Abstract
During protein synthesis, translating ribosomes encounter many challenges imposed by various types of defective mRNAs that can lead to reduced cellular fitness and, in some cases, even threaten cell viability. Aberrant translation leads to activation of one of several quality control pathways depending on the nature of the problem. These pathways promote the degradation of the problematic mRNA as well as the incomplete translation product, the nascent polypeptide chain. Many of these quality control systems feature critical roles for specialized regulatory factors that work in concert with conventional factors. This review focuses on the mechanisms used by these quality control pathways to recognize aberrant ribosome stalling and discusses the conservation of these systems.
INTRODUCTION
Accurate gene expression is the basis of life. Accumulation of abnormal proteins causes various cellular dysfunctions. Hence, maintenance of normal cell function requires recognition and elimination of abnormal proteins via homeostatic mechanisms. Abnormal proteins can be generated via post-translational changes in their three-dimensional structures mediated by various factors, by abnormal mRNAs, or by incorrect translation. The ribosome plays crucial roles in protein synthesis, in formation of properly folded three-dimensional structures, and in transport to organelles (1). Recent studies have demonstrated crucial roles of the ribosome in protein quality control systems and revealed the mechanisms by which they recognize aberrant translations and eliminate abnormal mRNAs and products (2–8). The conservation of quality control mechanisms for abnormal translation between prokaryotes and eukaryotes highlights their importance in maintaining protein homeostasis in cells.
RESCUE OF STALLED RIBOSOMES AT THE 3′ END OF NONSTOP mRNAs
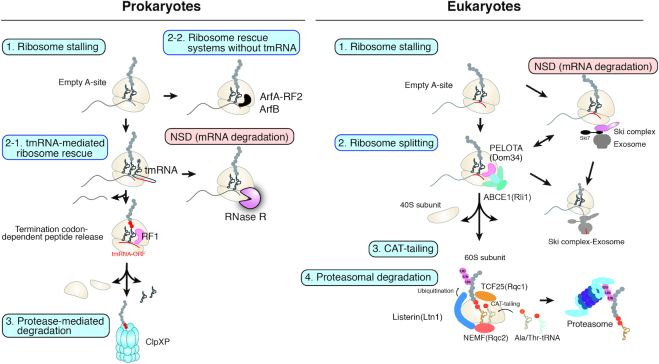
Aberrant mRNAs lacking a termination codon, known as nonstop mRNAs, are produced by errors during mRNA processing or cleavage. Ribosomes stall at the 3′ end of nonstop mRNAs due to no codon in the A site. However, prokaryotes and eukaryotes have developed quality control systems to rescue stalled ribosomes and recycle them for re-use (Figure 1). Recycling of the ribosome results in release of the truncated mRNA and the incomplete protein, thereby facilitating their degradation and inhibition of the next round of translation. In prokaryotes, transfer-messenger RNA (tmRNA) is primarily responsible for rescuing stalled ribosomes. In eukaryotes, the nonstop mRNA decay (NSD) system recognizes and eliminates nonstop mRNAs, which are mainly produced by aberrant polyadenylation within the open reading frame (ORF) (9–13).
Figure 1.
Rescue systems for ribosomes stalled at the 3′ end of nonstop mRNAs. Ribosomes that translate aberrant mRNAs lacking stop codons become stalled at the 3′ end of the mRNA. Left: In prokaryotes, three systems are responsible for rescue of the stalled ribosome. The tmRNA–SmpB complex recruits the highly conserved 3′–5′ RNase R, which is responsible for targeted degradation of nonstop mRNAs (21). Right: In eukaryotes, Dom34 recognizes the empty A-site of ribosomes that are stalled at the 3′ end of nonstop mRNAs. ABCE1-mediated subunit dissociation of the stalled 80S ribosome results in rapid degradation of the released nonstop mRNA by the Ski complex and the exosome. Association with the stalled ribosome facilitates the ATPase activity of the Ski complex and degradation of the nonstop mRNA. Pelota (Dom34)-mediated dissociation of the stalled ribosome and association of the Ski complex–exosome with the ribosome may act cooperatively to ensure processive degradation of the nonstop mRNA from the 3′ end.
Rescue of stalled ribosomes at the 3′ end of mRNAs in prokaryotes
In bacteria, truncated mRNAs are produced mainly via exonucleolytic degradation of the 3′-ends of mRNAs or endonucleolytic cleavage by RNases such as RNase III. Truncation of mRNAs within the coding region (ORF) results in stop codon loss and stalling of ribosomes at the 3′ end of the resulting nonstop mRNAs. Accumulation of stalled ribosomes is detrimental to cells, and recycling stalled ribosomes is essential for cell viability (14,15). Three ribosome rescue systems, trans-translation and alternative rescue factors A and B (ArfA and ArfB), are responsible for rescuing stalled ribosomes in prokaryotes (reviewed in (16)) (Figure 1, Table 1). In addition, the bacterium Francisella tularensis uses a fourth mechanism in which ArfT acts as an alternative ribosome rescue factor via a mechanism that is distinct from those of ArfA and ArfB (17).
Table 1.
The three ribosome rescue systems in prokaryotes
| tmRNA-SmpB | ArfA-RF2 | ArfB | |
|---|---|---|---|
| Ribosome rescue | Yes | Yes | Yes |
| Degradation of aberrant mRNA (NSD) | Yes (RNase R) | Yes? | Yes? |
| Degradation of nonstop products | Yes (Ala-Ala tag) | No | No |
Trans-translation by tmRNA is a quality control system that is primarily responsible for coping with nonstop mRNAs in prokaryotes. As a back-up system for the absence of tmRNA, two other rescue systems present in some bacteria, which employ either ArfA and RF2 or ArfB, rescue stalled ribosomes on truncated mRNAs by hydrolyzing the peptidyl-tRNA (14). Trans-translation induces the rapid degradation of nonstop mRNAs and truncated products. By contrast, the ArfA/RF2 and ArfB systems rescue the ribosome but do not induce degradation of the aberrant products; however, it is unknown whether these two systems can induce mRNA decay.
Trans-translation as a quality control system for nonstop mRNAs
The trans-translation system, which is mediated by tmRNA and its binding partner small protein B (SmpB), recycles stalled ribosomes at the 3′-ends of truncated mRNAs. This is followed by degradation of both the incomplete mRNA and the protein. The tmRNA contains a domain similar to the acceptor arm of tRNAAla as well as an mRNA-like domain (14,15). The tmRNA–SmpB complex is delivered to an empty A-site by GTP-bound EF-Tu as a quaternary complex, although it does not decode the Ala codon in the mRNA. Translation elongation factor EF-G then translocates the tmRNA–SmpB complex through the ribosome in a manner similar to the translocation that occurs during canonical translation. The ribosome swaps messages, and the subsequent tmRNA-templated translation results in addition of a polypeptide tag to the nascent chain, which guarantees that the ribosome will terminate at a stop codon and be released. Even though trans-translation efficiently adds a degradation tag to the C-termini of truncated polypeptide chains, thereby marking them for degradation, the inactivation of trans-translation is not lethal. This discrepancy is due to the presence of back-up systems mediated by two alternative rescue factors, ArfA and ArfB. These factors recycle stalled ribosomes but allow synthesis of truncated protein variants encoded by stop-less mRNAs (Table 1). These observations indicate that the essential function of trans-translation is to recycle stalled ribosomes rather than to degrade aberrant products (18,19).
Biochemical and structural analyses have revealed the mechanism underlying the addition of the C-terminal degradation tag to aberrant nonstop products via trans-translation and how the tmRNA moves through the ribosome (reviewed in (16,20)) (Figure 2). When the 70S ribosome stalls at the 3′-end of a nonstop mRNA, EF-Tu delivers Ala–tmRNA–SmpB to the ribosome, where the C-terminal tail of SmpB forms a α-helix in the downstream mRNA channel to form a pre-accommodation state. EF-Tu then leaves, and tmRNA–SmpB accommodates into the A-site in a conformation such that the SmpB tail remains α-helical. The tRNAAla-like domain of tmRNA–SmpB directs the alanine on its 3′-CCA into the peptidyl-transferase center (PTC) to form a peptide bond with the nascent peptide. EF-G then translocates tmRNA–SmpB from the A-site into the P-site to release the original mRNA and tRNA. During the translocation step, the SmpB tail flips to the opposite side of the mRNA channel, where it binds in the E-site. The mRNA-like domain then passes through the A-site latch and moves into the space in the mRNA channel. Ala-tRNAAla decodes the first codon of the tmRNA ORF, and a peptidyl-transferase reaction transfers the peptide from tmRNA to tRNAAla. EF-G then translocates the peptidyl-tRNAAla into the P-site, after which the mRNA-like domain is again loaded into the mRNA channel via a latch, this time in the E-site. Translation of the ORF continues until the tmRNA reaches a stop codon, and the peptide is then targeted for degradation by proteases that recognize the polypeptide tag. Together with EF-G, ribosome recycling factor recycles the 70S ribosome in a post-termination state. Therefore, it is likely that ribosome recycling factor and EF-G also dissociate the stalled ribosome into the 30S and 50S subunits and facilitate decay of the nonstop mRNA from the 3′-end. For rapid degradation of the nonstop mRNA, tmRNA–SmpB recruits the highly conserved 3′–5′ exoribonuclease ribonuclease R (RNase R), which is responsible for targeted degradation of nonstop mRNAs after rescue of the stalled ribosome (21,22) (Figure 1).
