Abstract
The winter tick Dermacentor albipictus (Packard) has a single-host life cycle that allows it to reach severe infestation levels on ungulates, particularly moose. Genotypic variation within these and related ticks has been a source of taxonomic confusion, although the continuity in their morphology and life history has generally been interpreted as indicating the existence of a single species. To further investigate this variation, we sequenced regions of two mitochondrial DNA (mtDNA) genes (COI and 16S rDNA), two nuclear genes (lysozyme and ITS-2), and two bacterial markers from Francisella-like endosymbionts found in these ticks (eubacterial mtDNA 16S rRNA and a homolog of Francisella tularensis [Dorofe’ev] 17-kDa lipoprotein). We sampled 42 D. albipictus individuals from whitetail and mule deer culled from three populations in east-central Alberta, as well as four D. albipictus and two Dermacentor variabilis (Say) from other locations. We then compared DNA sequence variation between the genes and related this to variation in the morphology of spiracle plates. Both mtDNA regions indicated two deeply diverged lineages (mean difference of 7.1% for COI and 4.5% for 16S) that would normally be considered diagnostic of distinct species in DNA barcoding studies. However, very little divergence was revealed by nuclear gene sequences, bacterial endosymbionts, and morphometric analyses, and any variation that did occur in these markers was not congruent with mtDNA divergences. We conclude that the sampled populations in Alberta represent a single species, D. albipictus, and reiterate the importance of integrative approaches in species delimitation.
Keywords: Dermacentor albipictus, DNA barcoding, integrative taxonomy, cryptic species
Dermacentor albipictus (Packard 1896) overwinters on its host and is commonly found on ungulates across forested areas of North America and South to Central America (Samuel 2004, Yunker et al. 1986). Severe infestations on moose may cause anemia, extensive epithelial damage, weight loss, and shedding of protective winter coats by hosts as a result of stress and excessive grooming (Anderson 2002, Glines and Samuel 1989, McLaughlin and Addison 1986). D. albipictus may also carry a variety of disease-causing organisms (Baldridge et al. 2009). Closely related tick species and even different populations within a tick species can differ in their ability to transmit pathogens (Anderson 2002, Baker 1998, Sonenshine 2009). Unfortunately, species delimitation among ticks can be difficult if only morphological traits are used. Hybridization experiments or molecular markers may be necessary to distinguish such tick species (Zahler and Gothe 1997, Zahler et al. 1995, Baker 1998, Fukunaga et al. 2000, Dergousoff and Chilton 2007).
The taxonomy of D. albipictus has been debated since Packard (1896) first formally described two forms of the winter tick as Ixodes albipictus and Ixodes nigrolineatus (later placed in the genus Dermacentor by Banks in 1907). Cooley (1938) did not consider the morphological difference between these two forms to be significant enough to warrant recognition as two species, with the difference being viewed as a result of more transparent cuticle in Dermacentor nigrolineatus.Cooley’s (1938) synonymy of D. nigrolineatus under D. albipictus was supported by their ecological similarity as one-host ticks that share the same host ranges and are active at the same time of year. Ernst and Gladney (1975) later showed that the two forms of D. albipictus could hybridize and produce viable offspring. However, recent studies of Dermacentor phylogeny based on mitochondrial 16S rDNA gene sequences have revealed variation among D. albipictus individuals that suggests the presence of a species complex (Crosbie et al. 1998).
For this project, we reassess the DNA variation reported by Crosbie et al. (1998) in the context of D. albipictus sampled primarily from east-central Alberta. We sequenced several tick genes (including mitochondrial 16s rDNA) and prokaryotic DNA markers from our specimens. We then compared DNA variation with morphometric variation in tick spiracle plates, which are structures that have typically been used for species identification in Dermacentor (Yunker et al. 1986, Keirans and Litwak 1989). We anticipated that if different species were present, then nuclear, mitochondrial, and endosymbiont DNA markers, as well as morphometric analyses, would exhibit congruent variation.
Materials and Methods
Animals.
The majority of tick specimens were collected from three localities in Alberta near the Saskatchewan border. Fourteen ticks were sampled from the vicinity of each of three areas: Chauvin (52° 41′N, 110° 08′W), Oyen (51° 21′N, 110° 29′W), and Dillberry Park (52° 36′N, 110° 05′W). These ticks were obtained from mule deer (Odocoileus hemionus [Rafinesque, 1817]) and white-tailed deer (Odocoileus virginianus [Zimmermann, 1780]), and only a single tick was used per host animal. These 42 deer were culled as part of a chronic wasting disease control program implemented by Alberta Fish and Wildlife Division during 6–22 March 2007 and 1–25 March 2008. Four other tick specimens from Edmonton (53° 31′N, 113° 30′W), one from nearby North Cooking Lake (53° 27′N, 112° 59′W) and one tick from ≈10 km east of Carmacks, Yukon (62° 05′N, 136° 05′W), were also sampled. All Dermacentor ticks were identified to species using morphological characters in the taxonomic key of Yunker et al. (1986). All ticks from Edmonton were collected off human patients that had not traveled outside the province recently. Of these four ticks, two were identified as Dermacentor variabilis.
