERS-Uganda chimera expressing RFP, we utilized the MERS-CoV backbone, replacing ORF5 with RFP as previously described (19). Synthetic constructions of chimeric mutant and full-length MERS-Uganda and HKU5-CoV were approved by the University of North Carolina Institutional Biosafety Committee.
Replication in Vero, Calu-3 2B4, Caco-2, Huh7, and HAE cells was performed as previously described (12, 52–54). Briefly, cells were washed with PBS and inoculated with virus or mock diluted in Opti-MEM for 60 minutes at 37°C. Following inoculation, cells were washed three times, and fresh medium with or without trypsin was added to signify time zero. Three or more biological replicates were harvested at each described time point. For HAE cultures, apical surfaces were washed with PBS containing 5 μg/ml trypsin at 0, 8, 18, 24, and 48 hours postinfection. Sample collections were not conducted in a blind manner nor were samples randomized. Microscopy photos were captured via a Keyence BZ-X700 microscope.
For antibody neutralization assays, MERS-CoV and MERS-Uganda stocks were grown in Opti-MEM both with and without trypsin. All stocks were quantified via plaque assay by overlaying cells with 0.8% agarose in Opti-MEM supplemented with 0.5 μg/ml trypsin and anti/anti. MERS-Uganda stocks grown without trypsin had low titers but were sufficient for neutralization assays.
For anti-DPP4 blocking experiments, Vero cells were preincubated with serum-free Opti-MEM containing 5 μg/ml anti-human DPP4 antibody (R&D systems, MN) for 1 hour. Medium was removed, and cells were infected for 1 hour with virus or mock inoculum at a multiplicity of infection of 0.1. The inoculum was removed, cells were washed three times with PBS, and medium was replaced.
RNA isolation and quantification.
RNA was isolated via TRIzol reagent (Invitrogen, CA) and Direct-zol RNA miniprep kit (Zymo Research, CA) according to the manufacturer’s protocol. MERS-CoV and MERS-Uganda genomic RNA (gRNA) was quantified via TaqMan fast virus 1-step master mix (Applied Biosystems, CA) using previously reported primers and probes targeting ORF1ab (54) and normalized to host 18S rRNA (Applied Biosystems). HKU5-CoV RNA was first reverse transcribed using SuperScript III (Invitrogen) and was then assayed using SsoFast EvaGreen supermix (Bio-Rad, CA) and scaled to host glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript levels. HKU5 gRNA was amplified with the following primers: forward, 5′-CTCTCTCTCGTTCTCTTGCAGAAC-3′; and reverse, 5′-GTTGAGCTCTGCTCTATACTTGCC-3′. GAPDH RNA was amplified with the following primers: forward, 5′-AGCCACATCGCTGAGACA-3′; and reverse, 5′-GCCCAATACGACCAAATCC-3′. Fold change was calculated using the threshold cycle (ΔΔCT) method and was scaled to RNA present at 0 hours postinfection.
Generation of VRP, polyclonal mouse antisera, and Western blot analysis.
Virus replicon particles (VRPs) expressing the MERS-CoV nucleocapsid, MERS-Uganda spike, or HKU5-5 CoV spike were constructed using a non-select agent biosafety level 2 (BSL2) Venezuelan equine encephalitis virus (VEEV) strain 3546 replicon system, as previously described (55). Briefly, RNA containing the nonstructural genes of VEEV and either MERS-CoV nucleocapsid or HKU5-5 CoV spike was packaged using helper RNAs encoding VEEV structural proteins as described previously (56). Six-week-old female BALB/c mice were primed and boosted with VRPs to generate mouse antisera toward either MERS-CoV nucleocapsid or HKU5-5 CoV spike. Following vaccination, mouse polyclonal sera were collected as described previously (57). For Western blotting, lysates from infected cells were prepared as described before in detail (58), and these blots were probed using the indicated mouse polyclonal sera. MERS-CoV N sera was able to detect to HKU5-CoV N protein via Western blot as previously described (7).
Virus neutralization assays.
