Abstract
Sexual reproduction generates genetic diversity and purges genomes of deleterious mutations, allowing organisms to adapt to changing environments and avoid extinction. Cryptococcus species utilize a variety of sexual reproduction mechanisms, which contribute to their ability to occupy myriad environmental niches and to exhibit a range of pathogenic potential in humans. Pathogenic Cryptococcus species can undergo both bisexual and unisexual reproduction when stimulated by properties associated with their environmental niches, which proceed through well-characterized signaling pathways and corresponding morphological changes. Genes governing mating are encoded within the mating-type (MAT) loci, and influence pathogenesis, population dynamics, and lineage divergence within each Cryptococcus species. Although the genes encoded by MAT remain largely conserved across these species, MAT has undergone several important evolutionary changes within the Cryptococcus genus. In Cryptococcus, MAT can exist as either two, genetically and physically unlinked loci in species with tetrapolar mating systems or as a single, large locus in bipolar mating systems; pathogenic Cryptococcus species employ bipolar systems for sexual reproduction, while their characterized nonpathogenic counterparts have tetrapolar mating systems. There is evidence that the transition from the ancestral tetrapolar state in nonpathogenic species to a bipolar mating system in pathogenic Cryptococcus species was mediated by intercentromeric recombination followed by multiple chromosomal rearrangements. In addition to the transition from tetrapolar to bipolar, the Cryptococcus MAT loci have experienced several internal reconfigurations. Due to the variety of established sexual reproduction mechanisms Cryptococcus species utilize and the robust characterization of the evolution of mating and MAT in this genus, Cryptococcus species provide key insights into the evolution of sexual reproduction.
I. Introduction
Sexual reproduction is ubiquitous throughout eukaryotic organisms. It involves mate recognition, zygote formation through gamete fusion, and then gamete or progeny formation through meiosis. In general, sexual reproduction allows reshuffling of genetic material from two parents, which generates recombinant progeny with variable adaptive potential and allows natural selection to act more efficiently to purge deleterious mutations that have accumulated in the parental genomes. While these basic characteristics are shared, there is great diversity in sexual reproduction strategies that have been adopted by different organisms (Goodenough and Heitman, 2014).
In fungi, sexual reproduction is governed by the mating-type locus (MAT), and modes of sexual reproduction differ across fungal species (Heitman et al., 2013). There are self-fertile, inbreeding species (homothallic), as well as outbreeding species where sexual reproduction only occurs between two individuals that are mating compatible (heterothallic). There are two primary mating-type systems that govern mating compatibility in fungi: bipolar mating systems (e.g. ascomycetes) in which mating compatibility is determined by a single MAT locus, and tetrapolar mating systems (e.g. most Basidiomycetous species) where mating compatibility is determined by two genetically and physically unlinked MAT loci. In addition to bipolar and tetrapolar mating systems, the number of functional alleles at the MAT loci also varies among different species, from bi-allelic to multi-allelic, which adds additional complexity to fungal mating systems. Moreover, different mating systems can often be identified in species that are closely related. Thus, the fungal kingdom provides a unique opportunity to study the underlying molecular mechanisms, as well as the evolutionary origins, dynamics, and consequences of sexual reproduction [reviewed in (Heitman, 2015; Heitman et al., 2017; Heitman et al., 2007; Kües et al., 2011; Lee et al., 2010; Ni et al., 2011)].
The human pathogenic Cryptococcus species complex belongs to the phylum Basidiomycota; based on genetic divergence and reproductive isolation identified through genetic crosses, there are currently seven different species defined in this complex (Hagen et al., 2015; Kwon-Chung et al., 2017; Sun and Xu, 2007, 2009). The Cryptococcus pathogenic species are major opportunistic human fungal pathogens that cause cryptococcal meningoencephalitis in both immunocompromised and immunocompetent individuals, and it has been estimated that Cryptococcus is responsible for 223,100 cases of meningoencephalitis annually, with an associated mortality rate of 20% to 70% (Rajasingham et al., 2017). Pathogenic Cryptococcus species have bipolar mating systems, with a single mating-type locus (MAT) and two alternative alleles that define the two compatible mating types: MATα and MATa. The MAT locus in Cryptococcus is large (~120 kb), contains more than 20 genes, and has been shown to be associated with virulence (Lengeler et al., 2002; Lin et al., 2008). Interestingly, comparative genomic studies of Cryptococcus and its closely related non-pathogenic tetrapolar sister species revealed that the MAT locus of the bipolar mating system evolved from fusion of the two MAT loci in the tetrapolar mating system through ectopic recombination likely mediated via inter-centromeric recombination (Sun et al., 2017).
While Cryptococcus species can outcross through bisexual reproduction between isolates of α and a mating types, MATα cells can also reproduce unisexually, which increases inbreeding potential and is an extreme form of inbreeding when it occurs through endoreplication of a solo MATα cell (Fu and Heitman, 2017; Kwon-Chung, 1975, 1976b; Lin et al., 2005). Indeed, evidence of both bisexual and unisexual reproduction has been found in population genetic and genomic studies of natural isolates (Chowdhary et al., 2011; Desjardins et al., 2017; Farrer et al., 2015; Hiremath et al., 2008; Lin et al., 2007).
Thus, the pathogenic Cryptococcus species have a unique MAT locus with diverse mating systems, which provide excellent opportunities for studying the evolution of mating-type loci and mating systems, as well as the evolutionary dynamics and consequences of sexual reproduction. In this review, we focus on: 1) recent advances in studies of bisexual and unisexual reproduction; 2) how sexual reproduction in Cryptococcus affects population structure and dynamics, lineage differentiation, and speciation; and 3) evolution of the bipolar mating system from its ancestral tetrapolar mating system in Cryptococcus and other basidiomycetous species.
II. Sexual reproduction of the pathogenic Cryptococcus species
Bisexual and Unisexual reproduction
Cryptococcus species occupy a number of known environmental niches including Mopane trees native to sub-Saharan Africa, oak trees in North Carolina, eucalyptus trees, and the soil surrounding these trees, but are also known to associate with pigeon guano and animals including many bird and mammal species (Chowdhary et al., 2012; Farrer et al., 2015; Litvintseva et al., 2005; Lugarini et al., 2008; Malik et al., 2003; O’Brien et al., 2004; Singer et al., 2014). Compounds present within several of these niches stimulate mating of Cryptococcus. For example, myo-inositol, a sugar produced by most plants, and the plant hormone indole acetic acid, pigeon guano and various copper concentrations all contribute to initiating mating responses in Cryptococcus (Xue, 2012; Xue et al., 2007). Media made with pigeon guano and various copper concentrations have also been shown to stimulate robust mating (Figure 1) (Chitty et al., 2019; Gyawali et al., 2017; Kent et al., 2008; Lin et al., 2006; Nielsen et al., 2007; Staib, 1981). A myriad of additional environmental stimuli can also trigger Cryptococcus mating. Nutritional cues such as overall nutrient concentration, the absence of glucose, or the presence of glucosamine or galactose are all known to stimulate filamentation (Alspaugh et al., 1997; Xu et al., 2017). Cell-cell interactions among Cryptococcus isolates are also of primary importance in mating. Recently the quorum sensing peptide produced by Cryptococcus cells, Qsp1, which is involved in density-dependent growth and virulence, has also been shown to be involved in the regulation of sexual development (Homer et al., 2016; Lee et al., 2007; Tian et al., 2018). Many other abiotic factors including humidity, CO2 concentration, light, and temperature all contribute to the complex and precise environmental state required to initiate mating in Cryptococcus (Figure 1) (Casadevall and Perfect, 1998; Heitman et al., 2011; Idnurm and Heitman, 2005a, b; Sia et al., 2000).
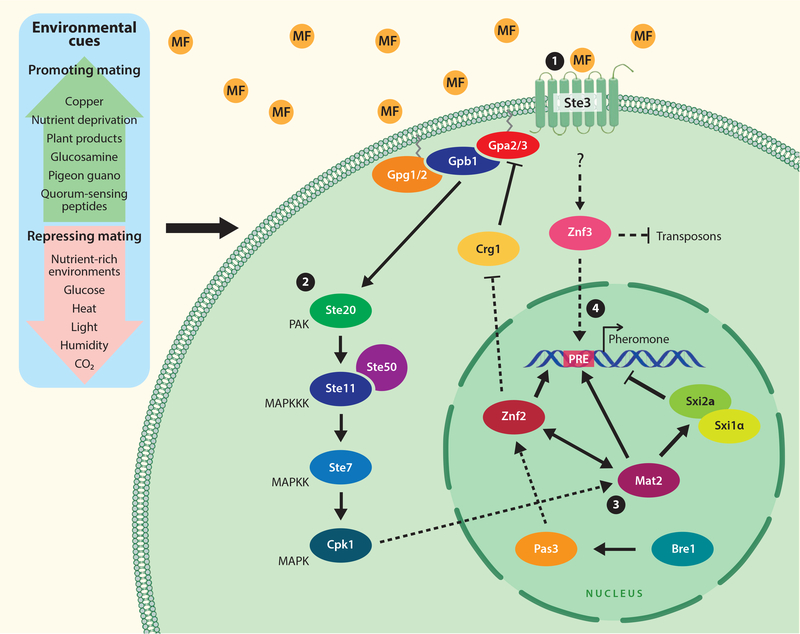
Figure 1. Signal transduction pathways involved in initiation of sexual development in pathogenic Cryptococcus species, as well as environmental factors that influence this process.
The various environmental cues that can either stimulate or repress mating are in the blue shaded box to the left. Schematic depicts the signaling pathway that is initiated when pheromone binds to the Ste3 GPCR. An identical signaling pathway is controlled by the constitutively activated pheromone-like receptor Cpr2. Following GPCR activation, signaling proceeds through a mitogen-activated protein kinase (MAPK) cascade, which activates the key high-mobility transcription factor Mat2. Mat2 signals in concert with other transcription factors and also directly binds the pheromone response element (PRE), initiating transcription of the pheromone-encoding gene MFa/α, and also STE3a/α, SXI1α, SXI2a, GPA2, GPA3, as well as many other genes (Kruzel et al., 2012). In addition to signaling mediated by the Mat2 transcription factor, pheromone-response genes are upregulated in response to signaling from the transcription factor Znf3.
In the proper environment, Cryptococcus cells begin secreting small mating-associated lipid-modified peptides, known as pheromones (Davidson et al., 2000; McClelland et al., 2002). These pheromones are unique to either one or the other of the two Cryptococcus mating types, mating-type α (MATα) and mating-type a (MATa), and are bound by the G-protein coupled receptors (GPCRs) Ste3α and Ste3a (Shen et al., 2002). Pheromone binding to this GPCR activates the pheromone response pathway of Cryptococcus, promoting expression of a variety of genes involved in mating including those encoded by the mating-type locus (MAT) (Figure 1). A constitutively active pheromone receptor-like GPCR, Cpr2, also activates the same signaling pathway (Hsueh et al., 2009). Following activation, the GPCR and its associated subunits signal through the PAK kinase Ste20 to a mitogen-activated protein (MAP)-kinase cascade, which includes Ste11, Ste7, Cpk1, and a recently identified Ste50 adaptor protein (Jung et al., 2011; Wang and Heitman, 1999). This signaling cascade activates the transcription factor Mat2, which directly and indirectly regulates expression of genes encoded by the MAT locus, orchestrating a complex signaling network through several other transcription factors, including Znf2, and the mating-type-specific homeodomain proteins Sxi1α and Sxi2a (Hull et al., 2005; Hull et al., 2002; Kruzel et al., 2012; Lin et al., 2010b; Mead and Hull, 2016). Recent identification of additional novel factors involved in regulating mating in Cryptococcus, including the PAS domain protein Pas3 and Bre1, a ubiquitin ligase required for histone modification, illustrate the important role of epigenetics in this process and identify an opportunity for further study (Zhao et al., 2018). A signaling pathway independent of Mat2 has also been discovered and is regulated, in part, by Znf3, which helps to defend the genome by inhibiting transposition during meiosis (Feretzaki et al., 2016; Feretzaki and Heitman, 2013). Transposition is further regulated during sexual reproduction by an RNAi-dependent process known as sex-induced silencing (SIS) (Wang et al., 2013; Wang et al., 2010).
Following activation of the pheromone response signaling cascade, Cryptococcus cells undergo a number of morphological and cellular changes to complete the sexual cycle, which vary based on the mode of reproduction and species involved (Figure 2). Initially during bisexual reproduction, one of the mating partners (typically the MATα parent) produces a conjugation tube that extends towards its mate, similar to the shmooing process in Saccharomyces cerevisiae. The plasma membranes of the mating partners eventually meet and fuse in a process mediated by the plasma membrane fusion protein Prm1 (Fu and Heitman, 2017). This cell-cell fusion generates a diploid zygote and the zygote subsequently produces a single, elongating hyphal filament, which typically initiates from the MATa cell. The hyphae are divided intermittently by septa and organelles, like mitochondria and nuclei, are appropriately partitioned. Like many eukaryotes, mitochondria are uniparentally inherited in Cryptococcus, typically from the MATa parent. While several genes involved in sexual reproduction, as well as certain environmental factors have been shown to influence the uniparental mitochondrial inheritance in Cryptococcus, the precise mechanism behind this process remains to be elucidated (Gyawali and Lin, 2011; Sun and Xu, 2007; Xu et al., 2000a; Yan et al., 2004; Yan et al., 2007a; Yan et al., 2007b; Yan and Xu, 2003). Once the growing hypha reaches an appropriate length, the hyphal tip differentiates into a terminal, globose basidium. Nuclear fusion and meiosis then typically occur within this structure, generating four recombinant products and marking the penultimate stage of sexual reproduction in Cryptococcus. Ultimately these meiotic products undergo repeated rounds of mitosis and bud from the basidium as four basipetal basidiospore chains (Kwon-Chung, 1975; Kwon-Chung, 1976a; Kwon-Chung, 1976b). Each of these processes that Cryptococcus undergoes during sexual reproduction are mediated by specific genetic regulation. Genes involved in cell-cell fusion (PRM1 and RSC9), filamentation (ZNF2, PUM1, FAS1, CFL1, PAS3, BRE1), sporulation (DDI1, DST1, TOP1, UBC5) and germination (IRR1, PRP11, PRP31, ISP2, EMC3, GRE202) have been identified, although the genetic regulation specific for conjugation tube development and basidium formation has not yet been characterized (Feretzaki and Heitman, 2013; Fu and Heitman, 2017; Huang et al., 2015; Lin et al., 2010a; Wang et al., 2014; Zhao et al., 2018).
