In citrus, PH4–Noemi, a regulatory complex, controls proanthocyanidin accumulation by activating the transcription of proanthocyanidin biosynthetic genes, and also regulates expression of Noemi, promoting proanthocyanidin accumulation by positive feedback.
Keywords: Citrus, positive feedback, proanthocyanidin biosynthesis, regulatory complex, transcriptional regulation
Abstract
Proanthocyanidins (PAs; or condensed tannins) are a major class of flavonoids that contribute to citrus fruit quality. However, the molecular mechanism responsible for PA biosynthesis and accumulation in citrus remains unclear. Here, we identify a PH4–Noemi regulatory complex that regulates proanthocyanidin biosynthesis in citrus. Overexpression of PH4 or Noemi in citrus calli activated the expression of PA biosynthetic genes and significantly increased the PA content. Interestingly, Noemi was also shown to be up-regulated in CsPH4-overexpressing lines compared with wild-type calli. Simultaneously, CsPH4 partially complemented the PA-deficient phenotype of the Arabidopsis tt2 mutant and promoted PA accumulation in the wild-type. Further analysis revealed that CsPH4 interacted with Noemi, and together these proteins synergistically activated the expression of PA biosynthetic genes by directly binding to the MYB-recognizing elements (MRE) of the promoters of these genes. Moreover, CsPH4 could directly bind to the promoter of Noemi and up-regulate the expression of this gene. These findings explain how the CsPH4–Noemi regulatory complex contributes to the activation of PA biosynthetic genes via a positive feedback loop and provide new insights into the molecular mechanisms underlying PA biosynthesis, which can be effectively employed for metabolic engineering to improve citrus fruit quality.
Introduction
Proanthocyanidins (PAs) are prominent polyphenolic secondary metabolites that are synthesized via the flavonoid biosynthesis pathway (Winkel-Shirley, 2001). These metabolites are usually present in different tissues of many plant species (Dixon et al., 2005; de Rezende et al., 2009; Barbehenn and Constabel, 2011). PAs play important roles in plant defence against various forms of stress (Peters and Constabel, 2002; Dixon et al., 2005; Koes et al., 2005; Shirley, 2008; Barbehenn and Constabel, 2011; Hichri et al., 2011), and in modulating seed dormancy, longevity, and dispersion (Debeaujon et al., 2000; Lepiniec et al., 2006; Jia et al., 2012; Oikawa et al., 2015). In addition, PAs act as potential dietary antioxidants that have beneficial effects on human health, including protection against free radical-mediated injury and cardiovascular disease (Bagchi et al., 2000; Ling et al., 2001; Goufo and Trindade, 2014).
PA biosynthesis is one branch of the flavonoid biosynthesis pathway, which also produces anthocyanins and flavonols (see Supplementary Fig. S1 at JXB online). The flavonoid biosynthesis pathway has been extensively elucidated in many plant species, such as maize (Zea mays), snapdragon (Antirrhinum majus), Arabidopsis, apple (Malus domestica), grapevine (Vitis vinifera), and poplar (Populus spp.) (Mol et al., 1998; Allan et al., 2008; Tohge et al., 2017; Wang et al., 2017a). Among flavonoid biosynthetic genes, those encoding anthocyanidin reductase (ANR) and leucoanthocyanidin reductase (LAR) are specific to the PA branch of the pathway and produce flavan-3-ols, typically epicatechin and catechin, respectively (Abrahams et al., 2003; Xie et al., 2003). Moreover, laccase 15 (TT10), MATE transporter (TT12), glutathione-S-transferase (TT19/GST) and H+-ATPase isoform 10 (AHA10) have been reported to be involved in PA modification, transport, and oxidation (Kitamura et al., 2004; Baxter et al., 2005; Pourcel et al., 2005; Marinova et al., 2007).
The regulation of PA biosynthesis is co-ordinately induced by many transcription factors (TFs), such as R2R3-MYB, basic helix–loop–helix (bHLH), WD40-repeat proteins (WDRs), MADS box, and WRKY (Johnson et al., 2002; Hichri et al., 2011; Xu et al., 2017). In Arabidopsis, the TRANSPARENT TESTA2 (TT2) gene, which encodes an R2R3-MYB protein, has been identified as a key regulator of the transcription of ANR, DFR, and AHA10 (Nesi et al., 2001; Sharma and Dixon, 2005; Lepiniec et al., 2006). In grapevine (V. vinifera), three MYB-like TFs, namely VvMYBPA1, VvMYBPA2, and VvMYB5b, are involved in the regulation of PA biosynthesis via activation of VvLAR1, VvANS, VvF3′5′H, or VvCHI in developing grape berries (Bogs et al., 2007; Deluc et al., 2008; Terrier et al., 2009). In poplar (P. tomentosa), a TT2-like gene, MYB115, which significantly enhanced the expression of ANR1 and LAR3, was isolated and characterized (Wang et al., 2017a). The MYB–bHLH–WD40 (MBW) complex, formed by MYB, bHLH, and WDR proteins, has been widely elucidated in regulating PA biosynthesis (Schaart et al., 2013; Xu et al., 2014). The bHLH and WDR cofactors are adaptable and are required for PA biosynthesis in many plant species, interacting with PA-specific R2R3-MYB proteins, such as TT8 and TTG1 from Arabidopsis, VvMYC1 from grapevine, MtTT8 and MtWD40-1 from alfalfa, and OsRc from rice (Walker et al., 1999; Nesi et al., 2000; Furukawa et al., 2007; Hichri et al., 2011; Li et al., 2016).
The genus Citrus, comprising some of the most widely cultivated fruit crops in the world, provides an abundance of natural variations in metabolites and an interesting system for analysis of the evolution of fruit quality (Huang et al., 2018; Wu et al., 2018). Similar to most fruit crops, citrus also accumulate large amounts of flavonoids, which have significant effects on quality (Kawaii et al., 1999, 2000; Chen et al., 2015; Wang et al., 2016, 2017b). Blood oranges (Citrus sinensis) and purple pummelo (C. grandis) accumulate considerable amounts of anthocyanins in mature fruits, which give the fruit a striking colour (Butelli et al., 2012; Huang et al., 2018). PAs, one of the most important determinants of quality in citrus fruit, are widely accumulated in the fruits, leaves, roots, and seeds (Wang et al., 2017b). Previous studies have reported that TFs such as Ruby, CsMYBF1 and Noemi play important roles in flavonoid biosynthesis in citrus (Butelli et al., 2012; Liu et al., 2016; Butelli et al., 2019; Huang et al., 2018). However, although our knowledge of flavonoid biosynthesis and accumulation in plants has increased substantially, there remains much to learn about the regulation of flavonoid biosynthesis in citrus, especially the molecular mechanism responsible for PA biosynthesis.
‘Anliu’ (C. sinensis Osbeck cv. Anliu), ‘Hong Anliu’ (C. sinensis Osbeck cv. Hong Anliu, a bud mutant of ‘Anliu’) and ‘Succari’ (C. sinensis Osbeck cv. Succari), all of which are sweet orange varieties, exhibit significant differences in major metabolite accumulation in fruit, providing an ideal set of resources for investigation of the regulatory mechanisms underlying the differences in flavonoid biosynthesis (Pan et al., 2014; Chen et al., 2015; Guo et al., 2016; Huang et al., 2016). ‘Hong Anliu’ and ‘Succari’ are two acidless varieties, with white seed, and these varieties do not accumulate PAs in seeds (Guo et al., 2016; Huang et al., 2016). In addition, ‘Hong Anliu’ exhibits high accumulation of lycopene in the fruit juice sacs (Liu et al., 2007). ‘Anliu’ accumulates certain organic acids, and the seeds of this variety accumulate PAs and are brown in colour (Liu et al., 2007). A number of studies have been performed at different levels to gain insight into high lycopene accumulation and the acidless phenotype using these three varieties (Liu et al., 2007; Guo et al., 2016; Huang et al., 2016), whereas very little attention has been paid to the molecular mechanism underlying PA modulation.
Here, based on the RNA-seq analysis, we identified differentially expressed genes putatively involved in PA metabolism. Subsequent experiments identified the CsPH4–Noemi regulatory complex as a key regulator of PA biosynthesis in citrus. Moreover, the CsPH4–Noemi regulatory complex also regulates the expression of Noemi, thereby promoting PA accumulation via a positive feedback loop. These results fill a gap in the molecular mechanisms underlying the regulation of PA accumulation in citrus.
Materials and methods
Plant materials and growth conditions
Fruit samples of ‘Anliu’, ‘Hong Anliu’, and ‘Succari’ were harvested from three mature trees, with 10 representative fruits from each tree at 90 days after flowering (DAF). The flavedos, pulps, and seeds were separately and immediately frozen in liquid nitrogen, and stored at −80 °C until analysis.
The citrus callus used for genetic transformation was derived from Marsh grapefruit (C. paradise Macf., ‘RM’), a citrus variety with low concentration of anthocyanin and PA. The citrus callus was subcultured on solid MT basal medium in darkness at 25 °C every 20 d. Tobacco (Nicotiana benthamiana) was planted in a growth chamber under a 14 h light–10 h dark photoperiod and 24 °C conditions. The wild-type (Col-0) and tt2 mutants (CS83) plants were planted in a growth chamber under a 16 h light–8 h dark photoperiod and 21 °C conditions.
RNA isolation, transcriptome profile analysis and real-time quantitative PCR analysis
All RNA samples were isolated as described by Lu et al. (2018). Three biological replicates of seeds and pulps from ‘Anliu’, ‘Hong Anliu’, and ‘Succari’ at 90 DAF were subjected to RNA-sequencing (RNA-seq). RNA-seq libraries were constructed and sequenced on Illumina HiSeqTM. The RNA-seq data were aligned to the sweet orange reference genome (http://citrus.hzau.edu.cn/orange/) using TopHat (v2.0.9). The mapped reads from each sample were normalized to fragments per kilobase of exon model per million reads mapped (FPKM) mapped for each predicted transcript using HTSeq v0.6.1 software in union mode (see Supplementary Tables S1, S2; Trapnell et al., 2010). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed with the GOSeq R package and KOBAS software using default parameters (Supplementary Tables S3, S4), respectively (Mao et al., 2005; Young et al., 2010).