All tick specimens were cut longitudinally, slightly off the midline, using sterilized razor blades. The larger halves of each sample were stored as vouchers in 95% EtOH at −20°C, whereas DNA was extracted from the smaller sections. Specimen remains have been deposited in the E. H. Strickland Entomological Museum, University of Alberta. Images and associated data can be viewed at http://www.biology.ualberta.ca/facilities/strickland/Vouchers/index.html.
Molecular.
Tick samples were ground up in buffer solution using autoclaved pestles. DNA was extracted using the QIAamp DNA mini kit (Qiagen, Valencia, CA).
We attempted to sequence several genetic markers, including two mitochondrial (16S rDNA and COI) and four nuclear genes: ITS-2, lysozyme (lys), ferritin (fer), and calreticulin (crt). COI is a mitochondrial DNA (mtDNA) protein-coding gene commonly used in insect molecular studies and has been proposed as an effective marker for species delimitation in animals (Hebert et al. 2003). However, ITS-2 (a nuclear gene spacer) and 16S rDNA (part of mtDNA) have previously been used in several other studies on Dermacentor phylogeny (Crosbie et al. 1998, Zahler and Gothe 1997, Zahler et al. 1995). Similarly, the nuclear protein-coding genes lys, crt, and fer have been suggested as potentially useful for phylogenetic reconstruction of Dermacentor ticks (Xu et al. 2004, 2005). We also sequenced two DNA markers from Francisella-like endosymbionts known to be present in some D. albipictus ticks (Scoles 2004, Baldridge et al. 2009): eubacterial mtDNA16S rDNA, and a homolog of Francisella tularensis 17-kDa lipoprotein. We wanted to see whether bacterial symbionts reflected tick DNA variation.
Polymerase chain reactions (PCRs) were carried out in either a 48-well Biometra T Personal Thermal Cycler (Montreal Biotech, Province of Quebec, Canada) or a 96-well 2720 Thermal Cycler (Applied Biosystems, Foster City, CA). Primers and annealing temperatures used in the PCRs followed: Murrell et al. (2001) and Folmer et al. (1994) for COI; Scoles 2004 for tick 16S rDNA, eubacterial 16S rDNA, and 17-kDa lipoprotein homolog; Dergousof and Chilton (2007) for ITS-2; Simser et al. (2004) for lys; and Xu et al. (2004, 2005) for fer and crt. The general PCR recipe was as follows: 35.5 μl of distilled water, 5 μl of 10× PCR buffer, 2 μl of MgCl2, 1 μl of dNTPs, 2 μl of each relevant primer, 2 μl of DNA template, and 0.5 μl of Taq polymerase. Variation in PCR recipe included the use of 5 μl of MgCl2 for fer amplification and 4 μl for 17-kDa lipoprotein and eubacterial 16S rDNA, accompanied by corresponding changes in the amount of distilled water to bring total PCR volume to 50 μl.
Amplified products were visualized in DNA agar or agarose gel stained with ethidium bromide and purified using the QIAgen purification kit (Qiagen, Valencia, CA). Cycle sequencing was performed using the Big Dye kit and protocol. Cycle-sequenced products were purified using sodium acetate-ethylenediaminetetraacetic acid ethanol precipitation (BigDye protocol; Applied Biosystems, Foster City, CA), and then visualized on an ABI3730 automated capillary sequencer (Applied Biosystems, Foster City, CA). All sequence chromatograms were viewed, and contigs created, using Sequencher 4.1.2 (Gene Codes, Ann Arbor, MI). Heterozygosity for lys was determined by subtracting known alleles from homozygotes, as determined by sequence chromatograms without double peaks, from heterozygous sequences that had double peaks in the sequence chromatograms (Abe et al. 2005).
Manual alignments, heuristic parsimony searches, and phylogenetic trees were generated using PAUP* version 4.0 (altivec) (Swofford 2002). Trees were rooted using outgroup sequences either from D. variabilis or more distant species obtained from GenBank (http://www.ncbi.nlm.nih.gov/Genbank/). Sequences taken from GenBank for use in our study were originally published in the studies cited above as well as in Black and Piesman (1994), Lysyk and Scoles (2008), and Dobec et al. (2009) for ticks, and Machado-Ferreira et al. (2009) for eubacterial DNA. One outgroup sequence from GenBank (AY008679) was originally reported as Anocentor nitens (Neumann), a species that is now placed in Dermacentor (Murrell et al. 2001). Unique tick haplotypes generated in our study were deposited on GenBank as accession GU968826–GU968843 for mtDNA COI, GU968848–GU968861 for 16S rDNA mtDNA, GU968847 for ITS-2 nuclear DNA, and GU968844–GU968846 for lys nuclear DNA. For bacterial symbionts, new sequences deposited on GenBank from our study include GU968873–GU968877 for F. tularensis 17-kDa lipoprotein homolog and GU968862–GU968872 for Francisella-like endosymbiont (FLE) eubacterial 16S rDNA.
