Abstract
The genetic background of Atopic Dermatitis (AD) with chronic pruritus is complex. Filaggrin (FLG) is an essential gene in the epidermal barrier formation s. Loss-of-function (LOF) variants in FLG associated with skin barrier dysfunction constitute the most well-known genetic risk factor for AD. In this study, we focused on the frequency and effect of FLG loss-of-function variants in association with self-reported age-of-onset of AD. The dataset consisted of 386 whole-genome sequencing (WGS) samples. We observe a significant association between FLG LOF status and age-of-onset, with earlier age of onset of AD observed in the FLG LOF carrier group (p-value 0.0003, Wilcoxon two-sample test). We first tested this on the two most prevalent FLG variants. Interestingly, the effect is even stronger when considering all detected FLG LOF variants. Having two or more FLG LOF variants associates with the onset of AD at 2 years of age. In this study, we have shown enrichment of rare variants in the EDC region in cases compared with controls. Age-of-onset analysis shows not only the effect of the FLG and likely EDC variants in terms of the heightened risk of AD, but foremost enables to predict early-onset, lending further credence to the penetrance and causative effect of the identified variants. Understanding the genetic background and risk of early-onset is suggestive of skin barrier dysfunction etiology of AD with chronic pruritus
Subject terms: Disease genetics, Skin diseases, Genetics research, Risk factors
Introduction
Atopic dermatitis (AD) is a chronic, inflammatory skin disease with an estimated prevalence of 7.3% in the US1. The genetic background of chronic pruritus in AD is complex. The heritability of AD is estimated to be around 75%2. Interestingly, when one or both parents have AD the risk of a child developing AD is higher than the risk of developing other atopic conditions, such as asthma and allergic rhinitis3. This suggests that there are genetic factors specific to AD beyond those for general atopy4.
One of the main functions of the skin is to act as a barrier between the individual and the environment, preventing water loss and at the same time preventing pathogen and allergen entry5. Skin barrier dysfunction is a key clinical feature of AD, as this facilitates penetration of allergens, immunological dysfunction, and consequently an increased risk of developing eczema5,6. The skin barrier dysfunction has been associated with the etiology of the itch-scratch cycle. Genes encoding skin barrier proteins have been shown to play a role in the heritability of AD7,8. FLG is the most studied gene in AD. Loss of function (LOF) variants resulting in aberrant FLG production, constitute the best-known AD gene-association and have been shown to predispose individuals to AD5,8. FLG initially synthesizes profilaggrin, which is then transformed to FLG monomers which interact with intermediate filaments in the stratum corneum (SC), causing such to aggregate into dense parallel arrays of macrofilaments. This promotes cellular compaction and keratin crosslinking in the SC, which forms a highly insoluble matrix that acts as a protective barrier5,9.
The disrupted skin barrier of individuals harboring FLG LOF variants is characterized by dry and fissured skin. LOF variants in FLG are associated with lower levels of natural moisturizing factors in AD10. This facilitates penetration of allergens, immunological dysfunction, and consequently an increased risk of developing eczema5,6. Two prevalent FLG LOF variants, p.R501* and p.S761fs were identified as causes of Ichthyosis vulgaris, (dry, thick, scaly skin), a common feature of moderate to severe AD11,12. The allelic frequency of these variants in European cohort with eczema were estimated to be 2.8% and 6.6% for p.R501* and p.S761fs respectively, with a combined frequency of 9.3%13,14. They demonstrate the strongest association with AD, at 18% and 48% for moderate and severe disease, respectively15. However, both the p.R501* and p.S761fs variants, as well as the other two prevalent variants in Europeans, p.S324* and p.R2447*, are highly uncommon in Asian populations16–18. Furthermore, most of the identified FLG null variants display a higher prevalence in individuals of Caucasian ancestry as compared to the African Americans (27.5% vs. 5.8%)19.
The genetic architecture of AD is likely a combination of common variants, but also rare LOF variants within the FLG and pathways involved12. Several rare FLG LOF variants have been identified across different populations. For example, two rare LOF variants not found in European groups were identified in Asian AD populations, p.S2554* and p.S1107fs (prevalence 4.2% and 1.4%, respectively)16. Two studies in Singaporean Chinese AD populations identified a total of 14 additional FLG LOF variants20,21.
Additional genes involved in skin barrier function are thought to have a potential role in AD. The epidermal differentiation complex (EDC) encodes proteins critical to the proper development of keratinocytes and normal formation of the skin barrier22. The proteins in the EDC come from three gene families with closely related functions: the cornified envelop precursor family, and the S100 protein family and the S100 fused type proteins (SFTP)22,23. FLG, which is located on chromosome 1q21, is a member of the SFTP family of the EDC5. Dysregulation of other EDC genes has also been implicated in AD24,25. Several profiling studies of the transcriptome have shown significant downregulation of EDC genes, such as ivolucrin, loricrin and late cornified envelop 2B, in AD24–26. For example, the deletion of the EDC member gene Small Proline-Rich Protein 3 was shown to be associated with AD27. Another large-scale genome-wide association study of AD patients also identified multiple risk loci in genes involved in epidermal proliferation and differentiation, in addition to a strong signal at the FLG locus28.
Our work aimed to investigate the frequency and effect of rare FLG and EDC LOF variants on the age of onset, on the severity of the phenotype itself and on clinically relevant measures in AD. We also evaluated other potential risk loci within the entire EDC in patients with AD, participants of a clinical study, VP-VLY-686-2102 (also referred to as AD1 throughout this paper). Study AD1 was a randomized, double-blind, placebo-controlled, multicenter study of 168 patients (placebo or treatment) with chronic pruritus associated with AD. The genetic results from this study were then replicated in a subset of a second ongoing clinical study, EPIONE (also referred to as AD2 throughout this paper). Study AD2 was also a randomized, double-blind, placebo-controlled, ongoing multicenter study of patients with chronic pruritus associated with AD. We evaluate the effect of these variants on the age-of-onset of AD to further understand the penetrance and the consequences of these variants in the phenotype context.
