Abstract
Over half of cutaneous melanoma tumors have BRAFV600E/K mutations. Acquired resistance to BRAF inhibitors (BRAFi) remains a major hurdle in attaining durable therapeutic responses. In this study we demonstrate that approximately 50–60% of melanoma cell lines with vemurafenib resistance acquired in vitro show activation of RhoA family GTPases. In BRAFi-resistant melanoma cell lines and tumors, activation of RhoA is correlated with decreased expression of melanocyte lineage genes. Using a machine learning approach, we built gene expression-based models to predict drug sensitivity for 265 common anti-cancer compounds. We then projected these signatures onto the collection of TCGA cutaneous melanoma and found that poorly differentiated tumors were predicted to have increased sensitivity to multiple Rho kinase (ROCK) inhibitors. Two transcriptional effectors downstream of Rho, MRTF and YAP1, are activated in the RhoHigh BRAFi-resistant cell lines, and resistant cells are more sensitive to inhibition of these transcriptional mechanisms. Taken together, these results support the concept of targeting Rho-regulated gene transcription pathways as a promising therapeutic approach to restore sensitivity to BRAFi-resistant tumors or as a combination therapy to prevent the onset of drug resistance.
Keywords: BRAF inhibitor, resistance, RhoA, MRTF, YAP1, Melanoma
Introduction
Most cutaneous melanomas have point mutations in V-Raf Murine Sarcoma Viral Oncogene Homolog B (BRAF), a serine/threonine kinase with the V600E/K point mutations being the most common15. These mutations result in constitutive BRAF activity and downstream Mitogen Activated Protein Kinase (MAPK) pathway activation, independent of upstream stimuli. Given the prevalence of these mutations in human melanoma tumors, several drugs have been developed which target mutant BRAF, including vemurafenib and dabrafenib. The most common class of BRAFi resistance mechanisms result in MAPK-reactivation17, 20, 67. This includes alterations in the BRAF gene itself such as BRAF amplification56, 57, or aberrant splice variants45, which can drive resistance to BRAFi therapy. Alterations in other genes in the MAPK pathway such as Mitogen-Activated Protein Kinase Kinase (MEK)7, Neuroblastoma RAS Viral Oncogene Homolog (NRAS)38, Neurofibromin 1 (NF1)70, and others19, 51, 55 also promote resistance to BRAF inhibitors. Receptor Tyrosine Kinase (RTK) activation is another mechanism by which cells can generate BRAFi resistance, at least partially through re-activation of the MAPK pathway, through either upregulation of the receptor itself1, 38, 63, or through increased expression of the RTK ligand71.
The combination of BRAF inhibitors with Mitogen-Activated Protein Kinase Kinase 1/2 (MEK1/2) inhibitors was proposed as an approach to overcome BRAF inhibitor resistance 38 and it is clinically superior to BRAF inhibitor monotherapy against BRAFV600-mutant tumors 4, 25, 47. However, acquired resistance to the BRAF and MEK inhibitor combination is still common20, consistent with non-MAPK pathway resistance mechanisms being important clinically 17, 60. Some MAPK-independent resistance mechanisms result from cancer cell intrinsic, epigenomically driven, adaptive responses to drug pressure early during therapy60. These may result in wide-ranging phenotypic switches resulting in MAPK inhibitor resistance in patients and ultimately relapse during therapy17. Melanoma cells grown without drug pressure stochastically switch between a rapid-cycling cell state and a rare slow-cycling cell state53. These cells are selected for during treatment with a BRAFi, ultimately giving rise to a stable population of resistant cells57, 60. These data are further supported by the observation that BRAFi/MEKi-resistant cells and tumors can be re-sensitized to treatment with BRAF or MEK inhibitors after a “drug holiday” 6, 16, 34.
Among the non-MAPK resistance mechanisms, compensatory activation of other GTPases may be important during the development of drug resistance. The RhoA subfamily (RhoA, RhoB, and RhoC) of GTPases act as molecular switches which regulate actin dynamics. The RhoA and RhoC isoforms are highly similar and often function redundantly in the cell, but in some contexts these two isoforms signal differently69. In melanoma the RhoA subfamily, especially RhoC, promotes invasion and metastasis 5, 22, 50, and inhibition of the RhoA isoform suppresses tumor growth 49. Canonically, the RhoA GTPases (encompassing RhoA, B, and C) promote the formation of actin stress fibers by stimulating G-actin polymerization and inhibiting F-actin depolymerization 14, 59, 61. Actin stress fibers have been shown to be increased in melanoma cells with acquired BRAFi resistance 21 and we confirm and extend that finding here.
In addition to regulating actin dynamics, RhoA GTPases also regulate gene transcription. This occurs, in part, through actin polymerization-dependent activation of Myocardin-Related Transcription Factor (MRTF) and YES Proto-Oncogene 1 (YAP1). MRTF and YAP1 are transcriptional co-activators which, upon activation, translocate into the nucleus and regulate gene transcription. Silencing of MRTF or Serum Response Factor (SRF), a transcription factor by which MRTF modulates gene expression, prevents melanoma metastasis 32. Previously, we have developed a series of MRTF-pathway inhibitors including CCG-203971 and CCG-222740 3, 13, 18 and demonstrated that CCG-203971 prevents melanoma metastasis, induces G1 cell cycle arrest, and reduces growth of melanoma cells 13. YAP1 promotes BRAFi/MEKi resistance in melanoma through suppression of apoptosis via BCL-xL and BIM dysregulation 10, 17, 21, 28. Accumulation of YAP1 protein and enrichment of a YAP1 gene signature has been documented in about 40% of clinical melanoma samples from patients who relapsed on MAPK inhibitor therapies 17.
Previous studies have demonstrated that non-mutational, acquired resistance mechanisms represent a major hurdle in maintaining a durable response to MAPK-directed therapeutics 17. We hypothesize that activation of the RhoA pathway is one such acquired resistance mechanism. In this study, we build upon existing literature to demonstrate that actin stress fiber accumulation and RhoA signaling are elevated in approximately half of the vemurafenib-resistant melanoma cell lines tested and that this mechanism is also active in a significant fraction of clinical tumors. RhoAHigh but not RhoALow-resistant lines are partially re-sensitized to vemurafenib by two structurally distinct ROCK inhibitors. We also demonstrate that RhoA activation is linked to loss of melanocyte lineage genes, a pattern also observed in human tumors. Finally, de-differentiated BRAFi-resistant cells have increased MRTF and YAP1 activation and these cells are more sensitive to pharmacological inhibition of these transcriptional mechanisms. De-differentiation of melanoma cells is a major mechanism of acquired BRAFi-resistance 9, 41, 53, 63, 66 and we have identified signaling alterations commonly associated with de-differentiation. This information is critical for developing therapeutic strategies to target this class of drug-resistant tumors.
Materials and Methods
Cell lines and culture
Selection of Vemurafenib-resistant cells
UACC62 and SK-Mel-19 cells were seeded into 10-cm tissue culture plates at ~30% confluence and grown in DMEM as described below. After the cells had adhered to the plate (~16 h), culture medium was supplemented with 2 μM vemurafenib. Medium was exchanged every 2–3 days for 10 mL of fresh media supplemented with 2 μM vemurafenib. Cells were split at a 1:3 ratio into a new 10-cm tissue culture plate when they reached ~75% confluence (approximately 3–4 weeks) and approximately weekly for each subsequent passage. After two months of selection, cell populations were expanded in vemurafenib-containing media and frozen.
Sources of cells
Three additional pairs of parental (P) and vemurafenib-resistant (R) melanoma lines, M229P/R, M238P/R, and M249P/R cells, were generously provided by Dr. Roger Lo at UCLA 38. SK-Mel-19 and UACC62 cells were obtained from Dr. Maria Soengas at The University of Michigan and were made resistant as described above.
Cells were cultured in DMEM (Gibco #11995-065) supplemented with 10% FBS (Gibco #10437-028) and 1% Antibiotic-Antimycotic reagent (ThermoFisher, Waltham, MA, USA #15240062). Vemurafenib-resistant cells were continuously cultured in the presence of 2 μM vemurafenib. Cells were split at ~75% confluence. Vemurafenib was removed from the culture medium when cells were seeded for experiments (e.g. immunofluorescence staining or qRT-PCR), except where otherwise indicated. Cells were routinely tested for mycoplasma contamination by DAPI staining. STR profiling on all cell lines was performed at the MSU genomics core. In all cases, isogenic pairs of cell lines had the same STR profile.
Cloning
For CRISPR experiments the sgRNA sequences are listed in (Table S1). These guide sequences were cloned into the pLentiCRISPRv2 vector (from Feng Zhang, Addgene plasmid #52961). All guide RNA sequences were confirmed by Sanger sequencing.