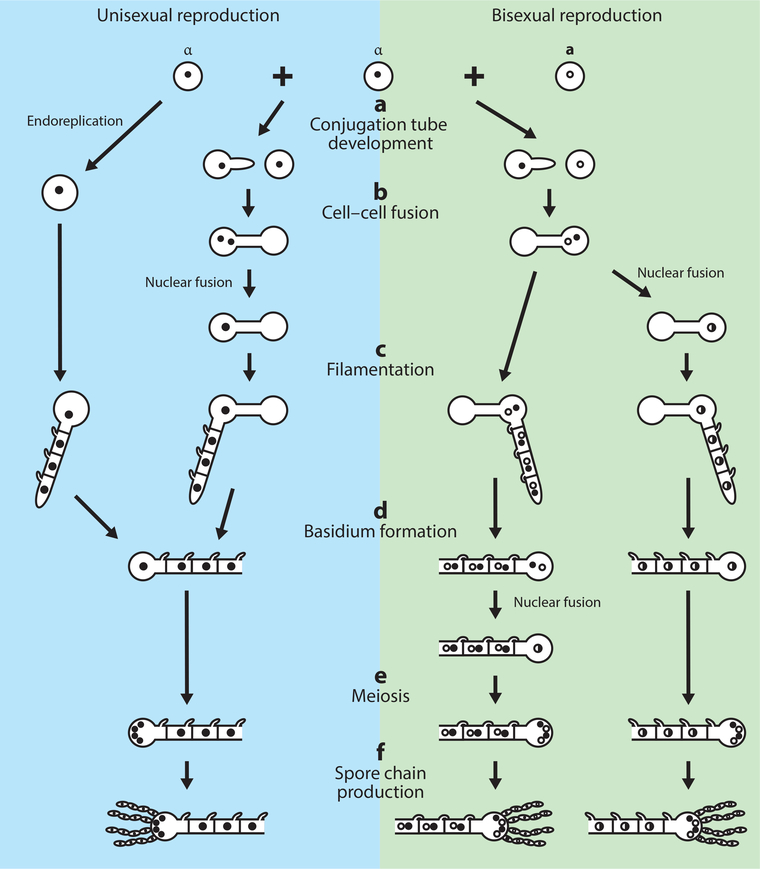
Figure 2. The sexual cycle of pathogenic Cryptococcus species.
Cryptococcus pathogenic species can undergo both unisexual (involving only one mating type) and bisexual reproduction (involving two opposite mating types), depicted in the blue and green shaded boxes, respectively. Following mate-recognition in the appropriate environment, one mating partner produces a conjugation tube that grows toward and eventually fuses with the mating partner. Following cell-cell fusion, a filament protrudes and continues to grow, eventually differentiating into a basidium at its terminus. Meiosis then occurs within the basidium and the meiotic products undergo repeated rounds of mitosis, budding from the basidium as four basidiospore chains. In bisexual reproduction, nuclear fusion can occur at various phases: in C. deneoformans unisex, nuclear fusion can occur post cell-cell fusion before filamentation; in C. deneoformans bisexual reproduction, nuclear fusion can occur post-cell-cell fusion prior to filamentation or within the terminal basidium; in C. neoformans bisexual reproduction, nuclear fusion occurs in the basidium. If nuclear fusion occurs post-cell-cell fusion, the subsequent filament is monokaryotic with unfused clamp cells. If nuclear fusion occurs within the basidium, the prior filament is dikaryotic with fused clamp cells. An alternative mechanism of C. deneoformans unisexual reproduction can occur within a single cell in which the nucleus undergoes endoreplication; this pathway is depicted on the far right.
During bisexual reproduction in C. deneoformans, nuclear fusion, also known as karyogamy, can occur at various stages following cell-cell fusion and is mediated by several karyogamy genes (Fu and Heitman, 2017; Lee and Heitman, 2012). C. deneoformans zygotes produce both dikaryotic hyphae, with two separate haploid nuclei, and monokaryotic hyphae, with a single migrating diploid nucleus, during bisexual reproduction. Conversely, a dikaryotic filament is maintained throughout bisexual reproduction in C. neoformans until nuclear fusion occurs in the basidium. In addition to bisexual reproduction, C. deneoformans can undergo unisexual reproduction, also known as haploid or monokaryotic fruiting (Fu et al., 2014; Lin et al., 2005; Sun and Heitman, 2015). Unlike bisexual reproduction, filaments produced during unisexual reproduction are exclusively monokaryotic and clamp cells along these monokaryotic filaments, which aid in nuclear segregation in dikaryotic hyphae, remain unfused (Lin et al., 2005). Diploidization during unisexual reproduction can occur through either fusion of two cells of the same mating type or through endoreplication (Feretzaki and Heitman, 2013; Fu and Heitman, 2017; Lin et al., 2005). Additionally, it appears that pheromones, the pheromone receptors, and the homeodomain proteins Sxi1α and Sxi2a are dispensable during unisexual reproduction in hyperfilamentous Cryptococcus strains (e.g. XL280) (Gyawali et al., 2017), although they may still be required for haploid fruiting in strains that are less filamentous (e.g. JEC21). Recently, it has been shown that the quorum sensing peptide, Qsp1, is involved in the initiation and coordination of unisexual reproduction in Cryptococcus, in a pheromone- and pheromone receptor-independent fashion. Additionally, an atypical zinc finger regulator Cqs2 has been identified as an important component of the Qsp1 signaling cascade (Tian et al., 2018).
Studies conducted on sexual reproduction in non-neoformans Cryptococcus species suggest that related species utilize similar mating mechanisms, but require different environmental conditions for success. For instance, mating in the pathogenic species C. deuterogattii is more efficient at lower temperatures and takes much longer to produce basidiospores than mating of the C. neoformans and C. deneoformans species (Fraser et al., 2003; Kwon-Chung, 1975; Kwon-Chung, 1976a; Kwon-Chung, 1976b). On the other hand, the closely related non-pathogenic C. depauperatus constitutively undergoes sexual reproduction regardless of environment and may be an example of an obligate sexual fungus (Findley et al., 2009; Kwon-Chung, 2011; Rodriguez-Carres et al., 2010).
Effects of Cryptococcus sexual reproduction on pathogenesis
The most widely acknowledged association between Cryptococcus sexual reproduction and pathogenesis is the generation of sexual basidiospores, which have been shown to be infectious propagules in both a murine inhalation virulence model and the invertebrate model host Galleria mellonella (Giles et al., 2009; Velagapudi et al., 2009). Compared to yeast cells, sexual spores are not only smaller in size, which makes them ideal for alveolar deposition, but also behave differently within the host (Alvarez and Casadevall, 2006; Botts et al., 2009). For example, while yeast cells cannot be phagocytosed by macrophages without opsonization, spores are readily phagocytosed in the absence of opsonins. In addition, spores within macrophages are able to germinate and grow into yeast cells that can withstand reactive oxygen and nitrogen intermediates produced during the macrophage killing response, enabling them to proliferate and eventually escape from the macrophages and disseminate (Alvarez and Casadevall, 2006; Giles et al., 2009; Velagapudi et al., 2009).
Another hallmark of Cryptococcus sexual reproduction is the transition of growth from budding yeast to hyphae, and studies have shown that this morphological change might also be intricately connected with pathogenesis (Lin et al., 2010b; Neilson et al., 1978; Wang et al., 2012; Zimmer et al., 1984). For example, it has been previously shown that one of the key regulators of the yeast-to-hyphal morphological transition is the transcription factor Znf2 (Lin et al., 2010b). Interestingly, cells in which the ZNF2 gene is deleted are locked in the yeast phase and exhibit elevated virulence in the murine inhalation model; on the other hand, cells in which the wild-type ZNF2 allele is replaced with an overexpression allele show elevated filamentation, and consequently, attenuated virulence (Lin et al., 2010b; Wang et al., 2012). Remarkably, a recent study showed that mice pre-treated with either live or heat-killed ZNF2-overexpression filamentous cells were protected against subsequent infection with Cryptococcus isolates that would have otherwise been highly virulent (Zhai et al., 2015). However, it should be noted that hyphae production is not necessarily always associated with attenuated virulence, as some highly self-filamentous isolates (XL280) are more virulent compared closely related isolates that are less filamentous (JEC21) (Feretzaki et al., 2014).
Pathogenic Cryptococcus species are opportunistic human pathogens, which means their primary niches are natural environments, and they spend most of their time interacting with non-human organisms in these natural environments. Thus, from an evolutionary point of view, virulence factors that contribute to pathogenesis in humans may be byproducts of long-term survival in harsh, natural environments and defense strategies developed to combat and compete against natural predators. For example, it has been shown that hyphae (as well as pseudohyphae produced under nutrient limitation conditions) could be beneficial for Cryptococcus to survive amoeba, which is presumably one of their major natural predators (Casadevall, 2012; Lin et al., 2015; Magditch et al., 2012; Steenbergen et al., 2001). Additionally, the natural habitats of Cryptococcus include trees and bird droppings that are normally nutrient limited (promoting mating) and harsh (e.g. temperature fluctuations and extremes, UV and oxidative stress), which could be more challenging to yeast cells than to spores. Thus, the production of spores, an infectious propagule causing human infections, could also be a byproduct of a long-term strategy adopted by Cryptococcus to survive in nature (Botts et al., 2009; Steenbergen et al., 2001).
Another unique morphotype of Cryptococcus, referred to as the titan cell, was recently identified in the lungs of infected patients and mice, and was later successfully induced in vitro (Crabtree et al., 2012; Dambuza et al., 2018; Gerstein et al., 2015; Hommel et al., 2018; Okagaki et al., 2010; Okagaki et al., 2011; Trevijano-Contador et al., 2018). Titan cells are large (>20 um in diameter compared to ~5 um for typical yeast cells), have polyploid genomes, and produce budding cells that are usually haploid, diploid, or aneuploid. Titan cells are resistant to phagocytosis by macrophages, and thus can better sustain attacks from the host immune system than yeast cells. Thus, they may play unique roles in latency, dissemination, and infection. The fact that titan cells are polyploid and produce haploid budding cells suggests that their cell cycle involves ploidy changes, mimicking what happens during sexual reproduction, undergoing 1N (parental cells) → 2N (zygotes) → 1N (meiotic progeny) transitions in ploidy. Although it is not known whether the formation of titan cells, as well as the subsequent generation of haploid/diploid/aneuploid daughter cells, involves sex or meiosis, these observations may reflect aspects of unisexual reproduction occurring during infection.
Effects of Cryptococcus sexual reproduction on population structure and dynamics in nature
Advances in sequencing technology have propelled the analyses of natural Cryptococcus isolates from population genetics to population genomics, allowing more comprehensive genomic studies to identify evolutionary signatures of clonal expansion, recombination, and gene flow, as well as signs of positive and/or purifying selection. Several recent studies of large collections of C. neoformans and C. gattii isolates showed that natural Cryptococcus populations are largely clonal, with signs of sexual reproduction and recombination occurring at low frequencies (Billmyre et al., 2014; Desjardins et al., 2017; Engelthaler et al., 2014; Farrer et al., 2015; Rhodes et al., 2017). Additionally, evidence of recombination has been detected in both VNBI/VNBII lineages that have both MATa and MATα isolates occurring at comparable frequencies, and the VNI lineage in which the vast majority of the isolates are MATα, consistent with models in which both bisexual and unisexual reproduction can promote recombination. This is in overall agreement with previous population genetics studies of Cryptococcus that identified clonal structures among natural Cryptococcus isolates (Hiremath et al., 2008; Randhawa et al., 2008). Interestingly, a recent population genomics study of different C. neoformans lineages revealed the a and α MAT loci have distinct evolutionary trajectories, and suggested introgression of the MATa allele from VNBI lineage to the VNI group (Desjardins et al., 2017). Specifically, the authors found that the MATα alleles display a phylogeny that overall matches the whole-genome phylogeny with one exception: VNBI MATα appeared to be paraphyletic with respect to VNBII, and has 2 distinct alleles – one comprised of some VNBI isolates and the other of the remaining VNBI isolate in addition to all of the VNBII isolates. The phylogeny of the MATa alleles also differed from whole-genome phylogeny in that the VNI and VNBI lineages are sister groups that are separated by a large distance from VNBII, suggesting that the extant MATa alleles of the VNI lineage originated from an introgression event from the VNBI group. Furthermore, the MATa allele transgressed into the VNI lineage carries ∼5.3 kb of VNBI sequence upstream of the 5′ end of the MAT locus, consistent with previous studies showing the presence of recombination hot spots flanking MAT in Cryptococcus (Hsueh et al., 2006; Sun and Heitman, 2016; Sun et al., 2012). We hypothesize that the most common global Cryptococcus lineage (serotype A VNI) lost the MATa locus and then regained it by introgression from the divergent VNBI lineage endemic to sub-Saharan Africa. Notably, the VNI lineage is almost exclusively α mating-type (99.9%), consistent with a genetic bottleneck while exiting Africa and transition to unisexual reproduction. Introgression of mating-type alleles by inter-specific hybridization has also been reported in other fungi, such as the Dutch elm disease fungus Ophiostoma novo-ulmi. It has been shown that when this plant pathogen first invaded Europe as a series of clonal populations, there was only one mating type (MAT-2) and a single vegetative incompatibility type. The populations then rapidly became diversified with the appearance of the MAT-1 mating type, which O. novo-ulmi likely acquired from another species, Ophiostoma ulmi, through inter-species hybridization. This inter-species gene transfer of the MAT-1 mating type has been hypothesized to have facilitated the rapid adaptation of O. novo-ulmi to a new environment after the initial invasion (Et-Touil et al., 1999; Paoletti et al., 2006).
