Chloroplast protein import systems have evolved to accommodate thousands of nuclear-encoded proteins. Diversification of import routes supports tissue- and development-specific plastid physiology in terrestrial plants.
Keywords: Chloroplast biogenesis, chloroplast protein import, protein targeting, protein quality control, transit peptide, endosymbiosis
Abstract
The evolution of chloroplasts from the original endosymbiont involved the transfer of thousands of genes from the ancestral bacterial genome to the host nucleus, thereby combining the two genetic systems to facilitate coordination of gene expression and achieve integration of host and organelle functions. A key element of successful endosymbiosis was the evolution of a unique protein import system to selectively and efficiently target nuclear-encoded proteins to their site of function within the chloroplast after synthesis in the cytoplasm. The chloroplast TOC–TIC (translocon at the outer chloroplast envelope–translocon at the inner chloroplast envelope) general protein import system is conserved across the plant kingdom, and is a system of hybrid origin, with core membrane transport components adapted from bacterial protein targeting systems, and additional components adapted from host genes to confer the specificity and directionality of import. In vascular plants, the TOC–TIC system has diversified to mediate the import of specific, functionally related classes of plastid proteins. This functional diversification occurred as the plastid family expanded to fulfill cell- and tissue-specific functions in terrestrial plants. In addition, there is growing evidence that direct regulation of TOC–TIC activities plays an essential role in the dynamic remodeling of the organelle proteome that is required to coordinate plastid biogenesis with developmental and physiological events.
Introduction
In vascular plants, 2000–3000 nuclear genes are required for plastid function, with only ~120 genes retained in the plastid genome (Jarvis and Lopez-Juez, 2013). The translocon at the outer chloroplast envelope (TOC)–translocon at the inner chloroplast envelope (TIC) system mediates the import of the vast majority of nuclear-encoded plastid proteins from the cytoplasm, and it is therefore referred to as the general import system (Richardson et al., 2017; Bölter, 2018; Day and Theg, 2018). Although many mechanistic details of the general import system remain to be defined, the known activities of the core TOC–TIC components fulfill all of the criteria for a functional protein import system (Fig. 1). Nuclear-encoded plastid proteins contain intrinsic targeting signals to direct them to the organelle. Cytosolic factors and protein import receptors at the chloroplast surface specifically recognize the targeting signals and direct preproteins to the organelle. Targeting is assisted by cytosolic chaperones that prevent misfolding or aggregation in the cytoplasm. The targeting receptors are coupled to membrane channels, which mediate transport across the organelle membrane. In turn, the membrane channels associate with an energy-dependent import motor that provides the driving force for transport from the cytoplasm into the stroma. In the case of plastids, membrane channels in the outer and inner membrane work in concert to facilitate transport across the envelope and prevent mistargeting to the intermembrane space.
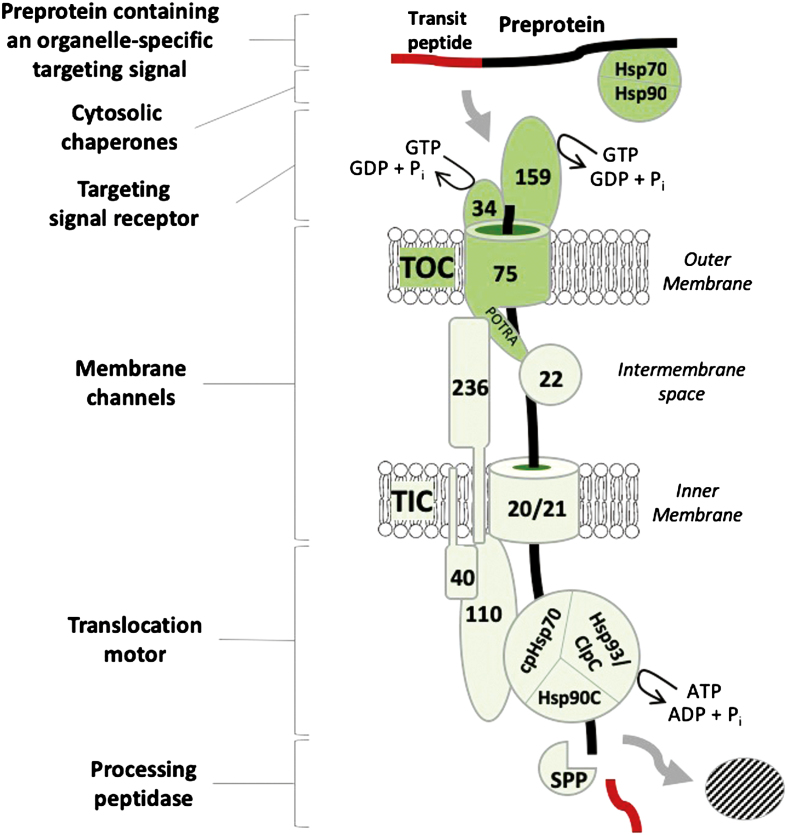
Fig. 1.
The core components of the TOC–TIC general import machinery of chloroplasts. The core components of the general import machinery are conserved across the green lineage. The newly synthesized preprotein is targeted to the TOC complex at the outer membrane by binding of its intrinsic transit peptide to the Toc34 (34) and Toc159 (159) receptors. Targeting is aided by cytosolic chaperone complexes of the Hsp70 and Hsp90 families. The GTPase activities of the receptors function as a checkpoint for the commitment of the preprotein to transport through the TOC and TIC membrane channels formed by Toc75 (75) and Tic20/21 (20/21), respectively. TOC–TIC supercomplexes formed by the binding of Tic236 (236) to Toc75 and Tic110 (110) at the TIC complex facilitates direct transport of the preprotein from the cytoplasm to the stroma. Mistargeting to the intermembrane space is avoided by the combined chaperone activities of the Toc75 POTRA domains and Tic22 (22). Tic20/21 form the major components of the TIC import channel, and they associate with Tic110 and Tic40 (40), which form a scaffold for the assembly of the ATP-dependent import motor in the stroma. The import motor drives unidirectional translocation of the preprotein via the combined activities of cpHsp70, Hsp93/ClpC, and Hsp90C chaperones. Upon import, the transit peptide is cleaved by the stromal processing peptidase (SPP). The reader is referred to other recent reviews for more comprehensive descriptions of accessory components associated with the general import machinery.
Protein import systems with functions comparable with TOC–TIC have not been identified in cyanobacteria or other Gram-negative bacteria (Day and Theg, 2018). Although the major bacterial protein targeting systems, including the Sec, Tat, and signal recognition particle (SRP) pathways, were conserved in chloroplasts, they do not participate in protein import, but were modified to participate in the biogenesis of internal membrane systems, such as the thylakoids and inner envelope membrane (Day and Theg, 2018). It therefore appears that the plastid protein import system was a novel adaptation to enable endosymbiosis. Despite their unique functions, we now know that core components of the TOC–TIC general import system were also derived from specialized bacterial protein targeting systems and repurposed to constitute the protein import apparatus. Here, we provide a brief overview of the mechanism of protein import, but our focus will be on highlighting the origins of the core, conserved components of the TOC–TIC systems that constitute the general import system. We will also highlight recent studies that reveal how diversification and regulation of the general protein import machinery in terrestrial plants contribute to the control and functional remodeling of plastids during developmental and physiological changes. The reader is referred to a number of recent reviews that focus on the mechanism of protein import for a more comprehensive description of additional components associated with the general import apparatus (Paila et al., 2015; Chotewutmontri et al., 2017; Lee et al., 2017; Richardson et al., 2017; Sjuts et al., 2017; Bölter, 2018; Day and Theg, 2018; Schwenkert et al., 2018).
Overview of TOC–TIC function
Nuclear-encoded plastid proteins are translated as preproteins on cytoplasmic ribosomes, containing intrinsic transit peptides that serve as the targeting signals for protein import (Fig. 1). Transit peptides are highly diverse in length and primary structure, and can range from 30 to 150 amino acids (Chotewutmontri et al., 2017; Lee and Hwang, 2018). Although there are limited exceptions, transit peptides are typically cleaved from the preprotein upon entry of the protein into the plastid stroma (Park et al., 2018). Like other organelle targeting systems, a number of cytosolic chaperone systems, including members of the heat shock protein (Hsp) 70 and Hsp90 families, are implicated in aiding transit of preproteins to the organelle surface (Fig. 1) (Kourtz and Ko, 1997; May and Soll, 2000; Qbadou et al., 2006; Flores-Pérez and Jarvis, 2013; Chotewutmontri and Bruce, 2015). These chaperone complexes probably interact with the unfolded or partially folded preprotein as it is synthesized, to prevent misfolding, aggregation, or mistargeting prior to recognition by the protein import machinery. Cytosolic chaperones are common features of all protein targeting systems and were probably recruited to the chloroplast protein import system from the host cell to fulfill a general chaperone function.
The recognition of plastid preproteins at the outer envelope membrane of plastids is mediated by binding of the transit peptide to the core TOC complex, composed of Toc34, Toc159, and Toc75 (Fig. 1) (Paila et al., 2015; Chotewutmontri et al., 2017). Toc34 and Toc159 are membrane-bound GTPases that function as the primary import receptors at the outer membrane (Kessler and Schnell, 2009; Schleiff and Becker, 2011; Chang et al., 2012). They initiate transit peptide binding to the TOC complex and control the early steps in import via their intrinsic GTPase activities, thereby constituting a checkpoint in the import reaction to ensure the fidelity of targeting (Chang et al., 2017; Richardson et al., 2018; Wiesemann et al., 2019). Toc75 is the major component of the protein import channel at the outer membrane (Fig. 1) (Ganesan and Theg, 2019). Toc75 also binds to transit peptides, and it functions in coordination with the GTPase receptors to mediate the insertion of the transit peptide across the outer envelope membrane (Schleiff et al., 2003; Kikuchi et al., 2006; Chen and Li, 2007; Koenig et al., 2010). Toc75 is essential in Arabidopsis (Jackson-Constan and Keegstra, 2001; Baldwin et al., 2005; Hust and Gutensohn, 2006), and site-specific cross-linking demonstrates its intimate association with preproteins at all stages in import (Ma et al., 1996; Richardson et al., 2018).