Figure 2.
Mechanism of trans-translation. During the first step of trans-translation, the 70S ribosome stalls at the 3′-end of a nonstop mRNA. In step 2, EF-Tu delivers Ala–tmRNA–SmpB to the ribosome, where the C-terminal tail of SmpB forms a α-helix in the downstream mRNA channel to form a pre-accommodation state. In step 3, EF-G translocates tmRNA–SmpB from the A-site into the P-site to release the original mRNA and tRNA. In step 4, the nonstop mRNA is switched for the ORF of the tmRNA and translation is resumed. In step 5, translation of the ORF of the tmRNA results in tagging of the aberrant polypeptide with a degradation tag that is recognized by proteases. In step 6, the ribosome is recycled by conventional translation factors.
Recycling of stalled ribosome complexes in the absence of trans-translation
The two additional rescue systems present in some bacteria, which employ either ArfA and release factor 2 (RF2) or ArfB, rescue stalled ribosomes on truncated mRNAs by hydrolyzing the peptidyl-tRNA (23). ArfA represents a back-up system for trans-translation and is required for cell survival in the absence of tmRNA. ArfA mediates ribosome rescue by recruiting the canonical termination factor RF2 to ribosomes stalled on truncated mRNAs. The C-terminal tail of ArfA probes the vacant mRNA channel and recruits RF2 to the nonstop ribosome to catalyze peptidyl-tRNA hydrolysis. ArfA cooperates with RF2, but does not interact with RF2 in solution (24,25). ArfA initially binds to the 30S subunit in the vicinity of the mRNA channel and recruits RF2 to nonstop ribosomes (25). Analyses of five cryo-electron microscopy (cryo-EM) structures of ArfA-RF2-stalled ribosome complexes revealed that ArfA interacts almost exclusively with the small subunit (16,26–28). In addition, tmRNA represses ArfA expression. A truncated arfA mRNA species is produced via endonucleolytic cleavage of a stem-loop structure in the 3′-end of the mRNA by RNase III (23,29,30). In the presence of tmRNA, the short ArfA protein produced from the truncated arfA mRNA is tagged by tmRNA and targeted for degradation. The full-length ArfA protein is 72 amino acids long and includes a C-terminal hydrophobic region that promotes aggregation of the protein in vivo (23). In the absence of tmRNA, the shorter form of ArfA can be expressed from the truncated arfA mRNA, and, as it lacks the terminal region, it is not degraded. Instead, the short ArfA protein product hydrolyzes the peptidyl-tRNA to rescue and recycle ribosomes stalled on truncated mRNAs (23,24,30,31).
ArfB also probes the vacant mRNA channel of nonstop ribosomes (32). ArfB overexpression can rescue cells lacking the tmRNA and ArfA rescue systems (32). In vivo, ArfB can efficiently catalyze the hydrolysis of peptidyl-tRNAs in ribosomes stalled at the 3′-ends of nonstop mRNAs (31–33). The N-terminal domain of ArfB contains the GGQ motif that catalyzes the hydrolysis of polypeptides from peptidyl-tRNAs. (16,33–36). Mutations in the ArfB GGQ motif impair the rescue activity of ArfB (32,33). Consistently, the N-terminal domain interacts with the large subunit such that the GGQ motif is positioned at the PTC (37). The C-terminal region of ArfB is crucial for ribosome binding and hydrolysis of the peptidyl-tRNA (32–34). The α-helix in the C-terminal region of ArfB binds in the mRNA entry tunnel downstream of the A-site (37), and this interaction distinguishes actively translating ribosomes from those stalled on truncated mRNAs. The N-terminal domain and C-terminal helix are connected by a 12-amino acid flexible linker that adopts an extended conformation on the ribosome. A deletion analysis of the ArfB linker suggested that it is important for positioning the N-terminal domain in the PTC of the ribosome (34).
Quality control of nonstop mRNAs in eukaryotes
The yeast Dom34 protein recognizes the empty A-site of ribosomes stalled at the 3′-end of the mRNA and plays a crucial role in NSD (Figure 1). Biochemical and genetic analyses demonstrated that yeast Dom34 and its human homolog Pelota promote the dissociation of the translation elongation complex into subunits (38–44). A cryo-EM analysis of the structure of yeast Dom34 bound to a yeast ribosome stalled at the 3′-end of a nonstop mRNA demonstrated that domain N of Dom34 competes with the mRNA in the A-site and part of the mRNA entry channel. The truncation of mRNAs within ORFs via self-cleavage of hammerhead ribozyme sequences results in stalled ribosomes with an empty A-site at the 3′-end of the mRNA. Dom34 monitors the mRNA channel of stalled ribosomes with an empty A-site and, along with ABCE1, facilitates dissociation of the ribosome from the mRNA, thereby inducing rapid degradation of nonstop mRNAs (38,45,46).
Dissociation of the subunits of stalled 80S ribosomes is a key step for the degradation of truncated mRNAs and their aberrant products. Pelota (Dom34) recruits ABCE1 to dissociate the subunits of stalled 80S ribosomes. ABCE1 (Saccharomyces cerevisiae Rli1) is a conventional recycling factor that is required for ribosome recycling after eukaryotic releasing factor 1 (eRF1) hydrolyzes the peptidyl-tRNA via a normal termination reaction (43,47–49). In NSD quality control, ABCE1-mediated dissociation of stalled 80S ribosome subunits results in rapid degradation of the released nonstop mRNA via the Ski complex and exosomes (50) (Figure 1). The Ski2–Ski3–Ski8 (Ski) complex is a helicase that functions alongside the RNA-degrading exosome to mediate 3′–5′ mRNA decay. A cryo-EM analysis of the structure of an endogenous ribosome–Ski complex revealed that ribosome binding displaces the autoinhibitory domain of the Ski2 helicase, positioning it in an open conformation near the ribosomal mRNA entry tunnel (51). It is thought that the ribosome stalls at the 3′ end of the nonstop mRNA and interacts with the Ski complex, thereby activating Ski2 helicase activity to facilitate unwinding of the aberrant mRNA (Figure 1).
CO-TRANSLATIONAL PROTEASOMAL DEGRADATION OF NASCENT POLYPEPTIDES INDUCED BY RIBOSOME STALLING
In addition to stalling at the 3′ end of nonstop mRNAs, ribosomes can also stall during translation elongation within the ORF. Emerging evidence demonstrates that ribosome stalling within the ORF induces co-translational degradation of the nascent peptide by the ubiquitin–proteasome pathway and mRNA quality control systems. This quality control mode is referred to as Ribosome-associated Quality Control (RQC), and its mechanism has been elucidated via identification of the essential factors required for each step of the pathway, from the recognition of stalled ribosomes to the degradation of their products (3–8,52–54). These findings clearly demonstrate that stalled ribosomes efficiently induce subunit dissociation and proteasomal degradation of peptidyl-tRNA in the 60S subunit.
Steps in RQC
RQC is a translation arrest-induced quality control system that leads to degradation of the arrest products via three steps (Figure 3): recognition of stalled ribosomes, subunit dissociation, and degradation of aberrant nascent polypeptides. In the first step, abnormal stalled ribosomes are recognized and ubiquitinated at one or more specific residues. In yeast, the E3 ubiquitin ligase Hel2 ubiquitinates uS10 at K6/8 and plays a crucial role in RQC (55). Zinc finger protein 598 (ZNF598), a mammalian homolog of Hel2, ubiquitinates the eS10 and uS10 ribosomal proteins at K138/139 and K4/8, respectively, to stimulate translational arrest and RQC triggered by poly-lysine mRNA sequences (56–58) (Table 2). In the second step of RQC, ribosomal ubiquitination induces subunit dissociation, which is a crucial event for Listerin (S. cerevisiae Ltn1)-dependent ubiquitination of the arrest products in the 60S large ribosomal subunit (39–43,53). The RQC-trigger (RQT) complex is required for RQC and is proposed to recognize ubiquitinated stalled ribosomes and induce subunit dissociation in yeast. The RQT complex is composed of three factors: the RNA helicase-family protein Slh1/Rqt2, the ubiquitin-binding protein Cue3/Rqt3, and Rqt4 (Table 2) (55,59). The RQT complex is conserved in mammalian cells; ASCC3 is a homolog of Slh1 (55) and ASCC2 is likely to be a homolog of Cue3. The ATPase activity of Slh1 and the ubiquitin-binding activity of Cue3 are crucial for RQC (55). In the third step of RQC, dissociation of the 40S subunit allows binding of 60S ribosome-nascent chain (RNC) complexes to nuclear export mediator factor (NEMF; S. cerevisiae Rqc2), which then recruits the E3 ubiquitin ligase Listerin (S. cerevisiae Ltn1). Next, Listerin ubiquitinates the nascent chains, thus targeting them for degradation (52,53,60–63). The subsequent proteasomal degradation of a ubiquitinated nascent chain requires its prior release from the 60S subunit. Several lines of evidence indicate that ANKZF1 (Vms1) is responsible for the release of the ubiquitinated polypeptide from the tRNA in the 60S subunit (64–68).
