Abstract
Autosis is a distinct form of cell death that requires both autophagy genes and the Na+,K+-ATPase pump. However, the relationship between the autophagy machinery and Na+,K+-ATPase is unknown. We explored the hypothesis that Na+,K+-ATPase interacts with the autophagy protein Beclin 1 during stress and autosis-inducing conditions. Starvation increased the Beclin 1/Na+,K+-ATPase interaction in cultured cells, and this was blocked by cardiac glycosides, inhibitors of Na+,K+-ATPase. Increases in Beclin 1/Na+,K+-ATPase interaction were also observed in tissues from starved mice, livers of patients with anorexia nervosa, brains of neonatal rats subjected to cerebral hypoxia-ischemia (HI), and kidneys of mice subjected to renal ischemia/reperfusion injury (IRI). Cardiac glycosides blocked the increased Beclin 1/Na+,K+-ATPase interaction during cerebral HI injury and renal IRI. In the mouse renal IRI model, cardiac glycosides reduced numbers of autotic cells in the kidney and improved clinical outcome. Moreover, blockade of endogenous cardiac glycosides increased Beclin 1/Na+,K+-ATPase interaction and autotic cell death in mouse hearts during exercise. Thus, Beclin 1/Na+,K+-ATPase interaction is increased in stress conditions, and cardiac glycosides decrease this interaction and autosis in both pathophysiological and physiological settings. This crosstalk between cellular machinery that generates and consumes energy during stress may represent a fundamental homeostatic mechanism.
Keywords: Cell Biology
Keywords: Autophagy, Cell stress
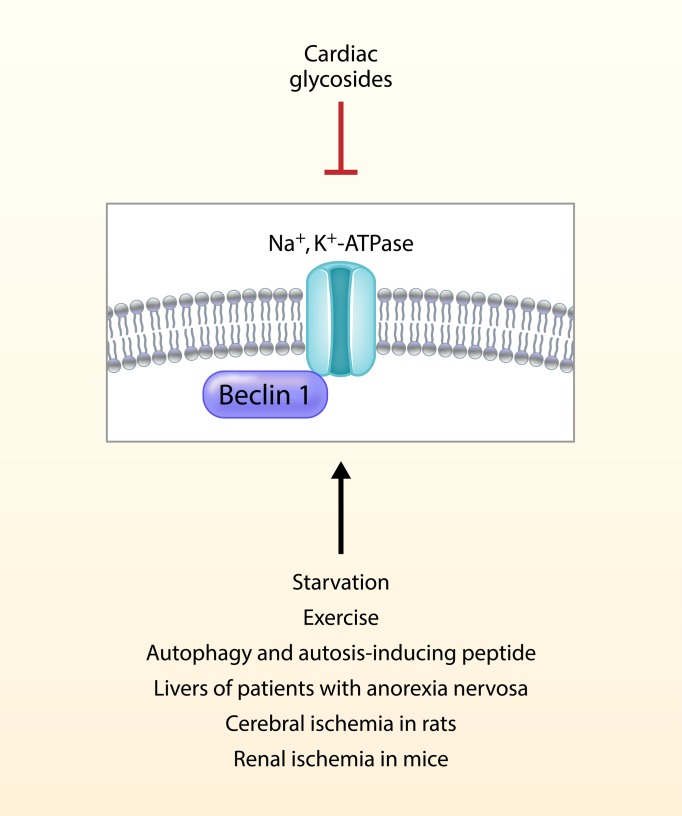
A nexus between the autophagy machinery and Na+,K+-ATPase is described during physiological and pathological conditions, including starvation, exercise, anorexia, and tissue ischemia.
Introduction
Eukaryotic cells are constantly exposed to different types of both extracellular and intracellular stress. To counteract such stress and maintain cellular homeostasis, cells trigger a tightly regulated network of prosurvival responses that include autophagy (1). Autophagy is an evolutionarily conserved pathway that mediates the transport of cytosolic components to the lysosome for degradation (2). Through this catabolic process, cells can survive a wide range of insults. For example, during energy deprivation, autophagy mediates the mobilization of different cellular energy stores, such as lipids or glycogen, providing a cell-autonomous mechanism for ATP generation (3). However, high levels of autophagy that occur during severe stress, especially tissue ischemia, can also be detrimental for the cell, leading to its death (4). The concept of autophagic cell death describes different types of cell death that typically exhibit massive formation of autophagic vacuoles and that are dependent on autophagy genes (4, 5).
Our group previously described autosis, a specific type of autophagic cell death that shows distinctive morphological features, including increased substrate adhesion and expansion of the perinuclear space (6). This unique pathway has been described under different stress conditions in vitro and in vivo (7–9), including in the livers of patients with severe anorexia nervosa (7) and a rat model of perinatal cerebral hypoxia-ischemia (6, 10). A growing number of studies suggest a dual role for autophagy in ischemia in different organs (11–15), with autophagy-dependent cell death partially contributing to adverse clinical outcomes (4, 16–19). However, the precise determinants of whether autophagy is detrimental during ischemic injury remain unclear. It is interesting to speculate that additional molecules may interface with the autophagy pathway, thereby regulating its switch from a protective process to a death pathway and providing potential therapeutic targets in ischemic disease.
Although the precise molecular mechanisms of autosis remain unknown, this pathway requires Na+,K+-ATPase (6). Genetic silencing of Na+,K+-ATPase or pharmacological inhibition of Na+,K+-ATPase by cardiac glycosides, natural inhibitors of Na+,K+-ATPase (20), blocks autosis in vitro, and the cardiac glycoside, neriifolin, decreases autosis and cerebral infarct size in a rat model of cerebral hypoxic-ischemic injury (6). Na+,K+-ATPase is a pump that is responsible for the ATP-dependent exchange of Na+ and K+ ions across membranes (21). Na+,K+-ATPase is also involved in the import of protons (22) and the initiation of different signaling pathways (23). Interestingly, Na+,K+-ATPase activity is markedly reduced during hypoxia or a single bout of exercise (24), and endogenous cardiac glycosides (synthesized by the adrenal gland and hypothalamus) are released into the circulation in such settings (25). These studies suggest the existence of a systemic regulatory mechanism characterized by the production of endogenous cardiac glycosides that may control potentially detrimental increases in Na+,K+-ATPase activity during specific contexts in vivo, thereby preventing autotic cell death in tissues.
Here, we report that Na+,K+-ATPase and the autophagy protein Beclin 1 interact in a wide range of conditions, including autophagy-inducing treatments in vitro, mice subjected to starvation, patients with anorexia nervosa, and animal models of cerebral and renal ischemia. Furthermore, cardiac glycosides decrease the Na+,K+-ATPase/Beclin 1 interaction, the numbers of autotic cells, and the severity of ischemic injury. Moreover, endogenous cardiac glycosides block the increased Na+,K+-ATPase/Beclin 1 interaction and autosis during exercise, a physiological inducer of autophagy. Taken together, our results show an unexpected link between 2 pathways oppositely involved in cellular energy balance whose regulation may define cell fate in different physiological and pathological conditions.
Results
Identification of autosis regulators in a genome-wide siRNA screen.
A previous chemical screen revealed that cardiac glycosides, antagonists of Na+,K+-ATPase, are inhibitors of autosis (6). To further characterize autotic cell death and identify potential mediators of the pathway, we performed a genome-wide siRNA screen using a Dharmacon Human siGENOME siRNA library. Following transfection with the siRNA library, HeLa cells were treated with the autophagy-inducing peptide Tat-Beclin 1 (26), and cell viability was assessed by measuring cellular ATP levels using the CellTiter-Glo assay (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.133282DS1). For the top 240 hits of the primary screen (defined as those with Z scores ≥ 3.0) (Supplemental Figure 1, B and C, and Supplemental Table 1), we performed a confirmation screen using the SYTOX Green assay for cell death (Supplemental Figure 1B). Those genes whose silencing resulted in a reduction of autotic cell death of more than 40% were chosen for further analysis (Supplemental Table 2). After eliminating genes that decreased Tat-Beclin 1 peptide entry into the cells, we performed a deconvolution screen using individual siRNAs. This screen identified 13 candidate regulators of autosis whose inhibition resulted in greater than 40% protection against autotic cell death. Notably, the strongest scoring hit was the α1 subunit of Na+,K+-ATPase (ATP1A1), showing an essential role for the Na+,K+-ATPase pump in autosis (Supplemental Table 3).
Beclin 1 and Na+,K+-ATPase interact during autophagy- and autosis-inducing conditions.
