Abstract
Celiac disease (CeD) is a common immune-mediated disease of the small intestine that is triggered by exposure to dietary gluten. While the HLA locus plays a major role in disease susceptibility, 39 non-HLA loci were also identified in a study of 24,269 individuals. We now build on this earlier study by adding 4125 additional Caucasian samples including an Argentinian cohort. In doing so, we not only confirm the previous associations, we also identify two novel independent genome-wide significant associations at loci: 12p13.31 and 22q13.1. By applying a genomics approach and differential expression analysis in CeD intestinal biopsies, we prioritize potential causal genes at these novel loci, including LTBR, CYTH4, and RAC2. Nineteen prioritized causal genes are overlapping known drug targets. Pathway enrichment analysis and expression of these genes in CeD biopsies suggest that they have roles in regulating multiple pathways such as the tumor necrosis factor (TNF) mediated signaling pathway and positive regulation of I-κB kinase/NF-κB signaling.
Subject terms: Coeliac disease, Genome-wide association studies, Disease genetics, Genetics research
Introduction
Celiac disease (CeD) is a common immune-mediated disease (IMD) present in ~1% of the Western population that is characterized by inflammation of the small intestine, villous atrophy, and crypt hyperplasia. CeD is caused by an interaction of environmental and genetic factors [1]. The main environmental factor is exposure to dietary gluten and the only available treatment is life-long adherence to a gluten-free diet. An estimation of the genetic component of CeD in twins is 75% [2]. The main genetic risk factors for the development of CeD are human leukocyte antigen (HLA) molecules, specifically the HLA-DQ2.5 and HLA-DQ2.2 haplotypes, which are responsible for 40% of disease heritability [3]. However, while the presence of the HLA-DQ2.5 and HLA-DQ2.2 haplotypes are necessary to develop the disease, they are not sufficient in themselves to promote the disease; many HLA carriers do not develop CeD, indicating that additional genetic factors may play a role.
Previous genome-wide association studies (GWAS) [4, 5] have identified 26 loci outside the HLA-region that increase the risk of developing CeD. In 2011, we fine-mapped more than 50% of these previously CeD-associated loci and identified 13 novel non-HLA loci using the Immunochip platform, which led to the identification of 57 independent SNPs in 39 loci. Although it remains a challenge to pinpoint the causal variants and genes at these 39 CeD-associated loci, much progress has been made using integrative functional genomic approaches that combine multiple layers of omics information. Analysis of the candidate causal genes for CeD from these loci has led to a better understanding of disease pathology and identified new causal pathways, such as interferon gamma signaling [6] and autophagy [7].
Bigger sample sizes and integrative omics approaches for other diseases have not only led to a better understanding of disease biology [8–10], they have also pinpointed new treatment options [11]. In the present study, we aimed to identify new loci contributing to CeD by increasing the sample size (adding >4000 new samples) and adding ethnic diversity of our patient cohort. We then used a system genetics approach to identify new pathways playing a role in disease pathogenesis.
Material and methods
Subjects
Aside from the two cohorts from Latin America of self-reported Argentinian origin, all individuals included in our analysis are Europeans from the Netherlands, Spain, Italy, Ireland, and Poland (Supplementary Table 1). All cases were diagnosed according to standard clinical criteria, positive tissue transglutaminase antibodies or endomysial antibodies and, in all cases, small intestinal biopsy with Marsh stages II or III. Written informed consent was obtained for all individuals and the study was approved by the ethics committee or institutional review board of all participating institutions. We used the British, Italian, Polish, Spanish, and Dutch cohorts that were included and described in our previous Immunochip analyses [3], and added additional samples to the Dutch, Spanish, Italian, and Polish cohorts (for a total of 3925 cases and 4743 controls) following the same inclusion criteria (Supplementary Table 1). The Indian cohort included in Trynka’s study was instead excluded being too genetically different from our cohorts. We also added an Irish cohort (393 cases and 455 controls) that was described previously by Coleman et al. [12], and two Argentinian cohorts specifically collected for this study. The Argentinian cases were included after diagnosis by the presence of tissue transglutaminase IgA or anti-deamidated gliadin peptide IgG antibodies and positive endomysial antibodies, and intestinal biopsy class March IIIA or above. Argentinian controls were unselected blood donors and population controls. Blood samples for DNA isolation were collected in the Gastroenterology hospital “Dr. C. Bonorino Udaondo” in Buenos Aires, Argentina and in the OSEP in Mendoza, Argentina after written informed consent was given. See Supplementary note 1 for more information.
Genotyping and quality control
DNA isolation of the Argentinian samples was carried out at the University Medical Center Groningen (UMCG) by the salting out procedure [13]. The additional samples were genotyped on the Immunochip at the UMCG, following Illumina’s standard protocols. Variant calling was performed using Genome Studio with the same cluster used by Trynka et al. [3]. All quality control (QC) checks and filters were performed per cohort using PLINK versions 1.07 [14] and 1.9 [15]. Specifically, non-polymorphic markers and markers with duplicated rs identifiers were removed and data mapped to the human reference 19 (build 37) using the LiftOver tool from UCSC (http://genome.ucsc.edu/cgi-bin/hgLiftOver) [16]. Samples with a call rate <98% and single nucleotide polymorphism (SNPs) with a call rate <99% or Hardy–Weinberg equilibrium exact test <0.001 were discarded.
Hidden relationships between samples within cohorts were identified by calculating identity by descent estimates, which were derived using 14,453 non-HLA independent variants (two rounds of linkage-disequilibrium (LD) pruning using the “indep” option (window size of 50, step ≥2 size, r2 > 0.2)). When we identified duplicated individuals, the sample with the best call rate was kept. Individuals with first and second degree of consanguinity (pihat > 0.2) were excluded. Population outliers within each cohort were detected by multi-dimensional scaling (MDS) plots using R Studio (http://www.rstudio.com/) with the previously described set of SNPs and excluded.
