Significance
Unlike other eukaryotic viruses, fungal viruses generally lack an extracellular phase in their life cycle. It thus remains unclear whether fungal viruses can spread beyond their native hosts. Herein, we investigated the infectivity of a fungal virus, Cryphonectria hypovirus 1 (CHV1) in Nicotiana tabacum plants. CHV1 is able to systemically infect plants after mechanical coinoculations with plant viruses such as tobacco mosaic virus (TMV) or when inoculated to transgenic plants expressing the TMV movement protein. In the fungal inoculation experiment, we demonstrated that plant virus infections in plants enable horizontal transfer of CHV1 from fungi to plants and to other heterologous fungal species. Hence, our results reveal a facilitative effect of plant viruses in spreading fungal viruses.
Keywords: plant virus, mycovirus, cross-kingdom, infection
Abstract
Plants and fungi are closely associated through parasitic or symbiotic relationships in which bidirectional exchanges of cellular contents occur. Recently, a plant virus was shown to be transmitted from a plant to a fungus, but it is unknown whether fungal viruses can also cross host barriers and spread to plants. In this study, we investigated the infectivity of Cryphonectria hypovirus 1 (CHV1, family Hypoviridae), a capsidless, positive-sense (+), single-stranded RNA (ssRNA) fungal virus in a model plant, Nicotiana tabacum. CHV1 replicated in mechanically inoculated leaves but did not spread systemically, but coinoculation with an unrelated plant (+)ssRNA virus, tobacco mosaic virus (TMV, family Virgaviridae), or other plant RNA viruses, enabled CHV1 to systemically infect the plant. Likewise, CHV1 systemically infected transgenic plants expressing the TMV movement protein, and coinfection with TMV further enhanced CHV1 accumulation in these plants. Conversely, CHV1 infection increased TMV accumulation when TMV was introduced into a plant pathogenic fungus, Fusarium graminearum. In the in planta F. graminearum inoculation experiment, we demonstrated that TMV infection of either the plant or the fungus enabled the horizontal transfer of CHV1 from the fungus to the plant, whereas CHV1 infection enhanced fungal acquisition of TMV. Our results demonstrate two-way facilitative interactions between the plant and fungal viruses that promote cross-kingdom virus infections and suggest the presence of plant–fungal-mediated routes for dissemination of fungal and plant viruses in nature.
Diverse fungal viruses (or mycoviruses) are widely distributed in all major groups of fungi (1, 2). Initially, most reported fungal viruses had double-stranded RNA (dsRNA) genomes, but a number of positive (+) and negative (–) sense, single-stranded RNA (ssRNA) viruses, as well as a circular single-stranded DNA virus, have also been identified (3). Fungal viruses are currently classified into 16 families and 24 genera, with a number of families sharing sequence similarities with plant or animal viruses (4). Most fungal virus infections are asymptomatic or cryptic, but many fungal viruses reduce the growth and/or attenuate the virulence of their fungal hosts (termed “hypovirulence”) or, conversely, enhance fungal virulence (hypervirulence) (5–8). Thus, it is possible that fungal viruses can serve as molecular tools for investigations of fungal pathogenicity. Numerous studies have identified fungal viruses that infect agriculturally important phytopathogenic fungi, so it is possible that some of these might provide biological control agents of fungal crop diseases (9, 10).
Unlike animal and nonpersistent plant viruses, fungal viruses generally lack an extracellular phase in their life cycle and are transmitted vertically through sporulation and horizontally via hyphal fusion (1). Except for a fungal DNA virus that uses insects as a transmission vector (11), no other evidence of a biological vector for fungal viruses has been reported. A significant number of fungal viruses lack capsids (3), suggesting that they have adapted to an intracellular life cycle in the host. Moreover, no viral-encoded proteins related to those encoded by animal and plant viruses that function in cell entry or spread in the host have been identified in fungal viruses (12, 13). Different fungal strains or species commonly exhibit vegetative incompatibility that hinders the spread of viruses via hyphal anastomosis (14), albeit some virus transmissions across vegetative incompatible strains or species have been observed in the laboratory or in nature (15, 16). However, fungal viruses often have diverse host ranges that include taxonomically distant fungal species; therefore, hitherto unknown routes for spread of fungal viruses across different strains and species may exist in nature.
Plants host various fungi, including phytopathogenic, mycorrhizal, and endophytic fungi (17). Plants and infecting fungi can bidirectionally exchange various molecules; for example, fungi absorb water and nutrients from the plant cell and also secrete enzymes and effector proteins that promote fungal proliferation or suppress host defense responses (18, 19). Transfer of small RNA molecules to plants by phytopathogenic fungi have also been found to serve as effector molecules that inhibit expression of plant defense-related genes (i.e., transkingdom RNAi phenomenon) (20–22). Additionally, some fungi can acquire small plant RNAs that mediate gene suppression (21). Interestingly, our recent studies have demonstrated transfer of a plant virus and viroids (the smallest known plant pathogens) from plants to fungi and in the opposite direction during fungal colonization of plants under laboratory condition (23, 24). Moreover, studies using artificial inoculation have demonstrated compatibility of some plant viruses and viroids with fungal hosts including yeast (23–28), suggesting that certain plant viruses and viroids have an inherent ability to replicate in fungi. Moreover, this raises the interesting question of whether fungal viruses can also be transmitted and spread to plants during fungal infection. Fungal viruses infecting a marine fungus (Penicillium aurantiogriseum var. viridicatum) associated with seaweed were shown to replicate in plant protoplasts (29), but infection of whole plants by fungal viruses was not reported. Unlike fungi, developed plants consist of various tissues and organs connected by the vascular system. To invade the whole plant, a virus must be able to circumvent the cell wall via plasmodesmata (PD) and spread systemically through the vasculature and adapt to these tissues while simultaneously countering plant defense responses (30–32). Thus, for successful plant infections, fungal viruses must overcome a number of challenging obstacles.
Fungal viruses that have ssRNA genomes currently consist of nine families: Hypoviridae, Endornaviridae, Narnaviridae, Barnaviridae, Alphaflexiviridae, Gammaflexiviridae, Deltaflexiviridae, Mymonaviridae, and Botourmiaviridae and one floating genus, Botybirnavirus (4). The family Hypovirudae contains only one genus (Hypovirus), with four virus species (Cryphonectria hypovirus 1 through 4, CHV1–4) that infect an ascomycete phytopathogenic fungus, the chestnut blight fungus Cryphonectria parasitica (Diaporthales, Sordariomycetes) (33). The members of this genus are characterized by their naked large (+)ssRNA genomes ranging from 9.1 to 12.7 kb that contain either a single, long open-reading frame (ORF) or two ORFs encoding polyproteins with a cis-acting papain-like cysteine protease at the N terminus (34). CHV1 infection attenuates the growth and virulence of C. parasitica and represents the first example of the successful use of a mycovirus as a biocontrol agent against a plant pathogen (35, 36). CHV1 is one of the most intensively studied fungal viruses, and several molecular aspects of fungal virology, such as virus replication (37–40), pathogenicity (41, 42), and RNA silencing-associated host immunity (43–46), have been investigated using the CHV1–C. parasitica pathosystem.
In this study, we observed facilitative and synergistic interactions between CHV1 and a well-known plant (+)ssRNA virus, tobacco mosaic virus (TMV, genus Tobamovirus, family Virgaviridae, a plant alpha-like virus) during mixed infections with their respective fungal and plant hosts. Using mechanical inoculation, coinfection with TMV enabled the systemic spread of CHV1 in plants, and TMV infection also promoted high levels of CHV1 accumulation in leaf tissue. Conversely, CHV1 infection enhanced TMV accumulation in the phytopathogenic ascomycete fungus, Fusarium graminearum (Hypocreales, Sordariomycetes). Using in planta fungal inoculations, we further demonstrated two-way interactions between CHV1 and TMV in facilitating cross-kingdom virus infection. These findings have implications for a more profound understanding of fungal and plant virus cooperation to support dissemination in nature.
Results
Coinfection with a Plant Virus Enables Systemic Infection of CHV1 in Plants.
