Abstract
Microglia are parenchymal macrophages of the central nervous system (CNS); as professional phagocytes they are important for maintenance of the brain’s physiology. These cells are generated through primitive hematopoiesis in the yolk sac and migrate into the brain rudiment after establishment of embryonic circulation. Thereafter, microglia develop in a stepwise fashion, reaching complete maturity after birth. In the CNS, microglia self-renew without input from blood monocytes. Recent RNA-seq studies have defined a molecular signature for microglia under homeostasis. During disease, microglia undergo remarkable phenotypic changes, which reflect the acquisition of specialized functions tailored to the pathological context. In addition to microglia, the brain-border regions host populations of extra-parenchymal macrophages with disparate origins and phenotypes that have recently been delineated. In this review we outline recent findings that provide a deeper understanding of both parenchymal microglia and extra-parenchymal brain macrophages in homeostasis and during disease.
1. Ontogeny and phenotypic maturation of microglia
1.1. Microglia stem from yolk sac-derived myeloid progenitors
Microglia, the parenchymal macrophages of the CNS, originate from cKit+CD31+CD41+CD45lo erythromyeloid precursor cells (EMPs) generated during embryonic primitive hematopoiesis (1, 2). EMPs bud from clusters of Tie2+CD34+Flk1+ hemogenic endothelial cells in the yolk sac (YS) as early as E7.5, and provide the first cohort of macrophages and erythrocytes to the developing embryo (3–5). EMP-derived macrophage precursors (CD45+CD115+F4/80+CX3CR1+) appear in the blood around E8.5, spread via the embryonic circulation and seed all the tissues (including the CNS), wherein they complete the differentiation process (3, 6, 7). EMP commitment towards macrophages is supported by a combination of transcription factors, especially Runx1, PU.1 and IRF8 (4, 6, 8–10). Notably, microglia do not require the transcription factor Myb (11), which is necessary for the bone marrow hematopoiesis (12). YS-derived EMPs also migrate into the fetal liver, thus giving rise to the transient-definitive hematopoiesis. During the third week of gestation, EMPs in the liver are progressively outcompeted by hematopoietic stem cells (HSCs), which eventually establish definitive hematopoiesis in the bone marrow (13). Microglia first appear in the neuroepithelium at E9.5 (6, 8) (Figure 1). However, invasion of the embryonic CNS ends around E14.5, after the formation of the blood-brain barrier (BBB) (14). Thereafter, microglia spread throughout the CNS parenchyma and undertake a stepwise differentiation process, reaching a final maturation state around the second postnatal week (15, 16).
Figure 1:
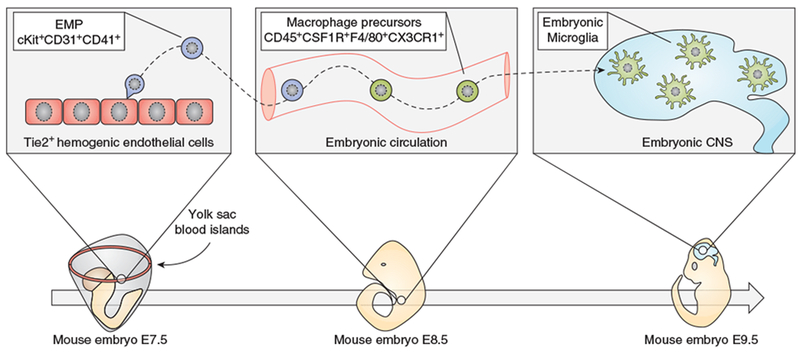
Key steps in the development of embryonic microglia
The concept that mouse microglia are solely derived from yolk sac EMPs dominates the field and is currently supported by several lines of evidence (3, 6, 17). However, a recent publication suggested that about 20-25% of brain microglia stem from Hoxb8-positive fetal liver-derived monocytes (FL-Mo), arising from fetal liver hematopoiesis around E12 (18). Thus, FL-Mo would enter the embryonic CNS and acquire phenotype and functions barely distinguishable from YS-derived microglia. Albeit highly provocative, this model might explain previous findings that contradict a sole YS origin for microglia. First, a transient increase in Ly6C+ monocytes was noted in the embryonic brain between E12 and E14 (19). Second, early depletion of YS-derived macrophages prevents the seeding of microglia in the brain rudiment at E10.5 and enhances the recruitment of monocytes at E14.5. Then, a microglial population reappears during the late gestational period, when EMPs are no longer present (19, 20). Third, the S100a4Cre line, supposedly specific for monocytes, engenders fluorescent labeling in roughly 20% of embryonic microglia, and this population is maintained in the adult brain (19, 21). Further studies are needed to determine whether multiple hematopoietic waves contribute to microglia in mice. Current fate-mapping tools may not accurately resolve partially overlapping populations such as YS-derived macrophages and FL-derived monocytes. In the near future, techniques involving photoinducible labeling, such as NICHE-seq (22), might be exploited to track the fate of macrophage-progenitors in vivo.
1.2. Brain cytokines are critical for microglial survival and phenotype
Microglial survival is dependent on the CSF1R ligands CSF1 and IL34 (23–27). In the brain, IL34 is produced by neurons, whereas CSF1 is produced by both neurons (26, 28, 29) and microglia themselves (15). Interestingly, these cytokines exhibit non-overlapping expression patterns. CSF1 is highly expressed in the cerebellum, corpus callosum and spinal cord, whereas IL34 is more abundant in the neocortex, olfactory bulb, striatum and hippocampus (25–27). Genetic ablation of either CSF1 (24, 26) or IL34 (25, 27) results in a partial reduction of microglia, estimated to be about 30% and 50%, respectively. Furthermore, it has recently been shown that deletion of CSF1 in the neuroectodermal lineage causes specific depletion of cerebellar microglia (30). At present, it is not known whether CSF1-dependent and IL34-dependent microglia differ in terms of phenotype and function.
TGFβ is another critical factor for microglial homeostasis. In the brain, TGFβ is produced by microglia, astrocytes and neurons (31–33), whereas TGFβR1 and TGFβR2 are chiefly expressed in microglia (32–34). TGFβ knock-out mice exhibit a deficit of microglia development (34). Likewise, ablation of TGFβR2 in microglia using either the Sall1CreER or the Cx3cr1CreER conditional deletion systems disrupts the homeostatic morphology and phenotype of microglia (35–37). Mice with TGFβR2-deficient microglia were also shown to develop white matter pathology and a progressive limb paralysis during adulthood (36). Similarly, mice deficient for Lrrc33, a critical molecule for TGFβ signaling in macrophages, develop microglia alterations, progressive paralysis, and die prematurely (38, 39). At present, which component(s) of the SMAD transcriptional machinery are crucial for TGFβ signaling in microglia is still not clear.
1.3. Microglia are long-lived cells with slow turnover
Several independent studies have shown that blood monocytes do not contribute to microglial turnover under homeostasis (3, 40, 41). However, monocytes can infiltrate the CNS and generate monocyte-derived macrophages in the presence of BBB damage (42–44). At steady state, microglia self-renew through a slow but constant process of apoptosis and cell division in a stochastic manner (45–48). By contrast, microglia undergo clonal expansion under pathological conditions (47, 48). Microglia are rather long-lived cells, with a turnover rate of a few months that varies depending on the brain region they occupy. For example, the median lifespan of mouse cortical microglia is about 15 months (48). One study attempted to determine microglial turnover in the human brain and, although limited to only two subjects, found that human microglia proliferate at a very slow rate, with 0.08% cells being replaced per day. Using a mathematical model, the average lifespan of human microglia was estimated to be about 4.2 years, although some cells may be older than 20 years. This means that the entire microglial population is probably renewed multiple times during a human life (49).
1.4. Competition between resident microglia and monocytes for tissue niches
Depletion of microglia, either by injection of diphtheria toxin into mice expressing diphtheria toxin-receptor (DTR) in macrophages or via chronic administration of CSF1R inhibitors, has provided deep insight into the dynamics of microglia turnover. Following depletion, microglia rapidly proliferate and repopulate the CNS in a few days (50–52). Determining the origin of repopulating microglia has been an active field of research for the past few years. Multiple studies determined that virtually all repopulating microglia originate from a few microglial cells that survive the depletion period, whereas blood monocytes do not contribute to repopulation, at least in the absence of BBB disruption due to irradiation or myeloablative chemotherapy (35, 53, 54). At present, the factors promoting microglia repopulation are obscure. However, it has been shown that deletion of either IL-1R1 or IKKβ (upstream kinase of the NF-kB cascade) in microglia significantly delays the repopulation (52, 54). In stark contrast to these studies, a recent work using the Cx3cr1CreER26-DTR mice concluded that Ly6Chi monocytes do engraft the brain parenchyma in the absence of head irradiation. Monocyte-derived microglia were identified as F4/80hiClec12a+, whereas resident microglia were F4/80loClec12a− (55). Similarly, another group used Cx3cr1CreERCsf1rflox/flox mice, which yielded partial depletion of microglia accompanied by monocyte recruitment into the CNS (56). It was also shown that irradiation-free transplantation of wild-type bone marrow into CSF1R-deficient pups (devoid of microglia) generates a massive invasion of donor-derived monocytes into the host CNS (57). Moreover, these studies have shown that resident and monocyte-derived microglia encompass phenotypically distinct populations. For example, unlike resident microglia, monocyte-derived counterparts do not express the transcriptional regulator Sall1 (35, 55, 57, 58). ATAC-sequencing revealed that certain loci typically open in resident microglia (like Zfp691 (59, 60)), are transcriptionally inaccessible in monocyte-derived microglia (58). Conversely, monocyte-derived microglia express high levels of various immune-related genes (like MHC-II chains, Lyz2, Clec12a, Ms4a7, ApoE, Cybb) that are typically silenced in resident microglia at steady state (55, 57, 58). Together, these data indicate that microglia and monocytes may contend for the colonization of empty niches in the brain. Under naïve conditions, microglia greatly outcompete monocytes, perhaps by being more suited to the brain environment, or simply because their homing into the CNS occurs during primitive hematopoiesis, long before adult hematopoiesis is established in bone marrow. Nevertheless, under conditions of microglia deficiency, monocytes may gain access to the CNS and differentiate into macrophages that partially resemble microglia (61).
1.5. Both ontogeny and environment sculpt the molecular fingerprint of microglia
Local environmental cues are critical for the final maturation and specialization of tissue-resident macrophages, including microglia (7, 62–66). Indeed, acutely isolated microglia lose their molecular identity within a few hours upon exposure to cell culture conditions (60, 67). Conversely, cultured microglia transplanted back into the mouse brain reacquired their original phenotype in about two weeks (57, 67). Similarly, transplantation of iPSC-derived macrophages into the post-natal mouse brain generated microglia-like cells fully integrated within the host tissue (68, 69). Overall, the brain environment is critical for the maturation and maintenance of the microglial phenotype. Nevertheless, the failure of monocytes to acquire a complete microglial signature suggests that origin from the yolk sac or bone marrow may imprint different repertoires of poised enhancers. For example, it has been shown that both miRNAs (70) and HDAC enzymes (71) critically shape microglia development during the embryonic stage, but not after birth. Altogether, we can hypothesize that yolk sac ontogeny dictates the epigenetic landscape in microglial progenitors, whereas the transcriptional signature is locally instructed within the CNS environment (57).