Results
Incidence of FLG variants in AD patients
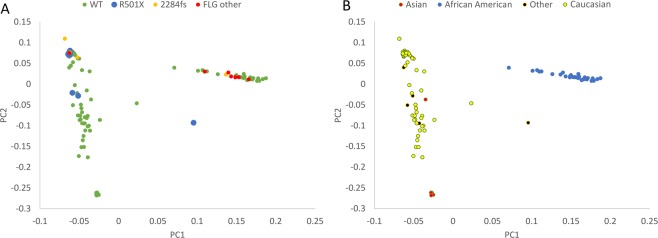
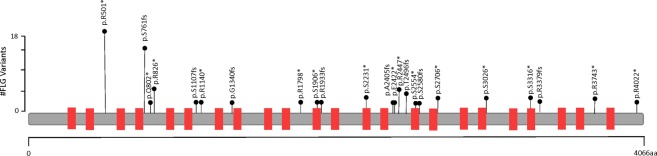
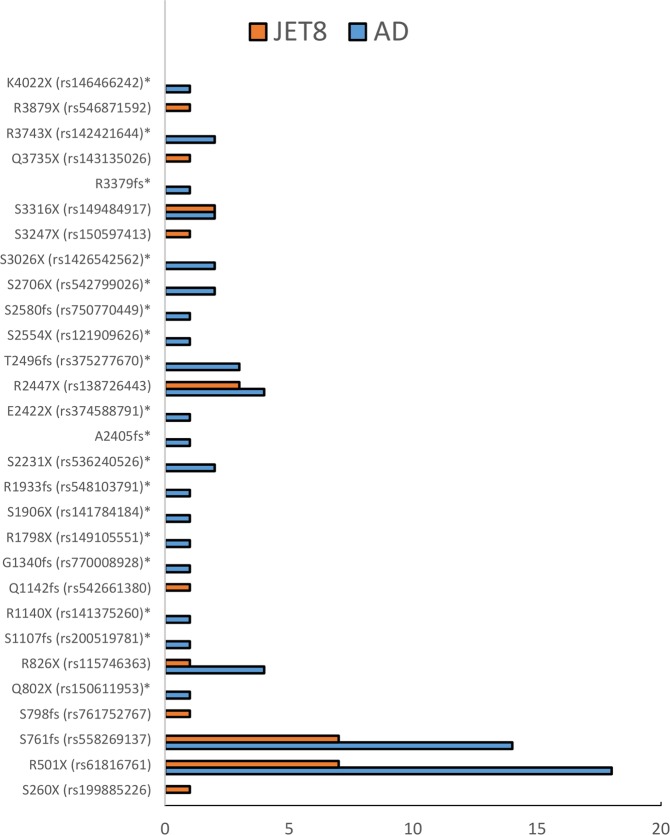
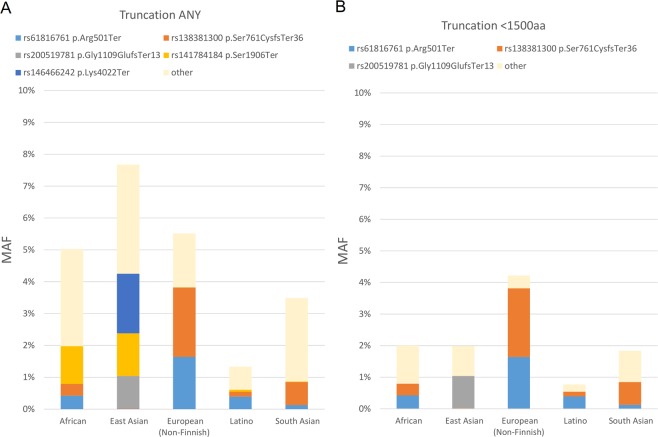
In clinical studyAD1, whole genome sequencing data was obtained from 116 subjects. In an ongoing study AD2 a subset of 270 patient samples were collected and whole genome sequenced. Figure 1 shows a PCA plot of the first study cohort 116/168, together with demographics and clinical characteristics of these patients provided in Supplementary Table 1. PCA was generated using WGS data, in order to examine population substructure of our studied cohort. Looking at panel B, we see enrichment of the two known variants in Caucasians and an interesting enrichment of other LOF FLG variants in the African American background. We investigated the incidence of all FLG LOF variants in the genomes of the clinical study patients, and compared them with the whole genome sequences of a control population of 316 healthy volunteers from clinical study 3107 (also referred to as JET8 throughout this paper). In AD1 study, 26 patients of the 116 (OR = 4.05, CI = 2.17–7.55, p < 0.0001) samples carried FLG LOF variant. In study AD2, 41 of the 270 sampled patients carried a FLG LOF variant (OR = 1.99, CI = 1.18–3.36, p = 0.0093). In the control population from the JET8 study, 21 of the 316 sampled patients harbored FLG LOF variants. The presence of FLG LOF variants in the combined population of AD1 and AD2 patients was significantly greater than the presence of these variants in the JET8 control population (OR = 2.95, CI = 1.76–4.93, p < 0.0001). The incidence of p.R501* variant alone was examined in the AD1 cohort, and was found in 10 of the 116 patients (MAF = 0.043). GnomAD’s allelic frequency total is 0.009 with the highest allelic frequency noted for European (Non-Finnish) population. The list of all the identified FLG LOF is present in Table 1 and displayed on Figs. 2 and 3. Figure 2 displays the location of individual LOF variant in both AD sets. We do observe a higher frequency of FLG LOF variants with many being rare singleton variants when compared to controls as displayed on Fig. 3. In addition, we have evaluated the presence of FLG population specific variants in gnomAD across ethnicities as shown on Fig. 4. Figure 4 shows the major frequency differences among populations especially when focusing the most actionable variants. Panel 4a. displays the frequent variants in the entire region whereas Panel B. displays truncating variants in the region <1500 aa. The motivation beyond examining truncation in the context of FLG LOFs was to explore physical truncation of the protein that would otherwise provide monomers as well as exploring the incidence of such.
