Significance
The opportunistic fungal pathogen Cryptococcus neoformans is a major threat to immunocompromised patients. Most fungal cell walls expose chitin, which is recognized by the human immune system, but C. neoformans is one of the few species that instead exposes chitosan, the deacetylated form of chitin. Chitosan is produced by chitin synthases and chitin deacetylases, the latter acting on chitin as a substrate. Here we describe the detailed functional analysis of CnCda4, a chitin deacetylase with unusual subsite specificity resulting in a preference for chitosan rather than chitin. By testing the effect of CnCda4 deacetylation products on human peripheral blood-derived macrophages, we show that the enzyme probably helps the pathogen to evade the host immune system by further deacetylating exposed chitosans.
Keywords: fungal pathogen, chitin, CnCda4/Fpd1/d25, degree of acetylation, pattern of acetylation
Abstract
Cryptococcus neoformans is an opportunistic fungal pathogen that infects ∼280,000 people every year, causing >180,000 deaths. The human immune system recognizes chitin as one of the major cell-wall components of invading fungi, but C. neoformans can circumvent this immunosurveillance mechanism by instead exposing chitosan, the partly or fully deacetylated form of chitin. The natural production of chitosans involves the sequential action of chitin synthases (CHSs) and chitin deacetylases (CDAs). C. neoformans expresses four putative CDAs, three of which have been confirmed as functional enzymes that act on chitin in the cell wall. The fourth (CnCda4/Fpd1) is a secreted enzyme with exceptional specificity for d-glucosamine at its −1 subsite, thus preferring chitosan over chitin as a substrate. We used site-specific mutagenesis to reduce the subsite specificity of CnCda4 by converting an atypical isoleucine residue in a flexible loop region to the bulkier or charged residues tyrosine, histidine, and glutamic acid. We also investigated the effect of CnCda4 deacetylation products on human peripheral blood-derived macrophages, leading to a model explaining the function of CnCda4 during infection. We propose that CnCda4 is used for the further deacetylation of chitosans already exposed on the C. neoformans cell wall (originally produced by CnChs3 and CnCda1 to 3) or released from the cell wall as elicitors by human chitinases, thus making the fungus less susceptible to host immunosurveillance. The absence of CnCda4 during infection could therefore promote the faster recognition and elimination of this pathogen.
The human immune system is well equipped to deal with fungal pathogens, partly because the fungal cell wall offers convenient pathogen-associated molecular patterns (PAMPs) such as chitin oligomers, which are set free by the action of human chitinases and subsequently recognized by pattern recognition receptors such as Toll-like receptors (TLRs) and C-type lectin-like receptors (CLRs) (1, 2). TLR2 was recently identified as the primary fungal chitin sensor in mammalian cells, recognizing chitin oligomers with a degree of polymerization (DP) ≥6 (3). However, a small number of fungal pathogens can evade the immune system by replacing the chitin in their cell walls with chitosan, a partially deacetylated derivative of chitin. These include fungal pathogens of plants (4) and some human pathogens such as Cryptococcus neoformans (5–7), both of which express enzymes known as chitin deacetylases (CDAs) to convert cell-wall chitin into chitosans.
CDAs (EC 3.5.1.41) (8) are amino hydrolases belonging to carbohydrate esterase family 4 (9, 10). They catalyze the hydrolysis of N-acetyl groups from N-acetylglucosamine (GlcNAc) units in chitin and chitosan oligomers and polymers (11–14). Although chitosans are rare in nature, CDAs have been identified in many fungi and insects as well as other organisms such as diatoms, bacteria, and even viruses. In fungi and insects, CDAs may play a role in morphogenesis, whereas bacterial CDAs may contribute to chitin degradation for nutritional purposes. Interestingly, CDAs differ in their regioselectivity when acting on chitin oligomers, yielding products with different degrees of acetylation (DAs) and patterns of acetylation (PAs) (4, 12, 15–22). This suggests that both the DA and PA may influence the biological activities of chitosan oligomers and, possibly, polymers (4, 23, 24). In fungi, different types of CDAs have been described based on their subcellular localization, which may also define different physiological roles (12). Some extracellular, secreted CDAs possess glycosylphosphatidylinositol (GPI) anchors that tether them to the outer surface of the plasma membrane, leading to a periplasmic localization. Their role is the deacetylation of nascent chitin polymer chains emerging from transmembrane chitin synthases in chitosan-containing cell walls. In contrast, nonanchored, secreted, extracellular CDAs often act on chitin oligomers released from the cell wall by host chitinases during host–pathogen interactions. In both cases, CDAs act as pathogenicity or virulence factors, helping the pathogen to evade the host immune system. CDAs are therefore promising targets for the development of antifungal drugs.
The targeting of CDAs has been proposed as a therapeutic strategy against C. neoformans, which infects ∼280,000 people every year, more than 180,000 of whom suffer fatal cryptococcal meningitis within 3 mo due to the low efficacy of current treatments (25–27). The pathogen infects humans via the lung and from there enters the bloodstream, allowing it to spread to other organs (28, 29). Ultimately, the central nervous system is infected, often leading to severe symptoms and death, particularly in immunocompromised patients (30–32). The C. neoformans genome contains four CDA-like genes, three of which (CDA1, CDA2, and CDA3) encode intracellular-type enzymes with GPI anchors, whereas there is evidence that CnCda4, also known as d25 (33) and fungal polysaccharide deacetylase 1 (Fpd1) (5), is a secreted enzyme. CnCda1 to 3 have been shown to account for all cell-wall chitosan produced during vegetative growth in yeast extract peptone dextrose medium, and each can compensate for the loss of the other two (5). In contrast, when C. neoformans was grown in RPMI-1640 medium (typically used for the cultivation of mammalian cells), CnCda1 alone was shown to be responsible for pathogenesis (34). The loss of CnCda1 to 3 activity strongly inhibits the virulence of C. neoformans. Mice infected with a mutant strain deficient for all three enzymes (C. neoformans cda1Δ, cda2Δ, cda3Δ) showed no symptoms of infection, and fungal cells were fully degraded within 24 h (6). Fungal clearance was closely associated with a spike in the concentration of proinflammatory molecules related to the Th1-type adaptive immune response (35). In contrast, the loss of CnCda4 had no effect on cell-wall composition or fungal virulence, hence its initial designation as Fpd1 rather than CnCda4. However, some studies have shown that all four CDAs seem to induce protective immune responses in mice, with Cda1, 2, and 3 being more effective than Fpd1/Cda4, thereby making promising drug targets (33, 34, 36).
To gain more insight into the role of CnCda4, we optimized its expression as a recombinant protein in Escherichia coli Lemo21 (DE3) and Rosetta2 (DE3) cells, followed by biochemical characterization and site-directed mutagenesis in the active groove in order to investigate in detail its substrate preferences and mode of action against chitin and chitosan oligomers. Finally, we evaluated its natural role in pathogenesis by examining the effect of the well-defined hexameric products of CnCda4 on human macrophages.
Results and Discussion
Heterologous Expression of CnCda4 in E. coli.
The codon-optimized CDA4 gene of C. neoformans was fused to a C-terminal Strep tag II sequence and expressed in E. coli Lemo21 cells. Optimal yields of soluble protein were achieved by including 1 mM α-l-rhamnose in the medium (SI Appendix, Fig. S1). CnCda4 was purified from the soluble fraction of bacterial lysates by affinity chromatography for further characterization. We found that the optimum temperature was 37 °C and the optimum pH was 7, and that 50 mM sodium bicarbonate was the most suitable buffer (SI Appendix, Fig. S2). The construct with the C-terminal Strep tag II achieved a moderate yield (24 µg/L culture medium) under optimal conditions. We therefore tested other constructs to see if this could be improved. The insertion of a linker sequence of 12 amino acids (mainly glycine and serine residues) between the C-terminal Strep tag II and protein did not have a significant effect. However, removing the 21-amino acid N-terminal signal peptide predicted by SignalP v4.1 (37) increased the yield to 137 µg/L. Attaching an additional N-terminal Strep tag II and expressing the protein in E. coli Rosetta (DE3) cells increased the yield even further, to 800 µg/L.
