Abstract
Background
Naturally occurring variation in Membrane-bound O-acyltransferase domain-containing 7 (MBOAT7), encoding for an enzyme involved in phosphatidylinositol acyl-chain remodelling, has been associated with fatty liver and hepatic disorders. Here, we examined the relationship between hepatic Mboat7 down-regulation and fat accumulation.
Methods
Hepatic MBOAT7 expression was surveyed in 119 obese individuals and in experimental models. MBOAT7 was acutely silenced by antisense oligonucleotides in C57Bl/6 mice, and by CRISPR/Cas9 in HepG2 hepatocytes.
Findings
In obese individuals, hepatic MBOAT7 mRNA decreased from normal liver to steatohepatitis, independently of diabetes, inflammation and MBOAT7 genotype. Hepatic MBOAT7 levels were reduced in murine models of fatty liver, and by hyper-insulinemia. In wild-type mice, Mboat7 was down-regulated by refeeding and insulin, concomitantly with insulin signalling activation. Acute hepatic Mboat7 silencing promoted hepatic steatosis in vivo and enhanced expression of fatty acid transporter Fatp1. MBOAT7 deletion in hepatocytes reduced the incorporation of arachidonic acid into phosphatidylinositol, consistently with decreased enzymatic activity, determining the accumulation of saturated triglycerides, enhanced lipogenesis and FATP1 expression, while FATP1 deletion rescued the phenotype.
Interpretation
MBOAT7 down-regulation by hyper-insulinemia contributes to hepatic fat accumulation, impairing phosphatidylinositol remodelling and up-regulating FATP1.
Funding
LV was supported by MyFirst Grant AIRC n.16888, Ricerca Finalizzata Ministero della Salute RF-2016–02,364,358, Ricerca corrente Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico; LV and AG received funding from the European Union Programme Horizon 2020 (No. 777,377) for the project LITMUS-“Liver Investigation: Testing Marker Utility in Steatohepatitis”. MM was supported by Fondazione Italiana per lo Studio del Fegato (AISF) ‘Mario Coppo’ fellowship.
Keywords: LPIAT1, NAFLD, Nash, Nonalcoholic fatty liver disease, Steatohepatitis, Phospholipid, Phosphatidylinositol
Abbreviations: ALD, alcoholic liver disease; APOB, Apolipoprotein B; BMI, Body Mass Index; CDP, Cytidine-Diphosphate; CI, Confidence Interval; CL, Cardiolipin; CPT1, Carnitine Palmitoyltransferase I; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; CXCL10, C-X-C Motif Chemokine 10; DAG, Diacylglycerol; FABP1, Fatty Acid-Binding Protein 1, FAS, Fatty Acid Synthase; FATP1, Fatty Acid Transport Protein 1; FOXO1, Forkhead Box protein O1; FOXA2, Forkhead Box A2; G3P, Glyceraldehyde-3-Phosphate; HDL, High Density Lipoproteins; hHEPS, Human Hepatocytes; hHSC, Human Hepatic Stellate Cells; HOMA-IR, homeostasis Model Assessment of Insulin Resistance; IFG, Impaired Fasting Glucose; i.p., Intraperitoneal; i.v., Intravenous; LDL, Low Density Lipoproteins; LPA, Lyso-Phosphatidic Acid; LPIAT1, Lysophosphatidylinositol Acyltransferase 1; Mboat7, Membrane bound O-acyltransferase domain containing 7; MCD, methionine choline deficient diet; MPO, morpholino; mTOR, mammalian target of Rapamycin; MTTP, Microsomal Triglyceride Transfer Protein; NAFLD, nonalcoholic fatty liver disease; NASH, Nonalcoholic Steatohepatitis; NHEJ, Non-Homologues End Joining; OA, Oleic Acid; PA, Palmitic Acid; PC, Phosphatidylcholine; ORO, Oil Red O Staining; PE, Phosphatidyl-Ethanolamine; PG, Phosphatidyl-Glycerol; PI, Phosphatidylinositol; PIP, Phosphatidyl-Inositol-Phosphate; PI3K, Phosphatidylinositol 3 Kinase; PNPLA3, Patatin-like Phospholipase Domain-containing-3; PPARα, Peroxisome Proliferator-Activated Receptor alpha; PS, Phosphatidyl-Serine; qRT-PCR, quantitative Real Time Polymerase Chain Reaction; SD, Standard Diet; SREBP1c, Sterol Regulatory Element-Binding Protein 1; TAG, Triglycerides; TGFβ, Transforming Growth Factor Beta; TM6SF2, Transmembrane 6 Superfamily Member 2; TMC4, Transmembrane Channel-Like 4; TNFα, tumor Necrosis Factor Alpha; T2DM, Type 2 Diabetes Mellitus; VLDL, Very Low Density Lipoprotein
Research in context.
Evidence before this study
Fatty liver, associated with obesity, type 2 diabetes and at-risk alcohol intake is the leading cause of liver disease worldwide. Inherited genetic factors play a major role in the development of fatty liver. Naturally occurring variation in the MBOAT7 gene, associated with reduced expression, have been linked to fatty liver and liver cancer. However, the mechanism linking MBOAT7 down-regulation with liver disease is still debated.
Added value of this study
We examined relationship between hepatic MBOAT7 down-regulation and fat accumulation in clinical samples, experimental models of NAFLD in vivo, and in genetically edited hepatocytes in vitro. We showed that, independently of the genetic background, down-regulation of Mboat7is a maladaptive response to hyper-insulinemia and facilitates intracellular fat accumulation in hepatocytes. The mechanism is related to impaired generation of arachidonoyl-phosphatidylinositol, leading to the conversion of saturated lyso-phosphatidylinositol to triglycerides, and enhanced lipogenesis.
Implications of all the available evidence
Down-regulation of MBOAT7 by hyper-insulinemia contributes to fatty liver independently of the genetic background. The underlying mechanism encompasses an impairment of intracellular phosphatidylinositol remodelling and lipid fluxes. These findings provide novel insight into the pathogenesis of fatty liver disease.
Alt-text: Unlabelled box
1. Introduction
Excessive hepatic fat accumulation, triggered by dysmetabolism and/or at-risk alcohol intake such as in nonalcoholic and alcoholic fatty liver disease (NAFLD and ALD), is the most frequent cause of liver disease worldwide, affecting more than one third of the general population. NAFLD is epidemiologically associated with obesity, metabolic syndrome features and type 2 diabetes (T2D) [1,2]. Fatty liver encompasses a wide spectrum of liver damage, ranging from isolated steatosis to nonalcoholic steatohepatitis (NASH), that more frequently evolves to cirrhosis and liver cancer (hepatocellular carcinoma: HCC) [3].
Fatty liver disease development is heavily influenced by inherited factors [4,5]. The main common genetic determinants of fatty liver are the variants in PNPLA3 (p.I148M) [6], and TM6SF2 (p.E167K) [7] genes. A genome-wide screening of the inherited genetic variants associated with alcoholic cirrhosis led to the identification of the common rs641738 C>T single nucleotide polymorphism at the Membrane bound O-acyltransferase domain-containing 7 – Transmembrane channel-like 4 (MBOAT7-TMC4) locus [[8], [9], [10]]. We showed that the minor rs641738 T allele is associated with increased predisposition towards hepatic fat accumulation and to the entire spectrum of liver damage related to NAFLD, including HCC [11–13]. Furthermore, the rs641738 variant has been associated with increased risk of liver fibrosis in patients with chronic viral hepatitis [14,15], suggesting that MBOAT7 variation is a common modifier of liver disease risk.
MBOAT7, also known as lyso-phosphatidylinositol (lyso-PI) acyltransferase 1 (LPIAT1), is involved in phospholipid acyl-chain remodelling of the membranes within the Lands’ cycle. It conjugates an acyl-CoA to the second acyl-chain of lyso-PI and other lyso-phospholipids, using as preferential substrate arachidonoyl-CoA. This process increases the desaturation of phospholipids using free arachidonic acid, a precursor of the proinflammatory eicosanoids [[16], [17], [18], [19], [20], [21]]. We previously reported that the likely mechanism behind the genetic association with liver damage is related to reduced hepatic expression of the MBOAT7 protein in carriers of 3′-untranslated region variants linked with rs641738 [11,13], leading to reduced hepatic PI species bound to polyunsaturated fatty acids [11,12].
Here we shown that, independently of the genetic background, down-regulation of hepatic MBOAT7 is a maladaptive response to hyper-insulinemia that induces fat accumulation in hepatocytes. We tested this hypothesis in clinical samples of patients, in experimental models of NAFLD and in response to normal physiological cues in vivo, and by genetic manipulation of hepatocytes in vitro.
2. Materials and methods
2.1. Patients
We evaluated hepatic and adipose tissue samples of 119 unselected severely obese individuals, who underwent percutaneous liver biopsy performed during bariatric surgery, and had no history of at-risk alcohol intake (>30/20 g/day in males/females) or other liver diseases (Bariatric surgery cohort). NASH was diagnosed when steatosis, lobular inflammation and ballooning were concomitantly present. Informed written consent was obtained from each patient and the study protocol was approved by the Ethical Committee of the Fondazione IRCCS Ca’ Granda and conforms to the ethical guidelines of the 1975 Declaration of Helsinki. The clinical characteristics of these patients are listed in Table S1.
Serum free fatty acids (FFAs) were measured in 72 NAFLD patients, of whom samples were available (Hepatology service cohort). Clinical characteristics are listed in Table S2.