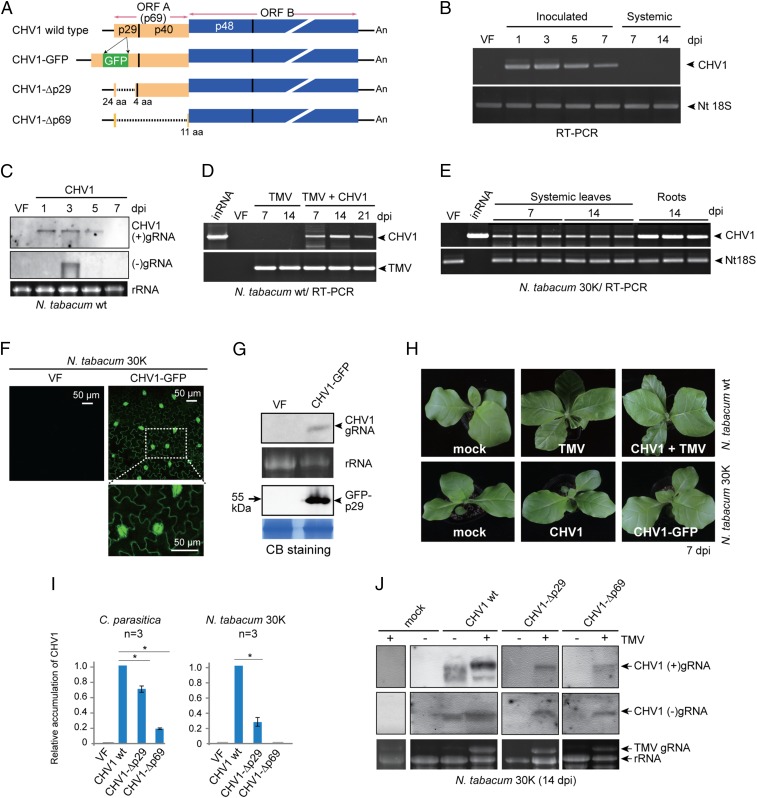
A previous report revealed replication of fungal viruses in plant cells (29), but the ability of fungal viruses to infect whole plants was not demonstrated. To investigate this issue, we selected CHV1 for artificial inoculation of Nicotiana tabacum plants because of the availability of an infectious full-length cDNA clone (Fig. 1A, CHV1-wild type (WT)) (38). Plus-strand in vitro CHV1 transcripts (45 ng/μL) were mechanically inoculated to leaves, and infection was tested by RT-PCR. Viral RNA was detected in inoculated leaves at 1 to 7 d postinoculation (dpi), but not in uninoculated upper leaves at 7 and 14 dpi (Fig. 1B). To confirm CHV1 replication in inoculated leaves, (+) genomic RNA (gRNA) was detected by RNA blotting at 1, 3, and 5 dpi but (–)gRNA accumulation was only detected at 3 dpi (Fig. 1C), indicating a short transient period of viral replication. Moreover, CHV1 (–)gRNA accumulation was reduced in inoculated leaves when lower concentrations of CHV1 transcripts were used for inoculation (SI Appendix, Fig. S1A). Thus it seems that inoculation with a high concentration of inoculum allows infection of a large number of cells. In a parallel experiment, when the total RNA fraction extracted from CHV1-infected C. parasitica was inoculated to leaves, both viral (+) and (–)gRNA accumulated in the inoculated leaves, with the most prominent accumulation of (–)gRNA at 3 dpi (SI Appendix, Fig. S1B), showing that total RNA from the infected fungus can also be used as inoculum. Together, these results indicate that CHV1 replicates in inoculated leaves but does not spread systemically to the upper leaves (Fig. 1B).
Fig. 1.
Infection of CHV1 in N. tabacum plants. (A) Schematic presentation of the CHV1 WT, CHV1-GFP, CHV1-Δp29, and CHV1-Δp69 genomes (not to scale). For CHV1-GFP, the gfp gene was inserted into the p29 gene. Colored boxes, lines, and dashed lines indicate the ORF, untranslated regions (UTRs), and deleted regions, respectively. (B) RT-PCR detection of CHV1 gRNA accumulation in inoculated and upper uninoculated leaves of N. tabacum plants. (C) RNA blotting analysis of CHV1 (+) and (–)gRNA accumulation in inoculated leaves. VF, virus free. The RNA gel was stained with ethidium bromide, and rRNA is shown as a loading control. (D) RT-PCR detection of viral gRNA accumulation in the upper leaves of plants inoculated with TMV alone or with TMV plus CHV1. inRNA, in vitro CHV1 transcripts. (E) RT-PCR detection of CHV1 RNA accumulation in uninoculated upper leaves and roots of transgenic N. tabacum plants expressing the TMV 30K movement protein. (F) Fluorescence microscopy observations of GFP expression in epidermal cells of CHV1-GFP–infected upper uninoculated leaves. (G) RNA and Western blot analyses of CHV-GFP (+)gRNA and GFP-p29 fusion protein accumulation in CHV1-GFP–infected upper uninoculated leaves. Coomassie brilliant blue (CB)-stained total proteins run on separate gels with the same sample batch are shown as loading controls. (H) Phenotypic growth of N. tabacum inoculated with TMV alone or with TMV plus CHV1, and transgenic N. tabacum 30K plants infected with CHV1 or CHV1-GFP. (I) Quantitative RT-PCR analysis of gRNA accumulation of CHV1 WT and CHV1 mutants in C. parasitica and in upper uninoculated N. tabacum 30K leaves. “*” indicates significant differences (P < 0.05, Student’s t test). (J) RNA blotting analysis of CHV1 (+) and (–)gRNA accumulation in N. tabacum 30K plants infected with TMV.
Considering that CHV1 may lack genes that are usually encoded by plant viruses for spread throughout the plant, we inoculated CHV1 to N. tabacum plants that had been previously inoculated with TMV. Interestingly, CHV1 was readily detected by RT-PCR in the upper leaves at 7, 14, and 21 dpi (Fig. 1D). Moreover, CHV1 systemic infections were also observed in plants infected with each of three other plant (+)ssRNA viruses, potato virus Y (PVY, family Potyviridae, a plant picorna-like virus), potato virus X (PVX, genus Potexvirus, family Alphaflexiviridae, a plant alpha-like virus), or cucumber mosaic virus (CMV, family Bromoviridae, a plant alpha-like virus) (SI Appendix, Fig. S2). Together, these results indicate that plant virus infections can facilitate CHV1 systemic infections in plants. Because viral movement proteins (MPs) are essential for cell-to-cell and long-distance transport of plant viruses (47, 48), we then inoculated CHV1 to a transgenic N. tabacum line expressing the TMV 30K MP protein (49). CHV1 RNA accumulation in uninoculated upper leaves and roots of the 30K plants was detected by RT-PCR (Fig. 1E and SI Appendix, Fig. S3A), indicating that the TMV MP facilitates systemic spread of CHV1. CHV1 RNA accumulation in N. tabacum 30K plants was considerably lower than in its natural host, C. parasitica, as compared by qRT-PCR (SI Appendix, Fig. S3B). To visually monitor CHV1 multiplication in the plants, CHV1 expressing GFP inserted within the p29 coding region (CHV1-GFP, Fig. 1A) (50) was inoculated to N. tabacum 30K plants. At 7 dpi, green fluorescence was observed in epidermal cells of uninoculated upper leaves using a fluorescence microscope (Fig. 1F), and accumulation of CHV1-GFP gRNA and the GFP-p29 fusion protein was detected by RNA and Western blotting (Fig. 1G).
CHV1 systemic infection did not induce visible symptoms on either the N. tabacum wild-type or the 30K-expresssing plants (Fig. 1H and SI Appendix, Fig. S2) and did not affect accumulation of the coinfecting plant viruses (SI Appendix, Fig. S4). Both sap extracts and total RNA derived from CHV1-infected systemic leaves were infectious when used as inoculum for mechanical inoculation (SI Appendix, Fig. S5). Moreover, gRNA of CHV1 that systemically infected the N. tabacum 30K plants for ∼40 d contained seven nucleotide changes, and three caused amino acid substitutions (SI Appendix, Fig. S6). These results clearly validate CHV1 infectivity in plants and suggest that genome evolution may occur during invasion.
