A discussion of recent discoveries in the etiolation/de-etiolation field, focusing on post-transcriptional processes and ultrastructural changes, along with comments on usage of the term ‘etiolation’, and of common etiolation/de-etiolation systems.
Keywords: chloroplast biogenesis, de-etiolation, etiolation, etioplast, prolamellar body, skotomorphogenesis
Abstract
The state of etiolation is generally defined by the presence of non-green plastids (etioplasts) in plant tissues that would normally contain chloroplasts. In the commonly used dark-grown seedling system, etiolation is coupled with a type of growth called skotomorphogenesis. Upon illumination, de-etiolation occurs, marked by the transition from etioplast to chloroplast, and, at the seedling level, a switch to photomorphogenic growth. Etiolation and de-etiolation systems are therefore important for understanding both the acquisition of photosynthetic capacity during chloroplast biogenesis and plant responses to light—the most relevant signal in the life and growth of the organism. In this review, we discuss recent discoveries (within the past 2–3 years) in the field of etiolation and de-etiolation, with a particular focus on post-transcriptional processes and ultrastructural changes. We further discuss ambiguities in definitions of the term ‘etiolation’, and benefits and biases of common etiolation/de-etiolation systems. Finally, we raise several open questions and future research possibilities.
Introduction: defining etiolation
Etiolation involves prolonged growth in the absence of light that results in the development of etioplasts in tissue that would have chloroplasts if subjected to light. Etioplasts do not contain chlorophyll or stacked thylakoid membranes, but rather have a paracrystalline lipid–pigment–protein structure known as the prolamellar body (PLB). The PLB consists largely of the plastid lipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG), and an association of the chlorophyll precursor protochlorophyllide (Pchlide), the light-dependent protochlorophyllide oxidoreductase (LPOR) that is responsible for its conversion, and the cofactor NADPH (Fig. 1; etioplast composition and structure reviewed, for example, in Kowalewska et al. (2019) and Pribil et al. (2014)).
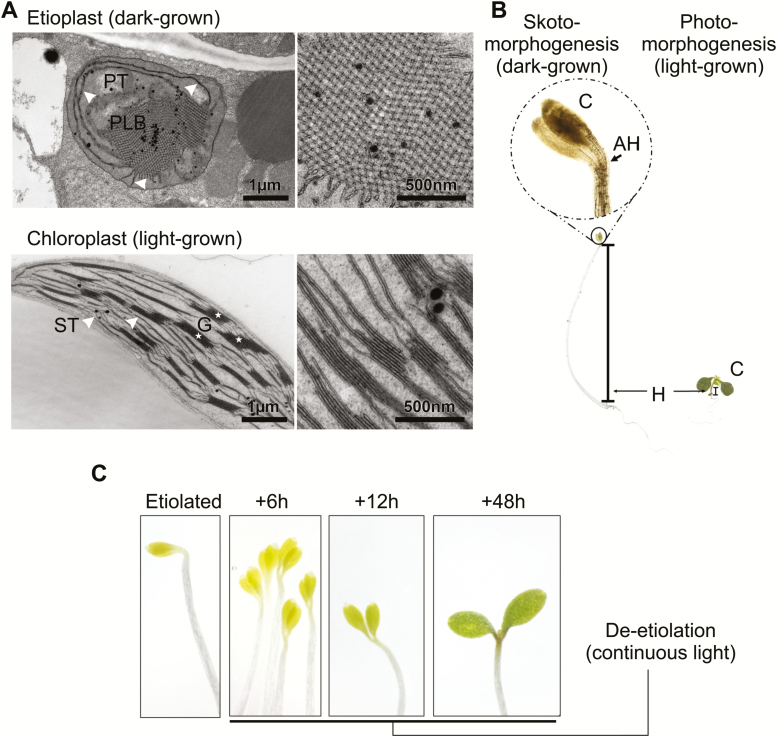
Fig. 1.
Etiolated phenotypes in plants (exemplified in Arabidopsis). (A) Plants grown in extended darkness develop etioplasts (upper panels). These plastids are physically defined by the presence of a paracrystalline membrane structure called prolamellar body (PLB), as well as prothylakoids (PT, indicated by white arrowheads). In the light, photosynthetic tissue develops chloroplasts (lower panels), which are defined structurally by thylakoid membranes that contain grana stacks (G, white asterisks) and stroma lamellae called stroma thylakoids (ST, white arrowheads). Images are from 6-day-old dark-grown Arabidopsis plant (upper panel), and a light-grown Arabidopsis plant at the rosette stage (lower panel). (B) Etiolation and de-etiolation studies generally involve germination and growth of seedlings in darkness, resulting in skotomorphogenic growth (left). This is defined by the presence of an apical hook (AH), closed and pale cotyledons, and an elongated hypocotyl. By contrast, plants grown the light (photomorphogenic conditions; right) have shorter hypocotyls, and open, green cotyledons. C, cotyledon; H, hypocotyls. Images taken from a 7-day-old dark-grown and a 9-day-old light-grown Arabidopsis seedling. (C) De-etiolation of dark-grown (etiolated) seedlings involves straightening of the apical hook, opening and greening of the cotyledons, as well as the transition from etioplast to chloroplasts (refer to Fig. 3). The etiolated seedlings were exposed to continuous white light (95 µmol photons m−2 s−1) for 6, 12, and 48 h.
As most scientifically observed etiolation systems involve (aseptic) germination and growth of seedlings in complete darkness, the term ‘etiolated’ is commonly defined additionally by the presence of a skotomorphogenic phenotype of elongated hypocotyls, shortened roots, and small, closed cotyledons (Fig. 1; reviewed in Josse and Halliday, 2008). In these systems, the light-driven etioplast-to-chloroplast transition is coupled to a transition from skotomorphogenic to photomorphogenic growth. These morphogenic traits are often portrayed in quantifiable and continuous terms, with variables of hypocotyl length, apical hook angle, and cotyledon angle considered. By these definitions, aberrant ‘photomorphogenic in darkness’ or ‘skotomorphogenic in light’ phenotypes have been utilized to identify multiple components involved in light sensing, signaling, or downstream responses. Many of these components have since been shown to have broad roles in non-etiolation-related light response.
The majority of the data discussed in this Expert View refer to work undertaken in such seedling-based etiolation/de-etiolation systems. The various limitation of these systems and possible alternative or complementary systems are also discussed (in the section ‘New systems required and new lessons learned’).
More broadly, the term ‘etiolated’, which has etymological roots in the French étiolier (i.e. straw), is still used as a descriptor for a range of pale or yellowing phenotypes. These include nitrogen-deficient rice (Oryza sativa; Sun et al., 2018a), graft-incompatible pomello (Citrus grandis; He et al., 2018), and heavy-metal-treated wheat (Triticum aestivum; Semenova et al., 2017). Similarly, a skotomorphogenic phenotype observed in infected light-grown creeping bentgrass (Agrostis stolonifera; Roberts et al., 2016) was recently termed ‘bacterial etiolation’. We consider these phenotypes to be largely outside our personal definition of etiolated tissues (i.e. having etioplasts), and will not discuss them within this work. Nonetheless, we note that in recent years, similar ‘etiolated’ phenotypes have been linked to pigment accumulation (Chen et al., 2018b) and light signaling defects (Peng et al., 2019). Furthermore, the pale barley (Hordeum vulgare L.) albostrians mutant (Muramoto et al., 1999), has been shown to contain structures in its albino sectors that are highly reminiscent of transforming PLBs (Li et al., 2019). As such, these ‘etiolated’ plants should be consdered a potential source of new players in the regulation of chloroplast development, particularly in non-model species. Finally, this review will not discuss etiolation-like responses in non-angiosperm species, a still under-represented and debated research field (reviewed in Mathews, 2006).
Recent developments in understanding etiolation and the etioplast-to-chloroplast transition
The response to light was one of the earliest phenomena observed in plants by naturalists, and much progress has been made in understanding both the perception of light by various photoreceptors, and the resultant signaling cascades that lead to transcriptional activation or repression of genes involved in de-etiolation. We will not discuss these processes, which have been recently reviewed (Casal et al., 2014; Huang et al., 2014; Casal and Qüesta, 2018; Pham et al., 2018; Podolec and Ulm, 2018), but rather focus here on breakthroughs in post-transcriptional regulation and ultrastructural changes during etiolation and de-etiolation (summarized in Box 1; Fig. 2).
