Abstract
In contrast to desiccation-tolerant ‘orthodox’ seeds, so-called ‘intermediate’ seeds cannot survive complete drying and are short-lived. All species of the genus Coffea produce intermediate seeds, but they show a considerable variability in seed desiccation tolerance (DT), which may help to decipher the molecular basis of seed DT in plants. We performed a comparative transcriptome analysis of developing seeds in three coffee species with contrasting desiccation tolerance. Seeds of all species shared a major transcriptional switch during late maturation that governs a general slow-down of metabolism. However, numerous key stress-related genes, including those coding for the late embryogenesis abundant protein EM6 and the osmosensitive calcium channel ERD4, were up-regulated during DT acquisition in the two species with high seed DT, C. arabica and C. eugenioides. By contrast, we detected up-regulation of numerous genes involved in the metabolism, transport, and perception of auxin in C. canephora seeds with low DT. Moreover, species with high DT showed a stronger down-regulation of the mitochondrial machinery dedicated to the tricarboxylic acid cycle and oxidative phosphorylation. Accordingly, respiration measurements during seed dehydration demonstrated that intermediate seeds with the highest DT are better prepared to cease respiration and avoid oxidative stresses.
Keywords: Coffea, dehydration, desiccation tolerance, intermediate seeds, late maturation, metabolic quiescence, respiration, seed development, transcriptome
Seeds of coffee species that are more tolerant to desiccation display a specific late maturation transcriptional program that primes them to better cease respiration and avoid oxidative stress upon dehydration.
Introduction
Desiccation tolerance (DT) is the ability of an organism or tissue to withstand removal of intracellular water whilst retaining structural integrity and viability, and then to resume normal metabolism upon rehydration (Crowe et al., 1992; Leprince and Buitink, 2015). The vast majority of flowering plants produce seeds that are desiccation tolerant. In a dry quiescent state at maturity, these seeds are endowed with an exceptional capacity to endure extreme adverse environmental conditions after dispersal (Sano et al., 2016). These so-called ‘orthodox’ seeds can survive ex situ storage for very long periods under conventional gene-bank conditions (Li and Pritchard, 2009). DT is a complex trait that is mostly genetically determined in the developing seed and is put in place during the late maturation program. This is achieved through the interplay of multiple cellular protectants and repair mechanisms designed to cope with the various desiccation-associated stresses including oxidation, hyperionicity, mechanical strain associated with cell shrinkage, protein misfolding/aggregation and phase transition of lipid membranes (Crowe et al., 1992; Leprince and Buitink, 2015). Coordinated repression of metabolism, stabilization of membranes and proteins against conformational change by chemical and protein chaperones, and the accumulation of efficient antioxidant, detoxication and repair systems appear to be among the essential attributes that contribute to seed DT (Hoekstra et al., 2001; Buitink and Leprince, 2008; Leprince and Buitink, 2010). Some of these protective mechanisms are shared across eukaryotic kingdoms since the accumulation of chemical and protein chaperones such as non-reducing sugars (e.g. trehalose), heat-shock proteins, and intrinsically disordered proteins (e.g. late embryogenesis abundant (LEA) proteins) have been recently confirmed to be of tremendous importance in conferring DT in unicellular yeasts, tardigrades, and nematodes (Erkut et al., 2011; Tapia et al., 2015; Boothby et al., 2017). The repertoire of DT-related genes, including key transcriptional regulators, has been extensively studied in orthodox seeds through transcriptome profiling and co-expression analyses during the acquisition of desiccation tolerance (e.g. Verdier et al., 2013; Righetti et al., 2015; González-Morales et al., 2016; for a review see Leprince et al., 2017).
Estimates suggest that about 8% of the world’s flowering plants (>40 000 species) produce desiccation-sensitive short-lived seeds (Tweddle et al., 2003; Wyse and Dickie, 2017). Two other categories of seeds—intermediate and recalcitrant—have been defined with respect to their storability in gene banks (Roberts, 1973; Ellis et al., 1990) and these terms are now used to describe their sensitivity to drying. Although desiccation tolerance may vary considerably within each of these two seed categories (Dussert et al., 1999; Berjak and Pammenter, 2008), it is commonly acknowledged that recalcitrant seeds are extremely sensitive to dehydration and do not survive if dried to about 90% relative humidity (RH), while intermediate seeds are able to withstand enforced drying to about 30% RH (Black and Pritchard, 2002). Seeds of many major tropical crops are recalcitrant (e.g. cocoa, coconut, rubber tree) or intermediate (e.g. coffee, tea, oil palm, citrus), which represents a major constraint for growers, seed companies, and germplasm repositories (Li and Pritchard, 2009; Walters et al., 2013).
Albuminous seeds of Coffea species are the model system for the ‘intermediate seed’ category (Ellis et al., 1990; Dussert et al., 1999). As in all fleshy fruits, coffee seeds are not subjected to dry ambient air during maturation (Dussert et al., 2000). Partial DT is acquired without dehydration in planta during the late stage of maturation, well after the onset of reserve deposition in the cellular endosperm, a living tissue that represents more than 98% of the mature coffee seed mass (Dussert et al., 2018). The desiccation-induced loss of seed viability is due to damage to the endosperm only, the embryo being much more tolerant to desiccation (Dussert et al., 2006). Transcriptomic analysis of developing C. arabica seeds revealed that the cellular and regulation processes that occur during late maturation in the coffee seed are strikingly similar to those known or thought to be involved in orthodox seed DT (Dussert et al., 2018). Briefly, acquisition of DT coincides with a dramatic transcriptional switch characterized by the repression of genes involved in the cell cycle, DNA processing, and primary metabolism, and the up-regulation of a large number of genes coding for proteins involved in cell protection and rescue, including reactive oxygen species (ROS)-scavenging enzymes, LEA proteins and heat-shock protein stress proteins, defense-, cold-, and drought-induced proteins, as well as many components of sugar and nutrient sensing systems. The DT-associated transcriptional switch also had the signature of a coordinated metabolic slow-down, with a marked decrease in the mRNA abundance of genes involved in protein fate and major energy processes such as photosynthesis, the tricarboxylic acid (TCA) cycle, respiratory electron transport, ATP synthase, and glycolysis. With regard to phytohormones, there was an up-regulation of genes for abscisic acid (ABA) synthesis, perception, and signaling during the late maturation and a drastic down-regulation of cytokinin biosynthetic genes associated with a large drop in cytokinin content. Finally, major transcription factors (TFs) homologous to those identified as associated with DT in orthodox seeds have been described as massively up-regulated during DT acquisition in coffee seeds, namely PLATZ, DOGL4, HSFA9, and DREB2G (Prieto-Dapena et al., 2006, 2008; Righetti et al., 2015; González-Morales et al., 2016). The late maturation program described in coffee seed is therefore qualitatively comparable to that of orthodox seeds. This led to the hypothesis that the differences observed in DT between orthodox and intermediate seeds, and between intermediate seeds displaying different DT levels, could be due to a quantitative variation in gene expression and metabolite accumulation. In the present work we therefore compared the seed maturation transcriptomes of three closely related Coffea species (Cenci et al., 2012) that were previously shown to differ in their DT (Dussert et al., 1999): C. arabica, C. eugenioides, and C. canephora.
Material and methods
Plant material
Seeds at three different maturation stages known to span the acquisition of desiccation tolerance in C. arabica (ST5, ST6, and ST7, Dussert et al., 2018) were collected from three trees of each species from the Biological Resource Center Coffea, Bassin Martin, Reunion Island. The average seed development duration being different for the three studied species, i.e. respectively ca. 7, 8, and 10 months for C. arabica, C. eugenioides, and C. canephora, respectively (Dussert et al., 2000). The developmental stages were selected based on marked anatomical and morphological seed and fruit traits that are shared across coffee species, as defined and described previously for C. arabica (Joët et al., 2009; Dussert et al., 2018). Briefly, stage 5 is the peak of reserve deposition and corresponds to endosperm hardening due to massive deposition of galactomannans in cell walls, stage 6 coincides with fruit veraison, and stage 7 corresponds to mature cherry fruits with red pericarp. After being cross-sectioned, the seed was separated from the pericarp and immediately frozen in liquid nitrogen and stored at −80 °C. The endosperm was separated from the perisperm and the embryo while frozen. To minimize the impact of genotypic effect and facilitate inter-species comparisons, developing seeds were collected (pools of ca. 50 seeds) on different wild accessions for each species (C. arabica accessions AR28-06, AR02-06, AR38b/05; C. eugenioides accessions DA71, DA78, DA78c; C. canephora accessions BD55, BD56, DAF71), and were considered biological replicates.