Figure 1.
PCA plot displaying the population structure of AD1 cohort. (A) PCA plot colored by variant status. (B) PCA plot (WGS data) colored by ethnicity.
Table 1.
All LOF in FLG identified in the AD set.
| Patient ID | Ethnicity | Gene | Transcript ID | NT change | AA change | RSziD | Chromosome | Start_Position | End_Position |
|---|---|---|---|---|---|---|---|---|---|
| 1011019 | Asian | FLG | NM_002016 | c.C8117G | p.S2706* | rs542799026 | 1 | 152279245 | 152279245 |
| 1081007 | Caucasian | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 1091012 | African American | FLG | NM_002016 | c.5799delG | p.R1933fs | rs548103791 | 1 | 152281563 | 152281563 |
| 1241019 | Asian | FLG | NM_002016 | c.C8117G | p.S2706* | rs542799026 | 1 | 152279245 | 152279245 |
| 1451005 | Asian | FLG | NM_002016 | c.7487delC | p.T2496fs | rs375277670 | 1 | 152279875 | 152279875 |
| 1471013 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 1531004 | Caucasian | FLG | NM_002016 | c.10137delG | p.R3379fs | . | 1 | 152277225 | 152277225 |
| 1601018 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 1601033 | Caucasian | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 1601033 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 1601038 | African American | FLG | NM_002016 | c.7740_7741del | p.S2580fs | rs750770449 | 1 | 152279621 | 152279621 |
| 1621005 | African American | FLG | NM_002016 | c.C2476T | p.R826* | rs115746363 | 1 | 152284886 | 152284886 |
| 1631028 | Caucasian | FLG | NM_002016 | c.C9077G | p.S3026* | rs1426542562 | 1 | 152278285 | 152278285 |
| 1631028 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 1711007 | African American | FLG | NM_002016 | c.C2404T | p.Q802* | rs150611953 | 1 | 152284958 | 152284958 |
| 1711027 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 1721001 | Caucasian | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 1721003 | Caucasian | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 1721003 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 42973718 | Caucasian | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 42978887 | Asian | FLG | NM_002016 | c.7487delC | p.T2496fs | rs375277670 | 1 | 152279875 | 152279875 |
| 42989053 | African American | FLG | NM_002016 | c.C2476T | p.R826* | rs115746363 | 1 | 152284886 | 152284886 |
| 42989254 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 43022549 | African American | FLG | NM_002016 | c.C7339T | p.R2447* | rs138726443 | 1 | 152280023 | 152280023 |
| 43022550 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 43050811 | African American | FLG | NM_002016 | c.C5717A | p.S1906* | rs141784184 | 1 | 152281645 | 152281645 |
| 43055206 | Caucasian | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 43062910 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 43079509 | African American | FLG | NM_002016 | c.C6692A | p.S2231* | rs536240526 | 1 | 152280670 | 152280670 |
| 43099438 | Caucasian | FLG | NM_002016 | c.C7339T | p.R2447* | rs138726443 | 1 | 152280023 | 152280023 |
| 43099528 | African American | FLG | NM_002016 | c.C6692A | p.S2231* | rs536240526 | 1 | 152280670 | 152280670 |
| 43109908 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 43116759 | Caucasian | FLG | NM_002016 | CNV_DELETION | CNV_DELETION | CNV_DELETION | 1 | 152274652 | 152297680 |
| 43132454 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 43134422 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 43135174 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 43148497 | Caucasian | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 43179517 | Caucasian | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 43198393 | Caucasian | FLG | NM_002016 | c.4020delA | p.G1340fs | rs770008928 | 1 | 152283341 | 152283342 |
| 43203074 | African American | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 43221146 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 43221147 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 43260548 | Caucasian | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 43297049 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 43306754 | African American | FLG | NM_002016 | c.C2476T | p.R826* | rs115746363 | 1 | 152284886 | 152284886 |
| 5500365697 | Caucasian | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 5502137729 | African American | FLG | NM_002016 | c.C9077G | p.S3026* | rs1426542562 | 1 | 152278285 | 152278285 |
| 5502362305 | African American | FLG | NM_002016 | c.C9947G | p.S3316* | rs149484917 | 1 | 152277415 | 152277415 |
| 5502362305 | African American | FLG | NM_002016 | c.C2476T | p.R826* | rs115746363 | 1 | 152284886 | 152284886 |
| 5502432960 | African American | FLG | NM_002016 | c.C5392T | p.R1798* | rs149105551 | 1 | 152281970 | 152281970 |
| 5504556880 | Caucasian | FLG | NM_002016 | c.C7339T | p.R2447* | rs138726443 | 1 | 152280023 | 152280023 |
| 5504556880 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 5505347456 | Caucasian | FLG | NM_002016 | c.C1501T | p.R501* | rs61816761 | 1 | 152285861 | 152285861 |
| 5505609600 | Asian | FLG | NM_002016 | c.C7661G | p.S2554* | rs121909626 | 1 | 152279701 | 152279701 |
| 5506294081 | Caucasian | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 5507409104 | African American | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 5508687105 | African American | FLG | NM_002016 | c.C9947G | p.S3316* | rs149484917 | 1 | 152277415 | 152277415 |
| 5508688512 | Caucasian | FLG | NM_002016 | c.7487delC | p.T2496fs | rs375277670 | 1 | 152279875 | 152279875 |
| 5512912848 | Caucasian | FLG | NM_002016 | c.C11227T | p.R3743* | rs142421644 | 1 | 152276135 | 152276135 |
| 5514977857 | African American | FLG | NM_002016 | c.2282_2285del | p.S761fs | rs558269137 | 1 | 152285077 | 152285077 |
| 5515794176 | Caucasian | FLG | NM_002016 | c.C7339T | p.R2447* | rs138726443 | 1 | 152280023 | 152280023 |
| 5515801488 | Asian | FLG | NM_002016 | c.G7264T | p.E2422* | rs374588791 | 1 | 152280098 | 152280098 |
| 5515801488 | Asian | FLG | NM_002016 | c.7214_7215del | p.A2405fs | . | 1 | 152280147 | 152280147 |
| 5515862528 | Asian | FLG | NM_002016 | c.A12064T | p.K4022* | rs146466242 | 1 | 152275298 | 152275298 |
| 5515996305 | Asian | FLG | NM_002016 | c.C3418T | p.R1140* | rs141375260 | 1 | 152283944 | 152283944 |
| 5516026753 | Asian | FLG | NM_002016 | c.C11227T | p.R3743* | rs142421644 | 1 | 152276135 | 152276135 |
| 5516124672 | Asian | FLG | NM_002016 | c.3321delA | p.S1107fs | rs200519781 | 1 | 152284041 | 152284041 |
Figure 2.
