The high complexity of in situ ecosystems renders it difficult to study marine microbial photoautotroph-heterotroph interactions. Two-member coculture systems of picocyanobacteria and single heterotrophic bacterial strains have been thoroughly investigated. However, in situ interactions comprise far more diverse heterotrophic bacterial associations with single photoautotrophic organisms. In the present study, combined metagenomic and metaproteomic data supplied the metabolic potentials and activities of uncultured dominant bacterial populations in the coculture system. The results of this study shed light on the nature of interactions between photoautotrophs and heterotrophs, improving our understanding of the complexity of in situ environments.
KEYWORDS: Synechococcus culture, interactions, high-molecular-weight DOM, low-molecular-weight DOM, metagenome, metaproteome, exoproteome, transporters
ABSTRACT
Microbial photoautotroph-heterotroph interactions underlie marine food webs and shape ecosystem diversity and structure in upper ocean environments. Here, bacterial community composition, lifestyle preference, and genomic- and proteomic-level metabolic characteristics were investigated for an open ocean Synechococcus ecotype and its associated heterotrophs over 91 days of cocultivation. The associated heterotrophic bacterial assembly mostly constituted five classes, including Flavobacteria, Bacteroidetes, Phycisphaerae, Gammaproteobacteria, and Alphaproteobacteria. The seven most abundant taxa/genera comprised >90% of the total heterotrophic bacterial community, and five of these displayed distinct lifestyle preferences (free-living or attached) and responses to Synechococcus growth phases. Six high-quality genomes, including Synechococcus and the five dominant heterotrophic bacteria, were reconstructed. The only primary producer of the coculture system, Synechococcus, displayed metabolic processes primarily involved in inorganic nutrient uptake, photosynthesis, and organic matter biosynthesis and release. Two of the flavobacterial populations, Muricauda and Winogradskyella, and an SM1A02 population, displayed preferences for initial degradation of complex compounds and biopolymers, as evinced by high abundances of TonB-dependent transporters (TBDTs), glycoside hydrolase, and peptidase proteins. Polysaccharide utilization loci present in the flavobacterial genomes influence their lifestyle preferences and close associations with phytoplankton. In contrast, the alphaproteobacterium Oricola sp. population mainly utilized low-molecular-weight dissolved organic carbon (DOC) through ATP-binding cassette (ABC), tripartite ATP-independent periplasmic (TRAP), and tripartite tricarboxylate transporter (TTT) transport systems. The heterotrophic bacterial populations exhibited complementary mechanisms for degrading Synechococcus-derived organic matter and driving nutrient cycling. In addition to nutrient exchange, removal of reactive oxygen species and vitamin trafficking might also contribute to the maintenance of the Synechococcus-heterotroph coculture system and the interactions shaping the system.
INTRODUCTION
Interactions between photoautotrophic and heterotrophic bacteria shape the structure and diversity of upper ocean ecosystems and are foundational to marine food webs (1–3). Phytoplankton are significant contributors to primary production in aquatic environments, and marine picocyanobacteria primarily comprising Prochlorococcus and Synechococcus species account for almost half of the total ocean primary production (4, 5). Marine bacteria take up labile dissolved organic carbon (DOC) that is released by phytoplankton into surrounding waters, and they transform a fraction of this DOC into recalcitrant forms via a series of metabolic processes collectively referred to as the microbial carbon pump (6). During DOC mineralization by heterotrophic bacteria, inorganic nutrients (e.g., NH4+, NO3−, PO43−, and Fe2+) that promote phytoplankton growth are regenerated. Thus, marine bacteria are important contributors to the global biogeochemical cycling of carbon, nitrogen, and phosphate (7, 8).
Interactions between photoautotrophs and heterotrophs in both natural environments and laboratory cultures have been extensively explored in recent decades (2, 8–10). Among these interactions, synergistic and antagonistic relationships have been commonly observed between photoautotrophs and heterotrophs. However, it is generally thought that photoautotrophs and heterotrophs develop mutualistic relationships through nutrient cycling. Specifically, photoautotrophs provide labile organic matter for heterotrophic growth, and in return, they benefit from the supply of essential micronutrients from heterotrophic bacteria, including vitamins and bioavailable trace metals (9, 11, 12), in addition to the removal of reactive oxygen species (ROS) (13, 14). In addition to mutualistic interactions, competition and antagonistic behaviors have also been observed between photoautotrophs and heterotrophs. For example, such interactions include competition for essential inorganic nutrients like phosphate and nitrate, in addition to the production of algicidal compounds by heterotrophic bacteria (15, 16).
Isolated marine unicellular cyanobacterial cultures, including those of Synechococcus and Prochlorococcus, typically contain coexisting heterotrophic bacterial partners (17, 18). Such associations between Synechococcus and heterotrophic bacteria have also been observed in natural environments (19). Although axenic cultures of Cyanobacteria can be achieved, Synechococcus and Prochlorococcus cultures with coexisting bacterial populations are more stable and have more robust and longer lifespans than axenic cultures (8, 13, 20). The interactions between picocyanobacteria and single heterotrophic bacterial strains (e.g., Synechococcus-Ruegeria pomeroyi DSS-3, Synechococcus-Shewanella, Synechococcus-Vibrio, and Prochlorococcus-Alteromonas) have been thoroughly investigated, yielding evidence for synergistic relationships among the two partners in coculture systems (8, 20–24). However, in situ interaction networks between photoautotrophs and heterotrophs are not only two-member systems but instead comprise diverse heterotrophic bacterial assemblies associated with a single photoautotrophic strain. These myriad relationships are not only synergistic in nature but also competitive and antagonistic.
The lifestyles and genomic characteristics of dominant heterotrophic bacteria associated with coastal Synechococcus sp. strain XM-24 cultures have been investigated in detail, but the associated heterotrophs also show geographic distribution patterns that are related to the environments of Synechococcus strain isolation (25, 26). Thus, several important aspects of these relationships remain unclear including (i) how open ocean Synechococcus culture ecotypes interact with their associated heterotrophic bacteria, (ii) how different heterotrophic bacterial populations interact with single cyanobacterial strains in coculture systems, and (iii) the nature of the roles of major bacterial populations in the Synechococcus culture systems. To address these gaps in knowledge, this study investigated (i) the assembly of culture-associated heterotrophic bacterial populations and their lifestyles across the growth period of an open ocean Synechococcus ecotype, (ii) the metabolic characteristics of the dominant bacteria in the coculture systems via metagenomic and metaproteomic analyses, and (iii) the potential associations of different bacterial populations in the context of the functioning and maintenance of coculture systems.
RESULTS AND DISCUSSION
Abundance variations and morphology of Synechococcus and associated heterotrophic bacteria.
Synechococcus strain YX04-3 was isolated from the South China Sea using PRO2 liquid medium and purified after ∼5 to 7 rounds of dilution until a single Synechococcus internal transcribed spacer (ITS) sequence was obtained (25). This strain belongs to the clade III, subcluster 5.1 group of Synechococcus (see Fig. S1 in the supplemental material), and represents an open ocean ecotype. Its associated heterotrophs were a self-selected natural assemblage. During the 91-day incubation, no extra inorganic nutrients were added, and nutrient recycling maintained the coculture for a long time. The Synechococcus strain was inoculated at an initial density of 1.43 × 107 cells ml−1 and plateaued at a population size of ∼1.60 × 108 cells ml−1 during the experiment. Heterotrophic bacterial abundances ranged from the inoculated abundance of 7.78 × 106 to 6.39 × 108 cells ml−1 over the experiment. The Synechococcus growth curve exhibited four distinct phases comprising the lag, exponential, stationary, and decline phases (Fig. 1A). Phosphate was an important factor controlling Synechococcus growth in the culture (Fig. S2).
FIG 1.
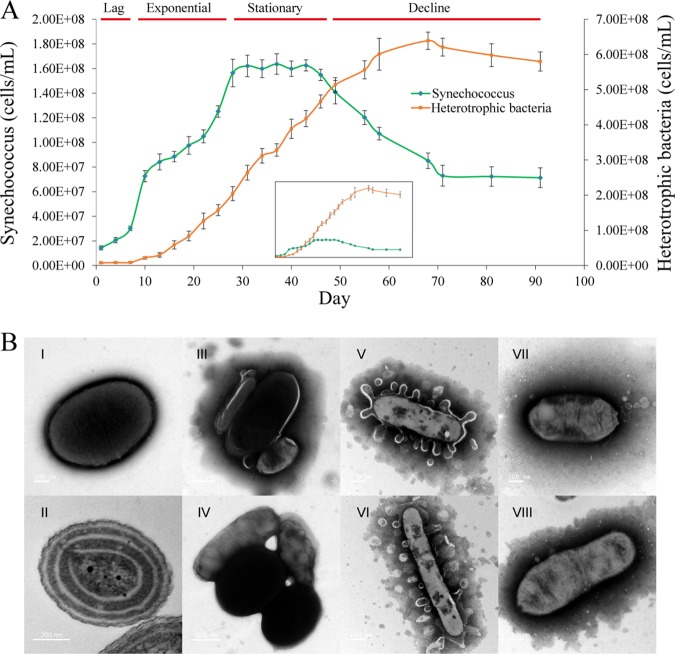
Abundance dynamics (A) and morphology (B) of Synechococcus and associated heterotrophic bacteria in the Synechococcus sp. strain YX04-3 coculture. The green curve represents Synechococcus cell abundances (left vertical axis) over time, while the orange curve shows heterotrophic bacterial cell abundances (right vertical axis) over time. The inset figure shows the dynamics among Synechococcus and heterotrophic bacterial abundances on the same vertical axis scale. Time (in days) is shown on the x axis. Error bars represent the ranges of values from triplicate measurements. The morphology of Synechococcus (I and II), attached cells or aggregates (III and IV), and associated heterotrophic bacteria (V to VIII) in the coculture system are depicted. Image scales are shown in the lower-left corner of the micrographs.