Desiccation tolerance assays and respiration measurement during desiccation
Mature seed lots (50 seeds) of C. arabica, C. canephora, and C. eugenioides were desiccated by equilibration over various saturated salt solutions (K-acetate (23% RH), K2CO3 (45% RH), NH4NO3 (62% RH), and (NH4)2SO4 (81% RH)) for 20 d at 27 °C in the dark, as previously described (Dussert et al., 2000). For germination tests, batches of nine seeds were placed on 18 g of vermiculite fully imbibed with 50 ml of sterile water in closed plastic boxes (Magenta, Chicago, IL, USA). After 6 weeks of culture at 27 °C in the dark, successful seed germination was scored (emergence of the hypocotyl and radicle geotropic growth). Desiccation sensitivity was quantified using the previously developed quantal response model (Dussert et al., 1999). For respiration measurements, fresh ST7 mature seeds were desiccated at 25 °C by equilibration over a saturated K2CO3 solution. During 4 d of desiccation, subsamples of four to five seeds were taken every 12 h for respiration measurement. For each replicate, seeds (the fresh weight was recorded) were placed in individual 10 ml hermetically sealed vials at 25 °C. CO2 release (µmol CO2 min−1 g−1 DM) was measured at 5 min intervals for 25 min by gas chromatography with an Agilent M200 apparatus (SRA, Marcy l’Etoile, France). Variations in gas concentration were corrected using the volume of the seeds in the vial.
Transcriptome analysis
For each of the 27 samples of the experimental design, a mix of >20 endosperms was ground to a fine powder in an analytical grinder (IKA A10, Staufen, Germany) and total RNA was extracted from 70 mg using the Qiagen RNeasy Lipid Tissue kit (Qiagen, Stanford, CA, USA). cDNA libraries were constructed using the TruSeqTM Stranded mRNA sample preparation kit (Illumina, USA) then sequenced on an Illumina HiSeq 2500 (single reads, 100 nt) on the MGX platform (Montpellier Genomix, http://www.mgx.cnrs.fr/). After quality filtering using Cutadapt (quality score>Q30 and removal of reads shorter than 60 bp or higher than 140 bp), a total of 875 million reads were retained (average of 31.95 million reads per library) (Supplementary Table S1 at JXB online). The entire dataset has been deposited at the European Nucleotide Archive (ENA) under the project number PRJEB32533. Owing to the very low genetic divergence between the three Coffea species (average of 1.3% gene sequence difference; Cenci et al., 2012), the trimmed reads (cutadapt, Genome Analysis Toolkit) of each library were mapped to the C. canephora coding transcriptome DNA reference sequence (25574 CDS) (Denoeud et al., 2014) using BWA MEM (Li, 2013, Preprint) with the default parameters. Reads were counted using IDXstats in SAMtools (Li et al., 2009) and counts normalized (RPKM; Supplementary Table S2). A total of 628 million reads were mapped (average of 23.3 million reads per library). Genes with extremely low expression (<20 counts in total) were removed for subsequent analyses (22 503 retained genes).
Differential expression analysis was performed using the DESeq2 package with default parameters (version 1.14.1, Love et al., 2014) in R (version 3.3.2). Hierarchical clustering of species stages was calculated using the Euclidean method after read count normalization of each sample. Each species-stage was contrasted to each other (36 pairwise comparisons) and P-values were adjusted using the Benjamini–Hochberg method (Supplementary Table S3). Combinations of Boolean operators were used as filters of differentially expressed genes (DEGs) to define different categories of DT- and desiccation sensitivity (DS)-related candidate genes (Table 1). Each candidate gene was manually curated, taking into account recent relevant literature and the predicted conserved domains (www.ncbi.nlm.nih.gov/Structure/cdd), and assigned to a functional group. Gene ontology (GO) annotation was performed using Blast2GO software (Götz et al., 2008) with default parameters and Mapman annotation (Klie and Nikoloski, 2012) was performed using Mercator (Lohse et al., 2014). Fisher’s exact test was used to evaluate the significance (P-value<0.05) of term enrichment in each category of candidates.
Table 1.
Categories of DT-related candidates and the filters used for each category
| Cluster | Filtersa | n | Uniques | Candidates | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | a | A5<A6 | E5<E6 | C6<A6 | C6<E6 | C5<A6b | C5<E6b | 49 | 17 | C1=150 |
| 1 | b | A5<A7 | E5<E7 | C7<A7 | C7<E7 | C5<A7b | C5<E7b | 133 | 101 | |
| 2 | a | A5>A6 | E5>E6 | C6>A6 | C6>E6 | C5>A6b | C5>E6b | 111 | 33 | C2=292 |
| 2 | b | A5>A7 | E5>E7 | C7>A7 | C7>E7 | C5>A7b | C5>E7b | 259 | 181 | |
| 3 | a | C5<C6 | C6>A6 | C6>E6 | C6>A5 | C6>E5 | 180 | 57 | C3=571 | |
| 3 | b | C5<C7 | C7>A7 | C7>E7 | C7>A5 | C7>E5 | 514 | 391 |
Cluster C1 is designed to comprise genes that are up-regulated during late maturation in both C. arabica and C. eugenioides desiccation-tolerant seeds, and whose expression is concomitantly lower in desiccation-sensitive C. canephora seeds (key positive effectors of DT). Cluster C2 groups genes that are down-regulated during late maturation in both C. arabica and C. eugenioides desiccation-tolerant seeds, and whose expression is concomitantly higher in desiccation-sensitive C. canephora seeds (negative effectors of seed DT). Cluster C3 is the opposite of C1; it includes genes that are up-regulated during late maturation in desiccation-sensitive C. canephora seeds, and whose expression is concomitantly lower in both C. arabica and C. eugenioides seeds. C3 thus comprises late maturation genes specific to a desiccation-sensitive context.
a An adjusted P-value cut-off of 0.05 was used during the filtering.
b For these filters a P-value cut-off was not imposed.
Proteome analysis
Soluble proteins were extracted from 500 mg of ground mature coffee endosperms by suspension and stirring in 10 ml buffer (50 mM HEPES pH 8.0, 1 mM EDTA, 5 mM DTT, 4% polyvinylpolypyrrolidone, 700 units of benzonase endonuclease, and 1% protease inhibitor cocktail) for 40 min at 4 °C. After centrifugation, proteins were precipitated from supernatant at 4 °C with trichloroacetic acid (10% v/v), washed twice with acetone, and air dried. Proteome analysis included protein fractionation by one-dimensional SDS-PAGE sliced into four bands, tryptic digestion, and analysis of samples by nano-LC-MS/MS using a QExactive Plus (ThermoFisher Scientific Inc), as described by Geiger et al. (2015). Raw files were analysed using Maxquant 1.5.5.1 on the predicted C. canephora peptide database (25 574 peptides, http://coffee-genome.org/) with a protein and peptide false discovery rate <1%, as described in Chen et al. (2019). Difference in protein abundance between species was tested by one-way ANOVA and post hoc Tukey test. Label-free data have been deposited at ProteomeXchange under the project number PXD015806.
Hormone analysis
Hormones in freeze-dried powder of endosperms (50 mg) were analysed by National Research Council, Canada. ABA and ABA metabolites, cytokinins, auxins, and gibberellins were quantified by ultra-performance liquid chromatography–electrospray ionization–tandem mass spectrometry as previously described (Chiwocha et al., 2003).
Promoter cis-element analysis
The 1000 bp sequences upstream of the start codon of the candidate genes and of a random selection of 500 C. canephora genes (control genes) were collected from the Coffee Genome Hub (coffee-genome.org) and submitted to the Genomatix MatInspector tool (Cartharius et al., 2005). The presence of seed and stress-related plant cis-elements was determined using the default parameters of MatInspector. The resulting contingency tables were submitted to enrichment analysis (hypergeometric test in R) by comparing the candidate gene cis-elements by cluster with those found in the control genes.
Results
The transcriptional program associated with late seed maturation shares common essential features among the different coffee species
Mature seeds of C. eugenioides and C. arabica were tolerant to relatively intense dehydration, with almost no loss of viability noticed when dried up to 23% RH (Fig. 1A). For both species, the equilibrium relative humidity at which 50% of the initial viability was lost, RH50, was lower than 10%. By contrast, C. canephora seeds displayed significantly higher desiccation sensitivity. They could only withstand mild drying without noticeable loss of viability, i.e. drying at 62% RH, and did not survive drying at 23% RH. The equilibrium RH50 estimated for C. canephora was around 50%. At stage (ST) 5, most seeds of the three species were already able to germinate and to develop into normal seedlings but displayed high mortality upon drying, at both 45% and 62% RH (Fig. 1B). The seeds of the three studied species acquired most of the capacity to be dried to 62% RH between ST5 and ST6 (Fig. 1B), demonstrating a conserved phenological sequence among the three coffee species for partial DT acquisition.
Fig. 1.
Characterization of seed desiccation tolerance in the three coffee species, Coffea arabica, C. canephora, and C. eugenioides. (A) Survival rate of mature (stage 7) coffee seeds after equilibration drying at various relative humidity (RH) conditions. The curves correspond to the fitted patterns using the quantal response model described in Dussert et al. (1999). (B) Changes in viability (%) after equilibration drying at various RH conditions during seed development (stage 5–6).
