Abstract
The metabolic characteristics of tumors present considerable hurdles to immune cell function and cancer immunotherapy. Using a glutamine antagonist, we metabolically dismantled the immunosuppressive microenvironment of tumors. We demonstrate that glutamine blockade in tumor-bearing mice suppresses oxidative and glycolytic metabolism of cancer cells, leading to decreased hypoxia, acidosis, and nutrient depletion. By contrast, effector T cells responded to glutamine antagonism by markedly up-regulating oxidative metabolism and adopting a long-lived, highly activated phenotype. These divergent changes in cellular metabolism and programming form the basis for potent antitumor responses. Glutamine antagonism therefore exposes a previously undefined difference in metabolic plasticity between cancer cells and effector T cells that can be exploited as a “metabolic checkpoint” for tumor immunotherapy.
Cancer cells have prodigious anabolic and energetic requirements and use distinct metabolic pathways for growth and survival (1, 2). Termed the Warburg effect, cancer cells are highly glycolytic, metabolizing glucose to lactic acid to produce adenosine triphosphate (ATP) and regenerate nicotinamide adenine dinucleotide (NAD+) (1, 3). In addition to glucose, glutamine plays an important role in providing both carbon and nitrogen necessary for anabolic metabolism (3, 4). As glucose is metabolized through glycolytic pathways to lactic acid, glutamine can fuel the tricarboxylic acid (TCA) cycle, generating metabolic intermediates to serve as building blocks for lipids, proteins, and nucleic acids that are crucial to anabolic growth and proliferation. Notably, these same metabolic programs have also been attributed to facilitating the tremendous growth associated with T cell activation and proliferation (5, 6). An important consequence of the specialized metabolism of cancer cells is the creation of a hypoxic, acidic, nutrient-depleted tumor microenvironment (TME) that is hostile to antitumor immune responses (7–14). In other words, the specialized metabolic programming of cancer cells not only promotes tumor growth but can also block the generation of an effective antitumor immune response.
We hypothesized that blocking glutamine metabolism would not only inhibit tumor growth but also relieve a master immunometabolic checkpoint and enable restoration of antitumor immunity. Glutaminase facilitates the conversion of glutamine to glutamate, and the subsequent conversion of glutamate to α-ketoglutarate represents a major pathway for glutamine carbons to enter the TCA cycle (1, 3). However, the therapeutic response of tumors in vivo to targeted blockade of glutaminase is generally limited (15–17). Instead of inhibiting a single enzyme, we chose to comprehensively inhibit glutamine metabolism by using drugs based on the glutamine antagonist 6-diazo-5-oxo-l-norleucine (DON). In addition to glutaminase, DON inhibits a broad range of glutamine-requiring enzymes (18). Consequently, DON exposure potently inhibits tumor cell viability, proliferation, and cell cycle progression (fig. S1, A to E). Indeed, DON has previously been evaluated in a number of clinical trials for cancer, but these pursuits were eventually abandoned secondary to unacceptable toxicity, particularly to the gastrointestinal tract (19–24). Given the extra-ordinary potential of DON to not only inhibit tumor growth but also condition the TME and enhance immunotherapy, we have developed a series of prodrugs of DON that are designed to circulate intact and inert but are activated in the TME upon enzymatic cleavage of the promoieties, thus mitigating the previously reported toxicity of glutamine antagonists on susceptible tissues such as the gut (fig. S2, A and B) (19, 25). Of note, although we do observe an enrichment of the active, parent compound, DON, in tumor over plasma in our mouse model (fig. S2C), these prodrugs are much more rapidly activated in rodents compared with larger animals (25–27). Nonetheless, we established a robust therapeutic window for in vivo mouse studies using the compound JHU083 to explore whether tumor glutamine metabolism can be targeted as an immunologic checkpoint (fig. S2, A to C) (19, 25, 28–30).
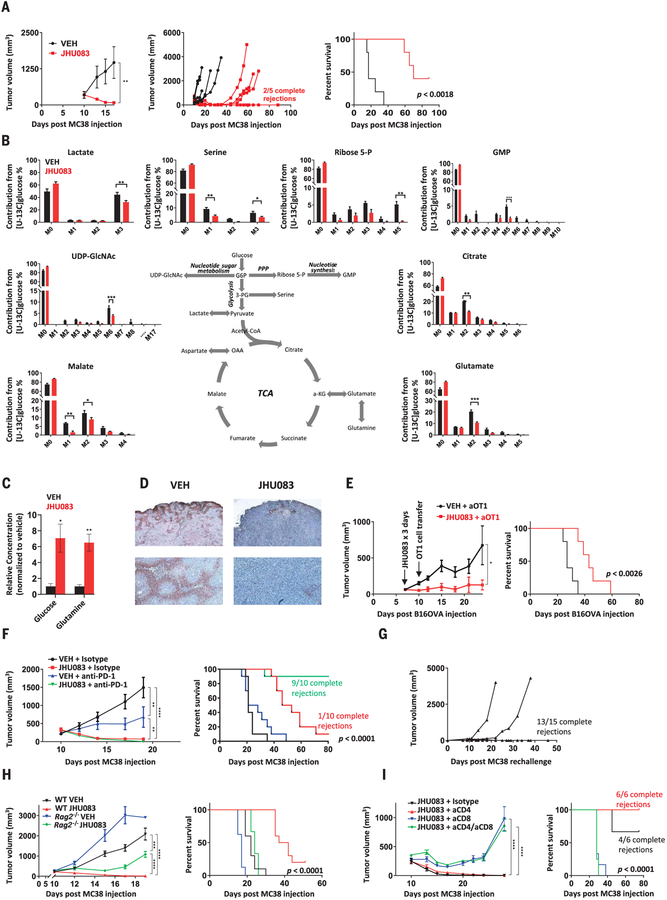
To examine the ability of JHU083 to inhibit tumor growth, we performed a series of experiments using syngeneic tumor models. Mice were injected subcutaneously with MC38 colon cancer, EL-4 lymphoma, CT26 colon cancer, or B16 melanoma. For each tumor type tested, treatment with JHU083 led to a marked decrease in tumor growth and improved survival (Fig. 1A and fig. S2, D to F). In the case of MC38, JHU083 monotherapy for 14 days led to a substantial proportion of durable responses. Given the ability of JHU083 to cause tumor regression, we used the MC38 in vivo model to examine the specific effects on tumor metabolism. Using 13C-glucose tracing with mass spectrometry, we observed differential enrichment of glucose metabolites in metabolic pathways. Blocking glutamine metabolism inhibited glucose metabolism through the TCA cycle, as well as through glycolysis and related pathways, thus disabling Warburg physiology (Fig. 1B and fig. S2G). We also noted a significant decline in glucose contribution to purine synthesis, the hexosamine pathway, and the pentose phosphate pathway (Fig. 1B). This broad suppression of metabolic activity was accompanied by a significant increase in glutamine and glucose content within tumors (Fig. 1C), as well as a decrease in tumor hypoxia (Fig. 1D). Such findings suggest that blocking glutamine metabolism can critically disrupt the metabolism of the tumor as a whole, with marked effects on the nutrient milieu within the TME.