Plaque reduction neutralization titer assays were performed with previously characterized antibodies against MERS-CoV, as previously described (25, 49). Briefly, antibodies were serially diluted 6- to 8-fold and incubated with 80 PFU of the indicated viruses for 1 h at 37°C. The virus and antibodies were then added to a 6-well plate of confluent Vero cells in triplicate. After a 1 hour incubation at 37°C, cells were overlaid with 3 ml of 0.8% agarose in Opti-MEM supplemented with 0.5 μg/ml trypsin and anti/anti. Plates were incubated for 2 or 3 days at 37°C for MERS-CoV or MERS-Uganda, respectively, and were then stained with neutral red for 3 h, and plaques were counted. The percentage of plaque reduction was calculated as (1 – [number of plaques with antibody/number of plaques without antibody]) × 100.
Biosafety and biosecurity.
Reported studies were initiated after the University of North Carolina Institutional Biosafety Committee approved the experimental protocols. All work for these studies was performed with approved standard operating procedures (SOPs) and safety conditions for MERS-CoV and other related CoVs. Our institutional CoV BSL3 facilities have been designed to conform to the safety requirements recommended by Biosafety in Microbiological and Biomedical Laboratories (BMBL), the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention (CDC), and the National Institutes of Health (NIH). Laboratory safety plans have been submitted, and the facility has been approved for use by the UNC Department of Environmental Health and Safety (EHS) and the CDC.
Accession number(s).
The nearly complete genome sequence for MERS-CoV EMC (GenBank accession number JX869059) and PREDICT/PDF-2180 (GenBank accession number KX574227) were previously deposited in GenBank (16, 59).
ACKNOWLEDGMENTS
The research described in this work was supported by grants from the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT project (cooperative agreement number GHN-A-OO-09-00010-00) and from the National Institute of Allergy and Infectious Disease and the National Institute of Aging of the NIH under awards U19AI109761 and AI110700 to R.S.B. and R00AG049092 to V.D.M. HAE cultures were supported by the National Institute of Diabetes and Digestive and Kidney Disease under award NIH DK065988 to S.H.R.
Trypsin-resistant Vero cells were kindly provided by Linda Saif. Monoclonal antibody LCA60 was provided by Davide Corti and Humabs Biomed SA.
The content described herein is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
REFERENCES
- 1.Reperant LA, Osterhaus A. 2017. AIDS, avian flu, SARS, MERS, Ebola, Zika … what next? Vaccine 35:4470–4474. doi: 10.1016/j.vaccine.2017.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morse SS, Mazet JA, Woolhouse M, Parrish CR, Carroll D, Karesh WB, Zambrana-Torrelio C, Lipkin WI, Daszak P. 2012. Prediction and prevention of the next pandemic zoonosis. Lancet 380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham AA, Daszak P, Wood JLN. 2017. One Health, emerging infectious diseases and wildlife: two decades of progress? Philos Trans R Soc Lond B Biol Sci 372:20160167. doi: 10.1098/rstb.2016.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chafekar A, Fielding BC. 2018. MERS-CoV: understanding the latest human coronavirus threat. Viruses 10:E93. doi: 10.3390/v10020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agnihothram S, Yount BL Jr., Donaldson EF, Huynh J, Menachery VD, Gralinski LE, Graham RL, Becker MM, Tomar S, Scobey TD, Osswald HL, Whitmore A, Gopal R, Ghosh AK, Mesecar A, Zambon M, Heise M, Denison MR, Baric RS. 2014. A mouse model for Betacoronavirus subgroup 2c using a bat coronavirus strain HKU5 variant. mBio 5:e00047-14. doi: 10.1128/mBio.00047-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agnihothram S, Gopal R, Yount BL Jr., Donaldson EF, Menachery VD, Graham RL, Scobey TD, Gralinski LE, Denison MR, Zambon M, Baric RS. 2014. Evaluation of serologic and antigenic relationships between middle eastern respiratory syndrome coronavirus and other coronaviruses to develop vaccine platforms for the rapid response to emerging coronaviruses. J Infect Dis 209:995–1006. doi: 10.1093/infdis/jit609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson BA, Graham RL, Menachery VD. 2018. Viral metagenomics, protein structure, and reverse genetics: key strategies for investigating coronaviruses. Virology 517:30–37. doi: 10.1016/j.virol.