Abstract
Many commercial plasticizers are toxic endocrine-disrupting chemicals that are added to plastics during manufacturing and may leach out once they reach the environment. Traditional phthalic acid ester plasticizers (PAEs), such as dibutyl phthalate (DBP) and bis(2-ethyl hexyl) phthalate (DEHP), are now increasingly being replaced with more environmentally friendly alternatives, such as acetyl tributyl citrate (ATBC). While the metabolic pathways for PAE degradation have been established in the terrestrial environment, to our knowledge, the mechanisms for ATBC biodegradation have not been identified previously and plasticizer degradation in the marine environment remains underexplored. From marine plastic debris, we enriched and isolated microbes able to grow using a range of plasticizers and, for the first time, identified the pathways used by two phylogenetically distinct bacteria to degrade three different plasticizers (i.e., DBP, DEHP, and ATBC) via a comprehensive proteogenomic and metabolomic approach. This integrated multi-OMIC study also revealed the different mechanisms used for ester side-chain removal from the different plasticizers (esterases and enzymes involved in the β-oxidation pathway) as well as the molecular response to deal with toxic intermediates, that is, phthalate, and the lower biodegrading potential detected for ATBC than for PAE plasticizers. This study highlights the metabolic potential that exists in the biofilms that colonize plastics—the Plastisphere—to effectively biodegrade plastic additives and flags the inherent importance of microbes in reducing plastic toxicity in the environment.
Introduction
Marine plastic pollution is both ubiquitous and well-acknowledged1−3 with estimates that 4.8 to 12.7 million tons of plastic waste enter the oceans every year.4 The ecotoxicological problems derived from environmental plastic pollution are direct, for example, through ingestion or entanglement,5 or indirect, for example, through the transfer of toxic chemicals to marine life.6,7 Plastics are known to harbor toxic chemicals in concentrations up to 106 times higher than in their surrounding environment,8 with up to 906 different chemicals associated with plastics.9 These chemical pollutants come from hydrophobic sorption to plastics, such as polychlorinated biphenyls (PCBs) and polyaromatic hydrocarbons (PAHs),10 or are directly added during manufacturing processes, such as stabilizers, flame retardants, and plasticizers.9
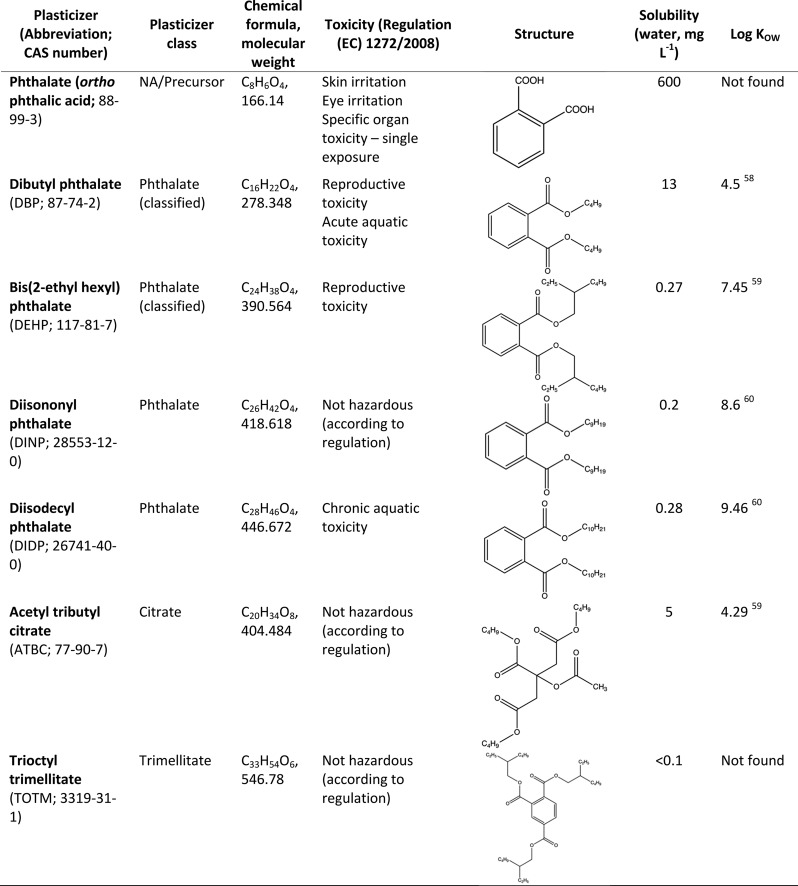
Plasticizers are added to plastics to change the physical and chemical properties of the material and can represent up to 10–70% of the material’s weight. They are used mainly in the production of plasticized or flexible poly(vinyl chloride) (PVC), a material which comprises 80% of the global use of plasticizers.11 There are several classes of plasticizers that can be used: phthalates, terephthalates, epoxies, aliphatics, trimellitates, and citrates. Phthalic acid ester plasticizers (PAEs; phthalates) represent ∼65% of the global plasticizer market share, with bis(2-ethyl hexyl) phthalate (DEHP) and dibutyl phthalate (DBP) being the two most widely used.12 Plasticizers are not chemically bound to the plastic and are therefore able to leach out of these materials, either during consumer use or on their subsequent release into the environment.11,13−15 Both DEHP and DBP, in addition to other plasticizers, have been found on both new plastics and plastic marine debris,14 as well as in a range of plastics ingested by marine wildlife.16,17 Concerns have been raised over the continued use of plasticizers as they may not only be highly toxic—DBP and DEHP have recently been identified as some of the most hazardous chemicals associated with plastics9—but many are also known endocrine disruptors and carcinogens, even at low concentrations.18 Due to their toxicity, there is now a push to replace the traditional PAEs with other less toxic alternatives, such as acetyl tributyl citrate (ATBC).19,20
ATBC is widely stated to be a nontoxic and biodegradable alternative to DEHP and other PAEs;19,21 however, to our knowledge, the catabolic pathway for ATBC biodegradation has not been experimentally shown. In fact, one study found that using ATBC as a plasticizer actually decreased enzymatic degradation rates of the biodegradable plastic poly(lactic acid).22 On the other hand, the pathways used for PAE biodegradation have been characterized to some extent in a number of terrestrial microorganisms.23,24,33−39,25−32 Esterases have been suggested to carry out the first step of PAE biodegradation by hydrolyzing the side chains and generating phthalate and short-chain alcohols and fatty acids.23,26,29,30,33−39 However, only two of these studies34,36 actually identified the specific esterases involved and, furthermore, β-oxidation of the side-chain fatty acids has even been suggested to occur directly on the PAE molecule.33,40 Hence, it is not known whether the mechanism for biodegradation is the same for different PAE plasticizers. The metabolic pathways for phthalate degradation are well established and follow one of the branches of the β-ketoadipate pathway.28,41,42 Despite the widespread distribution of plastics and, consequently, plasticizers in the oceans, the degradation of these additives by marine bacteria has been less extensively studied15,28,37 and the catabolic pathways are unknown.
Here, a natural microbial community obtained from marine plastic debris was enriched with each of six different plasticizers. Forty-two strains were isolated and tested for their ability to grow on each of the six plasticizers. A detailed proteogenomic and metabolomic analysis was then performed on two selected strains, Halomonas sp. ATBC28 and Mycobacterium sp. DBP42, allowing the identification of the enzymes and pathways involved in the biodegradation of phthalate, two of the most widely used plasticizers (DBP and DEHP), as well as a new alternative plasticizer ATBC.
Materials and Methods
All culturing and incubations described here were carried out using one carbon source only, that is, plasticizers were not mixed. A schematic depicting all experimental setups is found in Supporting Information Figure S1.
Enrichments and Microbial Isolation from Marine Plastic Debris
Tissue culture flasks (25 cm2) containing 25 mL of Bushnell–Haas mineral medium,43 supplemented with 0.52 M NaCl, 0.005% (w/v) yeast extract (as a supplement of growth factors; Merck KGaA, Germany), and each one of six plasticizers (0.1% v/v of DBP, DEHP, DINP, DIDP, ATBC, or TOTM; Sigma-Aldrich, U.K.; Table 1), were inoculated with 1 mL of a natural microbial community obtained from bulk marine plastic debris (Supporting Information Figure S2) collected during boat tows from both Plymouth Sound (Devon, U.K.; June 2016) and Portaferry (Northern Ireland, U.K.; August 2016) and detached by thorough vortexing. Cultures were incubated at 30 °C in the dark with shaking at 150 rpm. When growth was apparent (after approximately 3 weeks and assessed by visual turbidity), 1 mL of these cultures was used to inoculate fresh tissue culture flasks (25 cm2) with 25 mL of custom media, as above, in a second enrichment step. Agar plates were made with the supplemented Bushnell–Haas mineral medium, containing 0.1% (v/v) of one of the six plasticizers listed above and 1.5% agar. Media were blended immediately before pouring the plates to ensure homogeneous dispersal of the hydrophobic plasticizers. Marine agar (BD Difco) plates were also used for isolation. One-hundred microliters of the second enrichment cultures was spread on to replicate plates and incubated for 3 weeks at 30 °C. Morphologically distinct colonies (n = 42) were picked and streaked onto fresh plates until isolates were obtained.
Table 1. Characteristics of Plasticizers Used in this Study58−60.
Screening and Selection of Isolates
Each one of the 42 isolates was grown in a six-well plate, each well containing 2.5 mL of supplemented Bushnell–Haas mineral media with 0.1% (v/v) of one of the six plasticizers, and incubated for 2 weeks at 30 °C in the dark. Growth was monitored weekly through visual assessment due to the heterogeneity caused by the hydrophobic substrates (Supporting Information Table S1). The ten isolates that showed the highest potential for growth with several of the plasticizers were identified by partial amplification (using primers 27F and 1492R and PCR conditions as described previously44) and sequencing (GATC BioTech, Germany) of the 16S rRNA gene. Isolates were grown on marine agar (BD Difco) plates and DNA extractions were performed using a modified DNeasy Plant Mini Kit (Qiagen) protocol.45 Purification of PCR products was performed using the QIAquick PCR purification kit (Qiagen), in accordance with the manufacturer’s instructions, prior to being sent for sequencing. BLASTN46 searches of the NCBI database were used to identify the resulting 16S rRNA gene sequences.