Proteins destined for the plastid interior must traverse both the outer and inner envelope membranes (Schnell and Blobel, 1993; Kouranov et al., 1998; Inoue and Akita, 2008). To achieve this, the TOC complex associates with a second transport system at the inner membrane, designated TIC, to form supercomplexes that facilitate direct translocation of the protein from the cytoplasm into the stroma (Fig. 1) (Kikuchi et al., 2013; Chen and Li, 2017; Richardson et al., 2018). The coordinate action of the TOC and TIC systems is facilitated by Tic236, a component that appears to form a physical link between TOC, the TIC membrane channel, and components of the ATP-dependent molecular motor responsible for driving protein import across the two membranes (Chen et al., 2018). Transit of preproteins through the intermembrane space is also facilitated by the Tic22 small chaperones (Kouranov et al., 1998; Qbadou et al., 2007; Kasmati et al., 2013; Rudolf et al., 2013).
Tic20 and Tic110 are proposed to constitute components of the TIC membrane channel at the inner membrane (Fig. 1) (Chen et al., 2002; Hirabayashi et al., 2011; Kasmati et al., 2011; Kovacs-Bogdan et al., 2011). The membrane channel activity of Tic20 has been demonstrated (Kovacs-Bogdan et al., 2011), and it is in close proximity to the transit peptide as the preprotein initiates translocation across the envelope (Richardson et al., 2018). Tic110 also contains a transit peptide-binding site, and this property facilitates the handoff of the protein import substrate to the chaperones to initiate ATP-dependent translocation through TOC–TIC supercomplexes (van den Wijngaard and Vredenberg, 1999; Inaba et al., 2003; Chou et al., 2006; Richardson et al., 2018). Tic110 and the membrane-bound co-chaperone, Tic40, also form a scaffold for the binding of chloroplast stromal chaperones (Kessler and Blobel, 1996; Lubeck et al., 1996; Nielsen et al., 1997; Jackson et al., 1998; Chou et al., 2003, 2006; Inaba et al., 2003, 2005; Kovacheva et al., 2005; Kao et al., 2012). The import-associated chaperone complex in the stroma includes members of the Hsp70 (cpHsp70), Hsp90 (Hsp90C), and Hsp100 (ClpC/Hsp93) families (Fig. 1) (Flores-Pérez and Jarvis, 2013). Although the precise role of each chaperone remains to be defined, the combination of molecular genetic, biochemical, and physiological data conclusively demonstrates that the import-associated chaperone complex acts as the ATP-dependent translocation motor for protein import across the envelope (Constan et al., 2004; Kovacheva et al., 2007; Su and Li, 2010; Inoue et al., 2013; Liu et al., 2014). Their activities also are likely to facilitate protein quality control, protein folding, or subsequent targeting events once the protein has completed import into the organelle.
Origins of plastid protein import
The origins of transit peptides remain a mystery, due in large part to their remarkable size and sequence diversity (Bruce, 2000; Lee and Hwang, 2018). However, similar extensions are absent in the corresponding ancestral cyanobacterial genes, indicating that transit peptides are probably of host origin and co-evolved with the TOC–TIC system. Regardless of the exact origin, recent studies suggest that transit peptides, particularly in vascular plants, have evolved to consist of tandemly arranged functional motifs that mediate the interactions of preproteins with various import components (Li and Teng, 2013; Lee and Hwang, 2018; Holbrook et al., 2016). This hypothesis is consistent with the demonstrated interactions of transit peptides with TOC and TIC proteins at each stage in import (Richardson et al., 2018), and it is one possible mechanism to facilitate sequential, unidirectional transport of the preprotein from the cytoplasm into the stroma. Furthermore, the modular arrangement of transit peptides provides the potential for evolving new or modified molecular recognition motifs to enhance the interactions with specific import components, and thereby regulate the import of specific proteins during plastid biogenesis or plastid-type transitions (see below).
Phylogenetic analyses of the general import machinery demonstrate that they are conserved across all lineages of the plant kingdom, providing compelling evidence that the primary endosymbiotic event that gave rise to chloroplasts was a rare, if not singular, event (McFadden, 2014; Zimorski et al., 2014; Garg and Gould, 2016). Toc75, the TOC channel component, is highly conserved, even amongst secondary endosymbiont lineages (Voulhoux et al., 2003; Inoue and Potter, 2004; Schleiff and Soll, 2005; Hsu and Inoue, 2009; Schleiff and Becker, 2011; Noinaj et al., 2013). Clues as to the origin of the Toc75 channel first emerged when sequence analysis demonstrated that it belongs to the outer membrane protein (OMP) 85 superfamily of β-barrel proteins that are found exclusively in the outer membranes of Gram-negative bacteria, mitochondria, and chloroplasts (Day et al., 2014; Simmerman et al., 2014; O’Neil et al., 2017). This family of proteins is characterized by the presence of a C-terminal β-barrel membrane domain, which typically functions in protein transport or insertion at the membrane, and a variable number of structurally related N-terminal polypeptide transport-associated (POTRA) domains, which interact with the protein substrates and accessory factors to facilitate protein transport (Paila et al., 2016; O’Neil et al., 2017).
The OMP85 family includes the membrane components of the β-barrel assembly machinery (BAM) and translocation and assembly module (TAM) complexes, BamA/TamA, that participate in the transport and assembly of bacterial OMPs. On this basis, it has been proposed that Toc75 in chloroplasts was derived from an ancestral OMP85 β-barrel membrane transporter gene related to the BAM/TAM systems of the original endosymbiont (Fig. 2) (Day et al., 2014; Heinz and Lithgow, 2014). In support of this hypothesis, a second TAM homolog, Tic236, was recently identified in chloroplasts. Tic236 is phylogenetically related to TamB, which links the bacterial inner membrane with BAM/TAM, thereby facilitating passage of substrates from the Sec secretion system to the BAM in the outer membrane (Chen et al., 2018; Schnell, 2018). Tic236 interacts with both Toc75 and Tic110, which suggests that it co-evolved with Toc75 to maintain a link between outer and inner membranes while simultaneously acquiring new interactions that established a link between TOC and TIC channels (Fig. 2) to facilitate preprotein import across both envelope membranes simultaneously. Both Toc75 and Tic236 are closely related to proteins found in the outer membrane of the cyanobacterium, Synechocystis sp. PCC 6803 (Bölter et al., 1998; Reumann et al., 1999; Chen et al., 2018). Although there is no evidence that the cyanobacterial proteins are functional orthologs of the chloroplast proteins, these observations are consistent with the proposal that the Toc75–Tic236 system arose from the duplication and/or adaptation of cyanobacterial genes.
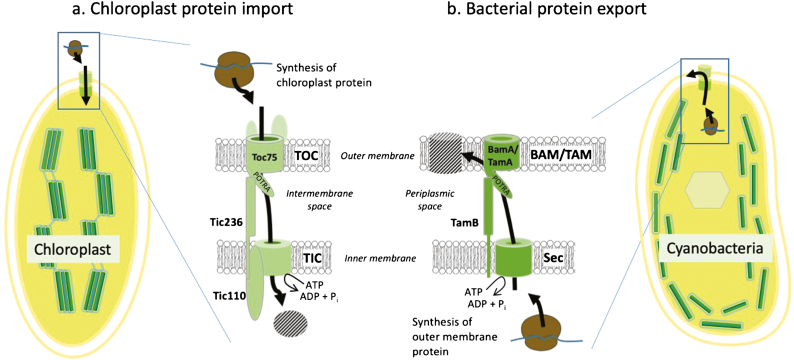
Fig. 2.
Core components of the chloroplast general protein import system evolved from bacterial protein export systems. (a) Protein import is mediated by TOC and TIC complexes in the outer and inner envelope membranes of plastids, respectively. Import occurs simultaneously through TOC and TIC supercomplexes to avoid mistargeting of preproteins to the intermembrane space. Supercomplexes appear to be assembled by the binding of Tic236 to the TOC import channel, Toc75, and the core TIC component, Tic110. Tic110 is associated with the TIC import channel and the ATP-dependent import motor that drives preprotein import. (b) Tic236 and its binding partner in the TOC complex, Toc75, are related to TamB and BamA/TamA of the BAM/TAM protein export complexes from Gram-negative bacteria, respectively. Therefore, it appears that key elements of the chloroplast protein import system were derived from the bacterial protein export system during the evolutionary assimilation of the photosynthetic bacterial ancestor by a host cell during endosymbiosis. The adaptation of the chloroplast import apparatus from BAM/TAM components required the acquisition of additional TOC and TIC components to reverse the directionality of protein translocation and confer specificity for the import of nuclear-encoded plastid preproteins.
In addition to interacting with Tic236 (Chen et al., 2018), the POTRA domains of Toc75 bind transit peptides and the small intermembrane space chaperones, Tic22-III and Tic22-IV (Fig. 1) (Paila et al., 2016). The POTRA domains also possess chaperone activity (Paila et al., 2016; O’Neil et al., 2017) and, together with the Tic22 proteins, are proposed to provide a chaperone complex to facilitate preprotein transit across the intermembrane space. These activities are analogous to the role of the POTRA domains of BamA, which constitute a platform for binding of the periplasmic chaperone, SurA, to facilitate passage of the secreted substrate from the Sec machinery in the inner membrane to the BamA membrane insertase in the outer membrane (Noinaj et al., 2015). Cyanobacteria also possess proteins related to the Tic22-III and Tic22-IV chaperones, but their function in bacteria is unknown (Tripp et al., 2012). Nonetheless, it suggests that the existing chaperone activity of these components was adapted for protein import into chloroplasts.
