Abstract
Staufen is a dsRNA-binding protein involved in many aspects of RNA regulation, such as mRNA transport, Staufen-mediated mRNA decay and the regulation of mRNA translation. It is a modular protein characterized by the presence of conserved consensus amino acid sequences that fold into double-stranded RNA binding domains (RBDs) as well as degenerated RBDs that are instead involved in protein–protein interactions. The variety of biological processes in which Staufen participates in the cell suggests that this protein associates with many diverse RNA targets, some of which have been identified experimentally. Staufen binding mediates the recruitment of effectors via protein–protein and protein–RNA interactions. The structural determinants of a number of these interactions, as well as the structure of full-length Staufen, remain unknown. Here, we present the first solution structure models for full-length hStaufen155, showing that its domains are arranged as beads-on-a-string connected by flexible linkers. In analogy with other nucleic acid-binding proteins, this could underpin Stau1 functional plasticity.
Keywords: SAXS, Staufen, dsRNA-binding domain, structural biology
INTRODUCTION
Staufen (Stau) is a dsRNA-binding protein originally identified in Drosophila melanogaster, where it plays an essential role in oocyte development (Schupbach and Wieschaus 1986; St Johnston et al. 1991). It is well conserved from nematodes to humans and, depending on the species, is composed of four or five dsRNA-binding domains (RBDs) (Wickham et al. 1999). In humans, there are two Staufen paralogs: hStau1 and hStau2, each present in several isoforms (Park et al. 2013). Much of our knowledge on human Stau1 is based on the study of isoform hStau155. hStau155 is associated with 40S and 60S ribosomal subunits and colocalizes with the rough endoplasmic reticulum (Marión et al. 1999; Wickham et al. 1999; Luo et al. 2002). hStau155 has also been characterized biochemically in the context of mRNA decay (Kim et al. 2005) and cell cycle control (Boulay et al. 2014). While Stau2 is expressed primarily in the neuromuscular system and is mostly involved in mRNA transport at particular sites of the post-synaptic muscles, Stau1 is ubiquitously expressed (Bélanger et al. 2003; Lebeau et al. 2008; Vessey et al. 2008; Ravel-Chapuis et al. 2012; Peredo et al. 2014). Even though Stau1 and Stau2 exhibit different tissue expression patterns, they have been shown to be involved in the same mechanisms of RNA regulation, such as mRNA transport (Martel et al. 2006, 2010; Ramasamy et al. 2006; Vessey et al. 2008; Ravel-Chapuis et al. 2012), Staufen-mediated mRNA decay (SMD) (Kim et al. 2005, 2014; Gong et al. 2009; Gong and Maquat 2011; Cho et al. 2012; Baker et al. 2013; Park and Maquat 2013) and regulation of mRNA translation (Ravel-Chapuis et al. 2012; Bonnet-Magnaval et al. 2016), myogenic differentiation (Ravel-Chapuis et al. 2014), stress granule formation(Ravel-Chapuis et al. 2016), regulation of adipogenesis (Cho et al. 2012; Baker et al. 2013), progression of the cell cycle (Boulay et al. 2014), and cellular differentiation (Gautrey et al. 2005, 2008; Kretz 2013; Peredo et al. 2014). They also are central players in virology, functioning in HIV infection by favoring viral RNA (vRNA) encapsidation (Mouland et al. 2000; Chatel-Chaix et al. 2004, 2008; Banerjee et al. 2014), in hepatitis C infection by transporting vRNA to the site of translation, in the replication of cellular DNA (Blackham and McGarvey 2013; Dixit et al. 2016) and as requirements for efficient influenza A virus propagation (de Lucas et al. 2010). The variety of cellular processes in which Stau1 is implicated suggests that it might adopt different binding modes with its diverse RNA targets and that structurally distinct RNA–Stau1 complexes mediate the recruitment of effectors via protein–protein and/or protein–RNA interactions.
Interactions between Stau1 and its RNA substrates were initially characterized for hStau155. Multiple copies of hStau155 can bind a single dsRNA. In cells, Stau1 binds intramolecular duplexes within the hARF1 mRNA (Martel et al. 2010). Furthermore, in vitro, multiple copies of Stau1 bind to mRNAs containing as many as 250 CUG repeats (Ravel-Chapuis et al. 2012). Additionally, the finding that hStau155 stabilizes imperfectly base-pairings formed between mRNAs and lncRNAs (Gong and Maquat 2011), suggests that multiple hStau1 molecules bind to the same dsRNA. Genome-wide analysis (Furic et al. 2008; Laver et al. 2013) and hiCLIP (RNA hybrid and individual-nucleotide resolution ultraviolet cross-linking and immunoprecipitation) (Fernandez Moya and Kiebler 2015; Sugimoto et al. 2015) of Stau-associated mRNAs identified secondary structures that confer binding specificity (Ricci et al. 2014). Nevertheless, what defines a Stau binding site remains unclear (de Lucas et al. 2014).
Stau proteins are characterized by two conserved consensus amino acid sequences that fold into dsRNA-binding domains (RBD3 and RBD4); hStau1 contains two other RBDs (RBD2 and RBD5) that are unable to bind RNA and, relative to hStau1, hStau2 has an additional RBD1 and only a partial RBD5 (Buchner et al. 1999; Wickham et al. 1999; Duchaine et al. 2002; Allison et al. 2004; Furic et al. 2008). hStau1 and hStau2 tubulin-binding domains (TBDs), which are involved in mRNA transport on the cytoskeleton, share only 18% identity. Functional activation of a number of dsRNA-binding proteins requires that they self-associate or associate with other dsRNA-binding proteins (Park et al. 2013). A Staufen swapping motif (SSM) has been identified to reside between TBD and RBD5. The SSM is necessary for the homodimeric or heterodimeric interactions between Stau1 and Stau2 (Park et al. 2013). This dimerization is critical for SMD (Martel et al. 2010; Gleghorn et al. 2013; Park et al. 2013). The amino-terminal α-helix of RBD5 was also identified as the major determinant for protein–protein interaction in vivo, intercalating with the two α-helices of the SSM. A recent SEC-MALLS report on purified protein also showed that, in the absence of RNA, SSM–RBD5 promotes dimerization (Lazzaretti et al. 2018). The importance of RBD2 in dimerization is less clear. BRET assays, aimed at the study of hStau155 multimerization, show that RBD2 (amino acids 37–79 of isoform hSTAU155) interacts with full-length hStau1 (Martel et al. 2010). On the other hand, recombinant purified hStau1-“RBD”2-RBD3 suggests that the contribution of RBD2 to hStau155 dimerization, while existing, is relatively minor (Martel et al. 2010; Park et al. 2013).
To date, analyses of the three-dimensional structure of Stau proteins have focused on studies of truncated versions of the protein, either in isolation or in complex with short RNA sequences or in complex with truncated versions of interacting proteins. The NMR structure of Drosophila RBD3 first confirmed that this construct is organized in the typical α-β-β-β-α fold (PDB ID: 1STU) (Bycroft et al. 1995). Mouse Stau2 RBD4, in the absence of dsRNA, also showed the α-β-β-β-α fold (PDB ID: 1UHZ). The structure of human Stau1 SSM–RBD5 solved by X-ray crystallography revealed a domain swapped dimer, which is responsible for mediating hStau1 dimerization (PDB ID: 4DKK) (Gleghorn et al. 2013). The X-ray crystal structure of the complex between Miranda and RBD5 showed two RBD5s symmetrically bound to the Miranda dimeric coiled-coil region through their exposed β-sheet faces, revealing a previously unrecognized protein interaction mode for RBDs (PDB ID: 5CFF) (Jia et al. 2015).