Lollipop plot displays the location and frequency of the identified variants in FLG. We observe an enrichment of rare LOF variants in FLG detected in AD patients, Red rectangles are FLG repeats.
Figure 3.
Comparison of variants detected in cases (AD – blue) and controls (JET8 - orange). We observe higher frequency of FLG variants in AD. The variants identified in AD cases not detected in controls are marked with a*.
Figure 4.
Prevalent FLG variants across ethnicities. MAF is displayed on the y-axis whereas the ethnicities are displayed on the x-axis. Panel A displays any truncation whereas Panel B displays truncation <1500 aa.
Enrichment of rare LOF variants in the EDC in AD patients
The incidence of rare LOF variants in the EDC was investigated. Specifically, evolutionarily related members of the S100 fused type proteins (SFTP) family were evaluated, such as CRNN, FLG2, HRNR and RPTN, among other EDC genes all listed Table 2 and LOF in EDC displayed on Fig. 5. We investigated the frequency and effect of rare LOF (stopgain, frameshift, splicing) variants in the SFTP gene family in the AD1 population compared to the JET8 controls, as defined by a MAF < 5%. In the AD1 cohort, 45 of the 116, 38% AD patients carry a LOF variant (Table 2). In the AD2 cohort 75 of the 270, 27.7% AD patients carry a LOF variant. In the JET8 controls, 55 of the 316, 17% patients carried these LOF variants. Comparing these two results, the presence of these variants is significantly higher in the AD patients than the controls (OR = 2.18, CI = 1.51–3.13, p < 0.0001). Cumulative risks (OR and RR) were calculated for these rare variants as well, based on the presence of at least one deleterious allele. The relative risk (RR) for the rare SFTP LOF SNPs (n = 13) was found to be 2.27 (p = 0.0005). The OR was found to be 2.66 (p = 0.0007). In addition we computed the genetic risk score of these top variants shown on S Fig. 2. with a p-value of 0.00001. We were able to replicate the effect in the AD2 study cohort, OR 2.11, p-value = 0.022.
Table 2.
ALL EDC LOFs identified in the AD set.
| Patient ID | Ethnicity | Gene | Transcript ID | NT change | AA change | RSziD | Chromosome | Start_Position | End_Position |
|---|---|---|---|---|---|---|---|---|---|
| 5500098961 | Caucasian | FLG2 | NM_001014342 | c.5475_5478del | p.T1825fs | . | 1 | 152324784 | 152324784 |
| 5508687105 | African American | FLG2 | NM_001014342 | c.C5399G | p.S1800* | rs1349434249 | 1 | 152324863 | 152324863 |
| 1641003 | African American | FLG2 | NM_001014342 | c.C5399G | p.S1800* | rs1349434249 | 1 | 152324863 | 152324863 |
| 1601037 | African American | FLG2 | NM_001014342 | c.3940_3941insTA | p.T1314fs | rs567184084 | 1 | 152326321 | 152326321 |
| 1771002 | Caucasian | FLG2 | NM_001014342 | c.C380A | p.S127* | rs373458772 | 1 | 152329882 | 152329882 |
| 43102554 | African American | FLG2 | NM_001014342 | c.3940_3941insTA | p.T1314fs | rs567184084 | 1 | 152326321 | 152326321 |
| 1601035 | African American | HRNR | NM_001009931 | c.8467delA | p.S2823fs | rs1441063152 | 1 | 152185638 | 152185638 |
| 5508393600 | Caucasian | HRNR | NM_001009931 | c.C2299T | p.R767* | rs148459733 | 1 | 152191806 | 152191806 |
| 5505310928 | Caucasian | HRNR | NM_001009931 | c.C2299T | p.R767* | rs148459733 | 1 | 152191806 | 152191806 |
| 5500363216 | Caucasian | HRNR | NM_001009931 | c.C2299T | p.R767* | rs148459733 | 1 | 152191806 | 152191806 |
| 1711017 | African American | HRNR | NM_001009931 | c.C2299T | p.R767* | rs148459733 | 1 | 152191806 | 152191806 |
| 1061004 | Caucasian | HRNR | NM_001009931 | c.C2299T | p.R767* | rs148459733 | 1 | 152191806 | 152191806 |
| 42989053 | African American | HRNR | NM_001009931 | c.C2299T | p.R767* | rs148459733 | 1 | 152191806 | 152191806 |
| 43306754 | African American | HRNR | NM_001009931 | c.C2299T | p.R767* | rs148459733 | 1 | 152191806 | 152191806 |
| 1721003 | Caucasian | LCE2C | NM_178429 | c.C96A | p.C32* | rs4119577 | 1 | 152648587 | 152648587 |
| 5512683728 | Caucasian | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 5509505040 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 5506327056 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 5502328576 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 5502137729 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 1681005 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 1651002 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 1641003 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 1601035 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 1511001 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 1431001 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 1371007 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 5506294081 | Caucasian | LCE4A | NM_178356 | c.245_254del | p.G82fs | rs763134811 | 1 | 152681796 | 152681796 |
| 42989053 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 43062909 | African American | LCE4A | NM_178356 | c.C9A | p.C3* | rs147765240 | 1 | 152681560 | 152681560 |
