Abstract
Background
Ginseng (Panax ginseng Meyer) is an essential source of pharmaceuticals and functional foods. Ginseng productivity has been compromised by high light (HL) stress, which is one of the major abiotic stresses during the ginseng cultivation period. The genetic improvement for HL tolerance in ginseng could be facilitated by analyzing its genetic and molecular characteristics associated with HL stress.
Methods
Genome-wide analysis of gene expression was performed under HL and recovery conditions in 1-year-old Korean ginseng (P. ginseng cv. Chunpoong) using the Illumina HiSeq platform. After de novo assembly of transcripts, we performed expression profiling and identified differentially expressed genes (DEGs). Furthermore, putative functions of identified DEGs were explored using Gene Ontology terms and Kyoto Encyclopedia of Genes and Genome pathway enrichment analysis.
Results
A total of 438 highly expressed DEGs in response to HL stress were identified and selected from 29,184 representative transcripts. Among the DEGs, 326 and 114 transcripts were upregulated and downregulated, respectively. Based on the functional analysis, most upregulated and a significant number of downregulated transcripts were related to stress responses and cellular metabolic processes, respectively.
Conclusion
Transcriptome profiling could be a strategy to comprehensively elucidate the genetic and molecular mechanisms of HL tolerance and susceptibility. This study would provide a foundation for developing breeding and metabolic engineering strategies to improve the environmental stress tolerance of ginseng.
Keywords: High light stress, Panax ginseng, Photoinhibition, Photoprotection, Transcriptome analysis
1. Introduction
Ginseng (Panax ginseng Meyer) is an important medicinal plant. Its roots have been widely used for thousands of years as a traditional medicine formulation for human health with various beneficial effects, such as antineoplastic, antistress, anti-inflammatory, antioxidative, and neuroprotective [1], [2].
Ginseng is a perennial shade plant. Shade plants, adapted to low light environments, have limited ability to cope with excess light and are more susceptible to photoinhibition than sun plants are [3]. The absorption of excessive light energy impairs photosynthetic machinery primarily by inactivating photosystem II (PSII) and eventually reduces photosynthetic capacity termed photoinhibition [4]. The proposed mechanism underlying photoinhibition is the disruption of oxygen-evolving complexes by excessive direct light, which inactivates the reaction center of PSII. Moreover, the excessive light-mediated production of reactive oxygen species (ROS) inhibits the repair of photodamaged PSII [4], [5], [6].
To minimize photoinhibition, all photosynthetic organisms have evolutionarily developed photoprotection and PSII repair mechanisms [4]. Plants can dissipate excess energy as heat, called nonphotochemical quenching, which is an efficient and relatively rapid response to sudden high light (HL) exposure [7]. Plants can also adjust light-harvesting capacity through leaf and chloroplast movement and reduce the light-harvesting antenna size as acclimation and relatively long-term response to HL stress [8]. Furthermore, detoxification of ROS is a key photoprotective response using antioxidants and ROS-scavenging enzymes [9]. Light-induced photoinhibition is considered to occur when the rate of PSII photodamage is higher than its rate of repair [4]. The PSII repair cycle includes successive steps of phosphorylation, dephosphorylation, disassembly of PSII components, proteolysis, de novo synthesis of PSII proteins (particularly D1 protein), and reconstitution of PSII complex [10], [11].
In ginseng, photosynthetic and physiological characteristics and growth performance have been studied in response to HL stress [12], [13], [14], [15]. Ginseng displays photoinhibitory symptoms including photobleaching and chlorosis followed by reduced photosynthetic efficiency when the photosynthetic photon flux density is > 500 μmol m−2 s−1 [13], which is approximately 25% of full sunlight (2,000 μmol m−2 s−1) [3]. Therefore, in the production of ginseng, modulating and optimizing the light intensity by artificial shading is one of the most important cultivation practices. Generally, the light transmission rate of artificial shading is in the range of 8–20% of full sunlight [12], [14], [15], which is comparable to 160–400 μmol m−2 s−1 light intensity.
Along with cultivation practices, genetic improvement of HL tolerance would be an alternative strategy to stabilize and further increase ginseng productivity. Genetic improvement via conventional breeding, metabolic engineering, or both could be promoted by elucidating genetic and molecular mechanisms underlying target traits. Currently, however, genetic and molecular characteristics associated with HL tolerance and susceptibility have been uncovered in ginseng. In this study, the level of photoinhibitory stress induced by HL treatment (800 μmol m−2 s−1) was estimated and quantified based on the chlorophyll (Chl) fluorescence in ginseng leaves. Then, genome-wide analysis of gene expression in response to HL stress was performed, for the first time, in ginseng using extensive parallel sequencing of RNA. Furthermore, we explored transcript dynamics under both HL and recovery treatments and identified HL responsive genes. Putative functions of the identified ginseng genes were also analyzed and discussed based on the comparative analysis of Arabidopsis thaliana orthologs. The identified ginseng genes might contribute to photoprotection and acclimation to HL stress. This study provides an overview and understanding of transcriptomic responses to HL stress in ginseng.
2. Materials and methods
2.1. Plant material and growth conditions
Roots of 1-year-old Korean ginseng (P. ginseng cv. Chunpoong) collected from the experimental field of Korea Ginseng Corporation were transplanted in 2-L pots (2 roots per pot) containing soil mixes (Shinsung Mineral, Goesan, Korea). Plants were germinated and grown in the indoor plant growth facility (KIST U-Farm, Gangneung, Korea) under 200 μmol m−2s−1 light intensity with a 16-h photoperiod. Full-spectrum light-emitting diode (LED) lights (Plant Growth LED, WAVESYSCOM, Gwangju, Korea) were used as a light source. The manufacturer-provided relative spectrum curve of the LED light was shown in suppl. Fig. 1. The light intensity was set by adjusting the distance between the plants and LED lights. The temperature was set at 24°C/20°C (day/night). Plants were irrigated once a week with 400-mL tap water, and the soil moisture (volumetric water content) was maintained at 18% before irrigation.