Because both a prior chemical screen (6) and our current genome-wide siRNA screen indicated that Na+,K+-ATPase is an essential effector of autosis, we further investigated the molecular link between Na+,K+-ATPase and autophagy during autotic cell death. A map of the human autophagy network previously suggested that Beclin 1 may bind the α subunit of Na+,K+-ATPase (27). Thus, we hypothesized that the interaction between Beclin 1 and Na+,K+-ATPase may be regulated during different conditions where autophagy is induced, including autosis.
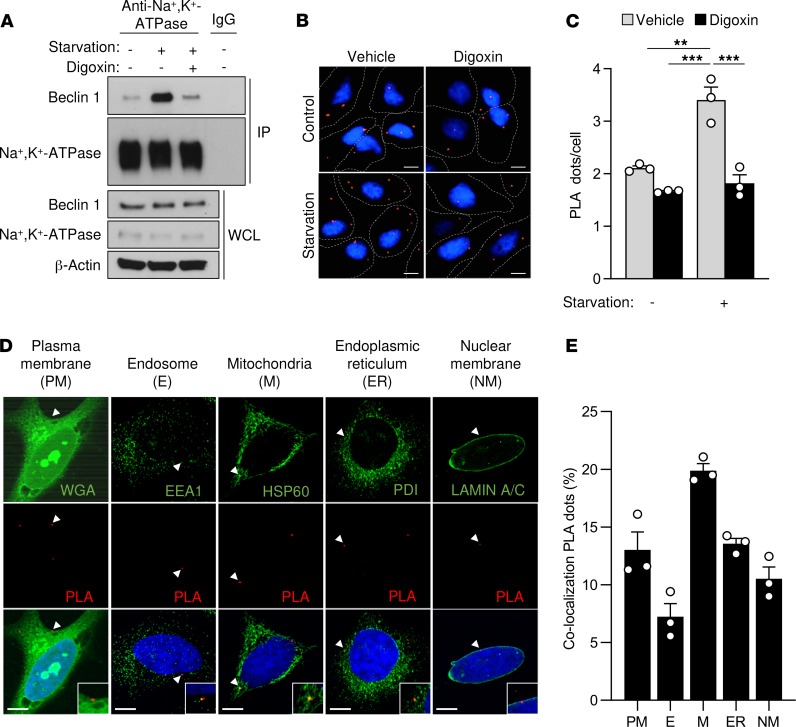
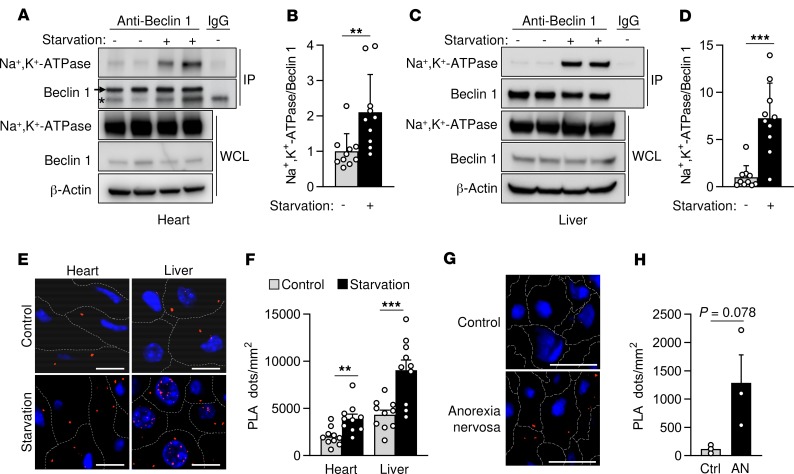
To investigate this hypothesis, we first used starvation or Tat-Beclin 1 peptide treatment to induce autophagy in HeLa cells in vitro. In both cases, the amount of Beclin 1 that immunoprecipitated with Na+,K+-ATPase increased (Figure 1A and Supplemental Figure 2A). However, this binding was reduced by treatment with the cardiac glycoside digoxin. Furthermore, using proximity ligase assays (PLAs), we observed increased Beclin 1/Na+,K+-ATPase interaction after starvation or Tat-Beclin 1 treatment that digoxin reduced (Figure 1, B and C, and Supplemental Figure 2, B and C). This interaction occurs not only at the plasma membrane but also at different intracellular compartments, such as the nuclear membrane, the endoplasmic reticulum, the mitochondria, and the early endosomes (Figure 1, D and E). Moreover, the increase in Beclin 1/Na+,K+-ATPase binding after prolonged starvation was more pronounced in autotic than in apoptotic cells (Supplemental Figure 3). Importantly, prolonged nutrient starvation in mice also led to enhanced Beclin 1/Na+,K+-ATPase interaction in mouse hearts and livers, as demonstrated both by coimmunoprecipitation and PLAs (Figure 2, A–F). Furthermore, livers from patients with anorexia nervosa that previously exhibited autotic cells (7) also showed a markedly increased binding of Beclin 1 to the Na+,K+-ATPase pump (Figure 2, G and H). Collectively, these results suggest that enhanced interaction between Beclin 1 and Na+,K+-ATPase occurs both in vitro during autophagy-inducing conditions and in vivo following extended nutrient deprivation.
Figure 1. Beclin 1 and Na+,K+-ATPase interact in cultured cells during starvation.
(A) Coimmunoprecipitation of Beclin 1 with the α subunit of Na+,K+-ATPase in HeLa cells after 3 hours of growth in normal medium (–) or HBSS starvation medium (+) treated with either vehicle or 10 μM digoxin. The same lysate from cells grown in normal medium without digoxin (lane 1) was used as a control for IgG immunoprecipitation. Similar results were observed in 3 independent experiments. (B and C) Representative images (B) and quantitation (C) of proximity ligase assays (PLAs) of Beclin 1 and Na+,K+-ATPase in the indicated conditions. In C, bars represent mean values ± SEM from 3 independent experiments (value for each experiment represents mean value of at least 100 cells per condition). (D) Representative images of PLAs of Beclin 1 and Na+,K+-ATPase costained with markers of plasma membrane (wheat germ agglutinin, WGA), endosomes (EEA1), mitochondria (HSP60), endoplasmic reticulum (PDI), and nuclear membrane (LAMIN A/C) in HeLa cells after 3 hours of starvation. The insets represent a 2-fold enlargement of the area of interest in the original image. (E) Quantitation of the percentage of PLA dots that colocalize with each indicated organelle marker. Bars represent mean values ± SEM for 3 experiments (each value represents percentage of PLA dots at indicated organelle using total number of PLA dots in >100 cells analyzed as the denominator). WCL, whole-cell lysate. Scale bars: 10 μm. **P < 0.01, and ***P < 0.001, 2-way ANOVA (C).
Figure 2. Beclin 1 and Na+,K+-ATPase interact in mouse and human tissues during starvation.
(A–D) Representative Western blots (A and C) and quantitation (B and D) of Na+,K+-ATPase coimmunoprecipitation with Beclin 1 from hearts (A and B) and livers (C and D) of mice after normal feeding or 48 hours of starvation. For A and C, the same tissue sample of the nonstarved group in lane 1 of the gel was used as a control for IgG immunoprecipitation. In B and D, bars represent mean ± SD (n = 10 mice). (E and F) Representative images (E) and quantitation (F) of proximity ligase assay of Beclin 1 and Na+,K+-ATPase in the indicated tissue and condition. For F, bars represent mean ± SEM (n = 10 mice, 10 randomly selected fields analyzed per mouse). (G and H) Representative images (G) and quantitation (H) of PLAs of Beclin 1 and the α subunit of Na+,K+-ATPase in livers from normal subjects (Ctrl) or patients with anorexia nervosa (AN). In H, bars represent mean ± SEM (n = 3 patients, at least 5 randomly selected fields analyzed per patient sample). Scale bars: 50 μm. Asterisk, nonspecific band. **P < 0.01, and ***P < 0.001, 2-tailed unpaired Student’s t test (B, D, F, and H).
Beclin 1 and Na+,K+-ATPase interact during ischemic injury.
