Abstract
Recent advances in technology over the last decades have strived to elucidate the diverse and abundant ecosystem of the human microbiome. The intestinal microbiota represents a densely inhabited environment that offers a plethora of beneficial effects to the host’s wellbeing. On the other hand, it can serve as a potential reservoir of Multi-Drug Resistant (MDR) bacteria and their antibiotic-resistant genes (ARgenes), which comprise the “gut resistome.” ARgenes, like antibiotics, have been omnipresent in the environment for billions of years. In the context of the gut microbiome, these genes may conflate into exogenous MDR or emerge in commensals due to mutations or gene transfers. It is currently generally accepted that Antimicrobial Resistance (AMR) poses a serious threat to public health worldwide. It is of paramount importance that researchers focus on, amongst other parameters, elaborating strategies to manage the gut resistome, particularly focusing on the diminution of AMR. Potential interventions in the gut microbiome field by Fecal Microbiota Transplant (FMT) or functional foods are newly emerged candidates for the uprooting of MDR strains and restoring dysbiosis and resilience. Probiotic nutrition is thought to diminish gut colonization from pathobionts. Yet only a few studies have explored the effects of antibiotics use on the reservoir of AR genes and the demanding time for return to normal by gut microbiota-targeted strategies. Regular administration of probiotic bacteria has recently been linked to restoration of the gut ecosystem and decrease of the gut resistome and AR genes carriers. This review summarizes the latest information about the intestinal resistome and the intriguing methods of fighting against AMR through probiotic-based methods and gut microbial shifts that have been proposed. This study contains some key messages: (1) AMR currently poses a lethal threat to global health, and it is pivotal for the scientific community to do its utmost in fighting against it; (2) human gut microbiome research, within the last decade especially, seems to be preoccupied with the interface of numerous diseases and identifying a potential target for a variety of interventions; (3) the gut resistome, comprised of AR genesis, presents very early on in life and is prone to shifts due to the use of antibiotics or dietary supplements; and (4) future strategies involving functional foods seem promising for the battle against AMR through intestinal resistome diminution.
Keywords: antimicrobial resistance, gut microbiome, resistome, antimicrobial resistance genes, probiotics, prebiotics
Introduction
The human gut microbiota constitutes a whole world of microbes, fungi, viruses, and parasites in an eternal interplay between them and their host. This abundant and diverse ecosystem, with more than 100 trillion of microorganisms, has created a unique niche and plays a pivotal role in its host’s physiology and metabolism by participating in digestion, innate and adaptive immunity, and protection from pathogen colonization. During the last decade, culture-independent techniques have set out to elucidate details for the dominant phyla: the external and internal factors influencing the intestinal microbiome, its “normal” and “abnormal” variations, and its relationship with health and disease (Rinninella et al., 2019).
On the other hand, antimicrobial resistance (AMR) has managed to besmear the miraculous antibiotics and endanger the achievements of modern medicine by nullifying the ability of antimicrobials to fight infections. The misuse and overuse of antibiotics promote bacterial resistance, and the malleable gut microbiota has consequently evolved into a reservoir of resistance genes, even from infancy (Francino, 2016).
Apart from strategies that involve Antibiotic Stewardship – the implementation of policies and new drugs in order to re-establish the therapeutic capability of antibiotics – a very promising solution has emerged from the field of functional foods. It’s becoming more than obvious that the gut resistome might be receptive to probiotic and prebiotic supplementation. The preferable result of this manipulation would be the elimination or even the uprooting of Multi-Drug Resistant (MDR) strains and restoration of dysbiosis and gut resilience (Wu et al., 2016; MacPherson et al., 2018).
In this paper, we aimed to provide an overview for the emergence and features of the gut resistome and the dynamics of prebiotics and probiotics administration targeting antibiotic-resistant genes (AR genes) as an alternative to tackling AMR today in the context of personalized medicine.
The Gut Microbiome in a Nutshell
Nowadays, it is generally accepted that microbial cells on and in our body outnumber our human cells, and the most densely populated site is the Gastrointestinal Tract (GI), which presents a gradual augmentation from 104/ml bacterial cells in the stomach and duodenum to almost 1011/ml in the colon (Sender et al., 2016). Either a “virtual organ” (Valdes et al., 2018) or a multiple set of “organs” (Cani, 2017) has become, in the last two decades, a very intriguing area of research, especially following the introduction of new technologies for analyzing DNA and RNA (16S rRNA sequencing–Shotgun metagenomics sequencing) directly from feces. The dominant phyla Firmicutes and Bacteroidetes represent 90% of gut microbiota (Arumugam et al., 2011), and Europeans are categorized to three enterotypes according to its variations (Zoetendal et al., 2008; Segata et al., 2012; Stavropoulou et al., 2019).
