Abstract
Cryptococcus neoformans and Cryptococcus gattii are two species complexes in the large fungal genus Cryptococcus and are responsible for potentially lethal disseminated infections. These two complexes share several phenotypic traits, such as production of the protective compound melanin. In C. neoformans, the pigment associates with key cellular constituents that are essential for melanin deposition within the cell wall. Consequently, melanization is modulated by changes in cell-wall composition or ultrastructure. However, whether similar factors influence melanization in C. gattii is unknown. Herein, we used transmission EM, biochemical assays, and solid-state NMR spectroscopy of representative isolates and “leaky melanin” mutant strains from each species complex to examine the compositional and structural factors governing cell-wall pigment deposition in C. neoformans and C. gattii. The principal findings were the following. 1) C. gattii R265 had an exceptionally high chitosan content compared with C. neoformans H99; a rich chitosan composition promoted homogeneous melanin distribution throughout the cell wall but did not increase the propensity of pigment deposition. 2) Strains from both species manifesting the leaky melanin phenotype had reduced chitosan content, which was compensated for by the production of lipids and other nonpolysaccharide constituents that depended on the species or mutation. 3) Changes in the relative rigidity of cell-wall chitin were associated with aberrant pigment retention, implicating cell-wall flexibility as an independent variable in cryptococcal melanin assembly. Overall, our results indicate that cell-wall composition and molecular architecture are critical factors for the anchoring and arrangement of melanin pigments in both C. neoformans and C. gattii species complexes.
Keywords: fungi, cell wall, polysaccharide, nuclear magnetic resonance (NMR), solid state NMR, Cryptococcus, melanin, molecular structure, opportunistic pathogen, virulence factor
Introduction
Unlike rigid walls made of bricks or stones, microbial cell walls are flexible and highly dynamic structures. Fungal cell walls undergo constant change during budding, cell growth, and yeast-to-hyphal cell transition. The fungal cell wall has such flexible viscoelastic properties that that it can allow the transit of liposomes (1). These external complex assemblies are sufficiently strong to protect the cell from the internal turgor pressure and from the outer environment, while also acting as molecular sieves that allow the intake of nutrients and the export of fungal products. Microbial cell walls are assembled from carbohydrates, proteins, and lipids in diverse architectures that depend on the fungal genus. In particular, fungal walls contain branched polysaccharides (mainly β-glucans) and mannoproteins, which are complexed with lipids, pigments, and inorganic salts (2, 3). Our current knowledge of fungal cell biology comes primarily from decades of research on model organisms such as Saccharomyces cerevisiae, Schizosaccharomyces pombe, Candida albicans, and Aspergillus fumigatus, which have laid the groundwork for studies on the cryptococcal cell wall (4–8). However, each of these organisms belongs to the ascomycetes, which have a cell-wall organization very different from that of the basidiomycetes (5, 6, 9).
Among the basidiomycetes, the genus Cryptococcus includes over 70 species of environmentally ubiquitous saprophytic encapsulated yeasts, of which two species complexes (10), Cryptococcus neoformans and Cryptococcus gattii, are relatively common causes of potentially fatal cryptococcal disease. C. neoformans is found worldwide in association with avian excreta (11, 12) or soil (13) and is the primary etiological agent of cryptococcal meningoencephalitis in immunocompromised patients (e.g. those infected with HIV/AIDS). On the other hand, C. gattii is found commonly in subtropical regions associated with various tree species, notably eucalyptus, and causes disease primarily in immunocompetent individuals (14). Infection by both cryptococcal species is acquired by inhalation of a spore or a desiccated yeast. Thus, whether an infection is cleared, becomes latent, or progresses to cryptococcosis depends on the integrity of an individual's immune system and the Cryptococcus species associated with the incursion (15). From an evolutionary perspective, these cryptococcal species are thought to have diverged from a common environmental saprophyte ancestor somewhere between 30–40 (16, 17) and 100 million years ago, a time that coincided with the breakup of the supercontinent Pangea that has been proposed to have contributed to cryptococcal speciation (18). Nevertheless, they have retained extraordinarily similar microbial traits in presumed response to selective pressures in both ecological and animal niches (19). A prime example of these traits is a robust cell wall coated by a polysaccharide capsule and reinforced with melanin pigments (20–22).
Melanins are complex heterogeneous polymers of phenolic and/or indolic origin characterized by a dark color, insolubility in most solvents, and the possession of a stable free radical signature (23, 24). These natural pigments are synthesized by members of all biological kingdoms and have a wide array of functions. In the environment, melanin provides Cryptococcus spp. with protection against desiccation, predation by microorganisms, and UV radiation; in mammalian hosts, it alters cytokine production, offers resistance to reactive oxygen species, and diminishes phagocytosis (25–27). Whereas cryptococcal melanin is known to be composed of oligomers or polymers of cyclized catecholamines such as 3,4-dihydroxyphenylalanine (l-DOPA)4 (28, 29), its insoluble and amorphous characteristics have hampered the elucidation of the pigment's detailed molecular architecture within the cellular milieu. Recent analyses using high resolution solid-state NMR (ssNMR) have revealed that aliphatic groups derived from cell-wall components such as polysaccharides (e.g. chitin) serve as a scaffold for the progressive incorporation of melanin pigments into the cryptococcal cell wall (30, 31). Furthermore, the identity of the melanin precursor (norepinephrine, epinephrine, methyl-l-DOPA, l-DOPA) alters the pigment structure as well as the polysaccharide- and lipid-based cellular scaffold associated with its deposition (32, 33). These analyses dovetail with previous reports in other fungi (34–36) as well as in insects (37), where an intimate association between melanin and the chitin polysaccharide was described. Conversely, several studies using both C. neoformans and C. gattii demonstrated that strains defective in the chitin/chitosan biosynthetic machinery are unable to retain melanin within the cell wall, thus yielding a “leaky melanin” phenotype characterized by visual detection of the pigment in the culture medium or agar surrounding the cells (38–40).
In the present work, we employ magic angle spinning (MAS) ssNMR assisted by isotopically labeled reagents and other supporting biochemical techniques to focus on the cell-wall structure of the two most clinically relevant cryptococcal isolates: C. neoformans H99 and C. gattii R265. Our results demonstrate that both polysaccharides, and particularly chitosan, together with lipids, play critical roles in anchoring and layering melanin within the cryptococcal cell wall.
Results
Melanized yeast cells of C. gattii R265 display notably augmented and condensed deposition of the pigment within the cell wall
Upon light microscopy examination of melanized cells from two isolates representing the C. neoformans and C. gattii species complexes, H99 and R265, respectively (Table 1), we observed that a subset of C. gattii R265 cells displayed highly pigmented cell walls, which were not observed for C. neoformans H99 (Fig. 1). The mean cell body and capsule sizes were also observed to differ between the two species (Fig. S1). Multiple studies have established that the molecular architecture of the C. neoformans cell wall directly influences the deposition and retention of melanin pigments (31, 38, 39, 41–43). Consequently, we tested the generality of cell-wall ultrastructural characteristics associated with melanin deposition by using transmission EM to further compare the melanized cell walls of C. neoformans H99 with C. gattii R265, which is a strain from the VGIIa molecular subtype reported to exhibit robust melanin production at 37 °C (44–46). We observed significant differences in the ultrastructural properties of C. neoformans H99 and C. gattii R265 (Fig. 2). Transmission EM revealed that the C. neoformans H99 melanized cell wall had a heterogeneous pigment distribution characterized by multiple layers of variable electron density (Fig. 2A, top), which is consistent with previous studies conducted by our group (42). In contrast, the melanin pigments embedded in C. gattii R265 cell walls were found to be more uniformly distributed, forming just two highly electron-dense layers that closely surrounded the cell body (Fig. 2A, bottom). The melanized cell walls of C. gattii R265 cells were additionally observed to have a significantly smaller thickness compared with C. neoformans H99 (Fig. 2B), a finding that could be attributed to the compact arrangement of their melanin pigments.
Table 1.
Cryptococcal strains used in this study
Figure 1.
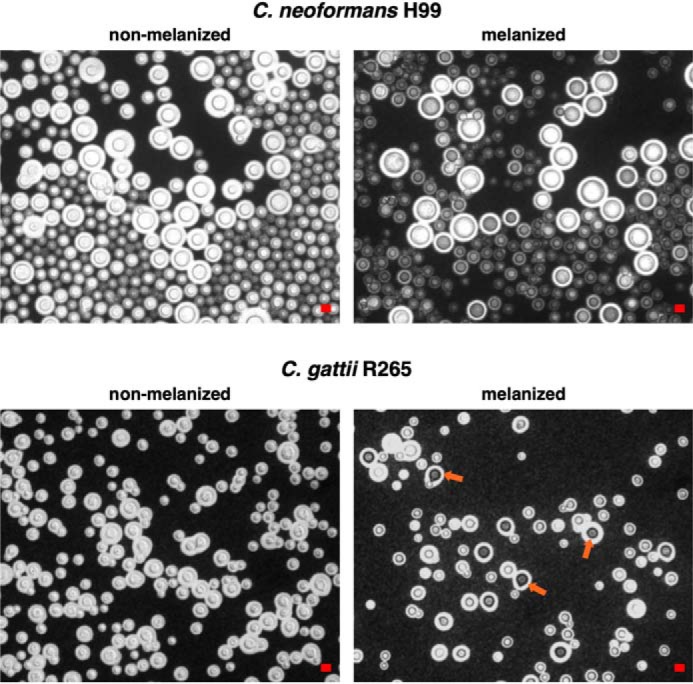
A subset of C. gattii R265 melanized cells exhibit highly pigmented cell walls. Shown are representative images of nonmelanized and melanized yeast cells of C. neoformans H99 and C. gattii R265 visualized by light microscopy using India ink counterstaining. Over 300 cells were counted at ×40 magnification in five different fields. Highly melanized C. gattii R265 cells showing enhanced cell-wall pigmentation (red arrows) corresponded to 7.5% of the total. Scale bar, 10 μm.
Figure 2.