Due to its confirmed association to CeD, extended LD and high complexity, we excluded the HLA region (chr6:19892021–39892022); we also excluded the X chromosome. After applying sample and SNP QC filters, we obtained 12,948 cases and 14,826 controls from eight different cohorts (Supplementary Table 1) and 127,855 SNPs for association analysis.
Statistical analysis
Logistic regression was implemented per cohort using PLINK 1.9 [15] including gender and three MDS components as covariates to correct for population stratification. We used a sample-size-weighted Z-score meta-analysis on the association results of the eight cohorts (Supplementary Table 1) in PLINK 1.9 [15]. The sample-phenotype information, summary statistics from the association analysis per cohort and the results from the meta-analysis have been deposited at the European Genome-phenome Archive, which is hosted by the EBI and the CRG, under accession number EGAS00001003805 (https://www.ebi.ac.uk/ega/studies/EGAS00001003805). Manhattan plots of −log10P and the QQ plot were generated using RStudio (http://www.rstudio.com/). The inflation factor in the non-CeD associated regions was 1.67 (Supplementary Fig. 1a) as expected by the Immunochip designed that included mainly immune-related genes, but similar to our previous Immunochip study [3] non-excess of associations was observed in three densely genotyped loci selected for bipolar disorder (λ = 1.055, Supplementary Fig. 1b). We used the standard genome-wide significance threshold of p < 5 × 10−8, as PLINK reports p-values below 2.22 × 10−16 as 0 for the meta-analysis, we assigned an arbitrary value of 9.99 × 10−17 for potting and results are indicate as p < 2.22 × 10−16 in the manuscript. Regional association plots for genome-wide significant loci were generated using LocusZoom (http://locuszoom.org/).
SNP annotation
We used the SNP2GENE function in FUMA (http://fuma.ctglab.nl/) to perform functional mapping and annotation of results from the association analysis. We selected the European population of the 1000 Genomes Project phase 3 to calculate LD and included all SNPs with a minor allele frequency (MAF) > 0.001. For gene mapping, we selected the eQTL option, including all databases except GTEx v6 (because it already includes the latest version (v7) and the samples overlap between the two databases). We included only significant eQTLs (FDR < 0.05). We used all genes in Ensembl version 92 for gene mapping.
Gene annotation
To explore tissue-specificity and biological context of identified genes and perform pathway enrichment analysis, we used the function GENE2FUNC. This function also allowed us to identify genes that are known targets of drugs in the DrugBank database (https://www.drugbank.ca/). We manually extracted the name of each of the target drugs and indications from the DrugBank.
We used Immunobase (www.immunobase.org) to explore if the associated regions were associated to other immune-mediated diseases (IMDs) or to CeD by other studies. To identify the closest gene, we annotated the strongest associated SNP at each locus (TopSNPs) using Haploreg v4 [17], and retrieved the effect on gene expression (eQTLs) from a selected list of 12 studies. We also extracted eQTL information from GTEx (http://www.gtexportal.org/home/) and mapped eQTLs using peripheral blood RNA-seq data from 2116 unrelated individuals, as described by Zhernakova et al. [18]. Finally, we used GeneNetwork (https://www.genenetwork.nl/) to predict the function of the genes.
Differential gene expression analysis in intestinal biopsies
We assessed the expression of the genes affected by the TopSNPs as detected by HaploReg and eQTL annotation, in intestinal biopsies of 12 celiac patients and 12 controls. The biopsies were selected according to United European Gastroenterology criteria; the biopsy sampling, RNA isolation [19] and microarray hybridization have been described previously [20]. The raw data has been deposited in EBI ArrayExpress with the accession ID “E-MTAB-4613” [20]. Expression data were quantile normalized using the Illumina Beadstudio program. Quantile-normalized and log2-transformed expression values were used for differential expression analysis and differences were assessed with a T-test. Significance was defined as a p < 0.05. Boxplots were generated using R Studio.
Protein-QTLs in plasma
We used existing imputed genotype data and LTBR concentrations from 1179 individuals from the Life lines-DEEP cohort generated in a previous study by Zhernakova et al. [21]. We performed Spearman correlation analysis between the SNP (rs2364484:C>R, hg19 chr12:g.6511996 C>R) dosage and LTBR levels to test the association between SNP genotypes and protein levels.
Results
Validation non-HLA loci previously associated to CeD
In a meta-analysis of the eight CeD-cohorts (Supplementary Table 1), we confirmed the association of all 38 previously reported autosomal non-HLA CeD loci [3]. In 35 of the associated loci, the previously reported TopSNP was also our most significantly associated SNP. In the other three loci, the previously reported TopSNP was excluded during QC in six of the cohorts, thus other SNPs in high LD with them showed the most significant associations. A Manhattan plot illustrating the results of the associations after excluding the HLA region is shown in Supplementary Fig. 2.
