Transient invaders can cause lasting shifts in community composition and function.
Abstract
Microbial dispersal often leads to the arrival of outsider organisms into ecosystems. When their arrival gives rise to successful invasions, outsider species establish within the resident community, which can markedly alter the ecosystem. Seemingly less influential, the potential impact of unsuccessful invaders that interact only transiently with the community has remained largely ignored. Here, we experimentally demonstrate that these transient invasions can induce a lasting transition to an alternative stable state, even when the invader species itself does not survive the transition. First, we develop a mechanistic understanding of how environmental changes caused by these transient invaders can drive a community shift in a simple, bistable model system. Beyond this, we show that transient invaders can also induce switches between stable states in more complex communities isolated from natural soil samples. Our results demonstrate that short-term interactions with an invader species can induce lasting shifts in community composition and function.
INTRODUCTION
Microbes are extremely adept at dispersal (1, 2), their migrations allowing them to colonize the most remote environments such as young ocean crust (3), medical implants (4), or even our space stations (5). Although this flow of dispersed microbes could homogenize microbial communities around the globe, countless microbial ecosystems nonetheless exhibit distinctive community stability (6). Not only this, but microbial communities can display different regimes known as alternative stable states (7–13), which, in turn, determine ecosystem functions such as host health. For example, the opportunistic pathogen Clostridium difficile can take over the gut microbiome of susceptible hosts, leading to an unhealthy, highly persistent community state (14). Moreover, microbial ecosystems often undergo transitions between alternative stable states in response to environmental disturbance (15–17), analogous to regime shifts observed in macroecosystems under stress (18–20). Among the many different stresses that can alter microbial ecosystems, however, there is still very little understanding of how microbial dispersal affects the stability of communities.
Much like other kinds of perturbations, the arrival of invader species (21) can markedly alter the structure of the resident community. Two major outcomes are typically analyzed in microbial invasions (22): establishment of the invader, which leads to changes in community composition (23), and community resilience, in which the invader is eliminated and the community remains unaltered (24). Beyond these two classical outcomes, however, the idea that transient microbiota can affect microbial ecosystems (25–27) has only recently been recognized. In the human gut microbiome, for example, many ingested microbes merely inhabit the gut temporarily, though, in some cases, they can potentially affect both the functionality and stability of the resident community (28). Nevertheless, the mechanisms allowing these transient microbes to interfere with the state of the resident community are very rarely understood (29)—specifically, whether community changes would last beyond the extinction of the invader has remained unexplored. This begs a better understanding of how transient community members affect microbial ecosystems in the long term.
RESULTS
Transient invaders can induce community shifts in a bistable model ecosystem
To explore how invasions affect multistable communities, we began by using a minimal model system in which two bacterial species mutually inhibit each other. As shown in our previous work, coculturing Corynebacterium ammoniagenes (Ca) and Lactobacillus plantarum (Lp) under a serial dilution protocol leads to two alternative outcomes. Lp can outcompete its partner by modifying the pH toward acidic values in which Ca cannot grow. Alternatively, Ca can induce a pH change toward alkaline values that inhibits Lp growth. In this way, either Ca or Lp can grow to dominate the ecosystem, depending on their initial relative abundance (30).
To determine whether both outcomes correspond to stable states of the system, we cocultured these two species under serial dilutions including a low migration rate, which we applied by adding fresh cells of each species into the ecosystem after every dilution cycle (Fig. 1A, fig. S1, and Materials and Methods). Despite this migration, the ecosystem still exhibited two alternative outcomes (Fig. 1B). This indicates that the two outcomes constitute alternative stable states of the ecosystem, each of them with a basin of attraction (Fig. 1B) that allows the system to resist some degree of stress or perturbation, such as moderate migration episodes.
Fig. 1. Transient invaders can induce shifts between alternative stable states in a laboratory ecosystem.
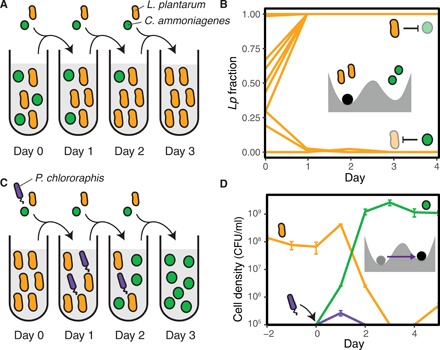
(A) We exposed cocultures of Lp and Ca to a serial dilution protocol that includes daily migration of fresh cells from both species. (B) Average fraction (three replicates, SE smaller than linewidth) of Lp cells in the community at the end of each dilution cycle, as described in (A). Depending on the initial species fraction, cocultures reach a different outcome in which either species grows to dominate the system. The inset cartoon shows a mechanical analog of the ecosystem: Each of the two basins of attraction can keep the marble (the community) in an alternative stable state. (C) We explored the effects of invasions into this bistable ecosystem. The cartoon shows an unsuccessful invasion by Pc that nevertheless induced a shift toward an alternative stable state. (D) Time series for the cell densities during an unsuccessful invasion by Pc (bars show the SE of three replicates). The inset cartoon depicts this invasion event as a perturbation that drives the system toward an alternative basin of stability, where it remains after the perturbation is gone.