2.2. HL treatment, recovery, sampling, and Chl fluorescence measurement
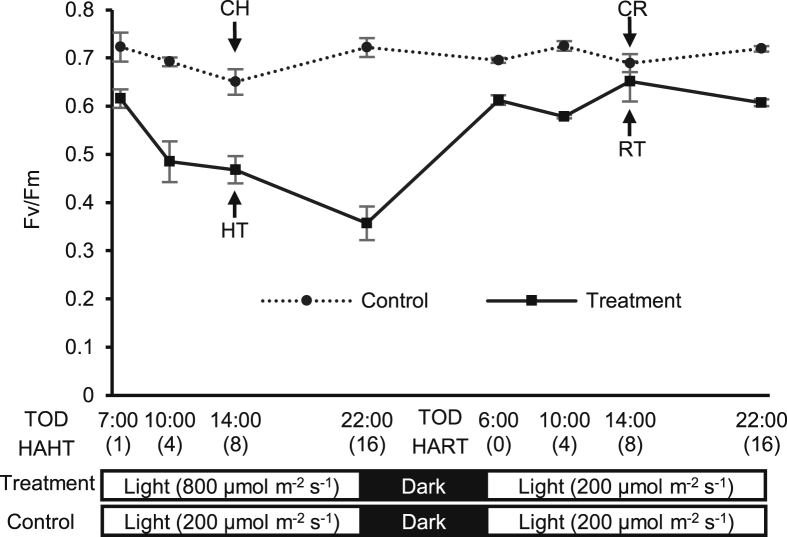
Seventy-five days after transplanting, plants were randomly placed under 200 or 800 μmol m−2 s−1 light intensity (control and HL treatments, respectively) on a 16-h photoperiod from 06:00 to 22:00. After an 8-h dark period from 22:00 to 06:00 (on the next day), HL-treated plants were placed under 200 μmol m−2 s−1 light intensity for recovery from 06:00 to 22:00 (16 h).
A pulse amplitude modulation fluorometer (Junior-PAM, Heinz Walz GmbH, Effeltrich, Germany) was used to measure Chl fluorescence and estimate the photosynthetic efficiency (the maximum quantum yield of PSII). The middle leaflet (the third of five leaflets of one compound leaf) of each plant was adapted to the dark for 20 min before the determination of basal (Fo) and maximum (Fm) Chl fluorescence. Fo was determined after exposure to a standard modulated excitation intensity of 0.5 μmol photon m−2 s −1, and then, Fm was measured after applying a saturating pulse of 2,400 μmol photons m−2 s −1. The maximum quantum yield of PSII was defined as Fv/Fm = (Fm-Fo)/Fm, and the value of Fv/Fm was used as a diagnostic indicator of photoinhibitory damage [16].
The Chl fluorescence of treated plants was measured with four biological replicates at 1 (07:00), 4 (10:00), 8 (14:00), and 16 (22:00) h after HL treatment (HAHT) and at 0 (06:00), 4(10:00), 8 (14:00), and 16 (22:00) h after recovery treatment (HART). The Chl fluorescence was also measured from corresponding control plants at each time point.
Samples for the transcriptome sequencing were collected from treated plants with two biological replicates at 8 HAHT and 8 HART and designated as HT (HL-treated sample) and RT (recovery-treated sample after HL treatment), respectively. Samples for the corresponding control plants were also collected at each time point with three biological replicates and designated as corresponding control for HT (CH) and corresponding control for RT (CR). Tissue specimens for the sequencing were from the middle leaflet (the third of five leaflets of one compound leaf) of each sample.
2.3. RNA extraction, library construction, transcriptome sequencing, and de novo assembly
Total RNA was extracted from HT, RT, CH, and CR samples using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). RNA integrity was confirmed using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
A total of 10 RNA-Seq paired-end libraries were prepared from each control and treated sample consisting of three and two biological replicates, respectively, using the Illumina TruSeq RNA Sample Preparation Kit v2 (Illumina, San Diego, CA, USA). The library was quantified using the KAPA Library Quantification Kit (Kapa Biosystems, Wilmington, MA, USA) following the manufacturer's instructions.
Each library was loaded onto the Illumina HiSeq 2000 platform (Illumina), and high-throughput sequencing was performed ensuring that each sample met the desired average sequencing depth. After sequencing, the duplicated reads produced by the polymerase chain reaction were filtered through in-house scripts. The sequence data were trimmed using DynamicTrim and LengthScort from SolexaQA package [17]. De novo assembly of the trimmed reads was performed using the Velvet (v1.2.10) [18] and Oases (v0.2.09) [19] based on the de Bruijn graph algorithm. To obtain the best de novo assembly, hash length (k-mer) optimization was performed, and the final assembly result was retrieved at k-mer = 65 (Table 1).
Table 1.
Summary statistics of transcriptome sequencing, assembly, and annotation.
| Sequencing results | ||
|---|---|---|
| Data | Number of reads | Total length (bp) |
| Raw data | 206,971,028 | 20,904,073,828 |
| Trimmed data | 184,040,366 | 16,034,478,225 (trimming rate: 76.7%) |
| Assembly results (best assembly at k-mer=65) | ||||||
|---|---|---|---|---|---|---|
| Data | Num. of transcripts | Length (bp) |
||||
| Total | Minimum | Maximum | Average | N50 | ||
| Total transcripts | 136,188 | 167,400,586 | 200 | 11,964 | 1,229 | 1,825 |
| Representative transcripts | 45,987 | 35,721,987 | 200 | 11,964 | 776 | 1,272 |
| Annotation | ||
|---|---|---|
| Data | Number of annotated transcripts | Annotation rate (%) |
| Total transcripts | 108,279 | 79.5 |
| Representative transcripts | 29,184 | 63.5 |
2.4. Read mapping, normalization, and identification of differentially expressed genes
Trimmed sequence reads were mapped to the assembled transcripts using the bowtie2 (v2.1.0) software [20], allowing a maximum of 2 bp mismatches per read. Representative transcripts were selected after the previous study [21]. The number of mapped clean reads for each transcript was calculated and normalized using the DESeq package in R [22].
The gene expression levels were compared between each sample, and differentially expressed genes (DEGs) were identified and selected based on a minimum twofold change and the number of DEseq-normalized counts (read counts > 500 in at least one library). The false discovery rate (FDR) was used to identify the threshold of the p-value in multiple tests (FDR < 0.01) using DESeq. All correlation analysis and clustering were performed using the AMAP library in R [23].
2.5. Functional annotation of assembled transcripts and functional enrichment analysis
Using the clean reads merged from each sample, transcripts were validated and annotated by direct comparison with gene sequences in the Phytozome, Uniprotkb database, and the NCBI NR Viridiplantae database using BLASTX algorithms with a significant threshold e-value ≤ 1e-10. The proteins with the highest sequence similarity were retrieved for analysis. All the transcripts were further annotated using Gene Ontology (GO) based on the similarity of protein sequences in the GO database [24]. GO term enrichment of DEGs with Arabidopsis ortholog's information was performed against the PANTHER database [25] using Fisher's exact test with Benjamini-Hochberg procedure (FDR < 0.05). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis for upregulated and downregulated transcripts was carried out using ThaleMine (https://apps.araport.org/thalemine/begin.do) with Benjamini-Hochberg test correction (FDR < 0.05). The identified transcripts in the enriched KEGG pathway were indicated on the corresponding pathway map from the KEGG database [26].
2.6. Sequence data availability
The sequence data generated for this study are accessible via the NCBI Sequence Read Archive under the accession ID of SRP148526.
