ABSTRACT
The presence of nanomaterials in our everyday life is ever increasing, and so too are concerns about the possible health consequences of exposure to them. While evidence of their biological activity is growing, there is still scant knowledge of the epigenetic mechanisms that could be at play in these processes. Moreover, the great variability in the chemical and physical structures of these compounds handicaps the study of their possible health risks. Here we have synthesized reduced graphene oxide (rGO) through the thermal exfoliation/reduction of graphite oxide, and characterized the resulting material. We have then made use of Illumina’s MethylationEPIC arrays and bisulphite pyrosequencing to analyse the genome-wide and global DNA methylation dynamics associated with the medium-term exposure of human lung epithelial cells to rGO at concentrations of 1 and 10 µg/mL. The results show no genome-wide or global DNA methylation changes associated with either condition. Our observations thus suggest that medium-term rGO exposure does not have significant effects on the DNA methylation patterns of human lung epithelial cells.
KEYWORDS: Epigenetics, DNA methylation, nanomaterials, graphene, exposure
Introduction
Nanomaterials, those with dimensions below the 100 nm limit, are a growing component of our everyday life. Emanating from sources such as food, cosmetics or the chemical and biomedical industries, human exposure to nanomaterials has substantially increased during recent decades. Some of the most fundamental players in the nanomaterial ‘revolution’ of recent times have been carbon-based materials, such as fullerenes, carbon nanotubes (CNTs) and graphene. Their unique electrical, mechanical and thermal properties have led to their use in the development of a wide spectrum of technologies, like electrochemical storage (e.g., solar cells, batteries), electronics (e.g., transistors, field emission) and even biological technologies such as photothermal therapy and drug delivery [1]. Thus, directly or indirectly, these components are, or will soon be, present in our everyday lives, and in tandem with this increasing exposure, concern about their potential threat to human health has risen, in the case of both naturally occurring and engineered nanomaterials (ENMs) [2].
The potential toxicity of nanomaterials can be influenced not only by their composition, but also their size, the dose involved, time of exposure and target tissue [3]. Although the interest in the potential adverse effects caused by nanomaterials is recent, there is already considerable evidence of their biological activity in both in vitro and in vivo models [3–5], while, however, more limited evidence exists regarding direct associations with human exposure [6].
Epigenetic mechanisms constitute molecular pathways which can integrate environmental inputs to produce genomic responses [7] and thus are interesting targets for the study of the biological effects of nanomaterial exposure. To date, research has been carried out on the main epigenetic marks: DNA methylation (the covalent addition of methyl groups to DNA cytosines), histone modification (various post-translational modifications such as acetylation or methylation that occur at histone tails) and non-coding RNAs. The observed effects are varied and depend on a multitude of variables, although general trends have been observed in some works, especially regarding DNA methylation, such as a tendency towards DNMT (DNA methyltransferase) downregulation and DNA hypomethylation [8,9]. However, most studies have focused on changes either from a local perspective or have looked at total cellular levels of epigenetic marks. While providing necessary and informative results, genome-wide studies using up-to-date technologies are essential in order to uncover the genomic dynamics of epigenetic processes such as DNA methylation, which is context-specific [10,11], and also to detect more subtle changes which could in fact be the consequences of nanomaterial exposure in real-life settings.
Graphene derivatives such as reduced graphene oxide (rGO) and graphene oxide (GO) are currently being investigated for their biomedical applications, which cover a wide range of technologies such as their biofunctionalization with proteins, their use as drug delivery carriers, and the development of graphene-based biosensors [12]. It is therefore important to characterize the possible biological effects that these materials could have on living beings, and particularly to study the impact of these nanomaterial in human contact applications. In the present study we have focused on rGO, which is a graphene-like material produced by the reduction of oxidized graphene/graphite [13]. The main advantages of this method are its low-cost and the high-yield obtained, meaning that, ideally, rGO could constitute an advantageous methodology for generating graphene-like materials. In general, rGO effects in various in vitro and in vivo settings are usually smaller as compared to GO, or indeed non-existent [14], indicating that rGO could be a more biocompatible material, and that its biological characterization is therefore important. All the same, although various biochemical parameters have been studied, epigenetic analyses, especially on a genome-wide scale, remain to be carried out.
In this study we have synthesized rGO by the thermal exfoliation/reduction of graphite oxide and characterized the resulting material. Subsequently we analysed the genome-wide DNA methylation dynamics associated with mediun-term exposure to different concentrations of rGO in human lung epithelial cells using the Illumina MethylationEPIC platform, which covers more than 800,000 CpG sites and serves to potentially identify DNA methylation changes across the genome. Furthermore, we have also analysed global levels of DNA methylation by bisulphite sequencing of representative repetitive DNA loci.
Materials and methods
Synthesis and characterization of reduced graphene oxide
The material was prepared from a commercial synthetic graphite (Sigma-Aldrich) by an initial oxidation using a previously described modified Hummers’ method [15] to obtain graphite oxide, followed by a thermal exfoliation/reduction process similar to that described by Álvarez P. and colleagues [16] (see Supplementary Methods for additional information).