Identification of two new loci associated to CeD
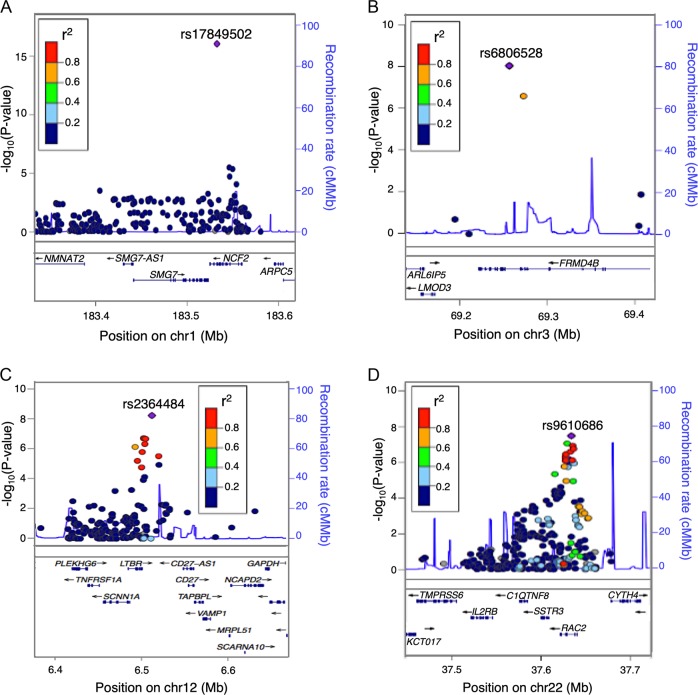
Four loci that were not reported in our previous Immunochip analysis reached genome wide significance (p < 5 × 10−8) in this analysis: 1q25.3, 3p14.1, 12p13.31, and 22q13.1 (Table 1). Results for each population can be found in Supplementary Table 2. The top SNP rs17849502 at the 1q25.3 locus (rs17849502:G>H, hg19 chr1:g.183532580G>H, Risk allele = T, OR = 1.43, p < 2.22 × 10−16, Table 1, Fig. 1a), is located in an exon of the Neutrophil Cytosolic Factor 2 gene (NCF2). The association to this missense variant (rs17849502:G>H) was missed in our previous Immunochip study [3] because it was a single association and there are not SNPs in LD with them within the locus. However, this low frequency variant (MAF = 0.07) was later identified in a resequencing study of CeD patients [22], and its association was replicated in a cross-disease meta-analysis of CeD and RA [23]. This region has also been associated to systemic lupus erythematosus [24]. The other three loci that reached genome-wide significance are all novel, with TopSNPs located in noncoding regions of the genome.
Table 1.
Genome-wide significant loci associate to celiac disease
| CHR | BP (hg19) | SNP | Risk allele | Other allele | N | OR | P(WZ) | MAF in CEU population | Closest gene | Is the region associated to other IMDs? | All eQTLs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 183532580 | rs17849502a | T | G | 7 | 1.43 | <2.22 × 10–16 | 0.06 | NCF2 | SLE, CeD | none |
| 3 | 69252899 | rs6806528 | T | C | 8 | 1.19 | 9.10 × 10−09 | 0.13 | FRMD4B | FRMD4B | |
| 12 | 6511996 | rs2364484 | C | A | 6 | 1.13 | 5.31 × 10−09 | 0.28 | RP1–102E24.8/ LTBR | AS, JIA, MS, PBC | LTBR |
| 22 | 37633851 | rs9610686 | C | T | 7 | 1.11 | 3.28 × 10−08 | 0.39 | RAC2 | T1D, VIT | C1QTNF6, SSTR3, CYTH4, RAC2 |
aMissense variant, BP base pairs, N number of cohorts, OR odds ratio, P(WZ) weighted P value, SLE systemic lupus erythematosus, AS ankylosing spondylitis, JIA juvenile idiopathic arthritis, MS multiple sclerosis, PBC primary biliary cirrhosis, T1D type 1 diabetes and VIT vitiligo
Fig. 1.
Regional plots of genome-wide significant loci. SNP with the strongest association in the region is shown in purple. SNPs in LD with the strongest associated SNP are shown in red (r2 < 1 and >0.8), orange (r2 < 0.8 and >0.6), green (r2 < 0.6 and >0.4), light blue (r2 < 0.4 and >0.2), and dark blue (r2 < 0.2). Lower panel shows the genes located within the region. a Association signals at the 1q25.3 locus. b Association signals at the 3p14.1 locus. c Association signals at the 12p13.31 locus. d Association signals at the 22q13.1 locus
The TopSNP rs6806528 (rs6806528:C>T, hg19 chr3>g.69252899C>T, Risk allele = T, OR = 1.18, p = 9.1 × 10−09, Table 1) at the 3p14.1 locus, which is located within an intron of the FERM domain containing the 4B gene (FRMD4B), was reported previously as a CeD-specific risk locus in a cross-disease meta-analysis [25] that increased the number of healthy controls included in the analysis to 22,308. In addition it showed modest association to CeD [26] in a cohort of 1550 North American CeD cases and 3084 controls (p = 0.0012). This has not been associated to any other IMD. This lack of association to other IMDs and in our previous study might be caused by the poor SNP coverage within the locus. For example, in the initial Immunochip analyses the 3p14.1 variants were excluded during the QC process, similar to the association identified to NCF2 in the 1q25.3 locus. We did not find any proxies of the TopSNP (r2 > 0.8) to be coding. The risk allele of rs6806528:C>T (T) increases the gene expression levels of FRMD4B in blood based on exon level eQTL analysis (p = 3.36 × 10−9, Fig. 2a, Table 1). The FRMD4B gene functions as a scaffolding protein [27] and is predicted by GeneNetwork to be involved in riboflavin metabolism, the Fc epsilon RI signaling pathway, the T cell receptor signaling pathway and axon guidance (Fig. 2b).
Fig. 2.