3. Results
3.1. Photoinhibition in HL-treated ginseng
The level of photoinhibitory stress induced by HL treatment (800 μmol m−2 s−1) was estimated using the Chl fluorescence parameter Fv/Fm. The Fv/Fm of the HL-treated ginseng decreased overtime during the 16-h photoperiod (Fig. 1). The Fv/Fm dropped from 0.616 to 0.357 over the HL treatment period (1–16 HAHT), whereas the Fv/Fm ranged from 0.651 to 0.722 for the corresponding control plants grown under relatively moderate light (200 μmol m−2 s−1).
Fig. 1.
Time-course changes in maximum potential quantum efficiency of photosystem II (Fv/Fm) for control and high light–treated plants. Light intensities for high light and recovery treatment were 800 and 200 μmol m−2 s−1, respectively (treatment), and control plants were under 200 μmol m−2s−1 light intensity during photoperiod (control). Numbers in parentheses represent hours after HL treatment (HAHT) or hours after recovery treatment (HART). Arrows indicate Fv/Fm values at sampling for transcriptome sequencing. TOD, time-of-day; HT, high light–treated sample; CH, corresponding control for HT; RT, recovery-treated sample after high light treatment; CR, corresponding control for RT. Vertical bars represent standard error (n = 4).
After an 8-h dark period followed by 16-h recovery photoperiod at 200 μmol m−2 s−1 light intensity, the HL-treated plants showed an increase in Fv/Fm from 0.357 to 0.652, indicating a recovery from photoinhibition. However, the Fv/Fm of recovered HL-treated ginseng was lower than that of the corresponding control plants at each time point (Fig. 1).
Samples for transcriptome sequencing were collected at 8 HAHT and 8 HART (Fig. 1). The Fv/Fm values of sequencing samples, CH vs. HT and CR vs. RT, were 0.651 vs. 0.468 and 0.689 vs. 0.652, respectively (Fig. 1).
3.2. Transcriptome sequencing, de novo assembly, and annotation
RNA sequencing was performed using 10 individual libraries, two biological replicates of HT or RT, and three biological replicates of CH or CR. Summary statistics of raw and trimmed sequencing data for each replicates were shown in Suppl. Table 1. A total of 184,040,366 clean reads (total length of 16,034,478,225 bp) were generated after trimming and performing a quality check, and it was assembled into 136,188 transcripts with maximum and average lengths of 11,964 bp and 1,229 bp (Table 1). Among the transcripts, 45,987 were representative, indicating an average of 2.96 transcripts per locus. The number of annotated transcripts was 108,279 (annotation rate: 79.5%), and 29,184 transcripts were annotated among 45,987 representative transcripts (annotation rate: 63.5%).
3.3. DEGs selection and expression dynamics in response to HL and recovery treatment
To investigate transcriptomic responses specific to HL stress, DEGs were analyzed using a pairwise comparison between HT and CH. Highly expressed DEGs were then selected and analyzed from the identified DEGs based on the number of DEseq-normalized read counts > 500 in at least one library. Among 438 highly expressed DEGs, 326 and 112 transcripts were upregulated and downregulated under the HL condition (HT vs. CH). To facilitate the functional classification and analysis, DEGs annotated with Arabidopsis orthologs were further selected, and 190 and 99 were upregulated and downregulated DEGs, respectively, (Suppl. Table 2).
To investigate expression patterns of the HL responsive transcripts under recovery conditions, pairwise comparisons of expression levels were performed between RT and HT (RT vs. HT), as well as RT and CR (RT vs. CR). Among 190 upregulated transcripts in HT vs. CH, most transcripts (91%, 173 transcripts) were downregulated after recovery treatment compared with HT (RT vs. HT; Cluster 1; Suppl. Fig. 2A and B), whereas the rest (9%, 27 transcripts) remained upregulated in the RT (Cluster 2; Suppl. Fig 2C). When the expression in the RT was compared with the corresponding control (RT vs. CR), 146 and 27 transcripts in Cluster 1 showed relatively higher and lower expression levels, respectively (grouped as Cluster 1A and 1B, respectively). All the transcripts in Cluster 2 were upregulated in RT vs CR (Suppl. Fig. 2A-C).
In 99 downregulated transcripts in HT vs. CH, 91% (90 transcripts) were upregulated in response to recovery treatment compared with HT (RT vs. HT; Cluster 3; Suppl. Fig. 2D and E), while 9% of transcripts remained downregulated in RT compared with HT samples (Cluster 4; Suppl. Fig 2F). In a pairwise comparison between RT and CR samples, 84 and six transcripts in Cluster 3 displayed relatively lower and higher expression in RT compared with CR samples, respectively (grouped as Cluster 3A and 3B, respectively). The expression levels of transcripts in Cluster 4 were lower in RT than they were in CR samples (Suppl. Fig. 2D-F).
3.4. GO term enrichment analysis
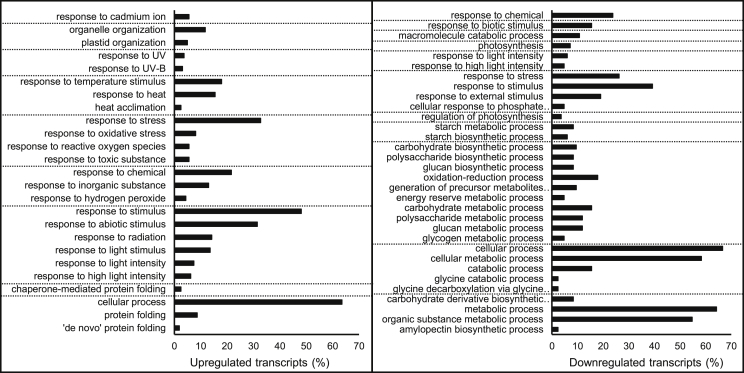
To identify overrepresented GO terms in DEGs in response to HL stress, GO term enrichment was performed against the Arabidopsis reference set. As shown in Fig. 2 and Suppl. Table 3, in the GO term “biological process,” most enriched GO terms were related to stresses such as response to “light intensity,” “abiotic stimulus,” “oxidative stress,” and “heat”. Particularly, the GO term “response to stress” was enriched with 33% upregulated transcripts. Furthermore, 6% of upregulated transcripts were involved in “response to HL intensity,” “response to ROS,” or both. GO terms for downregulated transcripts were largely different from those for upregulated transcripts. The enriched GO terms were mostly associated with “cellular metabolic process” and “carbohydrate/polysaccharide catabolic/biosynthetic process,” whereas these GO terms were not enriched in upregulated transcripts. Stress-related GO terms were also identified for downregulated transcripts, and 26% and 5% of downregulated transcripts were involved in “response to stress” and “response to HL intensity,” respectively.
Fig. 2.