Real-time quantitative PCR (qRT-PCR) was conducted according to Bustin et al. (2009). Single-strand cDNA was synthesized using the HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme). The primers used for qRT-PCR were from published articles (Huang et al., 2018; Butelli et al., 2019; Strazzer et al., 2019), and listed in Supplementary Table S5. The citrus Actin gene was used as an internal control. qRT-PCR was performed with a Roche LightCycler® 480 system using the 2×LightCycler 480 SYBR Green Master Mix (Roche) and a three-step programme: pre-incubation at 95 °C for 10 min; 40 cycles of amplification at 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 20 s; followed by a melting curve at 95 °C for 5 s, 65 °C for 1 min, then ramping at 0.11 °C s−1 to 97 °C with continuous fluorescence measurement. Fluorescence was measured at each extension step. Each run contained a negative control (water in place of cDNA) and each reaction was performed in triplicate. The reaction specificity was confirmed by the negative control and a Tm Calling analysis. The qRT-PCR data were analysed using 2−ΔΔCt method.
Isolation and sequence analysis of CsPH4 and Noemi
The full-length coding sequences (CDs) of CsPH4 and Noemi were amplified. The conserved domains were analysed using the NCBI Conserved Domain Database (CDD, https://www.ncbi.nlm.nih.gov/cdd) and the ExPASy PROSITE Database (http://prosite.expasy.org/) (Liu et al., 2014b). Multiple sequence alignments were performed using the Clustal W program and GeneDoc software. The neighbour-joining phylogenetic tree was constructed using MEGA6.0 software with bootstrap values from 1000 replicates.
Plasmid construction and plant transformation
The CDs of CsPH4 and Noemi were amplified and inserted into the PE3C vector and subsequently recombined into the binary overexpression vectors PK7WG2D (kanamycin resistance) and PH7WG2D (hygromycin resistance) to construct plasmids with a 3×HA tag fusion in the C-terminus. Then, the plasmids were transformed into Agrobacterium strain GV3101 and EH105, respectively. Plant transformation was performed using Agrobacterium-mediated methods described previously (Clough and Bent, 1998; Lu et al., 2016). Citrus calli and Arabidopsis were infected by recombinant A. tumefaciens and then putative transgenic plants were selected on medium supplemented with 50 mg l−1 hygromycin and 50 mg l−1 kanamycin, respectively. Seed phenotypes were observed in progeny from T2 transformants with a single copy of the transgene and further screened for homozygotes after germination.
Subcellular localization assay
The CDs of CsPH4 and Noemi without the stop codon were amplified and cloned into the PK7CWG2.0 vector, in frame with the cyan fluorescent protein (CFP) gene. For transient expression analysis, the fusion constructs (CsPH4–CFP and Noemi–CFP) were co-transformed with a plasmid coding for a nuclear marker, VirD2NLS, fused to mCherry into tobacco (N. benthamiana) leaves by A. tumefaciens infiltration based on a previous description (Kumar and Kirti, 2010). Fluorescence signals were observed with a confocal laser scanning microscope (Leica TCS SP2, Leica) 60 h after infiltration.
Transient expression assay
A transient expression assay was performed as previously described (Han et al., 2016; Lu et al., 2018). Briefly, for transcriptional activity assay, the CDs of CsPH4 and Noemi were inserted into the pBD vector to generate the effector vectors pBD-CsPH4 and pBD-Noemi. The empty vector was used as the negative control (pBD), while vector containing the VP16 activation domain was used as the positive control (pBD-VP16). The reporter vector contains a GAL4–LUC and an internal control REN driven by the 35S promoter; GAL4–LUC contains five copies of GAL4-binding element and TATA-box in front of the LUC.
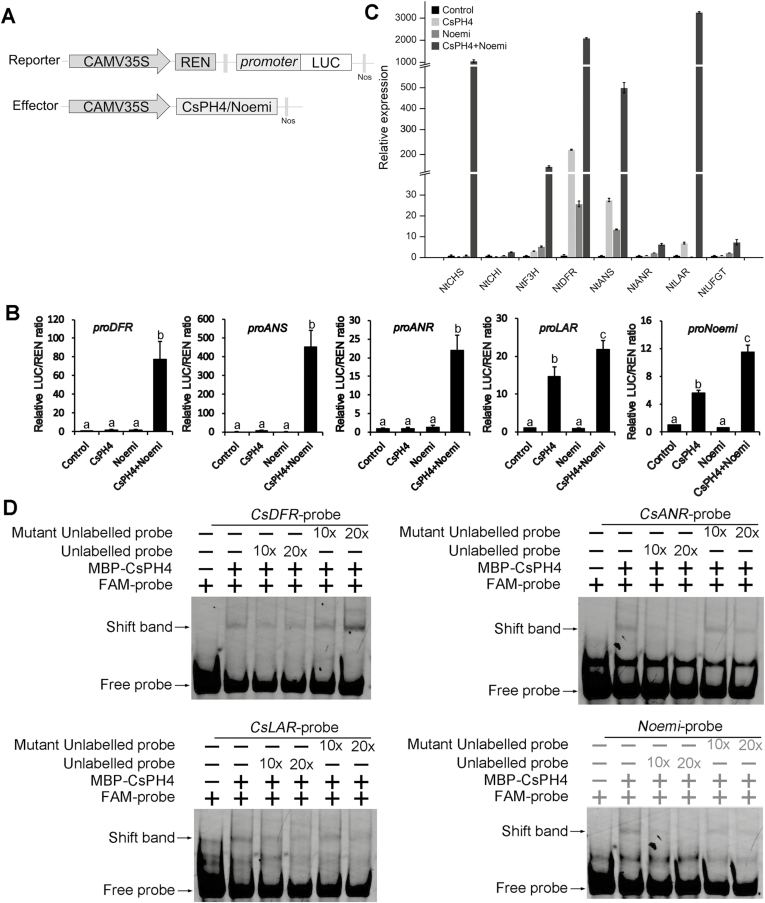
For the DNA-promoter interaction assay, the promoters of DFR, ANS, ANR, LAR, UFGT2, and Noemi were amplified and cloned into the pGreenII 0800-LUC vector to yield reporter. The PK7–HA–CsPH4 and PK7–HA–Noemi constructs were used as effectors. The empty vector pK7WG2D was used as the negative control (see Fig. 7A). Tobacco (N. benthamiana) leaves were infiltrated by Agrobacterium cells containing the effector and reporter using the agroinfiltration method described by Hellens et al. (2005). Luciferase activity was detected 72 h after infiltration using the Dual-Luciferase Reporter Assay System (Promega) with an Infinite200 Promicroplate reader (Tecan). Relative luciferase activity was calculated as the ratio of LUC/REN.
Fig. 7.
The CsPH4–Noemi complex activates the promoters of PA biosynthetic genes and Noemi by binding to these promoters. (A) Schematic diagrams of vectors used for the dual-luciferase assay. The reporter vector contained the promoter of CsDFR, CsANS, CsANR, CsLAR, and Noemi fused to LUC. (B) Transient promoter activity assays were carried out using LUC reporter gene under the control of the promoters of CsDFR, CsANS, CsANR, CsLAR, or Noemi, along with effectors (CsPH4+Noemi) and the empty vector as an internal control. Error bars represent the mean ±SD of eight biological replicates. Different letters indicate a significant difference using Duncan’s test: P<0.01. (C) qRT-PCR analysis of flavonoid biosynthetic genes (NtCHS, NtCHI, NtF3H, NtDFR, NtANS, NtANR, NtLAR, and NtUFGT) in tobacco leaves overexpressing CsPH4, Noemi, and CsPH4 plus Noemi and leaves infiltrated with the empty vector control. All transient overexpression experiments were conducted three times. The gene expression data in ‘Control’ were normalized to 1. Error bars represent the mean ±SD of three biological replicates. (D) EMSAs showing the binding of CsPH4 to the MREs of the CsDFR, CsANR, CsLAR, and Noemi promoters. For competition, 10- and 20-fold excess of non-labelled probes or mutant unlabelled probes were used. ‘+’ and ‘−’ indicate the presence and absence, respectively, of the indicated probe or protein.
Yeast two-hybrid analysis
Yeast two-hybrid (Y2H) assays were performed according to the manufacturer’s instructions (Clontech, Palo Alto, CA, USA). The CDs of CsPH4 and Noemi and the truncated CsPH4 peptide sequences (CsPH4ΔC1, amino acids 1–326; CsPH4ΔC2, amino acids 1–276) were amplified and inserted into pGBKT7 and pGADT7 to construct BD–CsPH4, DB–CsPH4ΔC1, DB–CsPH4ΔC2, DB–CsPH4ΔC3, BD–Noemi, and AD–Noemi. The AD and BD fusion constructs were transformed or co-transformed into yeast strain AH109 to examine self-activation and the interaction between CsPH4 and Noemi, respectively. Transformants were then screened on selection medium supplemented with SD base/−Trp/−Leu/−His/−Ade in the presence of 5-bromo-4-chloro-3-indolyl-α-D-galactopyranoside (X-α-Gal) to determine the interaction of CsPH4 with Noemi.
Bimolecular fluorescence complementation assay
For bimolecular fluorescence complementation (BiFC) assays, the CDs of CsPH4 without the stop codons were cloned into L101YNE to generate the CsPH4–nYFP construct, and Noemi without the stop codons was introduced into L101YCE to generate the Noemi–cYFP construct. The resulting constructs were introduced into A. tumefaciens strain GV3101 and then infiltrated into tobacco leaf epidermal cells according to the previous description with appropriate modification (Walter et al., 2004). After infiltration, plants were incubated for at least 48 h before observation. The YFP fluorescence was imaged using a Confocal Spectral Microscope Imaging System (Leica TCS SP5).
Recombinant protein purification and in vitro pull-down analysis
The CDs of CsPH4 and Noemi were amplified and inserted into pGEX-6P-1 to generate glutathione S-transferase (GST)-tagged recombinant protein and into pET32a to generate His-tagged recombinant protein. Pull-down assays were conducted according to the Pierce® GST Spin Purification Kit protocol (Pierce, Rockford, USA). The protein concentration of the eluted fractions was determined using the Pierce BCA Protein Assay Kit (product no. 23227). GST-tagged protein was eluted by adding elution buffer that contained glutathione, and the eluted proteins were analysed by western blotting using anti-His and anti-GST antibodies (Sangon Biotech, Shanghai, China).