All cloning primers are listed in (Table S1). Human RhoAG12V was amplified and N-terminal HA-tagged. This PCR product was used as a template for a second round of PCR amplification to add the Gateway adapter sequences. Human MRTFA was amplified out of the p3xFLAG-MRTFA vector (Addgene plasmid#11978) and tagged with gateway adapters which preserve the N-terminal 3x FLAG tag from the vector. The RhoA and MRTFA PCR products were first cloned into pDONR221 using the Gateway BP Clonase II Enzyme Mix from ThermoFisher (#11789020) using the manufacturer’s protocol. RhoA, MRTFA, and Gus (which is included in the BP reaction kit) were subcloned into the pLX301 lentiviral expression vector (from David Root, Addgene plasmid #25895) using the Gateway LR Clonase II Enzyme mix from ThermoFisher (#11791020). The presence of the correct insert in the final plasmid was confirmed by Sanger sequencing.
Virus Preparation and Infection
HEK-293T cells were seeded into 10-cm plates and were allowed to attach overnight. The next day at approximately 60–70% confluence, the cells were transfected with a plasmid cocktail containing 5000 ng of the pLentiCRISPRv2 or pLX301 plasmid, 3750 ng of psPAX2 (Addgene plasmid #12260), 1250 ng of pMD2.G (Addgene plasmid #12259), and 20 uL of Lipofectamine 2000 in 400 uL of OptiMEM. The next morning the medium was changed to 10 mL of fresh culture medium, and the next day each plate was supplemented with an additional 5 mL of culture medium. After 24 h, the culture medium was harvested and filtered through a 0.45-micron syringe filter. Virus was stored at 4 C and was used within 2 weeks.
Target melanoma cells (e.g. UACC62P/R) were seeded into 10-cm plates and were allowed to attach overnight. The next afternoon at approximately 30% confluence the medium was changed to 10-mL of complete medium and was supplemented with 1 mL of viral supernatant. The next morning, the medium was changed and the cells were incubated an additional 24 h. The cells were then treated with 10 μg/mL puromycin until all the untransformed cells died (approximately 72 h). For all virus experiments, the cells were used within 1–2 passages and each biological replicate for each experiment used a different batch of cells. We did not pick individual clones for the CRISPR cell lines, but instead used a pooled infection approach. Validation of CRISPR knockout efficiency was done by immunoblotting for the target protein.
Compounds and Antibodies
Vemurafenib (#S1267), Y-27632 (#S1049), fasudil (#S1573), and dasatinib (#S1021) were purchased from Sellekchem, Houston, TX, USA. Latrunculin B (#10010631), cytochalasin D (#11330), and erlotinib (#10483) were purchased from Cayman Chemical, Ann Arbor, MI, USA. Rho Inhibitor I (#CT04-A) was purchased from Cyoskeleton Inc, Denver, CO, USA. CCG-222740 18 was synthesized in the lab of Dr. Scott Larsen at the University of Michigan. All compounds were diluted in DMSO to a stock concentration of 10 mM. Compound stock solutions were frozen at −20 °C. Antibodies against YAP1 (#14074), MLC2 (#3672), pMLC2 (#3674), Sox10 (#89356), and pEGFR (#3777) were purchased from Cell Signaling, Danvers, MA, USA. Antibodies against MRTF-A (#sc21558), MRTF-B (#sc98989), and Actin (#sc1616) were purchased from Santa Cruz, Dallas, TX, USA. Donkey anti-Mouse800 (#926-32212), Donkey anti-Goat680 (#926-68074), and Donkey anti-Rabbit680 (#926-68073) immunoblotting secondary antibodies were purchased from LI-COR, Lincoln, NE, USA. Alexa Fluor goat anti-rabbit488 (#A11034) and donkey anti-goat488 (#A11055) were purchased from Invitrogen. Alexa Fluor546 Phalloidin (#A22263) was purchased from ThermoFisher.
qRT-PCR
Cells were cultured and treated as indicated, rinsed once in PBS, and total cellular RNA was harvested with the RNeasy kit purchased from the Qiagen, Hilden, Germany (#74104). RNA was eluted in nuclease-free H2O. cDNA was synthesized using the High-Capacity cDNA RT kit from ThermoFisher (#4368814) from 1000 ng of total RNA, according to the manufacturer’s protocol. qPCR was performed using the SYBR Green PCR Master Mix (#4309155) from ThermoFisher according to the manufacturer’s protocol using an Agilent Mx3000P qPCR instrument. Primers were purchased from Integrated DNA Technologies, San Jose, CA, USA and the primer sequences are included in (Table S1). Primers were designed using the Harvard Primer Bank tool (https://pga.mgh.harvard.edu/primerbank/). Fold-change analysis was performed using the ΔΔCT method.
RNA-Seq sample preparation and data processing
Total cellular RNA was extracted from UACC62P and UACC62R cells (two biological replicates per cell line) using the same method which was used for qPCR experiments. RNA concentration was measured by Qubit and quality control was performed on an Agilent 2100 Bioanalyzer in the MSU Genomics Core. All RNA samples had a RIN score > 8. Barcoded libraries were prepared using the Illumina TruSeq Stranded mRNA Library Preparation Kit on a Perkin Elmer Sciclone G3 robot following manufacturer’s recommendations. Completed libraries were QC’d and quantified using a combination of Qubit dsDNA HS and Caliper LabChipGX HS DNA assays. Libraries were pooled and run on two lanes, and sequencing was performed in a 1×50 bp single-end read format using HiSeq 4000 SBS reagents. Base calling was done by Illumina Real Time Analysis, RTA_ v2.7.7 and output of RTA was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v2.19.0. Sequencing was performed at a depth of >30M reads/sample. Quality control was performed on the FastQ files using FastQC v0.11.5, and reads were trimmed using Trimmomatic v0.33. Reads were mapped using HISAT2 v2.1.0 and analyzed using HTSeq v0.6.1. Differential gene expression was calculated using edgeR. Raw RNA-Seq reads and processed HTSeq read counts are available on GEO under GSE115938.
Immunoblotting
Cells were cultured and treated as indicated, placed on ice, and rinsed once in cold PBS. Cells were lysed in 2x Laemmli Sample Buffer (Biorad, #1610737). Samples were sonicated with a probe sonicator for approximately 5 sec, then boiled at 100 °C for 10 min. Samples were loaded onto a 12% polyacrylamide gel and transferred to Immobilon-FL PVDF Membrane (Millipore Sigma, Burlington, MA, USA, #IPFL00010). Membranes were blocked in 5% BSA + TBS-Tween (1:1000) for 1 h, then incubated in primary antibody overnight at 4 °C. Membranes were washed 3x in TBS-Tween and were then incubated in the appropriate secondary antibody at a 1:20000 dilution for 1 h at room temperature. All antibodies were diluted in blocking buffer. Membranes were washed 3x in TBS-Tween then dried and imaged on a LI-COR Odyssey FC imaging system.
Immunofluorescence staining
Cells were seeded into 8-well chamber slides and were treated as indicated in the figure legends. Cells were fixed with 3.7% formaldehyde for 15 min then blocked in 2% BSA PBS-Triton (0.1%) for 1 h at room temperature. Cells were incubated overnight at 4 °C in primary antibody at a 1:100 (MRTF-A or MRTF-B) or 1:500 (YAP1) dilution in blocking buffer. Cells were washed 3x in PBS then were incubated in the appropriate secondary antibody at a 1:1000 dilution for 1 h at room temperature. Cells were washed 3x in PBS then were mounted in ProLong Gold Antifade + DAPI (ThermoFisher, #P36935). Slides were cured overnight at room temperature and were then imaged on a Nikon TE2000-U Fluorescence Microscope at 20x magnification.
Cells were stained with Alexa Fluor546 Phalloidin (#A22263) to visualize F-Actin. For these experiments, cells were fixed and blocked as described above. Cells were then incubated in Phalloidin diluted 1:100 in blocking buffer for 1 h at room temperature before being washed and mounted. For all immunofluorescence experiments, images were blinded by an independent party or an automated R script before quantification. For a cell to be considered as stress fiber-positive, the cell was required to contain at least one stress fiber which spanned >90% the length of the cell. We repeated all staining experiments at least 3 times and typically analyzed at least 10 fields per biological replicate. In total we analyzed at least 400 cells per experimental group, but in most cases over 1000 cells per experimental group. For subcellular localization experiments, data are represented as a stacked bar graph wherein the fraction of cells that have predominantly nuclear, pan-cellular, or cytosolic localization is plotted as a fraction of the total cells. A cell was considered to have “cytosolic” localization if there was clear nuclear exclusion. Inversely a cell was described as having “nuclear” localization if the staining intensity was appreciably higher than in the cytosol. If there was no apparent difference between the nuclear and cytosolic staining, then the cell was described as having “pan-cellular” distribution of the protein being assessed.
Cell viability experiments
Cells were seeded into 384-well tissue culture plates (PerkinElmer, Waltham, MA, USA, #6007689) at a density of 1000 cells/well in 20 uL of media and were allowed to attach overnight. The next day, drugs were pre-diluted at 4x final concentration in culture medium then added to the 384-well plates so that the final volume was 40 μL/well. For the single compound dose response experiments, the compound was pre-diluted at 2x the final concentration and 20 uL was added to each well. A PBS or growth medium barrier was added to the outer wells of the plate to limit evaporation. Cells were cultured under these conditions for 72 h. To assess viability, 10 μL of CellTiter-Glo (Promega, Madison, WI, USA, #G7573) was added to each well. Plates were incubated for 5 min at room temperature then briefly centrifuged (4000 rpm, 60 seconds) before being read on a Bio-Tek Synergy Neo plate reader. Viability signal is plotted versus log(Vemurafenib concentration) for each treatment condition. The Area Under the Curve (AUC) was calculated for each curve using GraphPad Prism for the range log concentration from −9 to −5.