The elemental composition of the sample was determined in a LECO-CHNS-932 micro-analyser and the oxygen content was determined directly in a LECO-TF-900 furnace coupled to the previous equipment [15]. The analyses were performed using 1 mg of ground sample. The results are quoted from an average of the values of four determinations. In all cases, experimental error was < 0.5 % of the absolute value.
X-ray diffraction (XRD) analysis of the powdered sample was performed using a Bruker D8 Advance diffractometer. The radiation frequency employed was the Kα1 line from Cu (1.5406 Å), with a power supply of 40 kV and 40 mA [15]. The interlaminar distances (d002) of sheets and the average crystallite size along the c-axis (Lc) were obtained from the (002) reflection of the XRD patterns, which were recorded at steps of 0.01° and intervals of 6 s per step, using the Bragg’s law and the Scherrer equation respectively.
X-ray photoelectron spectroscopy (XPS) analyses were carried out in a VG-Microtech Mutilab 3000 device. The XPS C1s peak was analysed using a peak synthesis procedure that employs a combination of Gaussian and Lorentzian functions [17] in order to identify the functional groups and their respective percentages. The binding energy profiles were deconvoluted as follows: undamaged structures of Csp2-hybridized carbon (284.5 eV), damaged structures or sp3-hybridized carbons (285 eV), C–OH groups (285.7 eV), C-O-C functional groups (287 eV), C = O functional groups (287.5 eV) and C(O)OH groups at 288.7 eV).
The specific surface area was calculated from the N2 adsorption isotherm at 77 K using the Brunauer–Emmett–Teller (BET) equation [18]. The isotherm was obtained using an ASAP 2020 Micromeritics equipment. The sample was outgassed at 300°C for 3 h under vacuum conditions prior to the test.
SEM (scanning electron microscopy) images were obtained using a field emission gun scanning electron microscope (QUAN-TAN FEG 650, FEI) operating at 5 kV, and TEM (transmission electron microscopy) observations were performed with a JEOL 2000 EX-II instrument operating at 160 keV [15].
Cell culture, nanomaterial preparation and exposure
Human airway epithelial BEAS-2B cells were provided by Dr. G.M. Albaiceta’s group (Instituto de Investigación Sanitaria del Principado de Asturias). The cells were cultured as a monolayer with serum-free BEGM medium (Bronchial epithelial cell growth medium, Lonza) in culture dishes. This medium is complemented with the following factors: retinoic acid, insulin, hydrocortisone, transferrin, epinephrine, triiodothyronine, Bovine Pituitary Extract, hEGF (human epidermal growth factor) and GA-1000 (Gentamicin and Amphotericin). Cultures were maintained at 37ºC in a humified atmosphere of 5% CO2.
The rGO stock solution was prepared by pre-wetting with ethanol followed by dispersion in BSA-water (Nanogenotox protocol 2011, see Supplementary Methods for additional information). For rGO exposure, the working dilutions of 1 and 10 µg/mL were prepared directly in cell culture medium. 150 cm2 dishes were seeded with 106 cells, which were left to attach for 4–8 hours prior to the addition of the nanomaterials. Exposure medium was renewed every 4–5 days.
Global and repetitive DNA methylation analyses
Bisulphite pyrosequencing was used for the evaluation of the DNA methylation status of 5 different families of repetitive DNA: LINE1 [19], D4Z4 and NBL2 [20], SATα and AluYb8 [21] (see primer sequences in Table S2). First, genomic DNA was isolated by standard phenol-chloroform extraction. Secondly, bisulphite conversion was performed with the EZ DNA methylation-gold kit (Zymo Research) following the manufacturer’s instructions. Thirdly, modified DNA was amplified by PCR and, finally, pyrosequenced using PyroMark 24 reagents and a vacuum prep workstation, equipment and software (Qiagen).
Genome-wide DNA methylation analyses
The Illumina Infinium MethylationEPIC BeadChip was used for the genome-wide interrogation of the DNA methylation status of more than 800,000 CpG sites in the human BEAS-2B cell line genome. A total of 12 samples were analysed, which consisted of 3 exposure groups (control, 1 and 10 µg/mL rGO) at 2 time intervals (15 and 30 days), each condition having 2 technical replicates. DNA samples were hybridized to the BeadChip following the Illumina Infinium HD methylation protocol, and this service was provided by the Centro Nacional de Genotipado (CEGEN-ISCIII, Spain, http://www.cegen.org).
Infinium MethylationEPIC BeadChip data preprocessing
All the data were preprocessed by means of the statistical software R (version 3.4.2). The IDAT files from the Illumina MethylationEPIC array were processed following a pipeline built on the R/Bioconductor package minfi (version 1.22.1) [22]. Probes which have been shown to be potentially crossreactive or multimapping [23], those overlapping genetic variants [24] and those with a detection p-value > 0.01 in at least one sample were filtered out. The methylation data was normalized in a two-step procedure: first using the Noob method [25] as implemented in minfi for background correction and subsequently using the BMIQ method [26] to correct for probe design bias, implemented in the R/Bioconductor package ChAMP (version 2.8.9) [27]. β-values were extracted with the getBeta minfi function and M-values were obtained by the logit transformation of the normalized β-values with the R/Bioconductor package lumi (version 2.28.0) [28].
