Abstract
Background:
This study was conducted to determine the presence and molecular identify of Acanthamoeba, Naegleria and Vermamoeba in unimproved hot springs.
Methods:
From Jul to Aug 2017, 54 water samples were collected from hot springs in different parts of the Guilan Province, North Iran. For the isolation of Acanthamoeba, Naegleria and Vermamoeba approximately 500 ml of the water samples were filtered through a cellulose nitrate membrane with a pore size of 0.45 μm. The filter was transferred onto non-nutrient agar plates seeded with Gram-negative bacteria (Escherichia coli) as a food source. The morphological key of page was used to identify free-living amoebae (FLA) using an inverted microscope, PCR amplification targeting specific genes for each genus and sequencing determined frequent species and genotypes base on NCBI database.
Results:
Fifteen of the 54 samples were positive by culture and/or PCR for Acanthamoeba and other FLA from unimproved hot springs. By sequencing the positive isolates, the strains were shown to belong to Acanthamoeba castellanii (12 case isolates belonged to T4 genotype), 4 cases of V. vermiformis, and 3 cases of N. australiensis, 2 cases of N. pagei and 1 cases of N. gruberi.
Conclusion:
Although FLA-mediated illnesses are not as high as in environmental distribution, but because of a poor prognosis, more investigations about FLA distribution in hot springs is critical. Hot spring may enhance exposure of the amoebae in individuals. Hence, more attention to unimproved hot springs is needed to prevent free-living amoebae mediated diseases.
Keywords: Acanthamoeba, Naegleria Vermamoeba, PCR/DNA sequencing, Hot springs, Iran
Introduction
Free-living amoebae (FLA) as amphizoic amoebae are a group of parasitic protozoa with the growth abilities in different natural environments such as soil and water. In appropriate conditions, they are pathogenic in animals and humans (1). FLA include many genera which cause serious diseases such as cutaneous ulcers, sight threatening keratitis and fatal encephalitis.
Acanthamoeba spp., Naegleria fowleri and Balamuthia mandrillaris are the most commonly reported causes in the world (2). Other genera in this group, including Sappinia diploidea, Vermamoeba and Vahlkampfia mix infection with other FLA could also lead to severe diseases with a lower incidence around the world (3–5). Acanthamoeba strains, especially ones that are potentially pathogenic, can tolerate extremes of temperature, osmolarity, and pH (6). Vahlkampfia and Naegleria genera are commonly found in warm freshwater.
N. australiensis and N. italica can cause infection in experimental animals but N. fowleri is able to infect animals and humans with possible life threatening contamination (7, 8). Acanthamoeba spp. Vahlkammpfids and B. mandrillaris can cause serious infections in humans (1). In addition to Vermamoeba have as a suitable hosts for pathogenic microorganisms such as Legionella pneumophila (9, 10). Other diseases related to FLA are Vermamoeba keratitis, and Vahlkampfia keratitis (3, 5, 11).
Hot springs are highly regarded for their therapeutic effects and how much of their use in Iran, both as a therapeutically use and as a tourist attraction, is growing; which will increase the chance of exposure to these amoebae (12–14). Guilan Province is a tourist areas of Iran which plays host to millions of Iranian and foreign travelers annually, and one of the tourist attractions of in this province is its hot springs (15). The presence of FLA in the hot springs of the provinces bordering Guilan has been confirmed (14, 16). Parasitological methods only recognize parasite contamination however, it is not possible to identify the exact species of parasite involved (17).
Hence, we aimed to survey the incidence of waterborne FLA belonging to Vermamoeba, Naegleria spp. and Acanthamoeba spp. isolated from unimproved therapeutic hot springs of Guilan Province using morphological and sequence based methods.
