Abstract
DNA damage response (DDR) is critically important for cell survival, genome maintenance, and its defect has been exploited therapeutically in cancer treatment. Many DDR-targeting agents have been generated and have entered the clinic and/or clinical trials. In order to provide a global and unbiased view of DDR network, we designed a focused CRISPR library targeting 365 DDR genes and performed CRISPR screens on the responses to several DDR inhibitors and DNA-damaging agents in 293A cells. With these screens, we determined responsive pathways enriched under treatment with different types of small-molecule agents. Additionally, we showed that POLE3/4-deficient cells displayed enhanced sensitivity to an ATR inhibitor, a PARP inhibitor, and camptothecin. Moreover, by performing DDR screens in isogenic TP53 wild-type and TP53 knock-out cell lines, our results suggest that the performance of our CRISPR DDR dropout screens is independent of TP53 status. Collectively, our findings indicate that CRISPR DDR screens can be used to identify potential targets of small-molecule drugs and reveal that TP53 status does not affect the outcome of these screens.
1. Introduction
Genomic instability is one of the hallmarks of cancer [1]. The DNA damage response (DDR) network includes multiple cellular pathways that sense, signal and repair DNA lesions, which are critically important for cell survival and genome maintenance. Elucidating the complexity of DDR network will contribute meaningfully and decisively to our understanding of cancer biology and cancer treatment. DDR network integrates the regulation of cell cycle progression and the repair of DNA damage caused by numerous endogenous factors or exogenous agents, which ensures genome integrity and cell viability [2–6]. The success of PARP inhibitors for the treatment of BRCA-deficient cancers highlights the importance of investigating DDR deficiencies to provide novel therapeutic opportunities [7, 8]. DDR targeting agents are being developed to target molecules involved in either DNA damage signaling or DNA repair, including homologous recombination (HR), mismatch repair (MMR), Fanconi anemia (FA), nonhomologous end-joining (NHEJ), base excision repair (BER), and nucleotide excision repair (NER) [3, 5, 9]. In addition to the discovery of proper patients for these DDR targeting agents, it is also critical to identify the right dose and maximize the therapeutic window for cancer patients. Understanding of the interactions between drugs and genes can provide additional rationales that facilitate the development of combination therapies [9, 10].
Large-scale loss-of-function screening using clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 is a powerful tool for the identification of genetic interactions in human cells [11, 12]. This approach is already widely adopted by various laboratories, and numbers of CRISPR libraries targeting either the entire genome or a specific class of genes are available [8, 13–16]. Although mechanisms of action of many drugs were identified by RNA-interference (RNAi) screens [17–19], CRISPR-Cas9, the novel gene editing technology that creates frameshift mutations shows more precision than RNAi to unravel the mechanism of action of a drug or to identify the target of a small molecule. The appreciation of a single small-molecule genetic target or mechanism of action is critically important to developing the correct drug combination and the patient selection strategy for future clinical trials. Over the past 10 years, a large number of agents have entered the clinic that target molecules related to DNA damage repair, replication stress, cell cycle checkpoints, and other aspects of DDR [3]. However, the knowledge of the therapeutic window and the potential genes and pathways that can be used to predict response to DDR-targeting agents, including PARP inhibitors [7], remains limited. Moreover, understanding of gene and drug interactions may enable new combination therapies that could elicit more effective response than monotherapy.
Here, we created a focused CRISPR library including 365 DDR genes and performed CRISPR screens on the responses to several DDR inhibitors and DNA-damaging agents in 293A cells. We uncovered many responsive pathways under treatment with different types of DDR agents. We found that loss of POLE3/4, two subunits of DNA polymerase epsilon [20, 21], increased cells’ sensitivity to an ATR inhibitor, a PARP inhibitor, and camptothecin. Moreover, our results highlighted the importance of TP53 in orchestrating a variety of DDR mechanisms to maintain genome stability. Furthermore, we showed that the performance of dropout CRISPR screens is independent of TP53, which allows this powerful technology to be used broadly without the concern of p53 status.
2. Materials and methods
2.1. Cell Lines, plasmids, and antibodies
293A, HCT116, and 293T cells were purchased from ATCC and cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum. 293A or HCT116 TP53 knockout cells were generated using pSpCas9(BB)-2A-Puro (PX459)-containing sgRNAs targeting TP53. Clones were isolated from 96-well plates and confirmed by Western blotting and sequencing. Sequences of sgRNAs used for generating knockout cells are provided below: sgTP53–1: 5’-ACTTCCTGAAAACAACGTTC-3’; sgTP53–2: 5’-CCCCGGACGATATTGAACAA-3’; sgTP53–3: 5’-GAGCGCTGCTCAGATAGCGA-3’; sgLuc: 5’-CCCGGCGCCATTCTATCCGC-3’; sgmCherry: 5’-GGCCACGAGTTCGAGATCGA-3’; sgPOLE3: 5’-GTACAGCACGAAGACGCTGG-3’; sgPOLE4–1: 5’-GCGAGTGAAGGCCTTGGTGA-3’; sgPOLE4–2: 5’-TGTCCCGCTAGCGTCACGTC-3’.
pSpCas9(BB)-2A-Puro (PX459) and lentiCRISPR v2 were gifts from Feng Zhang (Addgene plasmid No. 52961) [22]. psPAX2 and pMD2G were used as lentiviral packaging plasmids.
Antibodies used in this study included those against POLE3 (A301–245A, Bethyl Laboratories), POLE4 (A9882, ABclonal Technology), vinculin (V9131, Sigma-Aldrich), and p53 (sc-126, Santa Cruz Biotechnology).
2.2. DDR inhibitors and DNA damaging agents
KU-55933, AZD6738, and LY2606368 were purchased from Selleck Chemicals. Olaparib was purchased from BioVision Inc. Hydroxyurea, cisplatin, mitomycin C (MMC), camptothecin, and etoposide were purchased from Sigma-Aldrich.