Genomic annotation of array probes
MethylationEPIC BeadChip probes were mapped to different CpG island-associated or gene-associated regions using the annotation of the R/Bioconductor package IlluminaHumanMethylationEPICanno.ilm10b2.hg19 (version 0.6.0). To annotate array probes to genomic repeats, because most of the probes do not cover repetitive elements, their locations were expanded to 100 bp windows and checked for overlaps with repetitive DNA families extracted from RepeatMasker libraries (hg19 – Feb 2009 – RepeatMasker open-4.0.5 – Repeat Library 20,140,131) [29]. This strategy, adapted from Lai et al. [30], uses close-proximity probes as surrogates for the methylation status of repeat elements.
Characterization of BEAS-2B’s DNA methylation patterns and definition of differentially methylated probes in the exposure experiments
To characterize the genome-wide DNA methylation patterns of the BEAS-2B cell line we collapsed the measurements for the 4 control samples used in the rGO exposure experiments, which allowed us to have highly reliable DNA methylation measurements for the CpG sites. To achieve this, we first filtered out CpG sites which varied more than 10% between the different samples (3% of all probes) and then averaged the DNA methylation values of the remaining sites (a total of 774,728) between the 4 controls.
To perform gene ontology analyses, the R/Bioconductor package missMethyl (version 1.10.0) [31] was used (see Supplementary Methods for additional information). To better visualize the results obtained, the significant ontologies detected were semantically summarized using the online tool ReViGO (http://revigo.irb.hr/) [32].
To study differentially methylated CpG sites between the conditions we collapsed the two technical replicates for each condition into a single measurement. In this case we first filtered out probes with differences in β-values between the pairs of technical replicates larger than 5% (on average, 5% of the probes) and then averaged the β-value of each remaining CpG site between the two replicates (a stricter threshold was used because only 2 samples were collapsed). Finally, we defined as differentially methylated those probes with absolute differences in β-value between different conditions equal to or over 30%. We further looked for lesser differences, such as 20% or 10%.
Results
Characterization of reduced graphene oxide
The oxidation of the synthesized graphene material (see Methods) was confirmed by XRD (Figure 1(a)); the interlayer distance (d002) was 0.35 nm, the average crystallite size along the c-axis (Lc) was 0.89 nm, and the average number of stacked layers, estimated by the equation (Lc/d) +1, ranged from 3 to 4. Representative SEM and TEM images showing the morphology of the rGO are shown in Figure 1(b). The rGO showed an oxygen content of up to 24 wt. %, and low contents of sulphur (1.1 wt. %), nitrogen (0.3 wt. %), and hydrogen (0.9 wt. %), determined by elemental analysis. This oxygen was mostly in the form of epoxy groups (11.3 at. %), followed by alcohols (8.6 at. %), carboxylic groups (6.4 at. %), and ketonic groups (4.8 at. %), according to XPS spectra (Figure 1(c)). The calculated BET surface area of the rGO was 397 m2 /g, and the isotherm (Figure 1(d)) showed a steady increase in nitrogen adsorption and hysteresis in the desorption branch, which is indicative of the presence of mesopores (mainly several tens nm), as a result of the mis-stacking of single layers (or small packs of a few layers).
Figure 1.
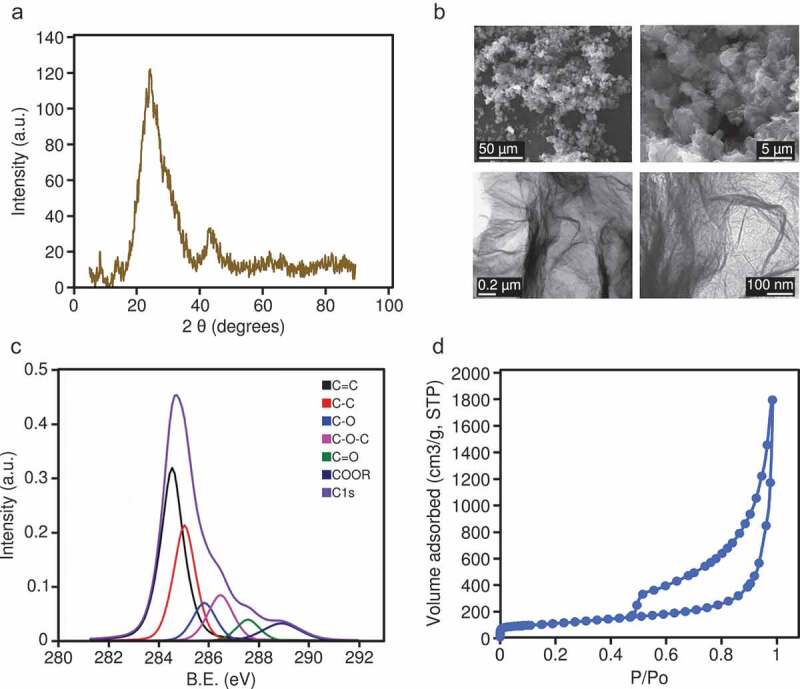
(a) XRD pattern for the rGO; intensity is expressed in arbitrary units (a.u.). (b) Representative SEM images (upper panels) and TEM images (lower panels) of the rGO. (c) High-resolution XPS spectra; intensity is expressed in arbitrary units (a.u.), binding energy (B.E.) is measured in electronvolts (eV). (d) Nitrogen adsorption/desorption isotherm of rGO; P/P0 is the relative pressure.