To investigate whether other fungal viruses are also able to infect N. tabacum 30K plants systemically, we mechanically inoculated the total RNA fraction extracted from a phytopathogenic ascomycete fungus, Alternaria alternata (Pleosporales, Dothideomycetes), infected with Alternaria alternata hypovirus 1 (AaHV1), a virus related to CHV1 (51). Accumulation of AaHV1 genomic RNA was detected in the uninoculated upper leaves at 7 dpi, but viral symptoms were not observed (SI Appendix, Fig. S7). This result suggests that certain fungal viruses (at least hypoviruses) can infect plants when the plant viral MP is provided in trans.
TMV Infection Enhances CHV1 Accumulation in Plants.
The CHV1 ORF A encodes p69, a polyprotein that is proteolytically processed into p29 and p40 (Fig. 1A) (33). The p29 protein is an RNA silencing (or RNAi) suppressor that functions by inhibition of the up-regulation of key RNAi genes (43, 52). A CHV1 p69 deletion mutant (CHV1-Δp69) retains replication competency in C. parasitica although gRNA accumulation is drastically reduced, as assessed by qRT-PCR (Fig. 1I) (37). We compared the infectivity of CHV1-Δp69 and a second CHV1 p29 deletion mutant (CHV1-Δp29) in N. tabacum 30K plants and used RT-PCR to determine that CHV1-Δp29, but not CHV1-Δp69, RNA accumulated in the inoculated leaves and the upper uninoculated leaves (SI Appendix, Fig. S8). Nevertheless, the CHV1-Δp29 RNA accumulation was markedly lower than that of the CHV1-WT in the uninoculated upper leaves (Fig. 1I). Thus, although p69 is dispensable for CHV1 replication in the fungus, it appears to be essential for CHV1 replication and/or accumulation in the plant.
Plant viruses often enhance the accumulation of coinfecting viruses (53). We inoculated CHV1-WT, CHV1-Δp29, and CHV1-Δp69 to N. tabacum 30K plants that were infected with TMV and evaluated virus accumulation in the upper leaves by RNA blotting. Both CHV1 (+) and (–)gRNA accumulation was higher in TMV-coinfected plants than in CHV1-singly infected plants (Fig. 1J). Strikingly, CHV1-Δp29 gRNA was readily detected by RNA blotting after coinfection with TMV (Fig. 1J), and CHV1-Δp69 gRNA was also detected in coinfected plants (Fig. 1J). These results indicate that TMV infection enhances CHV1 accumulation and rescues CHV1-Δp69 accumulation in tobacco.
CHV1 Infection Enhances TMV Accumulation in Fungi.
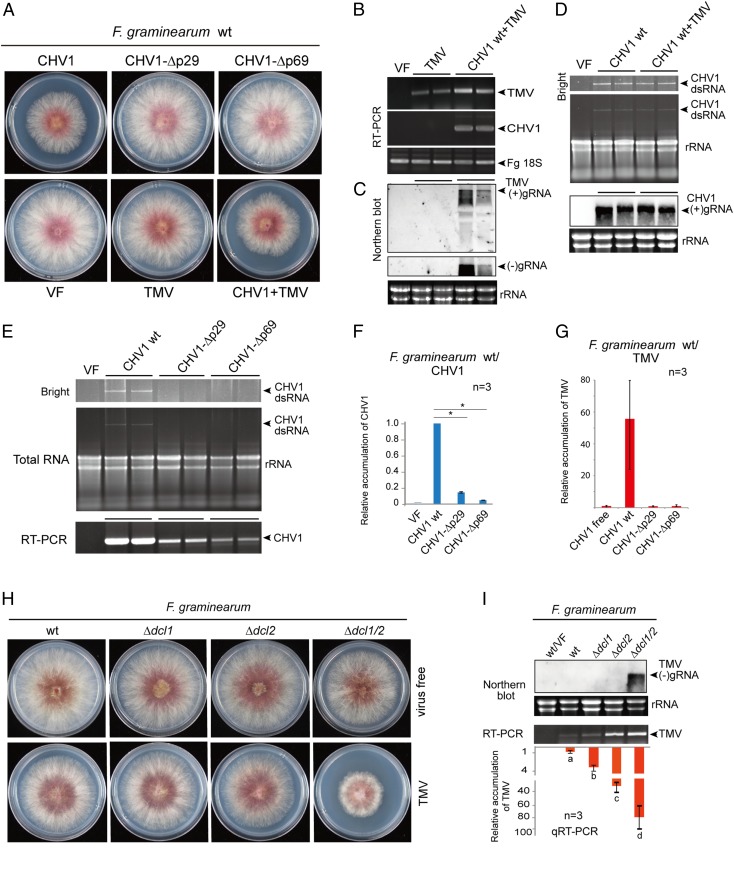
The synergism between the plant and fungal viruses in plants prompted us to explore whether such interactions occur in the fungal system. We used F. graminearum for this investigation owing to our initial findings that the fungus is a compatible host of CHV1 and TMV. Hence, TMV, CHV1, and CHV1 mutants were introduced into F. graminearum by protoplast transfection. After fungal growth and sporulation, both CHV1 and TMV were transmitted vertically through asexual spores (conidia), and coinfection with CHV1 enhanced TMV vertical transmission (SI Appendix, Fig. S9 A and B). In addition, infection with CHV1, but not the CHV1 mutants or TMV, reduced F. graminearum mycelial growth on potato dextrose agar (Fig. 2A and SI Appendix, Fig. S9C). TMV RNA accumulation in F. graminearum was relatively low and was detectable by RT-PCR (Fig. 2B), but not by RNA blotting (Fig. 2C). Strikingly, coinfection with CHV1 considerably elevated accumulation of TMV RNA (Fig. 2C), whereas accumulation of CHV1 RNA was not affected by coinfection with TMV (Fig. 2D). Moreover, TMV coat protein (CP) was detected in CHV1-coinfected fungi but not in fungi singly transfected with TMV (SI Appendix, Fig. S10). Thus, CHV1 confers a one-way synergistic effect on TMV replication and/or accumulation in F. graminearum.
Fig. 2.
CHV1 and TMV RNA accumulation in F. graminearum. (A) Phenotypic growth of fungal colonies that were virus-free (VF) or infected with WT CHV1, CHV1-Δp29, CHV1-Δp69, TMV, or CHV1 WT plus TMV. Fungi were grown on PDA medium (10-cm plates) for 3 d and then photographed. (B) RT-PCR detection of CHV1 and TMV RNA accumulation. (C) RNA blotting analysis of TMV (+) and (–)gRNA accumulation. (D) CHV1 dsRNA and (+)gRNA accumulation by gel electrophoresis and RNA blotting analyses. (E) dsRNA accumulation and RT-PCR detection of CHV1 WT and CHV1 mutants. (F) qRT-PCR analysis of CHV1 and CHV1 mutant RNA accumulation in F. graminearum. “*”indicates significant differences (P < 0.05, Student’s t test). (G) qRT-PCR analysis of TMV RNA accumulation in F. graminearum coinfected with CHV1 WT or CHV1 mutants. (H) Phenotypic growth of F. graminearum dcl mutant (Δdcl1, Δdcl2, and Δdcl1/2) fungal colonies in virus-free fungi or fungi infected with TMV. Fungi were grown on PDA medium (10-cm plates) for 3 d and then photographed. (I) RNA blotting and qRT-PCR analyses of TMV gRNA accumulation in F. graminearum dcl mutants. Different letters indicate significant differences (P < 0.05, one-way ANOVA).
Compared to CHV1, the accumulation of the CHV1-Δp29 and CHV1-Δp69 mutants in F. graminearum was markedly reduced (Fig. 2 E and F) and was quite similar to that of C. parasitica, the CHV1 natural fungal host (Fig. 1I), suggesting that p29 is essential for the high accumulation of CHV1 in F. graminearum. Moreover, coinfected CHV1-Δp29 and CHV1-Δp69 did not exhibit enhancing effects on TMV accumulation in F. graminearum (Fig. 2G).
An RNA Silencing Mechanism Suppresses TMV Accumulation in Fungi.
