Abstract
Natural products account for more than 50% of all small-molecule pharmaceutical agents currently in clinical use. However, low availability often becomes problematic when a bioactive natural product is promising to become a pharmaceutical or leading compound. Advances in synthetic biology and metabolic engineering provide a feasible solution for sustainable supply of these compounds. In this review, we have summarized current progress in engineering yeast cell factories for production of natural products, including terpenoids, alkaloids, and phenylpropanoids. We then discuss advanced strategies in metabolic engineering at three different dimensions, including point, line, and plane (corresponding to the individual enzymes and cofactors, metabolic pathways, and the global cellular network). In particular, we comprehensively discuss how to engineer cofactor biosynthesis for enhancing the biosynthesis efficiency, other than the enzyme activity. Finally, current challenges and perspective are also discussed for future engineering direction.
Subject Areas: Biological Sciences, Bioengineering, Metabolic Engineering, Biotechnology, Microbial Biotechnology
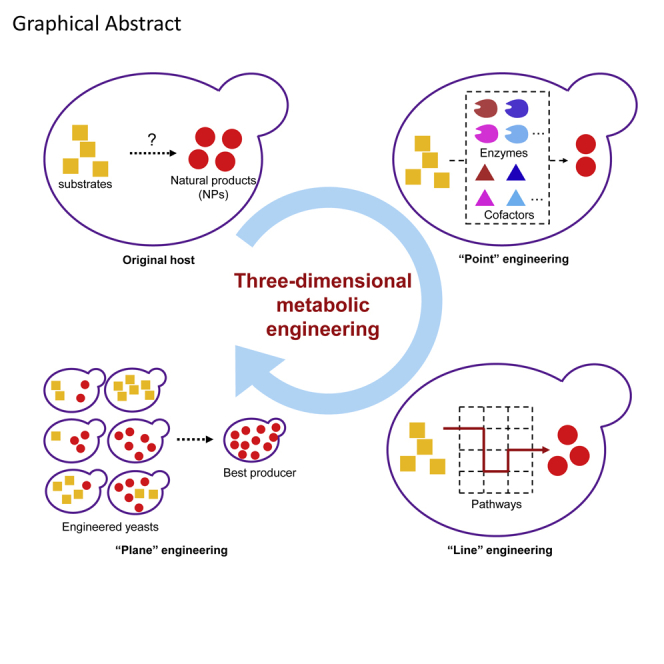
Graphical Abstract
Biological Sciences; Bioengineering; Metabolic Engineering; Biotechnology; Microbial Biotechnology
Introduction
Natural products, either directly or as inspiration, account for more than 50% of all small-molecule pharmaceutical agents in current clinical use (Patridge et al., 2016). By the end of 2013, natural products and derivatives possess about 38% of all US Food and Drug Administration-approved new molecular entities. However, the limited availability always becomes problematic when a bioactive natural product comes to a promising pharmaceutical (Atanasov et al., 2015). Metabolic engineering endows cells with the ability of overproduction of new products, which provides a feasible approach for supplying these precious molecules (Nielsen and Keasling, 2016).
Advances in synthetic biology accelerated metabolic engineering toward more complex synthesis and sustainable supply of limited natural resources in the past 10 years. Successful heterogeneous synthesis of artemisinic acid (Paddon et al., 2013) and resveratrol (Li et al., 2015a) highlighted the commercial feasibility of synthetic biology. Recently, the rapid development of synthetic biology enabled the microbial synthesis of more complex natural products, such as medicinal scutellarin (Liu et al., 2018b), alkaloids (Li et al., 2018, Srinivasan and Smolke, 2019, Stephanie et al., 2015), and cannabinoids (Luo et al., 2019). These successes were achieved through several rounds of “Design-Build-Test-Learn (DBTL),” in which a lot of knowledge was obtained for next round. However, several urgent challenges remain in advancing microbial engineering as a general approach for the biosynthesis of natural products, including (1) the difficulty in identification of biosynthetic pathways, (2) the lack of suitable environment for heterologous expression of enzyme with best activity, (3) the disruption of intracellular homeostasis with heterologous complex biosynthetic pathways, and (4) the low metabolic flux toward biosynthetic pathway.
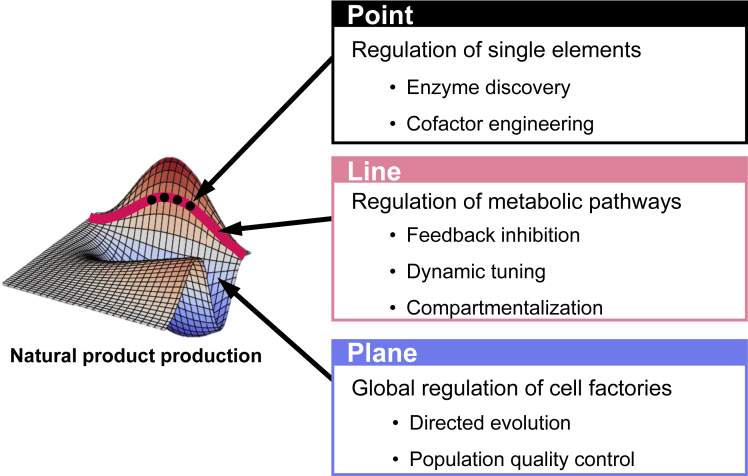
In this review, we first summarize the achievements made so far in the synthesis of natural products in yeast. Then we introduce the current advances in synthetic biology tools and metabolic engineering strategies at three dimensions (point, line, and plane), which are consistent with the general steps in building a cell factory. Point, line, and plane refer to individual optimization of engineering blocks (e.g., enzymes and cofactors), dynamic optimization of metabolic fluxes, and global optimization of cell robustness, respectively (Figure 1). In particular, we here discuss in detail the cofactor supply issue for retaining high activities of key enzymes other than their expression level. Meanwhile, we also present some perspectives to address the problems possibly encountered in the future.
Figure 1.
Metabolic Engineering Strategies for natural Product Production in Three Dimensions
Point, line, and plane represent the individual enzymes and cofactors, metabolic pathways, and the global cellular network, respectively.
Production of Natural Products in Yeast
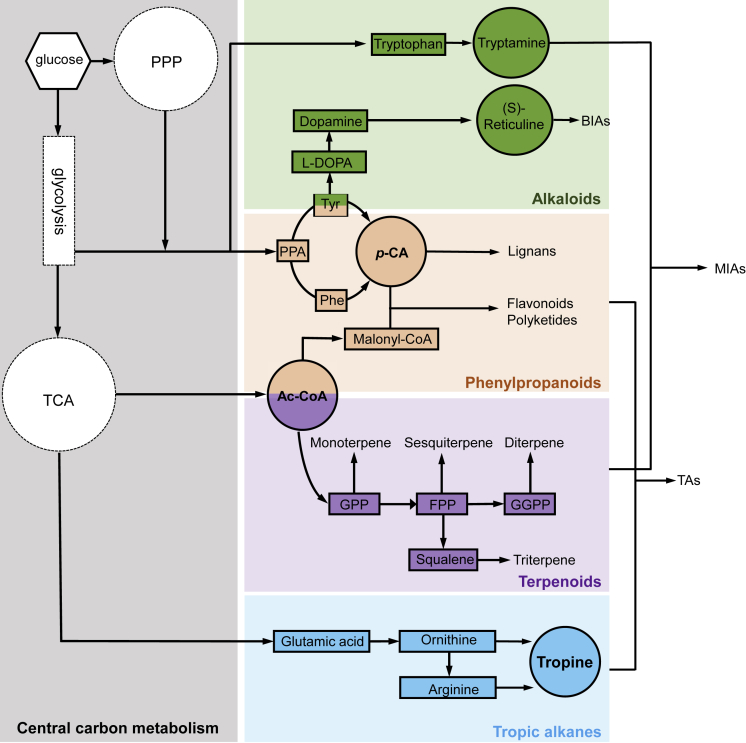
With the recent advances of functional genomics, metabolic engineering, systems and synthetic biology, a plenty of natural products or their precursors have been heterologously synthesized in microbes especially in the yeast cell factory. Here, we briefly introduce the yeast-based production of three types of most representative natural products: terpenoids, phenylpropanoids, and alkaloids (Figure 2).
Figure 2.
Simplified Biosynthetic Pathways for Three Types of Natural Product
Different colors represent different types of natural products. The colored circles mean key precursors, and the colored rectangles mean intermediates. TCA, tricarboxylic acid cycle; PPP, pentose phosphate pathway; PPA, prephenate; Ac-CoA, acetyl coenzyme A; malonyl-CoA, malonyl coenzyme A; Phe, phenylalanine; Tyr, tyrosine; p-CA, p-coumaric acid; FPP, farnesyl diphosphate; GPP, geranyl diphosphate; GGPP, geranylgeranyl pyrophosphate; BIAs, benzylisoquinoline alkaloids; MIAs, monoterpene indole alkaloids; TAs, tropane alkaloids.
Terpenoids
Terpenoids represent the largest class of natural products, which can be resources for pharmaceuticals, fragrances, and biofuels. The high-level production of artemisinic acid, the precursor of antimalarial artemisinin, showed the high potential of yeast Saccharomyces cerevisiae as a terpenoid cell factory (Paddon et al., 2013). The terpenoids have conserved skeletons with different amounts of isoprene units (C5H8), whose biosynthesis has been extensively reviewed elsewhere (Kirby and Keasling, 2009). We here classified the current progress in engineering yeast for overproduction of terpenoids into two parts, including (1) engineering central metabolism for sufficient supply of the key shared precursor acetyl-CoA and pyrophosphate precursors: isopentenyl pyrophosphate/dimethylallyl pyrophosphate (IPP/DMAPP), geranyl pyrophosphate (GPP) for monoterpene, farnesyl pyrophosphate (FPP) for sesquiterpene, geranylgeranyl pyrophosphate (GGPP) for diterpene, etc., and (2) engineering downstream decorating pathways for diverse functional terpenoids.
In S. cerevisiae, the main cytosolic acetyl-CoA was biosynthesized from pyruvate dehydrogenase (PDH) bypass pathway (or pyruvate decarboxylation pathway), which, however, is strongly competed by ethanol fermentation. Thus a bacterial PDH was overexpressed in S. cerevisiae for direct conversion of pyruvate into acetyl-CoA in the cytosol (Cardenas and Da Silva, 2016, Lian et al., 2014). However, the lack of cofactors such as lipoic acid (please refer to the section Engineering the Equilibrium State of Cofactors) for activating PDH (Lian and Zhao, 2016) limits the cytosolic PDH activity and the derived acetyl-CoA flux (Kozak et al., 2014). Alternatively, a non-native xylulose-5-phosphate-derived pathway was constructed for rewiring yeast central carbon metabolism to acetyl-CoA, which significantly boosted the farnesene production (130 g/L) with reduced requirement of ATP and oxygen (Meadows et al., 2016). The mitochondrial acetyl-CoA level is estimated to be 20- to 30-fold higher than that of the cytoplasm. Mitochondrial compartmentalization resulted >2-fold higher production of amorpha-4,11-diene and isoprene production than the cytosolic biosynthetic pathway (Lv et al., 2016, Yuan and Ching, 2016).
With regard to enhancing pyrophosphate precursor supply, the limiting steps of mevalonate (MVA) pathway were enhanced by overexpression of the catalyzing enzymes or corresponding mutants with higher activity. For example, a truncated tHMG1 or a variant HMG2K6R was overexpressed for enhancing the activity of rate-limiting 3-hydroxy-3-methylglutaryl-coenzyme reductase (HMG-R) (Ignea et al., 2015, Zhou et al., 2012). Furthermore, overexpression of ERG10 (encoding acetoacetyl-CoA thiolase), MK/ERG12 (encoding mevalonate kinase), and MVD1 (encoding mevalonate pyrophosphate decarboxylase) were shown to be helpful in enhancing MVA flux for terpene biosynthesis (Yao et al., 2018).
The 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway synthesizes IPP by condensation of the glycolysis intermediates glyceraldehyde-3-phosphate (G3P) and pyruvate, which has higher theoretical yield of IPP than that of the yeast MVA pathway (1 versus 0.66 mol IPP per mol glucose) (Li et al., 2010). However, the lack of cytosolic [4Fe-4S] clusters for functional MEP key enzymes 1-hydroxy-2-methyl-butenyl-4-diphosphate (HMBPP) synthase (IspG) and HMBPP reductase (IspH), makes it challenging to reconstruct fully functional MEP pathway in yeast cytosol (Kirby et al., 2016). Alternatively, non-natural isoprenoid alcohol (IPA) pathway (Clomburg et al., 2019) and isopentenol utilization pathway (Chatzivasileiou et al., 2019) enabled biosynthesis of IPP and DMAPP without requiring the [4Fe-4S] clusters, which, however, have not been verified in yeast yet.
Monoterpene overproduction in S. cerevisiae is always challenging, because there is no specific GPP synthase. In S. cerevisiae, GPP is synthesized by a bifunctional enzyme ERG20p (farnesyl diphosphate synthase [FPPS]) and is rapidly utilized toward FPP by ERG20p, which limits the C10 precursor pool. Enzyme engineering created ERG20p mutants for specific GPP synthase activity (Fischer et al., 2011, Ignea et al., 2014), which, however, hampered cell growth as the normal ERG20p is essential for the biosynthesis of essential cellular components. Construction of an orthogonal neryl diphosphate biosynthetic pathway enabled high-level production of monoterpenes without retarding cell growth (Cheng et al., 2019, Ignea et al., 2019). Recently, a plethora of C11 terpenes were synthesized in S. cerevisiae by construction of chimeric pathway harboring GPP methyltransferase and monoterpene synthase mutants that prefer C11 pyrophosphate (Ignea et al., 2019). This non-natural terpene biosynthesis can expand chemical space and diversity of natural products for pharmaceutical development (Zhou, 2018).
Once terpene scaffolds are produced, multiple decoration steps, including oxidation, glycosyl transfer, and methyl transfer, work together to produce structurally diverse terpenoids (Kirby and Keasling, 2008). The yeasts have been harnessed for heterologous synthesis of a variety of bioactive terpenoids, such as limonene, artemisinic acid, hydrocortisone, glycyrrhetinic acid, and ginsenosides (Table 1). Although systems biology approach can narrow the target genes, it is still challenging in identification and characterization of decoration enzymes. The yeast cells can be used for characterizing the enzymes in the biosynthesis of natural products such as tashinones (Guo et al., 2013, Guo et al., 2016) and ginsenosides (Yan et al., 2014) because of their clear backgrounds.
Table 1.