Figure 3.
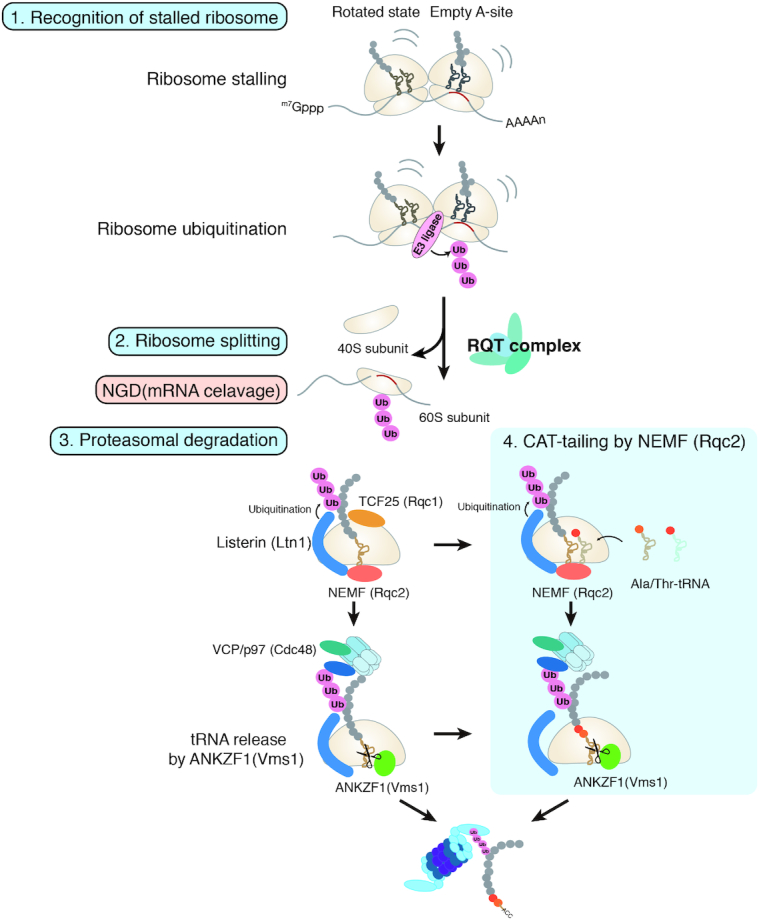
Quality control systems induced by ribosome stalling. RQC has three steps. Step 1: The abnormal stalled ribosome is recognized and ubiquitinated at one or more specific residues. Step 2: Ribosome ubiquitination induces subunit dissociation, which is a crucial event for Listerin (S. cerevisiae Ltn1)-dependent ubiquitination of the arrest products on the 60S large ribosomal subunit. The RQT complex is thought to recognize the ubiquitinated stalled ribosome and induce subunit dissociation in yeast. Step 3: Dissociation of the 40S subunits allows binding of 60S RNCs to NEMF (Rqc2), which recruits the E3 ubiquitin ligase Listerin (Ltn1). Subsequently, Listerin ubiquitinates the nascent polypeptide chains, targeting them for degradation. Rqc2 attaches CAT-tails (C-terminal alanyl/threonyl sequences) to stalled polypeptides. The ATPase VCP/p97 (Cdc48) forms a complex with the cofactors UFD1 (Ufd1) and NPLOC4 (Npl4) and unfolds ubiquitinated polypeptides, then extracts the peptidyl-tRNA from the 60S, thereby recruiting it to the 26S proteasome for degradation. Novel roles of ANKZF1 (Vms1)-mediated polypeptide release for proteasomal degradation: Rqc2 catalyzes the C-terminal extension of the stalled tRNA-bound peptides with CAT-tails through a non-canonical elongation reaction without mRNA. CAT-tails functionalize the carboxy termini of stalled polypeptides to drive their degradation on and off the ribosome. Vms1 interacts with the ribosomal 60S subunit to compete with Rqc2 and catalyze peptidyl-tRNA cleavage. Subsequently, tRNA nucleotidyl transferase 1 (TRNT1) is responsible for recycling of ANKZF1-cleaved tRNA fragments.
Table 2.
Conservation of ribosome ubiquitination and RQC-trigger factors in eukaryotes
| Ubiquitination of colliding ribosomes | Yeast | Mammals |
| E3 ligase | Hel2 | ZNF598 |
| Ubiquitination sites | uS10 | uS10, eS10 |
| Ubiquitin chain | K63-linked polyubiquitination | Mono/di-ubiquitination |
| RQC-trigger factors | Yeast | Mammals |
| RNA helicase | Shl1/Rqt2 | ASCC3 |
| Ubiquitin-binding protein | Cue3/Rqt3 | ASCC2 |
| Rqt4 | Rqt4 | ASCC1 or TRIP4 ? |
Top: The disomes formed by the leading stalled ribosome and colliding ribosomes. The critical E3 ubiquitin ligase Hel2 and its mammalian homolog ZNF598 recognize the colliding disome and ubiquitinate uS10 in yeast and uS10, eS10 and uS3 in mammals. Bottom: In yeast, ubiquitination of uS10 at the K6 or K8 residue is required for subunit dissociation by the RQT complex. The factors comprising the RQT complex are conserved in mammals, strongly suggesting that ribosome ubiquitination is required for RQT complex-mediated subunit dissociation in mammalian cells.
Recognition of aberrant stalled ribosomes: ribosome ubiquitination
Translation of the poly(A) tails of nonstop mRNAs or poly(A) sequences within ORFs results in strong translation arrest and co-translational proteasomal degradation in yeast (69,70), and mammals (4,5,35,42,57,60,71–73). Lysine-encoding poly(A) stretches control translation and induce ribosomes to slide along the mRNA in various organisms (74–76). Specific basic amino acid sequences within ORFs also induce co-translational proteasomal degradation in yeast (70,77), and certain combinations of rare arginine-coding codons are very potent in promoting ribosomal stalling (78) and RQC (55).
An important question in RQC is how ZNF598 (S. cerevisiae Hel2) recognizes and ubiquitinates its specific substrate. The first observation regarding the substrate specificity of Hel2 in yeast was that ribosomes co-purified with tagged Hel2 were mainly in the rotated state with hybrid tRNAs (55). In yeast, translation of tandem rare codons results in formation of di-ribosome (disome) units that consist of the stalled leading ribosome and the following colliding ribosome. In in vitro mammalian translation systems, disomes are formed in the presence of a mutant form of eRF1 that is defective in peptide-releasing activity (73). A cryo-EM analysis of disomes revealed that the colliding ribosomes are positioned in a rotated form (73,79) and recent studies using an in vitro translation system demonstrate that disomes are preferred targets of ZNF598 (Hel2)-dependent ubiquitination (73,79). Recognition of disomes containing colliding ribosomes in the rotated state by ZNF598 (Hel2) (73,79) leads to ubiquitination of uS10 in yeast and uS10, eS10 and uS3 in mammals (Figure 3, Table 2). Subsequently, the RQT complex specifically targets the ubiquitinated ribosomes in the hybrid state to allow dissociation into their respective subunits (55).
The potential connection between RQC and translation initiation repression
ZNF598 and Grb10-interacting GYF protein 2 (GIGYF2) are components of the eIF4E homologous protein (4EHP) complex, which is thought to repress translation by competing with eIF4E for binding to the 5′-cap structure of specific mRNAs. Because 4EHP does not interact with eIF4G, it fails to initiate translation (80). GIGYF2 recruits decapping and deadenylation complexes to 4EHP-containing ribonucleoproteins to induce the translational repression and degradation of target mRNAs (81). In addition, 4EHP-GIGYF2 acts as a cofactor of the zinc finger protein tristetraprolin (TTP), which promotes translational repression and degradation of mRNAs containing AU-rich elements (82). Moreovre, ZNF598 interacts with GIGYF1 via three proline-rich motifs and regulates known TTP targets, thereby regulating the inflammatory potential of cytokine-producing cells (83). Finally, a recent study proposed that ZNF598 recognizes ribosome stalling and inhibits translation initiation by recruiting translational repressor GIGYF2 (84).
The role of the RQT complex in subunit dissociation
The RQT complex is proposed to recognize ubiquitinated stalled ribosomes and induce subunit dissociation (55,59) (Figure 3, Table 2). The ubiquitin-binding activity of Cue3 is crucial for RQC but is not required for the interaction between the RQT complex and ubiquitinated ribosomes. The ATPase activity of Slh1 is also crucial for RQC, and the proposed model is that the interaction between the RQT complex and K63-linked polyubiquitinated ribosomes and ATP hydrolysis by Slh1 play essential roles in subunit dissociation (55). Although the mechanism by which the RNA helicase activity of Slh1 induces subunit dissociation remains unknown, the structure of ubiquitinated ribosomes may be modulated by Slh1 in an RNA helicase activity-dependent manner. Biochemical evidence is needed to demonstrate that the RQT complex is responsible for the dissociation of stalled ribosomes into their subunits.