Cofactor Analysis.
The catalytic mechanism of CDAs requires a divalent metal ion bound to the active site, coordinated by a conserved His-His-Asp triad (38). CDA reactions therefore tend to be inhibited by the metal-chelating compound EDTA and enhanced by the presence of Co2+, Ca2+, or Zn2+ (BRENDA:EC3.5.1.41). Accordingly, we found that the incubation of CnCda4 with 20 mM EDTA strongly inhibited its activity, but the subsequent addition of Zn2+ (and, to a lesser extent, Co2+ or Mn2+) restored activity to normal (SI Appendix, Fig. S3). However, none of the ions increased the activity of CnCda4 above the level achieved in the absence of EDTA. Therefore, no metal ions were added in subsequent activity tests.
CnCda4 Activity Against Polymeric Chitinous Substrates.
Zymography experiments revealed that CnCda4 showed high activity against glycol-chitin, but low activity against colloidal chitin in solution, only reducing the initial DA by ∼1% (SI Appendix, Fig. S3). In comparison, Podospora anserina CDA (PaCDA) (21), Puccinia graminis CDA (PgtCDA) (17), and PesCDA (7) reduced the initial DA by 12%, 6%, and 7%, respectively. CnCda4 did not show any detectable activity against the crystalline substrates α-chitin and β-chitin, which are very poor substrates for all CDAs described thus far. In contrast, partially acetylated chitosan polymers were good substrates for CnCda4. The enzyme reduced the DA of DA50% chitosan to 19% and that of DA30% chitosan to 15% within 24 h, corresponding to 15-fold (DA30%) and 32-fold (DA50%) higher activity against chitosans compared to colloidal chitin (SI Appendix, Fig. S2). Under similar conditions, PesCDA also showed high activity against chitosan polymers, albeit less activity than CnCda4, reducing the DA of DA50% chitosan to 34% and that of DA30% chitosan to 22% within 24 h (7), corresponding to 1.1-fold (DA30%) and 2.3-fold (DA50%) higher activity against chitosans compared to colloidal chitin. The higher preference of CnCda4 for chitosans in comparison to other CDAs is probably related to the unusual abundance of chitosan (at least twice as much chitosan compared to chitin) in the C. neoformans cell wall during vegetative growth (7).
CnCda4 Mode of Action Against Oligomeric Substrates.
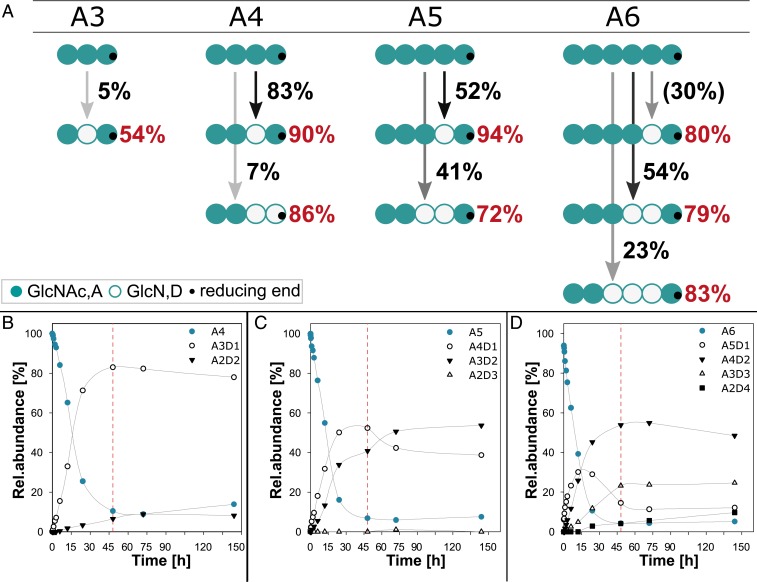
For brevity, partially acetylated chitosan oligosaccharides (paCOSs) are described hereafter using a single-letter code from the nonreducing to the reducing end: A, N-acetylglucosamine (GlcNAc); and D, glucosamine (GlcN). When chitin oligomers (DP 1 to 6) were used as substrates, CnCda4 was inactive against GlcNAc (A) and chitobiose (A2), slightly active against chitotriose (A3), and increasingly active against the larger oligomers (A4 to A6). The complete time course for the deacetylation of A4, A5, and A6 are shown in Fig. 1 B–D. Fig. 1A shows the mode of action of CnCda4 against the chitin oligomers by summarizing the results of our quantitative MS1 and MS2 experiments to determine the PA. After 48 h, CnCda4 modified only 5% of the chitotriose (A3), whereas more than 90% of the larger oligomers were converted to paCOS varying in DA. CnCda4 modified 90% of the chitin tetramer (A4), and most (83%) of the products were deacetylated on one unit to yield a rather pattern-pure oligomer (90% AADA). Only 7% of the tetrameric products were deacetylated at two units, with the predominant pattern AADD (86%). When presented with the chitin pentamer (A5), CnCda4 produced single and double deacetylated products, the former at a slightly higher proportion. The single deacetylated products were remarkably pattern-pure (94% AAADA) whereas the double deacetylated products had the predominant pattern AADDA (72%). Under the same conditions, the chitin hexamer (A6) was fully processed to A4D2 and A3D3, hence the single deacetylated product A5D1 was only detected at shorter incubation times. In all cases, there was a predominant pattern (A5D1, 80% AAAADA; A4D2, 79% AAADDA; and A3D3, 83% AADDDA). In the A5 and A6 substrates, the first attack was always at the sugar unit located adjacent to the reducing end, and deacetylation then continued toward the nonreducing end until only two adjacent acetylated units were left at the nonreducing end. By increasing the amount of enzyme or the reaction time, one further deacetylation step occurred, leaving only the reducing and nonreducing end units acetylated, thus yielding the patterns ADDDA and ADDDDA (Fig. 1 C and D). These data indicated that several factors affect the PA, including the DP and thereby the positioning of the substrate in the active site, ultimately revealing that certain subsite specificities play a key role during the deacetylation of chitin and chitosan oligomers by CnCda4. Given that we never observed deacetylation at the nonreducing end, whereas deacetylation at the reducing end was rare but did occur on occasion, we concluded that the catalytic activity of CnCda4 requires subsite −1 to be occupied. Therefore, we next investigated the subsite specificities in the substrate-binding cleft of CnCda4 and their potential influence on the mode of deacetylation when the enzyme acts on paCOS instead of chitin.
Fig. 1.
Mode of action and activity of CnCda4 on chitin oligomers. (A) Scheme to visualize the activity and mode of action of CnCda4 against oligomeric chitin substrates ranging from DP3 (A3) to DP6 (A6). The intensity of the arrows and the adjacent values represent the proportion of paCOS produced with the given DA, whereas red values represent the proportion of paCOS with the indicated PA among all paCOS with the same DA. All data were obtained after incubation for 48 h except for the data for A5D1, which were obtained after 15 h. (B–D) Time course of deacetylation on (B) the chitin tetramer (A4), (C) pentamer (A5), and (D) hexamer (A6), with red lines highlighting samples taken after 48 h as visualized in A. We incubated 0.5 µg CnCda4 with the chitin oligomers (1 mg/mL final concentration) in 50 mM NaHCO3 (pH 7) for up to 144 h (n = 3). Samples were taken after 0, 5, 15, and 30 min and after 1, 2, 3, 6, 12, 24, 48, 72, and 144 h by stopping the reaction with 0.5% (final concentration) formic acid. The profile of partially acetylated chitosan oligomers in all samples was determined by LC-MS1 and LC-MS2 analysis.