Bioinformatics
Dataset Processing
Cancer Cell Line Encyclopedia (CCLE) gene expression Affymetrix CEL files (Version 19-Mar-2013) were downloaded from the Broad Institute CCLE data portal. CEL files were processed using Affymetrix Expression Console (Build 1.4.0.38). Probe IDs were collapsed to gene names using the CollapseDataset function on GenePattern. The TCGA RNA-Seq dataset for Skin Cutaneous Melanoma (SKCM) was downloaded from the UCSC Cancer Genome Browser portal. No further data processing was performed prior to analysis.
RNA-Seq data for 62 human tumors paired for pre- and post- MAPK inhibitor resistance was downloaded from GSE6518517. Analysis of these data was performed on the pre-processed CuffnormFPKM dataset included in this series. RNA-Seq data for in vitro generated vemurafenib-resistant M229P/R and M238P/R cells was downloaded from GSE7531360. These data were processed using the above described RNA-Seq data processing pipeline.
Melanoma scRNA-Seq data was downloaded from GSE72056 and filtered to include only melanoma cells. Missing values were imputed with the MAGIC algorithm68.
Data for the M229 cells treated with vemurafenib for different times was downloaded from GSE110054. No further processing was performed on this dataset prior to ssGSEA analysis.
Gene Ontology/KEGG pathway analysis
Using the CCLE dataset, 38 adherent cell lines with BRAFV600 mutations were identified. For all cell lines, PLX4720 (activity area) was correlated with gene expression. A definition of Activity Area can be found in this study2. Genes highly expressed in resistant cells (genes with a Pearson correlation coefficient < −0.5 when correlated with PLX4720 sensitivity) and genes weakly expressed in resistant cells (Pearson correlation coefficient > 0.5) were identified. Gene ontology and KEGG pathway analysis was performed on the gene sets using GATHER (http://changlab.uth.tmc.edu/gather/gather.py) with network inference.
GSEA/ssGSEA
GSEA (v19.0.24) was performed using GenePattern (http://software.broadinstitute.org/cancer/software/genepattern/) with ‘number of permutations’ = 1000, and ‘permutation type’ = phenotype. All other parameters were left as default. ssGSEA (9.0.9) was performed on GenePattern with all parameters left as default. The ssGSEA output values were z-score normalized.
A RhoA/C gene signature was generated by using all genes which are upregulated > 2-fold by overexpression of either RhoA or RhoC from the GSE5913 dataset in NIH-3T3 cells. These two lists were merged and duplicates were removed. This resulted in a list of 79 genes (Table S1).
The melanocyte lineage signature included all genes in the GO_MELANIN_METABOLIC_PROCESS (GO: 0006582) and GO_MELANOCYTE_DIFFERENTIATION (GO: 0030318) MSigDB signatures. The combined list was filtered to remove duplicate genes.
The YAP1 signature used was the CORDENONSI_YAP_CONSERVED_SIGNATURE in the C6 collection on MSigDB. The MRTF signature is comprised of all genes downregulated > 2-fold upon MRTF knockdown in B16F2 melanoma cells 32 (Table S1).
Drug Response Signatures
The correlated gene expression profiling and drug IC50 values were downloaded from the GDSC data portal (https://www.cancerrxgene.org/downloads). Gene expression data was median centered so that the median expression of each gene across the cell lines was equal to 0. Data was randomly divided into a training (80%) and test (20%) set. A predictive model was built on the training set for each compound (n = 265 compounds) using a random forest algorithm (randomForest package in R) with ntrees = 500 and mtry = sqrt(#genes). Each model was validated on the test dataset by calculating the Pearson correlation coefficient between the predicted and actual IC50s. Models with a Pearson correlation coefficient > 0.3 were considered predictive. A full table of these results is included as (Table S2). To use gene expression data to predict drug response on clinical tumors, the TCGA SKCM data were median-centered using the same method used on the GDSC training data. Since the TCGA and GDSC datasets were collected on different gene expression analysis platforms, the two datasets were filtered to include only overlapping genes. Models from GDSC which were deemed predictive for a drug response were then projected onto the TCGA dataset. Melanocyte Lineage signature scores of TCGA samples were negatively skewed from a normal distribution (corrected z3 = −1.94). Of the 473 tumors, 70 were > 2 SD below the mean and none > 2 SD above the mean. Consequently, samples at least 2 SD below the mean are considered “lineage low” and all other tumor samples are considered “lineage high”. The average predicted IC50 for the Lineage low and Lineage high tumors was calculated by averaging the predicted log(IC50) for each sample class.
Statistical Analysis
Most bioinformatics analysis was performed using R v3.3.0. Data analysis and statistics were performed using GraphPad Prism v6 or v7. Dose response curves were fit using nonlinear least square regression [log(agonist) vs. response – Variable slope (four parameters)]. The AUC was calculated for each dose response curve in GraphPad Prism over a vemurafenib concentration range of 10−9 to 10−5. Datasets with two groups were analyzed by unpaired two-tailed t-tests. Pearson correlation coefficients were calculated in R (for drug response signatures) or GraphPad Prism (for all other analysis). Data are presented as mean ± S.E.M, and a p-value < 0.05 was considered statistically significant.
Results
RhoA activation in BRAFi-resistant melanoma cells and tumors
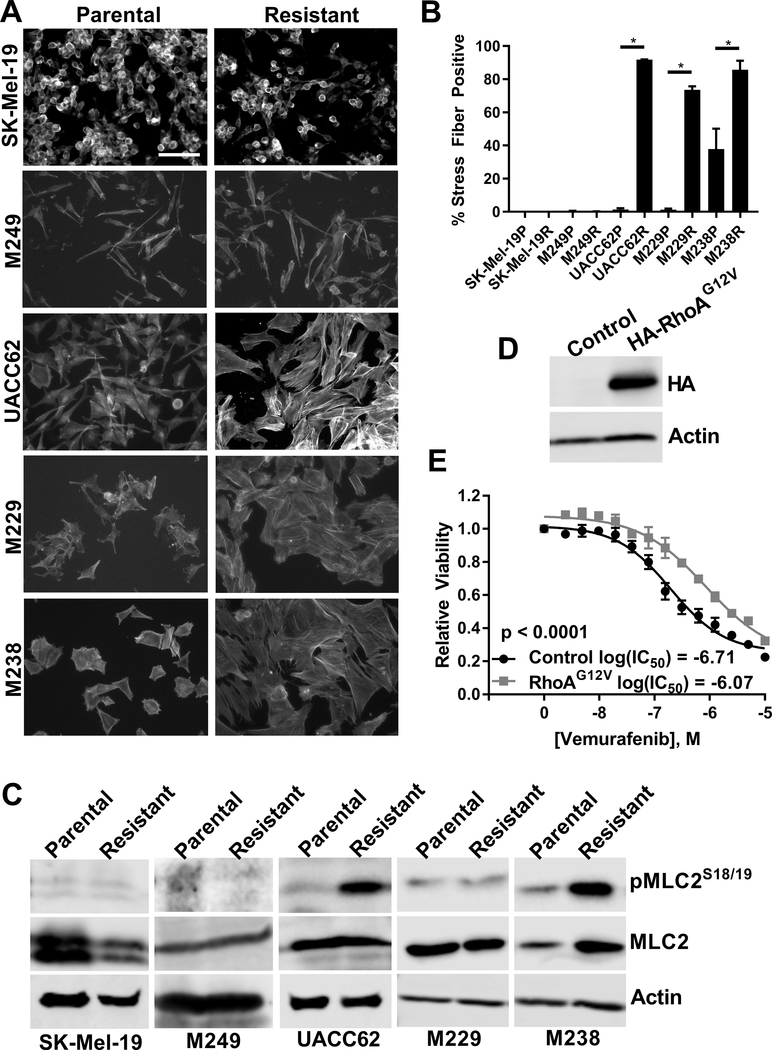
We analyzed a panel of matched parental (denoted by a P at the end of the cell line name) and BRAFi-resistant (denoted by an R at the end of the cell line name) melanoma cell lines and found that three of the resistant cell lines (UACC62R, M229R, and M238R) assumed a fibroblast-like morphology, while there was no overt change in the other two resistant cell lines (SK-Mel-19R and M249R). Since cell shape is controlled through modulation of the actin cytoskeleton, we examined F-actin structure by staining the cells with fluorescently labeled phalloidin. There was an increase in the number of actin stress fiber-positive UACC62R, M229R, and M238R cells compared to matched parental control cell lines; there was no overt change in stress fiber levels in the SK-Mel-19R and M249R cells (Fig. 1A/B). Since an increase in stress fibers would suggest that Rho activation is altered, we also analyzed Myosin Light Chain 2 (MLC2) phosphorylation in the matched parental and resistant cell lines. MLC2 is a RhoA effector so MLC2 phosphorylation is a readout for increased RhoA activation. MLC2 phosphorylation is increased in the stress fiber-positive UACC62R and M238R cell lines, but not in stress fiber-negative SK-Mel-19R or M249R cells (Fig. 1C). Interestingly, there was no change in MLC2 phosphorylation in the M229R cells despite the fact that they are stress fiber positive, which may suggest that these cells utilize an alternative signaling mechanism to activate RhoA and increase stress fibers or that RhoA may utilize different effector pathways in different cells.