Neighbor-joining phylogenetic tree based on 16S-23S rRNA ITS sequences of representatives from cultured lineages of Synechococcus. Bootstrap percentages from neighbor-joining (1,000 replications) are shown. The scale bar represents a distance of 0.05 substitutions per site. The Synechococcus strains shown in red were isolated and collected by our lab. Download FIG S1, PDF file, 0.3 MB (312.9KB, pdf) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Variation in phosphate (A) and organic carbon (B) concentrations in the coculture system over the incubation period. DOC, dissolved organic carbon; POC, particulate organic carbon. Error bars represent the ranges of values from triplicate measurements. Download FIG S2, PDF file, 0.3 MB (299.7KB, pdf) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transmission electron microscopy (TEM) was also used to investigate the morphological variation of Synechococcus and the heterotrophic bacterial cells, in addition to possible associations of the taxa in the coculture system. TEM revealed that Synechococcus cells exhibited short rod shapes (1.10 ± 0.10 μm by 0.85 ± 0.10 μm) with relatively strong electron densities (Fig. 1B, panels I and II). In addition, aggregated or attached cells with different morphologies were frequently observed in the coculture system. It seems that some aggregates comprised Synechococcus-like cells in the centers and attached heterotrophic bacteria at the periphery (Fig. 1B, panels III and IV). Bacteria exhibiting long rod shapes (0.20 ± 0.05 μm by 2.0 ± 0.50 μm) released considerable numbers of vesicles (Fig. 1B, panels V and VI). In addition, other bacteria with short rod shapes (0.40 ± 0.10 μm by 1.20 ± 0.20 μm) exhibited mouth-like pit structures at cell ends that could be channels related to the uptake of macromolecular organic matter (Fig. 1B, panels VII and VIII) (27, 28).
Microbial community assembly in the Synechococcus sp. strain YX04-3 coculture.
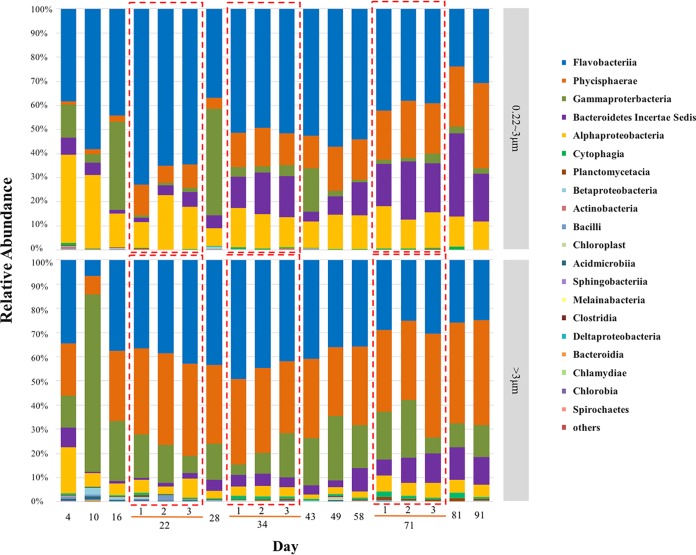
To evaluate the prevalence of different lifestyles (i.e., free-living versus attached/aggregated) among cells in the Synechococcus sp. strain YX04-3 coculture, 16S rRNA amplicons from 36 samples were amplified and sequenced over the duration of the culture incubation from two different size fractionation groups. A total of 1,271,032 16S rRNA gene sequences (617,028 sequences from the 0.22-to-3-μm size fractions and 654,004 sequences from the >3-μm size fractions) were generated after quality control. The average relative abundances of Synechococcus in the total 16S rRNA sequences for each size fraction represented 30.2% and 12.9% of the total bacteria in the 0.22-to-3-μm and >3-μm size fractions, respectively, over the entire culture growth period (Fig. 2 and 3). However, it should be noted that the relative abundances of Synechococcus are heavily underestimated due to multiple 16S rRNA gene copies in the genomes of the dominant heterotrophic bacteria in the coculture system (see Table S1A in the supplemental material). Consequently, we primarily focus on the distributional patterns among the different size fractions. After removal of Synechococcus sequences, a total of 430,701 and 569,501 heterotrophic bacterial sequences were obtained from the 0.22- and 3-μm size fractions, respectively. The heterotrophic bacterial 16S rRNA gene sequences primarily constituted five classes: Flavobacteriia (31.4% of the total 16S rRNA gene sequences), Phycisphaerae (20.7%), Gammaproteobacteria (11.1%), Bacteroidetes (6.8%), and Alphaproteobacteria (6.6%) (Fig. 2).
FIG 2.
Heterotrophic bacterial community structure from the 0.22-to-3-μm (top panel) and >3-μm (bottom panel) size fractions over the duration of the cultivation experiment. The relative abundances of different bacterial classes are based on the total heterotrophic bacterial 16S rRNA sequence abundances after removing Synechococcus sequences for each time point from the size-fractionated samples. Triplicate samples from the 0.22-to-3- μm and >3-μm size fractions were sequenced for the 22nd, 34th, and 71st day of cultivation (shown in red boxes).
FIG 3.
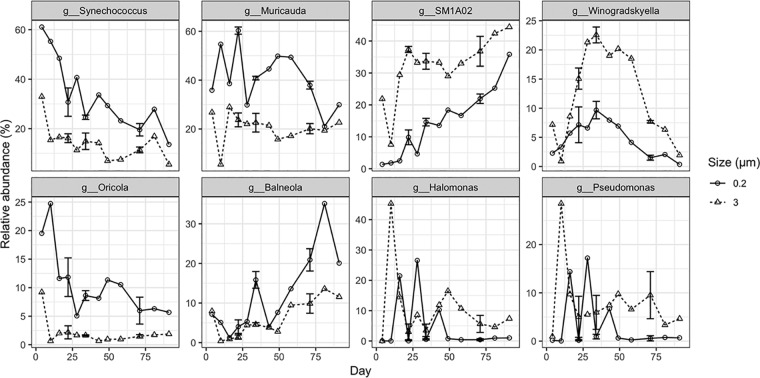
Relative abundances of Synechococcus (g_Synechococcus) in the total bacterial 16S rRNA sequence data sets and the seven dominant heterotrophic bacterial taxa among the total heterotrophic bacterial sequences from the 0.22-to-3-μm and >3-μm size fractions over the cultivation experiment. g_Synechococcus, genus Synechococcus, g__Muricauda, genus Muricauda, g__Winogradskyella, genus Winogradskyella, g__SM1A02, group SM1A02 belonging to family Phycisphaeraceae, g__Oricola, genus Oricola, g__Balneola, genus Balneola, g__Halomonas, genus Halomonas, g__Pseudomonas, genus Pseudomonas. Detailed descriptions of the lifestyle strategies of the dominant bacterial populations of the coculture are provided in Text S1 in the supplemental material.
(A) 16S rRNA gene copy numbers in the complete genomes of close relatives of the strains whose genomic bins were recovered in this study. (B) Proteins identified in the proteomes of each coculture population. Download Table S1, DOC file, 0.04 MB (44KB, doc) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic characteristics and lifestyle strategies of the dominant bacterial populations of the coculture, Synechococcus sp. YX04-3 coculture system exoproteome, supplemental Materials and Methods, and supplemental references. Download Text S1, DOC file, 0.08 MB (85.5KB, doc) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The seven most abundant heterotrophic genera/taxa, including Muricauda, SM1A02 group, Winogradskyella, Oricola, Balneola, Halomonas, and Pseudomonas, comprised more than 90% of the total heterotrophic bacterial 16S rRNA gene sequences within each size fraction. Comparison of taxonomic abundances across time points and size fractions revealed distinct patterns of lifestyle preference (free-living versus attached) and variation with Synechococcus growth phases (Fig. 3). The relative abundances of the Muricauda and Oricola genera from the Flavobacteriia and Alphaproteobacteria classes, respectively, were significantly higher in the 0.22-to-3-μm size fractions than in the >3-μm size fraction (t test, P < 0.01) (Fig. 3 and Table 1). In contrast, the relative abundances of the SM1A02 group and the Winogradskyella genus that belong to the Phycisphaerae and Flavobacteriia classes, respectively, were significantly higher in the >3-μm size fraction than in the 0.22-to-3-μm size fraction (P < 0.01) (Fig. 3 and Table 1). The Balneola genus exhibited increased relative abundances after the 50th day of cultivation (Fig. 3 and Table 1). In contrast, the gammaproteobacterial genera Halomonas and Pseudomonas did not exhibit lifestyle preferences or significant responses to Synechococcus growth phases (Fig. 3).
TABLE 1.
Characteristics of reconstructed genome bins and their relative abundances
| Bin IDa | Taxonomy (class) |
No. of contigs |
Assem. sizeb (Mbp) |
GC content (%) |
Est. comp.c (%) |
Est. cont.d (%) |
% relative abundance based on 16S rRNA gene sequencese
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.22 μm (D22) |
3 μm (D22) |
0.22 μm (D71) |
3 μm (D71) |
0.22 μm (avg) |
3 μm (avg) |
|||||||
|
Synechococcus sp. YX04-3 (Bin1) |
Cyanobacteria | 16 | 2.46 | 59.3 | 99.50 | 0.00 | 30.73 | 16.14 | 19.56 | 11.40 | 34.00 | 14.15 |
|
Muricauda sp. Bin2 |
Flavobacteria | 5 | 3.1 | 42 | 99.00 | 1.54 | 41.83 | 19.94 | 30.51 | 17.82 | 26.91 | 17.47 |
|
Winogradskyella sp. Bin3 |
Flavobacteria | 10 | 3.15 | 33.8 | 99.20 | 2.47 | 5.12 | 12.61 | 1.20 | 6.81 | 3.17 | 10.80 |
|
Phycisphaera sp. Bin4 |
Phycisphaerae | 1 | 3.26 | 61.2 | 98.90 | 0.00 | 6.93 | 31.36 | 17.68 | 32.57 | 10.41 | 27.57 |
|
Oricola sp. Bin5 |
Alphaproteo- bacteria |
7 | 3.63 | 64.8 | 99.20 | 0.39 | 8.03 | 1.82 | 4.77 | 1.31 | 6.49 | 1.67 |
|
Balneola sp. Bin6 |
Flavobacteria | 11 | 3.65 | 40 | 100 | 0.00 | 2.70 | 1.19 | 16.87 | 8.75 | 8.46 | 5.09 |
ID, identifier.
Assem. size, assembly size.
Est. comp., estimated completeness.
Est. cont., estimated contamination.
Percent relative abundance based on 16S rRNA gene sequences from 0.22-to-3-μm and >3-μm size fractions on day 22 (D22) and 71 (D71) of cultivation. The average information (avg) for 36 samples collected from two size fractions, 0.22-to-3-μm and >3-μm size fractions are shown in the two rightmost columns.
Metagenomic analysis of coculture communities.
Two samples from the exponential (22nd day) and decline (71st day) phases of the Synechococcus sp. YX04-3 culture were subjected to shotgun metagenomics sequencing on the Illumina MiSeq platform. Assembly of the metagenomic reads yielded 155 and 3,395 contigs (>1 kbp), comprising total assembly sizes of 15.7 and 24.2 Mbp, respectively. Six high-quality genomes (estimated genome completeness of >95% and estimated contamination of <5%) from the Synechococcus coculture were reconstructed from the assembled contigs, and are referred to as genomes Bin1 to Bin6 (Table 1). Five heterotrophic bacterial genomes (Muricauda sp. Bin2, Winogradskyella sp. Bin3, Phycisphaera sp. Bin4, Oricola sp. Bin5, and Balneola sp. Bin6) represented three bacterial phyla and corresponded to the dominant bacterial genera/groups of the cultivation community, with the exception of two gammaproteobacterial genera that were not represented.