RNA silencing mechanisms have been shown to operate against fungal viruses (44, 54–56) and viroids (24) in fungi, so we investigated whether similar mechanisms affect plant virus infection in fungi. The Dicer-like (DCL) protein is one of the key components in the RNA silencing pathway and functions through the generation of small interfering RNAs (57). Ascomycete fungi commonly encode two dcl genes, dcl1 and dcl2 (58). To dissect possible effects of these genes, TMV was introduced by protoplast transfections to F. graminearum single dcl knockout mutants (Δdcl1 and Δdcl2) and a double dcl knockout mutant (Δdcl1/2). TMV infection strongly reduced the growth of the Δdcl1/2 mutant but not the Δdcl1 and Δdcl2 mutants (Fig. 2H). Analyses of TMV RNA accumulation using RNA blotting and qRT-PCR indicated that both DCL proteins contribute to suppression of TMV accumulation, although DCL2 appeared to have a more pronounced effect than DCL1, and both dcl gene deletions supported drastically enhanced TMV accumulation (Fig. 2I and SI Appendix, Fig. S10). These findings demonstrate that RNA silencing has a substantial effect on suppression of TMV replication in F. graminearum, and this finding agrees with our previous observation that the silencing suppressor activity of CHV1 p29 is responsible for the synergistic effects on TMV accumulation in F. graminearum (Fig. 2G).
Plant Virus Infection Facilitates Transfer of CHV1 from Fungi to Plants.
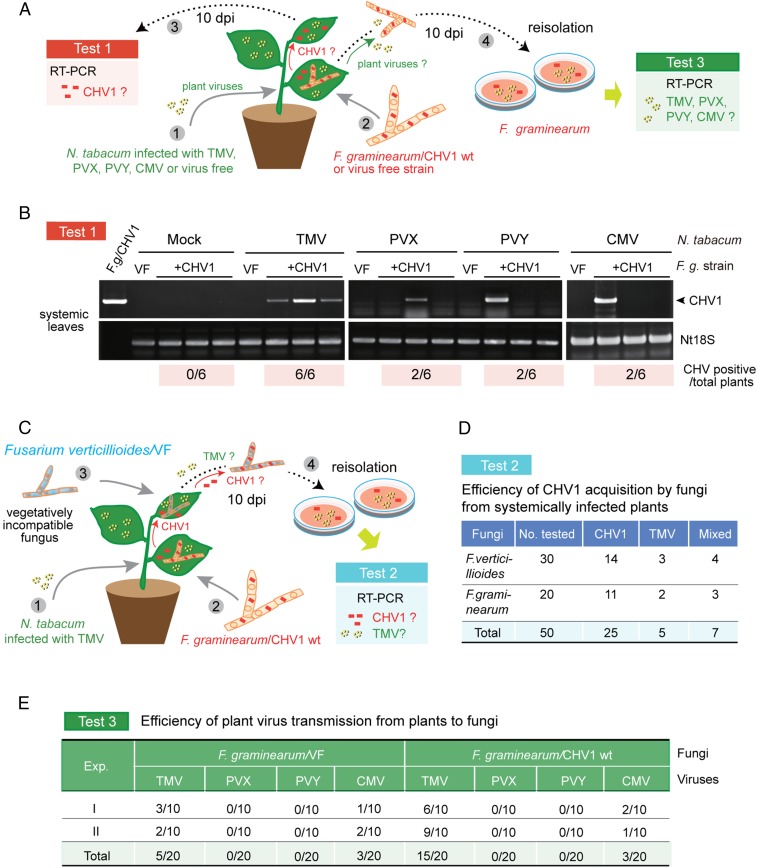
We previously demonstrated that a plant virus and viroids could be bidirectionally transferred between plants and fungi during colonization of plant tissue by fungal pathogens (23, 24). Our artificial inoculation experiments showing that plant virus infection of N. tabacum can facilitate the spread of CHV1 stimulated additional experiments to investigate whether such effects might enhance transmission of CHV1 from infected fungi to plants. First, we designed a fungal inoculation experiment in which a F. graminearum strain carrying CHV1 (Fig. 2A) was inoculated to N. tabacum plants infected with TMV, PVX, PVY, or CMV or that were virus free (mock, Fig. 3A). At 10 d after fungal inoculation, uninoculated upper leaves were tested for the presence of CHV1 by RT-PCR (Fig. 3A, test 1). The amplified bands showed that all six plants infected with TMV were also infected with CHV1, whereas two of six plants that were infected with PVX, PVY, or CMV were coinfected with CHV1 (Fig. 3B). In contrast, none of the six virus-free plants were CHV1 positive (Fig. 3B). This result demonstrates that the plant virus infections facilitate transmission of CHV1 from fungi to N. tabacum. The result also indicates that among the plant virus infections tested, TMV was the most effective in promoting CHV1 transfers to the plant. In the second fungal inoculation experiment, a F. graminearum strain carrying both CHV1 and TMV (Fig. 2A) was inoculated to virus-free plants, and at 10 d after inoculation, virus infections in the upper uninoculated leaves were detected by RT-PCR (SI Appendix, Fig. S11A). All six plants tested were infected with both CHV1 and TMV (SI Appendix, Fig. S11B). Thus, in this situation, TMV was transmitted efficiently from the fungus to the plants and facilitated systemic spread of CHV1 within the plant. Absence of F. graminearum in the upper leaves was confirmed by RT-PCR detection (SI Appendix, Fig. S11B) and growth of fungal mycelia was not evident from the portions of upper leaves placed on the medium (SI Appendix, Fig. S11C).
Fig. 3.
Transmission of CHV1 from fungi to plants with the help of plant virus. (A) Experimental procedure for investigating horizontal transfer of CHV1 from F. graminearum to N. tabacum plants infected with different plant viruses (test 1) and acquisition of plant viruses by F. graminearum (test 3). (B) RT-PCR detection of CHV1 RNA accumulation in uninoculated upper leaves after systemic infection with TMV, PVX, PVY, CMV, or virus-free plants, as described in A (test 1). The numbers of CHV1-positive plants out of six tested plants are shown below the gel images. (C) Experimental procedure for investigating CHV1 acquisition by vegetatively incompatible F. verticillioides from TMV-infected plants systemically infected with CHV1 (test 2) after colonization by CHVI-carrying F. graminearum. (D) Efficiency of CHV1 and TMV acquisition by F. verticillioides or F. graminearum in the experiment described in C (test 2). (E) Efficiency of TMV, PVX, PVY, and CMV acquisition by F. graminearum infected with CHV1 or a virus-free fungal isolate in the experiment described in A (test 3). Viruses were detected by RT-PCR.
We envisage that in nature, once fungal viruses are transferred from an infecting fungus to a plant, they can spread systemically in the plant with the assistance of a plant virus. Fungal viruses can then be acquired by similar or different fungal species that concurrently colonize other parts of the plant. This conceivably can enable transmission of fungal viruses to vegetatively incompatible strains or to different fungal species. To simulate this scenario, we inoculated F. graminearum carrying CHV1 to the leaves of TMV-infected N. tabacum plants and, simultaneously, the upper leaves of the same plant were inoculated with Fusarium verticillioides, a vegetatively incompatible virus-free fungus (Fig. 3C and SI Appendix, Fig. S12). At 10 d after fungal inoculation, F. verticillioides was cultured from the inoculated leaves and tested for the presence of the viruses (Fig. 3C, test 2 and SI Appendix, Fig. S13A). RT-PCR detection showed that 14 of 30 F. verticillioides isolates tested carried CHV1, 3 isolate carried TMV, and 4 isolates were coinfected with both viruses (Fig. 3D and SI Appendix, Fig. S13B). These numbers are quite similar to the acquisition of CHV1 and TMV by F. graminearum in a parallel experiment (Fig. 3D). Hence, the results suggest that systemic spread of fungal viruses in plants may culminate in transmission across diverse fungi in nature.
CHV1 Infection Enhances the Transmission Efficiency of TMV from Plant to Fungi.
In the fungal inoculation experiment described in Fig. 3A, we carried out an investigation about acquisition of plant viruses by F. graminearum. At 10 d after inoculation of virus-free and CHV-infected fungal strains, the fungus was reisolated from each infected plant and then cultured prior to virus detection (Fig. 3A, test 3). RT-PCR detection indicated that F. graminearum could acquire TMV and CMV, but not PVX or PVY (Fig. 3E). However, F. graminearum may not be a compatible host for PVX and PVY because these viruses cannot be transfected into F. graminearum protoplasts (SI Appendix, Fig. S14). Interestingly, a markedly higher proportion of CHV1-infected fungal strains acquired TMV compared with CHV1-free strains (15 versus 5 out of 20 isolates examined for each) (Fig. 3E). This suggests that CHV1 infection of fungi promotes TMV acquisition.