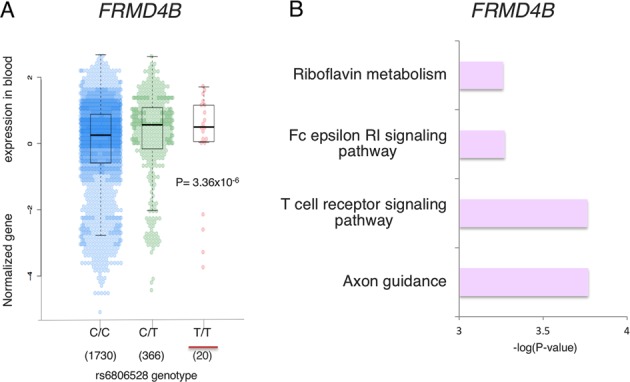
FRMD4B (3p14.1) locus. a The risk allele (underlined in red) of the Top-SNP rs6806528:C>T increases the expression of the FRMD4B gene (p = 3.36 × 10−6). The number of individuals analyzed is shown under each genotype. b FRMD4B gene function predictions based on GeneNetwork, a co-expression-based network
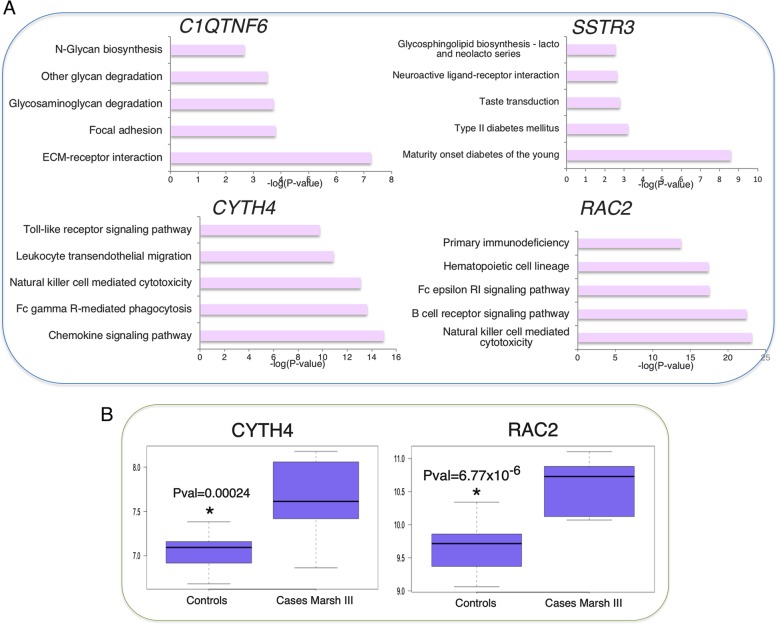
The TopSNP rs9610686 (rs9610686:C>T, hg19 chr2: g.37633851C>T, Risk allele = C, OR = 1.107, p = 3.28 × 10−09, Table 1) of the 22q13.1 locus (Fig. 1d) is a common variant located in an intron of the Ras-related C3 botulinum toxin substrate 2 gene (RAC2). This locus has previously been associated to type 1 diabetes [28] and vitiligo [29]. TopSNP rs9610686:C>T affects gene expression of multiple nearby genes: C1QTNF6, CYTH4, RAC2, and SSTR3 (Table 1). In artery aorta, blood, breast mammary tissue, and skin, it affects the gene expression of Complement C1q and tumor necrosis factor-Related protein 6 (C1QTNF6). C1QTNF6 modulates inflammation and insulin sensitivity in obese and diabetic mice and humans [30], and is predicted to be mainly involved in glycan-related processes and cell adhesion (Fig. 3a). The somatostatin receptor 3 gene (SSTR3), which regulates antiproliferative signaling and apoptosis [31] and is predicted to be involved in glycophospiloid biosynthesis and diabetes (Fig. 3a), is expressed in several tissues, including brain, ovary, pituitary, uterus, blood, and testis. SSTR3 expression level is affected by the rs9610686:C>T genotype only in testis, where its expression is higher than in the other tissues. In addition, the risk allele of rs9610686:C>T decreases the levels of expression in blood of two other genes, cytohesin 4 (CYTH4) and RAC2, both involved in immune-related processes (Fig. 3a). CYTH4’s strongest pathway predictions are for toll-like receptor signaling, leukocyte transendothelial migration, natural killer cell meditated cytotoxicity, Fc gamma R-mediated phagocytosis, and chemokine signaling. Mutations in RAC2 cause neutrophil immunodeficiency syndrome [32], which is characterized by severe bacterial infections and poor wound healing. RAC2 is involved in actin-based cellular functions of phagocyte cells, as well as cell proliferation and cell survival [33]. It is also predicted to be involved in primary immunodeficiency, hematopoietic cell lineages, the Fc epsilon RI signaling pathway, the B cell receptor signaling pathway, and natural killer cell mediated cytotoxicity. B cells [34] and natural killer cells [35] are important players in CeD pathogenesis. In addition to their immunity-related functions, CYTH4 and RAC2 are overexpressed in intestinal biopsies of celiac patients (p = 0.00024 and p = 6.77 × 10−6, respectively, Fig. 3b), further suggesting they have a role in the disease.
Fig. 3.
