Flaviviruses naturally cycle between the mosquito vector and vertebrate hosts. The disparate hosts provide selective pressures that drive virus genome evolution to maintain efficient replication during host alteration. Host adaptation may occur at different stages of the viral life cycle, since host-specific viral protein processing and virion conformations have been reported in the individual hosts. However, the viral determinants and the underlying mechanisms associated with host-specific functions remain obscure. In this study, using Zika virus, we found that the DAR-mediated genome cyclization regulates viral replication differently and is under different selection pressures in mammalian and mosquito cells. A more constrained complementarity of the DAR is required in mosquito cells than in mammalian cells. Since the DAR element is stably maintained among mosquito-borne flaviviruses, our findings could provide new information for understanding the role of flavivirus genome cyclization in viral adaptation and RNA evolution in the two hosts.
KEYWORDS: DAR element, flavivirus, genome cyclization, host-specific replication, Zika virus
ABSTRACT
Mosquito-borne flaviviruses, which include many important human pathogens, such as West Nile virus (WNV), dengue virus (DENV), and Zika virus (ZIKV), have caused numerous emerging epidemics in recent years. Details of the viral genome functions necessary for effective viral replication in mosquito and vertebrate hosts remain obscure. Here, using ZIKV as a model, we found that the conserved “downstream of AUG region” (DAR), which is known to be an essential element for genome cyclization, is involved in viral replication in a host-specific manner. Mutational analysis of the DAR element showed that a single-nucleotide mismatch between the 5′ DAR and the 3′ DAR had little effect on ZIKV replication in mammalian cells but dramatically impaired viral propagation in mosquito cells. The revertant viruses passaged in mosquito cells generated compensatory mutations restoring the base pairing of the DAR, further confirming the importance of the complementarity of the DAR in mosquito cells. We demonstrate that a single-nucleotide mutation in the DAR is sufficient to destroy long-range RNA interaction of the ZIKV genome and affects de novo RNA synthesis at 28°C instead of 37°C, resulting in the different replication efficiencies of the mutant viruses in mosquito and mammalian cells. Our results reveal a novel function of the circular form of the flavivirus genome in host-specific viral replication, providing new ideas to further explore the functions of the viral genome during host adaptation.
IMPORTANCE Flaviviruses naturally cycle between the mosquito vector and vertebrate hosts. The disparate hosts provide selective pressures that drive virus genome evolution to maintain efficient replication during host alteration. Host adaptation may occur at different stages of the viral life cycle, since host-specific viral protein processing and virion conformations have been reported in the individual hosts. However, the viral determinants and the underlying mechanisms associated with host-specific functions remain obscure. In this study, using Zika virus, we found that the DAR-mediated genome cyclization regulates viral replication differently and is under different selection pressures in mammalian and mosquito cells. A more constrained complementarity of the DAR is required in mosquito cells than in mammalian cells. Since the DAR element is stably maintained among mosquito-borne flaviviruses, our findings could provide new information for understanding the role of flavivirus genome cyclization in viral adaptation and RNA evolution in the two hosts.
INTRODUCTION
The genus Flavivirus includes many important human pathogens, such as West Nile virus (WNV), dengue virus (DENV), Zika virus (ZIKV), yellow fever virus (YFV), and tick-borne encephalitis virus (TBEV), which are endemic in different regions and have caused large public health burdens worldwide (1). Since 2015, ZIKV has drawn global attention due to large epidemics in Latin America and its possible association with high rates of severe neurological disorders, such as fetal microcephaly and Guillain-Barré syndrome (2–5). As an emerging virus, ZIKV represents one of the significant arthropod-borne viral pathogens in humans.
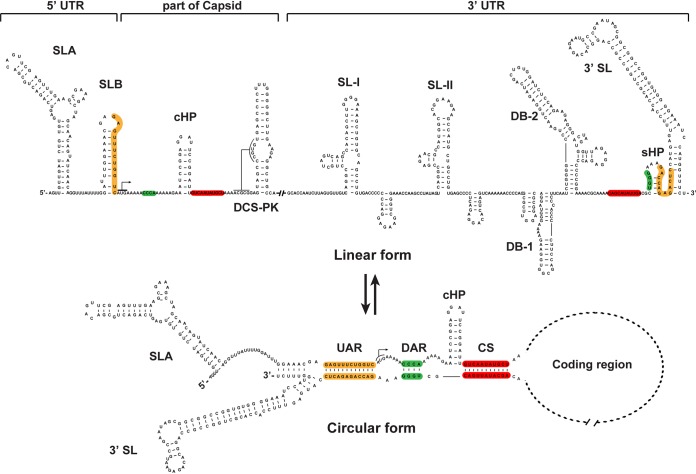
Flaviviruses are enveloped, positive-sense, single-stranded RNA viruses that are typically transmitted between arthropod vectors and vertebrate hosts (6). The genome of flaviviruses is around 11 kb and contains a single open reading frame (ORF) flanked by a 5ʹ untranslated region (UTR) and a 3ʹ UTR. The ORF encodes a polyprotein that can be processed into three structural (C, prM, and E) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins for viral particle formation and replication, respectively. Both UTRs contain conserved RNA sequences and highly structured RNA elements (7, 8) and play various roles in viral translation (9, 10), replication (11–17), pathogenesis (18–21), and host adaptation (20, 22–26). The 5ʹ UTR consists of two conserved stem-loop structures, SLA and SLB (Fig. 1). SLA serves as a promoter for viral RNA synthesis by binding the RNA-dependent RNA polymerase (RdRp) domain of NS5 (27, 28). The 5ʹ upstream AUG region (UAR) flanking stem (UFS) element within SLB (11); the local hairpin, known as the capsid-coding region hairpin element (cHP) (29); and the downstream of 5ʹ cyclization sequence (CS) pseudoknot (DCS-PK) (30), in the capsid-coding region, are necessary for efficient RNA replication. The 3ʹ UTR of mosquito-borne flaviviruses (MBFVs) can be sequentially divided into three domains: the stem-loop (SL) domain, the dumbbell (DB) domain, and the 3ʹ stem-loop domain, which includes a small hairpin (sHP) and 3ʹ SL structures (sHP-3ʹ-SL) (31, 32) (Fig. 1). The SL and DB structures are normally duplicated and function as host-specific RNA structures that have different evolution patterns in mammalian and mosquito hosts and facilitate viral fitness during host switching (22, 33, 34); sHP-3ʹ-SL is essential for viral replication (35–37).
FIG 1.
Structures of the linear and circular forms of the ZIKV genome. Based on the predictions and structural models identified by Li et al. in a recent study (59), the conserved RNA structures in the 5ʹ and 3ʹ ends of the genome are indicated, and the UAR, DAR, and CS sequences are highlighted in orange, green, and red, respectively, in the linear genome. The pairwise hybridizations between the 5ʹ UAR and the 3ʹ UAR, the 5ʹ DAR and the 3ʹ DAR, and the 5ʹ CS and the 3ʹ CS are shown in the circular form of the genome. The cis-acting element sequence present in the capsid-coding region (cHP) is present only in the linear form. The structures of the SLB, the sHP, and the bottom of the 3ʹ SL are changed in the circular form of the genome. The bent arrows denote the translation start site.
For positive-sense RNA viruses, several lines of evidence have demonstrated that genome cyclization also regulates viral RNA synthesis (15, 38, 39). Different strategies have been developed to bring the 5′ and 3′ ends of viral genomic RNA together during viral replication (40). In the case of flaviviruses, genome cyclization is mediated by direct, long-distance RNA-RNA interactions between different complementary sequences (Fig. 1) at the 5′ and 3′ ends of the viral genome (11–15, 17, 41). For MBFVs, at least three pairs of complementary interactions are necessary for genome cyclization (Fig. 1), i.e., the base pairing of 5′ CS-3′ CS, 5′ UAR-3′ UAR, and 5′ DAR-3′ DAR sequences (41). The CS sequences are highly conserved among flaviviruses and are thought to be the primary sites of cyclization (42). The 5ʹ-to-3ʹ UAR interaction is relatively weak and is dependent upon 5ʹ-3ʹ CS base pairing (42). Mutations in both the CS and UAR regions disrupting the base pairing abolish or severely affect viral replication (14–16, 43, 44). The DAR motif has fewer complementary bases and resides between the CS and UAR elements in the circular form of the genome (Fig. 1). It has been speculated that the interaction between 5ʹ and 3ʹ DARs takes place after the 5ʹ-3ʹ CS base pairing and then facilitates the 5ʹ-to-3ʹ UAR interaction (16). Mutagenesis analysis revealed that the complementarity between the 5ʹ DAR and 3ʹ DAR is necessary for genome cyclization and viral replication of DENV and WNV (16, 17). However, extensive analysis of the DAR element for viral replication and genome cyclization is still lacking.
In the present study, we demonstrate that 5′ DAR-3′ DAR-mediated genome cyclization is also essential for ZIKV replication. Importantly, it was found that different degrees of base pairing between the 5′ DAR and the 3′ DAR are required for viral replication in mammalian and mosquito cells. A single-nucleotide mismatch between the 5′ DAR and the 3′ DAR could dramatically impair ZIKV propagation in C6/36 cells, whereas it had slight or no effect on viral replication in Vero cells. Such a strict requirement for 5′ DAR-3′ DAR base pairing for ZIKV replication in mosquito cells was also confirmed by the recovery of revertant viruses, which contained mutations restoring both 5′ DAR-3′ DAR base pairing and viral replication in C6/36 cells. Further biochemical analysis indicated that the impaired genome cyclization caused by disrupted 5′ DAR-3′ DAR base pairing significantly decreased de novo RNA synthesis at 28°C instead of 37°C, resulting in the different replication efficiencies in C6/36 and Vero cells. These results suggest that flavivirus DAR-mediated genome cyclization is under different selection pressures in mosquito and mammalian cells, highlighting their roles in host adaptation and viral evolution.
RESULTS
Uncoupling cis-acting elements from capsid-coding sequences.
