Abstract
Objective
In these studies, we examined the association of single nucleotide polymorphisms (SNPs) of the IL1RN gene with radiographic severity of symptomatic knee osteoarthritis (SKOA) and the risk of incident OA. We also explored these genetic polymorphisms in patients with new onset rheumatoid arthritis (RA).
Methods
Over 1000 subjects who met American College of Rheumatology criteria for tibiofemoral OA were selected from three independent, National Institute of Health (NIH)-funded cohorts. CTA and TTG haplotypes formed from three SNPs of the IL1RN gene (rs419598, rs315952, rs9005) were assessed for association with radiographic severity, and risk for incident radiographic OA (rOA) in a nested case–control cohort. These IL1RN haplotypes were also assessed for association with disease activity (DAS28) and plasma inflammatory markers in patients with RA.
Results
Carriage of the IL1RN TTG haplotype was associated with increased odds of more severe rOA compared with age-matched, sex-matched and body mass index-matched individuals. Examination of the osteoarthritis initiative Incidence Subcohort demonstrated that carriage of the TTG haplotype was associated with 4.1-fold (p=0.001) increased odds of incident rOA. Plasma IL-1Ra levels were lower in TTG carriers, while chondrocytes from TTG carriers exhibited decreased secretion of IL-1Ra. In patients with RA, the TTG haplotype was associated with increased DAS28, decreased plasma IL-1Ra and elevations of plasma inflammatory markers (hsCRP, interleukin 6 (IL-6)).
Conclusion
Carriage of the IL1RN TTG haplotype is associated with more severe rOA, increased risk for incident OA, and increased evidence of inflammation in RA. These data suggest that the IL1RN TTG risk haplotype, associated with decreased IL-1Ra plasma levels, impairs endogenous ‘anti-inflammatory’ mechanisms.
Keywords: osteoarthritis, interleukins, inflammation, genetics, rheumatoid arthritis
Key messages.
What is already known about this subject?
Prior genetic studies have not identified any single causal locus with large effects on osteoarthritis (OA), but rather support the polygenic nature of the disease, consistent with the contribution of multiple variants with small effect sizes to variation in OA susceptibility or severity. The IL-1 gene cluster region has been associated with susceptibility to OA in various joints, but the results have been inconsistent.
What does this study add?
➢The IL1RN associations that we describe in over 1000 patients with symptomatic knee OA are compelling because the risk haplotype is highly prevalent and has a large, biologically consistent effect on age-dependent radiographic severity or risk of incident disease.
Our demonstration that the IL1RN risk haplotype is associated with more severe rheumatoid arthritis (RA) extends the biological implications to other chronic inflammatory conditions.
From a pathogenic perspective, the association of the IL1RN TTG risk haplotype with decreased plasma IL-1Ra and increased IL-6/hsCRP suggests that carriers of the IL1RN TTG haplotype experience more severe and earlier disease due to genetically determined impaired ‘anti-inflammatory’ mechanisms.
Key messages.
How might this impact on clinical practice or future developments?
Drug development in OA would benefit from genetic biomarkers that identify individuals at greater risk for more severe or incident OA.
Stratification by IL1RN risk haplotype in future clinical trial design and personalised medicine strategies could identify subsets of anti-IL1 responders/non-responders based on IL1RN risk haplotypes, as has been described in juvenile systemic arthritis.
Finally, the understanding of the pathogenic mechanisms of IL1RN variants that impair effective endogenous anti-inflammatory mechanisms in OA and RA could lead to the identification of novel targets for treatment.
Introduction
Osteoarthritis (OA) is characterised by focal loss of joint articular cartilage, osteophyte formation and subchondral bone remodelling. The production of interleukin 1 (IL-1β) and other mediators produced by cartilage and synovium induce a state of chronic low-grade inflammation that has been suggested to contribute to disease pathogenesis.1–4 Multiple genome-wide associations and candidate gene studies have identified genetic variants involved in the pathogenesis of OA,5–9 including variants in ALDH1A2, GDF5, VDR, IGF-1, COL11A1 and VEGF. However, genetic studies have not identified any single causal locus, but rather are consistent with the contribution of multiple variants with small effect sizes to variation in OA susceptibility or severity.10–12
We have previously examined 15 single nucleotide polymorphisms (SNPs) in six inflammatory response genes, including those for IL-1α, IL-1β, IL-1RN, TNFα, IL-10, oestrogen receptor 1 (ESR1) and determined whether polymorphisms of these genes could predict risk for radiographic knee OA severity. We found that radiographic severity was associated only with a three SNP haplotype (rs419598, rs315952 and rs9005) of IL1RN, the product of which is IL-1Ra.13 The goal of this study was to validate these findings in over 1000 additional individuals with or at risk for knee OA and to determine whether the findings extended to patients with rheumatoid arthritis (RA).
Methods
Participants with symptomatic knee OA
We assembled 1066 subjects from three independent cohorts of individuals with or at risk for knee OA. Participants met clinical (American College of Rheumatology) and radiographic criteria for tibiofemoral OA (Kellgren-Lawrence (KL) score ≥1); all had body mass index (BMI) <33 kg/m2 (see online supplementary file 1). Using these eligibility criteria, we established a study population by including 300–400 subjects from each cohort with the goal of reducing phenotypic heterogeneity across populations. Radiographs were scored for tibiofemoral KL grade (0–4) and minimal medial joint space width (mJSW).1 14–16
annrheumdis-2019-216055supp001.pdf (87.4KB, pdf)
New York University OA cohort
To validate our original observation linking IL1RN haplotypes to OA severity from the ‘founding’ cohort of 80 New York University (NYU) and 50 Duke symptomatic knee osteoarthritis (SKOA) patients,13 17 we recruited and followed 372 additional SKOA patients between 2008 and 2016. Individuals who comprised the ‘founding’ cohort are not included in this study (NYUSoM IRB approved no: # i05-131 and i12-03682).