The nature and origins of the TIC channel have been intensely investigated for many years. The accumulated evidence demonstrates that Tic20 plays a central role in membrane transport of preproteins at the inner membrane in chloroplasts (Fig. 1) (Ma et al., 1996; Kouranov and Schnell, 1997; Teng et al., 2006; Kovacs-Bogdan et al., 2011; Kikuchi et al., 2013; Richardson et al., 2018). Tic20 and a second structurally similar protein, Tic21 (Teng et al., 2006), are differentially expressed and probably have overlapping functions as central components of the import channel. Like Toc75 and Tic236, these core components of the TIC preprotein channel also appear to have been adapted from cyanobacterial genes. However, they are not derived from known bacterial protein transport systems. Proteins with ~30% sequence identity to both Tic20 and Tic21 have been identified in cyanobacteria (Reumann and Keegstra, 1999; Lv et al., 2009). The Tic21-like protein from Synechocystis sp. PCC 6803, SynTic21, complements the phenotypes of a null mutant lacking Tic21 in Arabidopsis, demonstrating that Tic21 from plants and cyanobacteria are orthologous (Lv et al., 2009). The cyanobacterial ortholog of Tic21 also possesses iron transport activity (Duy et al., 2007), raising the possibility that the components of the TIC channel were adapted from existing nutrient transporters in the cyanobacterial inner membrane.
One critical adaptation during the evolution of the chloroplast TOC–TIC system was the apparent reversal in transport function relative to the bacterial export systems (Fig. 2) (Day and Theg, 2018). BAM/TAM mediate export of proteins from the periplasmic space into the outer membrane, whereas TOC–TIC functions in the opposite direction to import proteins from the cytoplasm to the stroma. The reversal was accomplished, in part, by a highly specific targeting system, consisting of the transit peptide and the corresponding transit peptide receptors, Toc34 and Toc159, which evolved to mediate delivery of the preprotein to the Toc75 channel. The TOC receptors are essential in Arabidopsis and are found in all plant lineages (Jarvis et al., 1998; Bauer et al., 2000; Constan et al., 2004; Ivanova et al., 2004; Kubis et al., 2004; McFadden and van Dooren, 2004). They belong to the translation factor (TRAFAC)-related superclass of GTPases that include many cellular regulatory proteins (Leipe et al., 2002), and they can be further subclassified into the GTPases activated by dimerization (GAD) subfamily (Wittinghofer and Vetter, 2011). This subfamily includes other protein targeting factors, most notably the SRP that mediates the targeting of secretory proteins to the endoplasmic reticulum (ER). Although a clear ancestral lineage for Toc34 and Toc159 has not been defined (Reumann et al., 2005), they are likely to be of host origin and evolved to take advantage of the robust and pervasive GTPase regulatory mechanism to control preprotein targeting to the TOC complex. Consistent with their inclusion in the GAD subfamily, Toc34 and Toc159 control transit peptide recognition via a cycle of receptor dimerization that regulates nucleotide exchange and hydrolysis (Oreb et al., 2011; Lumme et al., 2014). In the case of the TOC receptors, the GTPase activators and effectors that are commonly associated with other regulatory GTPases to control their activities appear to be replaced by the transit peptide. Binding of the transit peptide triggers changes in receptor dimerization to allow nucleotide exchange and hydrolysis (Oreb et al., 2011; Wiesemann et al., 2019). Inhibiting this GTPase cycle prevents stable association of the transit peptide with the TIC channel and engagement of the import-associated chaperone motor, thereby blocking protein import (Perry and Keegstra, 1994; Ma et al., 1996; Kouranov and Schnell, 1997; Oreb et al., 2011; Chang et al., 2017; Richardson et al., 2018). As such, the TOC receptors have evolved to couple transit peptide recognition with a GTPase-driven checkpoint to ensure the fidelity of protein import and initiate directional transport across the outer membrane.
The acquisition of Tic110 was a second key element in establishing the directionality of import. It constitutes a critical link between the TOC and TIC import channels and the import motor via its association with Tic236, Tic20, and the stromal chaperones (Fig. 2). Tic110 also interacts with transit peptides and Tic40, a stromal co-chaperone associated with the inner membrane (Chou et al., 2003, 2006). Tic110 (Tsai et al., 2013) and Tic40 (Chou et al., 2003; Kao et al., 2012) contain conserved sequence motifs that mediate their interactions with stromal chaperones and stimulate chaperone ATP hydrolysis. By acting as a scaffold for chaperone organization, Tic110 and Tic40 tether the ATP-dependent preprotein binding and release cycle of the chaperones to the site of import, thereby providing the unidirectional driving force for translocation of preproteins from the cytoplasm to the stroma. Consistent with these roles, phylogenetic analysis demonstrates Tic110’s and Tic40’s universal presence in the green lineage (McFadden, 2014; Zimorski et al., 2014; Garg and Gould, 2016). Like the TOC receptors, evolution probably took advantage of ubiquitous structural and functional motifs to assemble and adapt Tic110 and Tic40 for preprotein and chaperone binding.
It is estimated that the protein import motor requires the hydrolysis of 650 ATP molecules per polypeptide to import the average preprotein (Shi and Theg, 2013). Although the relative contributions of each chaperone have not been fully defined, it is now generally accepted that the ATP-dependent import motor is composed of the stromal cpHsp70, Hsp90C, and Hsp93/ClpC chaperones (Kessler and Blobel, 1996; Akita et al., 1997; Nielsen et al., 1997; Shi and Theg, 2010; Su and Li, 2010; Inoue et al., 2013). Each of the chloroplast chaperones plays multiple roles in protein homeostasis within the organelle, and therefore they are not specialized to function in protein import alone. Albeit somewhat more complex, the import-associated motor is similar to the Hsp70-based motors that function in protein import or membrane translocation in mitochondria and the ER (Flores-Pérez and Jarvis, 2013). Thus, a similar mechanism has been adapted in numerous protein import systems to catalyze membrane transport.
Recently, complexes containing the plastid-encoded hypothetical chloroplast ORF (Ycf) 1 (recently named Tic214) and Ycf2 proteins of the inner envelope have been proposed to function as components of the TIC channel and import motor, respectively, in coordination with Tic20 (Kikuchi et al., 2013, 2018). Components of the Tic214 and Ycf2 complexes are absent from major algal and plant lineages (Glaucophyta, Rhodophyta, the Poaceae, and some other dicot species) (Huang et al., 2013; Kikuchi et al., 2013; de Vries et al., 2015), and an evolutionary survey of Ycf1/Tic214 also demonstrates a high degree of structural variability across species, a trait uncharacteristic of the high degree of conservation in confirmed TOC and TIC components (de Vries et al., 2015). Furthermore, studies from other groups demonstrate limited reduction in protein import activity when Tic214 complex components were depleted in Arabidopsis (Köhler et al., 2016; Agne et al., 2017; Bölter and Soll, 2017). Therefore, the precise roles of the Tic214 and Ycf2 complexes in protein import remain to be clarified, and more research is necessary to determine how these new components can be integrated into current, established models of protein import. As discussed below, there is growing evidence that the protein import apparatus plays a direct role in the regulation of plastid biogenesis and homeostasis. With this in mind, it will be important that investigators look beyond the constraints of current models of the import mechanism and consider alternative functions for these new components, including TIC assembly, protein quality control, and TOC–TIC regulation, or as mediators of substrate- or species-specific import (Paila et al., 2015; Sjuts et al., 2017; Bölter, 2018).
Regulation of chloroplast protein import during development and stress
Expansion and functional diversification of TOC complexes
The evolution of land plants resulted in expansion of the plastid family beyond chloroplasts to fulfill specialized metabolic, physiological, and signaling activities in distinct cell types, organs, and tissues (de Vries et al., 2016). More than a dozen plastid types are now recognized, and include proplastids, the undifferentiated precursors for all plastid types found in meristematic tissue; starch-storing amyloplasts, which are also important in gravitropism; pigment-containing chromoplasts such as those found in tomato fruit; and gerontoplasts, which function in nutrient re-mobilization in senescing tissues (Kiss et al., 1989; Lopez-Juez and Pyke, 2005; Jarvis and Lopez-Juez, 2013; Pinard and Mizrachi, 2018). All of these plastid types have unique proteomes, resulting from developmentally or environmentally triggered changes in gene expression. For example, as much as one-third of the cellular transcriptome in seedlings changes in response to light during photomorphogenesis (Ma et al., 2001; Tepperman et al., 2001). Many of the transcriptome changes lead to the synthesis of plastid proteins that are critical for the transition of proplastids and etioplasts to photosynthetically competent chloroplasts. It is now recognized that the TOC–TIC general protein import system has also diversified during evolution to accommodate the changes in the organelle proteome that are required to maintain basic organelle activities while transitioning to perform specialized functions (Bauer et al., 2000; Kubis et al., 2003, 2004; Inoue et al., 2010). Furthermore, additional evidence indicates that the import apparatus is directly regulated as part of key transitions in plastid type and function (Ling et al., 2012, 2019; Chu and Li, 2018).
The TOC GTPase receptor families have expanded and diversified in parallel with the expansion of the plastid family in the green lineage (Reumann et al., 2005; Kalanon and McFadden, 2008). Toc159 and Toc34 are encoded by small gene families in vascular plants, and the receptor variants exhibit distinct spatial and temporal expression patterns. In Arabidopsis, null mutants in different Toc159 or Toc34 isoforms impact plastid biogenesis in distinct ways (Bauer et al., 2000; Ivanova et al., 2004; Kubis et al., 2004; Inoue et al., 2010; Infanger et al., 2011; Dutta et al., 2014). These receptor isoforms assemble with a single Toc75 protein to form TOC complexes that mediate the selective import of subsets of precursor proteins (Fig. 3a). For example, atToc159 and atToc33 are the major isoforms in green tissues in Arabidopsis, and both play central roles in the import of photosynthetic proteins and other processes required for chloroplast biogenesis. AtToc132/120 and atToc34, other Toc159 and Toc34 family members, respectively, are expressed at similar levels in most tissues and appear to be essential for the maintenance of metabolic functions that are common to different plastid types (Fig. 3a) (Bauer et al., 2000; Kubis et al., 2003, 2004; Inoue et al., 2010; Bischof et al., 2011). These observations provided the first evidence that the general import apparatus plays a direct role in controlling the changes in the plastid proteome that are required for plastid-type specific functions. The co-evolution of distinct transit peptide classes and import complexes with correspondingly distinct specificities was probably a key adaptation that enabled the proteome changes necessary for functional or plastid-type transitions while maintaining organelle homeostasis.