In contrast to previous beliefs that newborns were free of bacteria, it is now a proven fact that bacteria are present in the endometrium, amniotic fluid, umbilical cord, and meconium (Amenyogbe et al., 2017), and different environmental factors, such as the delivery mode (cesarean or vaginal), feeding style (breastfeeding or formula), gestational age, administration of probiotics or prebiotics, diet, exercise, geography, genetics, and air pollution are the most pivotal determinants for gut microbiota modifications that lead to the development of a mature and healthy gut (Bezirtzoglou and Stavropoulou, 2011; Koren et al., 2013; Tanaka and Nakayama, 2017; Thursby and Juge, 2017; Angelakis and Raoult, 2018; Avelar Rodriguez et al., 2018; Ficara et al., 2018; Leoni et al., 2019). It’s quite controversial whether the human placenta includes microbes. Aagaard et al. (2014) studied placental specimens collected under sterile conditions from 320 subjects. Authors have characterized a unique placental microbiome niche, composed of non-pathogenic commensal microbiota from the Firmicutes, Tenericutes, Proteobacteria, Bacteroidetes, and Fusobacteria phyla. The placental microbiome was closely related to the human oral microbiome (Bray–Curtis dissimilarity <0.3) (Aagaard et al., 2014). On the other hand, de Goffau et al. (2019) as well as McDonald and McCoy (2019) in their respective studies, concluded that the human placenta is microbe-free. Studies have demonstrated that diversity and richness increases during life until adulthood when the intestinal microbiota stabilizes and, later on, reduces and adopts a poorer phenotype until the onset of old age (Belizário and Napolitano, 2015). A mutually beneficial relationship is established between the gut microflora and human host. This is called “eubiosis,” and, among other things, it aids in regulating digestion, immune function, resistance against pathogens, and the maintenance of an intact alimentary track epithelium (Brestoff and Artis, 2013; Avelar Rodriguez et al., 2018). Any disruption of this balance leads to changes in composition and diversity, called “dysbiosis,” and this has a negative impact on host health. Although the causality of the microbiome shifts has not yet been fully proven, a plethora of studies implicate this imbalance in multiple intestinal and extra-intestinal diseases, namely obesity, autoimmunity, cancer, allergies, neurological disorders, autism, etc. (Gao et al., 2017; Tanaka and Nakayama, 2017; Castaner et al., 2018; Clemente et al., 2018).
The restoration of dysbiosis by different kinds of approaches, such as neutraceuticals, diet interventions, and FMT, is a whole new scientific field, and, though poorly understood, it is very compelling and full of promise.
Antimicrobial Resistance and the Rise of Another –OME: the Resistome
Even though the emergence of antibiotics in the 1930s signified a new era in fighting infections, the truth is that they have been present in nature for eons. Unfortunately, their widespread usage has contributed to the growing global threat of AMR, which has resulted in the reduction of therapeutic options as well as a rise in lethal infections from MDR or PDR (Pan Drug Resistant) bacteria and “superbugs” in healthcare settings worldwide.
Bacteria have dwelled on the Earth for ages and have evolved the capability to avoid antibiotic attack through an accumulation of mutations or by a scattering of AR genes, using methods including horizontal gene transfer (transduction, conjugation, and transformation), toxin–antitoxin systems, and Mobile Genetic Elements (MGEs) (transposons, plasmids, integrons, and integrative–conjugative elements) (Burrus et al., 2002; Carattoli, 2013; Iyer et al., 2013; Sultan et al., 2018; Xie et al., 2018).
Antibiotic-resistant genes are mostly harbored by strictly anaerobic commensals in the gut, but, when a transfer of genetic material occurs by chance to pathobionts, it is conferred to the proliferation of MDR strains (van Schaik, 2015). Collectively all the AR genes and their precursors of gut bacteria pathogens and non-pathogens make up the resistome, as proposed by Gerard D. Wright in 2006 and 2007 (D’Costa et al., 2006; Wright, 2007). The intestine is considered a huge reservoir for antibiotic resistance determinants. The preponderance of determinants can, however, be considered inherent, and they are rarely shared with opportunistic pathogens (Ruppé et al., 2019). This is possibly due to the presence of unknown obstacles for this transfer from gut anaerobic eubionts (mainly Bacteroides) to Gram-negative facultative anaerobic pathobionts (e.g., Enterobacteriaceae) (van Schaik, 2015). Less is known concerning the role of Firmicutes in this procedure. Furthermore, the MGEs form the “mobilome” as a vehicle of transferring AR genes amongst gut microbes (Frost et al., 2005; Ravi et al., 2015, 2017; Pärnänen et al., 2018).
Over the last decades, opportunistic pathogens, such as Escherichia coli, Klebsiella pneumoniae, Enterococcus faecalis, and Enterococcus faecium from the Enterobacteriaceae and Enterococcaceae groups, represent very important MDR strains in hospitals that may cause serious infections when they translocate across the gut barrier. From the 1970s and 1980s, microbial “culturomics” have shown that Bacteroidales and Clostridiales may bear and transfer AR genes, not only among themselves but to pathobionts as well (Guiney and Davis, 1978; Privitera et al., 1979; Wüst and Hardegger, 1983; van Schaik, 2015). Metagenomic research suggested that effective antibiotic stewardship as a “One Health” policy (humans, animals, and the environment) may contribute to great changes regarding the abundance of AR genes in the gut microbiome of different countries’ dwellers and that resistance genes show specific geography clusters (Forslund et al., 2013; Hu et al., 2013). Likewise, hospitalized patients comprise a particular population under intense and (usually) prolonged antibiotics therapy; metagenomics revealed an increase in relative abundance of resistance genes, such as aminoglycosides (de Smet et al., 2009; Buelow et al., 2014).