DT acquisition was concomitant with a major transcriptional switch observed during late maturation in all three species (Fig. 2A). When species were analysed separately, most transcriptional changes appear to occur in the transition between the ST5 and ST6 with 2837, 3622, and 4109 DEGs identified in C. eugenioides, C. arabica, and C. canephora, respectively (11.3–16% of the reference genes). The ST5–ST7 comparison, i.e. taking the whole late maturation phase into account, identified a larger number of DEGs than the ST5–ST6 transition, specifically 6285, 5506, and 5535, in C. canephora, C. arabica, and C. eugenioides, respectively (21.5–24.6% of the reference genes). By contrast, the late maturation stages ST6 and ST7 were transcriptionally similar, with very few DEGs between them (16, 8, and 82 DEGs in C. canephora, C. arabica, and C. eugenioides, respectively). It is worth noting that down-regulated genes represented a slightly higher fraction than the up-regulated genes in these comparisons. Hierarchical clustering analysis of the global expression dataset grouped the vast majority of ST5 samples on one major branch of the dendogram (group I, Fig. 2B), while late stages ST6 and ST7 grouped together, by species, on another branch (group II). This suggests a close transcriptional program between the three species, with 2533 genes being commonly up- or down-regulated in the three coffee species (Fig. 2C; Supplementary Table S4).
Fig. 2.
Analysis of the transcriptional program associated with seed late maturation in the three coffee species Coffea arabica, C. canephora, and C. eugenioides. (A) Numbers of differentially expressed genes (adjusted P-value<0.05) in the three Coffea species between the consecutive developmental stages 5 and 6 (ST5–6 transition), stages 6 and 7 (ST6–7 transition), as well as over the entire late maturation period (ST5–7). Up- and down-regulated genes are labelled in red and blue, respectively. (B) Hierarchical clustering of normalized RNA-seq libraries based on Euclidian distance matrix; similar samples are darker blue. (C) Venn diagrams of the total up- and down-regulated genes between all stage comparisons, of each species studied (Ara, C. arabica; Eug, C. eugenioides; Can, C. canephora). Numbers in parentheses indicate the percentage of the DEGs that are found in each Venn category for a given species. The order is always Can, Eug, Ara.
Mapman-based DEG enrichment analysis showed a clear overview of molecular processes involved in the late maturation program, with a total of 94 functional categories detected as significantly enriched at least in one species (Fig. 3; Supplementary Tables S5, S6). More categories (62) were significantly over-represented among the down-regulated genes than among the up-regulated genes (30), and only two categories displayed contrasting behavior among different species (minor CHO metabolism and redox processes, mapman bin codes 3 and 21, respectively, which were enriched in C. canephora up-regulated genes and in C. eugenioides down-regulated genes). Most importantly, 24 functional categories (25.5%) were shared among the three species for at least one of the two comparisons (ST5–6 or ST5–7, labeled by red stars in Fig. 3) while each species also revealed a similar number (14–15) of specific categories. Transcriptional processes associated with C. canephora during the ST5–6 transition appear to be the most dissimilar compared with other stages and species, since only nine out of the 24 shared functional categories are represented during this early transition, whereas 23/24 were identified within the ST5–7 comparison. By contrast, most of these shared functional categories were already detected during the early ST5–6 transition among C. arabica and C. eugenioides. This important result indicates that the late maturation transcriptional program is delayed in the desiccation-sensitive C. canephora compared with species with desiccation-tolerant seeds.
Fig. 3.
Mapman functional class enrichment (Fisher enrichment test, adjusted P-value<0.05) for DEGs of ST5–6 and ST5–7 for each Coffea species. Up- and down-regulated genes are colored in shades of orange and blue, respectively; only significant enrichments are colored. Mapman functional classes which are found to be enriched in all three species are indicated by a red star. FDR, false discovery rate.
The composition for the 18 common categories shared between species for down-regulated genes strongly suggests a transcriptional co-ordination of metabolic slow-down during late maturation, with a significant decrease in mRNA abundance of genes involved in key energetic processes such as respiration (including the TCA cycle, mitochondrial electron transport and ATP synthase) and photosynthesis (including light reactions and photosystem I components). Genes encoding components of cytoskeleton, membrane transporters, or components of the machinery for cell wall precursors were also found to be significantly down-regulated (Fig. 3). GO enrichment analysis revealed similar features, but also pointed out a general decrease of carbohydrate metabolic process, and intracellular trafficking, including endomembrane systems and Golgi apparatus (Supplementary Table S7).
With regard to functional classes associated with late maturation up-regulated genes, only six were found to be shared among the three species. These mostly concerned RNA processing (splicing and RNA binding) and the translational machinery (i.e. ribosome biogenesis, including pre-rRNA processing and modifications). Surprisingly, functional enrichment analysis did not identify any functional class that would be specifically induced during late maturation in desiccation-tolerant seeds only. By contrast, it revealed several specific features of desiccation-sensitive C. canephora seeds. Indeed, functional processes such as gluconeogenesis and carbohydrate metabolism, flavonoid metabolism, redox homeostasis, and hormone metabolism (gibberellin synthesis and auxin-regulated genes) were significantly over-represented among the C. canephora genes up-regulated during late maturation (Fig. 3).
DT-related candidate genes and processes identified through inter-specific comparative transcriptomics
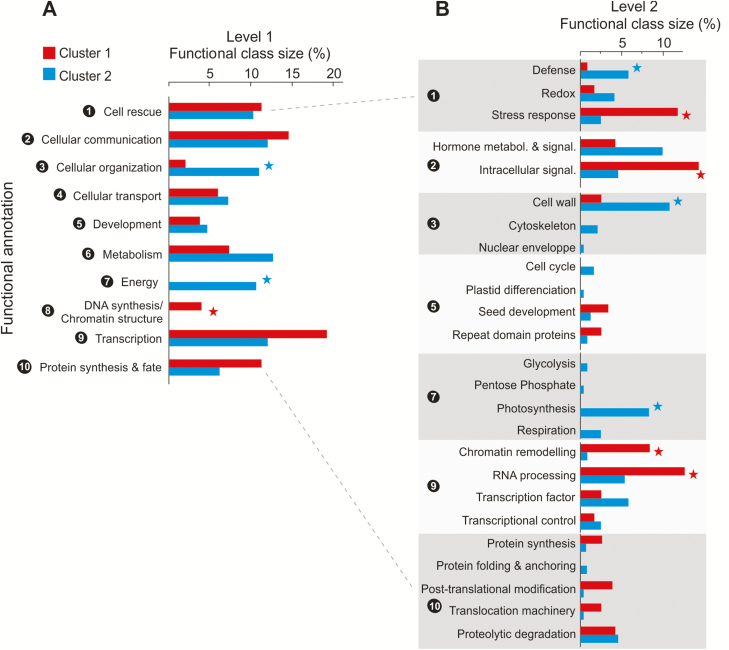
The DEGs between species/stages were filtered by using combinations of Boolean operators to identify candidate genes related to different categories of DT (Table 1; Fig. 4). The first cluster (C1; 150 genes), designed to encompass positive effectors of DT, groups genes that are up-regulated during late maturation in both C. arabica and C. eugenioides seeds, and whose expression is concomitantly lower in desiccation-sensitive C. canephora seeds (Fig. 4; Supplementary Tables S7, S8). Interestingly, the 1000 bp promoter regions of C1 candidate genes were significantly enriched in four cis-elements (Supplementary Table S9), including CGCG, which is bound by Calmodulin-binding transcription activator and known to play a key role in plant stress response (Galon et al., 2008; Doherty et al., 2009). Among the 120 genes with expert-assigned putative functions, the most-represented functional classes were transcriptional activities (19.2% of the 120 candidate genes), cellular communication (14.6%), cell rescue (11.3%), and protein synthesis and fate (11.3%) (Fig. 5A).
Fig. 4.
Candidate gene clusters related to desiccation tolerance. The standardized log2 of the gene expression is plotted against the developmental stage of each Coffea species. The average gene expression for each cluster is traced in a dark line, while individual gene expressions are plotted in thin gray lines.
Fig. 5.
The percentage of candidate genes in clusters 1 (red) and 2 (blue) that belong to each functional class. Class sizes that are significantly different between the two clusters (χ 2 test, P-value<0.05) are indicated with asterisks. (A) The first level of functional terms; (B) the second level of functional terms. Functional terms that are child terms of the first level are separated and numbered for ease of visualization.
The second cluster (C2), designed to comprise negative effectors of seed DT, was composed of genes that are down-regulated during late maturation in both C. arabica and C. eugenioides seeds, and whose expression is concomitantly higher in desiccation-sensitive C. canephora seeds (Fig. 4). Cluster C2 contains 292 genes, including 250 with a putative function assigned, with primary metabolism (12.7% of the candidate genes), cellular communication (12%), cellular organization (11%), transcriptional activities (11.6%), and energetic processes (10.6%) being the most represented functional classes in this group (Fig. 5A; Supplementary Table S7). Generation of precursor metabolites and energy, photosynthesis, and TCA cycle were among the biological processes significantly enriched using GO or Mapman ontologies (Supplementary Table S8). This result suggests that over the course of seed maturation differences in DT between species may be based on quantitative differences at the level of transcriptional control of the metabolic slow-down associated with seed quiescence.