Human hepatocytes and hepatic stellate cells (hHSC) were isolated from non-neoplastic tissue obtained from explanted or resected livers of 17 patients, while peripheral blood lymphocytes and monocytes were isolated from 16 healthy subjects. The detailed isolation protocols, and enrolment criteria are described in the Supplementary Methods.
All patients were genotyped for PNPLA3 rs738409 (p.I148M) and MBOAT7 rs641738 by TaqMan 5′-nuclease assays (Life Technologies, Carlsbad, CA) [11,22].
2.2. Experimental models
C57Bl/6 mice (Charles River, Calco, Italy) were housed at constant room temperature (23 °C) under 12-hour light/dark cycles with ad libitum access to water in compliance with the Principles of Laboratory Animal Care (NIH publication 86–23). C57Bl/6 Lepob/ob mice were purchased from Charles Rivers (Calco, Italy). Insulin receptor haplo-insufficient (InsR+/-) mice on C57Bl/6 background were kindly provided by Prof. Accili, Columbia University, NY, USA [23].
Wild-type and InsR+/- male mice, which are more susceptible to develop liver damage and fibrosis compared to females, were fed either methionine choline deficient diet (MCD: 14.8% protein, 12.4% fat and 72.8% carbohydrates; Test Diet, London, UK) or regular chow (standard diet: SD): 15.1% protein, 12.4% fat and 72.4% carbohydrates Test Diet, London, UK) for 6 weeks, starting at 6 weeks of age. Experiments were conducted in 10 mice per group. Before sacrifice, mice were fasted for 16 h and the interventions were done during the light cycle. Metabolic and biochemical features of InsR+/+ and InsR +/- mice upon SD or MCD feeding and Lepob/ob mice upon standard diet are shown in Table S3. In order to evaluate the physiological regulation of Mboat7 in response to the nutritional state, 6 weeks old wild-type male mice were fasted for 16 h (T0), refed, giving them ad libitum access to SD and then sacrificed at the following time points (before refeeding (T0), 15 min (T15’), 1hr (T1h), 4 hrs (T4h), 8 hrs (T8h)). In another experimental setting, wild-type male mice fasted for 16 h, injected with i.p. insulin (0.6 U/Kg) and then sacrificed at the following time points (before insulin (T0), 1hr (T1h) and 4 hrs (T4h)). These experiments were conducted in 7 mice per group per each time point.
The experimental protocol was approved by the University of Milan and the Italian Ministry of Health Review Boards (protocols 8/14 and 295/2012–A, received on 12/20/2012).
2.3. Gene silencing
To silence Forkhead box protein O1 (FoxO1) and/or Forkhead Box A2 (FoxA2), key mediators of insulin signalling at transcriptional level [[24], [25], [26], [27]], we exploited specific antisense oligonucleotides conjugated with morpholinos (MPO) that block protein translation or the “Standard control”. The respective sequences were: 5′-CCACCAGCAGAGAAGTACCGGGAGA-3′, 5′-TCCATCTTCACGGCTCCCAGCATAC-3′ and 5′-CCTCTTACCTCAGTTACAATTTATA-3′ (Gene tools, www.gene-tools.com). Primary mouse hepatocytes were isolated as previously described [28] and incubated with FoxO1 and/or FoxA2-specific or scramble MPO (10 μM) for 24 h then exposed to 0.33 μM insulin (Insuman Rapid, Sanofi Aventis) for 6 h.
In order to acutely silence in vivo Mboat7, wild-type male mice were treated with 12.5 mg/kg of Mboat7-specific MPO (5′-ATATGTCCATTCTTCGGGTGTCAT-3′) or scramble administered i.v. daily for 4 days (n = 6 per group). These mice were fed SD and fasted for 16 h before the sacrifice. Metabolic and biochemical features of mice treated with Scramble and anti-Mboat7-MPO are shown in Table S4.
2.4. Lipidomics
For lipidomic analysis, 106 hepatocytes were homogenized with 300 µl of methanol and 20 µl of a mix of internal standards (Avanti Polar Lipids, Alabaster, AL and Larodan, Solna, SE) were added to samples, which were then put on an oscillating plate for 30 min at 4 °C and centrifuged at 14,000 rpm. Afterwards, the methanol supernatant was analysed by liquid chromatography / mass spectrometry – quadrupole time of flight (LC/MS-QTOF; Agilent UHPLC 1290 Infinity-6540 QTOF, Santa Clara, CA). Separation of lipids obtained from mice livers is described in the Supplementary Methods.
Lipid classes were separated with ultra-high pressure liquid chromatography (UHPLC) equipped with ZORBAX Eclipse Plus C18 2.1 × 100 mm 1.8 µm columns (Agilent, Santa Clara CA), and lipids concentrations measured by MS-QTOF in both positive (for ceramides, sphyngomyelins, phosphatidyl-ethanolamines, phosphatidyl-cholines (PC), Lyso-PCs, diacylglycerols (DAG), triglycerides (TAG)) and negative electrospray ionization (for PI). To measure the contribution of de novo lipogenesis to intracellular fat accumulation, 10% deuterium (D2O) was added to the culture media (DMEM supplemented with 10% FBS, 1% l-Glu, 1% P/S) for 24 h [29]. The contribution of de novo lipogenesis of DAG/TAG synthesis was calculated by D2O enrichment divided by the number of hydrogens that can exchange D2O with the water pool and by%D2O added to the medium (10%) [29]. The detailed protocol is shown in the Supplementary Methods.
2.5. MBOAT7 deletion in HepG2 hepatocytes
MBOAT7−/− knockout clones were generated by exploiting the CRISPR-Cas9 technology following non-homologues end joining (NHEJ) in HepG2 hepatoma cells (ATCC—HB-8065), which are homozygous for the PNPLA3 I148M variant. Mboat7 variation has additive effect with the I148M PNPLA3 variant on the risk of NAFLD [11], [12], [13], making HepG2 an ideal model to test the impact of Mboat7 down-regulation on lipid metabolism. The detailed protocol of CRISPR-Cas9 pipeline, the characterization of MBOAT7−/− cells carrying different deletions (Δ31, Δ101 and Δ917) in homozygosis, and their sequences are summarized in the Supplementary Methods and in Fig. S1. In another experimental setting, we managed to achieve restoration of basal levels of FATP1 expression generating a double stable model of MBOAT7−/−/FATP1+/- hepatocytes (Fig. S2).
To induce lipid accumulation, visualized by Oil Red O (ORO) staining [30], cells were exposed for 24 h to a combination of palmitic (PA) and oleic (OA) acids (ratio 1:2), at the final concentration of 125 μM (Sigma-Aldrich, St Louis, MO). ORO positive areas were quantified by ImageJ software in 10 random micrographs (magnification 200x). To mimick hyper-insulinemia and insulin resistance and study the activation of mTOR signalling, cells were incubated with 0.33 μM insulin (Sanofi Aventis) alone or in combination with 0.33 mM glucose (Sigma-Aldrich, St Louis, MO) for 6 h, in DMEM supplemented with 0.5% BSA, 1% l-Glu, 1% P/S.
2.6. Statistics
For descriptive statistics, continuous variables were reported as mean and standard deviation or median and interquartile range for highly skewed biological variables. Variables with skewed distributions were logarithmically transformed before analyses. Differences between groups were calculated by two-tailed ANOVA; which was followed by post hoc t-tests adjusted for the number of comparisons when multiple groups were involved (Bonferroni correction). For lipidomic analyses, p values were adjusted by the Dunn's multiple comparisons test. Independent determinants of MBOAT7 and FATP1 mRNA levels, and of circulating fatty acids, were assessed by multivariate generalized linear model analysis. P values <0.05 (two-tailed) were considered statistically significant. Statistical analyses were performed using JMP 14.0 (SAS, Cary, NC) and Prism software (version 6, GraphPad Software). Software packages for the prediction analysis of Mboat7 promoter are listed in the Supplementary Methods section.
3. Results
3.1. Hepatic MBOAT7 down-regulation is associated with steatohepatitis and insulin resistance in obese individuals
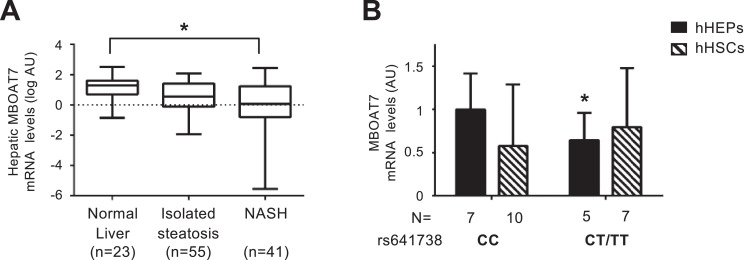
In severely obese individuals, hepatic MBOAT7 mRNA levels progressively decreased from normal liver to isolated steatosis and steatohepatitis (NASH); (p = 0.01 at ANOVA; adjusted p value=0.012 for NASH vs. normal liver; Fig. 1A). At multivariate analysis, impaired fasting glucose/T2D (IFG/T2D; p = 0.04), lobular inflammation (p = 0.005), and the rs641738 T allele (p = 0.001) were independently associated with reduced MBOAT7 (Table S5; Fig. S3A-B). In the visceral adipose tissue, MBOAT7 expression was 9-fold lower than in the liver, but IFG/T2D (p = 0.01) and the rs641738 T allele were similarly independently associated with reduced expression (p = 0.02; Table S5). These data suggest that down-regulation of hepatic MBOAT7 is associated with hepatic inflammation and insulin resistance independently of MBOAT7 genotype.