12p13.31 locus. a Functional predictions based on GeneNetwork for genes affected by the most-associated SNP in the locus (rs9610686:C>T). b Expression of the CYTH4 and RAC2 genes is significantly higher in CeD cases with a Marsh III diagnosis, as compared to healthy controls
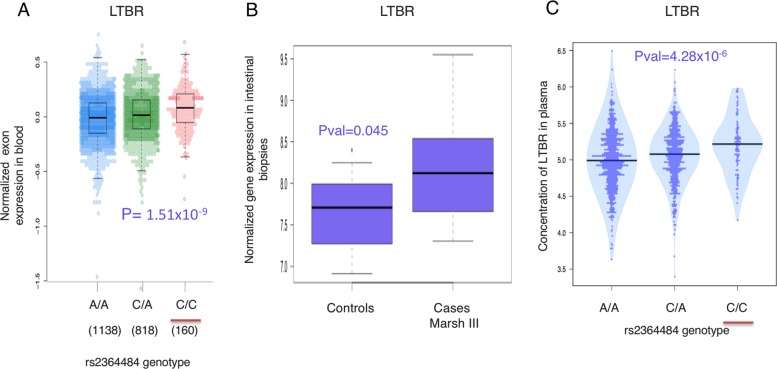
At the chromosome 12p13.31 locus, the TopSNP rs2364484 (rs2364484:C>R, hg19 chr12:g.6511996 C>R, Risk allele = C, OR = 1.13, p = 5.31 × 10−09, Table 1) is an intergenic variant between the Lymphotoxin Beta Receptor gene (LTBR) and the CD27 antisense RNA 1 gene (CD27-AS1) (Fig. 1c). This locus has previously been associated to ankylosing spondylitis [36], juvenile idiopathic arthritis [37], multiple sclerosis (MS) [10], and primary biliary cirrhosis [38]. The risk allele (C) of the TopSNP rs2364484: C>R increases the expression of LTBR in multiple tissues including blood (Fig. 4a), brain, stomach, testis, adipose, artery, breast, colon, esophagus, and pancreas. TopSNP rs2364484:C>R has also been shown to affect LTBR expression in ileal biopsies of 173 individuals [39] (p = 1.22 × 10−12), and the expression of LTBR was increased in intestinal biopsies of 12 CeD patients compared to controls (p = 0.045, Fig. 4b). Furthermore, the risk allele C of rs2364484:C>R increased the concentration of LTBR in plasma of 1179 healthy individuals from the Lifelines-DEEP cohort (p = 4.28 × 10−6, Fig. 4c). LTBR is involved in cell death, chemokine release, and inflammation [40], all important pathways in CeD, and the role of LTBR in the noncanonical NFκB activation cascade is well established [41, 42].
Fig. 4.
Functional annotation of the 12p13.31 locus containing LTBR. a The risk allele (underlined in red) of Top-SNP rs2364484:C>R, the strongest association in the 12p13.31 locus, increases the expression of the LTBR gene (p = 1.51 × 10−9). Number of individuals analyzed is shown under each genotype. b The expression of the LTBR gene is significantly higher in CeD cases with a Marsh III diagnosis, compared with healthy controls. c The risk allele of Top-SNP rs2364484:C>R (underlined in red) significantly increases the concentration of LTBR in plasma of healthy individuals
Functional annotation and pathway enrichment analyses on all CeD loci
To explore the functional impact of all CeD-associated loci, we performed functional annotation of significant loci and gene-mapping using FUMA. The SNP2GENE function identified 34 loci reaching genome-wide significance in this study, as some loci from our previous Immunochip study were only suggestive here (Supplementary Fig. 4a), comprising 4045 candidate SNPs including our TopSNPs and SNPs in high LD with them (r2 > 0.8). Thirty-six candidate SNPs were exonic within coding genes, 45 were exonic within ncRNAs, 53 were located in 3′ UTR and 19 in 5′ UTR (Supplementary Fig. 4b). Using multiple independent eQTLs datasets, FUMA mapped the candidate SNPs to 212 genes. The expression of these candidate genes was analyzed with MAGMA tissue expression analysis implemented in FUMA using 30 general tissue types from GTEx v7. We found significant enrichment of candidate genes to be expressed in blood, spleen and small intestine (Supplementary Fig. 5). It has been established that cells present in blood are important players in CeD [43], and that the disease leads to small-intestinal mucosal injury [1]. One third of CeD patients have defective spleen function and the prevalence of this dysfunction increases to 80% as the severity of the disease increases [44], indicating that the factors causing CeD also affect the spleen.
We explored if the results from the meta-analysis would lead to the discovery of new treatment options for CeD. While there are still no reported drugs for the treatment of CeD, 19 of the 212 candidate genes prioritized in the FUMA analysis are reported drug targets, including RAC2 from the novel 22q13.1 locus (Supplementary Table 3). Some of these drugs reduce inflammation or are immune-suppressants, and they are indicated for use in IMDs. Reported drugs include vedolizumab and CCX282 for the treatment for inflammatory bowel disease (IBD), natalizumab for multiple sclerosis (MS), abatacept for rheumatoid arthritis (RA) and juvenile idiopathic arthritis, galiximab for RA and psoriasis, 2-Methoxyestradiol for RA, and INCB3284 that is being investigated for use/treatment in inflammatory disorders, which might indicate a potential effect in CeD, but this requires follow-up study.
Using the 212 candidate genes as input, we looked for gene enrichment in multiple data sets and found enrichment of 286 Gene Ontology biological terms (Supplementary Table 4). We were able to confirm the enrichment to many well-known CeD pathways (Supplementary Table 4), including regulation of alpha beta T cell activation and proliferation, regulation of cell-cell adhesion, regulation of lymphocytes and leukocytes, production of multiple cytokines including interferon gamma (INF-γ), regulation of inflammatory response and regulation of B cell mediated immunity Table 1. Some of the pathways that contain novel associated genes (Supplementary Table 5) popped up for the first time, including TNF-mediated signaling, response to TNF regulation of I-κB kinase/NF-κB signaling, positive regulation of I-kappaB kinase/NF-κB signaling and apoptotic signaling.