These five dominant heterotrophic bacterial populations were closely related to strains reported to have close associations with marine phytoplankton or that otherwise exhibited the capacity to degrade algal polysaccharides. Muricauda can be abundant within diverse phytoplankton blooms or cultures, such as those produced by diatoms and dinoflagellates (29, 30). Further, Muricauda ruestringensis has been reported to utilize diverse carbohydrates and amino acids for growth (31). The flavobacterial genus Winogradskyella has been shown to associate with Cochlodinium polykrikoides and is a dominant particle-attached bacterial taxa (32). Winogradskyella rapida SCB36T was isolated from a seawater mesocosm experiment after the addition of protein and exhibited the most pronounced positive growth response to protein enrichment (33). Moreover, planctomycetes have frequently been observed to associate with marine phytoplankton blooms (34–39). Planctomycetes can exhibit diverse lifestyle strategies, including motility, unicellular stages, and aggregate formation with variable numbers of cells that are stacked with a fibrillar glycoproteic material known as the holdfast (40). Oricola cellulosilytica CC-AMH-0T is the only known member of the genus and has been previously shown to degrade cellulose and become particularly active under nutrient-limited conditions (41). Balneola has also been frequently isolated from phytoplankton cultures and particularly from those of Emiliania huxley and Cochlodinium polykrikoides (42, 43).
Detailed descriptions of genomic characteristics and lifestyle strategies of the dominant bacterial populations of the coculture are provided in Text S1 in the supplemental material.
Cellular metaproteomic analyses and inference of metabolic potential.
The metagenomic data could be used to predict the metabolic potentials of dominant bacterial populations, while the metaproteomic data could offer insights into the real metabolic activities occurring in the coculture system, improving our understanding of photoautotroph-heterotroph interactions. A total of 2,047 proteins were identified through cellular metaproteomic analyses (Tables S1B and S2). Proteins from the Synechococcus (Bin1) and Oricola (Bin5) populations comprised 43.2% and 43.8% of the total coculture proteomic profile, respectively. The considerable differences in protein abundances from Synechococcus (Bin1) and Oricola (Bin5) populations compared to the three Flavobacteria and Phycisphaera populations reflect differences in their trophic and lifestyle strategies during the exponential phase. A very low level of proteins was recovered from the Bin6 population, and thus, the other five bins were primarily used for further analysis. To constrain our investigation to the potential interactions within the coculture system, functional proteins that are involved in transmembrane transport and utilization of nutrients and in the storage of carbon and energy sources were specifically analyzed.
Cellular protein information for the proteomes of the five population bins of this study. Download Table S2, XLS file, 0.6 MB (634.5KB, xls) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proteomic profile of Synechococcus sp. YX04-3 (Bin1).
A total of 884 proteins that were attributable to Synechococcus in the coculture system were identified. Among these proteins, 27.48% of Synechococcus proteome (73 proteins) was dedicated to CO2 fixation and photosynthesis. A considerable relative abundance of the Synechococcus proteome was assigned to cell division (2.27% of Synechococcus proteome, 24 proteins), tRNA aminoacylation (1.26%, 21 proteins), ribosomal proteins (4.61%, 33 proteins), and translation factors (1.19%, 11 proteins). These recoveries were likely due to sampling during the exponential phase and the high metabolic and growth activity of Synechococcus during this time.
(i) Two-component regulatory systems and nutrient uptake. Bacteria typically use two-component regulatory systems comprising a sensor kinase and a response regulator to sense and respond to their environments (44). Three sensor histidine kinases and six response regulators were present in the proteomic profile of Synechococcus. Two of these were predicted to be involved in the phosphate sensor-regulatory (PhoRB) system. A total of 8 sensor histidine kinases and 11 response regulators were detected in the Synechococcus sp. YX04-3 genome, which is intermediate between those in the genomes of the oligotrophic Synechococcus strain WH8102 and the coastal strain CC9311 (45, 46). A lower abundance of sensors than response regulators is commonly observed in the genomes of Synechococcus and Prochlorococcus strains, which may represent an efficient regulation strategy wherein one sensor transmits signals to multiresponse regulators (44, 45).
Synechococcus requires sufficient nitrogen and phosphorus nutrients for growth. Accordingly, nitrate/nitrite transporters and reductases were identified in the Synechococcus proteome. Further, two abundant proteins, glutamine synthetase (0.43%) and glutamate synthase (0.02%), that are components of the primary bacterial pathway of ammonium assimilation were also identified. In addition, proteins that conferred the capacity for cyanate hydrolysis, including an ABC transporter system and a cyanase that catalyzes the metabolism of cyanide to CO2 and NH3, were recovered. Interestingly, several proteins involved in urea transport (UrtABCDE) and assimilation (UreCG) were detected, and they represented 0.84% of the Synechococcus proteome. Despite the fact that nitrate was the major N source provided in the medium, the urea ABC transporter protein was the most abundant inorganic nitrogen transporter protein observed, which is consistent with previous results (11, 47). Accordingly, a full complement of genes encoding a urea transporter system (urtABCDE) and assimilation pathway (ureABCDEFG) were present in the Synechococcus genome. The urea cycle is an important pathway of N recycling in Synechococcus cells, wherein urea, and eventually cyanate, are generated from arginine degradation (46).
In addition to nitrogen, phosphorus is a critical limiting nutrient for microbial growth in marine environments. High-affinity phosphate transporter (PstBS) and regulatory system (PhoBR) proteins were detected, and they accounted for 0.51% of the Synechococcus proteome. In addition, two phosphate starvation-inducible proteins (PhoH) were also observed on the 22nd day of cultivation, despite the fact that there was enough phosphate to support Synechococcus growth (Fig. S1). Iron (Fe) is another important limiting nutrient in the open ocean, and its transporter, one iron-binding ABC transport protein, was also identified in the proteomic profile of Synechococcus.
Amino acid and oligopeptide ABC transporters were also detected in the Synechococcus proteomic profile, indicating that the Synechococcus strain could utilize some low-molecular-weight (LMW) organic matter compounds (48). In addition, the genome of strain YX04-3 encoded proteins that could degrade chemically stable C-P bond-containing phosphonates (PhnCDE).
(ii) Photosynthesis and glycogen metabolism. Approximately 10% of the recovered Synechococcus proteins comprising 27.48% of the Synechococcus proteome were associated with CO2 fixation (5.75% of Synechococcus proteome, 10 proteins) and photosynthesis, including photosystem I (1.67%, 9 proteins) and II (3.22%, 13 proteins) assembly, chlorophyll (0.67%, 11 proteins) and carotenoid (0.36%, 7 proteins) biosynthesis, and phycobilisome (15.82%, 23 proteins) assembly (Fig. S3). A previous analysis of a close relative of strain YX04-3, Synechococcus sp. strain WH8102, under low light conditions indicated that it increased the expression of photosynthesis genes involved in light harvesting, phycobilisome assembly, and carbon fixation under these conditions (47). In addition to proteins involved in photosynthesis, seven proteins involved in glycogen biosynthesis and degradation (GlgABCDPX-MalZ) were detected and represented 0.87% of the Synechococcus proteome. Synechococcus can accumulate surplus photosynthetically fixed carbon to synthesize glycogen, which has been observed as granules in their cytoplasm (49).
Overview of the dominant metabolic processes of Synechococcus sp. YX04-3 including transporter systems and carbon, nitrogen, and phosphorus metabolisms in the proteome. The primary metabolic processes observed for strain YX04-3 were involved in inorganic nutrient (e.g., NO3−, PO43−, and cyanate) uptake, photosynthesis, organic matter (e.g., glycogen) biosynthesis, and organic compound [e.g., polysaccharides, lipopolysaccharide, lipids and (oligo)peptide] release. VB12, vitamin B12; SOD, superoxide dismutase. The background arrows indicate the potential material and energy flows detected in the proteomic data. Download FIG S3, PDF file, 0.4 MB (391.5KB, pdf) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(iii) Export and secretion systems. Several proteins were identified in high abundance (0.48% of the Synechococcus proteome) within the Synechococcus proteome that are involved in the secretion of organic compounds like lipids, polysaccharides, and proteins (Fig. S3). For example, proteins responsible for lipid A biosynthesis (LpxABCD, KtdA, and KdsD) and export (MsbA) were present in its proteomic profile. In addition, polysaccharide/lipopolysaccharide biosynthesis and export proteins were also abundant (0.5%) in its proteomic profile. Further, protein-export-related membrane proteins (SecADF) were abundant in the Synechococcus proteomic data, and especially the ATP-hydrolyzing SecA protein (0.11%). The release of organic matter is critical for heterotrophic bacterial growth that is associated with Synechococcus growth. Several proteins in the ABC and major facilitator superfamily group transporters were identified that are involved in the export of metals (e.g., lead, cadmium, zinc, and mercury), multidrugs, and toxins. These efflux transporter proteins could be involved in antagonistic interactions with cocultured heterotrophic bacteria via the export of toxins (44, 50).
Flavobacterial proteomic profiles (bins 2 and 3).
A total of 71 and 22 proteins were identified as belonging to the Flavobacteria Bin2 and Bin3 populations, respectively (Tables S1B and S2 ). These proteins were primarily involved in the transport and metabolism of complex high-molecular-weight (HMW) organic compounds in addition to supporting attached/aggregate lifestyles (Fig. S4).
Overview of the dominant metabolic processes associated with the two Flavobacteria population bins in their proteomes. The primary observed processes were involved in the degradation and transport of high-molecular-weight (dissolved) organic matter in Bin2 (left) and Bin3 (right). These activities were mediated by TonB-dependent transport systems and polysaccharide (or polymer) utilization loci (PUL) complexes. The polysaccharide utilization system of the figure was modified from Flint et al. (88). SOD, superoxide dismutase. The background arrows indicate the potential material and energy flows detected in the proteomic data. Download FIG S4, PDF file, 0.4 MB (467.6KB, pdf) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(i) Biopolymer transport and utilization. TonB-dependent transporters (TBDTs) were abundant proteins identified in the proteomic profiles of Bin2 (9 proteins) and Bin3 (5 proteins), comprising 10.95% and 19.37% of their proteomes, respectively. In particular, three SusCD protein complexes (defined here as A, B, and C) were detected in the Bin2 profile that are core components of TBDT systems and are responsible for the binding and uptake of biopolymers. In addition, three other SusC-like proteins were identified in the Bin2 proteome. A single SusCD protein complex (defined as D) that is encoded by the α-1,4-glucan utilization gene cluster was present in the Bin3 proteome, in addition to three other SusC-like proteins. Two glycohydrolase (GH) proteins and four peptidase proteins that are involved in the degradation and utilization of complex organic compounds were identified in the Bin2 proteome, with three and two such proteins identified in the Bin3 proteome, respectively.