Box 1. Key developments in understanding de-etiolation.
Small regulatory RNAs are highly dynamic during greening
Recent large-scale studies of small regulatory RNA (sRNA) changes during greening in Arabidopsis (Lin et al., 2017), rice, and maize (Xu et al., 2017) provide pioneer datasets, suggest new roles for several sRNAs, and demonstrate the power of de-etiolation systems in investigating pairwise relationships.
TOR connects light and nutrient signaling
The indirect activator of translation, target of rapamycin (TOR), acts downstream of the COP1–auxin cascade during de-etiolation (Chen et al., 2018a), but is also involved in light-independent developmental regulation in response to sugars (Mohammed et al., 2018). The complex demand/supply of resources associated with establishing photosynthesis has implications for the regulation and kinetics of chloroplast development, and for currently used etiolation systems.
Availability, not just abundance, counts for transcripts and proteins
Thousands of mRNA species are present yet translationally repressed by sequestration to processing bodies (P-bodies) in the dark (Jang et al., 2019). For plastid-encoded thylakoid membrane proteins, association of respective mRNA to ribosomes localizes them to membranes, but the membrane to soluble mRNA fraction changes little during greening (Legen and Schmitz-Linneweber, 2017). Soluble versus membrane localization of glutamyl-tRNA reductase (GluTR) does change with lighting, and the soluble (active) fraction shows early correlation with chlorophyll content (Schmied et al., 2018).
Singlet oxygen causes PSII damage and acts as a retrograde signal during de-etiolation
The early assembly of the PSII oxygen evolving complex results in the (damaging) formation of singlet oxygen (1O2; Shevela et al., 2019). 1O2 retrograde signaling mediates de-etiolation via the EXECUTER1 pathway (Chen et al., 2015; Carmody et al., 2016). A de-etiolation system was recently used to assign function to the elusive integrator of retrograde signalling, GUN1 (Wu et al., 2018).
Finally looking at membrane lipids (and how they get there)
Three recent studies investigated the effect of decreased MGDG (Fujii et al., 2017) and DGDG (Fujii et al., 2018) content on etioplast formation and greening (Fujii et al., 2019). They emphasize the role of DGDG in the dynamics of tubular-lamellar transformation occurring during PLB–thylakoid membrane transition as well as the crucial role of both neutral galactolipids in the membrane-associated steps of Chl biosynthesis. Future studies, using diverse systems and 3D imaging techniques, are suggested to further this developing field.
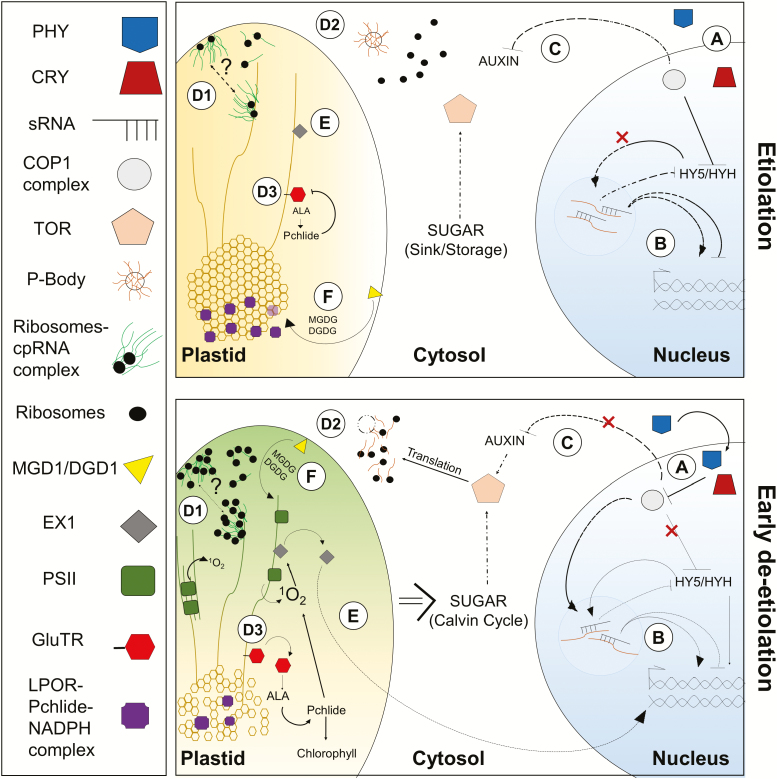
Fig. 2.
Signaling cascade and recently described players in de-etiolation. This simplified model shows a basic overview of (A) the PHY/CRY-mediated light-responsive signaling cascade, and (B–F) recent discoveries in the field discussed in this review. The upper panel shows the etiolated state, the lower panel shows the changes that occur early upon de-etiolation. (A) Light is perceived by photoreceptors such as phytochromes (PHY) and cryptochromes (CRY), resulting in indirect activation of the expression of Elongated Hypocotyl5/HY5 Homolog (HY5/HYH)-dependent photomorphogenesis-related genes by repression of the COP1 complex. (B) Small RNAs (sRNA) modulate transcript accumulation of both light-signaling molecules and ownstream effector genes, and the sRNA pathway itself is also controlled via light signaling pathways. (C) TOR indirectly activates translation via auxin, and is itself stimulated by light as well as by sugars. (D) Physical sequestration can limit functionality. (D1) Increased translation in the plastid is likely linked to increased ribosome density, as opposed to occupancy. (D2) Cytosolic transcripts are sequestered in processing bodies (P-bodies) during etiolation, with release allowing their translation. (D3) GluTR is soluble and active in the light, with the soluble form correlating with chlorophyll content during greening. (E) Retrograde signalling mediated by 1O2 produced by the early assembly of the oxygen evolving complex of PSII might contribute to the EXECUTER1 signaling pathway. (F) MGDG and DGDG, produced in the envelopes by monogalactosyldiacylglycerol synthase 1 (MGD1) and digalactosyldiacylglycerol synthase 1 (DGD1), are the primary plastid lipids, and have crucial and disparate roles in PLB formation and etioplast-to-chloroplast transition, but more research is required to understand the role of both lipids and proteins in membrane biogenesis.
Small RNAs fine-tune temporal and spatial expression of genes during de-etiolation
Small regulatory RNAs (sRNAs) are 20–24 nt-long molecules that regulate gene expression via RNA-dependent DNA methylation, translation inhibition, or mRNA cleavage (reviewed in Borges and Martienssen, 2015; Singh et al., 2018). Several important studies have highlighted the control of canonical light reception and response pathway factors by sRNAs, and the reciprocal light-based regulation not just of certain sRNA, but of the sRNA biogenesis process itself via these factors (Sorin et al., 2005; Zhang et al., 2011; Cho et al., 2014; Tsai et al., 2014; Achkar et al., 2018; Sun et al., 2018b). We refer the reader to two recent reviews (Sánchez-Retuerta et al., 2018; Manavella et al., 2019) for more details.
Recently, sRNAs were implicated in defining seedling tissue- or position- dependent greening responses: differential accumulation of certain sRNAs, and certain groups of sRNAs, was observed in different tissue types (Li et al., 2014). Most recently, two large-scale studies were undertaken: Lin et al. (2017) profiled sRNAs during Arabidopsis de-etiolation, while Xu and colleagues (2017) undertook comparative miRNA profiling in rice and maize (Zea mays) to understand the establishment of photosynthesis in C3 versus C4 species. These studies, which defined several specific sRNA roles, such as the repression of photomorphogenic growth by miR396 via members of the Growth Regulating Factors family (Lin et al., 2017), provide important pioneer work that defines global sRNA responses to greening (Fig. 2B). Furthermore, they demonstrate the use of de-etiolating systems—in which large scale yet highly temporally controlled changes occur—as a powerful tool for investigating pairwise relationships, for example, between regulators and their targets (Xu et al., 2016; Page et al., 2017; Xu et al., 2017).