Genome-wide DNA methylation characterization of human lung epithelial cells
We used the Illumina MethylationEPIC platform to characterize the genome-wide DNA methylation patterns of BEAS-2B human lung epithelial cells, and observed the expected bimodal distribution of DNA methylation (Figure 2(a), upper panel) whereby most of the probes where either unmethylated or highly methylated. To better characterize the epigenomic patterns observed, we calculated gene ontology enrichments for the genes associated with highly methylated (β-values > 0.8) or unmethylated (β-values < 0.2) CpG sites.
Figure 2.
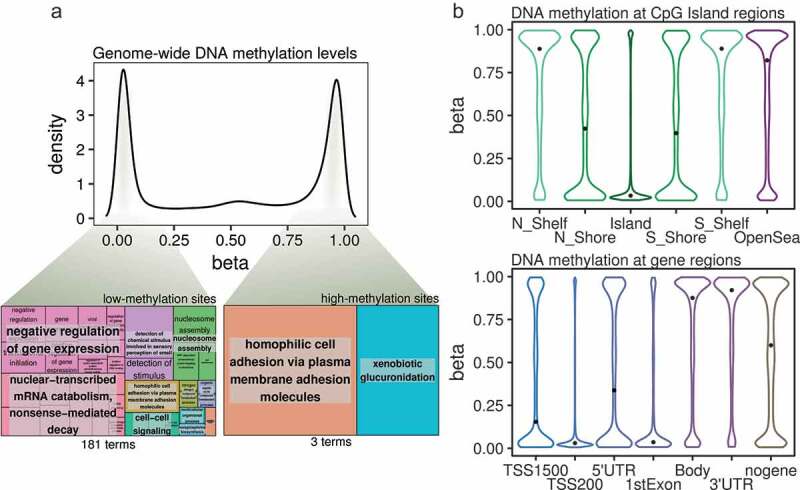
(a) Density plot showing DNA methylation β-values from all of the CpG sites analysed by the Illumina MethylationEPIC platform in the BEAS-2B control cells. Highlighted from the density plot are two treemap plots indicating the semantic summaries of gene ontologies found for those genes containing either low- or high-methylation CpG sites. (b) Violin plots indicating the DNA methylation β-value distribution of CpGs mapped to CpG Island-related regions (upper panel) or gene-related regions (lower panel). The median values of the distributions are highlighted by a black dot. (N_Shelf: north shelf, N_Shore: north shore, Island: CpG Island, S_Shelf: south shelf, S_Shore: south shore; TSS1500, TSS200: 1500 or 200 bp from transcription start site, nogene: intergenic).
DNA methylation can have different values within one particular gene and, specifically, we found that as much as 75% of the genes interrogated by the array contained both highly- and lowly-methylated probes in the BEAS-2B cells, and therefore we focused on genes which exclusively contained high- or low-methylation CpG sites (a total of 3357 and 2491 genes, respectively, see Supplementary Methods). Interestingly, while lowly-methylated genes were associated with several distinct pathways, some relating to the negative regulation of biological processes (Figure 2(a), lower left panel), highly-methylated genes were associated with many fewer pathways (Figure 2(a), lower right panel; see Table S1 for full information), suggesting that genes with low levels of DNA methylation are associated with specific functional pathways in the BEAS-2B cell line.
Next, we studied the DNA methylation state of the lung epithelial cells at different genomic sites. Regarding CpG island membership (Figure 2(b), upper panel), the lowest DNA methylation values were found at CpG islands, followed by immediate border elements (island shores), while high methylation values were found at farther elements (island shelves) and open sea sites. Furthermore, when looking at gene-region classifications (Figure 2(b), lower panel), low DNA methylation was found at transcription start sites and first exons – but not at 5ʹUTR regions – while high DNA methylation values were mainly observed at gene bodies and 3ʹUTR regions, and intermediate-high values at intergenic regions (the median methylation of all of the interrogated probes was 0.59). Thus, DNA methylation levels are correlated with CpG-density and specific genic regions at a genome-wide level in the BEAS-2B cell line.
Genome-wide DNA methylation changes upon reduced graphene oxide exposure
In order to characterize DNA methylation changes associated with rGO exposure in human lung epithelial cells, we designed an experimental setup with 2 different nanomaterial concentrations and 2 different exposure times (Figure 3(a), left panel): 1 or 10 μg/mL for either 15 or 30 days (as well as control groups with no exposure). To capture DNA methylation states at a genome-wide level, we used the Illumina MethylationEPIC platform on 2 technical replicates per condition, for a total of 12 arrays. We collapsed the replicates in each condition to define site specific DNA methylation values by retaining sites with differences between measurements of less than 5% (Figure 3(a), right panel; see methods), resulting in a final tally of 696,365 evaluated CpG sites across all conditions.