(ii) Proteins involved in adhesion or aggregation. Two important outer membrane porin proteins, OmpA and OmpH (51, 52), were detected in the Bin2 proteomic profile, while only OmpA was observed in the Bin3 proteomic profile. OmpA is thought to specifically interact with host receptor molecules (53) and exhibits important roles in bacterial adhesion and aggregation (52). Tetratricopeptide repeat proteins (TRP) that are involved in protein-protein interactions were also present in both flavobacterial proteomes, and could be involved in the adhesion or aggregation of Flavobacteria in the coculture system (54, 55). Although 15 gliding motility genes were present in both flavobacterial genomes, their encoded proteins were not identified in the proteomic data.
Phycisphaera sp. Bin4 proteomic profile.
The proteomic profile corresponding to Phycisphaera sp. Bin4 comprised 173 proteins, with 15.6% assigned to unknown function (i.e., hypothetical proteins). High abundances of peptidases and type IV pilus proteins in the Bin4 profile indicated the physiological potential for biopolymer degradation and motility. Seventeen peptidases, including a serine protease, glutamic protease, metalloprotease, and asparaginase, were identified and represented 6.74% of the Phycisphaera sp. Bin4 proteome, suggesting an enhanced ability to hydrolyze peptide bonds of proteins. Some secreted proteases can degrade extracellular proteins into amino acids that are important in nitrogen acquisition from proteins. The Phycisphaera sp. Bin4 population exhibited a preference for the utilization of peptide polymers and could mediate the supply of LMW DOC such as amino acids to the Synechococcus and Oricola sp. Bin5 populations.
Oricola sp. Bin5 proteomic profile.
A total of 896 proteins were identified that were produced by the Oricola sp. Bin5 population, of which 555 were involved in biological processes, 196 were associated with cellular components, and 618 were involved in molecular functions under the Gene Ontology (GO) category. That indicates Oricola sp. Bin5 was the most active heterotrophic bacteria in the coculture system, although its abundance was not the highest on day 22. A previous study also proved that alphaproteobacteria, especially members of Roseobacter, were better “helper” for Synechococcus growth than other heterotrophic bacteria (8). Remarkably, approximately 100 transporters that are involved in the uptake of LMW DOC and inorganic nutrient uptake/transport were present in the Bin5 proteomic profile (Fig. S5).
Overview of the dominant metabolic processes associated with the Oricola sp. Bin5 population in the proteome. The dominant processes identified were associated with transporter systems and carbon, nitrogen, and phosphorus metabolisms. Oricola sp. Bin5 primarily imported low-molecular-weight organic substrates, including amino acids, di-/oligopeptides, nucleosides, ribose, maltose, xylose, dihydroxyacetone, C4-dicarboxylates, pyruvate, and sialic acids, as well as inorganic nutrients like NO3− and PO43− using ATP-binding cassette (ABC), tripartite ATP-independent periplasmic (TRAP), and tripartite tricarboxylate transporters (TTT) transport systems. VB12, vitamin B12; SOD, superoxide dismutase. The background arrows indicate the potential material and energy flows detected in the proteomic data. Download FIG S5, PDF file, 0.3 MB (338.7KB, pdf) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(i) Polyhydroxyalkanoate metabolism. Polyhydroxyalkanoate (PHA) is an intracellular carbon and energy storage polyester, and its accumulation and degradation are typically related to microbial growth in response to various environmental conditions like nutrient levels (56, 57). The key enzymes for PHA biosynthesis (i.e., PHA polymerase, PhaC) and degradation (i.e., PHA depolymerase, PhaZ) were present in the proteomic profile of Oricola sp. Bin5. Thus, Oricola sp. Bin5 cells were likely accumulating intracellular PHA in the rich organic matter environment of the coculture system.
(ii) Transport systems and nutrient uptake. A considerable number of transport proteins, including ATP-binding cassette (ABC) transporters and tripartite ATP-independent periplasmic (TRAP) transporters, were identified in the proteomic profile of Oricola sp. Bin5. A total of 87 ABC transporter proteins were identified, and they comprised 11.54% of the Oricola sp. Bin5 proteome, in which ∼80% were involved in the transport of LMW organic matter compounds, including amino acids, di-/oligopeptides, and carbohydrates (including glycerol-3-phosphate) (Table S2). Among these proteins, ABC transporter proteins involved in carbohydrate transport and uptake were the most abundant (36 proteins, 5.47% of the Oricola sp. Bin5 proteome). In particular, transporters of LMW carbohydrates, including xylose, fructose, nucleoside, ribose, maltose, polyols, dihydroxyacetone, and glycerol-3-phosphate were prevalent. In addition, nine proteins (1.68%) were identified in the Bin5 proteome that are associated with general ABC transporters and those for polar and branched-chain amino acids, in addition to 15 ABC transporters (1.92%) involved in di-/oligopeptide transport. An additional nine ABC transporter proteins with inferred roles in transporting putrescine, pyrimidine, and thiamine were also observed in the proteome. Further, 10 TRAP transporter proteins (2.22%) involved in transporting C4-dicarboxylates, pyruvate, sialic acid, and other unknown substrates were found in the proteomic profile of Bin5. The different substrate preference between two dominant groups, flavobacteria and alphaproteobacteria, could reduce their competition in the coculture system.
Exoproteome information for the five population bins of this study. Download Table S3, XLS file, 0.1 MB (96KB, xls) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A large complement of genes involved in the uptake and metabolism of inorganic nitrogen and phosphate compounds were also observed in the Oricola sp. Bin5 genome. In addition, three proteins (GlnAE and GltB) involved in ammonia assimilation were identified in the proteomic profile of Oricola sp. Bin5. Further, a urea ABC transporter (UrtA) protein and two proteins involved in urea decomposition (UreAC) were present in its proteome. The Oricola sp. Bin5 proteomic profile also contained two high-affinity phosphate ABC transporter proteins (PstSB) and an associated regulatory protein (PhoU). Last, two ABC transporter proteins involved in binding ferric iron were observed in the Bin5 proteome. Thus, the import of extracellular organic and inorganic nutrients was critical for Oricola sp. Bin5 growth. That also indicates Oricola sp. Bin5 may compete inorganic nutrients with Synechococcus, especially phosphate, in the coculture system.
Exoproteome of Synechococcus sp. YX04-3 coculture.
A total of 150 proteins were identified (Tables S1B and S3) from the Synechococcus sp. YX04-3 coculture exoproteome which exhibited similar metabolic characteristics with cellular metaproteome (Text S1).
Utilization of HMW complex biopolymers by Flavobacteria.
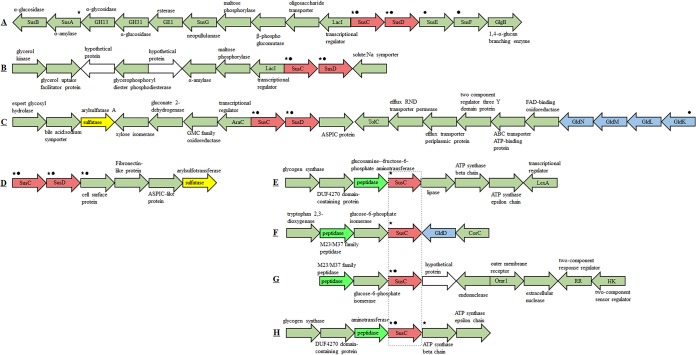
TBDT systems are outer membrane proteins involved in the uptake and utilization of HMW DOC molecules that cannot diffuse across membranes via porins. TBDTs targeting biopolymers (e.g., polysaccharides, proteins, and proteoglycan) are usually coupled to macromolecule substrate degradative enzymes (e.g., GHs and peptidases) to facilitate the efficient use of complex compounds (58, 59). SusC is a member of the TonB receptor family and is primarily responsible for importing oligosaccharides and polymers from the outer membrane into the periplasm, while SusD is an extracellular lipoprotein that is primarily involved in binding polysaccharides or other polymers at the outer cell membrane (60, 61). In addition to SusCD protein complexes, other SusC proteins were identified in the two flavobacterial proteomic profiles, suggesting that individual SusC proteins may independently mediate the transport of specific polymer types. Their upstream and downstream genes were next to genes involved in polymer degradation (e.g., GHs, amylases, peptidases, and sulfatases), motility, and attachment (Fig. 4).
FIG 4.
Multigene clusters encoding SusCD protein complexes (A, B, C, and D) and SusC-like proteins (E, F, G, and H) in the proteomic profiles of Bin3 and Bin2. A small solid black circle indicates that the proteins were detected in the cellular proteome, and a black star indicates that proteins were found in exoproteomes. (A and B) PULs targeting α-1,4-glucans; (C) PUL involved in the degradation of xylose-containing sulfated polysaccharides; (D) the PUL containing potential capacity for the degradation of sulfated polysaccharides/biopolymers; (E to H) gene clusters involved in the degradation of oligo- and polypeptides or proteins.
The identified SusCD protein complex in the Bin3 proteome was encoded by genes associated with a polysaccharide utilization locus (PUL) (here defined as PUL01) that was predicted to target α-1,4-glucans like starch and glycogen (Fig. 4A and Fig. S4). The TonB-dependent receptor SusC operates in concert with the glucan-binding lipoproteins SusDEFG. SusDEF are active in the glucan-binding process, while SusG (GH13) catalyzes the initial degradation, SusC imports oligosaccharides into the periplasm, and SusA (GH13, also known as neopullulanse) and SusB (GH97, an α-glucosidase) further degrade oligosaccharides to glucose (62) (Fig. S4). The SusCD protein complex A in the Bin2 proteome, their encoding genes were adjacent to those coding for a maltose phosphorylase (GH65) and an alpha-amylase (GH13) (PUL02) (Fig. 4B), and therefore was also predicted to target α-1,4-glucans (61). The identified SusCD protein complex B encoding gene cluster in the Bin2 genome was located with genes upstream that encode a xylose isomerase and an arylsulfatase, suggesting the involvement of the former in the degradation of xylose-containing sulfated polysaccharides. In addition, genes located downstream encoded outer membrane efflux proteins and gliding motility proteins (GldKLMN) (Fig. 4C). Further, the SusCD protein complex C encoded in the Bin2 genome was associated with genes encoding cell surface proteins and a sulfatase, indicating the potential capacity for the degradation of sulfated polysaccharides/biopolymers (Fig. 4D). All of the identified individual SusC protein-coding genes in the Bin2 genome, except for one located at the end of the contig, were always adjacent to peptidases, suggesting that they specialized in the degradation of oligo- and polypeptides or proteins (Fig. 4E to H).