Fig. 1.
Hepatic MBOAT7 expression decreases with liver damage severity and MBOAT7 rs641738 C>T affects MBOAT7 expression in human hepatocytes.
Hepatic MBOAT7 mRNA levels were evaluated by qRT-PCR in 119 severely obese patients stratified by liver damage severity (A). Demographic, anthropometric and clinical characteristics of these patients are shown in Table S1. Boxes span from 25° to 75° percentile, while whiskers indicate the 10° and 90° percentile. Data is normalized to β-actin, log-transformed and expressed as fold increase (Arbitrary Units - AU). * adjusted p<0.05. MBOAT7 mRNA levels were evaluated by qRT-PCR in freshly isolated human hepatocytes (hHEPS) and activated human hepatic stellate cells (hHSCs, at third passage) stratified by the presence of MBOAT7 rs641738 T allele. The mRNA levels were normalized for β-actin and expressed as fold increase (Arbitrary Units - AU), as compared to CC genotype. (n = 17 hHEPs; n = 12 hHSCs) *p<0.05 compared to hHEPs or hHSCs carrying the protective CC genotype by two-tailed Student's t-tests (B).
We then surveyed the expression and regulation of MBOAT7 in different cell types. MBOAT7mRNA levels were similar between hepatocytes and HSCs. However, the rs641738 variant was associated with reduced MBOAT7 expression only in hepatocytes (p<0.05 for T+ vs. CC genotype; Fig. 1B). Additional data on MBOAT7 expression in inflammatory cells are reported in the Supplementary results and Fig. S3C.
3.2. Hepatic Mboat7 expression is down-regulated in experimental models of NAFLD
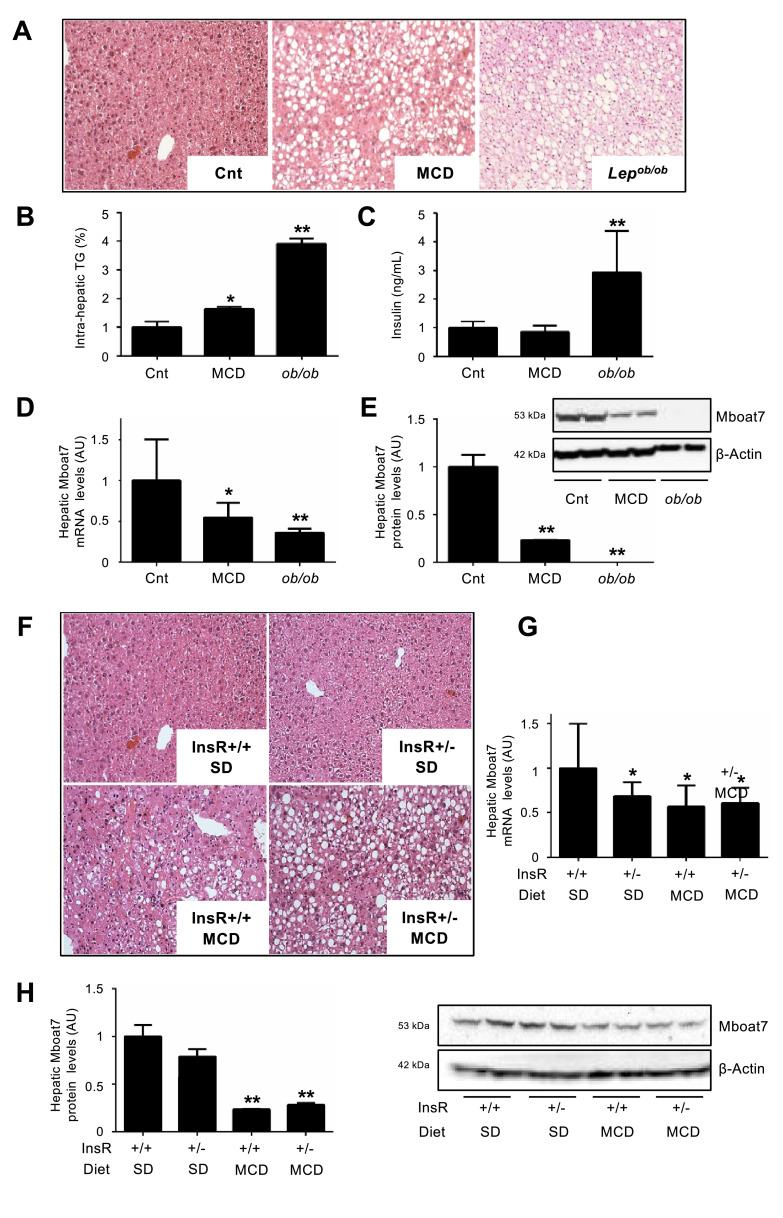
We next evaluated hepatic Mboat7 expression in two complementary mouse models of NAFLD, that is MCD, which induces NASH, but not insulin resistance, and genetically obese Lepob/ob mice, which develop severe insulin resistance without fibrosing NASH. The metabolic and biochemical features of wild-type mice on SD or MCD feeding and Lepob/ob mice are shown in Table S3. As expected, hepatic TAG content was higher in mice fed MCD compared to standard diet (p = 0.005 at ANOVA; adjusted p<0.05; Fig. 2A-B), but more so in Lepob/ob mice (adjusted p<0.01 vs. wild-type mice on SD; Fig. 2A-B). Fasting insulin, glucose and insulin resistance after insulin tolerance test were increased in Lepob/ob mice compared to wild-type mice fed standard diet or MCD (all p<0.05 at ANOVA; all adjusted p<0.05; Fig. 2C and Fig. S4A-B). In Lepob/ob mice, hyper-insulinemia was paralleled by lower hepatic protein levels of FoxA2, a key regulator of lipid metabolism, that is transcriptionally active in the fasted state (p = 0.0002 at ANOVA; adjusted p<0.01 vs. wild-type mice on SD; Fig. S4C). Hepatic Mboat7 mRNA and protein levels were reduced in MCD-fed mice (p<0.0001 and p = 0.0016, respectively at ANOVA; adjusted p<0.05 vs. SD; Fig. 2D-E), but more so in Lepob/ob mice (adjusted p<0.01 vs. SD; Fig. 2D-E). These data suggest that inflammation and hyper-insulinemia are independently associated with hepatic Mboat7 down-regulation.
Fig. 2.
Hepatic Mboat7 expression is down-regulated in experimental models of NAFLD/NASH and hyper-insulinemia.
Hematoxylin and Eosin (H&E) stain of hepatic specimens (A) and hepatic triglyceride (TG) content (B) from wild-type mice fed standard diet (referred to as Cnt) or Methionine choline deficient diet (MCD) and from Lepob/ob mice. Original magnification: 200X. Serum insulin was measured in mice fasted for 16 h (n = 10 per group) (C). Hepatic Mboat7 mRNA levels were evaluated by qRT-PCR, and mRNA levels were normalized for β-actin and are expressed as fold increase (Arbitrary Units - AU), as compared to Cnt (D). Western blot and quantification of Mboat7 protein levels in livers from Cnt, MCD and Lepob/ob mice, fasted 16 h (n = 10 per group) (E). * adjusted p<0.05, ** adjusted p<0.01 compared to Cnt. H&E stain of hepatic specimens from wild-type and InsR+/- mice fed standard diet (SD) or MCD (F). Original magnification: 200X. Hepatic Mboat7 mRNA levels were evaluated by qRT-PCR. mRNA levels were normalized for β-actin and expressed as fold increase (Arbitrary Units - AU), as compared to wild-type SD (InsR+/+ SD) (G). Western blot and quantification of Mboat7 protein levels in livers pooled from wild-type and InsR+/- MCD mice, fasted for 16 h (n = 10 per group) (H). * adjusted p<0.05; ** adjusted p<0.01 compared to InsR+/+ SD. All reactions were performed in duplicate in the same gel.
3.3. Hepatic Mboat7 down-regulation is linked to hyper-insulinemia
To evaluate the independent contribution of the metabolic state to Mboat7 regulation, we next examined a genetic model of insulin resistance. Haplo-insufficiency for Insulin receptor (InsR+/-) causes insulin resistance and hyper-insulinemia, characterized by tonic activation of the Akt signalling pathway in the liver. Metabolic and biochemical features of InsR+/+ and +/- mice on SD or MCD feeding are shown in Table S3. InsR+/- mice developed more severe steatosis compared to wild-type mice when fed MCD, whereas no macroscopic steatosis was detected on SD (Fig. 2F) [28]. Mboat7 mRNA levels were reduced in InsR+/- compared to wild-type mice, independently of the diet (adjusted p<0.05; Fig. 2G). However, Mboat7 protein levels were not significantly decreased in InsR+/- compared to wild-type mice fed a standard diet (p = 0.1; Fig. 2H), whereas they were down-regulated in both wild-type and InsR+/- mice fed MCD (p = 0.0011 at ANOVA; adjusted p <0.01 vs wild-type fed SD; Fig. 2H), suggesting that Mboat7 protein stability is modulated by steatosis/inflammation.