Fig. 1. Glutamine blockade suppresses cancer cell metabolic programs and enhances antitumor immune response.
(A) Average tumor growth curve (left), spider plots (center), and survival curve (right) from vehicle (VEH)– and JHU083-treated MC38-bearing mice. (B) In vivo 13C-glucose tracing experiment in MC38 tumor–bearing mice. M, unlabeled mass of isotope; M+n, native metabolite mass (M) plus number of isotopically labeled carbons (n); UDP-GlcNAc, uridine diphosphate N-acetylglucosamine; G6P, glucose-6-phosphate; GMP, guanosine monophosphate; 3-PG, d-glycerate 3-phosphate; OAA, oxaloacetate; α-KG, α-ketoglutarate. (C) Relative mass spectrometric quantification of glucose and glutamine in MC38 tumors from vehicle- and JHU083-treated mice (per milligram tumor tissue, normalized to vehicle group). (D) Pimonidazole immunohistochemistry staining for hypoxia in tumor sections from vehicle- and JHU083-treated mice. (E) B16OVA-bearing C57BL/6 mice treated with JHU083 or vehicle on days 7 to 9 after tumor inoculation received 1.5 × 106 activated OTI T cells on day 10. Tumor growth curve (left) and survival curve (right) are shown. (F) MC38-bearing C57BL/6 mice treated with vehicle, JHU083, anti– PD-1, or combination JHU083 and anti–PD-1 beginning on day 10 after tumor inoculation. Tumor growth curve (left) and survival curve (right) are shown. (G) Mice initially cured with 14 days of JHU083 treatment were rechallenged ≥30 days after last dose of JHU083; spider plots of tumor volume are shown. (H) MC38-bearing C57BL/6 wild-type (WT) and Rag2−/− mice treated with 14 days of vehicle or JHU083. Average tumor volume (until first sacrifice in WT VEH group) (left) and survival curve (right) are shown. (I) MC38-bearing C57BL/6 mice treated with JHU083 after depletion of CD8 cells, CD4 cells, or both compared with isotype control. Tumor growth curve (left) and survival curve (right) are shown. Error bars represent SEM. Data are representative of one (B), three [(A), (E) and (F)], or five (D) independent experiments with n = 3 to 10 mice per group. Tumor growth curves were assessed by two-way analysis of variance (ANOVA). Log-rank (Mantel-Cox) tests were performed for survival data. Metabolite data assessed with two-tailed Student’s t test for multiple comparisons. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 using two-tailed Student’s t test.
Given the pronounced changes observed in the TME, we asked whether targeting glutamine metabolism would enhance the efficacy of immunotherapy. We were initially concerned that blocking glutamine metabolism would severely impair lymphocyte function, thus metabolic treatment was administered before initiating adoptive cell transfer (ACT) immunotherapy. Mice harboring ovalbumin (OVA)– expressing B16 melanoma (B16OVA) were pretreated with JHU083 for 3 days before adoptive transfer of activated OVA-specific CD8+ T cells (OT1 cells). Treated mice showed improvement in tumor control and survival (Fig. 1E), suggesting that glutamine blockade can condition the TME to enhance ACT. Next, we combined glutamine targeting with programmed cell death protein 1 (PD-1) checkpoint immunotherapy. Because it is possible that glutamine antagonism could suppress antitumor immune cells, we designed our initial treatment regimens to use sequential dosing of JHU083 for a defined period of several days, followed by anti–PD-1 treatment. However, we found that the most efficacious regimens for inducing antitumor response required concurrent, not sequential, dosing of JHU083 and anti–PD-1. To this end, we found that tumor-bearing mice treated with concurrent dosing of the glutamine antagonist and anti–PD-1 showed dramatically improved antitumor effects compared with anti–PD-1 therapy alone, with complete response rates approaching 100% in the MC38 model (versus no complete response in mice receiving anti–PD-1 monotherapy) (Fig. 1F and fig. S2H). Importantly, these findings indicate that blockade of glutamine metabolism, despite the established role of glutamine for activated lymphocyte proliferation and function, does not metabolically disable antitumor immune cells, but to the contrary, appears to robustly enhance their function and effectiveness.
These findings, in addition to the observed ability of glutamine antagonism to promote durable tumor remissions as a single agent (Fig. 1A), led us to investigate the intriguing hypothesis that blocking glutamine metabolism alone could enhance endogenous antitumor responses, even in the absence of additional immunotherapy. To test this, we rechallenged mice that had been cured with JHU083 monotherapy with an equal burden of the same tumor injected on the opposite flank. Notably, almost all of the animals cured by single-agent JHU083 completely rejected the tumor upon rechallenge (Fig. 1G), suggesting the establishment of immunologic memory. To confirm the immunologic basis of this phenomenon, we compared JHU083 treatment of MC38-bearing Rag2−/− mice and wild-type mice. RAG2-deficient mice lack the V(D)J recombinase machinery necessary for rear-ranging antigen receptor genes and, as such, do not produce mature B or T lymphocytes and are incapable of mounting an adaptive immune response. Although glutamine blockade with JHU083 had some initial effect on tumor growth in the Rag2−/− mice, tumor growth rates recovered after several days and the animals succumbed to the disease in a similar time frame to untreated wild-type mice (Fig. 1H). Using T cell depleting anti-bodies (31), we were able to demonstrate that the efficacy of glutamine antagonist therapy was completely dependent upon the activity of the CD8+ T cell subset of the adaptive immune response (Fig. 1I). These observations demonstrate that glutamine blockade, without additional immunotherapy, markedly enhances endogenous antitumor immunity and, more generally, that tumor immune rejection and adaptive immune memory can be triggered solely through this metabolic intervention.