TOR connects light and nutrient signaling to activate translation
Target of rapamycin (TOR) is an evolutionarily conserved protein kinase that acts as a central hub to control cellular- and organism-level development (reviewed in Caldana et al., 2019; Xiong and Sheen, 2014). Disruption of TOR results in plants with reduced chloroplast size and number, poorly developed thylakoid membranes, and decreased expression of key photosynthesis-related proteins (Xiong et al., 2017). Furthermore, TOR (i) is required for proper regulation of photomorphogenic growth via regulation of translation and brassinosteroid signaling (Xiong et al., 2017), (ii) acts as an indirect positive regulator of chlorophyll biosynthesis and photosynthesis-related genes (Li et al., 2015), and (iii) is involved in the accumulation of the MGDG and DGDG synthases (Sun et al., 2016). Thus, TOR positively contributes to plastid development. Nonetheless, seedlings with repressed TOR activity were recently reported to undergo more rapid accumulation of chlorophyll, PS-related transcripts, and plastid membrane lipids during de-etiolation—surprising results that the authors attributed to altered nutrient content of TOR-repressed seeds (Zhang et al., 2018). Indeed, recent research underlines the essential role of TOR in sugar-status response during early development. This includes (indirect) positive control of cell elongation in dark-grown seedlings (Zhang et al., 2016), and de-repression of shoot apical meristem growth in the dark via sugar-induced TOR activity (Li et al., 2017b; Mohammed et al., 2018). In light of a recently clarified position for TOR in the constitutively photomorphogenic 1 (COP1)–auxin cascade (Chen et al., 2018a), these findings suggest that TOR balances light and sugar signaling to control plant and plastid development both at near-instantaneous and at more gradual time scales (Fig. 2C).
Recent studies have suggested that chloroplast protein production represents ~70% of the ATP cost of total cellular protein synthesis (Li et al., 2017a), and two-thirds of the cellular nitrogen budget (Evans and Clarke, 2019). The need for greening seedlings to balance the cost of photosynthesis with its ultimate reward may therefore define (i) the control of gene expression that exerts control primarily at the (costly) translational stage (Shen et al., 2009; Ning et al., 2016); and (ii) the recently observed multi-phase accumulation of photosynthesis-related products and activities (Dubreuil et al., 2018; Armarego-Marriott et al., 2019). We note that, in addition to defining greening, the availability of resources like carbon (Kósa et al., 2015) and nitrogen (Vitányi et al., 2013) influences etioplast formation. Therefore, these recent works highlight the importance of considering resource availability in studying all aspects of etiolation and de-etiolation. Given that these resources arise from both (exhaustible) seed storage tissues and medium supplementation, it is clear that the choice of experimental system can largely influence observations.
Control by location: where is as important as when
As well as massive transcriptional changes (Ma et al., 2001), greening can result in a global 2-fold increase in translational activity, and altered translation of ~1/3 of all transcripts (Liu et al., 2012). Translation of cytosolic mRNAs can increase due to changes in the number of ribosomes on individual transcripts (ribosome density) or changes in the proportion of transcripts occupied by ribosomes (ribosome occupancy) (Liu et al., 2013). In the plastid, transcripts are sequestered to membrane fractions in a ribosome-dependent manner, but membrane association of transcripts changes only minimally during maize leaf greening, suggesting that ribosome density, and not occupancy, drives greening-induced translation (Legen and Schmitz-Linneweber, 2017) (Fig. 2D1).
Within the cytosol, light-stimulated translation has been linked to processing bodies (P-bodies): RNA–protein complexes that are conserved in eukaryotes and regulate gene expression by degradation or translational arrest of mRNA (reviewed in Xu and Chua, 2011; Maldonado-Bonilla, 2014). Dark-grown seedlings of a P-body defective mutant (Xu and Chua, 2009) displayed prematurely opened apical hooks and augmented translation of thousands of transcripts, including those involved in the chlorophyll biosynthesis pathway (Jang et al., 2019). Despite previous links between sRNA-mediated mRNA cleavage and P-bodies (Pomeranz et al., 2010), Jang et al. (2019) noted limited overlap between mRNA cleavage and sequestration-induced translational ‘pausing’ (Fig. 2D2). Recently, physical sequestration has also been implicated in post-translational regulation. Localization of glutamyl-tRNA reductase (GluTR) to the chloroplast stroma, but not to the membrane, was associated with its enzymatic activity, and was shown to correlate with accumulation of chlorophyll during the early hours of greening (Schmied et al., 2018). Interestingly, GluTR partitioning also changes following dark exposure of light-grown plants, suggesting that this regulation has relevance beyond the etioplast-to-chloroplast transition (Schmied et al., 2018) (Fig. 2D3). Together, these recent studies underline that, in addition to cellular abundance of proteins and mRNAs, subcellular localization also needs to be taken into consideration.
Retrograde signaling: coupling the import and assembly of photosystems
Communication from the chloroplast to the nucleus, known as retrograde signaling, is a critical step during chloroplast biogenesis and maintenance (reviewed in Hernández-Verdeja and Strand, 2018; Rochaix and Ramundo, 2018; Leister, 2019; Pesaresi and Kim, 2019). Of six early identified Genomes uncoupled (gun) mutants defective in plastid-to-nucleus retrograde signaling (Susek et al., 1993), five (gun2–6) have defects in genes for enzymes involved in tetrapyrrole biosynthesis. More recently, a role for the enigmatic GUN1 in regulating protein import via the cytosolic heat shock protein 90 (HSP90) chaperone was clarified using a de-etiolation system (Wu et al., 2019). This followed observations that the GUN1 protein accumulates primarily during early chloroplast development (Wu et al., 2018) and that gun1 mutants showed retarded de-etiolation (Mochizuki et al., 1996). The early flowering phenotype observed in GUN1 overexpressing plants has led to the proposal that the protein may play a role in developmental phase transitions beyond chloroplast biogenesis (Wu et al., 2018).
Singlet oxygen (1O2) is produced early during greening as a by-product of tetrapyrrole biosynthesis (Zhang et al., 2015; Wang and Apel, 2019) and via early photosystem II (PSII) oxygen evolving complex activity (Zavafer et al., 2015). In addition to potentially causing significant harm to the developing chloroplast, including damage to emerging PSII complexes prior to their protective incorporation into grana stacks (Shevela et al., 2019), singlet oxygen may act in retrograde signaling via the Filamentation temperature sensitive H (FtsH; a membrane metalloprotease)-activated EXECUTER1 (EX1) pathway (Dogra et al., 2017). Previous research suggests that the 1O2-mediated EXECUTER pathway primes etioplasts to develop into chloroplasts (Kim et al., 2009), and also mediates high-light responses in the chloroplast, by regulation of multiple nucleus-encoded stress related transcripts (Carmody et al., 2016). Localization of EXECUTER proteins to grana margins (Wang et al., 2016b) further supports a potential role during PSII repair. Recently, Dogra et al. (2019) showed that the oxidation of a specific tryptophan residue (Trp643) in the singlet oxygen sensor domain contained in EX1 is essential for membrane localization and protein stability, and is also required for FtsH2-mediated EX1 degradation and further, as yet undefined, signaling to the nucleus (Dogra et al., 2019). Interestingly, EX1 is also involved in carbon/nitrogen partitioning during light acclimation (Uberegui et al., 2015), supporting a strong link between nutrient regulation and controlled chloroplast development (Fig. 2E).
Structural and functional membrane dynamics: recent focus on lipids in the regulation of membrane rearrangements
Although thylakoid membranes and etioplast internal membranes are both primarily composed of the galactolipids MGDG and DGDG, the lipid to lipid ratios (MGDG:DGDG) and lipid to protein ratios change with greening (Selstam and Sandelius, 1984). The role of lipid composition and content in plastid membrane structure has been studied extensively for several decades, but has recently returned to the spotlight with the publication of several studies involving disruption of galactolipid synthesis enzymes. Studies with mutants having slight decreases in galactolipid content and showing disrupted membranes in fully developed chloroplasts (Mazur et al., 2019) display limited or no structural disruptions in etioplasts (Jarvis et al., 2000), an effect attributable to the lower absolute requirement for lipids in etioplasts (Fujii et al., 2014, 2017). In recent work, etiolated plants with severe MGDG and DGDG deficits were shown to accumulate less photoactive Pchlide, LPOR, and carotenoids compared with respective wild types (Fujii et al., 2017, 2018). The decrease in photoactive Pchlide levels in a MGDG-deficient mutant observed under sugar-supplemented growth conditions (Fujii et al., 2017) contrasts with previous Pchlide increases seen in soil-grown mutants (Aronsson et al., 2008), again underlining the role of resource availability on plastid development. The decrease in DGDG content also resulted in significant structural PLB lattice perturbations, strong reduction of prothylakoid number, and retarded PLB disassembly in the light (Fujii et al., 2019). Furthermore, while MGDG- and DGDG-deficient plants showed impairment in accumulation of Chl and the light-harvesting complex II protein LHCB1 during greening, changes in photosynthesis-related gene transcript accumulation were, relatively, delayed (Fujii et al., 2019), suggesting that lipid status is sensed indirectly (e.g. via disrupted protein insertion or function).