2. Microglial phenotypes during homeostasis and disease
2.1. Developmental and regional heterogeneity of microglia
Bulk RNAseq studies identified a number microglia-specific genes, like Crybb1, Fcrls, Gpr34, Gpr84, Hexb, Olfml3, P2ry12, P2yr13, Rnase4, Sall1, Siglech, Slc2a5, and Tmem119 (16, 32, 34, 59, 72). Nevertheless, caution should be used because evolving technologies for multidimensional analysis and deep sequencing always reveal previously unappreciated subpopulations. For example, Siglech is also a marker for plasmacytoid dendritic cells (pDC) (73). Although pDCs represent a minor population of meningeal DCs under homeostasis, encompassing about 1.5% of the total MHC-II+ cells of the brain (74, 75), their number expands significantly during disease (76). Moreover, recent studies showed that Fcrls is broadly expressed in multiple brain macrophage subsets (76, 77), whereas Sall1 is apparently expressed in a population of macrophages within the apical choroid plexus epithelium (77).
More recently, single-cell RNAseq enabled more in-depth characterization of the transcriptional landscape in microglia at different stages of development. Embryonic microglia are enriched for various lysosomal enzymes, ApoE and Ms4a7 (78, 79). By contrast, early postnatal microglia abundantly expressed Igf1, Spp1, Gpnmb, and Clec7a. Interestingly, Spp1+Gpnmb+Clec7a+ postnatal microglia were primarily found within heavily myelinated regions like the corpus callosum and cerebellum (78, 80). Of note, these studies consistently found a cluster of microglia particularly enriched for immediate early genes like Fos, Jun and Egr1 (77–80). It was, however, acknowledged that this cluster may have been artificially generated due to microglia activation during sample preparation (77, 80, 81).
One month after birth mouse microglia are phenotypically mature with transcriptomes prominently enriched for homeostatic genes, such as Tmem119, P2ry12, Slc2a5, Selplg, Cst3, Sparc, Tgfbr, Malat1 and others (78–80). Nonetheless, a region-dependent heterogeneity of microglia can be appreciated. For example, one study showed that microglia may vary phenotypically, depending on their topological distribution within the CNS (82). In particular, cerebellar microglia appeared skewed towards a more “immune alerted” and “metabolically demanding” phenotype, possibly because of the higher content of white matter as compared to other brain regions. By contrast, microglia in the cortex and striatum appeared in a more “quiescent” state, while hippocampal microglia had an intermediate phenotype. Another report identified transcriptomic heterogeneity in microglia selectively isolated from the cortex, nucleus accumbens, ventral tegmental area and substantia nigra (83). Ingenuity pathway analysis identified the highest variability in pathways for vesicle release, mitochondrial function, cell metabolism, oxidative stress, lysosomal activity and transport of metal ions. More recently, distinct patterns of gene-expression and epigenetic signatures were identified in cerebellar and striatal microglia. In particular, cerebellar microglia were highly enriched for genes linked to phagocytosis of apoptotic cells, while striatal microglia were more enriched for genes involved in immunological surveillance (84).
Lastly, molecular heterogeneity was recently described in human microglia. A mass cytometry study identified multiple region-specific subsets of microglia from postmortem brains (85). Consistently, single-cell RNAseq of microglia from healthy human brains formed multiple clusters with varying enrichment for Tmem119, Cx3cr1, P2ry12, Slc2a5, Cst3, Ccl2 and Ccl4 (79). Understanding the functional implications of such microglial phenotypes will be an important challenge for the years to come.
2.2. The DAM signature during pathology
Broad changes in the transcriptomic profile of microglia have been found in mouse models of amyloid pathology (86–88), Tau pathology (89), and Experimental Autoimmune Encephalomyelitis (EAE) (74, 76, 88). Altogether, pathological conditions cause downregulation of the microglial homeostatic genes (including Tmem119, P2ry12, Selplg, Cx3cr1, Tgfbr1, Sall1), whereas other genes are upregulated. For example, Trem2, Tyrobp, and ApoE were consistently found overexpressed in microglia in different neurodegeneration mouse models (86–92). Moreover, microglia exhibited ApoE upregulation during EAE (76, 88), and in the cuprizone model of toxic demyelination (79). The exact functions of TREM2 and ApoE during brain diseases are still controversial, and this topic has already been addressed elsewhere (93–96). Interestingly, independent studies identified a conserved molecular signature of microglia in models of amyloid pathology and neurodegeneration (86–88). This specific microglial phenotype has been termed “Disease-Associated Microglia” (DAM) (86).
In mouse, the DAM signature is characterized by higher expression of genes involved in lysosomal functions (Cst7, Ctsb/d), phagocytosis (Axl), antigen presentation (H2-Aa, H2-Ab), lipid transport (Lpl, Apoe), matrix remodeling (Spp1, Gpnmb), complement binding (CD11c), anti-microbial activity (Lyz2, Dectin-1), immune modulation (Lilrb4), and cell survival (Csf1, Igf1) (74, 77, 86–88) (Figure 2).
Figure 2:
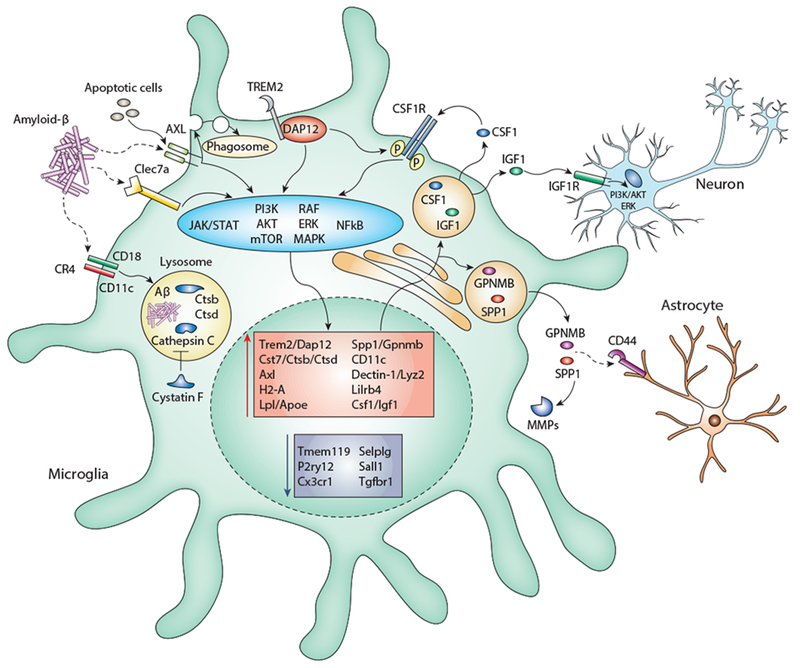
Possible mechanisms involving the DAM signature genes.
Cystatin F (Cst7) is a lysosomal cysteine protease inhibitor, which targets the lysosomal enzyme cathepsin C (97). Inhibition of Cst7 expression correlates with reduced amyloid pathology (98), indicating that Cst7 may reduce the capacity of microglia to degrade Aβ. Alongside, DAM signature is also characterized by higher expression of some cathepsins, especially Ctsb and Ctsd (86). Cathepsins are cysteine proteases important for lysosomal degradation of aggregated proteins (99). Additionally, cathepsins can be secreted, and therefore may play a role in cell migration (100).
Axl and Mertk belong to a family of TAM tyrosine kinases receptors mainly involved in the phagocytosis of dead cells (101). Axl is upregulated in microglia during neurodegeneration and neuroinflammatory diseases (86–88, 102), whereas Mertk is downregulated (74, 86, 88). Microglia lacking Axl and Mertk exhibit deficient phagocytosis of apoptotic cells and display reduced migration towards laser-induced injury (102, 103). In the EAE model, Axl-deficient mice show a more severe pathology and fewer macrophages infiltrating the spinal cord (104). Whether Axl or Mertk are directly involved in the uptake of protein aggregates (such as Aβ and α-synuclein) remains obscure.
Secreted Phosphoprotein-1 (Spp1, Osteopontin), is one of the most upregulated genes in the DAM signature (77, 86–88) and significantly increased levels of Spp1 were found in the CSF of AD and FTD patients (105, 106). In the periphery, Spp1 is highly expressed by both osteoblasts and osteoclasts and is important for the bone mineralization (107, 108). Spp1 is also secreted by different leukocytes including Th1 cells, macrophages and DCs (109). Spp1 induces IL12 production in DCs, thus promoting type-I immunity (110, 111). Moreover, Spp1 was shown to improve survival of autoreactive T-cells in EAE (112). However, it was also suggested that Spp1 regulates inflammatory reactions (113), for example via inhibition of NO production in macrophages (114). Spp1 is also known to bind CD44 (110, 115), which, in the brain, is primarily expressed by astrocytes (32, 33, 116), while the expression in microglia is negligible (117). Spp1 and CD44 may then form a communication axis between microglia and astrocytes under neurodegeneration. Additionally, secreted Spp1 represents a substrate for the activation of metalloproteinases (118), suggesting that it may play some role in microglia migration towards the injury site. Further studies on conditional Spp1-deficient mice are required to better understand the function of this protein in microglia. Similar to Spp1, Osteoactivin (Gpnmb) represents another possible ligand for CD44 (119). It was indeed suggested that Gpnmb may dampen astrocyte activation via CD44 signaling (120).
Clec7a (Dectin-1) is a C-type lectin serving as pattern-recognition receptor against fungi and bacteria (121). Clec7a contains an ITAM motif promoting a Syk-dependent signaling, which elicits an anti-microbial response in macrophages (122, 123). Albeit not expressed under homeostasis, Clec7a+ microglia were found in proximity of amyloid plaques (86, 88), as well as in mice with microglia-specific BRAF mutation, which causes microgliosis and late-onset neurodegeneration (124). In vitro, microglial metabolic activity is boosted by zymosan, which is a known ligand for Clec7a (125). Possibly, Clec7a may help mount the immune-activation state of plaque-associated microglia. Moreover, Aβ is known to induce microglia activation via various pattern-recognition receptors (126). It remains a question whether Clec7a is involved in a similar mechanism.
CD11c (Itgax), a prototypical DC marker, has repeatedly been found upregulated in activated microglia and represents one of the most consistent DAM signature genes (74, 77, 86, 88). CD11c and CD18 form the complement-receptor 4, which is important for the engulfment and fragmentation of complement-opsonized particles (127). It is then tempting to speculate that CD11c in plaque-associated microglia may help recognize or phagocytose Aβ aggregates. A conditional knock-out mouse model is needed to better investigate the role of CD11c in microglia under pathology.
Leukocyte immunoglobulin-like receptor B4 (Lilrb4) belongs to a family of ITIM-bearing inhibitory receptors widely expressed in different leukocytes (128). At present, the ligand as well as the exact function of mouse Lilrb4 is unknown. A recent study showed that conditional ablation of Lilrb4 exacerbates steatosis and systemic inflammation in mice under hyper-fat diet (129), suggesting that this receptor may play important immunomodulatory functions. Whether Lilrb4 could restrain the microglia activation state under brain pathology remains hypothetical.