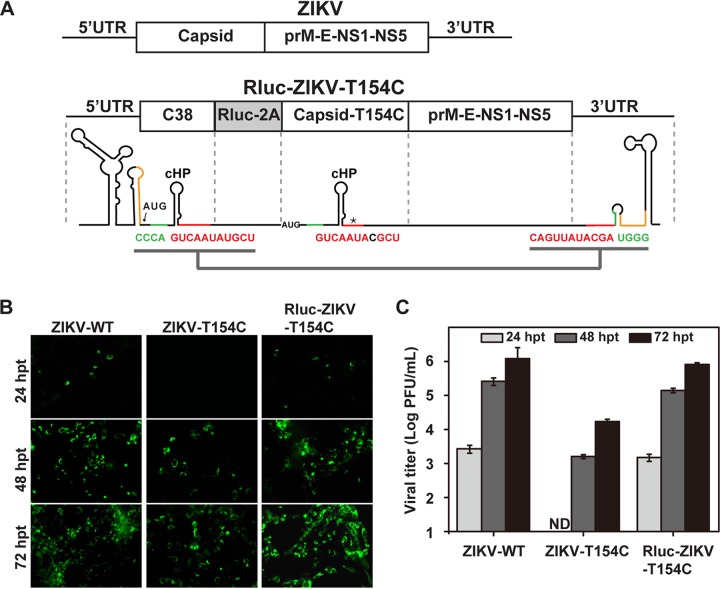
To investigate the function of the DAR during viral replication, we employed a Renilla luciferase (Rluc) reporter virus, ZIKV strain SZ-WIV01, with a T154C nucleotide substitution in the capsid (Rluc-ZIKV-T154C). Several genetic modifications were made to generate Rluc-ZIKV-T154C (Fig. 2A). The coding sequence of the N-terminal 38 amino acids of the capsid gene (designated C38), containing the intact RNA elements in the capsid, including the 5ʹ DAR, 5ʹ CS, cHP (29), DCS-PK (30), and C1 regions (12), was duplicated and placed upstream of the Rluc gene fused to a foot-and-mouth disease virus (FMDV) 2A protease sequence, followed by the complete ZIKV ORF. In the full-length capsid-coding region, we introduced a nucleotide substitution, T154C, within the 5ʹ CS sequence, leading to a mismatch between the 5ʹ and 3ʹ CS. These modifications were supposed to allow the nucleotide sequences of C38 preceding Rluc to act as the cis-acting RNA elements for viral replication. To test this hypothesis, we constructed a ZIKV-T154C mutant by introducing this T154C nucleotide mutation into the full-length infectious ZIKV clone and compared the replication levels of ZIKV-T154C and Rluc-ZIKV-T154C RNAs in transfected cells. As shown in Fig. 2B and C, compared to wild-type ZIKV (ZIKV-WT), the viral protein synthesis and virus production of ZIKV-T154C were significantly inhibited at each time point posttransfection, whereas Rluc-ZIKV-T154C exhibited replication patterns similar to those of the WT, indicating that the C38 sequence preceding Rluc works as a cis element to support viral replication of Rluc-ZIKV-T154C. This result confirmed that Rluc-ZIKV-T154C was a suitable tool to uncouple the cis-acting RNA elements from the capsid-coding region.
FIG 2.
Characterization of Rluc-ZIKV-T154C reporter virus uncoupling the DAR element from the capsid-coding region. (A) Schematic representation of the Rluc-ZIKV-T154C construct. The N-terminal 38 amino acids of the capsid (C38) were duplicated downstream of the 5ʹ UTR, and the Rluc gene fused with FMDV 2A sequence (Rluc-2A) was inserted at the C38 and capsid junction. In the full-length capsid, a nucleotide mutation, T154C, within the 5ʹ CS sequence was introduced to limit the interaction between the 5ʹ and 3ʹ CS. (B) IFA analysis of ZIKV-T154C and Rluc-ZIKV-T154C. Equal amounts of the in vitro-transcribed RNAs of Rluc-ZIKV-T154C, ZIKV-T154C, and ZIKV-WT were transfected into Vero cells, and IFA analysis was performed at the indicated time points posttransfection. The polyclonal antibody against NS3 and FITC-conjugated goat anti-mouse IgG were used as primary and secondary antibodies, respectively. (C) Production of ZIKV-WT, ZIKV-T154C, and Rluc-ZIKV-T154C at 24, 48, and 72 hpt. The data are representative of three independent transfections, and the error bars represent standard deviations.
Different degrees of DAR base pairing are required for ZIKV replication in Vero and C6/36 cells.
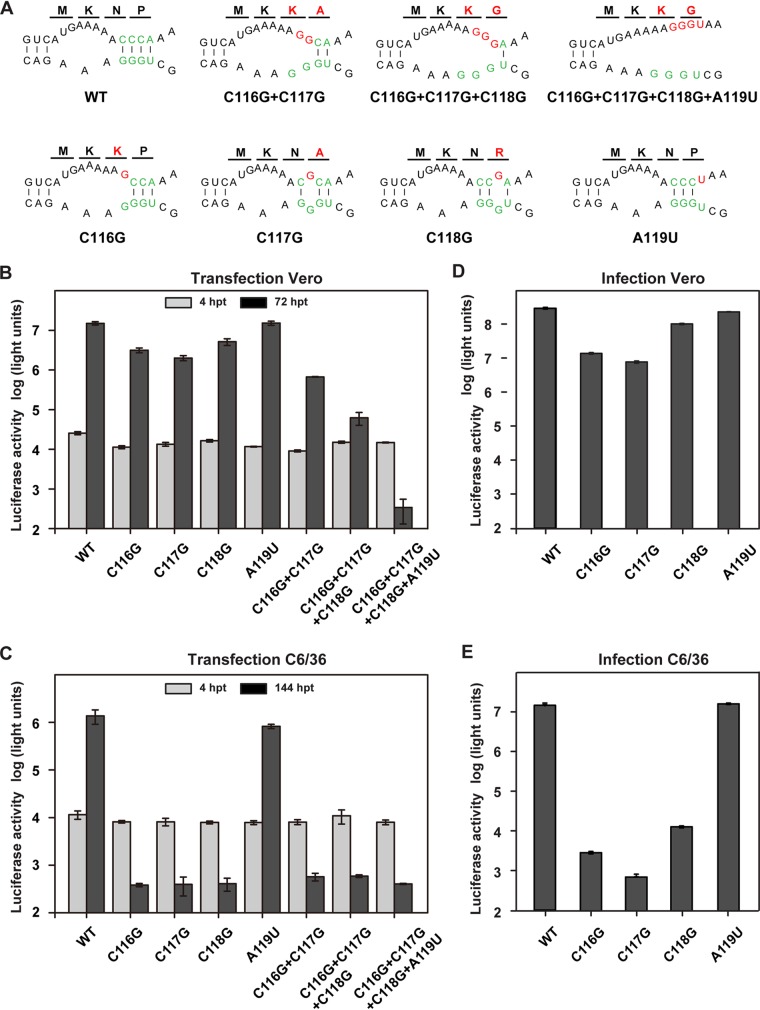
The 3ʹ DAR sequence overlaps the stem of the 3ʹ sHP structure (Fig. 1), and 3ʹ sHP integrity has been found to be essential for the replication of WNV (17) and DENV (35). To avoid destroying the stem of the 3ʹ sHP, a panel of mutations causing different degrees of mismatches in 5ʹ-3ʹ DAR base pairing were designed and introduced into the 5ʹ DAR motif 5ʹ -CCCA-3ʹ (Fig. 3A) within C38 of Rluc-ZIKV-T154C. To investigate the function of 5ʹ-to-3ʹ DAR hybridization in ZIKV replication, equal amounts of WT and mutant Rluc-ZIKV-T154C RNAs were transfected into mammalian Vero cells or mosquito C6/36 cells. Luciferase activity was monitored at different times posttransfection. All of the 5ʹ DAR mutants showed luciferase levels indistinguishable from that of WT Rluc-ZIKV-T154C at 4 h posttransfection (hpt) in both Vero and C6/36 cells, suggesting that the DAR base pairing is unnecessary for genome translation. However, these mutations had different impacts on viral RNA replication in Vero and C6/36 cells, leading to different levels of luciferase signals at 72 hpt in Vero cells and at 144 hpt in C6/36 cells (Fig. 3B and C). In Vero cells (Fig. 3B), a single-nucleotide mismatch had little or no effect on viral replication; the double- and triple-nucleotide mismatches (C116G plus C117G and C116G plus C117G plus C118G) impaired viral replication, with about 100-fold and 1,000-fold reductions in luciferase activity, respectively; the C116G-C117G-C118G-A119U mutant, which destroyed all base pairing of the DAR, was lethal, yielding a background luciferase signal. In C6/36 cells (Fig. 3C), however, even a single-nucleotide mismatch was able to dramatically impair viral replication, as only background luciferase signal was detectable at later time points posttransfection for all the mutations except A119U.
FIG 3.
Functional analysis of complementarity between DAR elements using the Rluc-ZIKV-T154C reporter virus. (A) Mutations introduced into theDAR sequences within the C38 region of Rluc-ZIKV-T154C. Predicted interaction between 5ʹ and 3ʹ DAR elements and their adjacent regions. The DAR sequences are labeled in green, and the mutations targeting the 5ʹ DAR are highlighted in red. The first four amino acids of the capsid encoded by each mutant RNA are shown above the nucleotide sequences, and the mutated amino acids are in red. (B and C) Translation and replication of the Rluc-ZIKV mutants in Vero (B) and C6/36 (C) cells. Equal amounts of WT and mutant RNAs were transfected into Vero or C6/36 cells, and the luciferase activity was measured at 4 hpt and 72 hpt in Vero cells and at 4 hpt and 144 hpt in C6/36 cells. (D and E) Infectivity of mutant reporter viruses in Vero (D) and C6/36 (E) cells. Viruses harvested from mutant-RNA-transfected Vero cells at 72 hpt were used to infect naive Vero and C6/36 cells at the same MOI of 0.1, and the luciferase activity was detected at 72 hpi in Vero cells and at 112 hpi in C6/36 cells. The data are representative of three independent experiments. Each experiment was performed in triplicate, and the error bars represent standard deviations.
To confirm the observation in mosquito cells, high titers of reporter viruses bearing the single-nucleotide substitution harvested from Vero cells were used to infect Vero (Fig. 3D) and C6/36 (Fig. 3E) cells at the same multiplicity of infection (MOI). Similar to the results obtained in the transfection assay, the A119U mutant replicated almost as efficiently as the WT in both Vero and C6/36 cells. The C116G, C117G, and C118G mutations showed slight replication delays in Vero cells, with an approximately 10-fold reduction in luciferase activity compared to the WT, whereas viral replication in C6/36 cells was dramatically inhibited, with more than a 1,000-fold reduction in luciferase signal. These data suggest that the mutations disrupting the complementarity of the 5ʹ-3ʹ DAR had more significant effect on viral replication in mosquito cells than in mammalian cells.