While these studies suggest differences in the roles of MGDG and DGDG during etiolation and de-etiolation, it is difficult to make concrete conclusions, due to the different reduction of galactolipid contents in each mutant and the inter-relationship between the lipids (DGDG is a downstream product of MGDG). These issues argue for alternative systems, such as the in vitro system recently used to show the requirement for MGDG and charged lipids in regulating LPOR complex formation and activity (Gabruk et al., 2017), and support a need for further biophysical studies that investigate the detailed distribution of lipid phases inside membranes (Garab et al., 2017; Ughy et al., 2019). In vivo time-resolved 3D techniques (e.g. Kowalewska et al. 2016), may be used to answer several open questions in the field, including how the PLB is formed and how and from where membrane components are recruited during the formation of grana stacks. On the latter topic, inner membrane-localized MGDG synthase has been suggested to be both a point of contact between thylakoids and the inner envelope membrane, and a supplier of lipids during thylakoid biogenesis (Rocha et al., 2018). We note that the nature of contact point(s), as being either direct or involving vesicles or tubules, remains debated (reviewed in Lindquist et al., 2016; Lindquist and Aronsson, 2018; Mechela et al., 2019). Notably, a recent 3D analysis of the proplastid-to-chloroplast transition (Liang et al., 2018) visualized direct connection points, which were proposed to both act as lipid transfer points and align growing thylakoids. Given that factors associated with these connections have been implicated in both thylakoid biogenesis and maintenance (e.g. Gao et al., 2006; Patil et al., 2018), understanding such connections is likely to bear importance throughout the lifetime of the plastids (Fig. 2F).
Etiolation studies and the future
New systems required and new lessons learned
To date, etiolation and de-etiolation work focused on the study of molecular processes has commonly been undertaken with dark-grown seedlings. The benefits of this system include that it (i) requires limited growth time and space yet provides sufficient material compared with other experimental systems such as the shoot apical meristem, and (ii) is highly customizable by use of different timing and lighting regimes and introduction of different substances to the growth medium (López-Juez et al., 2008; Mohammed et al., 2018; Dóczi et al., 2019). Nonetheless, there are limitations to this system, which should not be overlooked. These include the difficulties in separating plastid development (i.e. etioplast-to-chloroplast transition) from general seedling development programs, as well as issues associated with observing chloroplast development only in cotyledons, which are programmed differently from true leaves (reviewed in Pogson et al., 2015). Some limitations of the present system may be overcome by using other species and systems, although we stress that both etioplast formation and light-induced de-etiolation may largely differ depending on the species, timing, and conditions used (Skupień et al., 2017), making cross-system comparisons difficult. For example, both runner bean (Phaseolus coccineus) and pea (Pisum sativum) (Kowalewska et al., 2016) show similar skotomorphogenic growth to Arabidopsis, yet develop true leaves in darkness (Fig. 3). PLBs have also been observed in non-seedling systems, both in young leaves of tobacco following extended dark treatment (Armarego-Marriott et al., 2019) and in the innermost leaf primordia of the closed and opening leaf buds of trees (Solymosi and Böddi, 2006; Solymosi et al., 2006, 2012). The problem of uneven lighting that arises from gradual cotyledon opening or seed-coat shading (e.g. Solymosi et al., 2007) was recently overcome by using duckweed (Landoltia punctate), a flat-leafed aquatic monocot (Monselise et al., 2015). More artificially, cell cultures (Dubreuil et al., 2018), and even a callus-based system (Schaub et al., 2018), have been used to investigate various aspects of plastid development, and may putatively be adapted for de-etiolation. Nonetheless, these experimental systems come with their own caveats, in particular multiple impacts of carbon supplementation on plastid development (Eckstein et al., 2012; Häusler et al., 2014). Such systems may help to address issues related to spatial diversity of plastid types, seen previously within the shoot apical meristem (Charuvi et al., 2012), in chloroplasts in different leaf regions (Gügel and Soll, 2017), and in etioplasts within different tissues (Kósa et al., 2015) or even single cells (Solymosi et al., 2012).
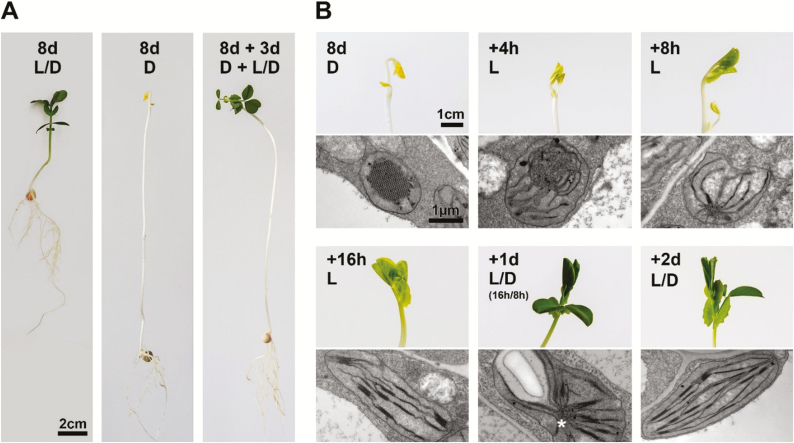
Fig. 3.
Pea (Pisum sativum) de-etiolating under light/dark conditions. (A) Pea seedlings grown for 8 d under light dark (L/D) conditions (16 h of light at 40 µmol photons m−2 s−1–8 h of darkness) (left panel), darkness (D) (middle panel), 8 d of darkness followed by 3 d of L/D (right panel). Pea, which develops true leaves in darkness, as well as other hypogeal germinating plants, may be used as an alternative system to epigeal germinating Arabidopsis plants, which only develop cotyledons in the dark. As ‘maternal tissue’, cotyledons are formed by and undergo different developmental programing from true leaves. (B) De-etiolating pea. The upper panels show seedling shoot apices; the lower panels show transmission electron micrograph. Plants grown in darkness for 8 d were de-etiolated under light–dark conditions (16 h of light at 40 µmol photons m−2 s−1–8 h of darkness). Note that following the first 24 h of growth there is partial reformation of the PLB, indicated by the white asterisk.
Curiously, while the etiolated state is largely defined by both the presence of a paracrystalline PLB and the absence of (stacked) thylakoid membranes, early studies in cucumber (Cucumis sativus; Ikeda, 1970) and avocado (Persea americana; Cran and Possingham, 1973), and more recent findings in bean (Phaseolus vulgaris) (Schoefs and Franck, 2008), various tree species (Solymosi et al., 2006), and tobacco (Nicotiana tabacum) (Armarego-Marriott et al., 2019), demonstrate that both structures can co-exist in a single plastid. Indeed, several studies indicate that PLB reformation may occur in young chloroplasts during extended darkness, or even during normal night periods during de-etiolation (see Fig. 3; Rudowska et al., 2012; Skupień et al., 2017; reviewed in Solymosi and Aronsson, 2013). These findings underscore the important influence of light regime, as well as light intensity, quality, and circadian-related effects (reviewed in Seluzicki et al., 2017) on greening, factors that must be considered when observing plastid development. We suggest PLB reformation as an interesting field for future study, and underline that the use of diverse systems may both further clarify current understandings of PLB formation and dissolution, and suggest new directions for future works.