Several recent studies have confirmed that the adult intestine has lower richness in AR genes than that of an infant, even if there is not prior exposure to antibiotics (Bäckhed et al., 2015; Moore et al., 2015; Gibson et al., 2016). There is conflicting evidence for the origins of these resistance genes, but most of it derives from mother’s gut as well as breast milk microbes (Zhang et al., 2011; Bode, 2012; Jost et al., 2014; Gosalbes et al., 2016). In this context, there is a hypothesis that this high percentage of AR genes, especially in preterm infants, originates from the high levels of Gammaproteobacteria among first commensals, which usually carry resistance genes (Hu et al., 2013; Moore et al., 2015; Gosalbes et al., 2016). Breastfeeding, apart from numerous other benefits, seems to suppress Gammaprotobacteria and promote Bifidobacteria, which are negatively related to resistance and consequently eliminate the antibiotics resistance burden of the infant gut, especially during the first 6 months (Bode, 2012; Moore et al., 2013; Moore et al., 2015; Pärnänen et al., 2018).
The impact of antibiotics upon the gut microbiota as a cause or promotor of dysbiosis has been widely investigated, and specific features, such as dosage and duration, have been implicated in the elimination of certain commensals that offer colonization resistance. Li et al. (2019) observed that even short-term antibiotic administration led to an individualized resistome shift and altered predominance of certain strains and their personal dynamics, thus indicating a new era in personalized therapy. The most investigated example is the Clostridium difficile as the cause of types of nosocomial diarrhea due to extended medication with broad-spectrum antibiotics (mainly clindamycin) (Casals-Pascual et al., 2018). The latest recommended therapy for recurrent C. difficile infection is FMT (Buffie and Pamer, 2013; Pamer, 2016). Moreover, there are studies showing the elimination of AR genes and MDR bacteria, indicating the potential use of the method for restoring colonization resistance in the gut, but this requires more elaboration (Francino, 2016; Marchesi et al., 2016). It is worth mentioning that the temporary use of probiotics (well-characterized beneficial gut microbes) in tandem with antibiotics therapy is correlated with lower risk of developing clinical overt C. difficile infection (Goldenberg et al., 2018).
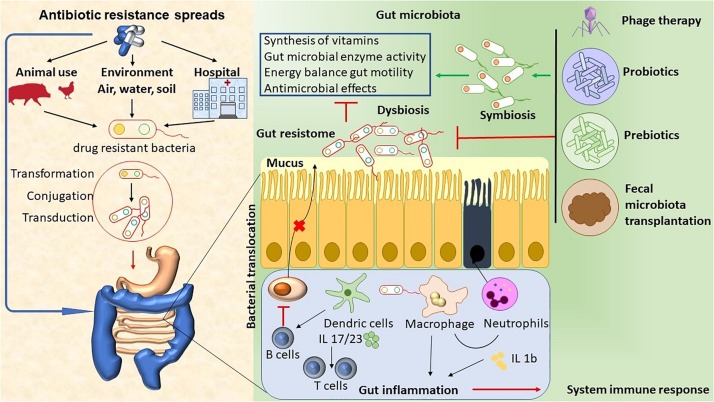
The abuse of antimicrobial agents (environmental, hospital, or animal) has lead to disturbances in the ecological community of the gut microbiota. This leads to mucus layer dysfunction and increased bacterial translocation. These bacteria promote the activation of DCs and macrophages by microorganism-associated molecular patterns (MAMPs) through Toll-like receptors (TLRs), and they induce neutrophil chemotaxis and interleukin (IL) production (Luca and Shoenfeld, 2019). Local inflammation can lead to damage to the epithelial cells. Antigen-presenting cells (APC) present antigens to prime and maintain T-cell responses, and B cells inhibit the production of IgA by plasma cells. Finally, the immune system’s activation promotes inflammation, which ultimately leads to an array of chronic diseases (Guven-Maiorov et al., 2017; Luca and Shoenfeld, 2019). Therapeutic approaches, like phage therapy, the use of functional foods with probiotics and prebiotics, and fecal microbiota transplantation (FMT), have the potential to target specific bacterial taxa, thus helping to re-establish homeostasis and a microbiome balance (Figure 1).
FIGURE 1.
The abuse of antimicrobial agents (environmental, hospital, or animal) has lead to disturbances in the ecological community of the gut microbiota. This leads to mucus layer dysfunction and increased bacterial translocation. These bacteria promote the activation of DCs and macrophages by microorganism- associated molecular patterns (MAMPs) through Toll-like receptors (TLRs), thus inducing neutrophil chemotaxis and IL production. Local inflammation leads to epithelial cells damage. APC present antigens to prime and maintain T-cell responses, and B cells inhibit the production of Ig A by plasma cells. Finally, immune system activation promotes inflammation, which ultimately leads to chronic diseases. The therapeutic approaches, such as phage therapy, the use of functional foods with probiotics and prebiotics, and fecal microbiota transplant (FMT), have the potential to target specific bacterial taxa, thus helping to re-establish homeostasis and microbiome balance.