Genetics of Generalized Osteoarthritis
We applied the same inclusion/exclusion criteria to select a subset of participants in the Genetics of Generalized Osteoarthritis (GOGO) study from Duke University,14 and identified 339 individuals who met the eligibility criteria. None of the GOGO patients selected for this study were among the participants included in the previously reported ‘founding’ cohort.13
Osteoarthritis initiative
We applied identical criteria to select a subject subset from the osteoarthritis initiative (OAI), an observational cohort study focused on identifying genetic and clinical risk factors, imaging and biochemical biomarkers for development and progression of knee OA. The OAI study recruited individuals divided into two subcohorts, ‘Progression’ and ‘Incidence’; inclusion and exclusion criteria for entry into the Progression and Incidence Subcohorts are available at http://oai.epi-ucsf.org/datarelease/.
Risk for SKOA
Using the OAI Incidence Subcohort, we performed a nested case–control study to assess the risk of incident disease. We identified 101 cases who developed either radiographic or symptomatic tibiofemoral radiographic knee OA within 2–4 years of baseline, and compared 101 controls who did not develop either frequent knee pain or radiographic tibiofemoral OA (>KL1) over a minimum of 4 years and for up to 96 months of follow-up, matched for gender, age and BMI at baseline visit (see online supplementary methods).
NYU new-onset RA cohort
All patients met the American College of Rheumatology/European League Against Rheumatism 2010 classification criteria for RA.18 Enrolled patients were seropositive: rheumatoid factor (95%); anti-citrullinated protein antibodies (100%). New-onset RA was defined as disease duration of a minimum of 6 weeks and up to 6 months since diagnosis, and absence of any treatment with disease-modifying anti-rheumatic drugs (DMARDs), biological therapy or steroids (ever) as we have described.19 Plasma samples from 145 RA subjects were selected for analysis. Clinical assessments included tender and swollen 28-joint counts, patient global disease activity assessment (0–100), and ESR to enable calculation of the DAS28-ESR.20 21
Haplotype determination
Since all three SNPs (rs419598, rs315952 and rs9005) are in the IL1RN gene, we evaluated haplotype effects on radiographic severity as described in our previous publication.13 All cases and controls were genotyped for the same set of SNP markers (rs419598, rs315952 and rs9005) in the IL1RN gene. Of the nine potential haplotypes that could be constructed from these three SNPs, two occurred with a frequency that were >1% (haplotypes CTA and TTG). Both CTA and TTG are found on the same locus. Specifically, 61.7% of subjects could be unambiguously inferred to carry 0, 1, or 2 copies of the TTG haplotype, and 12% of subjects could be unambiguously inferred to carry 1 or 2 copies of the CTA haplotype. Throughout this report, we denote the TTG-0 or TTG-1 or TTG-2 haplotype groups, to represent carriers of 0, 1, or 2 copies of the IL1RN TTG haplotype generated from the 3 IL1RN SNPs (rs419598, rs315952 and rs9005). The linkage disequilibrium parameters D′ and r2 for IL1RN SNPs rs419598, rs315952 and rs9005 are shown in online supplementary table 1 for all three cohorts. In the GOGO cohort, rs9005 was not directly genotyped but was imputed with high quality (INFO >0.8). For consistency, we used the most probable imputed genotypes for all three SNPs to generate IL1RN haplotypes. For both rs315952 and rs419598 SNPs genotype concordance was excellent (r2=0.981 and 0.976, respectively).
annrheumdis-2019-216055supp002.pdf (103.8KB, pdf)
Genetics and molecular analysis
Genotyping, cell culture assays and ELISA were performed as described in online supplementary file 1.
Statistical analyses
Primary analyses evaluated associations between haplotypes and radiographic severity. Genotype associations with radiographic severity were determined using Fisher’s exact test, adjusted using the false discovery rate, where appropriate.
For a continuous trait outcome, mJSW versus age, we used a regression model. Age and mJSW correlation were plotted, and at each age interval, the likelihood of mJSW was calculated with a 95% CI (for additional information see online supplementary file 1).
Results
Frequency of IL1RN haplotypes
The clinical, genetic and demographic parameters in the three cohorts are shown in table 1. The frequencies of the IL1RN TTG haplotypes, based on SNPs rs419598, rs315952 and rs9005, were similar across the three cohorts. For the combined cohort of 1066 participants, the frequencies of TTG-0, TTG-1 and TTG-2 were 26.3%, 42.2% and 19.5%, respectively. The overall frequency of the CTA-1 or CTA-2 haplotype was 12.0% but varied across cohorts (NYU 7%; GOGO 20%; OAI 9%). Approximately 30% of the TTG-0 haplotype subjects in the combined cohort were CTA carriers.
Table 1.
Demographic and radiographic characteristic features of symptomatic knee osteoarthritis patients from NYU, GOGO and OAI cohorts
| Cohort | Age in years | Sex (% females) | BMI | Ethnicity (% Caucasians) | KL 3/4 (%) |
mJSW in mm | Haplotype % frequencies | ||||
| TTG-0 | CTA-1/2 (TTG-0) | TTG-1 (CTA-0) | TTG-2 | CTA-1 (TTG-1) | |||||||
| NYU (n=372) | 61.4 (±10.4) | 62.7 | 26.41 (±3.52) | 64 | 48.9 | 3.21 (±1.53) | 22.3 | 8.0 | 46.0 | 15.3 | 10.5 |
| GOGO (n=339) | 67.0 (±8.2) | 75.5 | 26.8 (±3.40) | 100 | 29.8 | 3.35 (±1.4) | 32.0 | 20.0 | 41.0 | 27.0 | 0 |
| OAI (n=355) | 61.6 (±9.0) | 56.9 | 29.95 (±4.90) | 80 | 57.7 | 3.54 (±1.65) | 24.2 | 9.3 | 40.0 | 17.2 | 16.4 |
| Combined (n=1066) | 62.7 (±9.9) | 64.4 | 27.36 (±4.15) | 68.5 | 43.2 | 3.24 (±1.61) | 26.3 | 12.0 | 42.2 | 19.5 | 9.0 |
Details are shown of the mean (±SD) age, BMI and mJSW in millimetres (mm), as well as percentage of females, Caucasians, radiographic KL score 3 or 4 distribution and IL1RN TTG haplotype frequency distribution. The TTG-0 groups also include CTA-1/2 haplotype group. Haplotypes TTG-0 or TTG-1 or TTG-2, respectively, represent carriers of 0 or 1 or 2 copies of IL1RN haplotype produced using 3 IL1RN single nucleotide polymorphisms (rs419598, rs315952 and rs9005).