| 43181943 | Caucasian | LCE5A | NM_178438 | c.C235T | p.R79* | rs2282298 | 1 | 152484245 | 152484245 |
| 5502328576 | African American | RPTN | NM_001122965 | c.C2227T | p.R743* | rs184952075 | 1 | 152127348 | 152127348 |
| 1631025 | African American | RPTN | NM_001122965 | c.C2227T | p.R743* | rs184952075 | 1 | 152127348 | 152127348 |
| 5511537345 | Caucasian | RPTN | NM_001122965 | c.C1861T | p.Q621* | rs751330515 | 1 | 152127714 | 152127714 |
| 5515993537 | Caucasian | S100A16 | NM_001317007 | c.C5G | p.S2* | rs137911276 | 1 | 153580623 | 153580623 |
| 43179516 | Caucasian | S100A3 | NM_002960 | c.208delG | p.V70fs | rs576022937 | 1 | 153520255 | 153520255 |
| 1511001 | African American | SPRR3 | NM_001097589 | c.189_205del | p.E63fs | rs746080074 | 1 | 152975685 | 152975685 |
| 1451006 | African American | SPRR3 | NM_001097589 | c.189_205del | p.E63fs | rs746080074 | 1 | 152975685 | 152975685 |
| 1371005 | African American | SPRR3 | NM_001097589 | c.189_205del | p.E63fs | rs746080074 | 1 | 152975685 | 152975685 |
| 43079509 | African American | SPRR3 | NM_001097589 | c.189_205del | p.E63fs | rs746080074 | 1 | 152975685 | 152975685 |
| 43128537 | African American | SPRR4 | NM_173080 | c.22_23insA | p.R8fs | rs201207143 | 1 | 152944388 | 152944388 |
| 43153173 | African American | SPRR4 | NM_173080 | c.22_23insA | p.R8fs | rs201207143 | 1 | 152944388 | 152944388 |
| 5508393600 | Caucasian | TCHH | NM_007113 | c.C4309T | p.Q1437* | rs377677960 | 1 | 152081384 | 152081384 |
| 1611010 | Caucasian | TCHH | NM_007113 | c.C991T | p.Q331* | rs201930497 | 1 | 152084702 | 152084702 |
| 43050811 | African American | TCHH | NM_007113 | c.1delA | p.M1fs | rs748146582 | 1 | 152086555 | 152086555 |
| 5508457168 | Caucasian | TCHHL1 | NM_001008536 | c.C2686T | p.Q896* | rs148113334 | 1 | 152057472 | 152057472 |
| 5516026753 | Asian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 5514715841 | Caucasian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 5511537345 | Caucasian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 5508719312 | Caucasian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 5508456912 | Caucasian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 5508456784 | Caucasian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 5507675024 | Asian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 5506589377 | Caucasian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 1711004 | Caucasian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 1601032 | Caucasian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 1481008 | Caucasian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 1301005 | African American | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 1131007 | Caucasian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 1011018 | Caucasian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 5508687105 | African American | TCHHL1 | NM_001008536 | c.C880T | p.Q294* | rs61749316 | 1 | 152059278 | 152059278 |
| 5502362305 | African American | TCHHL1 | NM_001008536 | c.C880T | p.Q294* | rs61749316 | 1 | 152059278 | 152059278 |
| 1371007 | African American | TCHHL1 | NM_001008536 | c.C880T | p.Q294* | rs61749316 | 1 | 152059278 | 152059278 |
| 43022549 | African American | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 43099438 | Caucasian | TCHHL1 | NM_001008536 | c.C1966T | p.Q656* | rs150014958 | 1 | 152058192 | 152058192 |
| 43079509 | African American | TCHHL1 | NM_001008536 | c.C880T | p.Q294* | rs61749316 | 1 | 152059278 | 152059278 |
| 43099528 | African American | TCHHL1 | NM_001008536 | c.C880T | p.Q294* | rs61749316 | 1 | 152059278 | 152059278 |
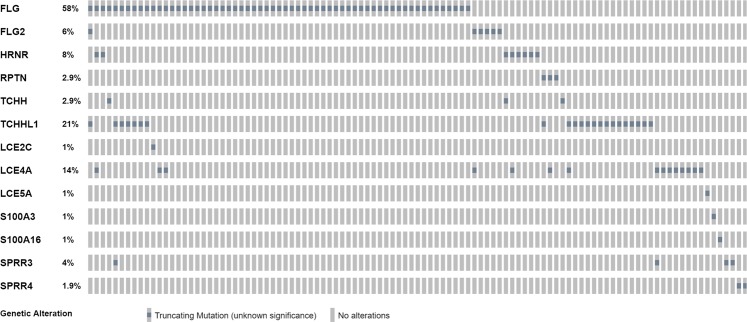
Figure 5.
Percentage of LOF variants identified in both AD cohorts in the EDC (with high incidence of LOFs identified in the SFTP family). The empty gray boxes indicate no alteration whereas the filled gray boxes indicate alteration (LOF) present in a particular subject in that particular gene. The spatial distribution displays all the patients carrying at least a single variant in the queried genes.
Regional enrichment analysis reiterates the significance of the locus
In order to study the LOF variant set of the whole SFTP family of the EDC, an optimal unified sequence kernel association test (SKAT-O) was used. This test was applied to compare the WGS’s of AD1 cases versus healthy controls. Overall, a significant accumulation of rare variants in the EDC was observed in AD cases when compared to controls (p = 4.7E-20). This value is notably much lower than the association of FLG alone (p = 4.5E-6). Specifically, en masse SKATO analysis showed highly significant association with AD in HRNR looking beyond mere FLG, genes. Accumulation of rare LOF variants in the EDC yields a p-value of 4.7e-20, much lower than for FLG alone p-value of 4.5e-6 (LOF set comparing AD with controls) and reiterates the importance of looking for LOF variants extending beyond FLG itself.