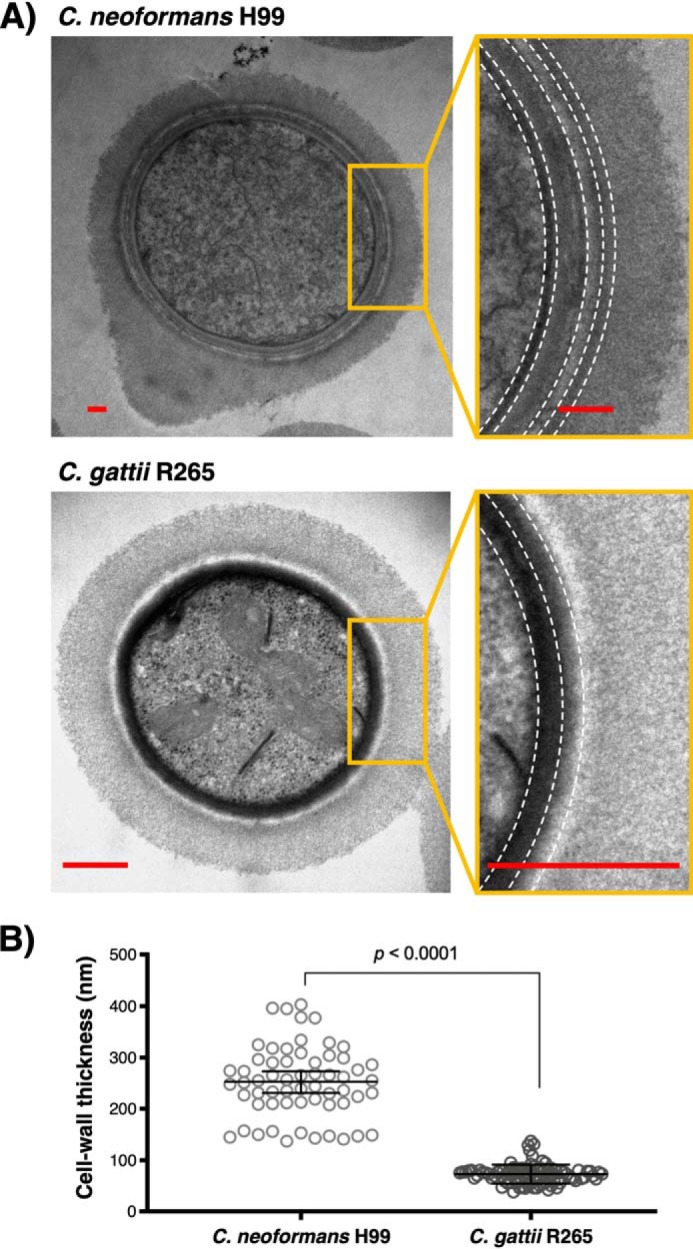
The melanized cell wall of C. gattii R265 reveals a compact and uniform deposition of melanin pigments. A, representative cross-sectional transmission electron micrographs of C. neoformans H99 (top) and C. gattii R265 (bottom) melanized yeast cells. The magnified images show a close-up view of the cell wall with dashed lines to illustrate the difference in layered melanin pigment arrangement between these two cryptococcal isolates, possibly attributable to their cell-wall chitinous compositions. Scale bar, 500 nm. B, measurement of C. neoformans H99 and C. gattii R265 melanized cell-wall thickness. Data were obtained by measuring 12–18 cells per strain (5 measurements/cell) and analyzed using the Mann–Whitney test, two-tailed p value with 99% confidence level. Error bars, 95% confidence interval of the median.
Interestingly, the phenotype displayed by C. gattii R265 cells was similar to that observed in C. neoformans H99 cells supplemented with GlcNAc (42). Prior work by our group demonstrated that C. neoformans H99 cells grown in the presence of 1 or 5 mm GlcNAc displayed a more uniform arrangement of cell-wall melanin pigments as well as diminished cell-wall and cell-body diameters compared with nonsupplemented control cells (42). We hypothesize that these morphological changes result from differences in the cell-wall composition of C. gattii, in analogy with GlcNAc-supplemented C. neoformans H99 cells that were found to have a relatively larger amount of cell-wall chitosan than controls. Therefore, our current results suggest that the chitinous composition of C. gattii R265 cell walls could favor enhancement of melanin deposition in a condensed and uniform manner.
The “leaky melanin” phenotype is associated with decreased levels of chitosan in both C. neoformans and C. gattii mutant strains
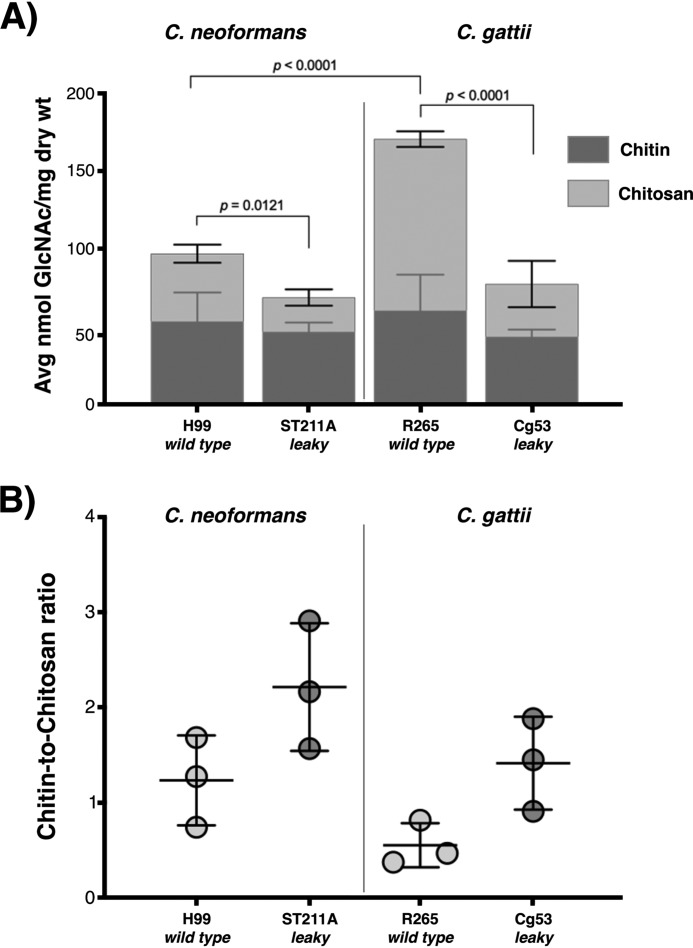
The leaky melanin phenotype refers to the observation that some Cryptococcus spp. mutants appear unable to retain melanin in the cell wall and instead release the pigment into the culture medium exterior to the cell (38, 39). To test the proposed correlation of melanin deposition and retention with chitinous molecular composition in C. gattii R265 melanized fungal cells, we conducted a biochemical measurement of chitin and chitosan content using the 3-methyl-2-benzothiazolone hydrazine hydrochloride (MBTH) colorimetric assay, as reported previously in GlcNAc-supplemented C. neoformans H99 cells (42). As hypothesized above, the cell-wall chitosan content of C. gattii R265 WT cells was found to be nearly 3-fold greater than in C. neoformans H99 WT cells (Fig. 3A). Although the change in the chitin/chitosan ratio associated with this increase was not statistically significant (Fig. 3B), C. gattii R265 cell walls also contained a relatively higher overall amount of total chitinous material (chitin plus chitosan) than C. neoformans H99 (Fig. 3A).
Figure 3.
Cryptococcal strains defective in the retention of cell-wall melanin exhibit decreased levels of cellular chitosan. A, biochemical determination of chitin and chitosan using the MBTH colorimetric reaction measured at 650 nm. Reduction in the cell-wall content of chitinous polysaccharides, primarily attributable to chitosan contribution, is associated with the leaky melanin phenotype in both C. neoformans ST211A and C. gattii Cg53 strains. Analysis of variance with a Bonferroni post hoc test indicated significant differences between groups. B, chitin/chitosan ratio. A boost in the chitin/chitosan ratio correlates with the leaky melanin phenotype independently of the Cryptococcus species. All data represent an average of three independent biological experiments. Error bars, S.D.
To further investigate whether a reduction in chitosan content was responsible for compromised pigment retention in both Cryptococcus species, we assessed the cell-wall chitin and chitosan content of two other leaky melanin strains previously identified for C. neoformans and C. gattii, ST211A and Cg53, respectively (40). The ST211A mutant has a frameshift in the CSR2 gene (GenBankTM accession number MK609896),5 along with a T-DNA insertion elsewhere in the genome. The Cg53 mutant has a T-DNA insertion in the CHS3 gene; the phenotype was complemented by transformation with a WT CHS3 gene. Chs3 is a chitin synthase that is an integral membrane protein, and Csr2 is an accessory protein to chitin synthase, which is membrane-associated likely via a C-terminal prenyl group (39). Both genes have been shown to be required for chitosan production in C. neoformans, and these two proteins have been hypothesized to form a membrane-bound complex that synthesizes the chitin that is then converted to chitosan (39). Whereas absolute levels of chitin were found to be similar for all four strains, including parent and leaky mutants, the chitosan absolute and proportional content for both the C. neoformans ST211A and C. gattii Cg53 mutants exhibited significant reductions compared with their respective parent strains (Fig. 3A) and, consequently, higher chitin/chitosan ratios (Fig. 3B). Interestingly, the lowered chitosan level of the C. gattii Cg53 mutant resulted in a chitin/chitosan ratio similar to that of C. neoformans H99, suggesting a dependence of aberrant pigment retention on other compositional or structural changes of the scaffold. These findings suggest that diminished cell-wall chitosan content may be a feature common to cells from leaky mutant strains regardless of the Cryptococcus species, underscoring the likely importance of this polysaccharide for anchoring of melanin to the cryptococcal cell wall. Moreover, these data support the contention that the phenotypic traits shared by melanized C. gattii R265 and GlcNAc-supplemented C. neoformans H99 cells are due to increased cell-wall chitosan content, which could promote uniform melanin pigment deposition and consequently result in diminished cell-wall thickness.