BMI, body mass index; GOGO, Genetics of Generalized Osteoarthritis study, Duke University; KL, Kellgren-Lawrence; mJSW, minimal medial joint space width; NYU, New York University School of Medicine; OAI, osteoarthritis initiative.
IL1RN TTG haplotype is associated radiographic severity
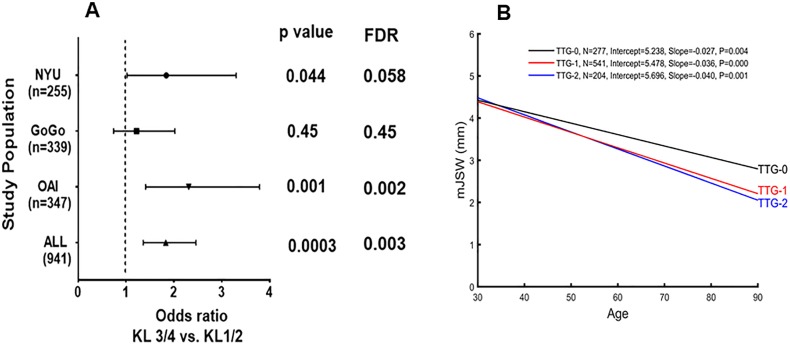
We first examined IL1RN TTG-0 versus TTG haplotypes (TTG-1 and TTG-2) for association with radiographic OA (rOA) severity as reported by KL scores and mean minimal medial radiographic joint space width (mJSW). As shown in a Forest plot (figure 1A), the IL1RN TTG haplotype was associated with an increased odds of more severe (KL 3/4 vs KL 1/2) radiographic knee OA compared with age-matched, sex-matched and BMI-matched knee OA patients with TTG-0 (OR of 1.83; 95% CI 1.36 to 2.46; p=0.0003). In the GOGO cohort, the TTG haplotype was associated with increased odds of KL 3/4 OA, which did not achieve significance. Of note, there was a lower percentage of participants with KL 3/4 OA severity (29.8%) in GOGO compared with the NYU (48.9%) and OAI (57.7%) cohorts, which may have reduced the statistical power of the test. We also note the higher frequency of the ‘protective’ CTA haplotype in the GOGO population (GOGO 20%; NYU 7%; OAI 9%), which could account for the smaller percentage of subjects with severe rOA, as we have reported.8
Figure 1.
Association of IL1RN TTG haplotypes with radiographic severity. (A) Forest plot displaying association of IL-1RN haplotypes (TTG-0 vs TTG-1/2) with radiographic severity in symptomatic knee osteoarthritis (OA) patients in three cohorts. Study-specific estimates of ORs with 95% CIs between severity of knee OA defined as Kellgren-Lawrence (KL) 1/2 versus KL 3/4 for haplotype rs419598, rs315952 and rs9005 ‘T-T-G’ are shown for three independent and all three combined cohort. (B) Association of radiographic minimal medial joint space width (mJSW), age and IL1RN haplotypes in NYU, OAI and GOGO cohorts. Influence of IL-1 receptor antagonist (IL1RN) haplotypes on the age relationship to mJSW of knee OA. Carriers of either TTG-1 or TTG-2 compared with TTG-0 had narrower JSW (mm) at each age (years) studied. The joint space width (JSW) of each knee in patients with knee OA who do not (TTG-0) or do carry the IL1RN TTG haplotype is plotted relative to age, and the regression line is shown for JSW relative to age. The figure shows the linear regression line for each of the IL1RN risk haplotypes. OA patients 982 out of 1066 from three cohorts are represented. GOGO, Genetics of Generalized Osteoarthritis; FDR, false discovery rate; NYU, New York University; OAI, osteoarthritis initiative.
We next assessed radiographic severity by minimal medial joint space width (mJSW). Table 2 shows that relative to TTG-0, carriage of TTG-1 or TTG-2 was associated with a TTG ‘dose-dependent’ decrease in mJSW in each individual population. Linear regression analysis confirmed that compared with TTG-0, the TTG-1 or TTG-2 haplotypes were significantly associated with decreased mJSW (online supplementary table 2A). This TTG dose effect was also observed for KL severity (online supplementary table 2B). The risk haplotype TTG carriers did not associate with either Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC) or Visual Analog Scale (VAS) pain in the NYU or OAI cohort.
Table 2.