Unfolding machinery for polypeptide ubiquitination and degradation
Stalled ribosomes are ubiquitinated and dissociated into their subunits, yielding 60S RNCs (55,57,58,73,79). The ubiquitin ligase Listerin (S. cerevisiae Ltn1) plays a crucial role in RQC (53,85). A forward genetics approach using a mouse model showed that mutation of the gene encoding Listerin can lead to neurodegeneration (86). NEMF (S. cerevisiae Rqc2) associates with 60S RNCs and recruits Listerin, which ubiquitinates peptidyl-tRNAs on dissociated 60S subunits (53). Rqc2 attaches C-terminal alanyl/threonyl sequences (CAT-tails) to stalled polypeptides (63,87). The ATPase valosin-containing protein (VCP), also known as p97 (S. cerevisiae Cdc48), is the unfolding machinery for ubiquitinated polypeptides. VCP forms a complex with its cofactors UFD1 (S. cerevisiae Ufd1) and NPLOC4 (S. cerevisiae Npl4) to promote the extraction of peptidyl-tRNAs from 60S subunits, and these peptidyl-tRNAs are then recruited to the 26S proteasome for degradation (4,54).
Vms1 connects polypeptide release and proteasomal degradation
In RQC pathways, the mechanism by which tRNA-linked nascent polypeptides are released from stalled 60S subunits prior to proteasomal degradation has been a long-standing puzzle. Several lines of evidence have demonstrated that mammalian ANKZF1 (S. cerevisiae Vms1) releases ubiquitinated nascent proteins from 60S ribosomal subunits for proteasomal degradation (Figure 3). Analysis of the crystal structure of Vms1 revealed that it is similar to eRF1 (65,88). Vms1 lacks the strict GGQ motif present in eRF1; however, it has a conserved glutamine (Q295) in a GxxQ context that can align with the catalytic glutamine in the eRF1 GGQ motif. The peptide release activity of Vms1 has been shown to depend on Q295 (64,65).
Pestova and co-workers reconstituted the entire mammalian RQC pathway in vitro, from ribosome stalling and recognition and processing of 60S complexes by the RQC ubiquitinating machinery, to ribosomal release of the ubiquitinated nascent polypeptides and their degradation by the 26S proteasome (68). In non-ubiquitinated nascent polypeptides on 60S subunits formed on nonstop mRNAs, the tRNAs are not firmly fixed in the P-site, which allows the peptidyl-tRNA hydrolase Ptrh1 to cleave the nascent peptidyl-tRNA in vitro. The release of non-ubiquitinated nascent polypeptides from the 60S subunit functions as an additional rescue pathway in cases where Listerin-dependent ubiquitination cannot be used. The 60S ribosome-nascent polypeptides are resistant to Ptrh1 due to their association with NEMF (Rqc2), and ubiquitination of nascent polypeptides results in the accommodation of nascent peptidyl-RNAs and susceptibility to ANKZF1. Subsequently, ANKZF1 induces specific cleavage in the tRNA acceptor arm to release ubiquitinated nascent polypeptides, which are then subject to proteasomal degradation. Shao and co-workers also investigated the fate of nascent polypeptide-tRNAs in 60S subunits using a eukaryotic cell-free system and found that ANKZF1 (Vms1) severs peptidyl-tRNAs in RQC complexes by precisely cleaving off the terminal 3′-CCA nucleotides, leaving a 2′,3′-cyclic phosphate (67). After removal of the cyclic phosphate, the sole CCA-adding enzyme tRNA nucleotidyl transferase 1 is responsible for recycling the ANKZF1-cleaved tRNA fragments, while also functioning in tRNA biogenesis in eukaryotes. Further studies may provide evidence whether the recycling of ANKZF1 (Vms1)-cleaved tRNAs is a crucial step to complete the quality control process without leaving behind any remnants. A more recent study by Beckmann and co-workers described the structural basis of the interaction between Vms1 and the ribosomal 60S subunit through which Vms1 competes with Rqc2 and catalyzes peptidyl-tRNA cleavage (66). Cryo-EM structures of native Vms1-60S ribosomal complexes revealed that Vms1 binds to the 60S subunit via the Vms1 release factor-like domain, the N-terminal zinc finger domain, and the ankyrin domain. The Vms1 release factor-like domain interacts with the ribosomal A-site and, like eRF1, projects its GSQ motif toward the PTC. This position overlaps with that observed for Rqc2 aminoacyl-tRNA (aa-tRNA), thus explaining the adversarial activity of Vms1. In the pre-hydrolysis state, the ABCF-type ATPase Arb1 accesses the ribosomal E-site, where it specifically interacts with the peptidyl-tRNA. Arb1 overexpression leads to partial suppression of the phenotypes resulting from Vms1 deletion, including the protein aggregation phenotype. Based on these observations, it is proposed that Arb1 plays a role in positioning peptidyl-tRNAs for cleavage by Vms1 and/or in dislocating peptidyl-tRNAs for tRNA extraction after cleavage (66).
Ribosome stalling often occurs during the translation of mitochondrially-targeted proteins. Nevertheless, mitochondrial polypeptides on stalled ribosomes are co-translationally imported into the mitochondria and are, therefore, inaccessible to ubiquitination due to the coupling of translation and translocation. Vms1 is a key component in a rescue pathway for ribosome-stalled mitochondrial polypeptides in yeast (89). Ltn1 and Vms1 protect cells against mitochondrial toxicity and maintain cell viability under respiratory conditions (89). These findings indicate that an ANKZF1 (Vms1)-mediated rescue pathway could be more crucial for maintaining protein homeostasis and ensuring cell survival. Under specific stress conditions, such as oxidative stress and rapamycin treatment, the rate of ribosome stalling can increase. Under these conditions, Vms1 accumulates on damaged mitochondria and recruits Cdc48 and Npl4 to facilitate proteasomal protein degradation of mitochondrial outer membrane proteins in yeast (90).
An alternative RQC pathway: CAT-tailing
Problematic nascent peptides remain bound to the 60S subunit, still connected to the tRNA in the P-site. In the 60S subunit, the Rqc2 protein catalyzes C-terminal extension of stalled tRNA-bound peptides with alanine and threonine residues (CAT-tails) via a non-canonical mRNA-independent elongation reaction (63,87) (Figure 3). CAT-tailing enables degradation of substrates that lack Ltn1p-accessible lysines by exposing lysines normally sequestered in the ribosome exit tunnel (87). This activity indicates that in the context of nascent chain degradation in budding yeast, CAT-tailing is a fail-safe mechanism that expands the range of RQC-degradable substrates (87). Brandman and co-workers developed quantitative techniques to assess how CAT-tails affect the degradation of stalled polypeptides, and found that in Saccharomyces cerevisiae, CAT-tails enhance the efficiency of the Ltn1 targeting of structured polypeptides, which are otherwise poor Ltn1 substrates (91). If Ltn1 fails to ubiquitinate stalled polypeptides or becomes limiting, CAT-tails act as degrons, marking proteins for proteasomal degradation. Joazerio and co-workers reported that RqcH extends the C-termini of incomplete nascent chains with polyalanine sequences to promote their degradation by the ClpXP protease in bacteria (92). RqcH protects cells against translational and environmental stresses in a redundant manner with tmRNA/ssrA (92).
Nuclear-encoded mitochondrial proteins are targets of CAT-tailing to prevent aggregation of the mitochondrial proteome (89). Vms1 plays a crucial role in RQC as it counteracts inhibitory CAT-tailing functions. Indeed, recent studies demonstrated that Vms1 terminates CAT-tail formation during RQC by acting as a general release factor (64–68).
NO-GO-DECAY (NGD) QUALITY CONTROL: MRNA ENDONUCLEOLYTIC CLEAVAGE INDUCED BY RIBOSOME STALLING
NGD quality control system triggers endonucleolytic cleavage of mRNAs when ribosomes encounter roadblocks such as stable RNA secondary structures, rare codons, or stretches of consecutive positively charged amino acids in the nascent chain, and responds to ribosome stalling (79,93–95,114,115). In contrast to the conventional mRNA degradation pathways, the NGD process is distinguished by endonucleolytic cleavage proximal to the ribosomal stalling site prior to degradation of the transcript by exonucleases (96). The endonucleolytic cleavage products produced during NGD quality control are also rapidly degraded by the 5′–3′-exoribonuclease Xrn1 and the 3′–5′-exoribonuclease Ski complex-exosome. The structural basis for the interaction between the mRNA decay machinery and ribosomes suggests that rapid mRNA degradation is efficiently coupled to endonucleolytic cleavage induced by translation arrest. Moreover, a recent study by Green and co-workers identified Cue2 as the endonuclease that may be responsible for the endonucleolytic cleavage induced by ribosome stalling in the NGD pathway (97).