The condensed pigment arrangement in melanized C. gattii R265 yeast cells does not result in increased melanin deposition
In our prior work, supplementation with GlcNAc was determined to augment the chitin/chitosan ratio of C. neoformans H99 cells, and increased levels of chitosan were correlated with greater cell-wall pigment deposition and retention, as evidenced by a greater mass yield of melanin ghosts with a relatively higher pigment content than nonsupplemented cells (42). Melanin ghosts are fungal pigment structures isolated via a degradative process that includes treatment with hot acid. Nevertheless, due to the insoluble and recalcitrant nature of melanin, a variety of cellular remnants remain associated with the pigment, resulting in structures that resemble hollowed-out yeast cells and thus are referred to as “melanin ghosts.” The relative pigment content of melanin ghost samples can be estimated using ssNMR spectroscopy and serves as a measure of fungal melanin deposition.
Thus, to determine whether the substantial chitosan content of C. gattii R265 is associated with increased cell-wall pigment deposition, quantitatively reliable 13C direct polarization magic angle spinning (DPMAS) spectra were acquired for melanin ghosts isolated from both C. neoformans H99 and C. gattii R265 cells. The relative content of indole-based pigment was estimated by comparing the aromatic region peak area (shaded) with the total signal area of each spectrum. Interestingly, aromatic signals comprised 35% of the C. neoformans total spectral area, whereas only 30% of the C. gattii melanin ghost total signal intensity was attributable to the pigment (Fig. 4A). Furthermore, C. neoformans H99 yielded a greater dry mass of melanin ghosts than C. gattii R265 (20.7 mg versus 16.6 mg, respectively) (Fig. 4B). In the context of our other data, this finding indicates that the condensed pigment deposition exhibited by C. gattii R265 melanized cell walls does not translate into an overall increase in its content.
Figure 4.
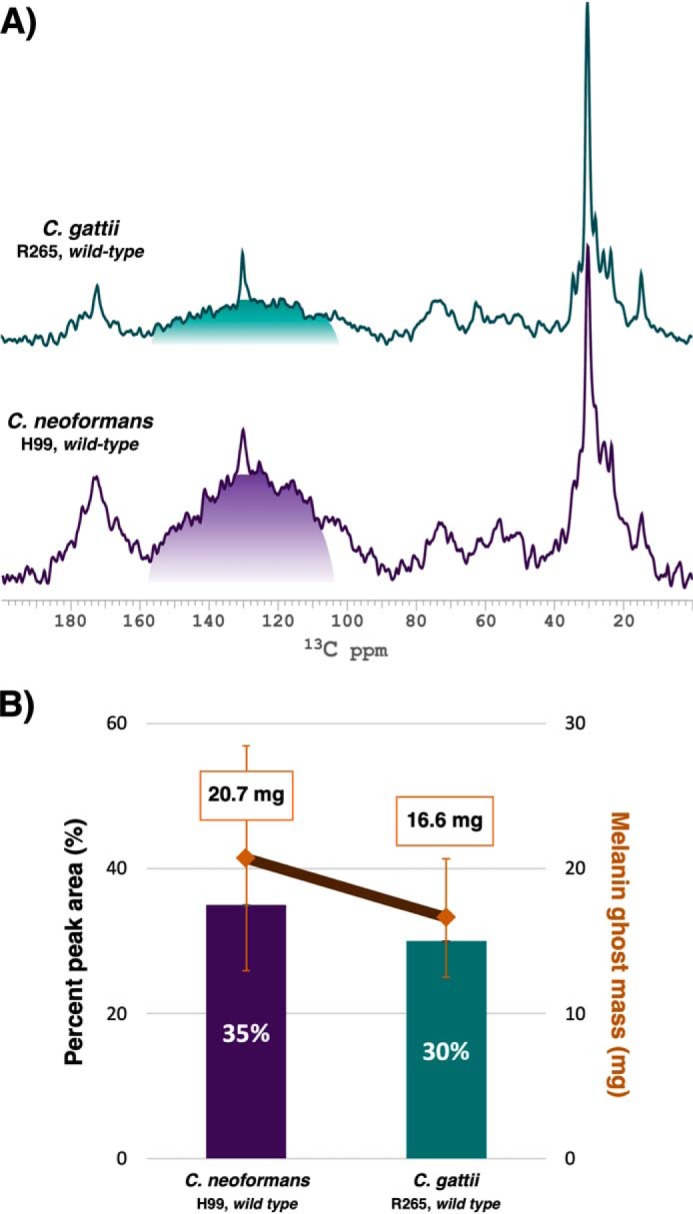
C. neoformans H99 yields a greater mass of melanin “ghosts” with a relatively higher pigment content than C. gattii R265. A, representative 1D 13C DPMAS spectra of melanin ghosts generated from C. neoformans H99 and C. gattii R265 parent strain cell cultures. The shaded region (110–160 ppm) represents the relative contribution of signal intensity attributable to the indole-based melanin pigment. The peak at ∼130 ppm arises from the unsaturated carbons of fatty acids and thus was excluded from the shaded region. B, estimation of pigment content relative to retained cellular constituents obtained from quantitatively reliable 13C DPMAS spectra (bar graph) and measurement of melanin ghost mass (line graph). The data represent the mean of results from three independent trials. The S.D. from the mean was less than 0.01% for the relative aromatic pigment content measurements, and thus the error bars are too small to be visible. Displayed error bars, S.D. from the mean melanin ghost mass generated from each isolate.
Lipid and polysaccharide constituents are retained differentially in leaky melanin ghosts and with species-specific relative compositions
In light of our finding that the C. gattii leaky mutant Cg53 had a chitin/chitosan ratio similar to that of the C. neoformans H99 strain, we sought a more comprehensive accounting of the compositional and architectural factors that distinguish the parent and leaky mutant strains of each Cryptococcus species. A quantitative 13C solid-state NMR analysis was carried out for melanin ghosts generated from cell cultures containing uniformly 13C-enriched glucose as the sole carbon source and natural-abundance l-DOPA as the obligatory melanization precursor. Because cryptococcal species are unable to synthesize melanin pigments from nutrient sources (47), the provision of [U-13C]glucose results in selective incorporation of the stable 13C isotope into glucose-derived nonpigment moieties. Thus, the resulting spectra display signals attributable solely to the aliphatic constituents that survive the degradative melanin ghost isolation protocol (Fig. 5). This strategy allowed us to estimate the relative amounts of long-chain lipid, polysaccharide, and “other” nonpigment components in these melanin ghost samples by acquiring quantitatively reliable 13C DPMAS spectra and comparing peak integrals expressed as fractions of the total integrated NMR signal intensity across the spectrum. The constituents retained by the ghosts should then reflect the adherence of the pigment and the architectural features that offer protection from chemical and enzymatic degradation.
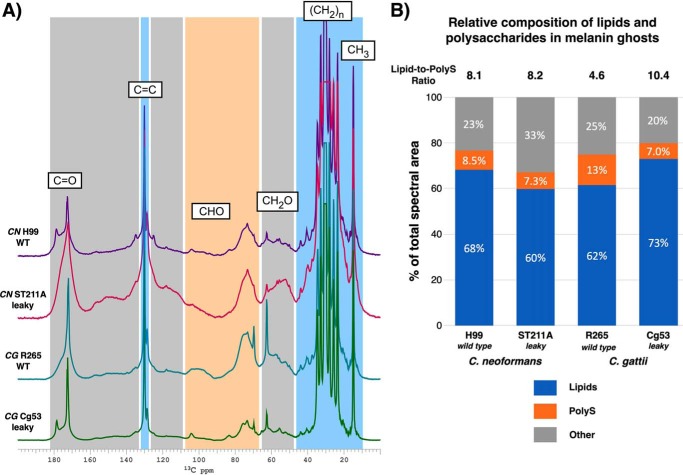
Figure 5.
The relative composition of retained cellular constituents differs between C. neoformans and C. gattii leaky mutant and parent strain melanin ghosts. A, 1D 13C DPMAS spectra of melanin ghosts generated from C. neoformans and C. gattii parent and leaky melanin strain cell cultures containing d-[U-13C6]glucose as the sole carbon source. B, measurement of each cellular constituent type relative to the total amount of retained nonpigment moieties obtained from quantitatively reliable 13C DPMAS spectra. PolyS, polysaccharide. Each spectrum is normalized by setting the largest peak to full scale. The top of the largest peak was cropped to improve the visibility of smaller peaks.
Upon visual inspection, the spectra of the four [U-13C]glucose-labeled melanin ghost samples appeared largely similar: they each displayed signals corresponding to cell-wall polysaccharides (∼55–105 ppm) as well as saturated and unsaturated long-chain methylene carbons (∼30 and 130 ppm, respectively) (Fig. 5A). However, quantitative analysis revealed considerable compositional differences among the four strains. Although resonances consistent with long-chain fatty acids ((CH2)n and CH3) that are primarily attributable to triacylglycerides6 accounted for the majority of signal intensity in each spectrum, their relative contributions in WT parent versus mutant strains exhibited opposite trends for the two Cryptococcus species. Whereas the melanin ghosts isolated from the C. neoformans ST211A leaky strain were slightly poorer in lipids than H99 (60% versus 68%), C. gattii Cg53 leaky ghosts had a greater proportion of lipids compared with the R265 sample (73% versus 62%) (Fig. 5B).
The peaks corresponding to polysaccharide carbons contributed only a minor proportion of the overall signal intensity in each spectrum. In C. neoformans, the relative polysaccharide content was essentially invariant for melanin ghosts isolated from H99 versus ST211A mutant cells (8.5% versus 7.3%). Both samples displayed similar lipid/polysaccharide ratios (8.1 versus 8.2 for parent and mutant, respectively) despite the diminished lipid percentage in the ST211A ghosts. Interestingly, the most substantial difference between the parent and leaky C. neoformans samples was the relative contribution of constituents other than long-chain methylenes or polysaccharides (23% versus 33%) (Fig. 5B). The ST211A melanin ghost sample displayed a visibly greater percentage of signal intensity in the “other” spectral region between ∼110 and 160 ppm than the H99 sample. Sharp peaks appearing in this chemical shift range are typically attributable to double-bonded carbons from unsaturated fatty acid chains, and thus, the ∼130 ppm resonances are categorized as lipids. However, the broad underlying components in this spectral region suggest a collection of relatively rigid, less motionally averaged conjugated moieties that are currently unassigned and under further investigation.