LTBR locus links NF-kB pathway to celiac disease
Pro-inflammatory cytokines, adhesion molecules and enzymes whose gene expression is known to be regulated by NFκB are involved in CeD [45]. There is also a deregulation of the NFκB -pathway in the intestine of CeD patients [46]. As mentioned before, LTBR is well known for its role in the NFκB -pathway. In addition to LTBR, three other genes involved in the NFκB pathway have also been prioritized as CeD genes: Receptor activator of nuclear factor kappa-Β ligand (RANKL), TNF Alpha Induced Protein 3 (TNFAIP3) and protein kinase C gamma (PRKCG). To formally test whether the NFκB -pathway was involved in CeD pathogenesis, we compared the expression of 95 genes involved in the NFκB signaling pathway according to the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg-bin/show_pathway?hsa04064) in intestinal biopsies of patients with active CeD with those of healthy controls. We observed that 37 of the 95 genes showed significant differences in their levels of expression (p < 0.05, Supplementary Fig. 6) as shown in Supplementary Fig. 6a, b. These differentially expressed genes are involved in both the canonical and noncanonical NFκB pathway.
Discussion
We report here the largest meta-analysis of celiac cases and controls analyzed to date (n = 27,774: 12,948 cases and 14,826 controls), adding 4125 new samples to our previous Immunochip study [3]. We identified four loci at genome-wide significance that were not reported in our previous Immunochip study and another 18 showing evidence for suggestive association. As expected, most of the genome-wide significant loci are within regions previously associated to other IMDs such as IBD, type 1 diabetes, psoriasis, and MS. These results may imply a high level of genetic sharing of IMDs, but could reflect the design of the Immunochip, which was designed to densely genotype regions associated by previous GWAS for fine-mapping purposes and suggestive variants for replication. To clarify on similarity and differences of CeD and other IMDs, association studies across the whole genome using large cohorts are needed. Such whole genome association approaches will also allow the discovery of additional new loci that are not fully covered on the Immunochip. These discoveries would lead to a better understanding of the disease-specific genetic and molecular mechanisms.
Our study implicates two new CeD-associated regions
The most plausible candidate genes in the 22q13.1 locus are CYTH4 and RAC2. While CYTH4 has been mainly associated with schizophrenia and bipolar disorder, an evolutionary analysis [47] looking at regulatory elements conserved across mammals within the RAC2 gene identified three major haplogroups present in the population. One of these was associated to an increased risk for MS and IBD, suggesting an important role for RAC2 in the pathogenesis of IMDs. RAC2 also activates T helper (Th) 1-specific signaling and IFN-γ gene expression [48]. In CeD, gliadin-specific CD4+T cells respond to gliadin peptides presented via HLA-DQ2 or HLA-DQ8, which represent the strongest genetic risk for the disease. Upon activation, gliadin-specific CD4+T cells polarize towards the Th1-type pathway and produce IFN-γ, whose expression is also upregulated in intestinal biopsies of untreated celiac patients [49], further implying a role for RAC2 in CeD.
The 12p13.31 locus containing LTBR has also been associated to ankylosing spondylitis [50], however through an independent variant that leads to the splicing of exon 6 of TNFRSF1A, resulting in loss of the transmembrane domain. Our CeD TopSNP was not in high LD with the ankylosing spondylitis variants. It is on the same haplotype as a nonsynonymous coding variant in LTBR associated to JIA [37] (rs2364480:C>W, hg19 chr12: g.6495275 C>W, r2 = 0.9, D′ = 0.96), which suggests that LTBR is an important causal gene for multiple autoimmune diseases. Furthermore, the CeD TopSNP increases the expression of the LTBR gene in blood and is differentially expressed in biopsies of celiac patients. LTBR is well known to be involved in multiple immune pathways, including the non-canonical NFκB pathway. Although the role of the NFκB pathway in CeD is well known and has been validated by experimental studies [45, 46], it was not clear whether the deregulation of this pathway is a cause or a consequence of CeD. Our study, however, suggest a causal role for NFκB in CeD pathogenesis as we find strong association of four NFκB genes and their differential expression in CeD intestinal biopsies.
Our systematic annotation of loci from the meta-analysis lead to the identification of drug targets for 19 prioritized genes. Some of these drugs reduce inflammation or are immune-suppressants, and they are indicated for use in RA, IBD, psoriasis, JIA and MS, while the reposition of such drugs to CeD may need further investigation, our results might help to prioritize drugs for further studies.
We acknowledge limitations of our study. Firstly, use of the Immunochip restricted our analysis to loci already implicated in autoimmune diseases, which could be one reason we did not discover novel non-immune pathways. Secondly, although we included a non-European population, the design of the Immunochip is based on the European population and does not include population-specific variants from other ethnicities, thus a more suitable platform should be used to study the Argentinian population.
In conclusion, we have shown that increasing the sample size of our previous study allowed us to not only map new regions associated to CeD, but also to identify new disease pathways. The integration of multiple layers of omics information provided more insight into the individual loci and into the pathways involved in disease pathogenesis.
Supplementary information
Acknowledgements
We thank the Argentinian clinicians for recruiting individuals with CeD to provide blood samples; the genotyping facility of the UMCG for help in generating the Immunochip data; Jeffrey Barrett, Rinse Weersma, and Ross McManus for providing genotypes from extra controls and the Irish cohort; all the participating CeD patients and controls; and Kate Mc Intyre for editing the manuscript.
Funding
This work was supported by an ERC Advanced grant [FP/2007-2013/ERC grant 2012-322698], an NWO Spinoza prize grant [NWO SPI 92-266], NWO-VIDI grants [864.13.013] to JF and [016.178.056] to AZ, Hypatia grant to VK from Radboud UMC, NWO Gravitation Netherlands Organ-on-Chip Initiative [024.003.001] to CW, a European Union Seventh Framework Programme grant (EU FP7) TANDEM project [HEALTH-F3-2012-305279] to CW and a ERC Starting Grant [715772] to AZ. GT is supported by the Wellcome Trust grant WT206194.