A dual role of Hel2 in two distinct NGD pathways
NGD and RQC require common factors and biochemical events, suggesting that these two quality control pathways are coupled (2,94). Ribosome profiling revealed that endonucleolytic cleavage occurs at sites of premature polyadenylation, and rescue of the ribosomes stalled at these sites is dependent on Dom34, suggesting that translation of poly(A) tails induces NGD as well as RQC (93). Both pathways are initiated by translation arrest and are dependent on 40S subunit-associated Asc1 in yeast (45,79,94,98). Moreover, Hel2-mediated K63-linked polyubiquitination is implicated in RQC after ribosome stalling at polybasic amino acid sequences and tandem CGA codons in yeast (99). Furthermore, ribosomal collisions trigger NGD-induced mRNA cleavage, and it is proposed that colliding ribosomes induce robust uS3 ubiquitination by Hel2 (95). Hel2-mediated uS10 ubiquitination is crucial for RQC; however, Hel2-mediated uS3 ubiquitination at lysine 212 is not essential for RQC or NGD (79); rather, it is involved in quality control for non-functional rRNA decay (100). Therefore, the role of Hel2-mediated uS3 ubiquitination remains to be elucidated.
As mentioned above, a disome unit consists of the leading stalled ribosome and the following colliding ribosome. In a recent study, endonucleolytic cleavage of an NGD-inducing reporter mRNA occurred at sites within disome units and depended on Hel2-mediated K63-linked polyubiquitination of uS10, demonstrating that NGD and RQC are coupled via this ubiquitination event (referred to as NGDRQC+) (79). Moreover, the ATPase activity of the RQT component Slh1 is required for cleavage within disomes, implying that subunit dissociation of stalled ribosomes is required for NGDRQC+. Further experiments revealed the existence of two interdependent branches of the NGD pathway: the RQC-coupled (NGDRQC+) and RQC-uncoupled (NGDRQC–) branches. In the NGDRQC– branch, endonucleolytic mRNA cleavage occurs upstream of stalled disomes when RQC is defective. Cleavage in the NGDRQC– branch requires K63-linked polyubiquitination of the eS7 ribosomal protein; this polyubiquitination also depends on Hel2 but is triggered by monoubiquitination by the E3 ligase Not4. Monoubiquitinated eS7 is a substrate for Hel2-mediated polyubiquitination, suggesting dual roles of Hel2 in two distinct NGD pathways, each of which requires specific ubiquitination events on stalled disomes.
Identification of Cue2 as a potential endonuclease responsible for NGD
In a recent study, Green and co-workers used constructs containing 12 tandem CGA rare codon repeats to screen for factors that reduced the mRNA level in a ribosome stalling-dependent manner, and identified the novel factor Cue2 in addition to the RQC factors Slh1 and Hel2 (97). Cue2 contains a ubiquitin-binding Cue domain at its N-terminus and an small MutS-related (SMR) hydrolase domain at its C-terminus. SMR domains show structural similarity to bacterial RNase E and function as DNA-nicking hydrolases (101). In addition, the SMR domain of SOT1 in plants exhibits RNA endonuclease activity (102). When aligned with the structure of the IF3-CTD on the ribosome, the conserved D348, H350 and R402 residues in the SMR domain of Cue2 are positioned along the mRNA channel. These residues are required for Cue2 function, and R402 has previously been implicated in the RNA cleavage activity of SOT1 in plants (102). Ribosome profiling and biochemical analyses have provided strong evidence that Cue2 cleaves mRNAs within the A-site of the colliding ribosome. The Cue2-dependent pathway becomes a major contributor to NGD in cells depleted of factors required for the resolution of stalled ribosome complexes. Further biochemical analyses will reveal the molecular mechanism by which Cue2 is recruited to the ubiquitinated ribosome and induces mRNA cleavage at the vicinity or inside the colliding ribosomes.
Interaction of the ribosome with mRNA decay machineries
Endonucleolytic cleavage during NGD results in the production of 5′-NGD and 3′-NGD mRNA intermediates that are further degraded by the Xrn1 exoribonuclease and the exosome, respectively (96). Xrn1 is the most prominent evolutionarily conserved 5′–3′-exoribonuclease, and it degrades substrates produced either by decapping or endonucleolytic cleavage. A recent study quantified the genome-wide abundance of 5′-phosphorylated mRNA degradation intermediates using a 5PSeq method (103) and found that Xrn1 interacts directly and specifically with 80S ribosomes for co-translational mRNA degradation. Analysis of the cryo-EM structure of the translating-degrading ribosome in complex with Xrn1 revealed that Xrn1 binds predominantly to the 40S ribosomal subunit at the mRNA exit site via interaction interfaces with numerous rRNA segments and ribosomal proteins, such as Asc1 (RACK1 in mammals) (104). These interfaces provide a channel for the passage of the mRNA directly from the ribosomal decoding site into the active center of the nuclease. In the NGDRQC+ pathway, endonucleolytic cleavage occurs mainly in the colliding ribosome. The leading stalled ribosome associates with 5′-NGD mRNA intermediates that will either undergo RQC or become substrates for Xrn1-dependent degradation. The Xrn1-ribosome structure led us to speculate that Xrn1 binds to the 40S subunit of the stalled ribosome to modulate the mRNA positioning for efficient access.
Several lines of evidence demonstrate the relevance of endonucleolytic cleavage events in a more general context (outside of NGD); for example, endogenous human mRNAs undergo repeated co-translational and ribosome-phased endonucleolytic cuts at the exit site of the ribosome mRNA channel (105–106). N6-methyladenosine (m6A) can also influence the fate of RNAs via proteins termed ‘readers’, which specifically recognize and bind to this modified nucleotide. YTHDC2 is a reader protein for m6A, and the YTHDC2 YTH domain preferentially binds m6A-containing RNAs. Interestingly, YTHDC2 interacts with the small ribosomal subunit and XRN1 via ankyrin repeats in an RNA-independent manner (107). It is proposed that YTHDC2 bridges interactions between m6A-containing mRNAs, ribosomes, and Xrn1 to regulate the stability and translation of the mRNAs.
The exosome acts as the exonuclease for 5′ NGD mRNA intermediate that is 5′ fragment produced by the endonucleolytic cleavage in NGD, and it cooperates with the Ski2–Ski3–Ski8 (Ski) helicase complex to mediate 3′–5′-mRNA decay. The structure of an endogenous ribosome–Ski complex generated via a cryo-EM analysis revealed that upon ribosome binding, the autoinhibitory domain of the Ski2 helicase relocates in an open conformation near the ribosomal mRNA entry tunnel (51). One possibility is that in the NGD pathway, ribosomes stalled at the 3′-end of 5′-NGD mRNA intermediates interact with the Ski complex, thereby activating the Ski2 helicase activity to unwind the aberrant mRNA.
QUALITY CONTROL OF NON-FUNCTIONAL RIBOSOMAL RNA
Non-functional ribosomes are subjected to quality control systems to avoid the production of potentially harmful products via abnormal translation. Moore and co-workers show that in yeast, a non-functional rRNA decay (NRD) quality control pathway can detect and eliminate mature rRNAs containing individual point mutations that adversely affect ribosome function, even when they are contained in fully assembled ribosomes and ribosomal subunits (108,109). Given the fundamental but differing functions of the 40S and 60S subunits during translation, NRD quality control pathways eliminate non-functional subunits via distinct pathways. The 25S NRD quality control pathway eliminates 25S rRNAs with mutations in the PTC, while the 18S NRD pathway eliminates mutated 18S rRNAs containing deleterious point mutations in the decoding center (108,109). The factors involved in RQC are also involved in 18S NRD (108,109), indicating that the stalling of non-functional ribosomes is recognized by ribosome ubiquitination-mediated subunit dissociation, leading to degradation of the non-functional rRNA. This section summarizes recent advances in our understanding of the mechanism by which the NRD pathway recognizes aberrant translation.
Ribosome ubiquitination plays a crucial role in 18S NRD
18S NRD is a quality control mechanism in which the base substitution mutant (A1492C) in the decoding center region of 18S rRNA constituting the 40S subunit is specifically degraded. The mechanisms involved in the 18S NRD pathway are still largely unknown; however, several lines of evidence suggest a connection between 18S NRD and RQC (Figure 4). For example, Asc1 is involved in 18S NRD (109,110) and is also required for RQC (57,58,94). The C-terminal 212–240 region of uS3 is essential for the 18S NRD pathway (110). The 212th lysine residue (K212) of uS3 is a target of Hel2-dependent ubiquitination (55), and sequential ubiquitination of uS3 at K212 triggers subunit dissociation in the 18S NRD pathway (100). It has been proposed that non-functional 80S ribosomes that stall due to decoding failures are sequentially ubiquitinated at the K212 residue of uS3. Mag2 mediates monoubiquitination of uS3 at K212, followed by Hel2- or Rsp5-mediated polyubiquitination (100). Subsequently, Slh1 stimulates subunit dissociation to promote Xrn1-dependent degradation of the non-functional 18S rRNA in the 40S subunit (100). Asc1 is also involved in uS3 ubiquitination and is thus required for subunit dissociation (100), suggesting that Asc1 is a platform for E3 ubiquitin ligases to recognize the ribosome stalled at the specific structure. Given the role of Slh1 in subunit dissociation in RQC, the crucial roles of ribosome ubiquitination and Slh1 are conserved in quality control pathways induced by ribosome stalling, as depicted in Figure 4.