Gene Ontology (GO) enrichment analysis for differentially expressed gene (DEG) set identified by pairwise comparison between high light–treated and corresponding control plants. GO-terms in the category of “biological process” are presented. Dashed lines indicate each subcategory, and terms in each subcategory are presented by the hierarchical order. Values on x-axis are percentages of upregulated or downregulated transcripts associated with each GO-term among all upregulated or downregulated transcripts in DEG set.
For the GO term “cellular component,” 18% of upregulated transcripts were related to “macromolecular complex” including “protein complex” and “chaperone complex,” which were not enriched in downregulated transcripts. In addition, 12% and 6% of upregulated transcripts were categorized into “chloroplast part” and “mitochondrial part,” respectively. For downregulated transcripts, 40 GO terms were significantly enriched, and 14 were related to the chloroplast. Among them, the GO terms “chloroplast,” “chloroplast envelope,” “chloroplast thylakoid,” and “photosystem” were enriched with 44%, 14%, 11%, and 6% of downregulated transcripts, respectively (Suppl. Table 4).
For the GO term “molecular function,” 59% of upregulated transcripts were associated with “binding” and subcategorized into “protein binding,” “nucleotide binding,” “organic cyclic compound binding,” or a combination of these terms. For downregulated transcripts, only the GO term “catabolic activity” and one subgroup of “transferase activity” were significantly enriched with 56% and 10% of transcripts, respectively (Suppl. Table 5).
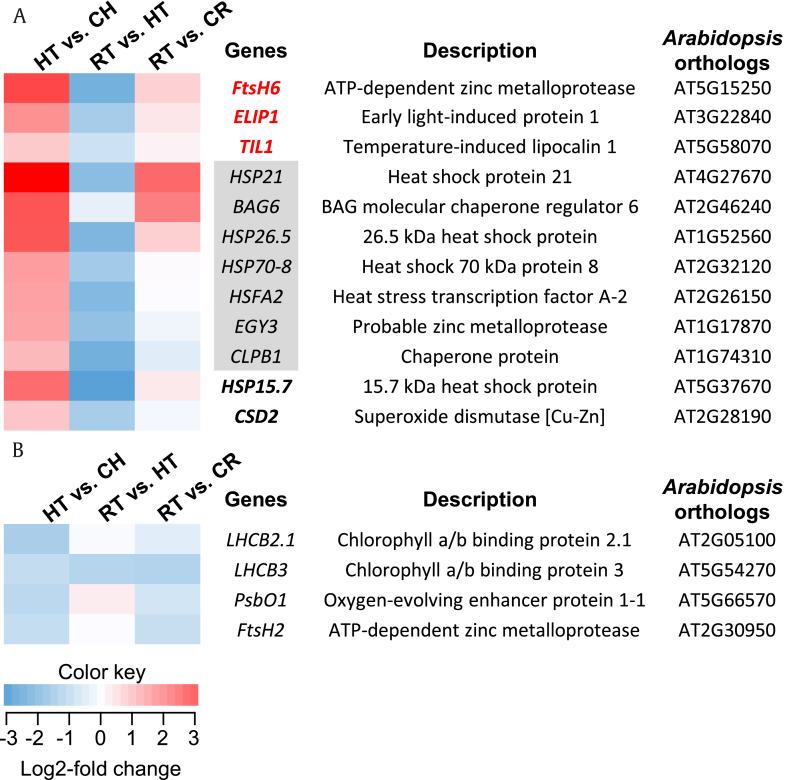
We further focused on the GO terms “response to HL intensity” and “response to ROS,” which were enriched with 10 and nine upregulated transcripts, respectively (Fig. 3A). Among the identified transcripts, three (FtsH6, ELIP1, and TIL1) and two (HSP 15.6 and CSD2) genes were particularly involved in “response to HL intensity” and “response to ROS,” respectively, whereas seven genes were redundant in both GO terms. All the upregulated transcripts in response to HL treatment were reversely downregulated after recovery treatment compared with HT samples (RT vs. HT; Fig. 3A). When the expression levels of the transcripts were compared with the corresponding control (RT vs. CR; Fig. 3A), they remained largely upregulated, except for EGY3, CLPB1, and CSD2.
Fig. 3.
Identified ginseng genes associated with Gene Ontology (GO)-terms, “response to high light intensity,” “response to reactive oxygen species (ROS),” or both and their expression profile under high light and recovery conditions. The color key indicates that color intensity represents the degree of log2-fold changes in gene expression in each pairwise comparison. (A) Upregulated genes in HT vs. CH and their expression profile in each pairwise comparison. Genes with bold red and bold black color were identified to be associated with “response to high light intensity” and “response to ROS,” respectively. Genes in grey box were involved in both GO-terms. (B) Downregulated genes in HT vs. CH identified to be associated with “response to high light intensity” and their expression profiles in each pairwise comparison.
HT, high light–treated sample; CH, corresponding control for HT; RT; recovery-treated sample after HL treatment; CR, corresponding control for RT.
Among downregulated transcripts, four (LHCB2.1, LHCB3, PsbO1, and FtsH2) were found to be involved in “response to HL intensity” (Fig. 3B). The expression levels of these transcripts after recovery treatment were slightly upregulated compared with the HT samples, except for LHCB3 (RT vs. HT; Fig. 3B). All transcripts remained downregulated after recovery treatment when their expression was compared with the corresponding control (RT vs. CR; Fig. 3B).
3.5. KEGG pathway enrichment analysis
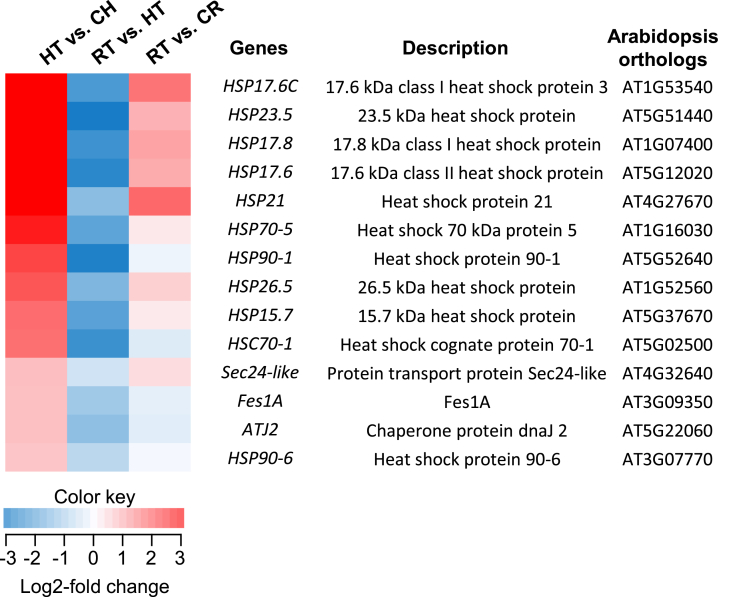
To obtain an overview of the biological functions of identified DEGs, KEGG pathway enrichment for upregulated and downregulated transcripts was performed against the Arabidopsis data set. For the upregulated transcripts in HT compared with CH samples, a KEGG pathway, “protein processing in the endoplasmic reticulum (ER; ath04141)” was significantly enriched with 14 transcripts (Fig. 4). The identified ginseng transcripts were heat shock proteins (HSPs), heat shock cognate protein, HSP binding proteins (HBP), and HSP-like chaperones, acting in protein recognition in ER, COPII-mediated protein transport, ER-associated degradation, and ubiquitin ligase complex (Fig. 4, Suppl. Fig. 3). The Arabidopsis orthologs of identified ginseng transcripts were mapped on the KEGG pathway (Suppl. Fig. 3). All those 14 upregulated transcripts in HT compared with CH were downregulated after recovery treatment (RT vs. HT; Fig. 4). In a pairwise comparison between RT and CR, nine transcripts remained upregulated, while five transcripts showed relatively lower expression in RT (RT vs. CR; Fig. 4).