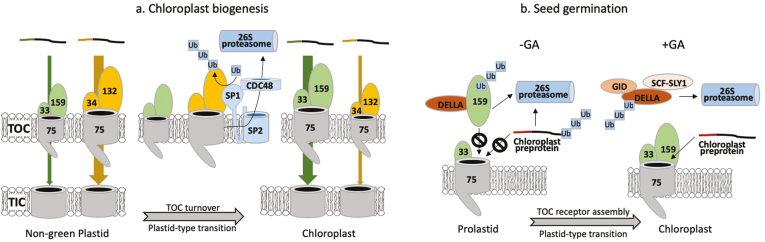
Fig. 3.
Regulation of the plastid general import machinery by the ubiquitin–proteasome system (UPS) during developmental transitions. Distinct TOC complexes mediate the import of specific classes (Groups 1 and 2) of nuclear-encoded preproteins into plastids. The specificity of the import complexes is determined by the Toc34–Toc159 family of transit peptide receptors. The relative abundance of distinct TOCs is regulated by the UPS and is required to balance the capacity of distinct import pathways with the changes in gene expression that accompany proteome remodeling during plastid-type transitions. (a) During the transition from non-green plastids to chloroplasts, the abundance of TOC complexes containing Toc132/Toc33 receptors (orange) relative to Toc159/Toc33 receptors (green) is altered to accommodate the import of proteins required for photosynthesis and the transition from chemoautotrophic to photoautotrophic metabolism. UPS-mediated turnover of TOC complexes involves a RING-type E3 ubiquitin ligase (SP1) that polyubiquitinates TOC components. The ubiquitinated TOC components appear to be extracted from the outer membrane and delivered to the 26S proteasome for degradation by the combined activities of the SP2 β-barrel channel protein and the cytosolic Cdc48 AAA+ ATPase. (b) Gibberellic acid (GA) plays an important part in preventing the premature biogenesis of chloroplasts during seed germination. The DELLA transcriptional regulators accumulate in the absence of GA and suppress the transcription of genes associated with germination, including genes required for chloroplast biogenesis. DELLAs also bind to the cytosolic form of the Toc159 protein import receptor, thereby promoting receptor degradation by the UPS and preventing the formation of TOC complexes required for the import and assembly of the photosynthetic machinery. A GA-regulated quality control system also targets cytosolic chloroplast preproteins for UPS degradation to prevent the accumulation of toxic preproteins in the cytosol. During germination, increased GA promotes UPS-mediated degradation of DELLAs via the GA–Gibberellin insensitive dwarf1 (GID) complex, the F-box protein SLY1, and the SCF E3 ligase, thereby inducing the expression of genes required for assembly of the photosynthetic machinery, while simultaneously allowing Toc159 to assemble with other TOC components to form complexes required for chloroplast biogenesis.
At least three distinct functional classes of transit peptides have also been identified that mediate preferential import into plastids at different developmental stages (Fig. 3a) (Teng et al., 2012). It has been proposed that the modular organization of transit peptides provides the platform for altering recognition by specific import components (e.g. distinct TOC receptors), while maintaining interactions with invariant import components (e.g. Toc75 or Tic110) (Li and Teng, 2013). In doing so, the import of functionally related subclasses of preproteins can be regulated in response to developmental or physiological signals that require changes in plastid function.
Regulation of protein import by the ubiquitin–proteasome system
The changes in the profiles of imported proteins during plastid-type transitions or organelle responses to physiological events require rapid modifications to the absolute and relative levels of specific TOC complexes. Two components involved in turnover of TOC complexes were discovered in a screen for suppressors of the pale phenotype of the atToc33 knockout mutant, ppi1. Suppressor of ppi1 locus 1 (SP1) is an E3 ubiquitin ligase that localizes to the chloroplast envelope and promotes turnover of atToc75, atToc159, and atToc33 via the ubiquitin–proteasome system (UPS) (Ling et al., 2012). The second component, SP2, is a β-barrel, OMP85 family member that is proposed to serve as a channel (retrotranslocon) for extraction of ubiquitinated TOC proteins from the membrane. SP1 and SP2 appear to associate with a cell division cycle protein (Cdc) 48 AAA+ ATPase in the cytosol, which is proposed to provide the pulling force for TOC extraction through SP2. Together, these components comprise a system hypothesized to function in a manner analogous to that which exists as part of the ER-associated degradation (ERAD) system, termed CHLORAD (Fig. 3a) (Ling et al., 2019). In fact, Cdc48 is a common component of the SP1–SP2 and ERAD systems. Knockout of SP1 or SP2 has no obvious growth defects under normal growth conditions; however, the sp1 mutant shows defects in chloroplast biogenesis during de-etiolation, and both sp1 and sp2 mutants attenuate photosynthetic decreases in mature leaves following premature dark treatment, mimicking leaf senescence. Conversely, SP1 or SP2 overexpression has an enhanced senescence phenotype, and SP1 overexpression shows enhanced de-etiolation (Ling et al., 2012, 2019). These observations are consistent with an important role for SP1 and SP2 during developmental transitions that involve plastid differentiation and/or changes in photosynthetic capacity (Fig. 3a) (Ling et al., 2012; Ling and Jarvis, 2015).
SP1 is a member of the expansive family of really interesting new gene (RING) E3 ubiquitin ligases in plants (Mazzucotelli et al., 2006). It was recently shown to also localize to peroxisomes and mitochondria in addition to chloroplasts (Pan et al., 2016; Pan and Hu, 2018). At peroxisomes, it is proposed that SP1 participates in turnover of the peroxisomal protein import machinery analogous to its role in chloroplasts, and negatively regulates peroxisome function (Pan et al., 2016). The co-localization of SP1 to chloroplasts, peroxisomes, and mitochondria may suggest the evolution of a common mechanism to coordinate the biogenesis of these three organelles by modulating protein import during developmental processes such as greening. This would serve as a mechanism to promote and regulate their coordinate metabolic functions, for example in photosynthesis, photorespiration, and during germination. Further work is needed to establish the targeting mechanism of SP1 to multiple organelles and its physiological relevance (Ling et al., 2017). Nonetheless, this observation may hint at coordinated regulation of the proteomes of chloroplasts and peroxisomes (and possibly mitochondria) during development by modulation of the levels and specificity of their protein import systems.
The ability to control plastid differentiation and chloroplast development during seedling establishment was also a key component of the adaptation of vascular plants to terrestrial life. Seeds are specially equipped to protect embryos from the harsh conditions of life on land, such as high light and an arid atmosphere, relative to the aquatic habitat of their ancestors (de Vries et al., 2016). In Arabidopsis (and most other dicots), proplastids are present in all cotyledon cells and develop into chloroplasts upon illumination, which involves extensive alterations of cellular and plastid proteomes. Hormone signaling pathways including gibberellic acid (GA), brassinosteroids (BRs), abscisic acid (ABA), and auxin also control chloroplast biogenesis during germination and early seeding development (reviewed in Pogson et al., 2015; Nee et al., 2017), and autophagy and protein turnover via the UPS contribute to remodeling of the cellular (and chloroplast) proteome during photomorphogenesis (Woodson, 2016; Aguilar-Hernandez et al., 2017).
It was recently reported that GA signaling regulates turnover of the major protein import receptor in Arabidopsis, atToc159, through the DELLA transcriptional regulators during seed germination (Fig. 3b) (Shanmugabalaji et al., 2018). GA promotes germination and chloroplast development by inducing degradation of DELLAs, which in turn induces expression of photosynthetic and chloroplast biogenesis genes. In the absence of GA, DELLAs accumulate and prevent this transcriptional response, suppressing chloroplast development in the dark to prevent premature greening and photooxidation caused by accumulation of chloroplast precursors (Cheminant et al., 2011; Pogson et al., 2015). In addition to transcriptional suppression of chloroplast biogenesis, the chloroplast protein import receptor atToc159 is degraded under low GA in a DELLA-dependent manner, and is stabilized in the presence of GA (Shanmugabalaji et al., 2018). Notably, atToc33 is also degraded under low GA, whereas atToc132 and atToc75 are not (Shanmugabalaji et al., 2018). This is consistent with the role of the atToc159 and atToc33 receptors in the import of a subset of preproteins that are critical for chloroplast biogenesis and assembly of the photosynthetic apparatus (Bauer et al., 2000; Kubis et al., 2003, 2004; Ivanova et al., 2004; Inoue et al., 2010). Low GA also promotes ubiquitination of the precursor to the Rubisco small subunit and reduces its accumulation in the cytosol, thereby preventing the toxic accumulation of the preprotein in the cytoplasm in the absence of a functional TOC import complex (Fig. 3b). atToc159 degradation in response to low GA appears to be independent of the SP1 E3 ligase, whereas Toc75 and atToc33 turnover is dependent upon the presence of SP1 (Shanmugabalaji et al., 2018). This suggests a novel mode of regulation for chloroplast biogenesis during seed dormancy and germination, in which the composition of the import apparatus is tightly regulated, and chloroplast precursor protein accumulation is minimized by a GA signaling mechanism to prevent premature greening. While this mechanism was studied only in germinating seeds, it is possible that similar mechanisms exist to regulate import during other key plastid developmental transitions.