In addition, a xylan utilization gene cluster was detected in the Muricauda sp. Bin2 genome (Fig. S6A). An arabinogalactan utilization gene cluster was also present in the Balneola sp. Bin6 genome that comprised genes encoding an arabinogalactanase (ganB), β-galactosidase, galactokinase, and a SusCD system (Fig. S6B) (63, 64). However, these two PULs were not detected in the proteomic profiles of Bin2 and Bin3. Thus, these overall results suggest that the three flavobacterial populations in the coculture system displayed different polysaccharide degradation abilities.
Operon structure and gene composition of two-polysaccharide utilization loci (PUL) identified in the Flavobacteria Bin2 (A) and Bin6 (B) genomes, respectively. Download FIG S6, PDF file, 0.3 MB (329.2KB, pdf) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The concentrations and organic molecular components of Synechococcus exudates vary over different growth phases or with physiological status (25, 26, 65). This variation in organic compound characteristics in turn influences the metabolic activities and population dynamics of heterotrophic bacteria in coculture systems. The two dominant flavobacterial populations, Bin2 and Bin3, should benefit from efficient strategies to metabolize specific HMW DOC compounds like glycogen, amylose, and maltodextrin that are released by Synechococcus in the exponential phase, relative to the flavobacterial Bin6 population. This is especially evident for the Winogradskyella sp. Bin3 population that could degrade α-1,4-glucans like glycogen that is biosynthesized by Synechococcus sp. YX04-3. This particular metabolic ability of the Bin3 population could partly explain its close association with Synechococcus cells.
Oxidative stress and SODs.
Superoxide is an important reactive oxygen species (ROS) encountered by cells that can cause cellular damage and is especially problematic for oxygenic photoautotrophs. Previous studies have suggested that the removal of ROS is a key interaction between Cyanobacteria and their associated heterotrophic bacterial cells (13, 14, 66). Superoxide dismutases (SODs) and catalase or catalase-peroxidase enzymes play important roles in antioxidant defenses via the catalysis of superoxide (O2−) radicals to either O2 and H2O. As observed for Prochlorococcus strains or its close relative Synechococcus sp. WH8102, strain Synechococcus sp. YX04-3 lacked catalase or catalase-peroxidase genes, indicating it needs “helper” bacteria to reduce the ROS (46, 67, 68).
The catalase-peroxidase protein KatG catalyzes the decomposition of H2O2 to H2O and O2 and was detected in the cellular proteome of both the Bin2 and Bin5 populations. Two additional ROS stress proteins, Fe-SOD and alkyl hydroperoxide reductase subunit C protein (AhpC), were also present in the cellular proteome of Bin5. Further, a thiol-specific antioxidant family protein associated with thiol-[disulfide interchange was identified in the proteome of Bin4. Last, two of the bacterial populations, Winogradskyella sp. Bin3 and Phycisphaera sp. Bin4, that displayed attached or aggregate lifestyle preferences, contained SOD proteins in their exoproteomes. Despite limited data, these overall observations indicate that the removal of ROS is an active and important process in the coculture system.
Vitamin B12 biosynthesis.
Many microbial species are auxotrophic for certain vitamins, lack specific vitamin biosynthetic pathways, and must therefore take up essential vitamins from their environments (69, 70). Vitamin B12 is an important cofactor for the activity of methionine synthase and is a core enzyme of cellular one-carbon metabolism, the production of the universal methyl donor S-adenosylmethionine, and for folate cycling required for DNA synthesis (71, 72).
Vitamin B12 is one of the most complex metabolites in natural environments and requires at least 19 separate enzymatic steps for its biosynthesis (69, 70). All of the heterotrophic bacterial genomic bins recovered, except the Oricola sp. Bin5 population, did not contain the key B12-independent methionine synthase (MetE) gene involved in biosynthesis, indicating that they all required exogenous vitamin B12. In contrast, the genomes of Synechococcus and Oricola sp. Bin5 contained all of the core genes for de novo cobalamin (vitamin B12) biosynthesis. Moreover, proteins involved in vitamin B12 biosynthesis (e.g., CobA, CobS, and CbiK) were observed in the proteomic profiles of Synechococcus sp. YX04-3 and Oricola sp. Bin5. Although the genes for vitamin B12 synthesis were not observed in the other genomes, genes involved in vitamin B12 transport were present in their genomes, including outer membrane receptors, binding proteins, permeases, and ATPase components. Two attached preferred bacterial populations, Winogradskyella sp. Bin3 and Phycisphaera sp. Bin4, lacked the core genes for thiamine (B1) and biotin (B7) biosynthesis, respectively. In addition, Oricola sp. Bin5 contained a set of ABC transport systems for importing thiamine instead of de novo biosynthesis in its genome. Thus, vitamin trafficking may contribute to interactions between populations in the coculture system and therefore partly account for the connectedness of microbial community networks in ocean environments (34, 73). These observations highlight the complex nature of interactions in these systems, as opposed to simple two-member interaction models.
Nutrient cycling in the coculture system.
The majority of organic matter in the coculture system was derived from photosynthetically fixed carbon by Synechococcus. Approximately 10% of the fixed carbon is released or leaked into the surroundings in the form of photosynthate (74–76) that then supports heterotrophic bacterial growth. The accumulation of photosynthate should thus become toxic to Synechococcus in nutrient-rich media and render inorganic nutrients unavailable within organic matter complexes (8). However, heterotrophs regenerate inorganic nutrients like NH4+, PO43−, and Fe2+/3+ via their metabolism of organic matter, that Synechococcus can use (Fig. 5). The close associations between Synechococcus and heterotrophic bacteria suggest that these synergistic interactions are important drivers of interactions in coculture ecosystems.
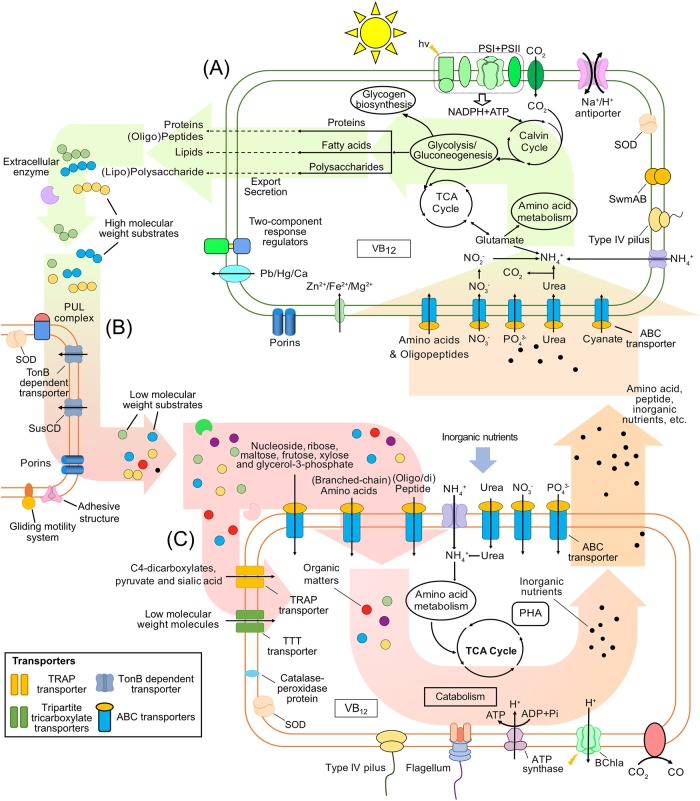
FIG 5.
Schematic of inorganic and organic nutrient cycling in the Synechococcus (A), Flavobacteria (B) and Alphaproteobacteria (C) coculture system. The dominant detected metabolic processes observed for bacterial populations in the system are shown. Synechococcus is the primary driver of the coculture system functioning, and its dominant metabolic processes comprise inorganic nutrient uptake, photosynthesis, and organic matter biosynthesis and release. Two flavobacterial populations within the system corresponding to Muricauda and Winogradskyella strains, in addition to a member of the SM1A02 group exhibited specific propensities toward the initial degradation of complex compounds and biopolymers, as indicated by abundant TBDT, GH, and peptidase proteins in their proteomes. The alphaproteobacterium Oricola sp. Bin5 population mainly utilizes low-molecular-weight dissolved organic matter, including by-products from flavobacterial population metabolism, via their use of ABC, TRAP, and TTT transport systems. Thus, the bacterial members of the coculture system exhibit complementary metabolisms that operate in the degradation of Synechococcus-derived organic matter and nutrient cycling within the coculture system. PSI, photosystem I; TCA cycle, tricarboxylic cycle; VB12, vitamin B12; SOD, superoxide dismutase. The background arrows indicate the potential material and energy flows detected in the proteomic data.
The uptake of inorganic nutrients (e.g., NO3−, PO43−, and Fe2+) is critical for photoautotrophic Synechococcus growth. Accordingly, transporter proteins for the above nutrients were abundant in both the cellular proteins and exoproteomes of Synechococcus (Fig. 5 and Fig. S3). In addition, approximately 10% of Synechococcus cellular proteins were associated with photosynthesis and CO2 fixation. Surplus photosynthetically fixed carbon could also be used to synthesize glycogen, a branched polysaccharide comprising glucose monomers that is an important mechanism for intracellularly storing carbon and energy reserves (49, 77). The multibranched glycogen biopolymer has a similar structure as amylopectin (a starch component) and is linked by α-1,4-glycosidic bonds on the stem chain and α-1,6-glycosidic bonds on branch chains. Many previous studies have shown that Synechococcus accumulates glycogen, although several Synechococcus strains harbor distinct α-polyglucans that have been termed semiamylopectin (78, 79). Proteins involved in the export of HMW biopolymers (e.g., polysaccharides, oligo/dipeptides, and lipids) were also identified in both the cellular and exoproteomic profiles of Synechococcus. Importantly, these compounds are also critical for the growth of coassociated heterotrophic bacterial populations.
The diverse nutrient transporters identified in the coculture system proteomes have different affinities and can be differentially regulated depending on nutrient concentrations and organic molecule complexity. Two of the flavobacterial strains appeared to be specialized in the degradation of complex HMW DOC molecules that exceed the typical 600- to 800-Da substrate range of normal porins (7, 80, 81) via the use of TBDT systems for direct attachment (Fig. S4). The alphaproteobacterial strain, Oricola sp. Bin5, appeared to be specialized in the uptake of labile LMW DOC including by-products of Synechococcus and flavobacterial growth via their expression of ABC and TRAP transporters and tripartite tricarboxylate transporters (TTTs) that were involved in monomer transport (Fig. S5). Thus, overall, it appeared that the flavobacterial and alphaproteobacterial strains acted synergistically to metabolize the organic matter released by Synechococcus (Fig. 5).