Biosynthesis of Terpenoids, Alkaloids, and Phenylpropanoids in Yeasts
| Compound Name | Activity | Host | Cultivation Condition | Titer (mg/L) | Reference |
|---|---|---|---|---|---|
| Terpenoids | |||||
| Limonene | Flavors, antibacterial and insecticide | S. cerevisiae | Semi-batch, CM | 166 | (Ignea et al., 2019) |
| Sabinene | Flavors, antibacterial and insecticide | S. cerevisiae | Semi-batch, CM | 113 | (Ignea et al., 2019) |
| Geraniol | Antimicrobial and antitumor | S. cerevisiae | Fed-batch, CM | 1680 | (Jiang et al., 2017) |
| α-Terpineol | Flavors and anti-fungal | S. cerevisiae | Fed-batch, CM | 21.9 | (Zhang et al., 2019a) |
| Artemisinic acid | Antimalarial | S. cerevisiae | Fed-batch, MM + glucose/ethanol | 25,000 | (Paddon et al., 2013) |
| Valencene | Flavors and fragrances | S. cerevisiae | Fed-batch, MM | 539 | (Chen et al., 2019a) |
| Valerenic acid | Anxiolytic and sedative | S. cerevisiae | Shake flask, CM | 4.0 | (Wong et al., 2018) |
| Patchoulol | Neuroprotection, anti-inflammatory, and anti-cancer | S. cerevisiae | Fed-batch, CM + glucose/glycerol/ethanol | 466.8 | (Ma et al., 2019a) |
| Sclareol | Antibacterial and fragrances | S. cerevisiae | Shake flask, MM | 403 | (Ignea et al., 2015) |
| Miltiradiene | Antibacterial, anti-inflammatory, and anti-cancer | S. cerevisiae | Fed-batch, SMM | 365 | (Zhou et al., 2012) |
| Hydrocortisone | Anti-inflammatory | S. cerevisiae | Shake flask, CM | 1060 | (Chen et al., 2019b) |
| Carnosic acid | Antioxidation, anti-inflammatory, and anti-tumor | S. cerevisiae | Shake flask, MM | 2.7 | (Scheler et al., 2016) |
| Ginsenoside Rh2 | Cancer prevention and therapy | S. cerevisiae | Fed-batch, CM | 2250 | (Wang et al., 2019a) |
| Ginsenoside Rg3 | Anti-tumor and anti-cancer | S. cerevisiae | Shake flask, CM | 49.8 | (Wang et al., 2015) |
| Glycyrrhetinic acid | Anti-viral, hepatoprotective, anti-allergic and antiulcer | S. cerevisiae | Fed-batch, CM | 18.9 | (Zhu et al., 2018) |
| Lycopene | Anti-tumor, cardiovascular protection, anti-cancer, antioxidation | S. cerevisiae | Fed-batch, CM + glucose/ethanol | 2370 | (Ma et al., 2019b) |
| (+)-Nootkatone | Anti-platelet aggregation and anti-proliferative | K. phaffii | Fed-batch, MM | 208 | (Wriessnegger et al., 2014) |
| (+)-Ambrein | Anti-nociceptive and aphrodisiac | K. phaffii | Fed-batch, MM | 105 | (Moser et al., 2018) |
| Lycopene | Anti-tumor, cardiovascular protection, anti-cancer, antioxidation | K. phaffii | Fed-batch, MM | 714 | (Zhang et al., 2019c) |
| α-Santalene | Antibacterial and diuretic | Y. lipolytica | Fed-batch, CM | 27.9 | (Jia et al., 2019) |
| Limonene | Flavors, antibacterial and insecticide | Y. lipolytica | Shake flask, CM | 23.6 | (Cao et al., 2016) |
| (+)-Nootkatone | Anti-platelet aggregation and anti-proliferative | Y. lipolytica | Shake flask, CM | 0.98 | (Guo et al., 2018) |
| Protopanaxadiol | Anti-cancer, anti-tumor, anti-viral, and antibiotic | Y. lipolytica | Fed-batch, CM + xylose | 301 | (Wu et al., 2019) |
| Campesterol | Anti-inflammatory | Y. lipolytica | Fed-batch, CM | 453 | (Du et al., 2016) |
| Ginsenoside K | Antitumor and anti-inflammatory | Y. lipolytica | Fed-batch, CM | 162 | (Li et al., 2019a) |
| Astaxanthin | Antioxidant | Y. lipolytica | Fed-batch, CM | 285 | (Tramontin et al., 2019) |
| Alkaloids | |||||
| Opioids (thebaine, hydrocodone) | Pain killer | S. cerevisiae | Shake flask, MM | 0.3–6.4*10−3 | (Galanie et al., 2015) |
| Noscapine | Anti-cancer and anti-tussive | S. cerevisiae | Shake flask, CM | 2.2 | (Li et al., 2018) |
| Strictosidine | Anti-cancer | S. cerevisiae | Shake flask, MM | 0.5 | (Brown et al., 2015) |
| Tropine | Treatment for neurological disorder | S. cerevisiae | Fed-batch, SM | 6.0 | (Srinivasan and Smolke, 2019) |
| Pseudotropine | Anticholinergic | S. cerevisiae | Shake flask, CM | 0.08 | (Ping et al., 2019) |
| Betanin | Food dye and spectrofluorometric probes | S. cerevisiae | Shake flask, MM | 16.8 | (Grewal et al., 2018) |
| Phenylpropanoids | |||||
| Resveratrol | Nutraceutical and antioxidation | S. cerevisiae | Fed-batch, MM + ethanol | 531 | (Li et al., 2015a) |
| Scutellarin | Cardio- and cerebrovascular diseases prevention | S. cerevisiae | Fed-batch, MM | 108 | (Liu et al., 2018b) |
| Anthocyanin (pelargonidin-3-O-glucoside, cyanidin-3-O-glucoside, delphinidin-3-O-glucoside) | Antioxidation, neuroprotection, vision improvement, cardiovascular protection, antidiabetic, anti-inflammatory | S. cerevisiae | Shake flask, MM | 0.85–1.86 | (Eichenberger et al., 2018) |
| 8-Prenylnaringenin | Potential treatment to adverse symptoms of menopause | S. cerevisiae | Shake flask, MM | 0.12 | (Levisson et al., 2019) |
| Kaempferol | Anti-cancer, anti-inflammatory effects | S. cerevisiae | Shake flask, CM | 86.8 | (Lyu et al., 2019) |
| Fisetin | Anti-cancer, cardio-protective, and anti-inflammatory | S. cerevisiae | Shake flask, MM | 2.3 | (Rodriguez et al., 2017b) |
| Quercetin | Cardiovascular protection | S. cerevisiae | Shake flask, MM | 20.4 | (Rodriguez et al., 2017b) |
| Dihydrochalcones (phlorizin, naringin dihydrochalcone, nothofagin) | Hypoglycemic agent, antioxidation | S. cerevisiae | Shake flask, MM | 11.6–65.0 | (Eichenberger et al., 2017) |
| Raspberry ketone | Flavoring agent | S. cerevisiae | Shake flask, CM | 3.5 | (Lee et al., 2016) |
| Naringenin | Neuroprotective and antioxidation | S. cerevisiae | Shake flask, MM | 100 | (Levisson et al., 2019) |
| Salidroside | Anti-cancer, cardiovascular, nerve, and brain cell protection | S. cerevisiae | Fed-batch, MM | 732 | (Jiang et al., 2018) |
| 3′-Hydroxygenistein | Antioxidant, antiproliferative, anti-inflammatory, and anti-melanogenesis | K. phaffii | Shake flask, CM | 23.0 | (Wang et al., 2016) |
| Naringenin | Neuroprotective and antioxidation | Y. lipolytica | Fed-batch, MM | 898 | (Palmer et al., 2020) |
| Eriodictyol | Antioxidant and antiaging | Y. lipolytica | Shake flask, MM | 134 | (Lv et al., 2019) |
| Taxifolin | Anticancer, anti-inflammatory, and antidiabetic | Y. lipolytica | Shake flask, MM | 111 | (Lv et al., 2019) |
MM, minimal media; SMM, semi-minimal media containing complex media components such as yeast extract; CM, complex media.
Other than the budding yeast S. cerevisiae, some other yeasts have also been harnessed for the production of terpenoids. Yarrowia lipolytica is a non-conventional oleaginous yeast, which is supposed to be a valuable host for the production of terpenoids due to its own endogenous MVA pathway and high acetyl-CoA biosynthesis with high oil production capacity (Ma et al., 2019c). Indeed, Y. lipolytica has been engineered for producing a plethora of bioactive terprnoids such as ginsenoside (Li et al., 2019a) and nootkatone (Guo et al., 2018). The methylotrophic yeast Komagataella phaffii (previously named as Pichia pastoris) has also been engineered for the production of lycopene (Bhataya et al., 2009) and nootkatone (Wriessnegger et al., 2014). However, the limited genetic tools hindered the extensive metabolic rewiring, which resulted in much lower production than that in S. cerevisiae (Table 1).
Alkaloids
Alkaloids are a class of natural products that mostly contain basic nitrogen atoms that include analgesic, sedative, and anti-cancer agents (Dubuisson and Dennis, 1977, Kemény-Beke et al., 2006, Orhan et al., 2007). Recently, it is attracting more attention to engineer yeast for the synthesis of alkaloids such as benzylisoquinoline, monoterpene indole, and tropane alkaloids (BIAs, MIAs and TAs) (Brown et al., 2015, Narcross et al., 2016, Srinivasan and Smolke, 2019).
BIAs biosynthetic pathway can be divided into two parts (Figure 2): the production of the key branchpoint metabolite (S)-reticuline and the downstream modification (Trenchard et al., 2015). Although high-level production of (S)-reticuline has been achieved in Escherichia coli (Minami et al., 2008, Nakagawa et al., 2011), it is still challenging in the efficient biosynthesis of (S)-reticuline in S. cerevisiae due to the difficulties in functional expression of bacterial tyrosinase that specifically catalyzes tyrosine hydroxylation toward L-3,4-dihydroxyphenylalanine (L-DOPA) (Deloache et al., 2015). Instead of tyrosinase, tyrosine hydroxylases (THs) and cytochrome P450 hydroxylase CYP76AD1 can serve as alternative options for L-DOPA production in S. cerevisiae (Narcross et al., 2016). Although TH has high specificity toward L-DOPA biosynthesis, it is heavily allosterically inhibited by substrate tyrosine and requires cofactor 5,6,7,8-tetrahydrobiopterin (BH4) (Daubner et al., 2011). A feedback inhibition-resistant mutant TH∗ (Li et al., 2018) and reconstruction of BH4 biosynthetic module (please refer to the section Reconstruction of Cofactor Biosynthesis) enabled efficient biosynthesis of (S)-reticuline. Similarly, expression of a CYP76AD1 mutant with abolished side activity enhanced L-DOPA biosynthesis in S. cerevisiae (Deloache et al., 2015, Hatlestad et al., 2012).
The (S)-reticuline-overproducing strain provides the chassis for the synthesis of versatile BIAs in S. cerevisiaec (Table 1). With integration of downstream modification enzymes, more and more BIAs were successfully de novo synthesized in S. cerevisiae, such as thebaine (Stephanie et al., 2015) and noscapine (Li et al., 2018, Stephanie et al., 2015). It is worthy to mention that up to 30 genes from various species were functionally expressed in S. cerevisiae for BIA biosynthesis, which demonstrated the high potential and capacity of yeast cell factories for natural product biosyntheses (Li et al., 2018). However, the yield of alkaloids in yeast remains low, which is commonly believed to be caused by insufficient core precursor (S)-reticuline (Deloache et al., 2015, Stephanie et al., 2015).
MIAs are a diverse family of complex nitrogen-containing plant-derived metabolites, which were composed of a secoiridoid moiety and an indole moiety with very diverse structures. Reconstruction of a seven-step pathway in S. cerevisiae enabled the production of anti-cancer drug precursor vindoline from tabersonine (Qu et al., 2015). Reconstruction of a plant-derived pathway with 15 genes, along with deletion of side pathways and enhancement of precursor supply, enabled for the first time de novo the biosynthesis of strictosidine in yeast (Brown et al., 2015).
TAs, containing common tropane ring, are a group of more than 200 specialized metabolites naturally produced in most solanaceous plants, which are widely used for treating nerve agent poisoning, gastrointestinal spasms, cardiac arrhythmia, and symptoms of Parkinson disease. With the characterized enzyme from a Chinese medical plant Anisodus acutangulus, the six-step biosynthetic pathway was reconstructed for de novo biosynthesis of tropine and pseudotropine (about 0.1 mg/L) in S. cerevisiae (Ping et al., 2019). Almost at the same time, the yeast central metabolism was extensively rewired for improving precursor supply and relieving the side pathway competition, which enabled much higher tropine production of 6 mg/L. With this chassis platform, the non-natural TA cinnamoyltropine was de novo synthesized by coupling biosynthetic modules from diverse plant lineages, which may accelerate the drug discovery pipeline (Srinivasan and Smolke, 2019).
Phenylpropanoids
The phenylpropanoids are commonly derived from L-tyrosine in microorganisms (Jendresen et al., 2015) and L-phenylalanine in plants (Havir and Hanson, 1968, Jun et al., 2018). The phenylpropanoid biosynthesis can be divided into two modules: the module for supplying precursor p-coumaric acid (p-CA) and the functional decoration module. Rewriting the central carbon metabolism, combined with the microbial tyrosine ammonia lyase and plant cinnamic acid hydroxylase branches, enabled high-level production of p-CA (12.5 g/L) in S. cerevisiae (Liu et al., 2019d, Rodriguez et al., 2015, Rodriguez et al., 2017a). Several flavonoids and simple polyphenols such as scutellarin, anthocyanin, and resveratrol have been successfully heterologously synthesized in yeast (Table 1). However, there is no report regarding engineering yeast for the synthesis of complex phenylpropanoids, such as active lignan clemastanin B (Zhang et al., 2016), and etoposide (Lau and Sattely, 2015), which mainly contributed to the complexity of downstream decoration steps. The very poor specificity of downstream enzymes can lead to accumulations of many by-products. For example, 4-coumaric acid:CoA ligase can in a promiscuous manner convert the p-CA and caffeic acid to the corresponding CoA molecules, which limits the pathway efficiency. Compartmentalization and co-cultivation may be feasible approaches to address this problem, which mimics spatiotemporal control in plants to avoid metabolic interference (Chen et al., 2017b, Wang et al., 2014). Recently, Y. lipolytica has been engineered as a phenylpropanoid-derived polyketide producer due to its high fluxes of acetyl and malonyl-CoA precursors, which were benefited from increased β-oxidation and peroxisome numbers, and overexpression of acetyl-CoA carboxylase (ACC1) (Palmer et al., 2020). Many attempts archived production of phenylpropanoid-derived polyketides in other non-conventional yeasts, such as flavonoids in Y. lipolytica (Lv et al., 2019) and K. phaffii (Wang et al., 2016) and triacetic acid lactone in Y. lipolytica (Liu et al., 2019a).
Enzyme Discovery
To date, more than 45,000 genomes are available in public databases, providing abundant resources for gene discovery. Even many functional genes and secondary metabolic pathways have been identified through the combination of computational mining and experimental confirmation (Ziemert et al., 2016), it is still challenging to identify suitable enzymes for the construction of biosynthetic pathways in specific hosts. Heterologous protein expression in yeast often results in reduced activity or undesired catalytic properties, because of incorrect folding, low expression levels, unsuitable microenvironments or feedback inhibition, etc. (Schuler and Werck-Reichhart, 2003). Here, we will discuss the advanced approaches to get suitable enzymes and regulation components for efficient biosynthetic pathways.
Multi-species Genomes Provide Clues for Enzyme Discovery
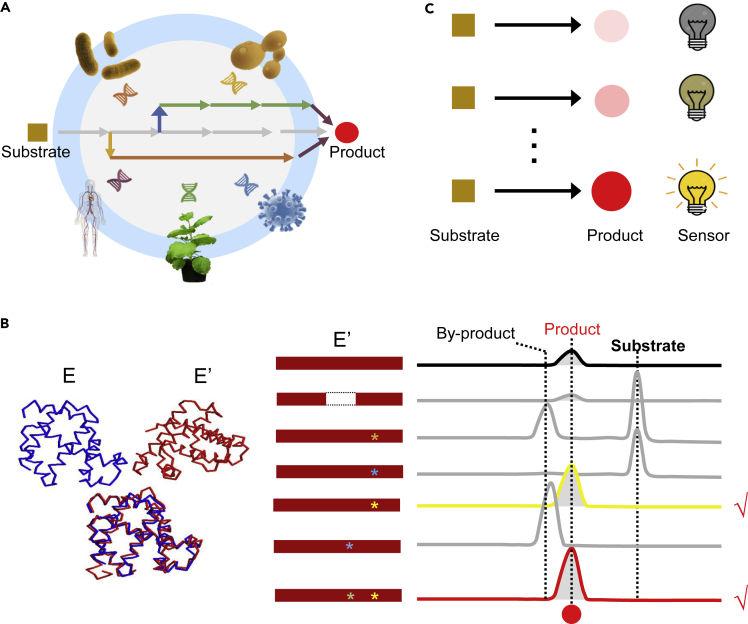
Specific biosynthesis can be conducted by different pathways (even different enzymes) among various species due to evolution diversity. There will be low efficiency when wholesale adopting inherent biosynthetic pathway from single species into a heterologous host. Alternatively, combinatorial optimization by using alternative steps or isoenzymes from multi-species genomes may result in efficient biosynthetic pathways (Stephanie et al., 2015) (Figure 3A). For example, non-natural IPA pathway was reconstructed for isoprenoid biosynthesis by combining multi-species genes from Staphylococcus aureus, Myxococcus xanthus, Clostridium beijerinckii, and E. coli. This novel IPA pathway is more energy efficient than native MVA and MEP pathways, resulting in the production of 0.6 g/L monoterpenoids from glycerol (Clomburg et al., 2019). As mentioned above, integration of four non-native metabolic reactions in S. cerevisiae improved acetyl-CoA supply and enabled high-level production of farnesene with reduced ATP requirement and carbon loss (Meadows et al., 2016). The production of caffeic acid from coumaric acid in plants requires multiple steps of oxidation and esterification, which are very weak in plant and yeast (Liu et al., 2019b, Ruben et al., 2013). A shorter and more efficient pathway catalyzed by HpaB (4-hydroxyphenylacetate 3-monooxygenase) and HpaC (NADPH-flavin oxidoreductase) from bacteria was constructed to replace the native plant pathway in E. coli and S. cerevisiae, which significantly improved conversion of coumaric acid toward caffeic acid (Eudes et al., 2014, Liu et al., 2019b, Wang et al., 2017a).
Figure 3.