Natural populations of Cryptococcus are dominated by isolates of the MATα mating type (Randhawa et al., 2008), and MATa strains have been primarily isolated from African countries, such as Botswana (Litvintseva et al., 2003). Indeed, population genomics studies have shown that isolates from Africa display the clearest signals of sexual reproduction and have increased genetic diversity. However, the discovery of unisexual reproduction among MATα cells, via either cell-cell fusion or endoreplication, suggests that the frequency of sexual reproduction in nature might have been underestimated (Fu and Heitman, 2017; Lin et al., 2005). Just as in bisexual reproduction, unisexual reproduction produces robust numbers of basidiospores, which could serve as infectious propagules in places where MATa cells are mostly absent. Additionally, it has been shown that compared to bisexual reproduction between MATα and MATa cells, unisexual reproduction between MATα cells has comparable frequencies of meiotic recombination, and consequently, has similar potential in reshuffling genetic materials from the two parental strains and generating progeny with unique genotypes (Desjardins et al., 2017; Farrer et al., 2015; Rhodes et al., 2017; Roth et al., 2018; Sun et al., 2014). Additionally, during unisexual reproduction, the two parental MATα alleles are collinear, and thus allow proper pairing and crossing over to occur within the MAT region, which can enable recombinational repair of the MAT genes (Sun et al., 2014). Diploid Cryptococcus isolates with two MATα alleles (e.g. αAAα, αDDα) have also been identified among natural isolates, providing further evidence that unisexual reproduction occurs in nature (Lin et al., 2007; Lin et al., 2009; Rhodes et al., 2017).
Effects of Cryptococcus sexual reproduction on lineage divergence and speciation
Sexual reproduction facilitates gene flow between diverging lineages, and therefore counteracts the establishment of species boundaries. Without sufficient interbreeding, when lineages of the same species diverge, the accumulation of genetic variants will reduce the viability and/or the fertility of progeny produced by inter-lineage sexual reproduction. Within the pathogenic C. neoformans/C. gattii species complex, there are currently as many as seven recognized species (Hagen et al., 2015). Additionally, within the C. neoformans species there are four lineages that are well-separated from each other phylogenetically (Farrer et al., 2015). It is most likely that the reproductive boundaries among the Cryptococcus species and lineages are post-zygotic, as inter-lineage and inter-species crosses often produce normal sexual structures, including hyphal growth, clamp cell formation, basidia formation, and basidiospore production. This is also consistent with studies showing that factors involved in pre-zygotic mating interactions, such as pheromones and pheromone receptor genes, are highly conserved and display mating-type specific topologies (Findley et al., 2009; Findley et al., 2012; Fraser et al., 2007).
Post-zygotic reproductive barriers are likely due to accumulated sequence divergence, including both sequence polymorphisms and gross chromosomal rearrangements that collectively compromise meiosis, particularly chromosomal mis-segregation during meiosis I. This typically increases the frequency of progeny with imbalanced genetic makeups that are either inviable or infertile. Indeed, during sexual reproduction between C. neoformans and C. deneoformans, progeny exhibit highly reduced viability, and are largely either diploid or aneuploid with various levels of heterozygosity throughout the genome, consistent with compromised meiosis with significantly reduced levels of recombination (Lengeler et al., 2001; Sun and Xu, 2007, 2009). Diploid serotype AD hybrid Cryptococcus isolates have also been frequently isolated from natural environments, suggesting either ongoing hybridization between C. neoformans and C. deneoformans, or that diploid hybrid progeny might be advantageous (hybrid vigor), and thus, persistent in natural environments (Lin et al., 2007; Lin et al., 2009; Xu et al., 2002; Xu et al., 2000b). Additionally, studies have shown that diploid and aneuploid Cryptococcus progeny produced from sexual reproduction can undergo LOH events that are either regional or across entire chromosomes through mitotic recombination and whole-chromosome loss and regain, respectively, which could provide additional sources of genetic and phenotypic diversity among meiotic progeny (Sun et al., 2014).
As mentioned earlier, post-zygotic reproductive isolation is likely due to genetic divergence accumulated between different lineages/species, including both nucleotide substitutions as well as chromosomal rearrangements such as inversions and translocations. It has been shown that during hybridization between different yeast (as well as bacteria) species, the DNA mismatch repair system functions to prevent recombination between homologous chromosomal regions with elevated nucleotide divergence. The resulting compromised chromosomal pairing and insufficient crossovers leads to chromosomal mis-segregation during meiosis I and results in progeny with imbalanced genetic material that are often inviable (Chambers et al., 1996; Greig et al., 2003; Hunter et al., 1996; Liti et al., 2006; Rayssiguier et al., 1989; Roeder, 1997). Thus, a similar mechanism may operate in Cryptococcus and contribute to the observed low spore viability from inter-lineage/inter-species hybridization. Interestingly, natural Cryptococcus isolates with null mutations in key mismatch repair genes have been identified in population genomic studies (Billmyre et al., 2017). These isolates have long branches in their associated phylogenies, consistent with accelerated rates of mutation accumulation. They also show elevated rates of mutation in the lab that lead to a high frequency of resistance to certain anti-fungal drugs. Future studies on the impact of these mismatch repair defects on relaxing reproductive isolation boundaries could yield interesting insights into the evolution of this species.
Another source of genetic variation that contributes to reproductive isolation is chromosomal rearrangements, which have been shown to have significant effects on speciation, or at least during the onset of these events (Delneri et al., 2003; Fischer et al., 2000; Hou et al., 2014). For example, it has been shown that chromosomal rearrangements alone could explain the observed hybrid sterility caused by meiosis I chromosome segregation between two closely related yeast species (Rogers et al., 2018). For Cryptococcus species, while it has been known that karyotypic variation and chromosomal rearrangements are present among natural isolates (Boekhout and Belkum, 1997; Dromer et al., 1994; Fries et al., 1996; Klepser and Pfaller, 1998; Perfect et al., 1993; Polacheck and Lebens, 1989; Sun and Xu, 2009; Wickes et al., 1994), most population genomic studies have focused on nucleotide polymorphisms (Desjardins et al., 2017; Farrer et al., 2015). This is partly due to the technical limitations of analyzing chromosomal rearrangements, which require comparison of high quality de novo genome assemblies that, until recently, have been difficult to generate. Improvements in long-read sequencing technologies, such as PacBio and Nanopore, as well as accompanying advancements in genome assembly algorithms, have allowed efficient de novo assembly of high quality, end-to-end chromosomal-level genome assemblies for Cryptococcus and closely related species (Passer et al., 2019; Sun et al., 2017; Yadav et al., 2018). These advances will enable analyses of chromosomal rearrangements events, including their origins, maintenance, and spread in the population, as well as their potential effects on fitness, pathogenesis, and lineage divergence, thus allowing a better understanding of the evolution and speciation of Cryptococcus.
Evolution of sexual reproduction in Cryptococcus and closely related species
All of the species in the C. neoformans/C. gattii complex have a bipolar mating system, in which the two mating types, α and a, are determined by a single MAT locus that is unusually large (~120 kb in size) and encompasses more than 20 genes (Fraser et al., 2004; Lengeler et al., 2002; Loftus et al., 2005). While recombination within the MAT locus is repressed during bisexual reproduction, crossover events within the MAT locus have been observed during unisexual reproduction between two MATα cells, and recombination and gene conversion hotspots have been identified in regions flanking and within the MAT locus, respectively (Hsueh et al., 2006; Sun et al., 2014; Sun and Heitman, 2015, 2016; Sun et al., 2012; Sun and Xu, 2007). Interestingly, the species that are most closely related to the C. neoformans/C. gattii complex, such as Cryptococcus amylolentus, Cryptococcus floricola, and Cryptococcus wingfieldii have tetrapolar mating systems, which is the ancestral configuration of the basidiomycetes (Findley et al., 2012; Passer et al., 2019; Sun et al., 2017). Genome comparison between C. neoformans and C. amylolentus showed that almost all of the genes that are present in the C. neoformans MAT locus are also present within the P/R and HD MAT loci in C. amylolentus. This comparison also revealed that the transition from the ancestral tetrapolar system to the extant bipolar mating system in C. neoformans is the result of a fusion between the ancestral P/R and HD loci that was likely mediated by inter-centromeric ectopic recombination (Sun et al., 2017). This fusion occurred once in the last shared common ancestor to the pathogenic species complex, concomitant with the emergence of this monophyletic group of highly successful human fungal pathogen.
So what could be the benefits of having a bipolar mating system for the pathogenic Cryptococcus species? One possible advantage of bipolar mating systems might be related to the ability of individuals to find suitable mating partners. For species with bipolar mating systems, the chance that two isolates in a random encounter are also mating compatible is 50% when the two mating types are at equilibrium in a population. This is always higher compared to tetrapolar species with low numbers of alleles at the P/R and HD loci when the two loci are in linkage equilibrium. In actuality, for tetrapolar species to achieve 50% mating compatibility between two random individuals, both P/R and HD loci must have more than two alleles and the total number of alleles of the two MAT loci must be equal to or greater than eight (e.g. 2/3 * 4/5 = 8/15, i.e. chance of compatibility is about 53%). While there are many examples of tetrapolar fungal species with large numbers of alleles at both P/R and HD loci that together constitute hundreds or even thousands of different mating types, such as Coprinus cinereus and Schizophyllum commune (Kothe, 1996; Kües et al., 2011; Raper, 1966), there could be scenarios where the numbers of mating-type alleles present in the population are significantly reduced or the two loci are in significant linkage disequilibrium (e.g. population bottlenecks and clonal expansions) which could then provide selective advantages for bipolar mating systems. Indeed, there are tetrapolar species that are bi-allelic at one of the two MAT loci, such as Ustilago maydis, which is bi-allelic at the a locus (P/R locus) and multi-allelic at the b locus (HD locus) (Kronstad and Leong, 1990). Interestingly, the closely related species Ustilago hordei has a bipolar mating system in which the P/R and HD loci show complete genetic linkage even though they are located far from each other on the same chromosome (Bakkeren and Kronstad, 1994). There are also species with pseudobipolar mating systems, in which the P/R and HD loci are located on the same chromosomes but are not fused, and populations where recombination between the two MAT loci has been detected, such as those in the Malassezia species complex (Coelho et al., 2013; Gioti et al., 2013). However, it is not yet clear in these cases how many different functional alleles are present at the two MAT loci, and how much recombination is occurring between them in natural populations. Nevertheless, conditions favoring the transition from tetrapolar to pseudobipolar or bipolar mating systems might be more prevalent in nature than currently appreciated.
The population of pathogenic Cryptococcus species in Africa has been hypothesized to be the origin of the global Cryptococcus population and shows a relatively balanced distribution of the two alleles, α and a, of the MAT locus. Interestingly though, the global Cryptococcus population has a highly skewed MAT allele distribution, with the vast majority of the isolates belonging to the α mating type. This disproportionate number of MATα isolates may have initially significantly reduced outcrossing potential. It is possible that this scenario provided selective pressure or an advantage for the evolution of unisexual reproduction, which allowed Cryptococcus to take advantage of the benefits of sex, even in the face of a population composed of individuals of the same mating type, by increasing the outcrossing potential to almost 100%.
Another possible advantage of transitioning to a bipolar mating system could be provided by selective pressures on chromosomal regions other than MAT. Cryptococcus amylolentus is the closest known sister species to the pathogenic Cryptococcus species complex and it has a tetrapolar mating system (Findley et al., 2012; Sun et al., 2017). While neither the P/R nor the HD MAT locus in C. amylolentus is tightly linked to its centromere physically, both loci show elevated genetic linkage to their respective centromere and repressed recombination within the inter-MAT-CEN regions. Thus, it is possible that in the intermediate pseudobipolar stage, in which the P/R and HD loci have moved onto the same chromosome via inter-centromeric ectopic recombination, the two MAT loci are located on opposite sides of the centromere and recombination is repressed across the entire region between the two MAT loci that encompasses the centromere. This situation may have provided selective pressure for the two MAT loci to be located closer together and to eventually fuse with each other to form the extant bipolar configuration in the pathogenic Cryptococcus species, such that the region that was originally trapped between the two MAT loci could be released to freely undergo recombination (Sun et al., 2017). If this is the case, one would expect reduced recombination frequencies in regions between the two MAT loci in species with pseudobipolar mating systems, like those in the Malassezia species complex, such as M. sympodialis and M. globosa (Gioti et al., 2013; Sankaranarayanan et al., 2019; Zhu et al., 2017).