Figure 1. RhoA is activated in BRAFi-resistant melanoma cells and tumors.
A) Cells were seeded into 8-well chamber slides and were allowed to attach overnight. The next day cells were fixed and stained with fluorescently labeled phalloidin. Representative images from n = 3 biological replicates and n = 1 technical replicate. Scale bar is 10 μm. B) Actin stress fiber positive cells were quantified using ImageJ. Statistical analysis was performed using unpaired t-tests to compare matched parental (denoted by a P at the end of the cell line name) and resistant (denoted by an R at the end of the cell line name) lines. * indicates that p < 0.05. C) MLC2S18/19 phosphorylation in the parental and resistant cells was assessed by immunoblotting. Total MLC2 and Actin were used as loading controls. Representative blots from n = 3 biological replicates and n = 1 technical replicate. D) UACC62P cells stably expressing HA-RhoAG12V were lysed and immunoblotted with anti-HA and anti-Actin antibodies. Representative images from n = 3 biological replicates and n = 1 technical replicate. E) UACC62P cells stably expressing Gus (control) or HA-RhoAG12V were seeded into 384-well plates and treated with a 14-point vemurafenib concentration gradient with a top dose of 10 μM as described in the materials and methods. Data is average from n = 3 biological replicates with n = 3 technical replicates.
These data suggested that RhoA was activated in the resistant cell lines, but it was not clear whether RhoA itself was functionally important in BRAFi resistance. To address this question, we generated UACC62P cells which stably express RhoAG12V (Fig. 1D). This specific mutation is not found in any Skin Cutaneous Melanoma (SKCM) tumors in the TCGA dataset, however, the constitutively active RhoAG12V model is a useful tool for studying mechanisms of Rho signaling since it is independent of upstream stimuli. Consistent with our observations suggesting that RhoA is activated in a subset of the resistant cell lines, overexpression of RhoAG12V reduced vemurafenib sensitivity by approximately 6-fold (Fig. 1E). To further confirm the role of RhoA in vemurafenib resistance, we pharmacologically inhibited the function of RhoA using the cell permeable Botulinum Exotoxin C3 (Rho Inhibitor I) to test whether RhoA inhibition reverses vemurafenib resistance (Fig. S1). M238R and UACC62R cells were more sensitive to single agent treatment with Rho Inhibitor I, suggesting that these cells are re-wired to depend on RhoA signaling for their survival. M229R cells did not have increased sensitivity to single agent treatment with Rho Inhibitor I, rather Rho Inhibitor I treatment increases vemurafenib sensitivity. As expected, Rho Inhibitor I was not selective for M249R cells over M249P cells, which is consistent with the idea that these cells do not develop resistance through RhoA activation. Since RhoA activation should result in an increase in actin polymerization, we next tested whether actin polymerization is functionally important for vemurafenib resistance. Similar to the findings with Rho Inhibitor I, cytochalasin D was more active against all three of the RhoAHigh cell lines as a single agent treatment and it partially re-sensitized M229R cells to vemurafenib (Fig. S2).
To more broadly confirm this finding, we correlated cell sensitivity to PLX4720 (a BRAF inhibitor which is structurally similar to vemurafenib) with the gene expression results for 38 BRAFV600-mutant cell lines from the Cancer Cell Line Encyclopedia (CCLE). Genes which are highly expressed in PLX4720-resistant cells (genes with a Pearson correlation of gene expression values vs drug activity area < −0.5) were analyzed by Gene Ontology and KEGG pathway analysis. One of the most statistically significant GO terms was “small GTPase mediated signal transduction” (Fig. S3A) and the most statistically significant KEGG pathway was “Regulation of actin cytoskeleton” (Fig. S3B). A RhoA/C gene signature was also inversely correlated (R = −0.42) with PLX4720 sensitivity (Fig. S4). Collectively, these data support the idea that RhoA activation is positively correlated with BRAFi resistance across a wide array of melanoma cell lines. To determine whether these cell line observations are applicable in the clinical context, we analyzed RNA-seq data from 41 tumors before and after development of resistance to BRAFi/MEKi 17. More than half of the resistant tumors (n = 24) had an increased RhoA/C signature score over the baseline tumor (Fig. S5). Taken together, these data suggest that RhoA is activated in approximately half of BRAFi-resistant cells and tumors and that RhoA activation is inversely correlated with BRAFi sensitivity.
Since the most common class of BRAFi resistance mechanisms is through MAPK re-activation we then wondered whether RhoA activation was mutually exclusive with MAPK reactivation-mediated resistance. If resistance is developed through MAPK re-activation then the resistant cells should retain ERK phosphorylation when treated with vemurafenib. As expected, vemurafenib inhibits ERK phosphorylation in all 5 parental cell lines. Vemurafenib fails to inhibit ERK phosphorylation in the two RhoALow resistant lines (SK-Mel-19R and M249R), which in the case of M249R is expected since these cells developed resistance by acquiring an NRASQ61K mutation. In the three RhoAHigh resistant cell lines, vemurafenib partially inhibited ERK phosphorylation in two (M229R and UACC62R) but failed to suppress ERK phosphorylation in the other (M238R) (Fig. S6). This finding is important since it suggests that Rho may be important even in cells which harbor MAPK-reactivating resistance mechanisms.
Resistant cell lines with a low level of melanocyte differentiation show high RhoA activity
We next wanted to understand mechanistically why the RhoA pathway is only activated in a subset of vemurafenib-resistant cells. We performed RNA-Seq on the UACC62P/R cell line pair (Fig. 2A), and also analyzed published RNA-Seq data for the M229P/R and M238P/R cells. The most striking finding was that a number of genes linked to the melanocyte lineage and pigment production were downregulated in all three of the RhoAHigh resistant cell lines. To more quantitively analyze this phenotype we generated a “Melanocyte Lineage” gene signature (Table S3) which is comprised of genes involved in pigment production and the melanocyte lineage. A majority of the signature genes are downregulated in all three of the RhoHigh resistant cell lines (Fig. 2B) which suggests that loss of melanocyte identity is associated with Rho activation in BRAFi-resistant cells. There is also a temporal association between expression of the melanocyte lineage genes and RhoA/C signature genes (Fig. S7). One of the most strongly downregulated genes, at the mRNA level, is the transcription factor Sox10 which is one of the “master regulators” of the melanocyte lineage; we confirmed that Sox10 is also downregulated at the protein level (Fig. 2C). Interestingly, there was no change in Sox10 protein expression in the M249P/R cells which did not have increased stress fibers (Fig. 2C). We also found that Sox9 is upregulated at the mRNA level in all three of the RhoAHigh resistant cell lines but not in the RhoALow resistant lines (Fig. S8). These results are consistent with previous findings which suggest that Sox10 suppresses Sox9 expression 54, and suggest that this switch in transcription factor expression may be reflective of the differentiation status of the resistant cells.
Figure 2. Melanoma differentiation status is inversely correlated with Rho activation.
A) RNA-Seq was performed on parental (UACC62P) and Vemurafenib-resistant (UACC62R) cells. Differential gene expression was visualized on a volcano plot. n = 2 biological replicates and n = 1 technical replicate per treatment condition. B) Heatmap of differential expression of Melanocyte Lineage signature genes in M229P/R, M238P/R, and UACC62P/R cells. Blue indicates that the gene is downregulated in the resistant cell line, and red indicates that the gene is upregulated in the resistant cell line. C) Sox10 protein expression was assessed across a panel of 4 parental and resistant melanoma cell lines. Actin was used as a loading control. Representative image from n = 3 biological replicates and n = 1 technical replicate. D) Histogram of Melanocyte Lineage signature scores for samples in the SKCM TCGA dataset (n = 473). Dotted line represents 2 SD below the mean of the Gaussian fit. Samples were stratified into Melanocyte Lineage high and Melanocyte lineage low samples as described in the Materials and Methods section. E) ssGSEA was used to calculate the RhoA/C signature score for each TCGA SKCM tumor sample. ssGSEA was performed as described in the Materials and Methods section. The output signature score from this analysis was not subjected to further processing. The tumors were stratified based on their Melanocyte Lineage signature score as described in panel D of this figure and in Materials and Methods. The average RhoA/C signature score for each class of tumor samples is plotted where tumor samples with a high Melanocyte Lineage score are in grey and tumor samples with a low Melanocyte Lineage score are in red. F) Predictive signatures were generated for 265 common anti-cancer compounds using a random forest algorithm. The models were made such that gene expression data was used to predict drug response. The drug response data was derived from the GDSC dataset73 and the numerical values for drug sensitivity are the IC50 values from this dataset. The samples were stratified into Melanocyte Lineage Low and Melanocyte Lineage High and the average predicted IC50 for each drug for each class of samples was calculated. The differential predicted IC50 was subsequently determined by calculating the differential in the average IC50 between the two classes of tumor samples. The values on the Y-axis of this plot are the fold change in predicted IC50 between the Melanocyte Lineage Low and Melanocyte Lineage High samples. A positive value means that the compound was predicted to be more effective against Melanocyte Lineage Low tumor samples, while a negative value means that the compound was predicted to be more effective against Melanocyte Lineage High tumor samples. G) Previously published single cell RNA-Seq data was used for this experiment 65. These data were downloaded and processed as described in Materials and Methods. The RhoA/C signature and the Melanocyte Lineage signatures were calculated using ssGSEA. Since this dataset is comprised of tumor cells derived from multiple different tumors the cells group together based upon tumor of origin. The cells are color coded based on the tumor from which they were derived from and the colors were chosen arbitrarily.