Association of IL1RN haplotype (TTG) with radiographic mean minimal medial joint space width (mJSW) in three combined (NYU, GOGO and OAI) cohorts of symptomatic knee osteoarthritis patients
| Cohorts | TTG-0 | TTG-1 | TTG-2 | Beta (95% CI) | P value | FDR |
| NYU (n=372) | 3.43 (±1.44) (n=83) |
3.28 (±1.45) (n=209) |
2.60 (±1.70) (n=57) |
−0.39 (−0.64 to −0.13) |
0.0030 | 0.0046 |
| GOGO (n=339) | 3.67 (±1.31) (n=111) |
3.08 (±1.51) (n=138) |
3.35 (±1.38) (n=90) |
−0.18 (−0.38 to 0.02) |
0.0049 | 0.0950 |
| OAI (n=355) | 3.40 (±1.46) (n=86) |
3.20 (±1.63) (n=200) |
3.09 (±1.81) (n=61) |
−0.16 (−0.42 to 0.10) |
0.4787 | 0.2364 |
| All (n=1066) | 3.52 (±1.40) (n=280) |
3.20 (±1.53) (n=547) |
3.07 (±1.63) (n=208) |
−0.23 (−0.37 to −0.10) |
0.0023 | 0.0021 |
| All meta-analysis (n=1066) |
– | – | – | −0.23 (−0.37 to −0.10) |
0.0008 | 0.0021 |
mJSW data are presented in millimetres as mean (±SD) unless otherwise indicated; number of subjects in each group is represented in square brackets below each value. Haplotypes TTG-0 or TTG-1 or TTG-2, respectively, represent carriers of 0 or 1 or 2 copies of IL1RN haplotype produced using 3 IL1RN single nucleotide polymorphisms (rs419598, rs315952 and rs9005). Linear regression model (beta and 95% CI) was performed and p value was adjusted by FDR. The last row indicates the meta-analysis of all three cohorts by including cohort as a covariate.
FDR, false discovery rate; GOGO, Genetics of Generalized Osteoarthritis; NYU, New York University; OAI, osteoarthritis initiative.
We next performed a more detailed analysis of the effects of different combinations of CTA and TTG haplotypes on mJSW. As shown in table 3, any combination of CTA-1 or CTA-2 was associated with wider mJSW compared with TTG-2 or TTG-1. For example, the mJSW for CTA-2 and CTA-1 or CTA-2 carriers was 3.67 (1.3) and 3.51 (1.31), respectively. In comparison, the mean mJSW for TTG-2 and TTG-1 or TTG-2 carriers was 3.07 (1.62) and 3.19 (1.57), respectively. The differences between CTA and TTG mJSW were significant after adjustment for common covariates age, sex and BMI (table 3).
Table 3.
IL1RN TTG-1/2 carriers consistently associated with narrower joint space width in combined cohorts
| CTA-2 | TTG-2 | P value | P value_ ASB_ Adj |
CTA-1/2 (TTG-0) | TTG-2 | P value | P value_ ASB_ Adj |
CTA-1/2 (TTG-0) | TTG-1/2 (CTA-0) | P value | P value_ ASB_ Adj |
|
| mJSW, mm | 3.67 (±1.30) |
3.07 (±1.62) |
0.0063 | 0.0072 | 3.51 (±1.31) |
3.07 (±1.62) |
0.0106 | 0.0124 | 3.51 (±1.31) |
3.19 (±1.57) |
0.0304 | 0.0123 |
| Age, years | 65.01 (±8.24) |
65.03 (±9.29) | 0.9881 | – | 64.97 (±8.95) | 65.03 (±9.29) | 0.9498 | – | 64.97 (±8.95) | 63.64 (±9.85) | 0.1571 | – |
| BMI | 27.15 (±4.02) |
27.74 (±3.98) | 0.2933 | – | 27.31 (±3.66) | 27.74 (±3.98) | 0.3272 | – | 27.31 (±3.66) | 27.55 (±4.20) | 0.5478 | – |
| Sex (% male) | 36.76% | 35.1% | – | – | 33.6% | 35.1% | – | – | 33.6% | 33.4% | – | – |
Details are shown of the mean (±SD) age, BMI and mJSW in millimetres (mm), as well as percentage of males in each haplotype group in combined cohort (including New York University, Genetics of Generalized Osteoarthritis and osteoarthritis initiative). Haplotypes TTG-0 or TTG-1 or TTG-2, respectively, represent carriers of 0 or 1 or 2 copies of IL1RN haplotype produced using 3 IL1RN single nucleotide polymorphisms (rs419598, rs315952 and rs9005). Data are presented as mean (±SD) unless otherwise indicated. P values were determined by two-sample t-test and adjusted for ASB.
Bold indicates significant p values.
ASB, age, sex and BMI; BMI, body mass index; mJSW, minimal joint space width.
IL1RN TTG haplotype predicts age-related rOA
We next evaluated the interaction between age, mJSW and IL1RN genotypes relative to radiographic severity. As shown in figure 1B, linear regression analysis demonstrated that carriers of either TTG-1 or TTG-2 compared with TTG-0 had narrower JSW (mm) at each age studied. At age 70, for example, mean mJSW was 3.34 mm in TTG-0 versus 2.86 mm in TTG-2 (p<0.005).
We analysed whether rOA was associated with IL1RN risk haplotype after adjustment for risk covariates (age, sex and BMI) in the regression model. As shown in online supplementary table 3, IL1RN risk haplotype carriers (TTG-1 or 2) had significantly narrower tibiofemoral mJSW compared with TTG-0 carriers; the association remained significant after adjustment for the covariates. In gender-specific analyses, we show that both male and female carriers of either TTG-1 or TTG-2 carriers had narrower mJSW compared with TTG-0 (online supplementary table 4). In addition, among Blacks and Hispanics TTG-1 or TTG-2 carriers had narrower mJSW compared with TTG-0 (online supplementary tables 5 and 6) with a p value of 0.05–0.10.