Strategies for Enzyme Discovery And Engineering
(A) Multi-species genomes are a treasure trove of versatile enzymes. Integration of enzymes from multiple species (bacteria, fungus, virus, plant, and animal) for the construction of chimeric pathway is beneficial for optimization of natural product biosynthesis.
(B) Knowledge-driven protein engineering is a promising approach to rational enzyme design for specific catalysis. The letters E and E′ indicate the structure comparison of the original protein and rationally engineered protein. The red rectangles represent the engineered proteins, and the asterisks represent the mutation sites. In the chromatogram the ticks represent the functional mutants.
(C) Biosensor is an ideal strategy for high-throughput screening of desired enzymes according to intuitive phenotypic changes. The brightness of bulbs and the circle color represent the response intensity of the biosensor and the yield of the product, respectively.
These studies clearly demonstrated that the construction of chimeric pathways, by integration of multiple species genes, is beneficial for efficient biosynthesis of molecules of interest, which, however, strongly relies on the identification of efficient functional genes from the avalanche gene databases. Fortunately, extensive bioinformatic tools and high-throughput experimental data were developed to elucidate biological function from multi-species genomes (Blaby-Haas and De, 2011, Medema and Fischbach, 2015, Ziemert et al., 2016), which should facilitate the identification of efficient enzymes for construction of biosynthetic pathways.
Knowledge-driven Protein Engineering
Other than gene excavation from multi-species genomes, knowledge-driven or rational protein engineering is a more targeted strategy to improve enzyme specificity and efficiency with the aid of protein crystal structure and computer simulation (Figure 3B). Furthermore, protein engineering can help to create efficient enzymes for specific reactions without available enzymes, by engineering the promiscuous enzymes from similar reaction.
In S. cerevisiae, ERG20p catalyzes consecutive reactions for the biosynthesis of GPP (C10) and FPP (C15) by consecutively condensing IPP and DMAPP (C5 units). To improve the monoterpene and diterpene production, ERG20p was engineered into a double-mutant GPP synthase (F96W-N127W) (Ignea et al., 2014) and a single-mutant GGPP synthase (F96C or Y95A) (Ignea et al., 2015), through protein engineering, when compared with previously identified crystal model of the avian FPPS (Stanley Fernandez et al., 2000, Tarshis et al., 1996). Similarly, ERG20p was specifically engineered to a non-natural C11 terpene synthase by a single-residue switch, which enabled the biosynthesis of a serious of non-natural C11 terpenes in S. cerevisiae (Ignea et al., 2018). With the development of structural biology, more and more crystal structures of catalytic enzyme have been resolved, which laid the foundation for the homology model construction and directed evolution. Knowledge-driven or rational protein engineering can provide more efficient enzymes for construction of robust yeast cell factories (Larue et al., 2016, Rodriguez et al., 2015, Zheng et al., 2004).
Biosensor for High-Throughput Screening
Biosensor is an ideal strategy for high-throughput screening of desired homologous proteins or mutants (Cirino and Arnold, 2002), which requires a certain correlation between biosensor and the enzyme activity with an easily observed signal, such as color, fluorescence, or growth state (Figure 3C).
Even the synthesis of (S)-reticuline from glucose was already achieved in E. coli (Nakagawa et al., 2011), non-functional TH limited the synthesis of BIAs in S. cerevisiae (Hawkins and Smolke, 2008). A small-molecule biosensor was developed by setting a negative correlation between the activity of TH and yellow by-product betaxanthin, which achieved high-throughput screening of TH mutants for the efficient synthesis of upstream intermediate L-DOPA and a 7.4-fold improvement of dopamine production with the best mutant (Deloache et al., 2015). Transcription factor-based biosensor is much more commonly used and can directly respond to molecules with conformational change of transcription factors (Williams et al., 2016). Using a bacterial transcription factor FapR and its corresponding operator fapO, a malonyl-CoA biosensor was constructed in S. cerevisiae to gauge and improve intracellular malonyl-CoA levels (Li et al., 2015b). The yeast-based biosensor was also successfully used in the screening of itaconic acid (Ekr et al., 2018) and short-/medium-chain fatty acids producer (Baumann et al., 2018).
Cofactor Engineering
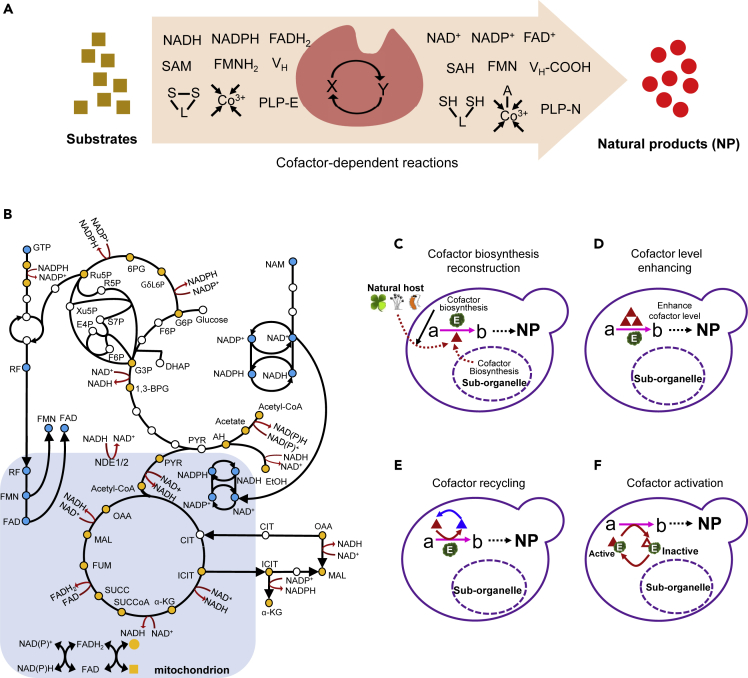
Although great progresses have been achieved in reconstruction of metabolic pathways for natural products, heterologous expression of enzymes from other species such as plants may encounter low-efficiency or inactive forms due to the lack of suitable cofactors (Figure 4A). In particular, eukaryotic organisms have compartmentalized cellular metabolism and distinct cofactor distribution in sub-organelles. For example, cytosolic reconstruction of cytochrome P450 enzyme-containing pathways may have insufficient cofactor heme (Michener et al., 2012, Szczebara et al., 2003), which is mainly localized in the mitochondria for maintaining efficient redox reactions. Figure 4B shows the biosynthetic pathways of redox-related cofactor (pairs) in S. cerevisiae and the mutual transformation in central carbon metabolism, which clearly demonstrates the complexity of cofactor regulation in eukaryotic cells. Although the role of NAD(P)+/NAD(P)H on regulating cellular redox balance and metabolic flux has been extensively reviewed elsewhere (Chen et al., 2014, Wang et al., 2013, Wang et al., 2017b), we here, for the first time, compressively review how the cofactors affect the biosynthetic pathways and cofactor regulation strategies for improved biosynthesis.
Figure 4.
Cofactor Engineering for Enhancing Biosynthetic Efficiency of Natural Products
(A) Representative cofactors involved in natural product biosynthesis. Two different states (X and Y) of cofactors keep cellular hemostasis. The squares and circles represent the substrates and natural products (NP), respectively.
(B) The biosynthetic pathways of redox-related cofactors in S. cerevisiae. Circles represent metabolites; yellow and blue circles represent metabolites involved in cofactor transformation and cofactor biosynthesis, respectively.
(C–F) Different regulation strategies of cofactors: Cofactor biosynthesis reconstruction (C), enhancing cofactor level (D), cofactor recycling (E), and cofactor activation (F). a denotes substrate, b denotes intermediates, and NP stands for natural products. Triangles of different colors represent cofactors of different forms. NAD+, nicotinamide adenine dinucleotide; NADP+, nicotinamide adenine dinucleotide phosphate; NADH, reduced from of NAD+; NADPH, reduced from of NADP+; FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide; FADH2, reduced from of FAD; FMNH2, reduced form of FMN; SAM, S-adenosyl methionine; SAH, S-Adenosyl-L-homocysteine; VH, biotin; VH-COOH, carboxylated biotin; S-L-S, lipoamide; SH-L-SH, dihydrolipoamide; =Co3+ = , 5′-deoxyadenosylcobalamin (vitamin B12); A-Co3+, inactive vitamin B12; PLP-E, pyridoxal-5-phosphate-protein complex; PLP-N, PLP aldimine; GTP, guanosine triphosphate; RF, riboflavin; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; G3P, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate; 1,3-BPG, 1,3- bisphosphoglycerate; PYR, pyruvate; AH, acetaldehyde; EtOH, ethanol; ACE, acetate; Ac-CoA, acetyl-CoA; GδL6P, 6-phosphoglucono-δ-lactone; 6PG, 6-phosphoglucononate; Ru5P, ribulose-5-phosphate; R5P, ribose-5-phosphate; Xu5P, xylulose-5-phosphate; S7P, sedoheptulose-7-phosphate; E4P, erythrose-4-phosphate; CIT, citrate; ICIT, isocitrate; α-KG, α-ketoglutarate; SUCCoA, succinyl-CoA; SUCC, succinate; FUM, fumarate; MAL, malate; OAA, oxaloacetate; NAM, nicotinamide.
Reconstruction of Cofactor Biosynthesis
Some enzymes often require special cofactors to achieve high catalytic activities. Heterologous expression of enzymes derived from plant and other species in yeast may suffer from insufficient or deficient cofactor supply, due to the different cellular environment between yeast and their natural hosts. Thus, reconstituting the cofactor biosynthesis should be helpful for maintaining the high enzyme activity for efficient natural product biosynthesis in yeast (Figure 4C).
Previously studies found that plant TH had low activity in S. cerevisiae (Deloache et al., 2015, Hawkins and Smolke, 2008), which limited the alkaloid biosynthesis in yeast. The main reason is the deficiency of its cofactor BH4 in yeast (Fitzpatrick, 1999). Thus, BH4 biosynthetic pathway was reconstituted and enabled the significant noscapine biosynthesis in S. cerevisiae (Li et al., 2018). We also found that reconstruction of bacterial NADPH-dependent electron transfer systems, Fd/Fnr (ferredoxin-ferredoxin reductase) and Fld/Fnr (putidaredoxin-putidaredoxin reductase), significantly improved the performance of bacterial alkane and 1-alkene biosynthetic pathways in yeast (Buijs et al., 2015, Zhou et al., 2018).
Biotin acts as a cofactor for biotin-dependent carboxylases involved in essential carboxylation and decarboxylation reactions (Streit and Entcheva, 2003). Most yeasts are deficient in de novo biosynthesis of biotin or have insufficient level for fast growth and product formation (Phalip et al., 1999) and thus require external addition of this expensive vitamin in cultures. Laboratory evolution rendered full biotin prototrophy of S. cerevisiae, and genome sequencing revealed causal mutations for biotin deficiency in S. cerevisiae (Bracher et al., 2017), which should be helpful for the construction of biotin-dependent biosynthetic pathways in yeast.
Pyridoxal 5′-phosphate (PLP), one of the most versatile cofactors, is essential for over 160 enzymes (di Salvo et al., 2012), and is synthetized through the ribose-5-phosphate-dependent de novo and salvage pathways. Therefore, PLP plays an important role in natural product biosynthesis (Schiroli and Peracchi, 2015), because PLP-dependent enzymes catalyze diverse chemical reactions, including decarboxylation, transamination, racemization, Cα-Cβ bond cleavage, and α, β-elimination reaction. Supplementation of PLP and eliminating its degradation significantly improved the whole-cell transamination for stereoselective production of (R)-1-phenylethylamine (>99% enantiomeric excess) in yeast S. cerevisiae (Weber et al., 2014, Weber et al., 2017). Similarly, using a PLP-dependent Rosa hybrid phenylacetaldehyde synthase along with internal de novo PLP synthesis produced 0.34 g/L 2-phenylethanol for the first time in E. coli (Achmon et al., 2014).
The eukaryotic cell metabolism is compartmentalized in sub-organelles, whose membranes are impermeable to various cofactors. Thus, it is necessary to reconstitute cofactor biosynthesis to drive the corresponding cofactor-dependent biosynthetic pathways in specific sub-organelles even if there is cofactor localization in other sub-organelles (Figure 4C). PDH complex consists of three catalytic subunits, PDH (E1), dihydrolipoyl transacetylase (E2), and dihydrolipoyl dehydrogenase (E3), which catalyze the efficient conversion of pyruvate to acetyl-CoA in yeast mitochondria. Reconstruction of PDH into cytosol is an ideal strategy for enhancing cytosolic acetyl-CoA supply without accumulation of by-product ethanol, which would be beneficial for biosynthesis of acetyl-CoA-derived molecules such as terpenoids and polyketides. However, it is challenging in cytosol reconstruction of PDH, because the biosynthesis of cofactor lipoic acid for E2 subunit activation localizes in mitochondria. Thus, external lipoic acid supplementation and reconstruction of cytosolic lipoic acid biosynthetic pathways are prerequisites for functional assembly of cytosolic PDH (Kozak et al., 2014, Lian and Zhao, 2016).
Enhancing the Cofactor Level
The cellular cofactor levels determine the metabolic flux, because cofactors involve enzyme reactions and thus regulate chemical equilibrium state. Therefore, enhancing cellular cofactor levels by engineering their biosynthetic pathway could drive the metabolic flux toward biosynthesis of the target product (Figure 4D).
S-adenosyl-L-methionine (SAM) is a central cofactor that participates in a variety of reactions and physiological processes mainly as methyl donor (Lieber and Packer, 2002, Malakar et al., 2006, Thomas and Surdin-Kerjan, 1997). Also, many natural product biosyntheses involve methylation steps catalyzed by SAM-dependent methyltransferases (Do et al., 2007, Zou et al., 2014). Many studies indicated that SAM level had a greater effect on methylation reactions than methyltransferase expression (Chen et al., 2017a, Kunjapur et al., 2016, Zhao et al., 2010). Although rarely reported in yeast metabolic engineering, it has been widely shown that up-regulation of SAM significantly improved the production of ephedrine (Morris et al., 2018), vanillin (Kunjapur et al., 2016), and pterostilbene (Heo et al., 2017) in E. coli.
Flavin is an active cofactor involved in maintaining redox homeostasis in mitochondria (Giancaspero et al., 2013), endoplasmic reticulum (ER) (Kim et al., 2018), and nucleus (Teresa Anna et al., 2013). Flavin adenine dinucleotide (FAD)-dependent monooxygenase (hpaB) enabled higher-level production of caffeic acid from p-CA than plant-specific cytochrome P450-dependent monooxygenase (Aymerick et al., 2013, Liu et al., 2019b, Wang et al., 2017a). It is expected that this pathway will consume a large amount of cytoplasmic FADH2 and disrupt intracellular FADH2/FAD homeostasis, and enhancing FAD(H2) level will further improve caffeic acid production in yeast.
Engineering the Equilibrium State of Cofactors
In addition to the cofactor levels, the cellular equilibrium states (the ratio between reducing and oxidation forms) affect the efficiency of the corresponding enzyme and even cellular redox hemostasis. The NAD+/NADH, NADP+/NADPH, FMN/FMNH2, and FAD/FADH2 cofactor pairs are important components of the electron transport chain and involved in at least 800 biochemical reactions (Chen et al., 2014). Thus, engineering the cellular redox balance is important for increasing the biosynthesis of target compounds and robustness of cell factories (Figure 4E).
We previously reprogrammed S. cerevisiae metabolism from ethanol fermentation toward lipogenesis. Ethanol fermentation (Crabtree effect) is mainly attributed to the high NADH/NAD+ from efficient glycolysis, and ethanol biosynthesis can re-oxidize cytosolic NADH. Enhancing fatty acid biosynthesis required high level of NADPH, and blocking ethanol fermentation disturbed the cellular redox hemostasis. Redirecting NADH to NADPH by engineered transhydrogenase and pentose phosphate pathway, and down-regulating glycolytic flux for reduced NADH generation, resulted a Crabtree-negative yeast for high-level production of fatty acids (Yu et al., 2018). It is worthy to mention that Crabtree effect can be relieved by enhancing the respiration of cytosolic NADH via external mitochondrial NADH dehydrogenases (NDE1/2) (Bakker et al., 2001, Luttik et al., 1998). Many natural products involve P450-catalyzed tailoring steps, which require NADPH for electron transfer. For example, noscapine biosynthesis involves five P450 enzymes (Vidal et al., 2017, Yan et al., 2015), which would consume large amount of NADPH for high-level production of noscapine and disturb the balance of NADPH/NADP+. Thus the NADPH was enhanced by overexpressing NADH kinase, three isocitrate reductases, prephenate dehydrogenase, and NADP-dependent glycerol dehydrogenase (GCY1p), leading to significant improvement of noscapine production by using glycerol as carbon source (Li et al., 2018). 3-Hydroxypropionic acid (3-HP) biosynthesis from acetyl-CoA requires extensive NADPH-dependent reduction, which deprives the cellular reducing power. Coupling the glycolysis with NADPH regeneration by expressing an NADP+-dependent G3P dehydrogenase, enabled a 2-fold improvement of 3-HP in S. cerevisiae (Chen et al., 2014). NADPH is the driving force for fatty acid biosynthesis; thus extensive redox engineering was implemented for converting NADH to NADPH in Y. lipolytica, which achieved a new redox equilibrium state and enabled a high-level lipid production of 90 g/L (Qiao et al., 2017).