3.4. Hepatic Mboat7 down-regulation depends on the nutritional state and insulin levels
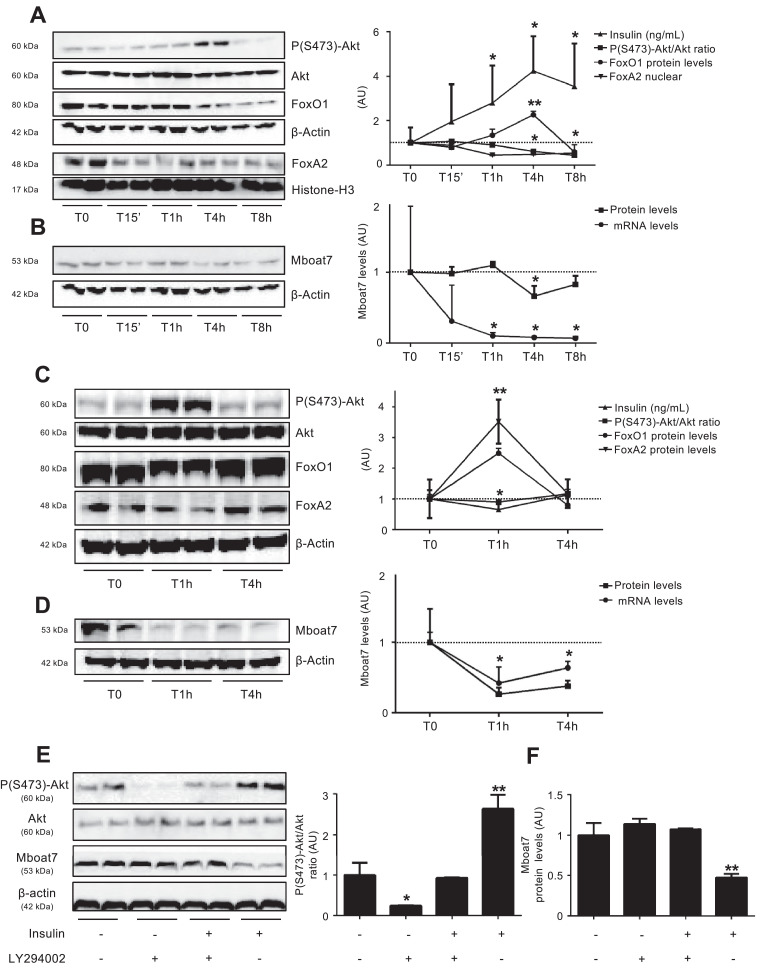
We next examined whether hepatic Mboat7 is physiologically regulated by excursions in insulin levels following fasting-refeeding cycles. Following refeeding, there was the expected rise of insulin levels (p <0.05 vs. T0; Fig. 3A) paralleled by activation of hepatic insulin signalling, as detected by an increased phospho (Ser473)-Akt/Akt ratio, (p<0.05 vs. T0; Fig. 3A) and by decreased protein levels of FoxO1/FoxA2 (p<0.05 vs. T0; Fig. 3A). This process was closely followed by down-regulation of both mRNA and protein levels of hepatic Mboat7 (p<0.05 vs. T0; Fig. 3B). After refeeding, Mboat7 mRNA levels were also down-regulated in the proximal intestine and the adipose tissue (p<0.05 vs. T0; Fig. S5A-B).
Fig. 3.
Hepatic Mboat7 expression is modulated by nutritional state and insulin.
Serum insulin (ng/mL) and Western blot and quantification of hepatic pSer473-Akt/Akt ratio, FoxO1, nuclear FoxA2 (A) and Mboat7 (B) protein levels evaluated in wild-type mice, fasted for 16 h (T0) or refed for 15 min (T15’), 1 h (T1h), 4 h (T4h) or 8 h (T8h) (n = 7 per group). FoxA2 nuclear fraction was normalized for Histone-H3 protein levels. Mboat7 mRNA levels were evaluated by qRT-PCR, normalized for β-actin and expressed as fold increase (Arbitrary Units - AU), as compared to T0. *p < 0.05, **p < 0.01 compared to T0. Western blot and quantification of hepatic pSer473-Akt/Akt ratio, FoxO1, FoxA2 (C) and Mboat7 (D) protein levels and serum insulin evaluated in wild-type mice, fasted for 16 h (T0) or 1 h (T1) and 4 h (T4) after insulin administration (0.6 U/Kg i.p, n = 7 per group). *p < 0.05; **p < 0.01 compared to T0.
Western blot and quantification of pSer473-Akt/Akt ratio (E) and Mboat7 protein levels (F) in hepatocytes isolated from wild-type mice (n = 3) and treated with insulin (0.33 μM), LY294002 (50 μM) or a combination of both for 6 h; * adjusted p < 0.05, ** adjusted p < 0.01 compared to wild-type untreated cells. At least three independent experiments were conducted. For each condition, freshly protein lysates were pooled. All reactions were performed in duplicate in the same gel.
Hepatic Mboat7 mRNA and protein down-regulation can be reproduced by insulin injection (0.6 U/Kg, by i.p. bolus) independently of feeding (p <0.05 vs. T0; Fig. 3C,D). Insulin downregulated Mboat7 also in the proximal intestine and in adipose tissue (p<0.05 vs. T0; Fig. S5C-D).
We next compared the regulation of Mboat7 to Mboat5, the closest paralogue that also catalyzes the incorporation of arachidonic acid into phospholipids, and has been involved in lipoprotein secretion [[31], [32], [33]]. Differently from Mboat7,Mboat5 was up-regulated during hyper-insulinemia, suggesting that Mboat7, but not Mboat5, down-regulation may be involved in hepatic fat accumulation in metabolic disorders (Supplementary Results and Fig. S6A–D).
3.5. Mboat7 is down-regulated by insulin signalling in primary mouse hepatocytes
To test whether Mboat7 down-regulation is a cell autonomous process dependant on activation of insulin signalling, we isolated primary hepatocytes from wild-type male mice and exposed them to insulin (0.33 μM) alone or in combination with a selective PI3K inhibitor LY294002 (50 μM). Insulin induced a 50% reduction in Mboat7 protein levels (p = 0.0009 at ANOVA; adjusted p <0.01 vs. untreated hepatocytes; Fig. 3E,F), which was completely abolished by LY294002 (adjusted p <0.01 vs. insulin-treated hepatocytes; Fig. 3E-F). Conversely, mammalian target of Rapamycin (mTOR) pharmacological inhibition did not restore Mboat7 expression (Fig. S7A). These data suggest that insulin down-regulates Mboat7 in hepatocytes in a PI3Kinase-depedent manner. Consistently, in InsR+/- hepatocytes insulin-dependant down-regulation of Mboat7 was lost (p<0.01 vs. untreated hepatocytes; Fig. S7B).
Analysis of the human Mboat7 human MBOAT7 promoter predicted the presence of FOXO1/FOXA2 binding sites, close to the transcription start site of the main MBOAT7 isoform. These binding sites are evolutionary conserved, but not in mice (Fig. S8A-B). Additional data on Mboat7 regulation by FoxO1/FoxA2 in murine hepatocytes, showing that these transcription factors are nonetheless required for insulin mediated down-regulation of Mboat7, are reported in the Supplementary Results and in Fig. S8C-F.
3.6. Acute hepatic Mboat7 down-regulation induces steatosis in mice
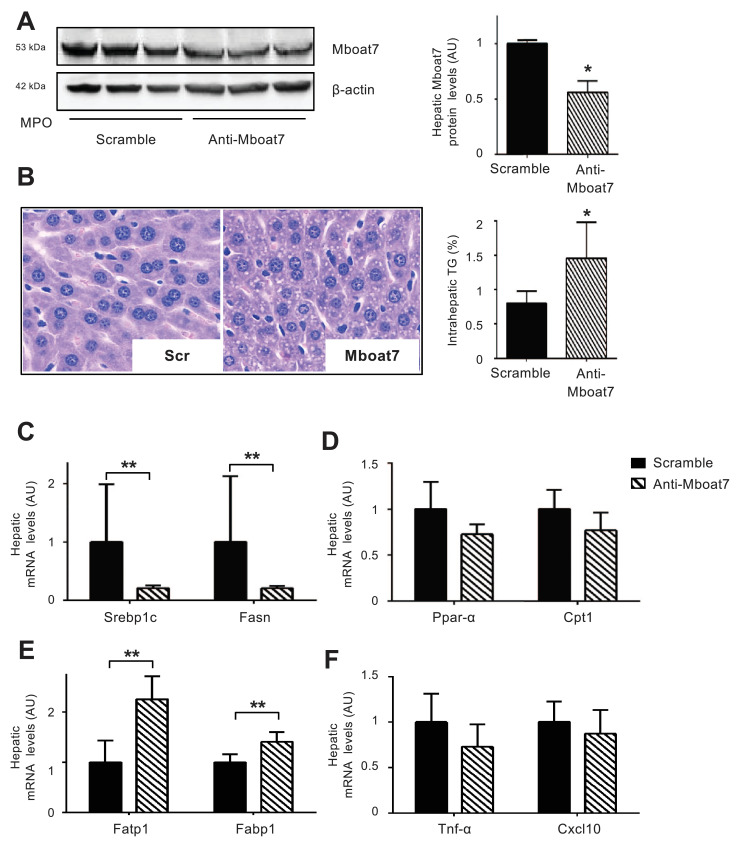
To ascertain the causal role of Mboat7 down-regulation on the development of fatty liver, we silenced Mboat7 in wild-type male mice by i.v. administration for 4 consecutive days of anti-Mboat7-MPO. The metabolic and biochemical features of mice are shown in Table S4. Anti-Mboat7-MPO induced a 45% silencing of hepatic Mboat7 (p <0.05 vs. scramble-MPO; Fig. 4A), which is comparable to that observed in individuals carrying the rs641738 risk variant. Acute down-regulation of Mboat7 led to an 80% increase in hepatic TAG content (p <0.05 vs. scramble-MPO; Fig. 4B), and to micro-vesicular steatosis development. This was associated with an alteration of the lipid species, consistent with decreased enzymatic activity (Supp. Results and Supp. File). Increased hepatic TAG concentration following Mboat7 down-regulation was not accompanied by detectable changes in circulating glucose, insulin, lipid levels, and ALT (Table S4).