The solution structure of Drosophila melanogaster Stau RBD3 bound to a 12-bp stem–loop RNA, determined by NMR spectroscopy, revealed the interaction of the canonical α-β-β-β-α RBD fold with dsRNA (PDB ID: 1EKZ) (Ramos et al. 1999, 2000). The crystallographic structure of the RBD3–RBD4 construct, bound to dsRNA as a dimer (monomers A and B), shows that the interaction surface with the RNA spans the major groove and the two adjacent minor groove surfaces. Furthermore, RBD3 from monomer B is bound on the opposite side of the RNA molecule, in an antiparallel orientation to RBD3A, whereas density for the sec RBD4 is missing (Lazzaretti et al. 2018). Human, Drosophila, and Caenorhabditis elegans Stau bind dsRNA without apparent sequence specificity in vitro (St Johnston et al. 1992; Marión et al. 1999; Wickham et al. 1999; Ramos et al. 2000; LeGendre et al. 2013; Wang et al. 2015). Bono and coworkers recently showed that, in addition to the interactions with the sugar-phosphate backbone previously identified for RBD3 (Ramos et al. 2000), both domains of hStau1 directly contact RNA bases in the minor groove of the dsRNA used. Indeed, they also show that specific base recognition is relevant in vivo and may therefore contribute to the overall sequence selectivity by Stau, possibly together with additional regions of the protein or with other regulators (Lazzaretti et al. 2018). The macromolecular interaction events that happen downstream from Stau RNA binding are still structurally unknown.
We used an integrated structural biology approach, combining homology modeling, small-angle X-ray scattering (SAXS), NMR, and hydrodynamic methods to characterize the structure of the human full-length Staufen1 protein. To interpret this structure, we also studied an hStau155_ΔRBD2 truncated variant, individual hStau155 domains, and several tandem multidomain Stau1 fragments. We confirmed that the deletion of RBD2 influences the oligomeric state of the protein, as well as reporting for the first time its effect on protein solubility. Our data show for the first time that hStau155 adopts an elongated conformation in solution. Furthermore, in the absence of RNA, RBD3 and RBD4 are connected by a linker that is very flexible in solution, and they do not interact with one another.
The reshaping or folding of flexible components in the presence of target nucleic acid in proteins has already been linked to multifunctionality. As an example, DNA and RNA nucleases (Tsutakawa et al. 2014) behave like molecular level transformers that can rebuild themselves by sometimes altering their protein conformations and typically sculpting the nucleic acid to control both their specificity and efficiency functions. We propose that the extreme flexibility and the independent movement of individual domains could also be the basis for the functional plasticity of Stau1 protein: Different relative movements of domains on themselves and/or on RNA can create multiple joint recognition surfaces, reshaping itself to elicit diverse RNA metabolism tasks.
RESULTS AND DISCUSSION
RBD2 influences both solubility and oligomeric state of Staufen1
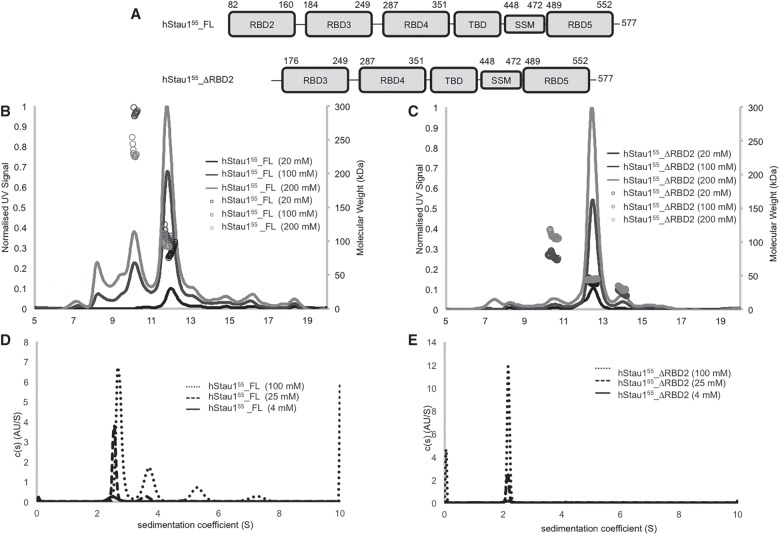
Stau proteins were purified to homogeneity by immobilized nickel chromatography followed by size exclusion chromatography. Additive screening to determine conditions that would allow the protein to achieve high concentration and good homogeneity for subsequent structural studies was performed using 10 K MWCO spin-concentrators (Rambo 2017). The addition of L-Arg HCl to the buffer proved necessary for maintaining the solubility of the full-length protein to enable further experiments. Interestingly, the solubility of hStau155_ΔRBD2 is not affected by the presence (or absence) of L-Arg HCl in the buffer. However, this additive was used for all constructs for consistency with the purification requirements of hStau155_FL. Size exclusion chromatography (SEC) traces show that the hydrodynamic volumes of hStau155_FL and of hStau155_ΔRBD2 (∼130–140 kDa) are higher than expected for globular proteins with corresponding molecular weights, suggesting that the two proteins might have an elongated shape or might form homomultimers. The domains organization of hStau155_FL and hStau155_ΔRBD2 is shown in Figure 1A. SEC-MALS analysis of hStau155_FL (Fig. 1B) shows the presence of multiple assemblies. In contrast to the recently published SEC profile for hStau163_FL (Lazzaretti et al. 2018), both SEC profiles for hStau155_FL and hStau155_ΔRBD2 described in this study present symmetrical peaks. MALS analysis of the eluting species highlights the different behavior of hStau155_FL and hStau155_ΔRBD2. Measurements were performed at three different concentrations (20, 100, and 200 μM). The samples used for this analysis had not been subject to the final SEC purification, hence small amounts of species other than hStau155_FL and hStau155_ΔRBD2 were seen to be present. However, the predominant peak in the hStau155_ΔRBD2 sample had a molecular weight consistent with that of a monomer (Fig. 1C). The main SEC peak for hStau155_FL appears with a molecular weight consistent with a dimer but the high polydispersity seen across the peak suggests that this is an equilibrium species between a monomer and higher order oligomers. This is consistent with the previous observation that RBD2 mediates hStau155_FL self-association (Martel et al. 2010; Lazzaretti et al. 2018), showing that its presence is fundamental for the formation of a stable oligomer in solution. The recent report describing hStau163 did not contain SEC-MALS analysis for the full-length protein (Lazzaretti et al. 2018).
FIGURE 1.
Architecture and biophysical characterization of Staufen proteins. (A) Domains organization of hStau155_FL and hStau155_ΔRBD2. (B) SEC-MALS of hStau155_FL at 20, 100, and 200 μΜ. (C) SEC-MALS of hStau155_ΔRBD2 at 20, 100, and 200 μΜ. (D) Analytical ultracentrifugation (AUC) of hStau155_FL at 4, 25, and 100 μΜ. (Ε) Analytical ultracentrifugation (AUC) of hStau155_ΔRBD2 at 4, 25, and 100 μΜ.
To resolve the oligomeric assemblies of the species eluting in the main SEC peak, we performed analytical ultracentrifugation (AUC) experiments. AUC analysis of the peak fraction from hStau155_FL SEC (Fig. 1D) confirms the coexistence of a number of species with molecular weights consistent with that of the monomer (major species in solution) and higher oligomers. Measurements were performed at three different concentrations (4, 25, and 100 μM). We chose to analyze a wide range of concentrations to address the role of concentration in the oligomeric state of the protein. The number of oligomers increases with increasing sample concentration, and the position of the peaks also shifts to a higher sedimentation coefficient. Both of these phenomena indicate concentration-dependent self-association equilibrium for hStau155_FL. On the other hand, both SEC-MALS (Fig. 1C) and AUC (Fig. 1E) analysis of hStau155_ΔRBD2 show that the truncated protein is only present in solution as monomer.