Functional Foods and Gut Microbiota Modifications
The gut microbiota ecosystem, in order to keep its role in the interface of inner and outer worlds, requires the security of the equilibrium state of eubiosis-homeostasis. The rather stable adult intestinal microbiome is characterized by resilience and plasticity, meaning that, after dietary changes, antibiotics, or infections, it has the ability to mostly restore its shifts in diversity and abundance within days or months (Turnbaugh et al., 2007; Lozupone et al., 2012). Several interventions for resurrecting the lost “healthy” gut microbiome encompass bacteriotherapy (by FMT), prebiotics (compounds that benefit microbes), and probiotics (certain beneficial gut strains) (Al-Kharousi et al., 2018).
Functional foods usually acquire additional functions (often related to health promotion or disease prevention) by adding new ingredients or more of the existing ingredients (Functional Foods and Nutraceuticals, 2008), ingredients like probiotics and prebiotics amongst others. The World Health Organization defines probiotics as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” (Food and Agriculture Organization of the United Nations, World Health Organization, 2006), and prebiotics are non-digestible food components that promote the growth and vivacity of commensals (Shafi et al., 2014). Prebiotics have the capacity to upgrade the expansion and metabolic activities of probiotics. Therapeutic synbiotics serve as a nutritional supplement, referred to as “combination of prebiotics and probiotics in the form of synergism that advantageously affect the host” (Gibson and Roberfroid, 1995; Bezirtzoglou and Stavropoulou, 2011).
Probiotic strains present antimicrobial activity, and they inhibit the growth and displace the adhesion of potential pathogens to human mucus. Collado et al. (2007), tested different probiotic strains against eight selected pathogens. The results indicated that all probiotic strains tested were able to inhibit and displace the adhesion of Bacteroides, Clostridium, Staphylococcus, and Enterobacter (Collado et al., 2007). Furthermore, probiotics restore mucosal homeostasis (Suchodolski and Jergens, 2016), and they also play a pivotal role in the prevention and treatment of H. pylori (Gotteland and Cruchet, 2003) and have a positive effect upon allergic conditions (Forsythe et al., 2007) etc. Prebiotics mainly yield a food source for the flourishing of probiotics and also increase glucose uptake (Breves et al., 2001) and the minerals’ bioavailability (Smiricky-Tjardes et al., 2003).
The exact short- and long-term effects of probiotics and prebiotics on intestinal microbiota are not yet fully elucidated, and conflicting evidence exist. Their impact does not fit into a “one-size-fits-all” model (Sanz et al., 2018) and seems to be unique for every individual for any period of their life.
Resistome Diminution Due to Dietary Supplementation – Is It Feasible?
Excessive antibiotics exposure, especially from early life or even in the uterus, leads to dysbiosis with numerous – yet not completely understood – consequences. Imbalances and disruptions of the intestinal microbiota stemming from antibiotics may decrease its efficiency in fighting infections, threaten immune homeostasis, decontrol metabolism, and gather AR genes that are capable of spreading across species (Francino, 2016). In this vein, the use of probiotic bacteria seems to be a very promising intervention either for preventing perturbations by synchronous administration with antibiotics or by tapping into their benefits for healing the “injured” microbiota and counteract the detrimental effects of antibiotics afterward.
But, as AMR currently addresses public health globally, efforts have been made, apart from the discovery of new drugs, to investigate alternative strategies to combat it. As the pipeline of research into antimicrobial agent, with a few exceptions, seems to dry up, dietary interventions are under active consideration for the time being. In Wu et al. (2016) showed in their study that a gut microbiota-targeted dietary intervention in obese children managed not only to alter the composition and functionality of the intestinal microbiome but, furthermore, to remarkably decrease their gut resistome. The diet that they had previously implemented (Zhang et al., 2015) included whole grains, traditional Chinese medicinal foods, and prebiotics (WTP diet), and with a genome-centric metagenomic approach, this resulted in a reduction of body weight and inflammation parameters. According to their data, other than weight loss and health improvements, they reported a decrease of the resistome richness, a reduction in the AR gene members, and the diminution of multiple-resistance AR gene carriers (reduced 69%), showcasing a possible novel perspective for the issue (Wu et al., 2016). Another important notion in this study is that, because of the diet type (high carbohydrate), Bifidobacterium was augmented and had few or no ARgenes, whereas the decreased bacteria resembled a minefield with their great burden combined with the final reduction of the resistome.
Researchers also demonstrated last year that MDR pathogens have different behaviors depending on the presence in their environment of co-pathogens, commensals, interactions with the host, and probiotics (Chan et al., 2018). Moreover, shifts in the gut microbiome and resistome after antibiotic administration succeeded in recovery within a week of the end of treatment, and this was probably because of the resilience and plasticity of the gut ecosystem, which acts in a very characteristic and predictable way (MacPherson et al., 2018). Evans et al. (2016) first introduced the presented evidence that L. helveticus R0052 and L. rhamnosus R0011 supplementation had a meaningful negative impact on the duration of diarrhea-like defecations following antibiotics.