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays (EMSAs) were conducted as described previously with some modifications (Huang et al., 2018; Lu et al., 2018). The CDs of CsPH4 without the stop codon was cloned into a double-tagged expression vector to generate the recombinant vector MBP–CsPH4–His. The 5′FAM-labelled oligonucleotide probes were directly synthesized and labelled. Unlabelled probes with the same or mutated oligonucleotides were used as cold competitors. The binding was implemented with EMSA/Gel-shift Binding Buffer (Beyotime Biotechnology, Shanghai, China, no. GS006). Purified protein and 1 μl of the FAM-labelled probe (10 μmol−1) were mixed together and incubated at 4 °C for 30 min. For the competition assays, the unlabelled probe was incubated with protein and binding buffer at 4 °C for 30 min. Next, 1 μl of the FAM-labelled probe (10 μmol−1) was added and incubated at 4°C for 30 min. The samples were loaded onto a pre-run 6% polyacrylamide gel. Electrophoresis was performed at 4 °C using 0.5× Tris–borate–EDTA buffer in the dark for 1 h. Gel images were acquired using the Amersham Imager 600 (GE Healthcare). Probes information is given in Supplementary Table S5.
DMACA staining and quantification of PAs
Extraction and quantification of total soluble and insoluble PAs were performed as previously described by Pang et al. (2008). Briefly, approximately 0.5 g of freeze-dried tissues was extracted using 5 ml of 70% acetone solution (v/v) containing 0.1% (w/v) ascorbic acid by vortexing, and then sonicated at 4 °C in the dark for 1 h. For quantification of soluble PAs, 1 ml of DMACA reagent (2% w/v p-dimethyl-amino-cinnamaldehyde (DMACA) in methanol–3 M HCl) was added to 1 ml of fluid under test. After reaction for 30 min, the total soluble PA levels was calculated spectrophotometrically at 640 nm, with (+)-catechin as the standard (Yuanye Bio-Technology, Shanghai, China). Quantification of insoluble PAs (with butanol–HCl) was performed as described previously (Pang et al., 2008). Absorbance values were converted into PA equivalents with procyanidin B1 as a standard (Yuanye Bio-Technology). The Arabidopsis seeds and citrus calli were stained with 0.2% DMACA in methanol: 6 M HCl (1:1) for at least 30 min and then washed in ethanol: acetic acid (75:25) for visualization of PAs under UV light on a universal fluorescence microscope (Olympus BX61, Tokyo, Japan).
Quantification of anthocyanins
Anthocyanin determinations were based largely on a previously reported protocol with modifications (Huang et al., 2018). The freeze-dried tissues were extracted using 2 ml of methanol and 1% (v/v) HCl for 30 min at 4 °C with ultrasonic vibration. The supernatant was calculated spectrophotometrically at 530 nm and 657 nm. The (A530–0.25×A657)/dry weight was considered anthocyanin content.
Accession numbers
The sequence data from this article can be found in the NCBI database or in the genome databases of Citrus (http://citrus.hzau.edu.cn/orange/) or Arabidopsis (http://www.arabidopsis.org/index.jsp). GenBank accession numbers of the proteins are listed in Supplementary Table S6.
Results
Analysis of PA levels ‘Anliu’, ‘Hong Anliu’, and ‘Succari’ sweet oranges
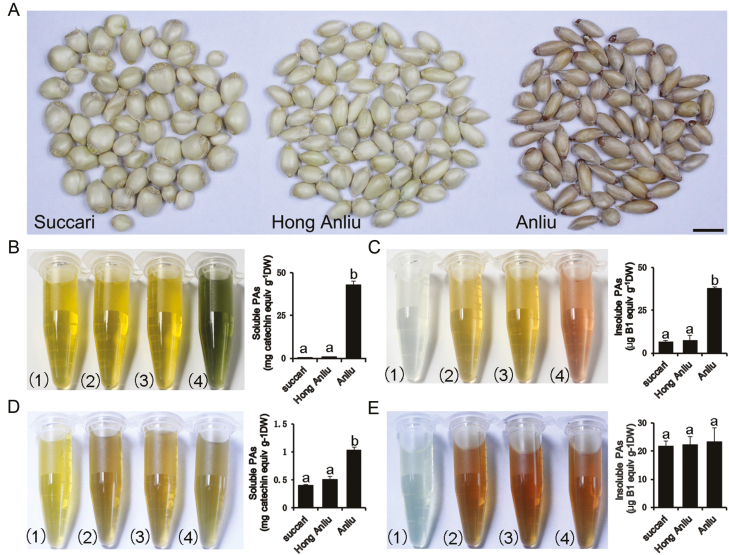
PAs, a polymer of catechin and epicatehin, are often deposited in the endothelial layer of the seed coat, leading to a brown coloration in many species. According to the degree of polymerization, PAs are divided into soluble (oligomeric, dimer and trimer polymerization) and insoluble (polymeric, tetramer or more polymerization). As shown in Fig. 1A, the ‘Anliu’, ‘Hong Anliu’, and ‘Succari’ seeds showed significant differences in colour. The juice of ‘Anliu’ fruits is more acidic (pH 3.59±0.13) than that of ‘Hong Anliu’ fruits (pH 5.64±0.06) and ‘Succari’ fruits (pH 5.60±0.01) (see Supplementary Fig. S2). The seed of ‘Anliu’ displayed higher concentrations of PA (soluble, 42.82±2.36 mg g−1 DW; insoluble, 38.05±0.39 µg g−1 DW) than the seed of the ‘Hong Anliu’ (soluble, 0.85±0.09 mg g−1 DW; insoluble, 7.46±2.69 µg g−1 DW) and ‘Succari’ (soluble, 0.45±0.06 mg g−1 DW; insoluble, 6.68±0.45 µg g−1 DW) (Fig. 1B, C). To further analyse the differences in PA content among the three varieties, we examined PA levels in the flavedos and pulps of these varieties. The soluble PA content in the pulps of ‘Anliu’ (1.03±0.06 mg g−1 DW) was higher than that in the pulps of ‘Hong Anliu’ (0.51±0.05 mg g−1 DW) and ‘Succari’ (0.41±0.01 mg g−1 DW) (Fig. 1D), while no significant differences were observed for the insoluble PA content (Fig. 1E). In addition, the three varieties contained comparable amounts of both soluble and insoluble PAs in their flavedos (Supplementary Fig. S3A, B).
Fig. 1.
Characterization of PA levels in ‘Anliu’, ‘Hong Anliu’, and ‘Succari’ sweet oranges. (A) Mature seeds without testae from three sweet orange varieties. Scale bar: 1 cm. (B) The contents and extracts after DMACA staining of soluble PAs in seeds. (C) The contents and extracts after DMACA staining of insoluble PAs in seeds. (D) The contents and extracts after DMACA staining of soluble PAs in pulps. (E) The contents and extracts after DMACA staining of insoluble PAs in pulps. (1) Blank control, (2) ‘Succari’, (3) ‘Hong Anliu’, (4) ‘Anliu’. DW, dry weight. Error bars represent the mean ±SD of three biological replicates. (This figure is available in colour at JXB online.)
Comparison of the transcriptomes of ‘Anliu’, ‘Hong Anliu’, and ‘Succari’ sweet oranges
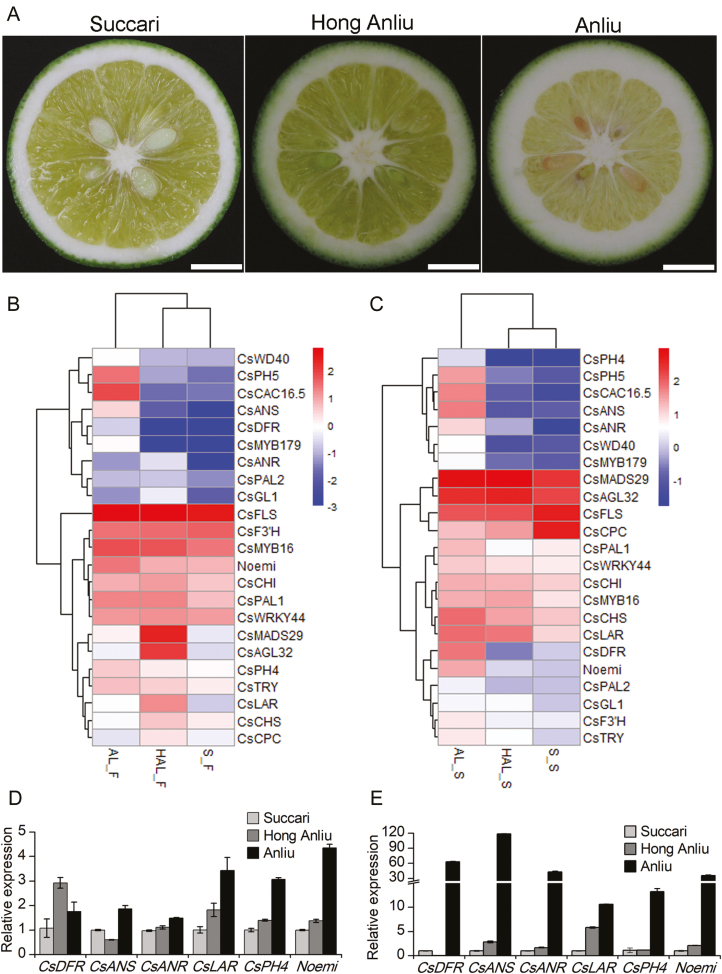
To identify the genes potentially associated with the phenotypic differences, seeds and pulps from the three varieties collected at 90 DAF (Fig. 2A) were subjected to RNA-seq. Given that the accumulation of PAs in ‘Anliu’ was significantly different from that in ‘Hong Anliu’ and ‘Succari’, while there was almost no difference between ‘Hong Anliu’ and ‘Succari’, the comparative transcriptomic analysis was performed between ‘Anliu’ and ‘Hong Anliu’, and between ‘Anliu’ and ‘Succari’. Comparison of the dataset from seed samples led to the identification of 329 (144 up-regulated and 185 down-regulated, ‘Anliu’ versus ‘Hong Anliu’, corrected P-value <0.05, fold change >1.5) and 2741 (1436 up-regulated and 1305 down-regulated, ‘Anliu’ versus ‘Succari’, corrected P-value <0.05, fold change >1.5) differentially expressed genes, among which, genes involved in cellulose metabolism, glucan metabolism, and TF activity were statistically over-represented. Meanwhile, 462 (140 up-regulated and 322 down-regulated, ‘Anliu’ versus ‘Hong Anliu’, corrected P-value <0.05, fold change >1.5) and 2457 (1414 up-regulated and 1043 down-regulated, ‘Anliu’ versus ‘Succari’, corrected P-value <0.05, fold change >1.5) differentially expressed transcripts were detected from the pulp comparison group (see Supplementary Fig. S4), and these genes showed marked enrichment of functions associated with lipid metabolism, stimulus response and TF activity (Supplementary Fig. S5). KEGG analysis revealed a high percentage of differentially expressed transcripts responsible for the biosynthesis of secondary metabolites, hormone signal transduction and phenylalanine metabolism (Supplementary Fig. S6).