Materials and Methods
Geographical area of study
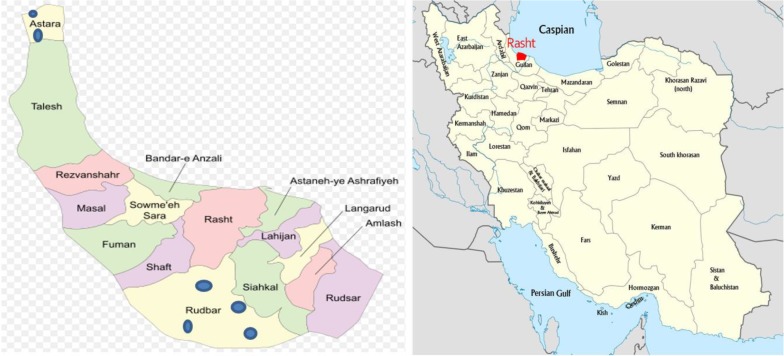
Guilan Provinces lies along the Caspian Sea. It has a plenty of annual rainfall, humid temperate weather and is known for its mild, moderate and Mediterranean-like climate (15) (Fig. 1).
Fig. 1:
Map of the investigated Guilan greater area (left) and its location in Iran (right) The sampling points are indicated by blue circles ( )
)
Sampling
This cross sectional study (Code of ethics: IR.AJUMS.REC.1396.501) was performed from July to August 2017. To run this study, 54 water samples were collected from hot springs in different parts of the Guilan Province, North Iran (Fig. 1). Six hot springs in Guilan, North of Iran were included in this study and nine samples per site (reservoir- margin- center) were obtained. Samples were collected from the surface of the waters (<5 cm below) (14).
Filtration, cultivation and cloning
Approximately 500 ml water samples were filtered using cellulose nitrate membranes with a pore size of 0.45 μm. Filters were transferred to non-nutrient agar (NNA) plates seeded with Escherichia coli as a food source (14, 15). The morphological key of page was use for the identification FLA using an inverted microscope. Cloning of the suspected amoebae was performed using culture replicates method (11, 18).
DNA extraction, PCR analysis and Sequencing
DNA was extracted using GeNet Bio kit, according to manufacturer's instructions (GeNet Bio, South Korea). Four sets of primers were used in order to detect various FLA shown in Table 1. To carry out the PCR reaction, 25 ml of red master mix (Denmark) was combined with DNA (10 ng), 0.1 μM of each primer and distilled water. The cycling condition was set as pre-denaturation step for 3 min at 94 °C, followed by 35 repetitions at 94 °C for 35 sec, annealing steps were at 56 °C, 56 °C, 56 °C and 58 °C for 1 min (for Acanthamoeba, Vahlkampfiids, N. fowleri and Vermamoeba, respectively), and 72 °C for 1 min. All sequences were edited manually and analyzed with reference sequences by Chromas software program. The sequences were submitted to gene bank under following accession numbers: MH347242-MH347263.
Table 1:
Primers used in this study
| FLA1 | Primer sequence | Reference |
|---|---|---|
| Acanthamoeba spp. | JDP15′-GGCCCAGATCGTTTACCGTGAA-3′ | 19 |
| JDP2 5′-TCTCACAAGCTGCTAGGGAGTCA-3′ | ||
| Vahlkampfiids | ITS1 F5′-GAACCTGCGTAGGGATCATTT- 3′ | 2 |
| ITS2 R 5′TTTCTTTTCCTCCCCTTATTA-3′ | ||
| N. fowleri 2 | F5′-GTGAAAACCTTTTTTCCATTTACA-3′ | 14 |
| R5′-AAATAAAAGATTGACCATTTGAAA-3′ | ||
| Vermamoeba | Hv1227F 5′- -TTA CGA GGT CAG GAC ACTGT-3′ | 2 |
| Hv1728R 5′-GAC CAT CCG GAG TTC TCG-3′ |
. Free living amoebae: FLA
. Naegleria fawleri
Results
Of 54 water samples, 15 (27.7%) cases were positive for outgrowth of free-living amoebae. Indeed, the positive samples included 7 cases of Acanthamoeba, 2 cases of Vahlkammpfids, 1 case of Vermamoeba, 2 mixed cases of Acanthamoeba, Vahlkammpfids and Vermamoeba, 2 mixed cases of Acanthamoeba and Vahlkammpfids, and 1 mixed case of Acanthamoeba and Vermamoeba. Accordingly, Acanthamoeba were found in 12 (54.5%) samples as the most prevalent amoebae in the tested samples (Tables 1, 2).