2.3. CRISPR DDR library design and construction
We designed a CRISPR DDR sub-library containing 4530 sgRNAs targeting 365 genes with a known or suspected DDR or DNA repair function, 45 core-essential genes, and 50 nonessential genes. We included 275 DDR genes described in the Knijnenburg paper [23], expanded the gene list based on the Pearl paper [5], and incorporated additional genes from other experts on core DDR pathways. We also included 45 out of 684 CEGv2 (i.e. core-essential) genes and 50 out of 927 NEGv1 (i.e. nonessential) genes as controls described in the Hart paper [16]. If possible, 10 sgRNAs were designed for each gene. Each oligo (77 nt) contained 20 nt sgRNA, 5’ universal flanking sequence: CTTGTGGAAAGGACGAAACACCG and 3’ flanking sequence: GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGG.
The oligo library was synthesized by Custom Array Inc. (Bothell, WA). The sequences of sgRNAs are provided in Supplementary Table S1. The DDR sub-library was amplified and cloned into lentiCRISPRv2 plasmid using Gibson Assembly Reaction Master Mix (NEB, E2611S).
2.4. CRISPR pooled screen and data processing
293A cells (2 × 107 cells per replicate) were infected with lentiviral CRISPR DDR library at a low multiplicity of infection (~ 0.3) and then selected with 2 μg/mL puromycin for 3 days. The infected cells were collected at the initial time point T0 and passaged every 3 days for 21 days and maintained at 1000-fold coverage.
Five million cells (1000-fold coverage) were harvested at T0 and the final time point at 21 days (T21) for genomic DNA extraction using a QIAamp DNA Blood Midi Kit (Qiagen). sgRNA inserts were amplified by PCR using NEBNext Q5 Hot Start HiFi PCR Master Mix (New England Biolabs). Illumina TruSeq adapters with i5 and i7 barcodes were added in a second round of PCR, and PCR products were sequenced on an Illumina NextSeq 500 High Output platform to determine sgRNA representations in each sample.
Next-generation sequencing reads were aligned to the DDR sub-library, and each sgRNA was counted using MAGeCK [24]. The sgRNA reads were processed with MAGeCK [24] and BAGEL [16, 25].
WebGestalt (http://www.webgestalt.org/) over-representation analysis [26] developed by Dr. Bing Zhang’s lab was used to profile DDR pathways based on top hits from MAGeCK analysis (false discovery rate < 0.1).
2.5. Cell viability assay
Cell viability was measured by a CellTiter-Glo luminescent kit (Promega, G7572). In brief, cells were seeded into 96-well plates at a density of 1000 cells per well. After 24 hours, cells were treated with different concentrations of DNA damage reagents or DMSO for 4 days. Cells were equilibrated at room temperature for approximately 15 minutes. CellTiter-Glo reagents were added into each well for 10 minutes on a shaker to induce cell lysis. Cell lysates were transferred into Corning 96-well white polystyrene microplates and subjected to luminescence detection using a BioTek Synergy 2 Multi-Mode Microplate Reader.
2.6. Statistical analysis
Statistical analyses were performed using GraphPad Prism 8 with two-way ANOVA followed with Bonferroni’s multiple comparisons test. The P-value less than 0.05 was considered statistically significant.
3. Results
3.1. CRISPR DDR screens with small-molecule agents
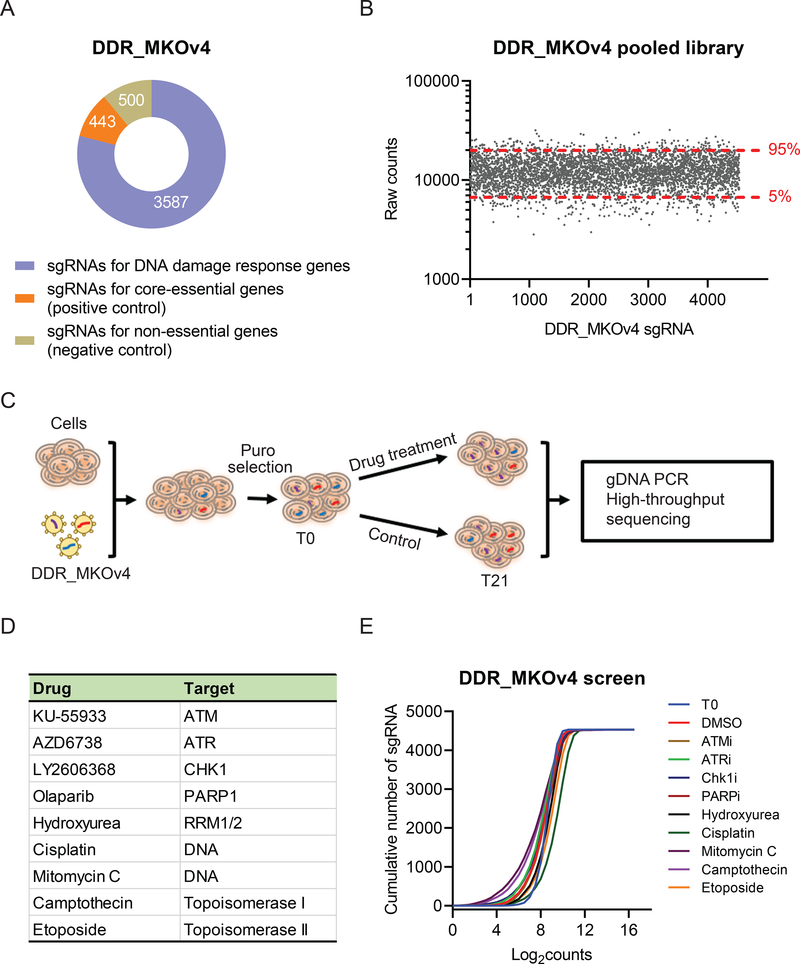
To understand the interplay between DDR pathways and DDR inhibitors or DNA-damaging agents, we performed CRISPR loss of function screens in 293A cells following treatment with a number of small-molecule drugs. We first designed a focused sgRNA library of 365 genes with known or suspected roles in DDR. We named this library DDR_MKOv4. The DDR_MKOv4 library harbors 4530 sgRNAs (up to 10 sgRNAs per gene) targeting 365 DDR genes, 50 nonessential genes as the negative control, and 45 core-essential genes as the positive control (Figure 1A, Supplementary Table 1). We cloned DDR_MKOv4 into lentiCRISPRv2 vector, and our library has more than 300-fold coverage of each sgRNA. We further verified successful library construction by sgRNA readout using Illumina sequencing (Figure 1B), which showed 100% recovery of input sgRNAs and 3-fold distribution of 90% of sgRNAs in this library (minimum reads: 2823; 5% reads: 6681; median reads: 11,978; 95% reads: 19,899; maximum reads: 32,023).