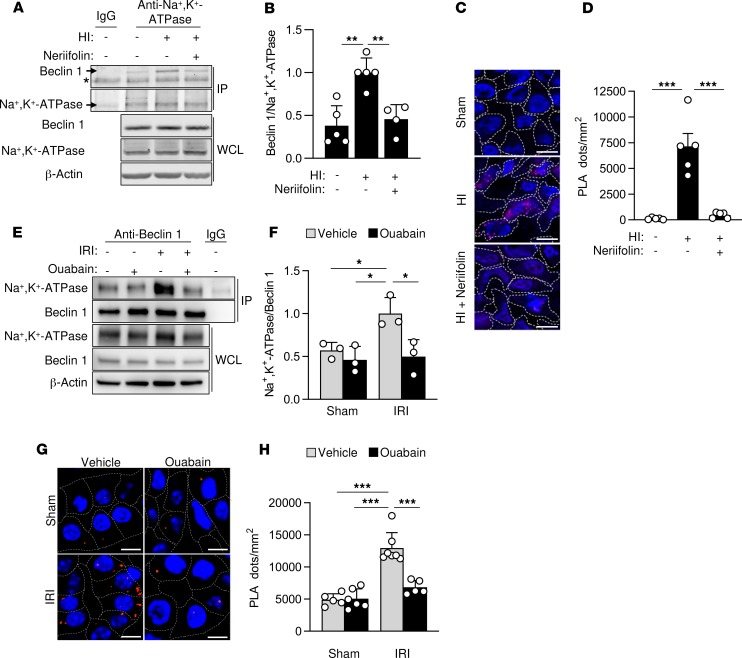
We next investigated whether the Beclin 1/Na+,K+-ATPase interaction increases in vivo during tissue ischemia. Previously, we described autotic cell death in neonatal rat brains subjected to hypoxia-ischemia (HI) injury and demonstrated that autotic death, as well as central nervous system (CNS) infarct size, were reduced by a CNS-penetrating cardiac glycoside, neriifolin (6). Using this same model of rat neonatal cerebral HI, we measured Beclin 1/Na+,K+-ATPase interaction by coimmunoprecipitation and PLAs. The brains of rats subjected to cerebral HI injury displayed an increase in Beclin 1/Na+,K+-ATPase association compared with the brains of control sham-operated animals (Figure 3, A–D) and neriifolin prevented this increase. Thus, Beclin 1/Na+,K+-ATPase interaction is enhanced in an established model of ischemia-induced autotic cell death (rat neonatal cerebral HI), and the interaction of these proteins is inhibited by a cardiac glycoside previously shown to exert beneficial effects in this model (6).
Figure 3. Beclin 1 and Na+,K+-ATPase interact in vivo during cerebral HI and renal IRI.
(A and B) Representative Western blots (A) and quantitation (B) of coimmunoprecipitation of Beclin 1 with the α subunit of Na+,K+-ATPase in the hippocampus of rat pups that were sacrificed 6 hours after receiving intraperitoneal vehicle or neriifolin (0.22 mg/kg) and subjected to sham operation or cerebral HI. In B, bars represent mean ± SD (n = 4–5 rats per group). (C and D) Representative images (C) and quantitation (D) of PLAs of Beclin 1 and the α subunit of Na+,K+-ATPase in the CA3 hippocampal region of rat pups that were sacrificed 6 hours after receiving intraperitoneal vehicle or neriifolin (0.22 mg/kg) and subjected to sham operation or cerebral HI. In D, bars represent mean ± SEM (n = 5 rats per group, 4 randomly selected fields analyzed per animal). (E and F) Representative Western blots (E) and quantitation (F) of coimmunoprecipitation of the α subunit of Na+,K+-ATPase with Beclin 1 in kidneys from mice that were subjected to sham operation or renal IRI after peritoneal administration with either vehicle or ouabain (0.25 mg/kg). In F, bars represent mean ± SD (n = 3 mice per group). (G and H) Representative images (G) and quantitation (H) of PLAs of Beclin 1 and the α subunit of Na+,K+-ATPase in kidneys from mice that were subjected to sham operation or renal IRI after peritoneal administration with either vehicle or ouabain (0.25 mg/kg). In H, bars represent mean ± SEM (n = 5–7 mice per group, 10 randomly selected fields analyzed per mouse). For A and E, the same tissue sample of the sham group (lane 1 of gel) was used as a control for IgG immunoprecipitation. Asterisk, nonspecific band. Scale bars: 10 μm (C), 50 μm (G). *P < 0.05, **P < 0.01, and ***P < 0.001; 1-way (B and D) or 2-way (F and H) ANOVA test.
We then examined whether increased Beclin 1/Na+,K+-ATPase interaction occurred in another rodent tissue ischemia model — renal ischemia/reperfusion injury (IRI) secondary to renal artery ligation. The kidneys of mice examined 48 hours after IRI displayed increased association between Beclin 1 and Na+,K+-ATPase (as demonstrated by coimmunoprecipitation and PLAs) compared with kidneys from control sham-operated mice (Figure 3, E–H). Moreover, this increase was blocked by the cardiac glycoside ouabain, further confirming that digoxin-like molecules regulate the interaction between Beclin 1 and Na+,K+-ATPase in ischemic injury.
Ouabain decreases autotic cell death and improves outcome in renal IRI.
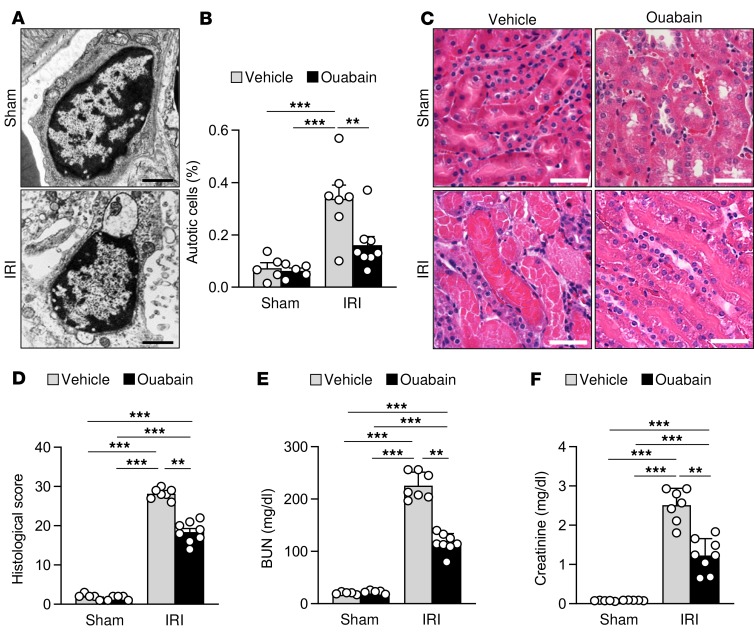
Analogous to observations in the rat neonatal cerebral HI model (ref. 6 and Figure 3, A–D), the increase in Beclin 1/Na+,K+-ATPase interaction in kidneys of mice subjected to IRI was associated with increased autotic death. Electron microscopic analyses of kidneys 48 hours after IRI revealed a marked increase in the number of cells that displayed the unique morphological features of autosis (including the expansion of the perinuclear space and a concave nucleus) (Figure 4, A and B). This increase in autotic cell death was blocked by ouabain. Autotic cell death was observed in renal peritubular capillary regions (mainly pericytes) and not in renal tubular cells. This observation is consistent with previous studies indicating that pericyte loss during acute kidney injury is associated with permanent peritubular capillary rarefaction, tubular damage, and renal fibrosis (28, 29). Notably, in parallel with reducing Beclin 1/Na+,K+-ATPase interaction and decreasing autotic cell death, treatment with ouabain also improved renal histopathology (Figure 4, C and D) and enhanced renal function as measured by serum levels of blood urea nitrogen (BUN) and creatinine after IRI (Figure 4, E and F). Moreover, although approximately 25% of the injured mice treated with vehicle died during the 48 hours of reperfusion, all the mice treated with ouabain survived (data not shown). Together with our previous study (6), these results demonstrate that autosis occurs during ischemic injury and that cardiac glycosides may be beneficial in reducing autosis and end-organ damage during tissue ischemia.
Figure 4. Ouabain decreases autotic cell death and improves outcome in renal IRI.
(A and B) Representative micrographs (A) and quantitation (B) of autotic pericytes in the kidneys from mice that were subjected to sham operation or renal IRI after peritoneal administration of either vehicle or ouabain (0.25 mg/kg). In B, bars represent mean ± SEM (n = 5–8 mice per group, more than 50 cells analyzed per mouse). (C and D) Representative H&E images (C) and histological score (D) of kidneys from mice that were subjected to sham operation or renal IRI after peritoneal administration of either vehicle or ouabain (0.25 mg/kg). In D, bars represent mean ± SEM. (n = 5–8 mice per group, 10 randomly selected fields analyzed per mouse). (E and F) Assessment of renal damage as measured by blood urea nitrogen (BUN) (E) and creatinine (F) levels in plasma from mice that were subjected to sham operation or renal IRI after peritoneal administration of either vehicle or ouabain (0.25 mg/kg). Bars represent mean ± SD (n = 5–8 mice per group). Scale bars: 1 μm (A), 50 μm (C). **P < 0.01; ***P < 0.001; 2-way ANOVA test.