Table 2:
Data regarding isolated of free-living amoebae in Guilan Hot Springs, and Samples Sites
| City | Sampling Site | No. of Samples | Number/positive samples | Acanthamoea | Vahlkam pfid | Vermamoeba | Mixed Acanthamoeba, Vahlkampfiid and Vermamoeba | Mixed Acanthamoeba and Vahlkampfi id | Mixed Acanthamoe a and Vermamoeba | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R | C | M | |||||||||
| Roud | Kolour | 3 | 3 | 3 | 9/2 | 1 | 1 | 0 | 0 | 0 | 0 |
| bar | Louye | 3 | 3 | 3 | 9/4 | 2 | 0 | 0 | 1 | 1 | 0 |
| Mastkhor | 3 | 3 | 3 | 9/4 | 3 | 0 | 0 | 0 | 1 | 0 | |
| Kalashtar | 3 | 3 | 3 | 9/2 | 1 | 0 | 0 | 0 | 0 | 1 | |
| Astara | Alidashi | 3 | 3 | 3 | 9/3 | 0 | 1 | 1 | 1 | 0 | 0 |
| Koutekoume | 3 | 3 | 3 | 9/0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total | 6 | 1 | 1 | 1 | 54/15(27.7%) | 7 | 2 | 1 | 2 | 2 | 1 |
| 8 | 8 | 8 | |||||||||
R: Reservoir C: Center M: Margin
For morphological determination of Acanthamoeba castellanii, the following characters were regarded; a double walled with a smooth wrinkled outer cyst wall and stellate endocyst (Figs. 2 and 3) for Vahlkammpfids round cysts with smooth wall and V. vermiforims round cysts with smooth wall but smaller Vahlkammpfids (Fig. 3 and 4).
Fig. 2:
Acanthamoeba castellanii cysts (400 X)
Fig. 3:
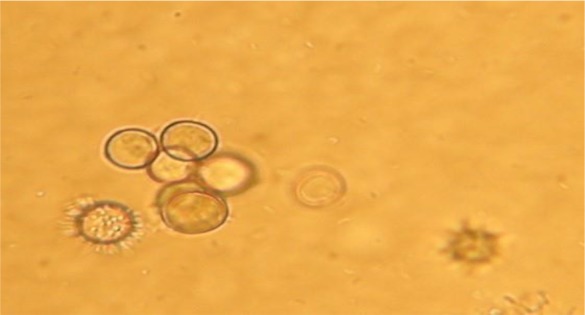
Mixed Vahlkammpfids cysts (1) (400 X) and Acanthamoeba castellanii trophozoite (2) (400 X)
Fig. 4:
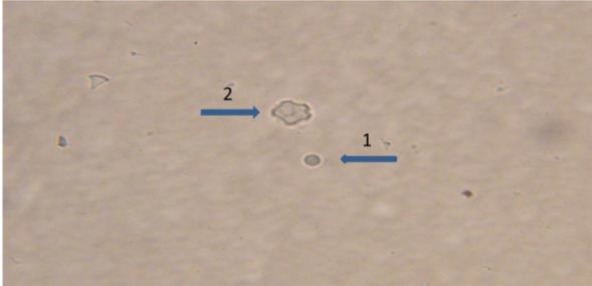
Mixed Vermamoeba vermiformis cysts (1) (400 X) and Acanthamoeba castellanii cysts (2) (400 X)
In electrophoresis of PCR products, twelve cases of Acanthamoeba demonstrated an approximately 500 bp band (Fig. 5). Sequencing analysis of 12 positive cases showed T4 (100%) genotypes with homology analysis of NCBI website revealing 95%-100% similarity.