Fig. 4.
In vivo silencing of hepatic Mboat7 induces TAG accumulation in hepatocellular lipid droplets Wild-type mice were treated with anti-Mboat7-MPO (12.5 mg/kg) or scramble-MPO (Scr) administered i.v. daily for 4 days (n = 6 per group) and fasted for 16 h before sacrifice. Western blot quantification of Mboat7 hepatic protein levels were normalized for β-actin; *p<0.05 compared to Scr, by two-tailed Student's t-tests. For each condition, freshly protein lysates were pooled. All reactions were performed in triplicate in the same gel (A). H&E staining and intrahepatic TAG content assessment in anti-Mboat7-MPO or scramble-MPO mice (B). Original magnification: 400X. Hepatic Srebp1 and Fasn (genes involved in de novo lipogenesis) (C), Ppar-α and Cpt1 (genes involved in β-oxidation) (D), Fatp1 and Fabp1 (fatty acids transporters) (E), Tnf-α and Cxcl10 (inflammatory markers) (F) expression was evaluated by qRT-PCR; mRNA levels were normalized to β-actin. Data are expressed as fold increase compared to Scr mice (n = 6 mice per group; *p<0.05, **p<0.01 by two-tailed Student's t-tests.
Steatosis development in mice silenced for Mboat7 was not associated with up-regulation of genes involved in lipogenesis. Indeed, despite enhanced hepatic phospho-mTOR and Raptor protein levels (p<0.01 vs. Scr-MPO; Fig. S9A), there was reduced phosphorylation of the downstream targets p70 s6 kinase and 4E-bp1 (p<0.01 vs. scramble-MPO; Fig. S9A), consistent with down-regulation of hepatic Srepb1c and Fasn mRNA levels (p<0.01 vs. scramble-MPO; Fig. 4C). On the other hand, the expression of genes regulating β-oxidation (Ppar-α and Cpt1) and VLDL export (Mttp and ApoB) was not affected (Fig. 4D; Fig. S10A). However, mRNA expression of fatty acids transporters (Fatp1 and Fabp1) was up-regulated in mice exposed to anti-Mboat7-MPO (p<0.01 and p<0.05 vs. scramble-MPO respectively; Fig. 4E). Mboat7silencing was not associated with alterations in the hepatic expression of inflammatory cytokines and chemokines (Fig. 4F). Furthermore, there was no modification in the expression of genes related to Mboat7 (Tmc4 at the same locus and Mboat5; Fig. S10B). This data supports the notion that reduced Mboat7 expression is responsible for fatty liver development in carriers of the rs641738 risk allele.
As Mboat7 was also expressed and downregulated in the adipose tissue, the impact of the decreased Mboat7 expression on circulating FFAs in vivo Mboat7 silencing and in patients carrying the rs641738 T allele is reported in the supplementary results, and Tables S4 and S6. In these experimental conditions and in selected patients, we could not rule out an impact of Mboat7 down-regulation in promoting adipose tissue lipolysis, which may contribute to hepatic fat accumulation.
3.7. MBOAT7 deletion increases intracellular fat in hepatocytes
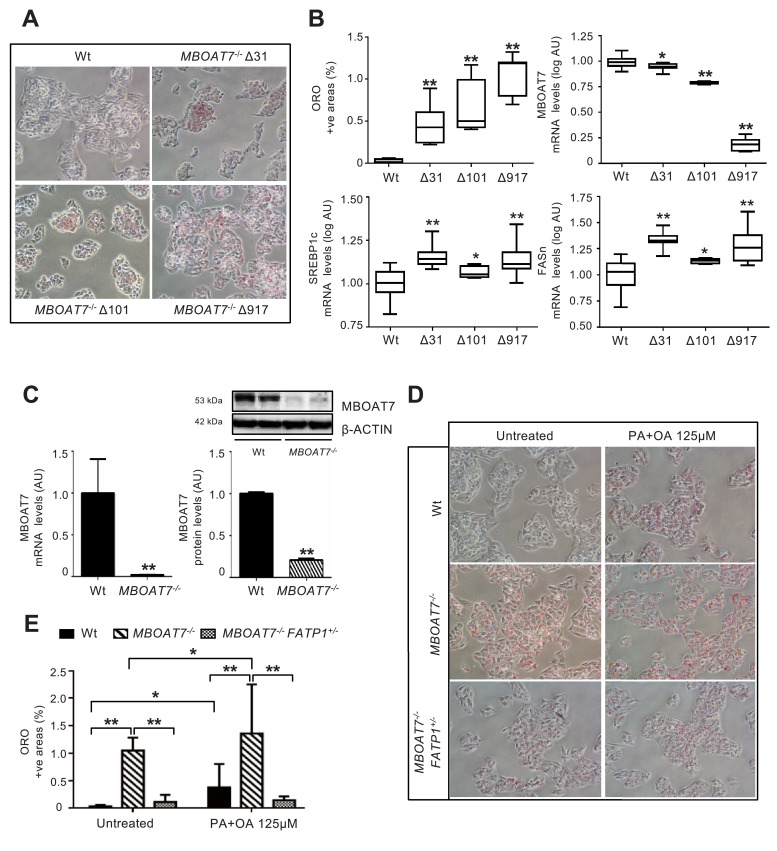
We characterized three independent HepG2 clones carrying homozygous MBOAT7 gene deletions (Δ31, Δ101 and Δ917; Fig. 5A-B and Fig. S1). Strikingly, they all displayed a spontaneous increase in intracellular fat content (p<0.0001 at ANOVA; adjusted p<0.01 vs. wild-type untreated; Fig. 5B), with the largest deletions being associated with reduced Mboat7 mRNA abundance (p<0.0001 at ANOVA; adjusted p<0.01 vs. wild-type untreated; Fig. 5B). In this model, intracellular fat accumulation was associated with induction of the lipogenic program, as detected by up-regulation of SREBP1c and FASn (p<0.0001 at ANOVA; adjusted p<0.05 vs. wild-type; Fig. 5B), and by activation of the mTOR pathway (Supp. results and Fig. S9B). In particular, MBOAT7 mRNA and protein levels were almost completely undetectable in the Δ917 clone, characterized by the most marked fat accumulation (p<0.01 vs. wild-type untreated; Fig. 5C). We therefore selected the Δ917 clone (henceforth MBOAT7−/−) for further experiments. Notably, MBOAT7−/− cells were also more susceptible to accumulate intracellular lipid droplets after exposure to a combination of fatty acids (PA+OA at a final concentration 125 μM; adjusted p<0.01 vs. wild-type exposed to PAOA 125 μM; Fig. 5D,E). These results suggest that genetically determined decrease in MBOAT7 expression/activity facilitates hepatocellular fat accumulation in a cell autonomous manner.
Fig. 5.
MBOAT7 deletion increases spontaneous fat accumulation in HepG2 hepatoma cells.
Spontaneous fat accumulation in untreated MBOAT7−/− cells carrying Δ31, Δ101 and Δ917 was evaluated by ORO staining (200X magnification) and quantified by ImageJ software in 10 random non-overlapping micrographs for each experimental condition, by calculating the percentage of pixels above the threshold value with respect to the total pixels per area. At least three independent experiments were conducted (A). Impact of MBOAT7 deletions on hepatic fat, and MBOAT7, SREBP1c, FASn mRNA, evaluated by qRT-PCR then normalized for β-actin (Arbitrary Units - AU). Boxes span from 25° to 75° percentile, while whiskers indicate the 10° and 90° percentile (B). * adjusted p<0.05, ** adjusted p<0.01 compared to wild-type (Wt) cells.
The Δ917 MBOAT7 deletion abrogates MBOAT7 expression, resulting in increased spontaneous fat accumulation, which was maintained after supplementation with palmitic and oleic acid (PA+OA 125 μM; ratio 1:2). mRNA levels were evaluated by qRT-PCR and protein levels by Western blot and then normalized for β-actin. For each condition, freshly protein lysates were pooled. All reactions were performed in duplicate in the same gel. **p<0.01 by two-tailed Student's t-tests (C). Lipid accumulation was evaluated by ORO staining (200X magnification) and quantified by ImageJ software in 10 random non-overlapping micrographs for each experimental condition, by calculating the percentage of pixels above the threshold value with respect to the total pixels per area. At least three independent experiments were conducted. * adjusted p<0.05, ** adjusted p<0.01 (D-E).