Fig. 4.
Identified ginseng genes associated with the Kyoto Encyclopedia of Genes and Genome pathway, “protein processing in endoplasmic reticulum” and their expression profile under high light and recovery conditions. HT, high light–treated sample; CH, corresponding control for HT; RT, recovery-treated sample after HL treatment; CR, corresponding control for RT. The color key indicates that color intensity represents the degree of log2-fold changes in gene expression in each pairwise comparison.
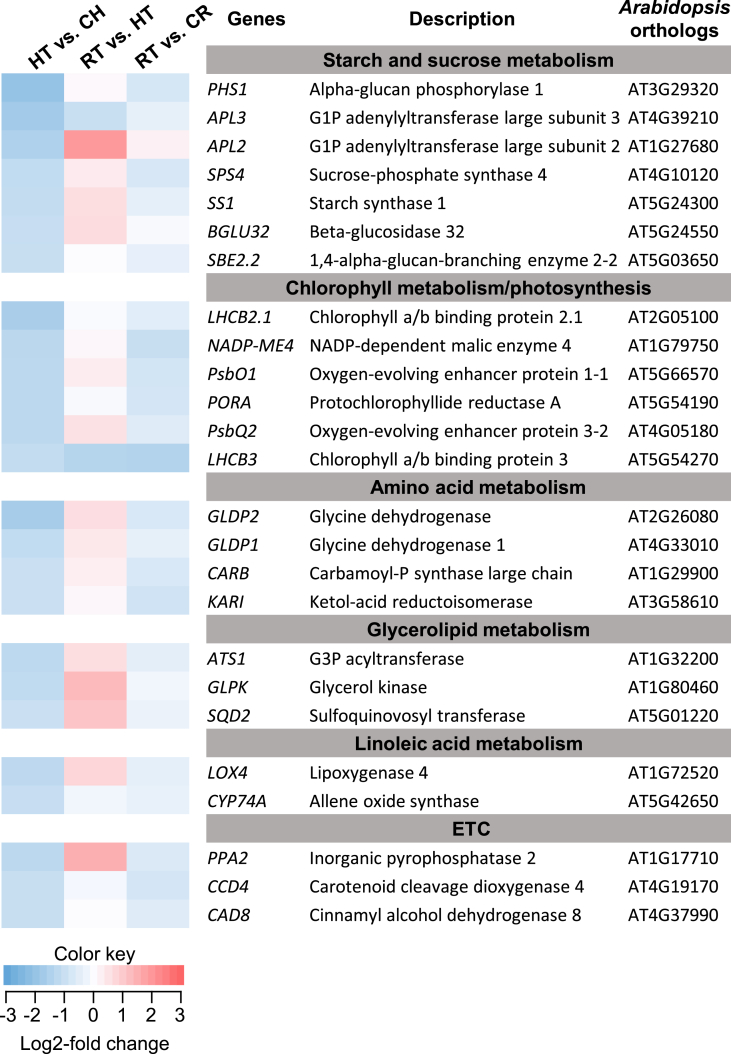
For the downregulated transcripts in the pairwise comparison between HT and CH samples, the KEGG pathways, “metabolic pathways (ath01100)” and “starch and sucrose metabolism (ath00500)” were significantly enriched with 25 and 7 transcripts, respectively. Among the 25 transcripts in “metabolic pathways,” seven were mapped onto “starch and sucrose metabolism” (Fig. 5; Suppl. Fig. 4). The remaining 18 transcripts in “metabolic pathways” were found to be involved in Chl metabolism/photosynthesis, amino acid metabolism, glycerolipid metabolism, linoleic acid metabolism, vitamin B6 metabolism, phenylpropanoid biosynthesis, and carotenoid biosynthesis (Fig. 5). Most downregulated transcripts (19 of 25 transcripts) in HT samples displayed an increased expression after recovery, whereas the remaining transcripts remained downregulated after recovery treatment (RT vs HT; Fig. 5). A comparison of the expression levels in RT with the corresponding control revealed that they were relatively lower in RT, except for that of the APL2 gene (RT vs. CR; Fig. 5).
Fig. 5.
Identified ginseng genes associated with the Kyoto Encyclopedia of Genes and Genome pathway, “metabolic pathways” and their expression profile under high light and recovery conditions. PPA2, CCD4, and CAD8 under the subgroup of ETC were identified to be involved in vitamin B6 metabolism, phenylpropanoid biosynthesis, and carotenoid biosynthesis, respectively. HT, high light–treated sample; CH, corresponding control for HT; RT, recovery-treated sample after HL treatment; CR, corresponding control for RT. The color key indicates that color intensity represents the degree of log2-fold changes in gene expression in each pairwise comparison.
4. Discussion
High-throughput transcriptome sequencing has been an effective strategy for understanding genetic and molecular mechanisms underlying complex biological processes and provides opportunities to discover genes of interests. Moreover, this strategy enables genome-wide studies of nonmodel plants even without prior genome and sequence information [27].
Considering the importance of ginsenosides as potential active compounds in ginseng, genome-wide transcriptome analysis has mostly focused on uncovering the ginsenoside biosynthetic pathways [28], [29], [30], [31], [32], [33]. However, transcriptome profiling associated with agricultural traits including a/biotic stress tolerance remains lacking. To our knowledge, this is the first study to investigate the transcriptomic responses of ginseng to HL stress, which is the major environmental stress compromising ginseng productivity.