Since Sox10 silencing results in activation of multiple RTKs, including EGFR, we sought to determine whether EGFR is activated in Sox10Low BRAFi-resistant cells. EGFR mRNA is upregulated approximately 8–40-fold in Sox10Low BRAFi-resistant cells (Fig. S9A) and this mRNA upregulation is accompanied by an increase in EGFR phosphorylation (Fig. S9B). We next tested whether EGFR was required for actin remodeling, however, treatment with the EGFR inhibitor erlotinib did not alter the assembly of actin stress fibers (Fig. S9C).
To determine whether this de-differentiation phenotype was also important in human SKCM tumors we projected the “Melanocyte Lineage” signature onto the SKCM TCGA dataset and then fit a Gaussian distribution to the signature scores. The distribution was skewed towards lower signature scores (corrected z3 = −1.94). While most of the tumors fell within 2 standard deviations of the mean, there was a subset of tumors (n = 70) which had low expression of melanocyte lineage genes (low was defined at being > 2SD below the mean) (Fig. 2D). There were no tumors which had a signature score > 2 SD above the mean. As expected, tumor purity was correlated with the expression of melanocyte lineage genes (Fig. S10), but this does not fully explain why these tumors have lower expression of these genes given the magnitude of the downregulation of the melanocyte lineage signature. Consistent with the finding that RhoA is activated in de-differentiated BRAFi-resistant cell lines, we also found that tumors with decreased expression of melanocyte lineage genes have increased expression of RhoA/C target genes (Fig. 2E).
The small fraction of tumors (n = 70 out of 473 total tumors) which have decreased expression of melanocyte lineage genes may be due to the fact that all of the tumors in this dataset were treatment-naïve with respect to BRAF inhibitors. Since the transcriptional profile of these lineage-low tumors is similar to that of the BRAFi-resistant cell lines, it is possible that these tumors may have intrinsic resistance to BRAF inhibitors. To test this hypothesis, we generated gene expression signatures from GDSC data to predict drug response for 265 common anti-cancer compounds using a random forest machine learning algorithm (see materials and methods). These signatures were then projected onto the TCGA dataset to predict drugs to which the de-differentiated tumors should be differentially sensitive to (Fig. 2F, Table S4). As expected, the de-differentiated tumors are predicted to be less sensitive to multiple BRAF and MEK inhibitors, including PLX4720 (a structurally similar vemurafenib analog). These predictions support the idea that the ~15% of treatment-naive melanoma tumors with a de-differentiated transcriptional signature are less sensitive to BRAF inhibition even before selection by BRAFi treatment. This supports what we observed in experimentally derived resistant cell line models. Also, de-differentiated tumors are predicted to have increased sensitivity to multiple ROCK inhibitors which is interesting since ROCK is one of the canonical RhoA effector proteins 12, 37.
The observation that RhoA activation is inversely correlated with differentiation status in human tumors could be marred by the contribution of non-malignant cells to the overall bulk gene expression profile of the tumor. For example, it is expected that in some cases cancer-associated fibroblasts or endothelial cells might have high RhoA activity 40, 75. To more directly address the hypothesis that differentiation status is inversely correlated with Rho activation in melanoma cells we used publicly available single cell RNA-Seq data 65 to correlate a RhoA/C signature and the Melanocyte Lineage signature. As expected, cells clustered together based on their tumor of origin which is due to the strong inter-tumor transcriptomic heterogeneity 65. Even within a single tumor, poorly differentiated cells have elevated RhoA activation (Fig. 2G). In total, these data suggest that tumors which acquire a de-differentiated phenotype have elevated RhoA activation and are predicted to be more sensitive to inhibition of RhoA signaling.
ROCK inhibition sensitizes RhoAHigh BRAFi-resistant melanoma cells
It is difficult to therapeutically target RhoA directly, so an alternative approach is to target downstream effector pathways. Since we predicted that poorly differentiated human melanoma tumors are more sensitive to ROCK inhibitors, it is possible that de-differentiated BRAFi-resistant cells are more sensitive to ROCK inhibitors. It is also possible that ROCK inhibition might re-sensitize the resistant cells to Vemurafenib. To test this hypothesis we used two ROCK inhibitors, Y-27623 and Fasudil, which have structurally distinct chemical scaffolds. We also confirmed that both Y-27632 and fasudil reduce actin stress fiber formation in M229R cells (Fig. S11). RhoAHigh BRAFi-resistant cells (but not RhoALow resistant cells) are more sensitive to either of the ROCK inhibitors as a single agent (Fig. S12). ROCK inhibition also re-sensitizes RhoAHigh (but not RhoALow) BRAFi-resistant cells to vemurafenib (Fig. 3A–H). Re-sensitization to vemurafenib was most pronounced in M229R cells (Fig. 3I–K) which is interesting since these cells do not have increased MLC2 phosphorylation. Since increased sensitivity to ROCK inhibitors alone, or the effect of ROCK inhibitors on re-sensitizing cells to vemurafenib, is only observed in cells which have increased stress fibers it suggests that this combination treatment may be specific for cells/tumors which activate this signaling mechanism.
Figure 3. ROCK inhibition reverses BRAFi resistance in RhoAHigh BRAFi-resistant melanoma cells.
Parental and Vemurafenib-resistant cell lines were seeded into 384-well plates at a density of 1,000 cells/well and cells were allowed to attach overnight. The next day, cells were treated with Vemurafenib at the indicated concentrations with or without the ROCK inhibitors Y-27632 (red) or Fasudil (blue) at 10 μM. Cells were grown for 72 h then viability was measured with CellTiter-Glo. Pooled viability data from n = 3 biological replicates and n = 1 technical replicate. A-H) Cell lines were treated as labeled with ROCK inhibitors (Y-27632 or Fasudil) along with Vemurafenib. I) Schematic of Area Under Curve (AUC) calculation. Larger AUC indicates lower sensitivity to the drug combination and smaller AUC indicates greater sensitivity to the drug combination. J) Heatmap of AUC values for the Vemurafenib/Y-27632 drug combination for four parental and resistant cell line pairs. K) Heatmap of AUC values for the Vemurafenib/Fasudil drug combination for four parental and resistant cell line pairs. Blue values indicate a high AUC and red values indicate a low AUC.
MRTF and YAP activation in RhoAHigh BRAFi-resistant cells
In addition to modulating cytoskeletal re-arrangement, RhoA also regulates gene expression. Two transcriptional co-activators downstream of RhoA are YAP1 and MRTF. MRTF and YAP1 have similar transcriptional outputs and can perform redundant functions in several contexts 11, 76. To determine whether YAP1 and MRTF are activated in RhoAHigh BRAFi-resistant cells, we measured the subcellular localization of YAP1 and MRTF-A (Fig. 4A/B). YAP1 nuclear localization is elevated in M229R and M238R cells compared to matched parental cell lines and is elevated to a lesser extent in UACC62R cells. The converse is true with respect to MRTF-A localization since nuclear MRTF-A is increased in UACC62R cells but not M229R or M238R cells (Fig. 4A/B). Expression of several MRTF/YAP1 target genes is also elevated in the cells which have increased nuclear MRTF/YAP1 localization (Fig. S13A). Several YAP1- and MRTF-related genes are highly expressed in BRAF-mutant cell lines with intrinsic BRAFi resistance (Fig. S14). These include the YAP1/MRTF target gene CYR61 and genes encoding proteins which activate RhoA (ARHGEF12, GNA11, GNA12, TGFβ1) as well as YAP1 and YES1.
Figure 4. YAP1 and MRTF-A are activated in de-differentiated BRAFi-resistant cells.
A) M229P/R, M238P/R, and UACC62P/R cells were seeded into 8-well chamber slides and were allowed to attach overnight. The next day, cells were fixed and stained with an anti-YAP1 or anti-MRTF-A antibodies. Representative images from n = 3 biological replicates and n = 1 technical replicate. Scale bar is 5 μm. B) Quantification of staining from panel ‘A’. Data are represented as a stacked bar graph wherein the fraction of cells that have predominantly nuclear, pan-cellular, or cytosolic localization is plotted as a fraction of the total cells. C) MRTF and YAP1 signatures were predicted for human melanoma tumor pairs which had an increase in RhoA/C signature score from (Fig. 1G). Change in MRTF and YAP1 signature score between baseline and resistant tumors is plotted.