Increase the Active Form of Cofactors
Some cofactors do not work alone and require activation through covalent binding to the protein (Figure 4F). As mentioned above, lipoic acid is necessary for functional reconstruction of PDH in yeast cytosol, but it cannot directly activate E2 subunit of PDH. Its active form lipoamide functions as an acyl carrier for E2 activation by covalent attachment of the lipoic acid to the ϵ-amino group of a specific lysine residue of E2 (Cronan et al., 2005, Rock, 2009). Therefore, in addition to reconstruction of cytosolic lipoic acid biosynthesis, a de novo synthetic lipoylation machinery is also necessary for functional reconstitution of PDH in the yeast cytosol (Lian and Zhao, 2016).
Cofactor B12 is very important for glycerol dehydratase activity in 1,3-propanediol (1,3-PDO) production, which becomes inactive (B12-inact) when tightly bound to the dehydratase after the reaction with glycerol (Sauvageot et al., 2002). A protein partner (pduO), glycerol dehydratase reactivase, facilitates the dissociation of the B12-inact from glycerol dehydratase. Also, B12-inact is reconverted to B12-act with the participation of flavoprotein and ATP (Ohnson et al., 2001). Thus, engineering rapid conversion of B12-inact into B12-act, in addition to increased B12 level, is very beneficial for enhancing glycerol dehydratase activity (Cervin et al., 2010, Nakamura and Whited, 2003).
Pathway Construction and Optimization
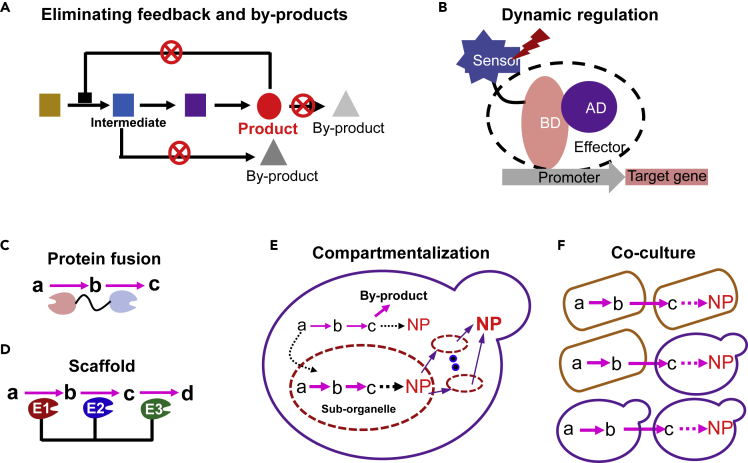
Despite the availability of versatile active enzyme elements and cofactors, it is still challenging in heterologous reconstruction of the efficient biosynthesis pathways, which is influenced by the complex interactions between metabolites and enzymes as well as the severely disturbing endogenous cellular metabolism. Several strategies, including relieving feedback inhibition, dynamic pathway regulation, and metabolon construction, were developed for improving the biosynthesis in yeast cell factories (Figure 5).
Figure 5.
Strategies to Enhance the Metabolic Flux of Natural Product Biosynthesis
(A) Eliminating feedback inhibition of key enzymes and blocking the side pathways for reducing by-product accumulation. Squares, circles, and triangles represent intermediate, target products, and by-products, respectively.
(B) Dynamic regulation of pathway activation through biosensor. BD and AD represent the binding domain and active domain of the transcription factor, respectively, which form the effector for gene activation.
(C) Protein fusion for substrate channeling.
(D) Scaffold enables boding enzymes for enhancing substrate channeling.
(E) Sub-organelle compartmentalization of heterogeneous pathways. Sub-organelle number and size can be engineered for enhancing biosynthesis.
(F) Co-culture engineering among multiple microorganisms, such as bacteria-bacteria, bacteria-yeast, and yeast-yeast, for improving natural products biosynthesis.
Eliminate Feedback Inhibition of Key Enzymes
The feedback inhibition mentioned here refers specifically to product inhibition at the enzyme level, not at the transcription level (Zhang and Liu, 2015) (Figure 5A). Introduction of feedback-insensitive amino acid biosynthetic genes (ARO4K229L and ARO7G141S) improved cellular aromatic compound levels by 200-fold in E. coli and S. cerevisiae (Billingsley et al., 2016, Rodriguez et al., 2015). Characterizing and expressing a non-feedback acetyl-CoA synthetase mutant from Salmonella enterica increased the production of acetyl-CoA/malonyl-CoA-derived products in yeast by enhancing acetyl-CoA supply (Cardenas and Silva, 2014). This strategy is generally based on a clear understanding of the enzyme structure and inhibition mechanism. Otherwise, biosensor-based enzyme engineering may be helpful for screening feedback-insensitive mutants (as mentioned in the section Biosensor for High-Throughput Screening).
Dynamic Regulation
Heterologous biosynthetic pathways may bring enormous stress on the cell fitness, such as the accumulation of toxic intermediates, the competition of carbon sources, and reducing power. Dynamic regulation of biosynthetic pathways may relieve the stresses by separating the cell growth and product biosynthesis. A dynamic control system comprises two elements: a sensor that perceives external environment change and an effector that can respond to the sensor (Qiu et al., 2019) (Figure 5B).
The carbon source sensor is extensively used for the construction of dynamic pathway, such as GAL and HXT systems. The promoters of galactose metabolism genes, such as GAL1p and GAL10p, are activated by galactose and repressed by glucose in S. cerevisiae (Ahn et al., 2013). With deletion of GAL80, these GAL promoters are constitutively transcribed at low glucose level and can be used for the construction of biosynthetic pathways responding to glucose level, which can divide fermentation process into cell growth and product biosynthesis phases. The GAL system has derived a variety of dynamic regulation strategies to meet different needs, which provides a tool for further refined control of metabolic engineering (Peng et al., 2018, Ryo et al., 2017). The promoter of hexose transporters (HXT1p, HXT6p, and HXT7p) were used to dynamically regulate by-product biosynthesis pathways (Scalcinati et al., 2012) and fatty alcohol transporter (Hu et al., 2018) with relieved yeast cell stresses.
Optogenetic regulation is an easily operational strategy for dynamic control of biosynthetic pathways, as light can be applied and removed easily without complex media changes (Zhao et al., 2018). A blue-light-responsive circuit was constructed by using a blue light transcription factor EL222 from the marine bacterium Erythrobacter litoralis (Nash et al., 2011), for dynamically activating cell growth upon blue light exposure and product biosynthesis in the dark (Zhao et al., 2018). This optogenetic regulation strategy successfully separated isobutanol biosynthesis from cell growth and improved the production of isobutanol by more than three times, which relieved the toxicity of isobutanol (Zhao et al., 2018). Although this strategy is easy operating, the permeability of light should be addressed for industrial high-density fermentation.
Metabolic Compartmentalization
Natural product biosynthesis always involves complex multi-step pathways, and reconstruction of yeast cell factory often encounters some common challenges, such as unsuitable physicochemical environments (pH and redox potential), insufficient supply of essential materials (substrates and cofactors), occurrence of undesired side reactions, toxicity of intermediates, and incompatibility with endogenous metabolism. Several spatialized metabolic engineering strategies including metabolon construction (protein level), sub-organelle engineering (subcellular level), and co-culture engineering (cell level) have been developed to address these problems.
Construction of metabolon is beneficial for substrate channeling by preventing unstable intermediates or relieving toxicity of some intermediates (Agapakis et al., 2012). There are two ways to construct a metabolon: protein fusion (Figure 5C) and protein scaffold (Figure 5D). Protein fusion is the most convenient strategy for enhancing substrate channeling (Zhou et al., 2012), which, however, may destroy the enzyme structure and is always limited to two enzymes (Figure 5C). Protein scaffold can gather several enzymes into proximity by affinity binding, which has been successfully applied in S. cerevisiae for improving resveratrol production (Wang and Oliver, 2012) and also xylose utilization with diminished the accumulation of by-product xylitol by co-localizing corresponding enzymes (Thomik et al., 2017).
Sub-organelle compartmentalization can provide suitable physicochemical environments and enough precursors/enzymes for complicated biosynthetic pathways (Figure 5E). The benefits of targeting biosynthetic pathways to subcellular compartments were reviewed previously, including mitochondria, peroxisomes, ER, Golgi, vacuoles, and cell wall, in different yeast species (Agapakis et al., 2012, Hammer and Avalos, 2017). The first example showed that mitochondrial compartmentalization of the tailoring P450 enzyme improved hydrocortisone production, because mitochondria are more suitable for maintaining P450 activity (Szczebara et al., 2003). Recently, mitochondrial compartmentalization was showed to be helpful for production of isoprenoids (Farhi et al., 2011, Yee et al., 2019, Yuan and Ching, 2016), short-chain alcohols (Avalos et al., 2013), and chemicals (acetoin and fumarate) (Chen et al., 2015, Li et al., 2014), because of sufficient supply of acetyl-CoA and tricarboxylic acid cycle intermediates. However, it may bring stresses when overexpressing enzymes in mitochondria that are crowded with respiration components. Some other sub-organelles, such as peroxisomes, lipid bodies, and vacuoles, are relatively orthogonal to the native essential process, making them ideal workhorses for metabolic compartmentalization. Furthermore, it has been showed that peroxisome compartmentalization of CoA-tailoring steps was required for efficient biosynthesis of penicillin (Meijer et al., 2010) and mycophenolic acid (Zhang et al., 2019b) in fungi, indicating that peroxisomes are ideal catalyzing houses for natural product biosynthesis. Peroxisomal targeting improved the production of fatty acid-derived molecules by up to 700% with relieved side pathway competition (Zhou et al., 2016, Zhu et al., 2017). Recently, peroxisome compartmentalization enabled high-level production of the triterpene squalene in S. cerevisiae, which suggested that peroxisomes were promising subcellular factories for terpene biosynthesis (Liu et al., 2020). Peroxisome compartmentalization of prodeoxyviolacein (PDV) biosynthetic pathway reduced the accumulation of by-product chromopyrrolic acid, although without improving PDV production (DeLoache et al., 2016), which indicated that the precursor might be insufficient. It is much more challenging to provide high flux of precursor for biosynthesis of heterologous natural products other than fatty acid or acetyl-CoA derivatives in peroxisome, which are the main organelles for fatty acid degradation in yeast. Similarly, compartmentalization of associated pathways in lipid droplet and other lipid-relevant organelles resulted in a 10 times higher production of fatty acid methyl esters when compared with the cytosolic pathway in Y. lipolytica (Yang et al., 2019). Recently, a synthetic non-endogenous nanoscale compartment was constructed in yeast by expressing prokaryotic encapsulins (Lau et al., 2018), which represented a strictly orthogonal compartment. Engineering organelle size and biogenesis can provide more space for enzyme encapsulation and product storage. Enlarging lipid droplets increased lycopene production by 25%, which might improve the storage of hydrophilic lycopene (Ma et al., 2019b). Similarly, expanding ER resulted in an 8-fold higher production of protopanaxadiol (Kim et al., 2019) and a 7.1-fold increase of protein secretion (Besada-Lombana and Da Silva, 2019), which demonstrated that ER space expansion increased the capacity of ER protein synthesis and folding and thus relieved metabolic constraints imposed by limited enzyme abundance.
Co-culture and Stepwise Culture Engineering
Non-linear biosynthetic pathways, such as diverging and converging pathways, are particularly challenging in pathway optimization in mono-culture system, because they require a delicate balancing between all interconnected pathway modules (Zhang and Wang, 2016). Furthermore, functional expression of multiple genes from various species may require different cellular environments, which is difficult in a single organism. The emergence of modular co-culture engineering offers a feasible approach for modularizing and balancing complicated biosynthetic pathways (Figure 5F), whose application in production of natural products has just been comprehensively reviewed (Wang et al., 2019b). The optimized multiple-strain culture of same species such as E. coli showed 38-fold higher production of rosmarinic acid over the mono-culture system (Li et al., 2019b). Construction of chimeric S. cerevisiae-E.coli co-cultures enabled high-level production of oxygenated terpenoids, by taking advantage of the high-efficiency terpene biosynthesis in E. coli and suitable environment of S. cerevisiae for P450 expression (Zhou et al., 2015). Engineering co-culture methylotrophic yeasts K. phaffii (two strains) enabled 50% to 70% higher production of polyketide drug monacolin J and lovastatin than that from mono-culture by relieving metabolic stress (Liu et al., 2018c). Stepwise fermentation is consequently culturing multiple strains that harbor divided modules of complicated biosynthetic pathways. The opiate biosynthetic pathway was divided into four modules and distributed into four E. coli strains (Nakagawa et al., 2016), and stepwise culture of four engineered strains produced 2.1 mg/L thebaine from glycerol, corresponding to a 300-fold increase from yeast system (Stephanie et al., 2015). This stepwise culture system can avoid tetrahydropapaveroline (THP) degradation by o-diphenolase activity of tyrosinase (Nakagawa et al., 2014).
Although modular co-culture has some unique advantages over monoculture, there are still some major obstacles to overcome for large-scale industrial applications. The biggest obstacle is coordinating multiple microbial groups into stable synthetic consortia. The strategies of commensalism-based or mutualistic synthetic consortia might be a way out (Sgobba et al., 2018, Zhou et al., 2015). However, little is known about the specific principles of cooperation and competition in these mutualistic models. When these interactions scale to higher dimensions, the behaviors of complex ecosystems are even more elusive (Mee et al., 2014). In addition, achieving a balance of metabolic ratios among different modules and efficient delivery of intermediates may also be obstacles for the application of co-culture strategy.
The Global Regulation
In addition to the pathway engineering and flux rewiring, enhancing the robustness of microbial cell factories is essential for industrial application with improved resistance to harsh industrial conditions and toxicity of natural products (Gong et al., 2017). Engineering cellular tolerance against harsh conditions has been discussed in detail in previous reviews by us and others (Gong et al., 2017, Zhu et al., 2012). Here we emphasize the global regulation of cellular performance for natural biosynthesis by directed evolution and population quality control.
Directed Evolution at the Level of the Whole Organism
Directed evolution at the cellular level emphasizes “collaboration” between scientists and microorganisms to achieve design goals, harnessing the initiative of the yeast rather than relying entirely on biologists' deliberate choices (Szymanski and Calvert, 2018). This global-directed evolution strategy often requires yeast to adapt to challenging environments by genomic flexibility and DNA recombination that are very difficult to realize through rational engineering. Similar to protein-directed evolution, cellular-directed evolution requires efficient mutagenesis and high-throughput screening. Adaptive laboratory evolution of S. cerevisiae toward enhanced resistance of periodic hydrogen peroxide shocking improved carotenoid production by exploiting the antioxidant properties of carotenoids (Reyes et al., 2014). Similarly, this “hydrogen peroxide-shocking” strategy also improved the production of antioxidant 3′-hydroxygenistein in recombinant K. phaffii (Wang et al., 2016). Incorporating the synthetic chromosome rearrangement and modification by loxP-mediated evolution into designed yeast cells with loxP sites enabled generation of genotype diversity for improved production of carotenoids, violacein, and penicillin in yeast (Blount et al., 2018, Jia et al., 2018, Liu et al., 2018a). Further high-throughput sequencing can map genotype-phenotype relationships, which is helpful for the construction of robust cell factories by reverse engineering (Yu et al., 2018). Together with random mutations, directed evolution can be combined with rational designs to form a loop, which in turn provides the targets for future rational design and regulation.
Population Quality Control
Nongenetic variation is naturally inherent and will lead to suboptimal performance at the ensemble level due to the subpopulations of low-performance variants consuming nutrients without efficiently synthesizing products. It was found that a minority (15%) of the total cell population produced more than half of the total free fatty acid in an engineered E. coli, which indicated that most cells performed very weakly (Xiao et al., 2016). To our knowledge, there is no report on engineering population quality control in yeast for production of natural products, which might be attributed to the challenge in coupling the cellular fitness with natural product biosynthesis, although genetic circuits such as dual feedback loops have been shown to suppress cellular heterogeneity in yeast (Ramsey et al., 2006).