The proteomic profile of Oricola sp. Bin5 indicated that a large amount of nitrogen-rich [e.g., amino acids, (oligo)peptides, and proteins] and phosphorus-rich (e.g., glycerol-3-phosphate) compounds were present in the coculture system. These substrates were likely originally released as complex HMW compounds from Synechococcus that were further degraded by Flavobacteria. Nitrogen or phosphorus containing dissolved organic matter (DOM) molecules, typically as labile substrates, are preferentially utilized by heterotrophic bacteria. Specifically, heterotrophic bacteria use labile DOM to synthesize their biomass and respire it during energy conservation. The regenerated inorganic N and P nutrients, and especially those with P, would thus be returned to the culture medium, allowing the maintenance of a stable, healthy, and long-lived coculture system. The phase of coculture growth with Synechococcus and heterotrophic bacterial abundance stability after the 70th day may represent a temporary steady state of environmental variation via the utilization and regeneration of PO43−, mitigation of oxidative stresses, and removal of ROS.
Conclusions.
A long-term, 91-day cultivation experiment of the open ocean ecotype Synechococcus and its associated dominant heterotrophic bacterial populations revealed dynamic variation in their abundances, lifestyles, and organic matter metabolisms over the growth period. The Synechococcus and heterotrophic bacterial strains primarily exhibited synergistic interactions via nutrient exchange, reduction of ROS stress, and vitamin trafficking. Specifically, Flavobacteria and Alphaproteobacteria strains synergistically metabolized HMW Synechococcus photosynthate to LMW DOM and helped recycle inorganic nutrients. Indeed, the activities of the dominant coculture populations all contributed to the maintenance of the Synechococcus and heterotroph system. Light availability, inorganic and organic nutrient availability, and viral lysis and predation, together shape the cycling of matter and energy between photoautotrophs and heterotrophs in the upper ocean. To better understand these complex relationships in situ, additional research is needed to identify top-down and bottom-up controls that impact photoautotroph-heterotroph interactions.
MATERIALS AND METHODS
Synechococcus culture and abundance measurement.
Synechococcus sp. strain YX04-3, and its associated heterotrophic bacterial species, were isolated from the South China Sea using PRO2 liquid medium (25). In the present study, the YX04-3 coculture was incubated in triplicate at 25°C with 10 μmol photons · m−2 · s−1 in SN medium (82). Synechococcus and heterotrophic bacterial cell numbers in the cocultures were measured by flow cytometry as described previously (25).
16S rRNA gene sequencing and metagenome sequencing.
DNA from the 36 size-fractionated samples of the Synechococcus strain YX04-3 coculture were subjected to PCR amplification of 16S rRNA genes using primers targeting the bacterial V3-V4 hypervariable regions (515F [5′-GTGCCAGCMGCCGCGGTAA-3′] and 907R [5′-CCGTCAATTCMTTT RAGTTT-3′]) (83, 84). Sequence libraries were constructed using the NEBNext Ultra DNA library prep kit for Illumina (New England Biolabs, USA). The detailed protocols are described in Zheng et al. (25). The 16S rRNA gene sequence data were deposited in the NCBI Sequence Read Archive under the BioProject accession no. PRJNA498017.
Shotgun metagenomic sequencing was also conducted on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) using the MiSeq V2 kit chemistry reagents with a 2 × 250 paired-end cycle sequencing run. Raw sequencing reads were trimmed and filtered and then de novo assembled by SPAdes (85). Assembled contig (>4 kbp) sequence properties, including GC content, tetranucleotide frequencies, and sequencing depth coverage were used to bin assembled contigs into draft genomes with the MaxBin 2.0 software program (86). Estimated genome sizes and completeness of the reconstructed genomes were then assessed using CheckM (87). The raw metagenomics sequence reads were deposited in the NCBI database under the BioProject accession no. PRJNA498190. The six metagenome bins were deposited in the NCBI database under the BioProject accession no. PRJNA497203.
Metaproteomic analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Metaproteomic analysis of the Synechococcus sp. YX04-3 coculture was conducted to elucidate the molecular mechanisms underlying population-level interactions. The 22nd day of cocultivation was chosen for targeted proteomic analysis because the number of Synechococcus cells was equivalent to the number of heterotrophic bacterial cells at this time point. To generate the metaproteomes, triplicate 120-ml liquid cultures were centrifuged (3,000 × g for 15 min at 4°C). The supernatant was then filtered through 0.22-μm-pore-size filter unit (Sterivex-GV, Millipore) and subjected to exoproteome analysis. The detailed protocol was conducted by the method of Christie-Oleza et al. (8) and supplied in the supplemental material. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD015067.
Data availability.
Sequence data were deposited in the NCBI Sequence Read Archive under BioProject no. PRJNA498017, PRJNA498190, and PRJNA497203.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (NSFC) project (91751207), the National Key Research Programs (2018YFA0605800), the Senior User Project of RV KEXUE (KEXUE2019GZ03), the NSFC project (41776145, 41876150, and 41861144018), the Fundamental Research Funds for the Central Universities (20720190095), and Natural Science Foundation of Fujian Province of China (2018J05072).
We declare that we have no conflicts of interest.
Q.Z., F.C., and N.J. conceived and designed the experiments. Q.Z., Y.W., J.L., and W.L. conducted the experiments. Q.Z., Y.W., and J.L. analyzed the data. All authors assisted in writing the manuscript, discussed the results, and commented on the manuscript.
Footnotes
This article is a direct contribution from Nianzhi Jiao, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by David Hutchins, University of Southern California, and Joseph Christie-Oleza, University of the Balearic Islands.
Citation Zheng Q, Wang Y, Lu J, Lin W, Chen F, Jiao N. 2020. Metagenomic and metaproteomic insights into photoautotrophic and heterotrophic interactions in a Synechococcus culture. mBio 11:e03261-19. https://doi.org/10.1128/mBio.03261-19.
REFERENCES
- 1.Azam F, Fenchel T, Field JG, Gray JS, Meyer-Reil LA, Thingstad F. 1983. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser 10:257–263. doi: 10.3354/meps010257. [DOI] [Google Scholar]
- 2.Buchan A, LeCleir GR, Gulvik CA, Gonzalez JM. 2014. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol 12:686–698. doi: 10.1038/nrmicro3326. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Meng Z, Liu X, Zhang X. 2019. Microbial assembly, interaction, functioning, activity and diversification: a review derived from community compositional data. Mar Life Sci Technol 1:112–128. doi: 10.1007/s42995-019-00004-3. [DOI] [Google Scholar]
- 4.Partensky F, Hess WR, Vaulot D. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev 63:106–127. doi: 10.1128/MMBR.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biller SJ, Berube PM, Lindell D, Chisholm SW. 2015. Prochlorococcus: the structure and function of collective diversity. Nat Rev Microbiol 13:13–27. doi: 10.1038/nrmicro3378. [DOI] [PubMed] [Google Scholar]
- 6.Jiao N, Herndl GJ, Hansell DA, Benner R, Kattner G, Wilhelm SW, Kirchman DL, Weinbauer MG, Luo T, Chen F, Azam F. 2010. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat Rev Microbiol 8:593–599. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
- 7.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang SX, Mann AJ, Waldmann J, Weber M, Klindworth A, Otto A, Lange J, Bernhardt J, Reinsch C, Hecker M, Peplies J, Bockelmann FD, Callies U, Gerdts G, Wichels A, Wiltshire KH, Glockner FO, Schweder T, Amann R. 2012. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 8.Christie-Oleza JA, Sousoni D, Lloyd M, Armengaud J, Scanlan DJ. 2017. Nutrient recycling facilitates long-term stability of marine microbial phototroph-heterotroph interactions. Nat Microbiol 2:17100. doi: 10.1038/nmicrobiol.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amin SA, Parker MS, Armbrust EV. 2012. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev 76:667–684. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azam F, Malfatti F. 2007. Microbial structuring of marine ecosystems. Nat Rev Microbiol 5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 11.Christie-Oleza JA, Armengaud J, Guerin P, Scanlan DJ. 2015. Functional distinctness in the exoproteomes of marine Synechococcus. Environ Microbiol 17:3781–3794. doi: 10.1111/1462-2920.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazamia E, Czesnick H, Thi TVN, Croft MT, Sherwood E, Sasso S, Hodson SJ, Warren MJ, Smith AG. 2012. Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ Microbiol 14:1466–1476. doi: 10.1111/j.1462-2920.2012.02733.x. [DOI] [PubMed] [Google Scholar]
- 13.Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. 2008. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol 74:4530–4534. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER. 2011. Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLoS One 6:e16805. doi: 10.1371/journal.pone.0016805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi RJ, Huang HH, Qi ZH, Hu WA, Tian ZY, Dai M. 2013. Algicidal activity against Skeletonema costatum by marine bacteria isolated from a high frequency harmful algal blooms area in southern Chinese coast. World J Microbiol Biotechnol 29:153–162. doi: 10.1007/s11274-012-1168-1. [DOI] [PubMed] [Google Scholar]
- 16.Crenn K, Duffieux D, Jeanthon C. 2018. Bacterial epibiotic communities of ubiquitous and abundant marine diatoms are distinct in short-and long-term associations. Front Microbiol 9:2879. doi: 10.3389/fmicb.2018.02879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore LR, Coe A, Zinser ER, Saito MA, Sullivan MB, Lindell D, Frois-Moniz K, Waterbury J, Chisholm SW. 2007. Culturing the marine cyanobacterium Prochlorococcus. Limnol Oceanogr Methods 5:353–362. doi: 10.4319/lom.2007.5.353. [DOI] [Google Scholar]
