Abstract
A new method for synthesizing deoxynucleic guanidine (DNG) oligonucleotides that uses iodine as a mild and inexpensive coupling reagent is reported. This method eliminates the need for the toxic mercury salts and pungent thiophenol historically used in methods aimed at preparing DNG oligonucleotides. This coupling strategy was readily translated to a standard MerMade 12 oligonucleotide synthesizer with coupling yields of 95% and has enabled the synthesis of a 20-mer DNG oligonucleotide, the longest DNG strand to date, in addition to mixed DNA–DNG sequences with 3–9 DNG inserts. Importantly, DNG oligonucleotides exhibit robust unaided cellular uptake as compared to unmodified oligonucleotides without apparent cellular toxicity. Taken together, these findings should greatly increase the accessibility of cationic backbone modifications and assist in the development of oligonucleotide-based drugs.
Graphical Abstract

INTRODUCTION
The programmable base-pairing of nucleic acids makes them attractive targets for biological applications such as diagnostic platforms, intracellular detection agents, drug delivery vehicles, and therapeutics.1,2 However, these applications are often limited by the polyanionic backbone of oligonucleotides, which suffers from high nuclease sensitivity and poor cellular uptake.3 Synthetic backbone modifications have been developed to address these challenges.4 Among them, phosphorothioate linkages have been widely adopted due to their ease of synthesis, increased nuclease stability, and moderate improvement in cellular uptake.5,6 While most backbone modifications enhance nuclease stability, increasing cellular uptake is more challenging. Charge alterations are a promising approach to address this problem, as demonstrated by uncharged peptide nucleic acid (PNA) linkages7 and through the incorporation of positively charged groups using phosphoramidate8,9 and amide backbone modifications.10–15 The variety of advantageous properties afforded by these modifications, including cell uptake, have been summarized in recent reviews16,17 and indicate that the continual development of backbone modifications expands the possibilities of this class of molecules. However, their synthesis, often incompatible with phosphoramidite methods, impedes their wide adoption by other groups.
Almost 30 years ago, Bruice and others introduced deoxyribonucleic guanidines (DNGs), oligonucleotides with guanidinium moieties in place of the phosphodiester backbone, as a strategy to increase cellular uptake, inspired by arginine-rich cell-penetrating peptides.18–21 DNGs are resistant to nuclease degradation and have been made with all four nucleobases.22–25 Short DNG sequences were reported to retain base-pairing fidelity over nonspecific electrostatic association, in contrast to findings with other positively charged oligonucleotides; however, duplex formation has not been studied for DNG sequences longer than eight bases.25–28 As opposed to alternative guanidine-containing backbone modifications, these oligonucleotides can be synthesized in the 3’ to 5’ direction using the same solid support and protection groups afforded by phosphoramidite chemistry, enabling their synthesis on standard instruments.23 However, current DNG production protocols require successive couplings with toxic mercury salts and pungent thiophenol washes, limiting the use of such structures and their broad scale adoption on oligonucleotide synthesizers (Scheme 1). For example, to date the interaction of DNGs with cells has yet to be explored, and therapeutically relevant lengths have not been attained.
Scheme 1. Solid-Phase DNG Synthesis Cyclea.

aB1 and B2, nucleobases; MMTr, monomethoxy trityl; TEA, triethylamine; TMP, 2,2,6,6-tetramethylpiperidine; DCA, dichloroacetic acid; NMI, N-methylimidazole; DMF, dimethylformamide.
*Indicates steps that are identical to standard phosphoramidite coupling chemistry.
Herein, we report a single-step coupling for a poly guanidine synthesis cycle that uses iodine as a mild oxidant to couple protected thiourea monomers to the 5’-amine of growing DNG strands anchored on a solid support. This method removes toxic reagents and increases access to this class of oligonucleotide modifications. Reaction conditions were initially identified and optimized through a solution-phase screen. Promising results from the screen were successfully translated to an automated oligonucleotide synthesizer and have enabled the synthesis of the longest DNG strand made to date. Moreover, for the first time since their development, we report that DNGs show rapid cellular uptake into mammalian cells without the need for auxiliary transfection reagents and do not affect cell viability. These findings highlight the potential of DNGs for further development as cationic self-transfecting gene regulation agents and as probes for making intracellular measurements.3
RESULTS AND DISCUSSION
Solution-Phase Screen for Coupling Conditions.