IL1RN TTG haplotype predicts the risk of incident rOA
We examined participants from the Incidence Subcohort of the OAI, selecting the subgroup without clinical or radiographic evidence of knee OA at baseline. We identified 101 cases who developed either radiographic or symptomatic tibiofemoral radiographic knee OA within 2–4 years of baseline. Using a nested case–control approach, we selected 101 controls from the OAI Incidence Subcohort who did not develop either pain or radiographic tibiofemoral OA (>KL1) over a similar period matched for age, sex and BMI. These subjects were followed for 2–8 years. Table 4 shows that the presence of the IL1RN TTG-2 haplotype significantly increased the risk of incident knee rOA (OR=4.13 (1.75–9.72); p=0.001). After adjustment for age, sex and BMI with a logistic regression model, carriage of the TTG haplotype remained positively and significantly associated with incident rOA (beta coefficient=1.38; 95% CI 0.48 to 2.28; p=0.002).
Table 4.
IL1RN TTG haplotype increases risk of incident osteoarthritis (OA)
| Age (years) | BMI | Sex | TTG-2 | TTG-0 | OR (95% CI); P value |
Beta ASB adjusted | |
| Cases | 62.6±8.9 | 26.4±3.3 | M=31; F=70 | 48 (M=14; F=34) |
16 (M=7; F=9) |
4.13 (1.75–9.72); 0.001 |
1.38 (0.48–2.28); 0.002 |
| Controls | 62.6±8.8 | 26.3±3.3 | M=31; F=70 | 16 (M=6; F=10) |
22 (M=0; F=22) |
Development of incident OA in cases was defined as development of frequent knee pain and radiographic OA (KL ≥1 or 2) in the same knee or in bilateral knees. Controls were individuals whose baseline Kellgren-Lawrence (KL)=0 or 1 did not change at follow-up AND who did not develop frequent pain in either knee at 24, 36, 48, 72 and 96 months. Cases and controls were matched for age, sex and BMI. Estimates of OR with 95% CIs between severity of knee OA defined as KL 1/2 versus KL 3/4 for haplotype rs419598, rs315952 and rs9005 ‘T-T-G’ are shown. Haplotypes TTG-0 or TTG-1 or TTG-2, respectively, represent carriers of 0 or 1 or 2 copies of IL1RN haplotype produced using 3 IL1RN SNPs (rs419598, rs315952 and rs9005). Data for age and BMI are presented as mean±SD; data for sex are presented as N of male and female. The ORs of patients falling into case or control groups versus IL1RN haplotype were calculated using Fisher’s exact test. Beta coefficient (and 95% CI) from logistic regressions were adjusted for ASB.
ASB, age, sex and BMI; BMI, body mass index.
TTG risk haplotype is associated with decreased plasma IL-1Ra levels in patients with OA
Genetic variants of IL1RN have been associated with plasma levels of IL-1Ra and may regulate intracellular IL-1Ra protein trafficking.22–25 In our studies, mean plasma IL1Ra protein concentrations in TTG-2 carriers were lower than in CTA-2 carriers (346.50 vs 479.45 pg/mL, p=0.05, respectively). In this subset of patients, carriers of the CTA-2 haplotype had wider mean (SD) mJSW than did TTG-2 carriers (3.28 (1.46) vs 2.60 (1.67) mm, p=0.046, age-adjusted, sex-adjusted and BMI-adjusted). This was despite the fact that CTA-2 carriers were a mean 6 years older than the TTG-2 carriers (68.94 (9.92) vs 62.33 (10.96) years, p=0.08), consistent with an age-evident ‘protective’ effect of CTA on rOA.
We also performed a causal analysis to determine relationships among IL1RN TTG, CTA haplotypes, age, sex, BMI, IL-1Ra and mJSW. As shown in online supplementary figure 2, the CTA haplotype and BMI, but not age, are independently associated with plasma IL-1Ra. The causal analysis also indicated that the TTG haplotype directly associated with mJSW. As expected, both age and BMI associated with mJSW, and these effects were independent of the TTG haplotype.
annrheumdis-2019-216055supp003.pdf (2.1MB, pdf)
IL1RN haplotypes in patients with RA
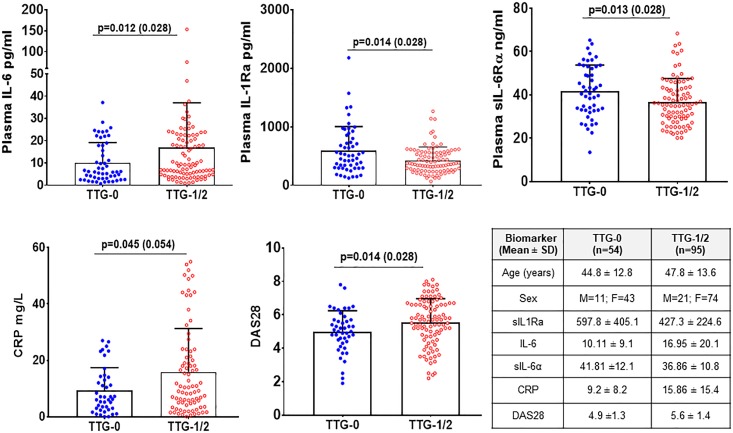
We examined plasma samples from new-onset, DMARD-untreated patients with RA, followed at NYU.20 21 As shown in figure 2, carriers of the TTG risk haplotype exhibited lower levels of plasma IL-1Ra and the soluble IL-6 receptor antagonist alpha (sIL-6Rα) than age-matched, BMI-matched and sex-matched individuals with RA. Conversely, in TTG carriers plasma IL-6 and hsCRP were higher. Clinically, carriers of the TTG haplotype exhibited greater disease activity (DAS28).
Figure 2.
IL1RN TTG-risk haplotype carriers have decreased IL-1Ra, soluble IL-6Rα and increased IL-6, CRP accompanied by increased disease activity (DAS28) in rheumatoid arthritis patients. Plasma levels of biomarkers were determined using ELISA as described in methods. The mean (SD) age, sex and biomarkers and number of subjects in each haplotype group are shown in the table. Each dot represents individual sample. The solid horizontal bar in each group represents the mean and the vertical bar represents the positive SD values. Mann-Whitney U test was used to analyse significance difference between specific haplotype groups (TTG-1/2) with haplotype of TTG-0 groups. The p value and false discovery rate adjusted values (in brackets) are shown in the figures. CRP, C‐reactive protein.