Despite the modest quantitative contributions of polysaccharide carbon signals apparent in both C. neoformans spectra, the polysaccharide proportion in C. gattii was nearly 2-fold greater in R265 ghosts compared with the Cg53 mutant (13% versus 7.0%). The compositional differences between the two C. gattii samples are further underscored by the significantly higher lipid/polysaccharide ratio of Cg53 (10.4, compared with 4.6 for the parent strain), which results from concurrently increased lipid and decreased polysaccharide levels. Thus, from a compositional perspective, our 13C NMR measurements revealed that leaky melanin phenotypes could result from a mix of constituent classes that is particular to the Cryptococcus species: elevated “other” materials comprising unidentified aromatic ring structures for C. neoformans or an enhanced lipid/polysaccharide ratio for C. gattii.
Two-dimensional (2D) 13C-15N NMR spectroscopy establishes chitin and chitosan as glucose-derived constituents retained by melanin ghosts
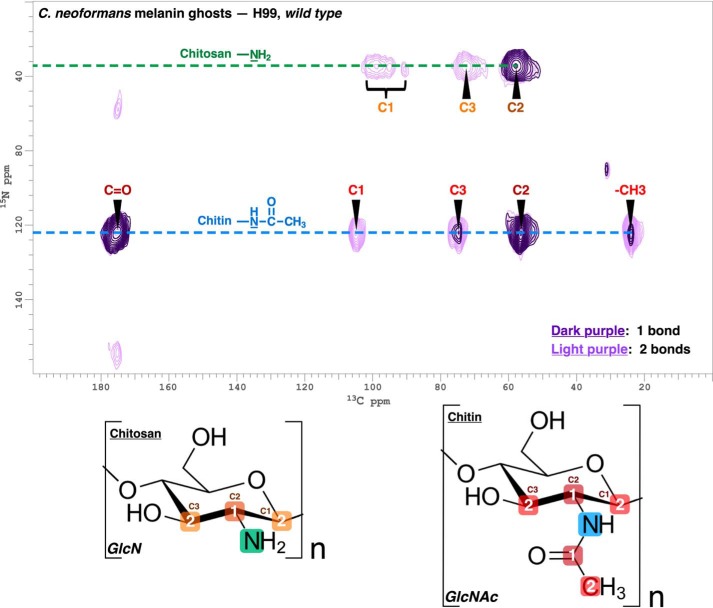
Although the MBTH assay provided a practical means to correlate changes in chitin/chitosan ratios with the leaky melanin phenotype in both Cryptococcus species, this approach is not practical for melanized fungal samples and falls short of revealing the macromolecular architecture of these polysaccharides within the underlying cellular scaffold. Moreover, the recalcitrant nature of melanin would require very harsh chemical or enzymatic procedures to isolate the pigment-associated cellular constituents from ghost samples, risking hydrolysis in an effort to achieve identification and characterization. Instead, we have previously used solid-state 2D 13C-13C NMR of [U-13C]glucose-labeled ghosts to obtain 13C chemical shifts and structural connections that implicate the acetyl group of chitin (31). Although chitosan lacks distinctive carbon-containing structural features that could distinguish it from the multitude of other moieties present in 13C NMR spectra of melanin ghosts, along with chitin it is unique among polysaccharides in possessing nitrogenous functional groups. Thus, we prepared melanized C. neoformans H99 cell cultures containing [U-13C]glucose and [15N]glycine as the sole carbon and nitrogen sources, respectively, to verify that chitosan and chitin were components of the melanized cell-wall scaffold. 15N NMR spectra of the ghosts (Fig. S2) displayed signals from chitin (123 ppm) and chitosan (35 ppm) (48, 49), and definitive identifications could be made from 2D 13C-15N transferred echo double resonance (TEDOR) experiments that identify spatially proximal 13C-15N pairs (Fig. 6) (50, 51).
Figure 6.
C. neoformans H99 melanin ghosts contain highly deacetylated chitosan. 2D 13C-15N TEDOR spectra of C. neoformans H99 melanin ghosts generated from cell cultures containing d-[U-13C6]glucose and [15N]glycine as the sole carbon and nitrogen sources, respectively. Coherence transfer periods of 1.40 and 4.00 ms were used in separate experiments to obtain proximal carbon-nitrogen pairs separated by distances corresponding to ∼1 bond length (dark purple contours) and ∼2 bond lengths (light purple contours). The boxed numbers displayed on the chitin GlcNAc and chitosan glucosamine (GlcN) monomeric units denote the number of bonds between each 13C-15N nuclear pair.
Depending on the experimental parameters, the TEDOR contour plot displays proximal 13C-15N pairs located within selected distance ranges up to 5–6 Å. We acquired two complementary spectra that distinguished correlations involving 13C-15N nuclei with intervening distances corresponding to 1–2 bonds from longer-range pairs at distances up to 3 bonds. The short-distance melanin ghost 2D TEDOR plot (Fig. 6, dark purple contours) displayed three prominent cross-peaks attributable to proximal 13C-15N pairs (that are also directly bonded in the structures) in chitinous polysaccharides: the amide nitrogen of chitin is covalently linked to the C2 ring carbon (55 ppm 13C × 123 ppm 15N) and also to the carbonyl carbon of the acetyl group (174 × 123), and the amine nitrogen of chitosan is linked to the C2 ring carbon (57 × 35). The correlations displayed in the long-distance TEDOR spectrum of melanin ghosts (Fig. 6, light purple contours) further substantiate identification of the two principal nitrogenous constituents as chitin and chitosan: three additional cross-peaks involving the 123-ppm chitin amide nitrogen are unambiguously identifiable as intramolecular correlations with the methyl carbon of the acetyl group (24 × 123), C1 (104 × 123), and C3 ring carbons (73 × 123), respectively, each located 2 bonds away. Similarly, correlations between the chitosan amino nitrogen at 35 ppm are observed with C3 (72 × 35) and with C1 carbons situated in slightly different chemical environments (∼90–100 × 35).
Although chitosan is often referred to as the deacetylated form of chitin, it would more accurately be classified as a binary heteropolysaccharide, because a percentage of the glucosamine residues remain acetylated (52). The C1 carbon participates in forming the glycosidic bond that links two adjacent monosaccharide units; consequently, its chemical shift is influenced by the acetylation state of the succeeding residue and can provide information about the number and distribution of acetylated units within a chitin/chitosan polymer. The chitosan C1 correlations we observe in the TEDOR spectrum have carbon chemical shift values that span a 10-ppm range of upfield values below 100 ppm (Fig. 6), consistent with a highly deacetylated chitosan in which the glucosamine units have slightly different magnetic environments, depending on their proximity to sparse acetylated units with a “nonsequential random distribution” (52–54). These findings imply that the chitinous material in C. neoformans melanin ghosts comprises distinct polymers of GlcNAc (chitin) and glucosamine (chitosan), suggesting the possibility that they originate from different regions of the cell wall and play contrasting roles in the deposition of melanin pigments.
Melanin retention is associated with the flexibility of the cell-wall polysaccharide network
Thus far, a substantial body of evidence from our group and others has indicated that the cell-wall balance between chitin and chitosan directly influences the retention of melanin pigments (38, 39, 42). However, the possible changes in macromolecular architecture or chitin-chitosan arrangements that could promote or inhibit pigment retention have not been explored. To compare the molecular-level structural features of the chitin-chitosan network within the melanized cell walls of C. neoformans and C. gattii, including cells from both parent and “leaky” mutant strains, we examined the [U-13C]glucose-enriched ghost samples using the 2D 13C-13C dipolar assisted rotation recoupling (DARR) solid-state NMR experiment. The parameters were optimized to detect proximal pairs of 13C-13C nuclei separated by distances corresponding to ∼1 bond length.
To analyze the DARR contour plots in Fig. 7, we followed a roadmap to key 13C chemical shift assignments that was provided by the 13C-15N TEDOR data of Fig. 6. Thus, the TEDOR spectra of U-13C,15N-enriched C. neoformans H99 melanin ghosts served to distinguish chitosan from the numerous other carbon-containing moieties in the [U-13C]glucose-enriched DARR spectra: whereas the majority of polysaccharides have C1 ring carbons that resonate at 100–105 ppm, the C1 carbon chemical shift observed for chitosan reflects the highly deacetylated form of this polymer. As noted above, this value can vary between 90 and 100 ppm according to the acetylation state of its adjacent sugar units. The corresponding cross-peaks in the 13C-13C DARR spectrum can then be taken to correlate with proximal ring carbons located 1 bond (C1–C2, 98 × 57 ppm) or 2 bonds (C1–C3, 98 × 73; C1–C5, 98 × 75 ppm) apart (Fig. 7, A–D).
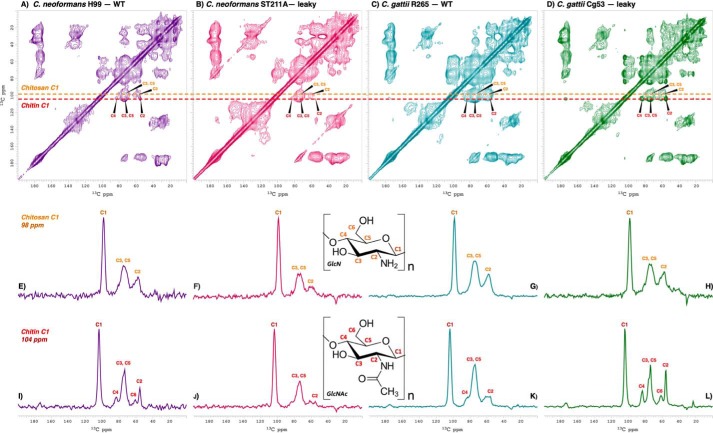
Figure 7.