Figure 1. CRISPR/Cas9 screens with DNA damage response (DDR) agents in 293A cells.
(A) Pie chart showing the distribution of sgRNA composition in the DDR_MKO4 library. Orange: 443 sgRNAs targeting core-essential genes as the positive control. Green: 500 sgRNAs targeting nonessential genes as the negative control. Blue: 3587 sgRNAs targeting the genes with known or potential DDR functions. (B) Validation of the quality of the DDR_MKO4 library. NextSeq data from DDR_MKOv4 plasmid showed 90% sgRNA counts a 3-fold distribution. (C) Workflow of CRISPR screens performed in 293A cells following treatment with DDR agents. (D) DDR agents used in this study and their targets. (E) Cumulative frequency of sgRNAs from T0 and T21 in 293A cells without treatment and treated with different DDR agents. Shifts among the curves represent the depletion of subset of sgRNAs under the indicated condition.
For DDR-focused screening, cells were infected with DDR_MKOv4 library at a low multiplicity of infection (< 0.3), and infected cells were selected with puromycin for 3 days. The infected 293A cells were treated with either small-molecule drugs or DMSO in 3 technical replicates for 21 days. We used the IC20 of drugs for the screens. Samples were collected at T0 and T21 for high-throughput sequencing (Figure 1C). To evaluate broader and effective mechanisms of drugs, we used 9 different types of agents (Figure 1D), which included ATM inhibitor KU55933, ATR inhibitor AZD6738, CHK1 inhibitor LY2606368, PARP inhibitor olaparib, hydroxyurea, cisplatin, MMC, and topoisomerase inhibitors camptothecin and etoposide. As expected, high-throughput sequencing data from multiple samples revealed significant alterations of sgRNA diversity among different treatments (Figure 1E).
3.2. Analysis of CRISPR DDR screens data
We performed bioinformatics analysis using the MAGeCK count algorithm and BAGEL fold-change algorithm for CRISPR DDR screens. Total counts and distributions of sgRNAs indicated the high depth of sequencing and adequacy for subsequent bioinformatic analysis (Supplementary Figure 1A). The quality control analysis showed the fold changes of sgRNAs between T21 and T0 of positive and negative controls (Supplementary Figure 1B), which indicated robust cell viability measurements during these screens.
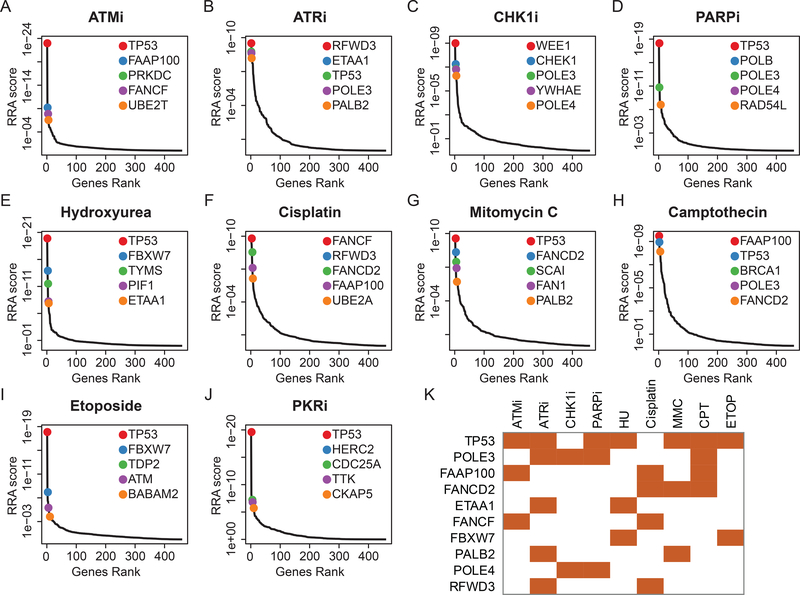
To identify the top hits, we used the MAGeCK RRA algorithm to compare drug-treated samples and DMSO-treated samples at T21. The genes were ranked by RRA scores (i.e., negative selection scores; Supplementary Table 2). Based on the threshold of false discovery rate < 0.1, we identified 25 genes whose loss sensitized cells to ATM inhibitor KU55933, 53 genes whose loss sensitized cells to ATR inhibitor AZD6738, 15 genes whose loss sensitized cells to CHK1 inhibitor, 48 genes whose loss sensitized cells to PARP inhibitor olaparib, 15 genes whose loss sensitized cells to hydroxyurea, 36 genes whose loss sensitized cells to cisplatin, 35 genes whose loss sensitized cells to MMC, 40 genes whose loss sensitized cells to camptothecin, and 20 genes whose loss sensitized cells to etoposide. The top 5 genes whose depletion sensitized cells to the indicated drugs are shown in Figure 2A–I. We included a non-DDR agent, the specific protein kinase R inhibitor (PKRi, C16), as the control and identified 36 genes whose loss sensitized cells to PKRi (Figure 2J).
Figure 2. Top 5 candidates when depletion could sensitize 293A cells to a number of small molecular agents based on MAGeCK RRA analysis.
Gene ranking is accomplished using a negative selection score (RRA score) from MAGeCK analysis under treatment with ATM inhibitor (ATMi) (A), ATR inhibitor (ATRi) (B), CHK1 inhibitor (CHK1i) (C), PARP inhibitor (PARPi) (D), hydroxyurea (E), cisplatin (F), mitomycin C (G), camptothecin (H), etoposide (I), or PKR inhibitor (PKRi) (J). Top 5 genes are highlighted with colored dots. (K) Crosstalk between top candidates and different DDR inhibitors and/or DNA-damaging agents. All listed genes appeared in at least 2 different DDR inhibitor and/or DNA-damaging agent treatment groups.