Lastly, the DAM signature shows an enrichment for Csf1 and Igf1. Interestingly, both genes are highly expressed in microglia at the early stages of brain development (15, 80, 130, 131). Csf1 is important for microglial survival and proliferation (51, 132), whereas microglia-derived Igf1 was shown to support the survival of newborn neurons (130). Csf1 could act on microglia in an autocrine/paracrine manner, thus promoting microglia proliferation/survival within the plaque-surrounding environment. RNAseq data indicate that expression of Igf1r in microglia is negligible, but detectable in neurons (32, 33). Microglia-derived Igf1 could then provide neurotrophic support to the neighboring neurons, thus protecting against Aβ cytotoxicity.
2.3. Similarities and discrepancies between mouse and human microglia
Our understanding of the molecular properties of microglia relies chiefly on mouse models. At present, only a few pioneering works have delineated the transcriptomic profiles of microglia from healthy subjects, as well as from patients with Alzheimer’s Disease (AD) and multiple sclerosis (MS) (60, 79, 116, 133). A seminal study showed that the transcriptomes of both mouse and human microglia are characterized by a dominant PU.1-dependent signature and about 50% of the microglia-specific genes (such as Cx3cr1, Tmem119, P2ry12, Trem2 and Sall1) are similarly expressed in both species. Using a cutoff of 10-fold, 2.5% of the transcripts were highly enriched in human microglia (like C3, SPP1, and APOE) and 1.9% appeared specific for mouse microglia (like Hexb, Sparc, and Sall3). Overall, these data indicate a substantial overlap between mouse and human microglia at the molecular level (60). Similar findings were reported in a following study that, however, highlighted increasing molecular disparity between mouse and human microglia during ageing (134). A mass cytometry study showed that human microglia express high levels of TMEM119 and P2RY12, which are absent in blood myeloid cells. Human microglia are also positive for EMR1 (F4/80), TREM2, CX3CR1, CD64 and CD115, whereas expression of CD45, CD44, CCR2, CD206 and CD163 is either low or negligible. This repertoire of surface markers closely resembles that of mouse microglia. However, unlike their mouse counterparts in the steady state, human microglia express CD11c, MHC-II, and relatively low levels of CD11b (85).
In AD patients, microglia upregulate CD74, HLA-DR, APOE, TREM2, C1Q and CD14. Interestingly, transcriptomic changes in microglia seemed to correlate with the severity of both amyloid and tau pathology (133). Given the difficulties of working with human brain samples, it has been suggested that iPSCs may be a powerful tool for modeling human microglia during brain diseases (68, 69, 135, 136). For example, a recent study showed that following transplantation into the brains of mice with amyloid pathology, iPSC-derived human microglia efficiently migrated towards amyloid plaques and, like their murine counterparts, up-regulated APOE, HLA-DR, LGALS3, MS4A7, ITGAX, and TREM2. Of note, some DAM signature genes like TYROBP, CST7, CLEC7A and CSF1 were not significantly altered in iPSC-derived human microglia (137). This suggests that mouse and human microglia mount similar immunological responses against amyloid pathology in vivo; however, certain pathways may not be conserved between species. Our own data support this view, as we detected a prominent IRF8-dependent signature in human microglia from AD patients, but not in mouse models (138). Further studies are required to better understand similarities and differences in the DAM signatures of human and mouse microglia.
3. Shared and distinct properties of microglia and Border Associated Macrophages (BAMs)
3.1. Ontogeny and phenotype of BAMs
Microglia are not the only brain-resident macrophages; indeed, populations of extra-parenchymal macrophage patrol the brain-blood interface in both mice and humans. Perivascular macrophages (pvMPs) are primarily found in the perivascular Virchow-Robin spaces of the cortical blood vessels. Meningeal macrophages (mMPs) are located within the meningeal membranes, either on the pia mater or within the dura. Lastly, choroid plexus macrophages (cpMPs) lie beneath the epithelial cell layer of the choroid plexus (139). These extra-parenchymal brain macrophages are collectively referred to as border associated macrophages (BAMs) (74). Although microglia and BAMs share the expression of several phenotypic markers (including Iba1, CD11b, CX3CR1, CD64, Mertk, CD115, and others), transcriptome studies identified a repertoire of molecules that are specifically expressed by each population (Figure 3). Like microglia, BAMs are dependent on CSF1R signaling for their survival (74, 77). However, it is still unknown whether anatomically distinct BAMs subsets are differentially dependent on CSF1 and IL34. Interestingly, genetic deletion of the super-enhancer fms-intronic regulatory element (FIRE), which is critical for the expression of CSF1R in YS-derived macrophages, causes complete depletion of microglia, but leaves pvMPs and mMPs seemingly unaffected. This may suggest that transcriptional and epigenetic mechanisms differentially regulate CSF1R in microglia and extra-parenchymal macrophages (140).
Figure 3:
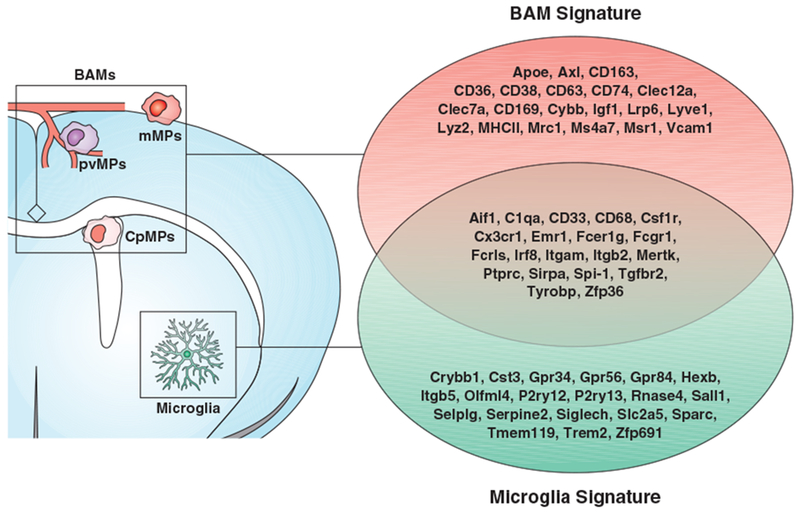
Molecular signature of microglia and BAMs under homeostasis.
It has long been thought that BAMs are derived from and constantly replaced by BM-derived monocytes. However, recent fate-mapping studies revealed that, like microglia, BAMs are generated by YS-derived progenitors. Embryonic-derived pvMPs and mMPs are long-lived cells, whereas cpMPs are rapidly replaced by BM-derived monocytes soon after birth (141). Of note, a recent paper described a population of BAMs localized on the apical choroid plexus epithelium (previously identified as epiplexus Kolmer’s cells) that share key features of microglia. These cells are embryonic derived, express Sall1 and can self-renew with no obvious input from the periphery. By contrast, stromal cpMPs are negative for Sall1 and undergo constant monocyte-mediated turnover (77). At present, it is unclear whether YS-derived myeloid progenitors are already committed to become either microglia or BAMs during the embryonic development, or whether the two differentiation pathways are locally instructed by environmental stimuli. The first case predicts the existence of a common myeloid progenitor (possibly downstream of the EMP stage) that generates two separate lineages. Alternatively, we may hypothesize that the local environment of the blood-brain interface guides the differentiation of embryonic macrophages towards a BAM phenotype, whereas the parenchymal environment supports the differentiation into microglia (Figure 4). Future studies will hopefully shed some light on this outstanding question.
Figure 4:
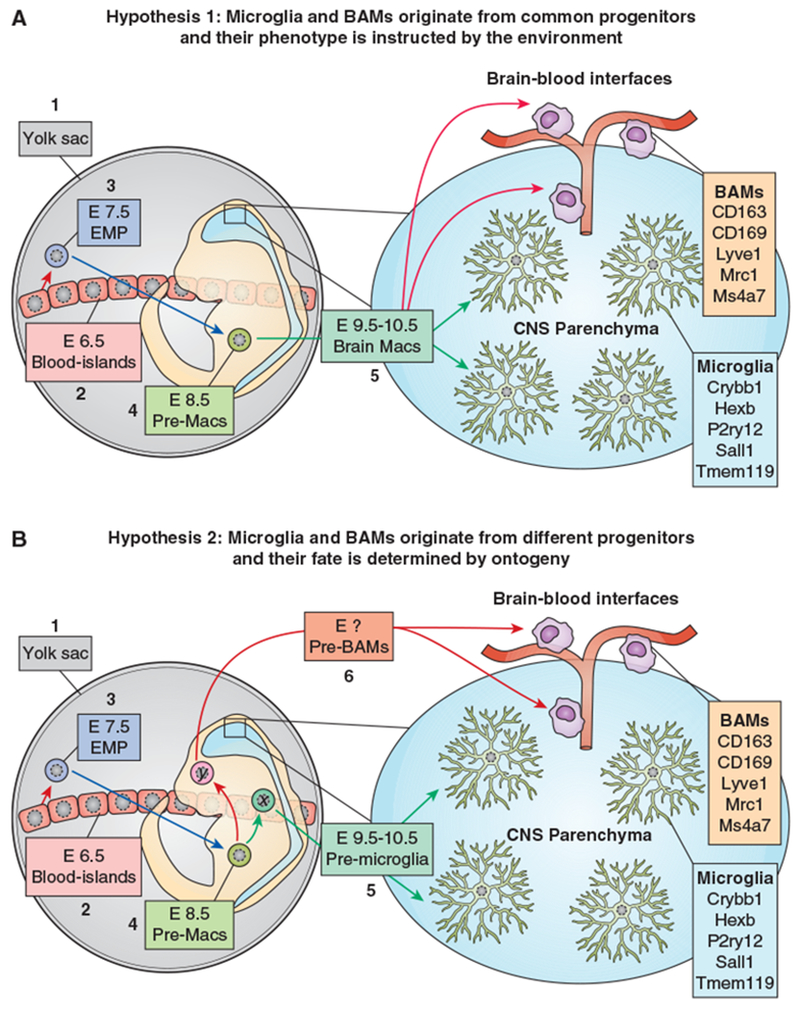
Two alternative hypotheses for the origin of microglia and BAMs.
a) Hypothesis 1 - EMPs arise from YS-blood islands around E7.5 (1) and at E8.5 migrate into the embryo proper where they differentiate into macrophage precursors (pre-Macs) (2). These cells seed all embryonic tissues including the brain (3). Depending on the local environment, they differentiate into either microglia (4) or BAMs (5). In this model, microglia and BAMs originate from common progenitors and the local environment plays a major role in determining their fate. The genes indicated in boxes 4 and 5 refer to transcriptional signatures identified in the adult mouse brain.
b) Hypothesis 2 - EMPs arise from YS-blood islands around E7.5 (1) and at E8.5 migrate into the embryo proper where they differentiate into macrophage precursors (pre-Macs) (2). These cells generate two separate lineages (x and y) giving rise to either pre-microglia (3) or pre-BAMs (4), which respectively colonize the CNS parenchyma and the brain-blood interfaces. The local environment dictates the final maturation into microglia (5) and BAMs (6). In this model, microglia and BAMs share a common ancestor cell (EMP), but eventually develop through distinct lineages. Therefore, a combination of ontogeny and environment is critical for the fate of both cell types. The genes indicated in boxes 5 and 6 refer to transcriptional signatures identified in the adult mouse brain.