These ten isolates were then narrowed down to two isolates, Mycobacterium sp. DBP42 and Halomonas sp. ATBC28, based on their robust growth on a variety of plasticizers and their diverse phylogenetic origin.
Characterization and Genome Sequencing of Two Selected Isolates
To determine appropriate substrates for use as reference conditions, the two selected isolates were further tested on different labile growth substrates, that is, glucose, sodium succinate, pyruvate, glycerol, N-acetyl-d-glucosamine, fructose, phthalate, and marine broth (BD Difco). Each condition was tested in duplicate in 96-well plates, with each well containing 200 μL of supplemented Bushnell–Haas mineral medium with 0.1% (w/v or v/v) substrate, and incubated at 30 °C with shaking at 150 rpm for 72 h. Growth was monitored by optical density measurements (600 nm; 30 min intervals using a Synergy HTX microplate reader). The substrates that produced the highest growth for each of the isolates (i.e., glycerol for Mycobacterium sp. DBP42 and pyruvate for Halomonas sp. ATBC28; Supporting Information Figure S3) were used as reference controls for subsequent incubations using plasticizers. To test the toxicity of phthalate to the bacteria, neither of which was able to grow with 0.1% phthalate (Supporting Information Figure S4), the two isolates were grown in triplicate in 3 mL of marine broth (BD Difco) in glass vials with 0, 0.02, 0.1, 0.4, and 1.6% (w/v) phthalate, with and without 0.1% (v/v) DBP as an inducer. These were incubated at 30 °C in the dark with shaking at 180 rpm and growth was measured by optical density (600 nm) on days 0, 1, 2, 3, and 4.
The two selected isolates were grown in triplicate in 22 mL glass vials containing 10 mL of supplemented Bushnell–Haas mineral medium with 0.1% (v/v) of each of the six plasticizers, 0.02% (w/v) phthalate (due to the toxicity of phthalate at higher concentrations; Supporting Information Figure S4), or 0.1% of the labile positive control (i.e., glycerol for Mycobacterium sp. DBP42 or pyruvate for Halomonas sp. ATBC28) and incubated at 30 °C in the dark with shaking at 150 rpm to confirm growth on these substrates. Growth was monitored by optical density measurements (600 nm) on days 1, 2, 4, and 7.
Whole cells were scraped from marine broth (BD Difco) agar plates and sent to MicrobesNG (Birmingham, U.K.) for whole genome sequencing. MicrobesNG performed the DNA extraction, library preparation, sequencing, and assembly of genomes. Assembled genomes were then annotated using Prokka version 1.1347 and KEGG Orthology And Links Annotation (KOALA48,49). The complete genome sequences of the two selected strains, that is, Mycobacterium sp. and DBP42 Halomonas sp. ATBC28 (Supporting Information Table S2), were deposited in the GenBank database under the BioProject nos. PRJNA525197 and PRJNA525098, respectively. Local BLAST searches with the proteins identified in Azoarcus sp. strain PA0150 and Aromatoleum aromaticum(51) as involved in anaerobic phthalate degradation were carried out on the genomes of both isolates.
Preparation of Cellular and Extracellular Samples for OMIC Analyses
The two selected isolates were grown in 100 mL Erlenmeyer flasks containing 40 mL of supplemented Bushnell–Haas mineral medium and 0.1% (v/v) of DBP, DEHP, or ATBC or 0.02% (w/v) of phthalate as sole carbon sources. Pyruvate and glycerol (0.1% w/v) were used as the control growth conditions for Halomonas sp. ATBC28 and Mycobacterium sp. DBP42, respectively. Three independent cultures for each condition were incubated at 30 °C in the dark with shaking at 150 rpm until absorbance (600 nm) reached 0.3, or large clumps of cells were visible (approximately 21 days for plasticizers and phthalate and 7 days for controls with pyruvate or glycerol) at which point cells were harvested by centrifugation (4000 rpm at 4 °C for 15 min) and supernatants were further filtered using 0.2 μm pore PTFE filters (Millex-LG; Millipore, Germany). After removing 1.5 mL for the metabolomic analyses, the extracellular proteomes within the supernatant filtrates (∼38 mL) were concentrated using a trichloroacetic acid protocol as previously described.52,53 Both cell pellets and exoproteome precipitates were dissolved in 1× LDS loading buffer (Invitrogen, USA), and the equivalent of 20 mL of culture was loaded on a precast 10% Tris-Bis NuPAGE gel (Invitrogen, USA) using 1× MOPS solution (Invitrogen, USA) as the running buffer. SDS-PAGE was performed for a short gel migration (5 min), allowing the removal of contaminants and purification of the polypeptides in the polyacrylamide gel.
Trypsin In-Gel Proteolysis, NanoLC-MS/MS, and Data Analysis of the Proteomes
Polyacrylamide gel bands containing the cellular and extracellular proteomes were excised and standard in-gel reduction with dithiothreitol, and alkylation with iodoacetamide was performed prior to trypsin (Roche, Switzerland) proteolysis.52 The resulting tryptic peptide mixture was extracted from the polyacrylamide gel bands and prepared for mass spectrometry as previously described.53 Samples were analyzed by means of nanoLC-ESI-MS/MS using an Ultimate 2000 LC system (Dionex-LC Packings) coupled to an Orbitrap Fusion mass spectrometer (Thermo Scientific, USA) using a 60 min LC separation (exoproteomes) or a 120 min LC separation (cellular proteomes) on a 25 cm column and settings as described previously.54 Raw proteomics files containing all mass spectral data were processed using MaxQuant version 1.5.5.155 for protein identification and quantification, using default parameters, match between runs and in-house protein databases obtained from the genome annotation for each bacterium. Matched proteins and corresponding label-free quantification were used for downstream analyses. The complete lists of detected peptides and polypeptides are provided in Supporting Information Tables S3 and S4 for Mycobacterium sp. DBP42 and Halomonas sp. ATBC28, respectively. The comparative proteomic analysis between samples was carried out using Perseus version 1.5.5.356 following the pipeline described previously.53 Data were then imported into Python where custom scripts (Python version 3.6.8 with modules numpy, os, csv, and math) were used to carry out two-sample Student’s t-tests for significance and calculate fold changes (Supporting Information Tables S5 and S6 for Mycobacterium sp. DBP42 and Halomonas sp. ATBC28, respectively). Fold changes were calculated for all proteins that were detected in all three replicates of at least one treatment. All custom analysis scripts can be found at https://github.com/R-Wright-1/Plasticizer-Degradation. Conserved domain searches were carried out for manual curation of the functions assigned to all key proteins identified and further mapped onto KEGG degradation pathways.49,57
Metabolomics for the Identification of Degradation Intermediates in Culture Supernatants (LC-MS)
Filtered supernatants (1.5 mL) were adjusted to pH 2.5 using 2 M HCl after which 3 mL of ethyl acetate was added for hydrophobic compound extraction and purification. Briefly, samples were vortexed for 10 s and left for 10 min for phase separation after which the organic fraction was collected into a new tube and evaporated using a GeneVac EZ-2 evaporator. Pellets were resuspended in 250 μL of 16 mM phosphate buffer (pH 2.5) containing 20% (v/v) of DMSO and filtered through a microcentrifugal tube with a 0.2 μm nylon membrane (Thermo Fisher Scientific, U.K.) before analysis. An UHPLC system (Ultimate 3000; Thermo Fisher Scientific, Waltham, MA, USA) coupled to a Q-Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific) operating with a heated electrospray interface (HESI) was employed, and the spectra were recorded in both positive and negative modes.
Chromatographic separation was accomplished using two strategies: (i) untargeted metabolomics were performed on a reversed-phase C18 core–shell (100 x 2.1 mm ID, 1.6 μm; Luna Omega) chromatographic column preceded by a C18-security guard ultracartridge (2.1 mm ID), both from Phenomenex (Torrence, CA); (ii) targeted metabolomics were performed on a reversed-phase C18 core–shell (250 x 4.6 mm ID, 5.0 μm; Ultrasphere) chromatographic column preceded by a C18-security guard cartridge (45 x 4.6 mm ID, 5.0 μm; Ultrasphere), both from Hichrom (Leicester, U.K.). Both strategies were used a mobile phase of (A) 0.1% (v/v) formic acid in water and (B) 0.1%(v/v) formic acid in acetonitrile at 0.2 mL min–1 (untargeted metabolomics) or 0.5 mL min–1 (targeted metabolomics). For untargeted metabolomics, the elution gradient started with 5% of solution B for 5 min and then increased to 55% over 5 min and was maintained at 55% for 3 min. Over 1 min, the proportion of solution B was reduced from 55 to 5% and it was maintained at 5% for 6 min. For targeted metabolomics, the elution gradient started with 5% of solution B for 6 min and then increased to 55% B over 8 min and was maintained at 55% for 4.25 min. Over 15 s, the proportion of solution B was reduced from 55 to 5% and it was maintained at 5% for 7 min. For both strategies, the injection volume was 5 μL. For the monitoring of plasticizer consumption, the method followed that of the targeted metabolomics, above, but the elution gradient had a constant flux of 0.2 mL min–1 and started with 40% of solution B for 0.5 min and then increased to 100% B in 24.5 min. It was maintained at 100% B for 10 min and then over 1 min, and then the proportion of B was reduced from 100 to 40% over 1 min and maintained at 40% for 5 min.
For MS and MS/MS acquisition, the temperature of ion transfer capillary, spray voltage, sheath gas flow rate, auxiliary gas flow rate, and S-lens RF level were set to 350 °C, 3.9 KV in positive mode and 3.1 kV in negative mode, 35 arbitrary units (AU), 10 AU, and 55 AU, respectively. Full scan acquisition over a range of 80–600 m/z was performed with a resolution of 70,000. During the MS/MS scans, precursors were fragmented with a normalized collisional energy of 50 AU. The top five ions were selected for MS/MS analysis with a dynamic exclusion of 1 s.