Discussion
Fungi are evolutionarily more closely related to animals than plants, but fungal viruses share more features with plant viruses than with animal viruses (59), reflecting the long-term intimate relationships between plants and fungi. Notably, the majority of fungal virus families also have members that infect plant species (4). In the case of the dsRNA virus families Chrysoviridae, Endornaviridae, and Partitiviridae, as well as ssRNA virus genera Ourmiavirus and Narnavirus, the phylogenetic relationships among plant and fungal members extend beyond their apparent host origins. In fact, some phylogenetic clades contain both plant and fungal viruses (60–63), and well-studied examples are the flexuous rod-shaped fungal viruses (botrexviruses) that highly resemble plant potexviruses (genus Potexvirus) (64–66). These observations suggest that despite the taxonomic diversity of fungi and plants, there have been relatively recent transmissions of these viruses across these taxa. In support of this hypothesis, our research group has provided direct evidence for horizontal transfer of a plant virus to a fungus in nature: CMV, an aphid-transmitted agriculturally important plant virus with a wide plant host range, was found to infect a soil-borne phytopathogenic fungus, Rhizoctonia solani (basidiomycete) isolated from a potato field (23). Moreover, by artificial inoculation, several plant RNA viruses in distinct taxonomic groups have been shown to replicate in various fungi, including yeast (23, 25, 27, 28), and an oomycete (67). In contrast, infection of fungal viruses in plants has not been widely explored. As fungal viruses have adapted to an intracellular life cycle, it is likely that they generally lack the ability for cell-to-cell movement or long-distance spread, as is commonly exhibited by plant viruses. Overall, our current findings provide direct evidence that mutualistic interactions between plant and fungal viruses can facilitate horizontal virus transfer between plants and fungi, and provide an additional mechanism whereby fungal viruses can be disseminated in nature.
Members of the Hypoviridae family have not been found in plants, but hypoviruses and plant (+)ssRNA viruses belonging to the family Potyviridae (picorna-like viruses supergroup) have similar characteristics, suggesting that they share a common ancestry (68). There is evidence for interspecies transmission of CHV1 within the genus Cryphonectria in nature (69), but CHV1 infection of other fungi has not been reported. An infectious cDNA clone of CHV1 was established in 1992 (38), thus enabling reverse genetic studies on gene expression and functional analyses (37, 40–42) and the introduction of CHV1 to heterologous fungal hosts (70). The papain-like protease p29 of CHV1 (encoded in ORF A) (Fig. 1A) is dispensable for viral replication and host virulence attenuation, but it contributes to the suppression of host pigmentation and conidiation (41). The p29 protein is associated with vesicles derived from the fungal trans-Golgi network that contain CHV1 dsRNA and RNA polymerase activity (71). The p29 protein also functions as an RNA silencing suppressor by interfering with up-regulation of transcripts of DCL2 and AGL2 (Argonaute-like protein 2), two key C. parasitica antiviral RNA silencing pathway proteins that are induced by virus infection (43, 52). CHV1 p40 (encoded by ORF A) (Fig. 1A) is also dispensable for CHV1 replication, but it largely contributes to viral RNA accumulation (37). Deletion of p29 drastically reduced CHV1 RNA accumulation in N. tabacum 30K plants (Fig. 1I, CHV1-Δp29), indicating that p29 is functional in plants, as has been previously shown (72), although it is possible that cis-acting RNA elements on the genome could affect virus replication in plants. It remains to be seen whether CHV1 infection in plants also induces up-regulation of RNA silencing-related genes (DCLs, AGOs, and RDRs), as observed in C. parasitica. Notably, CHV1-Δp69 was unable to accumulate in N. tabacum 30K plants (SI Appendix, Fig. S8), suggesting that p69 or possibly p40 may have an important role in CHV1 replication in plants, which is in line with the notion that p40 provides an accessory function in CHV1 RNA amplification (37). Interestingly, TMV infection rescued the replication of CHV1-Δp69 mutant virus (Fig. 1J), suggesting that TMV proteins could support CHV1-Δp69 replication. The silencing suppressor activities of the TMV replicase are an obvious candidate for such support (34).
The systemic spread of viruses in plants is a complex process but basically consists of two consecutive steps: virus cell-to-cell movement through PD and long-distance moment through the vasculature (30). Plant viruses establish movement in plants through the support of virally encoded MPs that usually bind viral gRNA and increase the PD size exclusion limits (47). Four different plant (+)ssRNA virus infections are able to facilitate systemic spread of CHV1 in N. tabacum and TMV appears to be the most effective helper virus for CHV1 spread (Fig. 3B). This observation resembles a coinfection experiment of two plant viruses, in which the helper virus facilitates the spread of a movement-defective virus (73). Notably, CHV1 spread is independent of the replication of the TMV helper plant viruses, because CHV1 can establish systemic infections in the N. tabacum 30K transgenic line (Fig. 1E). Previously, an insect RNA virus, flock house virus (a nodavirus) was shown to spread systemically in transgenic Nicotiana benthamiana expressing plant viral MP genes (74). These results support a model whereby MP functions help facilitate nonplant virus movement in plants. TMV, PVX, PVY, and CMV belong to different virus families with distinct, specialized viral MPs (75). However, as (+)ssRNA viruses, it is generally thought that they are transported through the PD in the form of ribonucleoprotein complexes containing viral RNA, movement protein, replicase and, in some cases, coat protein (30). The movement of TMV and other (+)ssRNA viruses is closely linked to their replication processes, where cell trafficking involves the viral replication complex, host proteins, and cellular membranes (76–78). CHV1 is a capsidless (+)ssRNA virus that forms replicase- and viral RNA-containing membranous vesicles that are thought to be replication complexes (39, 79). We plan to carry out additional studies to investigate how CHV1 establishes replication complexes in plants and whether these complexes are involved in CHV1 cell trafficking.
Synergistic interactions between coinfecting viruses have been observed in both plants and fungi (5, 53, 80–82). One classical example is coinfection of PVY and PVX in N. tabacum in which PVY enhances PVX accumulation (83). In most cases, suppression of RNA silencing-mediated defenses are underlying mechanisms for viral synergism (53). Interestingly, TMV and CHV1 have synergistic effects only in their respective plant and fungal hosts (Figs. 1J and 2C). TMV infection is thought to interfere with the antiviral RNA silencing response targeting CHV1 in plants and to lead to increased CHV1 accumulation. The replicase encoded by TMV and other tobamoviruses functions as an RNA silencing suppressor, most likely through direct binding or modification of small interfering RNA duplexes (34, 84, 85). In F. graminearum, TMV accumulation is strongly suppressed by antiviral RNA silencing (Fig. 2I), and CHV1 infection drastically elevated TMV accumulation (Fig. 2C), TMV transmission through spores (SI Appendix, Fig. S9), and TMV acquisition by the fungus (Fig. 3E). Moreover, the CHV1 p29 suppressor mediates enhanced mycoreovirus 1 (a dsRNA virus, family Reoviridae) accumulation in a coinfection experiment (86), and p29 appears to function in increasing TMV accumulation in F. graminearum (Fig. 2G). Thus, TMV silencing suppression activities in fungi may not be as effective as in plants; likewise, p29 appears not to be fully functional in plants. These phenomena point to possible mechanistic differences between antiviral RNA silencing in plants and fungi (45, 46, 52); therefore, plant and fungal viruses appear to have adapted to the antiviral mechanisms of their conventional host.