We next studied the effects of introducing an invader species into our bistable microbial ecosystem. For this purpose, we first mixed Lp and Ca cells at a 95:5 ratio in replicate cocultures and applied two daily cycles of dilution and migration, allowing the system to reach one of its stable states. During the third dilution cycle, we inoculated an invader species into the coculture, simulating a single shot invasion (Fig. 1C). As expected, we observed a variety of invasion outcomes out of seven different invader species. Only one invader candidate, namely, Bacillus cereus (Bc), succeeded at establishing itself in the community (fig. S2), while the other six invader species performed unsuccessful invasions (fig. S3). Escherichia coli (Ec) or Pseudomonas veronii (Pv), for example, both mirrored classical unsuccessful invasions, as these invader species neither survived nor significantly affected the ecosystem.
We also identified unsuccessful invasions that induced long-term changes in the resident community, which is an invasion outcome beyond the two classical ones. In particular, an invasion by Pseudomonas chlororaphis (Pc) led to a shift toward the stable state governed by Ca. Pc reaches extinction (falls below the detection limit) within 48 hours following its arrival into the community (Fig. 1D). Despite remaining for only a short time in the system (fig. S4), Pc affects the competition outcome between Lp and Ca, eventually leading to an increase in the Ca fraction that allows the community to reach its alternative stable state. This kind of unsuccessful invasion can be seen as a perturbation that drives the resident community toward the basin of attraction of an alternative stable state, where it remains after the perturbation ceases.
Environmental modulation by transient invaders can drive shifts between alternative stable states
In addition to a change in species abundances, we observed a rapid pH shift in the microbial environment following the invader inoculation (Fig. 2A). pH measurements at the end of each daily cycle revealed consistent acidification of the media in the initial community state: The pH dropped from 6.5 to 3.7 (±0.1) during each 24-hour cycle before the invasion. In contrast, the system persistently reached highly alkaline values (pH 9.2 ± 0.1) in every cycle following the invader inoculation. We hypothesized that the invader could have induced the rapid shift in pH, this environmental change driving the transition between alternative states.
Fig. 2. Feedback loops between microbial growth and pH can determine the community impact of transient invaders.
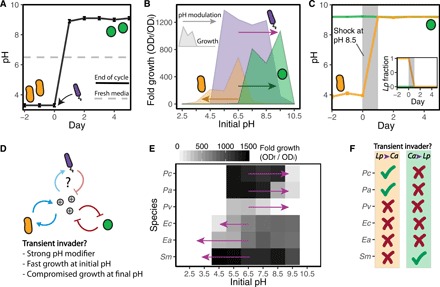
(A) Observed shift in pH after a Pc invasion into the stable state governed by Lp. The solid line stands for the pH at the end of each daily cycle. During each cycle, microbes induce changes in the pH of the fresh medium (dashed line) in which they were diluted into. (B) pH range in which Lp (in orange), Pc (purple), and Ca (green) exhibit growth, indicated by the fold growth in OD after monocultures spent 24 hours in highly (100 mM phosphate) buffered media. Arrows indicate how each species modifies the pH in standard (10 mM phosphate) buffer conditions. The head of the arrow points toward the pH value reached after a 24-hour culture that started at pH 6.5. (C) A temporary shock in which cells were transferred to alkaline medium during a single daily cycle (gray area) induced a transition from the Lp state to the Ca state (orange), while cocultures in the Ca state (green) remained unaltered. (D) Three main features observed in species that can act as transient invaders, as predicted by a minimal model that considers feedbacks between microbial growth and pH. (E) Fold growth in highly buffered media for monocultures from six different species, and pH modification induced by those species in standard buffer conditions (arrows). (F) Ticks and crosses indicate which species acted as transient invaders inducing community switches: Pa and Pc induced switches toward the alkaline state, and Sm was able to cause a switch toward the acidic state. Data in (B) and (D) correspond to average from four replicates (fold growth SE ≤ 2, pH modification SE ≤ 0.1).
Feedback between microbial growth and pH (30) was previously shown to drive the interaction between the resident species, Ca and Lp. Measuring the pH change after culturing Ca for 24 hours showed that this species was able to remarkably alkalize the environment (Fig. 2B). Moreover, culturing Ca at different pH values revealed that its growth is strongly favored in alkaline environments, showing that Ca modifies the environment in a way that promotes its own growth. Similarly, Lp is able to acidify the environment in a way that is sustainable for itself but not for Ca. The invader species Pc displays a different pattern, since Pc induces a pH increase that it is not beneficial for itself (Fig. 2B). Instead, Pc inhibits its own growth by driving the pH toward highly alkaline values, where Ca grows optimally. We also studied the effects of externally applied pH shocks, which revealed that temporary perturbations in the pH were sufficient to induce transitions between the Ca and Lp stable states (Fig. 2C and fig. S5). Together, these results show that pH modification by an invader species, namely, Pc, can trigger switches from the stable state governed by Lp toward the one governed by Ca.