For species with bipolar mating systems, progeny will be mating compatible with 50% of their siblings, a high fraction relative to progeny of a tetrapolar species regardless of the numbers of alleles at the P/R and HD loci in the population. Inbreeding among these progeny could be detrimental in the long-term. However, the long-term cost could be offset if sexual structures (e.g. hyphae and spores) provided immediate benefits in the context of nutrient acquisition, predator defense, or stress tolerance. Additionally, if two sibling progeny are sufficiently genetically divergent, consecutive rounds of inter-sibling mating could also break up allele combinations in the parental strains and generate a progeny pool that represents extensive reshuffling of the parental genetic material, and thus, increase the genetic and phenotypic diversity of the progeny. Additionally, both bisexual and unisexual reproduction can generate phenotypic and genotypic diversity de novo, in many cases as a result of aneuploidy (Ni et al., 2013).
For microorganisms that can also reproduce mitotically, sexual reproduction is not necessarily always the optimal form of reproduction. Sexual reproduction produces pools of progeny that are genetically and phenotypically diverse, and only some will have higher fitness. Progeny with higher fitness will likely undergo clonal expansion, generating linkage disequilibrium, which could in turn be broken up during subsequent rounds of sexual reproduction when conditions favoring sexual reproduction arise again. Thus, a possible reproductive strategy for microorganisms could involve rare sexual reproduction interrupted by long epochs of asexual clonal expansion (Heitman et al., 2013).
III. Evolution of the mating-type locus (MAT) in Cryptococcus
MAT locus structure
Mating-type identity in fungi is controlled by genes at the mating-type (MAT) locus. This region of the genome is unique in that it differs considerably between mating partners of the same species. The mating type has been hypothesized to be a virulence factor in C. neoformans based on disease epidemiology and animal studies (Kwon-Chung et al., 1992; Nielsen et al., 2005; Nielsen et al., 2003), the infectious properties of basidiospores, and the prevalence of the α mating type in clinical and environmental isolates (Kwon-Chung and Bennett, 1978). Given these intriguing characteristics, differences between MATa and MATα cell types as well as the genes encoded by the MAT locus and their targets became a great source of interest. Therefore, extensive work has been conducted focusing on this unique genomic region of C. neoformans.
Early studies by Moore and Edman used a difference cloning procedure to isolate sequences of the α-genome that were absent in the a-genome, identifying a 35-kb region, which included the C. neoformans pheromone gene (MFα1) and co-segregated with the α-mating type (Moore and Edman, 1993). Subsequent efforts analyzing cosmid subclones containing α-specific DNA defined the MATα locus boundaries to a ~50-kb region specific to MATα strains (Karos et al., 2000; Moore and Edman, 1993). This region contained multiple copies of MFα and one copy of a gene encoding the GPCR Ste3α. Additionally, three homologs of the S. cerevisiae pheromone response pathway (STE11α, STE12α, and STE20α) were identified within 24 kb of MFα1 (Karos et al., 2000), already hinting at the possibility that other genes involved in mating or sexual reproduction would likely also be located within the MAT locus, indicative of selection favoring linkage of genes with related functions into co-segregating units.
To understand the organization of the C. neoformans MAT locus further, Lengeler et al. carried out sequencing analysis of both MATα and MATa regions in C. neoformans (serotype A, strains H99α and 125.91a) and C. deneoformans (serotype D, strains JEC21α and JEC20a) strains of opposite mating types using genomic BAC libraries (Lengeler et al., 2002). Alignment of the nucleotide sequences revealed identical sequences flanking a region of non-identical DNA sequences between mating types that spanned 105 to 125 kb, demonstrating the MAT locus was larger than previously hypothesized (Lengeler et al., 2002) (Figure 3). Additional analyses identified the homeodomain gene SXI1α (Sex Inducer 1α) of the HD1 class at the left end of the MATα locus (Lengeler et al., 2002). SXI1α was later shown to be a key factor for establishing α-cell identity and progression through the sexual cycle, and interestingly, was sufficient to drive sexual development when expressed in a cells (Hull et al., 2002). However, it was obvious that a factor from MATa cells was also required for completion of the sexual cycle. Using a combination of molecular genetics and sequence analysis approaches, a homeodomain gene counterpart (SXI2a) of the HD2 class specific to a cells was successfully identified, which likewise forced α cells to adopt an a/α cell fate when introduced into α cells, resulting in production of hyphae, basidia, and basidiospores (Hull et al., 2005).
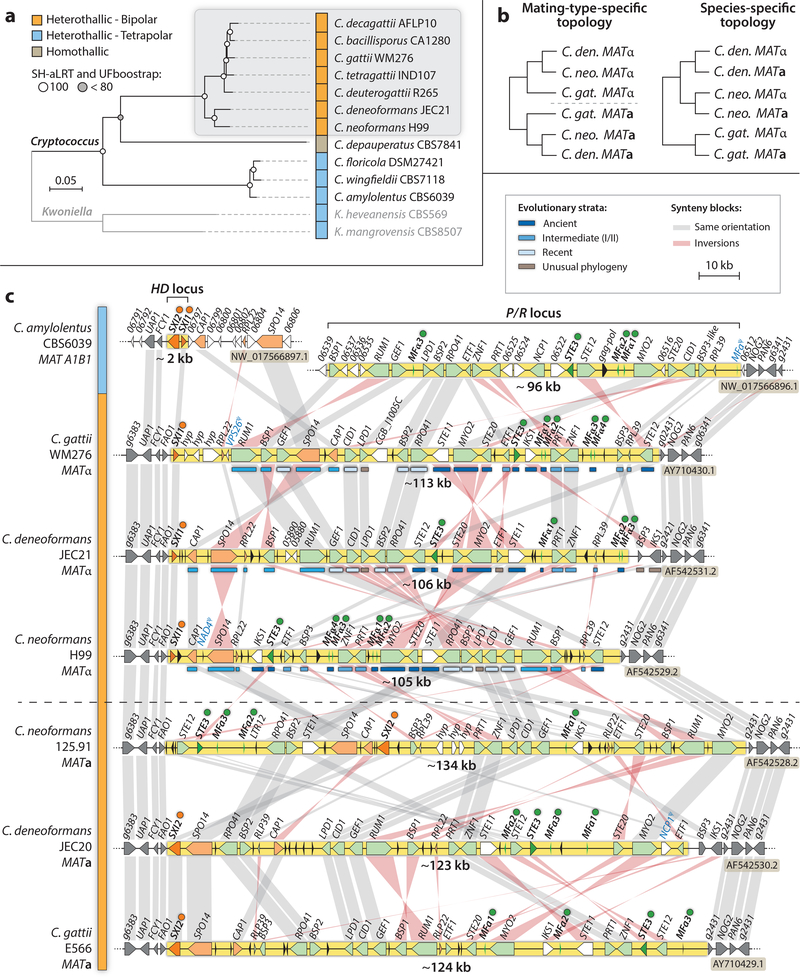
Figure 3. Cryptococcus phylogeny and comparative analyses of MAT alleles from pathogenic and closely related non-pathogenic Cryptococcus species.
(a) Seven species are currently recognized among the pathogenic C. neoformans/C. gattii species complex (shadowed in grey), all of which have linked P/R and HD mating-type loci (bipolar). Conversely, C. amylolentus, C. floricola, C. wingfieldii, and the two Kwoniella species (outgroup) are tetrapolar, with mating-type genes at two genetic loci on different chromosomes representing the ancestral configuration of the MAT loci. The tree was inferred from 734 single copy orthologs (149,332 informative amino acid sites), using maximum likelihood and the LG+F+R4 model of amino acid substitution in IQ-TREE. Branch support was assessed by ultrafast bootstrap (UFboot) and SH-aLRT methods. Scale bar indicates the number of substitutions per site. (b) Example of tree topologies observed for genes anciently (left) or recently (right) recruited to MAT, representing different evolutionary strata. (c) The MAT alleles of the pathogenic species are highly rearranged between species and mating types and result from linkage between P/R and HD gene clusters located on different chromosomes in C. amylolentus. The nonrecombining MAT region is depicted in yellow (with the number beneath indicating the approximate size), and is bordered by ~10 kb of common flanking regions depicted on each side (4 genes in dark grey), except in C. deneoformans where the IKS1 and BSP3 genes were evicted from MAT and fixed in the flanking region by inversion and recombination. Common genes found in the HD and P/R regions of C. amylolentus are colored in orange and green, respectively, in the extant MAT locus of the pathogenic species. Genes encoding homeodomain transcription factors (SXI1 and SXI2), pheromones (MF), and pheromone receptors (STE3) are bulleted. Blue bars below the genes indicate the evolutionary strata class they belong to as shown in the key. Genes in both the “Ancient” and the “Intermediate” strata classes have mating-type specific phylogenies (they entered MAT prior to speciation), while genes in the “Recent” strata class display species-specific phylogenies (they began diverging only after speciation). Genes with unusual phylogenies represent cases where the gene underwent gene conversion in one or the other lineage, thereby fixing one of the two alleles in both mating types and with concomitant loss of the alternative allele. Grey and pink bars connect orthologs with the same or inverted orientation, respectively. Genes depicted in white are not conserved in all species. Pseudogenes are labeled in blue, and black arrows represent transposable elements or their remnants.
Subsequent comparative analyses showed that the C. neoformans MAT locus is a unique genomic region because it differs considerably between mating types and encompasses more than 20 genes (Fraser et al., 2004; Lengeler et al., 2002), including those encoding mating pheromones (MFα or MFa) and their cognate receptors (STE3α or STE3a), as well as homeodomain transcription factors (HD1 or SXI1α and HD2 or SXI2a) that together govern cell-type identity. Of the 20 genes, 5 are essential based on gene disruption studies (MYO2, PRT1, RPL39, RPL22, and RPO41) (Fraser et al., 2004), while others have diverse functions during mating (STE11, STE12, and STE20), sporulation (SPO14 and RUM1), and in virulence (CAP1). It is also noteworthy that in contrast to other basidiomycetes, where a divergently transcribed HD1/HD2 gene pair is present in each mating type (Coelho et al., 2017), the C. neoformans MATα locus encodes only Sxi1α (HD1), while the MATa locus encodes only Sxi2a (HD2). This is, however, an exception because for almost every other MAT locus gene, there are similar, yet non-identical, alleles in both MATα and MATa in C. neoformans (Fraser et al., 2004).
Although MATa is clearly allelic to MATα, the organization of MATa reveals extensive rearrangement of the genes compared to MATα, a feature associated with suppression of recombination within MAT (Figure 3) (Fraser et al., 2004). This represents a scenario where recessive losses of genes from one mating type that do not play an essential role in haploid cells, as shown for SXI1α and SXI2a (Hull et al., 2005), could likely occur because each is sheltered in a permanent hemizygous state. These losses may constitute a first sign of mating-type chromosome degeneration as expected for longstanding, non-recombining regions (Bergero and Charlesworth, 2009). However, the presence of essential genes spaced throughout the MAT locus (Fraser et al., 2004) may, in contrast, pose an evolutionary constraint ensuring that large regions of the MAT locus are not lost through sexual recombination. Additionally, it has been shown that recombination hotspots associated with the presence of GC-rich motifs are located in the regions that flank the MAT locus in Cryptococcus (Hsueh et al., 2006), which potentially function as repressors of crossover within the MAT locus due to interference (Berchowitz and Copenhaver, 2010; Sun et al., 2017).
To gain further insight into the evolutionary trajectory of the MAT locus, Lengeler et al. and Fraser et al. (Fraser et al., 2004; Lengeler et al., 2002) cloned and sequenced extant MAT alleles in C. neoformans and C. deneoformans, which diverged ~20 million years ago (mya), and in C. gattii, which diverged from the C. neoformans/C. deneoformans common ancestor approximately ~40 mya (Casadevall et al., 2017; D’Souza et al., 2011; Sharpton et al., 2008; Xu et al., 2000b) (Figure 3a). This study revealed that the MAT gene cohort has been dramatically rearranged during evolution of the three lineages (Fraser et al., 2004) (Figure 3c). The subsequent completion of the genome sequences of representatives of three species (i.e. C. deneoformans JEC21, C. neoformans H99 and C. gattii WM276) (D’Souza et al., 2011; Janbon et al., 2014; Loftus et al., 2005) has broadened and accelerated Cryptococcus research. For instance, sequencing confirmed that the MAT locus occupies ~6% of a 1.8-Mb chromosome in an ~19-Mb genome (excluding the rDNA repeats that constitute ~5% of the genome) (Loftus et al., 2005). It also became clear that rearrangements at the MAT locus are highly atypical compared to non-MAT regions of the genome, and likely resulted from intra-allelic recombination between transposable elements and their remnants, which are enriched in the MAT locus relative to the rest of the genome (excluding the transposon-rich centromeric regions) (Fraser et al., 2004; Janbon et al., 2014; Loftus et al., 2005; Yadav et al., 2018).
Phylogenetic analyses of the alleles of MAT genes across these three Cryptococcus species demonstrated that the MAT locus is composed of four gene strata of distinct evolutionary ages, including: (i) a set of ancestral genes (STE3, MF and genes of the pheromone-sensing MAPK pathway) with mating-type-specific phylogenies; (ii) two strata of more intermediate evolutionary origin (e.g. RPL22, SPO14, RUM1, BSP1) and (iii) a set of recently acquired genes that have species-specific tree topologies, similar to genes outside MAT (Fraser et al., 2004) (Figure 3b,c). However, genes in the Cryptococcus MAT locus are no longer organized by age, as they been heavily reshuffled during speciation and mating-type divergence (Figure 3c). Furthermore, some genes seem to have been punctuated by more recent gene conversion events, resetting their molecular clock (e.g. IKS1 in C. deneoformans, Figure 3c), and direct experimental evidence of such phenomena has been more recently obtained (Sun et al., 2012). Nevertheless, by plotting the synonymous divergence level of each protein coding gene in MAT it was possible to infer an ancestral state where one gene cluster contained the pheromone and pheromone receptor genes (P/R cluster), and the other cluster contained the homeodomain-encoding gene (HD cluster). Together, this led to a model where two ancestral unlinked P/R and HD loci regions were juxtaposed by a chromosomal translocation, giving rise to the extant bipolar Cryptococcus MAT locus (Fraser et al., 2004).