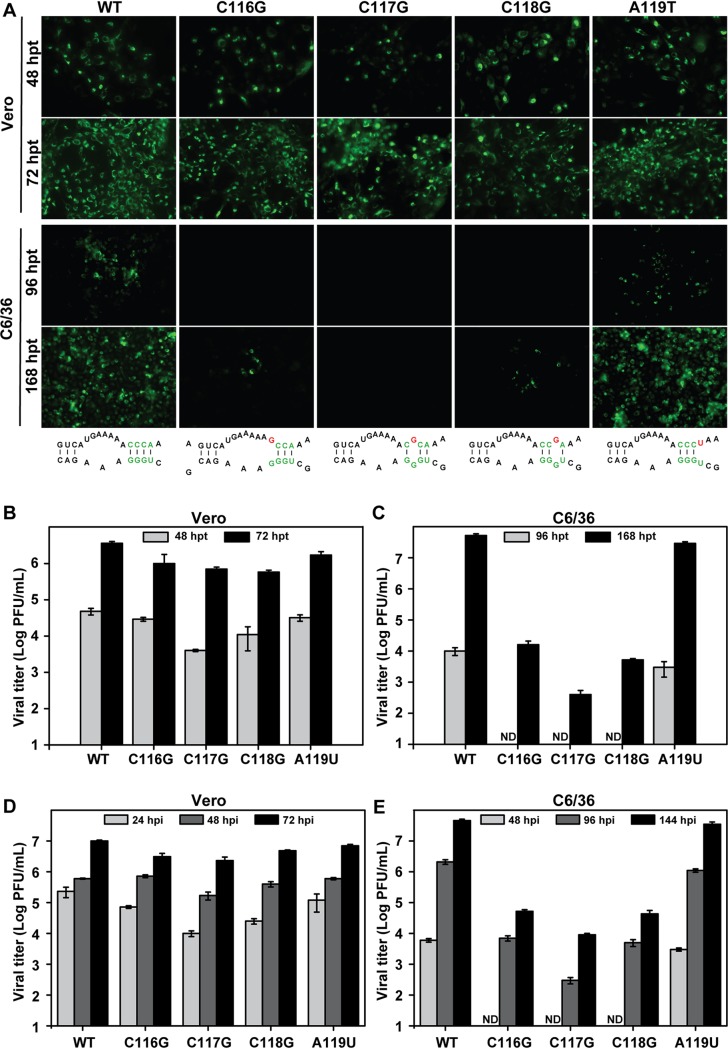
To further confirm the different functions of the 5ʹ-3ʹ DAR element in mammalian and mosquito cells, we introduced the single-nucleotide substitutions into a full-length ZIKV infectious clone. It should be noted that most of these nucleotide substitutions also inevitably led to a 1-amino-acid replacement in the capsid protein in the context of the infectious ZIKV clone (Fig. 3A). Equal amounts of in vitro-transcribed RNAs were transfected into Vero and C6/36 cells, and immunofluorescence assays (IFA) and plaque assays were performed to analyze RNA replication and virus production. Levels of IFA-positive cells (Fig. 4A) and virus production (Fig. 4B) comparable to or slightly lower than those of the WT were detected in the mutant-RNA-transfected Vero cells, whereas the C116G, C117G, and C118G mutants exhibited apparent replication defects compared to the WT and the A119U mutant in C6/36 cells (Fig. 4A and C). To further compare their growth kinetics in Vero and C6/36 cells, the viruses harvested from RNA-transfected Vero cells were used to infect both cell lines at an MOI of 0.1, and viral titers at different time points postinfection were quantified by plaque assay. The mutants with a single-nucleotide substitution showed a minor effect on viral growth in Vero cells (Fig. 4D), but the viral titers of the C116G, C117G, and C118G mutants were reduced sharply in C6/36 cells (Fig. 4E). These results further confirmed different requirements for DAR base pairing for viral replication in mammalian and mosquito cells. Moreover, the consistent data based on the Rluc-ZIKV-T154C reporter virus cDNA clone and the full-length infectious ZIKV cDNA clone also indicate that the disruption of DAR complementarity, rather than the changes of amino acid in the capsid, leads to differential replication of the DAR mutants in mammalian and mosquito cells.
FIG 4.
Characterization of DAR mutants using the ZIKV infectious clone. (A) IFA analysis of viral protein expression in mutant-RNA-transfected Vero and C6/36 cells. Equal amounts of the in vitro-transcribed RNAs were transfected into Vero and C6/36 cells, and IFA was performed at 48 and 72 hpt in Vero cells and at 96 and 168 hpt in C6/36 cells. (B and C) Viral production of supernatants from mutant-RNA-transfected Vero (B) and C6/36 (C) cells. The supernatants harvested from the two cell types at the indicated time points were subjected to plaque assays to determine viral titers. (D and E) Growth curves of mutant viruses in Vero (D) and C6/36 (E) cells. The mutant viruses collected from Vero cells at 72 hpt were used to infect naive Vero and C6/36 cells at an MOI of 0.1, and the virus titers at the indicated time points after infection were determined by plaque assay. ND, not detectable. The data are representative of three independent experiments. Each experiment was performed in triplicate, and the error bars represent standard deviations.
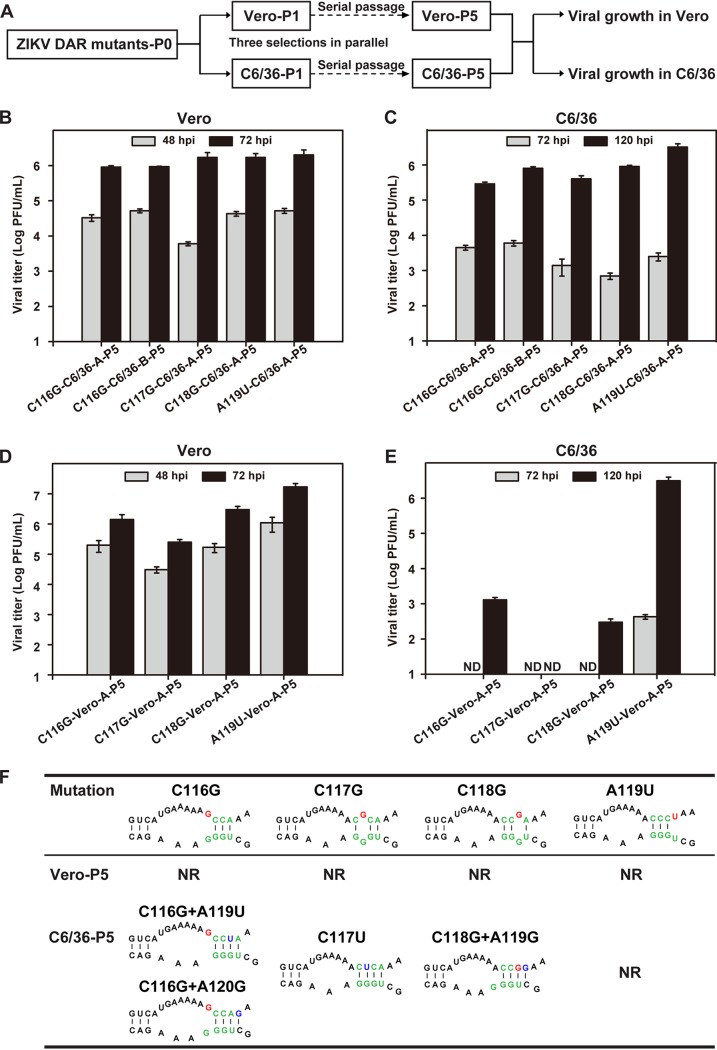
Compensatory mutations of C116G, C117G, and C118G mutants are specifically recovered in C6/36 cells.
To further verify the different requirements for DAR base pairing in mammalian and mosquito cells, the mutant viruses containing the C116G, C117G, or C118G mutation were passaged in C6/36 and Vero cells (Fig. 5A). Three independent selections (designated A, B, and C) were performed for each mutant. After 5 rounds of extensive passage in C6/36 cells, adaptive viruses (designated C6/36-P5) that replicated efficiently in C6/36 cells were recovered (Fig. 5B). Complete genome sequencing of passage 5 (P5) viruses revealed that the compensatory mutations occurred exclusively within the 5ʹ DAR region (Fig. 5F). For C116G and C118G mutants, additional adaptive mutation was found, while the original engineered mutation was maintained. Specifically, a single-nucleotide change of A119U or A120G was recovered from C116G-C6/36-P5, and A119G was identified from C118G-C6/36-P5. For C117G-C6/36-P5, a consistent pseudoreversion of 117U was found in all three independent selections. These identified compensatory mutations are predicted to promote the base pairings between the 5ʹ-3ʹ DAR or adjacent nucleotides (Fig. 5F), which were confirmed by RNA binding assays (see Fig. 8). At the same time, there were no adaptive mutations identified for C116G, C117G, and C118G mutant viruses in Vero cells after 5 rounds of passage (designated Vero-P5) (Fig. 5F). All three mutants of Vero-P5 still could not propagate efficiently in C6/36 cells (Fig. 5D), although they could replicate efficiently in Vero cells (Fig. 5E). For the A119U mutant, no compensatory mutations were found in both Vero cells and C6/36 cells after serial passages. These results demonstrated that the DAR mutants undergo different selections in mammalian and mosquito cells, as the compensatory mutations restoring the base pairing of the DAR specifically appear in mosquito cells, but not in Vero cells.
FIG 5.
Identification of revertant viruses of DAR mutants. (A) Flowchart of successive passages of DAR mutant viruses in Vero and C6/36 cells. (B and C) Growth of P5 revertant viruses derived from C6/36 cells in C6/36 (B) and Vero (C) cells. The C6/36-P5 viruses were used to infect C6/36 and Vero cells at an MOI of 0.1, and the titers of the viruses collected at the indicated time points after infection were determined. (D and E) Propagation of Vero-P5 viruses in C6/36 (D) and Vero (E) cells. ND, not detectable. The data are representative of three independent experiments. Each experiment was performed in triplicate, and the error bars represent standard deviations. (F) Genome sequencing of the C6/36-P5 and Vero-P5 viruses. The compensatory mutations identified in the C6/36-P5 revertant viruses were predicted to restore base pairing between DAR sequences. The predicted structures are shown, with the DAR sequences in green, the original mutations in red, and the compensatory mutations in blue. NR, not reverted.