3.2. Antigen-presentation capacity of BAMs and microglia during neuroinflammatory diseases
The composition of BAMs during neuroinflammatory disease has recently been investigated by single-cell RNAseq and multi-dimensional immunophenotyping techniques. During EAE, for example, expression of several activation markers (MHC-II, CD74, CD44, Sca-1, and CD11c) was increased, whereas the BAM homeostatic markers Lyve-1, Fcrls, and CD206 were downregulated. Interestingly, expression of other markers like Ms4a7, CD38, CD169 and CD86 remained stable during disease (74, 76). Meningeal myeloid cells have often been implicated in the re-activation of auto-aggressive T-cells during EAE (142–145). Nonetheless, experimental proof that BAMs license T-cell entry into the CNS is lacking. A landmark publication used intravital two-photon imaging to illustrate the path of autoreactive T-cells in a rat model of transfer EAE (146). Both MBP and MOG reactive T-cells enter the CNS by crawling along the leptomeninges, where they randomly encounter mMPs. Given that the leptomeninges represent a main gateway for infiltrating T-cells, mMPs probably constitute the first line of antigen-presenting cells (APCs) at the brain/blood interface. TCR engagement by meningeal APCs induces nuclear translocation of NFAT in antigen-competent T-cells, thereby eliciting their encephalitogenic potential (146). Intrathecal administration of blocking antibody for LFA1/VLA4 integrins augmented the number of MBP-reactive T-cells in the CSF, but dramatically reduced their infiltration into the underlying parenchyma (146). Consistent with this, in a rat model of gray matter inflammation, the number of synuclein-reactive T-cells was diminished in both the parenchyma and meninges after intrathecal injection of anti-MHC-II antibody. In addition, this treatment significantly attenuated expression of inflammatory cytokines (IFNγ and IL17) as well as the EAE clinical score (147). Altogether, these findings indicate that the meningeal myeloid compartment represents a critical checkpoint for the re-stimulation of auto-reactive T-cells before their invasion of the CNS parenchyma. To more precisely interrogate the antigen-presentation capacity of microglia and BAMs during EAE, three independent groups studied the effect of MHC-II deletion in microglia using either the Cx3cr1CreER or the Sall1CreER inducible systems. EAE pathology was unaffected in mice with MHC-II deficient microglia in all three studies (75, 76, 148). By contrast, deletion of MHC-II using the CD11cCre line (which targets both DCs and activated macrophages) completely abolished the onset of paralysis, CNS infiltration and demyelination (76). Additional studies are now required to clarify whether meningeal DCs are uniquely competent APCs in the brain, or whether other BAMs also play a role in this context. Nevertheless, these data strongly indicate that microglia are probably dispensable for the reactivation of T-cells during EAE.
3.3. Microglia promote both tissue damage and repair during neuroinflammation
Under neuroinflammation, microglia might significantly contribute to tissue damage by releasing inflammatory cytokines. A mass cytometry study in EAE identified seven distinct microglial populations expressing variable levels of pro-inflammatory (mostly TNF-α, IL-6, GM-CSF) and anti-inflammatory (TGF-β and IL-10) cytokines (149). This may indicate that microglia both promote and help resolve neuroinflammation during EAE. Furthermore, a recent study based on two-photon intravital microscopy during EAE suggested that CNS phagocytes shift from a pro-inflammatory to a wound-healing phenotype, depending on the lesion context (150). A seminal study on Cx3cr1CreERTak1flox/flox mice showed that deletion of the NF-kB activator Tak1 in microglia remarkably ameliorated EAE pathology in mice (41), which implies that microglia can be gravely detrimental in neuroinflammatory conditions. However, like microglia, meningeal and perivascular macrophages are CX3CR1+ and undergo very limited turnover (141). Therefore, the observed protective effect could partially stem from Tak1 deletion in BAMs, rather than microglia. In contrast, deletion of NF-kB negative regulators (like AHR or A20) in microglia was shown exacerbate neuroinflammation and EAE pathology (151, 152). Similarly, the Ubiquitin Specific Peptidase 18 (USP18) was shown to act as a negative regulator of the Stat1 pathway in white matter microglia, thus dampening the type-I interferon response (153). Therefore, microglia lacking USP18 constitutively upregulate IFN-dependent genes resulting in white-matter pathology and behavioral defects (154).
Besides their pro-inflammatory function at the peak of the disease, microglia may play a critical role in the resolution of inflammation and tissue repair. Indeed, microglia were repeatedly shown to be important for the clearance of myelin debris and ensuing remyelination both in EAE (155) and toxic-induced demyelination models (156–160). This suggests that microglia can support the healing of the white matter after demyelination. Indeed, a recent study showed that a population of CD11c+Clec7a+Gpnmb+ microglia promote remyelination via secretion of Igf1 during EAE (131). Additionally, microglia were shown to upregulate PD-L1 in the EAE model (74), whereas a microglial subset expressing Galectin-1 was found in post-mortem brain samples from MS patients (79). Both of these genes critically modulate the activation of CD8+ lymphocytes (161–164). It is then tempting to speculate that microglia could help dampen the T-cell mediated cytotoxic response during MS. Of note, microglia depletion approaches have generated conflicting results in the context of EAE, resulting in either beneficial, detrimental or no effects (165–167). We would hypothesize that non-selective depletion of both microglia and BAMs may produce unpredictable confounding effects.
Altogether, microglia may promote tissue damage during neuroinflammation via cytokine and ROS production. However, microglia are also critical for efficient scavenging of cellular debris and tissue regeneration. Environmental cues that selectively promote these functions are currently under investigation.
4. Conclusions
Brain macrophages encompass multiple populations characterized by different anatomical distribution, phenotype, ontogeny/turnover, and, very likely, different functions. Growing evidence suggests that molecular signature of microglia and BAMs is instructed by a combination of local environment and ontogeny. However, whether microglia and BAMs arise from a unique YS-derived progenitor or develop through independent pathways is still unknown (see also 168). Additionally, we just started to scratch the surface of the transcriptomic changes in brain macrophages during disease. For example, a number of independent works provided a list of candidate genes which identifies the DAM signature of plaque-associated microglia during amyloid pathology. Nevertheless, the exact function of these genes remains to be determined. This information may help us harness specific DAM genes to either reduce amyloid burden or improve viability of neighboring brain cells in Alzheimer Disease.
Acknowledgments
We warmly thank Dr. Susan Gilfillan for the helpful suggestions during the preparation of the present manuscript.
This work was supported by NIH grants AG051485, AG059176, AG059082 and by the Cure Alzheimer Fund
Glossary
- CNS
Central Nervous System
- EMPs
Erythromyeloid Precursor Cells
- YS
Yolk sac
- HSCs
Hematopoietic Stem Cells
- BBB
Blood-Brain Barrier
- FL-Mo
Fetal Liver-derived Monocytes
- DTR
Diphtheria Toxin-Receptor
- DAM
Disease-Associated Microglia
- AD
Alzheimer Disease
- MS
Multiple sclerosis
- BAMs
Border Associated Macrophages
- pvMPs
Perivascular macrophages
- mMPs
Meningeal macrophages
- cpMPs
Choroid plexus macrophages
- EAE
Experimental Autoimmune Encephalomyelitis
References
- 1.Sieweke MH, and Allen JE. 2013. Beyond stem cells: self-renewal of differentiated macrophages. Science 342: 1242974. [DOI] [PubMed] [Google Scholar]
- 2.Ginhoux F, and Jung S. 2014. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol 14: 392–404. [DOI] [PubMed] [Google Scholar]
- 3.Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, and Rodewald H-R. 2014. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518: 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGrath KE, Frame JM, Fegan KH, Bowen JR, Conway SJ, Catherman SC, Kingsley PD, Koniski AD, and Palis J. 2015. Distinct Sources of Hematopoietic Progenitors Emerge before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell Rep. 11: 1892–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginhoux F, and Guilliams M. 2016. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 44: 439–449. [DOI] [PubMed] [Google Scholar]
- 6.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, and Merad M. 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330: 841–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mass E, Ballesteros I, Farlik M, Halbritter F, Günther P, Crozet L, Jacome-galarza CE, Händler K, Klughammer J, Kobayashi Y, Gomez E, Schultze JL, Beyer M, Bock C, and Geissmann F 2016. Specification of tissue-resident macrophages during organogenesis. Science 353(6304). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Gomez Perdiguero E, Wieghofer P, Heinrich A, Riemke P, Hölscher C, Müller DN, Luckow B, Brocker T, Debowski K, Fritz G, Opdenakker G, Diefenbach A, Biber K, Heikenwalder M, Geissmann F, Rosenbauer F, and Prinz M. 2013. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci 16: 273–280. [DOI] [PubMed] [Google Scholar]
- 9.Crotti A, and Ransohoff RM. 2016. Microglial Physiology and Pathophysiology: Insights from Genome-wide Transcriptional Profiling. Immunity 44: 505–515. [DOI] [PubMed] [Google Scholar]
- 10.Hagemeyer N, Kierdorf K, Frenzel K, Xue J, Ringelhan M, Abdullah Z, Godin I, Wieghofer P, Costa Jordão MJ, Ulas T, Yorgancioglu G, Rosenbauer F, Knolle PA, Heikenwalder M, Schultze JL, and Prinz M. 2016. Transcriptome‐based profiling of yolk sac‐derived macrophages reveals a role for Irf8 in macrophage maturation. EMBO J. e201693801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SEW, Pollard JW, Frampton J, Liu KJ, and Geissmann F. 2012. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336: 86–90. [DOI] [PubMed] [Google Scholar]
- 12.Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, and Potter SS. 1991. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65: 677–689. [DOI] [PubMed] [Google Scholar]
- 13.Hoeffel G, and Ginhoux F. 2018. Fetal monocytes and the origins of tissue-resident macrophages. Cell. Immunol 330:5–15. [DOI] [PubMed] [Google Scholar]