We found that TP53 was among the top 5 genes enriched by 7 of the 9 drugs (Figure 2K). The increased sensitivity of TP53-deficient cells to DDR inhibitors or DNA-damaging agents is consistent with the multitasking functions of p53 in controlling DNA damage checkpoints, which may impact the outcomes of various DNA repair processes [4, 27]. We also revealed the effect of TP53 status on CRISPR screen in subsequent experiments (please see below). Cells with POLE3/POLE4 loss were more sensitive to ATR, CHK1, and PARP inhibitors and camptothecin. The loss of core components of Fanconi anemia (FA) pathways, including FAAP100, FANCD2, FANCF, and PALB2, also endowed sensitivity to at least 2 different DDR agents (Figure 2K).
3.3. KEGG pathway profiling under treatment with different small-molecule agents
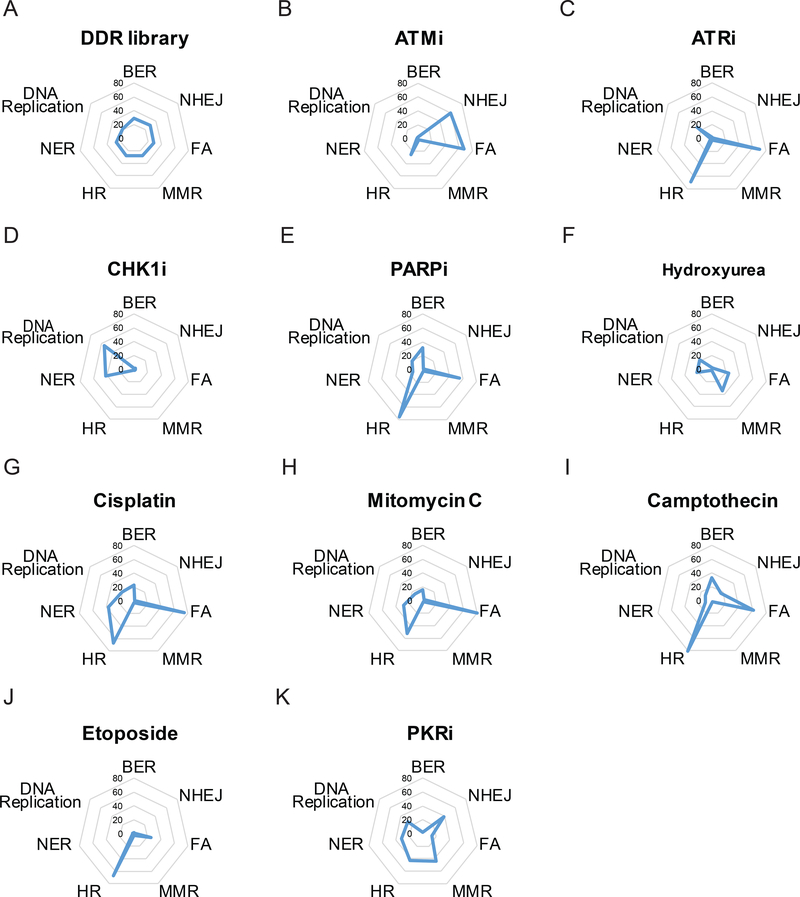
To determine pathways that affect the outcome of treatment with small-molecule agents, we performed WebGestalt over-representation analysis on genes that were depleted in response to different drug treatments. The DDR_MKOv4 library showed equal distribution of KEGG pathways, including HR, MMR, FA, NHEJ, BER, DNA replication, and NER (Figure 3A).
Figure 3. KEGG pathway profiling diagrams for individual agents.
(A) Radial plot of frequency of different KEGG pathways in the DDR_MKOv4 library. Kyoto Encyclopedia of Genes and Genomes pathway enrichment ratio was analyzed from WebGestalt. All the input genes were shown in the top list with false discovery rate < 0.1. Values in radial plot indicate the enrichment ratio calculated using WebGestalt KEGG. (B-K) Radial plots of KEGG pathway enrichment for indicated agents. Please be aware that since these were negative screens, the higher enrichment ratio indicates that a given KEGG pathway is more likely to be depleted following treatment with these agents. HR, homologous recombination; MMR, mismatch repair; FA, Fanconi anemia; NHEJ, nonhomologous end-joining; BER, base excision repair; NER, nucleotide excision repair. ATMi, ATM inhibitor; ATRi, ATR inhibitor; CHK1i, CHK1 inhibitor; PARPi, PARP inhibitor; PKRi, PKR inhibitor.
Upon treatment with different small-molecule agents, multiple pathways were depleted (Figure 3B–J). As shown in Figure 3B, genes related to NHEJ and FA pathways were largely lost under ATM inhibitor treatment. Inhibitors of ATR (Figure 3C) or PARP (Figure 3E) and camptothecin (Figure 3I) trigged cells more sensitive to loss of numerous genes involved in the HR or FA pathways. For DNA cross-linking agents such as cisplatin (Figure 3G) and MMC (Figure 3H), our results showed highly correlated profiling, with mainly depletion of the HR and FA pathways; this finding agrees with the fact that cisplatin and MMC have similar mechanisms of action. As the negative control, the PKR inhibitor showed similar ratios among many DDR pathways (Figure 3K), except the BER and FA pathways. Taken together, our results can be used to further understand the similarities and differences among the mechanisms of action of different small-molecule agents.