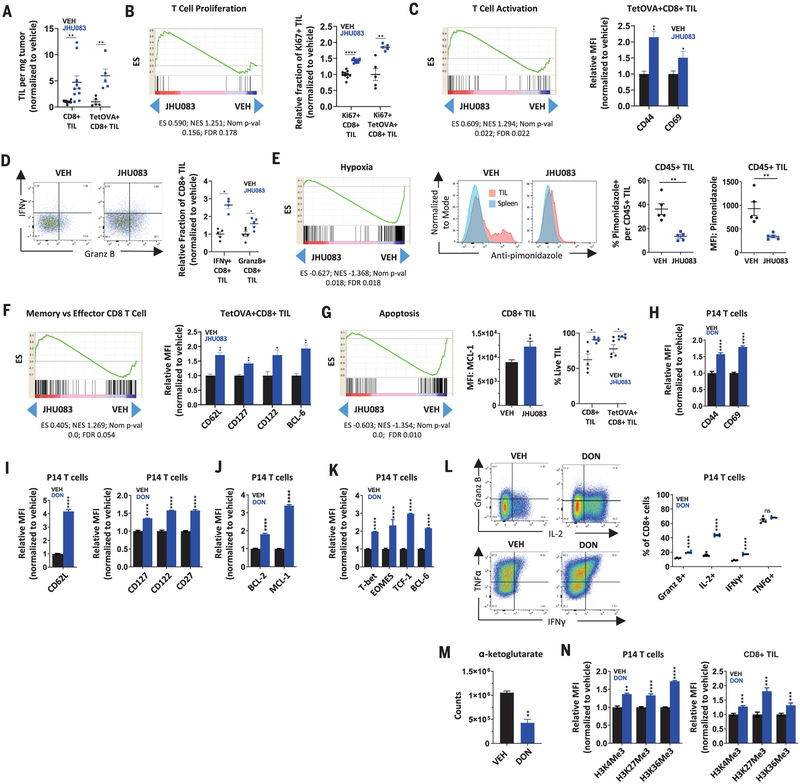
To further investigate the mechanism of this enhanced antitumor response to JHU083 monotherapy, we examined tumor-infiltrating lymphocytes (TILs) from vehicle- and JHU083-treated mice bearing either MC38 or OVA-expressing MC38 tumors. OVA-expressing MC38 tumors (M38OVA) allow identification and study of antigen specific CD8+ T cells by tetramer staining. Consistent with our depletion studies, we observed a marked increase in infiltrating CD8+ T cells with JHU083 treatment, and a greater percentage of these TILs were antigen specific (Fig. 2A). To explore the mechanism of the enhanced endogenous antitumor immunity, we performed RNA sequencing (RNA-seq) on CD8+ TILs sorted by fluorescence-activated cell sorting (FACS). RNA-seq revealed distinct differences between TILs from treated and untreated mice (fig. S3, A and B), with 4313 genes differentially expressed in a statistically significant manner. Gene set enrichment analyses (GSEA) showed that CD8+ TILs from treated mice had transcriptional programs consistent with a highly proliferative (Fig. 2B), robustly activated (Fig. 2C), less exhausted and less anergic phenotype (fig. S3C). These transcriptional changes were also reflected at the protein level. FACS analyses of CD8+ TILs from MC38 tumors and of tetramer-positive CD8+ TILs from OVA-expressing MC38 tumors showed increased proliferation and activation markers (Fig. 2, B and C, and fig. S3D), a decreased percentage of PD-1+LAG-3+ double-positive TILs (fig. S3E), and enhanced interferon-g (IFN-g) (Fig. 2D), granzyme B (Fig. 2D), and interleukin-2 (IL-2) (fig. S3F) production upon ex vivo stimulation. As was anticipated from our preliminary TME analysis, GSEA also suggested that CD8+ TILs in treated mice experience less hypoxia and oxidative stress (Fig. 2E and fig. S3G), which was corroborated by flow cytometry showing decreased pimonidazole (a marker of exposure to low oxygen tension) staining on CD45+ TILs (Fig. 2E). Furthermore, we observed a relative up-regulation of transcripts associated with long-lived memory T cells (Fig. 2F) and a marked down-regulation of apoptotic transcriptional programs in TILs from mice treated with glutamine blockade (Fig. 2G and fig. S3, H and I). Indeed, increased expression of memory markers (CD62L, CD127, CD122, and BCL-6) (Fig. 2F and fig. S3, J and K) and increased expression of anti-apoptotic protein MCL-1 (Fig. 2G) were also observed by flow cytometry. We also confirmed by reverse transcription polymerase chain reaction (RT-PCR) that apoptotic genes uncovered through leading edge GSEA analysis (fig. S3, H and I) were indeed down-regulated in CD8+ TILs from JHU083-treated mice (fig. S3L). This down-regulation has a tangible effect, as demonstrated by the fact that CD8+ TILs, or tetramer-OVA+CD8+ TILs, were less susceptible to intratumoral cell death in JHU083-treated mice (Fig. 2G, right). In accord with recent reports, these findings suggest that an important mechanism of CD8+ T cell dysfunction and tumor immune evasion may be apoptosis of tumor-specific T cells within the TME (32). Blocking glutamine metabolism led to similar changes when combined with anti–PD-1 treatment (fig. S3, D, E, J, and K). Taken together, our results demonstrate that glutamine blockade conditions TILs toward a long-lived, memory-like phenotype that is highly proliferative, markedly activated, and capable of enhanced effector function.
Fig. 2. Glutamine blockade conditions CD8+ TILs toward a highly proliferative, activated, and long-lived phenotype.