- 14.Daneman R, Zhou L, Kebede AA, and Barres BA. 2010. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 468: 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada Gonzalez F, Perrin P, Keren-Shaul H, Gury M, Lara-Astaiso D, Thaiss CA, Cohen M, Bahar Halpern K, Baruch K, Deczkowska A, Lorenzo-Vivas E, Itzkovitz S, Elinav E, Sieweke MH, Schwartz M, and Amit I. 2016. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353: aad8670. [DOI] [PubMed] [Google Scholar]
- 16.Bennett ML, Bennetta C, Liddelowa SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, Weissman IL, Chang EF, Gordon L, Grant GA, Hayden Gephart MG, and Barres BA. 2016. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci 113(12):E1738–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheng J, Ruedl C, and Karjalainen K. 2015. Most Tissue-Resident Macrophages Except Microglia Are Derived from Fetal Hematopoietic Stem Cells. Immunity 43: 382–393. [DOI] [PubMed] [Google Scholar]
- 18.De S, Van Deren D, Peden E, Hockin M, Boulet A, Titen S, and Capecchi MR. 2018. Two distinct ontogenies confer heterogeneity to mouse brain microglia. Development 145(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, Beaudin AE, Lum J, Low I, Forsberg EC, Poidinger M, Zolezzi F, Larbi A, Ng LG, Chan JKY, Greter M, Becher B, Samokhvalov IM, Merad M, and Ginhoux F. 2015. C-Myb+ Erythro-Myeloid Progenitor-Derived Fetal Monocytes Give Rise to Adult Tissue-Resident Macrophages. Immunity 42: 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Squarzoni P, Oller G, Hoeffel G, Pont-Lezica L, Rostaing P, Low D, Bessis A, Ginhoux F, and Garel S. 2014. Microglia Modulate Wiring of the Embryonic Forebrain. Cell Rep. 8: 1271–1279. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, Greter M, Mortha A, Boyer SW, Forsberg EC, Tanaka M, van Rooijen N, García-Sastre A, Stanley ER, Ginhoux F, Frenette PS, and Merad M. 2013. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38: 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medaglia C, Giladi A, Stoler-Barak L, De Giovanni M, Salame TM, Biram A, David E, Li H, Iannacone M, Shulman Z, and Amit I. 2017. Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science 358:1622–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wȩgiel J, Wiśniewski HM, Dziewiątkowski J, Tarnawski M, Kozielski R, Trenkner E, and Wiktor-Jȩdrzejczak W. 1998. Reduced number and altered morphology of microglial cells in colony stimulating factor-1-deficient osteopetrotic op / op mice. Brain Res. 804: 135–139. [DOI] [PubMed] [Google Scholar]
- 24.Erblich B, Zhu L, Etgen AM, Dobrenis K, and Pollard JW. 2011. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One 6(10):e26317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, Kündig TM, Frei K, Ginhoux F, Merad M, and Becher B. 2012. Stroma-Derived Interleukin-34 Controls the Development and Maintenance of Langerhans Cells and the Maintenance of Microglia. Immunity 37: 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nandi S, Gokhan S, Dai XM, Wei S, Enikolopov G, Lin H, Mehler MF, and Richard Stanley E. 2012. The CSF-1 receptor ligands IL-34 and CSF-1 exhibit distinct developmental brain expression patterns and regulate neural progenitor cell maintenance and maturation. Dev. Biol 367: 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, Barrow AD, Diamond MS, and Colonna M. 2012. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol 13: 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan Z, Kuhn JA, Wang X, Colquitt B, Solorzano C, Vaman S, Guan AK, Evans-Reinsch Z, Braz J, Devor M, Abboud-Werner SL, Lanier LL, Lomvardas S, and Basbaum AI. 2015. Injured sensory neuron-derived CSF1 induces microglial proliferation and DAP12-dependent pain. Nat. Neurosci 19(1):94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chitu V, Gokhan Ş, Nandi S, Mehler MF, and Stanley ER. 2016. Emerging Roles for CSF-1 Receptor and its Ligands in the Nervous System. Trends Neurosci. 39: 378–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kana V, Desland FA, Casanova-Acebes M, Ayata P, Badimon A, Nabel E, Yamamuro K, Sneeboer M, Tan I-L, Flanigan ME, Rose SA, Chang C, Leader A, Le Bourhis H, Sweet ES, Tung N, Wroblewska A, Lavin Y, See P, Baccarini A, Ginhoux F, Chitu V, Stanley ER, Russo SJ, Yue Z, Brown BD, Joyner AL, De Witte LD, Morishita H, Schaefer A, and Merad M. 2019. CSF-1 controls cerebellar microglia and is required for motor function and social interaction. J. Exp. Med jem.20182037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bialas AR, and Stevens B. 2013. TGF-β signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat. Neurosci 16: 1773–1782. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, and Wu JQ. 2014. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J. Neurosci 34: 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, Goeva A, Nemesh J, Kamitaki N, Brumbaugh S, Kulp D, and McCarroll SA. 2018. Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174(4):1015–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, and Weiner HL. 2014. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci 17: 131–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buttgereit A, Lelios I, Yu X, Vrohlings M, Krakoski NR, Gautier EL, Nishinakamura R, Becher B, and Greter M. 2016. Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol 17: 1397–1406. [DOI] [PubMed] [Google Scholar]
- 36.Lund H, Pieber M, Parsa R, Grommisch D, Ewing E, Kular L, Han J, Zhu K, Nijssen J, Hedlund E, Needhamsen M, Ruhrmann S, Guerreiro-Cacais AO, Berglund R, Forteza MJ, Ketelhuth DFJ, Butovsky O, Jagodic M, Zhang XM, and Harris RA. 2018. Fatal demyelinating disease is induced by monocyte-derived macrophages in the absence of TGF-β signaling letter. Nat. Immunol 19: 435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zöller T, Schneider A, Kleimeyer C, Masuda T, Potru PS, Pfeifer D, Blank T, Prinz M, and Spittau B. 2018. Silencing of TGFβ signalling in microglia results in impaired homeostasis. Nat. Commun 9(1):4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong K, Noubade R, Manzanillo P, Ota N, Foreman O, Hackney JA, Friedman BA, Pappu R, Scearce-Levie K, and Ouyang W. 2017. Mice deficient in NRROS show abnormal microglial development and neurological disorders. Nat. Immunol 18(6):633–641 . [DOI] [PubMed] [Google Scholar]
- 39.Qin Y, Garrison BS, Ma W, Wang R, Jiang A, Li J, Mistry M, Bronson RT, Santoro D, Franco C, Robinton DA, Stevens B, Rossi DJ, Lu C, and Springer TA. 2018. A Milieu Molecule for TGF-β Required for Microglia Function in the Nervous System. Cell 174(1):156–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ajami B, Bennett JL, Krieger C, Tetzlaff W, and V Rossi FM. 2007. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci 10: 1538–43. [DOI] [PubMed] [Google Scholar]
- 41.Goldmann T, Wieghofer P, Muller PF, Wolf Y, Varol D, Yona S, Brendecke SM, Kierdorf K, Staszewski O, Datta M, Luedde T, Heikenwalder M, Jung S, and Prinz M. 2013. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci 16: 1618–1626. [DOI] [PubMed] [Google Scholar]
- 42.Priller J, a Flügel, Wehner T, Boentert M, a Haas C, Prinz M, Fernández-Klett F, Prass K, Bechmann I, a de Boer B, Frotscher M, Kreutzberg GW, a Persons D, and Dirnagl U. 2001. Targeting gene-modified hematopoietic cells to the central nervous system: use of green fluorescent protein uncovers microglial engraftment. Nat. Med 7: 1356–61. [DOI] [PubMed] [Google Scholar]
- 43.Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch U-K, Mack M, Heikenwalder M, Brück W, Priller J, and Prinz M. 2007. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat. Neurosci 10: 1544–53. [DOI] [PubMed] [Google Scholar]
- 44.Ajami B, Bennett JL, Krieger C, McNagny KM, and V Rossi FM. 2011. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat. Neurosci 14: 1142–1149. [DOI] [PubMed] [Google Scholar]
- 45.Lawson LJ, Perry VH, and Gordon S. 1992. Turnover of resident microglia in the normal adult mouse brain. Neuroscience 48: 405–415. [DOI] [PubMed] [Google Scholar]
- 46.Askew K, Li K, Olmos-Alonso A, Garcia-Moreno F, Liang Y, Richardson P, Tipton T, Chapman MA, Riecken K, Beccari S, Sierra A, Molnár Z, Cragg MS, Garaschuk O, Perry VH, and Gomez-Nicola D. 2017. Coupled Proliferation and Apoptosis Maintain the Rapid Turnover of Microglia in the Adult Brain. Cell Rep. 18: 391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tay TL, Mai D, Dautzenberg J, Fernández-Klett F, Lin G, Sagar, Datta M, Drougard A, Stempfl T, Ardura-Fabregat A, Staszewski O, Margineanu A, Sporbert A, Steinmetz LM, Pospisilik JA, Jung S, Priller J, Grün D, Ronneberger O, and Prinz M. 2017. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci 20(6):793–803. [DOI] [PubMed] [Google Scholar]
- 48.Füger P, Hefendehl JK, Veeraraghavalu K, Wendeln AC, Schlosser C, Obermüller U, Wegenast-Braun BM, Neher JJ, Martus P, Kohsaka S, Thunemann M, Feil R, Sisodia SS, Skodras A, and Jucker M. 2017. Microglia turnover with aging and in an Alzheimer’s model via long-term in vivo single-cell imaging. Nat. Neurosci 20: 1371–1376. [DOI] [PubMed] [Google Scholar]
- 49.Réu P, Khosravi A, Bernard S, Mold JE, Salehpour M, Alkass K, Perl S, Tisdale J, Possnert G, Druid H, and Frisén J. 2017. The Lifespan and Turnover of Microglia in the Human Brain. Cell Rep. 20: 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, Lafaille JJ, Hempstead BL, Littman DR, and Gan WB. 2013. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155: 1596–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elmore MRP, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, Kitazawa M, Matusow B, Nguyen H, West BL, and Green KN. 2014. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82: 380–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bruttger J, Karram K, Wörtge S, Regen T, Marini F, Hoppmann N, Klein M, Blank T, Yona S, Wolf Y, Mack M, Pinteaux E, Müller W, Zipp F, Binder H, Bopp T, Prinz M, Jung S, and Waisman A. 2015. Genetic Cell Ablation Reveals Clusters of Local Self-Renewing Microglia in the Mammalian Central Nervous System. Immunity 43: 92–107. [DOI] [PubMed] [Google Scholar]
- 53.Huang Y, Xu Z, Xiong S, Sun F, Qin G, Hu G, Wang J, Zhao L, Liang YX, Wu T, Lu Z, Humayun MS, So KF, Pan Y, Li N, Yuan TF, Rao Y, and Peng B. 2018. Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat. Neurosci 21: 530–540. [DOI] [PubMed] [Google Scholar]