IL1RN haplotypes affect chondrocytes production of IL-1Ra
We next examined the relationship between TTG haplotypes and IL-1Ra production by chondrocytes. Chondrocytes were isolated from patients undergoing total joint replacement surgery at NYU, as described.26 Cell lysates and matched supernatants were analysed for IL-1Ra protein concentrations after 24 hours culture in the presence or absence of IL-1β. As shown in online supplementary table 7, following exposure to IL-β, basal levels of secreted IL-1Ra did not increase in TTG carriers, whereas intracellular concentrations of IL-1Ra in TTG chondrocytes were markedly increased. In contrast, chondrocytes obtained from TTG-0 individuals significantly increased the production of both intracellular and extracellular IL-1Ra following stimulation with IL-1β.
Discussion
The IL-1 gene cluster region has been associated with susceptibility to OA in various joints, but the results have been inconsistent.27–32 In this study of more than 1000 individuals with SKOA, we show that carriers of the IL1RN CTA haplotype (rs419598, rs315952 and rs9005) exhibit decreased age-dependent radiographic severity. Conversely, the TTG haplotype is associated with more severe rOA. Moreover, we demonstrate that the IL1RN TTG haplotype significantly increased the risk for incident tibiofemoral knee OA.
These results are consistent with our previous report that CTA in a large meta-analysis associated with less severe radiographic severity of knee OA.17 We note that in the genome-wide association study of OA using the UK Biobank, individual associations of each IL1RN SNP did not associate with knee OA at the genome-wide threshold (p<5×10−8).33 Similarly, in our study, IL1RN individual SNPs did not associate with knee OA (either KL or JSW).17 However, only the IL1RN haplotype, not tested in the UK Biobank study, was associated with more severe rOA in our studies. Another difference is the use of patient and/or hospital reported OA knee cases in the UK Biobank study, rather than radiographically confirmed SKOA as in our cohort, that may have resulted in a more heterogenous population of OA cases in the UK cohort.6 33
We also tested for association between the IL1RN haplotypes and radiographic progression, but neither of these associations were statistically significant. This is in contrast to the studies by Wu et al, who reported that the IL1RN TTG haplotype associated with change in KL over 4–11 years).34 Therefore, the lack of an association with progression in our studies could be a consequence of low power or insufficient years of follow-up. Alternatively, it is possible that exposures (eg, genotype) that increase disease susceptibility may also promote progression but such an association could be hard to detect because both progressors and non-progressors may already be enriched for the susceptibility genotype. This is a form of selection bias known as ‘collider bias’.
The association of IL1RN haplotypes with increased rOA at earlier age and the risk of incident disease may have clinical implications. Drug development in OA would benefit from genetic biomarkers that identify individuals at greater risk for more severe or incident OA.35 Stratification by IL1RN risk haplotype in future clinical trial design could identify subsets of anti-IL1 responders/non-responders based on IL1RN risk haplotypes, as has been described in juvenile systemic arthritis.36
What might be the biological explanation for the ‘yin/yang’ genetic effects of CTA versus TTG on rOA? We have previously shown that individuals carrying the IL1RN CTA (TTG-0) haplotype had significantly lower synovial fluid levels of IL-10 and showed a trend towards lower levels of IL-1β and IL-6.13 We here report that in patients with both OA and RA, carriers of the TTG haplotype exhibit reduced plasma levels of IL-1Ra compared with CTA carriers. We provide evidence in chondrocytes that this may result from decreased secretion of IL-1Ra protein. Although our studies focused on cartilage, the source of IL-1Ra in the synovial joint fluid could be from various tissues in the joint, including inflamed synovium. We postulate that the greater severity of rOA in carriers of the TTG haplotype results from impaired antagonism of chronic inflammatory IL-1β-driven processes.1 2
In these studies, we also asked whether the association of the TTG haplotype with more severe disease was limited to OA, or could be demonstrated in patients with new onset RA. We found that in RA, as in OA, plasma levels of IL-1Ra were decreased in TTG carriers, and this was accompanied by increased plasma IL-6 and hsCRP in association with increased clinical disease activity (DAS28).
With regard to limitations, our studies were restricted to SKOA, since standardised radiographs of other joints were not available in each of the study cohorts. Therefore, the risk conferred by the IL1RN risk haplotypes to individuals with OA of the hip, hands, and/or the spine will need assessment in future studies. In addition, we note that participants enrolled in our three cohorts were predominantly North American Caucasian. However, although the numbers were small, subset analysis of Black and Hispanic subjects indicated a trend towards increased rOA severity in each subset (online supplementary tables 5 and 6). Thus, confirmation of these findings in Black, Asian and Hispanic populations will require future studies.
Conclusion
In summary, we demonstrate that the IL1RN TTG haplotype identifies a subset of individuals with knee OA who are at increased risk for age-dependent rOA and increased risk for incident OA. Evidence for increased serological and clinical markers of disease activity in TTG carriers is also provided in new onset RA. We postulate that carriers of the IL1RN TTG haplotype experience more severe disease due to genetically determined impaired ‘anti-inflammatory’ mechanisms.