Biotic and abiotic stress, including light, salt, drought, and osmotic stress, result in high levels of photooxidative damage to chloroplast proteins, and protein turnover is an important means by which plants contend with oxidative damage (Li et al., 2009; Nelson et al., 2014; Jarvi et al., 2015; Woodson, 2016; Otegui, 2018). Consequently, the import of photosynthetic proteins and balancing the stoichiometry of nuclear- and chloroplast-encoded subunits of the multisubunit photosynthetic apparatus is critical for maintenance and repair of the photosynthetic apparatus under stress. This was recently underscored by the finding that loss or overexpression of SP1 or SP2 renders Arabidopsis plants hypersensitive or resistant to salt, osmotic, and oxidative stresses, respectively. These observations were linked to the ability of SP1 to control the levels of specific TOC components, and thereby the import of photosynthetic proteins, which ultimately controls the levels of photooxidation and reactive oxygen species (ROS) production (Ling and Jarvis, 2015; Ling et al., 2019). It has also been proposed that manipulation of the SP1–SP2 protein import control system could be exploited by pathogens to minimize host responses to infection (Sowden et al., 2018). It is possible that modulation of protein import into peroxisomes (and mitochondria) additionally contributes to the stress-tolerant phenotype of the SP1 overexpression plants, since both organelles are also involved in ROS generation and signaling (Hu et al., 2012).
The TOC receptors are also phosphorylated, although the physiological significance of this post-translational modification is still somewhat unclear (Fulgosi and Soll, 2002; Aronsson et al., 2006; Oreb et al., 2008; Agne et al., 2010; Zufferey et al., 2017). The atToc159 receptors are phosphorylated in an ABA-dependent manner; atToc159 by the kinase SnRK2, and atToc132 and Toc120 in an SnRK2-independent manner (Wang et al., 2013). The chloroplast outer envelope kinase, KOC1, also phosphorylates Toc159, and koc1 mutants show defects in chloroplast protein import, demonstrating that phosphorylation by this kinase is an important regulator of import (Zufferey et al., 2017). It has also been proposed that phosphorylation regulates the dimerization capabilities of the Toc GTPase receptors, and negatively regulates their interaction with preproteins, which may also be phosphorylated (Fulgosi and Soll, 2002; Oreb et al., 2008). Although direct evidence is lacking (Aronsson et al., 2006), these observations suggest that phosphorylation plays a role in regulating the TOC complex, and one possibility is that phosphorylation is tied to UPS-mediated degradation of TOC components.
Coordinate regulation of protein import with plastid–nucleus communication
Protein import and chloroplast to nucleus retrograde signaling
Chloroplasts have also evolved important roles in sensing physiological and environmental changes as part of the organelle–nucleus communication networks that were critical for the integration of host and endosymbiont. Complex chloroplast to nucleus signaling pathways have been identified that relay the status of the chloroplast to the nucleus to control expression of nuclear-encoded chloroplast genes. The most well studied signals involve tetrapyrroles produced in the chloroplast, plastid gene expression, ROS, and chloroplast metabolites (Chi et al., 2015; Pogson et al., 2015; Chan et al., 2016; Kleine and Leister, 2016). A chloroplast unfolded protein response was also recently shown to trigger a nuclear response upon disruption of protein homeostasis within the chloroplast. Chloroplast signals result in the up-regulation of nuclear-encoded chloroplast chaperones (Llamas et al., 2017).
Perturbance of chloroplast protein import also elicits a nuclear response. Plants that lack the major chloroplast protein import receptor Toc159 (plastid protein import 2; ppi2) have an albino phenotype and are defective in the import of several photosynthetic proteins but accumulate many other non-photosynthetic proteins normally (Bauer et al., 2000; Bischof et al., 2011). Expression of nuclear-encoded photosynthetic genes is down-regulated in ppi2 (Bauer et al., 2000; Kakizaki et al., 2009; Lee et al., 2009), suggesting that a plastid to nucleus signaling pathway exists to attenuate photosynthetic gene expression in the absence of Toc159. This signal does not appear to involve Mg-Protoporphyrin IX (Mg-ProtoIX), a chlorophyll intermediate that is proposed to be a chloroplast retrograde signaling molecule, as ppi2 does not accumulate Mg-ProtoIX (Kakizaki et al., 2009; Chi et al., 2015; Chan et al., 2016; Kleine and Leister, 2016). Plastid to nucleus signaling in ppi2 does involve GUN1, a plastid protein known to be involved in multiple plastid–nucleus signaling pathways, and the transcription factor Golden-like 1 (GLK1), a key regulator of chloroplast biogenesis (Fitter et al., 2002; Kakizaki et al., 2009). There appear to be (at least) two plastid signals that dampen the GLK1 transcriptional response in ppi2; one that down-regulates transcription of GLK1, and the other that results in degradation of GLK1 via the UPS in a GUN1-independent mechanism (Kakizaki et al., 2009; Tokumaru et al., 2017). There are still many remaining questions about how the nucleus senses defects in protein import, and whether this response is important during normal plant growth and development. Nonetheless, these observations point toward a system for sensing disruptions in protein import that is distinct from the known pathways that monitor chloroplast damage. The identification of key signaling molecules/proteins specific to protein import stress will be important for understanding protein import regulation during development and environmental stress.
Inhibition of protein import and the cytosolic unfolded protein response
When chloroplast protein import is down-regulated or perturbed, accumulating chloroplast precursors in the cytosol can have toxic effects on the cell. In addition to plastid signals that down-regulate nuclear gene expression of photosynthetic proteins, the cell also has at least one known mechanism for removing accumulating chloroplast precursors from the cytosol. In ppi2, chloroplast precursors that are unable to be imported efficiently due to the absence of Toc159 are degraded in the cytosol via the UPS, which involves the activity of the E3 ligase C-terminus of Hsp70-interacting protein (CHIP) (Lee et al., 2009). The cytosolic chaperone Hsp70-4, which is one of at least five cytosolic Hsp70 chaperones in Arabidopsis (Lin et al., 2001; Sung et al., 2001; Lee et al., 2009), is highly up-regulated in ppi2 and binds to the transit peptide of cytosolic chloroplast precursor proteins. Hsp70-4 recruits CHIP, which results in ubiquitination of accumulating precursors that are subsequently degraded by the proteasome (Lee et al., 2009). The inability of Hsp70-deficient plants to remove accumulating precursors leads to cytotoxic effects that interfere with normal growth and development of plants, highlighting the importance of this proteotoxic stress response in plant growth and development (Lee et al., 2009). In tomato, Hsp70 and Hsp90 are also known to be part of a general response to accumulating chloroplast precursors (Tillmann et al., 2015).
In Arabidopsis, similar to in non-plant species, components of the heat shock response are important for mitigating the effects of protein unfolding in the cytosol (Sugio et al., 2009; Lin et al., 2018). In response to heat stress, expression of Hsps is induced by a network of transcription factors called heat shock factors (HSFs). In non-plant species such as yeast and Drosophila, a single HSF regulates heat shock response genes, and in mammals the HSF gene family is limited to four members (Akerfelt et al., 2010). However, Arabidopsis (and other plant species) has an expanded repertoire of HSF genes, forming a gene regulatory network that is involved in the response to various biotic and abiotic stresses including pathogenS, salinity, drought, and cold stress (Swindell et al., 2007; von Koskull-Doring et al., 2007; Guo et al., 2016). In Arabidopsis, part of this network mediates a general cytoplasmic unfolded protein response that also appears to be up-regulated when protein import is impaired (Lee et al., 2009; Sugio et al., 2009; Gladman et al., 2016; Lin et al., 2018). These observations fit nicely with the recently shown GA-dependent degradation of chloroplast precursors in the cytosol during seed germination via the UPS, and are consistent with a carefully regulated system to prevent premature accumulation of chloroplast preproteins in the cytosol, which can lead to cellular toxicity.
Conclusions
The evolution of the TOC–TIC general import apparatus was essential to the integration of host and bacterial functions that accompanied endosymbiosis and the establishment of the plant kingdom. The evolution of core TOC–TIC components from bacterial protein export systems and the addition of novel components to confer directionality on targeting is a remarkable example of molecular adaptation to provide a novel function that was required for the assimilation of the new organelle. For many years, protein import was viewed simply as a basic protein trafficking pathway. We now know that the general import system plays a dynamic role in controlling and responding to changes in the flux and profiles of nuclear-encoded proteins that are required for plastid-type transitions during development and to control the flux of import in response to stress. The emergence of the import machinery as a site of regulation opens up a new phase in studies of the TOC–TIC machinery that shifts the focus from the basic mechanism to understanding how the regulation of import contributes to organelle homeostasis, developmental transitions, and plastid–nucleus signaling. Undoubtedly, recent discoveries, including the role of the UPS system in regulating import in response to developmental and hormonal signals, are only the tip of the iceberg in this new chapter of understanding the contributions of the TOC–TIC general import machinery in plastid biogenesis and function.
Acknowledgements
This work was supported by National Institutes of Health grant 2RO1-GM061893 to DJS. LGLR is a recipient of a Postdoctoral Fellowship from the Natural Sciences and Engineering Research Council of Canada.