- 54.Zhan L, Krabbe G, Du F, Jones I, Reichert MC, Telpoukhovskaia M, Kodamaid L, Wangid C, Cho SH, Sayed F, Li Y, Leid D, Zhou Y, Shen Y, West B, and Gan L. 2019. Proximal recolonization by self-renewing microglia re-establishes microglial homeostasis in the adult mouse brain. PLoS Biol. 17(2):e3000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lund H, Pieber M, Parsa R, Han J, Grommisch D, Ewing E, Kular L, Needhamsen M, Espinosa A, Nilsson E, Överby AK, Butovsky O, Jagodic M, Zhang XM, and Harris RA. 2018. Competitive repopulation of an empty microglial niche yields functionally distinct subsets of microglia-like cells. Nat. Commun 9(1):4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cronk JC, Filiano AJ, Louveau A, Marin I, Marsh R, Ji E, Goldman DH, Smirnov I, Geraci N, Acton S, Overall CC, and Kipnis J. 2018. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J. Exp. Med 215(6):1627–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bennett FC, Bennett ML, Yaqoob F, Mulinyawe SB, Grant GA, Hayden Gephart M, Plowey ED, and Barres BA. 2018. A Combination of Ontogeny and CNS Environment Establishes Microglial Identity. Neuron 98(6):1170–1183.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shemer A, Grozovski J, Tay TL, Tao J, Volaski A, Süß P, Ardura-Fabregat A, Gross-Vered M, Kim JS, David E, Chappell-Maor L, Thielecke L, Glass CK, Cornils K, Prinz M, and Jung S. 2018. Engrafted parenchymal brain macrophages differ from microglia in transcriptome, chromatin landscape and response to challenge. Nat. Commun 9(1):5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma’ayan A, Chua W-J, Hansen TH, Turley SJ, Merad M, and Randolph GJ. 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol 13: 1118–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O’Connor C, Fitzpatrick C, Pasillas MP, Pena M, Adair A, Gonda DD, Levy ML, Ransohoff RM, Gage FH, and Glass CK. 2017. An environment-dependent transcriptional network specifies human microglia identity. Science 356: 1248–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guilliams M, and Scott CL. 2017. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol 17(7):451–460. [DOI] [PubMed] [Google Scholar]
- 62.Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, and Glass CK. 2014. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159: 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, and Amit I. 2014. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159: 1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van de Laar L, Saelens W, De Prijck S, Martens L, Scott CL, Van Isterdael G, Hoffmann E, Beyaert R, Saeys Y, Lambrecht BN, and Guilliams M. 2016. Yolk Sac Macrophages, Fetal Liver, and Adult Monocytes Can Colonize an Empty Niche and Develop into Functional Tissue-Resident Macrophages. Immunity 44: 755–768. [DOI] [PubMed] [Google Scholar]
- 65.Scott CL, Zheng F, De Baetselier P, Martens L, Saeys Y, De Prijck S, Lippens S, Abels C, Schoonooghe S, Raes G, Devoogdt N, Lambrecht BN, Beschin A, and Guilliams M. 2016. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun 7: 10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gundra UM, Girgis NM, Gonzalez MA, San Tang M, Van Der Zande HJP, Lin J-D, Ouimet M, Ma LJ, Poles J, Vozhilla N, Fisher EA, Moore KJ, and Loke P. 2017. Vitamin A mediates conversion of monocyte-derived macrophages into tissue-resident macrophages during alternative activation. Nat. Immunol 18(6):642–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bohlen CJ, Bennett FC, Tucker AF, Collins HY, Mulinyawe SB, and Barres BA. 2017. Diverse Requirements for Microglial Survival, Specification, and Function Revealed by Defined-Medium Cultures. Neuron 94: 759–773.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takata K, Kozaki T, Lee CZW, Thion MS, Otsuka M, Lim S, Utami KH, Fidan K, Park DS, Malleret B, Chakarov S, See P, Low D, Low G, Garcia-Miralles M, Zeng R, Zhang J, Goh CC, Gul A, Hubert S, Lee B, Chen J, Low I, Shadan NB, Lum J, Wei TS, Mok E, Kawanishi S, Kitamura Y, Larbi A, Poidinger M, Renia L, Ng LG, Wolf Y, Jung S, Önder T, Newell E, Huber T, Ashihara E, Garel S, Pouladi MA, and Ginhoux F. 2017. Induced-Pluripotent-Stem-Cell-Derived Primitive Macrophages Provide a Platform for Modeling Tissue-Resident Macrophage Differentiation and Function. Immunity 47: 183–198.e6. [DOI] [PubMed] [Google Scholar]
- 69.Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, Yeromin AV, Scarfone VM, Marsh SE, Fimbres C, Caraway CA, Fote GM, Madany AM, Agrawal A, Kayed R, Gylys KH, Cahalan MD, Cummings BJ, Antel JP, Mortazavi A, Carson MJ, Poon WW, and Blurton-Jones M. 2017. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 94: 278–293.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varol D, Mildner A, Blank T, Shemer A, Barashi N, Yona S, David E, Boura-Halfon S, Segal-Hayoun Y, Chappell-Maor L, Keren-Shaul H, Leshkowitz D, Hornstein E, Fuhrmann M, Amit I, Maggio N, Prinz M, and Jung S. 2017. Dicer Deficiency Differentially Impacts Microglia of the Developing and Adult Brain. Immunity 46: 1030–1044.e8. [DOI] [PubMed] [Google Scholar]
- 71.Datta M, Staszewski O, Raschi E, Frosch M, Hagemeyer N, Tay TL, Blank T, Kreutzfeldt M, Merkler D, Ziegler-Waldkirch S, Matthias P, Meyer-Luehmann M, and Prinz M. 2018. Histone Deacetylases 1 and 2 Regulate Microglia Function during Development, Homeostasis, and Neurodegeneration in a Context-Dependent Manner. Immunity 48: 514–529.e6. [DOI] [PubMed] [Google Scholar]
- 72.Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang L, Means TK, and El Khoury J. 2013. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci 16: 1896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Swiecki M, and Colonna M. 2015. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol 15(8):471–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mrdjen D, Pavlovic A, Hartmann FJ, Schreiner B, Utz SG, Leung BP, Lelios I, Heppner FL, Kipnis J, Merkler D, Greter M, and Becher B. 2018. High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity 48: 380–395.e6. [DOI] [PubMed] [Google Scholar]
- 75.Mundt S, Mrdjen D, Utz SG, Greter M, Schreiner B, and Becher B. 2019. Conventional DCs sample and present myelin antigens in the healthy CNS and allow parenchymal T cell entry to initiate neuroinflammation. Sci. Immunol 4(31). pii: eaau8380. [DOI] [PubMed] [Google Scholar]
- 76.Jordão MJC, Sankowski R, Brendecke SM, Sagar, Locatelli G, Tai YH, Tay TL, Schramm E, Armbruster S, Hagemeyer N, Groß O, Mai D, Çiçek Ö, Falk T, Kerschensteiner M, Grün D, and Prinz M. 2019. Neuroimmunology: Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363(6425). pii: eaat7554. [DOI] [PubMed] [Google Scholar]
- 77.Van Hove H, Martens L, Scheyltjens I, De Vlaminck K, Pombo Antunes AR, De Prijck S, Vandamme N, De Schepper S, Van Isterdael G, Scott CL, Aerts J, Berx G, Boeckxstaens GE, Vandenbroucke RE, Vereecke L, Moechars D, Guilliams M, Van Ginderachter JA, Saeys Y, and Movahedi K. 2019. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci 22(6):1021–1035. [DOI] [PubMed] [Google Scholar]
- 78.Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, Marsh SE, Saunders A, Macosko E, Ginhoux F, Chen J, Franklin RJM, Piao X, McCarroll SA, and Stevens B. 2019. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50(1):253–271.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Masuda T, Sankowski R, Staszewski O, Böttcher C, Amann L, Scheiwe C, Nessler S, Kunz P, van Loo G, Coenen VA, Reinacher PC, Michel A, Sure U, Gold R, Priller J, Stadelmann C, and Prinz M. 2019. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566(7744):388–392. [DOI] [PubMed] [Google Scholar]
- 80.Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, Gulati G, Bennett ML, Sun LO, Clarke LE, Marschallinger J, Yu G, Quake SR, Wyss-Coray T, and Barres BA. 2019. Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron 101(2):207–223.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haimon Z, Volaski A, Orthgiess J, Boura-Halfon S, Varol D, Shemer A, Yona S, Zuckerman B, David E, Chappell-Maor L, Bechmann I, Gericke M, Ulitsky I, and Jung S. 2018. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol 19: 636–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK, Stevens MP, Freeman TC, Summers KM, and McColl BW. 2016. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci 19: 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Biase LM, Schuebel KE, Fusfeld ZH, Jair K, Hawes IA, Cimbro R, Zhang HY, Liu QR, Shen H, Xi ZX, Goldman D, and Bonci A. 2017. Local Cues Establish and Maintain Region-Specific Phenotypes of Basal Ganglia Microglia. Neuron 95: 341–356.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ayata P, Badimon A, Strasburger HJ, Duff MK, Montgomery SE, Loh YHE, Ebert A, Pimenova AA, Ramirez BR, Chan AT, Sullivan JM, Purushothaman I, Scarpa JR, Goate AM, Busslinger M, Shen L, Losic B, and Schaefer A. 2018. Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci 21(8):1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Böttcher C, Schlickeiser S, Sneeboer MAM, Kunkel D, Knop A, Paza E, Fidzinski P, Kraus L, Snijders GJL, Kahn RS, Schulz AR, Mei HE, Hol EM, Siegmund B, Glauben R, Spruth EJ, de Witte LD, and Priller J. 2019. Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat. Neurosci 22(1):78–90. [DOI] [PubMed] [Google Scholar]
- 86.Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, Itzkovitz S, Colonna M, Schwartz M, and Amit I. 2017. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169: 1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- 87.Mathys H, Adaikkan C, Gao F, Young JZ, Manet E, Hemberg M, De Jager PL, Ransohoff RM, Regev A, and Tsai LH. 2017. Temporal Tracking of Microglia Activation in Neurodegeneration at Single-Cell Resolution. Cell Rep. 21: 366–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, Greco DJ, Smith ST, Tweet G, Humulock Z, Zrzavy T, Conde-Sanroman P, Gacias M, Weng Z, Chen H, Tjon E, Mazaheri F, Hartmann K, Madi A, Ulrich JD, Glatzel M, Worthmann A, Heeren J, Budnik B, Lemere C, Ikezu T, Heppner FL, Litvak V, Holtzman DM, Lassmann H, Weiner HL, Ochando J, Haass C, and Butovsky O. 2017. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47: 566–581.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shi Y, Yamada K, Liddelow SA, Smith ST, Zhao L, Luo W, Tsai RM, Spina S, Grinberg LT, Rojas JC, Gallardo G, Wang K, Roh J, Robinson G, Finn MB, Jiang H, Sullivan PM, Baufeld C, Wood MW, Sutphen C, McCue L, Xiong C, Del-Aguila JL, Morris JC, Cruchaga C, Fagan AM, Miller BL, Boxer AL, Seeley WW, Butovsky O, Barres BA, Paul SM, and Holtzman DM. 2017. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549(7673):523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parhizkar S, Arzberger T, Brendel M, Kleinberger G, Deussing M, Focke C, Nuscher B, Xiong M, Ghasemigharagoz A, Katzmarski N, Krasemann S, Lichtenthaler SF, Müller SA, Colombo A, Monasor LS, Tahirovic S, Herms J, Willem M, Pettkus N, Butovsky O, Bartenstein P, Edbauer D, Rominger A, Ertürk A, Grathwohl SA, Neher JJ, Holtzman DM, Meyer-Luehmann M, and Haass C. 2019. Loss of TREM2 function increases amyloid seeding but reduces plaque-associated ApoE. Nat. Neurosci 22(2):191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tay TL, Sagar, Dautzenberg J, Grün D, and Prinz M. 2018. Unique microglia recovery population revealed by single-cell RNAseq following neurodegeneration. Acta Neuropathol. Commun 6(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH, Holtzman DM, Cirrito JR, and Colonna M. 2015. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160(6):1061–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yeh FL, Hansen DV, and Sheng M. 2017. TREM2, Microglia, and Neurodegenerative Diseases. Trends Mol. Med 23: 512–533. [DOI] [PubMed] [Google Scholar]