CnCda4 Activity Against Defined paCOS with Specific Acetylation Patterns.
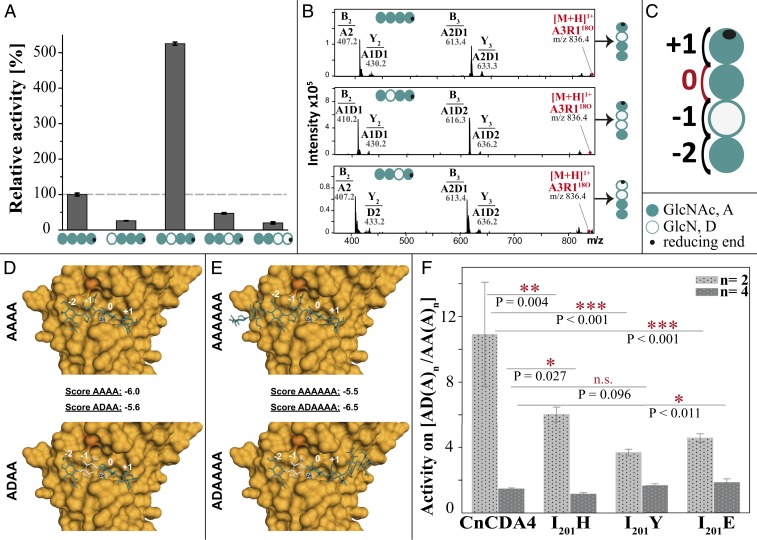
When the pattern-pure chitosan tetramers DAAA, ADAA, AADA, and AADD were used as substrates, CnCda4 showed 5 to 10 times higher activity against the single deacetylated tetramer ADAA than the fully acetylated A4, and even lower activity against the other three chitosan tetramers (Fig. 2A). This suggests that the natural substrates of CnCda4 are partially acetylated chitosans rather than fully acetylated chitin. To further understand the mode of action, the PA of the resulting products was analyzed by MS2 (Fig. 2B) to propose a model explaining the binding preferences of CnCda4 (Fig. 2C). The activity of CnCda4 against the substrates DAAA and AADD was too low to analyze the PA of the products, but, when presented with the substrates ADAA and AADA, CnCda4 preferred to deacetylate the neighboring unit downstream (further toward the reducing end) of the already deacetylated unit, yielding the products ADDA and AADD, respectively (Fig. 2B). Given that CnCda4 shows atypical activity against the chitin tetramer A4 in comparison to A5 and A6, we investigated its activity and mode of action against ADAAAA and A6 (Fig. 2F). Again, CnCda4 showed a significant (1.6-fold) preference for the partially deacetylated substrate, and, interestingly, the preferred deacetylation site was again the position downstream of the already deacetylated unit, yielding ADDAAA as the main product (53%). ADAADA, the product that would result if the four subsites of CnCda4 bound to the fully acetylated part of the substrate ADAAAA, was less frequent (24%). This confirmed that CnCda4 prefers the −1 subsite to be occupied by GlcN rather than GlcNAc, which explains its preference for partially deacetylated chitosan substrates rather than chitin. This substrate preference sets CnCda4 apart from already characterized CDAs of other organisms, all of which act on both chitin and chitosans. We therefore propose that CnCda4 is a “chitosan deacetylase” differing from known “chitin deacetylases.”
Fig. 2.
Regioselectivity and regiospecificity of CnCda4 against partially acetylated chitosan tetramers and hexamers. (A) Relative activity of CnCda4 against five tetrameric substrates differing in DA and PA, as determined by quantitative MS1 (performed with four independent enzyme batches of CnCda4; values are means ± SD). (B) MS2 spectra of the deacetylation products of A4, ADAA, and AADA after fragmentation, showing the fragment masses and composition of b-ions (nonreducing end) and y-ions (reducing end). (C) Model representing the binding preferences of CnCda4 at four subsites (−2, −1, 0, +1) where subsite 0 is the catalysis site. (D) Homology model of CnCda4 (based on PDB ID code 2IW0), with Zn2+ (gray) and I201 in dark orange, showing docking results for AAAA and ADAA in the active site. Subsites −2, −1, 0, and +1 are indicated in white. Docking scores are given for all substrates. (E) The same homology model showing docking results for AAAAAA and ADAAAA. (F) Activity ratio of wild-type CnCda4 and three mutants against partially deacetylated AD(A)n vs. fully acetylated AA(A)n, where n = 2 for tetrameric substrates and n = 4 for hexameric substrates (values are means of four replicates each ±SD).
Homology Modeling and Molecular Docking.
To gain insight into the substrate-binding behavior of CnCda4, we modeled the enzyme using the structure of Colletotrichum lindemuthianum CDA (ClCDA; PDB ID code 2IW0) as a template for the four subsites ranging from −2 to +1. The chitin substrates A4 and A6, and corresponding single deacetylated variants with GlcN adjacent to the nonreducing end (ADAA and ADAAAA), were generated using GLYCAM-Web. All four substrates were successfully docked into the active site using AutoDock VinaCarb v1.0 (39), resulting in docking scores of −6.0 (AAAA), −5.6 (ADAA), −5.5 (AAAAAA), and −6.5 (ADAAAA; Fig. 2 D and E). The high docking scores were thus in agreement with the experimental data shown in Fig. 2 A and F.
Site-Directed Mutagenesis.
To understand the CnCda4–substrate interactions at a molecular level, the amino acid sequence of CnCda4 was compared to the well-characterized CDAs PesCDA and ClCDA, which strongly prefer A4 as a substrate over any partially deacetylated tetramer. Given that CnCda4 strongly prefers a deacetylated unit at subsite −1, whereas PesCDA and ClCDA both prefer an acetylated unit, the amino acid residues surrounding subsite −1 were selected for mutagenesis. CnCda4 has an isoleucine residue at position 201 (I201; dark orange in Fig. 2 D and E), which is, according to the model, part of a flexible loop close to subsite −1, whereas PesCDA and ClCDA feature a histidine residue at the corresponding position. In addition to the I201H substitution, we also created the mutants I201Y and I201E, which replace the isoleucine residue in CnCda4 with less flexible (tryptophan) or bulky and negatively charged (glutamic acid) residues. The three mutants were then compared to wild-type CnCda4 in terms of their activity against the single deacetylated substrates ADAA and ADAAAA and the fully acetylated counterparts A4 and A6 (Fig. 2F). All three mutants still preferred ADAA over A4, but not as strongly as the wild-type CnCda4 (Fig. 2F), indicating that there was a greater acceptance of acetylated units at subsite −1 in all of the mutants. In contrast, the mutants showed no significant changes in preference for the hexameric substrates. Interestingly, whereas the wild-type enzyme converts ADAAAA mostly into ADDAAA (as detailed earlier), all three mutants produced significantly more ADAADA than the wild-type enzyme, indicating a reduced preference for GlcN at subsite −1. We hypothesize that, by changing I201 to the less flexible tyrosine or the larger and charged histidine or glutamic acid residues, we have weakened the preference for a deacetylated unit at the −1 subsite of CnCda4.