Another target group that is relatively vulnerable to factors attributed to dysbiosis are infants, particularly preterm ones. As their microbiome is unstable and less diverse, probiotics administration may promote colonization resistance, protection from serious conditions such as necrotizing enterocolitis, and mitigate the negative impact of antibiotics (Esaiassen et al., 2018). Limitations to the procedures have to do with sampling, the duration of the probiotic supplementation, small number of participants, etc., but it is a very promising intervention for dealing with AMR nowadays.
Breastfeeding in Early Life and Gut Microbiota
The gut microbiome is established during the newborn period. Different factors during prenatal, perinatal, and postnatal life may affect the gut microbial composition (Chong et al., 2018). Breastfeeding in early life configurates the gut microbiota, both directly by exposure of the neonate to the milk microbiota and indirectly via maternal milk factors, such as milk oligosaccharides, secretory IgA, and anti-microbial peptides that affect bacterial growth (van den Elsen et al., 2019). On the other hand, the infant gut microbiota has a high abundance of ARgenes, even in the absence of antibiotics exposure. Pärnänen et al. (2018), studied potential sources of infant gut ARgenes by performing metagenomic sequencing of breast milk as well as infant and maternal gut microbiomes. The results of this study suggested that fecal ARgenes of infants resembled those of their own mothers. The infants inherit the legacy of the past antibiotic consumption of their mothers via the transmission of genes (Pärnänen et al., 2018).
Conclusion and Future Perspectives
The emergence of AMR jeopardizes all the achievements of medical science, and it is of paramount importance to urgently raise awareness of this fact and promote the need for the implementation of effective strategies. In this notion, the gut resistome represents a continuous menace for the host’s wellbeing and public health. Different approaches strive to eliminate it, and, amongst other proposed methods, the exploitation of functional foods is strongly considered. It is indeed a very intriguing and promising intervention, though little is yet known with regards to the real impact it can have upon the ARgenes and the potential diminution and restoration of dysbiosis. Further research and more detailed and personal data are essential in order to evaluate and standardize manipulations and functionalities of diet components in a more personalized approach.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Aagaard K., Ma J., Antony K. M., Ganu R., Petrosino J., Versalovic J. (2014). The placenta harbors a unique microbiome. Sci. Transl. Med. 6:237ra65. 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Kharousi Z., Guizani N., Al-Sadi A., Al Bulushi I. (2018). “Fresh fruit and vegetable bacteria: Diversity, antibiotic resistance and their possible contribution to gut microbiota,” in Fruit and Vegetable Consumption and Health: New Research, ed. Jongen W. (Hauppauge, NY: Nova Science Publishers, Inc; ), 39–66. [Google Scholar]
- Amenyogbe N., Kollmann T. R., Ben-Othman R. (2017). Early-life host–microbiome interphase: the key frontier for immune development. Front. Pediatr. 5:111. 10.3389/fped.2017.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelakis E., Raoult D. (2018). Gut microbiota modifications and weight gain in early life. Hum. Microbio. J. 7 10–14. 10.1016/j.humic.2018.01.002 [DOI] [Google Scholar]
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473 174–180. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avelar Rodriguez D., Peña Vélez R., Toro Monjaraz E. M., Ramirez Mayans J., Ryan P. M. (2018). The gut microbiota: a clinically impactful factor in patient health and disease. SN Compr. Clin. Med. 1 188–199. 10.1007/s42399-018-0036-1 [DOI] [Google Scholar]
- Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microb. 17:852 10.1016/j.chom.2015.05.012 [DOI] [PubMed] [Google Scholar]
- Belizário J. E., Napolitano M. (2015). Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front. Microbiol. 6:1050. 10.3389/fmicb.2015.01050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezirtzoglou E., Stavropoulou E. (2011). Immunology and probiotic impact of the newborn and young children intestinal microflora. Anaerobe 17 369–374. 10.1016/j.anaerobe.2011.03.010 [DOI] [PubMed] [Google Scholar]
- Bode L. (2012). Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22 1147–1162. 10.1093/glycob/cws074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff J. R., Artis D. (2013). Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 14 676–684. 10.1038/ni.2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breves G., Szentkuti L., Schröder B. (2001). Effects of oligosaccharides on functional parameters of the intestinal tract of growing pigs. DTW Dtsch. Tierarztl. Wochenschr. 108 246–248. [PubMed] [Google Scholar]
- Buelow E., Gonzalez T. B., Versluis D., Oostdijk E. A. N., Ogilvie L. A., van Mourik M. S. M., et al. (2014). Effects of selective digestive decontamination (SDD) on the gut resistome. J. Antimicrob. Chemother. 69 2215–2223. 10.1093/jac/dku092 [DOI] [PubMed] [Google Scholar]