Glossary
Abbreviations
- BAM
β-barrel assembly machinery
- Cdc48
cell cycle division 48
- CHIP
C-terminus of Hsp70-interacting protein
- ERAD
endoplasmic reticulum-associated degradation
- GA
gibberellic acid
- GAD
GTPases activated by dimerization
- HSF
heat shock factor
- Hsp
heat shock protein
- KOC
kinase of the outer chloroplast membrane
- Mg-ProtoIX
Mg-Protoporphyrin IX
- OMP
outer membrane protein
- POTRA
polypeptide transport-associated
- ppi
plastid protein import mutant
- PUB4
U-box domain-containing protein 4
- ROS
reactive oxygen species
- SP
suppressor of ppi1
- SRP
signal recognition particle
- TAM
translocation and assembly module
- TIC
translocon at the inner chloroplast envelope
- TOC
translocon at the outer chloroplast envelope
- UPS
ubiquitin–proteasome system
- Ycf
hypothetical chloroplast ORF
References
- Agne B, Andrès C, Montandon C, Christ B, Ertan A, Jung F, Infanger S, Bischof S, Baginsky S, Kessler F. 2010. The acidic A-domain of Arabidopsis TOC159 occurs as a hyperphosphorylated protein. Plant Physiology 153, 1016–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agne B, Köhler D, Baginsky S. 2017. Protein import-independent functions of Tic56, a component of the 1-MDa translocase at the inner chloroplast envelope membrane. Plant Signaling & Behavior 12, e1284726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Hernández V, Kim DY, Stankey RJ, Scalf M, Smith LM, Vierstra RD. 2017. Mass spectrometric analyses reveal a central role for ubiquitylation in remodeling the Arabidopsis proteome during photomorphogenesis. Molecular Plant 10, 846–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerfelt M, Morimoto RI, Sistonen L. 2010. Heat shock factors: integrators of cell stress, development and lifespan. Nature Reviews. Molecular Cell Biology 11, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akita M, Nielsen E, Keegstra K. 1997. Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. Journal of Cell Biology 136, 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson H, Combe J, Patel R, Jarvis P. 2006. In vivo assessment of the significance of phosphorylation of the Arabidopsis chloroplast protein import receptor, atToc33. FEBS Letters 580, 649–655. [DOI] [PubMed] [Google Scholar]
- Baldwin A, Wardle A, Patel R, Dudley P, Park SK, Twell D, Inoue K, Jarvis P. 2005. A molecular–genetic study of the Arabidopsis Toc75 gene family. Plant Physiology 138, 715–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F. 2000. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403, 203–207. [DOI] [PubMed] [Google Scholar]
- Bischof S, Baerenfaller K, Wildhaber T, et al. 2011. Plastid proteome assembly without Toc159: photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. The Plant Cell 23, 3911–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölter B. 2018. En route into chloroplasts: preproteins’ way home. Photosynthesis Research 138, 263–275. [DOI] [PubMed] [Google Scholar]
- Bölter B, Soll J. 2017. Ycf1/Tic214 is not essential for the accumulation of plastid proteins. Molecular Plant 10, 219–221. [DOI] [PubMed] [Google Scholar]
- Bölter B, Soll J, Schulz A, Hinnah S, Wagner R. 1998. Origin of a chloroplast protein importer. Proceedings of the National Academy of Sciences, USA 95, 15831–15836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce BD. 2000. Chloroplast transit peptides: structure, function and evolution. Trends in Cell Biology 10, 440–447. [DOI] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. 2016. Learning the languages of the chloroplast: retrograde signaling and beyond. Annual Review of Plant Biology 67, 25–53. [DOI] [PubMed] [Google Scholar]
- Chang JS, Chen LJ, Yeh YH, Hsiao CD, Li HM. 2017. Chloroplast preproteins bind to the dimer interface of the Toc159 receptor during import. Plant Physiology 173, 2148–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WL, Soll J, Bölter B. 2012. The gateway to chloroplast: re-defining the function of chloroplast receptor proteins. Biological Chemistry 393, 1263–1277. [DOI] [PubMed] [Google Scholar]
- Cheminant S, Wild M, Bouvier F, Pelletier S, Renou JP, Erhardt M, Hayes S, Terry MJ, Genschik P, Achard P. 2011. DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. The Plant Cell 23, 1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KY, Li HM. 2007. Precursor binding to an 880-kDa Toc complex as an early step during active import of protein into chloroplasts. The Plant Journal 49, 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LJ, Li HM. 2017. Stable megadalton TOC–TIC supercomplexes as major mediators of protein import into chloroplasts. The Plant Journal 92, 178–188. [DOI] [PubMed] [Google Scholar]
- Chen X, Smith MD, Fitzpatrick L, Schnell DJ. 2002. In vivo analysis of the role of atTic20 in protein import into chloroplasts. The Plant Cell 14, 641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Chen LJ, Chu CC, Huang PK, Wen JR, Li HM. 2018. TIC236 links the outer and inner membrane translocons of the chloroplast. Nature 564, 125–129. [DOI] [PubMed] [Google Scholar]
- Chi W, Feng P, Ma J, Zhang L. 2015. Metabolites and chloroplast retrograde signaling. Current Opinion in Plant Biology 25, 32–38. [DOI] [PubMed] [Google Scholar]
- Chotewutmontri P, Bruce BD. 2015. Non-native, N-terminal Hsp70 molecular motor recognition elements in transit peptides support plastid protein translocation. Journal of Biological Chemistry 290, 7602–7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P, Holbrook K, Bruce BD. 2017. Plastid protein targeting: preprotein recognition and translocation. International Review of Cell and Molecular Biology 330, 227–294. [DOI] [PubMed] [Google Scholar]
- Chou ML, Chu CC, Chen LJ, Akita M, Li HM. 2006. Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. Journal of Cell Biology 175, 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Fitzpatrick LM, Tu SL, Budziszewski G, Potter-Lewis S, Akita M, Levin JZ, Keegstra K, Li HM. 2003. Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. The EMBO Journal 22, 2970–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CC, Li HM. 2018. Developmental regulation of protein import into plastids. Photosynthesis Research 138, 327–334. [DOI] [PubMed] [Google Scholar]
- Constan D, Froehlich JE, Rangarajan S, Keegstra K. 2004. A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiology 136, 3605–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day PM, Potter D, Inoue K. 2014. Evolution and targeting of Omp85 homologs in the chloroplast outer envelope membrane. Frontiers in Plant Science 5, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day PM, Theg SM. 2018. Evolution of protein transport to the chloroplast envelope membranes. Photosynthesis Research 138, 315–326. [DOI] [PubMed] [Google Scholar]
- de Vries J, Sousa FL, Bölter B, Soll J, Gould SB. 2015. YCF1: a green TIC? The Plant Cell 27, 1827–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Stanton A, Archibald JM, Gould SB. 2016. Streptophyte terrestrialization in light of plastid evolution. Trends in Plant Science 21, 467–476. [DOI] [PubMed] [Google Scholar]
- Dutta S, Teresinski HJ, Smith MD. 2014. A split-ubiquitin yeast two-hybrid screen to examine the substrate specificity of atToc159 and atToc132, two Arabidopsis chloroplast preprotein import receptors. PLoS One 9, e95026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy D, Wanner G, Meda AR, von Wirén N, Soll J, Philippar K. 2007. PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. The Plant Cell 19, 986–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. 2002. GLK gene pairs regulate chloroplast development in diverse plant species. The Plant Journal 31, 713–727. [DOI] [PubMed] [Google Scholar]
- Flores-Pérez Ú, Jarvis P. 2013. Molecular chaperone involvement in chloroplast protein import. Biochimica et Biophysica Acta 1833, 332–340. [DOI] [PubMed] [Google Scholar]
- Fulgosi H, Soll J. 2002. The chloroplast protein import receptors Toc34 and Toc159 are phosphorylated by distinct protein kinases. Journal of Biological Chemistry 277, 8934–8940. [DOI] [PubMed] [Google Scholar]
- Ganesan I, Theg SM. 2019. Structural considerations of folded protein import through the chloroplast TOC/TIC translocons. FEBS Letters 593, 565–572. [DOI] [PubMed] [Google Scholar]
- Garg SG, Gould SB. 2016. The role of charge in protein targeting evolution. Trends in Cell Biology 26, 894–905. [DOI] [PubMed] [Google Scholar]
- Gladman NP, Marshall RS, Lee KH, Vierstra RD. 2016. The proteasome stress regulon is controlled by a pair of NAC transcription factors in Arabidopsis. The Plant Cell 28, 1279–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Liu JH, Ma X, Luo DX, Gong ZH, Lu MH. 2016. The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Frontiers in Plant Science 7, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz E, Lithgow T. 2014. A comprehensive analysis of the Omp85/TpsB protein superfamily structural diversity, taxonomic occurrence, and evolution. Frontiers in Microbiology 5, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Kikuchi S, Oishi M, Nakai M. 2011. In vivo studies on the roles of two closely related Arabidopsis Tic20 proteins, AtTic20-I and AtTic20-IV. Plant & Cell Physiology 52, 469–478. [DOI] [PubMed] [Google Scholar]
- Holbrook K, Subramanian C, Chotewutmontri P, Reddick LE, Wright S, Zhang H, Moncrief L, Bruce BD. 2016. Functional analysis of semi-conserved transit peptide motifs and mechanistic implications in precursor targeting and recognition. Molecular Plant 9, 1286–1301. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Inoue K. 2009. Two evolutionarily conserved essential beta-barrel proteins in the chloroplast outer envelope membrane. Bioscience Trends 3, 168–178. [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK. 2012. Plant peroxisomes: biogenesis and function. The Plant Cell 24, 2279–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Friso G, Nishimura K, Qu X, Olinares PD, Majeran W, Sun Q, van Wijk KJ. 2013. Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships; the concept of orthoproteomics. Journal of Proteome Research 12, 491–504. [DOI] [PubMed] [Google Scholar]
- Hust B, Gutensohn M. 2006. Deletion of core components of the plastid protein import machinery causes differential arrest of embryo development in Arabidopsis thaliana. Plant Biology 8, 18–30. [DOI] [PubMed] [Google Scholar]
- Inaba T, Alvarez-Huerta M, Li M, Bauer J, Ewers C, Kessler F, Schnell DJ. 2005. Arabidopsis tic110 is essential for the assembly and function of the protein import machinery of plastids. The Plant Cell 17, 1482–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T, Li M, Alvarez-Huerta M, Kessler F, Schnell DJ. 2003. atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. Journal of Biological Chemistry 278, 38617–38627. [DOI] [PubMed] [Google Scholar]
- Infanger S, Bischof S, Hiltbrunner A, Agne B, Baginsky S, Kessler F. 2011. The chloroplast import receptor Toc90 partially restores the accumulation of Toc159 client proteins in the Arabidopsis thaliana ppi2 mutant. Molecular Plant 4, 252–263. [DOI] [PubMed] [Google Scholar]