Endogenous cardiac glycosides regulate Beclin 1/Na+,K+-ATPase binding and autosis during exercise.
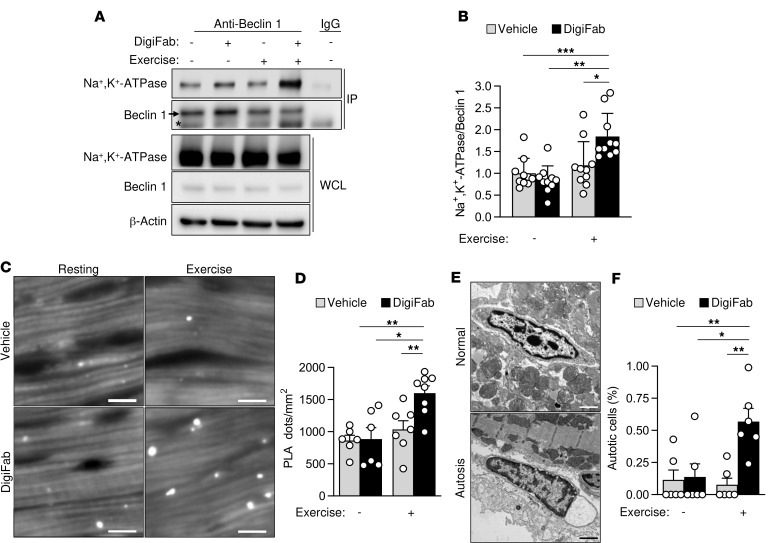
Previous studies have identified the presence of endogenous cardiac glycosides in mammals (30) and have shown that their plasma concentrations increase during exercise (25). Given the regulation of the Beclin 1/Na+,K+-ATPase interaction by cardiac glycoside drugs, we asked whether endogenous cardiac glycosides might prevent the increase in Beclin 1/Na+,K+-ATPase interaction during exercise. Even though exercise is a well-established physiological inducer of autophagy (31, 32), it did not increase Beclin 1/Na+,K+-ATPase binding in hearts from mice after they ran for 80 minutes on a treadmill, as shown by coimmunoprecipitation and PLAs (Figure 5, A–D). However, this interaction increased when mice were treated with DigiFab, an antibody that effectively blocks endogenous digoxin-like molecules (33), before running on the treadmill (Figure 5, A–D). Moreover, the hearts from mice that were subjected to both DigiFab treatment and exercise showed an increase in detectable autotic cells (Figure 5, E and F). Notably, only nonmyocytes (i.e., fibroblasts) were found to die by autosis, resembling the cell type–specific susceptibility to autosis that we observed in the kidney. Taken together, these results indicate that endogenous cardiac glycosides may regulate Beclin 1/Na+,K+-ATPase interaction during exercise while preventing autotic cell death.
Figure 5. Endogenous cardiac glycosides regulate Beclin 1/Na+,K+-ATPase binding and autosis during exercise.
(A and B) Representative Western blots (A) and quantitation (B) of coimmunoprecipitation of Beclin 1 with the α subunit of Na+,K+-ATPase in hearts from mice treated with either intraperitoneal vehicle or cardiac glycoside–blocking antibody DigiFab (10 mg/kg) before 80 minutes of resting or exercise. For A, the same issue sample of the resting group without DigiFab (lane 1) was used as a control for IgG immunoprecipitation. In B, bars represent mean ± SD (n = 10 mice per group). (C and D) Representative images (C) and quantitation (D) of PLAs of Beclin 1 and the α subunit of Na+,K+-ATPase in hearts from mice treated with either intraperitoneal vehicle or cardiac glycoside–blocking antibody DigiFab (10 mg/kg) before 80 minutes of resting or exercise. In D, bars represent mean ± SEM (n = 6–8 mice per group, 10 randomly selected fields analyzed per mouse). (E and F) Representative micrographs (E) and quantitation (F) of autotic noncardiomyocyte cells in hearts from mice treated with either intraperitoneal vehicle or cardiac glycoside blocking antibody DigiFab (10 mg/kg) before resting or running until exhaustion. In F, bars represent mean ± SEM (n = 6 randomly selected areas from 3 mice per condition, more than 1,000 analyzed cells per area). Asterisk, nonspecific band. Scale bars: 10 μm (B), 1 μm (C). *P < 0.05, **P < 0.01, and ***P < 0.001; 2-way ANOVA test.
Discussion
Our study defines an essential role of Na+,K+-ATPase in autotic cell death and uncovers unexpected crosstalk between Na+,K+-ATPase and the autophagy protein Beclin 1 during physiological and pathophysiological stress. Specifically, we show that Na+,K+-ATPase and Beclin 1 interact in cells during nutrient starvation, in rodent tissues during starvation and ischemic injury, and in livers of patients with severe anorexia nervosa. Cardiac glycosides, antagonists of Na+,K+-ATPase, inhibit this interaction during neonatal rat cerebral HI injury and mouse renal ischemia in parallel with improving clinical outcomes and decreasing autotic cell death. Moreover, blockade of endogenous cardiac glycosides during exercise increases Na+,K+-ATPase/Beclin 1 interaction and autotic cell death in the heart. This crosstalk between the cell’s major consumer of ATP (Na+,K+-ATPase) and a key generator of ATP during nutrient stress (autophagy) represents a newly described nexus for the regulation of cell survival.
Further studies are required to definitively determine whether, and if so how, the Na+,K+-ATPase/Beclin 1 interaction causes autotic cell death. In theory, the interaction could (a) alter Na+,K+-ATPase ion exchange or Na+,K+-ATPase ion exchange–independent effects on cellular signaling, (b) alter functions of Beclin 1 in autophagy or related membrane trafficking events, or (c) exert effects on the cell not directly related to known functions of Na+,K+-ATPase or Beclin 1. The robust induction of autophagy during tissue starvation (32, 34) suggests that enhanced Na+,K+-ATPase/Beclin 1 interaction does not inhibit the function of Beclin 1 in autophagy. Of note, the unique morphology of autosis involves endoplasmic reticulum (ER) and focal perinuclear space expansion, phenotypes that have been observed in cells with mutations in genes that affect ER membrane properties, including transport or channel conductance (35). It is thus relevant to note that we observed Na+,K+-ATPase/Beclin 1 interaction not only at the plasma membrane but also (and more frequently) on intracellular membranes, such as the ER and perinuclear membrane, as well as mitochondria and endosomes. Although best known for its plasma membrane localization, Na+,K+-ATPase has also been reported to localize to the perinuclear membrane, mitochondria, and endosomes (36–38), with some evidence for nuclear envelope Na+,K+-ATPase–dependent regulation of local Na+/K+ gradients and calcium signaling (36). Thus, delineation of the effects of Na+,K+-ATPase and Beclin 1 interaction on ion, osmolyte, and fluid homeostasis across intracellular membranes may help elucidate the basis for the distinct morphological features of autosis as well as potential mechanisms whereby the interaction between these proteins could lead to cell death.
In addition to Na+,K+-ATPase, our genome-wide siRNA screen identified other potential mediators of autosis (Supplemental Tables 1–3). Related to our hypothesis that autosis may involve a dysregulation of ion transport, other confirmed hits included genes encoding the intermediate conductance calcium-activated potassium channel protein 4 (KCNN4) and the potassium voltage-gated channel subfamily KQT member 2 (KCNQ2). In addition, a key feature that distinguishes autosis from other forms of cell death is the increased cellular substrate adherence (6); accordingly, the identification of genes involved in cell-to-matrix adhesion, such as adhesion G protein-coupled receptor F1 (ADGRF1) and thrombospondin 3 (THBS3), may indicate a requirement for increased cell-to-matrix adhesion in the autotic death pathway. Further studies are needed to investigate the role of these, and additional genes found in our genome-wide screen, in autosis.