Effect of Early and Late CnCda4 Products on Macrophages.
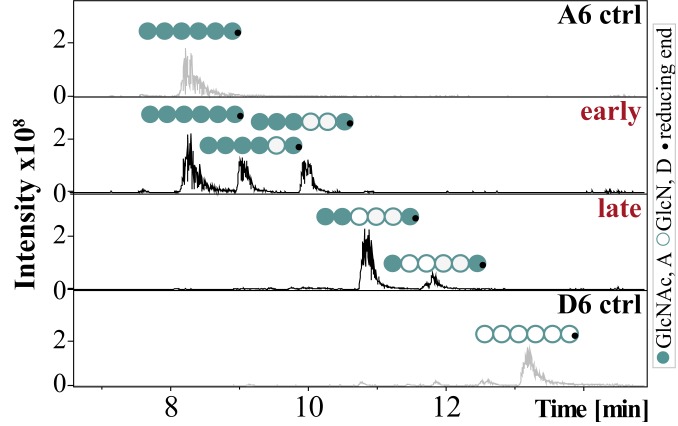
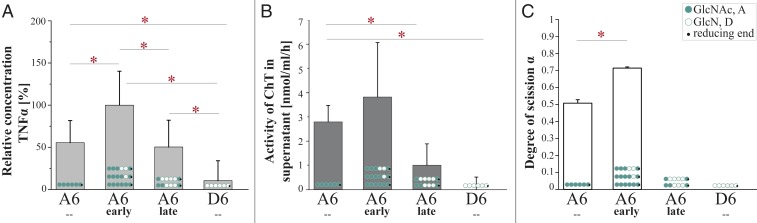
CnCda4 is known to stimulate CD4+ T cells, causing them to secrete higher levels of IFNγ and IL-2. It also induces a delayed-type hypersensitive reaction in mice infected with C. neoformans (33). CnCda1 to 3 are required for the synthesis of cell-wall chitosans (40) and thus confer virulence (6), but no natural function has yet been assigned to the secreted enzyme CnCda4. However, a CDA secreted by the plant endophytic fungus Pestalotiopsis sp. may help to inactivate elicitor-active chitin oligomers (4). We therefore tested the CnCda4 reaction products for their ability to induce the proinflammatory marker tumor necrosis factor α (TNFα) in human peripheral blood-derived macrophages (HPBMs), anticipating low activity. The fully acetylated chitin hexamer A6 is a potent activator of macrophages, but the fully deacetylated chitosan hexamer D6 is not (41, 42). A6 also induces the secretion of human chitotriosidase (ChT), which cleaves A6 into smaller, elicitor-inactive oligomers. In humans, two functional chitinases are known: ChT and the acetic mammalian chitinase (43). Monocytes express high levels of ChT during their differentiation into macrophages, whereas the expression levels of acetic mammalian chitinase remain very low (44). We therefore treated HPBMs with the control oligomers A6 and D6 (both incubated with heat-inactivated CnCda4) as well as the early and late products of CnCda4 (Fig. 3). MS analysis revealed the early fraction comprised A6, A5D1 (AAAADA), and A4D2 (AAADDA), whereas the late fraction comprised A3D3 (AADDDA) and A2D4 (ADDDDA). After removing the enzyme, all samples were tested for their ability to elicit the release of TNFα and ChT from HPBMs (Fig. 4 A and B and SI Appendix, Fig. S4). As anticipated, A6 elicited the release of TNFα and ChT into the supernatant, whereas D6 did not. Interestingly, the early products of CnCda4 showed even stronger elicitor activity than A6, whereas the late products were weaker. Given that, in contrast to most fungi, the C. neoformans cell wall contains more chitosan than chitin, the availability of a chitosan deacetylase rather than a chitin deacetylase may help to eliminate weakly deacetylated chitosan oligomers with strong elicitor activity by converting them into more completely deacetylated products with weak elicitor activity. We therefore propose that the initial deacetylation steps may inhibit hydrolysis by ChT but not yet prevent recognition by a potential chitin receptor, thereby increasing elicitor activity. The detection of chitin and, possibly, sufficiently acetylated chitosan oligomers by the human immune system may involve different subsets of pattern recognition receptors such as TLR2 or Dectin-1 (45). Further deacetylation of the chitosan oligomers by CnCda4 may prevent molecular recognition and thus reduce elicitor activity. To test this hypothesis, we next investigated the ability of purified ChT to hydrolyze the hexameric CnCda4 products (Fig. 4C).
Fig. 3.
LC-MS chromatograms of the oligomeric fractions tested for their ability to stimulate macrophages. Chitin hexamer (600 µg) was incubated in 50 mM NaHCO3 (pH 7) at 37 °C for either 1 h (early) or 90 h (late) with 100 µL (116 µg) of purified CnCda4. A heat-inactivated batch of CnCda4 was used for the two controls (A6 and D6). The reaction was stopped by passing the sample through a 3-kDa filter, removing the enzyme.
Fig. 4.
The effect of early and late CnCda4 products and A6/D6 controls on the secretion of TNFα by macrophages and the cleavage products generated by recombinant human chitotriosidase (ChT). (A) Activation of human peripheral blood-derived macrophages (HPBMs) by different chitin and chitosan hexamers (1 µg/mL). Secretion of TNFα into the supernatant was measured 6 h after stimulation. Data from five independent experiments with different batches of macrophages were normalized to the negative control (starvation medium, 0%) and to the positive control (10 ng/mL LPS, 100%). Data are presented as means ± SD (B) Activity of the human ChT measured in the supernatant after stimulation with the different chitin and chitosan hexamers (1 µg/mL). The concentration of ChT in 5 µL supernatant was determined by adding 20 mM of the ChT-specific substrate 4-methylumbelliferyl-β-d-N,N′-diacetylchitobioside hydrate in 100 mL McIlvain buffer (100 mM citric acid, 200 mM sodium phosphate, pH 5.2). The enzyme reaction was stopped after 20 min at 37 °C by adding 200 mL of 300 mM glycine-NaOH buffer, pH 10.5. Converted substrate was measured by fluorimetry at 450 nm and related to a standard of recombinant ChT. Values are means of three independent experiments ±SD. (C) Relative ChT activity on the oligomeric substrates GlcNAc6 (A6), GlcN6 (D6), and the early and late hexameric products of CnCda4. Isolated ChT (50 ng) was incubated with the oligomeric substrates (2.5 µg) for 24 h at 37 °C in 50 mM ammonium acetate (pH 4.5). Products were detected and quantified by HPLC-MS analysis, and ChT activity was quantified by the amount of hydrolyzed substrate, described as the degree of scission (α), where 100% equals complete degradation of the substrate. All data are based on five independent experiments (n = 5) and are given as means ± SD. Statistical analysis was performed using the t test (*P < 0.5).
Effect of Recombinant ChT on Early and Late CnCda4 Products.
The early and late hexameric products of CnCda4 (and the controls A6 and D6) were incubated with purified recombinant ChT, and the resulting products were quantified by LC-MS1. ChT activity was presented as the degree of scission (α), defined as the number of substrate cleavages relative to theoretical complete hydrolysis (Fig. 4C). The complete product profiles are shown in SI Appendix, Fig. S5. ChT hydrolyzed ∼50% of the A6 substrate, but D6 and the late deacetylation products of CnCda4 were untouched. Interestingly, and counter to our expectations, the early deacetylation products were more efficiently hydrolyzed than the fully acetylated chitin hexamer A6. ChT is a chitinase belonging to glycosyl hydrolase family 18 (GH18) with an absolute specificity for acetylated units at subsites −2 and −1 and a preference for an acetylated unit at subsite +1, although a deacetylated unit can also be accepted (46). As a consequence, A6 should be the preferred substrate in the early CnCda4 product mixture, but the partially deacetylated products AAAADA and AAADDA can probably also be hydrolyzed, although this should be a slower reaction. Taking a closer look at the hydrolysis profile of the early CnCda4 products (SI Appendix, Fig. S5), ChT indeed appears to mainly attack A6, but also A5D1 (AAAADA). Based on these data, the subsite specificity of ChT should be investigated in more detail.