Xcalibur 4.1, Trace Finder 4.1 SP2, and Compound Discoverer 2.0 software (Thermo Fisher Scientific) were used for LC-MS and LC-MS/MS acquisition, targeted metabolomics, and untargeted metabolomics data processing, respectively. Mass tolerance (Δ) was set for the analysis with Trace Finder 4.1 SP2 and Compound Discoverer 2.0 to 14 and 10 ppm, respectively, according to data obtained with external standards analyzed in negative mode using the FreeStyle 1.3 software (Thermo Fisher Scientific): salicylic acid (targeted, RT 21.02 min, Δ 10.22 ppm; untargeted, RT 15.11 min, Δ 6.46 ppm), phthalic acid (targeted, RT 18.12 min, Δ 8.67 ppm; untargeted, RT 12.08 min, Δ 4.99 ppm), catechol (targeted, RT 18.14 min, Δ 13.75 ppm; untargeted, RT 7.7 min, Δ 2.1 ppm), protocatechuate (3,4-dihydroxy benzoate; targeted, RT 16.7 min, Δ 8.49 ppm; untargeted, RT 12.23 min, Δ 6.18 ppm), and citric acid (targeted, RT 6.97 min, Δ 5.76 ppm; untargeted, RT 1.87 min, Δ 2.86 ppm). Additional basic parameters used for analysis were as follows: Trace Finder 4.1 SP2, negative mode, Genesis as the detection algorithm, and the highest peak as the peak detection strategy; Compound Discoverer 2.0, minimum peak intensity of 5 × 106, signal–noise ratio of 5, and retention time tolerance of 0.075 min. Full details of all detected compounds are in Supporting Information Table S7.
Results
Enrichment, Isolation, and Selection of Plasticizer-Degrading Bacteria
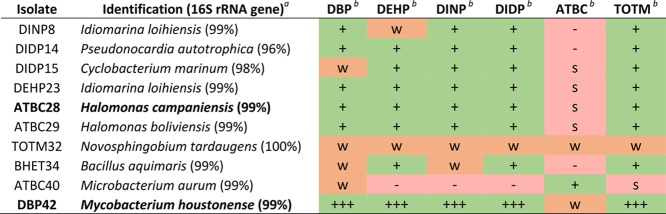
Only 17 out of the 42 isolates achieved considerable growth with at least one of the plasticizers (i.e., DBP, DEHP, DINP, DIDP, ATBC, and TOTM; Table 1) in liquid cultures, whereas others only produced weak growth (n = 15) or emulsified the plasticizers (n = 10; Supporting Information Table S1). Interestingly, while each one of the PAEs and TOTM could be used by 11 to 14 of the isolates, clear growth on ATBC was only observed with one of the strains (i.e., isolate 40; Supporting Information Table S1). Those ten isolates that showed growth on a range of plasticizers were identified by partial sequencing of their 16S rRNA gene (Table 2). Isolates 28 and 42, identified as Halomonas campaniensis (99% 16S rRNA gene sequence identity) and Mycobacterium houstonense (99% 16S rRNA gene sequence identity), respectively (hereafter, named as Halomonas sp. ATBC28 and Mycobacterium sp. DBP42), were selected for further genomic characterization based on their diverse phylogenetic origin (i.e., Gammaproteobacteria and Actinomycetales, respectively) as well as their robust growth on and consumption of a variety of plasticizers (Table 2 and Figure 1; growth on all six plasticizers as well as phthalate is found in Supporting Information Figure S5). Phthalate was toxic to both microbes at 0.1% (w/v). Toxicity testing showed that the maximum concentration of phthalate that either isolate could grow with was 0.02% (w/v; Supporting Information Figure S4). Interestingly, the addition of 0.1% DBP to the cultures (an inducer of the biodegradation pathways as shown below) increased the toxicity of phthalate possibly by solubilizing the crystals as assessed visually.
Table 2. Ten Isolates That Showed the Best Growth on a Wide Range of Plasticizers.
Isolates were identified through partial sequencing of their 16S rRNA gene and subsequent BLAST searches of the NCBI database with this sequence.
+ indicates growth on the corresponding substrate, +++ indicates strong growth, – indicates no growth, w indicates weak growth, and s indicates that there was evidence that a surfactant was produced, but no growth was obvious.
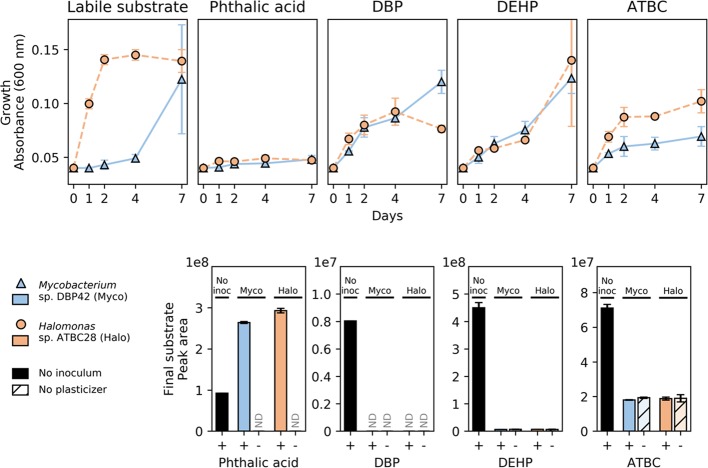
Figure 1.
Growth (top panels) and final substrate concentration (bottom panels) of the reference labile substrate (0.1% w/v pyruvate and 0.1% v/v glycerol, respectively, as tested in Supporting Information Figure S3) and three different plasticizers (0.1% v/v), as well as phthalate (0.02% w/v) by Mycobacterium sp. DBP42 and Halomonas sp. ATBC28. Points and error bars show means and standard deviations, respectively, of three (top) and two (bottom) biological replicates. Substrate consumption graphs also show controls with either no bacterial inoculum (solid black bars) or no plasticizer (“-“; colored bars with hatched lines). ND denotes that the compound was below the limit of detection. Growth of Mycobacterium sp. DBP42 and Halomonas sp. ATBC28 on three additional plasticizers, DINP, DIDP, and TOTM, is shown in Supporting Information Figure S5.
A subset of three of the plasticizers were chosen to further characterize the mechanisms of plasticizer degradation by Mycobacterium sp. DBP42 and Halomonas sp. ATBC28: DBP and DEHP, two of the most abundantly used PAE plasticizers,12 both with differing chain lengths and degrees of branching, and ATBC, a less toxic plasticizer for which no biodegradation pathway has been described to date. Curiously, while the plasticizers DBP, DEHP, and ATBC were depleted when incubated in the presence of both microorganisms (i.e., DBP was degraded to values below the limit of detection, and DEHP and ATBC to the background values obtained when these substrates were not added to the cultures; Figure 1), phthalate detection increased by the end of the incubation period, possibly due to the increase in solubilization of the phthalate crystals in the presence of the microbes.
Genome Analysis of Halomonas sp. ATBC28 and Mycobacterium sp. DBP42
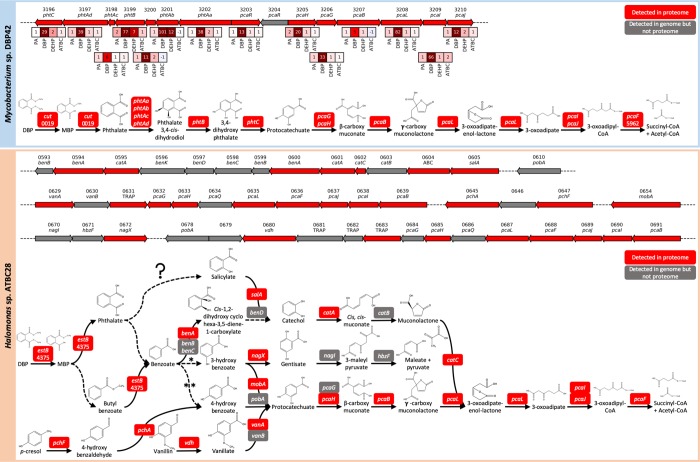
Genome sequencing revealed that Mycobacterium sp. DBP42 had a genome size of 6.52 Mb with 6316 coding sequences, 1581 of which were annotated as hypothetical proteins, and a GC content of 66.15%. Halomonas sp. ATBC28 had a genome size of 5.47 Mb, with 5111 coding sequences, 2367 of which were annotated as hypothetical proteins, and a GC content of 54.6%. As expected, the genomes of both organisms revealed a large potential for PAE biodegradation (Supporting Information Table S2): (i) both strains encoded genes involved in phthalate degradation and (ii) a number of esterases, cutinases, and lipases or enzymes from the β-oxidation of fatty acids, necessary for the removal of the ester side chains from the phthalate molecule, were also encoded. A comprehensive exo- and cellular proteomic (Supporting Information Tables S3–S6) and metabolomic analysis (Supporting Information Table S7) was performed with Mycobacterium sp. DBP42 and Halomonas sp. ATBC28 to ascertain the pathways and enzymes used for degrading the plasticizers DBP, DEHP, and ATBC, as summarized in Figure 2 and detailed below. Both isolates also encoded genes involved in the anaerobic degradation of phthalate (Supporting Information Table S2);50,51 however, they were either not detected or were not upregulated in the proteomes, as expected from the aerobic conditions of the cultures..
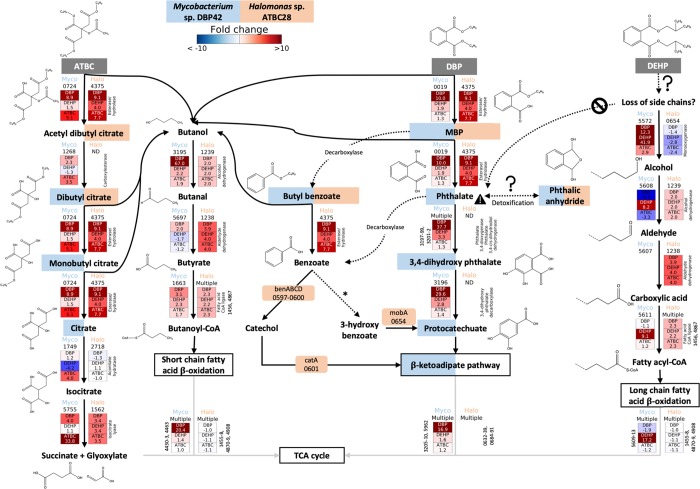
Figure 2.