Perspective
Non-conventional yeast refers to more than 2,000 identified yeast species except for two model yeasts, S. cerevisiae and Schizosaccharomyces pombe (Radecka et al., 2015). Several non-conventional yeasts have attracted more interest in metabolic engineering applications because of their unique advantages and characteristics (Wagner and Alper, 2015). In contrast to the Crabtree yeast S. cerevisiae, many con-conventional yeasts are Crabtree negative, have much higher biosynthetic capacity, and use low-cost or market-surplus carbon sources, such as methanol or glycerol (Czajka et al., 2018, Mayer et al., 1999, Wriessnegger et al., 2014). K. phaffii has been known as a powerful protein expression system because of its capacity to perform complex post-translational modifications (Yang and Zhang, 2018). Ogataea polymorpha and Ogataea thermomethanolica are thermo-tolerant yeast and have high potential for industrial application along with saving cooling cost (Ryabova et al., 2003). These characteristics and advantages of non-conventional yeasts make them more suitable as hosts than S. cerevisiae for specific production (Duan et al., 2018). It should be emphasized that fungi such as Aspergillus are much more suitable as cell factories for heterologous production of complicated antibiotics that are derived from fungi (Skellam, 2019). However, lack of genetic tools and inefficient homologous recombination (HR) make it challenging in engineering these microbes (Cai et al., 2019).
Engineering natural product production always involves construction of complex and long pathways with multiple genes (Li et al., 2018, Luo et al., 2019), which requires efficient gene editing platform for high-fidelity assembly of long DNA fragments. With the rapid development of DNA chemical synthesis services, a series of long DNA fragment assembly techniques have been introduced, including polymerase cycling assembly (PCA) (Smith et al., 2003), emulsion PCA (Plesa et al., 2018), ligase cycling assembly (De et al., 2014), Gibson assembly (Gibson et al., 2009), and golden gate assembly (Engler and Marillonnet, 2014). However, sequence mismatch or errors always happen at special DNA assembly, such as long repeat sequence and hairpin loop structure, which hinder the precise construction of biosynthetic pathways. Therefore, bioinformatics and sequence optimization might be promising solutions.
Recently, the CRISPR-Cas9 system has emerged as a powerful gene editing tool for metabolic engineering and synthetic biology, which has worked well in organisms including model microorganism S. cerevisiae and E. coli. A vast number of Cas9-mediated genetic tools enabled efficient and rapid genetic editing in S. cerevisiae (Lian et al., 2018). Furthermore, numerous genetic manipulation toolboxes were developed for the construction of heterologous pathways, including promoters (Sun et al., 2012), terminators (Curran et al., 2013, Yamanishi et al., 2013), and neutral sites (Mikkelsen et al., 2012). These genetic toolboxes make S. cerevisiae the most attractive cell factory for the production of natural products. As mentioned above, most of non-conventional yeasts display a stronger preference for non-homologous end joining (NHEJ) when exogenous DNA is introduced, which hampers the precise genetic editing in metabolic engineering (Vogl et al., 2013). Although this problem has been alleviated to some extent through the regulation of the HR- or NHEJ-related proteins such as Rad52p (Shao et al., 2017), Ku70p (Liu et al., 2019c), and Ku80p (Schwartz et al., 2015), the HR efficiency is still far less for precise genetic engineering, in particular multiple DNA fragment assembly. Besides efficient genetic editing, non-conventional yeast systems require an expanded synthetic biology toolbox (such as highly controllable promoters and enough terminators without homologous sequences) for pathway construction.
Current cell factory construction mainly focuses on enhancing precursor supply, enzyme engineering, and flux balancing. However, the cofactors that are essential for enzyme activity have always been ignored. In particular, many catalytic enzymes of natural product biosynthesis require the specific cofactors, but heterologous hosts such yeasts have insufficient cofactor supply. Although some strategies have been developed for cofactor engineering, we still know very little about the regulation mechanism of cellular cofactor equilibrium. On the other hand, the variation of cofactor biosynthesis and regulation among different organisms poses a huge challenge for the cofactor engineering in cell factories. Thus, it is essential to illuminate the cellular cofactor biosynthesis, distribution and transportation in yeast for cofactor regulation. Artificial intelligence (AI) provides an opportunity for speeding up DBTL cycles by computer automatic design and optimization. The computer-aided design (CAD) systems have greatly improved the design quality and documentation communications, and created shareable databases for manufacturing. Recently, deep neural networks were trained from published reactions in organic chemistry, which enabled the discovery of retrosynthetic routes and resulted in faster and more selective synthesis than traditional methods (Segler et al., 2018). Some attempts have also been made in synthetic biology. An automated DBTL pipeline succeeded in improving the production of flavonoid (2S)-pinocembrin in E. coli (Carbonell et al., 2018). Another computational platform ClusterCAD formalized a paradigm for the design of PKS (type I modular polyketide synthase) chimeras and streamlined the process of designing experiments to test strategies for engineering PKS variants (Eng et al., 2018). However, the complexity and nonlinearity of biological systems makes it challenging in precise modeling and operation and thus needs novel and robust computational tools. The current protein design platforms rely on the homology of existing proteins at different resolutions and accurately predicting the structure of a novel protein is still challenging so far (Alford et al., 2017, Davey et al., 2017), not to mention the design of complex biological systems. Building an interoperable and reliable standard database is a prerequisite in biological engineering, which will be beneficial for rapid integration of data, continuous self-renewing of biological DBTL cycles, and automated design of metabolic pathways and molecules.
In summary, the yeast cells are ideal workhorses for overproduction of natural products, and advanced synthetic biology and systems biology can accelerate the rising of yeast cell factory for industrial application.
Acknowledgments
Authors acknowledge funds from the National Key Research and Development Program of China (2018YFA0900300), National Natural Science Foundation of China (Grant nos. 21922812 and 31970316), LiaoNing Revitalization Talents Program (XLYC1807191), the DICP and QIBEBT program (DICP & QIBEBT UN201706), and the Shanghai Sail Program (19YF1459300).
Author Contributions
R.C. and S.Y. wrote and revised the manuscript. L.Z. revised and provided feedback on the manuscript. Y.J.Z. conceived this study and wrote the manuscript.
Footnotes
My lab website: www.synbc.dicp.ac.cn
Dedicated to the 70th anniversary of Dalian Institute of Chemical Physics, Chinese Academy of Sciences
References
- Achmon Y., Zelas B.B., Fishman A. Cloning Rosa hybrid phenylacetaldehyde synthase for the production of 2-phenylethanol in a whole cell Escherichia coli system. Appl. Microbiol. Biotechnol. 2014;98:3603–3611. doi: 10.1007/s00253-013-5269-z. [DOI] [PubMed] [Google Scholar]
- Agapakis C.M., Boyle P.M., Silver P.A. Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat. Chem. Biol. 2012;8:527–535. doi: 10.1038/nchembio.975. [DOI] [PubMed] [Google Scholar]
- Ahn J., Park K.M., Lee H., Son Y.J., Choi E.S. GAL promoter-driven heterologous gene expression in Saccharomyces cerevisiae Δ strain at anaerobic alcoholic fermentation. FEMS Yeast Res. 2013;13:140–142. doi: 10.1111/1567-1364.12009. [DOI] [PubMed] [Google Scholar]
- Alford R., Leaver-Fay A., Jeliazkov J., O'Meara M., DiMaio F., Park H., Shapovalov M., Renfrew P., Mulligan V., Kappel K. The rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theor. Comput. 2017;13:3031–3048. doi: 10.1021/acs.jctc.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasov A., Waltenberger B., Pferschy-Wenzig E., Linder T., Wawrosch C., Uhrin P., Temml V., Wang L., Schwaiger S., Heiss E. Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnol. Adv. 2015;33:1582–1614. doi: 10.1016/j.biotechadv.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos J.L., Fink G.R., Stephanopoulos G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat. Biotechnol. 2013;31:335–341. doi: 10.1038/nbt.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymerick E., Darmawi J., Baidoo E.E.K., William Collins F., Keasling J.D., Dominique L. Correction: production of hydroxycinnamoyl anthranilates from glucose in Escherichia coli. Microb. Cell. Fact. 2013;12:62. doi: 10.1186/1475-2859-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker B.M., Overkamp K.M., van Maris A.J., Kötter P., Luttik M.A., van Dijken J.P., Pronk J.T. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001;25:15–37. doi: 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- Baumann L., Rajkumar A., Morrissey J., Boles E., Oreb M. A yeast-based biosensor for screening of short- and medium-chain fatty acid production. ACS Synth. Biol. 2018;7:2640–2646. doi: 10.1021/acssynbio.8b00309. [DOI] [PubMed] [Google Scholar]
- Besada-Lombana P., Da Silva N. Engineering the early secretory pathway for increased protein secretion in Saccharomyces cerevisiae. Metab. Eng. 2019;55:142–151. doi: 10.1016/j.ymben.2019.06.010. [DOI] [PubMed] [Google Scholar]
- Bhataya A., Schmidt-Dannert C., Lee P.C. Metabolic engineering of Pichia pastoris X-33 for lycopene production. Process. Biochem. 2009;44:1095–1102. [Google Scholar]
- Billingsley J.M., Denicola A.B., Tang Y. Technology development for natural product biosynthesis in Saccharomyces cerevisiae. Curr. Opin. Biotechnol. 2016;42:74–83. doi: 10.1016/j.copbio.2016.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby-Haas C.E., De C.-L.V. Mining high-throughput experimental data to link gene and function. Trends Biotechnol. 2011;29:174–182. doi: 10.1016/j.tibtech.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount B.A., Gowers G.-O.F., Ho J.C.H., Ledesma-Amaro R., Jovicevic D., McKiernan R.M., Xie Z.X., Li B.Z., Yuan Y.J., Ellis T. Rapid host strain improvement by in vivo rearrangement of a synthetic yeast chromosome. Nat. Commun. 2018;9:1932. doi: 10.1038/s41467-018-03143-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracher J.M., De H.E., Koster C.C., Van D.B.M., Daran J.G., Aja V.M., Pronk J.T. Laboratory evolution of a biotin-requiring Saccharomyces cerevisiae strain for full biotin prototrophy and identification of causal mutations. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.00892-17. e00892–00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S., Clastre M., Courdavault V., O'Connor S.E. De novo production of the plant-derived alkaloid strictosidine in yeast. Proc. Natl. Acad. Sci. U S A. 2015;112:3205–3210. doi: 10.1073/pnas.1423555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijs N.A., Zhou Y.J., Siewers V., Nielsen J. Long-chain alkane production by the yeast Saccharomyces cerevisiae. Biotechnol. Bioeng. 2015;112:1275–1279. doi: 10.1002/bit.25522. [DOI] [PubMed] [Google Scholar]
- Cai P., Gao J., Zhou Y. CRISPR-mediated genome editing in non-conventional yeasts for biotechnological applications. Microb. Cell. Fact. 2019;18:63. doi: 10.1186/s12934-019-1112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Lv Y.B., Chen J., Imanaka T., Wei L.J., Hua Q. Metabolic engineering of oleaginous yeast Yarrowia lipolytica for limonene overproduction. Biotechnol. Biofuels. 2016;9:214. doi: 10.1186/s13068-016-0626-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell P., Jervis A.J., Robinson C.J., Yan C., Dunstan M., Swainston N., Vinaixa M., Hollywood K.A., Currin A., Rattray N.J.W. An automated Design-Build-Test-Learn pipeline for enhanced microbial production of fine chemicals. Commun. Biol. 2018;1:66. doi: 10.1038/s42003-018-0076-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas J., Da Silva N.A. Engineering cofactor and transport mechanisms in Saccharomyces cerevisiae for enhanced acetyl-CoA and polyketide biosynthesis. Metab. Eng. 2016;36:80–89. doi: 10.1016/j.ymben.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Cardenas J., Silva N.A.D. Metabolic engineering of Saccharomyces cerevisiae for the production of triacetic acid lactone. Metab. Eng. 2014;25:194–203. doi: 10.1016/j.ymben.2014.07.008. [DOI] [PubMed] [Google Scholar]
- Cervin, M.A., Soucaille, P., and Valle, F. (2010). Process for the biological production of 1,3-propanediol with high yield. US. 7745184B2.