The analysis of the relative size and composition of the different gene functional classes (Fig. 5B) revealed striking differences between DT-associated (C1) and DS-related genes (C2), with some functional classes being almost exclusively represented in one group. Specifically, C1 alone includes representatives of the functional class related to the synthesis and repair of DNA and chromatin structure (Fig 5A). Similarly, the relative size of the functional class related to transcriptional processes was highest for DT-associated genes, with many genes predicted to be implicated in RNA processing or encoding many chromatin remodeling factors involved in regulation of transcription, but with very few TFs (three) including GATA and MADS factors (Fig 5B). In this context, it is worth noting that C1 contains multiple genes encoding putative chromatin remodeling factors/complex subunits, including MRG1 (Cc07_g06470), SWC6 (Cc07_g10850), SWI3A (Cc07_g20270), and SWIB complex BAF60b domain-containing protein (Cc02_g18450). In Arabidopsis, MRG1 has been shown to be involved in increasing the transcriptional levels of the flowering time genes FLC and FT (Bu et al., 2014) while SWC6 is a component of the SWR1 complex that mediates transcriptional regulation of selected genes such as FLC (Choi et al., 2007). By contrast, C2 does not contain chromatin remodeling factors; instead it contains many TFs (14) related to key developmental activities and cell differentiation, including homologs of HOX, B3, LOB, MADS, and MYB TFs. This result suggests DT acquisition could be associated with intense chromatin remodeling and repression of specific developmental TFs.
Moreover, C2 displays a higher number of genes involved in redox homeostasis and defense against biotic stress while C1 includes many genes with a direct role in the regulation of abiotic stress response (Fig 5B). C1 genes include key stress proteins such as LEA (EM6, Cc05_g00080), co-chaperone (JAC1, Cc10_g03210; Cc08_g03570), cold-stress (COR28, Cc02_g18840), and drought-stress proteins (ERD4, Cc11_g12570) (Supplementary Table S7). C1 also includes mediators of ABA and jasmonate stress response (AFP2, ABO5, NHL6, MKS1), a regulator of ROS homeostasis (APP1, Cc07_g09130; Yu et al., 2016), a modulator of unfolded protein response (CPR5, Cc07_g16050), and proteins involved in plastid stress response mediated by effector nucleotides (ppGpp synthetase, Cc10_g06010)(Supplementary Table S7).
With regard to cellular communication processes, while C1 presents mainly genes implicated in signal transduction processes (Fig 5B), such as components of receptor-like kinase signaling pathways, protein kinase cascades, and protein phosphatases (subunits of PP2A and PP2C), C2 mostly contains genes implicated in hormone metabolism and signaling, a majority of them being putatively dedicated to the metabolism, transport, and perception of auxin, and, to a lesser extent, cytokinin and gibberellins (Supplementary Table S7).
Finally, functional classes related to energetic processes appear mostly restricted to DS-related genes (Fig 5). Many C2 genes are associated with key energetic processes such as respiration (including molecular components of the TCA cycle, respiratory electron transfer chain and ATP synthase), photosynthesis, and glycolysis, as well as key enzymes of the pentose phosphate pathway. These candidates point to the existence of a transcriptional orchestration of metabolic quiescence that differs between desiccation-tolerant and desiccation-sensitive seeds. This could lead to different levels of repression of energetic processes and basal cellular processes when entering quiescence during seed desiccation.
Desiccation-sensitive specific genes expressed during late maturation
Finally, a third cluster (C3) included 571 genes specific to a desiccation-sensitive context, i.e. genes that are up-regulated during late maturation in C. canephora seeds, and whose expression is concomitantly lower in both C. arabica and C. eugenioides desiccation-tolerant seeds (Fig. 4). The Mapman functional classes over-represented within this cluster are mainly related to nitrogen metabolism, tocopherol biosynthesis, and translational control (Supplementary Table S8). As shown for genes that were up-regulated during C. canephora late seed maturation (Fig. 3), auxin-regulated genes were also enriched in C3. Accordingly, the analysis of promoter regions for C3 candidate genes revealed significant enrichment for three cis-elements (Supplementary Table S9), including the auxin response factor ARF3 cis-element. This motif is bound by auxin-regulated TFs (Chandler, 2016), probably including the ARF homolog gene (Cc08_g16330) detected in C3. The cis-element GCCF (GCC-box bound by TF of the AP2/ERF family) was shown to be enriched in both C3 and C1 genes, suggesting this cis-element is an important driver of gene expression at the late maturation stage, independently of the DT level. The most common GCCF cis-element signatures found in these two clusters are those of the AtERF-1 and of the RAP2.6 TF, known to be involved in jasmonate- and ethylene-dependent stress response, and in ABA and salt stress response signaling, respectively (Lorenzo et al., 2003; Pré et al., 2008; Zhu et al., 2010).
A comparison of the functional classes’ relative size between C1 and C3 also highlighted striking differences in late maturation transcripts specific to C. arabica and C. eugenioides compared with the desiccation-sensitive C. canephora seeds (Fig. 6). Interestingly, genes of C1 are enriched in functions related to stress response, chromatin remodeling, and RNA processing when compared with C3, as seen in the C1–C2 comparison. By contrast, C3 contains many genes involved in defense against biotic stress or associated with energetic processes (cytosolic and plastidial glycolysis, control of the redox poise in plastid and mitochondrial respiratory electron transporters), and cell cycle or organelle differentiation (Fig. 6). In addition to many TFs, C3 also contains numerous genes encoding members of the Pentatricopeptide repeat-containing (PPR) protein family. To date, a majority of characterized PPRs are predicted or shown to localize to chloroplasts or mitochondria where they play essential roles in gene expression, mRNA editing, mRNA processing, and protein synthesis (Hayes et al., 2015; Liu et al., 2016).
Fig. 6.
The percentage of candidate genes in clusters 1 (red) and 3 (green) that belong to each functional class. Class sizes that are significantly different between the two clusters (χ 2 test, P-value<0.05) are indicated with asterisks. (A) The first level of functional terms; (B) the second level of functional terms. Functional terms that are child terms of the first level are separated and numbered for ease of visualization.
Comparative proteomics validates processes and candidate genes inferred from transcriptome analysis
The soluble proteome of mature coffee seeds was analysed among the three species in order to ascertain the differential functions inferred from transcriptomic analysis (Supplementary Table S10; Supplementary Fig. S1). Among the 1389 proteins accurately detected in at least one species, quantification revealed 95 proteins whose abundance is higher in desiccation-tolerant C. arabica and C. eugenioides seeds compared with C. canephora (Supplementary Table S11). Mapman functional classes over-represented within this group are mainly related to calcium signaling, protein post-translational modification, and synthesis of amino acids of the serine–glycine–cysteine group (Supplementary Table S12). This proteomic analysis also revealed 99 proteins over-accumulated in desiccation-sensitive C. canephora mature seeds. Hormone metabolism related to auxin, secondary metabolism associated with flavonoids, and biotic stress-associated PR proteins were among the biological processes significantly enriched using Mapman ontologies (Supplementary Table S12). All these functional classes were also detected at the transcriptome. Besides, the expert analysis of the relative size and composition of the different functional classes revealed striking differences between DT- and DS-associated proteins, with some functional classes such as those related to glycolysis, photosynthesis, hormone signaling, and defense against biotic stress being higher in desiccation-sensitive C. canephora seeds as observed with transcriptomic data (Supplementary Fig. S2). Finally, DT candidates detected at both transcript and protein levels included the LEA EM6 in desiccation-tolerant seeds and the glycolytic GAPC2c (Cc07_g00460) or the redox-involved NRX1 (Cc11_g14760) for desiccation-sensitive seeds (Supplementary Table S13).
Hormone sensing and signaling pathways are differentially expressed between desiccation-tolerant and -sensitive coffee species
Phytohormone profiling detected only faint amounts of active molecules in mature seeds and no significant differences among the three species studied (Supplementary Table S14). However, our survey highlighted key genes involved in hormone sensing, predominantly in desiccation-sensitive seeds (Supplementary Table S7). Indeed, in the DT-repressed genes (C2) we found homologs of major hormone transporters (auxin, PIN1, Cc06_g19880; ABA, ABCG40, Cc11_g01420; cytokinin, ABCG14, Cc08_g05640; Zhang et al., 2014; Kang et al., 2015), several predicted auxin response regulators (Phabulosa, Cc09_g08040; LBD16, Cc01_g18520; MIZU-KUSSEI-like protein, Cc04_g07700; Lee et al., 2009; Moriwaki et al., 2011; Müller et al., 2016), and a cytokinin signaling inhibitor that mediates auxin–cytokinin crosstalk (AHP6, Cc04_g04370; Bishopp et al., 2011). Furthermore, C3 (DS-specific late maturation genes) contained predicted hormone receptors (ABA, PYL8, Cc08_g15960; auxin, AFB5, Cc10_g15520; Prigge et al., 2016), and other potential auxin response proteins (ARF9, Cc08_g16330, IAA1, Cc04_g03620). This gene composition fingerprint suggests increased hormone sensitivity in desiccation-sensitive seeds, especially for auxin and cytokinin, while desiccation-tolerant seeds displayed higher expression (C1) for NHL6 (Cc05_g06810), a mediator of ABA signaling in Arabidopsis (Bao et al., 2016). Differentially accumulated proteins associated with auxin signaling were also detected in the proteome. Similar levels of phytohormones in mature seeds do not exclude the possibility that desiccation-tolerant and -sensitive species integrate and respond to the hormone signals differently.