Catabolic pathways informed by genomic, proteomic, and metabolomic analyses for DBP, DEHP, and ATBC degradation by Mycobacterium sp. DBP42 and Halomonas sp. ATBC28. Initial plasticizer substrates are shown in dark gray boxes, while degradation intermediates that were detected by metabolomics are shown with blue and orange boxes if they were present in the Mycobacterium sp. DBP42 (Myco) or Halomonas sp. ATBC28 (Halo) treatments, respectively. General multistep pathways are outlined in boxes. Dashed arrows show reactions inferred by metabolomics or KEGG degradation pathways, although no enzyme catalyzing the reaction could be confidently assigned by proteogenomics. Solid arrows indicate reactions catalyzed by detected enzymes. Enzyme ID number for both Mycobacterium sp. DBP42 and Halomonas sp. ATBC28 is shown for each reaction. The fold change in protein abundance in each treatment (DBP, DEHP, and ATBC vs labile substrate control, determined in Supporting Information Figure S3) is shown where this enzyme was detected in all three biological replicates of at least one treatment. All enzymes shown here were detected in the cellular proteome, aside from the isocitrate lyase 5755 in Mycobacterium sp. DBP42 and the esterase/hydrolase 4375 in Halomonas sp. ATBC28, which were detected in the exoproteome. Dashed lines in chemical structures indicate uncertainty on composition. * denotes that this step is described in KEGG, but no enzyme capable of this reaction is currently known. Full pathways are shown for Mycobacterium sp. DBP42 in Supporting Information Figure S6 and Halomonas sp. ATBC28 in Supporting Information Figure S7.
Proteomic and Metabolomic Identification of the Pathways Used by Mycobacterium sp. DBP42 for Plasticizer Degradation
For Mycobacterium sp. DBP42, it was confirmed by metabolomics that DBP is degraded through a sequential removal of the ester side chains, first producing monobutyl phthalate and then phthalate, as well as two butanol molecules (Figure 2 and Supporting Information Figure S6). The strong proteomic upregulation of the cutinase 0019 in the presence of DBP (i.e., 10.0-fold) suggests that this enzyme may be involved in the cleavage of these side chains. Phthalate is then converted to protocatechuate and further processed via the β-ketoadipate pathway, as indicated by the detection of the expected intermediates by targeted metabolomics (Figure 2 and Supporting Information Figure S6 and Table S7) and the upregulation of all proteins encoded within the gene cluster 3196-3210 in the presence of DBP (Figures 2 and 3). Interestingly, the metabolomic analysis flagged the possible accumulation of compounds derived from side reactions, that is, butyl benzoate and phthalic anhydride (Figure 2 and Supporting Information Table S7). While butyl benzoate may come as a consequence of monobutyl phthalate decarboxylation (and it is unknown whether it is then further processed into benzoate as this intermediate was not detected), phthalic anhydride may function as a detoxification strategy during the build-up of phthalate. Phthalic anhydride was previously detected during the degradation of the long-chain PAEs DINP and DIDP,29,30 as well as dipentyl phthalate.61 A dihydroxy acid dehydratase (3668) that could be responsible for catalyzing this reaction was slightly upregulated in both the presence of phthalate and DBP (i.e., 1.9-fold; Supporting Information Table S5); however, it is also possible that this could occur through a hydrolysis reaction for which a number of enzymes could be responsible or spontaneous decay of phthaloyl-CoA as suggested during anaerobic biodegradation of phthalate.42,49 The hydrolyzed DBP side chains, that is, two butanol molecules, are further metabolized through the action of alcohol and aldehyde dehydrogenases (3195 and 5697, respectively) and a fatty acid-CoA ligase (1663) before entering the short-chain fatty acid β-oxidation pathway (4429–4434), a pathway previously suggested,39 and for which all enzymes were strongly upregulated in this treatment (Figure 2).
Figure 3.
Gene clusters within the genomes of Mycobacterium sp. DBP42 and Halomonas sp. ATBC28 that are used for the degradation of phthalate and other related aromatic compounds. Arrows represent coding domain sequences that were detected in the genomes and proteomes (red) or were detected in the genomes but not the proteomes (gray). All genes are drawn to scale and arrows represent the direction of transcription. Numbers above the arrows indicate the gene ID within each genome. Boxes showing the fold change in protein abundance in phthalate (PA), DBP, DEHP, and ATBC treatments relative to the control growth (labile substrate) are shown for Mycobacterium sp. DBP42, where all proteins within this cluster were present in all three biological replicates in at least one treatment. Annotations are shown for ABC (ABC transporter), benA (benzoate/toluate 1,2-dioxygenase alpha subunit), benB (benzoate/toluate 1,2-dioxygenase beta subunit), benC (benzoate/toluate 1,2-dioxygenase reductase component), benD (dihydroxycyclohexadiene carboxylate dehydrogenase), benK (benzoate transport protein), catA (catechol 1,2-dioxygenase), catB (muconate cycloisomerase), catC (muconolactone D-isomerase), cut (cutinase), hbzF (maleyl pyruvate hydrolase), mobA (molybdenum cofactor guanyltransferase), nagI (gentisate 1,2-dioxygenase), nagX (3-hydroxybenzoate 6-monooxygenase), pcaB (3-carboxy-cis, cis-muconate cycloisomerase), pcaD (3-oxoadipate enol-lactonase), pcaF (histone acetyltransferase), pcaG (protocatechuate 3,4-dioxygenase alpha subunit), pcaH (protocatechuate 3,4-dioxygenase beta subunit), pcaI (3-oxoadipate CoA transferase alpha subunit), pcaJ (3-oxoadipate CoA transferase beta subunit), pcaL (3-oxoadipate enol-lactonase/4-carboxymuconolactone decarboxylase), pcaQ (pca operon transcriptional regulator), pcaR (pca operon regulatory protein), pchA (salicylate biosynthesis isochorismate synthase), pchF (pyochelin synthetase), phtAa (phthalate 3,4-dioxygenase alpha subunit), phtAb (phthalate 3,4-dioxygenase beta subunit), phtAc (phthalate 3,4-dioxygenase ferredoxin component), phtAd (phthalate 3,4-dioxygenase ferredoxin reductase component), phtB (phthalate 3,4-cis-dihydrodiol dehydrogenase), phtC (3,4-dihydroxyphthalate decarboxylase), pobA (p-hydroxybenzoate 3-monooxygenase), salA (salicylate 1-monooxygenase/salicylate hydroxylase), TRAP (TRAP transporter), vanA (vanillate monooxygenase), vanB (vanillate monooxygenase ferredoxin subunit), and vdh (vanillin dehydrogenase). Details of how the function of each protein was assigned are in Supporting Information Table S2 and full details of all proteins detected by proteomics are in Supporting Information Tables S3–S6.
Curiously, while the presence of DBP strongly induced the catabolic pathway for phthalate, this was not the case when Mycobacterium sp. DBP42 was grown in the presence of phthalate or DEHP (Figure 2). The transcriptional regulator encoded within the gene cluster (3203) increased 13.5-fold in the presence of DBP as opposed to just 1.2- to 1.4-fold in the phthalate, DEHP, and ATBC treatments (Figure 3). Furthermore, metabolic intermediates of phthalate and protocatechuate degradation were only detected in the DBP treatment and not in the presence of phthalate or DEHP. All of this evidence in conjunction with (i) the reduced growth of this strain with phthalate (Figure 1) and lack of growth at higher concentrations of phthalate, even when an alternative carbon source was supplied (Supporting Infomation Figure S4), (ii) the build-up of phthalic anhydride in the phthalate treatment (Supporting Infomation Table S7), and (iii) the higher relative abundance of proteins involved in DNA protection (5706; 18.0-fold increase), potential antigens (2974; 14.6-fold increase), and stress proteins (3075; 8.8-fold increase; Supporting Infomation Table S5) suggests that phthalate is toxic for this strain.
The degradation of DEHP by Mycobacterium sp. DBP42 did not seem to follow the same pathway as observed for DBP. The lack of detection of the expected degradation intermediates (e.g., mono(2-ethyl hexyl)phthalate, phthalate, or phthalic anhydride; Supporting Infomation Table S7) and lower abundance of the cutinase 0019, involved in hydrolyzing the ester side chains, suggest that DEHP degradation does not occur via sequential cleavage of ester side chains. The data indicate that degradation may occur as suggested previously,40 that is, via the β-oxidation of the fatty acid side chains directly on the DEHP molecule. In fact, enzymes from the long-chain fatty acid β-oxidation pathway were strongly induced by the presence of DEHP (an average of 17.2-fold; Figure 2 and Supporting Infomation Figure S6). We hypothesize that DEHP degradation is initiated through hydroxylation of the ester side chains by a monooxygenase (5572; 42-fold protein increase in the DEHP treatment) and enters the β-oxidation pathway as suggested in Figure 2.
ATBC degradation in Mycobacterium sp. DBP42 occurs through a sequential removal of the ester side chains, generating citrate that is then funneled into the central metabolism (Figure 2). The metabolomic analysis confirmed the presence of the ATBC degradation intermediates dibutyl citrate, monobutyl citrate, and citrate (Supporting Information Table S7 and Figure 2). This suggests that ATBC degradation initiates by the removal of acetic acid and butanol, possibly by the carboxylesterase 1268 (3.5-fold increase) or hydrolase 0724 (5.1-fold increase), respectively. As indicated in Figure 2, we tentatively suggest that acetyl dibutyl citrate is generated first because this intermediate was detected in the Halomonas sp. ATBC28 metabolome (see below). The other two butanol molecules are then successively removed, presumably by the hydrolase 0724, to produce citrate. While the degradation pathway for butanol was not highly upregulated in the presence of ATBC, the two-step conversion of citrate to succinate and glyoxylate was shown in Figure 2. Curiously, the isocitrate lyase (5755), presumably responsible for the conversion of isocitrate to succinate and glyoxylate, with no prediction for secretion,62 was detected in high abundance in the exoproteome of Mycobacterium sp. DBP42 only in this treatment (24.7% of the exoproteome and 33.8-fold upregulated; Supporting Information Table S5).