- Inoue H, Akita M. 2008. The transition of early translocation intermediates in chloroplasts is accompanied by the movement of the targeting signal on the precursor protein. Archives of Biochemistry and Biophysics 477, 232–238. [DOI] [PubMed] [Google Scholar]
- Inoue H, Li M, Schnell DJ. 2013. An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proceedings of the National Academy of Sciences, USA 110, 3173–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Rounds C, Schnell DJ. 2010. The molecular basis for distinct pathways for protein import into Arabidopsis chloroplasts. The Plant Cell 22, 1947–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Potter D. 2004. The chloroplastic protein translocation channel Toc75 and its paralog OEP80 represent two distinct protein families and are targeted to the chloroplastic outer envelope by different mechanisms. The Plant Journal 39, 354–365. [DOI] [PubMed] [Google Scholar]
- Ivanova Y, Smith MD, Chen K, Schnell DJ. 2004. Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Molecular Biology of the Cell 15, 3379–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DT, Froehlich JE, Keegstra K. 1998. The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. Journal of Biological Chemistry 273, 16583–16588. [DOI] [PubMed] [Google Scholar]
- Jackson-Constan D, Keegstra K. 2001. Arabidopsis genes encoding components of the chloroplastic protein import apparatus. Plant Physiology 125, 1567–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Aro EM. 2015. Photosystem II repair in plant chloroplasts—regulation, assisting proteins and shared components with photosystem II biogenesis. Biochimica et Biophysica Acta 1847, 900–909. [DOI] [PubMed] [Google Scholar]
- Jarvis P, Chen LJ, Li H, Peto CA, Fankhauser C, Chory J. 1998. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282, 100–103. [DOI] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E. 2013. Biogenesis and homeostasis of chloroplasts and other plastids. Nature Reviews. Molecular Cell Biology 14, 787–802. [DOI] [PubMed] [Google Scholar]
- Kakizaki T, Matsumura H, Nakayama K, Che FS, Terauchi R, Inaba T. 2009. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiology 151, 1339–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalanon M, McFadden GI. 2008. The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: a bioinformatic comparison of Toc and Tic components in plants, green algae and red algae. Genetics 179, 95–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao YF, Lou YC, Yeh YH, Hsiao CD, Chen C. 2012. Solution structure of the C-terminal NP-repeat domain of Tic40, a co-chaperone during protein import into chloroplasts. Journal of Biochemistry 152, 443–451. [DOI] [PubMed] [Google Scholar]
- Kasmati AR, Töpel M, Khan NZ, Patel R, Ling Q, Karim S, Aronsson H, Jarvis P. 2013. Evolutionary, molecular and genetic analyses of Tic22 homologues in Arabidopsis thaliana chloroplasts. PLoS One 8, e63863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasmati AR, Töpel M, Patel R, Murtaza G, Jarvis P. 2011. Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. The Plant Journal 66, 877–889. [DOI] [PubMed] [Google Scholar]
- Kessler F, Blobel G. 1996. Interaction of the protein import and folding machineries of the chloroplast. Proceedings of the National Academy of Sciences, USA 93, 7684–7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F, Schnell D. 2009. Chloroplast biogenesis: diversity and regulation of the protein import apparatus. Current Opinion in Cell Biology 21, 494–500. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Asakura Y, Imai M, et al. 2018. A Ycf2–FtsHi heteromeric AAA-ATPase complex is required for chloroplast protein import. The Plant Cell 30, 2677–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M. 2013. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339, 571–574. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Hirohashi T, Nakai M. 2006. Characterization of the preprotein translocon at the outer envelope membrane of chloroplasts by blue native PAGE. Plant & Cell Physiology 47, 363–371. [DOI] [PubMed] [Google Scholar]
- Kiss JZ, Hertel R, Sack FD. 1989. Amyloplasts are necessary for full gravitropic sensitivity in roots of Arabidopsis thaliana. Planta 177, 198–206. [PubMed] [Google Scholar]
- Kleine T, Leister D. 2016. Retrograde signaling: organelles go networking. Biochimica et Biophysica Acta 1857, 1313–1325. [DOI] [PubMed] [Google Scholar]
- Koenig P, Mirus O, Haarmann R, Sommer MS, Sinning I, Schleiff E, Tews I. 2010. Conserved properties of polypeptide transport-associated (POTRA) domains derived from cyanobacterial Omp85. Journal of Biological Chemistry 285, 18016–18024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler D, Helm S, Agne B, Baginsky S. 2016. Importance of translocon subunit Tic56 for rRNA processing and chloroplast ribosome assembly. Plant Physiology 172, 2429–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ. 1998. Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. Journal of Cell Biology 143, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A, Schnell DJ. 1997. Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. Journal of Cell Biology 139, 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtz L, Ko K. 1997. The early stage of chloroplast protein import involves Com70. Journal of Biological Chemistry 272, 2808–2813. [DOI] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Patel R, Dudley P, Twell D, Ríos G, Koncz C, Jarvis P. 2005. In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. The Plant Journal 41, 412–428. [DOI] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Wardle A, Patel R, Jarvis P. 2007. Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. The Plant Journal 50, 364–379. [DOI] [PubMed] [Google Scholar]
- Kovács-Bogdán E, Benz JP, Soll J, Bölter B. 2011. Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biology 11, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S, Baldwin A, Patel R, Razzaq A, Dupree P, Lilley K, Kurth J, Leister D, Jarvis P. 2003. The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. The Plant Cell 15, 1859–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S, Patel R, Combe J, et al. 2004. Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. The Plant Cell 16, 2059–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Hwang I. 2018. Evolution and design principles of the diverse chloroplast transit peptides. Molecules and Cells 41, 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Lee J, Hwang I. 2017. Sorting of nuclear-encoded chloroplast membrane proteins. Current Opinion in Plant Biology 40, 1–7. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee DW, Lee Y, Mayer U, Stierhof YD, Lee S, Jürgens G, Hwang I. 2009. Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin–26S proteasome system in Arabidopsis. The Plant Cell 21, 3984–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L. 2002. Classification and evolution of P-loop GTPases and related ATPases. Journal of Molecular Biology 317, 41–72. [DOI] [PubMed] [Google Scholar]
- Li HM, Teng YS. 2013. Transit peptide design and plastid import regulation. Trends in Plant Science 18, 360–366. [DOI] [PubMed] [Google Scholar]
- Li Z, Wakao S, Fischer BB, Niyogi KK. 2009. Sensing and responding to excess light. Annual Review of Plant Biology 60, 239–260. [DOI] [PubMed] [Google Scholar]
- Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M. 2001. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress & Chaperones 6, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KF, Tsai MY, Lu CA, Wu SJ, Yeh CH. 2018. The roles of Arabidopsis HSFA2, HSFA4a, and HSFA7a in the heat shock response and cytosolic protein response. Botanical Studies 59, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Q, Broad W, Trosch R, Topel M, Demiral Sert T, Lymperopoulos P, Baldwin A, Jarvis RP. 2019. Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science 363, eaav4467. [DOI] [PubMed] [Google Scholar]
- Ling Q, Huang W, Baldwin A, Jarvis P. 2012. Chloroplast biogenesis is regulated by direct action of the ubiquitin–proteasome system. Science 338, 655–659. [DOI] [PubMed] [Google Scholar]
- Ling Q, Jarvis P. 2015. Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Current Biology 25, 2527–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Q, Li N, Jarvis P. 2017. Chloroplast ubiquitin E3 ligase SP1: does it really function in peroxisomes? Plant Physiology 175, 586–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, McNeilage RT, Shi LX, Theg SM. 2014. ATP requirement for chloroplast protein import is set by the Km for ATP hydrolysis of stromal Hsp70 in Physcomitrella patens. The Plant Cell 26, 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas E, Pulido P, Rodriguez-Concepcion M. 2017. Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PLoS Genetics 13, e1007022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Juez E, Pyke KA. 2005. Plastids unleashed: their development and their integration in plant development. International Journal of Developmental Biology 49, 557–577. [DOI] [PubMed] [Google Scholar]
- Lübeck J, Soll J, Akita M, Nielsen E, Keegstra K. 1996. Topology of IEP110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. The EMBO Journal 15, 4230–4238. [PMC free article] [PubMed] [Google Scholar]
- Lumme C, Altan-Martin H, Dastvan R, et al. 2014. Nucleotides and substrates trigger the dynamics of the Toc34 GTPase homodimer involved in chloroplast preprotein translocation. Structure 22, 526–538. [DOI] [PubMed] [Google Scholar]
- Lv HX, Guo GQ, Yang ZN. 2009. Translocons on the inner and outer envelopes of chloroplasts share similar evolutionary origin in Arabidopsis thaliana. Journal of Evolutionary Biology 22, 1418–1428. [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. 2001. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. The Plant Cell 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Kouranov A, LaSala SE, Schnell DJ. 1996. Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. Journal of Cell Biology 134, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T, Soll J. 2000. 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. The Plant Cell 12, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucotelli E, Belloni S, Marone D, De Leonardis A, Guerra D, Di Fonzo N, Cattivelli L, Mastrangelo A. 2006. The E3 ubiquitin ligase gene family in plants: regulation by degradation. Current Genomics 7, 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI. 2014. Origin and evolution of plastids and photosynthesis in eukaryotes. Cold Spring Harbor Perspectives in Biology 6, a016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI, van Dooren GG. 2004. Evolution: red algal genome affirms a common origin of all plastids. Current Biology 14, R514–R516. [DOI] [PubMed] [Google Scholar]
- Née G, Xiang Y, Soppe WJ. 2017. The release of dormancy, a wake-up call for seeds to germinate. Current Opinion in Plant Biology 35, 8–14. [DOI] [PubMed] [Google Scholar]