Fig. 2.
Transcriptomic differences among the three sweet orange varieties. (A) Fruits from the three citrus varieties 90 DAF. Scale bar: 1 cm. (B) Cluster heat map based on the expression of phenylpropanoid-related genes in the pulps of the three citrus varieties. AL_F, ‘Anliu’ pulps; HAL_F, ‘Hong Anliu’ pulps; S_F, ‘Succari’ pulps. (C) Cluster heat map based on the expression of phenylpropanoid-related genes in the seeds of the three citrus varieties. AL_S, ‘Anliu’ seeds; HAL_S, ‘Hong Anliu’ seeds; S_S, ‘Succari’ seeds. (D) Relative expression patterns of PA biosynthetic genes (CsDFR, CsANS, CsANR, CsLAR), CsPH4, and Noemi in the pulps of the three citrus varieties. (E) Relative expression patterns of PA biosynthetic genes (CsDFR, CsANS, CsANR, CsLAR), CsPH4, and Noemi in the seeds of the three citrus varieties. The gene expression data for ‘Succari’ were normalized to 1. Error bars represent the mean ±SD of three biological replicates. (This figure is available in colour at JXB online.)
To further investigate the molecular processes and regulatory mechanism underlying PA accumulation in citrus, we focused on genes associated with the flavonoid biosynthesis pathway. Interestingly, the expression of the four PA biosynthetic genes, namely, DFR, ANS, ANR, and LAR, showed a close association with PA accumulation in seeds and pulps (Fig. 2B, C). The TFs CsPH4 (Cs9g03070) and Noemi (Cs5g31400) were also co-ordinately expressed with PA accumulation in both seeds and pulps (Fig. 2B, C; Supplementary Table S7). Further, qRT-PCR analysis was performed to verify the results. Consistent with the prediction, the five flavonoid biosynthetic genes (CHS, DFR, ANS, ANR, and LAR) and two TFs were expressed at significantly higher levels in ‘Anliu’ than in ‘Hong Anliu’ and ‘Succari’ (Fig. 2D, E; Supplementary Fig. S7). These results indicated the regulatory potential of CsPH4 and Noemi in citrus PA biosynthesis.
Identification of CsPH4 and Noemi
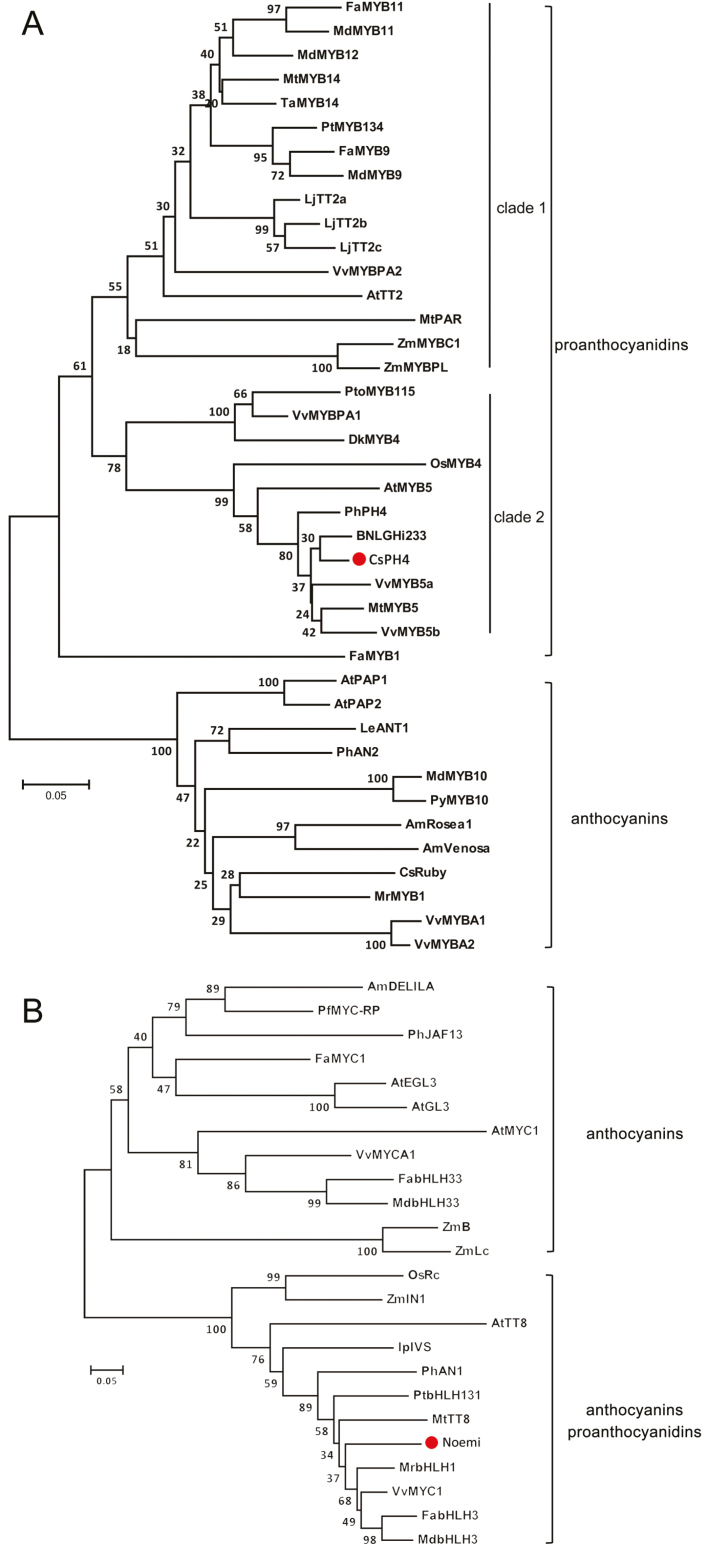
To verify the two candidate genes controlling PA biosynthesis in citrus, we isolated CsPH4 and Noemi from the seed cDNA library of ‘Anliu’ by PCR using gene-specific primers (see Supplementary Table S5). CsPH4, like other putative PA regulators such as VvMYB5a, VvMYB5b, and MtMYB5, contained the conserved R2R3 repeat domain, C1 and C3 motifs (Supplementary Fig. S8A). Compared with other MYB proteins, CsPH4 shared the highest sequence homology with VvMYB5b (61%), which was confirmed to regulate anthocyanin and PA biosynthesis during grape berry development (Deluc et al., 2008). A phylogenetic analysis of CsPH4 and other plant R2R3-MYB proteins associated with regulation of anthocyanidin and PA biosynthesis indicated that CsPH4 was closely related to PA regulators and distinct from anthocyanin regulators (Fig. 3A). The PA regulator clades consisted of two subclades, PA clades 1 and 2, while CsPH4, VvMYB5a, VvMYB5b, and MtMYB5 belonged to clade 2 (Fig. 3A). Compared with other bHLH proteins, Noemi, containing a conserved bHLH domain and an MYB-interacting region, shared the highest sequence homology with VvMYC1 (70% identity, 79% similarity) (Supplementary Fig. S8B). Phylogenetic analysis indicated that Noemi was closely related to PA and anthocyanin regulators and distinct from anthocyanin-specific regulators (Fig. 3B).
Fig. 3.
Phylogenetic analysis of CsPH4 and Noemi. (A) Phylogenetic analysis of predicted peptide sequences of CsPH4 and related genes from other plants. (B) Phylogenetic analysis of predicted peptide sequences of Noemi and related genes from other plants. Scale bar represents 0.05 substitutions per site and numbers next to the nodes are bootstrap values from 1000 replicates. Phylogenetic trees were constructed using the neighbour-joining method of MEGA v.5.1 software. Putative regulatory functions of most of the proteins in the control of flavonoid biosynthesis are indicated. (This figure is available in colour at JXB online.)
Transient expression of CsPH4–CFP and Noemi–CFP fusion protein in tobacco (N. benthamiana) epidermal cells demonstrated that both the CsPH4–CFP and Noemi–CFP proteins were localized in the nucleus (see Supplementary Fig. S9A). Furthermore, the transcriptional activity assay indicated that both CsPH4 and Noemi acted as transcriptional activators (Supplementary Fig. S9B, C).