- 18.Cole JK, Hutchison JR, Renslow RS, Kim YM, Chrisler WB, Engelmann HE, Dohnalkova AC, Hu DH, Metz TO, Fredrickson JK, Lindemann SR. 2014. Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: model systems for the study of autotroph-heterotroph interactions. Front Microbiol 5:109. doi: 10.3389/fmicb.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malfatti F, Samo TJ, Azam F. 2010. High-resolution imaging of pelagic bacteria by atomic force microscopy and implications for carbon cycling. ISME J 4:427–439. doi: 10.1038/ismej.2009.116. [DOI] [PubMed] [Google Scholar]
- 20.Biller SJ, Coe A, Chisholm SW. 2016. Torn apart and reunited: impact of a heterotroph on the transcriptome of Prochlorococcus. ISME J 10:2831–2843. doi: 10.1038/ismej.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tai V, Paulsen IT, Phillippy K, Johnson DA, Palenik B. 2009. Whole-genome microarray analyses of Synechococcus-Vibrio interactions. Environ Microbiol 11:2698–2709. doi: 10.1111/j.1462-2920.2009.01997.x. [DOI] [PubMed] [Google Scholar]
- 22.Beliaev AS, Romine MF, Serres M, Bernstein HC, Linggi BE, Markillie LM, Isern NG, Chrisler WB, Kucek LA, Hill EA, Pinchuk GE, Bryant DA, Wiley HS, Fredrickson JK, Konopka A. 2014. Inference of interactions in cyanobacterial-heterotrophic co-cultures via transcriptome sequencing. ISME J 8:2243–2255. doi: 10.1038/ismej.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aharonovich D, Sher D. 2016. Transcriptional response of Prochlorococcus to co-culture with a marine Alteromonas: differences between strains and the involvement of putative infochemicals. ISME J 10:2892–2906. doi: 10.1038/ismej.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaur A, Hernandez-Fernaud JR, Aguilo-Ferretjans MD, Wellington EM, Christie-Oleza JA. 2018. 100 days of marine Synechococcus-Ruegeria pomeroyi interaction: a detailed analysis of the exoproteome. Environ Microbiol 20:785–799. doi: 10.1111/1462-2920.14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Q, Wang Y, Xie R, Lang AS, Liu YT, Lu JY, Zhang XD, Sun J, Suttle CA, Jiao NZ. 2018. Dynamics of heterotrophic bacterial assemblages within Synechococcus cultures. Appl Environ Microbiol 84:e01517-17. doi: 10.1128/AEM.01517-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Q, Lu J, Wang Y, Jiao N. 2019. Genomic reconstructions and potential metabolic strategies of generalist and specialist heterotrophic bacteria associated with an estuary Synechococcus culture. FEMS Microbiol Ecol 95:fiz017. doi: 10.1093/femsec/fiz017. [DOI] [PubMed] [Google Scholar]
- 27.Momma K, Hashimoto W, Miyake O, Yoon H, Kawai S, Mishima Y, Mikami B, Murata K. 1999. Special cell surface structure, and novel macromolecule transport/depolymerization system of Sphingomonas sp A1. J Ind Microbiol Biotechnol 23:425–435. doi: 10.1038/sj.jim.2900719. [DOI] [PubMed] [Google Scholar]
- 28.Momma K, Okamoto M, Mishima Y, Mori S, Hashimoto W, Murata K. 2000. A novel bacterial ATP-binding cassette transporter system that allows uptake of macromolecules. J Bacteriol 182:3998–4004. doi: 10.1128/jb.182.14.3998-4004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolas JL, Corre S, Cochard JC. 2004. Bacterial population association with phytoplankton cultured in a bivalve hatchery. Microb Ecol 48:400–413. doi: 10.1007/s00248-003-2031-6. [DOI] [PubMed] [Google Scholar]
- 30.Lawson CA, Raina JB, Kahlke T, Seymour JR, Suggett DJ. 2018. Defining the core microbiome of the symbiotic dinoflagellate, Symbiodinium. Environ Microbiol Rep 10:7–11. doi: 10.1111/1758-2229.12599. [DOI] [PubMed] [Google Scholar]
- 31.Bruns A, Rohde M, Berthe-Corti L. 2001. Muricauda ruestringensis gen. nov., sp. nov., a facultatively anaerobic, appendaged bacterium from German North Sea intertidal sediment. Int J Syst Evol Microbiol 51:1997–2006. doi: 10.1099/00207713-51-6-1997. [DOI] [PubMed] [Google Scholar]
- 32.Park BS, Guo RY, Lim WA, Ki JS. 2017. Pyrosequencing reveals specific associations of bacterial clades Roseobacter and Flavobacterium with the harmful dinoflagellate Cochlodinium polykrikoides growing in culture. Mar Ecol 38:e12474. doi: 10.1111/maec.12474. [DOI] [Google Scholar]
- 33.Pinhassi J, Nedashkovskaya OI, Hagstrom A, Vancanneyt M. 2009. Winogradskyella rapida sp. nov., isolated from protein-enriched seawater. Int J Syst Evol Microbiol 59:2180–2184. doi: 10.1099/ijs.0.008334-0. [DOI] [PubMed] [Google Scholar]
- 34.Fukunaga Y, Kurahashi M, Sakiyama Y, Ohuchi M, Yokota A, Harayama S. 2009. Phycisphaera mikurensis gen. nov., sp nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov and Phycisphaerae classis nov in the phylum Planctomycetes. J Gen Appl Microbiol 55:267–275. doi: 10.2323/jgam.55.267. [DOI] [PubMed] [Google Scholar]
- 35.Morris RM, Longnecker K, Giovannoni SJ. 2006. Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ Microbiol 8:1361–1370. doi: 10.1111/j.1462-2920.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- 36.Bengtsson MM, Ovreas L. 2010. Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol 10:261–272. doi: 10.1186/1471-2180-10-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burke C, Thomas T, Lewis M, Steinberg P, Kjelleberg S. 2011. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J 5:590–600. doi: 10.1038/ismej.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lachnit T, Meske D, Wahl M, Harder T, Schmitz R. 2011. Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ Microbiol 13:655–665. doi: 10.1111/j.1462-2920.2010.02371.x. [DOI] [PubMed] [Google Scholar]
- 39.Lage OM, Bondoso J. 2011. Planctomycetes diversity associated with macroalgae. FEMS Microbiol Ecol 78:366–375. doi: 10.1111/j.1574-6941.2011.01168.x. [DOI] [PubMed] [Google Scholar]
- 40.Lage OM. 2013. Characterization of a planctomycete associated with the marine dinoflagellate Prorocentrum micans Her. Antonie Van Leeuwenhoek 104:499–508. doi: 10.1007/s10482-013-9991-4. [DOI] [PubMed] [Google Scholar]
- 41.Hameed A, Shahina M, Lai WA, Lin SY, Young LS, Liu YC, Hsu YH, Young CC. 2015. Oricola cellulosilytica gen. nov., sp. nov., a cellulose-degrading bacterium of the family Phyllobacteriaceae isolated from surface seashore water, and emended descriptions of Mesorhizobium loti and Phyllobacterium myrsinacearum. Antonie Van Leeuwenhoek 107:759–771. doi: 10.1007/s10482-014-0370-6. [DOI] [PubMed] [Google Scholar]
- 42.Taniguchi A, Tada Y, Hamasaki K. 2011. Seasonal variations in the community structure of actively growing bacteria in neritic waters of Hiroshima Bay, Western Japan. Microbes Environ 26:339–346. doi: 10.1264/jsme2.me11212. [DOI] [PubMed] [Google Scholar]
- 43.Rosana ARR, Orata FD, Xu Y, Simkus DN, Bramucci AR, Boucher Y, Case RJ. 2016. Draft genome sequences of seven bacterial strains isolated from a polymicrobial culture of coccolith-bearing (C-type) Emiliania huxleyi M217. Genome Announc 4:e00673-16. doi: 10.1128/genomeA.00673-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palenik B, Brahamsha B, Larimer FW, Land M, Hauser L, Chain P, Lamerdin J, Regala W, Allen EE, McCarren J, Paulsen I, Dufresne A, Partensky F, Webb EA, Waterbury J. 2003. The genome of a motile marine Synechococcus. Nature 424:1037–1042. doi: 10.1038/nature01943. [DOI] [PubMed] [Google Scholar]
- 45.Palenik B, Ren Q, Dupont CL, Myers GS, Heidelberg JF, Badger JH, Madupu R, Nelson WC, Brinkac LM, Dodson RJ, Durkin AS, Daugherty SC, Sullivan SA, Khouri H, Mohamoud Y, Halpin R, Paulsen IT. 2006. Genome sequence of Synechococcus CC9311: insights into adaptation to a coastal environment. Proc Natl Acad Sci U S A 103:13555–13559. doi: 10.1073/pnas.0602963103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post AF, Hagemann M, Paulsen I, Partensky F. 2009. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mackay RA, Henderson W. 2017. Introduction to modern inorganic chemistry. CRC Press, Boca Raton, FL. [Google Scholar]
- 48.Willey J, Waterbury J. 1989. Chemotaxis toward nitrogenous compounds by swimming strains of marine Synechococcus spp. Appl Environ Microbiol 55:1888–1894. doi: 10.1128/AEM.55.8.1888-1894.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carrieri D, Paddock T, Maness PC, Seibert M, Yu JP. 2012. Photo-catalytic conversion of carbon dioxide to organic acids by a recombinant cyanobacterium incapable of glycogen storage. Energy Environ Sci 5:9457–9461. doi: 10.1039/c2ee23181f. [DOI] [Google Scholar]
- 50.Long RA, Azam F. 2001. Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol 67:4975–4983. doi: 10.1128/AEM.67.11.4975-4983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marandi MV, Mittal KR. 1997. Role of outer membrane protein H (OmpH)- and OmpA-specific monoclonal antibodies from hybridoma tumors in protection of mice against Pasteurella multocida. Infect Immun 65:4502–4508. doi: 10.1128/IAI.65.11.4502-4508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Confer AW, Ayalew S. 2013. The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet Microbiol 163:207–222. doi: 10.1016/j.vetmic.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 53.Selvaraj SK, Periandythevar P, Prasadarao NV. 2007. Outer membrane protein A of Escherichia coli K1 selectively enhances the expression of intercellular adhesion molecule-1 in brain microvascular endothelial cells. Microbes Infect 9:547–557. doi: 10.1016/j.micinf.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gómez-Pereira PR, Schüler M, Fuchs BM, Bennke C, Teeling H, Waldmann J, Richter M, Barbe V, Bataille E, Glöckner FO, Amann R. 2012. Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean. Environ Microbiol 14:52–66. doi: 10.1111/j.1462-2920.2011.02555.x. [DOI] [PubMed] [Google Scholar]
- 55.Woyke T, Xie G, Copeland A, González JM, Han C, Kiss H, Saw JH, Senin P, Yang C, Chatterji S, Cheng J-F, Eisen JA, Sieracki ME, Stepanauskas R. 2009. Assembling the marine metagenome, one cell at a time. PLoS One 4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ackermann JU, Babel W. 1997. Growth-associated synthesis of poly(hydroxybutyric acid) in Methylobacterium rhodesianum as an expression of an internal bottleneck. Appl Microbiol Biotechnol 47:144–149. doi: 10.1007/s002530050903. [DOI] [Google Scholar]