To better understand switches between alternative stable states that result from environmentally mediated microbial interactions, we developed a theoretical model incorporating the main features of our experimental system (fig. S6) including daily dilutions, migration, and pH modification by microbes. This minimal model uses simple step functions to determine (i) the pH range in which a species can actively modify the pH, and (ii) the pH range in which each species either grows or is harmed. This simple model is able to recapitulate the observed outcomes for our bistable community, including invader-induced transitions between stable states (fig. S6). Furthermore, we identified three specific features allowing unsuccessful invaders to cause switches between alternative stable states in in silico communities (Fig. 2D). To disrupt the initial stable state, the invader first has to be able to overcome the resident species at modifying the environment. This can be achieved through a combination of (i) fast growth in the existing pH and (ii) a high ability to modify the pH. As a consequence of these two requirements, the model predicts a minimum inoculum size for the invader to cause shifts in the community, a prediction that we further validated experimentally (fig. S6). Last, losing the competition against the resident species once the environment has changed makes the invader unsuccessful. This leads to the third requirement: (iii) Transient invaders exhibit compromised growth in the final pH. As shown in Fig. 2B, Pc exhibits these three generic features that enable it to act as a transient invader that alters the state of the resident community.
To determine whether these three generic features predict which invaders can trigger switches between alternative stable states, we characterized the feedback between microbial growth and pH for the six unsuccessful invaders mentioned previously (fig. S3). Among those candidates, the species Pseudomonas aurantiaca (Pa) exhibited very similar features to those of the transient invader Pc (Fig. 2E). An invasion by Pa into the Lp stable state also induced a community switch in the bistable ecosystem (Fig. 2F and fig. S3). In contrast, an invasion by Pv did not result in a community switch. While Pv also alkalizes the environment, its lower growth rate potentially compromises its ability to overcome the initially abundant Lp species, as predicted by the model (Fig. 2D and fig. S6). The last three unsuccessful invaders, namely, Enterobacter aerogenes (Ea), Ec, and Serratia marcescens (Sm), modify the pH toward acidic values, and as expected based on the direction of the pH change, they were not able to induce a shift toward the alkaline (Ca) state. Instead, when the ecosystem was initially in the Ca stable state, we observed transient invasions by Sm that can induce a transition toward the acidic state governed by Lp (fig. S7), showing that the direction of the switch can be controlled by inoculating different invader species. Analogous to the case of Pa and Pc, the transient invader Sm also exhibits the highest growth rates among the three species that induce acidification (Fig. 2E). Together, these results show that how invaders modify the environment and react to it can determine the outcome of microbial invasions in a relatively simple bistable community.
Transient invaders can induce lasting shifts in complex microbial communities
Given that we used a minimal model system that exhibits bistability between two laboratory strains, the question arises of whether analogous community dynamics could unfold from transient invasions in more complex microbial ecosystems. To address this question, we propagated a natural soil sample in the laboratory under serial dilution and looked for signatures of multistability. After nine daily cycles, 74 of 88 replicate cultures originating from the same soil sample exhibited optical densities above the detection limit. Analysis of the time series for the pH (Fig. 3A), as well as optical density (OD) and plating on agar suggested that the different replicates reached a wide variety of community states (fig. S8) exhibiting different species composition. These results are in agreement with recent findings showing that soil communities can relax into alternative community compositions when propagated in laboratory environments (31).
Fig. 3. Transient invaders can drive transitions between stable states of a community isolated from the soil.
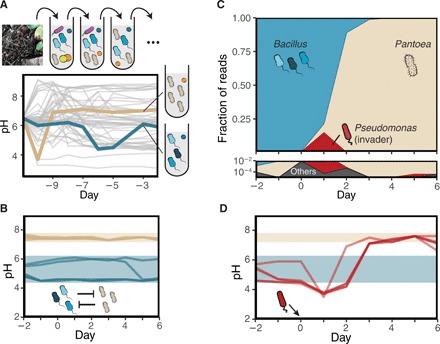
(A) Time series for the pH of 39 replicates of a soil community exposed to serial dilutions. At the end of nine cycles, these replicates showed signs of pH stabilization (fig. S8). The colored (blue and cream) lines correspond to two cultures in which the community composition was also stable. 16S sequencing revealed that the community in cream was highly dominated by a Pantoea genus, while the one in blue was governed by Bacillus (blue). (B) Time series for the pH as the Pantoea and Bacillus communities were exposed to migration from each other. Measures for six replicates for each stable community are shown. (C) Time series for the community composition during an invasion by Pc into the Bacillus community as revealed by 16S amplicon sequencing. The Bacillus community was exposed to a daily dilution protocol including migration from the Pantoea community as in (B). (D) Time series for the pH during the same invasion presented in (C), along with three additional replicates. For reference, shaded areas indicate the observed pH range for each community in the presence of migration [as in (B)].
We identified two community states that were consistently able to resist low doses of migration from one another (Fig. 3B and fig. S9). Analogous to the results in Fig. 1, A and B, this indicated the presence of alternative stable states in the microcosms originated from the soil community. Sequencing revealed that one community was dominated by a Pantoea and the other was dominated by closely related Bacillus strains that display a wide range of pH-growth dynamics and colony morphologies (fig. S10).