Cell-wall chitin flexibility is augmented in C. neoformans and C. gattii leaky mutant strain melanin ghosts. Top row, 2D 13C-13C DARR spectra (50-ms mixing time) of melanin ghosts generated from cell cultures containing d-[U-13C6]glucose as the sole carbon source. Cross-peaks represent proximal carbon pairs separated by distances corresponding to ∼1 bond length. Middle row, 98-ppm cross-sections displaying chitosan C1 correlations. Bottom row, 104-ppm cross-sections displaying chitin C1 correlations. A, E, and I, C. neoformans H99 (WT). B, F, and J, C. neoformans ST211A (leaky). C, G, and K, C. gattii R265 (WT). D, H, and L, C. gattii Cg53 (leaky). The vertical scale of the 2D spectra was adjusted such that all significant cross-peaks are visible, and the same numbers of contours are displayed for the carbonyl resonance at 173 ppm. The 1D cross-sections were adjusted by setting the largest peak to full scale.
Although a common set of intramolecular correlations involving the chitosan C1 ring carbon was displayed in the 13C-13C DARR plots of the melanin ghost samples isolated from all strains, the relative signal intensities of the cross-peaks with respect to their parent diagonal peaks varied noticeably among the four spectra. Strong signals signify large dipole-dipole interactions between a pair of carbon nuclei; for interactions within a sugar ring that has defined intramolecular distances, they report on the degree to which the structure is well-defined. One-dimensional 98-ppm “slices” from the 2D DARR plots of ghost samples from each leaky strain were found to display significantly attenuated chitosan correlations relative to those observed in the spectra of their respective parent strains, demonstrating partial averaging of the pairwise dipolar interactions and attributable to molecular disorder and/or flexibility in the solid state (Fig. 7, E–H). The reduction in chitosan cross-peak intensity was especially pronounced in the spectrum of the C. neoformans ST211A leaky strain (Fig. 7F). Overall, however, the chitosan-rich composition of the C. gattii parent strain produced more robust chitosan correlations for R265 ghosts (Fig. 7G) compared with H99 samples (Fig. 7E). As a result, the chitosan cross-peak signal intensities were rendered similar for C. neoformans H99 (Fig. 7E) and C. gattii leaky Cg53 ghosts (Fig. 7H), suggesting that the degree of disorder associated with the leaky phenotype is strain-specific with respect to this polysaccharide constituent.
In contrast to the broad and poorly resolved signals of chitosan, the cross-peaks involving chitin were sharp and well-resolved, exhibiting numerous proximal 13C-13C interactions that were most evident in comparisons of the representative one-dimensional (1D) cross-sections (Fig. 7, I–L). Strikingly, the C1 carbon of chitin resonates at ∼104 ppm; the peaks displayed in the corresponding 1D cross-section of the DARR spectrum are assigned to ring carbons located at distances corresponding to as many as 3 bonds apart (C1–C2, 104 × 56 ppm; C1–C3, 104 × 74; C1–C4, 104 × 84; C1–C5, 104 × 76). In comparison, the 98-ppm cross-section representative of the correlations involving the chitosan C1 ring carbon displays relatively broader peaks and carbon pairs located at most 2 bonds apart. These spectral variations are likely due to differences in molecular mobility between the two polysaccharides; chitin has a semicrystalline structure and is intrinsically more rigid than chitosan (55).
As observed for the chitosan constituent, variations in the 13C-13C DARR results for chitin were observed between the Cryptococcus strains and as a function of whether the melanin ghosts displayed the leaky phenotype. However, the trends were opposite for the C. neoformans and C. gattii leaky mutants: attenuation for C. neoformans ST211A (Fig. 7J) and augmentation for C. gattii Cg53 (Fig. 7L). Once again, the degree of disorder associated with the leaky phenotype for this polysaccharide constituent was found to be strain-specific. Thus, the leaky phenotype is found to occur when chitin is either anomalously disordered (C. neoformans) or unusually ordered (C. gattii). Nonetheless, a common hallmark of leaky mutants from both strains is the observation of disparate line widths and cross-peak intensities for the chitin and chitosan constituents, suggesting a mismatch in their degrees of crystallinity or propensity for molecular flexibility.
Discussion
This work was conceived following a curious observation under the light microscope: a portion of melanized cells from C. gattii R265 appeared to have strikingly pigmented cell walls, a peculiarity not witnessed in melanized cells from C. neoformans (Fig. 1). Further comparison of these two strains revealed that C. gattii R265 melanized cell walls had a relatively homogeneous distribution of melanin pigments throughout and are structurally more compact than those of C. neoformans H99 (Fig. 2, A and B). We can attribute this finding to the substantially higher chitosan content of the R265 strain in analogy with prior findings by our group: C. neoformans H99 cells grown in the presence of GlcNAc had increased chitosan levels compared with nonsupplemented control cells and concurrently exhibited a more uniform arrangement of cell-wall pigments, possibly due to an increase in electrostatic charge interactions (56, 57) and/or covalent linkage (58, 59) between chitosan and the melanin polymer. However, in GlcNAc-supplemented C. neoformans H99 cells, these features were found to correlate with an increase in melanization, whereas the C. gattii R265 strain actually retained a slightly smaller amount of cell-wall melanin pigments than C. neoformans H99 (Fig. 4). Thus, it is likely that chitosan promotes the uniform distribution of pigments, in turn leading to a more melanin-dense cell wall, but that other factors are involved in determining the overall extent of melanin deposition.
In the first reports linking cell-wall morphology and integrity with melanization, Banks et al. (39) and Baker et al. (38) determined that, of the eight chitin synthase proteins (Chs1–8) coded for by C. neoformans, the chitin synthase 3 enzyme (Chs3) and its chitin synthase regulator protein (Csr2) are responsible for the production of cell-wall chitin, a substantial portion of which is deacetylated to chitosan by one or more of three chitin deacetylase proteins (Cda1, Cda2, and Cda3) with redundant activity. Cells from C. neoformans chs3Δ, csr2Δ, and cda1Δ/cda2Δ/cda3Δ deletion strains were found to have normal or increased chitin levels but significantly diminished chitosan content. The mutant strains also displayed phenotypic traits such as incomplete cell separation during budding, increased sensitivity to certain cell-wall stressors, and, most notably, the inability to retain cell-wall melanin pigments, suggesting that chitosan deficiency could result in aberrant melanization.
In contrast, there is only one report of a C. gattii leaky melanin strain, designated Cg53, which was generated by Walton et al. (40) using insertional mutagenesis and subsequently determined to bear a mutation in the gene that encodes for Chs3. Nevertheless, no follow-up studies were reported that examine the effect of this genetic mutation on C. gattii cell-wall structure or the composition of chitinous polysaccharides. The C. neoformans ST211A leaky strain was also identified by Walton et al. (40) and found to bear a frameshift mutation in the CSR2 gene (GenBankTM accession number MK609896). Thus, ST211A is likely genetically equivalent to the csr2Δ strain first reported by Banks et al. (39), and Cg53 is the C. gattii analog of the C. neoformans chs3Δ mutant, both of which have previously been determined to display strikingly similar phenotypic traits, including decreased production of chitosan. Our biochemical determination of chitinous polysaccharides revealed that both of these mutants indeed have significantly lower chitosan levels than their respective parent strains (Fig. 3, A and B). Although anticipated for C. neoformans ST211A, this is the first demonstration that the deletion of CHS3 in C. gattii results in diminished chitosan content. Therefore, our findings indicate that chitosan deficiency contributes to the leaky melanin phenotype displayed by Cg53 and that the role of chitosan in the deposition and retention of cell-wall melanin pigments is conserved in these two Cryptococcus species.
A recent study of chitin and chitosan and the role of the chitin deacetylases in C. gattii (60) confirms our finding that there is increased chitosan in C. gattii compared with C. neoformans, and our results go further to demonstrate that the amount of chitin in both species is similar. What is puzzling from the C. gattii deacetylase study is that the R265 strain deleted for all three CDA genes did not leak melanin, despite having almost no chitosan, running counter to the interpretations in this paper. However, in C. neoformans, when the three deacetylases were deleted, the amount of chitin was substantially increased (38). The difference between the mutants with defects in CHS3 or CSR2 and a strain without deacetylases is that the deacetylase mutant strain still has its full complement of functional chitin synthases; the deacetylase mutant produces the same amount of chitin, but none of the chitin is deacetylated to chitosan, thus increasing the overall amount of chitin in the cell wall compared with WT. Therefore, it is possible that there are substantially increased chitin or other components in the R265 cda1Δcda2Δcda3Δ strain that change the architecture and/or the flexibility of the cell wall and thus allow the melanin to be retained.
Despite the undeniable importance of chitosan in the anchoring and arrangement of cell-wall melanin pigments, other nonpolysaccharide constituents are also likely to be involved. Prior work by our group has demonstrated that the fungal pigments present in C. neoformans “melanin ghost” samples are complexed with an intricate aliphatic scaffold that includes both polysaccharides and lipids (31, 32). The fact that these relatively labile moieties can withstand the enzymatically and chemically degradative purification process used to generate melanin ghosts suggests they have become intimately associated with the pigment during its transport and/or deposition into the cell wall and thus play an integral role in melanin assembly.
In the current work, our spectroscopic data verified that the melanin pigments produced by C. gattii are tightly associated with an aliphatic framework composed of the same cellular constituents as in C. neoformans. However, quantitative ssNMR analysis of [13C]glucose-enriched melanin ghosts isolated from the leaky and parent strains of both species revealed significant disparities in the relative amounts of retained constituent types. Notably, the relative proportions of polysaccharides estimated by ssNMR for ghosts (Fig. 5B) follow the same trend as the total amounts of chitin plus chitosan determined by the MBTH assay in unmelanized cells (Fig. 3A). Thus, cells that produce less chitinous material do not seem to compensate for this “loss” by retaining a greater percentage of their polysaccharides when in their melanized state. Instead, C. gattii Cg53 is found to retain more lipids, such as long-chain fatty acids, compared with its parent strain, whereas C. neoformans ST211A retains relatively more “other” components compared with H99. A possible explanation for this finding is that C. gattii and C. neoformans leaky mutant strains activate different compensatory mechanisms leading to increased production of nonpolysaccharide constituents as a response to the reduced production of cell-wall chitin and chitosan. Nevertheless, because Cg53 and ST211A bear mutations in different genes (CHS3 and CSR2, respectively), it is uncertain whether these outcomes are species-specific or due to different genetic mutations.