C57BL/6 mice with MC38 or MC38OVA tumors were treated on days 14 to 18 with vehicle or JHU083 (0.3 mg per kilogram of body weight per day) and sacrificed on day 18 for analysis. (A) Percentage of live CD8+ or TetOVA+CD8+ TILs per tumor weight for MC38 and MC38OVA models, respectively. TetOVA, tetramer-OVA. (B) GSEA tracing for positive regulation of αβ T cell proliferation (left) and percentage of Ki67+ cells per CD8+ in MC38 model or TetOVA+CD8+ in MC38OVA model (right). ES, enrichment score; NES, normalized enrichment score; FDR, false discovery ratio. (C) GSEA tracing for αβ T cell activation (left) and mean fluorescence intensity (MFI) of CD44 and CD69 of TetOVA+CD8+ TILs in MC38OVA model (right). (D) Relative fraction (normalized to vehicle) of IFN-g+ and granzyme B+ per CD8+ TILs from the MC38OVA model after ex vivo stimulation with SIINFEKL peptide for 4 hours. (E) GSEA tracing for hypoxic exposure (left) and FACS plots (center) and data summary (right) showing percentage of pimonidazole positive and pimonidazole MFI of CD45+ TILs in the MC38 model. (F) GSEA tracings for memory versus KLRG1high effector CD8+ T cells (left) and relative MFI of CD62L, CD127, CD122, and BCL-6 of TetOVA+CD8+ TILs (right). (G) GSEA tracing for apoptosis (left), MFI of MCL-1 on CD8+ in MC38 model (center), and live-cell percentage of CD8+ TILs in MC38 model and TetOVA+CD8+ in MC38OVA model (right). (H to K) Naïve P14 T cells activated in the presence of vehicle or DON (1 mM) for 2 days, rested in IL-2 for two additional days with vehicle or DON, and analyzed by FACS for activation markers (H), memory markers (I), survival markers (J), and transcription factors (K). (L) P14 T cells activated for 2 days as described and rested for 4 days in the presence of vehicle or DON before restimulation (with no drug present) for 4 hours. Flow plots (left) and data summary (right) for intracellular cytokines are shown. (M) Relative α-ketoglutarate levels at 36 hours post–P14 activation. (N) H3 histone trimethylation levels in P14 T cells 3 days post-activation (left) and CD8+ TILs (right). H3K4Me3, trimethylated histone H3 lysine 4; H3K27Me3, trimethylated histone H3 lysine 27; H3K36Me3, trimethylated histone H3 lysine 36. Error bars represent SEM. For MC38OVA experiments, FACS data are representative of two or three independent experiments with n = 5 mice per group. For MC38 experiments, FACS summary plots are combination data from two independent experiments with n = 5 mice per group. For RNA-seq, data are from treated and untreated groups of five mice in each group. In vitro experiments are representative of three to five independent experiments with n = 3 to 6. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and ns is not significant using two-tailed Student’s t test.
Our initial studies were motivated by the hypothesis that targeting tumor metabolism would alter the TME to enhance antitumor immune responses. Indeed, the decreased exposure of TILs to hypoxia in treated mice directly supports this mechanism (Figs. 1D and 2E). However, the notable changes observed in the activation and differentiation states of the TILs from treated and untreated mice led us to question whether glutamine antagonism was also directly affecting T cell activation and function. To assess the cell-intrinsic nature of these findings, we activated and expanded naïve CD8+ T cells with anti-CD3, anti-CD28, and IL-2 in the presence or absence of DON. (Because JHU083 requires in vivo enzymatic cleavage of promoieties, DON is used for all in vitro studies.) Much like our in vivo TIL analysis, in vitro activated T cells, in the presence of DON, showed robust up-regulation of activation markers (CD44 and CD69), memory markers (CD62L, CD127, CD122, and CD27), anti-apoptotic proteins (BCL-2 and MCL-1), and transcription factors (T-bet, EOMES, TCF-1, and BCL-6) (Fig. 2, H to K). Moreover, restimulation of DON-treated CD8+ T cells reveals an increased capacity to produce cytokines and granzyme B (Fig. 2L). These phenotypic changes are also observed when glutamine metabolism is inhibited in the context of acute infection with lymphocytic choriomeningitis virus (LCMV) Armstrong (fig. S3, M to O). Transcriptional changes associated with a memory T cell phenotype have previously been reported in response to modulation of α-ketoglutarate–dependent demethylases, including critical epigenetic remodeling enzymes (33, 34). α-ketoglutarate is a proximal metabolite of glutamine metabolism. We observed significant attenuation of α-ketoglutarate levels in DON-treated CD8 T cells 36 hours post-activation (Fig. 2M), with increased methylation (corresponding to decreased activity of histone demethylase enzymes) on a broad range of histone methylation sites (Fig. 2N, left). Importantly, identical changes in histone methylation are evident on TILs from explanted tumors (Fig. 2N, right). These findings demonstrate that glutamine blockade conditions, even in the absence of tumor, activated T cells in a cell-intrinsic manner toward a highly activated, long-lived, memory-like differentiation state.