Using etiolated systems and knowledge to go ‘beyond the darkness’
The benefits of the standard seedling etiolation and/or de-etiolation systems means that they have been used often in recent years to study diverse topics including gravitropism (Yamamoto et al., 2017), phototropism (Sullivan et al., 2019), resource limitation (Avin-Wittenberg et al., 2015; Kósa et al., 2015), and metabolite or hormone signaling (Gupta et al., 2015). Furthermore, etiolated growth can promote development of (i) certain tissue and organ types (e.g. adventitious roots; Sorin et al., 2005; da Costa et al., 2018; Trinh et al., 2018), (ii) certain growth types (e.g. growth by cellular expansion in hypocotyls; Sinclair et al., 2017; Ilias et al., 2019), and (iii) specific responses (e.g. ethylene ‘triple response’; Guzmán and Ecker, 1990; Ma et al., 2018) that cannot be easily observed in light-grown plants. Growth in darkness can also induce arrest of the shoot apical meristem, and thus de-etiolation can be used to observe shoot apical meristem development (López-Juez et al., 2008; Mohammed et al., 2018; Dóczi et al., 2019).
Beyond the practicality of the system itself, the greatest value of etiolation/de-etiolation studies lies in the central role of light signaling in plant life. Indeed, the overlap between factors involved in light responses with those involved in other response and growth processes has allowed basic knowledge from etiolation studies to be used to understand diverse plant processes (reviewed in Liu et al., 2017; Hsieh and Okamoto, 2014; Casal and Qüesta, 2018). In the applied sector, associations have been made between light receptors or responses and desirable crop attributes such as dwarfism (Hou et al., 2017), fruit or flower chromoplast development (Pankratov et al., 2016), and abiotic stress response (Zhou et al., 2018). Shade avoidance responses bear similarity to etiolation (Wang et al., 2016a), while ‘photobiotechnology’, in which modulated expression results in improved crop yield and resistance, has recently been proposed for improved food security (Ganesan et al., 2017). Clearly, future attempts to improve photosynthesis will require a detailed understanding of the chloroplast membrane structures and their biogenesis, as well as a thorough understanding of the processes involved in regulating the expression of photosynthesis-related genes (Ort et al., 2015). Taken together, while there is still much more to be learnt about de-etiolation itself, it is also clear that etiolation and de-etiolation systems provide the ideal environments to gain insight into the establishment of one of the most important processes for plant growth.
Acknowledgements
We thank Ralph Bock for his support in writing this paper. Transmission electron microscopy images were performed in the Laboratory of Electron Microscopy, Nencki Institute of Experimental Biology of PAS (Warsaw, Poland), using a JEM 1400 electron microscope (Jeol).
Glossary
Abbreviations:
- COP1
constitutively photomorphogenic 1
- DGDG
digalactosyldiacylglycerol
- EX1
Executer 1
- FtsH
Filamentation temperature sensitive H
- GluTR
Glutamyl-tRNA reductase
- GUN1
Genomes Uncoupled 1
- HSP90
heat shock protein 90 (chaperone)
- LPOR
light dependent protochlorophyllide oxidoreductase
- MGDG
monogalactosyldiacylglycerol
- Pchlide
protochlorophyllide
- PLB
prolamellar body
- PSII
photosystem II
- P-bodies
processing bodies
- sRNA
small RNA
- TOR
target of rapamycin
References
- Achkar NP, Cho SK, Poulsen C, et al. 2018. A quick HYL1-dependent reactivation of microRNA production is required for a proper developmental response after extended periods of light deprivation. Developmental Cell 46, 236–247.e6. [DOI] [PubMed] [Google Scholar]
- Armarego-Marriott T, Kowalewska Ł, Burgos A, et al. 2019. Highly resolved systems biology to dissect the etioplast-to-chloroplast transition in tobacco leaves. Plant Physiology 180, 654–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson H, Schöttler MA, Kelly AA, Sundqvist C, Dörmann P, Karim S, Jarvis P. 2008. Monogalactosyldiacylglycerol deficiency in Arabidopsis affects pigment composition in the prolamellar body and impairs thylakoid membrane energization and photoprotection in leaves. Plant Physiology 148, 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avin-Wittenberg T, Bajdzienko K, Wittenberg G, Alseekh S, Tohge T, Bock R, Giavalisco P, Fernie AR. 2015. Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. The Plant Cell 27, 306–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Martienssen RA. 2015. The expanding world of small RNAs in plants. Nature Reviews. Molecular cell biology 16, 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Martins MCM, Mubeen U, Urrea-Castellanos R. 2019. The magic ‘hammer’ of TOR: the multiple faces of a single pathway in the metabolic regulation of plant growth and development. Journal of Experimental Botany 70, 2217–2225. [DOI] [PubMed] [Google Scholar]
- Carmody M, Crisp PA, d’Alessandro S, Ganguly D, Gordon M, Havaux M, Albrecht-Borth V, Pogson BJ. 2016. Uncoupling high light responses from singlet oxygen retrograde signaling and spatial-temporal systemic acquired acclimation. Plant Physiology 171, 1734–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Candia AN, Sellaro R. 2014. Light perception and signalling by phytochrome A. Journal of Experimental Botany 65, 2835–2845. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Qüesta JI. 2018. Light and temperature cues: multitasking receptors and transcriptional integrators. New Phytologist 217, 1029–1034. [DOI] [PubMed] [Google Scholar]
- Charuvi D, Kiss V, Nevo R, Shimoni E, Adam Z, Reich Z. 2012. Gain and loss of photosynthetic membranes during plastid differentiation in the shoot apex of Arabidopsis. The Plant Cell 24, 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GH, Liu MJ, Xiong Y, Sheen J, Wu SH. 2018a TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. Proceedings of the National Academy of Sciences, USA 115, 12823–12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NG, Wang PR, Li CM, et al. 2018. b A single nucleotide mutation of the IspE gene participating in the MEP pathway for isoprenoid biosynthesis causes a green-revertible yellow leaf phenotype in rice. Plant and Cell Physiology 59, 1905–1917. [DOI] [PubMed] [Google Scholar]
- Chen S, Kim C, Lee JM, Lee HA, Fei Z, Wang L, Apel K. 2015. Blocking the QB-binding site of photosystem II by tenuazonic acid, a non-host-specific toxin of Alternaria alternata, activates singlet oxygen-mediated and EXECUTER-dependent signalling in Arabidopsis. Plant, Cell & Environment 38, 1069–1080. [DOI] [PubMed] [Google Scholar]
- Cho SK, Ben Chaabane S, Shah P, Poulsen CP, Yang SW. 2014. COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nature Communications 5, 5867. [DOI] [PubMed] [Google Scholar]
- Cran DG, Possingham JV. 1973. The fine structure of avocado plastids. Annals of Botany 37, 993–997. [Google Scholar]
- da Costa CT, Gaeta ML, de Araujo Mariath JE, Offringa R, Fett-Neto AG. 2018. Comparative adventitious root development in pre-etiolated and flooded Arabidopsis hypocotyls exposed to different auxins. Plant Physiology and Biochemistry 127, 161–168. [DOI] [PubMed] [Google Scholar]
- Dóczi R, Hatzimasoura E, Farahi Bilooei S, Ahmad Z, Ditengou FA, López-Juez E, Palme K, Bögre L. 2019. The MKK7-MPK6 MAP kinase module is a regulator of meristem quiescence or active growth in Arabidopsis. Frontiers in Plant Science 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra V, Duan J, Lee KP, Lv S, Liu R, Kim C. 2017. FtsH2-dependent proteolysis of EXECUTER1 is essential in mediating singlet oxygen-triggered retrograde signaling in Arabidopsis thaliana. Frontiers in Plant Science 8, 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra V, Li M, Singh S, Li M, Kim C. 2019. Oxidative post-translational modification of EXECUTER1 is required for singlet oxygen sensing in plastids. Nature Communications 10, 2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil C, Jin X, Barajas-López JD, et al. 2018. Establishment of photosynthesis through chloroplast development is controlled by two distinct regulatory phases. Plant Physiology 176, 1199–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein A, Zieba P, Gabrys H. 2012. Sugar and light effects on the condition of the photosynthetic apparatus of Arabidopsis thaliana cultured in vitro. Journal of Plant Growth Regulation 31, 90–101. [Google Scholar]