Figure 4.
Ribosome ubiquitination as a trigger of quality control systems induced by ribosome stalling. Left: In RQC, the critical E3 ubiquitin ligase Hel2 and its mammalian homolog ZNF598 recognize disomes containing colliding ribosomes in the rotated state (73,79), leading to ubiquitination of uS10 in yeast and uS10, eS10, and uS3 in mammals. Right: In 18S NRD, non-functional 80S ribosomes containing the A1492C mutation in the decoding center stall due to decoding failures and are sequentially ubiquitinated at the K212 residue of uS3. Mag2 monoubiquitinates uS3 at K212, followed by Hel2-mediated polyubiquitination. Subsequently, the Ski2-like RNA helicase Slh1 in the RQT complex stimulates subunit dissociation to promote Xrn1-dependent degradation of the non-functional 18S rRNA in the 40S subunit (100). In these quality control systems, the ribosome is stalled at the specific conformation, and the specific E3 ligases recognize and ubiquitinate particular sites to induce the subsequent quality control steps.
These findings suggest that the ubiquitination of specific residues of ribosomal proteins is a universal molecular mechanism for determining the fate of ribosomes. Accurate gene expression is guaranteed by the quality control mechanism that recognizes and eliminates ribosomes that are abnormally stalled during translation elongation. Elucidation of the molecular mechanism of 18S NRD, the quality control mechanism of ribosomes lacking function, will form the molecular basis for understanding diseases caused by ribosome defects.
Mechanism of 25S NRD
In NRD, the dissociation of the subunits of non-functional 80S ribosomes is a prerequisite step for the degradation of the non-functional subunit. The 25S NRD pathway requires an E3 ubiquitin ligase complex, and proteins associated with non-functional ribosomes are ubiquitinated in an Rtt101-Mms1-dependent manner (111), with Crt10 being responsible for substrate recognition by the Rtt101-Mms1-containing E3 ligase complex (112). The components of cullin-RING ubiquitin ligase (CRL) complexes are essential for the selective ubiquitination of ribosomal particles containing non-functional 25S rRNAs (113). CRL complexes constitute a large family of E3 ubiquitin ligases, including the SCF (Skp1–cullin–F-box) complex. Cullin is a scaffold component of the CRL complex, which is connected by Skp1 to an F-box protein, a substrate recognition subunit of the ligase. In the 25S NRD pathway, Rtt101 is thought to recognize the non-functional ribosome. The mechanism by which the E3 ubiquitin ligase complex recognizes its substrate and ubiquitinates the ribosome, as well as the role of ribosome ubiquitination in subunit dissociation, remain to be elucidated. Non-functional and ubiquitinated 60S subunits are dissociated from their 40S subunits in a Cdc48–Npl4–Ufd1 complex (Cdc48 complex)-dependent manner before being attacked by the proteasome (113).
CONCLUSION AND PERSPECTIVES
Recent advances in our understanding of quality control systems in cells have revealed that E3 ligases recognize and ubiquitinate stalled ribosomes. Each E3 ligase recognizes the specific structure of the stagnant ribosome and ubiquitinates the lysine residue at the specific site, thereby recruiting subsequent quality control factors. This molecular mechanism is a ubiquitin code in a broad sense and can be called a ribosome ubiquitin code. It is rational to speculate that putative quality controls for aberrant translation may be similarly induced by other molecular modifications, such as phosphorylation, methylation and acetylation. The modification sites could be rRNAs in addition to ribosomal proteins, and the levels of modification could be induced or increased by aberrant translation during stress conditions. Given that ribosome modification can play an essential function in translational control, there is no doubt that understanding the mechanisms of ribosome modification and recognition will become an increasingly important research subject in the future. Elucidation of the molecular mechanisms involved in translation quality control will not only lead to a better understanding of how it ensures accurate translation, but also will contribute to our understanding of diseases related to translational abnormalities and the development of therapeutic methods.
FUNDING
Japan Society for the Promotion of Science (JSPS) [JP18H03977]; Japan Agency for Medical Research and Development (AMED) [JP19gm1110010]. Funding for open access charge: JSPS [18H03977]; AMED [19gm1110010]
Conflict of interest statement. None declared.
REFERENCES
- 1. Pechmann S., Willmund F., Frydman J.. The ribosome as a hub for protein quality control. Mol. Cell. 2013; 49:411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shoemaker C.J., Green R.. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012; 19:594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Inada T. Quality control systems for aberrant mRNAs induced by aberrant translation elongation and termination. Biochim. Biophys. Acta. 2013; 1829:634–642. [DOI] [PubMed] [Google Scholar]
- 4. Brandman O., Hegde R.S.. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 2016; 23:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shao S., Hegde R.S.. Target selection during protein quality control. Trends Biochem. Sci. 2016; 41:124–137. [DOI] [PubMed] [Google Scholar]
- 6. Inada T. The ribosome as a platform for mRNA and nascent polypeptide quality control. Trends Biochem. Sci. 2017; 42:5–15. [DOI] [PubMed] [Google Scholar]
- 7. Joazeiro C.A.P. Mechanisms and functions of ribosome-associated protein quality control. Nat. Rev. Mol. Cell Biol. 2019; 20:368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Joazeiro C.A.P. Ribosomal stalling during translation: providing substrates for ribosome-associated protein quality control. Annu. Rev. Cell Dev. Biol. 2017; 33:343–368. [DOI] [PubMed] [Google Scholar]
- 9. Frischmeyer P.A., van Hoof A., O’Donnell K., Guerrerio A.L., Parker R., Dietz H.C.. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002; 295:2258–2261. [DOI] [PubMed] [Google Scholar]
- 10. van Hoof A., Frischmeyer P.A., Dietz H.C., Parker R.. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002; 295:2262–2264. [DOI] [PubMed] [Google Scholar]
- 11. van Hoof A., Parker R.. Messenger RNA degradation: beginning at the end. Curr. Biol. 2002; 12:R285–287. [DOI] [PubMed] [Google Scholar]
- 12. Wilson M.A., Meaux S., Parker R., van Hoof A.. Genetic interactions between [PSI+] and nonstop mRNA decay affect phenotypic variation. PNAS. 2005; 102:10244–10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meaux S., Van Hoof A.. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA. 2006; 12:1323–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Keiler K.C. Mechanisms of ribosome rescue in bacteria. Nat. Rev. Microbiol. 2015; 13:285–297. [DOI] [PubMed] [Google Scholar]
- 15. Himeno H., Nameki N., Kurita D., Muto A., Abo T.. Ribosome rescue systems in bacteria. Biochimie. 2015; 114:102–112. [DOI] [PubMed] [Google Scholar]
- 16. Huter P., Muller C., Arenz S., Beckert B., Wilson D.N.. Structural basis for ribosome rescue in bacteria. Trends Biochem. Sci. 2017; 42:669–680. [DOI] [PubMed] [Google Scholar]
- 17. Goralski T.D.P., Kirimanjeswara G.S., Keiler K.C.. A new mechanism for ribosome rescue can recruit RF1 or RF2 to nonstop ribosomes. mBio. 2018; 9:e02436-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moore S.D., Sauer R.T.. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 2007; 76:101–124. [DOI] [PubMed] [Google Scholar]
- 19. Huang C., Wolfgang M.C., Withey J., Koomey M., Friedman D.I.. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J. 2000; 19:1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rae C.D., Gordiyenko Y., Ramakrishnan V.. How a circularized tmRNA moves through the ribosome. Science. 2019; 363:740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Richards J., Mehta P., Karzai A.W.. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol. Microbiol. 2006; 62:1700–1712. [DOI] [PubMed] [Google Scholar]
- 22. Venkataraman K., Guja K.E., Garcia-Diaz M., Karzai A.W.. Non-stop mRNA decay: a special attribute of trans-translation mediated ribosome rescue. Front. Microbiol. 2014; 5:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chadani Y., Matsumoto E., Aso H., Wada T., Kutsukake K., Sutou S., Abo T.. trans-translation-mediated tight regulation of the expression of the alternative ribosome-rescue factor ArfA in Escherichia coli. Genes Genet. Syst. 2011; 86:151–163. [DOI] [PubMed] [Google Scholar]
- 24. Chadani Y., Ito K., Kutsukake K., Abo T.. ArfA recruits release factor 2 to rescue stalled ribosomes by peptidyl-tRNA hydrolysis in Escherichia coli. Mol. Microbiol. 2012; 86:37–50. [DOI] [PubMed] [Google Scholar]
- 25. Kurita D., Chadani Y., Muto A., Abo T., Himeno H.. ArfA recognizes the lack of mRNA in the mRNA channel after RF2 binding for ribosome rescue. Nucleic Acids Res. 2014; 42:13339–13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. James N.R., Brown A., Gordiyenko Y., Ramakrishnan V.. Translational termination without a stop codon. Science. 2016; 354:1437–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma C., Kurita D., Li N., Chen Y., Himeno H., Gao N.. Mechanistic insights into the alternative translation termination by ArfA and RF2. Nature. 2017; 541:550–553. [DOI] [PubMed] [Google Scholar]