Next, we inoculated different invader species into the two stable states governed by either Bacillus or Pantoea. Pc was consistently able to cause a switch toward the more alkaline stable state, the one dominated by Pantoea. Figure 3C shows the time series for a transient invasion by Pc as revealed by 16S amplicon sequencing. Before the invasion, the community is dominated by the Bacillus genus, but after inoculation of Pc on day 0, there is an increase of Pc accompanied by an increase in the abundance of Pantoea. This increase in Pantoea abundance continues even as the transient invader Pc is outcompeted. At the end of the experiment, Pantoea was dominant, Pc was extinct, and the abundance of Bacillus reads was consistent with the low migration rate applied at each dilution cycle. We performed three additional replicates of Pc invasions, all of them exhibiting analogous dynamics for both the pH (Fig. 3D) and community composition (Fig. 3C and fig. S11). As before, Pa was also able to act as a transient invader that induces a transition to the more alkaline community state (fig. S11). Notably, none of the considered invader species was able to induce a shift toward the stable state dominated by Bacillus, indicating a stronger stability of the Pantoea stable state against these invaders. Summarizing, these results show that transient invaders can induce shifts between alternative stable states in more complex microbial ecosystems derived from natural communities.
DISCUSSION
Our work demonstrates that microbial communities can experience lasting shifts in community composition induced by transient members. Analyzing both theoretically and experimentally a minimal bistable community, we have shown that feedback between microbial growth and environmental conditions (specifically, the pH) can mechanistically drive these transitions. While our results enhance the relevance of environmentally mediated interactions (30, 32, 33) in the microbial world, phenomenological models such as the generalized Lotka-Volterra model (34) and the stable marriage problem (27) indicate that analogous community dynamics could unfold from different microbial interactions. Interactions with the host are of special relevance in the case of gut microbiomes, where pathogenic invaders can induce systemic immune responses, such as bacterial adherence blocking and inflammation, that, in turn, affect the resident bacterial community (35). By selectively targeting specific bacterial taxa, phages (36) are exceptional candidates to act as transient invaders that reshape bacterial communities. Analogous boom-and-bust invasions (37) have also been reported as a main cause of macroecosystem disruption. Invasive herbivores, for example, can markedly change island landscapes before risking extinction as they deplete their main resources. Together, these findings suggest that transient invaders could frequently have a lasting impact also in natural microbiomes.
To date, a few longitudinal studies have linked microbial community shifts to transient invaders. Mallon et al. proposed that unsuccessful invaders can steer the community away from the invader’s niche (25) in a soil microbiome, although it remained unclear whether those changes lead the system to an alternative stable state. Notably, our analysis of the pH preference of the different laboratory strains is consistent with a transient invader that drives the community to an alternative niche. Although lacking strong evidence of underlying mechanisms, long-term community changes following transient invasions have also been observed in predator-prey microcosms (26), freshwater phytoplankton communities (38), as well as after infections in the human gut microbiome (39). Our work suggests that community switches after unsuccessful invasions may not be rare, as three of six different unsuccessful invaders induced a community switch in the minimal bistable community (two of six in the case of the soil communities). Given the currently increasing availability of temporal datasets for natural microbiomes, we expect that future analyses will frequently reveal transitions between alternative community states induced by microbial invaders.
In biomedical research, the frequent failure of probiotics (28) at establishing in the gut community has generated much skepticism about their efficacy. Our results show, however, that the manipulation of transient interactions has tremendous potential to control the long-term dynamics of microbial communities. Thus, exploring new avenues in which a set of transient microorganisms can steer the community into a healthy state (40) could lead to less intrusive interventions for the host. Beyond host-associated microbiota, future work could apply similar treatments to different microbial ecosystems (e.g., agricultural soil) and also explore the effects of coinvasions in which multiple invader species are involved.
MATERIALS AND METHODS
Media, buffer, cultures, and plating
Overnight precultures of laboratory strains were performed in nutrient media (NM) (30): yeast extract (10 g/liter) and soytone (10 g/liter) (both Becton Dickinson, Franklin Lakes, USA) and 10 mM sodium phosphate (pH 7). The experiments were performed in base media (30) (hereafter, BM) supplemented with glucose and urea. The stock of BM was prepared as follows: yeast extract (1 g/liter), soytone (1 g/liter), 10 mM sodium phosphate buffer, 0.1 mM CaCl2, 2 mM MgCl2, NiSO4 (4 mg/liter), MnCl2 (50 mg/liter), and 1X Trace Metals Mixture (Teknova, Hollister, CA), pH 6.5. A standard concentration of 10 mM sodium phosphate in BM was used in all the experiments, with the exception of experiments measuring the growth dependence on the pH (Fig. 2D) of monocultures, in which BM contained 100 mM sodium phosphate. Supplemented base media (SBM) for the experiments was prepared daily by adding glucose (10 g/liter) and urea (8 g/liter) to BM. All media were filter-sterilized using VWR Bottle Top Filtration Unit (VWR, Radnor, USA).