Consortium for the study of genetic associations of celiac disease in Latin-America
Emilia Sugai, María L. Moreno, Horacio Vázquez, Elsa Barrientos, Eduardo Mauriño, Martin Talamo, Analia Villarruel.
Data availability
The sample-phenotype information, summary statistics from the association analysis per cohort, and the results from the meta-analysis have been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001003805.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Javier Gutierrez-Achury, Ana Florencia Costa, Patrick Deelen
These authors contributed equally: Julio Cesar Bai, Vinod Kumar, Cisca Wijmenga
Contributor Information
Cisca Wijmenga, Email: c.wijmenga@umcg.nl.
Consortium for the study of genetic associations of celiac disease in Latin-America:
Emilia Sugai, María L. Moreno, Horacio Vázquez, Elsa Barrientos, Eduardo Mauriño, Martin Talamo, and Analia Villarruel
Supplementary information
The online version of this article (10.1038/s41431-019-0520-4) contains supplementary material, which is available to authorized users.
References
- 1.Tack GJJ, Verbeek WHMHM, Schreurs MWJWJ, Mulder CJJJJ. The spectrum of celiac disease: epidemiology, clinical aspects and treatment. Nat Rev Gastroenterol Hepatol. 2010;7:204–13. doi: 10.1038/nrgastro.2010.23. [DOI] [PubMed] [Google Scholar]
- 2.Kuja-Halkola R, Lebwohl B, Halfvarson J, Wijmenga C, Magnusson PKE, Ludvigsson JF. Heritability of non-HLA genetics in coeliac disease: a population- based study in 107 000 twins. Gut. 2016;65:1793–8. doi: 10.1136/gutjnl-2016-311713. [DOI] [PubMed] [Google Scholar]
- 3.Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubois P, Trynka G, Franke L, Hunt K, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–9. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar V, Westra HJ, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B et al. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet. 2013; 8. 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed]
- 7.Ricaño-Ponce I, Zhernakova DV, Deelen P, Luo O, Li X, Isaacs A, et al. Refined mapping of autoimmune disease associated genetic variants with gene expression suggests an important role for non-coding RNAs. J Autoimmun. 2016;68:62–74. doi: 10.1016/j.jaut.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–8. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jostins L, Ripke S, Weersma R, Duerr RH, Dermot P, Hui K, et al. Host-microbe413 interactions shape genetic risk for inflammatory bowel disease. Nature. 2012;49:119–24. doi: 10.1038/nature11582. [DOI] [Google Scholar]
- 10.Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman C, Quinn EM, Ryan AW, Conroy J, Trimble V, Mahmud N, et al. Common polygenic variation in coeliac disease and confirmation of ZNF335 and NIFA as disease susceptibility loci. Eur J Hum Genet. 2015;353:1–7. doi: 10.1038/ejhg.2015.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015; 4. 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed]
- 16.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward LD, Kellis M. HaploRegv4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–81. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhernakova DV, Deelen P, Vermaat M, van Iterson M, van Galen M, Arindrarto W, et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat Genet. 2016;49:139–45. doi: 10.1038/ng.3737. [DOI] [PubMed] [Google Scholar]
- 19.Diosdado B, Wapenaar MC, Franke L, Duran KJ, Goerres MJ, Hadithi M, et al. A microarray screen for novel candidate genes in coeliac disease pathogenesis. Gut. 2004;53:944–51. doi: 10.1136/gut.2003.018374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt KA, Zhernakova A, Turner G, Heap GAR, Franke L, Bruinenberg M, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhernakova DV, Le TH, Kurilshikov A, Atanasovska B, Bonder MJ, Sanna S, et al. Individual variations in cardiovascular-disease-related protein levels are driven by genetics and gut microbiome. Nat Genet. 2018;50:1524–32. doi: 10.1038/s41588-018-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt KA, Mistry V, Bockett NA, Ahmad T, Ban M, Barker JN, et al. Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature. 2013;498:232–5. doi: 10.1038/nature12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutierrez-Achury J, Zorro MM, Ricaño-Ponce I, Zhernakova DV, Diogo D, Raychaudhuri S, et al. Functional implications of disease-specific variants in loci jointly associated with coeliac disease and rheumatoid arthritis. Hum Mol Genet. 2016;25:180–90. doi: 10.1093/hmg/ddv455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bentham J, Morris DL, Cunninghame Graham DS, Pinder CL, Tombleson P, Behrens TW, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–64. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Márquez A, Kerick M, Zhernakova A, Gutierrez-Achury J, Chen W-M, Onengut- Gumuscu S, et al. Meta-analysis of Immunochip data of four autoimmune diseases reveals novel single-disease and cross-phenotype associations. Genome Med. 2018;10:97. doi: 10.1186/s13073-018-0604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garner C, Ahn R, Ding YC, Steele L, Stoven S, Green PH et al. Genome-wide association study of celiac disease in North America confirms FRMD4B as new celiac locus. PLoS ONE. 2014; 9. 10.1371/journal.pone.0101428. [DOI] [PMC free article] [PubMed]
- 27.Klarlund JK, Holik J, Chawla A, Park JG, Buxton J, Czech MP. Signaling complexes of the FERM domain-containing protein GRSP1 bound to ARF exchange factor GRP1. J Biol Chem. 2001;276:40065–70. doi: 10.1074/jbc.M105260200. [DOI] [PubMed] [Google Scholar]