We hypothesized that the mercury salts used during DNG synthesis could be replaced by less toxic reagents (Scheme 1). Bruice’s coupling chemistry relies on mercury-mediated sulfur abstraction from the thiourea and addition of an amine to form a guanidine.29 Therefore, we reviewed alternative reagents that similarly activate thioureas for a single step synthesis of carbodiimides and guanidines. Among them are Mukaiyama’s reagent (2-chloro-1-methylpyridinium iodide, CMPI),30,31 Sanger’s reagent (1-fluoro-2,4-dinitrobenzene, DNFB),31,32 hypervalent iodine (e.g., [hydroxy(tosyloxy)-iodo]benezene, HTIB),33,34 and molecular iodine,35–38 all in the presence of a nitrogenous base. We began exploring alternative chemistry by coupling P6 and I3-TIPS under reported conditions (Figure 1A). The two coupling partners were synthesized according to published protocols with minor variations (Schemes S1 and S2). To expedite the screening process, we carried out these reactions in solution phase with TIPS-protected 3’-hydoxyl of 5’-deoxyamino thymidine (I3-TIPS), which would usually be attached to the growing strand on a solid support. This allowed for rapid reaction screening in a 96-well format.
Figure 1.
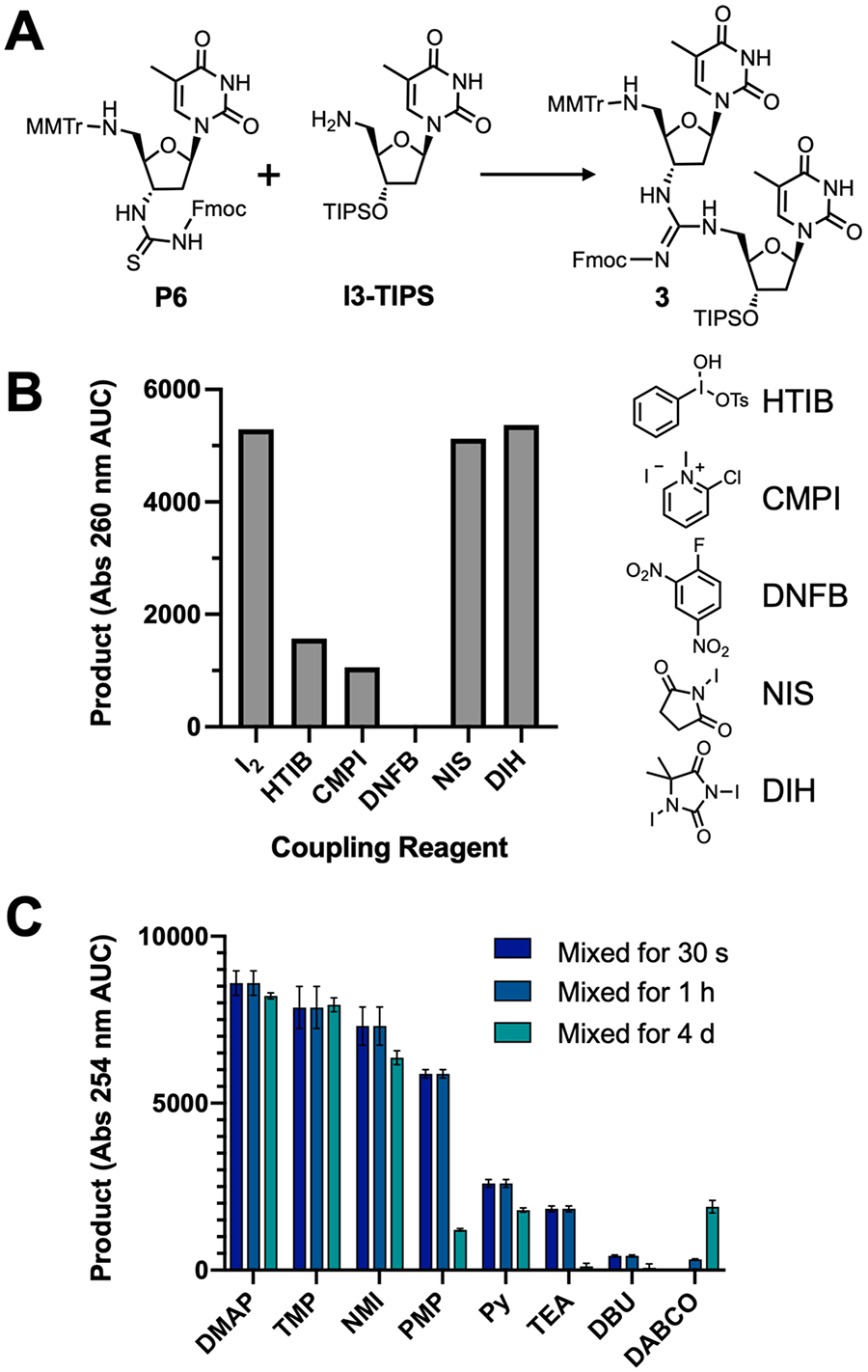
Solution-phase screen for coupling reagents and conditions. (A) Solution-phase reaction between Fmoc protected thiourea P6 and 3’-TIPS protected amine I3-TIPS to give the Fmoc-protected guanidine product 3. (B) Area under the curve of the UV trace of the product peak from UPLC analysis. Reactions contained P6, I3-TIPS, TMP, and indicated coupling reagent in 1:3 dichloromethane (DCM):acetonitrile (ACN). The structures of some of the coupling reagents used in the screen are shown. (C) Effects of base identity and storage in solution with iodine after 30 s, 1 h, and 4 days. Note: Error bars indicate the standard deviation of three reactions.
Each of these coupling reagents was incubated with thiourea P6 in the presence of 2,2,6,6-tetramethylpiperidine (TMP) as the base, chosen due to its similarity to other bases commonly used with these reactions (i.e., triethylamine) and its stability under the reaction conditions (vide infra). Coupling product 3 could be detected in all reactions tested; however, iodine resulted in far greater and faster product yields than did CMPI, DNFB, and HTIB (Figure 1B). We suspected that the active reagent responsible for the coupling in the iodine