Proteomic and Metabolomic Identification of the Pathways Used by Halomonas sp. ATBC28 for Plasticizer Degradation
The metabolic pathways used for the degradation of plasticizers and phthalate were less clear in Halomonas sp. ATBC28 (Figure 2). Interestingly, this bacterium encodes an array of enzymes predicted to be involved in the biodegradation of many aromatic compounds, as well as numerous transporters, within a tight genomic cluster (i.e., genes 0593-0691; Figure 3 and Supporting Information Figures S7 and S8 and Table S2), many of which were detected by proteomics (Supporting Information Tables S4 and S6). Also, in contrast to Mycobacterium sp. DBP42, Halomonas sp. ATBC28 did not build up any detectable degradation intermediates in the culture supernatant and, hence, we were unable to confirm the exact pathway used to degrade phthalate and phthalate-based plasticizers (Figure 2 and Supporting Information Table S7). Nonetheless, based on the genomic and proteomic data for Halomonas sp. ATBC28, we propose that the hydrolysis of the butyl side chains of DBP is catalyzed by the esterase 4375, a serine hydrolase with a signal peptide for secretion,62 which showed a 9.1-fold increase in the exoproteome under this treatment (Figure 2). The esterase 4375 also showed an increased abundance in the presence of other plasticizers (4- and 7.7-fold in the DEHP and ATBC treatments, respectively), suggesting that it may also be induced by other plasticizers. As observed for Mycobacterium sp. DBP42, the side chains of the plasticizers are likely to enter the β-oxidation pathway for fatty acid degradation; however, in Halomonas sp. ATBC28, there was no clear differentiation between the short- and long-chain pathways when growing with either DBP or DEHP (Figure 2). DBP is possibly catabolized via benzoate, as shown in Figure 2 and previously suggested.63−65 Benzoate could then be processed via three different pathways (Figure 3): (1) it is converted to maleate and pyruvate via 3-hydroxy benzoate, gentisate, and 3-maleyl pyruvate, although no enzyme has been described in KEGG for the conversion of benzoate to 3-hydroxy benzoate, (2) it follows the same pathway for protocatechuate degradation as seen in Mycobacterium sp. DBP42, via an initial conversion of the benzoate to either 3- (as above) or 4-hydroxy benzoate (by a cytochrome P450 monooxygenase; not detected in the genome), and then protocatechuate, or (3) it is converted to cis-1,2-dihydroxycyclohexa-3,5-diene-1-carboxylate and then catechol by benzoate 1,2-dioxygenases and dihydroxycyclohexadiene carboxylate dehydrogenase after which it is funneled back into the β-ketoadipate pathway via cis,cis-muconate and muconolactone. There are genes coding for almost all of these steps in Halomonas sp. ATBC28 and most were detected by proteomics (Figure 3) although in similar abundances to the reference culture (grown with pyruvate).
For ATBC, the intermediates acetyl dibutyl citrate and dibutyl citrate were detected in the metabolome of Halomonas sp. ATBC28 in this treatment (Figure 2), confirming the ability of this strain to metabolize ATBC. We again suggest esterase 4375 to be involved in hydrolyzing the side chains of ATBC, although the promiscuity of this enzyme for other substrates requires further confirmation.
The strong increase of 20 transporters and efflux pumps in Halomonas sp. ATBC28 in one or more of the treatments, when compared with growth on pyruvate (Supporting Information Figure S8 and Supporting Information Table S6), is also remarkable. The tripartite ATP-independent periplasmic (TRAP) transporter 0631, which is encoded within the phthalate degradation gene cluster (Figure 3), was strongly upregulated in the exoproteomes of all treatments, and a second TRAP transporter (0264) was strongly upregulated in the cellular proteomes of the plasticizer treatments (Supporting Information Figure S8). Also, upregulated more strongly in the plasticizer treatments were a cation/acetate symporter previously suggested to be involved in phthalate degradation27 and a sodium/hydrogen ion antiporter (2103). The strong upregulation of these proteins in Halomonas sp. ATBC28 opens the question as to whether these transporters are maybe exporting phthalate to reduce its cellular toxicity or importing the cleaved side chains or other intermediates into the cell. Other interesting cellular responses of Halomonas sp. ATBC28 to the presence of the different plasticizers were the strong increase in iron acquisition (e.g., bacterioferritin 2469 and three different iron binding transport systems) and an extracellular superoxide dismutase (4604) to deal with oxidative stress (Supporting Information Table S6). We also found an ompA-like protein (3348; Supporting Information Table S2) that is strongly upregulated in the exoproteomes of the phthalate, DBP, DEHP, and ATBC treatments (49, 204, 90, and 264-fold increases, respectively; Supporting Information Figure S8 and Table S6) that could be used as a surfactant to solubilize the plasticizers and aid in their detoxification, as previously suggested.66
Discussion
The microbial isolation and integrated multi-OMIC characterization that we present here using model conditions has allowed a comprehensive understanding of plasticizer biodegradation in bacteria obtained from marine plastic debris. Through the enrichment and isolation of 42 bacterial isolates capable of plasticizer degradation we found that, while the ability to grow using PAE plasticizers (e.g., DBP and DEHP) was fairly widespread, surprisingly, ATBC was the plasticizer that the lowest number of isolates was able to use for growth (Table 2; Supporting Information Table S1). ATBC is sold as a biodegradable and less toxic alternative to DEHP and other PAE plasticizers;19,21 however, to our knowledge, there are no studies that have reported microbial degradation of ATBC or identified the enzymes involved. The side chains of the ATBC molecules can hydrolyze spontaneously; however, this only occurs at high temperatures and ATBC has a predicted half-life of 2 years under the conditions applied here, that is, pH 7.67 Here, we suggest that ATBC side-chain hydrolysis is facilitated by encoded hydrolases in a reduced number of bacteria as only a small number of isolates could grow in the presence of this plasticizer. The use of ATBC in industry is more recent and it is therefore present in a smaller fraction of manufactured materials; consequently, environmental microbes have been exposed to this new compound for less time.
The biodegradation of most plasticizers requires an initial hydrolysis of the ester side chains, generating intermediates that are then funneled through known catabolic pathways into the central metabolism.23,37−39,41 The proteogenomic approach used here helped us to identify these esterases (i.e., cutinase 0019 and hydrolase 0724 for DBP and ATBC hydrolysis, respectively, in Mycobacterium sp. DBP42, and the possibly more promiscuous esterase 4375 in Halomonas sp. ATBC28) and to flag these enzymes as candidates for future biochemical characterization. Although cutinases are enzymes that were initially described for the hydrolysis of the plant polyester cutin,68 a previously identified cutinase-like enzyme was also shown to hydrolyze the PAE dipentyl phthalate to phthalate.61 Esterases acting on PAEs can catalyze a wide range of compounds, for example, p-nitrophenols61 and triacylglycerols,69 and hence, the promiscuity of the enzymes identified in this study is not surprising but needs to be further tested. While for DBP and ATBC, we observed a sequential removal of the ester-bound side chains by metabolomics (i.e., producing phthalate and citrate, respectively, as well as butanol), and DEHP presumably follows a sequential shortening of the side chains directly on the molecule by enzymes involved in the β-oxidation pathway for fatty acid degradation, as previously suggested.33
The use of two phylogenetically distinct bacteria to characterize the biodegradation of different plasticizers has highlighted the biological complexity behind the metabolic regulation of the different pathways. In Mycobacterium sp. DBP42, the enzymes for PAE degradation were only highly upregulated under the DBP treatment, suggesting that either DBP or monobutyl phthalate may act as inducers of this pathway. This stresses the fact that more microbes may be capable of biodegrading recalcitrant compounds, although their pathways might be silenced or only induced by very specific chemicals. The conversion of phthalate into protocatechuate is the most common pathway for phthalate degradation.41,42 Interestingly, in Mycobacterium sp. DBP42, we found these genes (i.e., phthalate dioxygenase and dihydroxy phthalate decarboxylase) clustered with the protocatechuate and benzoate degradation pathway (Figure 3), as reported previously in Mycobacterium vanbaalenii (M. vanbaalenii) PYR-1,70 although in M. vanbaalenii, the decarboxylase (responsible for the conversion of 3,4-dihydroxy phthalate to protocatechuate) was located elsewhere in the genome. The fact that we found that all genes clustered in Mycobacterium sp. DBP42 suggests a higher level of specialization in degrading this aromatic compound. Halomonas sp. ATBC28, which potentially catabolizes DBP via benzoate (Figure 2) and shows an enormous potential to degrade aromatic xenobiotic compounds condensed in a tight genomic cluster (Figure 3), does not show the strong gene regulation observed in Mycobacterium sp. DBP42, and most of these catabolic enzymes appear to be constitutively expressed. This raises the question of how both organisms would behave in the environment, where concentrations of these compounds may well be below the induction threshold.
There is relatively little data available on the concentrations of plasticizers in marine debris, but DBP and DEHP can be present in plastic waste at concentrations of up to 360 and 2700 μg g–1, respectively.71 The proportion of these PAEs that leach from the plastics and become bioavailable to microorganisms is not clear; however, DBP and DEHP are typically found in surface waters at concentrations of approximately <0.1 to 50 μg L–1 and have even been found at concentrations as high as 1.4 and 0.8 g L–1, respectively.72 Therefore, the plasticizer concentrations used in the study (0.1%; 1 g L–1) may well be within the upper limits found in polluted environments or at least those that microbes encounter at the surfaces of plastics containing them. Microbes enhance the solubilization of these compounds and may even facilitate their leaching from the materials that they colonize. In this study, we used standard laboratory conditions to reduce complexity and facilitate the understanding of the molecular mechanisms used by the marine isolates to degrade the plasticizers. Future research requires determining how these biodegrading organisms behave in complex biofilms, that is, in the presence of an array of nondegrading organisms and having access to more labile organic compounds produced by primary producers.