- 28.Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, et al. Meta- analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40:1399–401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben S, Riccardi SL, et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet. 2012;44:676–80. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei X, Seldin MM, Little HC, Choy N, Klonisch T, Wong GW. C1q/TNF-related protein 6 (CTRP6) links obesity to adipose tissue inflammation and insulin resistance. J Biol Chem. 2017;292:14836–50. doi: 10.1074/jbc.M116.766808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma K, Patel YC, Srikant CB. Subtype-selective induction of wild-type p53 and apoptosis, but not cell cycle arrest, by human somatostatin receptor 3. Mol Endocrinol. 1996;10:1688–96. doi: 10.1210/mend.10.12.8961277. [DOI] [PubMed] [Google Scholar]
- 32.Ambruso DR, Knall C, Abell AN, Panepinto J, Kurkchubasche A, Thurman G, et al. Human neutrophil immunodeficiency syndrome is associated with an inhibitory Rac2 mutation. Proc Natl Acad Sci USA. 2000;97:4654–9. doi: 10.1073/pnas.080074897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang FC, Kapur R, King AJ, Tao W, Kim C, Borneo J, et al. Rac2 stimulates Akt activation affecting BAD/Bcl-XL expression while mediating survival and actin function in primary mast cells. Immunity. 2000;12:557–68. doi: 10.1016/S1074-7613(00)80207-1. [DOI] [PubMed] [Google Scholar]
- 34.Mesin L, Sollid LM, Niro RDi. The intestinal B-cell response in celiac disease. Front Immunol. 2012;3:313. doi: 10.3389/fimmu.2012.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marafini I, Imeneo MG, Monteleone G. The role of natural killer receptors in celiac disease. Immunome Res. 2017;13:1–2. doi: 10.4172/1745-7580.10000129. [DOI] [Google Scholar]
- 36.Cortes A, Hadler J, Pointon JP, Robinson PC, Karaderi T, Leo P, et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45:730–8. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinks A, Cobb J, Marion MC, Prahalad S, Sudman M, Bowes J, et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet. 2013;45:664–9. doi: 10.1038/ng.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–32. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabakchiev B, Silverberg MS. Expression quantitative trait loci analysis identifies associations between genotype and gene expression in human intestine. Gastroenterology. 2013;144:1488–96. doi: 10.1053/j.gastro.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang Y-H, Hsieh S-L, Chen M-C, Lin W-W. Lymphotoxin beta receptor induces interleukin 8 gene expression via NF-kappaB and AP-1 activation. Exp Cell Res. 2002;278:166–74. doi: 10.1006/excr.2002.5573. [DOI] [PubMed] [Google Scholar]
- 41.VanArsdale TL, VanArsdale SL, Force WR, Walter BN, Mosialos G, Kieff E, et al. Lymphotoxin-beta receptor signaling complex: role of tumor necrosis factor receptor-associated factor 3 recruitment in cell death and activation of nuclear factor kappaB. Proc Natl Acad Sci USA. 1997;94:2460–5. doi: 10.1073/pnas.94.6.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li C, Norris PS, Ni C-Z, Havert ML, Chiong EM, Tran BR, et al. Structurally distinct recognition motifs in lymphotoxin-β receptor and CD40 for tumor necrosis factor receptor-associated factor (TRAF)-mediated signaling. J Biol Chem. 2003;278:50523–9. doi: 10.1074/jbc.M309381200. [DOI] [PubMed] [Google Scholar]
- 43.Meresse B, Malamut G, Cerf-Bensussan N. Celiac disease: an immunological jigsaw. Immunity. 2012;36:907–19. doi: 10.1016/j.immuni.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Di Sabatino A, Brunetti L, Carnevale Maffè G, Giuffrida P, Corazza GR. Is it worth investigating splenic function in patients with celiac disease? World J Gastroenterol. 2013;19:2313–8. doi: 10.3748/wjg.v19.i15.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maiuri MC, De Stefano D, Mele G, Iovine B, Bevilacqua MA, Greco L, et al. Gliadin increases iNOS gene expression in interferon-γ-stimulated RAW 264.7 cells through a mechanism involving NF-κB. Naunyn Schmiedebergs Arch Pharm. 2003;368:63–71. doi: 10.1007/s00210-003-0771-y. [DOI] [PubMed] [Google Scholar]
- 46.Fernandez-Jimenez N, Castellanos-Rubio A, Plaza-Izurieta L, Irastorza I, Elcoroaristizabal X, Jauregi-Miguel A, et al. Coregulation and modulation of NFκB-related genes in celiac disease: uncovered aspects of gut mucosal inflammation. Hum Mol Genet. 2014;23:1298–310. doi: 10.1093/hmg/ddt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sironi M, Guerini FR, Agliardi C, Biasin M, Cagliani R, Fumagalli M, et al. An evolutionary analysis of RAC2 identifies haplotypes associated with human autoimmune diseases. Mol Biol Evol. 2011;28:3319–29. doi: 10.1093/molbev/msr164. [DOI] [PubMed] [Google Scholar]
- 48.Li B, Yu H, Zheng W, Voll R, Na S, Roberts AW, et al. Role of the guanosine triphosphatase Rac2 in T helper 1 cell differentiation. Science. 2000;288:2219–22. doi: 10.1126/science.288.5474.2219. [DOI] [PubMed] [Google Scholar]
- 49.Nilsen EM, Jahnsen FL, Lundin KE, Johansen FE, Fausa O, Sollid LM, et al. Gluten induces an intestinal cytokine response strongly dominated by interferon gamma in patients with celiac disease. Gastroenterology. 1998;115:551–63. doi: 10.1016/S0016-5085(98)70134-9. [DOI] [PubMed] [Google Scholar]
- 50.Braun J, Sieper J, Devlam K, Calin A, Braun J, et al. Ankylosing spondylitis. Lancet. 2007;369:1379–90. doi: 10.1016/S0140-6736(07)60635-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sample-phenotype information, summary statistics from the association analysis per cohort, and the results from the meta-analysis have been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001003805.