Whereas changes in cell-wall composition can have a drastic impact on melanization, the possible influence of altered cell-wall architecture, even among strains with similar chitin content, had been unexplored. Our 2D ssNMR spectroscopic data support the premise that the macromolecular arrangement and structure of the constituents comprising the cell-wall framework play key roles in cryptococcal melanin deposition.
First, 2D 13C-15N TEDOR spectra of C. neoformans H99 melanin ghosts displayed 13C chemical shifts that implicated a highly deacetylated form of chitosan (Fig. 6), for which the few remaining acetylated units are distributed in a nonsequential manner (52–54). This carbon chemical shift information then allowed us to infer the presence of highly deacetylated chitosan in the 2D 13C-13C DARR spectra of melanin ghosts from all four fungal strains. In contrast, the chitosan produced by most organisms is only partially deacetylated, resulting in a copolymer that confers both rigidity and flexibility to cellular structures in which it is incorporated (61, 62). The fact that both C. neoformans and C. gattii were found to produce highly deacetylated chitosan could mean that, with hydrogen bonding between chitin and chitosan precluded, both of these Cryptococcus species achieve strengthening of their cell walls via an architecture that promotes pigment deposition.
Second, the spectral differences observed between the 2D 13C-13C DARR plots of leaky and parent strain melanin ghosts cannot be accounted for simply by disparities in cell-wall composition. Notably, the peaks corresponding to the intramolecular 13C-13C contacts within chitin were found to differ in line width and intensity between the leaky and parent strain of each species (Fig. 7). By comparing the 1D cross-sectional slices that display correlations involving the chitin C1 carbon, it is evident that the signal intensities observed for C. neoformans ST211A were significantly diminished compared with those of C. neoformans H99. Given similar chitin levels, we cannot attribute this observation to a reduction in chitin content. Instead, the diminished peak intensities likely arise from partial averaging of dipolar interactions between chitin carbons, signifying an increase in the flexibility and/or disorder of the chitin polymers. The spectral features corresponding to chitin carbons were additionally found to differ between the C. gattii Cg53 leaky mutant and its parent strain R265 despite their similar chitin content. In contrast to our findings with C. neoformans, the chitin peaks of Cg53 were observed to have greater intensities and reduced line widths compared with those displayed by R265, indicating an increase in rigidity and/or molecular order. Moreover, it is noteworthy that chitin correlations displayed by the parent type of each species have intermediate intensities and line widths, which suggests the presence of polymers that are both amorphous and crystalline to some degree.
The simplest explanation for these results is that the genetic mutation carried by each leaky strain has a direct influence on the ultrastructure of the chitin produced. Importantly, although the two leaky strains have mutations in two distinct genes, the proteins encoded by these genes have been hypothesized to form a complex that produces the chitin that can be converted to chitosan (39). Therefore, the two leaky strains are likely interrupting the same biological process and should have very similar phenotypes. For example, it is likely that the mutation of CHS3 in Cg53 results in increased production of chitin by one of the other chitin synthases it encodes for, as seen in C. neoformans chs3Δ mutants (39), which could in turn produce chitin polymers that have a greater propensity to form rigid and/or well-ordered crystalline microfibrils.
An alternative explanation is that the disparate compositional changes in nonchitin constituents exhibited by the respective leaky mutant strains impact chitin ultrastructure differentially. For example, the increased proportion of nonpolysaccharide and nonlipid “other” constituents retained by ST211A melanin ghosts, which are likely composed of bulky and rigid aromatic ring systems, could potentially disrupt the hydrogen bonding between individual chitin polymers and thus produce a relatively disordered architecture. In either case, the fact that the chitin produced by both C. neoformans and C. gattii parent strains appears to have a macromolecular arrangement with intermediate properties suggests a Goldilocks principle, whereby a “just right” balance of flexibility/disorder and rigidity/order is required for optimal cell-wall melanin pigment deposition and retention.
In summary, this study provides the first detailed compositional and structural analysis of melanized cell walls from the two most clinically relevant isolates of the Cryptococcus species complex: C. neoformans H99 and C. gattii R265. By leveraging the dependence of cryptococcal species on exogenous precursors for melanin synthesis and implementing ssNMR spectroscopy, we generated isotopically enriched melanin ghosts for which it could be determined that pigments produced by each C. neoformans and C. gattii preferentially associate with different proportions of chitinous polysaccharides and lipids during melanin assembly. In addition, we verified the role of chitosan as a critical component of the cell-wall scaffold that governs melanin deposition and retention in both cryptococcal species and represents a major insight for this field. Nonetheless, further studies using multiple strains from these two cryptococcal species are required for validation. Altogether, a better understanding of these fungal melanized cell-wall structures could support the development of new and highly selective antifungal drugs.
Experimental procedures
Cryptococcus strains and culture conditions
All strains used in the current study are listed in Table 1. Yeast cells were kept frozen in 20% glycerol stocks and subcultured into Sabouraud dextrose broth for 48 h at 30 °C prior to each experiment. The yeast cells were washed three times with Dulbecco's PBS (DPBS) with calcium and magnesium (Corning, Inc.) and counted using a hemocytometer. Cultures inoculated with 1 × 106 cells ml−1 (end concentration) were grown in minimal medium (MM) (29.4 mm KH2PO4, 10 mm MgSO4, 13 mm glycine, 3 μm thiamine, 15 mm glucose, pH 5.5) at 30 °C and shaken at moderate speed (120 rpm) for periods of 4–10 days in separate experiments. Melanized yeast cells were produced by inoculating cells in MM containing 1 mm l-DOPA as the obligatory melanization precursor and using the same culture conditions listed above. In designated experiments, glucose and/or glycine were replaced by [U-13C6]glucose (catalog no. GLC-082, Omicron Biochemical, Inc.) and [15N]glycine (catalog no. 299294, Sigma-Aldrich), respectively. Aliquots of each culture were collected for light microscopy and transmission EM.
Microscopic studies for cell body and capsule measurements
C. neoformans H99 and C. gattii R265 nonmelanized and melanized cells, grown for 4 or 7 days in separate experiments, were washed three times with DPBS. To determine cell body and capsule sizes by light microscopy, washed yeast cells were suspended in India ink and visualized under an Olympus AX70 microscope (Olympus America) using a ×40 objective. The cell body and capsule dimensions were calculated as done previously (42). For each condition, 150 cells were measured in two independent experiments.
Assessment of chitinous cell-wall constituents
Cellular chitin and chitosan of nonmelanized yeast cells were estimated as described previously (42). Briefly, each alkaline-extracted sample was divided into two aliquots: one aliquot was deacetylated with hydrochloric acid (HCl) to measure the total amount of glucosamine arising from chitin plus chitosan, and the second aliquot remained untreated to measure the glucosamine from chitosan only. The amount of chitin-derived glucosamine was determined by taking the difference between the two measurements. Each aliquot was treated with MBTH, which forms a complex with hexosamines (amino sugars) under mildly acid conditions. The intense blue color yielded by this reaction was recorded at 650 nm. The amount of hexosamine in each sample was determined by comparison with a standard curve prepared from a 1 mm GlcNAc stock.
Transmission EM
Aliquots from C. neoformans H99 and C. gattii R265 melanized yeast cells were washed, adjusted to 1 × 106 cell ml−1 in DPBS, and fixed overnight with the same volume of 2.5% (v/v) glutaraldehyde, 3 mm MgCl2 in 0.1 m sodium cacodylate buffer (pH 7.2) at 4 °C. After rinsing with buffer, cells were processed with 0.8% potassium ferrocyanide reduced with 1% OsO4, serially dehydrated in a graded series of ethanol, and embedded in Eponate 12 (Ted Pella) resin. Ultrathin sections were obtained and stained with 2% uranyl acetate in 50% methanol, followed by lead citrate for examination with a Philips/FEI Bio Twin CM120 transmission electron microscope at 80 kV. Images were captured with an AMT XR80 high-resolution (16-bit) 8-megapixel camera. Using ImageJ software (Fiji) (63), micrograph analyses of 10–18 cells (5 calculations/cell) per Cryptococcus strain were performed to determine the melanized cell wall dimensions.
Melanin isolation for solid-state NMR
Cryptococcal cells cultured in MM containing the melanization precursor l-DOPA for 10 days were subjected to acid digestion in a procedure known as “melanin ghost” isolation, where cellular material is digested but the acid-resistant melanin remains assembled into a structure with the dimensions of the original cell (33). The protocol involved progressive enzymatic and chemical removal of cell-wall polysaccharides, proteins, and lipids. All samples were extensively dialyzed against distilled water and subsequently lyophilized for 3 days before use.
ssNMR spectroscopy
Solid-state NMR measurements were carried out on a Varian (Agilent) DirectDrive2 (DD2) instrument operating at a 1H frequency of 600 MHz. All data were acquired on ∼5–10 mg of powdered samples using an MAS rate of 15.00 ± 0.02 kHz at a spectrometer-set temperature of 25 °C and processed with 100–150 Hz of line broadening. The 1D 13C DPMAS and 2D 13C-13C DARR experiments were conducted on a 1.6-mm T3 HXY fastMAS probe (Agilent Technologies, Santa Clara, CA) with 90° pulse lengths of 1.2 and 1.4 μs for 1H and 13C, respectively, and 104-kHz heteronuclear decoupling using the small phase incremental alternation pulse sequence (SPINAL) during signal acquisition. To estimate the relative amounts of carbon-containing constituents in melanin ghost samples, 1D 13C DPMAS experiments were performed with long (50-s) recycle delays to obtain quantitatively reliable signal intensities that allowed the integration of defined spectral regions using the GNU image manipulation program (GIMP). In 2D 13C-13C DARR experiments, the initial magnetization transfer from 1H to 13C was accomplished during a 1.5-ms cross-polarization period with a 10% linearly ramped radiofrequency field (rf) of 191 kHz for 1H and a 179-kHz constant rf for 13C. Proton irradiation with an rf of 16 kHz was applied during the 50-ms field-assisted diffusion mixing period. The 1D 15N cross-polarization magic angle spinning (CPMAS) and 2D 13C-15N TEDOR experiments were run on a 3.2-mm T3 HXY MAS probe (Agilent Technologies, Santa Clara, CA) with 90° pulse lengths of 2.6, 4.0, and 4.5 μs for 1H, 13C, and 15N, respectively, and 48-kHz SPINAL decoupling during acquisition. In the 1D 15N CPMAS experiments, 1H-15N transfer was accomplished during a 2-ms period with a 10% linearly ramped rf of 66 kHz for 1H and a 56-kHz constant rf for 15N. In the 2D 13C-15N TEDOR experiments, the initial 1H to 13C transfer was accomplished during a 1.0-ms cross-polarization period with a 10% linearly ramped rf of 85 kHz for 1H and a 63-kHz constant rf for 13C. Coherence transfer periods of 1.40 and 4.00 ms were used in separate experiments to obtain 13C-15N correlations corresponding to approximately 1- and 2-bond distances, respectively.