Staufen adopts distinct elongated structures in solution
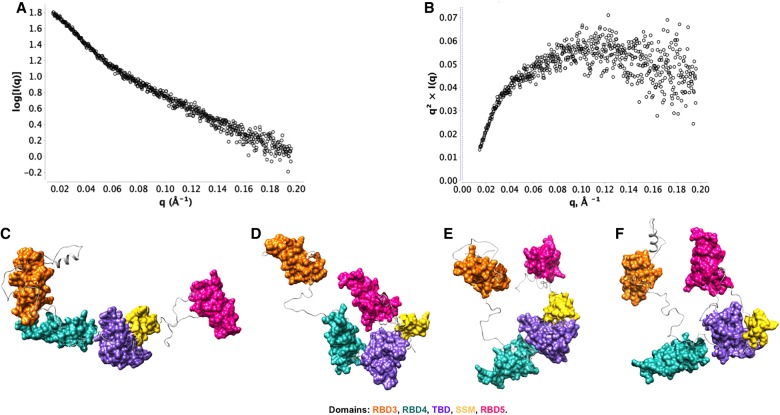
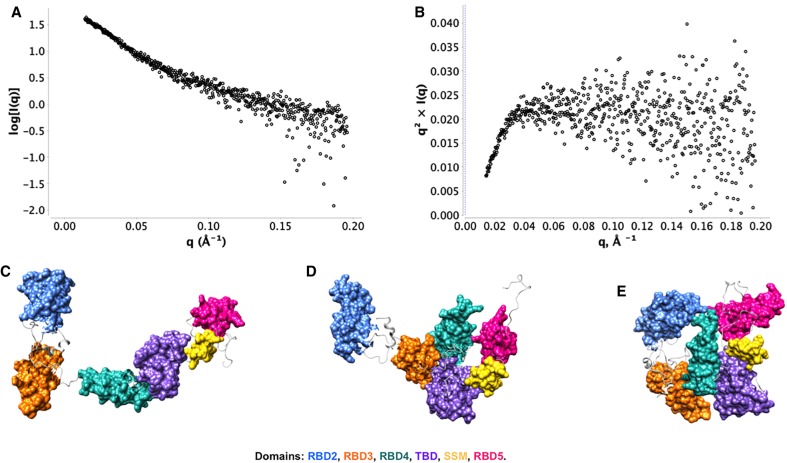
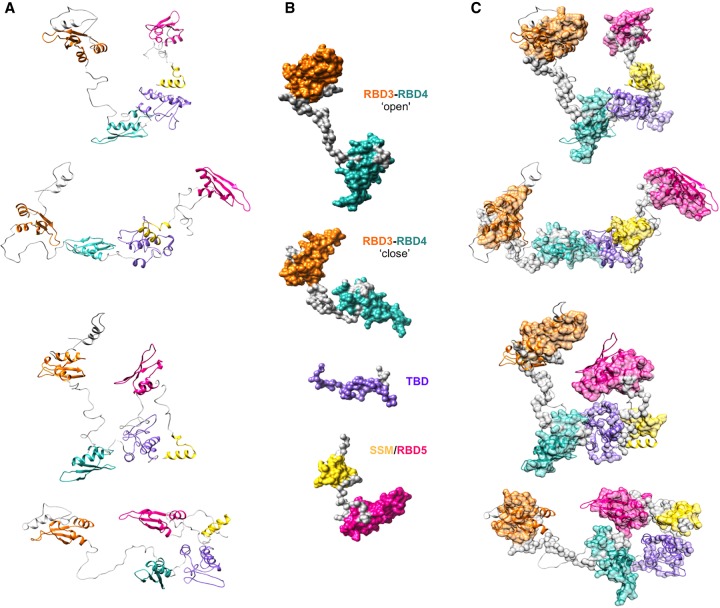
The first indication that Staufen protein and its hStau155_ΔRBD2 mutant adopt elongated structures in solution is given by their average hydrodynamic volume, which is much higher than expected for the estimated molecular weight of the monomeric protein. This is in agreement with the Rg and Dmax obtained from SAXS measurements for both the full-length (Rg ensemble = 48.11 Å, Dmax ensemble = 155.2 Å) and truncated ΔRBD2 (Rg ensemble = 50.26 Å, Dmax ensemble = 166.96 Å) proteins. The higher Rg and Dmax for hStau155_ΔRBD2 can be explained by a higher degree of conformational heterogeneity in the FL protein. Importantly, SAXS shows that both these systems are characterized by a high degree of flexibility, as shown by their Kratky plot in Figure 2. The three-dimensional models of hStau155_FL protein and of its truncation mutant hStau155_ΔRBD2 were obtained by combining homology modeling analysis and SAXS data. The models obtained for hStau155_ΔRBD2 show that the protein adopts a range of conformations (Fig. 2), from highly extended to more compact, where RBD3 and RBD5 are in closer proximity. The relative positions of RBD4, TBD, and SSM show only minor differences among the models obtained, due to the flexibility of the loops. On the other hand, the high level of flexibility of the loops between RBD3 and RBD4 and between SSM and RBD5 seems to be the main factor that contributes to the coexistence of a more distended and a more closed conformation of hStau155_ΔRBD2. These models show that all the individual domains do not coalesce to form a compact structure. The models obtained for hStau155_FL, represented in Figure 3, show more interdomain flexibility, resulting in the presence of elongated, as well as more compact, conformations. The major differences between the coexisting conformations are due to the disordered loops between RBD2 and RBD3, between RBD3 and RBD4 and between RBD4 and TBD. Thanks to the malleability of these linkers, hStau155_FL seems to be able to transition in solution from more elongated to more closed conformations, but still not globular.
FIGURE 2.
Small-angle X-ray scattering (SAXS) of Staufen proteins. (A) SAXS intensity curve; (B) Kratky analysis; (C–F) EOM models generated for hStau155_ΔRBD2 at 60 μΜ [Rg ensemble = 50.26 Å, Dmax ensemble = 166.96 Å, Rsigma = ∼80.2% (∼85.68%)].
FIGURE 3.
Small-angle X-ray scattering (SAXS) of Staufen proteins. (A) SAXS intensity curve; (B) Kratky analysis; (C–E) EOM models generated for hStau155_FL at 40 μΜ [Rg ensemble = 48.11 Å, Dmax ensemble = 155.2 Å, Rsigma = ∼74.45% (∼86.17%)].