FIG 8.
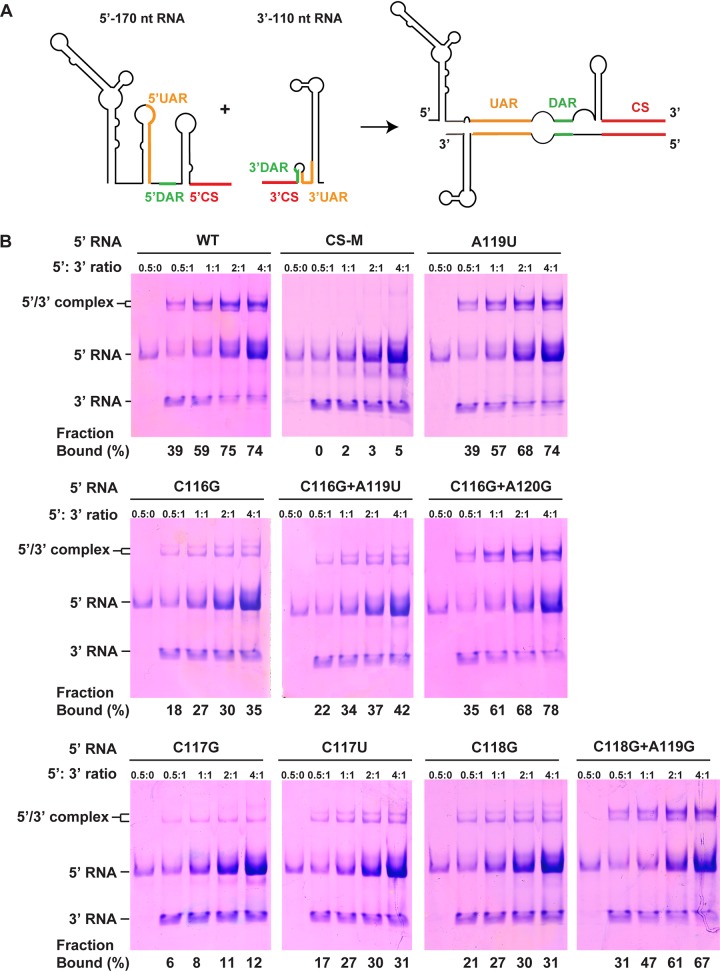
Effects of DAR mutations on interaction between terminal sequences of ZIKV RNA. (A) Schematic representation of RNA molecules used for binding assays. The structures in the 5ʹ RNA and 3ʹ RNA are indicated; the 5ʹ-3ʹ RNA complex is formed by hybridization between the cyclization sequences when the two RNAs are incubated together. (B) RNA binding assay of the DAR mutants. The 3ʹ RNA was incubated at different ratios with WT and DAR mutant 5ʹ RNAs, and the formation of RNA complexes was then analyzed by electrophoresis using a native polyacrylamide gel. The percentages of the bound 3ʹ RNAs, which were calculated by quantification of the band intensities of the 5ʹ-3ʹ RNA complex and the free 3ʹ RNA using the ImageJ software, are shown below the gels.
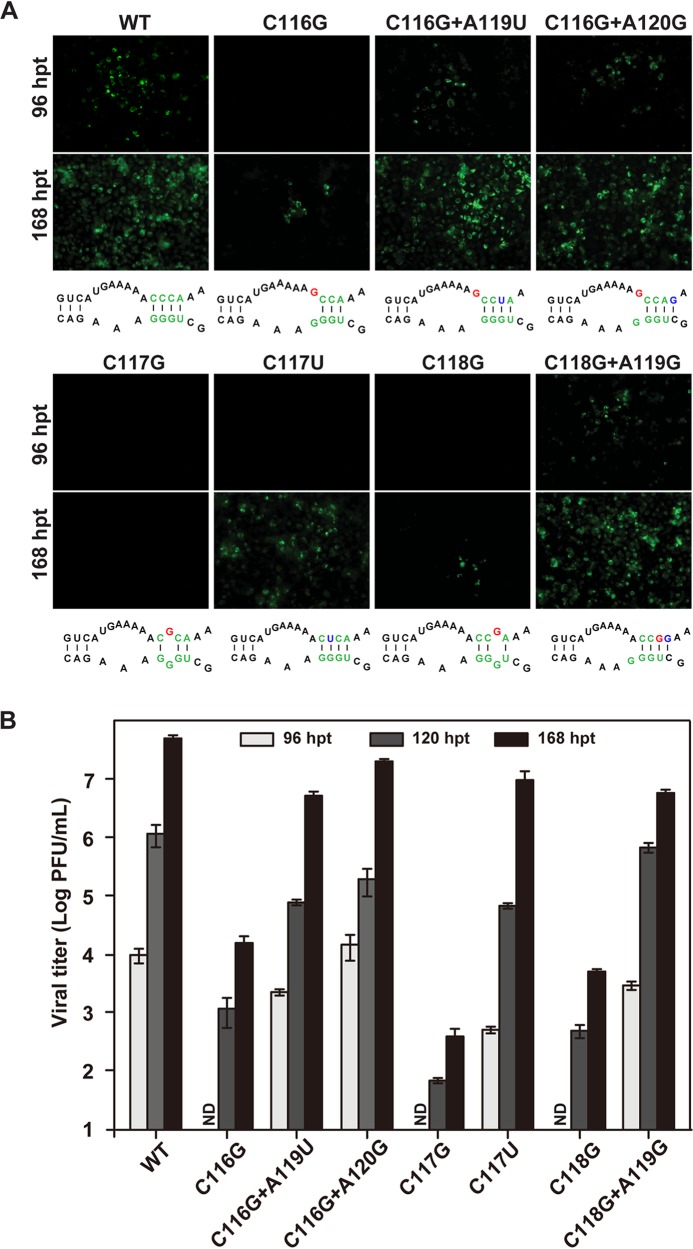
Compensatory mutations specifically rescue viral replication defects of DAR mutants in C6/36 cells.
To validate the roles of compensatory mutations of the 5ʹ DAR mutants in viral replication, we engineered the identified mutations back to the full-length infectious clones of their corresponding mutants. The same analyses were performed to compare viral replication efficiencies in mosquito cells, as mentioned above. In contrast to the poor replication of the C116G, C117G, and C118G mutants, the C116G-plus-A119U, C116G-plus-A120G, C117U, and C118G- plus-A119G mutants replicated efficiently, as comparable amounts of IFA-positive cells and WT cells were produced (Fig. 6A). All of the compensatory mutations could increase viral titers by about 1,000-fold relative to the respective parental mutants in C6/36 cells (Fig. 6B). Our results suggested that the compensatory mutations, which are predicted to restore the original number of base pairings in the DAR or adjacent sequences, could specifically rescue the viral replication defects of the DAR mutants in mosquito cells. These results also indicated that the secondary structure of the DAR, rather than its particular sequence, is important for viral replication.
FIG 6.
Genetic analysis of compensatory mutations of DAR mutants in C6/36 cells. (A) IFA detection of C6/36 cells transfected with RNAs bearing various mutations at 96 and 168 hpt. (B) Viral titers of transfected cells at 96, 120, and 168 hpt monitored by plaque assay. The data are representative of three independent experiments. Each experiment was performed in triplicate, and the error bars represent standard deviations.
Growth of the DAR mutants is temperature dependent.
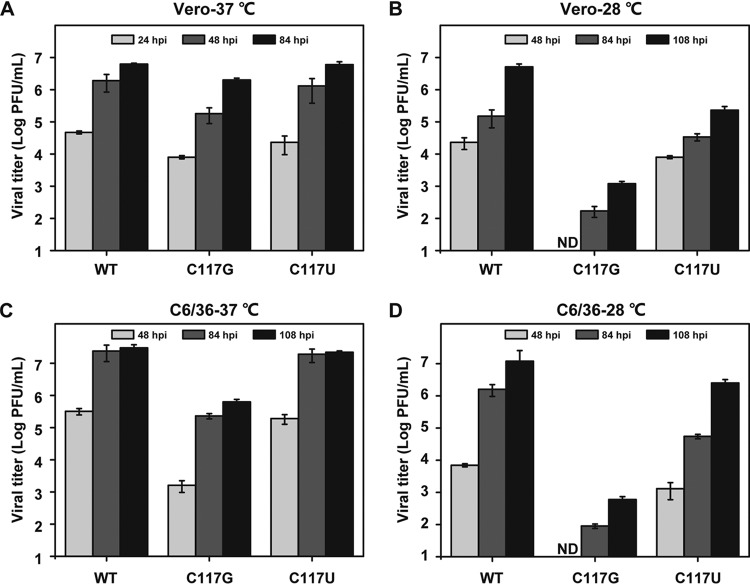
Since the mammalian and mosquito cells were grown at different temperatures, we wondered whether it was the temperature that caused such DAR-involved differences of viral replication between the two hosts. To this end, we chose the most severely replication-defective mutant in C6/36 cells, the C117G mutant, and its pseudoreversion, C117U, for investigation of their growth kinetics within the same host background at different temperatures. As stated above, in Vero cells, all the viruses replicated to high titers at 37°C (Fig. 7A). At 28°C, the WT and C117U viruses showed a modest delay in replication but still reached high titers of 5.1 × 106 PFU/ml and 4.3 × 105 PFU/ml, respectively, by 108 h postinfection (hpi). In contrast, the C117G mutant replicated very poorly, yielding a 3- to 4-log-unit reduction in viral titers compared to the viral titers generated at 37°C (Fig. 7B). In parallel, the same assay was performed in C6/36 cells. All of the viruses showed more efficient replication at 37°C than at 28°C in C6/36 cells. The WT and the C117U and C117G mutant viruses generated approximately 2-, 10-, and 1,000-fold increases, respectively, in viral titers at 37°C compared to titers at 28°C at 108 hpi (Fig. 7C and D). These results suggested that the temperature predominantly determined the different viral growth rates of DAR mutants in mammalian and mosquito cells, and more restricted DAR complementarity may be required for viral replication at a lower growth temperature.