- Nelson CJ, Alexova R, Jacoby RP, Millar AH. 2014. Proteins with high turnover rate in barley leaves estimated by proteome analysis combined with in planta isotope labeling. Plant Physiology 166, 91–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K. 1997. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. The EMBO Journal 16, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK. 2013. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N, Rollauer SE, Buchanan SK. 2015. The β-barrel membrane protein insertase machinery from Gram-negative bacteria. Current Opinion in Structural Biology 31, 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil PK, Richardson LGL, Paila YD, Piszczek G, Chakravarthy S, Noinaj N, Schnell D. 2017. The POTRA domains of Toc75 exhibit chaperone-like function to facilitate import into chloroplasts. Proceedings of the National Academy of Sciences, USA 114, E4868–E4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreb M, Höfle A, Koenig P, Sommer MS, Sinning I, Wang F, Tews I, Schnell DJ, Schleiff E. 2011. Substrate binding disrupts dimerization and induces nucleotide exchange of the chloroplast GTPase Toc33. Biochemical Journal 436, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreb M, Höfle A, Mirus O, Schleiff E. 2008. Phosphorylation regulates the assembly of chloroplast import machinery. Journal of Experimental Botany 59, 2309–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS. 2018. Vacuolar degradation of chloroplast components: autophagy and beyond. Journal of Experimental Botany 69, 741–750. [DOI] [PubMed] [Google Scholar]
- Paila YD, Richardson LG, Inoue H, Parks ES, McMahon J, Inoue K, Schnell DJ. 2016. Multi-functional roles for the polypeptide transport associated domains of Toc75 in chloroplast protein import. eLife 5, e12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paila YD, Richardson LGL, Schnell DJ. 2015. New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. Journal of Molecular Biology 427, 1038–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R, Hu J. 2018. The Arabidopsis E3 ubiquitin ligase SP1 targets to chloroplasts, peroxisomes, and mitochondria. Plant Physiology 176, 480–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R, Satkovich J, Hu J. 2016. E3 ubiquitin ligase SP1 regulates peroxisome biogenesis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 113, E7307–E7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Zhong R, Lamppa G. 2018. Chloroplast stromal processing peptidase activity is modulated by transit peptide determinants that include inhibitory roles for its N-terminal domain and initial Met. Biochemical and Biophysical Research Communications 503, 3149–3154. [DOI] [PubMed] [Google Scholar]
- Perry SE, Keegstra K. 1994. Envelope membrane proteins that interact with chloroplastic precursor proteins. The Plant Cell 6, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinard D, Mizrachi E. 2018. Unsung and understudied: plastids involved in secondary growth. Current Opinion in Plant Biology 42, 30–36. [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Ganguly D, Albrecht-Borth V. 2015. Insights into chloroplast biogenesis and development. Biochimica et Biophysica Acta 1847, 1017–1024. [DOI] [PubMed] [Google Scholar]
- Qbadou S, Becker T, Bionda T, Reger K, Ruprecht M, Soll J, Schleiff E. 2007. Toc64—a preprotein-receptor at the outer membrane with bipartide function. Journal of Molecular Biology 367, 1330–1346. [DOI] [PubMed] [Google Scholar]
- Qbadou S, Becker T, Mirus O, Tews I, Soll J, Schleiff E. 2006. The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. The EMBO Journal 25, 1836–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Davila-Aponte J, Keegstra K. 1999. The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: identification of a cyanobacterial homolog. Proceedings of the National Academy of Sciences, USA 96, 784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Inoue K, Keegstra K. 2005. Evolution of the general protein import pathway of plastids (review). Molecular Membrane Biology 22, 73–86. [DOI] [PubMed] [Google Scholar]
- Reumann S, Keegstra K. 1999. The endosymbiotic origin of the protein import machinery of chloroplastic envelope membranes. Trends in Plant Science 4, 302–307. [DOI] [PubMed] [Google Scholar]
- Richardson LGL, Singhal R, Schnell DJ. 2017. The integration of chloroplast protein targeting with plant developmental and stress responses. BMC Biology 15, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson LGL, Small EL, Inoue H, Schnell DJ. 2018. Molecular topology of the transit peptide during chloroplast protein import. The Plant Cell 30, 1789–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf M, Machettira AB, Groß LE, et al. 2013. In vivo function of Tic22, a protein import component of the intermembrane space of chloroplasts. Molecular Plant 6, 817–829. [DOI] [PubMed] [Google Scholar]
- Schleiff E, Becker T. 2011. Common ground for protein translocation: access control for mitochondria and chloroplasts. Nature Reviews. Molecular Cell Biology 12, 48–59. [DOI] [PubMed] [Google Scholar]
- Schleiff E, Soll J. 2005. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Reports 6, 1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E, Soll J, Küchler M, Kühlbrandt W, Harrer R. 2003. Characterization of the translocon of the outer envelope of chloroplasts. Journal of Cell Biology 160, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell DJ. 2018. Exit route evolved into entry path in plants. Nature 564, 45–46. [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Blobel G. 1993. Identification of intermediates in the pathway of protein import into chloroplasts and their localization to envelope contact sites. Journal of Cell Biology 120, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenkert S, Dittmer S, Soll J. 2018. Structural components involved in plastid protein import. Essays in Biochemistry 62, 65–75. [DOI] [PubMed] [Google Scholar]
- Shanmugabalaji V, Chahtane H, Accossato S, Rahire M, Gouzerh G, Lopez-Molina L, Kessler F. 2018. Chloroplast biogenesis controlled by DELLA–TOC159 interaction in early plant development. Current Biology 28, 2616–2623. [DOI] [PubMed] [Google Scholar]
- Shi LX, Theg SM. 2010. A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. The Plant Cell 22, 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LX, Theg SM. 2013. Energetic cost of protein import across the envelope membranes of chloroplasts. Proceedings of the National Academy of Sciences, USA 110, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmerman RF, Dave AM, Bruce BD. 2014. Structure and function of POTRA domains of Omp85/TPS superfamily. International Review of Cell and Molecular Biology 308, 1–34. [DOI] [PubMed] [Google Scholar]
- Sjuts I, Soll J, Bölter B. 2017. Import of soluble proteins into chloroplasts and potential regulatory mechanisms. Frontiers in Plant Science 8, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowden RG, Watson SJ, Jarvis P. 2018. The role of chloroplasts in plant pathology. Essays in Biochemistry 62, 21–39. [DOI] [PubMed] [Google Scholar]
- Su PH, Li HM. 2010. Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. The Plant Cell 22, 1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A, Dreos R, Aparicio F, Maule AJ. 2009. The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. The Plant Cell 21, 642–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung DY, Vierling E, Guy CL. 2001. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiology 126, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Huebner M, Weber AP. 2007. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng YS, Chan PT, Li HM. 2012. Differential age-dependent import regulation by signal peptides. PLoS Biology 10, e1001416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng YS, Su YS, Chen LJ, Lee YJ, Hwang I, Li HM. 2006. Tic21 is an essential translocon component for protein translocation across the chloroplast inner envelope membrane. The Plant Cell 18, 2247–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. 2001. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proceedings of the National Academy of Sciences, USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann B, Röth S, Bublak D, Sommer M, Stelzer EH, Scharf KD, Schleiff E. 2015. Hsp90 is involved in the regulation of cytosolic precursor protein abundance in tomato. Molecular Plant 8, 228–241. [DOI] [PubMed] [Google Scholar]
- Tokumaru M, Adachi F, Toda M, Ito-Inaba Y, Yazu F, Hirosawa Y, Sakakibara Y, Suiko M, Kakizaki T, Inaba T. 2017. Ubiquitin–proteasome dependent regulation of the GOLDEN2-LIKE 1 transcription factor in response to plastid signals. Plant Physiology 173, 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp J, Hahn A, Koenig P, et al. 2012. Structure and conservation of the periplasmic targeting factor Tic22 protein from plants and cyanobacteria. Journal of Biological Chemistry 287, 24164–24173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JY, Chu CC, Yeh YH, Chen LJ, Li HM, Hsiao CD. 2013. Structural characterizations of the chloroplast translocon protein Tic110. The Plant Journal 75, 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Wijngaard PW, Vredenberg WJ. 1999. The envelope anion channel involved in chloroplast protein import is associated with Tic110. Journal of Biological Chemistry 274, 25201–25204. [DOI] [PubMed] [Google Scholar]
- von Koskull-Döring P, Scharf KD, Nover L. 2007. The diversity of plant heat stress transcription factors. Trends in Plant Science 12, 452–457. [DOI] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265. [DOI] [PubMed] [Google Scholar]
- Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Tao WA, Zhu JK. 2013. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proceedings of the National Academy of Sciences, USA 110, 11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesemann K, Simm S, Mirus O, Ladig R, Schleiff E. 2019. Regulation of two GTPases Toc159 and Toc34 in the translocon of the outer envelope of chloroplasts. Biochimica et Biophysica Acta 1867, 627–636. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A, Vetter IR. 2011. Structure–function relationships of the G domain, a canonical switch motif. Annual Review of Biochemistry 80, 943–971. [DOI] [PubMed] [Google Scholar]
- Woodson JD. 2016. Chloroplast quality control—balancing energy production and stress. New Phytologist 212, 36–41. [DOI] [PubMed] [Google Scholar]
- Zimorski V, Ku C, Martin WF, Gould SB. 2014. Endosymbiotic theory for organelle origins. Current Opinion in Microbiology 22, 38–48. [DOI] [PubMed] [Google Scholar]
- Zufferey M, Montandon C, Douet V, Demarsy E, Agne B, Baginsky S, Kessler F. 2017. The novel chloroplast outer membrane kinase KOC1 is a required component of the plastid protein import machinery. Journal of Biological Chemistry 292, 6952–6964. [DOI] [PMC free article] [PubMed] [Google Scholar]