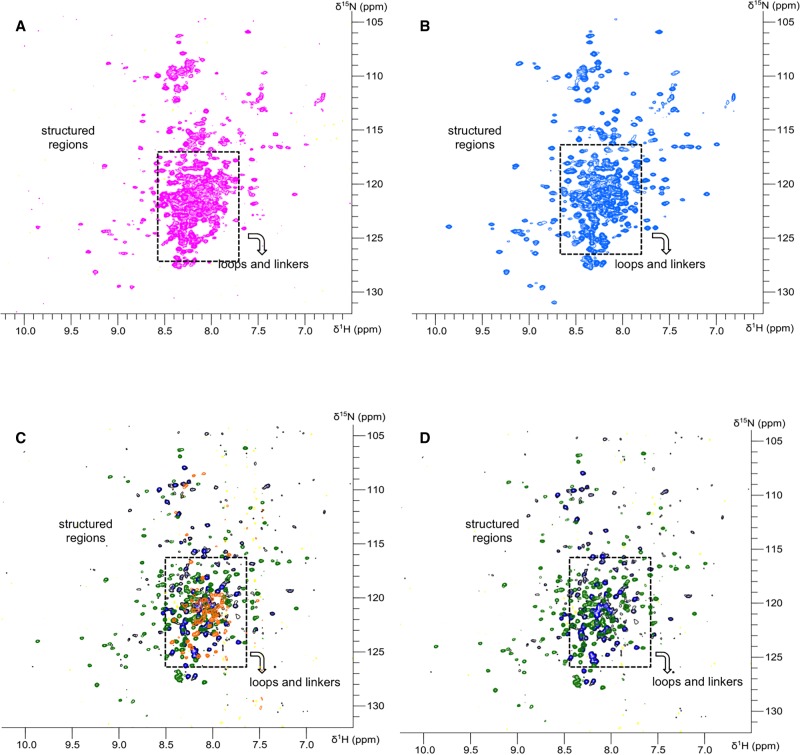
The structural information gathered from the SAXS models for hStau155_FL and hStau155_ΔRBD2 were validated using 2D NMR (Fig. 4). 15N,1H-TROSY spectra were recorded for individual and tandem domains, as well as for the full-length protein and its truncation mutant ΔRBD2. The large number of peaks in the central area of the spectra of both hStau155_FL and hStau155_ΔRBD2 indicates the presence of a large number of amino acids in disordered regions, this agrees well with the presence of long unstructured linkers that confer flexibility on the proteins. Moreover, the similar lineshapes and lack of significant chemical shift perturbations between the spectra recorded for individual and tandem domains (Supplemental Figs. S4, S5) show that any interactions between adjacent domains are very limited and that each of the domains constituting the tandem constructs tumbles independently in solution, thanks to the intervening linkers flexibility. In addition, it is possible to reconstitute the spectra for hStau155_FL and hStau155_ΔRBD2 almost entirely by overlaying those obtained for individual domains, showing that the domains tumble independently in the full-length protein.
FIGURE 4.
NMR analysis of Staufen proteins. (A) NMR spectra of hStau155_FL. (B) NMR spectra of hStau155_DRBD2. (C) Reconstitution of hStau155_FL spectra by overlapping of RBD2, RBD3–RBD4, and TBD–SSM/RBD5 NMR spectra. (D) Reconstitution of hStau155_DRBD2 spectra by overlapping of RBD2, RBD3–RBD4, and TBD–SSM/RBD5 NMR spectra.
Together, our data suggest that hStau155 is an extremely flexible protein and its domains can adopt several positions relative to each other, without interdomain interactions. Thanks to the flexibility of the linkers, the protein adopts an elongated conformation in solution and its domains behave as beads on a string. Connector regions are crucial players in Staufen allostery and conformational changes, in line with recent studies on the role of the dynamic linker in the modulation of protein function (Papaleo et al. 2016).
Linker flexibility mediates RBD3 and RBD4 rearrangement
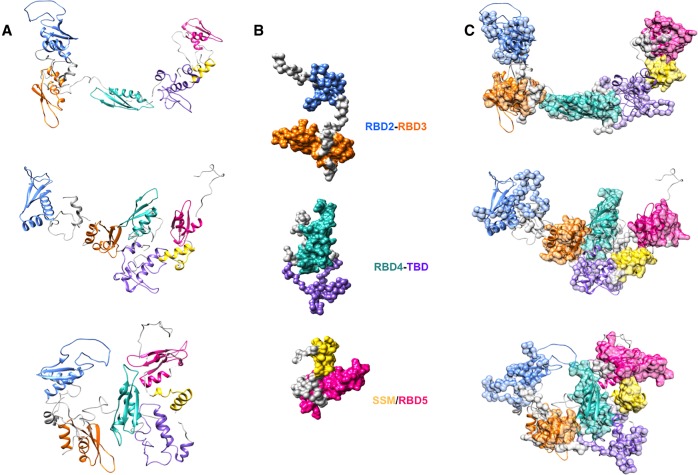
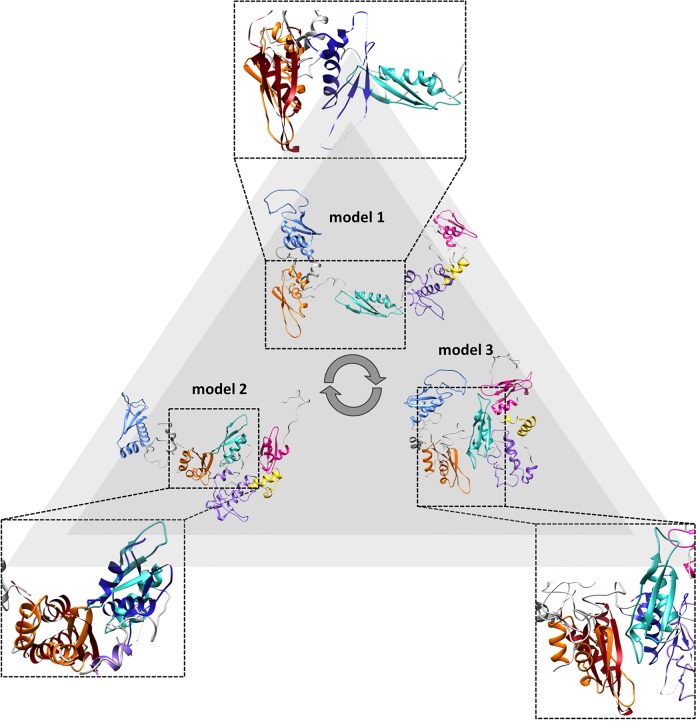
The fitting of a representative subset of SAXS models for individual or tandem domains in the SAXS models of hStau155_ΔRBD2 and hStau155_FL is shown in Figures 5, 6, respectively. The SAXS data and models obtained for individual and tandem domains are shown in Supplemental Figures S6–S14. A representative subset of solution scattering models of individual and tandem domains (Fig. 5B) was chosen in order to interpret the domains rearrangement observed in the models corresponding to the different conformations that hStau155_ΔRBD2 adopts in solution (Fig. 5A). From the fitting proposed in Figure 5C, it is possible to observe that in hStau155_ΔRBD2 the linker connecting RBD3 to RBD4 can be completely or partially distended. SAXS models obtained for the construct RBD3–RBD4 well describe the behavior of these two domains also when they belong to the truncated protein, showing that the presence of TBD and SSM/RBD5 does not have a great impact on RBD3–RBD4 rearrangements. On the contrary, the models obtained for the construct RBD3–RBD4 cannot be used for the interpretation of domains rearrangements in hStau155_FL (Fig. 6A) and a different subset of solution scattering models (Fig. 6B) needs to be used in the fitting to describe the conformational changes of the full-length protein (Fig. 6C). In fact, the solution models obtained for the tandem domain RBD3–RBD4 (Supplemental Fig. S11) show that a long linker, which is extremely elongated, connects these two domains. On the other hand, our hStau155_FL models show the coexistence of three main conformations in the solution for which it is interesting to notice the relative movement of RBD3 and RBD4 and their closer proximity, possibly in order to elicit the binding of RNA targets (Fig. 7). In the more elongated model of the full-length protein (model 1), RBD3 and RBD4 are in an “open” conformation that resembles the one assumed by the tandem domain on its own. However, in the other two models (2 and 3), RBD3 and RBD4 are “pulled” toward each other by conformational changes of the connecting linker, interestingly resembling the recently deposited structure of the hStau163 RBD3–RBD4 construct bound to dsRNA (Lazzaretti et al. 2018) (represented in red and blue in Fig. 7).
FIGURE 5.
Fitting of SAXS models for Staufen1 domains in the SAXS models obtained for hStau155_DRBD2. (A) EOM models generated for hStau155_DRBD2. (B) Subset of selected domains and tandem domains models. (C) Fitting of representative models for domains and tandem domains in the EOM models generated for hStau155_DRBD2.