FIG 7.
Temperature-dependent growth of DAR mutants. (A and B) Growth curves of DAR mutants in Vero cells at 37°C (A) and 28°C (B). Naive Vero cells were infected with WT, C117G, and C117U viruses at an MOI of 0.1 and maintained at 37°C or 28°C. The viral titers of the supernatants harvested at the indicated time points were determined by plaque assay. The same strategy was used to compare propagation rates of the viral mutants in C6/36 cells at 37°C (C) and 28°C (D). The data are representative of three independent experiments. Each experiment was performed in triplicate, and the error bars represent standard deviations.
Effects of DAR mutations on RNA-RNA interaction between the 5ʹ and 3ʹ ends of the ZIKV genome.
RNA binding assays were used to investigate the direct 5ʹ-3ʹ-end interactions of the ZIKV genome (15–17). The 170-nucleotide (nt) RNA of the 5ʹ end of the ZIKV genome carrying the specific DAR mutations and the last 116-nt RNA of the 3ʹ UTR (Fig. 8A) were prepared by in vitro transcription. The formation of the 5ʹ-3ʹ RNA complex was detected by electrophoretic mobility shift analysis. A 5ʹ RNA containing double mutations (underlined) in the 5ʹ CS sequence (5ʹ- GUCAAUAUGCU-3ʹ to 5ʹ- GUCUAUUUGCU-3ʹ), which had been described as abrogating the interaction of WNV 5ʹ-3ʹ RNA (17), was named CS-M and used as a negative control. As shown in Fig. 8B, the WT 5ʹ RNA was able to bind 3ʹ RNA to form the 5ʹ-3ʹ RNA complex, with an estimated dissociation constant (Kd) of 0.47 μM, whereas CS-M completely abolished the 5ʹ-to-3ʹ RNA interaction. It suggested that the RNA binding assay is a reliable method to study the long-range RNA interaction of the ZIKV genome, as described for DENV and WNV in previous studies (15–17). Unlike the A119U mutation, which had no influence on the interaction between 5ʹ and 3ʹ RNAs, the C116G, C117G, and C118G mutations significantly decreased the formation of 5ʹ-3ʹ RNA complexes, with estimated Kds of 2.87, 12.82, and 2.42 μM, respectively. However, the compensatory mutations, C116G plus A119U, C116G plus A120G, C117U, and C118G plus A119G, could partially restore the RNA binding affinity of their parental mutations, with Kds of 2.05, 0.51, 3.12, and 0.52 μM, respectively (Fig. 8B). Our data demonstrated that the single-nucleotide mismatch between the 5ʹ DAR and the 3ʹ DAR is sufficient to affect viral genome cyclization, which is in agreement with previous findings in DENV (16) and WNV (17).
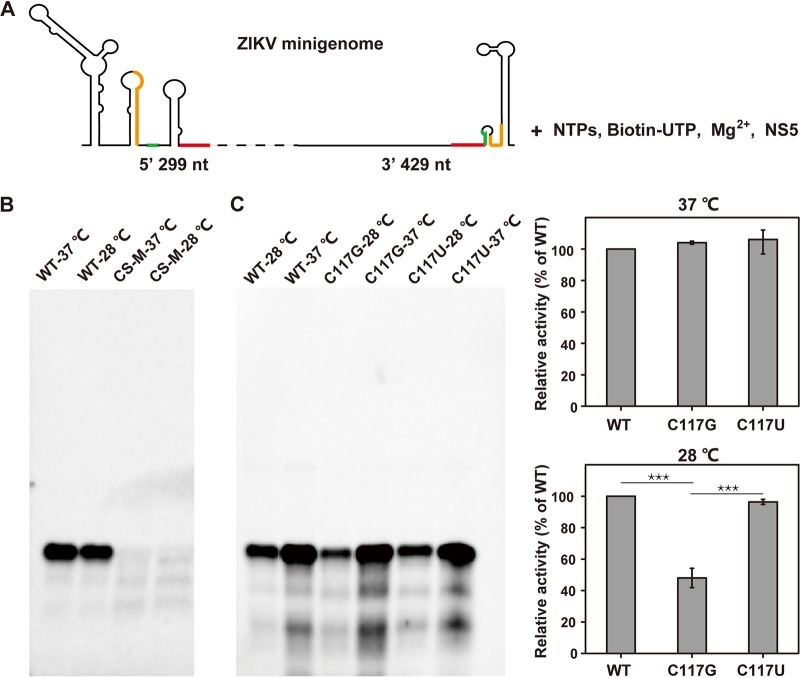
The DAR mutations have a more pronounced influence on de novo RNA synthesis at 28°C than at 37°C.
To investigate the effects of the DAR mutations on viral RNA synthesis, we employed a de novo RdRp assay of ZIKV NS5 using the ZIKV minigenome as the RNA template (Fig. 9A). We first used the WT minigenome and the minigenome bearing the CS-M mutation to test the feasibility of this assay in our hands. The assay was conducted at 37°C and 28°C. As expected, the synthesized RNA product was detectable using the WT template, in contrast to undetectable RdRp activity using the CS-M RNA template at either assay temperature. It should be noted that the efficiency of de novo RNA synthesis at 28°C was a little lower than that at 37°C (Fig. 9B), which may account for the modest delay in replication of the WT virus at the lower temperature (Fig. 7). We next compared the effects of C117G and its C117U pseudoreversion mutation on RNA synthesis. As shown in Fig. 9C, the de novo products in reactions using either the C117G or the C117U minigenome as the template were indistinguishable from that generated by using the WT minigenome as the template at 37°C. However, when the RdRp reaction mixtures were incubated at 28°C, the efficiency of RNA synthesis using the C117G mutant template was significantly reduced, generating 50% less reaction product than the WT, whereas the pseudoreversion C117U could restore the efficiency of RNA synthesis to almost the WT level. These results correlated well with viral replication at 28°C (Fig. 7B and D) between the C117G and C117U mutants. In addition, it indicated that a more adequate genome cyclization is required for de novo initiation of RNA synthesis to support viral replication at mosquito cell growth temperature (28°C). Taken together, these results demonstrated that the DAR-mediated RNA-RNA interaction had a more pronounced influence on de novo RNA synthesis at 28°C than at 37°C, resulting in efficient growth of the mutant viruses in mammalian cells but poor replication in mosquito cells.
FIG 9.
Temperature-dependent effects of DAR mutants on de novo RNA synthesis. (A) Schematic view of the reaction setup of the in vitro RdRp assay. The ZIKV minigenome RNA, which contains the 5ʹ 299-nt and 3ʹ UTR sequences of the genome, was used as the template; the UAR, DAR, and CS elements are labeled in orange, green, and red, respectively. The minigenome RNA template was incubated with a nucleoside triphosphate (NTP) mixture, biotin-UTP, and purified ZIKV NS5 protein in a reaction buffer containing 5 mM MgCl2, as described in Materials and Methods. (B) RdRp activities of NS5 at 37°C and 28°C. The WT minigenome was used as the template in the RdRp reaction; the reaction mixture was incubated at 37°C and 28°C for 90 min. The products were analyzed by formaldehyde-agarose gel electrophoresis, followed by blot analysis, as described in Materials and Methods. The minigenome containing the CS-M mutation was used as a negative control. (C) Effects of C117G and C117U mutations on de novo RNA synthesis at 37°C and 28°C. The WT and the C117G and C117U mutant minigenomes were used for RdRp reactions at 37°C and 28°C. The results were analyzed with ImageJ software, and the activity for each mutant is presented as the percentage relative to WT at each temperature. The data are representative of three independent experiments, and the error bars indicate standard deviations. The statistical analysis was performed using t test analysis with GraphPad Prism software. ***, P < 0.001.
DISCUSSION
Most flaviviruses cycle between insects and vertebrate hosts. Based on their host ranges, they are divided into four phylogenetic groups: MBFVs, tick-borne flaviviruses (TBFVs), insect-specific flaviviruses (ISFVs), and not-known-vector flaviviruses (NKVs) (11). Host range restriction of flaviviruses may occur at different points of the viral life cycle and is regulated by different host/viral factors (45). Increasing evidence indicates that some specific RNA elements in UTRs are under different selective pressures during host adaptation (22, 24) and regulate host-specific replication (22, 23, 46–49).
In this study, it was demonstrated that the DAR element was also able to regulate ZIKV replication by facilitating genome cyclization, as previously reported in WNV and DENV type 2 (DENV-2) (16, 17). In addition, we found that in mosquito cells, the replication of ZIKV required more stringent complementarity of the DAR than in mammalian cells. In mosquito cells, one base-pairing loss between 5ʹ DAR and 3ʹ DAR, except for the one caused by A119U nucleotide substitution, almost completely abolished viral replication, while in mammalian cells, viral replication was not significantly affected until more than one 5ʹ-3ʹ DAR base pairing was impaired. Further experiments indicated that the influence of DAR-mediated genome cyclization on viral RNA synthesis was temperature dependent, based on interpreting the different requirements of the complementarity of the DAR motif for viral replication in mammalian and mosquito cells.
Genome cyclization mediated by long-range RNA-RNA interactions has been demonstrated to be necessary for flavivirus replication. Following the binding of NS5 to the 5′ SLA element, it brings the 3′ SL of genomic RNA close to the NS5-SLA complex to facilitate the initiation of negative-strand RNA synthesis (Fig. 1) (50, 51). Our in vitro RdRp assay revealed that the single-nucleotide mutations affecting 5′-to-3′ DAR interaction impaired RNA synthesis at 28°C instead of 37°C. These data explained why differential complementarity between the 5′ DAR and the 3′ DAR is required for viral replication in mosquito and mammalian cells. Consistently, we also observed such temperature-dependent effects on viral replication capability in both mammalian and mosquito cells (Fig. 7). At 28°C in Vero cells, the C117G mutant virus showed rather poor replication capability in contrast to only slight reduction in viral titers at 37°C. In parallel, C117G mutant virus replicated efficiently in C6/36 cells when the temperature was shifted from 28°C to 37°C. The growth temperature is one of the most important factors impacting viral growth by regulating protein conformation, enzyme activity, and infection dynamics during viral replication (52). Viruses transmitted between different hosts have to resist the stresses of the different hosts’ growth temperatures. A previous study identified a conformational transition of DENV structure when the temperature changed from 37°C to 28°C (53), providing a possible tactic for viral adaption to different host environments. Here, we substantiated that the ZIKV DAR element has different effects on viral replication in mammalian and mosquito cells, suggesting a host-specific function of the DAR element that enables ZIKV to resist the temperature stresses of different hosts.