Statistical analyses
Statistical analyses were performed using GraphPad Prism version 8.00 for Mac OS X (GraphPad Software, San Diego, CA). The 90–95% confidence interval was determined for each set of results.
Author contributions
C. C., E. C., R. P.-R., J. K. L., A. C., and R. E. S. conceptualization; C. C., E. C., M. S. F., R. P.-R., S. C., R. J. C., J. K. L., A. C., and R. E. S. formal analysis; C. C., E. C., M. S. F., R. P.-R., S. C., R. J. C., J. K. L., A. C., and R. E. S. validation; C. C., E. C., M. S. F., R. P.-R., S. C., and R. J. C. investigation; C. C., E. C., J. K. L., A. C., and R. E. S. visualization; C. C., E. C., M. S. F., R. P.-R., S. C., R. J. C., J. K. L., A. C., and R. E. S. methodology; C. C., E. C., J. K. L., A. C., and R. E. S. writing-original draft; C. C., E. C., J. K. L., A. C., and R. E. S. project administration; C. C., E. C., M. S. F., R. P.-R., S. C., R. J. C., J. K. L., A. C., and R. E. S. writing-review and editing; J. K. L., A. C., and R. E. S. supervision.
Supplementary Material
Acknowledgments
We thank Alexander Idnurm (Mycology Laboratory, University of Melbourne) for the kind gift of C. neoformans strain ST211A and Barbara Smith (Microscopy Facility, School of Medicine, Johns Hopkins University) for her expertise and technical assistance with transmission EM. We also thank Hsin Wang (City College, CUNY) and Van Chanh Phan (Hostos College, CUNY) for the development of NMR programs and technical support. The 600-MHz NMR facilities used in this work are operated by City College and the CUNY Institute for Macromolecular Assemblies, with additional infrastructural support provided by National Institutes of Health Grant 3G12MD007603-30S2 from the National Institute on Minority Health and Health Disparities of the National Institutes of Health.
This work was supported by National Institutes of Health Grant R01-AI052733 (to A. C. and R. E. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2.
A. Idnurm, personal communication.
C. Chrissian, unpublished observations.
- l-DOPA
- 3,4-dihydroxyphenylalanine
- ssNMR
- solid-state NMR
- MAS
- magic angle spinning
- DPMAS
- direct polarization magic angle spinning
- CPMAS
- cross-polarization magic angle spinning
- TEDOR
- transferred echo double resonance
- DARR
- dipolar-assisted rotational resonance
- MBTH
- 3-methyl-2-benzothiazolone hydrazine hydrochloride
- DPBS
- Dulbecco's PBS
- MM
- minimal medium
- SPINAL
- small phase incremental alternation pulse sequence
- rf
- radiofrequency field
- 1D and 2D
- one- and two-dimensional, respectively.
References
- 1. Walker L., Sood P., Lenardon M. D., Milne G., Olson J., Jensen G., Wolf J., Casadevall A., Adler-Moore J., and Gow N. A. R. (2018) The viscoelastic properties of the fungal cell wall allow traffic of AmBisome as intact liposome vesicles. MBio 9, e02383–17 10.1128/mBio.02383-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Latgé J. P., and Beauvais A. (2014) Functional duality of the cell wall. Curr. Opin. Microbiol. 20, 111–117 10.1016/j.mib.2014.05.009 [DOI] [PubMed] [Google Scholar]
- 3. Perez P., and Ribas J. C. (2013) Fungal cell wall analysis. In Laboratory Protocols in Fungal Biology (Gupta V., Tuohy M., Ayyachamy M., Turner K., and O'Donovan A., eds) pp. 175–196, Springer, New York [Google Scholar]
- 4. Lesage G., and Bussey H. (2006) Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 317–343 10.1128/MMBR.00038-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gow N. A. R., Latge J. P., and Munro C. A. (2017) The fungal cell wall: structure, biosynthesis, and function. Microbiol. Spectr. 5 10.1128/microbiolspec.FUNK-0035-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Free S. J. (2013) Fungal cell wall organization and biosynthesis. Adv. Genet. 81, 33–82 10.1016/B978-0-12-407677-8.00002-6 [DOI] [PubMed] [Google Scholar]
- 7. Agustinho D. P., Miller L. C., Li L. X., and Doering T. L. (2018) Peeling the onion: the outer layers of Cryptococcus neoformans. Mem. Inst. Oswaldo Cruz 113, e180040 10.1590/0074-02760180040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perez P., and Ribas J. C. (2017) Fission yeast cell wall analysis. Cold Spring Harb. Protoc. 2017, pdb.top079897 10.1101/pdb.top079897 [DOI] [PubMed] [Google Scholar]
- 9. Doering T. L. (2009) How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annu. Rev. Microbiol. 63, 223–247 10.1146/annurev.micro.62.081307.162753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kwon-Chung K. J., Bennett J. E., Wickes B. L., Meyer W., Cuomo C. A., Wollenburg K. R., Bicanic T. A., Castaneda E., Chang Y. C., Chen J., Cogliati M., Dromer F., Ellis D., Filler S. G., Fisher M. C., et al. (2017) The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. mSphere 2, e00357–16 10.1128/mSphere.00357-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chowdhary A., Rhandhawa H. S., Prakash A., and Meis J. F. (2012) Environmental prevalence of Cryptococcus neoformans and Cryptococcus gattii in India: an update. Crit. Rev. Microbiol. 38, 1–16 10.3109/1040841X.2011.606426 [DOI] [PubMed] [Google Scholar]
- 12. Ellis D. H., and Pfeiffer T. J. (1990) Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 28, 1642–1644 10.1128/JCM.28.7.1642-1644.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emmons C. W. (1955) Saprophytic sources of Cryptococcus neoformans associated with the pigeon (Columba livia). Am. J. Hyg. 62, 227–232 10.1093/oxfordjournals.aje.a119775 [DOI] [PubMed] [Google Scholar]
- 14. Sorrell T. C., Brownlee A. G., Ruma P., Malik R., Pfeiffer T. J., and Ellis D. H. (1996) Natural environmental sources of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 34, 1261–1263 10.1128/JCM.34.5.1261-1263.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. May R. C., Stone N. R., Wiesner D. L., Bicanic T., and Nielsen K. (2016) Cryptococcus: from environmental saprophyte to global pathogen. Nat. Rev. Microbiol. 14, 106–117 10.1038/nrmicro.2015.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Findley K., Rodriguez-Carres M., Metin B., Kroiss J., Fonseca A., Vilgalys R., and Heitman J. (2009) Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the Tremellales. Eukaryot. Cell 8, 353–361 10.1128/EC.00373-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu J., Vilgalys R., and Mitchell T. G. (2000) Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9, 1471–1481 10.1046/j.1365-294x.2000.01021.x [DOI] [PubMed] [Google Scholar]
- 18. Casadevall A., Freij J. B., Hann-Soden C., and Taylor J. (2017) Continental drift and speciation of the Cryptococcus neoformans and Cryptococcus gattii species complexes. mSphere 2, e00103–17 10.1128/mSphere.00103-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Freij J. B., Fu M. S., De Leon Rodriguez C. M., Dziedzic A., Jedlicka A. E., Dragotakes Q., Rossi D. C. P., Jung E. H., Coelho C., and Casadevall A. (2018) Conservation of intracellular pathogenic strategy among distantly related cryptococcal species. Infect. Immun. 86, e00946–17 10.1128/IAI.00946-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gerstein A. C., and Nielsen K. (2017) It's not all about us: evolution and maintenance of Cryptococcus virulence requires selection outside the human host. Yeast 34, 143–154 10.1002/yea.3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eisenman H. C., and Casadevall A. (2012) Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 93, 931–940 10.1007/s00253-011-3777-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zaragoza O., Rodrigues M. L., De Jesus M., Frases S., Dadachova E., and Casadevall A. (2009) The capsule of the fungal pathogen Cryptococcus neoformans. Adv. Appl. Microbiol. 68, 133–216 10.1016/S0065-2164(09)01204-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. White L. P. (1958) Melanin: a naturally occurring cation exchange material. Nature 182, 1427–1428 10.1038/1821427a0 [DOI] [PubMed] [Google Scholar]
- 24. Enochs W. S., Nilges M. J., and Swartz H. M. (1993) A standardized test for the identification and characterization of melanins using electron paramagnetic resonance (EPR) spectroscopy. Pigment Cell Res. 6, 91–99 10.1111/j.1600-0749.1993.tb00587.x [DOI] [PubMed] [Google Scholar]
- 25. Nosanchuk J. D., and Casadevall A. (2006) Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob. Agents Chemother. 50, 3519–3528 10.1128/AAC.00545-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nosanchuk J. D., and Casadevall A. (2003) The contribution of melanin to microbial pathogenesis. Cell Microbiol. 5, 203–223 10.1046/j.1462-5814.2003.00268.x [DOI] [PubMed] [Google Scholar]
- 27. Mednick A. J., Nosanchuk J. D., and Casadevall A. (2005) Melanization of Cryptococcus neoformans affects lung inflammatory responses during cryptococcal infection. Infect. Immun. 73, 2012–2019 10.1128/IAI.73.4.2012-2019.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Williamson P. R., Wakamatsu K., and Ito S. (1998) Melanin biosynthesis in Cryptococcus neoformans. J. Bacteriol. 180, 1570–1572 10.1128/JB.180.6.1570-1572.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu L., Wakamatsu K., Ito S., and Williamson P. R. (1999) Catecholamine oxidative products, but not melanin, are produced by Cryptococcus neoformans during neuropathogenesis in mice. Infect. Immun. 67, 108–112 10.1128/IAI.67.1.108-112.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhong J., Frases S., Wang H., Casadevall A., and Stark R. E. (2008) Following fungal melanin biosynthesis with solid-state NMR: biopolymer molecular structures and possible connections to cell-wall polysaccharides. Biochemistry 47, 4701–4710 10.1021/bi702093r [DOI] [PubMed] [Google Scholar]