Fig. 5:
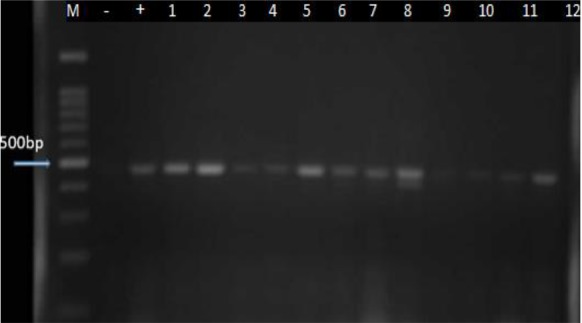
PCR amplification of the isolated Acanthamoeba strains. M marker, -=Neg Control, +=Pos Control, 1–12 samples
Taken together, 6 cases of Vahlkammpfids (2 single and 4 mixed with Acanthamoeba) and 4 cases of Vermamoeba (1 single and 3 mixed with Acanthamoeba) were determined. Six cases of Vahlkammpfids exhibited an approximately 400 bp band, and 4 samples of Vermamoeba demonstrated a nearly 500bp band during electrophoresis (Figs. 6 and 7).
Fig. 6:

PCR amplification of the isolated naegleria strains. M marker, -=Neg Control, +=Pos Control, (2, 7,8 and 10 samples)
Fig. 7:
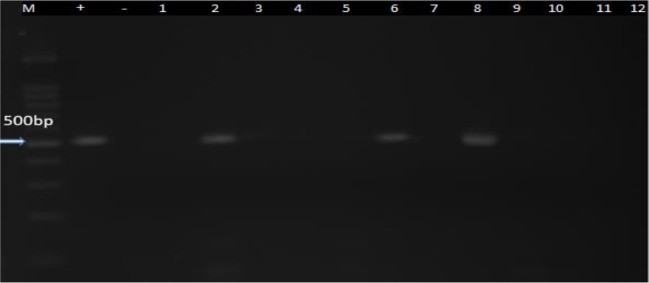
PCR amplification of the isolated Vermamoeba. M marker, - = Neg Control, + = Pos Control, (2,6 and 8 samples)
Water PH and temperature were assessed in situ by using a portable pH meter (Digital tester DMT-20), so that these parameters of hot springs were respectively measured as 23.4%-31.6 °C and 6.5–7.2 (Table 3).
Table 3:
Data of the free living amoebae from unimproved hot springs, Guilan Province, Iran
| Name Hot Spring | Isolate code | Morphology | PH | Temperature °C | PCR (JDP1,2) | PCR (ITS1, 2) | PCR for N. fowleri | PCR (Hv1227F, Hv1728R) | Sequencing | Accession Number |
|---|---|---|---|---|---|---|---|---|---|---|
| Kolour | RN2 | Acanthamoeba | 7.1 | 31.6* | + | - | - | - | T4 genotype | MH347242 |
| RN3 | Vahlkamfiid | 6.8 | 31.2 | - | + | - | - | N. pagei | MH347254 | |
| Louye | RN14 | Acanthamoeba | 6.5* | 30.3 | + | - | - | - | T4 genotype | MH347243 |
| RN16 | Acanthamoeba | 7 | 30 | + | - | - | - | T4 genotype | MH347244 | |
| RN17 |
Acanthamoeba,Vahlkamfiid Vermamoeba |
6.8 | 30.5 | + | + | - | + | T4 genotype N. Pagei V.vermiformis |
MH347245 MH347255 MH347260 |
|
| RN18 | Acanthamoeba, Vahlkamfiid | 7.1 | 23.4* | + | + | - | - | T4 genotype N. gruberi |
MH347246 MH347256 |
|
| Mastkhor | RN20 | Acanthamoeba | 6.6 | 23.6 | + | - | - | - | T4 genotype | MH347247 |
| RN21 | Acanthamoeba, Vahlkamfiid | 6.8 | 23.4 | + | + | - | - | T4 genotype N. australiensis |
MH347248 MH347257 |
|
| RN22 | Acanthamoeba | 7.2* | 30.6 | + | - | - | - | T4 genotype | MH347249 | |
| RN23 | Acanthamoeba | 6.6 | 30.1 | + | - | - | - | T4 genotype | MH347250 | |
| Kalashtar | RN30 | Acanthamoeba | 7 | 28.4 | + | - | - | - | T4 genotype | MH347251 |
| RN36 | Acanthamoeba,Vermamoeba | 6.8 | 27.6 | + | - | - | + | T4 genotype V.vermiformis |
MH347252 MH347261 |
|
| Alidashi | AN43 |
Acanthamoeba.Vahlkamfiid Vermamoeba |
6.6 | 27.8 | + | + | - | + | T4 genotype N. australiensis V.vermiformis |
MH347253 MH347258 MH347262 |
| AN44 | Vahlkamfiid | 7 | 30.4 | - | + | - | - | N. australiensis | MH347259 | |
| AN45 | Vermamoeba | 7.2 | 28.9 | - | - | - | + | V.vermiformis | MH347263 |
= maximum and minimum water PH and temperature