Additionally, we noticed that even at 37°C, where the C117G mutant virus showed a great increase in replication in C6/36 cells, it still replicated less efficiently than in Vero cells (an ∼100-fold decrease versus an ∼10-fold decrease in viral titers relative to the corresponding WT) (Fig. 7A and C). This result implied that, besides the temperature, some unique host factors in mammalian and mosquito cells might function differently to regulate viral replication. Recently, a number of host factors that interact with flavivirus UTRs have been identified in either mammalian (54–56) or mosquito (57, 58) cells, and one mammalian protein, AUF 1, has even been demonstrated to rearrange stem structures at both ends of the flavivirus genome, assisting genome cyclization and concurrently enabling the RdRp to initiate RNA synthesis. Our future work will focus on elucidating how the host factors regulate viral replication in these two host cells.
The 3′ DAR sequence overlaps the stem of the 3′ sHP element that is present in the linear form of the flavivirus genome. A previous study showed that the mutations that do not alter 3′ sHP structure in DENV were tolerated in mammalian cells but were lethal in mosquito cells, implying that the sequence of the DENV 3′ sHP is host specific (23). The overlapping sequences of the 3′ DAR and the sHP element provide the possibility that the disruption of base pairing of the 5′-3′ DAR is responsible for the replication defect of the reported sHP mutants in mosquito cells. Here, we confirmed that the ZIKV DAR-mediated long-range RNA-RNA interaction has different effects on viral replication in Vero and C6/36 cells. Therefore, we hypothesize that the flavivirus genome cyclization differentially affects viral replication in mosquito and mammalian cells. Additionally, previous studies demonstrating that the interaction between the capsid-coding sequence and DB1 in the 3′ UTR (12) and a pseudoknot formed within DB2 (DB2-PK) in the 3′ UTR (33) are involved in DENV genome cyclization and have more pronounced effects on viral replication in mosquito cells than in mammalian cells also support our hypothesis.
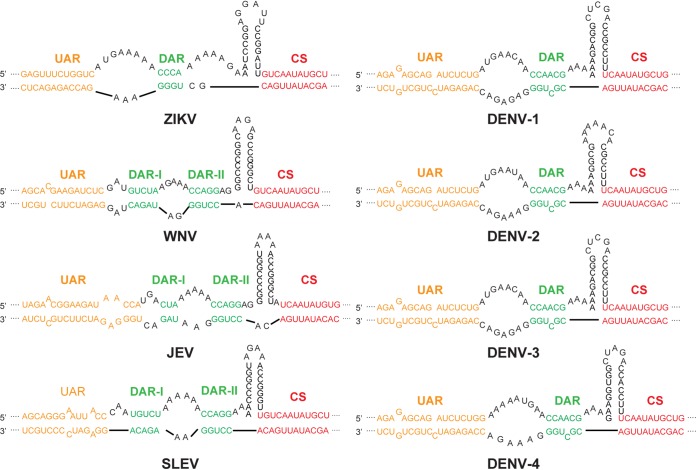
By summarizing the results from previous studies (59, 60) and performing Mfold secondary-structure prediction (61), we analyzed the nucleotide sequences and the structures of the cyclization elements in different MBFVs. In contrast to the high conservation rate of the CS sequences, the DAR sequences were not conserved, but the complementarity between the 5′ DAR and 3′ DAR was maintained in all MBFVs (Fig. 10). Furthermore, it was demonstrated that the compensatory mutations in the revertant viruses of the DAR mutants could restore both DAR base pairing and viral replication (Fig. 6), providing direct evidence that efficient viral replication relies on the complementarity of the DAR, rather than the specific sequence of the DAR. As more stringent DAR complementarity is required for viral replication in C6/36 cells, we suppose that different selective pressures are imposed on the DAR element during host switching and viral RNA evolution, resulting in the stable maintenance of the base pairing of the DAR element but varying primary RNA sequences of DARs among MBFVs. Since the DAR secondary structure is highly conserved among MBFVs, the DAR-mediated genome cyclization is possibly a general strategy for effective viral replication in both mosquito and mammalian hosts. Our findings provide new insights into the functions of the circular form of flavivirus and other positive-strand RNA virus genomes and suggest an evolutionary role for DAR-mediated genome cyclization during host adaption of flaviviruses.
FIG 10.
Long-range RNA-RNA interactions in the MBFV genome. Shown are schematic representations of the UAR, DAR, and CS sequences of different MBFVs based on the predictions of and the structures identified by recent studies (59, 60). The UAR, DAR, and CS elements are labeled in orange, green, and red, respectively. The sequences corresponding to the 5ʹ termini and 3ʹ UTRs of ZIKV (GenBank accession no. KU963796), WNV (EF657887.1), Japanese encephalitis virus (JEV) (U14163), Saint Louis encephalitis virus (SLEV) (NC_007580), DENV-1 (AY277666.2), DENV-2 (HQ541799.1), DENV-3 (AF309641.1), and DENV-4 (JN638571.1) were analyzed with the Mfold Web server (61).
MATERIALS AND METHODS
Cell culture.
Vero (African green monkey kidney) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. The mosquito C6/36 cells were grown in RPMI 1640 medium supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Vero and C6/36 cells were maintained in 5% CO2 at 37°C and 28°C, respectively.
Plasmid construction.
The infectious clone of the ZIKV strain, SZ-WIV01 (GenBank accession no. KU963796) (62), which is driven by a T7 promoter designated pACYC-ZIKV, was used to construct the infectious clone of the reporter virus containing a T154C nucleotide substitution (Rluc-ZIKV-T154C). First, the fragments covering KpnI-T7-5′ UTR-C38 and C-prM-E-AvrII were produced using pACYC-ZIKV as a template, and the Rluc-2A fragment was amplified using pACYC-Rluc-JEV (63) as a template. The three fragments were fused together by two-step overlap PCR, generating the fragment KpnI-T7-5′ UTR-C38-Rluc-2A-C-prM-E-AvrII. The sequence from KpnI and AvrII was inserted into the corresponding site in pACYC-ZIKV to produce pACYC-Rluc-ZIKV. Finally, a capsid fragment with a T154C nucleotide substitution was produced by overlap PCR and replaced with the full-length sequence of the capsid in pACYC-Rluc-ZIKV at PacI and AvrII restriction sites, generating the infectious clone pACYC-Rluc-ZIKV-T154C. The various DAR mutations were generated by overlap PCR, and the fragments with the introduced mutations were engineered into pACYC-Rluc-ZIKV-T154C at KpnI and AscI restriction sites. To construct the infectious clones of the DAR mutants, fragments with the desired DAR mutations were engineered into pACYC-ZIKV at KpnI and AvrII restriction sites.
To construct the cDNA clone of the ZIKV minigenome, the fragment containing the T7 promoter and the 5ʹ-terminal 299 nt and the fragment including the entire 3ʹ UTR (429 nt) were amplified using pACYC-ZIKV as a template. The two fragments were fused by overlap PCR. The resulting fragment was inserted into the pACYC vector by digestion with KpnI and XhoI, generating the ZIKV minigenome plasmid. The minigenome vectors bearing different mutations in the DAR or CS region were constructed by introducing the corresponding mutations into the ZIKV minigenome plasmid. All the plasmids were validated by DNA sequencing analysis before the subsequent experiments.
In vitro RNA transcription.
Following linearization by XhoI and purification with phenol-chloroform, viral genomic RNAs were transcribed from the corresponding linearized cDNA plasmids using a mMessage mMachine T7 transcription kit (Invitrogen) according to the manufacturer’s protocols. To produce ZIKV minigenome RNAs, PCR was performed to amplify the minigenome cDNAs. After purification of the PCR products by phenol-chloroform extraction, the minigenome RNAs were synthesized through in vitro transcription using a MEGAscript T7 transcription kit (Invitrogen). To generate 5ʹ and 3ʹ RNAs for the RNA binding assay, the fragment containing the T7 promoter plus the 5ʹ-terminal 170 nt of the ZIKV genome and the fragment containing the last 116 nt of the genome were amplified by PCR and purified by polyacrylamide gel electrophoresis. The purified products were used as templates for in vitro transcription of the 5ʹ RNA and 3ʹ RNA, respectively, using a MEGAshortscript T7 transcription kit (Invitrogen). The transcribed RNAs were quantified by spectrophotometry and analyzed by agarose gel electrophoresis or PAGE to examine the quality.
Transfection, luciferase assay, and virus recovery.
To obtain the Rluc-ZIKV-T154C mutant viruses, approximately 0.5 μg of the Rluc-ZIKV-T154C mutant RNAs was transfected into 1 × 105 Vero or 3 × 105 C6/36 cells in a 12-well plate using DMRIE-C reagent (Invitrogen). The supernatants of the transfected cells were harvested, and the cell lysates were reclaimed at different time points. Luciferase activity was measured in a microplate reader (Varioskan Flash; Thermo Fisher, Finland) after mixing 20 μl of the lysates with 50 μl of substrate (Promega). The WT ZIKV and the DAR mutant viruses were rescued by transfecting 1 μg of RNAs into 2 × 105 Vero or 6 × 105 C6/36 cells in 35-mm dishes using DMRIE-C reagent. The supernatants of the transfected cells were collected at different time points after transfection, and the viruses were aliquoted and stored at −80°C for use in all experiments.
Immunofluorescence assay (IFA) and plaque assay.
Vero or C6/36 cells were seeded into 35-mm dishes with three preplaced coverslips, followed by transfection with ZIKV genomic RNA. At different time points after transfection, the coverslips containing the transfected cells were fixed in cold (−20°C) 5% acetic acid in methanol for 10 min at room temperature. The fixed cells were washed with phosphate-buffered saline (PBS) three times and incubated with mouse polyclonal antibody against ZIKV NS3 (1:200 dilution with PBS) for 1 h. After washing with PBS three times, the cells were incubated with goat anti-mouse IgG conjugated with fluorescein isothiocyanate (FITC) (1:125 dilution with PBS) at room temperature for another hour. Following three PBS washes, the slides were mounted with 95% glycerol and analyzed under a fluorescence microscope. The virus titer was determined by single-layer plaque assay with a standard procedure as described previously (64).