- 31. Chatterjee S., Prados-Rosales R., Itin B., Casadevall A., and Stark R. E. (2015) Solid-state NMR reveals the carbon-based molecular architecture of Cryptococcus neoformans fungal eumelanins in the cell wall. J. Biol. Chem. 290, 13779–13790 10.1074/jbc.M114.618389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chatterjee S., Prados-Rosales R., Tan S., Phan V. C., Chrissian C., Itin B., Wang H., Khajo A., Magliozzo R. S., Casadevall A., and Stark R. E. (2018) The melanization road more traveled by: precursor substrate effects on melanin synthesis in cell-free and fungal cell systems. J. Biol. Chem. 293, 20157–20168 10.1074/jbc.RA118.005791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chatterjee S., Prados-Rosales R., Frases S., Itin B., Casadevall A., and Stark R. E. (2012) Using solid-state NMR to monitor the molecular consequences of Cryptococcus neoformans melanization with different catecholamine precursors. Biochemistry 51, 6080–6088 10.1021/bi300325m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Z., Zheng L., Hauser M., Becker J. M., and Szaniszlo P. J. (1999) WdChs4p, a homolog of chitin synthase 3 in Saccharomyces cerevisiae, alone cannot support growth of Wangiella (Exophiala) dermatitidis at the temperature of infection. Infect. Immun. 67, 6619–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bull A. T. (1970) Chemical composition of wild-type and mutant Aspergillus nidulans cell walls: the nature of polysaccharide and melanin constituents. J. Gen. Microbiol. 63, 75–94 10.1099/00221287-63-1-75 [DOI] [PubMed] [Google Scholar]
- 36. Walker C. A., Gómez B. L., Mora-Montes H. M., Mackenzie K. S., Munro C. A., Brown A. J., Gow N. A., Kibbler C. C., and Odds F. C. (2010) Melanin externalization in Candida albicans depends on cell wall chitin structures. Eukaryot. Cell 9, 1329–1342 10.1128/EC.00051-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stavenga D. G., Leertouwer H. L., Hariyama T., De Raedt H. A., and Wilts B. D. (2012) Sexual dichromatism of the damselfly Calopteryx japonica caused by a melanin-chitin multilayer in the male wing veins. PLoS ONE 7, e49743 10.1371/journal.pone.0049743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baker L. G., Specht C. A., Donlin M. J., and Lodge J. K. (2007) Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell 6, 855–867 10.1128/EC.00399-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Banks I. R., Specht C. A., Donlin M. J., Gerik K. J., Levitz S. M., and Lodge J. K. (2005) A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 4, 1902–1912 10.1128/EC.4.11.1902-1912.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walton F. J., Idnurm A., and Heitman J. (2005) Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol. Microbiol. 57, 1381–1396 10.1111/j.1365-2958.2005.04779.x [DOI] [PubMed] [Google Scholar]
- 41. Baker L. G., Specht C. A., and Lodge J. K. (2009) Chitinases are essential for sexual development but not vegetative growth in Cryptococcus neoformans. Eukaryot. Cell 8, 1692–1705 10.1128/EC.00227-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Camacho E., Chrissian C., Cordero R. J. B., Liporagi-Lopes L., Stark R. E., and Casadevall A. (2017) N-Acetylglucosamine affects Cryptococcus neoformans cell-wall composition and melanin architecture. Microbiology 163, 1540–1556 10.1099/mic.0.000552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tsirilakis K., Kim C., Vicencio A. G., Andrade C., Casadevall A., and Goldman D. L. (2012) Methylxanthine inhibit fungal chitinases and exhibit antifungal activity. Mycopathologia 173, 83–91 10.1007/s11046-011-9483-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fraser J. A., Giles S. S., Wenink E. C., Geunes-Boyer S. G., Wright J. R., Diezmann S., Allen A., Stajich J. E., Dietrich F. S., Perfect J. R., and Heitman J. (2005) Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437, 1360–1364 10.1038/nature04220 [DOI] [PubMed] [Google Scholar]
- 45. Ngamskulrungroj P., and Meyer W. (2009) Melanin production at 37 ºC is linked to the high virulent Cryptococcus gattii Vancouver Island outbreak genotype VGIIa. Aust. Mycol. 28, 9–15 [Google Scholar]
- 46. Ngamskulrungroj P., Serena C., Gilgado F., Malik R., and Meyer W. (2011) Global VGIIa isolates are of comparable virulence to the major fatal Cryptococcus gattii Vancouver Island outbreak genotype. Clin. Microbiol. Infect. 17, 251–258 10.1111/j.1469-0691.2010.03222.x [DOI] [PubMed] [Google Scholar]
- 47. Polacheck I., Platt Y., and Aronovitch J. (1990) Catecholamines and virulence of Cryptococcus neoformans. Infect. Immun. 58, 2919–2922 10.1128/IAI.58.9.2919-2922.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Watson H. R., Apperley D. C., Dixon D. P., Edwards R., and Hodgson D. R. (2009) An efficient method for 15N-labeling of chitin in fungi. Biomacromolecules 10, 793–797 10.1021/bm8012814 [DOI] [PubMed] [Google Scholar]
- 49. Heux L., Brugnerotto J., Desbrières J., Versali M. F., and Rinaudo M. (2000) Solid state NMR for determination of degree of acetylation of chitin and chitosan. Biomacromolecules 1, 746–751 10.1021/bm000070y [DOI] [PubMed] [Google Scholar]
- 50. Jaroniec C. P., Filip C., and Griffin R. G. (2002) 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly 13C,15N-labeled solids. J. Am. Chem. Soc. 124, 10728–10742 10.1021/ja026385y [DOI] [PubMed] [Google Scholar]
- 51. Hing A. W., Vega S., and Schaefer J. (1993) Measurement of heteronuclear dipolar coupling by transferred-echo double-resonance NMR. J. Magn. Reson. Ser. A 103, 151–162 10.1006/jmra.1993.1146 [DOI] [Google Scholar]
- 52. Vårum K. M., Anthonsen M. W., Grasdalen H., and Smidsrød O. (1991) Determination of the degree of N-acetylation and the distribution of N-acetyl groups in partially N-deacetylated chitins (chitosans) by high-field n.m.r. spectroscopy. Carbohydr. Res. 211, 17–23 10.1016/0008-6215(91)84142-2 [DOI] [PubMed] [Google Scholar]
- 53. Vårum K. M., Anthonsen M. W., Grasdalen H., and Smidsrød O. (1991) 13C-n.m.r. studies of the acetylation sequences in partially N-deacetylated chitins (chitosans). Carbohydr. Res. 217, 19–27 10.1016/0008-6215(91)84113-S [DOI] [PubMed] [Google Scholar]
- 54. Weinhold M. X., Sauvageau J. C. M., Kumirska J., and Thöming J. (2009) Studies on acetylation patterns of different chitosan preparations. Carbohydr. Polym. 78, 678–684 10.1016/j.carbpol.2009.06.001 [DOI] [Google Scholar]
- 55. Oh D. X., Cha Y. J., Nguyen H. L., Je H. H., Jho Y. S., Hwang D. S., and Yoon D. K. (2016) Chiral nematic self-assembly of minimally surface damaged chitin nanofibrils and its load bearing functions. Sci. Rep. 6, 23245 10.1038/srep23245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nosanchuk J. D., and Casadevall A. (1997) Cellular charge of Cryptococcus neoformans: contributions from the capsular polysaccharide, melanin, and monoclonal antibody binding. Infect. Immun. 65, 1836–1841 10.1128/IAI.65.5.1836-1841.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Quemeneur F., Rinaudo M., and Pépin-Donat B. (2008) Influence of molecular weight and pH on adsorption of chitosan at the surface of large and giant vesicles. Biomacromolecules 9, 396–402 10.1021/bm700943j [DOI] [PubMed] [Google Scholar]
- 58. Zhang Y., Thomas Y., Kim E., and Payne G. F. (2012) pH- and voltage-responsive chitosan hydrogel through covalent cross-linking with catechol. J. Phys. Chem. B 116, 1579–1585 10.1021/jp210043w [DOI] [PubMed] [Google Scholar]
- 59. Zhu X., and Williamson P. R. (2004) Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res. 5, 1–10 10.1016/j.femsyr.2004.04.004 [DOI] [PubMed] [Google Scholar]
- 60. Lam W. C., Upadhya R., Specht C. A., Ragsdale A. E., Hole C. R., Levitz S. M., and Lodge J. K. (2019) Chitosan biosynthesis and virulence in the human fungal pathogen Cryptococcus gattii. mSphere 4, e00644–19 10.1128/mSphere.00644-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Grifoll-Romero L., Pascual S., Aragunde H., Biarnés X., and Planas A. (2018) Chitin deacetylases: structures, specificities, and biotech applications. Polymers (Basel) 10, E352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cui J., Yu Z., and Lau D. (2016) Effect of acetyl group on mechanical properties of chitin/chitosan nanocrystal: a molecular dynamics study. Int. J. Mol. Sci. 17, E61 10.3390/ijms17010061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Toffaletti D. L., Rude T. H., Johnston S. A., Durack D. T., and Perfect J. R. (1993) Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175, 1405–1411 10.1128/JB.175.5.1405-1411.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fraser J. A., Subaran R. L., Nichols C. B., and Heitman J. (2003) Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2, 1036–1045 10.1128/EC.2.5.1036-1045.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.