Moreover, sequencing analysis revealed 95%–100 % similarity with Vahlkammpfids and V. vermiformis. Accession numbers of nucleotide sequences were deposited in the GenBank database, and have been demonstrated in Table 3. Moreover, six hot springs, 83.4% (n=5), 66.6% (n=4) and 50% (n=3) were respectively positive for Acanthamoeba, Vahlkamfiids, and Vermamoeba amoebae.
Discussion
The present study is the first study on un-improved hot springs of Guilan Province, Northern Iran to determine the pathogenic free-living amoeba via molecular methods. In this investigation waterborne free living amoebae belonging to the Acanthamoeba T4 genotype, Naegleria (N. pagei, N. australiensis and N. gruberi) and Vermamoeba verformis were found in the unimproved hot springs of Guilan Province, Northern Iran and the present study is the second report of N. gruberi in the country. Acanthamoeba was detected in surface water of Guilan, previously (15).
No significant differences were shown between pH value (and temperature) and the presence/absence of Acanthamoeba, Naegleria, Vermamoeba. In previous studies, the T4 genotype was reported to be isolated from samples such as soil, hospital wards, surface waters, recreational water areas, dust sources and also hot springs in Iran (13, 19–23). In contrast with results reported by other studies that reported T15 and T3 as predominate genotypes in waters surveyed (11, 24), the founding of PCR analysis and sequencing in the present work confirmed that the T4 genotype was the predominant type (Table 3).
Among the 47 known species of Naegleria, only N. fowleri has been reported to be pathogenic for human (8). An investigation on hot springs sources in Iran reported an increased occurrence of Naegleria genus in the tested samples (14). The pathogenic N. fowleri was not found in this study. To our knowledge, so far, there is no report on the presence of pathogenic N. fowleri in environmental sources of Iran.
However, a clinical case of N. fowleri has been reported in the country (25).
In the present study, N. australiensis was the most prevalent species, which can be pathogenic to mouse (26). V. vermiforims was also one of the most detected FLA, based on molecular assays (Table 3). A case of Vermamoeba keratitis and a case of mix infection of V. vermiforims and Acanthamoeba were reported during previous studies (4, 5, 27).
To prevent infection and diseases related to free-living amoebae, hot springs should be periodically checked, in particular, during the summer season, when these surface water are used by thousands of tourists (28). The disease could originate to possess a seasonal mode of frequency in the region and serious monitoring for proper preparation against the disease should be in place (29).
Conclusion
Although FLA-mediated illnesses is not as high as in their environmental distribution, because of a poor diagnosis, more investigations about FLA distribution in hot springs is critical. Hot spring may enhance exposure of the amoebae to individuals. Hence, more attention to unimproved hot springs is needed to prevent free-living amoebae mediated diseases.