and TMP reaction was an electrophilic iodine source. When iodine and TMP are mixed in acetonitrile, a TMP-I2 complex crystallizes from the solution (Figure S1 for crystal structure), which resembles other electrophilic iodine reagents. To confirm the hypothesis that electrophilic iodine was responsible for the sulfur abstraction, we attempted the reaction with two frequently used sources of electrophilic iodine: N-iodosuccinimide (NIS)39 and 1,3-diiodo-5,5-dimiethylhydantoin (DIH). When either reagent was used, equally high product yields were observed (Figure 1B). On the basis of these results, we proceeded to further study a molecular iodine-based “activator”, due to its low cost and availability.
To preserve the standard operation of the oligonucleotide synthesizer, we constrained ourselves to storing the iodine and base in a single bottle, and the thiourea monomer in a separate bottle, to be delivered simultaneously during the coupling step. Therefore, it was important to identify a base that could accomplish the coupling, but also remain stable in solution with iodine for the duration of the synthesis. For this purpose, we explored a variety of alkyl and aryl nitrogen bases: 4-dimethylaminopyridine (DMAP), TMP, TEA, NMI, 1,2,2,6,6-pentamethylpiperidine (PMP), pyridine (Py), 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU), and 1,4-diazabicyclo[2.2.2]octane (DABCO). We also measured the amount of product formed in the solution-phase coupling reaction with the various iodine–base mixes after they had remained in the dark at room temperature for 30 s, 1 h, and 4 d.
DMAP, TMP, and NMI all afforded similar amounts of product. However, DMAP formed a precipitate under these conditions, while NMI began to afford less product at the longer time point. Surprisingly, TEA did not perform well in this screen, although it had been commonly reported as a base for carbodiimide and guanidine formation from thiour-eas.32–36,38–40 Similarly, PMP, pyridine, DBU, and DABCO only afforded the product in low amounts. Most poorly performing activator solutions changed color over time or formed a precipitate (Figure S2). These observations suggest that these formulations may decompose, possibly via a previously reported iodine-mediated oxidation at the α position.41 Because TMP did not exhibit any decrease in product yield over time nor precipitation, it showed promise for translation to automated synthesis.