Overexpression of CsPH4 or Noemi induces PA accumulation in citrus callus
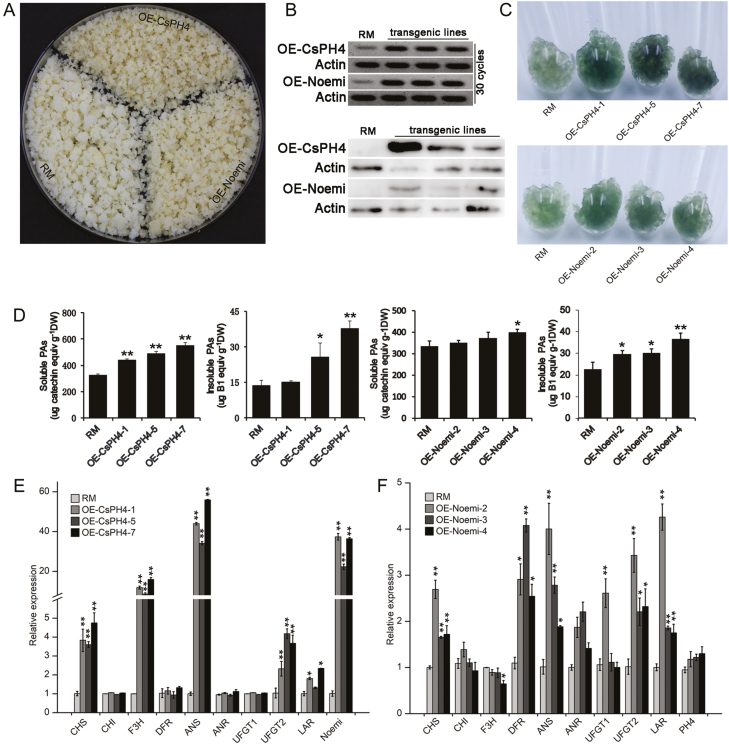
To investigate the functions of CsPH4 and Noemi, the PK7-HA-CsPH4 and PK7-HA-Noemi constructs were introduced into citrus calli (Fig. 4A). Three independent lines with high transcript and protein levels of CsPH4 and Noemi were chosen for further analysis (Fig. 4B). To visualize differences in PA levels, the transgenic calli and wild-type were stained with DMACA reagent. As expected, histochemical staining showed that the PA levels were higher in the CsPH4-overexpressing (OE-CsPH4) and Noemi-overexpressing (OE-Noemi) calli than in the wild-type (RM) (Fig. 4C). Quantitative measurements showed that the levels of PAs in the OE-CsPH4 (soluble, 1.36~1.69-fold changes; insoluble, 1.89~2.77-fold changes) and OE-Noemi (soluble, approximately 1.2-fold changes; insoluble, 1.31~1.62-fold changes) calli were significantly higher than those in the wild-type (Fig. 4D), but the content of anthocyanins did not change significantly (see Supplementary Fig. S10A).
Fig. 4.
Functional characterization of CsPH4 and Noemi overexpression in citrus calli. (A) Phenotypes of transgenic citrus calli. RM, wild-type citrus callus; OE-CsPH4, CsPH4-overexpressing callus; OE-Noemi, Noemi-overexpressing callus. (B) Semi-quantitative RT-PCR and western blotting analysis of CsPH4 and Noemi transcript and protein levels. An anti-HA antibody was used for immunoblotting. Actin, Actin gene (internal control). (C) DMACA staining of transgenic citrus calli. (D) Quantification of soluble and insoluble PA levels in transgenic citrus calli. DW, dry weight. (E) Relative expression of flavonoid biosynthetic genes in CsPH4-overexpressing calli. (F) Relative expression of flavonoid biosynthetic genes in Noemi-overexpressing calli. After several rounds of subculture, stable transgenic callus lines were established on selectable media. Calli grown for 15 d were collected for each assay. The gene expression data in ‘RM’ were normalized to 1. Error bars represent the mean ±SD of three biological replicates. Asterisks indicate significant differences using Student’s t-test: *P<0.05; **P<0.01. (This figure is available in colour at JXB online.)
To analyse the effects of CsPH4 overexpression on the transcription of flavonoid biosynthetic genes, qRT-PCR analysis was performed. OE-CsPH4 calli exhibited significantly up-regulated expression of CHS, F3H, ANS, LAR, and UFGT2 compared with the wild-type (Fig. 4E). Interestingly, the expression of Noemi was also significantly up-regulated in the OE-CsPH4 calli (Fig. 4E). Similarly, the expression of the flavonoid biosynthetic genes, including CHS, DFR, ANS, ANR, LAR, and UFGT2, was also up-regulated in OE-Noemi calli compared with the wild-type (Fig. 4F). Overall, these results demonstrated that overexpression of CsPH4 and Noemi in citrus calli could induce partially the expression of flavonoid biosynthetic genes and promote the accumulation of PAs.
CsPH4 partially complements the Arabidopsis tt2 mutant phenotype and promotes the accumulation of PAs in the Arabidopsis wild-type
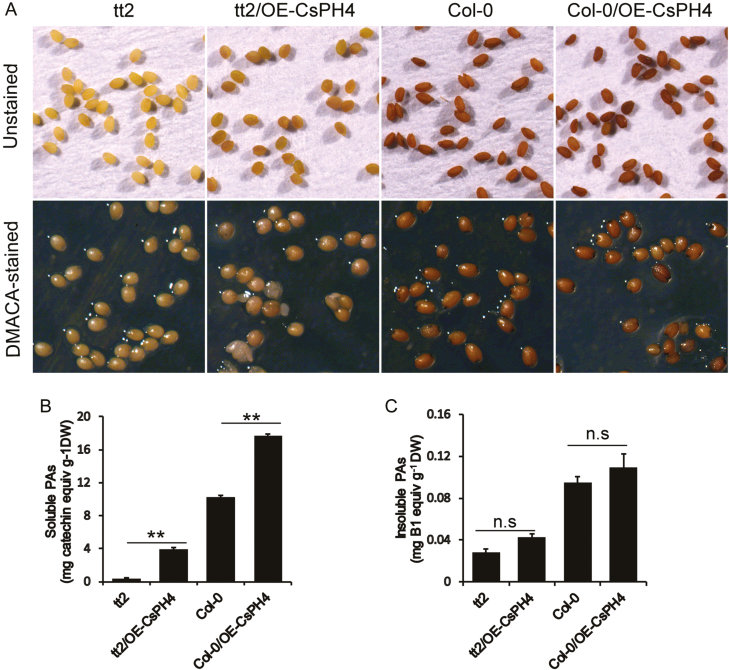
AtMYB123 (TT2) was reported to regulate PA biosynthesis in the seed coat of Arabidopsis (Nesi et al., 2001). To determine whether CsPH4 had a similar function to TT2, the PK7–HA–CsPH4 construct was introduced into the Arabidopsis tt2 mutant. The seeds of the tt2 mutant appeared bright yellow due to the lack of PAs, while the seeds of OE-CsPH4 lines with restored PA production (approximately 9.3-fold changes) were similar to those of the wild-type (Fig. 5A). The complementary phenotype of Arabidopsis seeds was further verified via DMACA staining (Fig. 5B). These results showed that CsPH4 could partially complement the PA-deficient phenotype of the tt2 mutant.
Fig. 5.
Functional characterization of CsPH4 overexpression in Arabidopsis. (A) Unstained and DMACA-stained seeds from tt2 mutants, tt2/OE-CsPH4 transformants, wild-type (Col-0), and Col-0/OE-CsPH4 transformants. Three independent transgenic lines were obtained and showed similar results. (B, C) Quantification of soluble and insoluble PAs in seed from tt2 mutants, tt2/OE-CsPH4 transformants, wild-type (Col-0), and Col-0/OE-CsPH4 transformants. DW, dry weight. Error bars represent the mean ±SD of three biological replicates. Asterisks indicate significant differences using Student’s t-test: **P<0.01; n.s., no significant difference. (This figure is available in colour at JXB online.)
To further analyse the function of CsPH4, we tested the effect of CsPH4 overexpression on PA biosynthesis in the Arabidopsis wild-type. Higher PA accumulation (approximately 1.7-fold change) was observed in the seeds of the OE-CsPH4 lines than in those of the wild-type (Fig. 5A–C). These results suggested that CsPH4 could promote the accumulation of PAs in the Arabidopsis wild-type.
CsPH4 interacts with Noemi to form a regulatory complex
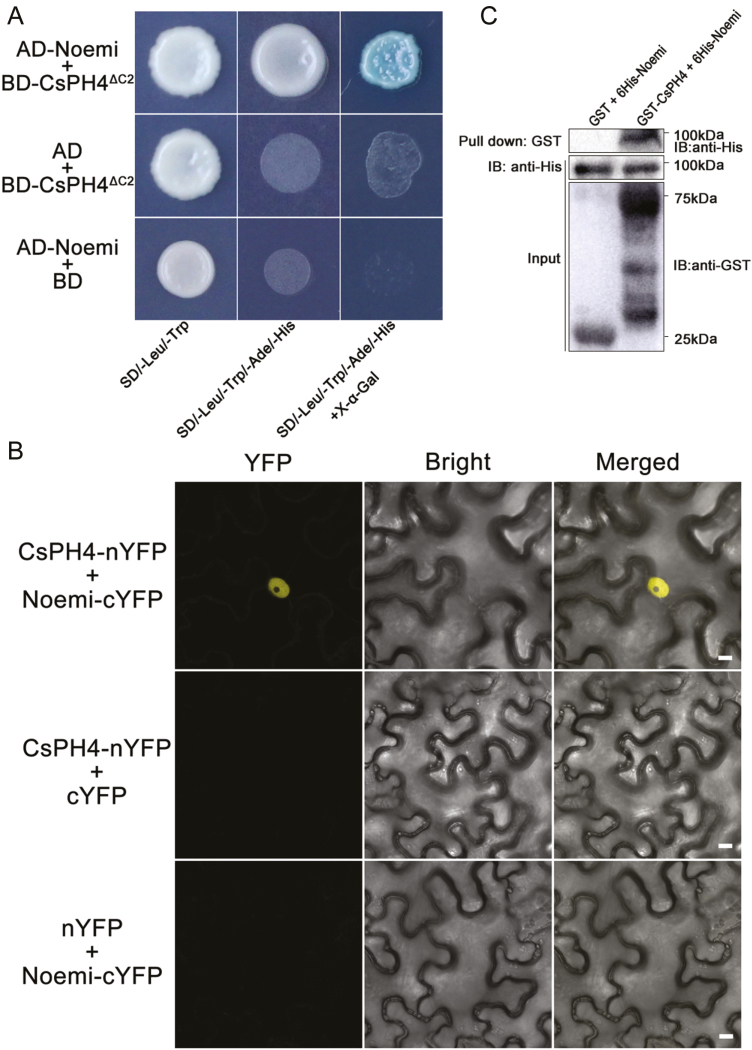
MYB TFs were reported to interact with bHLH TFs to form a regulatory complex (Allan et al., 2008). To determine whether CsPH4 and Noemi form a regulatory complex, Y2H assays were performed. Because BD–CsPH4 exhibited transcriptional self-activation in a prior investigation, the truncated CsPH4ΔC2 was used (see Supplementary Fig. S11). The Y2H assays showed that CsPH4ΔC2 could interact with Noemi (Fig. 6A).
Fig. 6.