FIGURE 6.
Fitting of SAXS models for Staufen1 domains in the SAXS models obtained for hStau155_FL. (A) EOM models generated for hStau155_FL. (B) Subset of selected domains and tandem domains models. (C) Fitting of representative models for domains and tandem domains in the EOM models generated for hStau155_FL.
FIGURE 7.
RBD3 and RBD4 rearrangements in hStau155_FL can explain its plasticity in the binding of diverse dsRNA targets. The SAXS models obtained for hStau155_FL show that RBD3 (orange) and RBD4 (cyan) transit from an “open” to a “more closed” conformation. A direct comparison of our models with the crystal structure of the hStau163 RBD3–RBD4 construct bound to Arf1 SBS43 (RBD3 displayed in red and RBD 4 in blue) shows that RBD3 and RBD4 are “pulled” toward each other by conformational changes of the connecting linker, possibly in order to elicit dsRNA binding.
Taken together, these data show that not only the presence of RBD2 triggers a spatial reorganization of RBD3 and RBD4, which is indeed mediated by the linker between these last two domains, but also the importance of RBD3–RBD4 relative position and rearrangements on dsRNA binding. Therefore, we propose that these two domains can change mutual orientation depending on the structure of the RNA target, in order to effectively bind different substrates in distinct biological contexts. Moreover, RBDs that are not involved in the binding of RNAs, such as RBD2 and RBD5, can adopt multiple conformations in the full-length protein, not only to elicit protein dimerization, but also to regulate hStau1 structural plasticity and multifunctionality in vivo. All in all, our solution studies demonstrate that Stau protein can adopt several conformations thanks to long linkers that facilitate domains rearrangements, providing a clue on the structural background for the role of Stau in multiple biological pathways. This would be reminiscent of the remodeling of flexible components in the presence of target nucleic acid, which has been seen in other DNA and RNA binding proteins (Tsutakawa et al. 2014) that can change overall shape by altering their protein conformations to switch among their multiple functions.
Conclusions
Canonical RBDs are composed of an α-β-β-β-α secondary structure that folds in three dimensions to recognize dsRNA. Recently, structural and functional studies of divergent RBDs revealed adaptations that include intra- and/or intermolecular protein interactions, sometimes in the absence of detectable dsRNA-binding ability (Krovat and Jantsch 1996; Gleghorn and Maquat 2014). The number of canonical RBDs per polypeptide is highly variable in RBD-containing proteins, ranging from one to five (Macrae et al. 2006; Barraud and Allain 2012; Thomas and Beal 2017). There is no clear correlation between the number of RBDs and dsRNA-binding affinity. Moreover, the extent of the contribution of specific RBDs to RNA substrate specificity is still an open question. Structural information reported on dsRBPs carrying one or more canonical RBD, such as Dicer (Macrae et al. 2006) and ADARs (Barraud and Allain 2012; Thomas and Beal 2017) highlights the importance of neighboring domains for substrate specificity and enzymatic activity. Conformational flexibility of Dicer protein is also proposed to play a central role in dsRNA recognition and processing (Macrae et al. 2006) and this could be extended to other RBD-containing proteins, such as hStau1. The lack of structural information on this protein, for which to date we only had three-dimensional models of truncation mutants, in isolation or in complex with short RNA sequences or truncated protein interactors (Ramos et al. 1999, 2000; Gleghorn et al. 2013; Gleghorn and Maquat 2014; Jia et al. 2015; Lazzaretti et al. 2018) made its functional understanding particularly challenging. Here, we provided for the first time structural information on the full-length hStau1 protein, using an integrated structural biology approach. Combination of hydrodynamic Materials and Materials and Methods, homology modeling, SAXS and NMR allowed us to show that Stau1 is a highly flexible protein, that recoils in solution from an elongated to a compact conformation in which the domains are in closer proximity but not interacting with each other. In this perspective, we propose that the flexible interdomain loops possess a regulatory role in hStau1 activity, allowing a high degree of freedom for recognition and binding of diverse RNA and protein targets and for the subsequent involvement of hStau1 in very diverse aspects of RNA metabolism and regulation. Interestingly, post-translational modifications have been identified in loops and linkers regions within hStau1 protein (Rigbolt et al. 2011; Zhou et al. 2013; Guo et al. 2014), which could be involved in the regulation of domains rearrangement or protein–protein interactions.
As far as the dimerization of hStau1 is concerned, we show that this is dependent on the presence of RBD2 and on protein concentration. Consistently with what has been shown crystallographically, the SSM–RBD5 construct dimerized in solution (Supplemental Fig. S2). SAXS experiments show that the full-length protein adopts at least three main conformations in solution, which therefore can explain its ability to bind diverse RNA targets and protein partners. Our data provide the first structural insight into the “Swiss knife” mechanism adopted by the Stau155 protein to elicit sometimes contrasting biological functions.
MATERIALS AND METHODS
Cloning of individual and tandem domains for NMR analysis
Individual and tandem hStau155 domains were amplified by PCR from pRSET-B-Stau155 vector (Kim et al. 2005) as described in Supplemental Table S1. Purified PCR products and pET28a were digested with NdeI and HindIIIHF (NEB) for 3 h at 37°C. Digested vector was purified from 1% agarose gel run in TBE using the MinElute Gel Extraction Kit (QIAGEN), whereas digested inserts were purified using the QIAquick PCR Purification Kit (QIAGEN). Ligations between the vector and the individual inserts (in ratio 1:3) were performed using the Quick Ligation Kit (NEB) for 5 min at room temperature (RT). Escherichia coli XL1-Blue cells were transformed by the heat-shock method with 2.5μL of the ligation reactions and plated in LB agar plates containing 50 μg/mL kanamycin. After overnight (o/n), for each of the transformations, 15 mL of LB supplemented with 50 μg/mL kanamycin were inoculated with a single colony and the cultures were grown at 37°C o/n. Subsequently, plasmid DNA was purified using the Wizard Plus SV Minipreps DNA Purification Systems and sequenced.
Protein overexpression and purification for SAXS, AUC, and EM experiments
Recombinant proteins were overexpressed in E. coli Rosetta pLysS cells transformed by heat-shock with pRSET-B vectors containing either hStau155_FL (Kim et al. 2005), hStau155_Δ RBD2 or pET28a containing individual domains (RBD2, RBD3, RBD4, TBD, SSM/RBD5) or tandem domains (RBD2–RBD3, RBD3–RBD4, RBD4–TBD, TBD–SSM/RBD5). Starting cultures were grown in LB medium containing 50 μg/mL ampicillin (for hStau155_FL and hStau155_ΔRBD2) or 50 μg/mL kanamycin (for individual and tandem domains) and 34 μg/mL chloramphenicol at 37°C o/n. One milliliter of overnight culture was inoculated in 1 L of terrific broth (TB) medium supplemented with antibiotics and cells were grown to an 0.6 OD600. Protein overexpression was induced by adding 0.5 mM IPTG and culturing the cells at 27°C overnight. Cells were harvested by centrifugation at 5000 rpm on a Beckman Avanti J-20 XP centrifuge with JLA 8.1000 rotor for 20 min and washed once with phosphate buffered saline (PBS) solution; cell pellets were aliquoted and stocked at −80°C. Frozen aliquots were thawed and lysed by sonication in 20 mL lysis buffer (25 mM HEPES pH 7.5, 1 M GndCl, 20 mM imidazole, 1% Triton X-100, 400 μL Complete EDTA-free protease inhibitors [50X] and 2 μL benzonase) followed by 30 min of incubation on ice. Soluble protein extracts were separated from cell pellets by centrifugation at 15,000 rpm for 30 min. 6-His tagged hStau1 proteins were purified by nickel chromatography using HisTrap FF columns (GE Healthcare) equilibrated in washing buffer (25 mM HEPES pH 7.5, 1 M GndCl, 20 mM imidazole). Elution was performed with 20 mM–1 M imidazole gradient. Fractions containing the protein of interest were pooled and concentrated to 10 mg/mL prior to size exclusion chromatography on Superdex200 (hStau155_FL and hStau155_ΔRBD2) or Superdex75 (individual and tandem domains) gel filtration columns (GE Healthcare) in buffer A (25 mM HEPES pH 7.5, 100 mM KCl, 10 mM MgCl2, 200 mM L-Arg HCl). Overexpression and purification efficiency were monitored by SDS–PAGE analysis on 12%, 15%, and 18% gels stained with SimpleBlue (Life Technologies). Fractions were also analyzed by western blot; the nitrocellulose membrane was incubated o/n at 4°C with anti-His antibody conjugated with alkaline phosphatase (1:4000) and developed using the SIGMA-FAST BCIP/NBT reagent. Size exclusion chromatography and SDS–PAGE analysis for hStau155_FL and hStau155_ΔRBD2 are shown in Supplemental Figure S1, those for individual and tandem domains are shown in Supplemental Figure S2.