Before plating for colony-forming units (CFU) counting, experimental cultures were diluted in phosphate-buffered saline (PBS; Corning, New York, USA). Plating was performed on tryptic soy broth (TSB; Teknova, Hollister, USA) with 2.5% agar (Becton Dickinson, Franklin Lakes, USA), in which we adjusted the pH to different values for selective plating (see below).
Overnight precultures took place in 5 ml of NM, inside 50-ml Falcon tubes for 24 hours in the case of Ca and Lp and 16 hours for the rest of strains, shaking at 250 rpm on a New Brunswick Innova 2100 shaker (Eppendorf, Hauppauge, NY, USA). Experimental cultures took place in 96-deepwell plates covered with AeraSeal adhesive sealing films (Excel Scientific, Victorville, USA), shaking at 1350 rpm using a Heidolph platform shaker (Titramax 100, Heidolph North America, Elk Grove Village, IL, USA). All precultures and cultures were incubated at 30°C, with a relative humidity of 50%.
Laboratory strains
Lp (ATCC 8014), Ca (ATCC 6871), Pc (ATTC 9446), Ea (ATCC 13048), Pa (ATCC 33663), Pv (ATCC 423 700474), and Sm (ATCC 13880) were obtained from the American Type Culture Collection. Bc was obtained from Ward’s Scientific Catalog. Ec MC4100 (CGSC #6152) was obtained from the E. coli Genetic Stock Center.
Soil microcosms
The soil was sampled from a lawn in Cambridge, Massachusetts, at a depth of ~15 cm. About 20 grains of soil (~0.5 g) were diluted into 20 ml of PBS, vortexed at intermediate speed for 30 s, and then incubated on a shaker at 250 rpm. After 30 min, the sample was allowed to settle for 5 min and the supernatant was transferred to a new Falcon tube. Aliquots (7 μl) of this supernatant were then transferred into 203 μl of SBM in a 96-deepwell plate, inoculating a total of 88 replicates. We applied seven daily dilution cycles to the 88 replicates as explained in the main text, and at the end of day 7 (which corresponds to day −4 in Fig. 3A), we froze the resulting communities (transferring them to a final concentration of 40% glycerol and then storing at −80°C).
Invasions to the soil communities were initiated by sampling the −80°C stocks to inoculate 203 μl of SBM in 96-deepwell plates. Then, we exposed these communities to daily dilution cycles with migration (see the “Migration” section below). To facilitate the thawed samples to recover stability after coming from −80°C, migration was not applied during the first daily cycle. Invaders were inoculated along the dilution protocol after the end of the fourth cycle. We used Ea, Ec, Pa, Pc, Pv, and Sm as candidate invader species for the soil communities.
Daily dilutions
At the end of every daily cycle, a 30-fold dilution was applied by transferring 7 μl of the experimental cultures into 203 μl of fresh SBM using a Viaflo 96-well pipettor (Viaflo 96, Integra Biosciences, Hudson, USA; Viaflo settings: pipet/mix program, aspirating 7 μl, three mixing cycles, mixing volume of 10 μl, speed of 6).
Migration
Overnight cultures of both Ca and Lp were washed in 15 ml of BM. To increase accuracy at adjusting the population densities of the migrant cells, we first adjusted both monocultures to an OD/cm of approximately 2.0 and then performed a second round of adjustment to a final OD/cm of 0.37 for Ca and 0.24 for Lp. We then mixed 10 ml of each monoculture, resulting in 20 ml of the migrant cells mix. Using a Viaflo 96-well pipettor, 3 μl of a fresh migrant mix was inoculated in the experimental cultures right after each dilution cycle (Viaflo settings: pipet/mix program, aspirating 3 μl, three mixing cycles, mixing volume of 10 μl, speed of 6). This resulted in a daily inoculation of (1.2 ± 0.1) × 105 fresh cells from each species into the culture (fig. S1).
To apply migrations between the stable states of the soil communities, at the beginning of the experiment, we initiated several control cultures from both stable states. These control cultures were exposed to 30-fold daily dilutions in parallel to the rest of experimental cultures. During each dilution cycle, samples of these control cultures were diluted by 100-fold in BM using a Viaflo 96-well pipettor. The resulting dilutions from each stable state were mixed at a 1:1 ratio so that the composition of migrant cells includes species from both stable states. Migration was applied by transferring 3 μl of the resulting mix into wells with fresh media, where the experimental cultures would also be propagated through the regular 30-fold dilution. This resulted in 4 × 10−3 volume ratio of migrant cultures relative to the regularly diluted community at the beginning of each daily cycle.
Invader inoculation
Overnight monocultures of the strains used as invaders were washed in 15 ml of BM. Then, we adjusted the OD/cm to obtain a final population density of approximately 3.3 × 108 cells/ml (OD versus CFU assays were performed previously to guide this adjustment). During the dilution cycle indicated in each figure caption, we inoculated 3 μl of the invader monocultures, resulting in an inoculation of approximately 105 invader cells (<10% of the total number of cells transferred at the beginning of the cycle; fig. S1). The same procedure was used for the experiments in fig. S6F, followed by a series of additional 10-fold dilutions to obtain the reported inoculum sizes. A higher inoculum size of 3 × 106 was used in fig. S7 for Sm to induce transitions toward the Lp state.