Based on the text mining of the annotated transcripts, GO term, and KEGG pathway enrichment analysis, a significant number of upregulated transcripts under HL conditions were stress-associated genes. In particular, 40 of the 190 upregulated genes (21%) were annotated as HSP (cognate), HBP, and (co)chaperones (Suppl. Table 2). In the enriched GO-terms “response to HL intensity” and “response to ROS,” six of the 12 identified genes were also HSPs and (co)chaperones (Fig. 3A). Furthermore, the KEGG pathway, “protein processing in ER” was enriched with 14 genes including 12 HSPs, one chaperone (ATJ2), one HSBP (Fes1A), and one transporter (Sec24-like) (Fig. 4; Suppl. Fig. 3). Consistent with our results, increased expression of HSPs and chaperones under photoinhibitory light conditions has been reported in a wide range of photosynthetic organisms, including cyanobacteria, algae, and various higher plants [9], [34], [35], [36], [37], [38]. HSPs and chaperones, evolutionary conserved and found in all living organisms, are responsible for proper protein (re)folding, assembly, translocation, and stabilization, as well as protein protection and degradation after various stressful conditions [39]. Considering previous reports of the biological functions of those proteins, identified ginseng HSPs and chaperones could have protective roles against HL stress. Particularly, as indicated in the enriched KEGG pathway, HSP-mediated protein recognition, translocation, ubiquitination, and degradation of photodamaged proteins would be crucial to photoprotection.
Any HL treatment consequently increases temperature, and HL treatment in our experimental setting increased the leaf temperature by 2.8°C (an average of 25.9 and 27.7°C for the control and HL treatments, respectively). Although HSPs were originally discovered and determined to be upregulated by heat stress [40], [41], temperatures ranging from 22°C to 31°C had no detectable effects on the expression of plant HSPs [9]. Therefore, the identified ginseng HSPs in this study would primarily be induced by HL treatment rather than heat.
Biotic and abiotic stresses including HL elevate intracellular levels of ROS, which leads to oxidative stress, impairing biomolecules and biological processes [5], [9]. Under excessive light conditions, ROS are proposed to induce oxidative damage to PSII and inhibit the repair of photodamaged PSII. Thus, plants use multiple antioxidants and ROS-scavenging enzymes to minimize photo-oxidative stress in the chloroplast [4], [9], [42], [43], [44]. Suppressed expression of CCD4 under HL stress (Fig. 5) might result in the reduced catabolism of carotenoids, which are main antioxidants [44]. Similar to the previous study on Arabidopsis [42], a chloroplastic superoxide-scavenging enzyme, CSD2, was identified as an upregulated gene under HL stress in ginseng (Fig. 3A). However, other HL-responsive ROS scavengers including ascorbate peroxidase and glutathione reductase [5] were not identified as DEGs (>twofold changes). Considering the sampling time (8 HAHT) in the present study, these genes might have different temporal expression patterns after HL stress, although they commonly protect against photo-oxidative stresses [5].
Along with HSPs, (co)chaperones, and ROS scavengers, several genes involved in photoprotection were identified, especially those functioning in PSII repair and chloroplast development/maintenance. Proteolysis and de novo synthesis of the PSII reaction center-binding protein (D1 protein) is a critical process during the repair of photodamaged PSII, while a zinc metalloprotease, FtsH, degrades the D1 protein [45], [46]. In Arabidopsis, 12 genes encode FtsHs, and the FtsH protease involved in D1 protein degradation is a heterohexamer composed of type A and B FtsH subunits encoded by four chloroplastic FtsH genes [45]. Highly increased expression of ginseng FtsH6 after HL stress (12-fold increase, Fig. 3A) suggests that this gene would be one of the subunits forming functional FtsH protease for PSII repair in ginseng. In contrast to ginseng FtsH6, FtsH2 showed decreased expression after HL treatment (Fig. 3A and B), indicating that ginseng FtsH2 might not be directly involved in D1 protein degradation.
ELIP1, EGY3, and TIL1 showed increased expression after HL stress in ginseng (Fig. 3A) and are known to have protective roles against photo-oxidative stress by functioning mainly in chloroplast development and maintenance [47], [48], [49]. ELIPs are transiently accumulated in response to HL and are a family of light-harvesting complexes, which bind Chl. Although the mode of action of ELIP in photoprotection is unclear, it is proposed to prevent the accumulation of free Chl, which produces singlet oxygen molecules causing oxidative damage [47], [50]. EGY is a metalloprotease, which is required for chloroplast development, especially for the formation of thylakoid grana and the lamellae system. EGY might participate in the metabolic process of chloroplast membrane protein formation as a chloroplastic membrane–bound protease. Furthermore, it is also thought to be involved in the PSII and PSI repair cycle [49]. TIL is one of the lipocalin family members that is induced by various stress conditions, such as cold, heat, salt, drought, and HL stress [48], [51], [52]. TIL plays pivotal roles in maintaining membrane stability by minimizing oxidative degradation of membrane lipids [51].
While a significant number of transcripts upregulated by HL stress treatment were stress-related genes, most downregulated transcripts were associated with the metabolic process as shown in the GO-term and KEGG pathway enrichment analysis (Fig. 2, Fig. 3, Fig. 5). Downregulation of those genes, especially those involved in collective carbohydrates, amino acids, and lipids metabolism, would be an acclimation process under HL conditions rather than a photoprotective response. Excessive light reduces photosynthetic capacity and directly limits carbon availability. Thus, distributing and controlling metabolic flux via transcriptional regulation would be a critical acclimation process to cope with changes in the carbon status [53], [54]. For instance, as shown in Fig. 4, starch synthesis could be suppressed primarily by deregulating AGPases (APL2 and APL3) and starch synthase (SS1) when carbon availability could decrease owing to HL stress. Furthermore, suppressing SPS4 would contribute to maintaining sucrose availability, thereby providing energy in the source tissues (mainly leaves) under the stress condition.
Transcriptional deregulation of some of the identified genes in Chl metabolism would be directly related to photoprotection. Notably, in this study and in agreement with previous findings [55], [56], [57], reduced expression of genes encoding the light-harvesting Chl a/b binding proteins of PSII (LHCII), LHCB2.1, and LHCB3 reflects the photoprotective reduction of the PSII antenna size, which downregulates the rate of PSII excitation. The PSII antenna size is adjusted by regulatory proteolysis of pre-existing LHCII proteins, and FtsH is proposed to be one of the candidate proteases [55], [58]. Furthermore, the expression of LHCII could be negatively regulated both transcriptionally and translationally in response to signaling molecules including H2O2 and ABA under HL conditions [57], [59].
In this study, the responses of ginseng transcriptomes to HL and recovery from photoinhibition were investigated. Most upregulated and downregulated transcripts under HL stress were reversely regulated after recovery treatment and were transiently regulated HL-responsive genes (categorized into clusters 1 and 3, respectively; Suppl. Fig. 2). DEGs maintaining their expression levels even after recovery (categorized into Cluster 2 and 4; Suppl. Fig. 2) would participate in relatively long-term acclimation process against HL stress.