Association of variant status with age-of-onset of AD
We collected age of onset of AD information for the whole genome sequencing (WGS) samples also genotyped for 2 most prevalent FLG variants (p.R501* (rs61816761), p.S761fs (rs558269137)). The average age of onset in this cohort is 23.2 years of age (S Fig. 3. Histogram of age of onset). We observe a significant association between FLG LOF status and age-of-onset, with earlier age of onset of AD observed in the FLG LOF carrier group (p-value 0.0003, Wilcoxon two-sample test).The median age of onset for WT is 20 years of age, and the mean age of onset is 4.
Interestingly, the effect is even stronger when considering all detected FLG LOF variants. Having two or more FLG LOF variants associates with onset of AD at 2 years of age (Wilcoxon Two-Sample Test). We observe a significant association between FLG LOF status and age-of-onset, with earlier age of onset of AD observed in the FLG LOF carrier group (z-score 3.95, p-value 0.00008) with the effect displayed on Fig. 6. The OR of having onset before 20 years of age in the AD population if the subject is a variant carrier is 8.9 (p-value 0.004). The OR of having onset before 5 years of age in the AD population if the subject is a variant carrier is 7.8 (p-value 0.0001). We have shown a significant enrichment of WGS for rare variants in the EDC region in cases compared with controls. Age-of-onset analysis shows not only the effect of the FLG and likely EDC variants in terms of heightened risk of AD, but foremost enables to predict early onset, lending further credence to the penetrance and causative effect of the identified variants.
Figure 6.
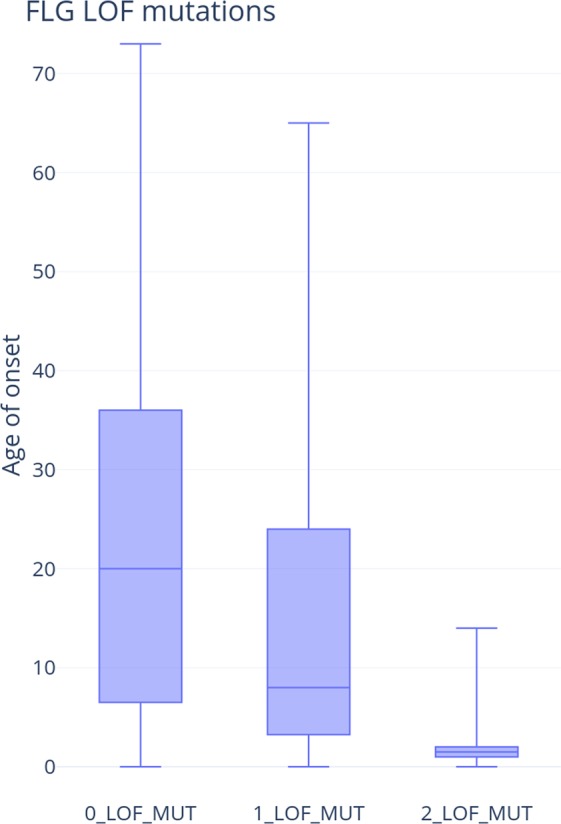
AD Age-of-onset box-plot of AD. We observe an earlier age-of-onset of AD in FLG LOF variant carriers. The effect is greater with higher frequency of FLG LOFs.
Discussion
Using screening methods that are dependent only on a subset of known FLG variants (p.R501*, p.S761fs etc.) we are missing out on ~30% of FLG LOF variation. Whole Genome Sequencing closes that gap of missing FLG impairment among AD patients. It is especially evident among non-European populations as shown in Figs. 1 and 4. With whole genome sequencing and focusing on all LOF variants we have shown that we can explain a higher proportion of patients as well as find other actionable variants (variants of well-established consequence on development and persistence of AD, that can guide diagnosis and likely therapeutic approaches) that are variable across populations. The same holds true when we expand to pathway based analyses and EDC complex, in example variants in HRNR and other SFTP family members.
It has been previously reported by Margolis et al., that subjects with an FLG variants were less likely (OR 0.54) to report as ‘symptom free’ in comparison with those without these variants. Interestingly, children in that cohort carrying the p.R501* variant (OR 0.44) were more likely to be non-responders to therapies19. Another interesting study reported by Koseki et al., investigating the effect of 8 LOF FLG variants, supports the notion that the effect of FLG LOFs variants is prominent during a very early stage of life29. Further evidence comes from another study focusing specifically on 2282del4 showing association with AD developed during infancy as reported by Rupnik et al.30. Furthermore, the authors showed an association with longer duration and more frequent hospitalization in this cohort of FLG variant carriers30. It is becoming more apparent that FLG and overall EDC, specifically SFTP variants predispose to risk of AD with effects differing across ages.
We used FLG and EDC status of variants and used that to correlate with age-of-onset and other clinical measures of AD and itch. The fact that age-of-onset is highly correlated with FLG status reflects upon the potential causes of the disorder itself, here likely skin barrier dysfunction as primary. The fact that frequency matters and amplifies the observed earlier onset lends credence to yet another hypothesis were the effect is intensified with number of causative variants.
There are several limitations of this work that may impact the conclusions as of course we extrapolate to EDC but there could be other ways of assigning pathway based categories that are involved in skin barrier function. Noteworthy is also the fact that we are not using controls evaluated for not having AD, so likely the magnitude of the effect would be even greater with such matched and screened for not having AD diagnosis set of controls. Nevertheless, it is likely that directionally our conclusions are valid and simply the magnitude may increase with differently matched controls. As we are accruing more subjects it will be very interesting to design a panel that would cover and work for different ethnicities as the majority of variants discussed in literature are applicable to Caucasian ancestry. It will also be relevant to quantify the effects of these variants on a translational basis looking at skin proteomics. In addition structural variants that are usually known to account for roughly 5% of the variation unexplained, should be evaluated, in this context to provide a more comprehensive outlook on the role of skin barrier genetics in AD.