Serial passage of the DAR mutants in Vero and C6/36 cells.
The supernatants collected from the DAR mutant-RNA-transfected Vero cells were designated P0 viruses and were subjected to three independent serial passages in Vero and C6/36 cells, respectively. For the first round of passage, 2 × 105 Vero cells or 6 × 105 C6/36 cells in 35-mm dishes were infected with P0 viruses at an MOI of 1. The supernatants from the Vero cells were collected at 3 days postinfection (dpi), while the viruses from the C6/36 cells were harvested at 7 dpi; they were designated Vero-P1 and C6/36-P1, respectively. The P1 viruses collected from different cells were blindly passaged in the corresponding cell lines. For the subsequent passages, 1 μl viruses was used to infect naive Vero cells and incubated for 3 days for each passage, whereas 500 μl viruses was used to infect naive C6/36 cells and incubated for 5 to 7 days for each passage. When the viruses were passaged to P5, the total RNA from the infected cells was extracted and used for reverse transcription (RT)-PCR for genome sequencing to analyze the nucleotide changes during viral passage.
RNA binding assay.
The interaction between 5ʹ and 3ʹ RNAs was analyzed with an RNA binding assay using in vitro-transcribed 5ʹ RNA (170 nt) and 3ʹ RNA (116 nt). First, 0.5 μg of 3ʹ RNA and different amounts of 5ʹ RNA (5ʹ/3ʹ RNA molar ratios were 0.5:1, 1:1, 2:1, and 4:1) were mixed in TE buffer (5 mM Tris-HCl, pH 8.0, 0.5 mM EDTA) in a volume of 10 μl. The samples were heat denatured at 95°C for 2 min and immediately placed on ice for 5 min. Then, 4.54 μl 3.3× RNA folding buffer (333 mM HEPES, pH 8.0, 20 mM MgCl2, 333 mM NaCl) was added to the samples and mixed by gentle pipetting. The final reactions were incubated for 30 min to form the 5ʹ-3ʹ RNA complex. The RNA-RNA interactions were analyzed by electrophoresis on native 8% polyacrylamide gels. The gels were prerun in 0.5× TBE buffer for 30 min on ice at 60 V, and then the samples were combined with 5 μl gel loading solution (Invitrogen) and loaded onto the gels. Electrophoresis was carried out at 60 V in 1× TBE buffer for 5 h on ice. The gels were stained with Stains-All (Sigma-Aldrich) for 1 h and scanned with an Epson scanner. Colored images obtained from the scanned gels were converted to grayscale images prior to quantification using ImageJ software. The percentage of bound 3ʹ RNA was calculated by quantifying the band intensities of the 5ʹ-3ʹ RNA complex and the free 3ʹ RNA. The dissociation constant (Kd) was estimated by nonlinear regression analysis (GraphPad Prism software) using the following equation: percent bound 3ʹ RNA = [x]/(Kd + [x]) (11, 15), in which [x] is the concentration of 5ʹ RNA.
In vitro RdRp assay.
The in vitro-transcribed minigenome RNAs were used as templates for the NS5-mediated RdRp reaction. About 1 μg of minigenome RNAs was mixed with 2 μg of the purified ZIKV NS5 protein (kindly provided by Peng Gong and Han-Qing Ye, Wuhan Institute of Virology, Chinese Academy of Science) in the reaction mixture. The standard reaction mixture contained 50 mM Tris-HCl, pH 7.9; 5 mM MgCl2; 50 mM NaCl; 500 μM (each) ATP, GTP, and CTP and 100 μM UTP; and 250 μM biotin-16-UTP (Roche, Switzerland), along with 4 U RNase inhibitor (Roche, Switzerland). The RdRp reaction was carried out at 37°C and 28°C for 90 min and terminated by acid phenol-chloroform extraction. The purified RNA pellets were dissolved in 17 μl RNA sample buffer containing 1.85% formaldehyde and 50% formamide in 1× MOPS buffer (20 mM MOPS [morpholinepropanesulfonic acid], pH 7.0, 5 mM sodium acetate [NaAc], 1 mM Na2-EDTA). The RNA samples were combined with 3 μl gel loading solution and loaded onto a 1.2% agarose gel with 2% formaldehyde. The electrophoresis proceeded at 90 V for 4 h at room temperature. The RNA in the gel was transferred to a Hybond N+ nylon membrane (GE Healthcare, Chicago, IL) by the capillary method and fixed by UV cross-linking. The membrane was then blocked, incubated with streptavidin-conjugated horseradish peroxidase (HRP), washed, and visualized using a chemiluminescent nucleic acid detection module kit (Thermo) and a chemiluminescence system (ChemiDoc; Bio-Rad) according to the manufacturer’s protocols. The image of the blot was analyzed with ImageJ software, and the activities of DAR mutants were expressed as a percentage of the corresponding WT activity.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2018YFA0507201) and the National Natural Science Foundation of China (81572003).
We are grateful to the Wuhan Key Laboratory of Special Pathogens and Biosafety for their helpful support during the course of the work.
REFERENCES
- 1.Fernández-Sanlés A, Ríos-Marco P, Romero-López C, Berzal-Herranz A. 2017. Functional information stored in the conserved structural RNA domains of flavivirus genomes. Front Microbiol 8:546. doi: 10.3389/fmicb.2017.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastere S, Valour F, Baudouin L, Mallet H, Musso D, Ghawche F. 2014. Zika virus infection complicated by Guillain-Barre syndrome—case report, French Polynesia. Euro Surveill 19:20720. [DOI] [PubMed] [Google Scholar]
- 3.Cao-Lormeau V-M, Blake A, Mons S, Lastère S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, Vial A-L, Decam C, Choumet V, Halstead SK, Willison HJ, Musset L, Manuguerra J-C, Despres P, Fournier E, Mallet H-P, Musso D, Fontanet A, Neil J, Ghawché F. 2016. Guillain-Barre syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuler-Faccini L, Ribeiro EM, Feitosa IM, Horovitz DD, Cavalcanti DP, Pessoa A, Doriqui MJ, Neri JI, Neto JM, Wanderley HY, Cernach M, El-Husny AS, Pone MV, Serao CL, Sanseverino MT, Brazilian Medical Genetics Society-Zika Embryopathy Task Force. 2016. Possible association between Zika virus infection and microcephaly—Brazil, 2015. MMWR Morb Mortal Wkly Rep 65:59–62. doi: 10.15585/mmwr.mm6503e2. [DOI] [PubMed] [Google Scholar]
- 5.Beckham JD, Pastula DM, Massey A, Tyler KL. 2016. Zika virus as an emerging global pathogen: neurological complications of Zika virus. JAMA Neurol 73:875–879. doi: 10.1001/jamaneurol.2016.0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ochsenreiter R, Hofacker IL, Wolfinger MT. 2019. Functional RNA structures in the 3′UTR of tick-borne, insect-specific and no-known-vector flaviviruses. Viruses 11:298. doi: 10.3390/v11030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gritsun TS, Gould EA. 2007. Origin and evolution of flavivirus 5′UTRs and panhandles: trans-terminal duplications? Virology 366:8–15. doi: 10.1016/j.virol.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Thurner C, Witwer C, Hofacker IL, Stadler PF. 2004. Conserved RNA secondary structures in Flaviviridae genomes. J Gen Virol 85:1113–1124. doi: 10.1099/vir.0.19462-0. [DOI] [PubMed] [Google Scholar]
- 9.Chiu WW, Kinney RM, Dreher TW. 2005. Control of translation by the 5′- and 3′-terminal regions of the dengue virus genome. J Virol 79:8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holden KL, Harris E. 2004. Enhancement of dengue virus translation: role of the 3′ untranslated region and the terminal 3′ stem-loop domain. Virology 329:119–133. doi: 10.1016/j.virol.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z-Y, Li X-F, Jiang T, Deng Y-Q, Ye Q, Zhao H, Yu J-Y, Qin C-F, Liu Z-Y, Li X-F, Jiang T, Deng Y-Q, Ye Q, Zhao H, Yu J-Y, Qin C-F. 2016. Viral RNA switch mediates the dynamic control. of flavivirus replicase recruitment by genome cyclization. Elife 5:e17636. doi: 10.7554/eLife.17636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Borba L, Villordo SM, Iglesias NG, Filomatori CV, Gebhard LG, Gamarnik AV. 2015. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J Virol 89:3430–3437. doi: 10.1128/JVI.02677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filomatori CV, Iglesias NG, Villordo SM, Alvarez DE, Gamarnik AV. 2011. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J Biol Chem 286:6929–6939. doi: 10.1074/jbc.M110.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khromykh AA, Meka H, Guyatt KJ, Westaway EG. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J Virol 75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol 79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friebe P, Harris E. 2010. Interplay of RNA elements in the dengue virus 5’ and 3’ ends required for viral RNA replication. J Virol 84:6103–6118. doi: 10.1128/JVI.02042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friebe P, Shi PY, Harris E. 2011. The 5’ and 3’ downstream AUG region elements are required for mosquito-borne flavivirus RNA replication. J Virol 85:1900–1905. doi: 10.1128/JVI.02037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan C, Muruato AE, Nunes BT, Luo H, Xie X, Medeiros DB, Wakamiya M, Tesh RB, Barrett AD, Wang T, Weaver SC, Vasconcelos PF, Rossi SL, Shi PY. 2017. A live-attenuated Zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat Med 23:763. doi: 10.1038/nm.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakai M, Yoshii K, Sunden Y, Yokozawa K, Hirano M, Kariwa H. 2014. Variable region of the 3’ UTR is a critical virulence factor in the Far-Eastern subtype of tick-borne encephalitis virus in a mouse model. J Gen Virol 95:823–835. doi: 10.1099/vir.0.060046-0. [DOI] [PubMed] [Google Scholar]
- 20.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, Khromykh AA. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Durbin AP, Karron RA, Sun W, Vaughn DW, Reynolds MJ, Perreault JR, Thumar B, Men R, Lai CJ, Elkins WR, Chanock RM, Murphy BR, Whitehead SS. 2001. Attenuation and immunogenicity in humans of a live dengue virus type-4 vaccine candidate with a 30 nucleotide deletion in its 3’-untranslated region. Am J Trop Med Hyg 65:405–413. doi: 10.4269/ajtmh.2001.65.405. [DOI] [PubMed] [Google Scholar]