YAP1 and MRTF gene signature activation is increased in the paired pre- and post-resistance human melanoma tumors which had an increase in RhoA/C signature gene expression (Fig. 4C). Out of this subset of tumors, only 3/24 failed to upregulate either YAP1 or MRTF target genes. Half (12/24) of the tumors had upregulation of both YAP1 and MRTF gene signatures, which could possibly result from the high degree of redundancy in the transcriptional output from YAP1 and MRTF. Another explanation is that this could result from the tumors consisting of a mixed population of YAP1High and MRTFHigh cells. Some tumors appeared to have selective activation of YAP or MRTF, which is interesting considering the apparent mutual exclusivity of MRTF-A/YAP1 activation in the experimentally derived cell line models. This is again consistent with the transcriptional alterations in the RhoAHigh BRAFi-resistant cell lines since MRTFA and YAP1 gene signatures are both increased in the poorly differentiated tumors (Fig. S15). Taken together, these data demonstrate that YAP1 and/or MRTF are activated in nearly all of the poorly differentiated BRAFi-resistant cells/tumors.
We hypothesized that since increased MRTF-A/YAP1 nuclear localization is only in RhoAHigh resistant cells that RhoA may be regulating their nuclear accumulation. To test this, we treated parental and resistant cells with Y-27632 and Fasudil and measured the subcellular localization of MRTF-A and YAP1 with immunofluorescent staining. Treatment with either ROCK inhibitor reduced YAP1 nuclear accumulation in M229R cells and reduced MRTF-A nuclear accumulation in UACC62R cells (Fig. S16)
Pharmacologically targeting MRTF/YAP-mediated gene transcription
Since our results indicated that YAP1 and MRTF are activated in de-differentiated BRAFi-resistant cells, we reasoned that pharmacologically targeting these transcriptional mechanisms would be sufficient to re-sensitize cells to vemurafenib. YAP1 is activated by YES1, a Src family kinase. Previous studies have used the Src family kinase inhibitor dasatinib to inhibit YES1, resulting in a downregulation of YAP1 activity 48. There is also evidence which suggests that other Src family kinases activate YAP1 24. Using Src inhibition as an approach to block YAP1 activity is also interesting since our bioinformatics analysis predicted that poorly differentiated human tumors are more sensitive to Src inhibitors, including dasatinib (Fig. 2F). To confirm this in the context of vemurafenib-resistant cells, we treated M229R and M238R cells with dasatinib and measured YAP1 nuclear localization. YAP1’s nuclear localization is decreased in both cell lines upon dasatinib treatment (Fig. 5A/B). While datastinib reduces nuclear accumulation of YAP1, which theoretically should reduce YAP1-mediated gene transcription, expression of several YAP1 target genes is not altered by dastatinib treatment (Fig. S13B–D).
Figure 5. De-differentiated BRAFi-resistant cells are more sensitive to dasatinib and CCG-222740.
A) Cells were seeded into 8-well chamber slides and were allowed to attach overnight. The next day, cells were treated with dasatinib (500 nM) for 16 h, then cells were fixed and stained with an anti-YAP1 antibody. Representative images from n = 3 biological replicates and n = 1 technical replicate. B) Quantification of YAP1 localization from panel “A”. Data are represented as a stacked bar graph wherein the fraction of cells that have predominantly nuclear, pan-cellular, or cytosolic localization is plotted as a fraction of the total cells. C-F) Parental and Resistant cell lines were seeded into 384-well plates at a density of 1,000 cells/well and cells were allowed to attach overnight. The next day, cells were treated in dose response with Vemurafenib at the indicated concentrations −/+ 100 nM dasatinib (red). After 72 h viability was measured with CellTiter-Glo. Dose response curves are viability data are from n = 3 biological replicates and n = 1 technical replicate. G) Heatmap of AUC values for the vemurafenib/dasatinib drug combination for four parental and resistant cell line pairs. H-K) Parental and Resistant cell lines were seeded into 384-well plates at a density of 1,000 cells/well and cells were allowed to attach overnight. The next day, cells were treated in dose response with Vemurafenib at the indicated concentrations −/+ 10 μM CCG-222740 (blue). After 72 h viability was measured with CellTiter-Glo. Dose response curves are viability data are from n = 3 biological replicates and n = 1 technical replicate. L) Heatmap of AUC values for the Vemurafenib/CCG-222740 drug combination for four parental and resistant cell line pairs. Blue values indicate a high AUC and red values indicate a low AUC. M) UACC62P cells were engineered to stably express Gus (Control) or MRTF-A as described in Materials and Methods. Immunoblots to measure MRTF-A levels were performed as described in Materials and Methods with β-Actin serving as a loading control. N) UACC62P cells stably expressing Gus (control) or MRTFA were seeded into 384-well plates and treated with a 14-point vemurafenib concentration gradient with a top dose of 10 μM as described in the materials and methods. Data is average from n = 3 biological replicates with n = 3 technical replicates.
We next wanted to determine whether dasatinib re-sensitizes de-differentiated BRAFi-resistant cells to vemurafenib. Dasatinib treatment has only a minor effect on potentiating the vemurafenib response in the parental UACC62P and M229P cells, however, the vemurafenib response is greatly potentiated in the resistant UACC62R and M229R cells (Fig. 5C–F). While UACC62R does not have as robust YAP1 activation as M229R and M238R, the minor increase in YAP1 nuclear localization could explain why these cells also respond to dasatinib. This effect is consistent across all three de-differentiated BRAFi-resistant cell lines (Fig. 5G). All three of the de-differentiated BRAFi-resistant cell lines also have increased sensitivity to dastatinib as a single agent (Fig. S17). Interestingly, in contrast with other similar experiments, we did not observe any change in vemurafenib sensitivity upon deletion of YAP1 with CRISPR (Fig. S18C/D)
Our lab has developed a series of MRTF pathway inhibitors, including CCG-222740 3, 13, 18, 29. We sought to determine whether this inhibitor can re-sensitize de-differentiated BRAFi-resistant cells to Vemurafenib. CCG-22740 has only a modest effect on re-sensitizing M229R or M238R cells, which have strong YAP1 but low MRTF-A activation and has the stronger re-sensitization effect in UACC62R cells (Fig. 5H–L) which was the only BRAFi-resistant cell line with strong nuclear localization of MRTF-A. Also, UACC62R cells are more sensitive to CCG-222740 as a single agent (Fig. S17). Interestingly, despite the effect of CCG-222740 on viability and vemurafenib re-sensitization, CCG-222740 (10 μM, 24 h) does not alter expression of several MRTF target genes at the mRNA level (Fig. S13B–D). To more directly determine the effect of MRTF-A on BRAFi resistance, we generated cells which stably express wildtype MRTF-A (Fig. 5M). Cells expressing MRTF-A are approximately 10-fold less sensitive to vemurafenib (Fig. 5N). Interestingly when we performed the inverse experiment, deletion of MRTF-A with CRISPR in resistant cells did not alter vemurafenib sensitivity (Fig. S18A/B). Although we did not observe any overt change in MRTF-B localization when parental and resistant cell lines were compared under basal conditions (Fig. S19), it is possible that MRTF-A depletion may induce MRTF-B activation. Taken together these data demonstrate that inhibition of RhoA-mediated gene transcription in de-differentiated melanoma cells, which can be mediated either by YAP1 or MRTF, re-sensitizes the melanoma cells to vemurafenib.
Discussion
In this study we sought to identify a pharmacological “Achilles heel” for BRAFi-resistant melanoma cells/tumors. In theory, if pathway-centric dependences can be identified for cells with acquired resistance, then co-targeting these resistance pathways concurrently with MAPK pathway inhibitors may delay, prevent, or reverse resistance. We found evidence for RhoA pathway activation in approximately half of BRAFi/MEKi-resistant human melanoma cells and tumors. In isogenic BRAFi-resistant cell lines, Rho pathway activation was accompanied by both an increase in actin stress fibers and usually MLC2 phosphorylation. These findings are consistent with previous reports which demonstrate that actin stress fibers are increased in cell line models of acquired BRAFi resistance 21. Building off these findings, we demonstrated that ROCK inhibition re-sensitizes RhoHigh BRAFi-resistant cells to vemurafenib, highlighting the importance of this signaling pathway in adaptive BRAFi resistance. This finding also supports our bioinformatics predictions, since multiple ROCK inhibitors were among the drugs predicted to be selective for poorly differentiated melanoma tumors.
We next wanted to identify signaling mechanisms which are associated with RhoA pathway activation. These signaling mechanisms could serve as biomarkers for RhoA activation or these pathways could directly promote RhoA activation. Upon acquisition of drug resistance all of the RhoAHigh cell lines downregulate an array of melanocyte lineage genes such as TYR, MLANA, and SOX10. This is accompanied by upregulation of multiple cancer invasion-associated genes including AXL and SOX9 as well as several collagen and integrin isoforms. De-differentiation of melanoma cells has previously been linked to drug resistance. For instance, a decrease in MITF/AXL gene expression ratio marks BRAFi resistance 23, 60, 65. In another study silencing of SOX10, which was one of the most downregulated genes in our analysis, promotes BRAFi resistance 63. But whether de-differentiation is directly inducing RhoA activation, or if RhoA activation is simply associated with de-differentiation is a question that still needs to be addressed.