Desiccation-tolerant and -sensitive coffee seeds display differences in transcriptional and metabolic control of respiratory processes
The three coffee species shared a transcriptional slow-down for energy metabolism during seed maturation with down-regulation of numerous mitorespiration-related transcripts, such as those encoding several subunits of ATP synthase, many components of the electron transfer chain (NADH dehydrogenase complex I, cytochrome bc1 complex, cytochrome c oxidase), as well as many enzymes of the TCA cycle (isocitrate dehydrogenase, pyruvate dehydrogenase, succinyl-CoA ligase, dihydrolipoyl dehydrogenase) (Supplementary Table S4). However, the interspecific transcriptome comparison revealed fundamental differences in the regulation of mitochondrial energy metabolism between desiccation-tolerant and desiccation-sensitive seeds. First, compared with C. canephora, desiccation-tolerant seeds displayed higher expression (C1 genes) of the translocase inner membrane subunit TIM44-2 (Cc01_g18340) and the mitochondrial splicing factor OTP439 (Cc01_g05040, Organelle transcript processing), involved in basal cellular processes such as protein import and organelle post-transcriptional processes, respectively (Murcha et al., 2014; Colas des Francs-Small et al., 2014). Secondly, there is a down-regulation in desiccation-tolerant seeds (C2 genes) of many genes encoding proteins pivotal for regulation of energetic processes, namely malate dehydrogenase MMDH1 (Cc02_g32320), a central enzyme in the TCA cycle (Sew et al., 2016), the voltage-dependent anion channel VDAC1 (Cc11_g13460), and the ATP synthase delta subunit (Cc02_g04940). VDAC1, an abundant protein of the outer mitochondrial membrane, contributes to the exchange of small metabolites essential for respiration and plays a role in redox control (Tateda et al., 2011; Robert et al., 2012). This transcriptional fingerprint suggests a slow-down of the machinery dedicated to oxidative phosphorylation, i.e. ATP production, without necessarily affecting the electron transfer chain operability. By contrast, desiccation-sensitive C. canephora seeds displayed late maturation induction (C3 genes) of several respiratory electron transfer chain complex components or genes involved in their assembly, namely complex I accessory subunits 6 and 7 (Cc02_g25880, Cc08_g10410) and complex I α subcomplex assembly factor 3 (Cc02_g25880), as well as the ubiquinone biosynthesis protein COQ4 (Cc06_g15640) and the cytochrome c-type biogenesis protein CcmH (Cc04_g07670). Desiccation-sensitive C. canephora seeds also displayed late maturation induction of genes involved in mitochondrial post-transcriptional and translational activities, such as elongation factors EFTu and EFTs (Cc03_g02960 and Cc05_g09920), and editing factors MEF10 and MEF11 (Cc04_g00410 and Cc11_g07090, respectively), the latter also being involved in ABA signaling (Sechet et al., 2015). The high expression level of these genes corresponds to the signature of organelles capable of maintaining high energy metabolism on demand. In this context, one may note the presence of a mitochondrial phosphate transporter, MPT3 (Cc07_g01200), which plays vital roles in phosphate homeostasis and ATP biosynthesis and whose expression levels modulate salt stress tolerance in Arabidopsis (Zhu et al., 2012). Finally, C3 contains two genes encoding proteins that may play a role in regulating mitochondrial metabolism under stress conditions: formate dehydrogenase (FDH, Cc07_g02270) and prohibitin PHB3 (Cc07_g00470). The mitochondrial FDH is a positive regulator of cell death and defense response, and confers tolerance to hypoxia, metal toxicity and low pH in Arabidopsis (Choi et al., 2014; Lou et al., 2016) while PHB3 functions in nitric oxide-mediated plant stress responses (Wang et al., 2010).
This transcriptional signature for differential energetic processes among desiccation-tolerant and desiccation-sensitive seeds led us to probe their respiratory activity during desiccation. Respiration rates, estimated as CO2 release, were measured on mature seeds of the three coffee species at various moisture contents upon dehydration (Fig. 7). Within the 0.6–0.8 g H2O g−1 DM hydration window, all seeds displayed roughly equivalent respiratory activities. Upon desiccation, C. arabica and C. eugenioides seeds displayed a smooth reduction in respiration rate until ceasing detectable respiration at approximately 0.3 g H2O g−1 DM. Desiccation-sensitive C. canephora seeds, however, had a biphasic mode of respiration, initially displaying rather stable respiration rates between 0.8 and 0.4 g H2O g−1 DM. Below this threshold C. canephora seeds experienced drastic reduction in respiration rate that remained significantly higher than that measured for C. arabica and C. eugenioides seeds (Fig. 7).
Fig. 7.
Respiration rates, measured as release of CO2 (µmol CO2 min−1 g−1 DM) as a function of the hydration state (g H2O g−1 DM) of fresh mature seeds (ST7) of the three coffee species during equilibrium drying.
Discussion
A late maturation program also occurs in desiccation-sensitive coffee seeds
Performed on developing seeds of three coffee species, the present transcriptome survey reveals the shared existence of a major transcriptional switch during the ST5–ST6 transition (Fig. 2A). This switch coincides with partial DT acquisition in the three species (Fig. 1), and is a mark of a late maturation program since it occurs well after transcriptional events associated with storage reserve deposition, as already described in C. arabica (Dussert et al., 2018). A large number of processes associated with late maturation are conserved among the three species, as demonstrated through functional enrichment analysis. This suggests a late maturation phase has been conserved during the course of Coffea evolution independently of DT levels. Among the 6285 DEGs (24.6% of the reference genes) identified in C. canephora, 40.3% were found to be commonly up- or down-regulated in the other coffee species (Fig. 2C), suggesting the late maturation phase serves the acquisition of several major functional traits other than DT. Initially thought to be restricted to orthodox seeds (reviewed in Leprince et al., 2017), a late maturation phase is presumably conserved in intermediate seeds, even those that display very low DT such as those of C. canephora. Interestingly, a recent proteomic and metabolomic survey of the recalcitrant cocoa developing seed also revealed important metabolic changes during the late phases of development, including accumulation of stress-related proteins, which occurred after the peak stage of reserve deposition, thus resembling a late maturation phase (Wang et al., 2016).
Desiccation-tolerant seeds harbor key stress proteins and integrators of stress response
Analysis of the DEGs identified relatively few DT-specific up-regulated genes (C1, 150 genes) compared with DT-specific down-regulated genes (C2, 292 genes), and identified many genes specifically induced in maturing desiccation-sensitive C. canephora seeds (C3, 571 genes; Fig. 4). Although transcription factors were under-represented among DT candidate genes (Figs 5, 6), three putative TFs were identified, a MADS (Cc09_g07070) and two GATA (Cc02_g34640; Cc01_g21240). These deserve further attention and validation of their potential role in orchestrating the DT-associated transcriptome during late maturation in coffee seeds.
In addition to these TFs, our survey did detect multiple genes related to cell rescue and stress response. Most notably, our survey revealed one LEA protein, EM6 (Cc05_g00080), whose gene expression and protein levels are significantly higher in desiccation-tolerant coffee seeds. EM6 protein accumulation has been previously observed temporally associated with DT acquisition during seed development, as well as with re‐induction of DT in germinated radicles of Medicago truncatula (Boudet et al., 2006; Chatelain et al., 2012). Due to their hydrophilic nature and hydration buffer capacity, certain LEA proteins may participate in the control of water loss during maturation. EM6 from Medicago was found in vitro to bind water much more efficiently at 75% RH compared with a control protein, acting as a molecular sponge (Boucher et al., 2010). Similarly, EM6 has been suggested to play a critical role in water binding during Arabidopsis maturation drying (Manfre et al., 2009). In addition to the EM6, two heat-shock protein 70 co-chaperones were identified among DT-associated genes, including JAC1 (Cc10_g03210), known to be involved in plastid movement in Arabidopsis (Takano et al., 2010). Since both heat-shock proteins and LEA proteins have been proposed to play important roles in DT through chaperone protection of membranes and/or proteins in plants and various other organisms (Tolleter et al., 2007; Chakrabortee et al., 2007; Erkut et al., 2011; Tapia et al., 2015; Boothby et al., 2017), it is tempting to attribute a quantitative role to these candidates in the seed DT level of coffee species. Furthermore, among stress-response genes with increased expression during DT acquisition, genes such as those for COR28, ERD4, and APP1 are worth noting, the latter playing a critical role in the control of H2O2 content and ROS homeostasis in Arabidopsis (Yu et al., 2016). ERD4 was shown to be an osmosensitive calcium-permeable cation channel (Hou et al., 2014), and therefore could play a central role in early salt and drought stress signaling through variation of cytosolic calcium fluxes. Because cytosolic calcium elevation is one of the earliest responses of plant cells to different stress stimuli (Knight et al., 1991), the presence in C1 of ERD4 and the calcium-binding protein Calmodulin-like38 (CML38, Cc07_g00660) is particularly attractive since they may act as sensors of early subtle variations of water potential upon dehydration and play a role in orchestrating metabolic quiescence. Interestingly, we also detected calcium signaling through proteomics, including calmodulin CAM7 (Cc04_g09200 and Cc06_g22690) involved in ABA responsiveness in Arabidopsis (Abbas and Chattopadhyay, 2014). Finally, concerning C1 genes involved in stress response, one may also note the presence of genes encoding DNA ligase (Cc00_g04310) and SMC5 (Cc02_g28540), which both have canonical functions in repair of damaged DNA and maintenance of genome integrity (Watanabe et al., 2009). Different DNA ligases were indeed shown to be involved in DNA repair during early imbibition, and were major determinants of Arabidopsis seed longevity (Waterworth et al., 2010).