- Chatzivasileiou A.O., Ward V., Edgar S.M., Stephanopoulos G. Two-step pathway for isoprenoid synthesis. Proc. Natl. Acad. Sci. U S A. 2019;116:506–511. doi: 10.1073/pnas.1812935116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Bao J., Kim I., Siewers V., Nielsen J. Coupled incremental precursor and co-factor supply improves 3-hydroxypropionic acid production in Saccharomyces cerevisiae. Metab. Eng. 2014;22:104–109. doi: 10.1016/j.ymben.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Chen X., Dong X., Wang Y., Zhao Z., Liu L. Mitochondrial engineering of the TCA cycle for fumarate production. Metab. Eng. 2015;31:62–73. doi: 10.1016/j.ymben.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhou H., Meng W., Tan T. Control of ATP concentration in Escherichia coli using an ATP-sensing riboswitch for enhanced S-adenosylmethionine production. RSC Adv. 2017;7:22409–22414. [Google Scholar]
- Chen Z., Sun X., Li Y., Yan Y., Yuan Q. Metabolic engineering of Escherichia coli for microbial synthesis of monolignols. Metab. Eng. 2017;39:102–109. doi: 10.1016/j.ymben.2016.10.021. [DOI] [PubMed] [Google Scholar]
- Chen H., Zhu C., Zhu M., Xiong J., Ma H., Zhuo M., Li S. High production of valencene in Saccharomyces cerevisiae through metabolic engineering. Microb. Cell. Fact. 2019;18:195. doi: 10.1186/s12934-019-1246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Fan F., Qu G., Tang J., Xi Y., Bi C., Sun Z., Zhang X. Identification of Absidia orchidis steroid 11beta-hydroxylation system and its application in engineering Saccharomyces cerevisiae for one-step biotransformation to produce hydrocortisone. Metab. Eng. 2019;57:31–42. doi: 10.1016/j.ymben.2019.10.006. [DOI] [PubMed] [Google Scholar]
- Cheng S., Liu X., Jiang G., Wu J., Zhang J.L., Lei D., Yuan Y.J., Qiao J., Zhao G.R. Orthogonal engineering of biosynthetic pathway for efficient production of limonene in Saccharomyces cerevisiae. ACS Synth. Biol. 2019;8:968–975. doi: 10.1021/acssynbio.9b00135. [DOI] [PubMed] [Google Scholar]
- Cirino P.C., Arnold F.H. Exploring the diversity of heme enzymes through directed evolution. In: Brakmann S., Johnsson K., editors. Vol. 10. Wiley-VCH Verlag GmbH & Co. KGaA; 2002. pp. 215–243. (Directed Molecular Evolution of Proteins: Or How to Improve Enzymes for Biocatalysis). [Google Scholar]
- Clomburg J.M., Qian S., Tan Z., Cheong S., Gonzalez R. The isoprenoid alcohol pathway, a synthetic route for isoprenoid biosynthesis. Proc. Natl. Acad. Sci. U S A. 2019;116:12810–12815. doi: 10.1073/pnas.1821004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan J.E., Zhao X., Jiang Y. Function, attachment and synthesis of lipoic acid in Escherichia coli. Adv. Microb. Physiol. 2005;50:103–146. doi: 10.1016/S0065-2911(05)50003-1. [DOI] [PubMed] [Google Scholar]
- Curran K.A., Karim A.S., Gupta A., Alper H.S. Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications. Metab. Eng. 2013;19:88–97. doi: 10.1016/j.ymben.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajka J., Nathenson J., Benites V., Baidoo E., Cheng Q., Wang Y., Tang Y. Engineering the oleaginous yeast Yarrowia lipolytica to produce the aroma compound β-ionone. Microb. Cell. Fact. 2018;17:136. doi: 10.1186/s12934-018-0984-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner S., Le T., Wang S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 2011;508:1–12. doi: 10.1016/j.abb.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J.A., Damry A.M., Goto N.K., Chica R.A. Rational design of proteins that exchange on functional timescales. Nat. Chem. Biol. 2017;13:1280–1285. doi: 10.1038/nchembio.2503. [DOI] [PubMed] [Google Scholar]
- De K.S., Stanton L.H., Slaby T., Durot M., Holmes V.F., Patel K.G., Platt D., Shapland E.B., Serber Z., Dean J. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth. Biol. 2014;3:97–106. doi: 10.1021/sb4001992. [DOI] [PubMed] [Google Scholar]
- Deloache W.C., Russ Z.N., Narcross L., Gonzales A.M., Martin V.J.J., Dueber J.E. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat. Chem. Biol. 2015;11:465–471. doi: 10.1038/nchembio.1816. [DOI] [PubMed] [Google Scholar]
- DeLoache W., Russ Z., Dueber J. Towards repurposing the yeast peroxisome for compartmentalizing heterologous metabolic pathways. Nat. Commun. 2016;7:11152. doi: 10.1038/ncomms11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Salvo M., Safo M., Contestabile R. Biomedical aspects of pyridoxal 5'-phosphate availability. Front. Biosci. 2012;4:897–913. doi: 10.2741/E428. [DOI] [PubMed] [Google Scholar]
- Do C.T., Pollet B.J., Sibout R., Denoue D., Barriere Y., Lapierre C., Jouanin L. Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta. 2007;226:1117–1129. doi: 10.1007/s00425-007-0558-3. [DOI] [PubMed] [Google Scholar]
- Du H.X., Xiao W.H., Wang Y., Zhou X., Zhang Y., Liu D., Yuan Y.J. Engineering Yarrowia lipolytica for campesterol overproduction. PLoS One. 2016;11:e0146773. doi: 10.1371/journal.pone.0146773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X.P., Gao J.Q., Zhou Y.J. Advances in engineering methylotrophic yeast for biosynthesis of valuable chemicals from methanol. Chin. Chem. Lett. 2018;29:681–686. [Google Scholar]
- Dubuisson D., Dennis S.G. The formalin test: a quantitative study of the analgesic effects of morphine, meperidine, and brain stem stimulation in rats and cats. Pain. 1977;4:161–174. doi: 10.1016/0304-3959(77)90130-0. [DOI] [PubMed] [Google Scholar]
- Eichenberger M., Lehka B.J., Folly C., Fischer D., Martens S., Simon E., Naesby M. Metabolic engineering of Saccharomyces cerevisiae for de novo production of dihydrochalcones with known antioxidant, antidiabetic, and sweet tasting properties. Metab. Eng. 2017;39:80–89. doi: 10.1016/j.ymben.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger M., Hansson A., Fischer D., Durr L., Naesby M. De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res. 2018;18:foy046. doi: 10.1093/femsyr/foy046. [DOI] [PubMed] [Google Scholar]
- Ekr H., Minton N.P., Malys N. A transcription factor-based biosensor for detection of itaconic acid. ACS. Synth. Biol. 2018;7:1436–1446. doi: 10.1021/acssynbio.8b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C.H., Backman T.W., Bailey C.B., Magnan C., García Martín H., Katz L., Baldi P., Keasling J.D. ClusterCAD: a computational platform for type I modular polyketide synthase design. Nucleic Acids Res. 2018;46:D509. doi: 10.1093/nar/gkx893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C., Marillonnet S. Golden gate cloning. Methods Mol. Biol. 2014;1116:119–131. doi: 10.1007/978-1-62703-764-8_9. [DOI] [PubMed] [Google Scholar]
- Eudes A., Juminaga D., Baidoo E.E., Collins F.W., Keasling J.D., Loqué D. Erratum to: production of hydroxycinnamoyl anthranilates from glucose in Escherichia coli. Microb. Cell. Fact. 2014;13:8. doi: 10.1186/1475-2859-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhi M., Marhevka E., Masci T., Marcos E., Eyal Y., Ovadis M., Abeliovich H., Vainstein A. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab. Eng. 2011;13:474–481. doi: 10.1016/j.ymben.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Fischer M.J., Meyer S., Claudel P., Bergdoll M., Karst F. Metabolic engineering of monoterpene synthesis in yeast. Biotechnol. Bioeng. 2011;108:1883–1892. doi: 10.1002/bit.23129. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick P.F. Tetrahydropterin-dependent amino acid hydroxylases. Annu. Rev. Biochem. 1999;68:355–381. doi: 10.1146/annurev.biochem.68.1.355. [DOI] [PubMed] [Google Scholar]
- Galanie S., Thodey K., Trenchard I.J., Interrante M.F., Smolke C.D. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancaspero T., Locato V., Barile M. A regulatory role of NAD redox status on flavin cofactor homeostasis in S. cerevisiae mitochondria. Oxid. Med. Cell. Longev. 2013;2013:612784. doi: 10.1155/2013/612784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D., Young L., Chuang R., Venter J., Hutchison C., Smith H. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gong Z., Nielsen J., Zhou Y.J. Engineering robustness of microbial cell factories. Biotechnol. J. 2017;12:1700014. doi: 10.1002/biot.201700014. [DOI] [PubMed] [Google Scholar]
- Grewal P.S., Modavi C., Russ Z.N., Harris N.C., Dueber J.E. Bioproduction of a betalain color palette in Saccharomyces cerevisiae. Metab. Eng. 2018;45:180–188. doi: 10.1016/j.ymben.2017.12.008. [DOI] [PubMed] [Google Scholar]
- Guo J., Zhou Y.J., Hillwig M.L., Shen Y., Yang L., Wang Y., Zhang X., Liu W., Peters R.J., Chen X. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. U S A. 2013;110:12108–12113. doi: 10.1073/pnas.1218061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Ma X., Cai Y., Ma Y., Zhan Z., Zhou Y.J., Liu W., Guan M., Yang J., Cui G. Cytochrome P450 promiscuity leads to a bifurcating biosynthetic pathway for tanshinones. New Phytol. 2016;210:525–534. doi: 10.1111/nph.13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Sun J., Li D., Lu W. Heterologous biosynthesis of (+)-nootkatone in unconventional yeast Yarrowia lipolytica. Biochem. Eng. J. 2018;137:125–131. [Google Scholar]
- Hammer S.K., Avalos J.L. Harnessing yeast organelles for metabolic engineering. Nat. Chem. Biol. 2017;13:823–832. doi: 10.1038/nchembio.2429. [DOI] [PubMed] [Google Scholar]
- Hatlestad G.J., Sunnadeniya R.M., Akhavan N.A., Gonzalez A., Goldman I.L., McGrath J.M., Lloyd A.M. The beet R locus encodes a new cytochrome P450 required for red betalain production. Nat. Genet. 2012;44:816–820. doi: 10.1038/ng.2297. [DOI] [PubMed] [Google Scholar]
- Havir E.A., Hanson K.R. L-phenylalanine ammonia-lyase. II. Mechanism and kinetic properties of the enzyme from potato tubers. Biochemistry. 1968;7:1904–1914. doi: 10.1021/bi00845a039. [DOI] [PubMed] [Google Scholar]
- Hawkins K.M., Smolke C.D. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat. Chem. Biol. 2008;4:564–573. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo K.T., Kang S.Y., Hong Y.S. De novo biosynthesis of pterostilbene in an Escherichia coli strain using a new resveratrol O-methyltransferase from Arabidopsis. Microb. Cell. Fact. 2017;16:30. doi: 10.1186/s12934-017-0644-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Zhu Z., Nielsen J., Siewers V. Heterologous transporter expression for improved fatty alcohol secretion in yeast. Metab. Eng. 2018;45:51–58. doi: 10.1016/j.ymben.2017.11.008. [DOI] [PubMed] [Google Scholar]
- Ignea C., Pontini M., Maffei M.E., Makris A.M., Kampranis S.C. Engineering monoterpene production in yeast using a synthetic dominant negative geranyl diphosphate synthase. ACS Synth. Biol. 2014;3:298–306. doi: 10.1021/sb400115e. [DOI] [PubMed] [Google Scholar]
- Ignea C., Trikka F.A., Nikolaidis A.K., Georgantea P., Ioannou E., Loupassaki S., Kefalas P., Kanellis A.K., Roussis V., Makris A.M. Efficient diterpene production in yeast by engineering Erg20p into a geranylgeranyl diphosphate synthase. Metab. Eng. 2015;27:65–75. doi: 10.1016/j.ymben.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Ignea C., Pontini M., Motawia M., Maffei M., Makris A., Kampranis S. Synthesis of 11-carbon terpenoids in yeast using protein and metabolic engineering. Nat. Chem. Biol. 2018;14:9. doi: 10.1038/s41589-018-0166-5. [DOI] [PubMed] [Google Scholar]
- Ignea C., Raadam M.H., Motawia M.S., Makris A.M., Vickers C.E., Kampranis S.C. Orthogonal monoterpenoid biosynthesis in yeast constructed on an isomeric substrate. Nat. Commun. 2019;10:3799. doi: 10.1038/s41467-019-11290-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendresen C.B., Stahlhut S.G., Li M., Gaspar P., Siedler S., Förster J., Maury J.M., Borodina I., Nielsen A.T., Pettinari M.J. Highly active and specific tyrosine ammonia-lyases from diverse origins enable enhanced production of aromatic compounds in bacteria and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2015;81:4458–4476. doi: 10.1128/AEM.00405-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B., Wu Y., Li B.-Z., Mitchell L.A., Liu H., Pan S., Wang J., Zhang H.-R., Jia N., Li B. Precise control of SCRaMbLE in synthetic haploid and diploid yeast. Nat. Commun. 2018;9:1933. doi: 10.1038/s41467-018-03084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Xu S., Sun J., Zhang C., Li D., Lu W. Yarrowia lipolytica construction for heterologous synthesis of alpha-santalene and fermentation optimization. Appl. Microbiol. Biotechnol. 2019;103:3511–3520. doi: 10.1007/s00253-019-09735-w. [DOI] [PubMed] [Google Scholar]
- Jiang G.Z., Yao M.D., Wang Y., Zhou L., Song T.Q., Liu H., Xiao W.H., Yuan Y.J. Manipulation of GES and ERG20 for geraniol overproduction in Saccharomyces cerevisiae. Metab. Eng. 2017;41:57–66. doi: 10.1016/j.ymben.2017.03.005. [DOI] [PubMed] [Google Scholar]
- Jiang J., Yin H., Wang S., Zhuang Y., Liu S., Liu T., Ma Y. Metabolic engineering of Saccharomyces cerevisiae for high-level production of salidroside from glucose. J. Agric. Food Chem. 2018;66:4431–4438. doi: 10.1021/acs.jafc.8b01272. [DOI] [PubMed] [Google Scholar]
- Jun S., Sattler S., Cortez G., Vermerris W., Sattler S., Kang C. Biochemical and structural analysis of substrate specificity of a phenylalanine ammonia-lyase. Plant Physiol. 2018;176:1452–1468. doi: 10.1104/pp.17.01608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemény-Beke á., Aradi J., Damjanovich J., Beck Z., Facskó A., Berta A., Bodnár A. Apoptotic response of uveal melanoma cells upon treatment with chelidonine, sanguinarine and chelerythrine. Cancer Lett. 2006;237:67–75. doi: 10.1016/j.canlet.2005.05.037. [DOI] [PubMed] [Google Scholar]
- Kim S., Kim C., Son Y., Choi J., Siegenthaler R., Lee Y., Jang T., Song J., Kang H., Kaiser C. Molecular basis of maintaining an oxidizing environment under anaerobiosis by soluble fumarate reductase. Nat. Commun. 2018;9:4867. doi: 10.1038/s41467-018-07285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Jang I., Son S., Ko Y., Cho B., Kim S., Lee J. Tailoring the Saccharomyces cerevisiae endoplasmic reticulum for functional assembly of terpene synthesis pathway. Metab. Eng. 2019;56:50–59. doi: 10.1016/j.ymben.2019.08.013. [DOI] [PubMed] [Google Scholar]
- Kirby J., Dietzel K.L., Wichmann G., Chan R., Antipov E., Moss N., Baidoo E.E.K., Jackson P., Gaucher S.P., Gottlieb S. Engineering a functional 1-deoxy-D-xylulose 5-phosphate (DXP) pathway in Saccharomyces cerevisiae. Metab. Eng. 2016;38:494–503. doi: 10.1016/j.ymben.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby J., Keasling J. Metabolic engineering of microorganisms for isoprenoid production. Nat. Prod. Rep. 2008;25:656–661. doi: 10.1039/b802939c. [DOI] [PubMed] [Google Scholar]
- Kirby J., Keasling J.D. Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu. Rev. Plant Biol. 2009;60:335–355. doi: 10.1146/annurev.arplant.043008.091955. [DOI] [PubMed] [Google Scholar]
- Kozak B., van Rossum H., Luttik M., Akeroyd M., Benjamin K., Wu L., de Vries S., Daran J., Pronk J., van Maris A. Engineering acetyl coenzyme A supply: functional expression of a bacterial pyruvate dehydrogenase complex in the cytosol of Saccharomyces cerevisiae. MBio. 2014;5 doi: 10.1128/mBio.01696-14. e01696–e01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunjapur A.M., Hyun J.C., Prather K.L.J. Deregulation of S -adenosylmethionine biosynthesis and regeneration improves methylation in the E. coli de novo vanillin biosynthesis pathway. Microb. Cell. Fact. 2016;15:61. doi: 10.1186/s12934-016-0459-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue K., Melgar M., Martin V.J.J. Directed evolution of a fungal β-glucosidase in Saccharomyces cerevisiae. Biotechnol. Biofuels. 2016;9:1–15. doi: 10.1186/s13068-016-0470-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau W., Sattely E. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science. 2015;349:1224–1228. doi: 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau Y.H., Giessen T.W., Altenburg W.J., Silver P.A. Prokaryotic nanocompartments form synthetic organelles in a eukaryote. Nat. Commun. 2018;9:1311. doi: 10.1038/s41467-018-03768-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Lloyd N.D., Pretorius I.S., Borneman A.R. Heterologous production of raspberry ketone in the wine yeast Saccharomyces cerevisiae via pathway engineering and synthetic enzyme fusion. Microb. Cell Fact. 2016;15:49. doi: 10.1186/s12934-016-0446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levisson M., Araya-Cloutier C., de Bruijn W.J.C., van der Heide M., Salvador Lopez J.M., Daran J.M., Vincken J.P., Beekwilder J. Toward developing a yeast cell factory for the production of prenylated flavonoids. J. Agric. Food Chem. 2019;67:13478–13486. doi: 10.1021/acs.jafc.9b01367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Cann A., Liao J. Biofuels: biomolecular engineering fundamentals and advances. Annu. Rev. Chem. Biomol. Eng. 2010;1:19–36. doi: 10.1146/annurev-chembioeng-073009-100938. [DOI] [PubMed] [Google Scholar]