We compared our 16S rDNA sequence results against those of Crosbie et al. (1998) by incorporating 16S rDNA sequences from their sequence figure and GenBank into a single file for heuristic parsimony analysis using PAUP* version 4.0 (altivec) (Swofford 2002). Matrices and the associated trees for each locus were submitted to TreeBASE (www.treebase.org; accession no. S2671).
Genetic structuring of mtDNA variation among sampling localities was evaluated using analysis of molecular variance (AMOVA) (nucleotide variation and F-statistics [Fst]) in Arlequin 3.1.1.1 (Excoffier et al. 2005).
Morphometrics.
Although scutum coloration was used by Packard (Cooley 1938) to distinguish D. albipictus from D. nigrolineatus, ethanol preservation of our specimens rendered color-based discrimination unreliable. We therefore based our morphometric analyses on spiracle plate shape and goblet cell sizes, which are characters widely used for identifying species in Dermacentor (Yunker et al. 1986, Keirans and Litwak 1989).
Forty-three tick specimens were classified into three groups depending on the shape of their spiracle plates. Round spiracle shape was defined as appearing round or oval in shape with no presence of prolongation on its posterior-dorsal region (Fig. 1). Intermediate spiracle plate shape was mostly round or oval, but had a slight prolongation on its posterior-dorsal region compared with a round spiracle plate. Prolonged spiracle plate shape had a distinct prolongation on its posterior-dorsal region compared with a round or intermediate spiracle plate. Digital images were taken using a Nikon COOLPIX 8400 digital camera mounted on a Wild Heerbrugg M5-40617 dissecting microscope (Switzerland). All images were taken at constant magnification. Images were not taken for three of the D. albipictus specimens as they were engorged and difficult to orient to obtain good images.
Fig. 1.
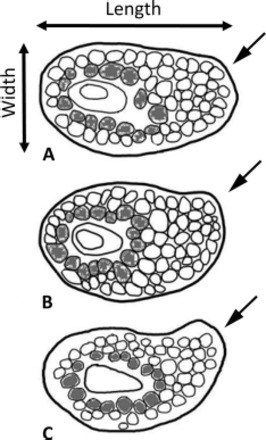
Representative examples of three categories of spiracle plate shape in D. albipictus: (A) round, (B) intermediate, and (C) prolonged. Plates are oriented so that the macula is horizontal. Arrows in the figure point to the region of the plate where prolongation is expected. Darker cells surrounding the macula indicate the cells counted to estimate goblet cell size.
The outlines of spiracle plates were traced using ImageJ (Abramoff et al. 2004), and the images were superimposed for comparison using the downloadable imaging freeware GNU Image Manipulation Program 2.6.4. (http://www.gimp.org/). Specimens were then scored on the basis of the shape of their spiracle plates (see Fig. 1). ImageJ (Abramoff et al. 2004) was also used to take the following measurements: 1) overall area of spiracle plate (measured in pixels at a uniform focal distance), 2) length of spiracle plate, and 3) width of spiracle plate. The ratio of length to width was used as a simplified quantification of the extent of prolongation in the spiracle plate. In addition, a count of the number of goblet cells in the first layer of cells immediately around the macula was used to assess relative goblet cell size, with a smaller number of cells usually indicating larger goblet cell size. Scatter plots were used to visually compare spiracle plate area and spiracle plate length with goblet cell counts, and linear regression analysis was used to observe whether overall spiracle plate sizes affect goblet cell counts. All quantitative data were input and analyzed using Microsoft Excel 2007 (Microsoft, Redmond, WA).
Results
Tick DNA.
DNA sequences for the 658-bp region of COI gave 18 unique COI haplotypes for the 46 D. albipictus specimens. A heuristic parsimony search of COI mtDNA sequences gave trees exhibiting two distinct lineages separated by deep divergence (COI lineages 1 and 2) (Fig. 2). COI lineage 2 was more closely related to Dermacentor nitens than it was to COI lineage 1 when rooted with D. variabilis and more distant Dermacentor species (Fig. 2). The average nucleotide difference between lineages 1 and 2 was 7.09% (with mean variation 3.23% between specimens within lineage 1 and 0.58% within lineage 2). When only unique haplotypes were considered, average nucleotide difference between the lineages was 7.48% (with 3.09% mean variation within lineage 1 and 0.92% within lineage 2). No haplotype was restricted to a single locality. Fst values for COI mtDNA sequences, obtained via AMOVA analysis, indicated that there was no genetic structuring between the different localities from which samples were obtained (Fst = −0.039, P = 0.55).
Fig. 2.