CsPH4 interacts with Noemi. (A) Yeast two-hybrid assay revealing an interaction between CsPH4ΔC2 and Noemi. The full-length coding sequences of Noemi and the truncated coding sequence of CsPH4ΔC2 were cloned into PGADT7 (AD–Noemi) and PGBKT7 (BD–CsPH4ΔC2), respectively. The interaction is indicated by yeast growth and X-α-Gal staining. Yeast grown in SD/−Trp/−Leu medium and SD/−Trp/−Leu/−His/−Ade medium is indicated. (B) BiFC assay of the interaction between CsPH4 and Noemi in epidermal cells of N. benthamiana. CsPH4–nYFP and Noemi–cYFP were used for the interaction assay, while nYFP plus Noemi–cYFP and CsPH4–nYFP plus cYFP were used as the controls. Yellow indicates a positive interaction signal. Scale bar: 10 μm. The experiment was repeated independently three times with similar results obtained each time. (C) Pull-down assays showing the interaction of CsPH4 and Noemi. The recombinant GST–CsPH4 or GST was incubated with 6His–Noemi. Blots were first probed with anti-His antibody and then with anti-GST antibody. (This figure is available in colour at JXB online.)
To further confirm the interaction between CsPH4 and Noemi, a BiFC assay was performed in tobacco (N. benthamiana) leaves. A yellow fluorescence signal was observed in the nuclei of tobacco cells co-transformed with CsPH4–nYFP and Noemi–cYFP (Fig. 6B). No fluorescence signal was observed in cells that were transformed with the empty vector nYFP plus CsPH4–cYFP or the empty vector cYFP plus Noemi–nYFP. These results indicated that CsPH4 interacted physically with Noemi in vivo (Fig. 6B).
In addition, an in vitro pull-down analysis was conducted sequentially to further verify the interaction results. The recombinant GST–CsPH4 protein and the GST control were incubated in vitro with the recombinant His–Noemi protein. The protein was eluted with glutathione and immunoblotted with anti-GST and anti-His antibodies. GST–CsPH4 was pulled down, but GST alone was not, suggesting that GST–CsPH4 interacted directly with the His–Noemi protein (Fig. 6C).
The CsPH4–Noemi complex activates the expression of PA biosynthetic genes and Noemi by binding to the promoters of these genes
As mentioned earlier, the transcript levels of flavonoid biosynthetic genes, including DFR, ANS, ANR, LAR, and UFGT2, were significantly enhanced by CsPH4 and Noemi. The expression of Noemi was significantly up-regulated in OE-CsPH4 citrus calli. To further test whether CsPH4 and Noemi directly regulated the expression of DFR, ANS, ANR, LAR, UFGT2, and Noemi, a transient expression assay was performed using the dual-luciferase system. We found that CsPH4 or Noemi alone could not activate the expression of DFR, ANS, and ANR, whereas co-expression of CsPH4 and Noemi significantly activated the expression of these genes (Fig. 7B). Transient expression assays also revealed that CsPH4 alone could activate the promoters of LAR and Noemi, while the ability to activate the promoters of LAR and Noemi was significantly enhanced after co-expression of CsPH4 and Noemi (Fig. 7B). However, UFGT2 was not activated in any way (see Supplementary Fig. S12). These results suggested that co-expression of CsPH4 and Noemi significantly activated the promoters of DFR, ANS, ANR, LAR, and Noemi but not UFGT2.
Tobacco leaves are widely used as a transient expression system for verifying gene function. To further determine whether the CsPH4–Noemi regulatory complex promoted the accumulation of PAs, CsPH4, Noemi, and CsPH4 plus Noemi were transiently overexpressed in tobacco leaves. Quantitative measurements showed that the soluble PA levels were significantly higher in tobacco leaves overexpressing CsPH4 plus Noemi than those overexpressing CsPH4, Noemi, or control, whereas no significant difference was detected in anthocyanin contents (see Supplementary Figs S10B, S13, S14). qRT-PCR analysis showed that transient co-expression of CsPH4 and Noemi significantly induced the expression of flavonoid biosynthetic genes (NtCHS, NtF3H, NtDFR, NtANS, and NtLAR) in tobacco leaves, while expression of the key anthocyanin biosynthetic gene NtUFGT was not significantly induced (Fig. 7C).
The regulation of PA biosynthesis by the MBW complex was mainly mediated by R2R3-MYB recognition of the consensus MYB-recognizing element (MRE) (Zhu et al., 2015; Zhai et al., 2016). By analysing the promoter sequences of citrus PA biosynthetic genes and Noemi, we found that the DFR, ANS, ANR, LAR, and Noemi promoters all contained the consensus MRE site. To test the ability of CsPH4 to bind the promoters of PA biosynthetic genes and Noemi, an EMSA assay was performed. The results confirmed that CsPH4 could bind to the MRE sites within the promoters of DFR, ANR, LAR, and Noemi but not ANS (Fig. 7D).
Discussion
The CsPH4–Noemi regulatory complex controls PA biosynthesis via a positive feedback loop
Fruit secondary metabolites, such as carotenoids, vitamins, and flavonoids, are important determinants of fruit quality (Goufo and Trindade, 2014; Nisar et al., 2015). The tissue specificity of secondary metabolite accumulation depends on the expression of structural genes and their transcriptional regulation level. In this study, the RNA-seq analysis showed that the differential expression of PA biosynthetic genes (DFR, ANS, ANR, and LAR) and several TFs (such as CsPH4, Noemi, CsWRKY44, CsTRY, and CsMYB179) were highly correlated with PA accumulation in both seeds and pulps (Fig. 2B, C). Previous studies have shown that CsWRKY44 (TTG2-like) and CsTRY (AtTRY-like) were involved in trichome and root hair patterning development (Johnson et al., 2002; Schellmann et al., 2002). The function of CsMYB196 (homologous to AtMYB6) has hardly been reported so far. Then, our sequence analysis revealed that CsPH4 has a high homology with VvMYB5a/b, AtMYB5, and MtMYB5, and Noemi shared the highest sequence homology with VvMYC1 (Fig. 3B). These homologous genes have been shown to modulate PA biosynthesis (Deluc et al., 2006; Deluc et al., 2008; Verdier et al., 2012; Liu et al., 2014a). Additionally, Butelli et al. (2019) reported that Noemi is essential for the production of citrus flavonoid pigments, based on the natural variations of gene sequence and flavonoid content among germplasms. These results indicated that CsPH4 and Noemi are the most likely candidate regulators of PA biosynthesis in citrus. Therefore, we choose these two genes for further analysis, with the purpose of elucidating the molecular mechanism underlying PA modulation.
Subsequent genetic evidence demonstrated that both CsPH4 and Noemi are positive regulators that are involved in PA accumulation by up-regulating flavonoid biosynthetic genes in citrus calli and Arabidopsis. Consistent with the findings in model species, CsPH4 interacts with Noemi to form a CsPH4–Noemi regulatory complex and synergistically activates the expression of PA biosynthetic genes in citrus. The regulatory complex also directly induced the expression of Noemi and further promoted the biosynthesis of PA via a positive feedback loop. Natural selection leads to increased precision of PA biosynthesis in citrus, and the tissue specificity of PA accumulation has obviously evolved. Our data indicated that PAs could be significantly accumulated in citrus only when CsPH4 and Noemi were both highly expressed with spatiotemporal specificity (e.g. in seeds of ‘Anliu’), while PAs could not be accumulated at low expression levels of CsPH4 and Noemi or without spatiotemporal specificity (e.g. in seeds and pulps of ‘Hong Anliu’ and ‘Succari’). Based on these results, we proposed a model that was consistent with the molecular mechanisms responsible for the regulation of PA biosynthesis in citrus (Fig. 8).
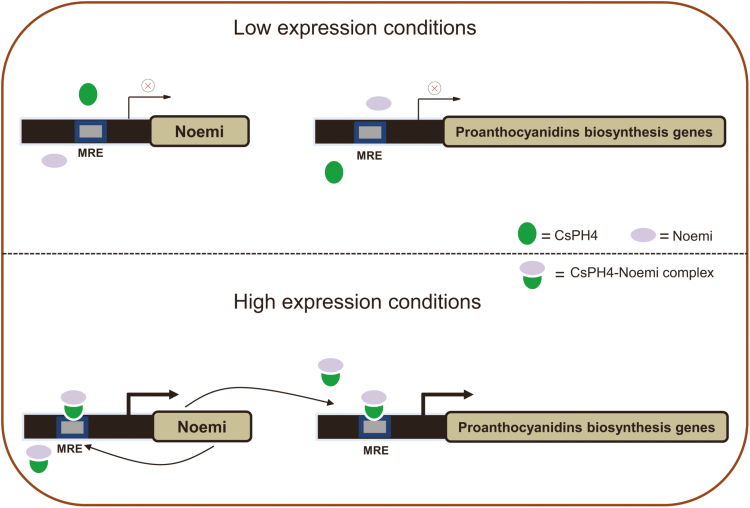
Fig. 8.
A proposed model of the mechanism by which the CsPH4–Noemi regulatory complex regulates PA biosynthesis. Top: under low-expression conditions, neither CsPH4 nor Noemi can be translated to the corresponding protein and form the complex, resulting in the inability to induce PA biosynthetic genes and Noemi expression; thus, PAs could not be effectively accumulated. Bottom: under high-expression conditions, CsPH4 interacts with the Noemi protein to form a regulatory complex, which is mediated by CsPH4 recognition by the consensus MRE site and induction of the expression of PA biosynthetic genes and Noemi. Simultaneously, the regulatory complex provides further positive feedback, which regulates the expression of Noemi, thereby enhancing the accumulation of PAs in citrus. The green parts represent the CsPH4 protein, and the purple parts represent the Noemi protein. (This figure is available in colour at JXB online.)
Bud mutants caused by point mutations, large deletions, and the insertions of retrotransposons are one of the most important breeding approaches for developing valuable fruit crops, such as citrus, grape, apple, and persimmon (Liu et al., 2007; Yamane et al., 2008; Pelsy, 2010). Using these mutations to develop molecular markers can effectively speed up the breeding process, especially for perennial fruit crops. Previous studies reported that the acidless phenotype of citrus is usually accompanied by the three traits of low fruit acidity, white flowers and green leaves, and seeds of light cream colour, and natural variation or low expression of Noemi is an important determinant of the phenotype (Butelli et al., 2019). Compared with ‘Anliu’, ‘Hong Anliu’ and ‘Succari’ are also two acidless varieties. Based on this and our results, we speculated that the sequence variation of Noemi or CsPH4 might be the primary cause leading to different PA accumulation among the three varieties. Nevertheless, in this study, we mainly focused on elucidating a common mechanism for the regulation of PA biosynthesis in citrus. No attention has been paid to revealing the mutations responsible for the phenotypes among these three genotypes. Further work needs to be done to determine the contribution of Noemi or CsPH4 sequence variants and molecular regulatory networks to both traits. The natural variation of CsPH4 or Noemi holds out the prospect of developing molecular markers for PA as well as acidless breeding of citrus fruits.