This study provides transcriptome profiling data on ginseng in response to HL stress and identifies potential candidate genes playing critical roles in photoprotection and acclimation processes against the stress. Furthermore, the identified genes could be rational candidates for metabolic engineering or germplasm screening to improve HL tolerance in ginseng. Ginseng, a shade plant, is particularly susceptible to HL stress. Comparative transcriptome analysis between sun and shade plants or among close relatives with different light tolerance would further facilitate the elucidation of genetic and molecular mechanisms underlying HL tolerance and susceptibility.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by intramural grants (2Z05310, 2Z05630, and 2E27380) from the Korea Institute of Science and Technology (KIST). Korea Ginseng Corporation (KGC) is gratefully acknowledged for providing ginseng materials.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2018.12.009.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kim J.H., Yi Y.-S., Kim M.-Y., Cho J.Y. Role of ginsenosides, the main active components of Panax ginseng in inflammatory responses and diseases. J Ginseng Res. 2017;41:435–443. doi: 10.1016/j.jgr.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park T.Y., Hong M., Sung H., Kim S., Suk K.T. Effect of Korean red ginseng in chronic liver disease. J Ginseng Res. 2017;41:450–455. doi: 10.1016/j.jgr.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powles S.B. Photoinhibition of photosynthesis induced by visible light. Annu Rev Plant Physiol. 1984;35:15–44. [Google Scholar]
- 4.Takahashi S., Badger M.R. Photoprotection in plants: a new light on photosystem II damage. Trends Plant Sci. 2011;16:53–60. doi: 10.1016/j.tplants.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Gururani Mayank A., Venkatesh J., Tran L.S.P. Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol Plant. 2015;8:1304–1320. doi: 10.1016/j.molp.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Murata N., Takahashi S., Nishiyama Y., Allakhverdiev S.I. Photoinhibition of photosystem II under environmental stress. Biochim Biophys Acta. 2007;1767:414–421. doi: 10.1016/j.bbabio.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Müller P., Li X.-P., Niyogi K.K. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001;125:1558–1566. doi: 10.1104/pp.125.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruban A.V. Evolution under the sun: optimizing light harvesting in photosynthesis. J Exp Bot. 2015;66:7–23. doi: 10.1093/jxb/eru400. [DOI] [PubMed] [Google Scholar]
- 9.Rossel J.B., Wilson I.W., Pogson B.J. Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol. 2002;130:1109–1120. doi: 10.1104/pp.005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samol I., Shapiguzov A., Ingelsson B., Fucile G., Crèvecoeur M., Vener A.V., Rochaix J.-D., Goldschmidt-Clermont M. Identification of a photosystem II phosphatase involved in light acclimation in Arabidopsis. Plant Cell. 2012;24:2596–2609. doi: 10.1105/tpc.112.095703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nath K., Jajoo A., Poudyal R.S., Timilsina R., Park Y.S., Aro E.-M., Nam H.G., Lee C.H. Towards a critical understanding of the photosystem II repair mechanism and its regulation during stress conditions. FEBS Letters. 2013;587:3372–3381. doi: 10.1016/j.febslet.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Jang I.-B., Lee D.-Y., Yu J., Park H.-W., Mo H.-S., Park K.-C., Hyun D.-Y., Lee E.-H., Kim K.-H., Oh C.-S. Photosynthesis rates, growth, and ginsenoside contents of 2-yr-old Panax ginseng grown at different light transmission rates in a greenhouse. J Ginseng Res. 2015;39:345–353. doi: 10.1016/j.jgr.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miskell J.-A., Parmenter G., Eaton-Rye J.J. Decreased Hill reaction rates and slow turnover of transitory starch in the obligate shade plant Panax quinquefolius L. (American ginseng) Planta. 2002;215:969–979. doi: 10.1007/s00425-002-0839-9. [DOI] [PubMed] [Google Scholar]
- 14.Cheon S.-G., Mok S.-G., Lee S.-S. Effect of light intensity and quality on the growth and quality of Korean ginseng (Panax ginseng C.A. Meyer) III. Effects of light intensity on the quality of ginseng plant. J Ginseng Res. 1991;15:144–151. [Google Scholar]
- 15.Parmenter G., Littlejohn R. Effect of shade on growth and photosynthesis of Panax ginseng. N Z J Crop Hortic Sci. 2000;28:255–269. [Google Scholar]
- 16.Maxwell K., Johnson G.N. Chlorophyll fluorescence—a practical guide. J Exp Bot. 2000;51:659–668. doi: 10.1093/jxb/51.345.659. [DOI] [PubMed] [Google Scholar]
- 17.Cox M.P., Peterson D.A., Biggs P.J. Solexaqa: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulz M.H., Zerbino D.R., Vingron M., Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J.-E., Choe J., Lee W., Kim S., Lee M., Kim T.-H., Jo S.-H., Lee J. De novo gene set assembly of the transcriptome of diploid, oilseed-crop species Perilla citriodora. J Plant Biotechnol. 2016;43:293–301. [Google Scholar]
- 22.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amap: Another multidimensional analysis package . 2014. R package version 0.8-12.http://CRAN.R-project.org/package=amap Available from: [Google Scholar]
- 24.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the panther classification system. Nat Protoc. 2013;8:1551. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M., Goto S. Kegg: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unamba C.I.N., Nag A., Sharma R.K. Next generation sequencing technologies: the doorway to the unexplored genomics of non-model plants. Front Plant Sci. 2015;6 doi: 10.3389/fpls.2015.01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo H., Sun C., Sun Y., Wu Q., Li Y., Song J., Niu Y., Cheng X., Xu H., Li C. Analysis of the transcriptome of Panax notoginseng root uncovers putative triterpene saponin-biosynthetic genes and genetic markers. BMC Genomics. 2011;12:S5. doi: 10.1186/1471-2164-12-S5-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun C., Li Y., Wu Q., Luo H., Sun Y., Song J., Lui E.M.K., Chen S. De novo sequencing and analysis of the American ginseng root transcriptome using a GS FLX Titanium platform to discover putative genes involved in ginsenoside biosynthesis. BMC Genomics. 2010;11:262. doi: 10.1186/1471-2164-11-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rai A., Yamazaki M., Takahashi H., Nakamura M., Kojoma M., Suzuki H., Saito K. RNA-seq transcriptome analysis of Panax japonicus, and its comparison with other Panax species to identify potential genes involved in the saponins biosynthesis. Front Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K., Jiang S., Sun C., Lin Y., Yin R., Wang Y., Zhang M. The spatial and temporal transcriptomic landscapes of ginseng, Panax ginseng C.A. Meyer. Sci Rep. 2015;5:18283. doi: 10.1038/srep18283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jayakodi M., Lee S.