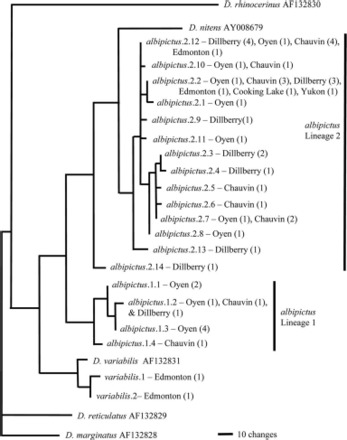
Parsimony tree showing relationship between unique COI haplotypes. The number of specimens for each combination of locality and haplotype is indicated in brackets.
A 408-bp region of 16S rDNA mtDNA sequenced from 18 D. albipictus specimens representing each of the 18 COI mtDNA haplotypes revealed 14 unique haplotypes (Fig. 7). A heuristic parsimony search of these sequences gave two distinct lineages, as for COI. The average nucleotide difference between the two 16S mtDNA lineages was 4.49%, with a mean variation of 0.00% within lineage 1 and 1.07% within lineage 2. Like COI mtDNA, AMOVA analysis for 16S rDNA indicated no genetic structuring between the different sampling localities (Fst = −0.10, P = 0.74).
Fig. 7.

Parsimony tree showing relationships between 16S rDNA haplotypes obtained for our study and those of Crosbie et al. (1998). Haplotype labels for sequences from our study are as in Fig. 2. Haplotypes from Crosbie et al. (1998) are labeled as in their study, with location in brackets. Additional sequences taken from GenBank show only the accession number.
A parsimony tree obtained from incorporation of our 16S mtDNA sequences with those of Crosbie et al. (1998) revealed two distinct lineages. Our 16S mtDNA lineage 1 sequences and the sequence from a D. albipictus tick from southeast Washington (Crosbie et al. 1998) formed a monophyletic group that was closely related to D. nitens. However, 16S mtDNA sequences from the other three D. albipictus from the study by Crosbie et al. (1998) were nested among our sequences for mtDNA lineage 2 (Fig. 7).
Sequences for lys and ITS-2 were obtained from the same 18 specimens for which 16S rDNA were sequenced. However, repeated attempts to obtain good sequences for crt and fer were unsuccessful, and these two markers were abandoned.
A single allele sequence was obtained for a 313-bp region of the ITS-2 gene for all 18 specimens of D. albipictus. This sequence was identical to one published on GenBank (accession M498348) (Fig. 3A). There was an interspecific divergence of 5.11% between ITS-2 sequences obtained for D. variabilis and D. albipictus.
Fig. 3.
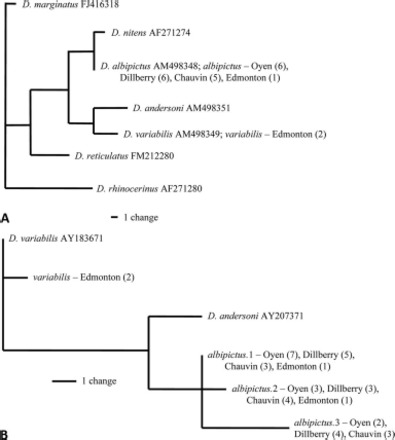
(A) Parsimony tree of ITS-2 nuclear sequences for 18 D. albipictus specimens and outgroups. (B) Parsimony tree for lys nuclear alleles in Dermacentor species. Numbers in brackets give the number of specimens with each haplotype.
The 512-bp lys nuclear gene sequences generated in our study were very similar to those from D. variabilis and Dermacentor andersoni available on GenBank (accession AY183671 and AY207371, respectively). We obtained three unique lys alleles in 18 D. albipictus specimens sequenced (giving 36 alleles for this diploid, nuclear locus), and heuristic parsimony analysis revealed no deep divergences among these alleles (Fig. 3B). The lys sequences obtained in our study included an insertion (370 bp in D. variabilis and 435 bp in D. albipictus) that was not present in the published lys sequences derived from cDNA for D. variabilis and D. andersoni. Of the 18 D. albipictus specimens, 10 were homozygous, whereas eight were heterozygous. There was one nucleotide substitution between lys alleles 1 and 2, three between lys alleles 1 and 3, and four between lys alleles 2 and 3.
Endosymbiont DNA.
Amplification of the 17-kDa lipoprotein gene homolog and 16S rDNA of FLE was attempted for all specimens available. Of the 46 D. albipictus surveyed, we obtained DNA sequences for 17-kDa lipoprotein homolog and eubacterial 16s rRNA from 16 specimens for each gene, but both genes were amplified in only 12 specimens. Five unique haplotypes were obtained for the F. tularensis 17-kDa lipoprotein homolog, whereas 11 unique haplotypes were obtained for FLE 16S rDNA. Unlike mtDNA, parsimony analysis of endosymbiont sequences (748-bp region of 17-kDa lipoprotein homolog, and 599-bp region of FLE 16S rDNA) revealed no major divergences in endosymbiont sequences (Figs. 4 and 5). The sequences also showed no relationship with mtDNA lineage. Furthermore, albipictus.1 haplotype for FLE was identical to FLE sequences obtained from D. variabilis and D. andersoni (GenBank accession AY375404 and AY375396, respectively) (Fig. 5), indicating no species-specific association between FLE sequences and the D. albipictus ticks surveyed in our study.