We discovered an important role for cardiac glycoside drugs (e.g., digoxin, neriifolin, and ouabain) and endogenous cardiac glycosides in regulating the interaction between Na+,K+-ATPase and Beclin 1 during starvation, cerebral HI injury, renal IRI, and exercise. Exogenous cardiac glycosides decrease the interaction and prevent autotic death during cellular starvation, rat cerebral HI injury, and renal IRI (the present study and ref. 6), whereas blockade of endogenous cardiac glycosides with DigiFab increases Na+,K+-ATPase/Beclin 1 interaction and autosis in cardiac tissue during exercise. Previously, we showed that neriifolin improves clinical outcomes in rats with neonatal cerebral HI injury (6), and in the current study, we found that ouabain improves clinical outcomes in mice with renal IRI. Thus, even though it is unknown whether Beclin 1/Na+,K+-ATPase interaction is a driving event in autosis, our findings raise the possibility that cardiac glycosides may be clinically useful in settings of tissue ischemia and autotic cell death. Moreover, we speculate that endogenous cardiac glycosides may represent an adaptive mechanism that regulates Na+,K+-ATPase activity, preventing its association with the autophagy machinery and allowing autophagy to be protective rather than pathological during stress.
An open question is how cardiac glycosides disrupt the interaction between Na+,K+-ATPase and Beclin 1. Cardiac glycosides bind to the extracellular domain of the α1 subunit of Na+,K+-ATPase and thereby inhibit Na+/K+ exchange as well as regulate diverse cellular signal transduction pathways (20). It is possible cardiac glycoside binding could trigger a conformational change in Na+,K+-ATPase that directly prevents its interaction with Beclin 1 on the cytoplasmic side of either the plasma membrane or intracellular membranes. It is also possible that one of the signaling cascades cardiac glycosides trigger, such as activation of EGFR tyrosine kinase (39) that phosphorylates Beclin 1 (40), indirectly alters the ability of Beclin 1 to interact with Na+,K+-ATPase. Interestingly, a previous study reported that EGFR functions as a switch between cell survival and cell death induced by autophagy in hypoxic cancer cells (8). Our detection of a Na+,K+-ATPase/Beclin 1 interaction at endosomes may indicate that its regulation occurs during the trafficking of Na+,K+-ATPase. However, the predominant localization of the interaction at intracellular membranes (e.g., mitochondria, ER/perinuclear membrane) suggests potential alternative mechanisms of regulation. Although cardiac glycosides are reported to enter cells through either passive diffusion (for those compounds such as digoxin that are hydrophobic) or uptake by drug transporters (41, 42), it is unclear how they would gain access to the extracellular domain of Na+,K+-ATPase (which would be on the luminal side of intracellular membranous compartments). Therefore, it seems more likely that interactions between Na+,K+-ATPase/Beclin 1 at the mitochondria and ER/perinuclear membrane are regulated indirectly via cardiac glycoside effects on ionic concentrations or signal transduction pathways. To unravel the precise mode of regulation of Na+,K+-ATPase/Beclin 1 interaction by cardiac glycosides, as well as the interaction’s significance in autotic cell death, an atomic understanding of the interaction will be essential.
Methods
Cell culture.
HeLa cells were obtained from American Type Culture Collection and cultured in DMEM supplemented with 10% (vol/vol) FBS, 1% l-glutamine, and 1% penicillin/streptomycin. For starvation experiments, cells were cultured in HBSS (MilliporeSigma, H4641) for the indicated time.
Chemical reagents and antibodies.
Digoxin was purchased from MilliporeSigma (D6003). The following antibodies were used for immunoblot analyses: anti–Na+,K+-ATPase α2 subunit (used in analyses of mouse hearts, livers, and kidneys; Proteintech, 18836-1-AP, 1:100 dilution), anti–Na+,K+-ATPase α3 subunit (used in analyses of rat brains, Abcam, ab182571 and ab2826, 1:1000 dilution), anti–Beclin 1 (Santa Cruz Biotechnology, sc-11427, 1:250 dilution), and anti-Actin (Santa Cruz Biotechnology, sc-47778, 1:5,000 dilution; and Invitrogen, Thermo Fisher Scientific, MA511869, 1:2000 dilution). The following antibodies were used for immunofluorescence analyses: anti–Na+,K+-ATPase α1 subunit (used in analyses of HeLa cells and human liver samples, Santa Cruz Biotechnology, sc-21712, 1:50 dilution), anti–Na+,K+-ATPase α2 subunit (used in analyses of mouse hearts, livers, and kidneys; Proteintech, 18836-1-AP, 1:100 dilution), anti–Na+,K+-ATPase α3 subunit (used in analyses of rat brains, Abcam, ab182571, 1:1000 dilution), anti–Beclin 1 (Santa Cruz Biotechnology, sc-11427 and sc-10086, 1:100 dilution), anti-EEA1 (Santa Cruz Biotechnology, sc-6415, 1:50 dilution), anti-HSP60 (Santa Cruz Biotechnology, sc-1052, 1:50 dilution), anti-PDI (Santa Cruz Biotechnology, sc-30932, 1:50 dilution), and anti–Lamin A/C (Santa Cruz Biotechnology, sc-6215, 1:50 dilution). Anti-WGA conjugated with Alexa Fluor 488 (Thermo Fisher Scientific, W11261, 1:200) was used to stain the plasma membrane according to the manufacturer’s instructions.
Genome-wide siRNA screen.
For the primary screen, 800 HeLa cells per well were transfected with 2 pmol Dharmacon Human siGENOME siRNA Library (Horizon Discovery, Inc.) using 0.1 μL Lipofectamine RNAiMAX (Thermo Fisher Scientific, 13778150) in 96-well plates (Greiner Bio-One, 655083). After 48 hours of transfection, cells were treated with 12.5 μM Tat-Beclin 1 peptide (YGRKKRRQRRRGGTNVFNATFEIWHDGEFGT; ref. 26) in 100 μL acidified OptiMEM (pH 6.8) for 4 hours and then lysed in 40 μL CellTiter-Glo solution (Promega, G8462). Cellular ATP levels were measured by an EnVision multimode plate reader (PerkinElmer, Inc.). Wells in the first column and the last column of each 96-well plate were transfected with noncoding (NC) siRNA control (Thermo Fisher Scientific, D-001210-02-20) and ATP1A1 siRNA (Invitrogen, Thermo Fisher Scientific, 4390824, CUCGCUCACUGGUGAAUCA), respectively. Each siRNA pool (4 siRNAs) was screened in triplicate. For the primary screen, we first fitted a liner model to compare the differences between the siRNA triplicates with negative controls on the same plate to estimate the Z score and fold change. Second, raw data from EnVision Reader were analyzed with Genedata Screener 11 software suite (Genedata AG). Data files (.txt or .csv) were loaded into the Assay Analyzer module of Genedata software and normalized by the test population (the transfecting siRNA library) in each plate, using the following equation (43): normalized values = ([raw values – median of all samples] / median of all samples) × 100.
An average Z′ factor of 0.54 was achieved in the entire screen. Normalized well values were then corrected using a group of reference plates (typically 3 plates with sequence-scrambled siRNA, placed at the first, middle, and last positions of a 30-plate run). A correction factor for each well was calculated using a proprietary pattern detection algorithm in the Assay Analyzer software (see Genedata user documentation and ref. 44). The corrected normalized activity values from replicates for each siRNA in each run were then condensed to a single value (condensed activity) using the Robust Condensing method in Genedata Screener. The condensed activity is the most representative single value of the triplicates. In general, the triplicates were precondensed into a pair of values as follows: values (X, Y) = (median of triplicates m) ± dispersion, where dispersion = median (|X1 – m|, |X2 – m|, |X3 – m|). The less X and Y differ (|X – Y|), the better the data quality. For data points where |X – Y| ≤ 30%, the condensed activity is equal to the median of the triplicate measurements. Otherwise, a condensing function, Max(X, Y), was used to estimate the condensed activity. Calculation of condensed activity values from triplicated measurements was implemented with a Pipeline Pilot protocol (BIOVIA). A robust Z score was calculated from the corrected normalized activity for each siRNA by using the following formula (44): robust Z score = (corrected activity – median of neutral controls) / robust SD of neutral controls.