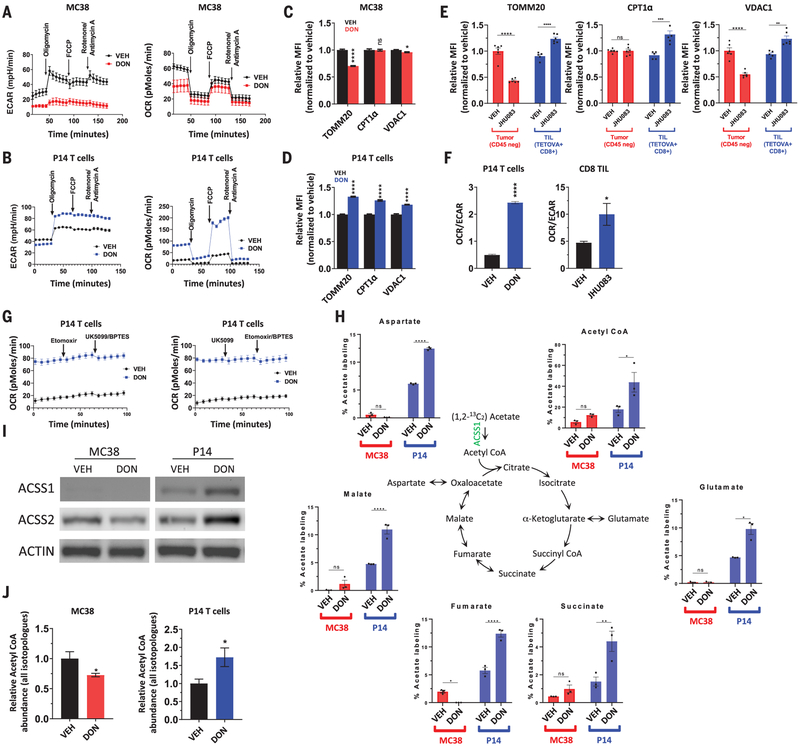
Given the differential ability of glutamine metabolic inhibition to disable tumor cells while enhancing T cell function, we were interested in defining the cell-specific metabolic adaptations of cancer cells and CD8+ effector T cells in response to glutamine blockade. Metabolic flux analyses revealed that glutamine blockade by DON suppresses aerobic glycolysis (i.e., Warburg physiology) in cancer cells and activated CD8+ T cells (Fig. 3, A and B, and fig. S4, A and B) as measured by extracellular acidification rate (ECAR). However, whereas DON profoundly suppresses oxidative phosphorylation (OXPHOS) in cancer cells, DON exposure during CD8+ T cell activation triggers a dramatic up-regulation of oxidative metabolism and spare respiratory capacity (SRC), defined as the ability of cells to increase OXPHOS above baseline (Fig. 3, A and B, and fig. S4C). In DON-treated T cells, but not in MC38 cancer cells, these changes in metabolic programs are associated with up-regulation of mitochondrial proteins TOMM20, CPT1α, and VDAC1 (Fig. 3, C and D). This is correlated with in vivo changes showing differential changes in expression of the same mitochondrial proteins in CD8+ TILs versus CD45-negative tumor cells in the TME (Fig. 3E). Functional changes favoring OXPHOS (with increased SRC) over Warburg physiology in T cells are consistent with metabolic characteristics of a T cell memory phenotype and can be approximated by the oxygen consumption rate (OCR)/ECAR ratio (35). In accord with memory-like phenotypic changes we observed in T cells exposed to JHU083 (Fig. 2, F, I, J, and K), we observed a markedly increased OCR/ECAR ratio in both in vitro T cell experiments as well as in CD8+ TILs sorted from treated and untreated MC38-bearing mice (Fig. 3F). To further explore the different metabolic responses in T cells and cancer cells, we were interested in determining what nutrient or nutrients were fueling increased OXPHOS in T cells undergoing glutamine blockade. Interestingly, we found that DON-treated T cells maintain dramatically increased levels of OXPHOS despite pharmacologic inhibition of mitochondrial transport of long-chain fatty acids, pyruvate, and glutamine with etomoxir, UK0599, and BPTES, respectively (Fig. 3G). Although metabolism of branched-chain amino acids does not appear to be a substantial source of fuel for DON-treated T cells (fig. S4D), 13C-tracing experiments show a dramatic ability of T cells to utilize acetate as a carbon source for the TCA cycle in the setting of glutamine blockade (Fig. 3H). The distinct ability of DON-treated T cells to metabolize acetate as a TCA fuel is supported by the up-regulation of acyl– coenzyme A (CoA) synthetase short-chain family member 1 (ACSS1) and ACSS2 in vitro and in vivo (Fig. 3I and fig. S4E). These findings are in accord with recent reports of the importance of acetate in CD8+ T cell function (36–38). By contrast, DON-treated MC38 cells down-regulate acetate metabolizing enzymes and show minimal incorporation of 13C-labeled acetate in TCA intermediates (Fig. 3, H and I). As such, acetyl-CoA generation is reciprocally affected in MC38 versus CD8+ T cells during glutamine blockade (Fig. 3J). Notably, down-regulation of ACSS1 and ACSS2 in response to glutamine blockade was observed in multiple cancer cell lines, suggesting a general effect of glutamine blockade on cancer cell metabolism (fig. S4F).
Fig. 3. Activated CD8+ T cells and MC38 tumor cells enact distinct metabolic programs in response to glutamine antagonist treatment.
(A to D) Differential metabolic characteristics of vehicle- versus DON-treated MC38 cells and vehicle- versus DON-treated activated P14 CD8+ T cells in vitro. ECAR and OCR from metabolic flux analyses [(A) and (B)]. FCCP, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone. Relative expression of mitochondrial proteins [(C) and (D)]. (E) Differential expression of mitochondrial proteins in CD8+ TILs and CD45-negative tumor cells from explanted MC38 tumors. (F) OCR/ECAR ratio in vehicle- and DON-treated activated P14 CD8+ T cells in vitro (left) and in CD8+ TILs harvested from MC38 murine tumors after vehicle or JHU083 treatment (right). (G) OCR response to etomoxir, UK5099, and BPTES in vehicle-versus DON-treated activated P14 CD8+ T cells in vitro. (H to J) Vehicle-versus DON-treated MC38 cells and vehicle- and DON-treated activated T CD8+ T cells in vitro. Liquid chromatography–mass spectrometry (LC-MS) analysis of TCA intermediates after stable isotope tracing with [1,2-13C] acetate (H). Western blot analysis of ACSS1 and ACSS2 expression. ACTIN is used as a loading control (I). LC-MS analysis of relative acetyl-CoA abundance (J). Error bars represent SEM. Data are representative of three [(A), (C), (D), (E), (G), and (I)] or six [(B) and (F)] independent experiments with n = 3 to 8 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and ns is not significant using two-tailed Student’s t test.