Conclusion
Understanding the genetic background and risk of early-onset is suggestive of skin barrier dysfunction etiology of the itch-scratch cycle present in early-onset AD. The pervasive effect of the variants would likely then manifest itself at an early age with AD phenotype of xerosis, and susceptibility to infection. Novel hence newly discovered LOF variants detected in this study expand the pool of risk loci. Whole-genome sequencing showed enrichment for rare variants in the EDC, specifically in the SFTP family, in AD patients compared to healthy controls. The strong association between these variants and AD suggests they may significantly affect a person’s risk of developing the disease. Further, these variants identified are often in genes that affect the skin barrier, lending evidence to the role of barrier dysfunction in the pathogenesis of AD. The novel LOF variants detected in this study help explain the missing heritability and etiology of AD and could serve as disease biomarkers, which could personalize treatment to improve disease outcomes. Adding the age component further reiterates the heritable component with an intensified effect and earlier onset observed at a young age and higher frequency of the LOF variants.
Methods
Clinical study design
The clinical studies VP-VLY-686-2102 and VP-VLY-686-3101 were multicenter, randomized, placebo-controlled, double-blind clinical studies in the United States. Participants provided written informed consent and were provided a copy of the signed consent form. All the studies were approved by the Advarra IRB located in Columbia, MD, USA and participants consented to participation in all the aspects of the studies. Methods were performed in accordance with the relevant guidelines and regulations, in adherence to the IRB approved protocol. Pharmacogenetic samples were taken at the screening visit for both studies. In EPIONE1, the self-reported age of onset was collected by the Medical History of Atopy questionnaire (developed by Vanda) at the first study visit. The study was divided into two phases: the screening phase and the evaluation phase (S Fig. 1).
Key study inclusion criteria included: chronic itch related to AD, defined as lasting 6 weeks or longer, that was refractory to previous treatment by patient history, average itch visual analog score (VAS) of greater than or equal to 70 mm out of 100 mm, and itch verbal response score (VRS) of greater than or equal to 3 on at least one of the past three days prior to randomization.
Study demographics were similar between treatment and placebo groups for sex, age, race and baseline itch and disease measures (S. Table 1).
DNA quantification
Incoming nucleic acid samples are quantified using fluorescent-based assays (PicoGreen) to accurately determine whether sufficient material is available for library preparation and sequencing.
DNA integrity
DNA sample size distributions are profiled by a Fragment Analyzer (Advanced Analytics) or BioAnalyzer (Agilent Technologies), to assess sample quality and integrity.
Genotyping
At the NYGC, we run the HumanCoreExome 24v1.3 array for all human DNA samples that we sequence.
WGS library preparation and sequencing, Truseq PCR-free (450 bp)
Whole genome sequencing (WGS) libraries were prepared using the Truseq DNA PCR-free Library Preparation Kit.
WGS Germline analysis part I
Whole Genome data were processed on NYGC automated pipeline. Paired-end 150 bp reads were aligned to the GRCh37 human reference (BWA-MEM v0.7.8) and processed with GATK best-practices workflow (GATK v3.4.0).
The mean coverage in the FLG region is 35.8, it reflects the samples average. The coverage for the FLG region tracks with the coverage for the whole genome. The coverage information per FLG region per sample is provided in the Supplementary File.
All high quality variants obtained from GATK were annotated for functional effects (intronic, intergenic, splicing, nonsynonymous, stopgain and frameshifts) based on RefSeq transcripts using Annovar (http://www.openbioinformatics.org/annovar/)31. Additionally, annovar was used to match general population frequencies from public databses (Exac, GnomAD, ESP6500, 1000 g) and was used to prioritize rare, loss-of-function variants.
Genomic data analysis part II
All variants in FLG and EDC were identified in whole genome sequencing data. The two most known genetic risk factors as defined by p.R501* (rs61816761), p.S761fs (rs558269137) were in addition verified with genotyping (S Fig. 4). All analyses were conducted in PLINK, MATLAB. In order to proceed with burden testing, we selected variants based on their quality, ancestry and pathogenicity, creating a subset of high-confidence variants. Variants that met these quality and pathogenicity filters were used for burden testing versus a. internal controls and b. Genome Aggregation Database (gnomAD).
Supplementary information
Acknowledgements
All participants of our clinical trials.
Author contributions
S.P.S. wrote the manuscript and conducted the analyses. S.W. edited the manuscript. D.X. and J.W. worked on the biostatistics section of the studies. G.B. and C.P. supervised and lead the clinical trials. M.P. mentorship, provided oversight for the analyses and is the P.I. for the study.
Competing interests
S.P.S., S.W., J.W., D.X., G.B., C.P., M.P. are all employees of Vanda Pharmaceuticals, There are no other interests to disclose.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-59627-7.