To provide experimental evidence to support this model, strains of C. neoformans were genetically engineered so that the homeodomain genes (SXI1α or SXI2a) were physically unlinked to the pheromone/pheromone receptor genes (Hsueh et al., 2008). In this way, the HD and P/R loci could segregate independently during meiosis. These strains mimicking a tetrapolar organization were able to mate and complete the sexual cycle, but when they were independently crossed with a strain harboring a contiguous MAT locus, only ~50% of the progeny were fertile. This provides evidence that the transitional “tripolar” state imposes disadvantages, which could in turn facilitate the transition from a tripolar to a bipolar state through recombination.
Evolution of the Cryptococcus MAT locus
Several studies support the hypothesis that tetrapolarity is the ancestral state of basidiomycetes. However, there have been multiple reports of transitions to bipolarity in the three major basidiomycete lineages ((Branco et al., 2017; Branco et al., 2018), reviewed in (Coelho et al., 2017)), including the human-pathogenic species of the C. neoformans/C. gattii complex as described above. Recent phylogenetic analyses resulted in the taxonomic reclassification of the Cryptococcus genus (Hagen et al., 2015; Liu et al., 2015). Several yeast species are now known to be closely related to the human-pathogenic clade, including the homothallic, strictly filamentous fungus Cryptococcus depauperatus, and a clade composed of three yeast species: C. amylolentus, C. wingfieldii, and the recently described Cryptococcus floricola (Passer et al., 2019) (Figure 3a).
To further understand how this unique bipolar mating system evolved in the pathogenic Cryptococcus species, studies have been conducted to investigate the MAT locus structures in non-pathogenic Cryptococcus species and also in more distantly related species of the sister genus Kwoniella (Findley et al., 2012; Guerreiro et al., 2013; Metin et al., 2010; Passer et al., 2019). In these studies, C. amylolentus, C. wingfieldii, C. floricola, Kwoniella heveanensis, and Kwoniella mangrovensis were shown to be tetrapolar, with P/R and HD loci located on different chromosomes (Figure 3a,b). The assignment of these species as tetrapolar is in accord with mating studies and analysis of F1 progeny (i.e. isolates need to differ at both the A and B MAT loci to be inter-fertile). Therefore, only in the lineage giving rise to the Cryptococcus human pathogens has there been a major genomic alteration, with the two MAT loci now residing on a single chromosome. Interestingly, in the non-pathogenic Cryptococcus species and in Kwoniella spp. the HD locus is restricted to an ~2 kb region, encodes two linked and divergently oriented homeodomain genes (HD1 and HD2) like most basidiomycetes, and is multiallelic, with allele numbers varying among species (e.g. K. heveanensis has at least 6 different HD alleles). This differs from the solo HD genes (SXI1α, SXI2a) of the pathogenic Cryptococcus complex and suggests that one or the other homeodomain gene was recently lost in C. neoformans/C. gattii in association with linkage of the two MAT loci. Compared to the HD locus, the P/R locus in the non-pathogenic species is expanded, spanning ~30 kb in Kwoniella spp. (Guerreiro et al., 2013; Metin et al., 2010) and up to 96 kb in C. amylolentus (Findley et al., 2012; Sun et al., 2017). These large P/R loci encode pheromone and pheromone receptor genes along with several other genes that are integrated within the MAT locus of the pathogenic Cryptococcus species. This indicates that some of these genes were already linked to the P/R locus prior to fusion of P/R and HD loci in the pathogenic Cryptococcus complex (Figure 3c). Therefore, the organization of MAT in other Cryptococcus species and in Kwoniella mirrors key aspects of the proposed intermediates in the evolution of MAT in the pathogenic Cryptococcus species.
Genome sequencing of hundreds of Cryptococcus isolates is currently taking place at an increasing scale and speed. While most of the data is still being generated on the Illumina platform, recent developments in long-read Pacific Biosciences and Oxford Nanopore Technologies sequencing technologies are providing more contiguous genome assemblies and are helping to resolve repetitive regions including the centromeres (Sun et al., 2017; Yadav et al., 2018). Indeed, a recent study by Sun et al. (Sun et al., 2017) revealed that, similar to the pathogenic Cryptococcus species (Janbon et al., 2014; Yadav et al., 2018), C. amylolentus has regional centromeres that are enriched with species-specific transposable elements. It was also shown that the MAT loci of C. amylolentus displays linkage with their respective centromeres despite overall collinearity of the intervening genomic regions (Sun et al., 2017). Suppression of recombination between each MAT locus and the centromere ensures segregation of mating-type in the first meiotic division, and thus, the generation of only two mating types per meiosis. This arrangement in a tetrapolar species therefore results in similar odds of compatibility between products of the same meiosis as that of progeny produced by linked P/R and HD loci in bipolar species. Similar instances of mating type loci evolution have been reported (e.g. in the anther-smut fungus Microbotryum lagerheimii) (Hood et al., 2015), indicating that equally beneficial phenotypes can emerge through different genomic changes in response to natural selection. Suppression of recombination without substantial rearrangements raises the hypothesis that genetic elements involved in epistatic interactions favor co-segregation of alleles from the same parent. Alternatively, recombination could be restricted in these regions due to the presence of epigenetic factors such as DNA methylation or histone modifications, which could potentially affect double-strand break formation and recombination initiation, as several examples have suggested (Maloisel and Rossignol, 1998; Termolino et al., 2016). Additional studies will be required to unravel the causes of recombination suppression in these regions and to define the role of suppression of recombination in the transition between unlinked and linked MAT loci.
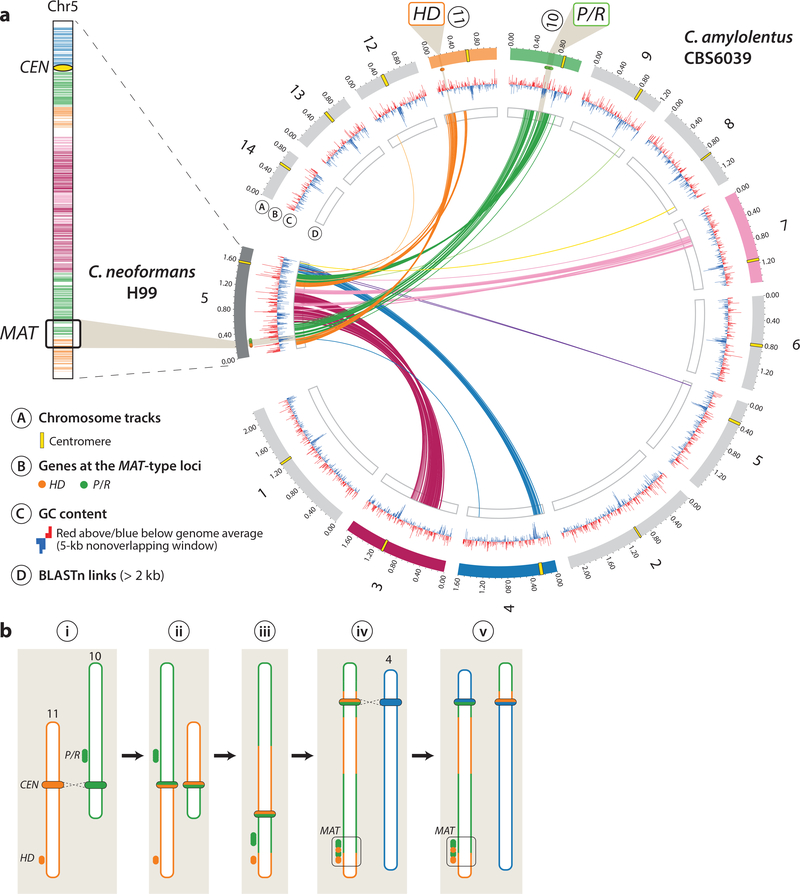
Finally, because centromeres are enriched with transposable elements and repetitive sequences, and some elements are shared among different chromosomes, Sun et al. proposed that the fusion of the ancestral P/R and HD loci that gave rise to the bipolar mating system in the pathogenic Cryptococcus species was initiated by ectopic intercentromeric recombination (Figure 4). Consistent with this, several lines of evidence now challenge the view that centromeres are typically recombination-free regions (Jaco et al., 2008; Shi et al., 2010; Symington and Petes, 1988). Recombination at these regions may lead to chromosomal translocations, which in this case would bring the ancestral P/R and HD loci onto the same chromosome, as the comparison of the extant MAT chromosome of C. neoformans with the chromosomes harboring the P/R and HD loci in C. amylolentus suggests (Figure 4).
Figure 4. Evolution of a bipolar mating system from a tetrapolar mating system via inter-centromeric ectopic recombination mediated by transposable elements shared between centromeres.
(a) Genome comparison between chromosome 5 of C. neoformans H99 and C. amylolentus CBS6039 chromosomes. Distribution of BLASTN hits showing that chromosome 5 of C. neoformans is made up of several C. amylolentus chromosome regions, implying multiple rearrangements along the evolutionary history of the two lineages. (b) Proposed model for the transition from tetrapolar to bipolar MAT organization. This model proposes intercentromeric recombination as a key mechanism bringing the two ancestrally unlinked P/R and HD loci into a single MAT locus, thus linking the two sets of genes determining mating type.
In conclusion, these analyses give rise to an evolutionary model in which the single MAT locus of the human-pathogenic Cryptococcus lineages evolved from an ancestral tetrapolar mating system via a series of steps, including gene acquisition, chromosomal translocations involving the centromere, and subsequent intrachromosomal rearrangements possibly mediated by transposable elements or repeat-rich regions.
MAT locus function
One distinct feature of the tetrapolar mating-system and consequently of mating in the Basidiomycota is that the genes encoding pheromones and pheromone receptors have acquired a mating-type defining role, in contrast to what occurs in the ascomycetes (Bennett and Turgeon, 2017; Coelho et al., 2017). The archetype of the P/R locus encodes pheromone-receptor genes homologous to the STE3 receptor gene from S. cerevisiae, a G-protein-coupled receptor with seven transmembrane domains, and pheromone precursor genes, which are homologous to the lipid-modified MFa pheromone gene of S. cerevisiae (Moore and Edman, 1993). Each of the mating types of Cryptococcus encodes a STE3 receptor gene that is activated by binding of its respective pheromone. It is not unusual for the MAT locus of basidiomycetes to encode more than one pheromone precursor gene, and in the case of Cryptococcus pathogenic species, each mating type has between three and four pheromone precursor genes (Figure 3c) (Coelho et al., 2017; Kües et al., 2011). Similarly to the pheromones of other basidiomycetes, Cryptococcus pheromones are first translated into pheromone precursor proteins which subsequently undergo post-translational processing at both the N- and C-terminal regions. A general feature of pheromone precursor proteins is the presence of a “CaaX” motif at the carboxyl terminus of the precursor, where “C” stands for cysteine, “a” for aliphatic amino acids and “X” stands for any residue (Raudaskoski and Kothe, 2010). The multistep maturation process of the pheromone precursors involves farnesylation, C-terminal proteolytic cleavage of the motif “aaX”, followed by carboxymethylation of the cysteine residue and proteolytic cleavage at the N-terminus, producing the mature lipopetide pheromones (Raudaskoski and Kothe, 2010). In addition to pheromones and pheromone receptors, the Cryptococcus MAT locus also encodes one homeodomain transcription factor gene in each of the two mating types, as mentioned previously SXI1α for MATα strains and SXI2a for MATa strains, each of which belongs to one of two distinct classes of homeodomain transcription factors based on structural homologies and distinct protein sequences of the DNA-binding motifs (Hull et al., 2005; Kües and Casselton, 1992; Kües et al., 2011). SXI1α belongs to the HD1 class of homeodomain transcription factors that are considered to have an atypical homeodomain, with three additional amino acids between helices one and two of the three helices comprising the homeodomain motif. Conversely, SXI2a belongs to the HD2 class of homeodomain transcription factors that have a canonical homeodomain, with 60 residues forming a three helical structure and a highly conserved DNA-binding region (Hull et al., 2005; Kües and Casselton, 1992; Kües et al., 2011). For these transcription factors to be functional in a heterothallic mating, it is necessary that they heterodimerize with each other after cell fusion in order to complete the mating process (Hull et al., 2005).
In addition to being key molecular determinants for mating, some of the MAT genes are also involved in other cellular processes of major importance for the cell. For example, SXI1 and SXI2 are not only important for determining mating-type identity and regulating sexual development, but are also indispensable for uniparental mitochondrial DNA inheritance (from the MATa cell) after cell fusion in heterothallic mating (Gyawali and Lin, 2013; Yan et al., 2007a). In C. amylolentus it was shown that mitochondrial inheritance is also uniparental and controlled by MAT-encoded genes, however in this case the P/R locus controls this process independently from the HD locus (Findley et al., 2012).