The Z scores estimated by linear model and robust Z score were averaged to get the final Z score. Primary hits were selected if the final Z score was equal to or greater than 3.0. For the confirmation screen, 1,000 HeLa cells were transfected with 2 pmol of the Dharmacon Human siGENOME siRNA Library using 0.1 μL Lipofectamine RNAiMAX (Thermo Fisher Scientific, 13778150) in a 96-well plate (Greiner Bio-One, 655090). Forty-eight hours after transfection, cells were treated with 18 μM Tat-Beclin 1 peptide in 100 μL acidified OptiMEM (pH 6.8) for 4 hours, and then 100 nM SYTOX Green (Invitrogen, Thermo Fisher Scientific, S7020) and 1 μg/mL Hoechst 33342 were added for 1 additional hour. Cells were then imaged using a BD Pathways 855 automated microscope. Cells in the first column and the last column of each 96-well plate were transfected with NC siRNA (Thermo Fisher Scientific, D-001210-02-20) and ATP1A1 siRNA (Invitrogen, Thermo Fisher Scientific, 4390824, CUCGCUCACUGGUGAAUCA), respectively. Each siRNA was screened in triplicate. Autotic cell death was counted as the percentage of SYTOX Green–positive cells. The protection of each siRNA was calculated as follows (“A” means the observed autotic cell death as a percentage of SYTOX Green–positive cells from triplicate plates): protection(siRNA) = ([A of NC – A of siRNA] / [A of NC – A of ATP1A1]) × 100%.
For the deconvolution screen, 1,000 HeLa cells were transfected with 2 pmol of each siRNA from the SMART pool for each gene (4 siRNAs per gene) in the Dharmacon Human siGENOME siRNA library or the Invitrogen Ambion Silencer Select siRNA using 0.1 μL Lipofectamine RNAiMAX (Thermo Fisher Scientific, 13778150) in 96-well plates (Greiner Bio-One, 655090). Forty-eight hours after transfection, cells were treated with 18 μM Tat-Beclin 1 in 100 μL acidified OptiMEM (pH 6.8) for 4 hours, and then 100 nM SYTOX Green and 1 μg/mL Hoechst 33342 were added for 1 additional hour. Cells were then imaged using a Bio-Tek Cytation 3 Cell Imaging Multi-Mode Reader. Cells in the first column and the last column of each 96-well plate were transfected with NC siRNA (Thermo Fisher Scientific, D-001210-02-20) and ATP1A1 siRNA (Invitrogen, Thermo Fisher Scientific, 4390824, CUCGCUCACUGGUGAAUCA), respectively. Each siRNA was screened in triplicate.
Peptide entry assay.
N-terminal biotinylated Tat-Beclin 1 peptide (>95% purity as determined by reverse-phase HPLC) was synthesized as described previously (26). For the peptide entry assay, siRNA-transfected cells were treated with 20 μM biotinylated Tat-Beclin 1 for 30 minutes, fixed with 4% paraformaldehyde at room temperature for 10 minutes, and permeabilized with Triton X-100 solution (0.5% Triton X-100, 20 mM HEPES at pH 7.4, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose) for 5 minutes on ice. After staining with Alexa Fluor 488–conjugated streptavidin for 1 hour, more than 400 cells per siRNA treatment were imaged using a PerkinElmer Ultraview spinning disk confocal microscope. Cells showing the typical intracellular staining pattern of Alexa Fluor 488 signal were counted as biotin-positive cells.
Renal IRI model.
Eight- to 12-week-old wild-type mice (C57BL/6J, The Jackson Laboratory) underwent bilateral ischemia for 45 minutes followed by 48 hours’ reperfusion. Laparotomy was conducted for sham treatment in control animals following our previously published protocol (45). Vehicle (0.5% ethanol/PBS) or ouabain (0.25 mg/kg diluted in 0.5% ethanol/PBS) (MilliporeSigma, O3125) was injected intraperitoneally 30 minutes prior to surgery. After 48 hours’ reperfusion, mice were euthanized under anesthesia and blood and kidneys were harvested. The kidney samples were either fixed in 4% paraformaldehyde for paraffin-embedded sections used for histopathological analyses or fixed in Karnovsky’s fixative (2.5% glutaraldehyde, 4% paraformaldehyde, 0.1 M cacodylate buffer, 8 mM CaCl2, pH 7.4) for ultrastructural analysis. The severity of kidney damage was semiquantified with a kidney histological score in a blinded manner as adapted from previous publications (45, 46). Briefly, the kidney injury was scored in H&E-stained kidney sections as a percentage of damaged tubules: 0, no damage; 1, damage in less than 25% of the field; 2, damage in 25%–50% of the field; 3, damage in 50%–75% of the field; and 4, damage in more than 75% of the field. Plasma BUN was analyzed using a Vitros Chemistry Analyzer (Ortho-Clinical Diagnosis). Plasma creatinine was measured using a P/ACE MDQ Capillary Electrophoresis System and photodiode detector (Beckman Coulter), as previously described (47).
In vivo starvation studies.
Eight-week-old wild-type mice (C57BL/6J, The Jackson Laboratory) were starved overnight and refed for 3 hours before sample collection (control group) or were starved for 48 hours (starved group) to minimize variability due to differences in food intake.
Mouse exercise studies.
Eight- to 12-week-old wild-type mice (C57BL/6J, The Jackson Laboratory) were acclimated to a 10° uphill Exer-3/6 Open Treadmill (Columbus Instruments) for 2 days. On day 1, mice ran for 5 minutes at 8 m/min, and on day 2, mice ran for 5 minutes at 8 m/min followed by another 5 minutes at 10 m/min. On day 4, mice were treated either with vehicle or with DigiFab (10 μg/kg) 30 minutes before being subjected to a single bout of running starting at the speed of 10 m/min. Forty minutes later, the treadmill speed was increased at a rate of 1 m/min every 10 minutes for 30 minutes and then increased at a rate of 1 m/min every 5 minutes until they ran for 80 minutes (for coimmunoprecipitations or PLAs, with sample collection immediately after exercise) or until they were exhausted (for electron microscopy analysis, with sample collection 6 hours later). Exhaustion was defined as the point at which mice spent more than 5 seconds on the electric shocker without attempting to resume running. Mice were euthanized and hearts were then harvested. The samples were frozen for Western blot analyses, fixed in 4% paraformaldehyde (embedded thereafter in 30% sucrose to prepare cryosections) for PLAs, or fixed in Karnovsky’s fixative (2.5% glutaraldehyde, 4% paraformaldehyde, 0.1 M cacodylate buffer, 8 mM CaCl2, pH 7.4) for electron microscopy analysis.
Rat model of neonatal cerebral HI.
Seven-day-old male rats (Sprague-Dawley, from Janvier Labs) underwent HI according to the Rice-Vannucci model (48) as described previously (49). Briefly, under isoflurane (3%) anesthesia, the right common carotid artery was isolated, double-ligated, and cut. One hour after carotid occlusion, neriifolin (0.22 mg/kg diluted in 0.5% ethanol/PBS) (MilliporeSigma, S961825) or vehicle (0.5% ethanol/PBS) was injected intraperitoneally. After 2 hours of recovery with the dam, the rat pups were exposed to 2 hours of systemic hypoxia at 8% of oxygen in a chamber maintained at around 35.5°C. Sham-operated animals (same anesthesia and surgery without cutting the artery) were used as controls. Hippocampal samples were microdissected and collected 6 hours after hypoxia.
Immunoprecipitation.