CsPH4 determines the functional specificity of the CsPH4–Noemi complex in regulating PA biosynthesis
In this study, both seed and pulp of ‘Anliu’ versus ‘Hong Anliu’ showed fewer differentially regulated genes compared with those of the comparison between ‘Anliu’ versus ‘Succari’, which suggest that the phenotypic variation among the three citrus varieties may be involved in the alteration of multiple biological processes and metabolic pathways. The bHLH proteins of the subgroup IIIf-1 (i.e. PhAN1, AtTT8, MtTT8, IpIVS, and VvMYC1) interact with various R2R3-MYBs to regulate anthocyanin and PA biosynthesis and cell development (Allan et al., 2008; Hichri et al., 2010, 2011; Nesi et al., 2000). Our analysis found that Noemi belonged to subgroup IIIf-1 and shared a highest sequence homology with VvMYC1 (Fig. 3B). Recent studies have shown that Noemi controls PA biosynthesis in seeds and is also essential for the regulation of fruit acidity (Butelli et al., 2019). These results indicated that Noemi not only regulate the biosynthesis of PAs, but also participate in other metabolic pathways. It is a possibility that Noemi forms complexes with different partners, such as CsPH4, Ruby, CsTRY (AtTRY-like), and CsWRKY44 (TTG2-like), to regulate different metabolic pathways and cell development (Johnson et al., 2002; Schellmann et al., 2002; Butelli et al., 2012). By contrast, the PA regulator clades of R2R3-MYB consisted of two subclades, PA clades 1 and 2, and clade 2 was subdivided into two distinct clusters (Fig. 3A), suggesting that members of R2R3-MYB responsible for PA regulation varied significantly among different species. PA biosynthesis is more specifically regulated by TT2, VvMYBPA2, MtPAR, and other genes, which clustered in clade 1. Conversely, VvMYB5a, AtMYB5, and MtMYB5, which belong to clade 2 (where CsPH4 clustered), are also in some way involved in accumulation of other metabolites.
As previously reported, the co-expression of CitPH4 and CitAN1 strongly induced CitPH1 and CitPH5 expression to lead to citric acid accumulation in citrus fruit (Li et al., 2015; Strazzer et al., 2019). Our data indicated that both CsCAC16.5 (a protease-like protein that may be involved in vacuolar acidification) and CsPH5 were consistently expressed at significantly lower levels in seeds and pulps of ‘Hong Anliu’ and ‘Succari’ than in ‘Anliu’. There was a tendency for co-expression between CsCAC16.5, CsPH5, and CsPH4 (Fig. 2A), suggesting that CsPH4 potentially regulates the expression of CsCAC16.5 and CsPH5. Thus, CsPH4 not only regulates PA biosynthesis but also participates in citric acid accumulation in citrus. Recent studies have shown that VvMYB5 has similar functions to PhPH4 and AtMYB5, and is also involved in controlling vacuolar hyper-acidification and trafficking in grapevine (Amato et al., 2019). These results suggested that the regulator involved in PA accumulation had obvious functional diversification, that is, the regulator of clade 1 more specifically regulated the biosynthesis of PAs, while the regulator of clade 2 not only regulated the biosynthesis of PAs but also regulated vacuolar acidification in plants.
The accumulation of anthocyanins and PAs shares the same upstream metabolic pathway. In most cultivated citrus varieties, the accumulation of anthocyanins is associated with the activity of the transcriptional activator Ruby (Butelli et al., 2017; Huang et al., 2018). However, our transcriptomic data showed no obvious difference in Ruby expression among the three varieties. Huang et al. (2018) overexpressed Ruby in Arabidopsis and found no significant changes in PA content (Huang et al., 2018). These results suggested that Ruby only participated in the biosynthesis of anthocyanins, not PAs. Meanwhile, the CsPH4 protein was verified to directly bind to MRE sites (Fig. 7) and provided DNA-binding specificity for the activation of target genes to regulate PA biosynthesis in citrus. Compared with the wild-type, the expression of UFGT2 in OE-CsPH4 calli was significantly up-regulated, whereas no significant increase of anthocyanin content detected. Also, the CsPH4–Noemi complex could not activate the expression of UFGT2 in the transient expression assays. Thus, the up-regulation of UFGT2 in transgenic calli is more likely to be the result of positive feedback regulation. Collectively, it can be concluded that CsPH4 determines the functional specificity of the CsPH4–Noemi complex in regulating PA biosynthesis, while Ruby is specifically involved in anthocyanins.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Simplified diagram of genes involved in flavonoid biosynthesis.
Fig. S2. pH in juice from breaker stage ‘Anliu’, ‘Hong Anliu’, and ‘Succari’ fruits.
Fig. S3. Characterization of PA levels of flavedos in ‘Anliu’, ‘Hong Anliu’, and ‘Succari’.
Fig. S4. Volcano plots showing the whole distribution of differentially expressed genes (DEGs).
Fig. S5. GO terms of DEGs.
Fig. S6. KEGG classifications of DEGs.
Fig. S7. Relative expression patterns of CsCHS and CsF3H in the seeds and pulps of three citrus varieties.
Fig. S8. Protein sequence alignment of CsPH4 and Noemi.
Fig. S9. Subcellular localization and transcriptional activity of CsPH4 and Noemi.
Fig. S10. The anthocyanin contents of transgenic citrus callus and the transient overexpression in tobacco (N. benthamiana) leaves.
Fig. S11. Amino acid sequence analysis of CsPH4 and transcriptional self-activation activity assay of CsPH4 and Noemi in yeast AH109.
Fig. S12. Transient promoter activity assays of CsUFGT2.
Fig. S13. Phenotype of the transient overexpression in tobacco (N. benthamiana) leaves.
Fig. S14. Characterization of PA levels of the transient overexpression in tobacco (N. benthamiana) leaves.
Table S1. Summary of read numbers based on the RNA-seq data.
Table S2. Transcriptome data of DEGs.
Table S3. Transcriptome data of KEGG classifications.
Table S4. Transcriptome data of GO enrichment.
Table S5. Primers used in this study.
Table S6. Accession numbers of MYB and bHLH transcription factors in the phylogenetic tree.
Table S7. Expression patterns of phenylpropanoid-related genes in the seeds and pulps of three citrus varieties.
Acknowledgements
We thank Prof. Wangjin Lu and Prof. Jianfei Kuang (South China Agricultural University) for providing vectors for the Dual Luciferase Assay; and Prof. Jihong Liu (Huazhong Agricultural University) for providing vectors for the subcellular localization and BiFC assay experiments. This project was supported by the National Key Research and Development Program of China (2018YFD1000200) and National Natural Science Foundation of China (No. 31630065).
Author contributions
XXD conceived of the project and supervised the research. YZ designed and performed the experiments. CYL was involved in the research design and the improvement of the manuscript. JLY and QX edited the manuscript. LCL provided samples of citrus fruits. YZ analysed the data and wrote the article.
Competing interests
The authors declare no competing interests.