- 28. Demo G., Svidritskiy E., Madireddy R., Diaz-Avalos R., Grant T., Grigorieff N., Sousa D., Korostelev A.A.. Mechanism of ribosome rescue by ArfA and RF2. eLife. 2017; 6:e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schaub R.E., Poole S.J., Garza-Sanchez F., Benbow S., Hayes C.S.. Proteobacterial ArfA peptides are synthesized from non-stop messenger RNAs. J. Biol. Chem. 2012; 287:29765–29775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garza-Sanchez F., Schaub R.E., Janssen B.D., Hayes C.S.. tmRNA regulates synthesis of the ArfA ribosome rescue factor. Mol. Microbiol. 2011; 80:1204–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shimizu Y. ArfA recruits RF2 into stalled ribosomes. J. Mol. Biol. 2012; 423:624–631. [DOI] [PubMed] [Google Scholar]
- 32. Chadani Y., Ono K., Kutsukake K., Abo T.. Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA- and ArfA-mediated pathways. Mol. Microbiol. 2011; 80:772–785. [DOI] [PubMed] [Google Scholar]
- 33. Handa Y., Inaho N., Nameki N.. YaeJ is a novel ribosome-associated protein in Escherichia coli that can hydrolyze peptidyl-tRNA on stalled ribosomes. Nucleic Acids Res. 2011; 39:1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kogure H., Handa Y., Nagata M., Kanai N., Guntert P., Kubota K., Nameki N.. Identification of residues required for stalled-ribosome rescue in the codon-independent release factor YaeJ. Nucleic Acids Res. 2014; 42:3152–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brown A., Shao S., Murray J., Hegde R.S., Ramakrishnan V.. Structural basis for stop codon recognition in eukaryotes. Nature. 2015; 524:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matheisl S., Berninghausen O., Becker T., Beckmann R.. Structure of a human translation termination complex. Nucleic Acids Res. 2015; 43:8615–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gagnon M.G., Seetharaman S.V., Bulkley D., Steitz T.A.. Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science. 2012; 335:1370–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsuboi T., Kuroha K., Kudo K., Makino S., Inoue E., Kashima I., Inada T.. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell. 2012; 46:518–529. [DOI] [PubMed] [Google Scholar]
- 39. Shoemaker C.J., Eyler D.E., Green R.. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010; 330:369–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shoemaker C.J., Green R.. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. PNAS. 2011; 108:E1392–E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pisareva V.P., Skabkin M.A., Hellen C.U., Pestova T.V., Pisarev A.V.. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011; 30:1804–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shao S., von der Malsburg K., Hegde R.S.. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell. 2013; 50:637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Becker T., Franckenberg S., Wickles S., Shoemaker C.J., Anger A.M., Armache J.P., Sieber H., Ungewickell C., Berninghausen O., Daberkow I. et al.. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012; 482:501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ikeuchi K., Yazaki E., Kudo K., Inada T.. Conserved functions of human Pelota in mRNA quality control of nonstop mRNA. FEBS Lett. 2016; 590:3254–3263. [DOI] [PubMed] [Google Scholar]
- 45. Ikeuchi K., Inada T.. Ribosome-associated Asc1/RACK1 is required for endonucleolytic cleavage induced by stalled ribosome at the 3′ end of nonstop mRNA. Sci. Rep. 2016; 6:28234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matsuda R., Ikeuchi K., Nomura S., Inada T.. Protein quality control systems associated with no-go and nonstop mRNA surveillance in yeast. Genes Cells. 2014; 19:1–12. [DOI] [PubMed] [Google Scholar]
- 47. Preis A., Heuer A., Barrio-Garcia C., Hauser A., Eyler D.E., Berninghausen O., Green R., Becker T., Beckmann R.. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 2014; 8:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Young D.J., Guydosh N.R., Zhang F., Hinnebusch A.G., Green R.. Rli1/ABCE1 Recycles terminating ribosomes and controls translation reinitiation in 3′UTRs In Vivo. Cell. 2015; 162:872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Franckenberg S., Becker T., Beckmann R.. Structural view on recycling of archaeal and eukaryotic ribosomes after canonical termination and ribosome rescue. Curr. Opin. Struct. Biol. 2012; 22:786–796. [DOI] [PubMed] [Google Scholar]
- 50. Kashima I., Takahashi M., Hashimoto Y., Sakota E., Nakamura Y., Inada T.. A functional involvement of ABCE1, eukaryotic ribosome recycling factor, in nonstop mRNA decay in Drosophila melanogaster cells. Biochimie. 2014; 106:10–16. [DOI] [PubMed] [Google Scholar]
- 51. Schmidt C., Kowalinski E., Shanmuganathan V., Defenouillere Q., Braunger K., Heuer A., Pech M., Namane A., Berninghausen O., Fromont-Racine M. et al.. The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex. Science. 2016; 354:1431–1433. [DOI] [PubMed] [Google Scholar]
- 52. Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C.C., Li G.W., Zhou S., King D., Shen P.S., Weibezahn J. et al.. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012; 151:1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bengtson M.H., Joazeiro C.A.. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010; 467:470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Defenouillere Q., Yao Y., Mouaikel J., Namane A., Galopier A., Decourty L., Doyen A., Malabat C., Saveanu C., Jacquier A. et al.. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. PNAS. 2013; 110:5046–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Matsuo Y., Ikeuchi K., Saeki Y., Iwasaki S., Schmidt C., Udagawa T., Sato F., Tsuchiya H., Becker T., Tanaka K. et al.. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 2017; 8:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Garzia A., Jafarnejad S.M., Meyer C., Chapat C., Gogakos T., Morozov P., Amiri M., Shapiro M., Molina H., Tuschl T. et al.. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat. Commun. 2017; 8:16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Juszkiewicz S., Hegde R.S.. Initiation of quality control during poly(A) Translation requires Site-Specific ribosome ubiquitination. Mol. Cell. 2017; 65:743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sundaramoorthy E., Leonard M., Mak R., Liao J., Fulzele A., Bennett E.J.. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol. Cell. 2017; 65:751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sitron C.S., Park J.H., Brandman O.. Asc1, Hel2, and Slh1 couple translation arrest to nascent chain degradation. RNA. 2017; 23:798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shao S., Hegde R.S.. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol. Cell. 2014; 55:880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lyumkis D., Oliveira dos Passos D., Tahara E.B., Webb K., Bennett E.J., Vinterbo S., Potter C.S., Carragher B., Joazeiro C.A.. Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. PNAS. 2014; 111:15981–15986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hilal T., Spahn C.M.. Ribosome rescue and protein quality control in concert. Mol. Cell. 2015; 57:389–390. [DOI] [PubMed] [Google Scholar]
- 63. Shen P.S., Park J., Qin Y., Li X., Parsawar K., Larson M.H., Cox J., Cheng Y., Lambowitz A.M., Weissman J.S. et al.. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science. 2015; 347:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Verma R., Reichermeier K.M., Burroughs A.M., Oania R.S., Reitsma J.M., Aravind L., Deshaies R.J.. Vms1 and ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes. Nature. 2018; 557:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zurita Rendon O., Fredrickson E.K., Howard C.J., Van Vranken J., Fogarty S., Tolley N.D., Kalia R., Osuna B.A., Shen P.S., Hill C.P. et al.. Vms1p is a release factor for the ribosome-associated quality control complex. Nat. Commun. 2018; 9:2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Su T., Izawa T., Thoms M., Yamashita Y., Cheng J., Berninghausen O., Hartl F.U., Inada T., Neupert W., Beckman R.. Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis. Nature. 2019; 570:538–542. [DOI] [PubMed] [Google Scholar]
- 67. Yip M.C.J., Keszei A.F.A., Feng Q., Chu V., McKenna M.J., Shao S.. Mechanism for recycling tRNAs on stalled ribosomes. Nat. Struct. Mol. Biol. 2019; 26:343–349. [DOI] [PubMed] [Google Scholar]