Acknowledgements
The present study was financially supported by Student Research Committee, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran (Grant No: 95 S 123). The authors are indebted to Dr. H. Hooshyar from the Department of Parasitology and Mycology, School of Medicine, Kashan University of Medical Sciences and to S. Ebadi from the Department of Parasitology, School of Medicine Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran for their technical assistance.
Footnotes
Conflict of interests
All the authors declare that they have no conflict of interest.
References
- 1.Saburi E, Rajaii T, Behdari A, et al. Free-living amoebae in the water resources of Iran: a systematic review. J Parasit Dis. 2017;41(4):919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Javanmard E, Niyyati M, Lorenzo-Morales J, et al. Molecular identification of waterborne free living amoebae (Acanthamoeba, Naegleria and Vermamoeba) isolated from municipal drinking water and environmental sources, Semnan province, north half of Iran. Exp Parasitol. 2017;183:240–244. [DOI] [PubMed] [Google Scholar]
- 3.Niyyati M, Lorenzo-Morales J, et al. First report of a mixed infection due to Acanthamoeba geno-type T3 and Vahlkampfia in a cosmetic soft contact lens wearer in Iran. Exp Parasitol. 2010;126(1):89–90. [DOI] [PubMed] [Google Scholar]
- 4.Lorenzo-Morales J, Martínez-Carretero E, Batista N, et al. Early diagnosis of amoebic keratitis due to a mixed infection with Acanthamoeba and Hartmannella. Parasitol Res. 2007;102(1):167–9. [DOI] [PubMed] [Google Scholar]
- 5.Abedkhojasteh H, Niyyati M, Rahimi F, et al. First report of Hartmannella keratitis in a cosmetic soft contact lens wearer in Iran. Iran J Parasitol. 2013;8(3):481–5. [PMC free article] [PubMed] [Google Scholar]
- 6.Khan NA. Acanthamoeba: biology and pathogenesis: Horizon Scientific Press; 2009. [Google Scholar]
- 7.Visvesvara GS. Chapter 10 - Infections with free-living amebae. In: Garcia HH, Tanowitz HB, Del Brutto OH, editors. Handb Clin Neurol. 114: Elsevier; 2013. p. 153–68. [DOI] [PubMed] [Google Scholar]
- 8.De Jonckheere JF. What do we know by now about the genus Naegleria? Exp Parasitol. 2014;145 Suppl:S2–9. [DOI] [PubMed] [Google Scholar]
- 9.Zbikowska E, Walczak M, Krawiec A. Distribution of Legionella pneumophila bacteria and Naegleria and Hartmannella amoebae in thermal saline baths used in balneotherapy. Parasitol Res. 2013;112(1):77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheid P. Relevance of free-living amoebae as hosts for phylogenetically diverse microorganisms. Parasitol Res. 2014;113(7):2407–14. [DOI] [PubMed] [Google Scholar]
- 11.Niyyati M, Lasgerdi Z, Lorenzo-Morales J. Detection and molecular characterization of potentially pathogenic free-living amoebae from water sources in Kish Island, Southern Iran. Microbiol Insights. 2015;8(Suppl 1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badirzadeh A, Niyyati M, Babaei Z, et al. Isolation of free-living amoebae from sarein hot springs in ardebil province, iran. Iran J Parasitol. 2011;6(2):1–8. [PMC free article] [PubMed] [Google Scholar]
- 13.Solgi R, Niyyati M, Haghighi A, et al. Occurrence of thermotolerant Hartmannella vermiformis and Naegleria spp. in hot springs of Ardebil Province, Northwest Iran. Iran J Parasitol. 2012; 7(2): 47–52. [PMC free article] [PubMed] [Google Scholar]