Figure 3.
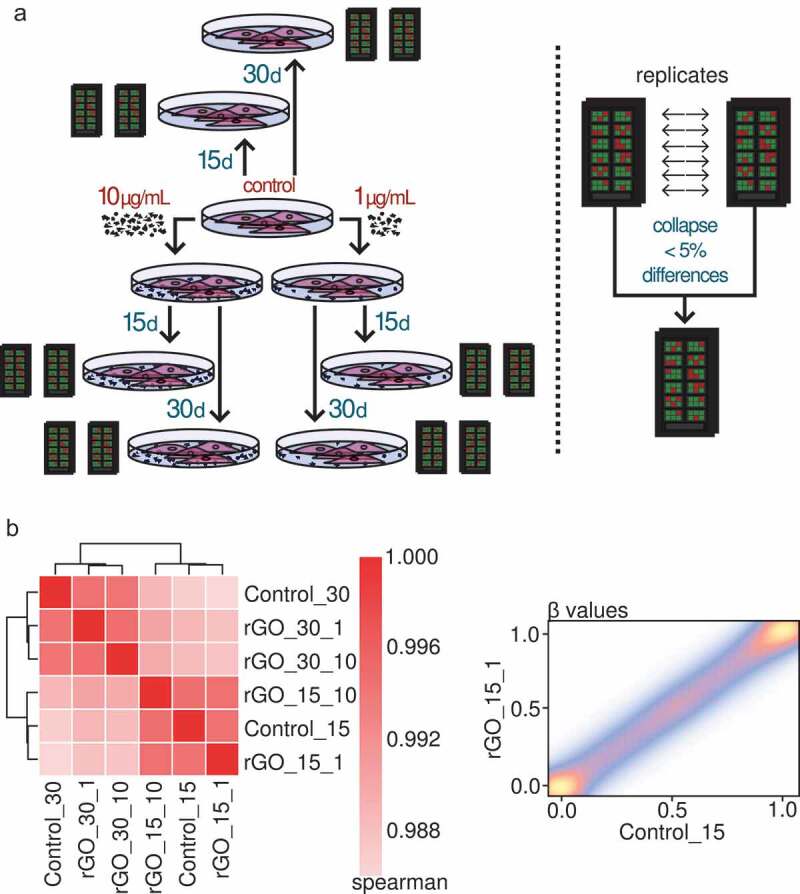
(a) Experimental set-up of the rGO exposure experiments and collapsing of the replicates. (b) Left panel: heatmap depicting Spearman correlation values for the correlation between the profiled β-values of the CpG sites of the different collapsed experimental conditions. Right panel: density scatter plot showing an example of the correlation between the β-values of the CpG sites of the rGO 15-day 1 μg/mL condition and the 15-day control condition. The colour indicates the density of the points, from low (blue) to high (yellow).
Next, we compared the different experimental conditions in order to look for differentially methylated sites. Interestingly, we found no DNA methylation changes associated with rGO exposure at concentrations of either 1 or 10 μg/mL at either 15 or 30 days of exposure. Correlation analyses (Figure 3(b)) showed a very strong epigenomic similarity between all of the conditions, especially within the same time groups. We lowered the threshold to 10% difference in methylation and still found no differentially methylated CpG sites. Our results therefore indicate that lung epithelial cells do not suffer significant DNA methylation changes when confronted with rGO during medium-term exposure.
Global and repetitive DNA methylation changes upon reduced graphene oxide exposure
Finally, we looked for global DNA methylation differences by performing bisulphite pyrosequencing on 5 different families of repetitive DNA: LINE1 [19], D4Z4 and NBL2 [20], SATα and AluYb8 [21] (see primer sequences in Table S2 and pyrosequencing measurements in Table S3). These multi-represented sequences cover large portions of the whole genome, and have been regularly used as proxies for the evaluation of the global levels of DNA methylation across the whole genome (e.g., LINE1, AluYb8), or at specific regions, e.g., subtelomeric D4Z4 repeat or the centromeric SATα repeats.
In consonance with the earlier genome-wide specific approach, we found no DNA methylation changes associated with rGO exposure at either concentration or exposure time for any repeat family (Figure 4). Furthermore, we made use of our previously generated genome-wide data to validate the bisulphite pyrosequencing observations by mapping the array probes to different repetitive DNA locations and looking at the DNA methylation β-values measured by the array (see methods). We again found no differences between conditions for a variety of genomic repeat families (see Figure S1). Thus our results suggest that lung epithelial cells do not experience extensive global and repetitive-associated DNA methylation changes when exposed to reduced graphene oxide.
Figure 4.