The MAT loci of the pathogenic Cryptococcus species also include genes that have been shown to be involved in virulence attributes, like the production of melanin, the formation of a polysaccharide capsule, and growth at 37°C (Kwon-Chung et al., 1992; Lengeler et al., 2002). For example, it has been reported that the MATα allele of C. deneoformans is associated with increased virulence given that MATα cells are more virulent than congenic MATa cells. However, this result was obtained only in certain genetic backgrounds (JEC21α/JEC20a and KN433α/KN433a) and not in others (KN3501α/KN3501a), and was also absent in C. neoformans congenic strains (H99α/KN99a), indicating that this phenotype might be not only species-specific but also strain-specific (Hsueh et al., 2011; Kwon-Chung et al., 1992; Nielsen et al., 2005; Nielsen et al., 2003). Additionally, the genes or molecular mechanisms responsible for the increase virulence observed in some of the MATα congenic strains of C. deneoformans have yet to be clearly elucidated (Wang et al., 2002). One gene hypothesized to be partially responsible for the increased virulence of C. deneoformans MATα is STE12, given that deletion or overexpression of this allele was shown to respectively, downregulate or upregulate the LAC1 gene, which encodes a laccase necessary for the production of the antioxidant pigment melanin, an important virulence factor of Cryptococcus (Chang et al., 2001; Chang et al., 2000; Kwon-Chung et al., 1992; Williamson, 1997). Additionally, both STE12 alleles influence the capsule size of cryptococcal cells in brain smears obtained from mice infected with C. deneoformans (Chang et al., 2001). In C. gattii, a ste12αΔ mutant showed a diminished production of melanin and attenuated virulence in mice models (Ren et al., 2006). However, C. neoformans ste12Δ mutants do not show reduced virulence (Yue et al., 1999), indicating that the relationship between STE12 and virulence may also be species-specific (Nielsen et al., 2003). The PAK kinase homologue STE20α encoded by the C. neoformans MAT locus is thought to play a role not only in the pheromone signal MAPK cascade, but also in virulence and thermotolerance (Nichols et al., 2004; Wang et al., 2002). Similarly, the MAT-encoded CAP1 gene is thought to encode a capsule-associated protein involved in the addition of xylose to the polysaccharide capsule of Cryptococcus, even though its biochemical functions remain unknown (Klutts et al., 2007; O’Meara and Alspaugh, 2012). Finally, other genes encoded at MAT do not appear to be directly involved in mating or virulence, including the genes thought to be essential based on genetic disruption assays, like the MYO2 gene that encodes a myosin homolog, the PRT1 gene that encodes a translation initiation factor, genes RPL39 and RPL22 that encode ribosomal proteins, and the RPO41 gene that encodes a mitochondrial RNA polymerase (Fraser et al., 2004).
Considering the speed and magnitude of the technical advances in the field of genome sequencing and analysis, we are likely at the cusp of understanding the differences observed between mating types in much more detail. Exploring the expression of all MAT-encoded transcripts (including long non-coding RNAs), the possible DNA or histone methylation states of this region at different stages of development, and the 3D interactions between the MAT locus and other regions of the genome, we are bound to gain a greater understanding of the factors that control mating and virulence in this important group of human fungal pathogens that are a cause of significant morbidity and mortality globally.
IV. Conclusion and future directions
We have gained great insights on the effects of sexual reproduction on many aspects of Cryptococcus, including its pathogenesis, evolution of the mating-type locus, and population structure and dynamics. Questions that remain to be investigated include:
What are the underlying mechanisms governing unisexual reproduction vs. bisexual reproduction in the distinct pathogenic Cryptococcus species?
What are the driving forces for the evolution of the bipolar mating system in Cryptococcus, as well as its large MAT locus and the genes resident therein? Are there other genes within the MAT locus other than the pheromone, pheromone receptor, and HD genes that play critical roles in sexual development in Cryptococcus species?
How much structural variation is present among the genomes of natural isolates, such as chromosomal translocations and inversions? Do these changes in genome structure have effects on fitness, including virulence? How do they affect sexual reproduction, and vice versa?
What are the underlying mechanisms of many cellular processes associated with Cryptococcus sexual reproduction, such as sex-induced genome defense against transposable elements as well as uniparental inheritance of mitochondria from the MATa parent?
What is the complete suite of genes and their networks that contribute to the pathogenesis of Cryptococcus? Taking advantage of sexual reproduction, combined with availability of high throughput genome sequencing and phenotyping, it is possible to perform high-resolution QTL analyses of virulence related factors, such as high-temperature tolerance and drug resistance, using segregants isolated from laboratory crosses. These analyses have the potential to identify new candidate loci/genes associated with Cryptococcus pathogenesis.
Acknowledgement
We thank Anna Floyd Averette, Ci Fu, and Vikas Yadav for comments and suggestions during the preparation of this review. This work was funded by the NIH/NIAID R37 MERIT award AI39115-21, R01 grant AI50113-15, and R01 grant AI133654-2 awarded to JH.
Reference
- Alspaugh JA, Perfect JR, and Heitman J (1997). Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev 11, 3206–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M, and Casadevall A (2006). Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol 16, 2161–2165. [DOI] [PubMed] [Google Scholar]
- Bakkeren G, and Kronstad JW (1994). Linkage of mating-type loci distinguishes bipolar from tetrapolar mating in basidiomycetous smut fungi. PNAS 91, 7085–7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, and Turgeon BG (2017). Fungal Sex: The Ascomycota In The Fungal Kingdom, Heitman J, Howlett BJ, Crous PW, Stukenbrock EH, James TY, and Gow NAR, eds. (Washington DC: ASM Press; ). [Google Scholar]
- Berchowitz LE, and Copenhaver GP (2010). Genetic interference: don’t stand so close to me. Curr Genomics 11, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, and Charlesworth D (2009). The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol 24, 94–102. [DOI] [PubMed] [Google Scholar]
- Billmyre RB, Clancey SA, and Heitman J (2017). Natural mismatch repair mutations mediate phenotypic diversity and drug resistance in Cryptococcus deuterogattii. eLife 6, e28802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billmyre RB, Croll D, Li W, Mieczkowski P, Carter DA, Cuomo CA, Kronstad JW, and Heitman J (2014). Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution. mBio 5, e01494–01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhout T, and Belkum AV (1997). Variability of karyotypes and RAPD types in genetically related strains of Cryptococcus neoformans. Curr Genet 32, 203–208. [DOI] [PubMed] [Google Scholar]
- Botts MR, Giles SS, Gates MA, Kozel TR, and Hull CM (2009). Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis. Eukaryot Cell 8, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco S, Badouin H, Rodríguez de la Vega RC, Gouzy J, Carpentier F, Aguileta G, Siguenza S, Brandenburg J-T, Coelho MA, Hood ME, et al. (2017). Evolutionary strata on young mating-type chromosomes despite the lack of sexual antagonism. PNAS 114, 7067–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco S, Carpentier F, Rodríguez de la Vega RC, Badouin H, Snirc A, Le Prieur S, Coelho MA, de Vienne DM, Hartmann FE, Begerow D, et al. (2018). Multiple convergent supergene evolution events in mating-type chromosomes. Nature Communications 9, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, ed. (2012). Amoeba provide insight into the origin of virulence in pathogenic fungi In: Mylonakis E, Ausubel F, Gilmore M., Casadevall A (eds) Recent Advances on Model Hosts. Advances in Experimental Medicine and Biology, vol 710 Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Freij JB, Hann-Soden C, and Taylor J (2017). Continental drift and speciation of the Cryptococcus neoformans and Cryptococcus gattii species complexes. mSphere 2, e00103–00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, and Perfect JR (1998). Cryptococcus neoformans (Washington, D.C.: ASM Press; ). [Google Scholar]
- Chambers SR, Hunter N, Louis EJ, and Borts RH (1996). The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol Cell Biol 16, 6110–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Penoyer LA, and Kwon-Chung KJ (2001). The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence. PNAS 98, 3258–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Wickes BL, Miller GF, Penoyer LA, and Kwon-Chung KJ (2000). Cryptococcus neoformans STE12alpha regulates virulence but is not essential for mating. The Journal of Experimental Medicine 191, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitty JL, Edwards DJ, Robertson AAB, Butler MS, Duley JA, Cooper MA, and Fraser JA (2019). Quantitation of purines from pigeon guano and implications for Cryptococcus neoformans survival during infection. Mycopathologia, 10.1007/s11046-11018-10315-11040. [DOI] [PubMed] [Google Scholar]
- Chowdhary A, Hiremath SS, Sun S, Kowshik T, Randhawa HS, and Xu J (2011). Genetic differentiation, recombination and clonal expansion in environmental populations of Cryptococcus gattii in India. Environ Microbiol 13, 1875–1888. [DOI] [PubMed] [Google Scholar]
- Chowdhary A, Randhawa HS, Prakash A, and Meis JF (2012). Environmental prevalence of Cryptococcus neoformans and Cryptococcus gattii in India: An update. Crit Rev Microbiol 38, 1–16. [DOI] [PubMed] [Google Scholar]
- Coelho MA, Bakkeren G, Sun S, Hood ME, and Giraud T (2017). Fungal Sex: The Basidiomycota In The Fungal Kingdom, Heitman J, Howlett BJ, Crous PW, Stukenbrock EH, James TY, and Gow NAR, eds. (Washington DC: ASM Press; ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MA, Sampaio JP, and Gonçalves P (2013). Living and thriving on the skin: Malassezia genomes tell the story. mBio 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree JN, Okagaki LH, Wiesner DL, Strain AK, Nielsen JN, and Nielsen K (2012). Titan cell production enhances the virulence of Cryptococcus neoformans. Infect Immun 80, 3776–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza CA, Kronstad JW, Taylor G, Warren R, Yuen M, Hu G, Jung WH, Sham A, Kidd SE, Tangen K, et al. (2011). Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. mBio 2, e00342–00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambuza IM, Drake T, Chapuis A, Zhou X, Correia J, Taylor-Smith L, LeGrave N, Rasmussen T, Fisher MC, Bicanic T, et al. (2018). The Cryptococcus neoformans Titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS Path 14, e1006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Moore TD, Odom AR, and Heitman J (2000). Characterization of the MFα pheromone of the human fungal pathogen Cryptococcus neoformans. Mol Microbiol 38, 1017–1026. [DOI] [PubMed] [Google Scholar]
- Delneri D, Colson I, Grammenoudi S, Roberts IN, Louis EJ, and Oliver SG (2003). Engineering evolution to study speciation in yeasts. Nature 422, 68–72. [DOI] [PubMed] [Google Scholar]
- Desjardins CA, Giamberardino C, Sykes SM, Yu C-H, Tenor JL, Chen Y, Yang T, Jones AM, Sun S, Haverkamp MR, et al. (2017). Population genomics and the evolution of virulence in the fungal pathogen Cryptococcus neoformans. Genome Res 27, 1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dromer F, Varma A, Ronin O, Mathoulin S, and Dupont B (1994). Molecular typing of Cryptococcus neoformans serotype D clinical isolates. J Clin Microbiol 32, 2364–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelthaler DM, Hicks ND, Gillece JD, Roe CC, Schupp JM, Driebe EM, Gilgado F, Carriconde F, Trilles L, Firacative C, et al. (2014). Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. mBio 5, e01464–01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Et-Touil A, Brasier CM, and Bernier L (1999). Localization of a pathogenicity gene in Ophiostoma novo-ulmi and evidence that it may be introgressed from O. ulmi. Mol Plant-Microbe Interact 12, 6–15. [Google Scholar]
- Farrer RA, Desjardins CA, Sakthikumar S, Gujja S, Saif S, Zeng Q, Chen Y, Voelz K, Heitman J, May RC, et al. (2015). Genome evolution and innovation across the four major lineages of Cryptococcus gattii. mBio 6, e00868–00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Billmyre RB, Clancey SA, Wang X, and Heitman J (2016). Gene network polymorphism illuminates loss and retention of novel RNAi silencing components in the Cryptococcus pathogenic species complex. PLoS Genet 12, e1005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Hardison SE, Wormley FL Jr., and Heitman J (2014). Cryptococcus neoformans hyperfilamentous strain is hypervirulent in a murine model of Cryptococcal meningoencephalitis. PLoS ONE 9, e104432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, and Heitman J (2013). Genetic circuits that govern bisexual and unisexual reproduction in Cryptococcus neoformans. PLoS Genet 9, e1003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K, Rodriguez-Carres M, Metin B, Kroiss J, Fonseca A, Vilgalys R, and Heitman J (2009). Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the Tremellales. Eukaryot Cell 8, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K, Sun S, Fraser JA, Hsueh Y-P, Averette AF, Li W, Dietrich FS, and Heitman J (2012). Discovery of a modified tetrapolar sexual cycle in Cryptococcus amylolentus and the evolution of MAT in the Cryptococcus species complex. PLoS Genet 8, e1002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, James SA, Roberts IN, Oliver SG, and Louis EJ (2000). Chromosomal evolution in Saccharomyces. Nature 405, 451–454. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, and Heitman J (2004). Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol 2, e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Hsueh Y-P, Findley K, and Heitman J (2007). Evolution of the mating-type locus: the basidiomycetes In Sex in Fungi, Heitman J, Kronstad JW,. Taylor JW, and Casselton LA, eds. (Washington, DC: ASM Press; ). [Google Scholar]