Fig. 4.
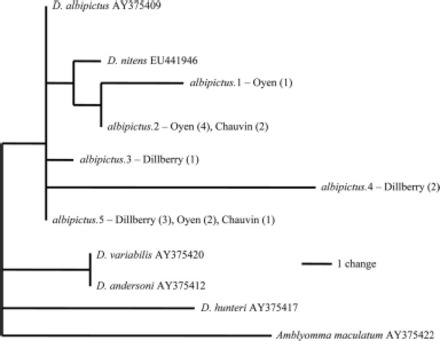
Parsimony tree showing relationship between five unique F. tularensis 17-kDa lipoprotein haplotypes. Numbers in brackets indicate number of specimens.
Fig. 5.

Parsimony tree of FLE eubacteria 16S rDNA from Dermacentor. Numbers in brackets indicate number of specimens.
Morphometrics.
The number of D. albipictus ticks that exhibited each of the three spiracle plate shapes is summarized in Fig. 6A. There was no observed association between spiracle plate shapes and tick gender. There was no significant difference in the proportions of mitochondrial lineage types relative to spiracle plate types (χ2 = 0.174, P = 0.917). A plot of mitochondrial lineage as a function of length/width ratios of spiracle plates against frequency revealed no obvious break in the distribution of ratios among the tick specimens (Fig. 6B). Similarly, spiracle plate area and the number of goblet cells immediately around the macula exhibited unimodal distributions that did not correspond with mtDNA lineages (Fig. 6, C and D). Linear regression analyses revealed no correlation between overall spiracle plate area (R2 = 0.023) and spiracle plate length with goblet cell counts (R2 = 0).
Fig. 6.
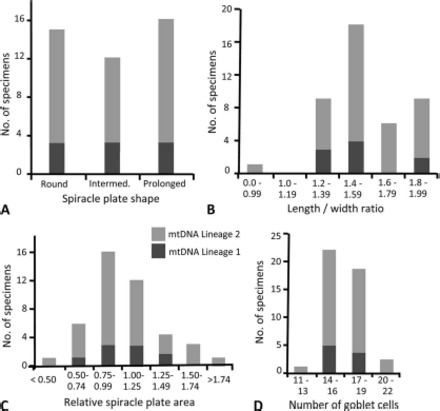
Counts of D. albipictus specimens with each mtDNA lineage as a function of (A) spiracle plate shape, (B) spiracle plate length: width ratio, (C) spiracle plate area, and (D) goblet cell number.
Discussion
Although previous studies of the morphology, life history, and geographical range of D. albipictus were taken as evidence for the conspecificity of forms that had originally been described as distinct species by Packard in 1896 (Cooley 1938, Ernst and Gladney 1975), a more recent survey of 16S mtDNA sequence variation in Dermacentor suggested that D. albipictus comprises at least two genetically distinct species (Crosbie et al. 1998). The mtDNA sequences generated in our study also showed two distinct lineages (Figs. 2 and 7), both of which were represented at each of our major sampling localities. Parsimony analysis of our 16S rDNA sequences combined with those of Crosbie et al. (1998) revealed the same two distinct 16S mtDNA lineages in both studies (Fig. 7). Thus, this divergent mtDNA variation in D. albipictus is geographically widespread, extending from Alberta to at least Washington state.
Unlike mtDNA, D. albipictus nuclear DNA sequences, endosymbiont gene sequences, and spiracle plate characters demonstrated little variation, and the minor variation that did exist showed no association with mitochondrial lineage differences. In a similar study on species limits of Ixodes didelphidis Fonseca & Aragão and Ixodes loricatus Neumann by Labruna et al. (2002), the authors concluded that the two tick species were conspecific because intraspecific ITS-2 sequence variation between these two putative species fell within 1.3-4.9% and there were no visually distinct differences in spiracle plate morphology. Analogously, taking all mitochondrial sequences, nuclear markers, bacterial sequences, and morphological analysis into account, we conclude that all D. albipictus sample in our study belong to a single species, despite the relatively high sequence divergence between the mitochondrial lineages 1 and 2 (COI = 7.48%, 16S rDNA = 4.49%).