-C., Park H.-S., Jang W., Lee Y.S., Choi B.-S., Nah G.J., Kim D.-S., Natesan S., Sun C. Transcriptome profiling and comparative analysis of Panax ginseng adventitious roots. J Ginseng Res. 2014;38:278–288. doi: 10.1016/j.jgr.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao H., Nuruzzaman M., Xiu H., Huang J., Wu K., Chen X., Li J., Wang L., Jeong J.-H., Park S.-J. Transcriptome analysis of methyl jasmonate-elicited Panax ginseng adventitious roots to discover putative ginsenoside biosynthesis and transport genes. Int J Mol Sci. 2015;16:3035–3057. doi: 10.3390/ijms16023035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hihara Y., Kamei A., Kanehisa M., Kaplan A., Ikeuchi M. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell. 2001;13:793–806. doi: 10.1105/tpc.13.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drzymalla C., Schroda M., Beck C.F. Light-inducible gene HSP70b encodes a chloroplast-localized heat shock protein in Chlamydomonas reinhardtii. Plant Mol Biol. 1996;31:1185–1194. doi: 10.1007/BF00040835. [DOI] [PubMed] [Google Scholar]
- 36.Barua D., Heckathorn S.A. The interactive effects of light and temperature on heat-shock protein accumulation in Solidago altissima (Asteraceae) in the field and laboratory. Am J Bot. 2006;93:102–109. [Google Scholar]
- 37.Schroda M., Vallon O., Wollman F.-A., Beck C.F. A chloroplast-targeted heat shock protein 70 (hsp70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell. 1999;11:1165–1178. doi: 10.1105/tpc.11.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stapel D., Kruse E., Kloppstech K. The protective effect of heat shock proteins against photoinhibition under heat shock in barley (Hordeum vulgare) J Photochem Photobiol B. 1993;21:211–218. [Google Scholar]
- 39.Wang W., Vinocur B., Shoseyov O., Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Ritossa F. A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia. 1962;18:571–573. [Google Scholar]
- 41.Lindquist S. Varying patterns of protein synthesis in Drosophila during heat shock: implications for regulation. Dev Biol. 1980;77:463–479. doi: 10.1016/0012-1606(80)90488-1. [DOI] [PubMed] [Google Scholar]
- 42.Abarca D., Roldán M., Martín M., Sabater B. Arabidopsis thaliana ecotype Cvi shows an increased tolerance to photo-oxidative stress and contains a new chloroplastic copper/zinc superoxide dismutase isoenzyme. J Exp Bot. 2001;52:1417–1425. doi: 10.1093/jexbot/52.360.1417. [DOI] [PubMed] [Google Scholar]
- 43.Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Havaux M., Niyogi K.K. The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc Natl Acad Sci U S A. 1999;96:8762–8767. doi: 10.1073/pnas.96.15.8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshioka-Nishimura M., Nanba D., Takaki T., Ohba C., Tsumura N., Morita N., Sakamoto H., Murata K., Yamamoto Y. Quality control of photosystem II: direct imaging of the changes in the thylakoid structure and distribution of FtsH proteases in spinach chloroplasts under light stress. Plant Cell Physiol. 2014;55:1255–1265. doi: 10.1093/pcp/pcu079. [DOI] [PubMed] [Google Scholar]
- 46.Adam Z., Frottin F., Espagne C., Meinnel T., Giglione C. Interplay between N-terminal methionine excision and FtsH protease is essential for normal chloroplast development and function in Arabidopsis. Plant Cell. 2011;23:3745–3760. doi: 10.1105/tpc.111.087239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hutin C., Nussaume L., Moise N., Moya I., Kloppstech K., Havaux M. Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc Natl Acad Sci U S A. 2003;100:4921–4926. doi: 10.1073/pnas.0736939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charron J.-B.F., Ouellet F., Houde M., Sarhan F. The plant apolipoprotein d ortholog protects Arabidopsis against oxidative stress. BMC Plant Biol. 2008;8:86. doi: 10.1186/1471-2229-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen G., Bi Y.R., Li N. EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J. 2005;41:364–375. doi: 10.1111/j.1365-313X.2004.02308.x. [DOI] [PubMed] [Google Scholar]
- 50.Tzvetkova-Chevolleau T., Franck F., Alawady A.E., Dall’Osto L., Carrière F., Bassi R., Grimm B., Nussaume L., Havaux M. The light stress-induced protein ELIP2 is a regulator of chlorophyll synthesis in Arabidopsis thaliana. Plant J. 2007;50:795–809. doi: 10.1111/j.1365-313X.2007.03090.x. [DOI] [PubMed] [Google Scholar]
- 51.Levesque-Tremblay G., Havaux M., Ouellet F. The chloroplastic lipocalin AtCHL prevents lipid peroxidation and protects Arabidopsis against oxidative stress. Plant J. 2009;60:691–702. doi: 10.1111/j.1365-313X.2009.03991.x. [DOI] [PubMed] [Google Scholar]
- 52.Abo-Ogiala A., Carsjens C., Diekmann H., Fayyaz P., Herrfurth C., Feussner I., Polle A. Temperature-induced lipocalin (TIL) is translocated under salt stress and protects chloroplasts from ion toxicity. J Plant Physiol. 2014;171:250–259. doi: 10.1016/j.jplph.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 53.Geigenberger P. Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiol. 2011;155:1566–1577. doi: 10.1104/pp.110.170399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walters R.G. Towards an understanding of photosynthetic acclimation. J Exp Bot. 2005;56:435–447. doi: 10.1093/jxb/eri060. [DOI] [PubMed] [Google Scholar]
- 55.Yang D.-H., Webster J., Adam Z., Lindahl M., Andersson B. Induction of acclimative proteolysis of the light-harvesting chlorophyll a/b protein of photosystem II in response to elevated light intensities. Plant Physiol. 1998;118:827–834. doi: 10.1104/pp.118.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Derks A., Schaven K., Bruce D. Diverse mechanisms for photoprotection in photosynthesis. Dynamic regulation of photosystem II excitation in response to rapid environmental change. Biochim Biophys Acta. 2015;1847:468–485. doi: 10.1016/j.bbabio.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 57.Borisova-Mubarakshina M.M., Ivanov B.N., Vetoshkina D.V., Lubimov V.Y., Fedorchuk T.P., Naydov I.A., Kozuleva M.A., Rudenko N.N., Dall'Osto L., Cazzaniga S. Long-term acclimatory response to excess excitation energy: evidence for a role of hydrogen peroxide in the regulation of photosystem II antenna size. J Exp Bot. 2015;66:7151–7164. doi: 10.1093/jxb/erv410. [DOI] [PubMed] [Google Scholar]
- 58.Luciński R., Jackowski G. Atftsh heterocomplex-mediated degradation of apoproteins of the major light harvesting complex of photosystem II (LHCII) in response to stresses. J Plant Physiol. 2013;170:1082–1089. doi: 10.1016/j.jplph.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 59.Liu R., Xu Y.-H., Jiang S.-C., Lu K., Lu Y.-F., Feng X.-J., Wu Z., Liang S., Yu Y.-T., Wang X.-F. Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function. J Exp Bot. 2013;64:5443–5456. doi: 10.1093/jxb/ert307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.