Phthalate and its isomers are known toxic compounds.41,73,74 Here, we show that phthalate is toxic to both organisms, even when an additional carbon source is present (Supporting Information Figure S4), but that there are a number of adaptations adopted by the two isolates to deal with phthalate toxicity, for example, via conversion into phthalic anhydride, as observed by metabolomics, or the production of stress response mechanisms and efflux pumps, as shown by proteomics. While phthalic anhydride was previously reported in other PAE biodegradation studies29,30,61 and is known to be less toxic than phthalic acid,75 the mechanisms or reasons for its production were not discussed in these studies. While it is possible that the dehydration of phthalate could occur spontaneously at high temperatures, many previous PAE degradation studies have not reported phthalic anhydride production.23,37−39 Carboxylic acid anhydride production usually requires the presence of a catalyst or an enzyme under the physiological conditions used here and, while we suggest a dihydroxy acid dehydratase as being responsible for phthalic anhydride production, this compound could also be produced by phthaloyl-CoA decay during the anaerobic degradation of phthalate,42,51 although the enzyme responsible for the production of phthaloyl-CoA (i.e., succinyl-CoA/phthalate-CoA transferase) was not found in either isolate (Supporting Information Tables S2, S5, and S6). This reaction deserves further biochemical characterization to confirm its involvement in phthalate detoxification.
Twenty active membrane transporters were upregulated in Halomonas sp. ATBC28 in the presence of one or more of the plasticizers, including two TRAP transporters (0264 and 0631), the latter of which is located within the phthalate degradation gene cluster (Figure 3). It is unknown though if these TRAP transporters are importing the ester side chains after removal from the phthalate and citrate molecules or the plasticizers themselves as suggested previously.26 The transporters used for importing PAEs are identified in very few studies to date26,27,70 and thus remain under studied.42 It is even possible that either the aforementioned TRAP or other transporters and efflux pumps76 (Supporting Information Figure S8 and Table S6) are being used as a detoxification mechanism, secreting the toxic phthalate out of the cell. In its environmental context, where these microbes grow in biofilms surrounded by other microorganisms, this transport may even promote an exchange of metabolites among the community and enhance the biodegradation of such compounds. Another protein that we believe could be involved in both detoxification and facilitating biodegradation is a membrane-linked ompA-like protein 3348, which was also strongly upregulated in the exoproteome of Halomonas sp. ATBC28 (Supporting Information Figure S8 and Table S6). OmpA proteins have been reported as active components of the biosurfactant alasan and implicated in the degradation of xenobiotic compounds such as alkanes in other Gammaproteobacteria.(77)
This study presents, for the first time, a comprehensive characterization of the plasticizer biodegrading potential extant in biofilms on marine plastic debris, from the enrichment and isolation of microbes to the proteogenomic and metabolomic analysis of degradation. The molecular analysis of the degradation of phthalate and three of the most widely used plasticizers (i.e., DBP, DEHP, and ATBC) in two newly isolated bacteria (i.e. Mycobacterium sp. DBP42 and Halomonas sp. ATBC28) has revealed (i) an array of esterases involved in the first steps of DBP and ATBC degradation, (ii) the use of different mechanisms for the removal of the ester side chains from different PAEs (DBP and DEHP), (iii) the complexity of induction of the catabolic pathways involved in the degradation of such compounds, and (iv) a number of strategies used by the microbes to deal with these toxic compounds. Noncovalently bound compounds linked to plastics are likely to be leached into the surrounding environment; microbial biofilms that grow on these materials are the first to encounter such chemicals and, as shown here, are likely to be able to catabolize them reducing their release into their aquatic surroundings.
Acknowledgments
We thank Maria del Mar Aguilo-Ferretjans for help with proteomics sample preparation, Danielle Sanger-Green for collecting the seawater from Portaferry, and the Christie-Oleza group (Vinko Zadjelovic, Audam Chhun and Gabriel Erni Cassola) for helpful discussions throughout the project. We also acknowledge technical support from the WPH Proteomic Facility at the University of Warwick and thank the Warwick Institute for Synthetic Biology for access to the GeneVac. Technical assistance in LC-Orbitrap-MS/MS data analysis provided by Dr. Gabriel Martorell and Dr. Rosa Gomila, both from the Scientific and Technical Center (SCT) of the University of the Balearic Islands, is also greatly appreciated.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.9b05228.
Supplementary files containing Figures S1–S8 (PDF)
Table S1, screening of isolates: details of all isolates, which treatment they were obtained from and whether they grew on the plasticizers (XLSX)
Table S2, genomic analysis of Halomonas sp. ATBC28 and Mycobacterium sp. DBP42: pathways identified in genomes, with all KEGG details and details of conserved domain searches for those identified here as being involved in plasticizer degradation (XLSX)
Table S3, Mycobacterium sp. DBP42 peptides and protein groups: all proteins from MaxQuant (XLSX)
Table S4, Halomonas sp. ATBC28 peptides and protein groups: all proteins from MaxQuant (XLSX)
Table S5, Mycobacterium sp. DBP42 proteomic analysis: relative abundances, t-tests, and details of all proteins suggested to be involved in degradation that are mentioned in the text (XLSX)
Table S6, Halomonas sp. ATBC28 proteomic analysis: relative abundances, t-tests, and details of all proteins suggested to be involved in degradation that are mentioned in the text (XLSX)
Table S7, metabolomic analyses: full results for targeted and untargeted metabolomics (XLSX)
Author Contributions
R.J.W. and J.C.-O. designed the study. R.J.W. performed all experiments and analysis with guidance from J.A.C.-O. and M.I.G. R.B. performed metabolomic analyses. R.J.W. and J.A.C.-O. wrote the first draft of the manuscript and all authors contributed substantially to revisions.
R.W. was supported by a Midlands Integrative Biosciences Training Partnership PhD scholarship via grant BB/M01116X/1. J.A.C.-O. by the National Environment Research Council Independent Research Fellowship NE/K009044/1, NERC research project NE/S005501/1, and Ramón y Cajal contract RYC-2017-22452 (funded by the Ministry of Science, Innovation and Universities, the National Agency of Research, and the European Social Fund). R.B. by the MINECO project CTM2015-70180-R (FEDER cofunding) and MIG by the European Research Council grant 638631.
The authors declare no competing financial interest.
Supplementary Material
References
- Barnes D. K. A.; Galgani F.; Thompson R. C.; Barlaz M. Accumulation and Fragmentation of Plastic Debris in Global Environments. Philos. Trans. R. Soc., B 2009, 364, 1985–1998. 10.1098/rstb.2008.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derraik J. G. B. The Pollution of the Marine Environment by Plastic Debris: A Review. Mar. Pollut. Bull. 2002, 44, 842–852. 10.1016/S0025-326X(02)00220-5. [DOI] [PubMed] [Google Scholar]
- Thompson R. C.; Moore C. J.; vom Saal F. S.; Swan S. H. Plastics, the Environment and Human Health: Current Consensus and Future Trends. Philos. Trans. R. Soc. B 2009, 364, 2153–2166. 10.1098/rstb.2009.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambeck J. R.; Geyer R.; Wilcox C.; Siegler T. R.; Perryman M.; Andrady A.; Narayan R.; Law K. L. Plastic Waste Inputs from Land into the Ocean. Science 2015, 347, 768–771. 10.1126/science.1260352. [DOI] [PubMed] [Google Scholar]
- Avio C. G.; Gorbi S.; Regoli F. Plastics and Microplastics in the Oceans: From Emerging Pollutants to Emerged Threat. Mar. Environ. Res. 2017, 128, 2–11. 10.1016/j.marenvres.2016.05.012. [DOI] [PubMed] [Google Scholar]