3.4. Comparison of CRISPR DDR screen and whole-genome screen
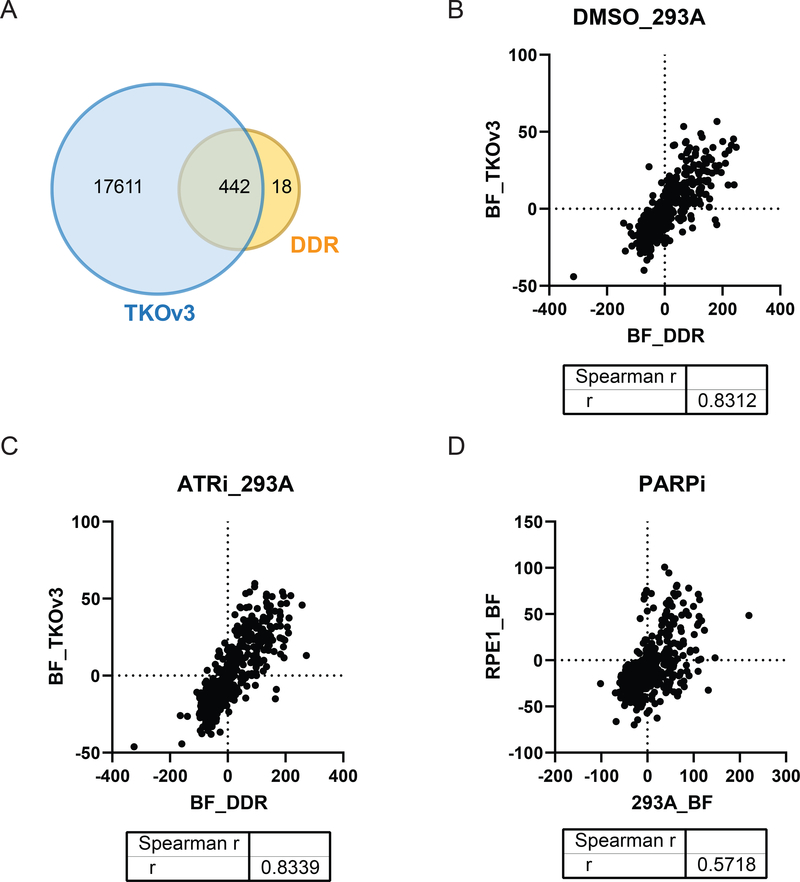
To ensure the quality of our CRISPR sub-library screens, we compared our data with publically available whole-genome screen data. As shown in Figure 4A, 442 of 460 genes in the DDR_MKOv4 library overlapped with the whole-genome library. According to BAGEL analysis, the fitness of these 442 overlapped genes displayed high correlation between the DDR_MKOv4 and TKOv3 screens in both DMSO-treated (Figure 4B, Spearman r = 0.8312) and ATR inhibitor-treated (Figure 4C, Spearman r = 0.8339) 293A cells. These correlations were higher than those between different cell lines (293A and RPE-1, Figure 4D, Spearman r = 0.5718). These analyses affirm the consistency of the whole-genome and sub-library screens for assessing gene functions but also point out that cellular genetic background may dramatically affect the results of these screens.
Figure 4. Correlation between screening results obtained from DNA damage response (DDR) sub-library and those obtained from the whole-genome library.
(A) Venn diagram showing the overlap genes presented in the DDR_MKOv4 library and the TKOv3 library. (B) Spearman correlation coefficient of Bayes factor (BF) distributions of the overlapped genes in 293A cells treated with DMSO. R = 0.8312. (C) Spearman correlation coefficient of BF distributions of the overlapped genes in ATR inhibitor (ATRi)-treated 293A cells. R = 0.8339. (D) Spearman correlation coefficient of BF distributions of the overlapped genes in PARP inhibitor (PARPi)-treated 293A cells (DDR_MKOv4) and in PARPi treated RPE-1 cells (TKOv1 [8]) is presented. R = 0.5718.
3.5. Deficiency in DNA polymerase epsilon subunits POLE3/POLE4 sensitize cells to ATR inhibitor, PARP inhibitor and camptothecin
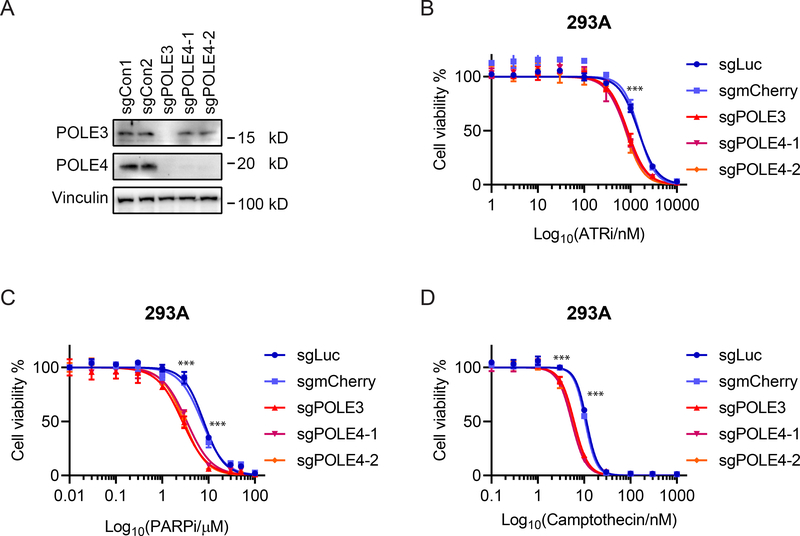
To further functionally validate the results, we next investigated the top candidates from our MAGeCK analysis. POLE3 and POLE4 are known subunits of DNA polymerase epsilon and more recently have been shown to form a newly identified histone H3-H4 chaperone complex that participates in the maintenance of chromatin integrity during DNA replication [20]. We knocked out POLE3 or POLE4 in 293A cells and found loss of POLE3 also decreased the stability of POLE4 (Figure 5A). We assessed the cells’ sensitivity to DDR targeting agents by CellTiter-Glo assay and observed that disruption of POLE3 or POLE4 caused hypersensitivity to ATR inhibition (Figure 5B), PARP inhibition (Figure 5C), and camptothecin treatment (Figure 5D) but had no effect on sensitivity to doxorubicin, cisplatin, MMC, and hydroxyurea (Supplementary Figure 2). We further validated some of these results using RPE-1 cells (Supplementary Figure 2).
Figure 5. POLE3/4-deficient cells are sensitive to ATRi, PARPi, and camptothecin.
(A) Confirmation of POLE3 and POLE4 knockout in 293A cells by Western blotting with the indicated antibodies. 293A cells were infected by lentivirus with either 2 control sgRNAs (sgLuciferase or sgmCherry) or sgRNAs targeting POLE3 or POLE4. Please note that POLE4 expression was abolished when POLE3 was knocked out. (B-D) CellTiter-Glo assay showed that POLE3- or POLE4-deficient cells were more sensitive to ATRi inhibitor (ATRi) (B), PARP inhibitor (PARPi) (C) or camptothecin (D) compared with control cells infected with lentivirus encoding sgLuc or sgmCherry. Cells were exposed to the indicated concentrations of inhibitors and cultured for 4 days. Data are from 3 technical replicates. *** indicates p value less than 0.001.