- Evans JR, Clarke VC. 2019. The nitrogen cost of photosynthesis. Journal of Experimental Botany 70, 7–15. [DOI] [PubMed] [Google Scholar]
- Fujii S, Kobayashi K, Nagata N, Masuda T, Wada H. 2017. Monogalactosyldiacylglycerol facilitates synthesis of photoactive protochlorophyllide in etioplasts. Plant Physiology 174, 2183–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Kobayashi K, Nagata N, Masuda T, Wada H. 2018. Digalactosyldiacylglycerol is essential for organization of the membrane structure in etioplasts. Plant Physiology 177, 1487–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Kobayashi K, Nakamura Y, Wada H. 2014. Inducible knockdown of MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE1 reveals roles of galactolipids in organelle differentiation in Arabidopsis cotyledons. Plant Physiology 166, 1436–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Nagata N, Masuda T, Wada H, Kobayashi K. 2019. Galactolipids are essential for internal membrane transformation during etioplast-to-chloroplast differentiation. Plant & Cell Physiology 60, 1224–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabruk M, Mysliwa-Kurdziel B, Kruk J. 2017. MGDG, PG and SQDG regulate the activity of light-dependent protochlorophyllide oxidoreductase. The Biochemical Journal 474, 1307–1320. [DOI] [PubMed] [Google Scholar]
- Ganesan M, Lee HY, Kim JI, Song PS. 2017. Development of transgenic crops based on photo-biotechnology. Plant, Cell & Environment 40, 2469–2486. [DOI] [PubMed] [Google Scholar]
- Gao H, Sage TL, Osteryoung KW. 2006. FZL, an FZO-like protein in plants, is a determinant of thylakoid and chloroplast morphology. Proceedings of the National Academy of Sciences, USA 103, 6759–6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garab G, Ughy B, Waard P, et al. 2017. Lipid polymorphism in chloroplast thylakoid membranes – as revealed by 31P-NMR and time-resolved merocyanine fluorescence spectroscopy. Scientific Reports 7, 13343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gügel IL, Soll J. 2017. Chloroplast differentiation in the growing leaves of Arabidopsis thaliana. Protoplasma 254, 1857–1866. [DOI] [PubMed] [Google Scholar]
- Gupta A, Singh M, Laxmi A. 2015. Multiple interactions between glucose and brassinosteroid signal transduction pathways in Arabidopsis are uncovered by whole-genome transcriptional profiling. Plant Physiology 168, 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR. 1990. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. The Plant Cell 2, 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler RE, Heinrichs L, Schmitz J, Flügge UI. 2014. How sugars might coordinate chloroplast and nuclear gene expression during acclimation to high light intensities. Molecular Plant 7, 1121–1137. [DOI] [PubMed] [Google Scholar]
- He W, Wang Y, Chen Q, Sun B, Tang HR, Pan DM, Wang XR. 2018. Dissection of the mechanism for compatible and incompatible graft combinations of Citrus grandis (L.) osbeck (“Hongmian Miyou’). International Journal of Molecular Sciences 19, E505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Verdeja T, Strand Å. 2018. Retrograde signals navigate the path to chloroplast development. Plant Physiology 176, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Niu H, Tao Q, Wang S, Gong Z, Li S, Weng Y, Li Z. 2017. A mutant in the CsDET2 gene leads to a systemic brassinosteriod deficiency and super compact phenotype in cucumber (Cucumis sativus L.). Theoretical and Applied Genetics 130, 1693–1703. [DOI] [PubMed] [Google Scholar]
- Hsieh HL, Okamoto H. 2014. Molecular interaction of jasmonate and phytochrome A signalling. Journal of Experimental Botany 65, 2847–2857. [DOI] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Deng XW. 2014. Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Current Opinion in Plant Biology 21, 96–103. [DOI] [PubMed] [Google Scholar]
- Ikeda T. 1970. Changes in fine structure of prolamellar body in relation to the formation of the chloroplast. The Botanical Magazine Tokyo 83, 1–9. [Google Scholar]
- Ilias IA, Negishi K, Yasue K, Jomura N, Morohashi K, Baharum SN, Goh HH. 2019. Transcriptome-wide effects of expansin gene manipulation in etiolated Arabidopsis seedling. Journal of Plant Research 132, 159–172. [DOI] [PubMed] [Google Scholar]
- Jang GJ, Yang JY, Hsieh HL, Wu SH. 2019. Processing bodies control the selective translation for optimal development of Arabidopsis young seedlings. Proceedings of the National Academy of Sciences, USA 116, 6451–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Dörmann P, Peto CA, Lutes J, Benning C, Chory J. 2000. Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proceedings of the National Academy of Sciences, USA 97, 8175–8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse EM, Halliday KJ. 2008. Skotomorphogenesis: the dark side of light signalling. Current Biology 18, R1144–R1146. [DOI] [PubMed] [Google Scholar]
- Kim C, Lee KP, Baruah A, Nater M, Göbel C, Feussner I, Apel K. 2009. 1O2-mediated retrograde signaling during late embryogenesis predetermines plastid differentiation in seedlings by recruiting abscisic acid. Proceedings of the National Academy of Sciences, USA 106, 9920–9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kósa A, Preininger É, Böddi B. 2015. Nitrogen deficiency hinders etioplast development in stems of dark-grown pea (Pisum sativum) shoot cultures. Physiologia Plantarum 155, 330–337. [DOI] [PubMed] [Google Scholar]
- Kowalewska Ł, Bykowski M, Mostowska A. 2019. Spatial organization of thylakoid network in higher plants. Botany Letters 166, 326–334. [Google Scholar]
- Kowalewska Ł, Mazur R, Suski S, Garstka M, Mostowska A. 2016. Three-dimensional visualization of the tubular-lamellar transformation of the internal plastid membrane network during runner bean chloroplast biogenesis. The Plant Cell 28, 875–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legen J, Schmitz-Linneweber C. 2017. Stable membrane-association of mRNAs in etiolated, greening and mature plastids. International Journal of Molecular Sciences 18, E1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. 2019. Piecing the puzzle together: the central role of reactive oxygen species and redox hubs in chloroplast retrograde signaling. Antioxidants & Redox Signaling 30, 1206–1219. [DOI] [PubMed] [Google Scholar]
- Li L, Nelson CJ, Trösch J, Castleden I, Huang S, Millar AH. 2017a Protein degradation rate in Arabidopsis thaliana leaf growth and development. The Plant Cell 29, 207–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LX, Song Y, Wang K, Dong P, Zhang XY, Li FG, Li ZG, Ren MZ. 2015. TOR-inhibitor insensitive-1 (TRIN1) regulates cotyledons greening in Arabidopsis. Frontiers in Plant Science 6, 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hensel G, Mascher M, et al. 2019. Leaf variegation and impaired chloroplast development caused by a truncated CCT domain gene in albostrians barley. The Plant Cell 31, 1430–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cai W, Liu Y, Li H, Fu L, Liu Z, Xu L, Liu H, Xu T, Xiong Y. 2017b Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proceedings of the National Academy of Sciences, USA 114, 2765–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Varala K, Hudson ME. 2014. A survey of the small RNA population during far-red light-induced apical hook opening. Frontiers in Plant Science 5, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Zhu N, Mai KK, Liu Z, Tzeng D, Osteryoung KW, Zhong S, Staehelin LA, Kang BH. 2018. Thylakoid-bound polysomes and a dynamin-related protein, FZL, mediate critical stages of the linear chloroplast biogenesis program in greening Arabidopsis cotyledons. The Plant Cell 30, 1476–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MC, Tsai HL, Lim SL, Jeng ST, Wu SH. 2017. Unraveling multifaceted contributions of small regulatory RNAs to photomorphogenic development in Arabidopsis. BMC Genomics 18, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist E, Aronsson H. 2018. Chloroplast vesicle transport. Photosynthesis Research 138, 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist E, Solymosi K, Aronsson H. 2016. Vesicles are persistent features of different plastids. Traffic 17, 1125–1138. [DOI] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Chen HM, Wu SH. 2012. Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Molecular Systems Biology 8, 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Wu JF, Lin WD, Wu YC, Tsai TY, Tsai HL, Wu SH. 2013. Translational landscape of photomorphogenic Arabidopsis. The Plant Cell 25, 3699–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XQ, Li Y, Zhong SW. 2017. Interplay between light and plant hormones in the control of Arabidopsis seedling chlorophyll biosynthesis. Frontiers in Plant Science 8, 1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juez E, Dillon E, Magyar Z, Khan S, Hazeldine S, de Jager SM, Murray JA, Beemster GT, Bögre L, Shanahan H. 2008. Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. The Plant Cell 20, 947–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Zhou Y, Chen H, et al. 2018. Membrane protein MHZ3 stabilizes OsEIN2 in rice by interacting with its Nramp-like domain. Proceedings of the National Academy of Sciences, USA 115, 2520–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. 2001. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. The Plant Cell 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Bonilla LD. 2014. Composition and function of P bodies in Arabidopsis thaliana. Frontiers in Plant Science 5, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Yang SW, Palatnik J. 2019. Keep calm and carry on: miRNA biogenesis under stress. The Plant Journal 99, 832–843. [DOI] [PubMed] [Google Scholar]
- Mathews S. 2006. Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Molecular Ecology 15, 3483–3503. [DOI] [PubMed] [Google Scholar]
- Mazur R, Mostowska A, Szach J, Gieczewska K, Wójtowicz J, Bednarska K, Garstka M, Kowalewska Ł. 2019. Galactolipid deficiency disturbs spatial arrangement of the thylakoid network in Arabidopsis thaliana plants. Journal of Experimental Botany 70, 4689–4704. [DOI] [PubMed] [Google Scholar]
- Mechela A, Schwenkert S, Soll J. 2019. A brief history of thylakoid biogenesis. Open Biology 9, 180237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Susek R, Chory J. 1996. An intracellular signal transduction pathway between the chloroplast and nucleus is involved in de-etiolation. Plant Physiology 112, 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed B, Bilooei SF, Dóczi R, Grove E, Railo S, Palme K, Ditengou FA, Bögre L, López-Juez E. 2018. Converging light, energy and hormonal signaling control meristem activity, leaf initiation, and growth. Plant Physiology 176, 1365–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monselise EB, Levkovitz A, Kost D. 2015. Ultraviolet radiation induces stress in etiolated Landoltia punctata, as evidenced by the presence of alanine, a universal stress signal: a 15N NMR study. Plant Biology 17 (Suppl 1), 101–107. [DOI] [PubMed] [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM. 1999. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. The Plant Cell 11, 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning DL, Liu KH, Liu CC, Liu JW, Qian CR, Yu Y, Wang YF, Wang YC, Wang BC. 2016. Large-scale comparative phosphoprotein analysis of maize seedling leaves during greening. Planta 243, 501–517. [DOI] [PubMed] [Google Scholar]
- Ort DR, Merchant SS, Alric J, et al. 2015. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proceedings of the National Academy of Sciences, USA 112, 8529–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MT, McCormac AC, Smith AG, Terry MJ. 2017. Singlet oxygen initiates a plastid signal controlling photosynthetic gene expression. New Phytologist 213, 1168–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratov I, McQuinn R, Schwartz J, Bar E, Fei Z, Lewinsohn E, Zamir D, Giovannoni JJ, Hirschberg J. 2016. Fruit carotenoid-deficient mutants in tomato reveal a function of the plastidial isopentenyl diphosphate isomerase (IDI1) in carotenoid biosynthesis. The Plant Journal 88, 82–94. [DOI] [PubMed] [Google Scholar]
- Patil M, Seifert S, Seiler F, Soll J, Schwenkert S. 2018. FZL is primarily localized to the inner chloroplast membrane however influences thylakoid maintenance. Plant Molecular Biology 97, 421–433. [DOI] [PubMed] [Google Scholar]
- Peng YL, Zou T, Li LM, Tang SW, Li Q, Zhang J, Chen YJ, Wang XC, Yang GT, Hu YG. 2019. Map-based cloning and functional analysis of YE1 in rice, which is involved in light-dependent chlorophyll biogenesis and photoperiodic flowering pathway. International Journal of Molecular Sciences 20, E758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Kim C. 2019. Current understanding of GUN1: a key mediator involved in biogenic retrograde signaling. Plant Cell Reports 38, 819–823. [DOI] [PubMed] [Google Scholar]
- Pham VN, Kathare PK, Huq E. 2018. Phytochromes and phytochrome interacting factors. Plant Physiology 176, 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolec R, Ulm R. 2018. Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Current Opinion in Plant Biology 45, 18–25. [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Ganguly D, Albrecht-Borth V. 2015. Insights into chloroplast biogenesis and development. Biochimica et Biophysica Acta 1847, 1017–1024. [DOI] [PubMed] [Google Scholar]
- Pomeranz MC, Hah C, Lin PC, Kang SG, Finer JJ, Blackshear PJ, Jang JC. 2010. The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiology 152, 151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribil M, Labs M, Leister D. 2014. Structure and dynamics of thylakoids in land plants. Journal of Experimental Botany 65, 1955–1972. [DOI] [PubMed] [Google Scholar]
- Roberts JA, Ritchie DF, Kerns JP. 2016. Plant growth regulator effects on bacterial etiolation of creeping bentgrass putting green turf caused by Acidovorax avenae. Plant Disease 100, 577–582. [DOI] [PubMed] [Google Scholar]
- Rocha J, Nitenberg M, Girard-Egrot A, Jouhet J, Marechal E, Block MA, Breton C. 2018. Do galactolipid synthases play a key role in the biogenesis of chloroplast membranes of higher plants? Frontiers in Plant Science 9, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J, Ramundo S. 2018. Chloroplast signaling and quality control. Essays in Biochemistry 62, 13–20. [DOI] [PubMed] [Google Scholar]
- Rudowska L, Gieczewska K, Mazur R, Garstka M, Mostowska A. 2012. Chloroplast biogenesis – correlation between structure and function. Biochimica et Biophysica Acta 1817, 1380–1387. [DOI] [PubMed] [Google Scholar]
- Sánchez-Retuerta C, Suaréz-López P, Henriques R. 2018. Under a new light: regulation of light-dependent pathways by non-coding RNAs. Frontiers in Plant Science 9, 962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub P, Rodriguez-Franco M, Cazzonelli CI, Álvarez D, Wüst F, Welsch R. 2018. Establishment of an Arabidopsis callus system to study the interrelations of biosynthesis, degradation and accumulation of carotenoids. PLoS ONE 13, e0192158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmied J, Hou Z, Hedtke B, Grimm B. 2018. Controlled partitioning of glutamyl-tRNA reductase in stroma- and membrane-associated fractions affects the synthesis of 5-aminolevulinic acid. Plant & Cell Physiology 59, 2204–2213. [DOI] [PubMed] [Google Scholar]
- Schoefs B, Franck F. 2008. The photoenzymatic cycle of NADPH: protochlorophyllide oxidoreductase in primary bean leaves (Phaseolus vulgaris) during the first days of photoperiodic growth. Photosynthesis Research 96, 15–26. [DOI] [PubMed] [Google Scholar]
- Selstam E, Sandelius AS. 1984. A comparison between prolamellar bodies and prothylakoid membranes of etioplasts of dark-grown wheat concerning lipid and polypeptide composition. Plant Physiology 76, 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluzicki A, Burko Y, Chory J. 2017. Dancing in the dark: darkness as a signal in plants. Plant, Cell & Environment 40, 2487–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova GA, Fomina IR, Kosobryukhov AA, Lyubimov VY, Nadezhkina ES, Balakhnina TI. 2017. Mesophyll cell ultrastructure of wheat leaves etiolated by lead and selenium. Journal of Plant Physiology 219, 37–44. [DOI] [PubMed] [Google Scholar]
- Shen Z, Li P, Ni RJ, et al. 2009. Label-free quantitative proteomics analysis of etiolated maize seedling leaves during greening. Molecular & Cellular Proteomics 8, 2443–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevela D, Ananyev G, Vatland AK, Arnold J, Mamedov F, Eichacker LA, Dismukes GC, Messinger J. 2019. ‘Birth defects’ of photosystem II make it highly susceptible to photodamage during chloroplast biogenesis. Physiologia Plantarum 166, 165–180. [DOI] [PubMed] [Google Scholar]