Conclusion
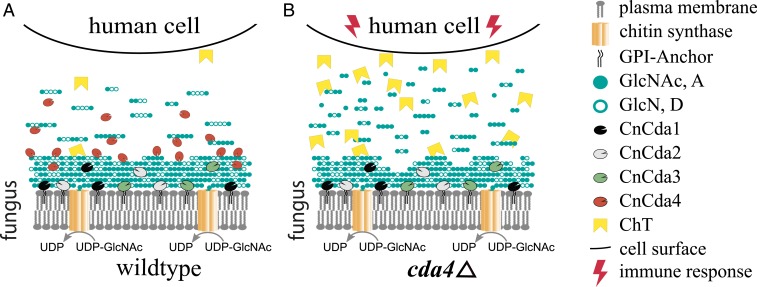
We have shown that CnCda4 is a functional enzyme that catalyzes the deacetylation of oligomeric and polymeric chitinous substrates. In contrast to other well-described CDAs, CnCda4 prefers partially deacetylated chitosans rather than fully acetylated chitin, making CnCda4 a chitosan deacetylase. Our data indicate that the final products of CnCda4 are not fully deacetylated but still possess a certain number of GlcNAc units, typically the terminal residues in longer oligomers. The physiological role of CnCda4 must be seen in the context of the other three C. neoformans CDAs, for which natural functions have been described. Cda1 is known to deacetylate cell-wall chitin produced by chitin synthase Chs3 and its regulator Csr2 (5, 7, 34) during vegetative growth, but, under certain conditions, CnCda2 and CnCda3 can compensate for the loss of CnCda1, thus ensuring that the predominance of chitosan over chitin in the cell wall is maintained (5). CnCda3 is known to mediate the adhesion of C. neoformans to epithelial lung cells, thereby also directly participating in the infection process (46). There is evidence that CnCda1 to 3 are mainly anchored to the plasma membrane and may also be associated with the cell wall (40) or even partly secreted (47), whereas CnCda4 is likely to be solely secreted into the medium or the glucuronoxylomannan (GXM) capsule surrounding the fungal cell. We hypothesize that CnCda4 supports the infection process by helping C. neoformans to evade the host immune system as summarized in Fig. 5, and that two main strategies may be involved. First, CnCda4 may further deacetylate cell-wall chitosans produced by the combined activities of Chs3, Csr2, Cda1, Cda2, and Cda3 (especially on the outer, exposed layers). Second, it may further deacetylate chitosan oligomers released as PAMPs from the fungal cell wall by host chitinases such as ChT so that they are trapped more efficiently in the negatively charged GXM capsule and/or no longer recognized by the host’s pattern recognition receptors (Fig. 5A). To evaluate this hypothesis and the proposed mechanisms, the phenotype of the existing CDA4-knockout mutant (5, 6) should be investigated during infection. Additionally, the influence of the GXM capsule of C. neoformans on the release of chitosan oligosaccharides should be investigated closely. Our data suggest that, in the absence of CnCda4, highly acetylated chitosan oligomers would be recognized by the host cell, initiating an early immune response involving the increased secretion of ChT, generating a positive feedback loop (Fig. 5B). Given that the C. neoformans cda1Δ, cda2Δ, cda3Δ strain is easily defeated by the host (35), it seems that CnCda4 alone, which is geared toward chitosan rather than chitin as a substrate, cannot achieve sufficient deacetylation of the cell-wall chitin to prevent recognition by the host. Rather, CnCda4 seems to act in concert with the other CDAs to amplify their stealth effect. This role of CnCda4 is similar to the proposed role of other secreted fungal CDAs such as PesCDA (4). In the case of C. neoformans, with its atypical chitosan-containing cell wall, this role is best performed by a chitosan deacetylase.
Fig. 5.
Proposed role of CnCda1 to 3 and CnCda4 during the infection of human cells by the opportunistic pathogen C. neoformans. (A) Infections by the wild-type pathogen. (B) Infection by a CnCda4-deficient mutant. Cell wall chitin produced by chitin synthase CnChs3 together with CnCsr2 is partially deacetylated by the cell wall-associated enzymes CnCda1 to 3, whereas CnCda4 further deacetylates the outer layers as well as the released oligomers and small polymers until only the terminal sugar units remain acetylated. (A) With all four CDAs present, C. neoformans can successfully hide its presence from the human immune system. (B) With CnCda4 absent, highly acetylated oligomers and small polymers are recognized by the host and defense responses are initiated, including secretion of chitotriosidase (ChT), attacking the fungal cell wall and thereby releasing more oligomers and small polymers, intensifying the host immune response.
Our study provides preliminary indications that the deacetylation of chitin oligosaccharides has a biological impact on the immune response to a cryptococcal infection, but more evidence is required. We therefore propose a two-step approach to develop an in vivo model. The first step would involve the evaluation of virulence in a CDA4-knockout strain of C. neoformans, such as the fpd1Δ strain that has already been described (5, 6). Removing CnCda4 from the system would prevent the inactivation of immunostimulatory chitosan oligomers with a high degree of acetylation, leading to reduced virulence. However, removing CnCda4 would also remove the immunostimulatory effect of the protein itself, consequently increasing virulence, and the two opposing effects might result in a noninformative phenotype. Therefore, the second step would involve the evaluation of virulence in a C. neoformans strain expressing a catalytically inactive form of CnCda4, which could be generated by site-directed mutagenesis. The loss of CnCda4 activity would again reduce virulence, but the protein would retain its immunostimulatory effect, and a more informative low-virulence phenotype would be anticipated.
Materials and Methods
Genes, Enzymes, and Plasmids.
The C. neoformans (formerly Filobasidiella neoformans) gene encoding CnCda4 (d25, Fpd1; UniProtKB Q96TR5_CRYNE, GenBank CAD10036.1, CNAG_06291) was codon-optimized for E. coli and synthesized by GeneArt. The gene was inserted into vector pET-22b(+) (Novagen), and variants were produced with the endogenous N-terminal signal peptide and a C-terminal Strep II tag [pET22b(+)::SP-CDA4-StrepII], with a C-terminal linker sequence inserted before the Strep II tag [pET22b(+)::SP-CDA4-linker-StrepII], without a linker but with the N-terminal signal peptide sequence removed [pET22b(+)::CDA4-strepII], and with Strep II tags at both termini [pET22b(+)::StrepII-CDA4-StrepII]. All sequences are shown in SI Appendix, Fig. S6. All constructs were introduced into E. coli Lemo21 (DE3) cells (New England Biolabs) by heat shock (48), and the fourth construct was also introduced into E. coli Rosetta2 (DE3) [pLysSRARE2] cells (Novagen). HPBM-derived cDNA representing the ChT gene was inserted into vector pDsRed-Monomer-N (Takara) in which the DsRed monomer cassette was replaced by a His6 tag. The ChT construct was introduced into HEK293 cells, and stably transfected cells were selected using geneticin (G418).
Recombinant Protein Expression.