Protein overexpression and purification for NMR analysis
E. coli Rosetta pLysS cells were transformed by heat-shock with pET-28 vectors carrying inserts for hStau155_FL, hStau155_ΔRBD2, individual domains (RBD2, RBD3, RBD4, TBD, SSM/RBD5), and tandem domains (RBD2–RBD3, RBD3–RBD4, RBD4–TBD, TBD-SSM/RBD5). Starting cultures were grown as described above and the following d cells were cultured to OD600 = 2 in TB medium supplemented with antibiotics. Cells were harvested by centrifugation at 5000 rpm on a Beckman AvantiTM J-20 XP centrifuge with JLA 8.1000 rotor for 20 min, resuspended in M9 minimal medium (1× M9, 2 mM MgSO4, 0.1 mM CaCl2) and grown for 30 min at 37°C. Successively, the minimal medium was supplemented with 400 μL thiamine [50 mg/mL], filter-sterilized glucose [3 g/L] and filter-sterilized ISOGRO-15N Powder-Growth Medium [1 g/L]. Protein overexpression was induced by adding 0.5 mM IPTG and culturing the cells at 27°C overnight. Purification of Staufen proteins was performed as described above, but replacing HEPES with 20 mM potassium phosphate buffer pH 7.6 in all buffers used. To prevent the formation of disulfide bonds, 2 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) was added to the purified proteins.
Analytical ultracentrifugation (AUC)
The experiments were performed at 40,000 rpm, using a Beckman XL-I analytical ultracentrifuge equipped with an An-50Ti rotor. Data were recorded using both absorbance (at 280 and 260 nm) and interference optical detection systems. The density and viscosity of the buffer were measured experimentally using a DMA 5000M densitometer equipped with a Lovis 200ME viscometer module. The partial specific volume for the protein was calculated using SEDNTERP from the amino acid sequence. Data were processed using SEDFIT, fitting to the c(s) model. Figures were made using GUSSI (Lebowitz et al. 2002).
SEC-MALS
SEC-MALS experiments were performed using a Superdex 200 10/300 Increase column (GE Healthcare) connected to an AktaPure 25 System (GE Healthcare). The protein sample (100 μL) was loaded onto the gel filtration column and eluted with one column volume (24 mL) of buffer A, at a flow rate of 0.7 mL/min. The eluting protein was monitored using a DAWN HELEOS-II 18-angle light scattering detector (Wyatt Technologies) equipped with a WyattQELS dynamic light scattering module, a U9-M UV/Vis detector (GE Healthcare), and an Optilab T-rEX refractive index monitor (Wyatt Technologies). Data were analyzed by using the Astra software (Wyatt Technologies) using a refractive increment value of 0.185 mL/g.
Small angle X-ray scattering and modeling
SAXS data for hStau155_FL and hStau155_ΔRBD2 were collected at B21, Diamond Light Source (Harwell, UK). Fifty-five microliters of each protein sample (∼10 mg/mL) were loaded onto a Superdex200 column (GE Healthcare), controlled by an Agilent HPLC system, coupled to an in-vacuum SAXS flow cell. HPLC-SAXS traces were processed using ScÅtter. High-resolution structures of individual domains were used as rigid bodies and constraints in the model generation. In our analysis, the modeled structures of the individual domains of Staufen1 were obtained using the Phyre2 web portal (Kelley et al. 2015). Human Staufen1 RBD3 was modeled by homology based on the NMR structure of D. melanogaster Staufen RBD3 (PDB ID: 1EKZ) (Ramos et al. 2000). The structure of human Staufen1 RBD4 was obtained by homology modeling based on the mouse RBD4 (PDB ID: 1UHZ). The structures of SSM and RBD5, were extracted from the structure solved by X-ray crystallography (PDB ID: 4DKK) (Gleghorn et al. 2013) and treated as two separate domains in this analysis, allowing complete interdomain loop flexibility. To obtain a more complete set of structural information to use as constraints for the interpretation of the SAXS data, the sequences of the 6-His + linker + RBD2 domain of hStau155_FL, the 6-His + linker of hStau155_ΔRBD2 and the TBD were modeled using the Phyre2 server (Kelley et al. 2015). The program EOM 2.0 (Tria et al. 2015) was used to obtain the models of hStau155_FL protein and deletion mutants. A pool of 10,000 independent models was generated, based on the sequence of hStau155_FL, or hStau155_ΔRBD2, and on constraints, we generated by homology modeling. After the creation of the pool of models, EOM (Tria et al. 2015) runs a genetic algorithm that compares the average theoretical scattering intensity from the ensemble of 10,000 conformations with the experimental scattering data and selects the models that best describe the experimental data, taking into account the constraints used as input (in this case, the homology models of individual domains).
SAXS data for individual domains and tandem domains were collected at B21, Diamond Light Source (Harwell, UK). Fifty-five microliters of each protein sample (∼10 mg/mL) were loaded onto a Superdex75 column (GE Healthcare), controlled by an Agilent HPLC system, coupled to an in-vacuum SAXS flow cell. HPLC-SAXS traces were processed using ScÅtter. Data were analyzed using different strategies depending on their flexibility level. Models for individual domains RBD3 and RBD4 were obtained using ScÅtter and DAMMIN and the tandem domain TBD_SSM/RBD5 was modeled using BUNCH (ATSAS). All the other domains, showing higher degree of flexibility, were modeled with EOM, as described above.
NMR spectroscopy
hStau155_FL, hStau155_ΔRBD2, individual and tandem domains were studied by NMR spectroscopy. Thirty microliters of D2O were added to 570 μL of protein in 20 mM potassium phosphate pH 7.5, 100 mM KCl, 10 mM MgCl2, 200 mM L-Arg HCl, 2mM TCEP at a suitable concentration for NMR experiments (Supplemental Table S2). 15N,1H-TROSY-HSQC spectra (Weigelt 1998) were acquired at 298 K using a Bruker AVANCE IIIHD 600 MHz spectrometer equipped with a 5 mm TCI cryoprobe. Data were processed using the Bruker TopSpin software and figures were generated using the CCPN analysis 2.4 software (Vranken et al. 2005).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We are grateful to Lynne Maquat for the kind gift of the pRSET-B-Stau155 vector (Kim et al. 2005). We acknowledge Eve Hartswood and Amy Whittaker for some initial protein production tests. We are grateful to Drs. Robert Rambo, Nathan Cowieson, Nikul Khunti, and Katsuaki Inoue (Diamond Light Source, UK) for assistance with data collection at B21, Diamond Light Source, Dr. Daniel Myatt (ISIS Pulsed Neutron and Muon Source) for help with the initial scattering experiments, and to Dr. David Scott for access to biophysical instrumentation at the Research Complex at Harwell. S.V. is supported by an ISIS Neutron Source/University of Edinburgh joint PhD Studentship (STFC/SA15). G.C. was supported by a Darwin Trust of Edinburgh PhD Studentship.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.072595.119.