Interestingly, C1 also included three genes coding for proteins that were shown to play important roles in ABA signaling, namely NHL6 (Cc05_g06810), AFP2 (Cc11_g11930) and ABO5 (Cc06_g12800). NHL6 has recently been demonstrated to act as positive regulator of ABA-mediated seed germination inhibition in Arabidopsis (Bao et al., 2016) while AFP2 acts epistatically to ABI5, a master regulator of ABA signaling and seed maturation in orthodox seeds (Garcia et al., 2008; Zinsmeister et al., 2016). Mutations in AFP2 result in increased sensitivity to ABA in Arabidopsis seeds (Garcia et al., 2008). Finally ABO5 mediates ABA response in organelles through splicing of key mitochondrial genes in Arabidopsis (Liu et al., 2010). Indeed, Arabidopsis abo5 mutants expressed lower transcripts of stress‐inducible genes as well as plastid-related genes involved in adaptation of photosynthesis to low ATP conditions (Liu et al., 2010). In genes specifically up-regulated in desiccation-sensitive seeds (C3, Fig. 6), we found PYL8 (Cc08_g15960), an ABA sensor in Arabidopsis seeds (Saavedra et al., 2010). The presence of these hormone-sensing and modulation genes in our candidate clusters suggests that a differential fine-tuning of ABA perception could be implicated in the differential acquisition and regulation of DT. Similarly, modulation of ABA sensitivity, rather than ABA content, was recently proposed to regulate the ability of DT re-induction and DT loss during Arabidopsis seed germination (Maia et al., 2014). Our survey also revealed DT and DS differences in the expression of nuclear-encoded mitochondrial genes dedicated to energetic processes. Recent evidence suggests hormones and the mitochondrial metabolism could be more closely linked than initially thought as ABA was shown to induce typical transcripts of the mitochondrial retrograde response while auxin has the inverse effect (Ivanova et al., 2014; Wagner et al., 2018).
Metabolic shut-down is better coordinated in desiccation-tolerant seeds
In coffee seeds, the major transcriptional switch observed during late maturation included several nuclear-encoded mitochondrial proteins that are markers of large reorganization of energy metabolism (Fig. 3), mimicking what is observed in orthodox seeds, such as down-regulation of genes dedicated to the TCA cycle (Logan et al., 2001; Howell et al., 2006). Furthermore, our dataset revealed numerous quantitative differences in transcript accumulation for key mitochondrial proteins between coffee seeds displaying different DT levels. Among them, it is worth noting that desiccation-sensitive C. canephora seeds displayed enhanced accumulation of transcripts for subunits of the complexes I and III (NADH dehydrogenase and cytochrome bc1), the major sites of superoxide production through one-electron reduction of molecular oxygen (Wagner et al., 2018). These transcriptomic differences observed between desiccation-tolerant and -sensitive coffee seeds appear to reflect the potential to slow down metabolism and respiration during dehydration rather than an actual slow-down phase during maturation since measured respiration rates in non-dehydrated mature seeds were similar among the three coffee species (Fig. 7). However, during their dehydration we observed striking differences in respiration rates. A better preparation or coordination of down-regulation of metabolism in desiccation-tolerant seeds during drying could play an important role in avoiding or limiting oxidative stress conditions and/or accumulation of by-products of metabolism to toxic levels. Seed respiration is generally detectable around 0.25 g H2O g−1 DM and respiration rates increase with increasing water contents above this threshold (Vertucci and Leopold, 1984; Vertucci, 1989). However, it is considered that at intermediate water contents the metabolism that occurs is unregulated, and there is evidence of damaging reactions that are probably free radical-mediated occurring in this water-content range (Leprince et al., 1990; Hendry et al., 1992; Hendry, 1993; Finch-Savage et al., 1994; Vertucci and Farrant, 1995). Interestingly, differences in the respiratory activities we observed between intermediate coffee seeds displaying different DT levels upon dehydration were similar to those observed between intermediate tea seeds and orthodox pea seeds (Walters et al., 2001), and between different recalcitrant Castanea seed tissues (axes and cotyledons) displaying differences in desiccation sensitivity (Leprince et al., 1999). The lack of coordinated repression of metabolism during drying is thought to play a predominant role in the oxidative burst that triggers lipid oxidation, membrane disruption, and ultimately the death of recalcitrant seeds (Leprince et al., 1999; Bailly, 2004; Kranner and Birtic, 2005). This mechanism could be shared with intermediate seeds displaying relatively low DT levels. Given that ROS‐provoked mitochondria‐dependent cell death has also been recently described for hydrated orthodox elm seeds under detrimental artificial ageing conditions (Wang et al., 2015), it defines mitochondria and the regulation of respiratory processes as universal targets of seed sensitivity to stress.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Protein abundance variation among mature coffee seeds of the three species.
Fig. S2. Functional categories of DT-related proteins.
Table S1. RNAseq statistics of samples used.
Table S2. Gene expression (RPKM) in each sample.
Table S3. Pairwise comparisons of gene expression.
Table S4. Genes commonly up- and down-regulated during late maturation.
Table S5. Mapman functional enrichment analysis of late seed maturation.
Table S6. GO-term enrichment analysis of late maturation.
Table S7. DT-related gene candidates of seed late maturation.
Table S8. Mapman and GO functional enrichment analysis of DT-related clusters.
Table S9. Promoter cis-element enrichment analysis.
Table S10. Proteome of mature coffee seeds.
Table S11. DT-related protein candidates.
Table S12. Mapman functional enrichment analysis of candidate proteins.
Table S13. Candidate genes detected at both transcript and protein levels.
Table S14. Phytohormone profiling in mature coffee seed.
Supplementary Material
Acknowledgements
AKS benefited from an EMBO long-term fellowship (ALTF 478-2017). The authors thank CIRAD for hosting visiting researchers at the Pôle de Protection des Plantes (3P, Unit PVBMT) and UMR Qualisud in Reunion Island, as well as the Mass Spectrometry Proteomics Platform (MSPP, Montpellier) for proteomic analysis. DS acknowledges financial support from France Génomique National infrastructure, funded as part of ‘Investissement d’avenir’ program managed by Agence Nationale pour la Recherche (contract ANR-10-INBS-09).
Glossary
Abbreviations:
- DEG
differentially expressed gene
- DS
desiccation sensitivity
- DT
desiccation tolerance
- ST
stage.