E. coli Lemo21 (DE3) cells carrying either of the plasmids were inoculated to an OD600 of 0.2 in lysogeny broth (LB) supplemented with 0 to 2 mM α-l-rhamnose and incubated at 37 °C shaking at 160 rpm until the OD600 reached 0.6 to 0.8. The expression of CnCda4 was then induced by adding IPTG to a final concentration of 400 µM and incubating the cells as described for a further 5 h. E. coli Rosetta2 (DE3) [pLysSRARE2] cells carrying pET22b(+)::StrepII-CDA4-StrepII were incubated in 500 mL autoinduction medium in a 2-L flask for 48 h at 26 °C, shaking at 160 rpm (49). Cells were harvested by centrifugation (4,000 × g, 20 min, 4 °C), resuspended in 30 mL of fast protein liquid chromatography (FPLC) washing buffer (20 mM TEA, 400 mM NaCl, pH 8.0), and stored at −20 °C. HEK293 cells carrying the ChT expression plasmid were cultivated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS), 1% penicillin/streptomycin, and 1% l-glutamine at 37 °C and 5% CO2. When the HEK293 cell layer reached a confluence of 80%, the culture medium was replaced with serum-free production medium (OptiPRO SFM; Gibco). After 3 d of culture, cell supernatants containing recombinant ChT were harvested and stored at −20 °C.
Enzyme Purification by Affinity Chromatography.
Cells were lysed by thawing and incubating at room temperature after adding lysozyme (1.5 g/L final concentration) and sodium chloride (∼10 g/L final concentration), followed by three 15-s pulses at 40% amplitude using a Branson Digital Sonifier model 250-D (Emerson). After centrifugation (60 min, 40,000 × g, 4 °C), CnCda4 was captured from the supernatant by Strep-Tactin affinity chromatography and eluted using 2.5 mM d-desthiobiotin in FPLC washing buffer. Recombinant ChT was captured from HEK293 cell supernatants by His6 tag affinity chromatography and eluted in phosphate-buffered saline (PBS; pH 7.4).
SDS/PAGE and Western Blotting.
The concentration of eluted protein was determined using the Bradford method (50). We then separated 3 to 6 µg per lane by SDS polyacrylamide gel electrophoresis (SDS/PAGE) (51). The gels were stained with 0.05% Coomassie Brilliant Blue G-250 in 40% methanol and 5% acetic acid. For Western blotting, proteins were transferred to a nitrocellulose membrane and detected using horseradish peroxidase (HRP)-conjugated Strep-Tactin and the chemiluminescent substrate luminol according to the manufacturer’s instructions (IBA).
Acetate Assay to Determine Enzyme Activity.
To determine the optimal pH, temperature, and buffer for CnCda4, as well as its activity against different polymeric substrates, we measured the amount of acetate released using the Acetate Assay Kit (r-Biopharm), adapted to a microtiter plate format (4).
LC-MS1 Analysis.
The activity of CnCda4 against oligomeric substrates was measured using the methods shown in Table 1 as previously described (52–55). Reaction products were separated on a Dionex Ultimate 3000RS UHPLC system (Thermo Fisher Scientific) coupled to an amaZon speed ESI-MSn detector (Bruker). Oligomers were separated by hydrophilic interaction chromatography (HILIC) using a VanGuard precolumn (1.7 µm, 2.1 × 35 mm) and an Acquity UHPLC BEH Amide column (1.7 µm, 2.1 × 150 mm; Waters). A gradient of solvent A (80% acetonitrile, 20% water) and solvent B (20% acetonitrile, 80% water), both containing 10 mM NH4HCO2 and 0.1% (vol/vol) formic acid, was used for separation. MS detection was performed in a positive mode and mass spectra acquired over a scan range of m/z of 50 to 2,000 in enhanced resolution scan mode. Data were processed using Data Analysis v4.1 software (Bruker). The oligomers were quantified by measuring peak areas.
Table 1.
LC-MS1 methods for the detection and quantification of partially acetylated chitosan oligomers
| Method | Column oven temperature, °C | Flow, mL/min | Isocratic 100% A, min | Linear gradient, min | Isocratic, min | Reequilibration, min | Isocratic 100% A, min | Duration, min |
| 1 | 35 | 0.4 | 0–2.5 | 2.5–12.5, 0–75% B | — | 12.5–13.5, 75–0% B | 13.5–15 | 15 |
| 2 | 75 | 0.8 | 0–0.8 | 0.8–3.3, 0–70% B | — | 3.3–4.3, 70–0% B | 4.3–5.5 | 5.5 |
| 3 | 75 | 0.8 | 0–0.8 | 0.8–4.8, 0–90% B | 4.8–6.8 | 6.8–7.3, 90–0% B | 7.3–8 | 8 |
LC-MS2 Analysis.
The PA of oligomer samples (5 to 10 µg) was determined by labeling the reducing ends with H218O (Euriso-top) in two steps (1 × 5 µL, 1 × 10 µL, 70 °C) as previously described (55). The samples were separated as described earlier for LC-MS1 analysis, and product ions of certain m/z values were selected (SI Appendix, Table S1). An isolation width of 1.0 m/z was used, and fragmentation was carried out using an amplitude gradient of 80 to 120% over 40 ms. MS/MS spectra were acquired over a scan range of m/z of 50 to 2,000 and analyzed using Data Analysis v4.1 software.
Mode of Action Against Chitin Oligomers.
CnCda4 (0.5 µg) was incubated with the chitin oligomers A1 to A6 (1 mg/mL final concentration) in 50 mM NaHCO3 (pH 7). Samples were taken after 0, 5, 15, and 30 min and after 1, 2, 3, 6, 12, 24, 72, and 144 h by stopping the reaction with 0.5% (final concentration) formic acid. The oligomer products in all samples were analyzed by LC-MS1 and LC-MS2 as described earlier.
Activity of CnCda4 Against Polymeric Substrates.
CnCda4 (1 µg) was incubated with different soluble and insoluble substrates (chitosan DA30% and DA50%, colloidal chitin, 1% [wt/vol] α-chitin, or 1% [wt/vol] β-chitin) for 24 h at 37 °C in 50 mM NaHCO3 (pH 7). Activity was determined by measuring the amount of acetate released as described earlier. Zymography experiments to determine activity on glycol chitin were performed according to the dot blot assay method (56), with glycol-chitin incorporated into the acrylamide gel. To detect CDA activity, the chitosan produced within the gel was depolymerized using a mixture of nitric and sulfuric acids (7.59 g NaNO2 dissolved in 20 mL H2O mixed with 550 µL concentrated H2SO4) for 10 min at room temperature before the gel was washed twice (10 min each) with water and illuminated under UV light. The α-chitin and β-chitin were purchased from Mahatani Chitosan. DA30% and DA50% chitosans were prepared by the chemical acetylation of DA0% chitosan (57). Colloidal chitin (58) and glycol chitin (59) were prepared as previously described.
Production of Defined paCOS.
Chitin oligomers A2, A3, A4, A5, and A6 were purchased from Megazyme. A4 and A6 were used as substrates for the preparation of pattern-specific paCOS by exposing them to the enzymes NodB [DAAA (52)], COD/VcCDA [ADAA, ADAAAA (52)], PesCDA [AADA (4)], and PgtCDA [AADD (17)].
Determining Regioselectivity and Regiospecificity.
Four independent batches of purified CnCda4, equally adjusted according to their activity against A4, were incubated with 1 mg/mL DAAA, ADAA, AADA, or AADD in 50 mM NaHCO3 (pH 7) at 37 °C. The reaction was stopped after 1 h or 72 h by adding formic acid (0.5% final concentration). The paCOS in all samples were analyzed by LC-MS1 and LC-MS2 as described earlier, and the relative activity at different sites was determined by counting the number of attacks.