Rapid coupling rates are crucial for translation to an automated system without prolonged cycle times. We found the solution-phase rate to increase with the number of equivalents of base added to the reaction (Figure S3). At low equivalencies of base, the reaction rate is markedly slower (amine consumed with a half-life of over 5 min), and the product degrades via MMTr removal; a large excess of base increases the reaction rate but can also remove the Fmoc protecting group. Optimal conditions were identified by varying the number of base equivalents required to maximize coupling. More than 2 equiv of base resulted in the highest product yields; 3 equiv was chosen for the fast rate of reaction, which reaches completion prior to the first measurement at 1 min. This illustrates the rapid coupling time that this chemistry can achieve. The extent of the reaction can also be approximated by the disappearance of the burgundy iodine color in solution, which occurs in seconds once the components are mixed together, but does not if iodine, base, or thiourea are not added (data not shown). Finally, the solution-phase screen was instrumental in determining solvent compatibility for this reaction and its impact on coupling efficiency (Figure S4). Among the five solvents tested (DMF, tetrahydrofuran, 25%v/v ACN in DCM, 25%v/v DCM in ACN, and 1-methyl-2-pyrrolidinone (NMP)), 25%v/v DCM in ACN generated the highest product yields.
Automated Synthesis on Solid Support.
We next translated the iodine coupling reaction to a MerMade 12 (BioAutomation) synthesizer. The synthesizer was minimally modified to accommodate both the iodine/TMP and the standard activators used for phosphoramidite chemistry, while the thiourea nucleoside monomer was placed into an empty amidite position (Figure S5). An additional wash step of neat DMF (vide infra) was incorporated into the cycle by following the manufacturer’s instructions. Following these modifications, the synthesizer could be used to make standard oligonucleotides as well as DNG strands (see the Supporting Information for automated cycle details).
The initial conditions identified by the solution-phase screen were used to perform 1–8 DNG dT couplings atop 19- to 11-mer dT standard DNA strands bearing a 5’-MMTr-protected amine through coupling with initiator unit I5 (Scheme S3). Following standard cleavage and deprotection with ammonium hydroxide and methylamine, the crude reaction afforded reverse phase chromatography (RP-HPLC) traces that clearly separated the failure strands lacking an MMTr group from the full-length strands retarded by the presence of the 5’-MMTr (Figure 2A). Another late eluting peak was also observed, which increased in area with more DNG insertions and is suspected to result from the electrostatic association of multiple strands. Both the second and the third peaks contained only the full-length product when analyzed by MALDI-TOF MS (Figure 2B), confirming successful purification. Coupling yield was estimated using the integrated peak areas of the UV absorption traces from batches of CPGs deprotected immediately prior to and following the addition of the three DNG bases. This method provided a simple way of comparing coupling conditions (Figure S6).
Figure 2.

Synthesis conditions and yield quantification of DNA–DNG chimeras. (A) Representative RP-HPLC trace of a 22-mer with three DNG linkages after cleavage and deprotection. (B) MALDI-TOF spectra of HPLC peaks containing the full-length peak (~22 min) and the assembly peak (~32 min). (C) Coupling yields of three DNG additions atop a DNA strand on a MerMade 12 automated synthesizer. (a) Coupling yields calculated per-coupling based on HPLC UV trace AUC of product and failure peaks. (b) Alternative wash contained ACN instead of DMF. (c) Iodine extracted with saturated NaHCO3 and dried with MgSO4 prior to making the activator solution. (d) Coupling yield measured by trityl absorbance of deprotection solution following each coupling cycle for 8 cycles atop an 11-mer DNA strand.
We found that the activator retained a constant coupling yield when stored on the synthesizer for up to 24 h but showed a significantly decreased coupling yield after 48 h (Figure 2C, entries 1–3). This suggests that the iodine–TMP solution should only be used for 1 day before being replaced, which is long enough to complete even extensive oligonucleotide syntheses.