Characterization of the soil Bacillus strains
To isolate the three bacillus strains used in fig. S10, we plated a diluted sample of the community (corresponding to day −4 in Fig. 3A, cream-colored curve) into a TSB agar plate. We picked each of the three colony morphologies and performed an overnight culture in NM, and then we stored the monocultures (−80°C, 40% glycerol). Plating samples of each overnight monoculture revealed a unique colony morphology for each monoculture, confirming a heritable colony morphology that is distinctive of each strain (fig. S10).
For the competition experiments between the isolated Bacillus strains (fig. S10), we grew colonies from the frozen isolates (1 day on TSB agar plates, 30°C). We then picked individual colonies to perform overnight cultures in NM. The next morning, we washed the monocultures in 15 ml of BM and then measured their OD and adjusted to a final population density of 3.3 × 108 CFU/ml (a previous independent CFU versus OD assay was performed to guide the population density adjustments). Then, we mixed the monocultures at the indicated ratios and inoculate 3 μl of the mixes into 207 μl of SBM to initiate the daily dilution cycles.
Estimation of population densities (CFU/ml)
For CFU counting, 10-μl droplets of PBS-diluted cultures were plated on agar. We used agar plates at pH 5 for selective plating of Lp, pH 10 to select for Ca colonies, and pH 7 to count the abundance of the rest of the strains (including those in the soil communities). Colonies from the different invaders generally grew faster than Ca and Lp on pH 7 agar, which allowed measurements of invader cell densities at very low abundances (down to relative fractions of approximately 10−4).
To prepare the 10-μl droplets, we serially diluted the experimental cultures via 10-fold dilutions (maximal dilution factor was 10−7) with a Viaflo 96-well pipettor using the program “pipet/mix” (pipetting volume, 20 μl; mixing volume, 180 μl; mixing cycles, 5; mixing and pipetting speed, 8). Droplets (10 μl) were then transferred to 150-mm-diameter agar plates with the 96-well pipettor (program “reverse pipette”; uptake volume, 20 μl; released volume, 10 μl; pipetting speed, 2). Droplets were allowed to dry, and the plates were incubated at room temperature for 1 to 2 days until colonies were visible. The different dilution steps allowed us to find a dilution at which colonies could be optimally counted with a Leica dissecting microscope (between ~5 and ~50 colonies). Up to three plating replicates per condition were performed to increase accuracy at measuring population densities.
pH measurement
To measure the pH of the microbial cultures, 150-μl samples were transferred into 96-well polymerase chain reaction plates (VWR, Radnor, USA), and the pH was measured using a pH microelectrode (Orion, PerpHecT, ROSS).
16S ribosomal RNA gene sequencing and analysis
The DNA extractions were performed using an Agencourt DNAdvance A48705 extraction kit (Beckman Coulter, Indianapolis, IN, USA) following the provided protocol. The obtained DNA was used for 16S amplicon sequencing targeting the V4-V5 region. The sequencing was done on an Illumina MySeq by the Center for Comparative Genomics and Evolutionary Bioinformatics–Integrated Microbiome Resource at the Dalhousie University, Halifax, NS, Canada.
We used the R package DADA2 (41) to obtain the amplicon sequence variants (ASVs) as described by Callahan et al. (42). Taxonomic identities were assigned to the ASVs by using the GreenGenes Database Consortium (version 13.8) (43) as a reference database.
Supplementary Material
Acknowledgments
We thank the members of the Gore laboratory for dedicated feedback. D.R.A. specially thanks J. Friedman, G. Leventhal, X. Yu, L. Seoane, and R. Solé for inspiring discussions. We also thank S. Umale for helping to collect the data in fig. S10. Funding: This research was funded by NIH (R01-GM102311). Author contributions: Conceptualization: D.R.A. and J.G.; data curation: D.R.A.; formal analysis: D.R.A., C.R., and J.G.; funding acquisition: J.G.; investigation: D.R.A.; methodology: D.R.A. and C.R.; project administration: D.R.A., C.R., and J.G.; resources: D.R.A.; software: D.R.A.; supervision: J.G.; validation: J.G.; visualization: D.R.A.; writing of original draft: D.R.A., C.R., and J.G. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The 16S sequencing dataset and the code used to obtain ASV abundances and taxonomy can be accessed at https://doi.org/10.5061/dryad.gb5mkkwkk. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/8/eaay8676/DC1
Fig. S1. Number of cells transferred to fresh media during daily dilution and migration steps.
Fig. S2. Successful invasion by Bc.
Fig. S3. Six different species performed unsuccessful invasions into the Ca and Lp ecosystem.
Fig. S4. The invader Pc thrives and decays within the 24 hours following inoculation into the Lp stable state.
Fig. S5. Temporary perturbations in pH induce shifts between the Lp and Ca states.
Fig. S6. Minimal model incorporating feedbacks between microbial growth and pH predicts a minimum inoculum size for the invasions to induce community shifts.