3.8. Impact of MBOAT7 deletion on intracellular lipid species in hepatocytes
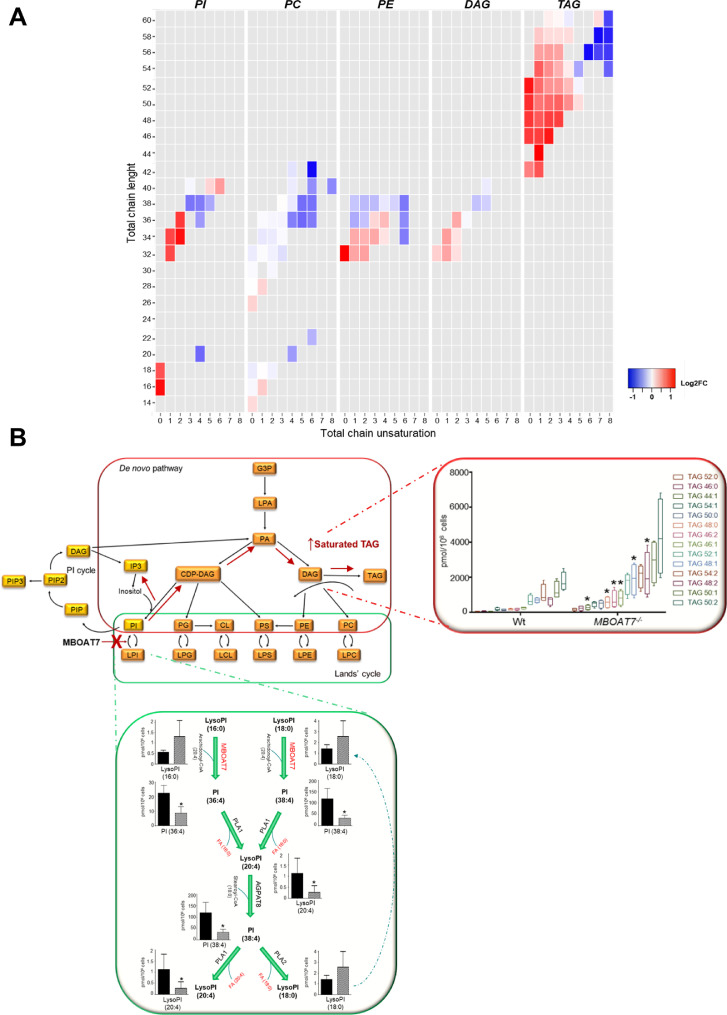
We next examined the impact of MBOAT7deletion on lipid species composition. MBOAT7−/− cells displayed a Lyso-PI/PI composition pattern suggestive of an impairment of MBOAT7 enzymatic activity (Fig. 6A, Fig. S11A-B and Supp. File). In particular, MBOAT7 deletion was associated with a pattern consistent with reduced concentration of Lyso-PI and PI conjugated with arachidonoyl-CoA (such as Lyso-PI 20:4, 36:4; 38:3; 38:5; 38:4) and increased PI (40:5) (adjusted p <0.05, Fig. 6A and Fig. S11A-B), but with a higher concentration of palmitoyl/oleyl-(Lyso) PI/(total) PI (i.e. Lyso-PI 16:0/18:0 and PI 36:2/34:1/34:2/32:1 Fig. 6A and Fig. S11A,B). In MBOAT7−/− cells there was also a reduced concentration of unsaturated phosphatidyl-ethanolamine (adjusted p<0.05 vs wild-type; Fig. 6A, Fig. S11C, and Supp. File).
Fig. 6.
Impact of MBOAT7 deletion on intracellular fat composition.
A)Heatmaps, which were generated calculating log2 fold change ratio between MBOAT7−/−/wt absolute quantification. Red shading indicates the induction and blue shading indicates repression relative to the average levels of each lipid species across all samples. PI: phosphatidyl-inositol, PC: phosphatidyl-choline, PE: phosphatidyl-ethanolamine, DAG: diacylglycerol and TAG: triacylglycerols.
B)Hypothetical model of the impact of reduced MBOAT7 activity on intracellular phospholipid metabolism in hepatocytes. The consequences of MBOAT7 downregulation are shown by the red cross and arrows. When MBOAT7 is not functional, LysoPI (20:4) cannot be generated from LysoPI (16:0) or LysoPI (18:0), which therefore accumulate, together with their precursors PI (36:4) and PI (38:4). Accumulating saturated-PIs are then shunted to the synthesis of TAGs.
CDP: cytidine-diphosphate, CL: cardiolipin, DAG: diacylglycerol, TAG: triacylglycerols, G3P: glyceraldehyde-3-phosphate, LPA: lyso-phosphatidic acid, PA: phosphatidic acid, PC: phosphatidyl-choline, PE: phosphatidyl-ethanolamine, PG: phosphatidyl-glycerol, PI: phosphatidyl-inositol, PIP: phosphatidyl-inositol-phosphate, PS: phosphatidyl-serine (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Besides the higher total DAG and TAG content, MBOAT7−/− cells displayed also a shift in their composition, with a higher concentration of saturated and mono-unsaturated TAG (i.e. TAG 50:0/50:1; adjusted p <0.05 vs. Wt; Fig. 6A, Fig. S11D and Supp. File). Similarly, in MBOAT7−/− cells we observed lower levels of unsaturated phosphatidyl-choline adjusted p<0.05 vs wild-type; Fig. 6A, Fig. S11E, and Supp. File).
To examine the relative contribution of de novo lipogenesis to the accumulation of TAG in MBOAT7−/− cells, 10% D2O was added to the culture media of wt and MBOAT7−/− cells. D2O incorporation in DAG and TAG species was higher in MBOAT7−/− cells compared to Wt (1.5-fold and 2.3-fold, respectively), especially for TAG with lower saturation (i.e. 54:2/54:1/50:2/50:1) (Fig. S12). This pattern is consistent with the heightened expression of lipogenic genes, and with reduced MBOAT7 enzymatic activity, due to the shunting of saturated-PI to the synthesis of DAG and TAG. A working model depicting the potential mechanism linking decreased MBOAT7 activity to altered intracellular lipid metabolism in hepatocytes is presented in Fig. 6B.
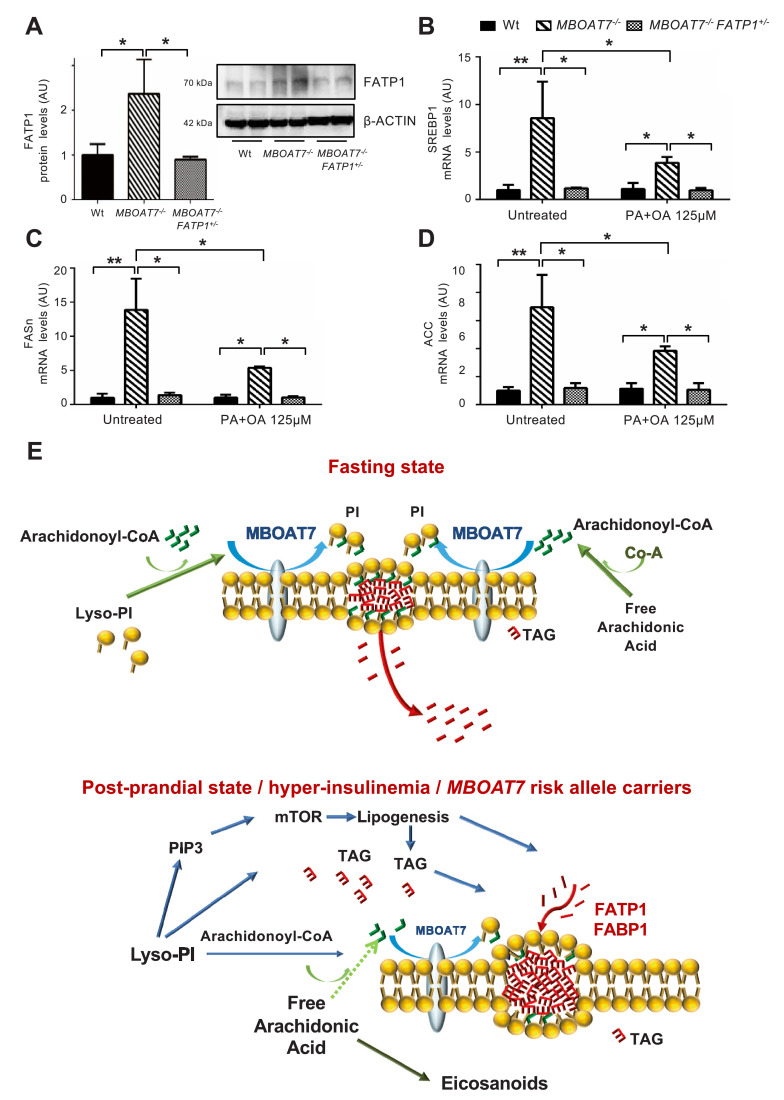
3.9. Role of FATP1 in hepatocellular fat accumulation associated with MBOAT7 down-regulation
Consistent with the results obtained following acute Mboat7 down-regulation in mice, FATP1 protein expression was up-regulated in MBOAT7−/− compared to wild-type cells (adjusted p <0.05; Fig. 7A). Therefore, we managed to achieve restoration of basal levels of FATP1 expression by hemizygous FATP1 deletion in MBOAT7−/−/FATP1+/− cells (adjusted p<0.05 vs Mboat7−/- cells; Fig. S2 and Fig. 7A). Remarkably, FATP1 down-regulation rescued the spontaneous increase in intracellular fat and reverted the induction of lipogenesis, despite persistent activation of the mTOR pathway (adjusted p<0.05 vs MBOAT7−/−; Figs. 5D,E, 7B–D, supplementary results and Fig. S11). In MBOAT7−/−/FATP1+/− cells, reduction of intracellular fat was maintained also in the presence of fatty acids that partially suppressed lipogenesis (adjusted p <0.05 vs. untreated MBOAT7−/−; Figs. 5D,E, 7B–D and Fig. S13A–C). These data suggest that FATP1 up-regulation is required to allow fat deposition related to MBOAT7 down-regulation.