- 57.Xiao N, Jiao NZ. 2011. Formation of polyhydroxyalkanoate in aerobic anoxygenic phototrophic bacteria and its relationship to carbon source and light availability. Appl Environ Microbiol 77:7445–7450. doi: 10.1128/AEM.05955-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang K, Jiao NZ, Liu KS, Zhang Y, Li SH. 2012. Distribution and functions of TonB-dependent transporters in marine bacteria and environments: implications for dissolved organic matter utilization. PLoS One 7:e41204. doi: 10.1371/journal.pone.0041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernández-Gómez B, Richter M, Schüler M, Pinhassi J, Acinas SG, González JM, Pedrós-Alió C. 2013. Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J 7:1026–1037. doi: 10.1038/ismej.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kappelmann L, Kruger K, Hehemann JH, Harder J, Markert S, Unfried F, Becher D, Shapiro N, Schweder T, Amann RI, Teeling H. 2019. Polysaccharide utilization loci of North Sea Flavobacteriia as basis for using SusC/D-protein expression for predicting major phytoplankton glycans. ISME J 13:76–91. doi: 10.1038/s41396-018-0242-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Degnan BA, Macfarlane GT. 1995. Arabinogalactan utilization in continuous cultures of Bifidobacterium longum - effect of co-culture with Bacteroides thetaiotaomicron. Anaerobe 1:103–112. doi: 10.1006/anae.1995.1005. [DOI] [PubMed] [Google Scholar]
- 64.Wu J, Deng A, Sun Q, Bai H, Sun Z, Shang X, Zhang Y, Liu Q, Liang Y, Liu S, Che Y, Wen T. 2018. Bacterial genome editing via a designed toxin–antitoxin cassette. ACS Synth Biol 7:822–831. doi: 10.1021/acssynbio.6b00287. [DOI] [PubMed] [Google Scholar]
- 65.van Rijssel M, Janse I, Noordkamp DJB, Gieskes W. 2000. An inventory of factors that affect polysaccharide production by Phaeocystis globosa. J Sea Res 43:297–306. doi: 10.1016/S1385-1101(00)00013-7. [DOI] [Google Scholar]
- 66.Lee MD, Walworth NG, McParland EL, Fu FX, Mincer TJ, Levine NM, Hutchins DA, Webb EA. 2017. The Trichodesmium consortium: conserved heterotrophic co-occurrence and genomic signatures of potential interactions. ISME J 11:1813–1824. doi: 10.1038/ismej.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grossart HP, Riemann L, Azam F. 2001. Bacterial motility in the sea and its ecological implications. Aquat Microb Ecol 25:247–258. doi: 10.3354/ame025247. [DOI] [Google Scholar]
- 68.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 69.Croft MT, Warren MJ, Smith AG. 2006. Algae need their vitamins. Eukaryot Cell 5:1175–1183. doi: 10.1128/EC.00097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giovannoni SJ. 2012. Vitamins in the sea. Proc Natl Acad Sci U S A 109:13888–13889. doi: 10.1073/pnas.1211722109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fenech M. 2012. Folate (vitamin B9) and vitamin B12 and their function in the maintenance of nuclear and mitochondrial genome integrity. Mutat Res 733:21–33. doi: 10.1016/j.mrfmmm.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 72.Helliwell KE, Lawrence AD, Holzer A, Kudahl UJ, Sasso S, Kräutler B, Scanlan DJ, Warren MJ, Smith AG. 2016. Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12. Curr Biol 26:999–1008. doi: 10.1016/j.cub.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walworth NG, Lee MD, Suffridge C, Qu PP, Fu FX, Saito MA, Webb EA, Sañudo-Wilhelmy SA, Hutchins DA. 2018. Functional genomics and phylogenetic evidence suggest genus-wide cobalamin production by the globally distributed marine nitrogen fixer Trichodesmium. Front Microbiol 9:189. doi: 10.3389/fmicb.2018.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kirchman DL, Suzuki Y, Garside C, Ducklow HW. 1991. High turnover rates of dissolved organic-carbon during a spring phytoplankton bloom. Nature 352:612–614. doi: 10.1038/352612a0. [DOI] [Google Scholar]
- 75.Burkill PH, Leakey RJG, Owens NJP, Mantoura R. 1993. Synechococcus and its importance to the microbial foodweb of the northwestern Indian Ocean. Deep Sea Res Part II 40:773–782. doi: 10.1016/0967-0645(93)90057-T. [DOI] [Google Scholar]
- 76.Richardson TL, Jackson GA. 2007. Small phytoplankton and carbon export from the surface ocean. Science 315:838–840. doi: 10.1126/science.1133471. [DOI] [PubMed] [Google Scholar]
- 77.Cano M, Holland SC, Artier J, Burnap RL, Ghirardi M, Morgan JA, Yu J. 2018. Glycogen synthesis and metabolite overflow contribute to energy balancing in Cyanobacteria. Cell Rep 23:667–672. doi: 10.1016/j.celrep.2018.03.083. [DOI] [PubMed] [Google Scholar]
- 78.Nakamura Y, Takahashi J-I, Sakurai A, Inaba Y, Suzuki E, Nihei S, Fujiwara S, Tsuzuki M, Miyashita H, Ikemoto H, Kawachi M, Sekiguchi H, Kurano N. 2005. Some cyanobacteria synthesize semi-amylopectin type α-polyglucans instead of glycogen. Plant Cell Physiol 46:539–545. doi: 10.1093/pcp/pci045. [DOI] [PubMed] [Google Scholar]
- 79.Aikawa S, Nishida A, Ho S-H, Chang J-S, Hasunuma T, Kondo A. 2014. Glycogen production for biofuels by the euryhaline cyanobacteria Synechococcus sp. strain PCC 7002 from an oceanic environment. Biotechnol Biofuels 7:88. doi: 10.1186/1754-6834-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rostovtseva TK, Nestorovich EM, Bezrukov SM. 2002. Partitioning of differently sized poly(ethylene glycol)s into OmpF porin. Biophys J 82:160–169. doi: 10.1016/S0006-3495(02)75383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krewulak KD, Vogel HJ. 2011. TonB or not TonB: is that the question? Biochem Cell Biol 89:87–97. doi: 10.1139/o10-141. [DOI] [PubMed] [Google Scholar]
- 82.Waterbury JB, Watson SW, Valois FW, Franks DG. 1986. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus, p 71–120. In Platt T, Li WKW (ed), Photosynthetic picoplankton. Department of Fisheries and Oceans, Ottawa, Canada. [Google Scholar]
- 83.Lane D. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, United Kingdom. [Google Scholar]
- 84.Stubner S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreenTM detection. J Microbiol Methods 50:155–164. doi: 10.1016/s0167-7012(02)00024-6. [DOI] [PubMed] [Google Scholar]
- 85.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu Y-W, Simmons BA, Singer SW. 2016. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 87.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neighbor-joining phylogenetic tree based on 16S-23S rRNA ITS sequences of representatives from cultured lineages of Synechococcus. Bootstrap percentages from neighbor-joining (1,000 replications) are shown. The scale bar represents a distance of 0.05 substitutions per site. The Synechococcus strains shown in red were isolated and collected by our lab. Download FIG S1, PDF file, 0.3 MB (312.9KB, pdf) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Variation in phosphate (A) and organic carbon (B) concentrations in the coculture system over the incubation period. DOC, dissolved organic carbon; POC, particulate organic carbon. Error bars represent the ranges of values from triplicate measurements. Download FIG S2, PDF file, 0.3 MB (299.7KB, pdf) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) 16S rRNA gene copy numbers in the complete genomes of close relatives of the strains whose genomic bins were recovered in this study. (B) Proteins identified in the proteomes of each coculture population. Download Table S1, DOC file, 0.04 MB (44KB, doc) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic characteristics and lifestyle strategies of the dominant bacterial populations of the coculture, Synechococcus sp. YX04-3 coculture system exoproteome, supplemental Materials and Methods, and supplemental references. Download Text S1, DOC file, 0.08 MB (85.5KB, doc) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cellular protein information for the proteomes of the five population bins of this study. Download Table S2, XLS file, 0.6 MB (634.5KB, xls) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overview of the dominant metabolic processes of Synechococcus sp. YX04-3 including transporter systems and carbon, nitrogen, and phosphorus metabolisms in the proteome. The primary metabolic processes observed for strain YX04-3 were involved in inorganic nutrient (e.g., NO3−, PO43−, and cyanate) uptake, photosynthesis, organic matter (e.g., glycogen) biosynthesis, and organic compound [e.g., polysaccharides, lipopolysaccharide, lipids and (oligo)peptide] release. VB12, vitamin B12; SOD, superoxide dismutase. The background arrows indicate the potential material and energy flows detected in the proteomic data. Download FIG S3, PDF file, 0.4 MB (391.5KB, pdf) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overview of the dominant metabolic processes associated with the two Flavobacteria population bins in their proteomes. The primary observed processes were involved in the degradation and transport of high-molecular-weight (dissolved) organic matter in Bin2 (left) and Bin3 (right). These activities were mediated by TonB-dependent transport systems and polysaccharide (or polymer) utilization loci (PUL) complexes. The polysaccharide utilization system of the figure was modified from Flint et al. (88). SOD, superoxide dismutase. The background arrows indicate the potential material and energy flows detected in the proteomic data. Download FIG S4, PDF file, 0.4 MB (467.6KB, pdf) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overview of the dominant metabolic processes associated with the Oricola sp. Bin5 population in the proteome. The dominant processes identified were associated with transporter systems and carbon, nitrogen, and phosphorus metabolisms. Oricola sp. Bin5 primarily imported low-molecular-weight organic substrates, including amino acids, di-/oligopeptides, nucleosides, ribose, maltose, xylose, dihydroxyacetone, C4-dicarboxylates, pyruvate, and sialic acids, as well as inorganic nutrients like NO3− and PO43− using ATP-binding cassette (ABC), tripartite ATP-independent periplasmic (TRAP), and tripartite tricarboxylate transporters (TTT) transport systems. VB12, vitamin B12; SOD, superoxide dismutase. The background arrows indicate the potential material and energy flows detected in the proteomic data. Download FIG S5, PDF file, 0.3 MB (338.7KB, pdf) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Exoproteome information for the five population bins of this study. Download Table S3, XLS file, 0.1 MB (96KB, xls) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Operon structure and gene composition of two-polysaccharide utilization loci (PUL) identified in the Flavobacteria Bin2 (A) and Bin6 (B) genomes, respectively. Download FIG S6, PDF file, 0.3 MB (329.2KB, pdf) .
Copyright © 2020 Zheng et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Sequence data were deposited in the NCBI Sequence Read Archive under BioProject no. PRJNA498017, PRJNA498190, and PRJNA497203.