Fig. S7. The invader Sm can induce transitions from the alkaline (Ca) to the acidic (Lp) stable state.
Fig. S8. Early dynamics of soil communities in laboratory environments.
Fig. S9. Mutual resilience against migration between the Bacillus and Pantoea soil communities.
Fig. S10. Different behavior in laboratory microcosms for three genetically similar soil isolates.
Fig. S11. Both Pc and Pa can induce transitions from the Bacillus to the Pantoea stable states.
Reference (44)
REFERENCES AND NOTES
- 1.Martiny J. B. H., Bohannan B. J. M., Brown J. H., Colwell R. K., Fuhrman J. A., Green J. L., Horner-Devine M. C., Kane M., Krumins J. A., Kuske C. R., Morin P. J., Naeem S., Øvreås L., Reysenbach A.-L., Smith V. H., Staley J. T., Microbial biogeography: Putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Finlay B. J., Global dispersal of free-living microbial eukaryote species. Science 296, 1061–1063 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Orcutt B. N., Bach W., Becker K., Fisher A. T., Hentscher M., Toner B. M., Wheat C. G., Edwards K. J., Colonization of subsurface microbial observatories deployed in young ocean crust. ISME J. 5, 692–703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renata Arciola C., Campoccia D., Montanaro L., Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 16, 397–409 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Lang J. M., Coil D. A., Neches R. Y., Brown W. E., Cavalier D., Severance M., Hampton-Marcell J. T., Gilbert J. A., Eisen J. A., A microbial survey of the International Space Station (ISS). PeerJ. 5, e4029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shade A., Peter H., Allison S. D., Baho D. L., Berga M., Bürgmann H., Huber D. H., Langenheder S., Lennon J. T., Martiny J. B. H., Matulich K. L., Schmidt T. M., Handelsman J., Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3, 417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vangay P., Johnson A. J., Ward T. L., Al-Ghalith G. A., Shields-Cutler R. R., Hillmann B. M., Lucas S. K., Beura L. K., Thompson E. A., Till L. M., Batres R., Paw B., Pergament S. L., Saenyakul P., Xiong M., Kim A. D., Kim G., Masopust D., Martens E. C., Angkurawaranon C., Mac Gready R., Kashyap P. C., Culhane-Pera K. A., Knights D., US immigration westernizes the human gut microbiome. Cell 175, 962–972.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J., Liu W., Deng Y., Jiang Y.-H., Xue K., He Z., Van Nostrand J. D., Wu L., Yang Y., Wang A., Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. MBio 4, e00584-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leventhal G. E., Boix C., Kuechler U., Enke T. N., Sliwerska E., Holliger C., Cordero O. X., Strain-level diversity drives alternative community types in millimetre-scale granular biofilms. Nat. Microbiol. 3, 1295–1303 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Friedman J., Higgins L. M., Gore J., Community structure follows simple assembly rules in microbial microcosms. Nat. Ecol. Evol. 1, 0109 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Lahti L., Salojärvi J., Salonen A., Scheffer M., de Vos W. M., Tipping elements in the human intestinal ecosystem. Nat. Commun. 5, 4344 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonnenburg E. D., Smits S. A., Tikhonov M., Higginbottom S. K., Wingreen N. S., Sonnenburg J. L., Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costea P. I., Hildebrand F., Arumugam M., Bäckhed F., Blaser M. J., Bushman F. D., de Vos W. M., Ehrlich S. D., Fraser C. M., Hattori M., Huttenhower C., Jeffery I. B., Knights D., Lewis J. D., Ley R. E., Ochman H., O’Toole P. W., Quince C., Relman D. A., Shanahan F., Sunagawa S., Wang J., Weinstock G. M., Wu G. D., Zeller G., Zhao L., Raes J., Knight R., Bork P., Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 3, 8–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seekatz A. M., Rao K., Santhosh K., Young V. B., Dynamics of the fecal microbiome in patients with recurrent and nonrecurrent Clostridium difficile infection. Genome Med. 8, 47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai L., Vorselen D., Korolev K. S., Gore J., Generic indicators for loss of resilience before a tipping point leading to population collapse. Science 336, 1175–1177 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Tropini C., Moss E. L., Merrill B. D., Ng K. M., Higginbottom S. K., Casavant E. P., Gonzalez C. G., Fremin B., Bouley D. M., Elias J. E., Bhatt A. S., Huang K. C., Sonnenburg J. L., Transient osmotic perturbation causes long-term alteration to the gut microbiota. Cell 173, 1742–1754.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dal Bello M., Rindi L., Benedetti-Cecchi L., Temporal clustering of extreme climate events drives a regime shift in rocky intertidal biofilms. Ecology 100, e02578 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Rocha J. C., Peterson G., Bodin Ö., Levin S., Cascading regime shifts within and across scales. Science 362, 1379–1383 (2018). [DOI] [PubMed] [Google Scholar]
- 19.M. Scheffer, Critical Transitions in Nature and Society (Princeton Univ. Press, 2009). [Google Scholar]
- 20.R. V. Solé, J. Bascompte, Self-organization in Complex Ecosystems (Princeton Univ. Press, 2006). [Google Scholar]
- 21.C. S. Charles S. Elton, The Ecology of Invasions by Animals and Plants (University of Chicago Press, 2000). [Google Scholar]