References
- 1.Chiesa Fuxench ZC, et al. Atopic Dermatitis in America Study: A Cross-Sectional Study Examining the Prevalence and Disease Burden of Atopic Dermatitis in the US Adult Population. J. Invest. Dermatol. 2019;139:583–590. doi: 10.1016/j.jid.2018.08.028. [DOI] [PubMed] [Google Scholar]
- 2.Elmose C, Thomsen SF. Twin Studies of Atopic Dermatitis: Interpretations and Applications in the Filaggrin Era. J. Allergy. 2015;2015:902359. doi: 10.1155/2015/902359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dold S, Wjst M, von Mutius E, Reitmeir P, Stiepel E. Genetic risk for asthma, allergic rhinitis, and atopic dermatitis. Arch. Dis. Child. 1992;67:1018–22. doi: 10.1136/adc.67.8.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes Kathleen C. An update on the genetics of atopic dermatitis: Scratching the surface in 2009. Journal of Allergy and Clinical Immunology. 2010;125(1):16-29.e11. doi: 10.1016/j.jaci.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandilands A, Sutherland C, Irvine AD, McLean WHI. Filaggrin in the frontline: role in skin barrier function and disease. J. Cell Sci. 2009;122:1285–94. doi: 10.1242/jcs.033969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao, P.-S. et al. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J. Allergy Clin. Immunol. 124, 507–13, 513.e1–7 (2009). [DOI] [PMC free article] [PubMed]
- 7.Czarnowicki T, Krueger JG, Guttman-Yassky E. Skin Barrier and Immune Dysregulation in Atopic Dermatitis: An Evolving Story with Important Clinical Implications. J. Allergy Clin. Immunol. Pract. 2014;2:371–379. doi: 10.1016/j.jaip.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Bin L, Leung DYM. Genetic and epigenetic studies of atopic dermatitis. Allergy, Asthma Clin. Immunol. 2016;12:52. doi: 10.1186/s13223-016-0158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elias PM. Stratum Corneum Defensive Functions: An Integrated View. J. Invest. Dermatol. 2005;125:183–200. doi: 10.1111/j.0022-202X.2005.23668.x. [DOI] [PubMed] [Google Scholar]
- 10.Engebretsen KA, et al. Concentration of filaggrin monomers, its metabolites and corneocyte surface texture in individuals with a history of atopic dermatitis and controls. J. Eur. Acad. Dermatology Venereol. 2018;32:796–804. doi: 10.1111/jdv.14801. [DOI] [PubMed] [Google Scholar]
- 11.Smith FJD, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat. Genet. 2006;38:337–342. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- 12.Sandilands A, et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat. Genet. 2007;39:650–654. doi: 10.1038/ng2020. [DOI] [PubMed] [Google Scholar]
- 13.Marenholz I, et al. Filaggrin loss-of-function mutations predispose to phenotypes involved in the atopic march. J. Allergy Clin. Immunol. 2006;118:866–871. doi: 10.1016/j.jaci.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 14.Irvine AD, Irwin McLean WH. Breaking the (Un)Sound Barrier: Filaggrin Is a Major Gene for Atopic Dermatitis. J. Invest. Dermatol. 2006;126:1200–1202. doi: 10.1038/sj.jid.5700365. [DOI] [PubMed] [Google Scholar]
- 15.O’Regan GM, Sandilands A, McLean WHI, Irvine AD. Filaggrin in atopic dermatitis. J. Allergy Clin. Immunol. 2009;124:R2–6. doi: 10.1016/j.jaci.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Nomura T, et al. Unique mutations in the filaggrin gene in Japanese patients with ichthyosis vulgaris and atopic dermatitis. J. Allergy Clin. Immunol. 2007;119:434–40. doi: 10.1016/j.jaci.2006.12.646. [DOI] [PubMed] [Google Scholar]
- 17.Rodríguez E, et al. Meta-analysis of filaggrin polymorphisms in eczema and asthma: robust risk factors in atopic disease. J. Allergy Clin. Immunol. 2009;123:1361–70.e7. doi: 10.1016/j.jaci.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 18.Brown SJ, McLean WHI. One remarkable molecule: filaggrin. J. Invest. Dermatol. 2012;132:751–62. doi: 10.1038/jid.2011.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margolis DJ, et al. The persistence of atopic dermatitis and filaggrin (FLG) mutations in a US longitudinal cohort. J. Allergy Clin. Immunol. 2012;130:912–7. doi: 10.1016/j.jaci.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colin Wong XC, et al. Array-based sequencing of filaggrin gene for comprehensive detection of disease-associated variants. J. Allergy Clin. Immunol. 2018;141:814–816. doi: 10.1016/j.jaci.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, et al. Wide spectrum of filaggrin-null mutations in atopic dermatitis highlights differences between Singaporean Chinese and European populations. Br. J. Dermatol. 2011;165:106–114. doi: 10.1111/j.1365-2133.2011.10331.x. [DOI] [PubMed] [Google Scholar]
- 22.Mischke, D., Korge, B. P., Marenholz, I., Volz, A. & Ziegler, A. Genes Encoding Structural Proteins of Epidermal Cornification and S100 Calcium-Binding Proteins Form a Gene Complex ("Epidermal Differentiation Complex") on Human Chromosome lq21. Journal of Investigative Dermatology106 (1996). [DOI] [PubMed]
- 23.Kypriotou M, Huber M, Hohl D. The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the ‘fused genes’ family. Exp. Dermatol. 2012;21:643–649. doi: 10.1111/j.1600-0625.2012.01472.x. [DOI] [PubMed] [Google Scholar]
- 24.Guttman-Yassky E, et al. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J. Allergy Clin. Immunol. 2009;124:1235–1244.e58. doi: 10.1016/j.jaci.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 25.Jensen J-M, et al. Impaired Sphingomyelinase Activity and Epidermal Differentiation in Atopic Dermatitis. J. Invest. Dermatol. 2004;122:1423–1431. doi: 10.1111/j.0022-202X.2004.22621.x. [DOI] [PubMed] [Google Scholar]
- 26.Sugiura H, et al. Large-scale DNA microarray analysis of atopic skin lesions shows overexpression of an epidermal differentiation gene cluster in the alternative pathway and lack of protective gene expression in the cornified envelope. Br. J. Dermatol. 2005;152:146–149. doi: 10.1111/j.1365-2133.2005.06352.x. [DOI] [PubMed] [Google Scholar]
- 27.Marenholz I, et al. Association screening in the Epidermal Differentiation Complex (EDC) identifies an SPRR3 repeat number variant as a risk factor for eczema. J. Invest. Dermatol. 2011;131:1644–9. doi: 10.1038/jid.2011.90. [DOI] [PubMed] [Google Scholar]
- 28.Paternoster L, et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat. Genet. 2012;44:187–192. doi: 10.1038/ng.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koseki R, et al. Effect of filaggrin loss-of-function mutations on atopic dermatitis in young age: a longitudinal birth cohort study. J. Hum. Genet. 2019;64:911–917. doi: 10.1038/s10038-019-0628-y. [DOI] [PubMed] [Google Scholar]
- 30.Rupnik H, Rijavec M, Korošec P. Filaggrin loss-of-function mutations are not associated with atopic dermatitis that develops in late childhood or adulthood. Br. J. Dermatol. 2015;172:455–461. doi: 10.1111/bjd.13477. [DOI] [PubMed] [Google Scholar]
- 31.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.