During the optimization process, we noticed that the drain line that transfers reagents from the column to the waste system would periodically become clogged, which did not occur during the standard phosphoramidite syntheses. We suspected that a precipitate, at times visible during the coupling step, was the cause of the clogs. The introduction of an additional DMF wash step to dissolve any precipitates prevented the clog from occurring since and did not impact the coupling efficiency (Figure 2C, entries 1 and 4).
In addition to testing the number of base equivalents in solution phase, we also tested this on a solid support. Three equivalents of base with respect to both thiourea and iodine resulted in the highest yield and did not remove the Fmoc protecting groups from the growing strand (Figure S7), while 2 or 4 equiv caused a decrease in coupling efficiency (Figure 2C, entries 6–8). Finally, increasing the amount of thiourea from 1 to 1.5 and 3 equiv with respect to iodine, while keeping the base constant at 3 equiv, caused a decreased yield (Figure 2C, entries 4, 5, and 7). This finding suggests that the excess unactivated thiourea may consume the activated thiourea monomer in a side reaction. Similar reactivity has been reported; however, we did not attempt to isolate these side products.42 The coupling yield of these optimized conditions was measured using the established trityl cation absorption protocol, and showed an average coupling efficiency of 95% over eight coupling cycles (Figure 2C, entry 5; Figure S8).43
Once a high yielding coupling was attained, a primarily positively charged strand was synthesized. Controlled pore glass (CPG) beads bearing a disulfide moiety, to provide a thiol handle for subsequent conjugation reactions, were coupled with initiator phosphoramidite I5 through a single phosphate linkage. Afterward, a 9-mer strand of DNG dT nucleosides was added to this nascent strand (sequence: 5’-(Tg)9(Tp)-OC3H6SSC3H6OH-3’, p is phosphate and g is guanidinium linkages). MALDI-TOF MS of the crude material showed the presence of the product peak along with a number of failure strands. The full-length strand was purified using a serendipitously discovered extraction procedure by washing the solid support with water and acetonitrile, which removed the failure strands, and then a 20% aqueous solution of acetic acid, which extracted the full-length product. HPLC purification of the strand proved elusive due to a strong and likely electrostatic attraction between the DNG strand and surfaces, and a general purification strategy is under development (Figures S9 and S10). The 10-mer DNG strand was characterized by SDS-PAGE and stained with SimplyBlue, a negatively charged protein stain that confirms the structure’s overall positive charge (Figure S11B). One main band was observed, attesting to the purity of the material. The purified 10-mer product showed a single primary mass peak when analyzed via MALDI-TOF MS, which matches the calculated mass (Figure 3A). To demonstrate the ability of this method to synthesize DNG strands of lengths relevant to therapeutic and detection applications, we also made a 20-mer DNG (Figure S11), the first example of a DNG strand of this length.
Figure 3.

Synthesis, cellular uptake, and toxicity of DNG (Tg)10 strands. (A) MALDI-TOF spectra of the crude and purified DNG strands, shot with DHAP matrix; (B) confocal fluorescence images of C166 cells after treatment with fluorophore-labeled DNA or DNG (red) with stained nuclei (blue); (C) uptake of fluorophore-labeled DNA and DNG strands into C166 cells as measured by flow cytometry; and (D) toxicity of DNA and DNG oligonucleotides toward C166 cells measured by Alamar Blue assay.
Cell-Penetrating Capabilities.
The improved coupling method and automated synthesis made the production of DNG strands at lengths not achieved to date practical and allowed us to finally test the 30-year hypothesis that the incorporation of guanidinium linkages as the backbone of oligonucleotides will affect their uptake into cells. We used AlexaFluor 647 to fluorophore-label the 10-mer DNG strand and a standard phosphate-backbone 10-mer DNA strand. Mouse endothelial C166 cells were separately treated with both labeled strands at a 600 nM concentration for 2.5 h to assess their uptake at concentrations relevant for oligonucleotide drug delivery. Confocal microscopy and flow cytometry were used to characterize the extent of uptake into cells (Figures 3B,C and S12–S15). Minimal fluorescence is visible in the DNA treatment group, attesting to the inefficient uptake of free oligonucleotides into cells. However, the DNG treated cells show intense fluorescence, both throughout the cytoplasm and in punctate areas arrayed inside the cell. Flow cytometry analysis further highlighted the high cellular uptake of DNGs, which is 38-fold higher than their DNA equivalent. To our delight, no toxicity was observed between 78 nM and 5 μM concentrations using an Alamar Blue cell viability assay (Figures 3D and S16).