Acknowledgments
The NYU and OAI cohort bio-specimens were collected as part of a grant to SBA from the US National Institutes of Health (R01AR052873). Ascertainment of GOGO study participants was made possible by funding from GlaxoSmithKline. The OAI is a public–private partnership comprised five contracts (N01-AR-2-2258; N01-AR-2-2259; N01-AR-2-2260; N01-AR-2-2261; N01-AR-2-2262) funded by the National Institutes of Health, a branch of the US Department of Health and Human Services, and conducted by the OAI Study Investigators. Private funding partners include Merck Research Laboratories; Novartis Pharmaceuticals Corporation, GlaxoSmithKline; and Pfizer. Private sector funding for the OAI was managed by the Foundation for the National Institutes of Health. This manuscript was prepared using an OAI public use data set and does not necessarily reflect the opinions or views of the OAI investigators, the NIH, or the private funding partners. This work was also supported by the generous contributions of the William and Lynda Steere Foundation. The authors would like to thank the staff and participants of the NYU Osteoarthritis Biomarkers Study.
Footnotes
Handling editor: Josef S Smolen
Contributors: MA, JS, SK, JUS, MH, JMJ, VBK and SBA conceived and designed the study. MA, JS, SK, JUS and JB acquired the data. All authors were involved in analysis and interpretation of the data. All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be submitted. MA, HZ, MY and SBA had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Funding: The study was funded in part by a grant to Dr Abramson from the US National Institutes of Health (R01AR052873), by funding from GlaxoSmithKline to support inclusion of GOGO study participants, and by generous contributions from the William and Lynda Steere Foundation. None of the funding sources had any involvement in the design, execution and reporting of this study.
Competing interests: MA and SBA have one provisional patent application and another approved patent for the use of inflammatory and genetic biomarkers in predicting at-risk knee OA patients.
Patient consent for publication: Not required.
Ethics approval: The current study was performed in accordance with the ethical standards of the Declaration of Helsinki 1975, as revised in 2000, and studies were approved by the Institutional Review Board (IRB) of NYU School of Medicine.
Provenance and peer review: Not commissioned; externally peer-reviewed.
Data availability statement: Patient deidentified- NYU data are freely available upon request. OAI data are available from OAI.epi-uscf.org
References
- 1. Attur M, Krasnokutsky S, Statnikov A, et al. Low-Grade inflammation in symptomatic knee osteoarthritis: prognostic value of inflammatory plasma lipids and peripheral blood leukocyte biomarkers. Arthritis Rheumatol 2015;67:2905–15. 10.1002/art.39279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Robinson WH, Lepus CM, Wang Q, et al. Low-Grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat Rev Rheumatol 2016;12:580–92. 10.1038/nrrheum.2016.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Watson Levings RS, Broome TA, Smith AD, et al. Gene therapy for osteoarthritis: pharmacokinetics of intra-articular self-complementary adeno-associated virus interleukin-1 receptor antagonist delivery in an equine model. Hum Gene Ther Clin Dev 2018;29:90–100. 10.1089/humc.2017.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Attur MG, Dave M, Cipolletta C, et al. Reversal of autocrine and paracrine effects of interleukin 1 (IL-1) in human arthritis by type II IL-1 decoy receptor. potential for pharmacological intervention. J Biol Chem 2000;275:40307–15. 10.1074/jbc.M002721200 [DOI] [PubMed] [Google Scholar]
- 5. Gonzalez A, Valdes AM. Big data boost for osteoarthritis genetics. Nat Rev Rheumatol 2018;14:387–8. 10.1038/s41584-018-0023-7 [DOI] [PubMed] [Google Scholar]
- 6. Zengini E, Hatzikotoulas K, Tachmazidou I, et al. Genome-Wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat Genet 2018;50:549–58. 10.1038/s41588-018-0079-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodriguez-Fontenla C, Calaza M, Evangelou E, et al. Assessment of osteoarthritis candidate genes in a meta-analysis of nine genome-wide association studies. Arthritis Rheumatol 2014;66:940–9. 10.1002/art.38300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yau MS, Yerges-Armstrong LM, Liu Y, et al. Genome-Wide association study of radiographic knee osteoarthritis in North American Caucasians. Arthritis Rheumatol 2017;69:343–51. 10.1002/art.39932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Styrkarsdottir U, Thorleifsson G, Helgadottir HT, et al. Severe osteoarthritis of the hand associates with common variants within the aldh1a2 gene and with rare variants at 1p31. Nat Genet 2014;46:498–502. 10.1038/ng.2957 [DOI] [PubMed] [Google Scholar]
- 10. Fernandez-Moreno M, Rego I, Carreira-Garcia V, et al. Genetics in osteoarthritis. Curr Genomics 2008;9:542–7. 10.2174/138920208786847953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loughlin J. Genetic contribution to osteoarthritis development: current state of evidence. Curr Opin Rheumatol 2015;27:284–8. 10.1097/BOR.0000000000000171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reynard LN, Loughlin J. The genetics and functional analysis of primary osteoarthritis susceptibility. Expert Rev Mol Med 2013;15:e2 10.1017/erm.2013.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Attur M, Wang H-Y, Kraus VB, et al. Radiographic severity of knee osteoarthritis is conditional on interleukin 1 receptor antagonist gene variations. Ann Rheum Dis 2010;69:856–61. 10.1136/ard.2009.113043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kraus VB, Jordan JM, Doherty M, et al. The genetics of generalized osteoarthritis (GOGO) study: study design and evaluation of osteoarthritis phenotypes. Osteoarthritis Cartilage 2007;15:120–7. 10.1016/j.joca.2006.10.002 [DOI] [PubMed] [Google Scholar]