References
- Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR. 2003. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. The Plant Journal 35, 624–636. [DOI] [PubMed] [Google Scholar]
- Allan AC, Hellens RP, Laing WA. 2008. MYB transcription factors that colour our fruit. Trends in Plant Science 13, 99–102. [DOI] [PubMed] [Google Scholar]
- Amato A, Cavallini E, Walker AR, et al. . 2019. The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. The Plant Journal 99, 1220–1241. [DOI] [PubMed] [Google Scholar]
- Bagchi D, Bagchi M, Stohs SJ, Das DK, Ray SD, Kuszynski CA, Joshi SS, Pruess HG. 2000. Free radicals and grape seed proanthocyanidin extract: importance in human health and disease prevention. Toxicology 148, 187–197. [DOI] [PubMed] [Google Scholar]
- Barbehenn RV, Constabel CP. 2011. Tannins in plant-herbivore interactions. Phytochemistry 72, 1551–1565. [DOI] [PubMed] [Google Scholar]
- Baxter IR, Young JC, Armstrong G, Foster N, Bogenschutz N, Cordova T, Peer WA, Hazen SP, Murphy AS, Harper JF. 2005. A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 102, 2649–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP. 2007. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiology 143, 1347–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, et al. . 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Butelli E, Garcia-Lor A, Licciardello C, et al. . 2017. Changes in anthocyanin production during domestication of Citrus. Plant Physiology 173, 2225–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Licciardello C, Ramadugu C, Durand-Hulak M, Celant A, Reforgiato Recupero G, Froelicher Y, Martin C. 2019. Noemi controls production of flavonoid pigments and fruit acidity and illustrates the domestication routes of modern citrus varieties. Current Biology 29, 158–164.e2. [DOI] [PubMed] [Google Scholar]
- Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C. 2012. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. The Plant cell 24, 1242–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang H, Pang Y, Cheng Y, Deng X, Xu J. 2015. Comparative study of flavonoid production in lycopene-accumulated and blonde-flesh sweet oranges (Citrus sinensis) during fruit development. Food Chemistry 184, 238–246. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M. 2000. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiology 122, 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Mérillon JM, Hamdi S. 2006. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiology 140, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Bogs J, Walker AR, Ferrier T, Decendit A, Merillon JM, Robinson SP, Barrieu F. 2008. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiology 147, 2041–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rezende AA, Graf U, Guterres Zda R, Kerr WE, Spanó MA. 2009. Protective effects of proanthocyanidins of grape (Vitis vinifera L.) seeds on DNA damage induced by Doxorubicin in somatic cells of Drosophila melanogaster. Food and Chemical Toxicology 47, 1466–1472. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Xie DY, Sharma SB. 2005. Proanthocyanidins—a final frontier in flavonoid research? New Phytologist 165, 9–28. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Maekawa M, Oki T, Suda I, Iida S, Shimada H, Takamure I, Kadowaki K. 2007. The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. The Plant Journal 49, 91–102. [DOI] [PubMed] [Google Scholar]
- Goufo P, Trindade H. 2014. Rice antioxidants: phenolic acids, flavonoids, anthocyanins, proanthocyanidins, tocopherols, tocotrienols, γ-oryzanol, and phytic acid. Food Science & Nutrition 2, 75–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LX, Shi CY, Liu X, Ning DY, Jing LF, Yang H, Liu YZ. 2016. Citrate accumulation-related gene expression and/or enzyme activity analysis combined with metabolomics provide a novel insight for an orange mutant. Scientific Reports 6, 29343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YC, Kuang JF, Chen JY, Liu XC, Xiao YY, Fu CC, Wang JN, Wu KQ, Lu WJ. 2016. Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and Expansins during fruit ripening. Plant Physiology 171, 1070–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V. 2011. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. Journal of Experimental Botany 62, 2465–2483. [DOI] [PubMed] [Google Scholar]
- Hichri I, Heppel SC, Pillet J, Léon C, Czemmel S, Delrot S, Lauvergeat V, Bogs J. 2010. The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Molecular Plant 3, 509–523. [DOI] [PubMed] [Google Scholar]
- Huang D, Wang X, Tang Z, Yuan Y, Xu Y, He J, Jiang X, Peng SA, Li L, Butelli E, Deng X, Xu Q. 2018. Subfunctionalization of the Ruby2–Ruby1 gene cluster during the domestication of citrus. Nature Plants 4, 930–941. [DOI] [PubMed] [Google Scholar]
- Huang D, Zhao Y, Cao M, Qiao L, Zheng ZL. 2016. Integrated systems biology analysis of transcriptomes reveals candidate genes for acidity control in developing fruits of sweet orange (Citrus sinensis L. Osbeck). Frontiers in Plant Science 7, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia LG, Sheng ZW, Xu WF, Li YX, Liu YG, Xia YJ, Zhang JH. 2012. Modulation of anti-oxidation ability by proanthocyanidins during germination of Arabidopsis thaliana seeds. Molecular Plant 5, 472–481. [DOI] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR. 2002. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. The Plant Cell 14, 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaii S, Tomono Y, Katase E, Ogawa K, Yano M. 1999. Quantitation of flavonoid constituents in citrus fruits. Journal of Agricultural and Food Chemistry 47, 3565–3571. [DOI] [PubMed] [Google Scholar]
- Kawaii S, Tomono Y, Katase E, Ogawa K, Yano M, Koizumi M, Ito C, Furukawa H. 2000. Quantitative study of flavonoids in leaves of citrus plants. Journal of Agricultural and Food Chemistry 48, 3865–3871. [DOI] [PubMed] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A. 2004. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. The Plant Journal 37, 104–114. [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science 10, 236–242. [DOI] [PubMed] [Google Scholar]
- Kumar KR, Kirti PB. 2010. A mitogen-activated protein kinase, AhMPK6 from peanut localizes to the nucleus and also induces defense responses upon transient expression in tobacco. Plant Physiology and Biochemistry 48, 481–486. [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M. 2006. Genetics and biochemistry of seed flavonoids. Annual Review of Plant Biology 57, 405–430. [DOI] [PubMed] [Google Scholar]
- Li P, Chen B, Zhang G, Chen L, Dong Q, Wen J, Mysore KS, Zhao J. 2016. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytologist 210, 905–921. [DOI] [PubMed] [Google Scholar]
- Li S-j, Liu X-j, Xie X-l, Sun C-d, Grierson D, Yin X-r, Chen K-s. 2015. CrMYB73, a PH-like gene, contributes to citric acid accumulation in citrus fruit. Scientia Horticulturae 197, 212–217. [Google Scholar]
- Ling WH, Cheng QX, Ma J, Wang T. 2001. Red and black rice decrease atherosclerotic plaque formation and increase antioxidant status in rabbits. The Journal of Nutrition 131, 1421–1426. [DOI] [PubMed] [Google Scholar]
- Liu C, Jun JH, Dixon RA. 2014a MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula. Plant Physiology 165, 1424–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Long J, Zhu K, Liu L, Yang W, Zhang H, Li L, Xu Q, Deng X. 2016. Characterization of a citrus R2R3-MYB transcription factor that regulates the flavonol and hydroxycinnamic acid biosynthesis. Scientific Reports 6, 25352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Wang X, Xu Y, Deng X, Xu Q. 2014b Genome-wide analysis of the R2R3-MYB transcription factor gene family in sweet orange (Citrus sinensis). Molecular Biology Reports 41, 6769–6785. [DOI] [PubMed] [Google Scholar]
- Liu Q, Xu J, Liu Y, Zhao X, Deng X, Guo L, Gu J. 2007. A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck). Journal of Experimental Botany 58, 4161–4171. [DOI] [PubMed] [Google Scholar]
- Lu S, Zhang Y, Zheng X, Zhu K, Xu Q, Deng X. 2016. Isolation and functional characterization of a lycopene β-cyclase gene promoter from citrus. Frontiers in Plant Science 7, 1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Zhang Y, Zhu K, Yang W, Ye J, Chai L, Xu Q, Deng X. 2018. The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiology 176, 2657–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Cai T, Olyarchuk JG, Wei L. 2005. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. [DOI] [PubMed] [Google Scholar]
- Marinova K, Pourcel L, Weder B, Schwarz M, Barron D, Routaboul JM, Debeaujon I, Klein M. 2007. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. The Plant cell 19, 2023–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R. 1998. How genes paint flowers and seeds. Trends in Plant Science 3, 212–217. [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. 2000. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. The Plant Cell 12, 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. 2001. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. The Plant Cell 13, 2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisar N, Li L, Lu S, Khin NC, Pogson BJ. 2015. Carotenoid metabolism in plants. Molecular Plant 8, 68–82. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Maeda H, Oguchi T, Yamaguchi T, Tanabe N, Ebana K, Yano M, Ebitani T, Izawa T. 2015. The birth of a black rice gene and its local spread by introgression. The Plant cell 27, 2401–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ZY, Li Y, Deng XX, Xiao SY. 2014. Non-targeted metabolomic analysis of orange (Citrus sinensis [L.] Osbeck) wild type and bud mutant fruits by direct analysis in real-time and HPLC-electrospray mass spectrometry. Metabolomics 10, 508–523. [Google Scholar]
- Pang YZ, Peel GJ, Sharma SB, Tang YH, Dixon RA. 2008. A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proceedings of the National Academy of Sciences, USA 105, 14210–14215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelsy F. 2010. Molecular and cellular mechanisms of diversity within grapevine varieties. Heredity 104, 331–340. [DOI] [PubMed] [Google Scholar]
- Peters DJ, Constabel CP. 2002. Molecular analysis of herbivore-induced condensed tannin synthesis: cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). The Plant Journal 32, 701–712. [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I. 2005. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. The Plant Cell 17, 2966–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaart JG, Dubos C, Romero De La Fuente I, van Houwelingen AM, de Vos RC, Jonker HH, Xu W, Routaboul JM, Lepiniec L, Bovy AG. 2013. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria×ananassa) fruits. New Phytologist 197, 454–467. [DOI] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M. 2002. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. The EMBO Journal 21, 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SB, Dixon RA. 2005. Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana. The Plant journal 44, 62–75. [DOI] [PubMed] [Google Scholar]
- Shirley BW. 2008. Flavonoids in seeds and grains: physiological function, agronomic importance and the genetics of biosynthesis. Seed Science Research 8, 415–422. [Google Scholar]
- Strazzer P, Spelt CE, Li S, Bliek M, Federici CT, Roose ML, Koes R, Quattrocchio FM. 2019. Hyperacidification of Citrus fruits by a vacuolar proton-pumping P-ATPase complex. Nature Communications 10, 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N, Torregrosa L, Ageorges A, Vialet S, Verriès C, Cheynier V, Romieu C. 2009. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiology 149, 1028–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, de Souza LP, Fernie AR. 2017. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. Journal of Experimental Botany 68, 4013–4028. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J, Zhao J, Torres-Jerez I, Ge S, Liu C, He X, Mysore KS, Dixon RA, Udvardi MK. 2012. MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula. Proceedings of the National Academy of Sciences, USA 109, 1766–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. 1999. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. The Plant Cell 11, 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, et al. . 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Wang L, Ran L, Hou Y, Tian Q, Li C, Liu R, Fan D, Luo K. 2017a The transcription factor MYB115 contributes to the regulation of proanthocyanidin biosynthesis and enhances fungal resistance in poplar. New Phytologist 215, 351–367. [DOI] [PubMed] [Google Scholar]
- Wang S, Tu H, Wan J, Chen W, Liu X, Luo J, Xu J, Zhang H. 2016. Spatio-temporal distribution and natural variation of metabolites in citrus fruits. Food Chemistry 199, 8–17. [DOI] [PubMed] [Google Scholar]
- Wang S, Yang C, Tu H, Zhou J, Liu X, Cheng Y, Luo J, Deng X, Zhang H, Xu J. 2017b Characterization and metabolic diversity of flavonoids in citrus species. Scientific Reports 7, 10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. 2001. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology 126, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GA, Terol J, Ibanez V, et al. . 2018. Genomics of the origin and evolution of Citrus. Nature 554, 311–316. [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA. 2003. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299, 396–399. [DOI] [PubMed] [Google Scholar]
- Xu W, Bobet S, Le Gourrierec J, et al. . 2017. TRANSPARENT TESTA 16 and 15 act through different mechanisms to control proanthocyanidin accumulation in Arabidopsis testa. Journal of Experimental Botany 68, 2859–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Grain D, Bobet S, Le Gourrierec J, Thévenin J, Kelemen Z, Lepiniec L, Dubos C. 2014. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis seed. New Phytologist 202, 132–144. [DOI] [PubMed] [Google Scholar]
- Yamane H, Ichiki M, Tao R, Esumi T, Yonemori K, Niikawa T, Motosugi H. 2008. Growth characteristics of a small-fruit dwarf mutant arising from bud sport mutation in Japanese persimmon (Diospyros kaki thunb.). HortScience 43, 1726–1730. [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biology 11, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai R, Wang Z, Zhang S, Meng G, Song L, Wang Z, Li P, Ma F, Xu L. 2016. Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (Pyrus bretschneideri Rehd.). Journal of Experimental Botany 67, 1275–1284. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Wang H, Wang Y, Guan S, Wang F, Tang J, Zhang R, Xie L, Lu Y. 2015. Characterization of the cis elements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation. Journal of Experimental Botany 66, 3775–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.