CONCLUSIONS
Herein, we have introduced a practical method for the automated synthesis of DNG oligonucleotides. This discovery is important for the following reasons: (1) it is an attractive, fully automated alternative to the previously reported mercury-based method, (2) it will accelerate the study of positively charged oligonucleotides, and (3) it will enable their translation as self-transfecting nucleic acid therapeutics. Taken together, we expect these findings will greatly improve the access to a previously underexplored class of oligonucleotides, which shows tremendous potential for biological and therapeutic applications.
Supplementary Material
ACKNOWLEDGMENTS
We would like to acknowledge support from the Vannevar Bush Faculty Fellowship program sponsored by the Basic Research Office of the Assistant Secretary of Defense for Research and Engineering and funded by the Office of Naval Research through grant N00014-15-1-0043; the Prostate Cancer Foundation and the Movember Foundation under award 17CHAL08; and the National Cancer Institute of the National Institutes of Health under award U54CA199091. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work made use of the IMSERC at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-1542205), the State of Illinois, and the International Institute for Nanotechnology (IIN). K.E.B. is supported by a Banting Fellowship from the Government of Canada. We would like to thank Professor Karl A. Scheidt for helpful and illuminating discussions.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.9b09937.
Detailed protocols for the synthesis of monomers I5, P6, and I3-TIPS; NMR spectra and HRMS data of monomers; crystal structure of iodine-TMP activator precipitate; further reaction characterization data; images of activator solutions over time; details of the automated synthesis cycle and synthesizer setup; flow cytometry gating strategy and results; confocal microscopy images and Z-stack analysis; and cell viability results (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Samanta D; Ebrahimi SB; Mirkin CA Nucleic-Acid Structures as Intracellular Probes for Live Cells. Adv. Mater 2019, 1901743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lundin KE; Gissberg O; Smith CIE Oligonucleotide Therapies: The Past and the Present. Hum. Gene Ther 2015, 26 (8), 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Deleavey GF; Damha MJ Designing Chemically Modified Oligonucleotides for Targeted Gene Silencing. Chem. Biol 2012, 19 (8), 937–954. [DOI] [PubMed] [Google Scholar]
- (4).Brad Wan W; Seth PP The Medicinal Chemistry of Therapeutic Oligonucleotides. J. Med. Chem 2016, 59 (21), 9645–9667. [DOI] [PubMed] [Google Scholar]
- (5).Eckstein F Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014, 24 (6), 374–87. [DOI] [PubMed] [Google Scholar]
- (6).Khvorova A; Watts JK The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol 2017, 35 (3), 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Nielsen PE; Egholm M; Berg RH; Buchardt O Sequence-Selective Recognition of DNA by Strand Displacement with a Thymine-Substituted Polyamide. Science 1991, 254 (5037), 1497–1500. [DOI] [PubMed] [Google Scholar]
- (8).Letsinger RL; Bach SA; Eadie JS Effects of pendant groups at phosphorus on binding properties of d-ApA analogues. Nucleic Acids Res. 1986, 14 (8), 3487–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Letsinger RL; Singman CN; Histand G; Salunkhe M Cationic oligonucleotides. J. Am. Chem. Soc 1988, 110 (13), 4470–4471. [Google Scholar]