- Rochman C. M.; Hoh E.; Kurobe T.; Teh S. J. Ingested Plastic Transfers Hazardous Chemicals to Fish and Induces Hepatic Stress. Sci. Rep. 2013, 3, 3263. 10.1038/srep03263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuten E. L.; Saquing J. M.; Knappe D. R. U.; Barlaz M. A.; Jonsson S.; Björn A.; Rowland S. J.; Thompson R. C.; Galloway T. S.; Yamashita R.; et al. Transport and Release of Chemicals from Plastics to the Environment and to Wildlife. Phil. Trans. R. Soc. B 2009, 364, 2027–2045. 10.1098/rstb.2008.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato Y.; Isobe T.; Takada H.; Kanehiro H.; Ohtake C.; Kaminuma T. Plastic Resin Pellets as a Transport Medium for Toxic Chemicals in the Marine Environment. Environ. Sci. Technol. 2001, 35, 318–324. 10.1021/es0010498. [DOI] [PubMed] [Google Scholar]
- Groh K. J.; Backhaus T.; Carney-Almroth B.; Geueke B.; Inostroza P. A.; Lennquist A.; Leslie H. A.; Maffini M.; Slunge D.; Trasande L.; Warhurst A. M.; Muncke J. Overview of Known Plastic Packaging-Associated Chemicals and Their Hazards. Sci. Total Environ. 2019, 651, 3253–3268. 10.1016/j.scitotenv.2018.10.015. [DOI] [PubMed] [Google Scholar]
- Bakir A.; Rowland S. J.; Thompson R. C. Competitive Sorption of Persistent Organic Pollutants onto Microplastics in the Marine Environment. Mar. Pollut. Bull. 2012, 64, 2782–2789. 10.1016/j.marpolbul.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Hahladakis J. N.; Velis C. A.; Weber R.; Iacovidou E.; Purnell P. An Overview of Chemical Additives Present in Plastics: Migration, Release, Fate and Environmental Impact during Their Use, Disposal and Recycling. J. Hazard. Mater. 2018, 344, 179–199. 10.1016/j.jhazmat.2017.10.014. [DOI] [PubMed] [Google Scholar]
- plasticizers.org
- Staples C. A.; Peterson D. R.; Parkerton T. F.; Adams W. J. The Environmental Fate of Phthalate Esters: A Literature Review. Chemosphere 1997, 35, 667–749. 10.1016/S0045-6535(97)00195-1. [DOI] [Google Scholar]
- Rani M.; Shim W. J.; Han G. M.; Jang M.; Al-Odaini N. A.; Song Y. K.; Hong S. H. Qualitative Analysis of Additives in Plastic Marine Debris and Its New Products. Arch. Environ. Contam. Toxicol. 2015, 69, 352–366. 10.1007/s00244-015-0224-x. [DOI] [PubMed] [Google Scholar]
- Paluselli A.; Fauvelle V.; Galgani F.; Sempéré R. Phthalate Release from Plastic Fragments and Degradation in Seawater. Environ. Sci. Technol. 2018, 53, 166–175. 10.1021/acs.est.8b05083. [DOI] [PubMed] [Google Scholar]
- Savoca D.; Arculeo M.; Barreca S.; Buscemi S.; Caracappa S.; Gentile A.; Persichetti M. F.; Pace A. Chasing Phthalates in Tissues of Marine Turtles from the Mediterranean Sea. Mar. Pollut. Bull. 2018, 127, 165–169. 10.1016/j.marpolbul.2017.11.069. [DOI] [PubMed] [Google Scholar]
- Vered G.; Kaplan A.; Avisar D.; Shenkar N. Using Solitary Ascidians to Assess Microplastic and Phthalate Plasticizers Pollution among Marine Biota: A Case Study of the Eastern Mediterranean and Red Sea. Mar. Pollut. Bull. 2019, 138, 618–625. 10.1016/j.marpolbul.2018.12.013. [DOI] [PubMed] [Google Scholar]
- Mathieu-Denoncourt J.; Wallace S. J.; de Solla S. R.; Langlois V. S. Plasticizer Endocrine Disruption: Highlighting Developmental and Reproductive Effects in Mammals and Non-Mammalian Aquatic Species. Gen. Comp. Endocrinol. 2015, 219, 74–88. 10.1016/j.ygcen.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Erceg M.; Kovačić T.; Klarić I. Thermal Degradation of Poly(3-Hydroxybutyrate) Plasticized with Acetyl Tributyl Citrate. Polym. Degrad. Stab. 2005, 90, 313–318. 10.1016/j.polymdegradstab.2005.04.048. [DOI] [Google Scholar]
- Van Vliet E. D. S.; Reitano E. M.; Chhabra J. S.; Bergen G. P.; Whyatt R. M. A Review of Alternatives to Di (2-Ethylhexyl) Phthalate-Containing Medical Devices in the Neonatal Intensive Care Unit. J. Perinatol. 2011, 31, 551–560. 10.1038/jp.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baiardo M.; Frisoni G.; Scandola M.; Rimelen M.; Lips D.; Ruffieux K.; Wintermantel E. Thermal and Mechanical Properties of Plasticized Poly(L-Lactic Acid ). J. Appl. Polym. Sci. 2003, 90, 1731–1738. 10.1002/app.12549. [DOI] [Google Scholar]
- Labrecque L. V.; Kumar R. A.; Davé V.; Gross R. A.; McCarthy S. P. Citrate Esters as Plasticizers for Poly(Lactic Acid). J. Appl. Polym. Sci. 1997, 66, 1507–1513. . [DOI] [Google Scholar]
- Ahuactzin-Pérez M.; Tlecuitl-Beristain S.; García-Dávila J.; González-Pérez M.; Gutiérrez-Ruíz M. C.; Sánchez C. Degradation of Di(2-Ethyl Hexyl) Phthalate by Fusarium Culmorum: Kinetics, Enzymatic Activities and Biodegradation Pathway Based on Quantum Chemical Modelingpathway Based on Quantum Chemical Modeling. Sci. Total Environ. 2016, 566-567, 1186–1193. 10.1016/j.scitotenv.2016.05.169. [DOI] [PubMed] [Google Scholar]
- Feng L.; Liu H.; Cheng D.; Mao X.; Wang Y.; Wu Z.; Wu Q. Characterization and Genome Analysis of a Phthalate Esters-Degrading Strain Sphingobium Yanoikuyae SHJ. Biomed Res. Int. 2018, 2018, 1–8. 10.1155/2018/3917054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselsoe M.; Boysen S.; Iversen N.; Jørgensen L.; Murrell J. C.; Mcdonald I.; Radajewski S.; Thestrup H.; Roslev P. Degradation of Organic Pollutants by Methane Grown Microbial Consortia. Biodegradation 2005, 16, 435–448. 10.1007/s10532-004-4721-2. [DOI] [PubMed] [Google Scholar]
- Kumar V.; Sharma N.; Maitra S. S. Comparative Study on the Degradation of Dibutyl Phthalate by Two Newly Isolated Pseudomonas Sp. V21b and Comamonas Sp. 51F. Biotechnol. Rep. 2017, 15, 1–10. 10.1016/j.btre.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V.; Maitra S. S. Biodegradation of Endocrine Disruptor Dibutyl Phthalate (DBP) by a Newly Isolated Methylobacillus Sp. V29b and the DBP Degradation Pathway. 3 BioTech 2016, 6, 200. 10.1007/s13205-016-0524-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D.-W.; Zhang T.; Fang H. H. P.; He J. Phthalates Biodegradation in the Environment. Appl. Microbiol. Biotechnol. 2008, 80, 183–198. 10.1007/s00253-008-1548-5. [DOI] [PubMed] [Google Scholar]
- Park J.-M.; Kim M.; Yoon J.; Kobayashi F.; Iwasaka Y.; Hong C.-S.; Min J.; Kim Y.-H. Biodegradation of Diisodecyl Phthalate (DIDP) by Bacillus Sp. SB-007. J. Basic Microbiol. 2009, 49, S31–S35. 10.1002/jobm.200800297. [DOI] [PubMed] [Google Scholar]
- Park J.-M.; Jeon M.; Lim E.-S.; Um H.-J.; Kim Y.-C.; Min J.; Kim Y.-H. Biodegradation of a Phthalate Plasticizer, Di-Isononyl Phthalate (DINP), by Sphingobium Chungbukense. Curr. Microbiol. 2008, 57, 515–518. 10.1007/s00284-008-9232-7. [DOI] [PubMed] [Google Scholar]
- Xie H.-J.; Shi Y.-J.; Zhang J.; Cui Y.; Teng S.-X.; Wang S.-G.; Zhao R. Degradation of Phthalate Esters (PAEs) in Soil and the Effects of PAEs on Soil microcosm Activity. J. Chem. Technol. Biotechnol. 2010, 85, 1108–1116. 10.1002/jctb.2406. [DOI] [Google Scholar]
- Yuan S. Y.; Liu C.; Liao C. S.; Chang B. V. Occurrence and Microbial Degradation of Phthalate Esters in Taiwan River Sediments. Chemosphere 2002, 49, 1295–1299. 10.1016/S0045-6535(02)00495-2. [DOI] [PubMed] [Google Scholar]
- Li F.; Liu Y.; Wang D.; Zhang C.; Yang Z.; Lu S.; Wang Y. Biodegradation of Di-(2-Ethylhexyl) Phthalate by a Halotolerant Consortium LF. PLoS One 2018, 13, e0204324 10.1371/journal.pone.0204324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislauskienė R.; Rudenkov M.; Karvelis L.; Gasparavičiu̅tė R.; Meškienė R.; Časaitė V.; Meškys R. Analysis of Phthalate Degradation Operon from Arthrobacter Sp. 68b. Biologija 2011, 57, 45–54. 10.6001/biologija.v57i2.1828. [DOI] [Google Scholar]
- Chen J.; Li X.; Li J.; Cao J.; Qiu Z.; Zhao Q.; Xu C.; Shu W. Degradation of Environmental Endocrine Disruptor Di-2-Ethylhexyl Phthalate by a Newly Discovered Bacterium, Microbacterium Sp. Strain CQ0110Y. Appl. Microbiol. Biotechnol. 2007, 74, 676–682. 10.1007/s00253-006-0700-3. [DOI] [PubMed] [Google Scholar]
- Yue F.; Lishuang Z.; Jing W.; Ying Z.; Bangce Y. E. Biodegradation of Phthalate Esters by a Newly Isolated Acinetobacter Sp. Strain LMB-5 and Characteristics of Its Esterase. Pedosphere 2017, 27, 606–615. 10.1016/S1002-0160(17)60355-2. [DOI] [Google Scholar]
- Yang T.; Ren L.; Jia Y.; Fan S.; Wang J.; Wang J.; Nahurira R.; Wang H.; Yan Y. Biodegradation of Di-(2-Ethylhexyl) Phthalate by Rhodococcus Ruber YC-YT1 in Contaminated Water and Soil. Int. J. Environ. Res. Public Health 2018, 15, 964. 10.3390/ijerph15050964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng N.-X.; Yu J.; Mo C.-H.; Zhao H.-M.; Li Y.-W.; Wu B.-X.; Cai Q.-Y.; Li H.; Zhou D.-M.; Wong M.-H. Biodegradation of Di-n-Butyl Phthalate (DBP) by a Novel Endophytic Bacillus Megaterium Strain YJB3. Sci. Total Environ. 2018, 616-617, 117–127. 10.1016/j.scitotenv.2017.10.298. [DOI] [PubMed] [Google Scholar]
- Ahuactzin-Pérez M.; Tlécuitl-Beristain S.; García-Dávila J.; Santacruz-Juárez E.; González-Pérez M.; Gutiérrez-Ruíz M. C.; Sánchez C. Kinetics and Pathway of Biodegradation of Dibutyl Phthalate by Pleurotus Ostreatus. Fungal Biol. 2018, 122, 991–997. 10.1016/j.funbio.2018.07.001. [DOI] [PubMed] [Google Scholar]