REFERENCES
- Allison R, Czaplinski K, Git A, Adegbenro E, Stennard F, Houliston E, Standart N. 2004. Two distinct Staufen isoforms in Xenopus are vegetally localized during oogenesis. RNA 10: 1751–1763. 10.1261/rna.7450204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KS, Chow EJ, Goodman PJ, Leisenring WM, Dietz AC, Perkins JL, Chow L, Sinaiko A, Moran A, Petryk A, et al. 2013. Impact of treatment exposures on cardiovascular risk and insulin resistance in childhood cancer survivors. Cancer Epidemiol Biomarkers Prev 22: 1954–1963. 10.1158/1055-9965.EPI-13-0610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Benjamin R, Balakrishnan K, Ghosh P, Banerjee S. 2014. Human protein Staufen-2 promotes HIV-1 proliferation by positively regulating RNA export activity of viral protein Rev. Retrovirology 11: 18 10.1186/1742-4690-11-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud P, Allain FH-T. 2012. ADAR proteins: double-stranded RNA and Z-DNA binding domains. Curr Top Microbiol Immunol 353: 35–60. 10.1007/82_2011_145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bélanger G, Stocksley MA, Vandromme M, Schaeffer L, Furic L, DesGroseillers L, Jasmin BJ. 2003. Localization of the RNA-binding proteins Staufen1 and Staufen2 at the mammalian neuromuscular junction. J Neurochem 86: 669–677. 10.1046/j.1471-4159.2003.01883.x [DOI] [PubMed] [Google Scholar]
- Blackham SL, McGarvey MJ. 2013. A host cell RNA-binding protein, Staufen1, has a role in hepatitis C virus replication before virus assembly. J Gen Virol 94: 2429–2436. 10.1099/vir.0.051383-0 [DOI] [PubMed] [Google Scholar]
- Bonnet-Magnaval F, Philippe C, Van Den Berghe L, Prats H, Touriol C, Lacazette E. 2016. Hypoxia and ER stress promote Staufen1 expression through an alternative translation mechanism. Biochem Biophys Res Commun 479: 365–371. 10.1016/j.bbrc.2016.09.082 [DOI] [PubMed] [Google Scholar]
- Boulay K, Ghram M, Viranaicken W, Trépanier V, Mollet S, Fréchina C, DesGroseillers L. 2014. Cell cycle-dependent regulation of the RNA-binding protein Staufen1. Nucleic Acids Res 42: 7867–7883. 10.1093/nar/gku506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner G, Bassi MT, Andolfi G, Ballabio A, Franco B. 1999. Identification of a novel homolog of the Drosophila staufen protein in the chromosome 8q13-q21.1 region. Genomics 62: 113–118. 10.1006/geno.1999.6015 [DOI] [PubMed] [Google Scholar]
- Bycroft M, Grünert S, Murzin AG, Proctor M, St Johnston D. 1995. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology with the N-terminal domain of ribosomal protein S5. EMBO J 14: 3563–3571. 10.1002/j.1460-2075.1995.tb07362.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Clement J-F, Martel C, Beriault V, Gatignol A, DesGroseillers L, Mouland AJ. 2004. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol 24: 2637–2648. 10.1128/MCB.24.7.2637-2648.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Boulay K, Mouland AJ, Desgroseillers L. 2008. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology 5: 41 10.1186/1742-4690-5-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim KM, Han S, Choe J, Park SG, Choi SS, Kim YK. 2012. Staufen1-mediated mRNA decay functions in adipogenesis. Mol Cell 46: 495–506. 10.1016/j.molcel.2012.03.009 [DOI] [PubMed] [Google Scholar]
- de Lucas S, Peredo J, Marión RM, Sanchez C, Ortin J. 2010. Human Staufen1 protein interacts with influenza virus ribonucleoproteins and is required for efficient virus multiplication. J Virol 84: 7603–7612. 10.1128/JVI.00504-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas S, Oliveros JC, Chagoyen M, Ortín J. 2014. Functional signature for the recognition of specific target mRNAs by human Staufen1 protein. Nucleic Acids Res 42: 4516–4526. 10.1093/nar/gku073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit U, Pandey AK, Mishra P, Sengupta A, Pandey VN. 2016. Staufen1 promotes HCV replication by inhibiting protein kinase R and transporting viral RNA to the site of translation and replication in the cells. Nucleic Acids Res 44: 5271–5287. 10.1093/nar/gkw312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Hemraj I, Furic L, Deitinghoff A, Kiebler MA, DesGroseillers L. 2002. Staufen2 isoforms localize to the somatodendritic domain of neurons and interact with different organelles. J Cell Sci 115: 3285–3295. [DOI] [PubMed] [Google Scholar]
- Fernández Moya SM, Kiebler MA. 2015. CLIPing Staufen to secondary RNA structures: size and location matter! Bioessays 37: 1062–1066. 10.1002/bies.201500052 [DOI] [PubMed] [Google Scholar]
- Furic L, Maher-Laporte M, DesGroseillers L. 2008. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA 14: 324–335. 10.1261/rna.720308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautrey H, McConnell J, Hall J, Hesketh J. 2005. Polarised distribution of the RNA-binding protein Staufen in differentiated intestinal epithelial cells. FEBS Lett 579: 2226–2230. 10.1016/j.febslet.2005.02.074 [DOI] [PubMed] [Google Scholar]
- Gautrey H, McConnell J, Lako M, Hall J, Hesketh J. 2008. Staufen1 is expressed in preimplantation mouse embryos and is required for embryonic stem cell differentiation. Biochim Biophys Acta 1783: 1935–1942. 10.1016/j.bbamcr.2008.05.017 [DOI] [PubMed] [Google Scholar]
- Gleghorn ML, Maquat LE. 2014. ‘Black sheep’ that don't leave the double-stranded RNA-binding domain fold. Trends Biochem Sci 39: 328–340. 10.1016/j.tibs.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleghorn ML, Gong C, Kielkopf CL, Maquat LE. 2013. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nat Struct Mol Biol 20: 515–524. 10.1038/nsmb.2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature 470: 284–288. 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. 2009. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev 23: 54–66. 10.1101/gad.1717309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, Yang V, Aguiar M, Kornhauser J, Jia X, et al. 2014. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol Cell Proteom 13: 372–387. 10.1074/mcp.O113.027870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia M, Shan Z, Yang Y, Liu C, Li J, Luo Z-G, Zhang M, Cai Y, Wen W, Wang W. 2015. The structural basis of Miranda-mediated Staufen localization during Drosophila neuroblast asymmetric division. Nat Commun 6: 8381 10.1038/ncomms9381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell 120: 195–208. 10.1016/j.cell.2004.11.050 [DOI] [PubMed] [Google Scholar]
- Kim MY, Park J, Lee JJ, Ha DH, Kim J, Kim CG, Hwang J, Kim CG. 2014. Staufen1-mediated mRNA decay induces Requiem mRNA decay through binding of Staufen1 to the Requiem 3'UTR. Nucleic Acids Res 42: 6999–7011. 10.1093/nar/gku388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M. 2013. TINCR, staufen1, and cellular differentiation. RNA Biol 10: 1597–1601. 10.4161/rna.26249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krovat BC, Jantsch MF. 1996. Comparative mutational analysis of the double-stranded RNA binding domains of Xenopus laevis RNA-binding protein A. J Biol Chem 271: 28112–28119. 10.1074/jbc.271.45.28112 [DOI] [PubMed] [Google Scholar]