As a result of modulating the actin cytoskeleton, Rho regulates gene transcription. Rho-induced F-actin polymerization allows for MRTF and YAP1 to translocate into the nucleus where they subsequently regulate gene transcription 8, 35, 36, 44, 52, 58. Interestingly, some reports suggest that MRTF and YAP1 physically interact and are present in close proximity on similar gene promoters 76, while others suggest more indirect mechanisms of shared gene expression control 11. While YAP1 has been previously demonstrated to promote BRAFi resistance in melanoma 10, 17, 21, 28, the role of MRTF in BRAFi resistance is unknown. In this study we demonstrate that nuclear accumulation of either MRTF-A or YAP1 is increased in RhoAHigh BRAFi-resistant cells. We also demonstrate that overexpression of MRTF-A induces vemurafenib resistance. Conversely, our data demonstrates that an MRTF pathway inhibitor increases vemurafenib sensitivity. Further work is required to determine how MRTF promotes BRAFi resistance and whether those signaling mechanisms are similar to the mechanisms by which YAP1 promotes BRAFi resistance. Interestingly, we observed YAP1 activation in 2 of 3 RhoAHigh resistant cell lines, and MRTF-A activation in the 3rd cell line. This may suggest that MRTF-A and YAP1 are acting redundantly in this context and that activation of either MRTF-A or YAP1 is sufficient to promote drug resistance.
Our data demonstrate that MRTF-A and YAP1 are activated in vemurafenib-resistant cells our data are conflicting on whether MRTF-A and YAP1 are required for the development of drug resistance. In this study deletion of YAP1 in M229R cells did not alter vemurafenib response, where other studies found that YAP1 silencing either partially reverses vemurafenib resistance or increases vemurafenib sensitivity17, 21, 28. One of these studies utilized M229, a cell line which we also used. In our study, cells were used within 14 days of viral CRISPR transduction. We hypothesize that YAP1 is promoting drug resistance by binding to transcription factors such as TEADs and regulating gene transcription. However, it is also possible that YAP1 may promote drug resistance via altering chromatin remodeling, in which case short-term deletion of YAP1 may not provide sufficient time for this process to take place.
Similarly, MRTF-A overexpression promotes vemurafenib resistance but we did not see reversal of resistance when MRTF-A was deleted. This could be because MRTF-A may promote drug resistance via altering chromatin remodeling, or it could be because MRTF-A deletion induces compensatory activation of other transcriptional mechanisms. We recently identified Pirin as a target for the CCG-222740 series of compounds29. Given the role of Pirin in melanoma 26, 27, 33, it is possible that compound effects on vemurafenib sensitivity may also involve Pirin actions that may or may not be directly related to MRTF-A.
This study focuses on MRTF/YAP1-mediated gene transcription downstream of RhoA, however there are other signaling mechanisms which may be important in RhoA-mediated drug resistance. Several studies have highlighted various signaling interactions between ERK and RhoA. For example RhoA increases ERK nuclear localization78 and RhoA silencing reduces ERK phosphorylation43. Other studies in different model systems observe the opposite effect since treatment with the ROCKi Y-27632 increases ERK phosphorylation and expression of constitutively active RhoA decreases ERK phosphorylation64. While these data are conflicting, these studies do raise the possibility that RhoA activation may modulate ERK activity in drug-resistant cells. RhoA activation could also promote BRAFi resistance through other transcriptional mechanisms such as AP-131 or NFκB42. Both AP-146 and NFκB62 promote BRAFi resistance, so transcriptional mechanisms other than MRTF/YAP1 may play a role in RhoA-mediated drug resistance. But the potential interaction between these transcription factors is especially interesting since AP-1 cooperates with TEAD30, 77 and NFκB74 to regulate gene transcription, suggesting that multiple RhoA effectors may work together to promote drug resistance.
This study demonstrates that RhoHigh BRAFi-resistant cells are re-sensitized to vemurafenib by ROCK inhibitors and that this RhoHigh phenotype is linked to de-differentiation. The direct signaling mechanisms which lead to Rho activation in melanoma cells are still unclear, but it is enticing to suggest that induction of TGFβ upon Sox10 loss 63 may lead to RhoA activation. However, it is possible that TGFβ may be inducing de-differentiation 39, 72 and RhoA activation simultaneously through different signaling mechanisms. Future studies will be necessary to elucidate details of these signaling networks. While it is already known that YAP1 promotes BRAFi resistance, these studies build upon that knowledge to demonstrate that dasatinib blocks the nuclear accumulation of YAP1 and enhanced drug sensitivity in BRAFi-resistant cells. Since dasatinib and other SRC kinase inhibitors are already FDA-approved for other indications, it highlights the potential of a re-purposing approach for treatment of BRAFi/MEKi-resistant melanomas. In this context, dasatinib may be most effective in combination with vemurafenib; in at least one resistant cell line vemurafenib potency was restored to that of parental cells. These studies also link MRTF-A activation to BRAFi resistance for the first time, highlighting the potential of targeting MRTF-mediated transcription to prevent or treat drug resistant melanoma. In total, these studies provide robust predictions of precision therapy approaches to prevent or treat clinical BRAFi resistance based on pharmacological inhibition of RhoA-mediated gene transcription.
Supplementary Material
Acknowledgements
This work was supported by R01 AR066049 (SDL), the MSU Gran Fondo Skin Cancer Research Fund (RRN), F31 CA232555 (SAM), MSUFCU Aitch Foundation Fellowship (SAM).
Financial support: MSU Gran Fondo Skin Cancer Research Fund (RRN), NIH R01 AR066049 (SDL), MSUFCU Aitch Foundation Fellowship (SAM), NIH F31 CA232555 (SAM)
Footnotes
Conflict of Interest Statement
Drs. Neubig and Larsen have intellectual property rights in the CCG-222740 compound used in this study. They are also founders and officers of FibrosIX Inc. which has an option for the IP for this and related compounds.
References
- 1.Abel EV, Basile KJ, Kugel CH 3rd, Witkiewicz AK, Le K, Amaravadi RK et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. J Clin Invest 2013; 123: 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell JL, Haak AJ, Wade SM, Kirchhoff PD, Neubig RR, Larsen SD. Optimization of novel nipecotic bis(amide) inhibitors of the Rho/MKL1/SRF transcriptional pathway as potential anti-metastasis agents. Bioorg Med Chem Lett 2013; 23: 3826–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011; 364: 2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature 2000; 406: 532–535. [DOI] [PubMed] [Google Scholar]
- 6.Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature 2013; 494: 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A 2009; 106: 20411–20416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N et al. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev 2014; 28: 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fallahi-Sichani M, Becker V, Izar B, Baker GJ, Lin JR, Boswell SA et al. Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state. Mol Syst Biol 2017; 13: 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher ML, Grun D, Adhikary G, Xu W, Eckert RL. Inhibition of YAP function overcomes BRAF inhibitor resistance in melanoma cancer stem cells. Oncotarget 2017; 8: 110257–110272. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 11.Foster CT, Gualdrini F, Treisman R. Mutual dependence of the MRTF-SRF and YAP-TEAD pathways in cancer-associated fibroblasts is indirect and mediated by cytoskeletal dynamics. Genes Dev 2017; 31: 2361–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujisawa K, Fujita A, Ishizaki T, Saito Y, Narumiya S. Identification of the Rho-binding domain of p160ROCK, a Rho-associated coiled-coil containing protein kinase. J Biol Chem 1996; 271: 23022–23028. [DOI] [PubMed] [Google Scholar]
- 13.Haak AJ, Appleton KM, Lisabeth EM, Misek S, Ji Y, Wade SM et al. Pharmacological inhibition of Myocardin-related transcription factor pathway blocks lung metastases of RhoC overexpressing melanoma. Mol Cancer Ther 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodge RG, Ridley AJ. Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol 2016; 17: 496–510. [DOI] [PubMed] [Google Scholar]
- 15.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP et al. A landscape of driver mutations in melanoma. Cell 2012; 150: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong A, Moriceau G, Sun L, Lomeli S, Piva M, Damoiseaux R et al. Exploiting Drug Addiction Mechanisms to Select against MAPKi-Resistant Melanoma. Cancer Discov 2018; 8: 74–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugo W, Shi H, Sun L, Piva M, Song C, Kong X et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell 2015; 162: 1271–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchings KM, Lisabeth EM, Rajeswaran W, Wilson MW, Sorenson RJ, Campbell PL et al. Pharmacokinetic optimitzation of CCG-203971: Novel inhibitors of the Rho/MRTF/SRF transcriptional pathway as potential antifibrotic therapeutics for systemic scleroderma. Bioorg Med Chem Lett 2017; 27: 1744–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 2010; 468: 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson DB, Menzies AM, Zimmer L, Eroglu Z, Ye F, Zhao S et al. Acquired BRAF inhibitor resistance: A multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur J Cancer 2015; 51: 2792–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim MH, Kim J, Hong H, Lee SH, Lee JK, Jung E et al. Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. EMBO J 2016; 35: 462–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein RM, Higgins PJ. A switch in RND3-RHOA signaling is critical for melanoma cell invasion following mutant-BRAF inhibition. Mol Cancer 2011; 10: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konieczkowski DJ, Johannessen CM, Abudayyeh O, Kim JW, Cooper ZA, Piris A et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov 2014; 4: 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamar JM, Xiao Y, Norton E, Jiang ZG, Gerhard GM, Kooner S et al. SRC tyrosine kinase activates the YAP/TAZ axis and thereby drives tumor growth and metastasis. J Biol Chem 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med 2014; 371: 1867–1876. [DOI] [PubMed] [Google Scholar]