3.6. Performance comparison of CRISPR DDR screens according to TP53 status
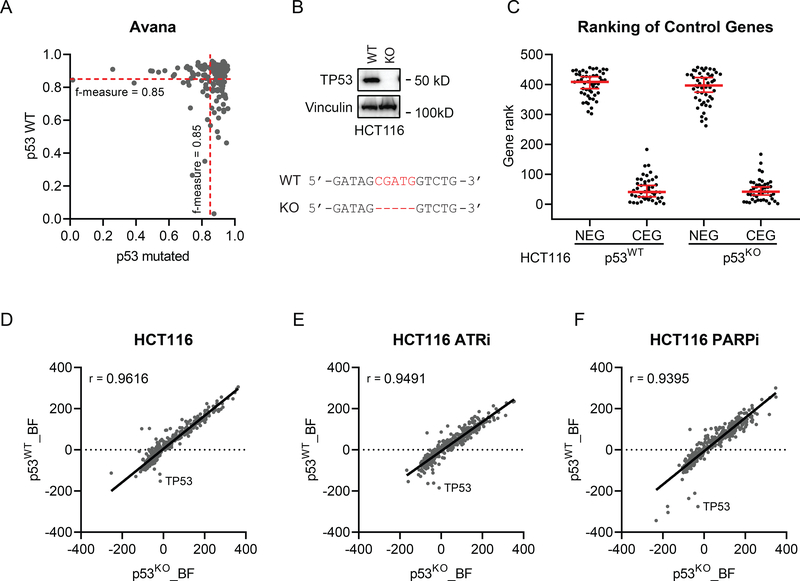
From our MAGeCK RRA analysis, we found loss of TP53 in cells trigged hypersensitivity to many DDR targeting drugs. Interestingly, recent studies showed discrepancy as to whether TP53 status would be a major concern during CRISPR screening because of TP53-mediated DDR [28–30]. We first examined the 514 CRISPR dropout screens performed in various cancer cell lines using the Avana whole-genome library [31]. The CRISPR screen performance calculated by F-measure based on precision and recall showed negligible difference across most of the cancer cell lines, regardless of TP53 status (Figure 6A). Second, we generated HCT116 TP53 knockout cells and 293A TP53 knockout cells (Figure 6B, Supplementary Figure 3A). Using the DDR_MKOv4 library, we performed CRISPR screens in HCT116 TP53 wild-type or TP53 knockout cells and 293A TP53 wild-type or TP53 knockout cells treated with DMSO, ATR inhibition, or PARP inhibition. For these screens, we adapted BAGEL analysis as previously described [16, 25] and calculated a Bayes factor (BF) for each gene. Based on BFs, genes were ranked from highest to lowest, and the gene ranking of core-essential genes was low as expected in both HCT116 wild-type cells and TP53 knockout cells (Figure 6C). However, there was no significant difference of core-essential gene rank between TP53 wild-type and TP53 knockout cells (Figure 6C). To evaluate the performance of all genes in HCT116 cells, we found that the BFs from TP53 wild-type cells highly correlated with those from TP53 knockout cells in the untreated group (Figure 6D, Spearman r = 0.9616), ATR inhibitor-treated group (Figure 6E, Spearman r = 0.9491), and PARP inhibitor-treated group (Figure 6F, Spearman r = 0.9395). Similar results from 293A cells are presented in Supplementary Figure 3.
Figure 6. Intact TP53 does not influence the performance of CRISPR screens.
(A) Similar CRISPR screen performance was noted in TP53 wild-type (WT) and TP53-mutated cancer cell lines. One hundred eighty-three Avana screens performed in TP53 WT cell lines were compared with randomly sampled same number of screens performed TP53 mutant cell lines. (B) Western blotting verified TP53 knockout (KO) in HCT116 cells. Vinculin was used as the loading control. Sanger sequencing showed that 5 nt (CGATG) of TP53 coding sequence was deleted in HCT116 KO cells. (C) The gene ranking of core-essential genes (CEG) and nonessential genes (NEG) in HCT116 WT and TP53 KO cells based on a log Bayes factor (BF) calculated using the BAGEL approach. Red line indicates median rank and 95% confidence interval. (D-F) Spearman correlation coefficient of BF distributions of DNA damage response genes in HCT116 WT and KO cells in mock treated (D), ATR inhibitor (ATRi)-treated (E), and PARP inhibitor (PARPi)-treated groups (F).
4. Discussion
In this study, we designed a focused sgRNA library, performed CRISPR DDR screenings in cells treated with different DDR inhibitors or DNA-damaging agents, and uncovered 64 genes that that affect the viability of 293A cells to two or more agents. Based on the genes with enhanced depletion under drug treatment, we determined KEGG pathways to highlight the potential synthetic lethetic pathways with these drugs. Our findings recovered many known synthetic lethal interactions. In addition, the use of our new DDR library allows further comparison of many DDR drug responses and may help the design of new combinations of these existing drugs.
It is possible that combination therapies using DDR inhibitors may provide better efficacy for cancer patients. Several clinical trials that combine PARP inhibitors with either an ATR inhibitor or WEE1 inhibitor are already ongoing. Our study showed the possible underlying mechanisms of different types of DDR agents, and these findings can be used to direct future studies of combination therapy. However, our current library can only be used for guiding the design of DDR gene-DDR targeting agent combinations. Development of DDR targeting agent combinations with other compounds, like agents targeting epigenetic regulation, will need additional CRISPR sub-libraries and/or other screening platforms [32]. It is anticipated that additional combination therapies based on DDR-targeting inhibitors with other agents that target different signaling pathways may reduce toxicity and show better efficacy. Moreover, the CRISPR DDR genetic screenings with DDR targeting agents performed in this study were carried out using in vitro cell culture; therefore, these results cannot be extended to the effects of these DDR agents in living subjects, as the agents may affect other physiological processes such as immune response. Further work by applying CRISPR DDR screens using cell line-derived xenograft [33] or genetically engineered mouse models will provide a better understanding of the mechanisms of action and their interaction with distinct genetic backgrounds in vivo, which would be important for selecting individual patients for these treatments.