- Li S., Liu L., Chen J. Compartmentalizing metabolic pathway in Candida glabrata for acetoin production. Metab. Eng. 2014;28:1–7. doi: 10.1016/j.ymben.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Li M., Kildegaard K.R., Chen Y., Rodriguez A., Borodina I., Nielsen J. De novo production of resveratrol from glucose or ethanol by engineered Saccharomyces cerevisiae. Metab. Eng. 2015;32:1–11. doi: 10.1016/j.ymben.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Li S., Si T., Wang M., Zhao H. Development of a synthetic malonyl-CoA sensor in Saccharomyces cerevisiae for intracellular metabolite monitoring and genetic screening. ACS Synth. Biol. 2015;4:1308–1315. doi: 10.1021/acssynbio.5b00069. [DOI] [PubMed] [Google Scholar]
- Li Y., Li S., Thodey K., Trenchard I., Cravens A., Smolke C.D. Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. U S A. 2018;115:E3922–E3931. doi: 10.1073/pnas.1721469115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Wu Y., Zhang C., Sun J., Zhou Z., Lu W. Production of triterpene ginsenoside compound K in the non-conventional Yeast Yarrowia lipolytica. J. Agric. Food Chem. 2019;67:2581–2588. doi: 10.1021/acs.jafc.9b00009. [DOI] [PubMed] [Google Scholar]
- Li Z., Wang X., Zhang H. Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering. Metab. Eng. 2019;54:1–11. doi: 10.1016/j.ymben.2019.03.002. [DOI] [PubMed] [Google Scholar]
- Lian J., Si T., Nair N.U., Zhao H. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metab. Eng. 2014;24:139–149. doi: 10.1016/j.ymben.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Lian J., Mishra S., Zhao H. Recent advances in metabolic engineering of Saccharomyces cerevisiae: new tools and their applications. Metab. Eng. 2018;50:85–108. doi: 10.1016/j.ymben.2018.04.011. [DOI] [PubMed] [Google Scholar]
- Lian J., Zhao H. Functional reconstitution of a pyruvate dehydrogenase in the cytosol of Saccharomyces cerevisiae through lipoylation machinery engineering. ACS Synth. Biol. 2016;5:689–697. doi: 10.1021/acssynbio.6b00019. [DOI] [PubMed] [Google Scholar]
- Lieber C.S., Packer L. S-Adenosylmethionine: molecular, biological, and clinical aspects–an introduction. Am. J. Clin. Nutr. 2002;76:1148S–1150S. doi: 10.1093/ajcn/76/5.1148S. [DOI] [PubMed] [Google Scholar]
- Liu W., Luo Z., Wang Y., Pham N., Tuck L., Pérez-Pi I., Liu L., Shen Y., French C., Auer M. Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods. Nat. Commun. 2018;9:1936. doi: 10.1038/s41467-018-04254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Cheng J., Zhang G., Ding W., Duan L., Yang J., Kui L., Cheng X., Ruan J., Fan W. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 2018;9:448. doi: 10.1038/s41467-018-02883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Tu X., Xu Q., Bai C., Kong C., Liu Q., Yu J., Peng Q., Zhou X., Zhang Y. Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol. Metab. Eng. 2018;45:189–199. doi: 10.1016/j.ymben.2017.12.009. [DOI] [PubMed] [Google Scholar]
- Liu H., Marsafari M., Wang F., Deng L., Xu P. Engineering acetyl-CoA metabolic shortcut for eco-friendly production of polyketides triacetic acid lactone in Yarrowia lipolytica. Metab. Eng. 2019;56:60–68. doi: 10.1016/j.ymben.2019.08.017. [DOI] [PubMed] [Google Scholar]
- Liu L., Liu H., Zhang W., Yao M., Li B., Liu D., Yuan Y. Engineering the biosynthesis of caffeic acid in Saccharomyces cerevisiae with heterologous enzyme combinations. Engineering. 2019;5:287–295. [Google Scholar]
- Liu Q., Shi X., Song L., Liu H., Zhou X., Wang Q., Zhang Y., Cai M. CRISPR-Cas9-mediated genomic multiloci integration in Pichia pastoris. Microb. Cell. Fact. 2019;18:1–11. doi: 10.1186/s12934-019-1194-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yu T., Li X., Chen Y., Campbell K., Nielsen J., Chen Y. Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat. Commun. 2019;10:4976. doi: 10.1038/s41467-019-12961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Reiter M.A., d'Espaux L., Wong J., Denby C.M., Lechner A., Zhang Y., Grzybowski A.T., Harth S., Lin W. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature. 2019;567:123–126. doi: 10.1038/s41586-019-0978-9. [DOI] [PubMed] [Google Scholar]
- Liu G.S., Li T., Zhou W., Jiang M., Tao X.Y., Liu M., Zhao M., Ren Y.H., Gao B., Wang F.Q. The yeast peroxisome: a dynamic storage depot and subcellular factory for squalene overproduction. Metab. Eng. 2020;57:151–161. doi: 10.1016/j.ymben.2019.11.001. [DOI] [PubMed] [Google Scholar]
- Luttik M.A., Overkamp K.M., Kötter P., de Vries S., van Dijken J.P., Pronk J.T. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH Dehydrogenases catalyzing the oxidation of cytosolic NADH. J. Biol. Chem. 1998;273:24529–24534. doi: 10.1074/jbc.273.38.24529. [DOI] [PubMed] [Google Scholar]
- Lv X., Wang F., Zhou P., Ye L., Xie W., Xu H., Yu H. Dual regulation of cytoplasmic and mitochondrial acetyl-CoA utilization for improved isoprene production in Saccharomyces cerevisiae. Nat. Commun. 2016;7:12851. doi: 10.1038/ncomms12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y., Marsafari M., Koffas M., Zhou J., Xu P. Optimizing oleaginous yeast cell factories for flavonoids and hydroxylated flavonoids biosynthesis. ACS Synth. Biol. 2019;8:2514–2523. doi: 10.1021/acssynbio.9b00193. [DOI] [PubMed] [Google Scholar]
- Lyu X., Zhao G., Ng K.R., Mark R., Chen W.N. Metabolic engineering of Saccharomyces cerevisiae for de novo production of kaempferol. J. Agric. Food Chem. 2019;67:5596–5606. doi: 10.1021/acs.jafc.9b01329. [DOI] [PubMed] [Google Scholar]
- Ma B., Liu M., Li Z.H., Tao X., Wei D.Z., Wang F.Q. Significantly enhanced production of patchoulol in metabolically engineered Saccharomyces cerevisiae. J. Agric. Food Chem. 2019;67:8590–8598. doi: 10.1021/acs.jafc.9b03456. [DOI] [PubMed] [Google Scholar]
- Ma T., Shi B., Ye Z., Li X., Liu M., Chen Y., Xia J., Nielsen J., Deng Z., Liu T. Lipid engineering combined with systematic metabolic engineering of Saccharomyces cerevisiae for high-yield production of lycopene. Metab. Eng. 2019;52:134–142. doi: 10.1016/j.ymben.2018.11.009. [DOI] [PubMed] [Google Scholar]
- Ma Y.-R., Wang K.-F., Wang W.-J., Ding Y., Shi T.-Q., Huang H., Ji X.-J. Advances in the metabolic engineering of Yarrowia lipolytica for the production of terpenoids. Bioresour. Technol. 2019;281:449–456. doi: 10.1016/j.biortech.2019.02.116. [DOI] [PubMed] [Google Scholar]
- Malakar D., Dey A., Ghosh A.K. Protective role of S-adenosyl-L-methionine against hydrochloric acid stress in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2006;1760:1298–1303. doi: 10.1016/j.bbagen.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Mayer A.F., Hellmuth K., Schlieker H., Lopez-Ulibarri R., Oertel S., Dahlems U., Strasser A.W., Van Loon A.P.G.M. A highly efficient and cost-effective process for phytase production by recombinant strains of Hansenula polymorpha. Biotechnol. Bioeng. 1999;63:373–381. doi: 10.1002/(sici)1097-0290(19990505)63:3<373::aid-bit14>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Meadows A.L., Hawkins K.M., Tsegaye Y., Antipov E., Kim Y., Raetz L., Dahl R.H., Tai A., Mahatdejkul-Meadows T., Xu L. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature. 2016;537:694–697. doi: 10.1038/nature19769. [DOI] [PubMed] [Google Scholar]
- Medema M., Fischbach M. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015;11:639–648. doi: 10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mee M.T., Collins J.J., Church G.M., Wang H.H. Syntrophic exchange in synthetic microbial communities. Proc. Natl. Acad. Sci. U S A. 2014;111:E2149–E2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer W.H., Gidijala L., Fekken S., Kiel J.A., van den Berg M.A., Lascaris R., Bovenberg R.A., van der Klei I.J. Peroxisomes are required for efficient penicillin biosynthesis in Penicillium chrysogenum. Appl. Environ. Microbiol. 2010;76:5702–5709. doi: 10.1128/AEM.02327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michener J.K., Nielsen J., Smolke C.D. Identification and treatment of heme depletion attributed to overexpression of a lineage of evolved P450 monooxygenases. Proc. Natl. Acad. Sci. U S A. 2012;109:19504–19509. doi: 10.1073/pnas.1212287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M.D., Buron L.D., Bo S., Olsen C.E., Hansen B.G., Mortensen U.H., Halkier B.A. Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab. Eng. 2012;14:104–111. doi: 10.1016/j.ymben.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Minami H., Kim J.S., Ikezawa N., Takemura T., Katayama T., Kumagai H., Sato F. Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. U S A. 2008;105:7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J., Groves R., Hagel J., Facchini P. An N-methyltransferase from Ephedra sinica catalyzing the formation of ephedrine and pseudoephedrine enables microbial phenylalkylamine production. J. Biol. Chem. 2018;293:13364–13376. doi: 10.1074/jbc.RA118.004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser S., Strohmeier G.A., Leitner E., Plocek T.J., Vanhessche K., Pichler H. Whole-cell (+)-ambrein production in the yeast Pichia pastoris. Metab. Eng. Commun. 2018;7:e00077. doi: 10.1016/j.mec.2018.e00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa A., Matsumura E., Koyanagi T., Katayama T., Kawano N., Yoshimatsu K., Yamamoto K., Kumagai H., Sato F., Minami H. Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli. Nat. Commun. 2016;7:10390. doi: 10.1038/ncomms10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa A., Minami H., Kim J., Koyanagi T., Katayama T., Sato F., Kumagai H. A bacterial platform for fermentative production of plant alkaloids. Nat. Commun. 2011;2:326. doi: 10.1038/ncomms1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa A., Matsuzaki C., Matsumura E., Koyanagi T., Katayama T., Yamamoto K., Sato F., Kumagai H., Minami H. (R,S)-Tetrahydropapaveroline production by stepwise fermentation using engineered Escherichia coli. Sci. Rep. 2014;4:6695. doi: 10.1038/srep06695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura C.E., Whited G.M. Metabolic engineering for the microbial production of 1,3-propanediol. Curr. Opin. Biotechnol. 2003;14:454–459. doi: 10.1016/j.copbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Narcross L., Fossati E., Bourgeois L., Dueber J.E., Martin V.J. Microbial factories for the production of benzylisoquinoline alkaloids. Trends Biotechnol. 2016;34:228–241. doi: 10.1016/j.tibtech.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Nash A.I., Reginald M.N., Mary Elizabeth S., Swartz T.E., Bogomolni R.A., Hartmut L., Gardner K.H. Structural basis of photosensitivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. Proc. Natl. Acad. Sci. U S A. 2011;108:9449–9454. doi: 10.1073/pnas.1100262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J., Keasling J.D. Engineering cellular metabolism. Cell. 2016;164:1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Ohnson C.L., Pechonick E., Park S.D., Havemann G.D., Leal N.A., Bobik T.A. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J. Bacteriol. 2001;183:1577–1584. doi: 10.1128/JB.183.5.1577-1584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orhan I., Özçelik B., Karaoğlu T., Şener B. Antiviral and antimicrobial profiles of selected isoquinoline alkaloids from fumaria and corydalis species. Z. Naturforsch. C. 2007;62:19–26. doi: 10.1515/znc-2007-1-204. [DOI] [PubMed] [Google Scholar]
- Paddon C.J., Westfall P.J., Pitera D.J., Benjamin K., Fisher K., McPhee D., Leavell M.D., Tai A., Main A., Eng D. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- Palmer C.M., Miller K.K., Nguyen A., Alper H.S. Engineering 4-coumaroyl-CoA derived polyketide production in Yarrowia lipolytica through a beta-oxidation mediated strategy. Metab. Eng. 2020;57:174–181. doi: 10.1016/j.ymben.2019.11.006. [DOI] [PubMed] [Google Scholar]
- Patridge E., Gareiss P., Kinch M.S., Hoyer D. An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discov. Today. 2016;21:204–207. doi: 10.1016/j.drudis.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Peng B., Wood R.J., Nielsen L.K., Vickers C.E. An expanded heterologous GAL promoter collection for diauxie-inducible expression in Saccharomyces cerevisiae. ACS. Synth. Biol. 2018;7:748–751. doi: 10.1021/acssynbio.7b00355. [DOI] [PubMed] [Google Scholar]
- Phalip V., Kuhn I., Lemoine Y., Jeltsch J.M. Characterization of the biotin biosynthesis pathway in Saccharomyces cerevisiae and evidence for a cluster containing BIO5, a novel gene involved in vitamer uptake. Gene. 1999;232:43–51. doi: 10.1016/s0378-1119(99)00117-1. [DOI] [PubMed] [Google Scholar]
- Ping Y., Li X., You W., Li G., Yang M., Wei W., Zhou Z., Xiao Y. De novo production of the plant-derived tropine and pseudotropine in yeast. ACS Synth. Biol. 2019;8:1257–1262. doi: 10.1021/acssynbio.9b00152. [DOI] [PubMed] [Google Scholar]
- Plesa C., Sidore A.M., Lubock N.B., Zhang D., Kosuri S. Multiplexed gene synthesis in emulsions for exploring protein functional landscapes. Science. 2018;359:343–347. doi: 10.1126/science.aao5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao K., Wasylenko T.M., Zhou K., Xu P., Stephanopoulos G. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat. Biotechnol. 2017;35:173–177. doi: 10.1038/nbt.3763. [DOI] [PubMed] [Google Scholar]
- Qiu C., Zhai H., Hou J. Biosensors design in yeast and applications in metabolic engineering. FEMS Yeast Res. 2019;19:foz082. doi: 10.1093/femsyr/foz082. [DOI] [PubMed] [Google Scholar]
- Qu Y., Easson M.L.A.E., Froese J., Simionescu R., Luca V.D. Completion of the seven-step pathway from tabersonine to the anticancer drug precursor vindoline and its assembly in yeast. Proc. Natl. Acad. Sci. U S A. 2015;112:6224–6229. doi: 10.1073/pnas.1501821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radecka D., Mukherjee V., Mateo R.Q., Stojiljkovic M., Thevelein J.M. Looking beyond Saccharomyces: the potential of non-conventional yeast species for desirable traits in bioethanol fermentation. FEMS Yeast Res. 2015;15:fov053. doi: 10.1093/femsyr/fov053. [DOI] [PubMed] [Google Scholar]
- Ramsey S.A., Smith J.J., Orrell D., Marelli M., Petersen T.W., de Atauri P., Bolouri H., Aitchison J.D. Dual feedback loops in the GAL regulon suppress cellular heterogeneity in yeast. Nat. Genet. 2006;38:1082–1087. doi: 10.1038/ng1869. [DOI] [PubMed] [Google Scholar]
- Reyes L.H., Gomez J.M., Kao K.C. Improving carotenoids production in yeast via adaptive laboratory evolution. Metab. Eng. 2014;21:26–33. doi: 10.1016/j.ymben.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Rock C.O. Opening a new path to lipoic acid. J. Bacteriol. 2009;191:6782–6784. doi: 10.1128/JB.01151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Kildegaard K.R., Li M., Borodina I., Nielsen J. Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab. Eng. 2015;31:181–188. doi: 10.1016/j.ymben.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Chen Y., Khoomrung S., Ozdemir E., Borodina I., Nielsen J. Comparison of the metabolic response to over-production of p-coumaric acid in two yeast strains. Metab. Eng. 2017;44:265–272. doi: 10.1016/j.ymben.2017.10.013. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Strucko T., Stahlhut S.G., Kristensen M., Svenssen D.K., Forster J., Nielsen J., Borodina I. Metabolic engineering of yeast for fermentative production of flavonoids. Bioresour. Technol. 2017;245:1645–1654. doi: 10.1016/j.biortech.2017.06.043. [DOI] [PubMed] [Google Scholar]
- Ruben V., Igor C., Katarzyna R., Yuguo X., Lisa S., Geert G., Hoon K., Joanna C., Kris M., Pedro A. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science. 2013;341:1103–1106. doi: 10.1126/science.1241602. [DOI] [PubMed] [Google Scholar]
- Ryabova O.B., Chmil O.M., Sibirny A.A. Xylose and cellobiose fermentation to ethanol by the thermotolerant methylotrophic yeast Hansenula polymorpha. FEMS Yeast Res. 2003;4:157–164. doi: 10.1016/S1567-1356(03)00146-6. [DOI] [PubMed] [Google Scholar]
- Ryo S., Ishii J., Matsuno T., Nakamura Y., Matsubara D., Tominaga M., Kondo A. Positive feedback genetic circuit incorporating a constitutively active mutant Gal3 into yeast GAL induction system. ACS Synth. Biol. 2017;6:928–935. doi: 10.1021/acssynbio.6b00262. [DOI] [PubMed] [Google Scholar]
- Sauvageot N., Muller C., Hartke A., Auffray Y., Laplace J.M. Characterisation of the diol dehydratase pdu operon of Lactobacillus collinoides. FEMS Microbiol. Lett. 2002;209:69–74. doi: 10.1111/j.1574-6968.2002.tb11111.x. [DOI] [PubMed] [Google Scholar]