References
- Abbas N, Chattopadhyay S. 2014. CAM7 and HY5 genetically interact to regulate root growth and abscisic acid responses. Plant Signaling & Behavior 9, e29763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C. 2004. Active oxygen species and antioxidants in seed biology. Seed Science Research 14, 93–107. [Google Scholar]
- Bao Y, Song WM, Pan J, et al. . 2016. Overexpression of the NDR1/HIN1-like gene NHL6 modifies seed germination in response to abscisic acid and abiotic stresses in Arabidopsis. PLoS ONE 11, e0148572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berjak P, Pammenter NW. 2008. From Avicennia to Zizania: seed recalcitrance in perspective. Annals of Botany 101, 213–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black M, Pritchard H. 2002. Desiccation and survival in plants. Drying without dying. Wallingford: CABI Publishing. [Google Scholar]
- Bishopp A, Help H, El-Showk S, Weijers D, Scheres B, Friml J, Benková E, Mähönen AP, Helariutta Y. 2011. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Current Biology 21, 917–926. [DOI] [PubMed] [Google Scholar]
- Boothby TC, Tapia H, Brozena AH, Piszkiewicz S, Smith AE, Giovannini I, Rebecchi L, Pielak GJ, Koshland D, Goldstein B. 2017. Tardigrades use intrinsically disordered proteins to survive desiccation. Molecular Cell 65, 975–984.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher V, Buitink J, Lin X, Boudet J, Hoekstra FA, Hundertmark M, Renard D, Leprince O. 2010. MtPM25 is an atypical hydrophobic late embryogenesis-abundant protein that dissociates cold and desiccation-aggregated proteins. Plant, Cell & Environment 33, 418–430. [DOI] [PubMed] [Google Scholar]
- Boudet J, Buitink J, Hoekstra FA, Rogniaux H, Larré C, Satour P, Leprince O. 2006. Comparative analysis of the heat stable proteome of radicles of Medicago truncatula seeds during germination identifies late embryogenesis abundant proteins associated with desiccation tolerance. Plant Physiology 140, 1418–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Z, Yu Y, Li Z, Liu Y, Jiang W, Huang Y, Dong AW. 2014. Regulation of Arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genetics 10, e1004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitink J, Leprince O. 2008. Intracellular glasses and seed survival in the dry state. Comptes Rendus Biologies 331, 788–795. [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942. [DOI] [PubMed] [Google Scholar]
- Cenci A, Combes MC, Lashermes P. 2012. Genome evolution in diploid and tetraploid Coffea species as revealed by comparative analysis of orthologous genome segments. Plant Molecular Biology 78, 135–145. [DOI] [PubMed] [Google Scholar]
- Chakrabortee S, Boschetti C, Walton LJ, Sarkar S, Rubinsztein DC, Tunnacliffe A. 2007. Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proceedings of the National Academy of Sciences, USA 104, 18073–18078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW. 2016. Auxin response factors. Plant, Cell & Environment 39, 1014–1028. [DOI] [PubMed] [Google Scholar]
- Chatelain E, Hundertmark M, Leprince O, Le Gall S, Satour P, Deligny-Penninck S, Rogniaux H, Buitink J. 2012. Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant, Cell & Environment 35, 1440–1455. [DOI] [PubMed] [Google Scholar]
- Chen Y, Rofidal V, Hem S, et al. . 2019. Targeted proteomics allows quantification of ethylene receptors and reveals SlETR3 accumulation in never-ripe tomatoes. Frontiers in Plant Science 10, 1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiwocha SD, Abrams SR, Ambrose SJ, Cutler AJ, Loewen M, Ross AR, Kermode AR. 2003. A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. The Plant Journal 35, 405–417. [DOI] [PubMed] [Google Scholar]
- Choi DS, Kim NH, Hwang BK. 2014. Pepper mitochondrial FORMATE DEHYDROGENASE1 regulates cell death and defense responses against bacterial pathogens. Plant Physiology 166, 1298–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Park C, Lee J, Oh M, Noh B, Lee I. 2007. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134, 1931–1941. [DOI] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Falcon de Longevialle A, Li Y, Lowe E, Tanz SK, Smith C, Bevan MW, Small I. 2014. The pentatricopeptide repeat proteins TANG2 and ORGANELLE TRANSCRIPT PROCESSING439 are involved in the splicing of the multipartite nad5 transcript encoding a subunit of mitochondrial complex I. Plant Physiology 165, 1409–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JH, Hoekstra FA, Crowe LM. 1992. Anhydrobiosis. Annual Review of Physiology 54, 579–599. [DOI] [PubMed] [Google Scholar]
- Denoeud F, Carretero-Paulet L, Dereeper A, et al. . 2014. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345, 1181–1184. [DOI] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. 2009. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. The Plant Cell 21, 972–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussert S, Chabrillange N, Engelmann F, et al. . 2000. Relationship between seed desiccation sensitivity, seed water content at maturity and climatic characteristics of native environments of nine Coffea L. species. Seed Science Research 10, 293–300. [Google Scholar]
- Dussert S, Chabrillange N, Engelmann F, Hamon S. 1999. Quantitative estimation of seed desiccation sensitivity using a quantal response model: application to nine species of the genus Coffea L. Seed Science Research 9, 135–144. [Google Scholar]
- Dussert S, Davey MW, Laffargue A, Doulbeau S, Swennen R, Etienne H. 2006. Oxidative stress, phospholipid loss and lipid hydrolysis during drying and storage of intermediate seeds. Physiologia Plantarum 127, 192–204. [Google Scholar]
- Dussert S, Serret J, Bastos-Siqueira A, Morcillo F, Déchamp E, Rofidal V, Lashermes P, Etienne H, Joët T. 2018. Integrative analysis of the late maturation programme and desiccation tolerance mechanisms in intermediate coffee seeds. Journal of Experimental Botany 69, 1583–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RH, Hong TD, Roberts EH. 1990. An intermediate category of seed storage behaviour? I. Coffee. Journal of Experimental Botany 41, 1167–1174. [Google Scholar]
- Erkut C, Penkov S, Khesbak H, Vorkel D, Verbavatz JM, Fahmy K, Kurzchalia TV. 2011. Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Current Biology 21, 1331–1336. [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Hendry GAF, Atherton NM. 1994. Free radical activity and loss of viability during drying of desiccation-sensitive tree seeds. Proceedings of the Royal Society of Edinburgh. Section B. Biological Sciences 102, 257–260. [Google Scholar]
- Galon Y, Nave R, Boyce JM, Nachmias D, Knight MR, Fromm H. 2008. Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Letters 582, 943–948. [DOI] [PubMed] [Google Scholar]
- Garcia ME, Lynch T, Peeters J, Snowden C, Finkelstein R. 2008. A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Molecular Biology 67, 643–658. [DOI] [PubMed] [Google Scholar]
- Geiger A, Hamidou Soumana I, Tchicaya B, Rofidal V, Decourcelle M, Santoni V, Hem S. 2015. Differential expression of midgut proteins in Trypanosoma brucei gambiense-stimulated vs. non-stimulated Glossina palpalis gambiensis flies. Frontiers in Microbiology 6, 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Morales SI, Chávez-Montes RA, Hayano-Kanashiro C, Alejo-Jacuinde G, Rico-Cambron TY, de Folter S, Herrera-Estrella L. 2016. Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 113, E5232–E5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Research 36, 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes ML, Dang KN, Diaz MF, Mulligan RM. 2015. A conserved glutamate residue in the C-terminal deaminase domain of pentatricopeptide repeat proteins is required for RNA editing activity. The Journal of Biological Chemistry 290, 10136–10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry GAF. 1993. Oxygen, free radical processes and seed longevity. Seed Science Research 3, 141–153. [Google Scholar]
- Hendry GAF, Finch-Savage WE, Thorpe PC, Atherton NM, Buckland SM, Nilsson KA, Seel WE. 1992. Free radical processes and loss of seed viability during desiccation in the recalcitrant species Quercus robur L. New Phytologist 122, 273–279. [DOI] [PubMed] [Google Scholar]
- Hoekstra FA, Golovina EA, Buitink J. 2001. Mechanisms of plant desiccation tolerance. Trends in Plant Science 6, 431–438. [DOI] [PubMed] [Google Scholar]
- Hou C, Tian W, Kleist T, He K, Garcia V, Bai F, Hao Y, Luan S, Li L. 2014. DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Research 24, 632–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell KA, Millar AH, Whelan J. 2006. Ordered assembly of mitochondria during rice germination begins with pro-mitochondrial structures rich in components of the protein import apparatus. Plant Molecular Biology 60, 201–223. [DOI] [PubMed] [Google Scholar]
- Ivanova A, Law SR, Narsai R, et al. . 2014. A functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates alternative oxidase1a expression in Arabidopsis. Plant Physiology 165, 1233–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joët T, Laffargue A, Salmona J, Doulbeau S, Descroix F, Bertrand B, de Kochko A, Dussert S. 2009. Metabolic pathways in tropical dicotyledonous albuminous seeds: Coffea arabica as a case study. New Phytologist 182, 146–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Yim S, Choi H, Kim A, Lee KP, Lopez-Molina L, Martinoia E, Lee Y. 2015. Abscisic acid transporters cooperate to control seed germination. Nature Communications 6, 8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klie S, Nikoloski Z. 2012. The choice between mapman and gene ontology for automated gene function prediction in plant science. Frontiers in Genetics 3, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ. 1991. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352, 524–526. [DOI] [PubMed] [Google Scholar]
- Kranner I, Birtic S. 2005. A modulating role for antioxidants in desiccation tolerance. Integrative and Comparative Biology 45, 734–740. [DOI] [PubMed] [Google Scholar]
- Lee HW, Kim NY, Lee DJ, Kim J. 2009. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiology 151, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince O, Bronchart R, Deltour R. 1990. Changes in starch and soluble sugars in relation to the acquisition of desiccation tolerance during maturation of Brassica campestris seed. Plant, Cell & Environment 13, 539–546. [Google Scholar]
- Leprince O, Buitink J. 2010. Desiccation tolerance: From genomics to the field. Plant Science 179, 554–564. [Google Scholar]