- 94.Song WM, and Colonna M. 2018. The identity and function of microglia in neurodegeneration. Nat. Immunol 19(10):1048–1058. [DOI] [PubMed] [Google Scholar]
- 95.Shi Y, and Holtzman DM. 2018. Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat. Rev. Immunol . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ulland TK, and Colonna M. 2018. TREM2 — a key player in microglial biology and Alzheimer disease. Nat. Rev. Neurol 14(11):667–675. [DOI] [PubMed] [Google Scholar]
- 97.Hamilton G, Colbert JD, Schuettelkopf AW, and Watts C. 2008. Cystatin F is a cathepsin C-directed protease inhibitor regulated by proteolysis. EMBO J. 27(3):499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ofengeim D, Mazzitelli S, Ito Y, DeWitt JP, Mifflin L, Zou C, Das S, Adiconis X, Chen H, Zhu H, Kelliher MA, Levin JZ, and Yuan J. 2017. RIPK1 mediates a disease-associated microglial response in Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A 114(41):E8788–E8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McGlinchey RP, and Lee JC. 2015. Cysteine cathepsins are essential in lysosomal degradation of α-synuclein. Proc. Natl. Acad. Sci 112(30):9322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mohamed MM, and Sloane BF. 2006. Cysteine cathepsins: Multifunctional enzymes in cancer. In Nature Reviews Cancer 6(10):764–75. [DOI] [PubMed] [Google Scholar]
- 101.Lemke G 2013. Biology of the TAM receptors. Cold Spring Harb. Perspect. Biol 5(11):a009076.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fourgeaud L, Través PG, Tufail Y, Leal-Bailey H, Lew ED, Burrola PG, Callaway P, Zagórska A, V Rothlin C, Nimmerjahn A, and Lemke G. 2016. TAM receptors regulate multiple features of microglial physiology. Nature 532: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tufail Y, Cook D, Fourgeaud L, Powers CJ, Merten K, Clark CL, Hoffman E, Ngo A, Sekiguchi KJ, O’Shea CC, Lemke G, and Nimmerjahn A. 2017. Phosphatidylserine Exposure Controls Viral Innate Immune Responses by Microglia. Neuron 93(3):574–586.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weinger JG, Brosnan CF, Loudig O, Goldberg MF, Macian F, Arnett HA, Prieto AL, Tsiperson V, and Shafit-Zagardo B. 2011. Loss of the receptor tyrosine kinase Axl leads to enhanced inflammation in the CNS and delayed removal of myelin debris during Experimental Autoimmune Encephalomyelitis. J. Neuroinflammation 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Comi C, Carecchio M, Chiocchetti A, Nicola S, Galimberti D, Fenoglio C, Cappellano G, Monaco F, Scarpini E, and Dianzani U. 2010. Osteopontin is increased in the cerebrospinal fluid of patients with Alzheimer’s disease and its levels correlate with cognitive decline. J. Alzheimer’s Dis 19(4):1143–8. [DOI] [PubMed] [Google Scholar]
- 106.Begcevic I, Brinc D, Brown M, Martinez-Morillo E, Goldhardt O, Grimmer T, Magdolen V, Batruch I, and Diamandis EP. 2018. Brain-related proteins as potential CSF biomarkers of Alzheimer’s disease: A targeted mass spectrometry approach. J. Proteomics 182:12–20. [DOI] [PubMed] [Google Scholar]
- 107.Yoshitake H, Rittling SR, Denhardt DT, and Noda M. 2002. Osteopontin-deficient mice are resistant to ovariectomy-induced bone resorption. Proc. Natl. Acad. Sci 96(14):8156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thurner PJ, Chen CG, Ionova-Martin S, Sun L, Harman A, Porter A, Ager JW, Ritchie RO, and Alliston T. 2010. Osteopontin deficiency increases bone fragility but preserves bone mass. Bone 46(6):1564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cantor H, and Shinohara ML. 2009. Regulation of T-helper-cell lineage development by osteopontin: The inside story. Nat. Rev. Immunol 9(2):137–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ashkar S, Weber GF, Panoutsakopoulou V, Sanchirico ME, Jansson M, Zawaideh S, Rittling SR, Denhardt DT, Glimcher MJ, and Cantor H. 2000. Eta-1 (osteopontin): An early component of type-1 (cell-mediated) immunity. Science. 287(5454):860–4. [DOI] [PubMed] [Google Scholar]
- 111.Shinohara ML, Jansson M, Hwang ES, Werneck MBF, Glimcher LH, and Cantor H. 2005. T-bet-dependent expression of osteopontin contributes to T cell polarization. Proc. Natl. Acad. Sci 102(47):17101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hur EM, Youssef S, Haws ME, Zhang SY, Sobel RA, and Steinman L. 2007. Osteopontin-induced relapse and progression of autoimmune brain disease through enhanced survival of activated T cells. Nat. Immunol 8(1):74–83. [DOI] [PubMed] [Google Scholar]
- 113.Denhardt DT, Noda M, O’Regan AW, Pavlin D, and Berman JS. 2001. Osteopontin as a means to cope with environmental insults: Regulation of inflammation, tissue remodeling, and cell survival. J. Clin. Invest 107(9):1055–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guo H, Cai CQ, Schroeder RA, and Kuo PC. 2001. Osteopontin Is a Negative Feedback Regulator of Nitric Oxide Synthesis in Murine Macrophages. J. Immunol 166(2):1079–86. [DOI] [PubMed] [Google Scholar]
- 115.Pietras A, Katz AM, Ekström EJ, Wee B, Halliday JJ, Pitter KL, Werbeck JL, Amankulor NM, Huse JT, and Holland EC. 2014. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 14(3):357–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schirmer L, Velmeshev D, Holmqvist S, Kaufmann M, Werneburg S, Jung D, Vistnes S, Stockley JH, Young A, Steindel M, Tung B, Goyal N, Bhaduri A, Mayer S, Engler JB, Bayraktar OA, Franklin RJM, Haeussler M, Reynolds R, Schafer DP, Friese MA, Shiow LR, Kriegstein AR, and Rowitch DH. 2019. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature doi: 10.1038/s41586-019-1404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Korin B, Ben-Shaanan TL, Schiller M, Dubovik T, Azulay-Debby H, Boshnak NT, Koren T, and Rolls A. 2017. High-dimensional, single-cell characterization of the brain’s immune compartment. Nat. Neurosci 20(9):1300–1309. [DOI] [PubMed] [Google Scholar]
- 118.Agnihotri R, Crawford HC, Haro H, Matrisian LM, Havrda MC, and Liaw L. 2001. Osteopontin, a Novel Substrate for Matrix Metalloproteinase-3 (Stromelysin-1) and Matrix Metalloproteinase-7 (Matrilysin). J. Biol. Chem 276(30):28261–7. [DOI] [PubMed] [Google Scholar]
- 119.Sondag GR, Mbimba TS, Moussa FM, Novak K, Yu B, Jaber FA, Abdelmagid SM, Geldenhuys WJ, and Safadi FF. 2016. Osteoactivin inhibition of osteoclastogenesis is mediated through CD44-ERK signaling. Exp. Mol. Med 48(9):e257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Neal ML, Boyle AM, Budge KM, Safadi FF, and Richardson JR. 2018. The glycoprotein GPNMB attenuates astrocyte inflammatory responses through the CD44 receptor. J. Neuroinflammation 15(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, and Brown GD. 2007. Dectin-1 is required for β-glucan recognition and control of fungal infection. Nat. Immunol 8(1):31–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shah VB, Huang Y, Keshwara R, Ozment-Skelton T, Williams DL, and Keshvara L. 2014. Beta-Glucan Activates Microglia without Inducing Cytokine Production in Dectin-1-Dependent Manner. J. Immunol 180(5):2777–85. [DOI] [PubMed] [Google Scholar]
- 123.Esteban A, Popp MW, Vyas VK, Strijbis K, Ploegh HL, and Fink GR. 2011. Fungal recognition is mediated by the association of dectin-1 and galectin-3 in macrophages. Proc. Natl. Acad. Sci 108(34):14270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mass E, Jacome-Galarza CE, Blank T, Lazarov T, Durham BH, Ozkaya N, Pastore A, Schwabenland M, Chung YR, Rosenblum MK, Prinz M, Abdel-Wahab O, and Geissmann F. 2017. A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature 549: 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ulland TK, Song WM, Huang SCC, Ulrich JD, Sergushichev A, Beatty WL, Loboda AA, Zhou Y, Cairns NJ, Kambal A, Loginicheva E, Gilfillan S, Cella M, Virgin HW, Unanue ER, Wang Y, Artyomov MN, Holtzman DM, and Colonna M. 2017. TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell 170(4):649–663.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Heneka MT, Kummer MP, and Latz E. 2014. Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol 14(7):463–77. [DOI] [PubMed] [Google Scholar]
- 127.Taborda CP, and Casadevall A. 2002. CR3 (CD11b/CD18) and CR4 (CD11c/CD18) are involved in complement-independent antibody-mediated phagocytosis of Cryptococcus neoformans. Immunity 16(6):791–802. [DOI] [PubMed] [Google Scholar]
- 128.Ravetch JV, and Lanier LL. 2000. Immune inhibitory receptors. Science 290(5489):84–9. [DOI] [PubMed] [Google Scholar]
- 129.Lu Y, Jiang Z, Dai H, Miao R, Shu J, Gu H, Liu X, Huang Z, Yang G, Chen AF, Yuan H, Li Y, and Cai J. 2018. Hepatic leukocyte immunoglobulin-like receptor B4 (LILRB4) attenuates nonalcoholic fatty liver disease via SHP1-TRAF6 pathway. Hepatology (4):1303–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, and Yamashita T. 2013. Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci 16: 543–51. [DOI] [PubMed] [Google Scholar]
- 131.Wlodarczyk A, Holtman IR, Krueger M, Yogev N, Bruttger J, Khorooshi R, Benmamar‐Badel A, de Boer‐Bergsma JJ, Martin NA, Karram K, Kramer I, Boddeke EW, Waisman A, Eggen BJ, and Owens T. 2017. A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. e201696056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.De I, Nikodemova M, Steffen MD, Sokn E, Maklakova VI, Watters JJ, and Collier LS. 2014. CSF1 overexpression has pleiotropic effects on microglia in vivo. Glia 62: 1955–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mathys H, Davila-velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L, Abdurrob F, Jiang X, Martorell AJ, Ransohoff RM, Hafler BP, Bennett DA, Kellis M, and Tsai LH. Single-cell transcriptomic analysis of Alzheimer ‘s disease. Nature 570(7761):332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Galatro TF, Holtman IR, Lerario AM, Vainchtein ID, Brouwer N, Sola PR, Veras MM, Pereira TF, Leite REP, Möller T, Wes PD, Sogayar MC, Laman JD, Den Dunnen W, Pasqualucci CA, Oba-Shinjo SM, Boddeke EWGM, Marie SKN, and Eggen BJL. 2017. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci 20: 1162–1171. [DOI] [PubMed] [Google Scholar]