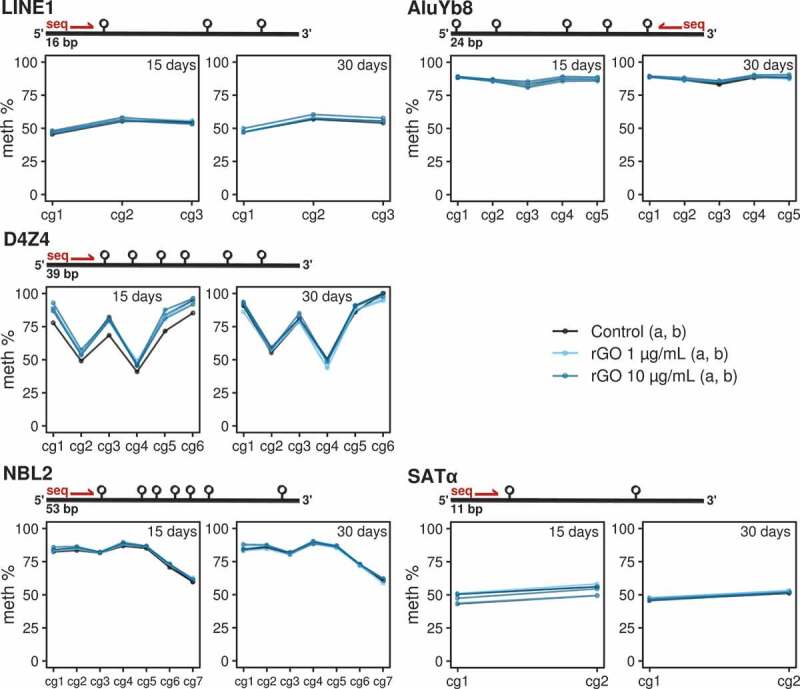
Line/point plots describing the DNA methylation values of different repeat elements measured by bisulphite pyrosequencing. For each experimental condition, the 2 technical replicates (‘a’ and ‘b’) are plotted as independent lines. For each DNA repeat region, a different number of CpGs were analysed. Values for the 15-day treatments at 0, 1 and 10 μg/mL concentrations and 30-day treatments at 0, 1 and 10 μg/mL concentrations are separated in 2 different plots.
Discussion
Interest in the potential consequences of the biological activities of nanomaterials has risen dramatically in the last few years. However, while biochemical and physiological studies are starting beginning to be more common, genomic studies, especially on a genome-wide scale are more scarce, not to mention those looking at epigenetic marks [8]. Thus there is a wide knowledge gap to fill in terms of the epigenetic mechanisms that might mediate the biological effects of nanomaterials. Of these, carbon-based materials are among the most important compounds application-wise, and we have previously studied the impact of MWCNTs on the epigenome of human lung epithelial cells [33] because inhalation is among the most common and most studied routes of exposure to nanomaterials We also focused on medium-term exposures at low concentrations, because these conditions might better mimic real human subchronic exposure.
In the present study we have focused on rGO because it is a low-cost and promising graphene-like derivative. However, the structure of rGO is far from that of perfect graphene sheets, and, importantly, oxygen is still present at relevant quantities in the structure of rGO, such that the hydrophilic groups present could alter its behaviour in biological environments and even provide functionalities different to those of GO.
We have investigated the genome-wide and global DNA methylation dynamics associated with rGO exposure in human lung epithelial cells, utilizing 2 different concentrations (1 and 10 µg/mL) with medium-term exposure times of 15 and 30 days. We have used concentrations in the range of those previously reported for in vitro studies using this and other graphene-based nanomaterials [14]. These studies, which evaluate physiological changes, usually choose shorter exposure windows (on a scale of hours). However, we sought to characterize altered epigenetic states which are maintained through larger time-scales because of the role they may play in the regulation of gene expression [34]. Nonetheless, because in general the influence of graphene-based nanomaterials (and in particular of rGO) on an epigenome-wide scale has not been studied, the exposure conditions (time and concentration) which can damage cells, at least on an epigenetic level, are still to be established.
We found no notable changes in DNA methylation associated with rGO exposure under any of the conditions evaluated, and regardless of the analysis method used (i.e., global or locus-specific approaches). Moreover, we used repetitive DNA-associated array probes to validate the lack of differences found in the analysis of repetitive DNA sequences in LINE1, D4Z4, NBL2, SATα and AluYb8, all of which have been widely used for the estimation of global levels of DNA methylation [19–21]. Our analyses thus indicate that medium-term in vitro exposure to rGO at concentrations up to 10 µg/mL has no apparent effects on the epigenome of human lung epithelial cells.
Interestingly, a recent in vitro study also found no adverse short-term cytotoxic or genotoxic effects induced by GO or rGO in murine lung cells at 5–200 μg/mL doses [35] and rGO at the same concentrations as used in our study did not cause any noticeable effects in murine spermatogonial stem cells [36]. On the other hand, rGO nanosheets were shown to have short-term cytotoxic effects on human lung cancer cells [37], pointing towards the choice of transformed versus normal in vitro models being an important variable. However, another recent study has found no effects of rGO nanosheet of up to 100 μg/mL concentrations on the cell viability of human lung cancer cells, although effects were found for a murine macrophage cell line [38]. These mixed findings point to the importance of standardizing the experimental assays used to evaluate cellular parameters. Our work provides insight into how doses similar to those used in other in vitro studies produce no effects even after up to 30 days of exposure.