- Leprince O, Buitink J. 2015. Introduction to desiccation biology: from old borders to new frontiers. Planta 242, 369–378. [DOI] [PubMed] [Google Scholar]
- Leprince O, Buitink J, Hoekstra FA. 1999. Axes and cotyledons of recalcitrant seeds of Castanea sativa Mill. exhibit contrasting responses of respiration to drying in relation to desiccation sensitivity. Journal of Experimental Botany 50, 1515–1524. [Google Scholar]
- Leprince O, Pellizzaro A, Berriri S, Buitink J. 2017. Late seed maturation: drying without dying. Journal of Experimental Botany 68, 827–841. [DOI] [PubMed] [Google Scholar]
- Li DZ, Pritchard HW. 2009. The science and economics of ex situ plant conservation. Trends in Plant Science 14, 614–621. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv, 1303.3997 [Preprint]. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, He J, Chen Z, Ren X, Hong X, Gong Z. 2010. ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. The Plant Journal 63, 749–765. [DOI] [PubMed] [Google Scholar]
- Liu JM, Zhao JY, Lu PP, Chen M, Guo CH, Xu ZS, Ma YZ. 2016. The E-subgroup pentatricopeptide repeat protein family in Arabidopsis thaliana and confirmation of the responsiveness PPR96 to abiotic stresses. Frontiers in Plant Science 7, 1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Millar AH, Sweetlove LJ, Hill SA, Leaver CJ. 2001. Mitochondrial biogenesis during germination in maize embryos. Plant Physiology 125, 662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Nagel A, Herter T, May P, Schroda M, Zrenner R, Tohge T, Fernie AR, Stitt M, Usadel B. 2014. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant, Cell & Environment 37, 1250–1258. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. 2003. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. The Plant Cell 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou HQ, Gong YL, Fan W, Xu JM, Liu Y, Cao MJ, Wang MH, Yang JL, Zheng SJ. 2016. A formate dehydrogenase confers tolerance to aluminum and low pH. Plant Physiology 171, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia J, Dekkers BJ, Dolle MJ, Ligterink W, Hilhorst HW. 2014. Abscisic acid (ABA) sensitivity regulates desiccation tolerance in germinated Arabidopsis seeds. New Phytologist 203, 81–93. [DOI] [PubMed] [Google Scholar]
- Manfre AJ, LaHatte GA, Climer CR, Marcotte WR Jr. 2009. Seed dehydration and the establishment of desiccation tolerance during seed maturation is altered in the Arabidopsis thaliana mutant atem6-1. Plant & Cell Physiology 50, 243–253. [DOI] [PubMed] [Google Scholar]
- Moriwaki T, Miyazawa Y, Kobayashi A, Uchida M, Watanabe C, Fujii N, Takahashi H. 2011. Hormonal regulation of lateral root development in Arabidopsis modulated by MIZ1 and requirement of GNOM activity for MIZ1 function. Plant Physiology 157, 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller CJ, Valdés AE, Wang G, Ramachandran P, Beste L, Uddenberg D, Carlsbecker A. 2016. PHABULOSA mediates an auxin signaling loop to regulate vascular patterning in Arabidopsis. Plant Physiology 170, 956–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha MW, Wang Y, Narsai R, Whelan J. 2014. The plant mitochondrial protein import apparatus – the differences make it interesting. Biochimica et Biophysica Acta 1840, 1233–1245. [DOI] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. 2008. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiology 147, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Dapena P, Castaño R, Almoguera C, Jordano J. 2006. Improved resistance to controlled deterioration in transgenic seeds. Plant Physiology 142, 1102–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Dapena P, Castaño R, Almoguera C, Jordano J. 2008. The ectopic overexpression of a seed-specific transcription factor, HaHSFA9, confers tolerance to severe dehydration in vegetative organs. The Plant Journal 54, 1004–1014. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Greenham K, Zhang Y, Santner A, Castillejo C, Mutka AM, O’Malley RC, Ecker JR, Kunkel BN, Estelle M. 2016. The Arabidopsis auxin receptor F-box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 6, 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Righetti K, Vu JL, Pelletier S, et al. . 2015. Inference of longevity-related genes from a robust coexpression network of seed maturation identifies regulators linking seed storability to biotic defense-related pathways. The Plant Cell 27, 2692–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert N, d’Erfurth I, Marmagne A, et al. . 2012. Voltage-dependent-anion-channels (VDACs) in Arabidopsis have a dual localization in the cell but show a distinct role in mitochondria. Plant Molecular Biology 78, 431–446. [DOI] [PubMed] [Google Scholar]
- Roberts E. 1973. Predicting the storage life of seed. Seed Science and Technology 1, 499–514. [Google Scholar]
- Saavedra X, Modrego A, Rodríguez D, González-García MP, Sanz L, Nicolás G, Lorenzo O. 2010. The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiology 152, 133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano N, Rajjou L, North HM, Debeaujon I, Marion-Poll A, Seo M. 2016. Staying alive: molecular aspects of seed longevity. Plant & Cell Physiology 57, 660–674. [DOI] [PubMed] [Google Scholar]
- Sechet J, Roux C, Plessis A, et al. . 2015. The ABA-deficiency suppressor locus HAS2 encodes the PPR protein LOI1/MEF11 involved in mitochondrial RNA editing. Molecular Plant 8, 644–656. [DOI] [PubMed] [Google Scholar]
- Sew YS, Ströher E, Fenske R, Millar AH. 2016. Loss of mitochondrial malate dehydrogenase activity alters seed metabolism impairing seed maturation and post-germination growth in Arabidopsis. Plant Physiology 171, 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Suetsugu N, Wada M, Kohda D. 2010. Crystallographic and functional analyses of J-domain of JAC1 essential for chloroplast photorelocation movement in Arabidopsis thaliana. Plant & Cell Physiology 51, 1372–1376. [DOI] [PubMed] [Google Scholar]
- Tapia H, Young L, Fox D, Bertozzi CR, Koshland D. 2015. Increasing intracellular trehalose is sufficient to confer desiccation tolerance to Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences, USA 112, 6122–6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateda C, Watanabe K, Kusano T, Takahashi Y. 2011. Molecular and genetic characterization of the gene family encoding the voltage-dependent anion channel in Arabidopsis. Journal of Experimental Botany 62, 4773–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D, Jaquinod M, Mangavel C, Passirani C, Saulnier P, Manon S, Teyssier E, Payet N, Avelange-Macherel MH, Macherel D. 2007. Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. The Plant Cell 19, 1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweddle JC, Dickie JB, Baskin CC, Baskin JM. 2003. Ecological aspects of seed desiccation sensitivity. Journal of Ecology 91, 294–304. [Google Scholar]
- Verdier J, Lalanne D, Pelletier S, et al. . 2013. A regulatory network-based approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiology 163, 757–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertucci CW. 1989. The effects of low water contents on physiological activities of seeds. Physiologia Plantarum 77, 172–176. [Google Scholar]
- Vertucci C, Farrant J. 1995. Acquisition and loss of desiccation tolerance. In: Kigel J, Galili G, eds. Seed development and germination. New York: Marcel Dekker, 237–271. [Google Scholar]
- Vertucci CW, Leopold AC. 1984. Bound water in soybean seed and its relation to respiration and imbibitional damage. Plant Physiology 75, 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Van Aken O, Elsässer M, Schwarzländer M. 2018. Mitochondrial energy signaling and its role in the low-oxygen stress response of plants. Plant Physiology 176, 1156–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters C, Berjak P, Pammenter N, Kennedy K, Raven P. 2013. Plant science. Preservation of recalcitrant seeds. Science 339, 915–916. [DOI] [PubMed] [Google Scholar]
- Walters C, Pammenter NW, Berjak P, Crane J. 2001. Desiccation damage, accelerated ageing and respiration in desiccation tolerant and sensitive seeds. Seed Science Research 11, 135–148. [Google Scholar]
- Wang L, Nägele T, Doerfler H, et al. . 2016. System level analysis of cacao seed ripening reveals a sequential interplay of primary and secondary metabolism leading to polyphenol accumulation and preparation of stress resistance. The Plant Journal 87, 318–332. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li Y, Xue H, Pritchard HW, Wang X. 2015. Reactive oxygen species-provoked mitochondria-dependent cell death during ageing of elm (Ulmus pumila L.) seeds. The Plant Journal 81, 438–452. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ries A, Wu K, Yang A, Crawford NM. 2010. The Arabidopsis prohibitin gene PHB3 functions in nitric oxide-mediated responses and in hydrogen peroxide-induced nitric oxide accumulation. The Plant Cell 22, 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Pacher M, Dukowic S, Schubert V, Puchta H, Schubert I. 2009. The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. The Plant Cell 21, 2688–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth WM, Masnavi G, Bhardwaj RM, Jiang Q, Bray CM, West CE. 2010. A plant DNA ligase is an important determinant of seed longevity. The Plant Journal 63, 848–860. [DOI] [PubMed] [Google Scholar]
- Wyse SV, Dickie JB. 2017. Predicting the global incidence of seed desiccation sensitivity. Journal of Ecology 105, 1082–1093. [Google Scholar]
- Yu Q, Tian H, Yue K, Liu J, Zhang B, Li X, Ding Z. 2016. A P-Loop NTPase regulates quiescent center cell division and distal stem cell identity through the regulation of ROS homeostasis in Arabidopsis root. PLoS Genetics 12, e1006175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Novak O, Wei Z, Gou M, Zhang X, Yu Y, Yang H, Cai Y, Strnad M, Liu CJ. 2014. Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nature Communications 5, 3274. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Zhang J, Gao X, Tong J, Xiao L, Li W, Zhang H. 2010. The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene 457, 1–12. [DOI] [PubMed] [Google Scholar]
- Zhu W, Miao Q, Sun D, Yang G, Wu C, Huang J, Zheng C. 2012. The mitochondrial phosphate transporters modulate plant responses to salt stress via affecting ATP and gibberellin metabolism in Arabidopsis thaliana. PLoS ONE 7, e43530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsmeister J, Lalanne D, Terrasson E, et al. . 2016. ABI5 is a regulator of seed maturation and longevity in legumes. The Plant Cell 28, 2735–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.