The large amount of intraspecific variation in D. albipictus mtDNA begs explanation. Previous studies of currently accepted Dermacentor species have shown that distinct tick species are nonetheless capable of interspecific hybridization (Dergousoff and Chilton 2007, Oliver et al. 1972, Ernst and Gladney 1975). However, if ongoing hybridization is the cause of the divergent mtDNA lineages in D. albipictus, we would expect to observe distinct heterogeneity in nuclear gene sequences and other markers (Labruna et al. 2002, Baker 1998, Zahler and Gothe 1997, Zahler et al. 1995). In our study, nuclear gene sequences for ITS-2 were uniform, and for lys showed relatively little variation in D. albipictus (Fig. 3, A and B). Structural variation in D. albipictus spiracles showed no relationship to mtDNA lineage variation (Fig. 6). Endosymbiont sequence variation revealed no consistent differences between D. albipictus mtDNA lineages or even between well-accepted species such as D. variabilis and D. andersoni (Figs. 4 and 5). These shared endosymbiont markers are most simply explained as being the result of cross-infection via shared vertebrate hosts rather than hybridization between species.
Introgression may potentially explain the much larger divergences in D. albipictus mtDNA compared with nuclear DNA. It is conceivable that mtDNA from another unidentified Dermacentor species may have been selectively incorporated into D. albipictus. One candidate for such a species is D. nitens, for which 16S sequences from Idaho (Crosbie et al. 1998) were most similar to D. albipictus mtDNA lineage 1 (Fig. 7). For COI, D. nitens COI sequences sampled from an unspecified location in a study by Murrell et al. (2001) were most similar to D. albipictus lineage 2 (Fig. 2). ITS-2 sequences for D. nitens were the most similar to D. albipictus among the seven Dermacentor species sampled (Fig. 3A). No lys sequence was available for D. nitens, and variation in the bacterial endosymbionts was not informative at this level (Figs. 4 and 5). However, variation in spiracle structure gives a very different picture, with D. nitens being characterized by such divergent spiracles and other morphological features that this species was placed in a separate genus, Anocentor, by Borges et al. (1998). All D. albipictus specimens in our study had at least 11 and usually >14 goblet cells surrounding the macula (Fig. 6D), whereas D. nitens typically has fewer and larger goblet cells in a round spiracle, and is found only in tropical or subtropical regions (Yunker et al. 1986). Nonetheless, it is intriguing to consider that D. albipictus and D. nitens share at least one important biological feature, their one-host life cycle, which is unusual within Dermacentor.
Retained ancestral polymorphism is a third potential explanation for the incongruent pattern of divergence between mtDNA, nuclear markers, and spiracle plate structures in D. albipictus. A gene phylogeny is not necessarily congruent with a species phylogeny, and so retained ancestral polymorphism may maintain divergent mtDNA lineages that are not reflected in any other traits, thereby also contributing to species misidentification (Funk and Omland 2003, Rich et al. 1997). However, this does not explain the strong similarity between D. nitens and D. albipictus in both mtDNA and ITS-2, a fast-evolving nuclear gene.
Because of the deep divergence present in the mtDNA sequences obtained, the ticks in our study would probably have been delimited as two different species if identification had been based solely on mtDNA sequences. Similarly, a study by Lysyk and Scoles (2008) revealed extensive mtDNA sequence variability between the prairie and montane populations of D. andersoni in western Canada. Yet, mating experiments carried out in the same study found little reproductive incompatibility between the two populations. As a result, the two populations of D. andersoni were not considered distinct species.
One of the challenges for delimiting species via molecular divergence is the difficulty in setting an overall threshold for species recognition. Hebert et al. (2003) considered any mtDNA COI sequences in Lepidoptera with 3% or more nucleotide divergence to indicate a distinct species. However, a survey of studies of inter- and intraspecific variation across several insect orders has shown that COI mtDNA sequence differences can vary substantially between closely related insect species, with up to 26.0% intraspecific compared with 0–30.7% interspecific sequence variation (Cognato 2006). For species delimitations, this demonstrates that reliance on a standard divergence threshold in a single genetic marker, particularly mtDNA (Galtier et al. 2009), is highly vulnerable to errors (Sperling and Roe 2009, Cognato 2006). Our study thus supports the value of integrative multicharacter approaches to species identification (Roe and Sperling 2007, Schlick-Steiner et al. 2009).
Based on the results from nuclear molecular markers and morphological analyses in our study, D. albipictus consists of a single species, not a species complex as Crosbie et al. (1998) suggested, although mtDNA sequence varies substantially among individuals. This mtDNA variation is most plausibly explained by introgression, but could also be a result of retained ancestral polymorphism. Improved identification and species delimitation of ticks are important because they can allow better understanding of variation in the effects of these arthropods on the health of their hosts.
Acknowledgements
We thank W. Samuel, M. Larsen, P. Merchant, G. Pohl, Beck Veterinary Clinic, the City of Edmonton Environmental Services Laboratory, and the Alberta Fish and Wildlife Chronic Wasting Disease field crew members for providing tick specimens. We thank S. Bromilow for providing training in molecular methods and analyses, D. Shpeley for assistance with morphometric imaging, T. Lysyk for advice on tick identification and literature, and the associate editor and an anonymous reviewer for constructive comments. Many thanks go to other members of the Sperling Laboratory for their assistance. This project was funded by a Natural Sciences and Engineering Research Council Discovery Grant to F.A.H.S.