Moreover, while in vivo studies do show adverse effects related to the exposure of graphene-based nanomaterials (GNMs), in general, rGO effects observed in various in vivo settings are usually smaller compared to GO or other types of GNMs [14,39]. This effect has also been observed in vitro: it has been hypothesized that the higher oxygenated functional group content of GO increases its cellular toxicity, as compared to rGO toxicity, through ROS pathways [40]. Nevertheless, a recent study using BEAS-2B cells has shown that GO compounds in general generate less genotoxicity than other graphene-derived materials, albeit they were found to cause slight DNA methylation changes at the global level [41].
Thus, our findings of no DNA methylation changes caused by rGO at low doses with medium-term exposures are not entirely surprising. Our observations, when accompanied by the aforementioned results on non-existent or generally lower cellular effects of rGO versus GO suggest that rGO could have considerable biocompatibility. It is important to stress that at different doses, or time exposures, to those used in this work it is possible that effects might be observed, and that our results would be strengthened if extended to other lung epithelial cell lines. Moreover, we have used an in vitro model of airway epithelium as an initial approximation, but our observations should be expanded upon with the use of in vivo models, which can more accurately model the process of exposure to nanomaterials through inhalation. We have also not looked at other epigenetic marks such as histone modifications, which are interrelated [42]. Our study is a starting point towards future analyses which should try to combine and integrate different technologies and, we think, focus on genome-wide approaches in order to uncover more subtle changes which could be associated with exposure to nanomaterials.
Funding Statement
This work has been financially supported by: The Plan Nacional de I+D+I co-funding FEDER (PI15/00892 and PI18/01527); the Government of the Principality of Asturias PCTI-Plan de Ciencia, Tecnología e Innovación de Asturias co-funding 2018-2022/FEDER (IDI/2018/146 and IDI/2018/121); AECC (PROYE18061FERN); FGCSIC (0348_CIE_6_E); IUOPA-ISPA-FINBA (The IUOPA is supported by the Obra Social Cajastur-Liberbank, Spain); A.F. Fernández is supported by a Miguel Servet II fellowship (contract CPII16/00007); R.F. Pérez is supported by ISPA-FINBA.
Acknowledgments
Special thanks go to Ronnie Lendrum, the English style editor, for her critical, constructive reading and invaluable comments. We thank Covadonga Huidobro for her help with the BEAS-2B cells.
Availability of data and materials
The raw IDAT and preprocessed data generated in this study by Illumina Infinium MethylationEPIC BeadChip technology are available in the ArrayExpress public repository under accession E-MTAB-7719.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
References
- [1].Bhushan B, Luo D, Schricker SR, et al., editors. Handbook of nanomaterials properties. Berlin: Springer; 2014. [Google Scholar]
- [2].MacPhail RC, Grulke EA, Yokel RA.. Assessing nanoparticle risk poses prodigious challenges: nanoparticle risk assessment. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2013;5:374–387. [DOI] [PubMed] [Google Scholar]
- [3].Yokel RA, MacPhail RC. Engineered nanomaterials: exposures, hazards, and risk prevention. J Occup Med Toxicol. 2011;6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Willhite CC, Karyakina NA, Yokel RA, et al. Systematic review of potential health risks posed by pharmaceutical, occupational and consumer exposures to metallic and nanoscale aluminum, aluminum oxides, aluminum hydroxide and its soluble salts. Crit Rev Toxicol. 2014;44:1–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yokel RA, Hussain S, Garantziotis S, et al. The yin: an adverse health perspective of nanoceria: uptake, distribution, accumulation, and mechanisms of its toxicity. Env Sci Nano. 2014;1:406–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pietroiusti A, Magrini A. Engineered nanoparticles at the workplace: current knowledge about workers’ risk. Occup Med. 2014;64:319–330. [DOI] [PubMed] [Google Scholar]
- [7].Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13:97–109. [DOI] [PubMed] [Google Scholar]
- [8].Sierra MI, Valdés A, Fernandez AF, et al. The effect of exposure to nanoparticles and nanomaterials on the mammalian epigenome. Int J Nanomedicine. 2016;11:6297–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stoccoro A, Karlsson HL, Coppedè F, et al. Epigenetic effects of nano-sized materials. Toxicology. 2013;313:3–14. [DOI] [PubMed] [Google Scholar]
- [10].Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. [DOI] [PubMed] [Google Scholar]
- [11].Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. [DOI] [PubMed] [Google Scholar]
- [12].Priyadarsini S, Mohanty S, Mukherjee S, et al. Graphene and graphene oxide as nanomaterials for medicine and biology application. J Nanostructure Chem. 2018;8:123–137. [Google Scholar]