- 135.Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, Bakiasi G, Tsai L-H, Aubourg P, Ransohoff RM, and Jaenisch R. 2016. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med 22: 1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, Kim G, Brown MA, Elkahloun AG, Maric D, Sweeney CL, Gossa S, Malech HL, McGavern DB, and Park JK. 2017. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat. Neurosci 20: 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hasselmann J, Coburn MA, England W, Gandhi SP, Spitale RC, Blurton-jones M, Hasselmann J, Coburn MA, England W, Velez DXF, Shabestari SK, Tu CH, Mcquade A, Kolahdouzan M, Echeverria K, Davtyan H, Glass CK, Healy LM, Gandhi SP, and Spitale RC. 2019. Development of a Chimeric Model to Study and NeuroResource Development of a Chimeric Model to Study and Manipulate Human Microglia In Vivo. Neuron 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Zaitsev K, Miller KR, Gilfillan S, Ulland TK, Beausoleil SA, Sainouchi M, Kakita A, Bennett DA, Schneider JA, Ulrich J, Holtzman DM, Artyomov M, Colonna M. Single-nucleus transcriptional mapping of mouse and human Alzheimer’s disease reveals TREM2-dependent and -independent cellular responses to pathology. In revision [Google Scholar]
- 139.Prinz M, Erny D, and Hagemeyer N. 2017. Ontogeny and homeostasis of CNS myeloid cells. Nat. Immunol 18: 385–392. [DOI] [PubMed] [Google Scholar]
- 140.Rojo R, Raper A, Ozdemir DD, Lefevre L, Grabert K, Caruso M, Gazova I, Zofia M, Alves J, Bradford B, Molina I, Davtyan H, Lodge RJ, Glover JD, Wallace R, David E, Amit I, Stephen J, Hardingham GE, Blurton-jones M, Mabbott NA, Kim M, Hohenstein P, Hume DA, Pridans C, Bush E, Unit T, Institutet K, and Queen T. 2019. Germ-line deletion of a highly-conserved intronic enhancer of the Csf1r locus has selective impacts on CSF1R expression and development of tissue macrophage populations. Nat. Commun 10(1):3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Goldmann T, Wieghofer P, Jordão MJC, Prutek F, Hagemeyer N, Frenzel K, Amann L, Staszewski O, Kierdorf K, Krueger M, Locatelli G, Hochgerner H, Zeiser R, Epelman S, Geissmann F, Priller J, V Rossi FM, Bechmann I, Kerschensteiner M, Linnarsson S, Jung S, and Prinz M. 2016. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol 17: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mues M, Bartholomäus I, Thestrup T, Griesbeck O, Wekerle H, Kawakami N, and Krishnamoorthy G. 2013. Real-time in vivo analysis of T cell activation in the central nervous system using a genetically encoded calcium indicator. Nat. Med 19(6):778–83. [DOI] [PubMed] [Google Scholar]
- 143.Bartholomäus I, Kawakami N, Odoardi F, Schläger C, Miljkovic D, Ellwart JW, Klinkert WEF, Flügel-Koch C, Issekutz TB, Wekerle H, and Flügel A. 2009. Effector T cell interactions with meningeal vascular structures in nascent autoimmune CNS lesions. Nature 462(7269):94–8. [DOI] [PubMed] [Google Scholar]
- 144.Kivisakk P, Imitola J, Rasmussen S, Elyaman W, Zhu B, Ransohoff RM, and Khoury SJ. 2009. Localizing central nervous system immune surveillance: Meningeal antigen-presenting cells activate T cells during experimental autoimmune encephalomyelitis. Ann. Neurol ;65(4):457–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lodygin D, Odoardi F, Schläger C, Körner H, Kitz A, Nosov M, Van Den Brandt J, Reichardt HM, Haberl M, and Flügel A. 2013. A combination of fluorescent NFAT and H2B sensors uncovers dynamics of T cell activation in real time during CNS autoimmunity. Nat. Med 19(6):784–90. [DOI] [PubMed] [Google Scholar]
- 146.Schläger C, Körner H, Krueger M, Vidoli S, Haberl M, Mielke D, Brylla E, Issekutz T, Cabanãs C, Nelson PJ, Ziemssen T, Rohde V, Bechmann I, Lodygin D, Odoardi F, and Flügel A. 2016. Effector T-cell trafficking between the leptomeninges and the cerebrospinal fluid. Nature 530(7590):349–53. [DOI] [PubMed] [Google Scholar]
- 147.Lodygin D, Hermann M, Schweingruber N, Flügel-Koch C, Watanabe T, Schlosser C, Merlini A, Körner H, Chang HF, Fischer HJ, Reichardt HM, Zagrebelsky M, Mollenhauer B, Kügler S, Fitzner D, Frahm J, Stadelmann C, Haberl M, Odoardi F, and Flügel A. 2019. β-Synuclein-reactive T cells induce autoimmune CNS grey matter degeneration. Nature 566(7745):503–508. [DOI] [PubMed] [Google Scholar]
- 148.Wolf Y, Shemer A, Levy-Efrati L, Gross M, Kim JS, Engel A, David E, Chappell-Maor L, Grozovski J, Rotkopf R, Biton I, Eilam-Altstadter R, and Jung S. 2018. Microglial MHC class II is dispensable for experimental autoimmune encephalomyelitis and cuprizone-induced demyelination. Eur. J. Immunol 48(8):1308–1318. [DOI] [PubMed] [Google Scholar]
- 149.Ajami B, Samusik N, Wieghofer P, Ho PP, Crotti A, Bjornson Z, Prinz M, Fantl WJ, Nolan GP, and Steinman L. 2018. Single-cell mass cytometry reveals distinct populations of brain myeloid cells in mouse neuroinflammation and neurodegeneration models. Nat. Neurosci 21: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Locatelli G, Theodorou D, Kendirli A, Jordão MJC, Staszewski O, Phulphagar K, Cantuti-Castelvetri L, Dagkalis A, Bessis A, Simons M, Meissner F, Prinz M, and Kerschensteiner M. 2018. Mononuclear phagocytes locally specify and adapt their phenotype in a multiple sclerosis model. Nat. Neurosci 21(9):1196–1208. [DOI] [PubMed] [Google Scholar]
- 151.Rothhammer V, Borucki DM, Tjon EC, Takenaka MC, Chao CC, Ardura-Fabregat A, De Lima KA, Gutiérrez-Vázquez C, Hewson P, Staszewski O, Blain M, Healy L, Neziraj T, Borio M, Wheeler M, Dragin LL, Laplaud DA, Antel J, Alvarez JI, Prinz M, and Quintana FJ. 2018. Microglial control of astrocytes in response to microbial metabolites. Nature 557(7707):724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Voet S, Mc Guire C, Hagemeyer N, Martens A, Schroeder A, Wieghofer P, Daems C, Staszewski O, Vande Walle L, Jordao MJC, Sze M, Vikkula H-K, Demeestere D, Van Imschoot G, Scott CL, Hoste E, Gonçalves A, Guilliams M, Lippens S, Libert C, Vandenbroucke RE, Kim K-W, Jung S, Callaerts-Vegh Z, Callaerts P, de Wit J, Lamkanfi M, Prinz M, and van Loo G. 2018. A20 critically controls microglia activation and inhibits inflammasome-dependent neuroinflammation. Nat. Commun 9(1):2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goldmann T, Zeller N, Raasch J, Kierdorf K, Frenzel K, Ketscher L, Basters A, Staszewski O, Brendecke SM, Spiess A, Tay TL, Kreutz C, Timmer J, Mancini GM, Blank T, Fritz G, Biber K, Lang R, Malo D, Merkler D, Heikenwalder M, Knobeloch K-P, and Prinz M. 2015. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J. 34(12):1612–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schwabenland M, Mossad O, Peres AG, Kessler F, Jose F, Maron M, Harsan L, Bienert T, Von Elverfeldt D, Knobeloch K, Staszewski O, Heppner FL, Meuwissen MEC, Mancini GMS, Prinz M, and Blank T. 2019. Loss of USP18 in microglia induces white matter pathology. Acta Neuropathol Commun. 7(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yamasaki R, Lu H, Butovsky O, Ohno N, Rietsch AM, Cialic R, Wu PM, Doykan CE, Lin J, Cotleur AC, Kidd G, Zorlu MM, Sun N, Hu W, Liu L, Lee J-C, Taylor SE, Uehlein L, Dixon D, Gu J, Floruta CM, Zhu M, Charo IF, Weiner HL, and Ransohoff RM. 2014. Differential roles of microglia and monocytes in the inflamed central nervous system. J. Exp. Med 211(8):1533–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Poliani PL, Wang Y, Fontana E, Robinette ML, Yamanishi Y, Gilfillan S, and Colonna M. 2015. TREM2 sustains microglial expansion during aging and response to demyelination. J. Clin. Invest 125(5):2161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cantoni C, Bollman B, Licastro D, Xie M, Mikesell R, Schmidt R, Yuede CM, Galimberti D, Olivecrona G, Klein RS, Cross AH, Otero K, and Piccio L. 2015. TREM2 regulates microglial cell activation in response to demyelination in vivo. Acta Neuropathol. 129(3):429–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Safaiyan S, Kannaiyan N, Snaidero N, Brioschi S, Biber K, Yona S, Edinger AL, Jung S, Rossner MJ, and Simons M. 2016. Age-related myelin degradation burdens the clearance function of microglia during aging. Nat. Neurosci 19: 995–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cantuti-Castelvetri L, Fitzner D, Bosch-Queralt M, Weil MT, Su M, Sen P, Ruhwedel T, Mitkovski M, Trendelenburg G, Lütjohann D, Möbius W, and Simons M. 2018. Defective cholesterol clearance limits remyelination in the aged central nervous system. Science 359(6376):684–688. [DOI] [PubMed] [Google Scholar]
- 160.Lloyd AF, Davies CL, Holloway RK, Labrak Y, Ireland G, Carradori D, Dillenburg A, Borger E, Soong D, Richardson JC, Kuhlmann T, Williams A, Pollard JW, des Rieux A, Priller J, and Miron VE. 2019. Central nervous system regeneration is driven by microglia necroptosis and repopulation. Nat. Neurosci 22(7):1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Perillo NL, Pace KE, Seilhamer JJ, and Baum LG. 1995. Apoptosis of T cells mediated by galectin−1. Nature 378(6558):736–9. [DOI] [PubMed] [Google Scholar]
- 162.Salama AD, Chitnis T, Imitola J, Ansari MJI, Akiba H, Tushima F, Azuma M, Yagita H, Sayegh MH, and Khoury SJ. 2003. Critical Role of the Programmed Death-1 (PD-1) Pathway in Regulation of Experimental Autoimmune Encephalomyelitis. J. Exp. Med 198(1):71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wilcox RA, Feldman AL, Wada DA, Yang ZZ, Comfere NI, Dong H, Kwon ED, Novak AJ, Markovic SN, Pittelkow MR, Witzig TE, and Ansell SM. 2009. B7-H1 (PD-L1, CD274) suppresses host immunity in T-cell lymphoproliferative disorders. Blood 114(10):2149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sundblad V, Morosi LG, Geffner JR, and Rabinovich GA. 2017. Galectin-1: A Jack-of-All-Trades in the Resolution of Acute and Chronic Inflammation. J. Immunol 199(11):3721–3730. [DOI] [PubMed] [Google Scholar]
- 165.Nissen JC, Thompson KK, West BL, and Tsirka SE. 2018. Csf1R inhibition attenuates experimental autoimmune encephalomyelitis and promotes recovery. Exp. Neurol 307:24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tanabe S, Saitoh S, Miyajima H, Itokazu T, and Yamashita T. 2019. Microglia suppress the secondary progression of autoimmune encephalomyelitis. Glia 67(9):1694–1704. [DOI] [PubMed] [Google Scholar]
- 167.Rubino SJ, Mayo L, Wimmer I, Siedler V, Brunner F, Hametner S, Madi A, Lanser A, Moreira T, Donnelly D, Cox L, Rezende RM, Butovsky O, Lassmann H, and Weiner HL. 2018. Acute microglia ablation induces neurodegeneration in the somatosensory system. Nat. Commun 9(1):4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kierdorf K, Masuda T, Jordão MJC, Prinz M. 2019. Macrophages at CNS interfaces: ontogeny and function in health and disease. Nat Rev Neurosci. doi: 10.1038/s41583-019-0201-x. [DOI] [PubMed] [Google Scholar]