In addition to help design of DDR gene-DDR targeting agent combinations, our work also validated a number of previous findings. BRCA1 or BRCA2 mutant cells show more sensitive to multiple drugs, including PARP inhibitor, cisplatin, MMC and camptothecin, as reported previously [18, 34, 35]. In Supplementary Table 2, we also reported FBXW7 depletion in 8 of 9 DDR inhibitors or DNA-damaging agents. Indeed, it was reported previously that FBXW7 is recruited to sites of DNA damage and enhances NHEJ repair [36]. Given that FBXW7 is known to be inactivated in human cancers, it is possible that loss of FBXW7 could be used as a potential biomarker for chemotherapy, radiation therapy, and others. Therefore, our dataset not only helps strengthen a number of previous observations, but also generate new biological hypotheses that need to be further tested. Our screen dataset can also be used to indicate the potential effect of genetic mutation and gene expression on drug activity and patient responsiveness. Our screens were performed in HEK93A cells, which are considered “normal” non-tumorigenic cells. Combined with the large cancer cell line databases such as the Cancer Cell Line Encyclopedia (CCLE), cBioPortal or DepMap portal, any investigator who is interested in our results can design new hypotheses in specific types of cancer based on the enriched pathways or genes. We highly recommend others to perform further validation studies using appropriate cancer cell lines with defined genetic and/or epigenetic alternations to test their drugs or genes of interest.
DDR pathways play important roles in genome maintenance and cancer development. Chemotherapy with cisplatin or mitomycin C are widely used for cancer treatments, which lead to a large amount of DNA lesions and result in cell death. Therefore, dissecting different DDR pathways is important for exploring new targets and/or strategies to increase efficacy of chemotherapy and expand clinical applications of these drugs. As shown in Figure 3, cells with HR, FA, NER, BER or DNA replication dysfunction show more sensitive to cisplatin or MMC comparing to cells with deficiency in other DDR pathways. Given that genetic testing for deficiency in various DNA damage related genes is becoming routine clinical practice, the information provided here may help us design clinical trials with new patient selection criteria.
Our data also suggest that TP53 status does not hamper the overall gene ranking of CRISPR screenings with ATR or PARP inhibition, which agrees with the recent finding by Brown and colleagues [28]. Our data from 2 pairs of cell lines, HCT116 and 293A, contributed to a clearer understanding that the performance of CRISPR loss-of-function screening is independent of TP53 status, although TP53 plays a prominent role in DDR [4]. There is no need to generate TP53-deficient cells prior to performing CRISPR screening because such manipulation not only would introduce extra work and cost but also might cause genomic instability due to TP53 loss and negatively affect the screening results.
More broadly, our study highlights the power of CRISPR screening to confirm or discover drugs’ mechanism of action, design new drug combinations, and characterize potential biomarkers for drug sensitivity. We anticipate that widely employing this powerful approach in preclinical studies will increase the successful applications of new drugs in clinical trials.
Supplementary Material
Supplementary Figure 1. Quality assessment of CRISPR/Cas9 DNA damage response (DDR) screens. (A) Deep-sequencing read distributions for each DDR agent treatment. The distributions of each sgRNA counts are presented. (B) Changes in sgRNAs targeting essential and nonessential genes during the CRISPR screens. Comparing the final time point T21 with baseline (T0), sgRNAs targeting essential genes changed dramatically, but no change in sgRNAs targeting nonessential genes was observed. NEG, non-essential genes; CEG, core-essential genes.
Supplementary Figure 2. Cell viability to doxorubicin, cisplatin, mitomycin C and hydroxyurea in POLE3/4-deficient cells. CellTiter-Glo assay showed that POLE3- or POLE4-deficient cells were not sensitive to doxorubicin (A), cisplatin (B), mitomycin C (C) or hydroxyurea (D) compared with control cells infected with lentivirus encoding sgLuc or sgmCherry. (E-I) Cell viability assay was repeated in RPE-1 cells. Cells were exposed to the indicated concentrations of inhibitors and cultured for 4 days. Data are from 3 technical replicates. * indicates p value less than 0.05 and *** indicates p value less than 0.001.
Supplementary Figure 3. (A) Western blotting and Sanger sequencing verified TP53 knockout (KO) in 293A cells. (B) The gene ranking of core-essential genes (CEG) and nonessential genes (NEG) in 293A WT and TP53 KO cells based on a log Bayes factor (BF) calculated using the BAGEL approach. Red line indicates median rank and 95% confidence interval. (C-E) Spearman correlation coefficient of BF distributions of DNA damage response genes in 293A WT and KO cells in mock treated (C), ATR inhibitor (ATRi)-treated (D), and PARP inhibitor (PARPi)-treated groups (E).
Highlights.
CRISRP DNA damage response (DDR) screens under DDR-related agents.
Consistency of the whole-genome and sub-library screens for assessing gene functions.
POLE3 or POLE4 deficiency caused cells hypersensitivity to ATRi, PARPi and CPT.
TP53 status does not hamper the power of CRISPR screens.
Acknowledgments
We thank all the members of the Chen laboratory for their help and constructive discussions. This work was supported in part by internal funding to J.C. In addition, J.C. received support from the Pamela and Wayne Garrison Distinguished Chair in Cancer Research. This work also benefited from the use of many reagents generated for studies funded by the Cancer Prevention & Research Institute of Texas (award RP160667 to J.C.) and NIH (award numbers CA210929, CA216911, and CA216437 to J.C.). We also thank The University of Texas MD Anderson Cancer Center Science Park Next-Generation Sequencing (NGS) Facility, which was supported by CPRIT Core Facility Support Grants RP120348 and RP170002. Editorial support was provided by Bryan Tutt in Scientific Publications Services, Research Medical Library, The University of Texas MD Anderson Cancer Center.