Fig. 7.
Fat accumulation in MBOAT7−/− hepatocytes requires FATP1.
FATP1 protein levels were evaluated in lysates from wild-type (Wt), MBOAT7−/− and MBOAT7−/−/FATP1+/- cells by Western blot and then normalized for β-actin. For each condition, freshly protein lysates were pooled. All reactions were performed in duplicate in the same gel (A). * adjusted p<0.05 compared to Wt or MBOAT7−/− cells. In Wt, MBOAT7−/− and MBOAT7−/−/FATP1+/- cells SREBP1 (B), FASn (C) and ACC (D) gene expressions were evaluated by qRT-PCR and normalized for β-actin. * adjusted p<0.05; compared to Wt or MBOAT7−/− cells. Hypothetical mechanism(s) linking down-regulation of MBOAT7 in hepatocytes with hepatic fat accumulation (E). During fasting, MBOAT7 localizes to the endoplasmic reticulum and lipid droplets, where it conjugates arachidonoyl-CoA to the second acyl-chain of Lyso-PI allowing the incorporation into cell membranes. During hyperinsulinemia and in carriers of the rs641738 risk variant, MBOAT7 is reduced, increasing the concentration of saturated PI, which accumulate and are diverted to TAG synthesis. This process requires up-regulation of the fatty acid transporter FATP1, and in vitro is associated with increased de novo lipogenesis.
To validate these findings in patients, we evaluated the independent determinants of hepatic FATP1 mRNA expression in the Bariatric surgery cohort (Table S7). FATP1 was independently associated with the presence of T2D (adjusted p = 0.01), induction of lipogenesis (SREBP1c; adjusted p = 0.001), and inflammation (TNF-α; adjusted p<0.0001). Viceversa, it was inversely associated with Mboat7 expression (adjusted p = 0.03).
4. Discussion
In this study, we examined the relationship between down-regulation of hepatic MBOAT7 and fat accumulation. We were prompted by previous evidence that genetically determined reduction in MBOAT7 expression is associated with increased hepatic fat [11] and liver disease development [13–15]. Here we show that in obese individuals hepatic MBOAT7 is down-regulated with the severity of liver damage and insulin resistance, independently of the genetic background. Furthermore, the MBOAT7 rs641738 T risk allele was associated with reduced MBOAT7 expression in human hepatocytes and inflammatory cells, consistently with a role in facilitating hepatic fat accumulation and in priming inflammation [34].
Down-regulation of hepatic Mboat7 was confirmed in experimental models of fatty liver. The reduction was more marked during obesity and hyperinsulinemia. Furthermore, during hyper-insulinemia in InsR+/- mice down-regulation of hepatic Mboat7 mRNA preceded the development of histological steatosis. We went on to show that hepatic Mboat7 mRNA and protein levels are physiologically down-regulated during fasting-feeding cycles and by hyperinsulinemia in vivo and in vitro, depending on the activation of insulin signalling [35].
We next sought independent confirmation that Mboat7 down-regulation is causally involved in the pathogenesis of hepatic fat accumulation. Acute silencing of hepatic Mboat7 to levels similar to those observed in carriers of the rs641738 risk variant increased hepatic fat leading to the development of steatosis. The onset of steatosis was linked to an alteration of hepatic lipid composition, and to the up-regulation of Fatp1 and Fabp1, fatty acid transporters in hepatocytes. Moreover, MBOAT7 deletion in HepG2 hepatoma cells caused a spontaneous increase in intracellular fat, coupled with an alteration of PI composition consistent with reduced total MBOAT7 enzymatic activity. MBOAT7 is involved in the re-acylation of phospholipids, transferring polyunsaturated acyl-CoAs, in particular arachidonoyl-CoA, to lyso-PI and other lyso-phospholipids. The prevalent accumulation of saturated and mono-unsaturated TAG is consistent with the increased expression of lipogenic genes, and with the reduced MBOAT7 enzymatic activity, due to the shunting of saturated-PI that cannot be inserted into the intracellular membranes, to the synthesis of DAG and TAG (Fig. 6B). In addition, the predicted accumulation of phosphatidyl-inositol-3-phosphate due to the alteration of lyso-PI fluxes may favour the tonic activation of insulin-PI3K-mTOR dependant activation of SREBP1c, contributing to the concurrent stimulation of lipogenesis that we observed in vitro. Strengthening the robustness of these findings, while this manuscript was under revision, Helsley et al. independently confirmed that MBOAT7 is suppressed during insulin resistance, and that Mboat7 silencing promotes the development of fatty liver mediated by the accumulation of saturated lyso-PI [36].
An intriguing finding was that MBOAT7deletion induced FATP1 up-regulation in hepatocytes, while FATP1 hemizygous deletion in the MBOAT7−/− genetic background ameliorated the spontaneous intracellular fat accumulation and increased lipogenesis. FATP1 is expressed in insulin-sensitive tissues and translocates from intracellular compartments to the plasma membrane in response to insulin, where it favours the uptake of long-chain fatty acids [37]. In addition, FATP1 is implicated in mTOR-dependant stimulation of intracellular fat accumulation [38]. Deletion of Fatp1 in mice impaired the regulation and redistribution of circulating FFAs following refeeding, protecting against diet-induced obesity and insulin resistance [37,39]. We also observed that in obese individuals down-regulation of hepatic MBOAT7 was associated with increased FATP1 mRNA. Therefore, although we cannot rule out that FATP1 is a marker of hepatocellular fat accumulation independently of MBOAT7 expression, FATP1 induction seems required to allow fat accumulation during MBOAT7 down-regulation.
In conclusion, hyper-insulinemia, a typical feature of metabolic syndrome and post-prandial state, reduces hepatic MBOAT7 expression via activation of the insulin signalling through the insulin receptor-PI3K pathway. Reduced MBOAT7 in turn favours hepatic fat accumulation. These data suggest that MBOAT7 down-regulation is implicated in mediating the effect of chronic hyperinsulinemia on liver disease development. A working model for the mechanisms through which MBOAT7 down-regulation induces hepatic fat accumulation is presented in Fig. 7E.
Acknowledgments
Grant support
LV was supported by MyFirst Grant AIRC n.16888, Ricerca Finalizzata Ministero della Salute RF-2016–02364358, Ricerca corrente Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, LV, AF and AG. received funding from the European Union (EU) Programme Horizon 2020 (under grant agreement No. 777377) for the project LITMUS-“Liver Investigation: Testing Marker Utility in Steatohepatitis”. MM was supported by Fondazione Italiana per lo Studio del Fegato (AISF) ‘Prof. Mario Coppo’ fellowship. AG received funding from the European Union (EU) Programme Horizon 2020 under grant agreement no. 634413 for the project EPoS - “Elucidating Pathways of Steatohepatitis”. AF was supported by Ricerca Finalizzata Ministero della Salute RF-2013–02358519.
Disclosures
The authors declare no conflict of interest relevant to the present manuscript.
Author contributions
The authors’ responsibilities were as follows: MMe, study design, in vivo and in vitro studies, genotyping, gene and protein expression analysis, data analysis and interpretation, manuscript drafting, wrote and edited the manuscript; PD study design, manuscript drafting, data analysis and interpretation, reviewed and edited the manuscript; ML in vitro studies, genotyping, gene and protein expression analysis, data analysis and interpretation; FC lipidomic analysis and data analysis and interpretation; GB, RR, PS, SB, MMa, ALF data and samples collection; MMe, PD, SR, SG, NOD, AG, LV contributed to discussion, manuscript revision, data interpretation; LV study design, manuscript drafting, data analysis and interpretation, study funding, supervision and has primary responsibility for final content. All authors read and approved the final manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102658.