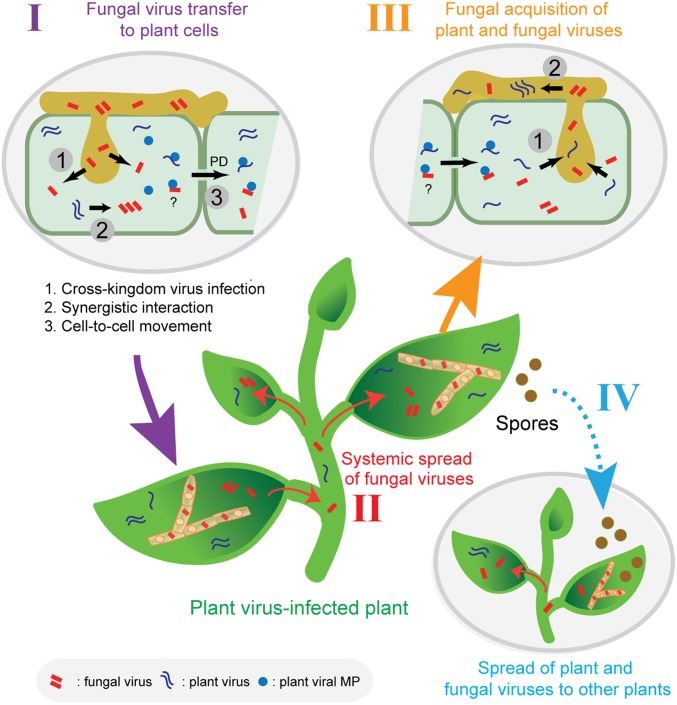
Studies about specific modes of fungal virus transmission are rudimentary. Some studies on filamentous phytophathogenic fungi have revealed virus transmission between vegetatively incompatible fungal hosts (16, 87–89), as well as virus transmission across different fungal species (15, 90–92). Additionally, a fungal virus has been shown to suppress nonself-recognition of the fungus, thus allowing horizontal transmission across vegetatively incompatible strains (93). Our data provide additional insights regarding possible routes of fungal virus transmission in nature (Fig. 4). This model is based on observations that plant virus infections can facilitate systemic spread of a fungal virus in plants after colonization with fungi harboring a fungal virus. As illustrated in Fig. 4, systemic spread of a fungal virus throughout the plant enhances accessibility of the fungal virus to other fungi that have colonized the same plant. We cannot rule out the possibility that some fungal viruses may be able to spread in particular plant tissues or organs without plant virus infections. Additionally, our findings also suggest the possible transmission of plant viruses by phytopathogenic fungi in nature. Indeed, biological vectors of many plant viruses such as tree-infecting viruses have not been investigated in much, if any, detail and merit intensive studies.
Fig. 4.
A proposed model for plant virus-facilitated spread of fungal viruses in nature. A fungal introduction of fungal viruses to plant cells during colonization of plant tissue (I). Fungal virus utilization of plant virus MPs for cell-to-cell and systemic spread in plants (II). Fungal virus acquisition by similar or different fungal species that concurrently colonize the plant. At the same time, the fungus may also acquire the plant virus (III). Fungal or plant virus vertical transmission to spores, and spread of fungal infections to other plants by virus infected spores (IV). These mechanisms permit fungal viruses to spread to vegetatively incompatible fungal strains or to different fungal species. In addition, fungi can also be plant virus vectors.
Materials and Methods
Fungal Strains, Viruses, Plasmids, and Plant Materials.
The C. parasitica strain EP155 (94) was kindly provided by Nobuhiro Suzuki, Okayama University, Kurashiki, Japan. F. graminearum PH-1, Δdcl1, Δdcl2, and Δdcl1/Δdcl2 mutants (95) were kindly provided Jinrong Xu, Northwest A&F University, Yangling, China. F. verticillioides YLM1 was isolated from maize plants in Yangling, Shaanxi Province, China. All fungal strains were grown on potato dextrose agar (PDA) media for 3 to 6 d at 24 °C to 26 °C for morphological observations or on cellophane-covered PDA medium for RNA and DNA extraction. The full-length cDNA clones of CHV1, CHV1-GFP, CHV1-Δp29, and CHV1-Δp69 (37, 38, 41, 50) were also provided by Nobuhiro Suzuki. An isolate of CMV-Fny was provided by Xianbing Wang, China Agricultural University, Beijing, China. PVX was derived from virus vector pGR106 (96) provided by David Baulcombe, Cambridge University, Cambridge, UK. PVY was from a laboratory collection that was isolated from a potato plant in Yulin, Shaanxi Province, China. TMV was from a laboratory collection that was isolated from a tobacco plant in Jingyang, Shaanxi Province, China. The N. tabacum 30K transgenic line (49) was a gift from Lesley Torrance, James Hutton Institute, Dundee, UK. All plants were grown in a plant-growth room at 22 °C ± 2 °C with a photoperiod of 16 h/8 h (day/night).
Fungal Protoplast Isolation and Virus Transfection.
Spheroplasts of F. graminearum strains were prepared by a previously described method (97). The cDNA clone plasmids were digested with SpeI and subjected to in vitro transcription with T7 RNA polymerase (RiboMAX Large Scale RNA Production Systems SP6 and T7, Promega) according to the manufacturer’s instructions. Transfection of fungal protoplasts with in vitro-transcribed RNA (1 µg) was performed as previously described (98). Vertical viral transmission assays through conidia were carried out as previously described (51).
RNA Extraction, RT-PCR, and RNA Blot Analysis.
Total RNA was extracted from leaves using TRIzol (Invitrogen). RT-PCR detection and qRT-PCR were carried out as described elsewhere (24). Total RNA was extracted from virus-infected fungal mycelia by phenol/chloroform extraction in a buffer containing 100 mM Tris/HCl (pH 8.0), 200 mM NaCl, 4 mM EDTA, 4% SDS followed by treatment of the nucleic acid with RQ1 DNase I to eliminate fungal DNA as previously described (86). RNA blot analysis was performed as previously described (23), and blots were hybridized with digoxigenin (DIG, Roche Diagnostics)-labeled RNA probes specific for CHV1 (p48 domain region in ORF B) (Fig. 1A), TMV (CP gene), PVX (CP gene), PVY (CP gene), and CMV (MP gene). Hybridization conditions and detection of viral RNAs were performed as described in the DIG Application Manual supplied by Roche. Chemiluminescent signals were visualized using a chemiluminometer (Clinx Science Instruments Co.). The experiments performed throughout this study were repeated at least twice. All primers used in the study are listed in SI Appendix, Table S1.
Virus and Fungal Inoculations to Plants.
Mechanical inoculation of viruses to plants, in vitro transcripts of CHV1 cDNA (45 ng/µL) or total RNA extracted from virus-infected fungal mycelia (5 µg/µL) was conducted by rubbing carborundum-dusted leaves. Inoculation of F. graminearum strains to plants was carried out as described elsewhere (24). To facilitate CHV1 spread in plants, N. tabacum leaves were first mechanically rub inoculated with the plant viruses TMV, PVX, PVY, or CMV, and at least 30 plants were inoculated in each experiment. Virus infection was subsequently confirmed by RT-PCR. At 5 d after plant virus inoculation, the plants were inoculated with F. graminearum strains. To examine virus acquisition by the fungus at 10 d after inoculation, virus-free F. graminearum and F. verticillioides were reisolated from the inoculated plants with a procedure similar to that described previously (23).
Fluorescence Imaging and Western Blot Analysis.
GFP fluorescence in CHV1-GFP–infected leaves was observed using a confocal laser‐scanning microscope (CLSM; Olympus FV3000). The protein sample preparation, SDS/PAGE, electroblotting, and immunodetection for Western blot analysis were carried out as previously described (99). GFP was detected using a primary anti-GFP monoclonal antibody (1:2,000) (Proteintech) and secondary polyclonal HRP-conjugated anti-mouse IgG (1:10,000) (Abcam). TMV CP was detected using anti-TMV CP polyclonal antibody (1:2,000) and secondary polyclonal HRP-conjugated anti-rabbit IgG (1:10,000) (Abcam). Protein bands were visualized with a Western blot ECL substrate kit (Bio-Rad).
Data Availability.
All of the materials, protocols, and data that were used or generated in this study are described and available in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
We deeply thank Drs. N. Suzuki, J. Xu, X. Wang, D. Baulcombe, and L. Torrance for providing research materials; Dr. Cheng-Gui Han for providing TMV CP antibody and helpful discussions; and Dr. Andrew O. Jackson for valuable comments and English editing of the manuscript. This work was supported in part by the National Natural Science Foundation of China (31970163), (U1703113) to L.S. and (31970159) to I.B.A., the National Key Research and Development Program of China (2017YFD0201100), the 111 Program (2016KW-069) to L.S., and Grants-in-Aid for Scientific Research on Innovative Areas from the Japanese Ministry of Education, Culture, Sports, Science and Technology (16H06436, 16H06429, and 16K21723) to H.K.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915996117/-/DCSupplemental.