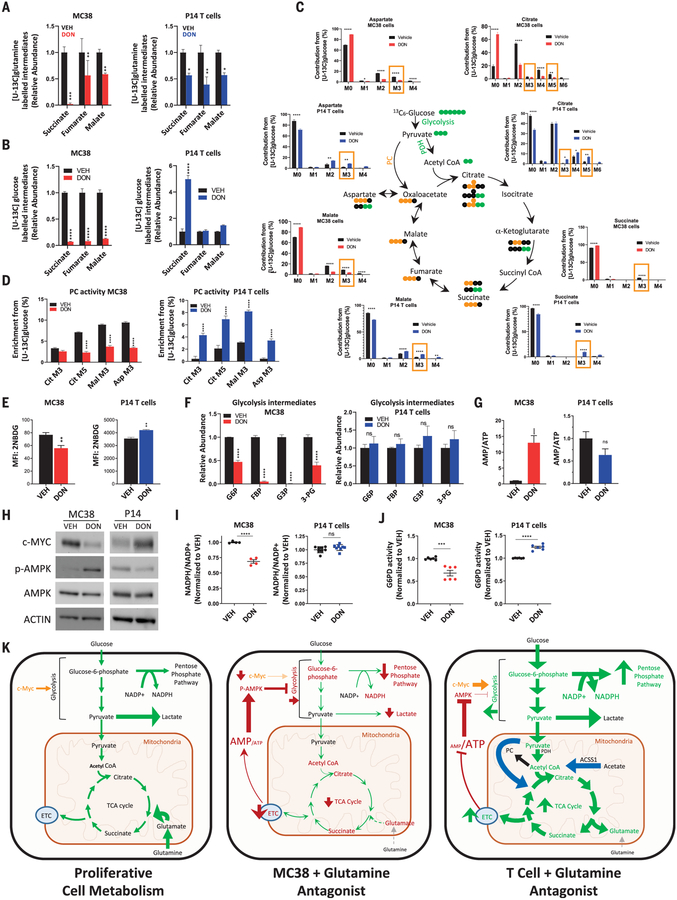
Oxidation of acetyl-CoA through the TCA cycle is a central process in cellular energy metabolism. However, highly proliferative cells, such as activated effector T cells, also use TCA cycle intermediates as building blocks for a range of biomolecules, including nonessential amino acids, lipids, and nucleic acids. Through a process called anaplerosis, these intermediates must be replenished to maintain TCA cycle function. Highly proliferative cells often fuel anaplerosis through glutamine metabolism, or glutaminolysis, which replenishes α-ketoglutarate in the TCA cycle through glutaminase (GLS) activity. Because glutamine contribution to TCA intermediates (as well as other glutamine-requiring processes) is suppressed in DON-treated T cells and MC38 cells (Fig. 4A and fig. S5), we hypothesized that T cells used an alternative anaplerotic source to maintain TCA activity. In this regard, pyruvate carboxylase (PC), which generates oxaloacetate from pyruvate, provides an important alternative anaplerotic pathway by allowing glucose to replenish TCA intermediates (39). As such, it has been reported that PC activity is required for cancer cell survival during targeted GLS inhibition (40). By performing stable-isotope tracing studies with glucose labeled with 13C at all six carbons, we found that DON-treated MC38 cells show a profound suppression of glucose-derived carbons contributing to the TCA cycle, an effect that is not apparent in DON-treated T cells nor in MC38 cells treated with the GLS inhibitor CB839 (Fig. 4B and fig. S6A). In sharp contrast, DON-treated T cells show considerable flexibility in glucose metabolism, glucose-derived carbons, both as acetyl-CoA as well as oxaloacetate through PC-mediated anaplerosis. As such, the pattern of glucose carbons entering the TCA cycle in DON-treated T cells (and GLS-inhibited MC38 cells)—including increased M+3 isotopologues in citrate, malate, and aspartate and M+5 isotopologue in citrate—is indicative of enhanced PC activity (Fig. 4, C and D, and fig. S6B) (41). Unlike DON-treated T cells, this signature is broadly attenuated in DON-treated MC38 cells (Fig. 4D). Notably, PC activity is critically dependent on activation by acetyl-CoA (42, 43). Accordingly, the differential activity of PC (which is expressed equally across each of these cell types and treatments) observed in these cells (fig. S6C) correlates with increased acetyl-CoA levels in DON-treated T cells and decreased acetyl-CoA in DON-treated MC38 cells (Fig. 3J).
Fig. 4. In response to glutamine blockade, activated CD8+ T cells, but not MC38 tumor cells, replenish TCA intermediates by up-regulating glucose anaplerosis.
(A to J) Metabolic characteristics of vehicle- versus DON-treated MC38 cells (red) and vehicle- versus DON-treated activated P14 CD8+ T cells (blue) in vitro. Relative abundance of labeled intermediates of the TCA cycle during [U-13C] glutamine (A) and [U-13C] glucose (B) tracing experiments. LC-MS analysis of TCA intermediates with [U-13C] glucose tracing (C). For (C), in the bar graphs, M3 and M5 values enclosed in yellow rectangles correspond to isotopologs indicative of PC activity, and in the TCA cycle diagram, PDH is pyruvate dehydrogenase; green and yellow circles indicate carbon atoms derived from PDH and PC activity, respectively; and black circles indicate unlabeled carbon atoms. Relative labeling of TCA isotopologues characteristic of PC activity (D). 2-NBDG uptake by flow cytometry analysis (E). Relative abundance of glycotic intermediates in MC38 (left) and P14 T cells (right) (F). G6P, glucose-6-phosphate; FDP, fructose-1,6-bis-phosphate; G3P, d-glyceraldehyde 3-phosphate; 3-PG, d-glycerate 3-phosphate. Relative AMP/ATP ratio (G). Western blot of c-MYC, phospho-AMPK, and total AMPK expression (H). ACTIN is used as a loading control. Relative NADP+/NADPH ratio (I). Relative G6PD activity (J). Error bars represent SEM. Data are representative of three or four independent experiments [(E), (H), (I), and (J)] with n = 3 or 4 [(E), (I), and (J)]. (G) is abundance data compiled from three independent tracing studies. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and ns is not significant using two-tailed Student’s t test. (K) Model depicting relative activity of fundamental metabolic pathways in highly proliferative cells (left), glutamine-inhibited MC38 cancer cells (center), and glutamine-inhibited effector CD8+ T cells (right). Highly proliferative cells in nutrient-rich microenvironments engage high levels of aerobic glycolysis (Warburg physiology), PPP activity, and glutaminolysis to maintain energy, redox, and metabolite homeostasis. Disruption of glutamine metabolism in MC38 cells leads to increased AMP/ATP ratio and decreased c-MYC, such that proximal glycolytic metabolism is suppressed and cells can no longer rely on Warburg physiology, the PPP, or TCA cycle activity. By contrast, activated T cells adapt to glutamine blockade and maintain redox and energy homeostasis by up-regulating OXPHOS through acetate catabolism, generating high levels of acetyl-CoA as a two-carbon source for the TCA cycle, and up-regulating PC activity for glucose anaplerosis. ETC, electron transport chain.