- Amir S.; Hafidi M.; Merlina G.; Hamdi H.; Jouraiphy A.; El Gharous M.; Revel J. C. Fate of Phthalic Acid Esters during Composting of Both Lagooning and Activated Sludges. Process Biochem. 2005, 40, 2183–2190. 10.1016/j.procbio.2004.08.012. [DOI] [Google Scholar]
- Vamsee-Krishna C.; Phale P. S. Bacterial Degradation of Phthalate Isomers and Their Esters. Indian J. Microbiol. 2008, 48, 19–34. 10.1007/s12088-008-0003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll M.; Geiger R.; Junghare M.; Schink B. Microbial Degradation of Phthalates: Biochemistry and Environmental Implications. Environ. Microbiol. Rep. 2019, 10.1111/1758-2229.12787. [DOI] [PubMed] [Google Scholar]
- Bushnell L. D.; Haas H. F. The Utilization of Certain Hydrocarbons by Microorganisms. J. Bacteriol. 1941, 41, 653–673. 10.1128/JB.41.5.653-673.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J. 16S/23S RRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt E., Goodfellow M., Eds.; Wiley: New York, 1991; pp 115–175. [Google Scholar]
- Wright R. J.; Gibson M. I.; Christie-Oleza J. A. Understanding Microbial Community Dynamics to Improve Optimal Microbiome Selection. Microbiome 2019, 7, 85. 10.1186/s40168-019-0702-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F.; Gish W.; Miller W.; Myers E. W.; Lipman D. J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Seemann T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Kanehisa M.; Sato Y.; Morishima K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Kanehisa M.; Furumichi M.; Tanabe M.; Sato Y.; Morishima K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghare M.; Spiteller D.; Schink B. Enzymes Involved in the Anaerobic Degradation of Ortho-Phthalate by the Nitrate-Reducing Bacterium Azoarcus Sp. Strain PA01. Environ. Microbiol. 2016, 18, 3175–3188. 10.1111/1462-2920.13447. [DOI] [PubMed] [Google Scholar]
- Mergelsberg M.; Egle V.; Boll M. Evolution of a Xenobiotic Degradation Pathway: Formation and Capture of the Labile Phthaloyl-CoA Intermediate during Anaerobic Phthalate Degradation. Mol. Microbiol. 2018, 108, 614–626. 10.1111/mmi.13962. [DOI] [PubMed] [Google Scholar]
- Christie-Oleza J. A.; Armengaud J. In-Depth Analysis of Exoproteomes from Marine Bacteria by Shotgun Liquid Chromatography-Tandem Mass Spectrometry: The Ruegeria Pomeroyi DSS-3 Case-Study. Mar. Drugs 2010, 8, 2223–2239. 10.3390/md8082223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A.; Hernandez-Fernaud J. R.; Aguilo-Ferretjans M. D. M.; Wellington E. M.; Christie-Oleza J. A. 100 Days of Marine Synechococcus–Ruegeria Pomeroyi Interaction: A Detailed Analysis of the Exoproteome. Environ. Microbiol. 2018, 20, 785–799. 10.1111/1462-2920.14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie-Oleza J. A.; Scanlan D. J.; Armengaud J. “You Produce While I Clean up”, a Strategy Revealed by Exoproteomics during Synechococcus–Roseobacter Interactions. Proteomics 2015, 15, 3454–3462. 10.1002/pmic.201400562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.; Mann M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Tyanova S.; Temu T.; Sinitcyn P.; Carlson A.; Hein M. Y.; Geiger T.; Mann M.; Cox J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat. Methods 2016, 13, 731–740. 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A.; Bo Y.; Han L.; He J.; Lanczycki C. J.; Lu S.; Chitsaz F.; Derbyshire M. K.; Geer R. C.; Gonzales N. R.; Gwadz M.; Hurwitz D. I.; Lu F.; Marchler G. H.; Song J. S.; Thanki N.; Wang Z.; Yamashita R. A.; Zhang D.; Zheng C.; Geer L. Y.; Bryant S. H. CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures. Nucleic Acids Res. 2017, 45, D200–D203. 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington J. J. Octanol/Water Partition Coefficients and Water Solubilities of Phthalate Esters. J. Chem. Eng. Data 1999, 44, 1414–1418. 10.1021/je990149u. [DOI] [Google Scholar]
- Bui T. T.; Giovanoulis G.; Cousins A. P.; Magnér J.; Cousins I. T.; de Wit C. A. Human Exposure, Hazard and Risk of Alternative Plasticizers to Phthalate Esters. Sci. Total Environ. 2016, 541, 451–467. 10.1016/j.scitotenv.2015.09.036. [DOI] [PubMed] [Google Scholar]
- Cousins I.; Mackay D. Correlating the Physical–Chemical Properties of Phthalate Esters Using the ‘three Solubility’ Approach. Chemosphere 2000, 41, 1389–1399. 10.1016/S0045-6535(00)00005-9. [DOI] [PubMed] [Google Scholar]
- Ahn J.-Y.; Kim Y.-H.; Min J.; Lee J. Accelerated Degradation of Dipentyl Phthalate by Fusarium Oxysporum f. Sp. Pisi Cutinase and Toxicity Evaluation of Its Degradation Products Using Bioluminescent Bacteria. Curr. Microbiol. 2006, 52, 340–344. 10.1007/s00284-005-0124-9. [DOI] [PubMed] [Google Scholar]
- Almagro Armenteros J. J.; Tsirigos K. D.; Sønderby C. K.; Petersen T. N.; Winther O.; Brunak S.; von Heijne G.; Nielsen H. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 2019, 37, 420–423. 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- Liu S. M.; Chi W. C. CO2–H2-Dependent Anaerobic Biotransformation of Phthalic Acid Isomers in Sediment Slurries. Chemosphere 2003, 52, 951–958. 10.1016/S0045-6535(03)00326-6. [DOI] [PubMed] [Google Scholar]
- Taylor B. F.; Ribbons D. W. Bacterial Decarboxylation of O-Phthalic Acids. Appl. Environ. Microbiol. 1983, 46, 1276–1281. 10.1128/AEM.46.6.1276-1281.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem R.; Hulshoff Pol L. W.; Lettinga G. Anaerobic Biodegradability of Phthalic Acid Isomers and Related Compounds. Biodegradation 1999, 10, 63–73. 10.1023/A:1008321015498.. [DOI] [PubMed] [Google Scholar]
- Shukla S. K.; Mangwani N.; Rao T. S.; Das S. Biofilm-Mediated Bioremediation of Polycyclic Aromatic Hydrocarbons; Das S., Ed.; Elsevier: Amsterdam, 2014, 10.1016/B978-0-12-800021-2.00008-X. [DOI] [Google Scholar]
- FR-MSCA . Analysis of the Most Appropriate Risk Management Option for Tributyl Citrate; ANSES, 2013. [Google Scholar]
- Carvalho C. M. L.; Aires-Barros M. R.; Cabral J. M. S. Cutinase Structure, Function and Biocatalytic Applications. Electron. J. Biotechnol. 1998, 1, 160–173. [Google Scholar]
- Martínez-Martínez M.; Lores I.; Peña-García C.; Bargiela R.; Reyes-Duarte D.; Guazzaroni M. E.; Peláez A. I.; Sánchez J.; Ferrer M. Biochemical Studies on a Versatile Esterase That Is Most Catalytically Active with Polyaromatic Esters. Microb. Biotechnol. 2014, 7, 184–191. 10.1111/1751-7915.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingley R. L.; Brezna B.; Khan A. A.; Cerniglia C. E. Novel Organization of Genes in a Phthalate Degradation Operon of Mycobacterium Vanbaalenii PYR-1. Microbiology 2004, 150, 3749–3761. 10.1099/mic.0.27263-0. [DOI] [PubMed] [Google Scholar]
- Pivnenko K.; Eriksen M. K.; Martín-Fernández J. A.; Eriksson E.; Astrup T. F. Recycling of Plastic Waste: Presence of Phthalates in Plastics from Households and Industry. Waste Manage. 2016, 54, 44–52. 10.1016/j.wasman.2016.05.014. [DOI] [PubMed] [Google Scholar]
- Net S.; Sempéré R.; Delmont A.; Paluselli A.; Ouddane B. Occurrence, Fate, Behavior and Ecotoxicological State of Phthalates in Different Environmental Matrices. Environ. Sci. Technol. 2015, 49, 4019–4035. 10.1021/es505233b. [DOI] [PubMed] [Google Scholar]
- Fajardo C.; Guyot J. P.; Macarie H.; Monroy O. Inhibition of Anaerobic Digestion by Terephthalic Acid and Its Aromatic By Products. Water Sci. Technol. 1997, 36, 83–90. 10.2166/wst.1997.0578. [DOI] [Google Scholar]
- Zhang Y.-M.; Sun Y.-Q.; Wang Z.-J.; Zhang J. Degradation of Terephthalic Acid by a Newly Isolated Strain of Arthrobacter Sp. 0574. S. Afr. J. Sci. 2013, 109, 1–4. 10.1590/sajs.2013/20120019. [DOI] [Google Scholar]
- Bang D.-Y.; Lee I.-K.; Lee B.-M. Toxicological Characterization of Phthalic Acid. Toxicol. Res. 2011, 27, 191–203. 10.5487/TR.2011.27.4.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D.; Wang-Kan X.; Neuberger A.; van Veen H. W.; Pos K. M.; Piddock L. J. V.; Luisi B. F. Multidrug Efflux Pumps: Structure, Function and Regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- Schneiker S.; dos Santos V. A. P. M.; Bartels D.; Bekel T.; Brecht M.; Buhrmester J.; Chernikova T. N.; Denaro R.; Ferrer M.; Gertler C.; Goesmann A.; Golyshina O. V.; Kaminski F.; Khachane A. N.; Lang S.; Linke B.; McHardy A. C.; Meyer F.; Nechitaylo T.; Pühler A.; Regenhardt D.; Rupp O.; Sabirova J. S.; Selbitschka W.; Yakimov M. M.; Timmis K. N.; Vorhölter F. J.; Weidner S.; Kaiser O.; Golyshin P. N. Genome Sequence of the Ubiquitous Hydrocarbon-Degrading Marine Bacterium Alcanivorax Borkumensis. Nat. Biotechnol. 2006, 24, 997–1004. 10.1038/nbt1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.