References
- 1.Ghabrial S. A., Suzuki N., Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47, 353–384 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Pearson M. N., Beever R. E., Boine B., Arthur K., Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 10, 115–128 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghabrial S. A., Castón J. R., Jiang D., Nibert M. L., Suzuki N., 50-plus years of fungal viruses. Virology 479–480, 356–368 (2015). [DOI] [PubMed] [Google Scholar]
- 4.King A. M. Q., et al. , Changes to taxonomy and the international code of virus classification and nomenclature ratified by the International Committee on Taxonomy of Viruses (2018). Arch. Virol. 163, 2601–2631 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Hillman B. I., Annisa A., Suzuki N., “Viruses of plant-interacting fungi” in Advances in Virus Research, Kielian M., Mettenleiter T. C., M. J. R., Eds. (Elsevier, 2018), vol. 100, pp. 99–116. [DOI] [PubMed] [Google Scholar]
- 6.Okada R., et al. , Molecular characterization of a novel mycovirus in Alternaria alternata manifesting two-sided effects: Down-regulation of host growth and up-regulation of host plant pathogenicity. Virology 519, 23–32 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Ahn I.-P., Lee Y.-H., A viral double-stranded RNA up regulates the fungal virulence of Nectria radicicola. Mol. Plant Microbe Interact. 14, 496–507 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Lau S. K. P., et al. , Novel partitivirus enhances virulence of and causes aberrant gene expression in Talaromyces marneffei. MBio 9, e00947-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie J., Jiang D., New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 52, 45–68 (2014). [DOI] [PubMed] [Google Scholar]
- 10.García-Pedrajas M. D., Cañizares M. C., Sarmiento-Villamil J. L., Jacquat A. G., Dambolena J. S., Mycoviruses in biological control: From basic research to field implementation. Phytopathology 109, 1828–1839 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Liu S., et al. , Fungal DNA virus infects a mycophagous insect and utilizes it as a transmission vector. Proc. Natl. Acad. Sci. U.S.A. 113, 12803–12808 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helenius A., Virus entry: Looking back and moving forward. J. Mol. Biol. 430, 1853–1862 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waigmann E., Ueki S., Trutnyeva K., Citovsky V., The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. Crit. Rev. Plant Sci. 23, 195–250 (2004). [Google Scholar]
- 14.Biella S., Smith M. L., Aist J. R., Cortesi P., Milgroom M. G., Programmed cell death correlates with virus transmission in a filamentous fungus. Proc. Biol. Sci. 269, 2269–2276 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melzer M. S., Ikeda S. S., Boland G. J., Interspecific transmission of double-stranded RNA and hypovirulence from Sclerotinia sclerotiorum to S. minor. Phytopathology 92, 780–784 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Hamid M. R., et al. , A novel deltaflexivirus that infects the plant fungal pathogen, Sclerotinia sclerotiorum, can be transmitted among host vegetative incompatible strains. Viruses 10, 295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selosse M.-A., Strullu-Derrien C., Martin F., Kamoun S., Kenrick P., Plants, fungi and oomycetes: A 400-million year affair that shapes the biosphere. New Phytol. 206, 501–506 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Lo Presti L., et al. , Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Giraldo M. C., Valent B., Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Weiberg A., et al. , Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua C., Zhao J.-H., Guo H.-S., Trans-kingdom RNA silencing in plant–fungal pathogen interactions. Mol. Plant 11, 235–244 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Hou Y., et al. , A Phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe 25, 153–165.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andika I. B., et al. , Phytopathogenic fungus hosts a plant virus: A naturally occurring cross-kingdom viral infection. Proc. Natl. Acad. Sci. U.S.A. 114, 12267–12272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei S., et al. , Symptomatic plant viroid infections in phytopathogenic fungi. Proc. Natl. Acad. Sci. U.S.A. 116, 13042–13050 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mascia T., et al. , Gene silencing and gene expression in phytopathogenic fungi using a plant virus vector. Proc. Natl. Acad. Sci. U.S.A. 111, 4291–4296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascia T., et al. , Infection of Colletotrichum acutatum and Phytophthora infestans by taxonomically different plant viruses. Eur. J. Plant Pathol. 153, 1001–1017 (2019). [Google Scholar]
- 27.Janda M., Ahlquist P., RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72, 961–970 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Nagy P. D., Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 46, 217–242 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Nerva L., Varese G. C., Falk B. W., Turina M., Mycoviruses of an endophytic fungus can replicate in plant cells: Evolutionary implications. Sci. Rep. 7, 1908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benitez-Alfonso Y., Faulkner C., Ritzenthaler C., Maule A. J., Plasmodesmata: Gateways to local and systemic virus infection. Mol. Plant Microbe Interact. 23, 1403–1412 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Ding S.-W., RNA-based antiviral immunity. Nat. Rev. Immunol. 10, 632–644 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Navarro J. A., Sanchez-Navarro J. A., Pallas V., Key checkpoints in the movement of plant viruses through the host. Adv. Virus Res. 104, 1–64 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Suzuki N., et al. ; Ictv Report Consortium , ICTV virus taxonomy profile: Hypoviridae. J. Gen. Virol. 99, 615–616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kubota K., Tsuda S., Tamai A., Meshi T., Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J. Virol. 77, 11016–11026 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rigling D., Prospero S., Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 19, 7–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anagnostakis S. L., Biological control of chestnut blight. Science 215, 466–471 (1982). [DOI] [PubMed] [Google Scholar]
- 37.Suzuki N., Nuss D. L., Contribution of protein p40 to hypovirus-mediated modulation of fungal host phenotype and viral RNA accumulation. J. Virol. 76, 7747–7759 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi G. H., Nuss D. L., Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257, 800–803 (1992). [DOI] [PubMed] [Google Scholar]
- 39.Fahima T., Wu Y., Zhang L., Van Alfen N. K., Identification of the putative RNA polymerase of Cryphonectria hypovirus in a solubilized replication complex. J. Virol. 68, 6116–6119 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo L. H., Sun L., Chiba S., Araki H., Suzuki N., Coupled termination/reinitiation for translation of the downstream open reading frame B of the prototypic hypovirus CHV1-EP713. Nucleic Acids Res. 37, 3645–3659 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Craven M. G., Pawlyk D. M., Choi G. H., Nuss D. L., Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 67, 6513–6521 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki N., Chen B., Nuss D. L., Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC-Pro protease. J. Virol. 73, 9478–9484 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Q., Choi G. H., Nuss D. L., A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. U.S.A. 106, 17927–17932 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segers G. C., Zhang X., Deng F., Sun Q., Nuss D. L., Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U.S.A. 104, 12902–12906 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andika I. B., Jamal A., Kondo H., Suzuki N., SAGA complex mediates the transcriptional up-regulation of antiviral RNA silencing. Proc. Natl. Acad. Sci. U.S.A. 114, E3499–E3506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andika I. B., Kondo H., Suzuki N., Dicer functions transcriptionally and posttranscriptionally in a multilayer antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 116, 2274–2281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lucas W. J., Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 344, 169–184 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Carrington J. C., Kasschau K. D., Mahajan S. K., Schaad M. C., Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8, 1669–1681 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deom C. M., et al. , Molecular characterization and biological function of the movement protein of tobacco mosaic virus in transgenic plants. Proc. Natl. Acad. Sci. U.S.A. 87, 3284–3288 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki N., Geletka L. M., Nuss D. L., Essential and dispensable virus-encoded replication elements revealed by efforts to develop hypoviruses as gene expression vectors. J. Virol. 74, 7568–7577 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., et al. , Identification of a novel hypovirulence-inducing hypovirus from Alternaria alternata. Front. Microbiol. 10, 1076 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiba S., Suzuki N., Highly activated RNA silencing via strong induction of dicer by one virus can interfere with the replication of an unrelated virus. Proc. Natl. Acad. Sci. U.S.A. 112, E4911–E4918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Syller J., Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 13, 204–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campo S., Gilbert K. B., Carrington J. C., Small RNA-based antiviral defense in the phytopathogenic fungus Colletotrichum higginsianum. PLoS Pathog. 