- [13].Pei S, Cheng H-M. The reduction of graphene oxide. Carbon. 2012;50:3210–3228. [Google Scholar]
- [14].Ema M, Gamo M, Honda K. A review of toxicity studies on graphene-based nanomaterials in laboratory animals. Regul Toxicol Pharmacol. 2017;85:7–24. [DOI] [PubMed] [Google Scholar]
- [15].Botas C, Álvarez P, Blanco C, et al. Critical temperatures in the synthesis of graphene-like materials by thermal exfoliation–reduction of graphite oxide. Carbon. 2013;52:476–485. [Google Scholar]
- [16].Álvarez P, Blanco C, Santamaría R, et al. Tuning graphene properties by a multi-step thermal reduction process. Carbon. 2015;90:160–163. [Google Scholar]
- [17].Yang D, Velamakanni A, Bozoklu G, et al. Chemical analysis of graphene oxide films after heat and chemical treatments by X-ray photoelectron and Micro-Raman spectroscopy. Carbon. 2009;47:145–152. [Google Scholar]
- [18].Roldán S, Villar I, Ruíz V, et al. Comparison between electrochemical capacitors based on NaOH- and KOH-activated carbons †. Energy Fuels. 2010;24:3422–3428. [Google Scholar]
- [19].Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. [DOI] [PubMed] [Google Scholar]
- [20].Ribel-Madsen R, Fraga MF, Jacobsen S, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS ONE. 2012;7:e51302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Choi SH, Worswick S, Byun H-M, et al. Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int J Cancer. 2009;125:723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinforma Oxf Engl. 2014;30:1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen Y, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Triche TJ, Weisenberger DJ, Van Den Berg D, et al. Low-level processing of Illumina Infinium DNA methylation beadarrays. Nucleic Acids Res. 2013;41:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinforma Oxf Engl. 2013;29:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Morris TJ, Butcher LM, Feber A, et al. ChAMP: 450k chip analysis methylation pipeline. Bioinforma Oxf Engl. 2014;30:428–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinforma Oxf Engl. 2008;24:1547–1548. [DOI] [PubMed] [Google Scholar]
- [29].Tejedor JR, Bueno C, Cobo I, et al. Epigenome-wide analysis reveals specific DNA hypermethylation of T cells during human hematopoietic differentiation. Epigenomics. 2018;10:903–923. [DOI] [PubMed] [Google Scholar]
- [30].Lai AY, Mav D, Shah R, et al. DNA methylation profiling in human B cells reveals immune regulatory elements and epigenetic plasticity at Alu elements during B-cell activation. Genome Res. 2013;23:2030–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinforma Oxf Engl. 2016;32:286–288. [DOI] [PubMed] [Google Scholar]
- [32].Supek F, Bošnjak M, Škunca N, et al. REVIGO summarizes and visualizes long lists of gene ontology terms. PloS One. 2011;6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sierra MI, Rubio L, Bayón GF, et al. DNA methylation changes in human lung epithelia cells exposed to multi-walled carbon nanotubes. Nanotoxicology. 2017;11:857–870. [DOI] [PubMed] [Google Scholar]
- [34].Almouzni G, Cedar H. Maintenance of epigenetic information. Cold Spring Harb Perspect Biol. 2016;8:a019372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bengtson S, Kling K, Madsen AM, et al. No cytotoxicity or genotoxicity of graphene and graphene oxide in murine lung epithelial FE1 cells in vitro: Graphene and Graphene Oxide in vitro. Environ Mol Mutagen. 2016;57:469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hashemi E, Akhavan O, Shamsara M, et al. Synthesis and cyto-genotoxicity evaluation of graphene on mice spermatogonial stem cells. Colloids Surf B Biointerfaces. 2016;146:770–776. [DOI] [PubMed] [Google Scholar]
- [37].Tabish TA, Pranjol MZI, Hayat H, et al. In vitro toxic effects of reduced graphene oxide nanosheets on lung cancer cells. Nanotechnology. 2017;28:504001. [DOI] [PubMed] [Google Scholar]
- [38].Esquivel-Gaon M, Nguyen NHA, Sgroi MF, et al. In vitro and environmental toxicity of reduced graphene oxide as an additive in automotive lubricants. Nanoscale. 2018;10:6539–6548. [DOI] [PubMed] [Google Scholar]
- [39].Bengtson S, Knudsen KB, Kyjovska ZO, et al. Differences in inflammation and acute phase response but similar genotoxicity in mice following pulmonary exposure to graphene oxide and reduced graphene oxide. Plos One. 2017;12:e0178355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Das S, Singh S, Singh V, et al. Oxygenated functional group density on Graphene Oxide: its effect on cell toxicity. Part Part Syst Charact. 2013;30:148–157. [Google Scholar]
- [41].Chatterjee N, Yang J, Choi J. Differential genotoxic and epigenotoxic effects of graphene family nanomaterials (GFNs) in human bronchial epithelial cells. Mutat Res Toxicol Environ Mutagen. 2016;798-799:1–10. [DOI] [PubMed] [Google Scholar]
- [42].Kundaje A, Meuleman W, Ernst J, et al.; Roadmap Epigenomics Consortium . Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw IDAT and preprocessed data generated in this study by Illumina Infinium MethylationEPIC BeadChip technology are available in the ArrayExpress public repository under accession E-MTAB-7719.