- (10).Meng M; Schmidtgall B; Ducho C Enhanced Stability of DNA Oligonucleotides with Partially Zwitterionic Backbone Structures in Biological Media. Molecules 2018, 23 (11), 2941–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Deglane G; Abes S; Michel T; Prevot P; Vives E; Debart F; Barvik I; Lebleu B; Vasseur JJ Impact of the guanidinium group on hybridization and cellular uptake of cationic oligonucleotides. ChemBioChem 2006, 7 (4), 684–92. [DOI] [PubMed] [Google Scholar]
- (12).Park M; Bruice TC Binding properties of positively charged deoxynucleic guanidine (DNG), AgTgAgTgAgT and DNG/DNA chimeras to DNA. Bioorg. Med. Chem. Lett 2008, 18 (12), 3488–3491. [DOI] [PubMed] [Google Scholar]
- (13).Schmidtgall B; Spork AP; Wachowius F; Hobartner C; Ducho C Synthesis and properties of DNA oligonucleotides with a zwitterionic backbone structure. Chem. Commun 2014, 50 (89), 13742–13745. [DOI] [PubMed] [Google Scholar]
- (14).Thomas SM; Sahu B; Rapireddy S; Bahal R; Wheeler SE; Procopio EM; Kim J; Joyce SC; Contrucci S; Wang Y; Chiosea SI; Lathrop KL; Watkins S; Grandis JR; Armitage BA; Ly DH Antitumor effects of EGFR antisense guanidine-based peptide nucleic acids in cancer models. ACS Chem. Biol 2013, 8 (2), 345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhou P; Wang M; Du L; Fisher GW; Waggoner A; Ly DH Novel binding and efficient cellular uptake of guanidine-based peptide nucleic acids (GPNA). J. Am. Chem. Soc 2003, 125 (23), 6878–9. [DOI] [PubMed] [Google Scholar]
- (16).Meng M; Ducho C Oligonucleotide analogues with cationic backbone linkages. Beilstein J. Org. Chem 2018, 14, 1293–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Jain ML; Bruice PY; Szabo IE; Bruice TC Incorporation of Positively Charged Linkages into DNA and RNA Backbones: A Novel Strategy for Antigene and Antisense Agents. Chem. Rev 2012, 112 (3), 1284–1309. [DOI] [PubMed] [Google Scholar]
- (18).Dempcy RO; Almarsson O; Bruice TC Design and synthesis of deoxynucleic guanidine: a polycation analogue of DNA. Proc. Natl. Acad. Sci. U. S. A 1994, 91, 7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Futaki S; Suzuki T; Ohashi W; Yagami T; Tanaka S; Ueda K; Sugiura Y Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem 2001, 276 (8), 5836–40. [DOI] [PubMed] [Google Scholar]
- (20).Bhadra J; Pattanayak S; Khan PP; Kundu J; Sinha S Internal Oligoguanidinium-Based Cellular Transporter Enhances Antisense Efficacy of Morpholinos in In Vitro and Zebrafish Model. Bioconjugate Chem. 2016, 27 (10), 2254–2259. [DOI] [PubMed] [Google Scholar]
- (21).Vandendriessche F; Voortmans M; Hoogmartens J; Van Aerschot A; Herdewijn P Synthesis of Novel N-Substituted Guanidine Linked Nucleoside Dimers and Their Incorporation into Oligonucleotides. Bioorg. Med. Chem. Lett 1993, 3 (2), 193–198. [Google Scholar]
- (22).Challa H; Bruice TC Deoxynucleic guanidine: synthesis and incorporation of purine nucleosides into positively charged DNG oligonucleotides. Bioorg. Med. Chem 2004, 12 (6), 1475–1481. [DOI] [PubMed] [Google Scholar]
- (23).Linkletter BA; Szabo IE; Bruice TC Solid-phase synthesis of oligopurine deoxynucleic guanidine (DNG) and analysis of binding with DNA oligomers. Nucleic Acids Res. 2001, 29 (11), 2370–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Szabo IE; Bruice TC DNG cytidine: Synthesis and binding properties of octameric guanidinium-linked deoxycytidine oligomer. Bioorg. Med. Chem 2004, 12 (15), 4233–4244. [DOI] [PubMed] [Google Scholar]