Comparative Activity Against AD(A)n and AA(A)n.
Wild-type CnCda4 and the corresponding mutants I201Y, I201H, and I201E were adjusted for their activity against A4 and incubated with 1 mg/mL of ADAA, A4, ADAAAA, or A6 in 50 mM NaHCO3 (pH 7) at 37 °C. The reaction was stopped after 45 min by adding formic acid (0.5% final concentration). The paCOS in all samples was analyzed by LC-MS1 and LC-MS2 as described earlier. In more challenging cases, MS2 analysis to determine the dominant PA was carried out using nonchemically acetylated samples. The relative activity was determined by quantifying the number of attacks, and the ratio of relative activities against the partially deacetylated and fully acetylated substrates was calculated. Significant differences in enzyme activities were determined by one-way analysis of variance (ANOVA).
Cofactor Analysis.
CnCda4 (1.2 M) was incubated with 20 mM EDTA in 50 mM NaHCO3 (pH 7) at room temperature for 2 h to remove all metal ions, before seven rounds of dialysis against 1,000 volumes of EDTA-free 50 mM NaHCO3 (90 h). EDTA levels were monitored by LC-MS. The mixture was aliquoted to add 1 mM ZnCl2, CoCl2, MnCl2, CaCl2, MgCl2, MgSO4, FeSO4, or CuCl2 in triplicate. After incubation at room temperature for 1 h, A4 was added to a final concentration of 0.5 mM. The samples were incubated at 37 °C for 2 h before the reaction was stopped by adding formic acid (0.5% final concentration). The activity was measured by LC-MS1 analysis as described earlier.
Homology Modeling and Substrate Docking.
A three-dimensional model of CnCda4 was generated using the online structure prediction tool SWISS-MODEL (60–62). The crystal structure of ClCDA (PDB ID code 2IW0; amino acid sequence identity 33%) was used as the template. The geometric accuracy of the model was evaluated with MolProbity (63). Because the models were created without a metal ion in the active site, the Zn2+ ion from the ClCDA crystal structure was inserted at the estimated location by aligning the two enzymes in PyMOL. Substrate docking was performed using two different chitin tetramers and two different chitin hexamers, all created using the carbohydrate builder from GLYCAM (64). All four substrates were docked using AutoDock VinaCarb v1.0 (39).
Sample Preparation for the Stimulation of HPBMs.
CnCda4 was heat-inactivated at 89 °C for 15 min before mixing with 600 µg D6 (Carbosynth) or 600 µg A6 at 37 °C for 90 h in 50 mM NaHCO3 (pH 7). In addition, 600 µg A6 was incubated under the same conditions with active CnCda4 for 1 h (to generate early products) and 90 h (to generate late products). The reaction was stopped by passage through a 3-kDa cutoff filter to remove the enzyme, and the sample composition was determined by LC-MS1 and LC-MS2 as described earlier.
Isolation and Stimulation of HPBMs.
HPBMs were isolated from buffy coats (German Red Cross) by two-step density centrifugation as previously described (65). Washed monocytes were seeded into a 24-well plate (200,000 cells per well) and cultivated for 7 d in 500 µL RPMI-1640 medium supplemented with 10% human serum, 1% nonessential amino acids, and 1% l-glutamine at 37 °C in a 5% CO2 atmosphere to differentiate them into macrophages. The cells were starved 24 h before stimulation with chitin and chitosan hexamers by switching to RPMI-1640 medium supplemented with 1% nonessential amino acids and 1% l-glutamine but no serum. HPBMs were stimulated with paCOS or lipopolysaccharides (LPSs) diluted in starvation medium (300 µL per well) for 6 h. TNFα and ChT activity were measured in cell supernatants by enzyme-linked immunosorbent assays (ELISAs) and enzyme activity assays, respectively, as previously described (41). Significant differences in the response to treatment were determined by one-way ANOVA.
Measurement of ChT Degree of Scission.
The hexameric samples used to stimulate HPBMs (2.5 µg) were incubated in vitro with 50 ng of purified human ChT at 37 °C for 24 h in 50 mM ammonium acetate (pH 4.5). Products were detected and quantified by HPLC-MS, and ChT activity was quantified by LC-MS1 and LC-MS2 analysis as described earlier. Significant differences in enzyme activities (degrees of scission, α) were determined by one-way ANOVA.
Data Availability.
All materials, data, and associated protocols will be made available upon request from the corresponding author.
Supplementary Material
Acknowledgments
We acknowledge Dr. Richard M. Twyman (Twyman Research Management) for editing the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915798117/-/DCSupplemental.
References
- 1.Mine S., Ikegami T., Kawasaki K., Nakamura T., Uegaki K., Expression, refolding, and purification of active diacetylchitobiose deacetylase from Pyrococcus horikoshii. Protein Expr. Purif. 84, 265–269 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Hoving J. C., Wilson G. J., Brown G. D., Signalling C-type lectin receptors, microbial recognition and immunity. Cell. Microbiol. 16, 185–194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs K., et al. , The fungal ligand chitin directly binds TLR2 and triggers inflammation dependent on oligomer size. EMBO Rep. 19, e46065 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cord-Landwehr S., Melcher R. L. J., Kolkenbrock S., Moerschbacher B. M., A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci. Rep. 6, 38018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker L. G., Specht C. A., Donlin M. J., Lodge J. K., Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell 6, 855–867 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker L. G., Specht C. A., Lodge J. K., Cell wall chitosan is necessary for virulence in the opportunistic pathogen Cryptococcus neoformans. Eukaryot. Cell 10, 1264–1268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banks I. R., et al. , A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 4, 1902–1912 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis L. L., Bartnicki-Garcia S., Chitosan synthesis by the tandem action of chitin synthetase and chitin deacetylase from Mucor rouxii. Biochemistry 23, 1065–1073 (1984). [Google Scholar]
- 9.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B., The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lombard V., et al. , A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 432, 437–444 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Caufrier F., Martinou A., Dupont C., Bouriotis V., Carbohydrate esterase family 4 enzymes: Substrate specificity. Carbohydr. Res. 338, 687–692 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y., Park R.-D., Muzzarelli R. A. A., Chitin deacetylases: Properties and applications. Mar. Drugs 8, 24–46 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kafetzopoulos D., Thireos G., Vournakis J. N., Bouriotis V., The primary structure of a fungal chitin deacetylase reveals the function for two bacterial gene products. Proc. Natl. Acad. Sci. U.S.A. 90, 8005–8008 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsigos I., Martinou A., Kafetzopoulos D., Bouriotis V., Chitin deacetylases: New, versatile tools in biotechnology. Trends Biotechnol. 18, 305–312 (2000). [DOI] [PubMed] [Google Scholar]
- 15.John M., Röhrig H., Schmidt J., Wieneke U., Schell J., Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc. Natl. Acad. Sci. U.S.A. 90, 625–629 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Wang L.-X., Wang X., Roseman S., The chitin catabolic cascade in the marine bacterium Vibrio cholerae: Characterization of a unique chitin oligosaccharide deacetylase. Glycobiology 17, 1377–1387 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Naqvi S., et al. , A recombinant fungal chitin deacetylase produces fully defined chitosan oligomers with novel patterns of acetylation. Appl. Environ. Microbiol. 82, 6645–6655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokuyasu K., Ohnishi-Kameyama M., Hayashi K., Mori Y., Cloning and expression of chitin deacetylase gene from a Deuteromycete, Colletotrichum lindemuthianum. J. Biosci. Bioeng. 87, 418–423 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Tokuyasu K., Ohnishi-Kameyama M., Hayashi K., Purification and characterization of extracellular chin deacetylase from Colletotrichum lindemuthianum. Biosci. Biotechnol. Biochem. 60, 1598–1603 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Martinou A., Bouriotis V., Stokke B. T., Vårum K. M., Mode of action of chitin deacetylase from Mucor rouxii on partially N-acetylated chitosans. Carbohydr. Res. 311, 71–78 (1998). [Google Scholar]