HeLa cells were collected and lysed in lysis buffer (20 mM Tris-HCl, 100 mM KCl, 4 mM MgCl2, 10% glycerol, 1% Triton X-100, protease inhibitor mixture from Roche Applied Sciences, and Halt Phosphatase Inhibitor Cocktail from Thermo Fisher Scientific, pH 7.4). Anti–human Na+,K+-ATPase α1 subunit antibody (Santa Cruz Biotechnology, sc-21712; 1:100 dilution) or control IgG (Santa Cruz Biotechnology, sc-2025) was then added to the lysate and incubated at 4°C for 1 hour. Protein G PLUS-Agarose (Santa Cruz Biotechnology, sc-2002) was then added (1:25 dilution), and samples were incubated at 4°C overnight. The beads were pelleted, washed, and boiled with 2× Laemmli buffer, and the supernatant was resolved on SDS-PAGE. For immunoprecipitation assays in mouse tissues, protein samples were obtained using a different lysis buffer, containing 250 mM HEPES, 250 mM KCl, 250 mM NaF, 25 mM Na4P2O7, 25 mM β-glycerophosphate, 5 mM EDTA, 5 mM EGTA, 200 mM vanadate, and 500 mM DTT and protease inhibitor mixture (Roche Applied Sciences). Samples were then incubated overnight with anti–mouse Beclin 1 antibody (Santa Cruz Biotechnology, sc-48341, 1:100) and protein-A Sepharose beads (MilliporeSigma, P-3391). The beads were pelleted, washed, transferred to a new tube, and boiled with 2× Laemmli buffer, and the supernatant was resolved on SDS-PAGE. For immunoprecipitation assays in rat brains, whole hippocampal tissues were collected in lysis buffer (20 mM HEPES at pH 7.4, 10 mM NaCl, 3 mM MgCl2, 2.5 mM EGTA, 0.1 mM DTT, 50 mM NaF, 1 mM Na3VO4, 1% Triton X-100, and a protease inhibitor cocktail [MilliporeSigma, 11873580001]), homogenized on ice, and rapidly sonicated. After protein concentration determination using a Bradford assay (Thermo Fisher Scientific), 500 μg of proteins were incubated in 300 μL of immunoprecipitation (IP) buffer (25 mM Tris, 150 mM NaCl, 1% NP-40, 1 mM Na3VO4, 20 mM NaF, and a protease inhibitor cocktail from MilliporeSigma) with 1–1.5 μg of antibody (anti–Na+,K+-ATPase α3 subunit rabbit monoclonal, ab182571, from Abcam, or anti–Beclin 1, sc-11427, rabbit polyclonal, or sc-48341, mouse monoclonal, from Santa Cruz Biotechnology) overnight at 4°C. Control IP was performed with an isotype control antibody (3900S) from Cell Signaling Technology or with an anti-GFP antibody (AB3080) from MilliporeSigma. After washing 3 times in IP buffer, 30–40 μL of Dynabeads Protein G (10004D) from Invitrogen (Thermo Fisher Scientific) was added to antibody/antigen complex and incubated 90 minutes at 4°C. Dynabeads were then washed 3 times in IP buffer, resuspended in 100 μL of IP buffer, and transferred to a new tube. Antibody/antigen complex was eluted by heating at 95°C for 10 minutes in 40 μL of 2× SDS loading buffer (125 mM Tris at pH 6.8, 4% SDS, 12.5% glycerol, 4% β-mercaptoethanol). The eluted samples were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, and analyzed by immunoblotting as previously described (50) except that IRDye 680RD detection reagent (catalog 926-68100) from LI-COR Biosciences or Fluorescent TrueBlot IgG DyLight secondary antibodies (catalogs 18-4516-32, 18-4417-32) from Rockland Immunochemicals, Inc. were used. Optical densities of bands were quantified using Odyssey v1.2 software (LI-COR Biosciences). See complete uncropped gels in the supplemental material.
PLA.
The PLA was performed according to the manufacturer’s instructions (MilliporeSigma, DUO92008). Briefly, cells were fixed with 3% paraformaldehyde for 10 minutes at room temperature and cold methanol for 20 minutes at 4°C and then blocked with 0.05% saponin and 1% BSA in PBS for 1 hour at room temperature. After incubation with primary antibodies overnight, cells were washed with 0.05% saponin and 1% BSA in PBS twice for 10 minutes. Cells were then incubated with Duolink In Situ PLA Probe Anti-Rabbit PLUS and Duolink In Situ PLA Probe Anti-Mouse MINUS (1:5 dilution) for 60 minutes at 37°C and then washed with buffer A twice for 10 minutes. Ligase was added at 1:40 dilution and incubated for 30 minutes at 37°C. After washing with buffer A twice for 5 minutes, PLA signal was amplified by polymerase (1:80) for 100 minutes at 37°C. Cells were then washed with 1× buffer B twice for 20 minutes and then 0.01× buffer B for 1 minute and mounted in Duolink In Situ Mounting Medium with DAPI for 15 minutes. Buffers A and B were part of the PLA kit. For tissues, frozen sections were incubated in L.A.B. solution (Polysciences, Inc., 24310-500) at 65°C for 5 minutes and then washed with PBS once. The samples were then incubated in 15 mL/L hydrogen peroxide in PBS for 15 minutes at room temperature and washed with PBS once. The standard PLA protocol was then performed starting from the blocking step.
Microscopy.
Immunofluorescent images were captured using either a Zeiss Axio Imager Z2 microscope or an LSM 780 Meta confocal microscope (Carl Zeiss). Electron microscopy images were captured using a JEOL 1200 EX II Electron Microscope (University of Texas [UT] Southwestern Electron Microscopy Core). SYTOX Green assay was analyzed using a BD Pathway 855 automated microscope.
Statistics.
Experimental data were analyzed using GraphPad Prism 8 software. P values were calculated using 2-tailed Student’s t test and 1- or 2-way ANOVA (for multiple-group analyses) followed by Tukey’s post hoc test. A P value of less than 0.05 was considered statistically significant. SEM was used when the individual data points represented mean values for a population of cells. SD was used when data points represented the value of an individual sample.
Study approval.
All human studies were conducted according to Declaration of Helsinki principles. The biopsies of human livers for research purposes and their analyses have been approved by the appropriate Institutional Review Board (Inserm Institutional Review Board, 00003888), and all participants gave written informed consent. All experiments involving mice were conducted following the Guide for the Care and Use of Laboratory Animals by the National Institutes of Health (NIH) (National Academies Press, 2011) and were approved by the Institutional Animal Care and Use Committee at UT Southwestern Medical Center. All experiments involving rat perinatal cerebral HI were performed in accordance with the Swiss laws for the protection of animals and were approved by the Vaud Cantonal Veterinary Office.
Author contributions
AFF, YL, VG, JP, MCH, and BL designed the study. AFF, YL, VG, and ZZ performed biochemical and fluorescence microscopic analyses. AFF and YL performed electron microscopy analysis. VG and MS performed animal surgeries. MCH performed histological analyses. AZ, BAP, and GX performed bioinformatics analyses. MT, VP, and PER assisted with studies of livers from patients. AFF, YL, VG, JN, JS, PER, JP, MCH, and BL discussed and analyzed data. AFF and BL wrote the manuscript.
Supplementary Material
Acknowledgments
The authors thank L. Nguyen for technical assistance and H. Smith for assistance with manuscript preparation. This work was supported by NIH grants R01 CA109618 (BL), U19 AI109725 (BL), U19 AI142784 (BL), R01 DK091392 (MCH), R01 DK092461 (MCH), 5P30 CA124543 (AZ and BAP), and R01 HL138720 (JS); Cancer Prevention Research Institute of Texas grant RP120718 (BL); Fondation Leducq grant 15CBD04 (JS and BL); the Pak Foundation for Endowed Professors Collaborative Research Support (MCH); the “Institut National de la Santé et de la Recherche Médicale” (ATIP AVENIR) (PER); the “Agence Nationale pour la Recherche” (ANR-14-CE12-0011, ANR-14-CE35-0022, ANR-18-CE14-0006-01) (PER); the Swiss National Foundation (310030, 163064, 310030, 182332) (JP); and a Fondation Motrice grant (JP).
Version 1. 01/16/2020
Electronic publication
Footnotes
Conflict of interest: BL is a scientific cofounder of Casma Therapeutics, Inc.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(1):e133282.https://doi.org/10.1172/jci.insight.133282.
Contributor Information
Álvaro F. Fernández, Email: alvaro.fernandez@utsouthwestern.edu.
Yang Liu, Email: Yang.Liu3@utsouthwestern.edu.
Vanessa Ginet, Email: vanessa.Ginetpuyal@unil.ch.
Mingjun Shi, Email: MINGJUN.SHI@UTSOUTHWESTERN.EDU.
Jihoon Nah, Email: gn111@njms.rutgers.edu.
Zhongju Zou, Email: zhongju.zou@utsouthwestern.edu.
Anwu Zhou, Email: Anwu.Zhou@UTSouthwestern.edu.
Guanghua Xiao, Email: GUANGHUA.XIAO@UTSOUTHWESTERN.EDU.
Marion Tanguy, Email: tanguymarion.mt@gmail.com.
Valérie Paradis, Email: valerie.paradis@aphp.fr.
Junichi Sadoshima, Email: sadoshju@njms.rutgers.edu.
Pierre-Emmanuel Rautou, Email: perautou@yahoo.fr.
Julien Puyal, Email: JulienPierre.Puyal@unil.ch.
Ming Chang Hu, Email: ming-chang.hu@utsouthwestern.edu.
Beth Levine, Email: beth.levine@utsouthwestern.edu.