- 26.Licciulli S, Luise C, Zanardi A, Giorgetti L, Viale G, Lanfrancone L et al. Pirin delocalization in melanoma progression identified by high content immuno-detection based approaches. BMC Cell Biol 2010; 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Licciulli S, Luise C, Scafetta G, Capra M, Giardina G, Nuciforo P et al. Pirin inhibits cellular senescence in melanocytic cells. Am J Pathol 2011; 178: 2397–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin L, Sabnis AJ, Chan E, Olivas V, Cade L, Pazarentzos E et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet 2015; 47: 250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lisabeth EM, Kahl D, Gopallawa I, Haynes SE, Misek SA, Campbell PL et al. Identification of Pirin as a Molecular Target of the CCG-1423/CCG-203971 Series of Anti-Fibrotic and Anti-Metastatic Compounds. bioRxiv 2018: 458554. [Google Scholar]
- 30.Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J et al. Tead and AP1 Coordinate Transcription and Motility. Cell Rep 2016; 14: 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marinissen MJ, Chiariello M, Tanos T, Bernard O, Narumiya S, Gutkind JS. The small GTP-binding protein RhoA regulates c-jun by a ROCK-JNK signaling axis. Mol Cell 2004; 14: 29–41. [DOI] [PubMed] [Google Scholar]
- 32.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol 2009; 11: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyazaki I, Simizu S, Okumura H, Takagi S, Osada H. A small-molecule inhibitor shows that pirin regulates migration of melanoma cells. Nat Chem Biol 2010; 6: 667–673. [DOI] [PubMed] [Google Scholar]
- 34.Moriceau G, Hugo W, Hong A, Shi H, Kong X, Yu CC et al. Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell 2015; 27: 240–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouilleron S, Guettler S, Langer CA, Treisman R, McDonald NQ. Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL. EMBO J 2008; 27: 3198–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mouilleron S, Langer CA, Guettler S, McDonald NQ, Treisman R. Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator. Sci Signal 2011; 4: ra40. [DOI] [PubMed] [Google Scholar]
- 37.Nakagawa O, Fujisawa K, Ishizaki T, Saito Y, Nakao K, Narumiya S. ROCK-I and ROCK-II, two isoforms of Rho-associated coiled-coil forming protein serine/threonine kinase in mice. FEBS Lett 1996; 392: 189–193. [DOI] [PubMed] [Google Scholar]
- 38.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010; 468: 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimura EK, Suzuki M, Igras V, Du J, Lonning S, Miyachi Y et al. Key roles for transforming growth factor beta in melanocyte stem cell maintenance. Cell Stem Cell 2010; 6: 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nobes CD, Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J Cell Biol 1999; 144: 1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan M, Reid MA, Lowman XH, Kulkarni RP, Tran TQ, Liu X et al. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat Cell Biol 2016; 18: 1090–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perona R, Montaner S, Saniger L, Sanchez-Perez I, Bravo R, Lacal JC. Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes Dev 1997; 11: 463–475. [DOI] [PubMed] [Google Scholar]
- 43.Pich C, Teiti I, Sarrabayrouse G, Gallardo F, Gence R, Tilkin-Mariame AF. Melanoma Expressed-CD70 Is Regulated by RhoA and MAPK Pathways without Affecting Vemurafenib Treatment Activity. PLoS One 2016; 11: e0148095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol 2006; 16: 588–596. [DOI] [PubMed] [Google Scholar]
- 45.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011; 480: 387–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramsdale R, Jorissen RN, Li FZ, Al-Obaidi S, Ward T, Sheppard KE et al. The transcription cofactor c-JUN mediates phenotype switching and BRAF inhibitor resistance in melanoma. Sci Signal 2015; 8: ra82. [DOI] [PubMed] [Google Scholar]
- 47.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med 2015; 372: 30–39. [DOI] [PubMed] [Google Scholar]
- 48.Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 2012; 151: 1457–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Routhier A, Astuccio M, Lahey D, Monfredo N, Johnson A, Callahan W et al. Pharmacological inhibition of Rho-kinase signaling with Y-27632 blocks melanoma tumor growth. Oncol Rep 2010; 23: 861–867. [PubMed] [Google Scholar]
- 50.Ruth MC, Xu Y, Maxwell IH, Ahn NG, Norris DA, Shellman YG. RhoC promotes human melanoma invasion in a PI3K/Akt-dependent pathway. J Invest Dermatol 2006; 126: 862–868. [DOI] [PubMed] [Google Scholar]
- 51.Saei A, Palafox M, Benoukraf T, Kumari N, Jaynes PW, Iyengar PV et al. Loss of USP28-mediated BRAF degradation drives resistance to RAF cancer therapies. J Exp Med 2018; 215: 1913–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seo J, Kim J. Regulation of Hippo signaling by actin remodeling. BMB Rep 2018; 51: 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 2017; 546: 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shakhova O, Cheng P, Mishra PJ, Zingg D, Schaefer SM, Debbache J et al. Antagonistic cross-regulation between Sox9 and Sox10 controls an anti-tumorigenic program in melanoma. PLoS Genet 2015; 11: e1004877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen CH, Kim SH, Trousil S, Frederick DT, Piris A, Yuan P et al. Loss of cohesin complex components STAG2 or STAG3 confers resistance to BRAF inhibition in melanoma. Nat Med 2016; 22: 1056–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun 2012; 3: 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 2014; 4: 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Si Y, Ji X, Cao X, Dai X, Xu L, Zhao H et al. Src inhibits the Hippo tumor suppressor pathway through tyrosine phosphorylation of Lats1. Cancer Res 2017. [DOI] [PubMed] [Google Scholar]
- 59.Sit ST, Manser E. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci 2011; 124: 679–683. [DOI] [PubMed] [Google Scholar]
- 60.Song C, Piva M, Sun L, Hong A, Moriceau G, Kong X et al. Recurrent Tumor Cell-Intrinsic and -Extrinsic Alterations during MAPKi-Induced Melanoma Regression and Early Adaptation. Cancer Discov 2017; 7: 1248–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spiering D, Hodgson L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh Migr 2011; 5: 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su Y, Wei W, Robert L, Xue M, Tsoi J, Garcia-Diaz A et al. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc Natl Acad Sci U S A 2017; 114: 13679–13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 2014; 508: 118–122. [DOI] [PubMed] [Google Scholar]
- 64.Taglietti V, Angelini G, Mura G, Bonfanti C, Caruso E, Monteverde S et al. RhoA and ERK signalling regulate the expression of the transcription factor Nfix in myogenic cells. Development 2018; 145. [DOI] [PubMed] [Google Scholar]
- 65.Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016; 352: 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsoi J, Robert L, Paraiso K, Galvan C, Sheu KM, Lay J et al. Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell 2018; 33: 890–904 e895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov 2014; 4: 94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ et al. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 2018; 174: 716–729 e727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vega FM, Fruhwirth G, Ng T, Ridley AJ. RhoA and RhoC have distinct roles in migration and invasion by acting through different targets. J Cell Biol 2011; 193: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov 2013; 3: 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 2012; 487: 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang G, Li Y, Nishimura EK, Xin H, Zhou A, Guo Y et al. Inhibition of PAX3 by TGF-beta modulates melanocyte viability. Mol Cell 2008; 32: 554–563. [DOI] [PubMed] [Google Scholar]
- 73.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013; 41: D955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ye S, Lawlor MA, Rivera-Reyes A, Egolf S, Chor S, Pak K et al. YAP1-Mediated Suppression of USP31 Enhances NFkappaB Activity to Promote Sarcomagenesis. Cancer Res 2018; 78: 2705–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yin L, Morishige K, Takahashi T, Hashimoto K, Ogata S, Tsutsumi S et al. Fasudil inhibits vascular endothelial growth factor-induced angiogenesis in vitro and in vivo. Mol Cancer Ther 2007; 6: 1517–1525. [DOI] [PubMed] [Google Scholar]
- 76.Yu OM, Benitez JA, Plouffe SW, Ryback D, Klein A, Smith J et al. YAP and MRTF-A, transcriptional co-activators of RhoA-mediated gene expression, are critical for glioblastoma tumorigenicity. Oncogene 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol 2015; 17: 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zuckerbraun BS, Shapiro RA, Billiar TR, Tzeng E. RhoA influences the nuclear localization of extracellular signal-regulated kinases to modulate p21Waf/Cip1 expression. Circulation 2003; 108: 876–881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.