- 22.Mallon C. A., Van Elsas J. D., Salles J. F., Microbial invasions: The process, patterns, and mechanisms. Trends Microbiol. 23, 719–729 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Kinnunen M., Dechesne A., Albrechtsen H.-J., Smets B. F., Stochastic processes govern invasion success in microbial communities when the invader is phylogenetically close to resident bacteria. ISME J. 12, 2748–2756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Roy K., Marzorati M., Negroni A., Thas O., Balloi A., Fava F., Verstraete W., Daffonchio D., Boon N., Environmental conditions and community evenness determine the outcome of biological invasion. Nat. Commun. 4, 1383–1385 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Mallon C. A., Le Roux X., Van Doorn G. S., Dini-Andreote F., Poly F., Salles J. F., The impact of failure: Unsuccessful bacterial invasions steer the soil microbial community away from the invader’s niche. ISME J. 12, 728–741 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olito C., Fukami T., Long-term effects of predator arrival timing on prey community succession. Am. Nat. 173, 354–362 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Goyal A., Dubinkina V., Maslov S., Multiple stable states in microbial communities explained by the stable marriage problem. ISME J. 12, 2823–2834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derrien M., van Hylckama Vlieg J. E. T., Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 23, 354–366 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Kleerebezem M., Binda S., Bron P. A., Gross G., Hill C., van Hylckama Vlieg J. E. T., Lebeer S., Satokari R., Ouwehand A. C., Understanding mode of action can drive the translational pipeline towards more reliable health benefits for probiotics. Curr. Opin. Biotechnol. 56, 55–60 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Ratzke C., Gore J., Modifying and reacting to the environmental pH can drive bacterial interactions. PLOS Biol. 16, e2004248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.C. Ratzke, J. Barrere, J. Gore, Strength of species interactions determines biodiversity and stability in microbial communities. bioRxiv 671008 [Preprint]. 13 June 2019. 10.1101/671008. [DOI] [PubMed]
- 32.Niehaus L., Boland I., Liu M., Chen K., Fu D., Henckel C., Chaung K., Miranda S. E., Dyckman S., Crum M., Dedrick S., Shou W., Momeni B., Microbial coexistence through chemical-mediated interactions. Nat. Commun. 10, 2052 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J., Prindle A., Humphries J., Gabalda-Sagarra M., Asally M., Lee D.-y. D., Ly S., Garcia-Ojalvo J., Süel G. M., Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 523, 550–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller T. E., terHorst C. P., Burns J. H., The ghost of competition present. Am. Nat. 173, 347–353 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Gopalakrishnan V., Helmink B. A., Spencer C. N., Reuben A., Wargo J. A., The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 33, 570–580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shkoporov A. N., Hill C., Bacteriophages of the human gut: The ‘Known Unknown’ of the microbiome. Cell Host Microbe 25, 195–209 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Simberloff D., Gibbons L., Now you See them, Now you don’t! – Population crashes of established introduced species. Biol. Invasions 6, 161–172 (2004). [Google Scholar]
- 38.Buchberger F., Stockenreiter M., Unsuccessful invaders structure a natural freshwater phytoplankton community. Ecosphere 9, e02158 (2018). [Google Scholar]
- 39.David L. A., Materna A. C., Friedman J., Campos-Baptista M. I., Blackburn M. C., Perrotta A., Erdman S. E., Alm E. J., Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao N., Cubillos-Ruiz A., Cameron D. E., Collins J. J., Probiotic strains detect and suppress cholera in mice. Sci. Transl. Med. 10, eaao2586 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P., DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Callahan B. J., Sankaran K., Fukuyama J. A., McMurdie P. J., Holmes S. P., Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. F1000Res. 5, 1492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., Andersen G. L., Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ratzke C., Denk J., Gore J., Ecological suicide in microbes. Nat. Ecol. Evol. 2, 867–872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/8/eaay8676/DC1
Fig. S1. Number of cells transferred to fresh media during daily dilution and migration steps.
Fig. S2. Successful invasion by Bc.
Fig. S3. Six different species performed unsuccessful invasions into the Ca and Lp ecosystem.
Fig. S4. The invader Pc thrives and decays within the 24 hours following inoculation into the Lp stable state.
Fig. S5. Temporary perturbations in pH induce shifts between the Lp and Ca states.
Fig. S6. Minimal model incorporating feedbacks between microbial growth and pH predicts a minimum inoculum size for the invasions to induce community shifts.
Fig. S7. The invader Sm can induce transitions from the alkaline (Ca) to the acidic (Lp) stable state.
Fig. S8. Early dynamics of soil communities in laboratory environments.
Fig. S9. Mutual resilience against migration between the Bacillus and Pantoea soil communities.
Fig. S10. Different behavior in laboratory microcosms for three genetically similar soil isolates.
Fig. S11. Both Pc and Pa can induce transitions from the Bacillus to the Pantoea stable states.
Reference (44)