- 14.Latifi AR, Niyyati M, Lorenzo-Morales J, et al. Occurrence of Naegleria species in therapeutic geothermal water sources, Northern Iran. Acta Parasitol. 2017;62(1):104–109. [DOI] [PubMed] [Google Scholar]
- 15.Mahmoudi M.R., Kazemi B., Haghighi A., Karanis P. Detection of Acanthamoeba and Toxoplasma in river water samples by molecular methods in Iran. Iran J Parasitol. 10(2), pp. 250–257. [PMC free article] [PubMed] [Google Scholar]
- 16.Solgi R, Niyyati M, Haghighi A, et al. Thermo-tolerant Acanthamoeba spp. isolated from therapeutic hot springs in Northwestern Iran. J Water Health. 2012;10(4):650–6. [DOI] [PubMed] [Google Scholar]
- 17.Feiz Haddad MH, Ghasemi E, Maraghi S, Tavala M. Identification of Leishmania Species Isolated from Human Cutaneous Leishmaniasis in Mehran, Western Iran Using Nested PCR. Iran J Parasitol. 2016;11(1):65–72. [PMC free article] [PubMed] [Google Scholar]
- 18.Page FC. A new key to freshwater and soil Gymnamoebae: with instructions for culture: Freshwater Biological Association; 1988. [Google Scholar]
- 19.Hooshyar Hossein, Hosseinbigi Bahram, Saraei Mehrzad, et al. Genotyping of Acanthamoeba isolated from surface and stagnant waters of Qazvin, Central Iran. Iran Red Crescent Med J. 2013;15(6): 536–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lasjerdi Z, Niyyati M, Haghighi A, et al. Potentially pathogenic free-living amoebae isolated from hospital wards with immunodeficient patients in Tehran, Iran. Parasitol Res. 2011;109(3):575–80. [DOI] [PubMed] [Google Scholar]
- 21.Nazar M, Haghighi A, Niyyati M, et al. Geno-typing of Acanthamoeba isolated from water in recreational areas of Tehran, Iran. J Water Health. 2011;9(3):603–8. [DOI] [PubMed] [Google Scholar]
- 22.Niyyati M, Lorenzo-Morales J, Rezaie S, et al. Genotyping of Acanthamoeba isolates from clinical and environmental specimens in Iran. Exp Parasitol. 2009;121(3):242–5. [DOI] [PubMed] [Google Scholar]
- 23.Feiz-Haddad MH, Shokri A, Habibpour H, Nejadi SM. A review of Acanthamoeba keratitis in the Middle East and Iran. Journal of Acute Disease. 2019;8(4):133–41. [Google Scholar]
- 24.Edagawa A, Kimura A, Kawabuchi-Kurata T, et al. Isolation and genotyping of potentially pathogenic Acanthamoeba and Naegleria species from tap-water sources in Osaka, Japan. Parasitol Res. 2009;105(4):1109–17. [DOI] [PubMed] [Google Scholar]
- 25.Movahedi Z, Shokrollahi MR, Aghaali M, et al. Primary amoebic meningoencephalitis in an Iranian infant. Case Rep Med. 2012;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.John DT, De Jonckheere JF. Isolation of Naegleria australiensis from an Oklahoma lake. J Protozool. 1985;32(4):571–5. [DOI] [PubMed] [Google Scholar]
- 27.Hajialilo E, Niyyati M, Solaymani M, Rezaeian M. Pathogenic free-living amoebae isolated from contact lenses of keratitis patients. Iran J Parasitol. 2015;10(4):541–6. [PMC free article] [PubMed] [Google Scholar]
- 28.Mahmoudi MR, Rahmati B, Seyedpour SH, Karanis P. Occurrence and molecular characterization of free-living amoeba species (Acanthamoeba, Hartmannella, and Saccamoeba limax) in various surface water resources of Iran. Parasitol Res. 2015;114(12):4669–74. [DOI] [PubMed] [Google Scholar]
- 29.Feiz-Haddad M-H, Kassiri H, Kasiri N, et al. Prevalence and epidemiologic profile of acute cutaneous leishmaniasis in an endemic focus, Southwestern Iran. J Acute Dis. 2015;4(4):292–7. [Google Scholar]