- Buffie C. G., Pamer E. G. (2013). Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13 790–801. 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V., Pavlovic G., Decaris B., Guédon G. (2002). Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46 601–610. 10.1046/j.1365-2958.2002.03191.x [DOI] [PubMed] [Google Scholar]
- Cani P. D. (2017). Gut microbiota - at the intersection of everything? Nat. Rev. Gastroenterol. Hepatol. 14 321–322. 10.1038/nrgastro.2017.54 [DOI] [PubMed] [Google Scholar]
- Carattoli A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. IJMM 303 298–304. 10.1016/j.ijmm.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Casals-Pascual C., Vergara A., Vila J. (2018). Intestinal microbiota and antibiotic resistance: perspectives and solutions. Hum. Microbio. J. 9 11–15. 10.3389/fmicb.2019.01704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaner O., Goday A., Park Y.-M., Lee S.-H., Magkos F., Shiow S. A. T. E., et al. (2018). The gut microbiome profile in obesity: a systematic review. Int. J. Endocrinol. 2018:4095789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. P., Choi Y., Brinkac L. M., Krishnakumar R., DePew J., Kim M., et al. (2018). Multidrug resistant pathogens respond differently to the presence of co-pathogen, commensal, probiotic and host cells. Sci. Rep. 8:8656. 10.1038/s41598-018-26738-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong C. Y. L., Bloomfield F. H., O’Sullivan J. M. (2018). Factors Affecting Gastrointestinal microbiome development in neonates. Nutrients 10:E274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente J. C., Manasson J., Scher J. U. (2018). The role of the gut microbiome in systemic inflammatory disease. BMJ 360:j5145. 10.1136/bmj.j5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M. C., Meriluoto J., Salminen S. (2007). Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Lett. Appl. Microbiol. 45 454–460. 10.1111/j.1472-765x.2007.02212.x [DOI] [PubMed] [Google Scholar]
- D’Costa V. M., McGrann K. M., Hughes D. W., Wright G. D. (2006). Sampling the antibiotic resistome. Science 311 374–377. 10.1126/science.1120800 [DOI] [PubMed] [Google Scholar]
- de Goffau M. C., Lager S., Sovio U., Gaccioli F., Cook E., Peacock S. J., et al. (2019). Human placenta has no microbiome but can contain potential pathogens. Nature 572 329–334. 10.1038/s41586-019-1451-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smet A. M. G. A., Kluytmans J. W., Cooper B. S., Mascini E. M., Benus R. F. J., van der Werf T. S., et al. (2009). Decontamination of the digestive tract and oropharynx in ICU patients. N. Engl. J. Med. 360 20–31. 10.1056/NEJMoa0800394 [DOI] [PubMed] [Google Scholar]
- Esaiassen E., Hjerde E., Cavanagh J. P., Pedersen T., Andresen J. H., Rettedal S. I., et al. (2018). Effects of probiotic supplementation on the gut microbiota and antibiotic resistome development in preterm infants. Front. Pediatr. 6:347. 10.3389/fped.2018.00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M., Salewski R. P., Christman M. C., Girard S.-A., Tompkins T. A. (2016). Effectiveness of Lactobacillus helveticus and Lactobacillus rhamnosus for the management of antibiotic-associated diarrhoea in healthy adults: a randomised, double-blind, placebo-controlled trial. Br. J. Nutr. 116 94–103. 10.1017/S0007114516001665 [DOI] [PubMed] [Google Scholar]
- Ficara M., Pietrella E., Spada C., Della Casa Muttini E., Lucaccioni L., Iughetti L., et al. (2018). Changes of intestinal microbiota in early life. J. Matern. Fetal Neonatal Med. 33 1036–1043. 10.1080/14767058.2018.1506760 [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations, World Health Organization, (eds) (2006). Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation. Rome: Food and Agriculture Organization. [Google Scholar]
- Forslund K., Sunagawa S., Kultima J. R., Mende D. R., Arumugam M., Typas A., et al. (2013). Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 23 1163–1169. 10.1101/gr.155465.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe P., Inman M. D., Bienenstock J. (2007). Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am. J. Respir. Crit. Care Med. 175 561–569. [DOI] [PubMed] [Google Scholar]
- Francino M. P. (2016). Antibiotics and the human Gut microbiome: dysbioses and accumulation of resistances. Front. Microbiol. 6:1543. 10.3389/fmicb.2015.01543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Leplae R., Summers A. O., Toussaint A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3 722–732. 10.1038/nrmicro1235 [DOI] [PubMed] [Google Scholar]
- Functional Foods and Nutraceuticals (2008). What are Functional Foods and Nutraceuticals. Available at: https://web.archive.org/web/20080607113000/http://www4.agr.gc.ca/AAFC-AAC/display-afficher.do?id=1171305207040&lang=e (accessed Feburary 27, 2019). [Google Scholar]
- Gao R., Gao Z., Huang L., Qin H. (2017). Gut microbiota and colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 36 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. R., Roberfroid M. B. (1995). Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125 1401–1412. 10.1093/jn/125.6.1401 [DOI] [PubMed] [Google Scholar]