- (25).Barawkar DA; Bruice TC Synthesis, biophysical properties, and nuclease resistance properties of mixed backbone oligodeox-ynucleotides containing cationic internucleoside guanidinium linkages: Deoxynucleic guanidine/DNA chimeras. Proc. Natl. Acad. Sci. U. S. A 1998, 95, 11047–11052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Schmidtgall B; Kuepper A; Meng M; Grossmann TN; Ducho C Oligonucleotides with Cationic Backbone and Their Hybridization with DNA: Interplay of Base Pairing and Electrostatic Attraction. Chem. - Eur. J 2018, 24 (7), 1544–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Linkletter BA; Szabo IE; Bruice TC Solid-phase synthesis of deoxynucleic guanidine (DNG) oligomers and melting point and circular dichroism analysis of binding fidelity of octameric thymidyl oligomers with DNA oligomers. J. Am. Chem. Soc 1999, 121 (16), 3888–3896. [Google Scholar]
- (28).Blaskó A; Dempcy RO; Minyat EE; Bruice TC Association of Short-Strand DNA Oligomers with Guanidinium-Linked Nucleosides. A Kinetic and Thermodynamic Study. J. Am. Chem. Soc 1996, 118 (34), 7892–7899. [Google Scholar]
- (29).Levallet C; Lerpiniere J; Ko SY The HgCl2-promoted guanylation reaction: The scope and limitations. Tetrahedron 1997, 53 (14), 5291–5304. [Google Scholar]
- (30).Yong YF; Kowalski JA; Lipton MA Facile and efficient guanylation of amines using thioureas and Mukaiyama’s reagent. J. Org. Chem 1997, 62 (5), 1540–1542. [Google Scholar]
- (31).Schneider SE; Bishop PA; Salazar MA; Bishop OA; Anslyn EV Solid phase synthesis of oligomeric guanidiniums. Tetrahedron 1998, 54 (50), 15063–15086. [Google Scholar]
- (32).Lammin SG; Pedgrift BL; Ratcliffe AJ Conversion of anilines to bis-Boc protected N-methylguanidines. Tetrahedron Lett. 1996, 37 (37), 6815–6818. [Google Scholar]
- (33).Zhu CJ; Xu D; Wei YY A New Synthetic Protocol for the Preparation of Carbodiimides Using a Hypervalent Iodine(III) Reagent. Synthesis 2011, 2011 (5), 711–714. [Google Scholar]
- (34).Singh CB; Ghosh H; Murru S; Patel BK Hypervalent iodine(III)-mediated regioselective N-acylation of 1,3-disubstituted thioureas. J. Org. Chem 2008, 73 (7), 2924–2927. [DOI] [PubMed] [Google Scholar]
- (35).Ali AR; Ghosh H; Patel BK A greener synthetic protocol for the preparation of carbodiimide. Tetrahedron Lett. 2010, 51 (7), 1019–1021. [Google Scholar]
- (36).Gaffney BL; Jones RA Synthesis of c-di-GMP analogs with thiourea, urea, carbodiimide, and guanidinium linkages. Org. Lett 2014, 16 (1), 158–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wang Z; Zhao Q; Hou J; Yu W; Chang J Iodine-mediated direct synthesis of multifunctional 2-aminobenzimidazoles from N-substituted o-diaminoarenes and isothiocyanates. Tetrahedron 2018, 74 (19), 2324–2329. [Google Scholar]
- (38).Duangkamol C; Pattarawarapan M; Phakhodee W Ultra-sonic-assisted synthesis of carbodiimides from N,N’-disubstituted thioureas and ureas. Monatsh. Chem 2016, 147 (11), 1945–1949. [Google Scholar]
- (39).Ohara K; Vasseur JJ; Smietana M NIS-promoted guanylation of amines. Tetrahedron Lett 2009, 50 (13), 1463–1465. [Google Scholar]
- (40).Yong YF; Kowalski JA; Lipton MA Facile and Efficient Guanylation of Amines Using Thioureas and Mukaiyama’s Reagent. J. Org. Chem 1997, 62 (5), 1540–1542. [Google Scholar]
- (41).Griffiths RJ; Burley GA; Talbot EPA Transition-Metal-Free Amine Oxidation: A Chemoselective Strategy for the Late-Stage Formation of Lactams. Org. Lett 2017, 19 (4), 870–873. [DOI] [PubMed] [Google Scholar]
- (42).Yella R; Khatun N; Rout SK; Patel BK Tandem regioselective synthesis of tetrazoles and related heterocycles using iodine. Org. Biomol. Chem 2011, 9 (9), 3235–3245. [DOI] [PubMed] [Google Scholar]
- (43).Eckstein F Oligonucleotides and Analogues: A Practical Approach; IRL Press: Oxford, NY, 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