References
- 1.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24(1):24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13(5):495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bialik S, Dasari SK, Kimchi A. Autophagy-dependent cell death - where, how and why a cell eats itself to death. J Cell Sci. 2018;131(18):jcs215152. doi: 10.1242/jcs.215152. [DOI] [PubMed] [Google Scholar]
- 5.Galluzzi L, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25(3):486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, et al. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci USA. 2013;110(51):20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kheloufi M, Boulanger CM, Codogno P, Rautou PE. Autosis occurs in the liver of patients with severe anorexia nervosa. Hepatology. 2015;62(2):657–658. doi: 10.1002/hep.27597. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, et al. Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia. Autophagy. 2016;12(6):1029–1046. doi: 10.1080/15548627.2016.1164357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang G, Luk BT, Hamidy M, Zhang L, Spector SA. Induction of a Na+/K+-ATPase-dependent form of autophagy triggers preferential cell death of human immunodeficiency virus type-1-infected macrophages. Autophagy. 2018;14(8):1359–1375. doi: 10.1080/15548627.2018.1476014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginet V, et al. Dying neurons in thalamus of asphyxiated term newborns and rats are autophagic. Ann Neurol. 2014;76(5):695–711. doi: 10.1002/ana.24257. [DOI] [PubMed] [Google Scholar]
- 11.Ravikumar B, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90(4):1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 12.Wang P, Shao BZ, Deng Z, Chen S, Yue Z, Miao CY. Autophagy in ischemic stroke. Prog Neurobiol. 2018;163-164:98–117. doi: 10.1016/j.pneurobio.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Kaushal GP, Shah SV. Autophagy in acute kidney injury. Kidney Int. 2016;89(4):779–791. doi: 10.1016/j.kint.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rautou PE, Mansouri A, Lebrec D, Durand F, Valla D, Moreau R. Autophagy in liver diseases. J Hepatol. 2010;53(6):1123–1134. doi: 10.1016/j.jhep.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Matsui Y, et al. Molecular mechanisms and physiological significance of autophagy during myocardial ischemia and reperfusion. Autophagy. 2008;4(4):409–415. doi: 10.4161/auto.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheng R, Qin ZH. The divergent roles of autophagy in ischemia and preconditioning. Acta Pharmacol Sin. 2015;36(4):411–420. doi: 10.1038/aps.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Descloux C, Ginet V, Clarke PG, Puyal J, Truttmann AC. Neuronal death after perinatal cerebral hypoxia-ischemia: focus on autophagy-mediated cell death. Int J Dev Neurosci. 2015;45:75–85. doi: 10.1016/j.ijdevneu.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Decuypere JP, et al. Autophagy and the kidney: implications for ischemia-reperfusion injury and therapy. Am J Kidney Dis. 2015;66(4):699–709. doi: 10.1053/j.ajkd.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 19.Nah J, Fernández ÁF, Kitsis RN, Levine B, Sadoshima J. Does autophagy mediate cardiac myocyte death during stress? Circ Res. 2016;119(8):893–895. doi: 10.1161/CIRCRESAHA.116.309765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diederich M, Muller F, Cerella C. Cardiac glycosides: from molecular targets to immunogenic cell death. Biochem Pharmacol. 2017;125:1–11. doi: 10.1016/j.bcp.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Clausen MV, Hilbers F, Poulsen H. The structure and function of the Na,K-ATPase isoforms in health and disease. Front Physiol. 2017;8:371. doi: 10.3389/fphys.2017.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vedovato N, Gadsby DC. Route, mechanism, and implications of proton import during Na+/K+ exchange by native Na+/K+-ATPase pumps. J Gen Physiol. 2014;143(4):449–464. doi: 10.1085/jgp.201311148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui X, Xie Z. Protein interaction and Na/K-ATPase-mediated signal transduction. Molecules. 2017;22(6):E990. doi: 10.3390/molecules22060990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clausen T. Na+-K+ pump regulation and skeletal muscle contractility. Physiol Rev. 2003;83(4):1269–1324. doi: 10.1152/physrev.00011.2003. [DOI] [PubMed] [Google Scholar]
- 25.Schoner W. Endogenous cardiac glycosides, a new class of steroid hormones. Eur J Biochem. 2002;269(10):2440–2448. doi: 10.1046/j.1432-1033.2002.02911.x. [DOI] [PubMed] [Google Scholar]
- 26.Shoji-Kawata S, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494(7436):201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeisberg M, Kalluri R. Physiology of the renal interstitium. Clin J Am Soc Nephrol. 2015;10(10):1831–1840. doi: 10.2215/CJN.00640114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramann R, Wongboonsin J, Chang-Panesso M, Machado FG, Humphreys BD. Gli1+ pericyte loss induces capillary rarefaction and proximal tubular injury. J Am Soc Nephrol. 2017;28(3):776–784. doi: 10.1681/ASN.2016030297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buckalew VM. Endogenous digitalis-like factors: an overview of the history. Front Endocrinol (Lausanne) 2015;6:49. doi: 10.3389/fendo.2015.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He C, Sumpter R, Levine B. Exercise induces autophagy in peripheral tissues and in the brain. Autophagy. 2012;8(10):1548–1551. doi: 10.4161/auto.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481(7382):511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMillin GA, et al. Comparable effects of DIGIBIND and DigiFab in thirteen digoxin immunoassays. Clin Chem. 2002;48(9):1580–1584. [PubMed] [Google Scholar]
- 34.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.e03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zwerger M, Kolb T, Richter K, Karakesisoglou I, Herrmann H. Induction of a massive endoplasmic reticulum and perinuclear space expansion by expression of lamin B receptor mutants and the related sterol reductases TM7SF2 and DHCR7. Mol Biol Cell. 2010;21(2):354–368. doi: 10.1091/mbc.e09-08-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garner MH. Na,K-ATPase in the nuclear envelope regulates Na+: K+ gradients in hepatocyte nuclei. J Membr Biol. 2002;187(2):97–115. doi: 10.1007/s00232-001-0155-5. [DOI] [PubMed] [Google Scholar]
- 37.Casciola-Rosen LA, Hubbard AL. Lumenal labeling of rat hepatocyte early endosomes. Presence of multiple membrane receptors and the Na+,K(+)-ATPase. J Biol Chem. 1992;267(12):8213–8221. [PubMed] [Google Scholar]
- 38.Chang CS, Kirk RG, Lee P. Presence of immunoreactive alpha 3 subunit isoform of Na,K-ATPase in mitochondria of kidney. Kidney Int. 1998;54(2):457–463. doi: 10.1046/j.1523-1755.1998.00031.x. [DOI] [PubMed] [Google Scholar]
- 39.Haas M, Askari A, Xie Z. Involvement of Src and epidermal growth factor receptor in the signal-transducing function of Na+/K+-ATPase. J Biol Chem. 2000;275(36):27832–27837. doi: 10.1074/jbc.M002951200. [DOI] [PubMed] [Google Scholar]
- 40.Wei Y, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154(6):1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Núñez-Durán H, Fernández P. Evidence for an intracellular site of action in the heart for two hydrophobic cardiac steroids. Life Sci. 2004;74(11):1337–1344. doi: 10.1016/j.lfs.2003.07.041. [DOI] [PubMed] [Google Scholar]
- 42.Mikkaichi T, et al. Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney. Proc Natl Acad Sci USA. 2004;101(10):3569–3574. doi: 10.1073/pnas.0304987101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 44.Wu Z, Liu D, Sui Y. Quantitative assessment of hit detection and confirmation in single and duplicate high-throughput screenings. J Biomol Screen. 2008;13(2):159–167. doi: 10.1177/1087057107312628. [DOI] [PubMed] [Google Scholar]
- 45.Bian A, et al. Downregulation of autophagy is associated with severe ischemia-reperfusion-induced acute kidney injury in overexpressing C-reactive protein mice. PLoS ONE. 2017;12(9):e0181848. doi: 10.1371/journal.pone.0181848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaushal GP. Autophagy protects proximal tubular cells from injury and apoptosis. Kidney Int. 2012;82(12):1250–1253. doi: 10.1038/ki.2012.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi M, et al. Cisplatin nephrotoxicity as a model of chronic kidney disease. Lab Invest. 2018;98(8):1105–1121. doi: 10.1038/s41374-018-0063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vannucci RC, Vannucci SJ. Perinatal hypoxic-ischemic brain damage: evolution of an animal model. Dev Neurosci. 2005;27(2-4):81–86. doi: 10.1159/000085978. [DOI] [PubMed] [Google Scholar]
- 49.Ginet V, Puyal J, Clarke PG, Truttmann AC. Enhancement of autophagic flux after neonatal cerebral hypoxia-ischemia and its region-specific relationship to apoptotic mechanisms. Am J Pathol. 2009;175(5):1962–1974. doi: 10.2353/ajpath.2009.090463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ginet V, et al. Involvement of autophagy in hypoxic-excitotoxic neuronal death. Autophagy. 2014;10(5):846–860. doi: 10.4161/auto.28264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.