Appendix. Supplementary materials
References
- 1.Marchesini G., Brizi M., Bianchi G., Tomassetti S., Bugianesi E., Lenzi M. Nonalcoholic fatty liver disease: a feature of the metabolic syndrome. Diabetes. 2001;50(8):1844–1850. doi: 10.2337/diabetes.50.8.1844. [DOI] [PubMed] [Google Scholar]
- 2.Marchesini G., Brizi M., Morselli-Labate A.M., Bianchi G., Bugianesi E., McCullough A.J. Association of nonalcoholic fatty liver disease with insulin resistance. Am J Med. 1999;107(5):450–455. doi: 10.1016/s0002-9343(99)00271-5. [DOI] [PubMed] [Google Scholar]
- 3.Marra F., Gastaldelli A., Svegliati Baroni G., Tell G., Tiribelli C. Molecular basis and mechanisms of progression of non-alcoholic steatohepatitis. Trends Mol Med. 2008;14(2):72–81. doi: 10.1016/j.molmed.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Dongiovanni P., Valenti L. Genetics of nonalcoholic fatty liver disease. Metab Clin Exp. 2016;65(8):1026–1037. doi: 10.1016/j.metabol.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 5.Dongiovanni P., Meroni M. miRNA signature in NAFLD: a turning point for a non-invasive diagnosis. 2018;19(12) doi: 10.3390/ijms19123966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L.A. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dongiovanni P., Petta S., Maglio C., Fracanzani A.L., Pipitone R., Mozzi E. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61(2):506–514. doi: 10.1002/hep.27490. [DOI] [PubMed] [Google Scholar]
- 8.Stickel F., Moreno C., Hampe J., Morgan M.Y. The genetics of alcohol dependence and alcohol-related liver disease. J. Hepatol. 2017;66(1):195–211. doi: 10.1016/j.jhep.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Buch S., Stickel F., Trepo E., Way M., Herrmann A., Nischalke H.D. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and Mboat7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015;47(12):1443–1448. doi: 10.1038/ng.3417. [DOI] [PubMed] [Google Scholar]
- 10.Meroni M., Longo M. Genetic and epigenetic modifiers of alcoholic liver disease. 2018;19(12) doi: 10.3390/ijms19123857. pii: E3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mancina R.M., Dongiovanni P., Petta S., Pingitore P., Meroni M., Rametta R. The Mboat7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology. 2016;150(5):1219–1230. doi: 10.1053/j.gastro.2016.01.032. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luukkonen P.K., Zhou Y., Hyotylainen T., Leivonen M., Arola J., Orho-Melander M. The Mboat7 variant rs641738 alters hepatic phosphatidylinositols and increases severity of non-alcoholic fatty liver disease in humans. J Hepatol. 2016;65(6):1263–1265. doi: 10.1016/j.jhep.2016.07.045. [DOI] [PubMed] [Google Scholar]
- 13.Donati B., Dongiovanni P., Romeo S., Meroni M., McCain M., Miele L. Mboat7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci Rep. 2017;7(1):4492. doi: 10.1038/s41598-017-04991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thabet K., Asimakopoulos A., Shojaei M., Romero-Gomez M., Mangia A., Irving W.L. Mboat7 rs641738 increases risk of liver inflammation and transition to fibrosis in chronic hepatitis c. Nat Commun. 2016;7:12757. doi: 10.1038/ncomms12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thabet K., Chan H.L.Y., Petta S., Mangia A., Berg T., Boonstra A. The membrane-bound O-acyltransferase domain-containing 7 variant rs641738 increases inflammation and fibrosis in chronic hepatitis b. Hepatology (Baltimore, Md) 2017;65(6):1840–1850. doi: 10.1002/hep.29064. [DOI] [PubMed] [Google Scholar]
- 16.Zarini S., Hankin J.A., Murphy R.C., Gijon M.A. Lysophospholipid acyltransferases and eicosanoid biosynthesis in zebrafish myeloid cells. Prostaglandins Other Lipid Mediat. 2014;113-115:52–61. doi: 10.1016/j.prostaglandins.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gijon M.A., Riekhof W.R., Zarini S., Murphy R.C., Voelker D.R. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J Biol Chem. 2008;283(44):30235–30245. doi: 10.1074/jbc.M806194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H.C., Inoue T., Sasaki J., Kubo T., Matsuda S., Nakasaki Y. LPIAT1 regulates arachidonic acid content in phosphatidylinositol and is required for cortical lamination in mice. Mol Biol Cell. 2012;23(24):4689–4700. doi: 10.1091/mbc.E12-09-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H.C., Inoue T., Imae R., Kono N., Shirae S., Matsuda S. Caenorhabditis elegans mboa-7, a member of the mboat family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol Biol Cell. 2008;19(3):1174–1184. doi: 10.1091/mbc.E07-09-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuda S., Inoue T., Lee H.C., Kono N., Tanaka F., Gengyo-Ando K. Member of the membrane-bound O-acyltransferase (MBOAT) family encodes a lysophospholipid acyltransferase with broad substrate specificity. Genes Cells Devoted Molecular Cellular Mech. 2008;13(8):879–888. doi: 10.1111/j.1365-2443.2008.01212.x. [DOI] [PubMed] [Google Scholar]
- 21.Hirata Y., Yamamori N., Kono N., Lee H.C., Inoue T., Arai H. Identification of small subunit of serine palmitoyltransferase a as a lysophosphatidylinositol acyltransferase 1-interacting protein. Genes Cells Devoted Molecul Cellular Mech. 2013;18(5):397–409. doi: 10.1111/gtc.12046. [DOI] [PubMed] [Google Scholar]
- 22.Dongiovanni P., Donati B., Fares R., Lombardi R., Mancina R.M., Romeo S. PNPLA3 I148M polymorphism and progressive liver disease. World J Gastroenterol. 2013;19(41):6969–6978. doi: 10.3748/wjg.v19.i41.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Accili D., Drago J., Lee E.J., Johnson M.D., Cool M.H., Salvatore P. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat Genet. 1996;12(1):106–109. doi: 10.1038/ng0196-106. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto M., Pocai A., Rossetti L., Depinho R.A., Accili D. Impaired regulation of hepatic glucose production in mice lacking the Forkhead transcription factor foxo1 in liver. Cell Metab. 2007;6(3):208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Nakae J., Kitamura T., Silver D.L., Accili D. The forkhead transcription factor foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J Clin Invest. 2001;108(9):1359–1367. doi: 10.1172/JCI12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfrum C., Asilmaz E., Luca E., Friedman J.M., Stoffel M. Foxa2 regulates lipid metabolism and Ketogenesis in the liver during fasting and in diabetes. Nature. 2004;432(7020):1027–1032. doi: 10.1038/nature03047. [DOI] [PubMed] [Google Scholar]
- 27.Wolfrum C., Besser D., Luca E., Stoffel M. Proceedings of the National Academy of Sciences of the United States of America. Vol. 100. 2003. Insulin regulates the activity of forkhead transcription factor hnf-3beta/foxa-2 by akt-mediated phosphorylation and nuclear/cytosolic localization; pp. 11624–11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dongiovanni P., Meroni M., Baselli G.A., Bassani G.A., Rametta R., Pietrelli A. Insulin resistance promotes lysyl oxidase like 2 induction and fibrosis accumulation in non-alcoholic fatty liver disease. Clin Sci. 2017;131(12):1301–1315. doi: 10.1042/CS20170175. [DOI] [PubMed] [Google Scholar]
- 29.Hackl M.T., Furnsinn C., Schuh C.M., Krssak M., Carli F., Guerra S. Brain leptin reduces liver lipids by increasing hepatic triglyceride secretion and lowering lipogenesis. Nat Commun. 2019;10(1):2717. doi: 10.1038/s41467-019-10684-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicoletti A., Kaveri S., Caligiuri G., Bariety J., Hansson G.K. Immunoglobulin treatment reduces atherosclerosis in APO E knockout mice. J Clin Invest. 1998;102(5):910–918. doi: 10.1172/JCI119892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hashidate-Yoshida T., Harayama T., Hishikawa D., Morimoto R., Hamano F., Tokuoka S.M. Fatty acid remodeling by LPCAT3 enriches arachidonate in phospholipid membranes and regulates triglyceride transport. eLife. 2015;4 doi: 10.7554/eLife.06328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabir I., Li Z., Bui H.H., Kuo M.S., Gao G., Jiang X.C. Small intestine but not liver lysophosphatidylcholine acyltransferase 3 (Lpcat3) deficiency has a dominant effect on plasma lipid metabolism. J Biol Chem. 2016;291(14):7651–7660. doi: 10.1074/jbc.M115.697011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rong X., Wang B., Dunham M.M., Hedde P.N., Wong J.S., Gratton E. Lpcat3-dependent production of arachidonoyl phospholipids is a key determinant of triglyceride secretion. eLife. 2015;4 doi: 10.7554/eLife.06557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takemasu S., Ito M., Morioka S., Nigorikawa K., Kofuji S., Takasuga S. Lysophosphatidylinositol-acyltrasferase-1 (LPIAT1) is involved in cytosolic Ca(2+) oscillations in macrophages. Genes Cells Devoted Molecul Cellular Mech. 2019 doi: 10.1111/gtc.12681. [DOI] [PubMed] [Google Scholar]
- 35.Pajvani U.B., Accili D. The new biology of diabetes. Diabetologia. 2015;58(11):2459–2468. doi: 10.1007/s00125-015-3722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helsley R.N., Venkateshwari V., Brown A.L., Gromovsky A.D., Schugar R.C., Ramachandiran I. Obesity-linked suppression of membrane-bound O-Acyltransferase 7 (Mboat7) drives non-alcoholic fatty liver disease. eLife. 2019;8 doi: 10.7554/eLife.49882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Q., Ortegon A.M., Tsang B., Doege H., Feingold K.R., Stahl A. FATP1 is an insulin-sensitive fatty acid transporter involved in diet-induced obesity. Mol Cell Biol. 2006;26(9):3455–3467. doi: 10.1128/MCB.26.9.3455-3467.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arif A., Terenzi F., Potdar A.A., Jia J., Sacks J., China A. EPRS is a critical mTORC1-S6K1 effector that influences adiposity in mice. Nature. 2017;542(7641):357–361. doi: 10.1038/nature21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J.K., Gimeno R.E., Higashimori T., Kim H.J., Choi H., Punreddy S. Inactivation of fatty acid transport protein 1 prevents fat-induced insulin resistance in skeletal muscle. J Clin Invest. 2004;113(5):756–763. doi: 10.1172/JCI18917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.