- Laver JD, Li X, Ancevicius K, Westwood JT, Smibert CA, Morris QD, Lipshitz HD. 2013. Genome-wide analysis of Staufen-associated mRNAs identifies secondary structures that confer target specificity. Nucleic Acids Res 41: 9438–9460. 10.1093/nar/gkt702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaretti D, Bandholz-Cajamarca L, Emmerich C, Schaaf K, Basquin C, Irion U, Bono F. 2018. The crystal structure of Staufen1 in complex with a physiological RNA sheds light on substrate selectivity. Life Sci Alliance 1: e201800187 10.26508/lsa.201800187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeau G, Maher-Laporte M, Topolnik L, Laurent CE, Sossin W, Desgroseillers L, Lacaille J-C. 2008. Staufen1 regulation of protein synthesis-dependent long-term potentiation and synaptic function in hippocampal pyramidal cells. Mol Cell Biol 28: 2896–2907. 10.1128/MCB.01844-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz J, Lewis MS, Schuck P. 2002. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci 11: 2067–2079. 10.1110/ps.0207702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGendre JB, Campbell ZT, Kroll-Conner P, Anderson P, Kimble J, Wickens M. 2013. RNA targets and specificity of Staufen, a double-stranded RNA-binding protein in Caenorhabditis elegans. J Biol Chem 288: 2532–2545. 10.1074/jbc.M112.397349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Duchaîne TF, DesGroseillers L. 2002. Molecular mapping of the determinants involved in human Staufen-ribosome association. Biochem J 365: 817–824. 10.1042/bj20020263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae IJ, Li F, Zhou K, Cande WZ, Doudna JA. 2006. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harb Symp Quant Biol 71: 73–80. 10.1101/sqb.2006.71.042 [DOI] [PubMed] [Google Scholar]
- Marión RM, Fortes P, Beloso A, Dotti C, Ortín J. 1999. A human sequence homolog of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol Cell Biol 19: 2212–2219. 10.1128/MCB.19.3.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Macchi P, Furic L, Kiebler MA, Desgroseillers L. 2006. Staufen1 is imported into the nucleolus via a bipartite nuclear localization signal and several modulatory determinants. Biochem J 393: 245–254. 10.1042/BJ20050694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Dugre-Brisson S, Boulay K, Breton B, Lapointe G, Armando S, Trepanier V, Duchaine T, Bouvier M, Desgroseillers L. 2010. Multimerization of Staufen1 in live cells. RNA 16: 585–597. 10.1261/rna.1664210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. 2000. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol 74: 5441–5451. 10.1128/JVI.74.12.5441-5451.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaleo E, Saladino G, Lambrughi M, Lindorff-Larsen K, Gervasio FL, Nussinov R. 2016. The role of protein loops and linkers in conformational dynamics and allostery. Chem Rev 116: 6391–6423. 10.1021/acs.chemrev.5b00623 [DOI] [PubMed] [Google Scholar]
- Park E, Maquat LE. 2013. Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA 4: 423–435. 10.1002/wrna.1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Gleghorn ML, Maquat LE. 2013. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself- and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci USA 110: 405–412. 10.1073/pnas.1213508110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peredo J, Villacé P, Ortín J, de Lucas S. 2014. Human Staufen1 associates to miRNAs involved in neuronal cell differentiation and is required for correct dendritic formation. PLoS ONE 9: e113704 10.1371/journal.pone.0113704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy S, Wang H, Quach HN, Sampath K. 2006. Zebrafish Staufen1 and Staufen2 are required for the survival and migration of primordial germ cells. Dev Biol 292: 393–406. 10.1016/j.ydbio.2006.01.014 [DOI] [PubMed] [Google Scholar]
- Rambo RP. 2017. Considerations for sample preparation using size-exclusion chromatography for home and synchrotron sources. Adv Exp Med Biol 1009: 31–45. 10.1007/978-981-10-6038-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Bayer P, Varani G. 1999. Determination of the structure of the RNA complex of a double-stranded RNA-binding domain from Drosophila Staufen protein. Biopolymers 52: 181–196. [DOI] [PubMed] [Google Scholar]
- Ramos A, Grünert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, Varani G. 2000. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J 19: 997–1009. 10.1093/emboj/19.5.997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel-Chapuis A, Bélanger G, Yadava RS, Mahadevan MS, DesGroseillers L, Côté J, Jasmin BJ. 2012. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative premRNA splicing. J Cell Biol 196: 699–712. 10.1083/jcb.201108113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel-Chapuis A, Crawford TE, Blais-Crépeau M-L, Bélanger G, Richer CT, Jasmin BJ. 2014. The RNA-binding protein Staufen1 impairs myogenic differentiation via a c-myc-dependent mechanism. Mol Biol Cell 25: 3765–3778. 10.1091/mbc.e14-04-0895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel-Chapuis A, Klein Gunnewiek A, Bélanger G, Crawford Parks TE, Côté J, Jasmin BJ. 2016. Staufen1 impairs stress granule formation in skeletal muscle cells from myotonic dystrophy type 1 patients. Mol Biol Cell 27: 1728–1739. 10.1091/mbc.e15-06-0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer EE, Ashar-Patel A, Peng L, Moore MJ. 2014. Staufen1 senses overall transcript secondary structure to regulate translation. Nat Struct Mol Biol 21: 26–35. 10.1038/nsmb.2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. 2011. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal 4: rs3 10.1126/scisignal.2001570 [DOI] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. 1986. Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev Biol 113: 443–448. 10.1016/0012-1606(86)90179-X [DOI] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nusslein-Volhard C. 1991. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66: 51–63. 10.1016/0092-8674(91)90138-O [DOI] [PubMed] [Google Scholar]
- St Johnston D, Brown NH, Gall JG, Jantsch M. 1992. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA 89: 10979–10983. 10.1073/pnas.89.22.10979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D'Ambrogio A, Luscombe NM, Ule J. 2015. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 26: 491–494. 10.1038/nature14280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JM, Beal PA. 2017. How do ADARs bind RNA? New protein–RNA structures illuminate substrate recognition by the RNA editing ADARs. Bioessays 39 10.1002/bies.201600187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tria G, Mertens HD, Kachala M, Svergun DI. 2015. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ 2: 207–217. 10.1107/S205225251500202X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Lafrance-Vanasse J, Tainer JA. 2014. The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once. DNA Repair (Amst) 19: 95–107. 10.1016/j.dnarep.2014.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JP, Macchi P, Stein JM, Mikl M, Hawker KN, Vogelsang P, Wieczorek K, Vendra G, Riefler J, Tubing F, et al. 2008. A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. Proc Natl Acad Sci USA 105: 16374–16379. 10.1073/pnas.0804583105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. 2005. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59: 687–696. 10.1002/prot.20449 [DOI] [PubMed] [Google Scholar]
- Wang X, Vukovic L, Koh HR, Schulten K, Myong S. 2015. Dynamic profiling of double-stranded RNA binding proteins. Nucleic Acids Res 43: 7566–7576. 10.1093/nar/gkv726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt J. 1998. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J Am Chem Soc 120: 10778–10779. 10.1021/ja982649y [DOI] [Google Scholar]
- Wickham L, Duchaîne T, Luo M, Nabi IR, DesGroseillers L. 1999. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol 19: 2220–2230. 10.1128/MCB.19.3.2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ, Mohammed S. 2013. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res 12: 260–271. 10.1021/pr300630k [DOI] [PubMed] [Google Scholar]