- Gibson M. K., Wang B., Ahmadi S., Burnham C.-A. D., Tarr P. I., Warner B. B., et al. (2016). Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat. Microbiol. 1:16024. 10.1038/nmicrobiol.2016.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg J. Z., Mertz D., Johnston B. C. (2018). Probiotics to prevent clostridium difficile infection in patients receiving antibiotics. JAMA 320 499–500. 10.1001/jama.2018.9064 [DOI] [PubMed] [Google Scholar]
- Gosalbes M. J., Vallès Y., Jiménez-Hernández N., Balle C., Riva P., Miravet-Verde S., et al. (2016). High frequencies of antibiotic resistance genes in infants’ meconium and early fecal samples. J. Dev. Orig. Health Dis. 7 35–44. 10.1017/s2040174415001506 [DOI] [PubMed] [Google Scholar]
- Gotteland M., Cruchet S. (2003). Suppressive effect of frequent ingestion of Lactobacillus johnsonii La1 on Helicobacter pylori colonization in asymptomatic volunteers. J. Antimicrob. Chemother. 51 1317–1319. 10.1093/jac/dkg227 [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Davis C. E. (1978). Identification of a conjugative R plasmid in Bacteroides ochraceus capable of transfer to Escherichia coli. Nature 274 181–182. 10.1038/274181a0 [DOI] [PubMed] [Google Scholar]
- Guven-Maiorov E., Tsai C.-J., Nussinov R. (2017). Structural host-microbiota interaction networks. PLoS Comput. Biol. 13:e1005579. 10.1371/journal.pcbi.1005579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Yang X., Qin J., Lu N., Cheng G., Wu N., et al. (2013). Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 4:2151. 10.1038/ncomms3151 [DOI] [PubMed] [Google Scholar]
- Iyer A., Barbour E., Azhar E., Salabi A. A. E., Hassan H. M. A., Qadri I., et al. (2013). Transposable elements in Escherichia coli antimicrobial resistance. Adv. Biosci. Biotechnol. 4 415–423. [Google Scholar]
- Jost T., Lacroix C., Braegger C. P., Rochat F., Chassard C. (2014). Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 16 2891–2904. 10.1111/1462-2920.12238 [DOI] [PubMed] [Google Scholar]
- Koren O., Knights D., Gonzalez A., Waldron L., Segata N., Knight R., et al. (2013). A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput. Biol. 9:e1002863. 10.1371/journal.pcbi.1002863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni C., Ceci O., Manzari C., Fosso B., Volpicella M., Ferrari A., et al. (2019). Human endometrial microbiota at term of normal pregnancies. Genes 10:E971. 10.3390/genes10120971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Rettedal E., van der Helm E., Ellabaan M. M. H., Panagiotou I., Sommer M. O. A. (2019). Antibiotic treatment drives the diversification of the human gut resistome. bioRxiv [Preprint]. 10.1016/j.gpb.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C. A., Stombaugh J. I., Gordon J. I., Jansson J. K., Knight R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489 220–230. 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F. D., Shoenfeld Y. (2019). The microbiome in autoimmune diseases. Clin. Exp. Immunol. 195 74–85. 10.1111/cei.13158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson C. W., Mathieu O., Tremblay J., Champagne J., Nantel A., Girard S. A., et al. (2018). Gut bacterial microbiota and its resistome rapidly recover to basal state levels after short-term amoxicillin-clavulanic acid treatment in healthy adults. Sci. Rep. 8:11192. 10.1038/s41598-018-29229-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi J. R., Adams D. H., Fava F., Hermes G. D. A., Hirschfield G. M., Hold G., et al. (2016). The gut microbiota and host health: a new clinical frontier. Gut 65 330–339. 10.1136/gutjnl-2015-309990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B., McCoy K. D. (2019). Maternal microbiota in pregnancy and early life. Science 6 984–985. 10.1126/science.aay0618 [DOI] [PubMed] [Google Scholar]
- Moore A. M., Ahmadi S., Patel S., Gibson M. K., Wang B., Ndao M. I., et al. (2015). Gut resistome development in healthy twin pairs in the first year of life. Microbiome 3:27. 10.1186/s40168-015-0090-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A. M., Patel S., Forsberg K. J., Wang B., Bentley G., Razia Y., et al. (2013). Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes. PLoS One 8:e78822. 10.1371/journal.pone.0078822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer E. G. (2016). Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 352 535–538. 10.1126/science.aad9382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pärnänen K., Karkman A., Hultman J., Lyra C., Bengtsson-Palme J., Larsson D. G. J., et al. (2018). Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat. Commun. 9:3891. 10.1038/s41467-018-06393-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privitera G., Dublanchet A., Sebald M. (1979). Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J. Infect. Dis. 139 97–101. 10.1093/infdis/139.1.97 [DOI] [PubMed] [Google Scholar]
- Ravi A., Avershina E., Foley S. L., Ludvigsen J., Storrø O., Øien T., et al. (2015). The commensal infant gut meta-mobilome as a potential reservoir for persistent multidrug resistance integrons. Sci. Rep. 5:15317. 10.1038/srep15317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi A., Estensmo E. L. F., Abée-Lund T. M. L., Foley S. L., Allgaier B., Martin C. R., et al. (2017). Association of the gut microbiota mobilome with hospital location and birth weight in preterm infants. Pediatr. Res. 82 829–838. 10.1038/pr.2017.146 [DOI] [PubMed] [Google Scholar]