- Fraser JA, Subaran RL, Nichols CB, and Heitman J (2003). Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver island, Canada. Eukaryot Cell 2, 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries BC, Chen F, Currie BP, and Casadevall A (1996). Karyotype instability in Cryptococcus neoformans infection. J Clin Microbiol 34, 1531–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, and Heitman J (2017). PRM1 and KAR5 function in cell-cell fusion and karyogamy to drive distinct bisexual and unisexual cycles in the Cryptococcus pathogenic species complex. PLoS Genet 13, e1007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Sun S, Billmyre RB, Roach KC, and Heitman J (2014). Unisexual versus bisexual mating in Cryptococcus neoformans: consequences and biological impacts. Fungal Genet Biol 78, 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Fu MS, Mukaremera L, Li Z, Ormerod KL, Fraser JA, Berman J, and Nielsen K (2015). Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. mBio 6, e01340–01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles SS, Dagenais TRT, Botts MR, Keller NP, and Hull CM (2009). Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect Immun 77, 3491–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioti A, Nystedt B, Li W, Xu J, Andersson A, Averette AF, Münch K, Wang X, Kappauf C, Kingsbury JM, et al. (2013). Genomic insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis. mBio 4, e00572–00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, and Heitman J (2014). Origins of eukaryotic sexual reproduction. Cold Spring Harbor Perspectives in Biology 6, a016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig D, Travisano M, Louis EJ, and Borts RH (2003). A role for the mismatch repair system during incipient speciation in Saccharomyces. J Evol Biol 16, 429–437. [DOI] [PubMed] [Google Scholar]
- Guerreiro MA, Springer DJ, Rodrigues JA, Rusche LN, Findley K, Heitman J, and Fonseca Á (2013). Molecular and genetic evidence for a tetrapolar mating system in the basidiomycetous yeast Kwoniella mangrovensis and two novel sibling species. Eukaryot Cell 12, 746–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyawali R, and Lin X (2011). Mechanisms of uniparental mitochondrial DNA inheritance in Cryptococcus neoformans. Mycobiology 39, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyawali R, and Lin X (2013). Prezygotic and postzygotic control of uniparental mitochondrial DNA inheritance in Cryptococcus neoformans. mBio 4, e00112–00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyawali R, Zhao Y, Lin J, Fan Y, Xu X, Upadhyay S, and Lin X (2017). Pheromone independent unisexual development in Cryptococcus neoformans. PLoS Genet 13, e1006772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, Falk R, Parnmen S, Lumbsch HT, and Boekhout T (2015). Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol 78, 16–48. [DOI] [PubMed] [Google Scholar]
- Heitman J (2015). Evolution of sexual reproduction: A view from the fungal kingdom supports an evolutionary epoch with sex before sexes. Fungal Biology Reviews 29, 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Howlett BJ, Crous PW, Stukenbrock EH, James TY, and Gow NAR (2017). The Fungal Kingdom (American Society for Microbiology). [Google Scholar]
- Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, and Casadevall A (2011). Cryptococcus (American Society for Microbiology). [Google Scholar]
- Heitman J, Kronstad JW, Taylor JW, and Casselton LA (2007). Sex in Fungi (Washington, DC: ASM Press; ). [Google Scholar]
- Heitman J, Sun S, and James TY (2013). Evolution of fungal sexual reproduction. Mycologia 105, 1–27. [DOI] [PubMed] [Google Scholar]
- Hiremath SS, Chowdhary A, Kowshik T, Randhawa HS, Sun S, and Xu J (2008). Long-distance dispersal and recombination in environmental populations of Cryptococcus neoformans var. grubii from India. Microbiology 154, 1513–1524. [DOI] [PubMed] [Google Scholar]
- Homer Christina M., Summers Diana K., Goranov Alexi I., Clarke Starlynn C., Wiesner Darin L., Diedrich Jolene K., Moresco James J., Toffaletti D, Upadhya R, Caradonna I, et al. (2016). Intracellular action of a secreted peptide required for fungal virulence. Cell Host & Microbe 19, 849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommel B, Mukaremera L, Cordero RJB, Coelho C, Desjardins CA, Sturny-Leclère A, Janbon G, Perfect JR, Fraser JA, Casadevall A, et al. (2018). Titan cells formation in Cryptococcus neoformans is finely tuned by environmental conditions and modulated by positive and negative genetic regulators. PLoS Path 14, e1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood ME, Scott M, and Hwang M (2015). Breaking linkage between mating compatibility factors: Tetrapolarity in Microbotryum. Evolution 69, 2561–2572. [DOI] [PubMed] [Google Scholar]
- Hou J, Friedrich A, de Montigny J, and Schacherer J (2014). Chromosomal rearrangements as a major mechanism in the onset of reproductive isolation in Saccharomyces cerevisiae. Curr Biol 24, 1153–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Fraser JA, and Heitman J (2008). Transitions in sexuality: recapitulation of an ancestral tri- and tetrapolar mating system in Cryptococcus neoformans. Eukaryot Cell 7, 1847–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Idnurm A, and Heitman J (2006). Recombination hotspots flank the Cryptococcus mating-type locus: implications for the evolution of a fungal sex chromosome. PLoS Genet 2, e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Metin B, Findley K, Rodriguez-Carres M, and Heitman J (2011). The mating type locus of Cryptococcus: evolution of gene clusters governing sex determination and sexual reproduction from the phylogenomic perspective In Cryptococcus: from human pathogen to model yeast, Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, and Casadevall A, eds. (Washington, DC: American Society for Microbiology; ), pp. 139–149. [Google Scholar]
- Hsueh YP, Xue C, and Heitman J (2009). A constitutively active GPCR governs morphogenic transitions in Cryptococcus neoformans. EMBO J 28, 1220–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Hebert AS, Coon JJ, and Hull CM (2015). Protein composition of infectious spores reveals novel sexual development and germination factors in Cryptococcus. PLoS Genet 11, e1005490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Boily MJ, and Heitman J (2005). Sex-specific homeodomain proteins Sxi1α and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot Cell 4, 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Davidson RC, and Heitman J (2002). Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes Dev 16, 3046–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Chambers SR, Louis EJ, and Borts RH (1996). The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J 15, 1726–1733. [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, and Heitman J (2005a). Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol 3, e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, and Heitman J (2005b). Photosensing fungi: phytochrome in the spotlight. Curr Biol 15, R829–832. [DOI] [PubMed] [Google Scholar]
- Jaco I, Canela A, Vera E, and Blasco MA (2008). Centromere mitotic recombination in mammalian cells. J Cell Biol 181, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G, Ormerod KL, Paulet D, Byrnes EJ III, Yadav V, Chatterjee G, Mullapudi N, Hon C-C, Billmyre RB, Brunel F, et al. (2014). Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet 10, e1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K-W, Kim S-Y, Okagaki LH, Nielsen K, and Bahn Y-S (2011). Ste50 adaptor protein governs sexual differentiation of Cryptococcus neoformans via the pheromone-response MAPK signaling pathway. Fungal Genet Biol 48, 154–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karos M, Chang YC, McClelland CM, Clarke DL, Fu J, Wickes BL, and Kwon-Chung KJ (2000). Mapping of the Cryptococcus neoformans MATα locus: presence of mating type-specific mitogen-activated protein kinase cascade homologs. J Bacteriol 182, 6222–6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent CR, Ortiz-Bermúdez P, Giles SS, and Hull CM (2008). Formulation of a defined V8 medium for induction of sexual development of Cryptococcus neoformans. Appl Environ Microbiol 74, 6248–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepser ME, and Pfaller MA (1998). Variation in electrophoretic karyotype and antifungal susceptibility of clinical isolates of Cryptococcus neoformans at a university-affiliated teaching hospital from 1987 to 1994. J Clin Microbiol 36, 3653–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klutts JS, Levery SB, and Doering TL (2007). A b-1,2-xylosyltransferase from Cryptococcus neoformans defines a new family of glycosyltransferases. J Biol Chem 282, 17890–17899. [DOI] [PubMed] [Google Scholar]
- Kothe E (1996). Tetrapolar fungal mating types: sexes by the thousands. FEMS Microbiol Rev 18, 65–87. [DOI] [PubMed] [Google Scholar]
- Kronstad JW, and Leong SA (1990). The b mating-type locus of Ustilago maydis contains variable and constant regions. Genes Dev 4, 1384–1395. [DOI] [PubMed] [Google Scholar]
- Kruzel EK, Giles SS, and Hull CM (2012). Analysis of Cryptococcus neoformans sexual development reveals rewiring of the pheromone response network by a change in transcription factor identity. Genetics 191, 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kües U, and Casselton LA (1992). Homeodomains and regulation of sexual development in basidiomycetes. Trends Genet 8, 154–155. [DOI] [PubMed] [Google Scholar]
- Kües U, James TY, and Heitman J (2011). Mating type in basidiomycetes: unipolar, bipolar, and tetrapolar patterns of sexuality In Evolution of fungi and fungal-like organisms, The Mycota XIV, Pöggeler S, and Wöstemeyer J, eds. (Springer Berlin Heidelberg; ), pp. 97–160. [Google Scholar]
- Kwon-Chung KJ (1975). A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67, 1197–1200. [PubMed] [Google Scholar]
- Kwon-Chung KJ (1976a). Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68, 821–833. [PubMed] [Google Scholar]
- Kwon-Chung KJ (1976b). A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68, 943–946. [PubMed] [Google Scholar]
- Kwon-Chung KJ (2011). Filobasidiella. In The Yeasts -- A Taxonomic Study, Kurtzman CP, Fell JW, and Boekhout T, eds. (Amsterdam, The Netherlands: Elsevier Science; ), pp. 1443–1455. [Google Scholar]
- Kwon-Chung KJ, and Bennett JE (1978). Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am J Epidemiol 108, 337–340. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE, Wickes BL, Meyer W, Cuomo CA, Wollenburg KR, Bicanic TA, Castañeda E, Chang YC, Chen J, et al. (2017). The case for adopting the “Species Complex” nomenclature for the etiologic agents of Cryptococcosis. mSphere 2, e00357–00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Edman JC, and Wickes BL (1992). Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun 60, 602–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Chang YC, Nardone G, and Kwon-Chung KJ (2007). TUP1 disruption in Cryptococcus neoformans uncovers a peptide-mediated density-dependent growth phenomenon that mimics quorum sensing. Mol Microbiol 64, 591–601. [DOI] [PubMed] [Google Scholar]
- Lee SC, and Heitman J (2012). Function of Cryptococcus neoformans KAR7 (SEC66) in karyogamy during unisexual and opposite-sex mating. Eukaryot Cell 11, 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Ni M, Li W, Shertz C, and Heitman J (2010). The evolution of sex: a perspective from the fungal kingdom. Microbiol Mol Biol Rev 74, 298–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Cox GM, and Heitman J (2001). Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect Immun 69, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Fox DS, Fraser JA, Allen A, Forrester K, Dietrich FS, and Heitman J (2002). Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot Cell 1, 704–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Idnurm A, and Lin X (2015). Morphology and its underlying genetic regulation impact the interaction between Cryptococcus neoformans and its hosts. Med Mycol 53, 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Huang JC, Mitchell TG, and Heitman J (2006). Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATalpha allele enhances filamentation. PLoS Genet 2, e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Hull CM, and Heitman J (2005). Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434, 1017–1021. [DOI] [PubMed] [Google Scholar]
- Lin X, Jackson JC, Feretzaki M, Xue C, and Heitman J (2010a). Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet 6, e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Jackson JC, Feretzaki M, Xue C, and Heitman J (2010b). Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet 6, e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, and Heitman J (2007). αADα hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet 3, 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Nielsen K, Patel S, and Heitman J (2008). Impact of mating type, serotype, and ploidy on the virulence of Cryptococcus neoformans. Infect Immun 76, 2923–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, and Heitman J (2009). Diploids in the Cryptococcus neoformans serotype A population homozygous for the α mating type originate via unisexual mating. PLoS Path 5, e1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Barton DB, and Louis EJ (2006). Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174, 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Kestenbaum L, Vilgalys R, and Mitchell TG (2005). Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J Clin Microbiol 43, 556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Marra RE, Nielsen K, Heitman J, Vilgalys R, and Mitchell TG (2003). Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot Cell 2, 1162–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Wang QM, Goker M, Groenewald M, Kachalkin AV, Lumbsch HT, Millanes AM, Wedin M, Yurkov AM, Boekhout T, et al. (2015). Towards an integrated phylogenetic classification of the Tremellomycetes. Stud Mycol 81, 85–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, Miranda M, Anderson IJ, Fraser JA, et al. (2005). The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307, 1321–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugarini C, Goebel CS, Condas LAZ, Muro MD, de Farias MR, Ferreira FM, and Vainstein MH (2008). Cryptococcus neoformans isolated from passerine and psittacine bird excreta in the state of Paraná, Brazil. Mycopathologia 166, 61–69. [DOI] [PubMed] [Google Scholar]
- Magditch DA, Liu T-B, Xue C, and Idnurm A (2012). DNA mutations mediate microevolution between host-adapted forms of the pathogenic fungus Cryptococcus neoformans. PLoS Path 8, e1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R, Krockenberger MB, Cross G, Doneley R, Madill DN, Black D, Mcwhirter P, Rozenwax A, Rose K, Alley M, et al. (2003). Avian cryptococcosis. Med Mycol 41, 115–124. [DOI] [PubMed] [Google Scholar]