- Scalcinati G., Knuf C., Partow S., Chen Y., Maury J., Schalk M., Daviet L., Nielsen J., Siewers V. Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquitepene alpha-santalene in a fed-batch mode. Metab. Eng. 2012;14:91–103. doi: 10.1016/j.ymben.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Scheler U., Brandt W., Porzel A., Rothe K., Manzano D., Bozic D., Papaefthimiou D., Balcke G.U., Henning A., Lohse S. Elucidation of the biosynthesis of carnosic acid and its reconstitution in yeast. Nat. Commun. 2016;7:12942. doi: 10.1038/ncomms12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiroli D., Peracchi A. A subfamily of PLP-dependent enzymes specialized in handling terminal amines. Biochim. Biophys. Acta. 2015;1854:1200–1211. doi: 10.1016/j.bbapap.2015.02.023. [DOI] [PubMed] [Google Scholar]
- Schuler M.A., Werck-Reichhart D. Functional genomics of P450s. Annu. Rev. Plant Biol. 2003;54:629–667. doi: 10.1146/annurev.arplant.54.031902.134840. [DOI] [PubMed] [Google Scholar]
- Schwartz C.M., Hussain M.S., Blenner M., Wheeldon I. Synthetic RNA polymerase III promoters facilitate high efficiency CRISPR-Cas9 mediated genome editing in Yarrowia lipolytica. ACS. Synth. Biol. 2015;5:356–359. doi: 10.1021/acssynbio.5b00162. [DOI] [PubMed] [Google Scholar]
- Segler M., Preuss M., Waller M. Planning chemical syntheses with deep neural networks and symbolic AI. Nature. 2018;555:604–610. doi: 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- Sgobba E., Stumpf A.K., Vortmann M., Jagmann N., Krehenbrink M., Dirks-Hofmeister M.E., Moerschbacher B., Philipp B., Wendisch V.F. Synthetic Escherichia coli-Corynebacterium glutamicum consortia for l-lysine production from starch and sucrose. Bioresour. Technol. 2018;260:302–310. doi: 10.1016/j.biortech.2018.03.113. [DOI] [PubMed] [Google Scholar]
- Shao S., Ren C., Liu Z., Bai Y., Chen Z., Wei Z., Wang X., Zhang Z., Xu K. Enhancing CRISPR/Cas9-mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52. Int. J. Biochem. Cell Biol. 2017;92:43–52. doi: 10.1016/j.biocel.2017.09.012. [DOI] [PubMed] [Google Scholar]
- Skellam E. Strategies for engineering natural product biosynthesis in fungi. Trends Biotechnol. 2019;37:416–427. doi: 10.1016/j.tibtech.2018.09.003. [DOI] [PubMed] [Google Scholar]
- Smith H.O., Hutchison C.A., Pfannkoch C., Venter J.C. Generating a synthetic genome by whole genome assembly: ϕX174 bacteriophage from synthetic oligonucleotides. Proc. Natl. Acad. Sci. U S A. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan P., Smolke C. Engineering a microbial biosynthesis platform for de novo production of tropane alkaloids. Nat. Commun. 2019;10:3634. doi: 10.1038/s41467-019-11588-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley Fernandez S.M., Kellogg B.A., Poulter C.D. Farnesyl diphosphate synthase. Altering the catalytic site to select for geranyl diphosphate activity. Biochemistry. 2000;39:15316–15321. doi: 10.1021/bi0014305. [DOI] [PubMed] [Google Scholar]
- Stephanie G., Kate T., Trenchard I.J., Maria F.I., Smolke C.D. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit W.R., Entcheva P. Biotin in microbes, the genes involved in its biosynthesis, its biochemical role and perspectives for biotechnological production. Appl. Microbiol. Biotechnol. 2003;61:21–31. doi: 10.1007/s00253-002-1186-2. [DOI] [PubMed] [Google Scholar]
- Sun J., Shao Z., Zhao H., Nair N., Wen F., Xu J.H., Zhao H. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2012;109:2082–2092. doi: 10.1002/bit.24481. [DOI] [PubMed] [Google Scholar]
- Szczebara F.M., Chandelier C., Villeret C., Masurel A., Bourot S., Duport C., Blanchard S., Groisillier A., Testet E., Costaglioli P. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat. Biotechnol. 2003;21:143–149. doi: 10.1038/nbt775. [DOI] [PubMed] [Google Scholar]
- Szymanski E., Calvert J. Designing with living systems in the synthetic yeast project. Nat. Commun. 2018;9:2950. doi: 10.1038/s41467-018-05332-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarshis L.C., Proteau P.J., Kellogg B.A., Sacchettini J.C., Poulter C.D. Regulation of product chain length by isoprenyl diphosphate synthases. Proc. Natl. Acad. Sci. U S A. 1996;93:15018–15023. doi: 10.1073/pnas.93.26.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teresa Anna G., Giovanni B., Concetta P., Claudia C., Angelica M., Grazia Maria L., Matilde C., Maria B. FAD synthesis and degradation in the nucleus create a local flavin cofactor pool. J. Biol. Chem. 2013;288:29069–29080. doi: 10.1074/jbc.M113.500066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomik T., Wittig I., Choe J., Boles E., Oreb M. An artificial transport metabolon facilitates improved substrate utilization in yeast. Nat. Chem. Biol. 2017;13:1158–1163. doi: 10.1038/nchembio.2457. [DOI] [PubMed] [Google Scholar]
- Tramontin L.R.R., Kildegaard K.R., Sudarsan S., Borodina I. Enhancement of astaxanthin biosynthesis in oleaginous yeast Yarrowia lipolytica via microalgal pathway. Microorganisms. 2019;7:472. doi: 10.3390/microorganisms7100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenchard I.J., Siddiqui M.S., Thodey K., Smolke C.D. De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast. Metab. Eng. 2015;31:74–83. doi: 10.1016/j.ymben.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal L.S., Kelly C.L., Mordaka P.M., Heap J.T. Review of NAD(P)H-dependent oxidoreductases: properties, engineering and application. Biochim. Biophys. Acta. 2017;1866:327–347. doi: 10.1016/j.bbapap.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Vogl T., Hartner F.S., Glieder A. New opportunities by synthetic biology for biopharmaceutical production in Pichia pastoris. Curr. Opin. Biotechnol. 2013;24:1094–1101. doi: 10.1016/j.copbio.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J.M., Alper H.S. Synthetic biology and molecular genetics in non-conventional yeasts: current tools and future advances. Fungal Genet. Biol. 2015;89:126–136. doi: 10.1016/j.fgb.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Wang Y., San K.Y., Bennett G.N. Cofactor engineering for advancing chemical biotechnology. Curr. Opin. Biotechnol. 2013;24:994–999. doi: 10.1016/j.copbio.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Wang J.P., Naik P.P., Chen H.-C., Shi R., Lin C.-Y., Liu J., Shuford C.M., Li Q., Sun Y.-H., Tunlaya-Anukit S. Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell. 2014;26:894–914. doi: 10.1105/tpc.113.120881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Wei Y., Fan Y., Liu Q., Wei W., Yang C., Zhang L., Zhao G., Yue J., Yan X. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab. Eng. 2015;29:97–105. doi: 10.1016/j.ymben.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Wang T.Y., Tsai Y.H., Yu I.Z., Chang T.S. Improving 3'-hydroxygenistein production in recombinant Pichia pastoris using periodic hydrogen peroxide-shocking strategy. J. Microbiol. Biotechnol. 2016;26:498–502. doi: 10.4014/jmb.1509.09013. [DOI] [PubMed] [Google Scholar]
- Wang J., Mahajani M., Jackson S.L., Yang Y., Chen M., Ferreira E.M., Lin Y., Yan Y. Engineering a bacterial platform for total biosynthesis of caffeic acid derived phenethyl esters and amides. Metab. Eng. 2017;44:89–99. doi: 10.1016/j.ymben.2017.09.011. [DOI] [PubMed] [Google Scholar]
- Wang M., Chen B., Fang Y., Tan T. Cofactor engineering for more efficient production of chemicals and biofuels. Biotechnol. Adv. 2017;35:1032–1039. doi: 10.1016/j.biotechadv.2017.09.008. [DOI] [PubMed] [Google Scholar]
- Wang P., Wei W., Ye W., Li X., Zhao W., Yang C., Li C., Yan X., Zhou Z. Synthesizing ginsenoside Rh2 in Saccharomyces cerevisiae cell factory at high-efficiency. Cell Discov. 2019;5:5. doi: 10.1038/s41421-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhao S., Wang Z., Koffas M. Recent advances in modular co-culture engineering for synthesis of natural products. Curr. Opin. Biotechnol. 2019;62:65–71. doi: 10.1016/j.copbio.2019.09.004. [DOI] [PubMed] [Google Scholar]
- Wang Y., Oliver Y. Synthetic scaffolds increased resveratrol biosynthesis in engineered yeast cells. J. Biotechnol. 2012;157:258–260. doi: 10.1016/j.jbiotec.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Weber N., Gorwa-Grauslund M., Carlquist M. Exploiting cell metabolism for biocatalytic whole-cell transamination by recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2014;98:10. doi: 10.1007/s00253-014-5576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber N., Gorwa-Grauslund M., Carlquist M. Improvement of whole-cell transamination with Saccharomyces cerevisiae using metabolic engineering and cell pre-adaptation. Microb. Cell. Fact. 2017;16:3. doi: 10.1186/s12934-016-0615-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T.C., Pretorius I.S., Paulsen I.T. Synthetic evolution of metabolic productivity using biosensors. Trends Biotechnol. 2016;34:371–381. doi: 10.1016/j.tibtech.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Wong J., d’Espaux L., Dev I., van der Horst C., Keasling J. De novo synthesis of the sedative valerenic acid in Saccharomyces cerevisiae. Metab. Eng. 2018;47:94–101. doi: 10.1016/j.ymben.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Wriessnegger T., Augustin P., Engleder M., Leitner E., Muller M., Kaluzna I., Schurmann M., Mink D., Zellnig G., Schwab H. Production of the sesquiterpenoid (+)-nootkatone by metabolic engineering of Pichia pastoris. Metab. Eng. 2014;24:18–29. doi: 10.1016/j.ymben.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Wu Y., Xu S., Gao X., Li M., Li D., Lu W. Enhanced protopanaxadiol production from xylose by engineered Yarrowia lipolytica. Microb. Cell. Fact. 2019;18:83. doi: 10.1186/s12934-019-1136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Bowen C.H., Liu D., Zhang F. Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat. Chem. Biol. 2016;12:339–344. doi: 10.1038/nchembio.2046. [DOI] [PubMed] [Google Scholar]
- Yamanishi M., Ito Y., Kintaka R., Imamura C., Katahira S., Ikeuchi A., Moriya H., Matsuyama T. A genome-wide activity assessment of terminator regions in Saccharomyces cerevisiae provides a ″terminatome″ toolbox. ACS. Synth. Biol. 2013;2:337–347. doi: 10.1021/sb300116y. [DOI] [PubMed] [Google Scholar]
- Yan X., Fan Y., Wei W., Wang P., Liu Q., Wei Y., Zhang L., Zhao G., Yue J., Zhou Z. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014;24:770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Hui L., Zhang X.-M., Gong J.-S., Li H., Rao Z.-M., Shi J.-S., Xu Z.-H. Improvement of NADPH-dependent P450-mediated biotransformation of 7α,15α-diOH-DHEA from DHEA by a dual cosubstrate-coupled system. Steroids. 2015;101:15–20. doi: 10.1016/j.steroids.2015.05.005. [DOI] [PubMed] [Google Scholar]
- Yang K., Qiao Y., Li F., Xu Y., Yan Y., Madzak C., Yan J. Subcellular engineering of lipase dependent pathways directed towards lipid related organelles for highly effectively compartmentalized biosynthesis of triacylglycerol derived products in Yarrowia lipolytica. Metab. Eng. 2019;55:231–238. doi: 10.1016/j.ymben.2019.08.001. [DOI] [PubMed] [Google Scholar]
- Yang Z., Zhang Z. Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris : a review. Biotechnol. Adv. 2018;36:182–195. doi: 10.1016/j.biotechadv.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Yao Z., Zhou P., Su B., Su S., Ye L., Yu H. Enhanced Isoprene production by reconstruction of metabolic balance between strengthened precursor supply and improved isoprene synthase in Saccharomyces cerevisiae. ACS Synth. Biol. 2018;7:2308–2316. doi: 10.1021/acssynbio.8b00289. [DOI] [PubMed] [Google Scholar]
- Yee D.A., DeNicola A.B., Billingsley J.M., Creso J.G., Subrahmanyam V., Tang Y. Engineered mitochondrial production of monoterpenes in Saccharomyces cerevisiae. Metab. Eng. 2019;55:76–84. doi: 10.1016/j.ymben.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T., Zhou Y.J., Huang M., Liu Q., Pereira R., David F., Nielsen J. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell. 2018;174:1549–1558. doi: 10.1016/j.cell.2018.07.013. [DOI] [PubMed] [Google Scholar]
- Yuan J., Ching C.-B. Mitochondrial acetyl-CoA utilization pathway for terpenoid productions. Metab. Eng. 2016;38:303–309. doi: 10.1016/j.ymben.2016.07.008. [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang X. Modular co-culture engineering, a new approach for metabolic engineering. Metab. Eng. 2016;37:114–121. doi: 10.1016/j.ymben.2016.05.007. [DOI] [PubMed] [Google Scholar]
- Zhang L., Chen J., Zhou X., Chen X., Li Q., Tan H., Dong X., Xiao Y., Chen L., Chen W. Dynamic metabolic and transcriptomic profiling of methyl jasmonate-treated hairy roots reveals synthetic characters and regulators of lignan biosynthesis in Isatis indigotica Fort. Plant Biotechnol. J. 2016;14:2217–2227. doi: 10.1111/pbi.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Li M., Zhao G.R., Lu W. Alpha-Terpineol production from an engineered Saccharomyces cerevisiae cell factory. Microb. Cell. Fact. 2019;18:160. doi: 10.1186/s12934-019-1211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du L., Qu Z., Zhang X., Li F., Li Z., Qi F., Wang X., Jiang Y., Men P. Compartmentalized biosynthesis of mycophenolic acid. Proc. Natl. Acad. Sci. U S A. 2019;116:13305–13310. doi: 10.1073/pnas.1821932116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang D., Duan Y., Zheng X., Lin Y., Liang S. Production of lycopene by metabolically engineered Pichia pastoris. Biosci. Biotechnol. Biochem. 2019;84:463–470. doi: 10.1080/09168451.2019.1693250. [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu C.J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant. 2015;8:17–27. doi: 10.1016/j.molp.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Zhao X., Gust B., Heide L. S-Adenosylmethionine (SAM) and antibiotic biosynthesis: effect of external addition of SAM and of overexpression of SAM biosynthesis genes on novobiocin production in Streptomyces. Arch. Microbiol. 2010;192:289–297. doi: 10.1007/s00203-010-0548-x. [DOI] [PubMed] [Google Scholar]
- Zhao E.M., Zhang Y., Mehl J., Park H., Lalwani M.A., Toettcher J.E., Avalos J.L. Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature. 2018;555:683–687. doi: 10.1038/nature26141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Baumann U., Reymond J.L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. Expanding the terpenoid kingdom. Nat. Chem. Biol. 2018;14:1069–1070. doi: 10.1038/s41589-018-0167-4. [DOI] [PubMed] [Google Scholar]
- Zhou Y.J., Gao W., Rong Q., Jin G., Chu H., Liu W., Yang W., Zhu Z., Li G., Zhu G. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J. Am. Chem. Soc. 2012;134:3234–3241. doi: 10.1021/ja2114486. [DOI] [PubMed] [Google Scholar]
- Zhou K., Qiao K., Edgar S., Stephanopoulos G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol. 2015;33:377–383. doi: 10.1038/nbt.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Buijs N., Zhu Z., Gómez D., Boonsombuti A., Siewers V., Nielsen J. Harnessing yeast peroxisomes for biosynthesis of fatty-acid-derived biofuels and chemicals with relieved side-pathway competition. J. Am. Chem. Soc. 2016;138:15368–15377. doi: 10.1021/jacs.6b07394. [DOI] [PubMed] [Google Scholar]
- Zhou Y.J., Hu Y., Zhu Z., Siewers V., Nielsen J. Engineering 1-alkene biosynthesis and secretion by dynamic regulation in yeast. ACS Synth. Biol. 2018;7:584–590. doi: 10.1021/acssynbio.7b00338. [DOI] [PubMed] [Google Scholar]
- Zhu L., Zhu Y., Zhang Y., Li Y. Engineering the robustness of industrial microbes through synthetic biology. Trends Microbiol. 2012;20:94–101. doi: 10.1016/j.tim.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Zhu M., Wang C., Sun W., Zhou A., Wang Y., Zhang G., Zhou X., Huo Y., Li C. Boosting 11-oxo-beta-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiae via pairing novel oxidation and reduction system from legume plants. Metab. Eng. 2018;45:43–50. doi: 10.1016/j.ymben.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Zhou Y.J., Kang M.K., Krivoruchko A., Buijs N.A., Nielsen J. Enabling the synthesis of medium chain alkanes and 1-alkenes in yeast. Metab. Eng. 2017;44:81–88. doi: 10.1016/j.ymben.2017.09.007. [DOI] [PubMed] [Google Scholar]
- Ziemert N., Alanjary M., Weber T. The evolution of genome mining in microbes–a review. Nat. Prod. Rep. 2016;33:988–1005. doi: 10.1039/c6np00025h. [DOI] [PubMed] [Google Scholar]
- Zou X., Liu Y., Hsu N., Huang C., Lyu S., Chan H., Chang C., Yeh H., Lin K., Wu C. Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase. Acta Crystallogr. D Biol. Crystallogr. 2014;70:1549–1560. doi: 10.1107/S1399004714005239. [DOI] [PubMed] [Google Scholar]