The inability of DON-treated MC38 cells to supply glucose-derived metabolites for the TCA cycle (Fig. 4C) or to maintain Warburg physiology (Fig. 3A) is likely due to the suppressive effects of DON on proximal glycolytic metabolism in these cells. Unlike DON-treated T cells and MC38 cells treated with CB839, DON-treated MC38 cancer cells exhibit decreased glucose uptake and diminished levels of early glycolytic intermediates (Fig. 4, E and F, and fig. S6D). We were interested in the mechanism of glycolysis suppression in MC38 cells treated with glutamine antagonism. Unlike T cells, wherein metabolic adaptations to DON treatment (including up-regulated acetate metabolism and PC activity) allow them to maintain the cellular adenosine monophosphate (AMP)/ATP ratio, MC38 cells are unable to compensate energetically, leading to dramatically increased AMP/ATP ratios and robust activation of AMP kinase (AMPK) (Fig. 4, G and H). Furthermore, MC38 cells exposed to glutamine blockade markedly down-regulate c-MYC (Fig. 4H), a phenomenon previously reported in cancer cells in response to glutamine deprivation (44). We observed this effect in a range of murine cancer cell lines (fig. S4F). As c-MYC and AMPK are critical regulators of glycolytic flux (45–47), the down-regulation of c-MYC and concurrent activation of AMPK play crucial roles in the suppression of glycolytic pathways in DON-treated MC38 cells. Like DON-treated T cells, however, CB839-treated MC38 cells maintain c-MYC expression and AMP/ATP ratios and show no increased AMPK signaling (fig. S6, C, E, and F).
Given the effects of glutamine antagonism on suppressing proximal glycolysis and Warburg physiology in MC38 cells, we hypothesized that parallel metabolic pathways may also be affected. We were particularly interested in the activity of the pentose phosphate pathway (PPP). The PPP is a branching pathway from glycolysis and is the major cellular source for reduced nicotinamide adenine dinucleotide phosphate (NADPH) production, which is vital for maintenance of redox homeostasis. In addition to their known role in regulating glycolytic flux, c-MYC and AMPK also regulate the activity of the PPP (46, 48, 49). Indeed, unlike DON-treated T cells and CB839-treated MC38 cells, DON-treated MC38 cells are unable to maintain NADP(H) homeostasis (Fig. 4I and fig. S6G), indicative of a defect in this pathway. This is further supported by measurement of the activity of the PPP rate-limiting enzyme glucose-6-phosphate dehydrogenase (G6PD), which is correspondingly suppressed in DON-treated MC38 cancer cells but increased in DON-treated CD8+ T cells. (Fig. 4J).
Although tracing experiments confirmed blockade of glutamine pathways during DON or JHU083 treatment (fig. S5), we were interested in assessing the specific contribution of key glutamine-utilizing enzymes to the observed metabolic phenotype (fig. S7A). We used short hairpin RNA (shRNA) lentiviral constructs to generate stable knockdowns of glutamine-utilizing enzymes in the MC38 cell line (fig. S7, B and C). We found that knocking down several of these genes individually (e.g., Gfpt1, Ctps1, and Asns) contributed to c-MYC down-regulation, reduced glucose uptake, and suppressed proliferation (fig. S7, D to F). However, when each of these lines was treated with DON, further down-regulation of c-MYC was observed (fig. S7D). That is, although several knockdown cell lines demonstrated suppressed glucose uptake, down-regulated c-MYC, and attenuated proliferation, no single gene completely accounted for the phenotype that was observed through inhibition of all glutamine-utilizing pathways simultaneously (with DON or JHU083) (fig. S7, D and G).
In contrast to cancer cells, activated CD8+ T cells adapt to glutamine blockade through the up-regulation of acetate metabolism, generating high levels of acetyl-CoA to fuel the TCA cycle directly (as a two-carbon source) and indirectly (as an activator of glucose anaplerosis through increased PC activity) (Fig. 4K). In addition to maintaining energy homeostasis (i.e., AMP/ATP ratio) (Fig. 4G), the ability of T cells to utilize acetate to fuel OXPHOS and produce ATP enables them to divert glucose into the PPP and maintain NADP(H) homeostasis (Fig. 4J). Overall, our studies demonstrate that for cancer cells, the interdependence of glycolysis, OXPHOS, and glutamine metabolism lacks plasticity, such that targeting glutamine metabolism leads to wide-ranging metabolic inhibition, disruption of NADP(H) homeostasis, and stymied growth. Conversely, targeting glutamine metabolism in T cells leads to adaptive metabolic reprogramming with enhanced survival, proliferation, and effector function.
From the time of the pioneering work of Otto Warburg, the study of immune cell metabolism has been linked to our understanding of cancer cell metabolism as a paradigm for the behavior of highly proliferative cells (5, 50–54). Although both activated T cells and cancer cells use Warburg physiology and glutamine metabolism to fuel notably high proliferative rates (2, 54, 55), the precise role of Warburg physiology, an inherently less efficient mode of ATP generation than oxidative respiration, has not been established. Recent theories have suggested that highly proliferative cells engage Warburg physiology to maintain the activity of critical pathways such as the PPP that emanate from the glycolytic pathway (56). We show here that, although increased glucose uptake may fuel increased PPP activity (as observed in DON-treated T cells), the function of this pathway does not require that glucose is metabolized to lactic acid, that is, Warburg physiology. In other words, the PPP runs in parallel with glycolysis but is not necessarily dependent on it. As such, in addition to establishing glutamine metabolism as a metabolic checkpoint for cancer immunotherapy, our studies suggest fundamental insights into the nature and role of Warburg physiology. Moreover, through the application of glutamine blockade, we demonstrate the possibility of differentially modulating the metabolism of cancer cells and antitumor immune cells by exploiting the differential metabolic plasticity of each cell type.
Supplementary Material
ACKNOWLEDGMENTS
We thank W. Xu, E. Thompson, S. Collins, R. Helms, M. Claiborne, and other members of the Powell lab for critical discussion of the manuscript.
Funding: This work was supported by the National Institutes of Health (R01CA226765 to J.D.P. and R01CA229451 to J.D.P. and B.S.S.) and The Bloomberg–Kimmel Institute for Cancer Immunotherapy.