- 21.Hoßbach J., et al. , A chitin deacetylase of Podospora anserina has two functional chitin binding domains and a unique mode of action. Carbohydr. Polym. 183, 1–10 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Shao Z., et al. , Comparative characterization of putative chitin deacetylases from Phaeodactylum tricornutum and Thalassiosira pseudonana highlights the potential for distinct chitin-based metabolic processes in diatoms. New Phytol. 221, 1890–1905 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Hayafune M., et al. , Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. U.S.A. 111, E404–E413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vander P., V rum K. M., Domard A., Eddine El Gueddari N., Moerschbacher B. M., Comparison of the ability of partially N-acetylated chitosans and chitooligosaccharides to elicit resistance reactions in wheat leaves. Plant Physiol. 118, 1353–1359 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park B. J., et al. , Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23, 525–530 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Kaplan J. E., et al. , Cryptococcal antigen screening and early antifungal treatment to prevent cryptococcal meningitis: A review of the literature. J. Acquir. Immune Defic. Syndr. 68 (suppl. 3), S331–S339 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Rajasingham R., et al. , Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 17, 873–881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benesová P., Buchta V., Cerman J., Zák P., Cryptococcosis–A review of 13 autopsy cases from a 54-year period in a large hospital. APMIS 115, 177–183 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Liaw Y. S., et al. , Direct determination of cryptococcal antigen in transthoracic needle aspirate for diagnosis of pulmonary cryptococcosis. J. Clin. Microbiol. 33, 1588–1591 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldmesser M., Kress Y., Novikoff P., Casadevall A., Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 68, 4225–4237 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deshaw M., Pirofski L.-A., Antibodies to the Cryptococcus neoformans capsular glucuronoxylomannan are ubiquitous in serum from HIV+ and HIV− individuals. Clin. Exp. Immunol. 99, 425–432 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L. C., Goldman D. L., Doering T. L., Pirofski La., Casadevall A., Antibody response to Cryptococcus neoformans proteins in rodents and humans. Infect. Immun. 67, 2218–2224 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biondo C., et al. , Identification and cloning of a cryptococcal deacetylase that produces protective immune responses. Infect. Immun. 70, 2383–2391 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Upadhya R., et al. , Cryptococcus neoformans Cda1 and its chitin deacetylase activity are required for fungal pathogenesis. MBio 9, 1–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Upadhya R., et al. , Induction of protective immunity to cryptococcal infection in mice by a heat-killed, chitosan-deficient strain of Cryptococcus neoformans. MBio 7, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Specht C. A., et al. , Vaccination with recombinant Cryptococcus proteins in glucan particles protects mice against cryptococcosis in a manner dependent upon mouse strain and cryptococcal species. MBio 8, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen H., “Predicting secretory proteins with SignalP” in Protein Function Prediction: Methods and Protocols, Kihara D., Ed. (Springer New York, 2017), pp. 59–73. [DOI] [PubMed] [Google Scholar]
- 38.Blair D. E., et al. , Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry 45, 9416–9426 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Nivedha A. K., Thieker D. F., Makeneni S., Hu H., Woods R. J., Vina-Carb: Improving glycosidic angles during carbohydrate docking. J. Chem. Theory Comput. 12, 892–901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilbert N. M., Baker L. G., Specht C. A., Lodge J. K., A glycosylphosphatidylinositol anchor is required for membrane localization but dispensable for cell wall association of chitin deacetylase 2 in Cryptococcus neoformans. MBio 3, 1–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorzelanny C., Pöppelmann B., Pappelbaum K., Moerschbacher B. M., Schneider S. W., Human macrophage activation triggered by chitotriosidase-mediated chitin and chitosan degradation. Biomaterials 31, 8556–8563 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Eide K. B., et al. , Human chitotriosidase-catalyzed hydrolysis of chitosan. Biochemistry 51, 487–495 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Boot R. G., et al. , Marked differences in tissue-specific expression of chitinases in mouse and man. J. Histochem. Cytochem. 53, 1283–1292 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Di Rosa M., et al. , Evaluation of AMCase and CHIT-1 expression in monocyte macrophages lineage. Mol. Cell. Biochem. 374, 73–80 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Elieh Ali Komi D., Sharma L., Dela Cruz C. S., Chitin and its effects on inflammatory and immune responses. Clin. Rev. Allergy Immunol. 54, 213–223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teixeira P. A. C., Penha L. L., Mendonça-Previato L., Previato J. O., Mannoprotein MP84 mediates the adhesion of Cryptococcus neoformans to epithelial lung cells. Front. Cell. Infect. Microbiol. 4, 106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levitz S. M., Nong S., Mansour M. K., Huang C., Specht C. A., Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T cell responses to Cryptococcus neoformans. Proc. Natl. Acad. Sci. U.S.A. 98, 10422–10427 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlegel S., et al. , Optimizing membrane protein overexpression in the Escherichia coli strain Lemo21(DE3). J. Mol. Biol. 423, 648–659 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Studier F. W., Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Bradford M. M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- 51.Laemmli U. K., Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970). [DOI] [PubMed] [Google Scholar]
- 52.Hamer S. N., et al. , Enzymatic production of defined chitosan oligomers with a specific pattern of acetylation using a combination of chitin oligosaccharide deacetylases. Sci. Rep. 5, 8716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haebel S., Bahrke S., Peter M. G., Quantitative sequencing of complex mixtures of heterochitooligosaccharides by vMALDI-linear ion trap mass spectrometry. Anal. Chem. 79, 5557–5566 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Pohlentz G., Egge H., [13] Neoglycolipids of 1-deoxy-1-phosphatidylethanolaminolactitol type: Synthesis, structure analysis, and use as probes for characterization of glycosyltransferases. Methods Enzymol. 242, 127–145 (1994). [DOI] [PubMed] [Google Scholar]
- 55.Cord-Landwehr S., et al. , Quantitative mass-spectrometric sequencing of chitosan oligomers revealing cleavage sites of chitosan hydrolases. Anal. Chem. 89, 2893–2900 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Govinda Rajulu M. B., et al. , Chitinolytic enzymes from endophytic fungi. Fungal Divers. 47, 43–53 (2011). [Google Scholar]
- 57.Lamarque G., Lucas J.-M., Viton C., Domard A., Physicochemical behavior of homogeneous series of acetylated chitosans in aqueous solution: Role of various structural parameters. Biomacromolecules 6, 131–142 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez-Kabana R., Godoy G., Morgan-Jones G., Shelby R. a., The determination of soil chitinase activity: Conditions for assay and ecological studies. Plant Soil 75, 95–106 (1983). [Google Scholar]
- 59.Trudel J., Asselin A., Detection of chitin deacetylase activity after polyacrylamide gel electrophoresis. Anal. Biochem. 189, 249–253 (1990). [DOI] [PubMed] [Google Scholar]
- 60.Arnold K., Bordoli L., Kopp J., Schwede T., The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T., The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 37, D387–D392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Biasini M., et al. , SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirschner K. N., et al. , GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 29, 622–655 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feige U., Overwien B., Sorg C., Purification of human blood monocytes by hypotonic density gradient centrifugation in Percoll. J. Immunol. Methods 54, 309–315 (1982). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All materials, data, and associated protocols will be made available upon request from the corresponding author.