- Maloisel L, and Rossignol JL (1998). Suppression of crossing-over by DNA methylation in Ascobolus. Genes Dev 12, 1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland CM, Fu J, Woodlee GL, Seymour TS, and Wickes BL (2002). Isolation and characterization of the Cryptococcus neoformans MATa pheromone gene. Genetics 160, 935–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead ME, and Hull CM (2016). Transcriptional control of sexual development in Cryptococcus neoformans. Journal of Microbiology 54, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metin B, Findley K, and Heitman J (2010). The mating type locus (MAT) and sexual reproduction of Cryptococcus heveanensis: insights into the evolution of sex and sex-determining chromosomal regions in fungi. PLoS Genet 6, e10000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TDE, and Edman JC (1993). The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol Cell Biol 13, 1962–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson JB, Ivey MH, and Bulmer GS (1978). Cryptococcus neoformans: Pseudohyphal forms surviving culture with Acanthamoeba polyphaga. Infect Immun 20, 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, Dietrich FS, and Heitman J (2013). Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol 11, e1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Feretzaki M, Sun S, Wang X, and Heitman J (2011). Sex in Fungi. Annu Rev Genet 45, 405–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CB, Fraser JA, and Heitman J (2004). PAK kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans. Mol Biol Cell 15, 4476–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Litvintseva AP, Mylonakis E, Malliaris SD, Benjamin DK Jr., Giles SS, Mitchell TG, Casadevall A, Perfect JR, et al. (2005). Cryptococcus neoformans α strains preferentially disseminate to the central nervous system during coinfection. Infect Immun 73, 4922–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, and Heitman J (2003). Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect Immun 71, 4831–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, De Obaldia AL, and Heitman J (2007). Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryot Cell 6, 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien CR, Krockenberger MB, Wigney DI, Martin P, and Malik R (2004). Retrospective study of feline and canine cryptococcosis in Australia from 1981 to 2001: 195 cases. Med Mycol 42, 449–460. [DOI] [PubMed] [Google Scholar]
- O’Meara TR, and Alspaugh JA (2012). The Cryptococcus neoformans capsule: a sword and a shield. Clin Microbiol Rev 25, 387–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chrétien F, Heitman J, Dromer F, and Nielsen K (2010). Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Path 6, e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Wang Y, Ballou ER, O’Meara TR, Bahn Y-S, Alspaugh JA, Xue C, and Nielsen K (2011). Cryptococcal titan cell formation is regulated by G-protein signaling in response to multiple stimuli. Eukaryot Cell 10, 1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M, Buck KW, and Brasier CM (2006). Selective acquisition of novel mating type and vegetative incompatibility genes via interspecies gene transfer in the globally invading eukaryote Ophiostoma novo-ulmi. Mol Ecol 15, 249–262. [DOI] [PubMed] [Google Scholar]
- Passer AR, Coelho MA, Billmyre RB, Nowrousian M, Mittelbach M, Yurkov AM, Averette AF, Cuomo CA, Sun S, and Heitman J (2019). Genetic and genomic analyses reveal boundaries between species closely related to Cryptococcus pathogens. bioRxiv, 557140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR, Ketabchi N, Cox GM, Ingram CW, and Beiser CL (1993). Karyotyping of Cryptococcus neoformans as an epidemiological tool. J Clin Microbiol 31, 3305–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck I, and Lebens GA (1989). Electrophoretic karyotype of the pathogenic yeast Cryptococcus neoformans. J Gen Microbiol 135, 65–71. [DOI] [PubMed] [Google Scholar]
- Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, and Boulware DR (2017). Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. The Lancet Infectious Diseases 17, 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa HS, Kowshik T, Chowdhary A, Preeti Sinha K, Khan ZU, Sun S, and Xu J (2008). The expanding host tree species spectrum of Cryptococcus gattii and Cryptococcus neoformans and their isolations from surrounding soil in India. Med Mycol 46, 823–833. [DOI] [PubMed] [Google Scholar]
- Raper JR (1966). Genetics of sexuality in higher fungi. (New York: Ronald Press Co.). [Google Scholar]
- Raudaskoski M, and Kothe E (2010). Basidiomycete mating type genes and pheromone signaling. Eukaryot Cell 9, 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayssiguier C, Thaler DS, and Radman M (1989). The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 23, 396–401. [DOI] [PubMed] [Google Scholar]
- Ren P, Springer DJ, Behr MJ, Samsonoff WA, Chaturvedi S, and Chaturvedi V (2006). Transcription factor STE12a has distinct roles in morphogenesis, virulence, and ecological fitness of the primary pathogenic yeast Cryptococcus gattii. Eukaryot Cell 5, 1065–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J, Desjardins CA, Sykes SM, Beale MA, Vanhove M, Sakthikumar S, Chen Y, Gujja S, Saif S, Chowdhary A, et al. (2017). Tracing genetic exchange and biogeography of Cryptococcus neoformans at the global population level. Genetics 207, 327–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Carres M, Findley K, Sun S, Dietrich FS, and Heitman J (2010). Morphological and genomic characterization of Filobasidiella depauperata: a homothallic sibling species of the pathogenic Cryptococcus species complex. PLoS ONE 5, e9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder GS (1997). Meiotic chromosomes: it takes two to tango. Genes Dev 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- Rogers DW, McConnell E, Ono J, and Greig D (2018). Spore-autonomous fluorescent protein expression identifies meiotic chromosome mis-segregation as the principal cause of hybrid sterility in yeast. PLoS Biol 16, e2005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C, Sun S, Billmyre RB, Heitman J, and Magwene PM (2018). A high resolution map of meiotic recombination in Cryptococcus deneoformans demonstrates decreased recombination in unisexual reproduction. Genetics 209, 567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan SR, Ianiri G, Reza MH, Thimmappa BC, Ganguly P, Coelho MA, Sun S, Siddharthan R, Tellgren-Roth C, Dawson TL, et al. (2019). Centromere-mediated chromosome break drives karyotype evolution in closely related Malassezia species. bioRxiv, 533794. [Google Scholar]
- Sharpton TJ, Neafsey DE, Galagan JE, and Taylor JW (2008). Mechanisms of intron gain and loss in Cryptococcus. Genome Biol 9, R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WC, Davidson RC, Cox GM, and Heitman J (2002). Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in Cryptococcus neoformans. Eukaryot Cell 1, 366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wolf SE, Burke JM, Presting GG, Ross-Ibarra J, and Dawe RK (2010). Widespread gene conversion in centromere cores. PLoS Biol 8, e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia RA, Lengeler KB, and Heitman J (2000). Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet Biol 29, 153–163. [DOI] [PubMed] [Google Scholar]
- Singer LM, Meyer W, Firacative C, Thompson GR, Samitz E, and Sykes JE (2014). Antifungal drug susceptibility and phylogenetic diversity among Cryptococcus isolates from dogs and cats in North America. J Clin Microbiol 52, 2061–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib F (1981). The perfect state of Cryptococcus neoformans, Filobasidiella neoformans, on pigeon manure filtrate agar. Zentralblatt für Bakteriologie, Mikrobiologie und Hygiene 1 Abt Originale A, Medizinische Mikrobiologie, Infektionskrankheiten und Parasitologie 248, 575–578. [PubMed] [Google Scholar]
- Steenbergen JN, Shuman HA, and Casadevall A (2001). Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. PNAS 98, 15245–15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Billmyre RB, Mieczkowski P, and Heitman J (2014). Unisexual reproduction drives meiotic recombination and phenotypic and karyotypic plasticity in Cryptococcus neoformans. PLoS Genet 10, e1004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, and Heitman J (2015). From two to one: unipolar sexual reproduction. Fungal Biology Reviews 29, 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, and Heitman J (2016). Running hot and cold: recombination around and within mating-type loci of fungi and other eukaryotes In Environmental and Microbial Relationships, Druzhinina SI, and Kubicek PC, eds. (Cham: Springer International Publishing; ), pp. 3–13. [Google Scholar]
- Sun S, Hsueh Y-P, and Heitman J (2012). Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet 8, e1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, and Xu J (2007). Genetic analyses of a hybrid cross between serotypes A and D strains of the human pathogenic fungus Cryptococcus neoformans. Genetics 177, 1475–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, and Xu J (2009). Chromosomal rearrangements between serotype A and D strains in Cryptococcus neoformans. PLoS ONE 4, e5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Yadav V, Billmyre RB, Cuomo CA, Nowrousian M, Wang L, Souciet J-L, Boekhout T, Porcel B, Wincker P, et al. (2017). Fungal genome and mating system transitions facilitated by chromosomal translocations involving intercentromeric recombination. PLoS Biol 15, e2002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, and Petes TD (1988). Meiotic recombination within the centromere of a yeast chromosome. Cell 52, 237–240. [DOI] [PubMed] [Google Scholar]
- Termolino P, Cremona G, Consiglio MF, and Conicella C (2016). Insights into epigenetic landscape of recombination-free regions. Chromosoma 125, 301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, He G-J, Hu P, Chen L, Tao C, Cui Y-L, Shen L, Ke W, Xu H, Zhao Y, et al. (2018). Cryptococcus neoformans sexual reproduction is controlled by a quorum sensing peptide. Nature Microbiology 3, 698–707. [DOI] [PubMed] [Google Scholar]
- Trevijano-Contador N, de Oliveira HC, García-Rodas R, Rossi SA, Llorente I, Zaballos Á, Janbon G, Ariño J, and Zaragoza Ó (2018). Cryptococcus neoformans can form titan-like cells in vitro in response to multiple signals. PLoS Path 14, e1007007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi R, Hsueh Y-P, Geunes-Boyer S, Wright JR, and Heitman J (2009). Spores as infectious propagules of Cryptococcus neoformans. Infect Immun 77, 4345–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tian X, Gyawali R, Upadhyay S, Foyle D, Wang G, Cai JJ, and Lin X (2014). Morphotype transition and sexual reproduction are genetically associated in a ubiquitous environmental pathogen. PLoS Path 10, e1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhai B, and Lin X (2012). The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Path 8, e1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, and Heitman J (1999). Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr Opin Microbiol 2, 358–362. [DOI] [PubMed] [Google Scholar]
- Wang P, Nichols CB, Lengeler KB, Cardenas ME, Cox GM, Perfect JR, and Heitman J (2002). Mating-type-specific and nonspecific PAK kinases play shared and divergent roles in Cryptococcus neoformans. Eukaryot Cell 1, 257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Darwiche S, and Heitman J (2013). Sex-induced silencing operates during opposite-sex and unisexual reproduction in Cryptococcus neoformans. Genetics 193, 1163–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hsueh Y-P, Li W, Floyd A, Skalsky R, and Heitman J (2010). Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev 24, 2566–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes BL, Moore TDE, and Kwon-Chung KJ (1994). Comparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans. Microbiology 140, 543–550. [DOI] [PubMed] [Google Scholar]
- Williamson PR (1997). Laccase and melanin in the pathogenesis of Cryptococcus neoformans. Front Biosci 2, e99–107. [DOI] [PubMed] [Google Scholar]
- Xu J, Ali RY, Gregory DA, Amick D, Lambert SE, Yoell HJ, Vilgalys RJ, and Mitchell TG (2000a). Uniparental mitochondrial transmission in sexual crosses in Cryptococcus neoformans. Curr Microbiol 40, 269–273. [DOI] [PubMed] [Google Scholar]
- Xu J, Luo G, Vilgalys RJ, Brandt ME, and Mitchell TG (2002). Multiple origins of hybrid strains of Cryptococcus neoformans with serotype AD. Microbiology 148, 203–212. [DOI] [PubMed] [Google Scholar]
- Xu J, Vilgalys R, and Mitchell TG (2000b). Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol Ecol 9, 1471–1481. [DOI] [PubMed] [Google Scholar]
- Xu X, Lin J, Zhao Y, Kirkman E, So Y-S, Bahn Y-S, and Lin X (2017). Glucosamine stimulates pheromone-independent dimorphic transition in Cryptococcus neoformans by promoting Crz1 nuclear translocation. PLoS Genet 13, e1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C (2012). Cryptococcus and beyond—inositol utilization and its implications for the emergence of fungal virulence. PLoS Path 8, e1002869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Tada Y, Dong X, and Heitman J (2007). The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host & Microbe 1, 263–273. [DOI] [PubMed] [Google Scholar]
- Yadav V, Sun S, Billmyre RB, Thimmappa BC, Shea T, Lintner R, Bakkeren G, Cuomo CA, Heitman J, and Sanyal K (2018). RNAi is a critical determinant of centromere evolution in closely related fungi. PNAS 115, 3108–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Hull CM, Heitman J, Sun S, and Xu J (2004). SXI1α controls uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr Biol 14, R743–744. [DOI] [PubMed] [Google Scholar]
- Yan Z, Hull CM, Sun S, Heitman J, and Xu JP (2007a). The mating-type specific homeodomain genes SXI1α and SXI2a coordinately control uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr Genet 51, 187–195. [DOI] [PubMed] [Google Scholar]
- Yan Z, Sun S, Shahid M, and Xu J (2007b). Environment factors can influence mitochondrial inheritance in the fungus Cryptococcus neoformans. Fungal Genet Biol 44, 315–322. [DOI] [PubMed] [Google Scholar]
- Yan Z, and Xu J (2003). Mitochondria are inherited from the MATa parent in crosses of the basidiomycete fungus Cryptococcus neoformans. Genetics 163, 1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue C, Cavallo LM, Alspaugh JA, Wang P, Cox GM, Perfect JR, and Heitman J (1999). The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics 153, 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B, Wozniak KL, Masso-Silva J, Upadhyay S, Hole C, Rivera A, Wormley FL, and Lin X (2015). Development of protective inflammation and cell-mediated immunity against Cryptococcus neoformans after exposure to hyphal mutants. mBio 6, e01433–01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Upadhyay S, and Lin X (2018). PAS domain protein Pas3 interacts with the chromatin modifier Bre1 in regulating Cryptococcal morphogenesis. mBio 9, e02135–02118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Engström PG, Tellgren-Roth C, Baudo CD, Kennell JC, Sun S, Billmyre RB, Schröder MS, Andersson A, Holm T, et al. (2017). Proteogenomics produces comprehensive and highly accurate protein-coding gene annotation in a complete genome assembly of Malassezia sympodialis. Nucleic Acids Res 45, 2629–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer BL, Hempel HO, and Goodman NL (1984). Pathogenicity of the basidiospores of Filobasidiella neoformans. Mycopathologia 85, 149–153. [DOI] [PubMed] [Google Scholar]