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker MM, Graham RL, Donaldson EF, Rockx B, Sims AC, Sheahan T, Pickles RJ, Corti D, Johnston RE, Baric RS, Denison MR. 2008. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc Natl Acad Sci U S A 105:19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rockx B, Baas T, Zornetzer GA, Haagmans B, Sheahan T, Frieman M, Dyer MD, Teal TH, Proll S, van den Brand J, Baric R, Katze MG. 2009. Early upregulation of acute respiratory distress syndrome-associated cytokines promotes lethal disease in an aged-mouse model of severe acute respiratory syndrome coronavirus infection. J Virol 83:7062–7074. doi: 10.1128/JVI.00127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheahan T, Rockx B, Donaldson E, Sims A, Pickles R, Corti D, Baric R. 2008. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J Virol 82:2274–2285. doi: 10.1128/JVI.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheahan T, Rockx B, Donaldson E, Corti D, Baric R. 2008. Pathways of cross-species transmission of synthetically reconstructed zoonotic severe acute respiratory syndrome coronavirus. J Virol 82:8721–8732. doi: 10.1128/JVI.00818-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menachery VD, Yount BL Jr., Sims AC, Debbink K, Agnihothram SS, Gralinski LE, Graham RL, Scobey T, Plante JA, Royal SR, Swanstrom J, Sheahan TP, Pickles RJ, Corti D, Randell SH, Lanzavecchia A, Marasco WA, Baric RS. 2016. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A 113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menachery VD, Yount BL Jr., Debbink K, Agnihothram S, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge XY, Donaldson EF, Randell SH, Lanzavecchia A, Marasco WA, Shi ZL, Baric RS. 2015. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I, Bannister R, Park Y, Babusis D, Mo C, Mackman RL, Spahn JE, Palmiotti CA, Siegel D, Ray AS, Cihlar T, Jordan R, Denison MR, Baric RS. 2017. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anthony SJ, Gilardi K, Menachery VD, Goldstein T, Ssebide B, Mbabazi R, Navarrete-Macias I, Liang E, Wells H, Hicks A, Petrosov A, Byarugaba DK, Debbink K, Dinnon KH, Scobey T, Randell SH, Yount BL, Cranfield M, Johnson CK, Baric RS, Lipkin WI, Mazet JA. 2017. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. mBio 8:e00373-17. doi: 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann M, Wyler R. 1988. Propagation of the virus of porcine epidemic diarrhea in cell culture. J Clin Microbiol 26:2235–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menachery VD, Dinnon KH III, Yount BL Jr., McAnarney ET, Gralinski LE, Hale A, Graham RL, Scobey T, Anthony SJ, Wang L, Graham B, Randell SH, Lipkin WI, Baric RS. 2019. Trypsin treatment unlocks barrier for zoonotic coronaviruses infection. bioRxiv 10.1101/768663. [DOI] [PMC free article] [PubMed]
- 19.Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, Menachery VD, Graham RL, Swanstrom J, Bove PF, Kim JD, Grego S, Randell SH, Baric RS. 2013. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J-E, Li K, Barlan A, Fehr AR, Perlman S, McCray PB Jr., Gallagher T. 2016. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci U S A 113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Wilde AH, Raj VS, Oudshoorn D, Bestebroer TM, van Nieuwkoop S, Limpens RWAL, Posthuma CC, van der Meer Y, Bárcena M, Haagmans BL, Snijder EJ, van den Hoogen BG. 2013. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J Gen Virol 94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banach BS, Orenstein JM, Fox LM, Randell SH, Rowley AH, Baker SC. 2009. Human airway epithelial cell culture to identify new respiratory viruses: coronavirus NL63 as a model. J Virol Methods 156:19–26. doi: 10.1016/j.jviromet.2008.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tekes G, Thiel HJ. 2016. Feline coronaviruses: pathogenesis of feline infectious peritonitis. Adv Virus Res 96:193–218. doi: 10.1016/bs.aivir.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song D, Park B. 2012. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corti D, Zhao J, Pedotti M, Simonelli L, Agnihothram S, Fett C, Fernandez-Rodriguez B, Foglierini M, Agatic G, Vanzetta F, Gopal R, Langrish CJ, Barrett NA, Sallusto F, Baric RS, Varani L, Zambon M, Perlman S, Lanzavecchia A. 2015. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc Natl Acad Sci U S A 112:10473–10478. doi: 10.1073/pnas.1510199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleine-Weber H, Elzayat MT, Hoffmann M, Pöhlmann S. 2018. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci Rep 8:16597. doi: 10.1038/s41598-018-34859-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biniossek ML, Nagler DK, Becker-Pauly C, Schilling O. 2011. Proteomic identification of protease cleavage sites characterizes prime and non-prime specificity of cysteine cathepsins B, L, and S. J Proteome Res 10:5363–5373. doi: 10.1021/pr200621z. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Liu C, Du L, Jiang S, Shi Z, Baric RS, Li F. 2015. Two mutations were critical for bat-to-human transmission of Middle East respiratory syndrome coronavirus. J Virol 89:9119–9123. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y, Shang J, Yang Y, Liu C, Wan Y, Geng Q, Wang M, Baric R, Li F, Zheng Y, Shang J, Yang Y, Liu C, Wan Y, Geng Q, Wang M, Baric R, Li F. 2018. Lysosomal proteases are a determinant of coronavirus tropism. J Virol 92:e01504-18. doi: 10.1128/JVI.01504-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Du L, Liu C, Wang L, Ma C, Tang J, Baric RS, Jiang S, Li F. 2014. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci U S A 111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham RL, Baric RS. 2010. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol 84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge X-Y, Li J-L, Yang X-L, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang Y-J, Luo C-M, Tan B, Wang N, Zhu Y, Crameri G, Zhang S-Y, Wang L-F, Daszak P, Shi Z-L. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo C-M, Wang N, Yang X-L, Liu H-Z, Zhang W, Li B, Hu B, Peng C, Geng Q-B, Zhu G-J, Li F, Shi Z-L, Luo C-M, Wang N, Yang X-L, Liu H-Z, Zhang W, Li B, Hu B, Peng C, Geng Q-B, Zhu G-J, Li F, Shi Z-L. 2018. Discovery of novel bat coronaviruses in South China that use the same receptor as Middle East respiratory syndrome coronavirus. J Virol 92:e00116-18. doi: 10.1128/JVI.00116-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klenk H-D, Rott R, Orlich M, Blödorn J. 1975. Activation of influenza A viruses by trypsin treatment. Virology 68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- 35.Luczo JM, Stambas J, Durr PA, Michalski WP, Bingham J. 2015. Molecular pathogenesis of H5 highly pathogenic avian influenza: the role of the haemagglutinin cleavage site motif. Rev Med Virol 25:406–430. doi: 10.1002/rmv.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark SM, Roth JR, Clark ML, Barnett BB, Spendlove RS. 1981. Trypsin enhancement of rotavirus infectivity: mechanism of enhancement. J Virol 39:816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roivainen M, Hovi T. 1987. Intestinal trypsin can significantly modify antigenic properties of polioviruses: implications for the use of inactivated poliovirus vaccine. J Virol 61:3749–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu H, Jung K, Vlasova AN, Chepngeno J, Lu Z, Wang Q, Saif LJ. 2015. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J Clin Microbiol 53:1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wicht O, Li W, Willems L, Meuleman TJ, Wubbolts RW, van Kuppeveld FJ, Rottier PJ, Bosch BJ. 2014. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J Virol 88:7952–7961. doi: 10.1128/JVI.00297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou P, Fan H, Lan T, Yang X-L, Shi W-F, Zhang W, Zhu Y, Zhang Y-W, Xie Q-M, Mani S, Zheng X-S, Li B, Li J-M, Guo H, Pei G-Q, An X-P, Chen J-W, Zhou L, Mai K-J, Wu ZX, Li D, Anderson DE, Zhang L-B, Li S-Y, Mi Z-Q, He T-T, Cong F, Guo P-J, Huang R, Luo Y, Liu X-L, Chen J, Huang Y, Sun Q, Zhang X-L-L, Wang Y-Y, Xing S-Z, Chen Y-S, Sun Y, Li J, Daszak P, Wang L-F, Shi Z-L, Tong Y-G, Ma J-Y. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li K, Wohlford-Lenane CL, Channappanavar R, Park J-E, Earnest JT, Bair TB, Bates AM, Brogden KA, Flaherty HA, Gallagher T, Meyerholz DK, Perlman S, McCray PB Jr.. 2017. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc Natl Acad Sci U S A 114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuyama S, Ujike M, Morikawa S, Tashiro M, Taguchi F. 2005. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc Natl Acad Sci U S A 102:12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng L-P, Gao Y-T, Ge X-Y, Zhang Q, Peng C, Yang X-L, Tan B, Chen J, Chmura AA, Daszak P, Shi Z-L. 2016. Bat severe acute respiratory syndrome-like coronavirus WIV1 encodes an extra accessory protein, ORFX, involved in modulation of the host immune response. J Virol 90:6573–6582. doi: 10.1128/JVI.03079-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Millet JK, Whittaker GR. 2015. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Earnest JT, Hantak MP, Li K, McCray PB Jr., Perlman S, Gallagher T. 2017. The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathog 13:e1006546. doi: 10.1371/journal.ppat.1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Li C, Zhao G, Chu H, Wang D, Yan HH, Poon VK, Wen L, Wong BH-Y, Zhao X, Chiu MC, Yang D, Wang Y, Au-Yeung RKH, Chan IH-Y, Sun S, Chan JF-W, To KK-W, Memish ZA, Corman VM, Drosten C, Hung IF-N, Zhou Y, Leung SY, Yuen K-Y. 2017. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci Adv 3:eaao4966. doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding Y, He L, Zhang Q, Huang Z, Che X, Hou J, Wang H, Shen H, Qiu L, Li Z, Geng J, Cai J, Han H, Li X, Kang W, Weng D, Liang P, Jiang S. 2004. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J Pathol 203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, Hasoksuz M, Spiro D, Halpin R, Wang S, Stollar S, Janies D, Hadya N, Tang Y, Ghedin E, Saif L. 2007. Complete genomic sequences, a key residue in the spike protein and deletions in nonstructural protein 3b of US strains of the virulent and attenuated coronaviruses, transmissible gastroenteritis virus and porcine respiratory coronavirus. Virology 358:424–435. doi: 10.1016/j.virol.2006.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, Cottrell CA, Becker MM, Wang L, Shi W, Kong W-P, Andres EL, Kettenbach AN, Denison MR, Chappell JD, Graham BS, Ward AB, McLellan JS. 2017. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A 114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallagher TM, Buchmeier MJ, Perlman S. 1992. Cell receptor-independent infection by a neurotropic murine coronavirus. Virology 191:517–522. doi: 10.1016/0042-6822(92)90223-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W, Hulswit RJG, Widjaja I, Raj VS, McBride R, Peng W, Widagdo W, Tortorici MA, van Dieren B, Lang Y, van Lent JWM, Paulson JC, de Haan CAM, de Groot RJ, van Kuppeveld FJM, Haagmans BL, Bosch B-J. 2017. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci U S A 114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sims AC, Tilton SC, Menachery VD, Gralinski LE, Schafer A, Matzke MM, Webb-Robertson B-J, Chang J, Luna ML, Long CE, Shukla AK, Bankhead AR III, Burkett SE, Zornetzer G, Tseng C-TK, Metz TO, Pickles R, McWeeney S, Smith RD, Katze MG, Waters KM, Baric RS. 2013. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J Virol 87:3885–3902. doi: 10.1128/JVI.02520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sims AC, Burkett SE, Yount B, Pickles RJ. 2008. SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium. Virus Res 133:33–44. doi: 10.1016/j.virusres.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Almazán F, DeDiego ML, Sola I, Zuñiga S, Nieto-Torres JL, Marquez-Jurado S, Andrés G, Enjuanes L. 2013. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio 4:e00650-13. doi: 10.1128/mBio.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agnihothram S, Menachery VD, Yount BL, Lindesmith LC, Scobey T, Whitmore A, Schafer A, Heise MT, Baric RS. 2018. Development of a broadly accessible Venezuelan equine encephalitis virus replicon particle vaccine platform. J Virol 92:e00027-18. doi: 10.1128/JVI.00027-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolles M, Deming D, Long K, Agnihothram S, Whitmore A, Ferris M, Funkhouser W, Gralinski L, Totura A, Heise M, Baric RS. 2011. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol 85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheahan T, Whitmore A, Long K, Ferris M, Rockx B, Funkhouser W, Donaldson E, Gralinski L, Collier M, Heise M, Davis N, Johnston R, Baric RS. 2011. Successful vaccination strategies that protect aged mice from lethal challenge from influenza virus and heterologous severe acute respiratory syndrome coronavirus. J Virol 85:217–230. doi: 10.1128/JVI.01805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, Nagel J, Johnson JB, Agnihothram S, Gates JE, Frieman MB, Baric RS, Donaldson EF. 2012. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol 86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]