- Rinninella E., Raoul P., Cintoni M., Franceschi F., Miggiano G. A. D., Gasbarrini A., et al. (2019). What is the healthy Gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 7:E14. 10.3390/microorganisms7010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppé E., Ghozlane A., Tap J., Pons N., Alvarez A.-S., Maziers N., et al. (2019). Prediction of the intestinal resistome by a three-dimensional structure-based method. Nat. Microbiol. 4:112. 10.1038/s41564-018-0292-6 [DOI] [PubMed] [Google Scholar]
- Sanz Y., Romaní-Perez M., Benítez-Páez A., Portune K. J., Brigidi P., Rampelli S., et al. (2018). Towards microbiome-informed dietary recommendations for promoting metabolic and mental health: opinion papers of the MyNewGut project. Clin. Nutr. Edinb. Scotl. 37(6 Pt A), 2191–2197. 10.1016/j.clnu.2018.07.007 [DOI] [PubMed] [Google Scholar]
- Segata N., Haake S. K., Mannon P., Lemon K. P., Waldron L., Gevers D., et al. (2012). Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13:R42. 10.1186/gb-2012-13-6-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafi A., Farooq U., Akram M., Hayat Z., Murtaza M. (2014). Prevention and control of diseases by use of pro- and prebiotics (Synbiotics). Food Rev. Int. 30 291–316. 10.1080/87559129.2014.929142 [DOI] [Google Scholar]
- Smiricky-Tjardes M. R., Grieshop C. M., Flickinger E. A., Bauer L. L., Fahey G. C. (2003). Dietary galactooligosaccharides affect ileal and total-tract nutrient digestibility, ileal and fecal bacterial concentrations, and ileal fermentative characteristics of growing pigs1. J. Anim. Sci. 81 2535–2545. 10.2527/2003.81102535x [DOI] [PubMed] [Google Scholar]
- Stavropoulou E., Tsigalou C., Bezirtzoglou E. (2019). Functions of the Human Intestinal Microbiota in Relation to Functional Foods. Available at: https://www.researchgate.net/publication/329729676_Functions_of_the_Human_Intestinal_Microbiota_in_Relation_to_Functional_Foods (accessed February 18, 2019). [Google Scholar]
- Suchodolski J. S., Jergens A. E. (2016). Recent advances and understanding of using probiotic-based interventions to restore homeostasis of the microbiome for the prevention/therapy of bacterial diseases. Microbiol. Spectr. 4:VMBF-0025-2015. 10.1128/microbiolspec.VMBF-0025-2015 [DOI] [PubMed] [Google Scholar]
- Sultan I., Rahman S., Jan A. T., Siddiqui M. T., Mondal A. H., Haq Q. M. R. (2018). Antibiotics, resistome and resistance mechanisms: a bacterial perspective. Front. Microbiol. 9:2066. 10.3389/fmicb.2018.02066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Nakayama J. (2017). Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. Off. J. JPN Soc. Allergol. 66 515–522. 10.1016/j.alit.2017.07.010 [DOI] [PubMed] [Google Scholar]
- Thursby E., Juge N. (2017). Introduction to the human gut microbiota. Biochem J. 474 1823–1836. 10.1042/BCJ20160510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C., Knight R., Gordon J. I. (2007). The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature 449 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes A. M., Walter J., Segal E., Spector T. D. (2018). Role of the gut microbiota in nutrition and health. BMJ 361:k2179 10.1136/bmj.k2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Elsen L. W. J., Garssen J., Burcelin R., Verhasselt V. (2019). Shaping the Gut microbiota by breastfeeding: the gateway to allergy prevention? Front. Pediatr. 7:47. 10.3389/fped.2019.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik W. (2015). The human gut resistome. Philos. Trans. R. Soc. B. Biol. Sci. 370:20140087. 10.1098/rstb.2014.0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G. D. (2007). The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5 175–186. 10.1038/nrmicro1614 [DOI] [PubMed] [Google Scholar]
- Wu G., Zhang C., Wang J., Zhang F., Wang R., Shen J., et al. (2016). Diminution of the gut resistome after a gut microbiota-targeted dietary intervention in obese children. Sci. Rep. 6:24030. 10.1038/srep24030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüst J., Hardegger U. (1983). Transferable resistance to clindamycin, erythromycin, and tetracycline in Clostridium difficile. Antimicrob. Agents Chemother. 23 784–786. 10.1128/aac.23.5.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Wei Y., Shen Y., Li X., Zhou H., Tai C., et al. (2018). TADB 2.0: an updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res. 46 D749–D753. 10.1093/nar/gkx1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Yin A., Li H., Wang R., Wu G., Shen J., et al. (2015). Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. eBio Med. 10 968–984. 10.1016/j.ebiom.2015.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Kinkelaar D., Huang Y., Li Y., Li X., Wang H. H. (2011). Acquired antibiotic resistance: are we born with it? Appl. Environ. Microbiol. 77 7134–7141. 10.1128/AEM.05087-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal E. G., Rajilic-Stojanovic M., de Vos W. M. (2008). High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57 1605–1615. 10.1136/gut.2007.133603 [DOI] [PubMed] [Google Scholar]