Footnotes
Conflict of Interest
The authors declare that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanahan D and Weinberg RA, Hallmarks of cancer: the next generation. Cell, 2011. 144(5): p. 646–74. [DOI] [PubMed] [Google Scholar]
- 2.Stingele J, Bellelli R, and Boulton SJ, Mechanisms of DNA-protein crosslink repair. Nat Rev Mol Cell Biol, 2017. 18(9): p. 563–573. [DOI] [PubMed] [Google Scholar]
- 3.Brown JS, et al. , Targeting DNA Repair in Cancer: Beyond PARP Inhibitors. Cancer Discov, 2017. 7(1): p. 20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams AB and Schumacher B, p53 in the DNA-Damage-Repair Process. Cold Spring Harb Perspect Med, 2016. 6(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearl LH, et al. , Therapeutic opportunities within the DNA damage response. Nat Rev Cancer, 2015. 15(3): p. 166–80. [DOI] [PubMed] [Google Scholar]
- 6.O’Connor MJ, Targeting the DNA Damage Response in Cancer. Mol Cell, 2015. 60(4): p. 547–60. [DOI] [PubMed] [Google Scholar]
- 7.Gourley C, et al. , Moving From Poly (ADP-Ribose) Polymerase Inhibition to Targeting DNA Repair and DNA Damage Response in Cancer Therapy. J Clin Oncol, 2019. 37(25): p. 2257–2269. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann M, et al. , CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature, 2018. 559(7713): p. 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nijman SM, Functional genomics to uncover drug mechanism of action. Nat Chem Biol, 2015. 11(12): p. 942–8. [DOI] [PubMed] [Google Scholar]
- 10.Lin A, et al. , Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci Transl Med, 2019. 11(509). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joung J, et al. , Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc, 2017. 12(4): p. 828–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T, Lander ES, and Sabatini DM, Viral Packaging and Cell Culture for CRISPR-Based Screens. Cold Spring Harb Protoc, 2016. 2016(3): p. pdb prot090811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, et al. , Genome-wide CRISPR screens reveal synthetic lethality of RNASEH2 deficiency and ATR inhibition. Oncogene, 2019. 38(14): p. 2451–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mengwasser KE, et al. , Genetic Screens Reveal FEN1 and APEX2 as BRCA2 Synthetic Lethal Targets. Molecular Cell, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munoz DM, et al. , CRISPR Screens Provide a Comprehensive Assessment of Cancer Vulnerabilities but Generate False-Positive Hits for Highly Amplified Genomic Regions. Cancer Discov, 2016. 6(8): p. 900–13. [DOI] [PubMed] [Google Scholar]
- 16.Hart T, et al. , High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell, 2015. 163(6): p. 1515–26. [DOI] [PubMed] [Google Scholar]
- 17.Nijman SM and Friend SH, Cancer. Potential of the synthetic lethality principle. Science, 2013. 342(6160): p. 809–11. [DOI] [PubMed] [Google Scholar]
- 18.Kaelin WG Jr., The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer, 2005. 5(9): p. 689–98. [DOI] [PubMed] [Google Scholar]
- 19.Elbashir SM, et al. , Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 2001. 411(6836): p. 494–8. [DOI] [PubMed] [Google Scholar]
- 20.Bellelli R, et al. , POLE3-POLE4 Is a Histone H3-H4 Chaperone that Maintains Chromatin Integrity during DNA Replication. Mol Cell, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Pursell ZF, and Linn S, Identification and cloning of two histone fold motif-containing subunits of HeLa DNA polymerase epsilon. J Biol Chem, 2000. 275(30): p. 23247–52. [DOI] [PubMed] [Google Scholar]
- 22.Sanjana NE, Shalem O, and Zhang F, Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods, 2014. 11(8): p. 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knijnenburg TA, et al. , Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep, 2018. 23(1): p. 239–254 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, et al. , MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol, 2014. 15(12): p. 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hart T and Moffat J, BAGEL: a computational framework for identifying essential genes from pooled library screens. BMC Bioinformatics, 2016. 17: p. 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao Y, et al. , WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res, 2019. 47(W1): p. W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kastenhuber ER and Lowe SW, Putting p53 in Context. Cell, 2017. 170(6): p. 1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown KR, et al. , CRISPR screens are feasible in TP53 wild-type cells. Mol Syst Biol, 2019. 15(8): p. e8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ihry RJ, et al. , p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med, 2018. [DOI] [PubMed] [Google Scholar]
- 30.Haapaniemi E, et al. , CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med, 2018. [DOI] [PubMed] [Google Scholar]
- 31.Meyers RM, et al. , Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet, 2017. 49(12): p. 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han K, et al. , Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol, 2017. 35(5): p. 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akcakaya P, et al. , In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendes-Pereira AM, et al. , Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med, 2009. 1(6–7): p. 315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farmer H, et al. , Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature, 2005. 434(7035): p. 917–21. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q, et al. , The WD40 domain of FBXW7 is a poly(ADP-ribose)-binding domain that mediates the early DNA damage response. Nucleic Acids Res, 2019. 47(8): p. 4039–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Quality assessment of CRISPR/Cas9 DNA damage response (DDR) screens. (A) Deep-sequencing read distributions for each DDR agent treatment. The distributions of each sgRNA counts are presented. (B) Changes in sgRNAs targeting essential and nonessential genes during the CRISPR screens. Comparing the final time point T21 with baseline (T0), sgRNAs targeting essential genes changed dramatically, but no change in sgRNAs targeting nonessential genes was observed. NEG, non-essential genes; CEG, core-essential genes.
Supplementary Figure 2. Cell viability to doxorubicin, cisplatin, mitomycin C and hydroxyurea in POLE3/4-deficient cells. CellTiter-Glo assay showed that POLE3- or POLE4-deficient cells were not sensitive to doxorubicin (A), cisplatin (B), mitomycin C (C) or hydroxyurea (D) compared with control cells infected with lentivirus encoding sgLuc or sgmCherry. (E-I) Cell viability assay was repeated in RPE-1 cells. Cells were exposed to the indicated concentrations of inhibitors and cultured for 4 days. Data are from 3 technical replicates. * indicates p value less than 0.05 and *** indicates p value less than 0.001.
Supplementary Figure 3. (A) Western blotting and Sanger sequencing verified TP53 knockout (KO) in 293A cells. (B) The gene ranking of core-essential genes (CEG) and nonessential genes (NEG) in 293A WT and TP53 KO cells based on a log Bayes factor (BF) calculated using the BAGEL approach. Red line indicates median rank and 95% confidence interval. (C-E) Spearman correlation coefficient of BF distributions of DNA damage response genes in 293A WT and KO cells in mock treated (C), ATR inhibitor (ATRi)-treated (D), and PARP inhibitor (PARPi)-treated groups (E).