References Cited
- Abe T. A., Spence J. R., Sperling F.A.H. 2005. Mitochondrial introgression is restricted relative to nuclear markers in a water strider (Hemiptera: Gerridae) hybrid zone. Can. J. Zool. 83: 432–444. [Google Scholar]
- Abramoff M. D., Magelhaes P. J., Ram S. J. 2004. Image processing with Image. J. Biophotonics Int. 11: 36–42. [Google Scholar]
- Anderson J. F. 2002. The natural history of ticks. Med. Clin. N. Am. 86: 205–218. [DOI] [PubMed] [Google Scholar]
- Baker S. C. 1998. Distinguishing species and populations of rhipicephaline ticks with ITS 2 ribosomal RNA. J. Parasitol. 84: 887–892. [PubMed] [Google Scholar]
- Baldridge G. D., Scoles G. A., Burkhardt N. Y., Schloeder B., Kurtti T. J., Munderloh U. G. 2009. Transovarial transmission of Francisella-like endosymbionts and Anaplasma phagocytophilum variants in Dermacentor albipictus (Acari: Ixodidae). J. Med. Entomol. 46: 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black W. C., IV, Piesman J. 1994. Phylogeny of hardand soft-tick taxa (Acari: Ixodida) based on mitochondrial 16S rDNA sequences. Proc. Natl. Acad. Sci. USA 91: 10034–10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges L.M.F., Labruna M. B., Linardi P. M., Ribeiro M.F.B. 1998. Recognition of the tick genus Anocentor Schultze, 1937 (Acari: Ixodidae) by numerical taxonomy. J. Med. Entomol. 35: 891–894. [DOI] [PubMed] [Google Scholar]
- Cognato A. I. 2006. Standard percent DNA sequence difference for insect does not predict species boundaries. J. Econ. Entomol. 99: 1037–1045. [DOI] [PubMed] [Google Scholar]
- Cooley R. A. 1938. The genera Dermacentor and Otocentor (Ixodidae) in the United States, with studies in variation. Natl. Inst. Health Bull. 171: 59–64. [Google Scholar]
- Crosbie P. R., Boyce W. M., Rodwell T. C. 1998. DNA sequence variation in Dermacentor hunteri and estimated phylogenies of Dermacentor spp. (Acari: Ixodidae) in the New World. J. Med. Entomol. 35: 277–288. [DOI] [PubMed] [Google Scholar]
- Dergousoff S. J., Chilton N. B. 2007. Differentiation of three species of Ixodid tick, Dermacentor andersoni, D. variabilis and D. albipictus, by PCR-based approaches using markers in ribosomal DNA. Mol. Cell. Probes 21: 343–348. [DOI] [PubMed] [Google Scholar]
- Dobec M., Golubic D., Punda-Polic V., Kaeppeli F., Sievers M. 2009. Rickettsia helvetica in Dermacentor reticulatus ticks. Emerg. Infect. Dis. 15: 98–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst S. E., Gladney W. J. 1975. Dermacentor albipictus: hybridization of the two forms of the winter tick. Ann. Entomol. Soc. Am. 68: 63–67. [Google Scholar]
- Excoffier L., Laval G., Schneider S. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3: 294–299. [PubMed] [Google Scholar]
- Fukunaga M., Yabuki M., Hamase A., Oliver J. H., Nakao M. 2000. Molecular phylogenetic analysis of Ixodid ticks based on the ribosomal DNA spacer, internal transcribed spacer 2, sequences. J. Parasitol. 86: 38–43. [DOI] [PubMed] [Google Scholar]
- Funk D. J., Omland K. E. 2003. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Syst. 34: 397–423. [Google Scholar]
- Galtier N., Nabholz B., Glemin S., Hurst G.D.D. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 18: 4541–4550. [DOI] [PubMed] [Google Scholar]
- Glines M. V., Samuel W. M. 1989. Effect of Dermacentor albipictus (Acari: Ixodidae) on blood composition, weight gain and hair coat of moose, Alces alces. Exp. Appl. Acarol. 6: 197–213. [DOI] [PubMed] [Google Scholar]
- Hebert P.D.N., Cywinska A., Ball S. L., deWaard J. 2003. Biological identifications through DNA barcodes. Proc. R. Soc. London Ser. B 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keirans J. E., Litwak T. R. 1989. Pictorial key to the adults of hard ticks, Family Ixodidae (Ixodida: Ix-odoidea), east of the Mississippi River. J. Med. Entomol. 26: 435–448. [DOI] [PubMed] [Google Scholar]
- Labruna M. B., Marrelli M. T., Heinemann M. B., Fava A. B., Cortez A., Soares R. M., Sakamoto S. M., Richtzenhain L. J., Marinotti O., Schumaker T.T.S. 2002. Taxonomic status of Ixodes didelphidis (Acari: Ixodidae). J. Med. Entomol. 39: 135–142. [DOI] [PubMed] [Google Scholar]