- 68. Kuroha K., Zinoviev A., Hellen C.U.T., Pestova T.V.. Release of ubiquitinated and non-ubiquitinated nascent chains from stalled mammalian ribosomal complexes by ANKZF1 and Ptrh1. Mol. Cell. 2018; 72:286–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Inada T., Aiba H.. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 2005; 24:1584–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ito-Harashima S., Kuroha K., Tatematsu T., Inada T.. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007; 21:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Akimitsu N., Tanaka J., Pelletier J.. Translation of nonSTOP mRNA is repressed post-initiation in mammalian cells. EMBO J. 2007; 26:2327–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shao S., Brown A., Santhanam B., Hegde R.S.. Structure and assembly pathway of the ribosome quality control complex. Mol. Cell. 2015; 57:433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Juszkiewicz S., Chandrasekaran V., Lin Z., Kraatz S., Ramakrishnan V., Hegde R.S.. ZNF598 is a quality control sensor of collided ribosomes. Mol. Cell. 2018; 72:469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Arthur L., Pavlovic-Djuranovic S., Smith-Koutmou K., Green R., Szczesny P., Djuranovic S.. Translational control by lysine-encoding A-rich sequences. Sci. Adv. 2015; 1:e1500154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Koutmou K.S., Schuller A.P., Brunelle J.L., Radhakrishnan A., Djuranovic S., Green R.. Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife. 2015; 4:e05534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Arthur L.L., Djuranovic S.. PolyA tracks, polybasic peptides, poly-translational hurdles. Wiley Interdiscip. Rev. RNA. 2018; 5:e1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dimitrova L.N., Kuroha K., Tatematsu T., Inada T.. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 2009; 284:10343–10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gamble C.E., Brule C.E., Dean K.M., Fields S., Grayhack E.J.. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell. 2016; 166:679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ikeuchi K., Tesina P., Matsuo Y., Sugiyama T., Cheng J., Saeki Y., Tanaka K., Becker T., Beckmann R., Inada T.. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 2019; 38:e100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Morita M., Ler L.W., Fabian M.R., Siddiqui N., Mullin M., Henderson V.C., Alain T., Fonseca B.D., Karashchuk G., Bennett C.F. et al.. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol. Cell Biol. 2012; 32:3585–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ruscica V., Bawankar P., Peter D., Helms S., Igreja C., Izaurralde E.. Direct role for the Drosophila GIGYF protein in 4EHP-mediated mRNA repression. Nucleic Acids Res. 2019; 47:7035–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fu R., Olsen M.T., Webb K., Bennett E.J., Lykke-Andersen J.. Recruitment of the 4EHP-GYF2 cap-binding complex to tetraproline motifs of tristetraprolin promotes repression and degradation of mRNAs with AU-rich elements. RNA. 2016; 22:373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tollenaere M.A.X., Tiedje C., Rasmussen S., Nielsen J.C., Vind A.C., Blasius M., Batth T.S., Mailand N., Olsen J.V., Gaestel M. et al.. GIGYF1/2-driven cooperation between ZNF598 and TTP in posttranscriptional regulation of inflammatory signaling. Cell Rep. 2019; 26:3511–3521. [DOI] [PubMed] [Google Scholar]
- 84. Hickey K.L., Dickson K., Cogan J.Z., Replogle J.M., Schoof M., D’Orazio K.N., Sinha N.K., Frost A., Green R., Kostova K.K. et al.. GIGYF2 and 4EHP Inhibit translation initiation of defective messenger RNAs to assist ribosome-associated quality control. 2019; bioRxiv doi:03 October 2019, pre-print: not peer reviewed 10.1101/792994. [DOI] [PMC free article] [PubMed]
- 85. Wilson M.A., Meaux S., van Hoof A.. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007; 177:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chu J., Hong N.A., Masuda C.A., Jenkins B.V., Nelms K.A., Goodnow C.C., Glynne R.J., Wu H., Masliah E., Joazeiro C.A. et al.. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. PNAS. 2009; 106:2097–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kostova K.K., Hickey K.L., Osuna B.A., Hussmann J.A., Frost A., Weinberg D.E., Weissman J.S.. CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science. 2017; 357:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Nielson J.R., Fredrickson E.K., Waller T.C., Rendon O.Z., Schubert H.L., Lin Z., Hill C.P., Rutter J.. Sterol oxidation mediates stress-responsive Vms1 translocation to mitochondria. Mol. Cell. 2017; 68:673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Izawa T., Park S.H., Zhao L., Hartl F.U., Neupert W.. Cytosolic protein vms1 links ribosome quality control to mitochondrial and cellular homeostasis. Cell. 2017; 171:890–903. [DOI] [PubMed] [Google Scholar]
- 90. Hey J.M., Nielson J.R., Dephoure N., Gygi S.P., Rutter J.. Intramolecular interactions control Vms1 translocation to damaged mitochondria. Mol. Biol. Cell. 2013; 24:1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sitron C.S., Brandman O.. CAT tails drive degradation of stalled polypeptides on and off the ribosome. Nat. Struct. Mol. Biol. 2019; 26:450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lytvynenko I., Paternoga H., Thrun A., Balke A., Muller T.A., Chiang C.H., Nagler K., Tsaprailis G., Anders S., Bischofs I. et al.. Alanine tails signal proteolysis in bacterial ribosome-associated quality control. Cell. 2019; 178:76–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Guydosh N.R., Green R.. Translation of poly(A) tails leads to precise mRNA cleavage. RNA. 2017; 23:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kuroha K., Akamatsu M., Dimitrova L., Ito T., Kato Y., Shirahige K., Inada T.. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 2010; 11:956–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Simms C.L., Yan L.L., Zaher H.S.. Ribosome collision is critical for quality control during no-go decay. Mol. Cell. 2017; 68:361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Doma M.K., Parker R.. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006; 440:561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. D’Orazio K.N., Wu C.C., Sinha N., Loll-Krippleber R., Brown G.W., Green R.. The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during no go decay. eLife. 2019; 8:e49117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tsuboi T., Yamazaki R., Nobuta R., Ikeuchi K., Makino S., Ohtaki A., Suzuki Y., Yoshihisa T., Trotta C., Inada T.. The tRNA splicing endonuclease complex cleaves the mitochondria-localized CBP1 mRNA. J. Biol. Chem. 2015; 290:16021–16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Saito K., Horikawa W., Ito K.. Inhibiting K63 polyubiquitination abolishes no-go type stalled translation surveillance in Saccharomyces cerevisiae. PLos Genet. 2015; 11:e1005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sugiyama T., Li S., Kato M., Ikeuchi K., Ichimura A., Matsuo Y., Inada T.. Sequential ubiquitination of ribosomal protein uS3 triggers the degradation of Non-functional 18S rRNA. Cell Rep. 2019; 26:3400–3415. [DOI] [PubMed] [Google Scholar]
- 101. Fukui K., Kuramitsu S.. Structure and function of the small MutS-Related domain. Mol. Biol. Int. 2011; 2011:691735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhou W., Lu Q., Li Q., Wang L., Ding S., Zhang A., Wen X., Zhang L., Lu C.. PPR-SMR protein SOT1 has RNA endonuclease activity. PNAS. 2017; 114:E1554–E1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pelechano V., Wei W., Steinmetz L.M.. Genome-wide quantification of 5′-phosphorylated mRNA degradation intermediates for analysis of ribosome dynamics. Nat. Protoc. 2016; 11:359–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tesina P., Heckel E., Cheng J., Fromont-Racine M., Buschauer R., Kater L., Beatrix B., Berninghausen O., Jacquier A., Becker T. et al.. Structure of the 80S ribosome-Xrn1 nuclease complex. Nat. Struct. Mol. Biol. 2019; 26:275–280. [DOI] [PubMed] [Google Scholar]
- 105. Ibrahim F., Mourelatos Z.. Capturing 5′ and 3′ native ends of mRNAs concurrently with Akron sequencing. Nat. Protoc. 2019; 14:1578–1602. [DOI] [PubMed] [Google Scholar]
- 106. Ibrahim F., Maragkakis M., Alexiou P., Mourelatos Z.. Ribothrypsis, a novel process of canonical mRNA decay, mediates ribosome-phased mRNA endonucleolysis. Nat. Struct. Mol. Biol. 2018; 25:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kretschmer J., Rao H., Hackert P., Sloan K.E., Hobartner C., Bohnsack M.T.. The m(6)A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′-3′ exoribonuclease XRN1. RNA. 2018; 24:1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. LaRiviere F.J., Cole S.E., Ferullo D.J., Moore M.J.. A late-acting quality control process for mature eukaryotic rRNAs. Mol. Cell. 2006; 24:619–626. [DOI] [PubMed] [Google Scholar]
- 109. Cole S.E., LaRiviere F.J., Merrikh C.N., Moore M.J.. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol. Cell. 2009; 34:440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Limoncelli K.A., Merrikh C.N., Moore M.J.. ASC1 and RPS3: new actors in 18S nonfunctional rRNA decay. RNA. 2017; 23:1946–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Fujii K., Kitabatake M., Sakata T., Miyata A., Ohno M.. A role for ubiquitin in the clearance of nonfunctional rRNAs. Genes Dev. 2009; 23:963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sakata T., Fujii K., Ohno M., Kitabatake M.. Crt10 directs the cullin-E3 ligase Rtt101 to nonfunctional 25S rRNA decay. Biochem. Biophys. Res. Commun. 2015; 457:90–94. [DOI] [PubMed] [Google Scholar]
- 113. Fujii K., Kitabatake M., Sakata T., Ohno M.. 40S subunit dissociation and proteasome-dependent RNA degradation in nonfunctional 25S rRNA decay. EMBO J. 2012; 31:2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Simms C.L., Hudson B.H., Mosior J.W., Rangwala A.S., Zaher H.S.. An active role for the ribosome in determining the fate of oxidized mRNA. Cell. 2014; 9:1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yan L.L., Simms C.L., McLoughlin F., Vierstra R.D., Zaher H.S.. Oxidation and alkylation stresses activate ribosome-quality control. Nature. 2019; 10:5611. [DOI] [PMC free article] [PubMed] [Google Scholar]