- 22.Villordo SM, Filomatori CV, Sanchez-Vargas I, Blair CD, Gamarnik AV. 2015. Dengue virus RNA structure specialization facilitates host adaptation. PLoS Pathog 11:e1004604. doi: 10.1371/journal.ppat.1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villordo SM, Gamarnik AV. 2013. Differential RNA sequence requirement for dengue virus replication in mosquito and mammalian cells. J Virol 87:9365–9372. doi: 10.1128/JVI.00567-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filomatori CV, Carballeda JM, Villordo SM, Aguirre S, Pallares HM, Maestre AM, Sanchez-Vargas I, Blair CD, Fabri C, Morales MA, Fernandez-Sesma A, Gamarnik AV. 2017. Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells. PLoS Pathog 13:e1006265. doi: 10.1371/journal.ppat.1006265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villordo SM, Carballeda JM, Filomatori CV, Gamarnik AV. 2016. RNA structure duplications and flavivirus host adaptation. Trends Microbiol 24:270–283. doi: 10.1016/j.tim.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, Harris E, Garcia-Blanco MA, Ooi EE. 2015. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lodeiro MF, Filomatori CV, Gamarnik AV. 2009. Structural and functional studies of the promoter element for dengue virus RNA replication. J Virol 83:993–1008. doi: 10.1128/JVI.01647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bujalowski PJ, Bujalowski W, Choi KH. 2017. Interactions between the dengue virus polymerase NS5 and stem-loop A. J Virol 91:e00047-17. doi: 10.1128/JVI.00047-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clyde K, Barrera J, Harris E. 2008. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology 379:314–323. doi: 10.1016/j.virol.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu ZY, Li XF, Jiang T, Deng YQ, Zhao H, Wang HJ, Ye Q, Zhu SY, Qiu Y, Zhou X, Qin ED, Qin CF. 2013. Novel cis-acting element within the capsid-coding region enhances flavivirus viral-RNA replication by regulating genome cyclization. J Virol 87:6804–6818. doi: 10.1128/JVI.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roby JA, Pijlman GP, Wilusz J, Khromykh AA. 2014. Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses 6:404–427. doi: 10.3390/v6020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Edmonds J, Dong H, Shi PY, Khromykh AA. 2010. RNA structures required for production of subgenomic flavivirus RNA. J Virol 84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Borba L, Villordo SM, Marsico FL, Carballeda JM, Filomatori CV, Gebhard LG, Pallares HM, Lequime S, Lambrechts L, Sanchez Vargas I, Blair CD, Gamarnik AV. 2019. RNA structure duplication in the dengue virus 3’ UTR: redundancy or host specificity?. mBio 10:e02506-18. doi: 10.1128/mBio.02506-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gritsun TS, Gould EA. 2007. Origin and evolution of 3’UTR of flaviviruses: long direct repeats as a basis for the formation of secondary structures and their significance for virus transmission. Adv Virus Res 69:203–248. [DOI] [PubMed] [Google Scholar]
- 35.Villordo SM, Alvarez DE, Gamarnik AV. 2010. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA 16:2325–2335. doi: 10.1261/rna.2120410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis WG, Basu M, Elrod EJ, Germann MW, Brinton MA. 2013. Identification of cis-acting nucleotides and a structural feature in West Nile virus 3’-terminus RNA that facilitate viral minus strand RNA synthesis. J Virol 87:7622–7636. doi: 10.1128/JVI.00212-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elghonemy S, Davis WG, Brinton MA. 2005. The majority of the nucleotides in the top loop of the genomic 3’ terminal stem loop structure are cis-acting in a West Nile virus infectious clone. Virology 331:238–246. doi: 10.1016/j.virol.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Neeleman L, Olsthoorn RC, Linthorst HJ, Bol JF. 2001. Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA. Proc Natl Acad Sci U S A 98:14286–14291. doi: 10.1073/pnas.251542798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newburn LR, White KA. 2015. cis-acting RNA elements in positive-strand RNA plant virus genomes. Virology 479-480:434–443. doi: 10.1016/j.virol.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 40.Nicholson BL, White KA. 2014. Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nat Rev Microbiol 12:493–504. doi: 10.1038/nrmicro3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang B, Dong H, Ye H, Tilgner M, Shi PY. 2010. Genetic analysis of West Nile virus containing a complete 3’CSI RNA deletion. Virology 408:138–145. doi: 10.1016/j.virol.2010.09.033. [DOI] [PubMed] [Google Scholar]
- 42.Polacek C, Foley JE, Harris E. 2009. Conformational changes in the solution structure of the dengue virus 5’ end in the presence and absence of the 3’ untranslated region. J Virol 83:1161–1166. doi: 10.1128/JVI.01362-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alvarez DE, Filomatori CV, Gamarnik AV. 2008. Functional analysis of dengue virus cyclization sequences located at the 5’ and 3’UTRs. Virology 375:223–235. doi: 10.1016/j.virol.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki R, Fayzulin R, Frolov I, Mason PW. 2008. Identification of mutated cyclization sequences that permit efficient replication of West Nile virus genomes: use in safer propagation of a novel vaccine candidate. J Virol 82:6942–6951. doi: 10.1128/JVI.00662-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Junglen S, Korries M, Grasse W, Wieseler J, Kopp A, Hermanns K, León-Juárez M, Drosten C, Kümmerer BM. 2017. Host range restriction of insect-specific flaviviruses occurs at several levels of the viral life cycle. mSphere 2:e00375-16 . doi: 10.1128/mSphere.00375-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cahour A, Pletnev A, Vazielle-Falcoz M, Rosen L, Lai CJ. 1995. Growth-restricted dengue virus mutants containing deletions in the 5’ noncoding region of the RNA genome. Virology 207:68–76. doi: 10.1006/viro.1995.1052. [DOI] [PubMed] [Google Scholar]
- 47.Blaney JE Jr, Sathe NS, Goddard L, Hanson CT, Romero TA, Hanley KA, Murphy BR, Whitehead SS. 2008. Dengue virus type 3 vaccine candidates generated by introduction of deletions in the 3’ untranslated region (3’-UTR) or by exchange of the DENV-3 3’-UTR with that of DENV-4. Vaccine 26:817–828. doi: 10.1016/j.vaccine.2007.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Men R, Bray M, Clark D, Chanock RM, Lai CJ. 1996. Dengue type 4 virus mutants containing deletions in the 3’ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J Virol 70:3930–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. 2005. Role of RNA structures present at the 3’UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 50.Nomaguchi M, Ackermann M, Yon C, You S, Padmanabhan R, Padmanbhan R. 2003. De novo synthesis of negative-strand RNA by Dengue virus RNA-dependent RNA polymerase in vitro: nucleotide, primer, and template parameters. J Virol 77:8831–8842. doi: 10.1128/jvi.77.16.8831-8842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ackermann M, Padmanabhan R. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem 276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- 52.Mojica KD, Brussaard CP. 2014. Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol Ecol 89:495–515. doi: 10.1111/1574-6941.12343. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. 2013. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc Natl Acad Sci U S A 110:6795–6799. doi: 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Friedrich S, Schmidt T, Geissler R, Lilie H, Chabierski S, Ulbert S, Liebert UG, Golbik RP, Behrens SE. 2014. AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. J Virol 88:11586–11599. doi: 10.1128/JVI.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Friedrich S, Engelmann S, Schmidt T, Szczepankiewicz G, Bergs S, Liebert UG, Kummerer BM, Golbik RP, Behrens SE. 2018. The host factor AUF1 p45 supports flavivirus propagation by triggering the RNA switch required for viral genome cyclization. J Virol 92:e01647-17. doi: 10.1128/JVI.01647-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA. 2011. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3’ UTR structures. RNA Biol 8:1173–1186. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yocupicio-Monroy M, Padmanabhan R, Medina F, del Angel RM. 2007. Mosquito La protein binds to the 3’ untranslated region of the positive and negative polarity dengue virus RNAs and relocates to the cytoplasm of infected cells. Virology 357:29–40. doi: 10.1016/j.virol.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 58.De Nova-Ocampo M, Villegas-Sepúlveda N, del Angel RM. 2002. Translation elongation factor-1alpha, La, and PTB interact with the 3’ untranslated region of dengue 4 virus RNA. Virology 295:337–347. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- 59.Li P, Wei Y, Mei M, Tang L, Sun L, Huang W, Zhou J, Zou C, Zhang S, Qin CF, Jiang T, Dai J, Tan X, Zhang QC. 2018. Integrative analysis of Zika virus genome RNA structure reveals critical determinants of viral infectivity. Cell Host Microbe 24:875–886 e5. doi: 10.1016/j.chom.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 60.Huber RG, Lim XN, Ng WC, Sim AYL, Poh HX, Shen Y, Lim SY, Sundstrom KB, Sun X, Aw JG, Too HK, Boey PH, Wilm A, Chawla T, Choy MM, Jiang L, de Sessions PF, Loh XJ, Alonso S, Hibberd M, Nagarajan N, Ooi EE, Bond PJ, Sessions OM, Wan Y. 2019. Structure mapping of dengue and Zika viruses reveals functional long-range interactions. Nat Commun 10:1408. doi: 10.1038/s41467-019-09391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng C, Liu S, Zhang Q, Xu M, Zhang H, Gu D, Shi L, He J, Xiao G, Zhang B. 2016. Isolation and characterization of Zika virus imported to China using C6/36 mosquito cells. Virol Sin 31:176–179. doi: 10.1007/s12250-016-3778-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li XF, Li XD, Deng CL, Dong HL, Zhang QY, Ye Q, Ye HQ, Huang XY, Deng YQ, Zhang B, Qin CF. 2017. Visualization of a neurotropic flavivirus infection in mouse reveals unique viscerotropism controlled by host type I interferon signaling. Theranostics 7:912–925. doi: 10.7150/thno.16615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng CL, Zhang QY, Chen DD, Liu SQ, Qin CF, Zhang B, Ye HQ. 2017. Recovery of the Zika virus through an in vitro ligation approach. J Gen Virol 98:1739–1743. doi: 10.1099/jgv.0.000862. [DOI] [PubMed] [Google Scholar]