- 15. Mercier C, Piperno M, Vignon E, et al. In normal knees, joint space width (JSW) is correlated with the intermargin distance (IMD), a measure of medial tibial plateau alignment. variations in IMD explain variability in JSW in serial radiographs. Joint Bone Spine 2013;80:183–7. 10.1016/j.jbspin.2012.07.006 [DOI] [PubMed] [Google Scholar]
- 16. Attur M, Statnikov A, Samuels J, et al. Plasma levels of interleukin-1 receptor antagonist (IL1Ra) predict radiographic progression of symptomatic knee osteoarthritis. Osteoarthritis Cartilage 2015;23:1915–24. 10.1016/j.joca.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kerkhof HJM, Doherty M, Arden NK, et al. Large-Scale meta-analysis of interleukin-1 beta and interleukin-1 receptor antagonist polymorphisms on risk of radiographic hip and knee osteoarthritis and severity of knee osteoarthritis. Osteoarthritis Cartilage 2011;19:265–71. 10.1016/j.joca.2010.12.003 [DOI] [PubMed] [Google Scholar]
- 18. Aletaha D, Neogi T, Silman AJ, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League against rheumatism collaborative initiative. Arthritis Rheum 2010;62:2569–81. 10.1002/art.27584 [DOI] [PubMed] [Google Scholar]
- 19. Scher JU, Sczesnak A, Longman RS, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013;2:e01202 10.7554/eLife.01202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jain M, Attur M, Furer V, et al. Increased plasma IL-17F levels in rheumatoid arthritis patients are responsive to methotrexate, anti-TNF, and T cell costimulatory modulation. Inflammation 2015;38:180–6. 10.1007/s10753-014-0020-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scher JU, Ubeda C, Equinda M, et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum 2012;64:3083–94. 10.1002/art.34539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rafiq S, Stevens K, Hurst AJ, et al. Common genetic variation in the gene encoding interleukin-1-receptor antagonist (IL-1ra) is associated with altered circulating IL-1ra levels. Genes Immun 2007;8:344–51. 10.1038/sj.gene.6364393 [DOI] [PubMed] [Google Scholar]
- 23. Matteini AM, Li J, Lange EM, et al. Novel gene variants predict serum levels of the cytokines IL-18 and IL-1RA in older adults. Cytokine 2014;65:10–16. 10.1016/j.cyto.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramírez-Pérez S, De la Cruz-Mosso U, Hernández-Bello J, et al. High expression of interleukine-1 receptor antagonist in rheumatoid arthritis: Association with IL1RN*2/2 genotype. Autoimmunity 2017;50:468–75. 10.1080/08916934.2017.1412431 [DOI] [PubMed] [Google Scholar]
- 25. Redlitz KH, Yamshchikov VF, Cominelli F. Differential contribution of IL-1ra isoforms to allele-specific IL-1ra mRNA accumulation. J Interferon Cytokine Res 2004;24:253–60. 10.1089/107999004323034123 [DOI] [PubMed] [Google Scholar]
- 26. Attur M, Yang Q, Shimada K, et al. Elevated expression of periostin in human osteoarthritic cartilage and its potential role in matrix degradation via matrix metalloproteinase-13. Faseb J 2015;29:4107–21. 10.1096/fj.15-272427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loughlin J, Dowling B, Mustafa Z, et al. Association of the interleukin-1 gene cluster on chromosome 2q13 with knee osteoarthritis. Arthritis Rheum 2002;46:1519–27. 10.1002/art.10260 [DOI] [PubMed] [Google Scholar]
- 28. Moos V, Rudwaleit M, Herzog V, et al. Association of genotypes affecting the expression of interleukin-1beta or interleukin-1 receptor antagonist with osteoarthritis. Arthritis Rheum 2000;43:2417–22. [DOI] [PubMed] [Google Scholar]
- 29. Smith AJP, Keen LJ, Billingham MJ, et al. Extended haplotypes and linkage disequilibrium in the IL1R1-IL1A-IL1B-IL1RN gene cluster: association with knee osteoarthritis. Genes Immun 2004;5:451–60. 10.1038/sj.gene.6364107 [DOI] [PubMed] [Google Scholar]
- 30. Kanoh T, Hasegawa Y, Masui T, et al. Interleukin-1Beta gene polymorphism associated with radiographic signs of osteoarthritis of the knee. J Orthop Sci 2008;13:97–100. 10.1007/s00776-007-1205-3 [DOI] [PubMed] [Google Scholar]
- 31. Moxley G, Han J, Stern AG, et al. Potential influence of IL1B haplotype and IL1A-IL1B-IL1RN extended haplotype on hand osteoarthritis risk. Osteoarthritis Cartilage 2007;15:1106–12. 10.1016/j.joca.2007.03.022 [DOI] [PubMed] [Google Scholar]
- 32. Moxley G, Meulenbelt I, Chapman K, et al. Interleukin-1 region meta-analysis with osteoarthritis phenotypes. Osteoarthritis Cartilage 2010;18:200–7. 10.1016/j.joca.2009.08.006 [DOI] [PubMed] [Google Scholar]
- 33. Tachmazidou I, Hatzikotoulas K, Southam L, et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat Genet 2019;51:230–6. 10.1038/s41588-018-0327-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu X, Kondragunta V, Kornman KS, et al. Il-1 receptor antagonist gene as a predictive biomarker of progression of knee osteoarthritis in a population cohort. Osteoarthritis Cartilage 2013;21:930–8. 10.1016/j.joca.2013.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karsdal MA, Christiansen C, Ladel C, et al. Osteoarthritis--a case for personalized health care? Osteoarthritis Cartilage 2014;22:7–16. 10.1016/j.joca.2013.10.018 [DOI] [PubMed] [Google Scholar]
- 36. Arthur VL, Shuldiner E, Remmers EF, et al. IL1RN Variation influences both disease susceptibility and response to recombinant human interleukin-1 receptor antagonist therapy in systemic juvenile idiopathic arthritis. Arthritis Rheumatol 2018;70:1319–30. 10.1002/art.40498 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
annrheumdis-2019-216055supp001.pdf (87.4KB, pdf)
annrheumdis-2019-216055supp002.pdf (103.8KB, pdf)
annrheumdis-2019-216055supp003.pdf (2.1MB, pdf)