- Sinclair SA, Larue C, Bonk L, et al. 2017. Etiolated seedling development requires repression of photomorphogenesis by a small cell-wall-derived dark signal. Current Biology 27, 3403–3418.e7. [DOI] [PubMed] [Google Scholar]
- Singh A, Gautam V, Singh S, Sarkar Das S, Verma S, Mishra V, Mukherjee S, Sarkar AK. 2018. Plant small RNAs: advancement in the understanding of biogenesis and role in plant development. Planta 248, 545–558. [DOI] [PubMed] [Google Scholar]
- Skupień J, Wójtowicz J, Kowalewska Ł, Mazur R, Garstka M, Gieczewska K, Mostowska A. 2017. Dark-chilling induces substantial structural changes and modifies galactolipid and carotenoid composition during chloroplast biogenesis in cucumber (Cucumis sativus L.) cotyledons. Plant Physiology and Biochemistry 111, 107–118. [DOI] [PubMed] [Google Scholar]
- Solymosi K, Aronsson H. 2013. Etioplasts and their significance in chloroplast biogenesis. In: Bisval B, Krupinska K, Biswal UC, eds. Plastid development in leaves during growth and senescence. Berlin, Heidelberg: Springer, 39− 71. [Google Scholar]
- Solymosi K, Böddi B. 2006. Optical properties of bud scales and protochlorophyll(ide) forms in leaf primordia of closed and opened buds. Tree Physiology 26, 1075–1085. [DOI] [PubMed] [Google Scholar]
- Solymosi K, Bóka K, Böddi B. 2006. Transient etiolation: protochlorophyll(ide) and chlorophyll forms in differentiating plastids of closed and breaking leaf buds of horse chestnut (Aesculus hippocastanum). Tree Physiology 26, 1087–1096. [DOI] [PubMed] [Google Scholar]
- Solymosi K, Morandi D, Bóka K, Böddi B, Schoefs B. 2012. High biological variability of plastids, photosynthetic pigments and pigment forms of leaf primordia in buds. Planta 235, 1035–1049. [DOI] [PubMed] [Google Scholar]
- Solymosi K, Vitányi B, Hideg E, Böddi B. 2007. Etiolation symptoms in sunflower (Helianthus annuus) cotyledons partially covered by the pericarp of the achene. Annals of Botany 99, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Bussell JD, Camus I, et al. 2005. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. The Plant Cell 17, 1343–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Kharshiing E, Laird J, Sakai T, Christie JM. 2019. Deetiolation enhances phototropism by modulating NON-PHOTOTROPIC HYPOCOTYL3 phosphorylation status. Plant Physiology 180, 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Yu Y, Hu W, Min Q, Kang H, Li Y, Hong Y, Wang X, Hong Y. 2016. Ribosomal protein S6 kinase1 coordinates with TOR-Raptor2 to regulate thylakoid membrane biosynthesis in rice. Biochimica et Biophysica Acta 1861, 639–649. [DOI] [PubMed] [Google Scholar]
- Sun YY, Zhu SC, Yang X, Weston MV, Wang K, Shen ZQ, Xu HW, Chen LS. 2018a Nitrogen diagnosis based on dynamic characteristics of rice leaf image. PLoS ONE 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZF, Li M, Zhou Y, Guo TT, Liu Y, Zhang H, Fang YD. 2018b Coordinated regulation of Arabidopsis microRNA biogenesis and red light signaling through Dicer-like 1 and phytochrome-interacting factor 4. PLoS Genetics 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. 1993. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799. [DOI] [PubMed] [Google Scholar]
- Trinh HK, Verstraeten I, Geelen D. 2018. In vitro assay for induction of adventitious rooting on intact Arabidopsis hypocotyls. Methods in Molecular Biology 1761, 95–102. [DOI] [PubMed] [Google Scholar]
- Tsai HL, Li YH, Hsieh WP, Lin MC, Ahn JH, Wu SH. 2014. HUA ENHANCER1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis. The Plant Cell 26, 2858–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberegui E, Hall M, Lorenzo Ó, Schröder WP, Balsera M. 2015. An Arabidopsis soluble chloroplast proteomic analysis reveals the participation of the Executer pathway in response to increased light conditions. Journal of Experimental Botany 66, 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ughy B, Karlický V, Dlouhý O, Javornik U, Materová Z, Zsiros O, Šket P, Plavec J, Špunda V, Garab G. 2019. Lipid-polymorphism of plant thylakoid membranes. Enhanced non-bilayer lipid phases associated with increased membrane permeability. Physiologia Plantarum 166, 278–287. [DOI] [PubMed] [Google Scholar]
- Vitányi B, Kósa A, Solymosi K, Böddi B. 2013. Etioplasts with protochlorophyll and protochlorophyllide forms in the under-soil epicotyl segments of pea (Pisum sativum) seedlings grown under natural light conditions. Physiologia Plantarum 148, 307–315. [DOI] [PubMed] [Google Scholar]
- Wang H, Wu GX, Zhao BB, Wang BB, Lang ZH, Zhang CY, Wang HY. 2016a Regulatory modules controlling early shade avoidance response in maize seedlings. BMC Genomics 17, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Apel K. 2019. Dose-dependent effects of 1O2 in chloroplasts are determined by its timing and localization of production. Journal of Experimental Botany 70, 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kim C, Xu X, Piskurewicz U, Dogra V, Singh S, Mahler H, Apel K. 2016b Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proceedings of the National Academy of Sciences, USA 113, E3792–E3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GZ, Chalvin C, Hoelscher M, Meyer EH, Wu XN, Bock R. 2018. Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiology 176, 2472–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GZ, Meyer EH, Richter AS, et al. 2019. Control of retrograde signalling by protein import and cytosolic folding stress. Nature Plants 5, 525–538. [DOI] [PubMed] [Google Scholar]
- Xiong F, Zhang R, Meng Z, Deng K, Que Y, Zhuo F, Feng L, Guo S, Datla R, Ren M. 2017. Brassinosteriod Insensitive 2 (BIN2) acts as a downstream effector of the Target of Rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. New Phytologist 213, 233–249. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. 2014. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiology 164, 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bräutigam A, Weber AP, Zhu XG. 2016. Systems analysis of cis-regulatory motifs in C4 photosynthesis genes using maize and rice leaf transcriptomic data during a process of de-etiolation. Journal of Experimental Botany 67, 5105–5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH. 2009. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. The Plant Cell 21, 3270–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH. 2011. Processing bodies and plant development. Current Opinion in Plant Biology 14, 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li Y, Wang Y, Liu X, Zhu XG. 2017. Altered expression profiles of microRNA families during de-etiolation of maize and rice leaves. BMC Research Notes 10, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto KT, Watahiki MK, Matsuzaki J, Satoh S, Shimizu H. 2017. Space-time analysis of gravitropism in etiolated Arabidopsis hypocotyls using bioluminescence imaging of the IAA19 promoter fusion with a destabilized luciferase reporter. Journal of Plant Research 130, 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavafer A, Cheah MH, Hillier W, Chow WS, Takahashi S. 2015. Photodamage to the oxygen evolving complex of photosystem II by visible light. Scientific Reports 5, 16363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW. 2011. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. The Plant Journal 65, 346–358. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang Y, McFarlane HE, Obata T, Richter AS, Lohse M, Grimm B, Persson S, Fernie AR, Giavalisco P. 2018. Inhibition of TOR represses nutrient consumption, which improves greening after extended periods of etiolation. Plant Physiology 178, 101–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu JY, Roh J, Marchive C, Kim SK, Meyer C, Sun Y, Wang W, Wang ZY. 2016. TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Current Biology 26, 1854–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZW, Zhang GC, Zhu F, Zhang DW, Yuan S. 2015. The roles of tetrapyrroles in plastid retrograde signaling and tolerance to environmental stresses. Planta 242, 1263–1276. [DOI] [PubMed] [Google Scholar]
- Zhou T, Meng L, Ma Y, Liu Q, Zhang Y, Yang Z, Yang D, Bian M. 2018. Overexpression of sweet sorghum cryptochrome 1a confers hypersensitivity to blue light, abscisic acid and salinity in Arabidopsis. Plant Cell Reports 37, 251–264. [DOI] [PubMed] [Google Scholar]