12, e1005640 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu J., Lee K.-M., Cho W. K., Park J. Y., Kim K.-H., Differential contribution of RNA interference components in response to distinct Fusarium graminearum virus infections. J. Virol. 92, e01756-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neupane A., Feng C., Mochama P. K., Saleem H., Lee Marzano S. Y., Roles of argonautes and dicers on Sclerotinia sclerotiorum antiviral RNA silencing. Front. Plant Sci. 10, 976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aliyari R., Ding S. W., RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol. Rev. 227, 176–188 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakayashiki H., RNA silencing in fungi: Mechanisms and applications. FEBS Lett. 579, 5950–5957 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Roossinck M. J., Evolutionary and ecological links between plant and fungal viruses. New Phytol. 221, 86–92 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Roossinck M. J., Lifestyles of plant viruses. Philos. Trans. R. Soc. B 365, 1899–1905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roossinck M. J., “Persistent plant viruses: Molecular hitchhikers or epigenetic elements?” in Viruses: Essential Agents of Life, Witzany Günther, Ed. (Springer, 2012), pp. 177–186. [Google Scholar]
- 62.Velasco L., Arjona-Girona I., Cretazzo E., López-Herrera C., Viromes in Xylariaceae fungi infecting avocado in Spain. Virology 532, 11–21 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Rastgou M., et al. , Molecular characterization of the plant virus genus Ourmiavirus and evidence of inter-kingdom reassortment of viral genome segments as its possible route of origin. J. Gen. Virol. 90, 2525–2535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howitt R. L., Beever R. E., Pearson M. N., Forster R. L., Genome characterization of Botrytis virus F, a flexuous rod-shaped mycovirus resembling plant ‘potex-like’ viruses. J. Gen. Virol. 82, 67–78 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Howitt R. L., Beever R. E., Pearson M. N., Forster R. L., Genome characterization of a flexuous rod-shaped mycovirus, Botrytis virus X, reveals high amino acid identity to genes from plant ‘potex-like’ viruses. Arch. Virol. 151, 563–579 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Xie J., et al. , Characterization of debilitation-associated mycovirus infecting the plant-pathogenic fungus Sclerotinia sclerotiorum. J. Gen. Virol. 87, 241–249 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Mascia T., Labarile R., Doohan F., Gallitelli D., Tobacco mosaic virus infection triggers an RNAi-based response in Phytophthora infestans. Sci. Rep. 9, 2657 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koonin E. V., Choi G. H., Nuss D. L., Shapira R., Carrington J. C., Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. U.S.A. 88, 10647–10651 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Y. C., Linder-Basso D., Hillman B. I., Kaneko S., Milgroom M. G., Evidence for interspecies transmission of viruses in natural populations of filamentous fungi in the genus Cryphonectria. Mol. Ecol. 12, 1619–1628 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Sasaki A., et al. , Extending chestnut blight hypovirus host range within diaporthales by biolistic delivery of viral cDNA. Mol. Plant Microbe Interact. 15, 780–789 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Jacob-Wilk D., Turina M., Van Alfen N. K., Mycovirus cryphonectria hypovirus 1 elements cofractionate with trans-Golgi network membranes of the fungal host Cryphonectria parasitica. J. Virol. 80, 6588–6596 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Segers G. C., van Wezel R., Zhang X., Hong Y., Nuss D. L., Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell 5, 896–904 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giesman-Cookmeyer D., Silver S., Vaewhongs A. A., Lommel S. A., Deom C. M., Tobamovirus and dianthovirus movement proteins are functionally homologous. Virology 213, 38–45 (1995). [DOI] [PubMed] [Google Scholar]
- 74.Dasgupta R., Garcia B. H. 2nd, Goodman R. M., Systemic spread of an RNA insect virus in plants expressing plant viral movement protein genes. Proc. Natl. Acad. Sci. U.S.A. 98, 4910–4915 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schoelz J. E., Harries P. A., Nelson R. S., Intracellular transport of plant viruses: Finding the door out of the cell. Mol. Plant 4, 813–831 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu C., Nelson R. S., The cell biology of Tobacco mosaic virus replication and movement. Front. Plant Sci. 4, 12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pitzalis N., Heinlein M., The roles of membranes and associated cytoskeleton in plant virus replication and cell-to-cell movement. J. Exp. Bot. 69, 117–132 (2017). [DOI] [PubMed] [Google Scholar]
- 78.Heinlein M., Plant virus replication and movement. Virology 479–480, 657–671 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Fahima T., Kazmierczak P., Hansen D. R., Pfeiffer P., Van Alfen N. K., Membrane-associated replication of an unencapsidated double-strand RNA of the fungus, Cryphonectria parasitica. Virology 195, 81–89 (1993). [DOI] [PubMed] [Google Scholar]
- 80.Mascia T., Gallitelli D., Synergies and antagonisms in virus interactions. Plant Sci. 252, 176–192 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Thapa V., Roossinck M. J., Determinants of coinfection in the mycoviruses. Front. Cell. Infect. Microbiol. 9, 169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aulia A., Andika I. B., Kondo H., Hillman B. I., Suzuki N., A symptomless hypovirus, CHV4, facilitates stable infection of the chestnut blight fungus by a coinfecting reovirus likely through suppression of antiviral RNA silencing. Virology 533, 99–107 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Vance V. B., Replication of potato virus X RNA is altered in coinfections with potato virus Y. Virology 182, 486–494 (1991). [DOI] [PubMed] [Google Scholar]
- 84.Kurihara Y., et al. , Binding of tobamovirus replication protein with small RNA duplexes. J. Gen. Virol. 88, 2347–2352 (2007). [DOI] [PubMed] [Google Scholar]
- 85.Vogler H., et al. , Modification of small RNAs associated with suppression of RNA silencing by tobamovirus replicase protein. J. Virol. 81, 10379–10388 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun L., Nuss D. L., Suzuki N., Synergism between a mycoreovirus and a hypovirus mediated by the papain-like protease p29 of the prototypic hypovirus CHV1-EP713. J. Gen. Virol. 87, 3703–3714 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Brusini J., Robin C., Mycovirus transmission revisited by in situ pairings of vegetatively incompatible isolates of Cryphonectria parasitica. J. Virol. Methods 187, 435–442 (2013). [DOI] [PubMed] [Google Scholar]
- 88.Yaegashi H., et al. , Appearance of mycovirus-like double-stranded RNAs in the white root rot fungus, Rosellinia necatrix, in an apple orchard. FEMS Microbiol. Ecol. 83, 49–62 (2013). [DOI] [PubMed] [Google Scholar]
- 89.Yang D., et al. , Sclerotinia minor endornavirus 1, a novel pathogenicity debilitation-associated mycovirus with a wide spectrum of horizontal transmissibility. Viruses 10, 589 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vainio E. J., Korhonen K., Tuomivirta T. T., Hantula J., A novel putative partitivirus of the saprotrophic fungus Heterobasidion ecrustosum infects pathogenic species of the Heterobasidion annosum complex. Fungal Biol. 114, 955–965 (2010). [DOI] [PubMed] [Google Scholar]
- 91.Vainio E. J., Piri T., Hantula J., Virus community dynamics in the conifer pathogenic fungus Heterobasidion parviporum following an artificial introduction of a partitivirus. Microb. Ecol. 65, 28–38 (2013). [DOI] [PubMed] [Google Scholar]
- 92.Khalifa M. E., MacDiarmid R. M., A novel totvirus naturally occurring in two different fungal genera. Front. Microbiol. 10, 2318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu S., et al. , Virus-mediated suppression of host non-self recognition facilitates horizontal transmission of heterologous viruses. PLoS Pathog. 13, e1006234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shapira R., Choi G. H., Nuss D. L., Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 10, 731–739 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zeng W., et al. , Dicer-like proteins regulate sexual development via the biogenesis of perithecium-specific microRNAs in a plant pathogenic fungus Fusarium graminearum. Front. Microbiol. 9, 818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu R., et al. , High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Churchill A., Ciuffetti L., Hansen D., Van Etten H., Van Alfen N., Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17, 25–31 (1990). [Google Scholar]
- 98.Hillman B. I., Supyani S., Kondo H., Suzuki N., A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the coltivirus genus of animal pathogens. J. Virol. 78, 892–898 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun L., Suzuki N., Intragenic rearrangements of a mycoreovirus induced by the multifunctional protein p29 encoded by the prototypic hypovirus CHV1-EP713. RNA 14, 2557–2571 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the materials, protocols, and data that were used or generated in this study are described and available in the manuscript and SI Appendix.