- Lysyk T. J., Scoles G. A. 2008. Reproductive compatibility of prairie and montane populations of Der, macentor andersoni. J. Med. Entomol. 45: 1064–1070. [DOI] [PubMed] [Google Scholar]
- Machado-Ferreira E., Piesman J., Zeidner N. S., Soares C. A. 2009. Francisella-like endosymbiont DNA and Francisella tularensis virulence-related genes in Brazilian ticks (Acari: Ixodidae). J. Med. Entomol. 46: 369–374. [DOI] [PubMed] [Google Scholar]
- McLaughlin R. F., Addison E. M. 1986. Tick (Dermacentor albipictus)-induced winter hair-loss in captive moose (Alces alces). J. Wildl. Dis. 22: 502–510. [DOI] [PubMed] [Google Scholar]
- Murrell A., Campbell N.J.H., Barker S. C. 2001. A total-evidence phylogeny of ticks provides insights into the evolution of life cycles and biogeography. Mol. Phylogenet. Evol. 21: 244–258. [DOI] [PubMed] [Google Scholar]
- Oliver J. H., Jr., Wilkinson P. R., Kohls G. M. 1972. Observations on hybridization of three species of North American Dermacentor ticks. J. Parasitol. 58: 380–384. [PubMed] [Google Scholar]
- Rich S. M., Rosenthal B. M., Telford S. R., Spielman A., Hartl D. L., Ayaia F. J. 1997. Heterogeneity of the internal transcribed spacer (ITS-2) region within individual deer ticks. Insect Mol. Biol. 6: 123–129. [DOI] [PubMed] [Google Scholar]
- Roe A. D., Sperling F.A.H. 2007. Population structure and species boundary delimitation of cryptic Dioryctria moths: an integrative approach. Mol. Ecol. 16: 3617–3633. [DOI] [PubMed] [Google Scholar]
- Samuel W. M. 2004. White as a ghost: winter ticks and moose. Federation of Alberta Naturalists, Edmonton, Alberta, Canada. [Google Scholar]
- Schlick-Steiner B. C., Steiner F. M., Seifert B., Stauffer C., Christian E., Crozier R. H. 2009. Integrative taxonomy: a multisource approach to exploring biodiversity. Annu. Rev. Entomol. 55: 421–435. [DOI] [PubMed] [Google Scholar]
- Scoles G. A. 2004. Phylogenetic analysis of the Francisellalike endosymbionts of the Dermacentor ticks. J. Med. Entomol. 41: 277–284. [DOI] [PubMed] [Google Scholar]
- Simser J. A., Macaluso K. R., Mulenga A., Azad A. F. 2004. Immune-responsive lysozymes from hemocytes of the American dog tick, Dermacentor variabilis and an embryonic cell line of the Rocky Mountain wood tick, D. andersoni. Insect Biochem. Mol. Biol. 34: 1235–1246. [DOI] [PubMed] [Google Scholar]
- Sonenshine D. E. 2009. Ticks, pp. 1003–1011. InEncyclopedia of insects. Elsevier, San Diego, CA. [Google Scholar]
- Sperling F.A.H., Roe A. D. 2009. Molecular dimensions of insect taxonomy, pp. 397–415. InFoottit R., Adler P. (eds.), Insect biodiversity: science and society. Blackwell Publishing, West Sussex, United Kingdom. [Google Scholar]
- Swofford D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland, MA. [Google Scholar]
- Xu G., Fang Q. Q., Keirans J. E., Durden L. A. 2004. Ferritin gene coding sequences are conserved among eight hard tick species (Ixodida: Ixodidae). Ann. Entomol. Soc. Am. 97: 567–573. [Google Scholar]
- Xu G., Fang Q. Q., Sun Y., Keirans J. E., Durden L. A. 2005. Hard tick calreticulin (CRT) gene coding regions have only one intron with conserved positions and variable sizes. J. Parasitol. 9: 1326–1331. [DOI] [PubMed] [Google Scholar]
- Yunker C. E., Keirans J. E., Clifford C. M., Easton E. R. 1986. Dermacentor ticks (Acari: Ixodoidea: Ixodidae) of the New World: a scanning electron microscope atlas. Proc. Entomol. Soc. Wash. 88: 609–627. [Google Scholar]
- Zahler M., Gothe R. 1997. Evidence for the reproductive isolation of Dermacentor marginatus and Dermacentor reticulatus (Acari: Ixodidae) ticks based on cross-breeding, morphology and molecular studies. Exp. Appl. Acarol. 21: 685–696. [PubMed] [Google Scholar]
- Zahler M., Gothe R., Rinder H. 1995. Genetic evidence against a morphologically suggestive conspecificity of Dermacentor reticulatus and D. marginatus (Acari: Ixodidae). J. Parasitol. 25: 1413–1419. [DOI] [PubMed] [Google Scholar]