Footnotes
Competing interests: J.D.P., B.S.S., and R.R. are scientific founders, paid consultants (J.D.P. and B.S.S.), and have equity in Dracen Pharmaceuticals. Technology arising in part from the studies described herein were patented by Johns Hopkins University and subsequently licensed to Dracen Pharmaceuticals (JHU083 is currently labeled as DRP-083). R.D.L., J.M.E., B.S.S., R.R., J.A., and J.D.P. are inventors for pending patent application no. PCT/US16/44829, submitted by Johns Hopkins University that covers the use of glutamine analogs, such as JHU083 (DRP-083), for cancer immunotherapy.
Data and materials availability: The chemical structure of JHU083 [ethyl 2-(2-amino-4-methylpentanamido)-DON] has previously been published as (25) and “compound 3” in (19) and “prodrug 4a” in (25). RNA-seq data have been deposited in the National Institutes of Health (NIH) Gene Expression Omnibus (GEO) repository, accession no. GSE120345.
SUPPLEMENTARY MATERIALS
science.sciencemag.org/content/366/6468/1013/suppl/DC1 Materials and Methods Figs. S1 to S7 References Data S1
REFERENCES AND NOTES
- 1.Pavlova NN, Thompson CB, Cell Metab 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolpaw AJ, Dang CV, Trends Cell Biol 28, 201–212 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altman BJ, Stine ZE, Dang CV, Nat. Rev. Cancer 16, 619–634 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Pavlova NN, Thompson CB, EMBO J 36, 1302–1315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frauwirth KA et al. , Immunity 16, 769–777 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Wang R et al. , Immunity 35, 871–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kouidhi S, Ben Ayed F, Benammar Elgaaied A, Front. Immunol 9, 353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kouidhi S, Noman MZ, Kieda C, Elgaaied AB, Chouaib S, Front. Immunol 7, 114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shevchenko I, Bazhin AV, Front. Immunol 9, 1816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prendergast GC, Malachowski WJ, Mondal A, Scherle P, Muller AJ, Int. Rev. Cell Mol. Biol 336, 175–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao E et al. , Nat. Immunol 17, 95–103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivadeneira DB, Delgoffe GM, Clin. Cancer Res 24, 2473–2481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang CH et al. , Cell 162, 1229–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong H, Bullock TN, Front. Immunol 5, 24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidson SM et al. , Cell Metab 23, 517–528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross M et al. , Cancer Res 76, 2329 (2016). [Google Scholar]
- 17.Hensley CT, Wasti AT, DeBerardinis RJ, J. Clin. Invest 123, 3678–3684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinkus LM, Methods Enzymol 46, 414–427 (1977). [DOI] [PubMed] [Google Scholar]
- 19.Lemberg KM, Vornov JJ, Rais R, Slusher BS, Mol. Cancer Ther 17, 1824–1832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch G, Kemeny N, Casper E, Am. J. Clin. Oncol 5, 541–543 (1982). [PubMed] [Google Scholar]
- 21.Earhart RH et al. , Invest. New Drugs 8, 113–119 (1990). [DOI] [PubMed] [Google Scholar]
- 22.Eagan RT, Frytak S, Nichols WC, Creagan ET, Ingle JN, Cancer Treat. Rep 66, 1665–1666 (1982). [PubMed] [Google Scholar]
- 23.Magill GB et al. , Cancer 10, 1138–1150 (1957). [DOI] [PubMed] [Google Scholar]
- 24.Krantz S et al. , J. Natl. Cancer Inst 22, 433–439 (1959).13631504 [Google Scholar]
- 25.Rais R et al. , J. Med. Chem 59, 8621–8633 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Fu J et al. , J. Pharm. Sci 105, 989–995 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Gelder J et al. , Int. J. Pharm 205, 93–100 (2000). [DOI] [PubMed] [Google Scholar]
- 28.JHU083 is referred to as “compound 3” in (19) and “compound 4a” in (25).
- 29.Zhu X et al. , Neuropsychopharmacology 44, 683–694 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nedelcovych MT et al. , J. Neuroimmune Pharmacol 14, 391–400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murter B et al. , Cancer Immunol. Res 7, 244–256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horton BL, Williams JB, Cabanov A, Spranger S, Gajewski TF, Cancer Immunol. Res 6, 14–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chisolm DA et al. , Immunity 47, 251–267.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyrakis PA et al. , Nature 540, 236–241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Windt GJ et al. , Immunity 36, 68–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balmer ML et al. , Immunity 44, 1312–1324 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Qiu J et al. , Cell Reports 27, 2063–2074.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vodnala SK et al. , Science 363, eaau0135 (2019).30923193 [Google Scholar]
- 39.Jitrapakdee S, Vidal-Puig A, Wallace JC, Cell. Mol. Life Sci 63, 843–854 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng T et al. , Proc. Natl. Acad. Sci. U.S.A 108, 8674–8679 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sellers K et al. , J. Clin. Invest 125, 687–698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.St. Maurice M et al. , Science 317, 1076–1079 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Utter MF, Keech DB, J. Biol. Chem 238, 2603–2608 (1963). [PubMed] [Google Scholar]
- 44.Dejure FR et al. , EMBO J 36, 1854–1868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faubert B et al. , Cell Metab 17, 113–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haikala HM, Anttila JM, Klefström J, Front. Cell Dev. Biol 5, 38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JW, Dang CV, Cancer Res 66, 8927–8930 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Kohan AB, Talukdar I, Walsh CM, Salati LM, Biochem. Biophys. Res. Commun 388, 117–121 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV, Cancer Discov 5, 1024–1039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warburg O, Gawehn K, Geissler AW, Z. Naturforsch. B 13, 515–516 (1958). [PubMed] [Google Scholar]
- 51.Bakker A, Klin. Wochenschr 6, 252–254 (1927). [Google Scholar]
- 52.Gerriets VA, Rathmell JC, Trends Immunol 33, 168–173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.MacIver NJ, Michalek RD, Rathmell JC, Annu. Rev. Immunol 31, 259–283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pearce EL, Poffenberger MC, Chang CH, Jones RG, Science 342, 1242454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siska PJ et al. , JCI Insight 2, e93411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liberti MV, Locasale JW, Trends Biochem. Sci 41, 211–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.