Abstract
Structural DNA nanotechnology is beginning to emerge as a widely accessible research tool to mechanistically study diverse biophysical processes. Enabled by scaffolded DNA origami in which a long single strand of DNA is weaved throughout an entire target nucleic acid assembly to ensure its proper folding, assemblies of nearly any geometric shape can now be programmed in a fully automatic manner to interface with biology on the 1–100-nm scale. Here, we review the major design and synthesis principles that have enabled the fabrication of a specific subclass of scaffolded DNA origami objects called wireframe assemblies. These objects offer unprecedented control over the nanoscale organization of biomolecules, including biomolecular copy numbers, presentation on convex or concave geometries, and internal versus external functionalization, in addition to stability in physiological buffer. To highlight the power and versatility of this synthetic structural biology approach to probing molecular and cellular biophysics, we feature its application to three leading areas of investigation: light harvesting and nanoscale energy transport, RNA structural biology, and immune receptor signaling, with an outlook toward unique mechanistic insight that may be gained in these areas in the coming decade.
Keywords: nanotechnology, synthetic structural biology, DNA origami, computational design, light harvesting, nanoscale energy transport, RNA structural biology, immunology
INTRODUCTION
The discovery of the double-helical structure of DNA arguably represents one of the most important discoveries in modern biology (52, 189, 191). Driving this landmark discovery was the close interaction between theory and experiment: Quantitative X-ray scattering data, generated by Franklin, Wilkins, and colleagues, were interpreted theoretically using a structural model of DNA proposed by Watson and Crick. Elucidation of the remarkably simple, elegant structure of DNA, which was confirmed only 20 years later by Rich and colleagues (142, 153), marked the first of several discoveries that enabled the rise of structural DNA nanotechnology to its prominence today, which has similarly been driven by a close interaction between theory and experiment.
Principal among these was the follow-on discovery of the Holliday junction used by cells in DNA recombination and repair, which confirmed that DNA may exist not only as a linear but also as a branched structure (75). This drove the remarkable conception by Ned Seeman (151) in 1982 that Watson-Crick base-pairing rules together with multiway branches may in principle be used to program synthetic DNA oligonucleotides to form complex, custom nanoscale materials. Chen & Seeman (30) subsequently applied these theoretical concepts to fabricate via self-assembly a synthetic, discrete DNA cube composed of six DNA strands, with 20 base pairs per cube edge, and an overall cube dimension of 7 nm, which arguably represents the birth of the field of structural DNA nanotechnology.
Lacking structural integrity and versatility in its branching capabilities, however, which limited this early design strategy from being generalized to more complex geometries, this architecture consisting of three-way vertices connected by single duplexes was soon supplanted by the introduction of the double-crossover (DX) motif (54). The DX motif displayed a twofold increase in stiffness over duplex DNA (144) and offered, in principle, the opportunity to program nearly arbitrarily complex nanostructures and arrays (99). Since then, the field of structural DNA nanotechnology has blossomed to include diverse strategies for programming two-dimensional (2D) and three-dimensional (3D) materials, which have recently been reviewed comprehensively (96, 133, 152, 154). In the present review, we highlight three discrete technological advances from the past decade that in our view pave the way for numerous high-impact applications in molecular and cellular biophysics, focusing on specific opportunities in the investigation of light harvesting and nanoscale energy transport, RNA structural biology, and immune receptor signaling.
In the first pivotal advance, Rothemund (143) introduced the concept of scaffolded DNA origami in which a long scaffold strand of DNA is folded into a complex, brick-like target shape via hybridization with hundreds of shorter oligonucleotide staple strands that stabilize the structure via Watson-Crick base-pairing. This 2D strategy was later generalized to rectilinear 3D structures by Shih and colleagues (42) and offered for the first time the ability to robustly synthesize discrete, structured DNA assemblies on the 10–100-nm scale. Importantly, in contrast to tile-based assembly (194), which was previously used to program higher-order nucleic acid assemblies, scaffolded DNA origami resulted in the quantitative yield of monodisperse structured DNA products. These DNA products can be used much like any discrete macromolecular assembly, such as a virus, polymer, or inorganic nanoparticle, with one important distinction: Scaffolded DNA origami is uniquely addressable at any distinct nucleotide position within the macromolecular assembly for purposes of molecular functionalization or templating.
In the second major advance, wireframe nucleic acid architectures were conceived (197) and applied using scaffolded DNA origami to program nearly arbitrarily complex lattice-like assemblies in two and three dimensions (201). In contrast to brick-like assemblies that are largely limited to acting as rectilinear pegboards, with or without variable degrees of bend and twist (39, 67), these assemblies now offer the facile generation of convex versus concave structured surfaces, internal and external positioning of molecules at arbitrary spatial positions, as well as access to a larger surface area per scaffold length. These assemblies are also compatible with a broader range of ionic conditions (186).
In the final major advance, top-down geometry-based computational design algorithms have generalized the wireframe scaffolded DNA origami approach to facilitate the automatic sequence design of 2D (17, 86; H. Jun, X. Wang & M. Bathe, unpublished information) and 3D (18, 85, 186) assemblies of nearly any shape and size on the 10–100-nm scale. These algorithms now enable any researcher to participate in the fabrication of these structured DNA assemblies for their own research applications of interest, without expertise in DNA origami design (see Figures 1 and 2, the sidebar titled Bottom-Up Computational Design of Scaffolded DNA Origami, and the sidebar titled Top-Down Computational Design of Wireframe Scaffolded DNA Origami).
Figure 1.
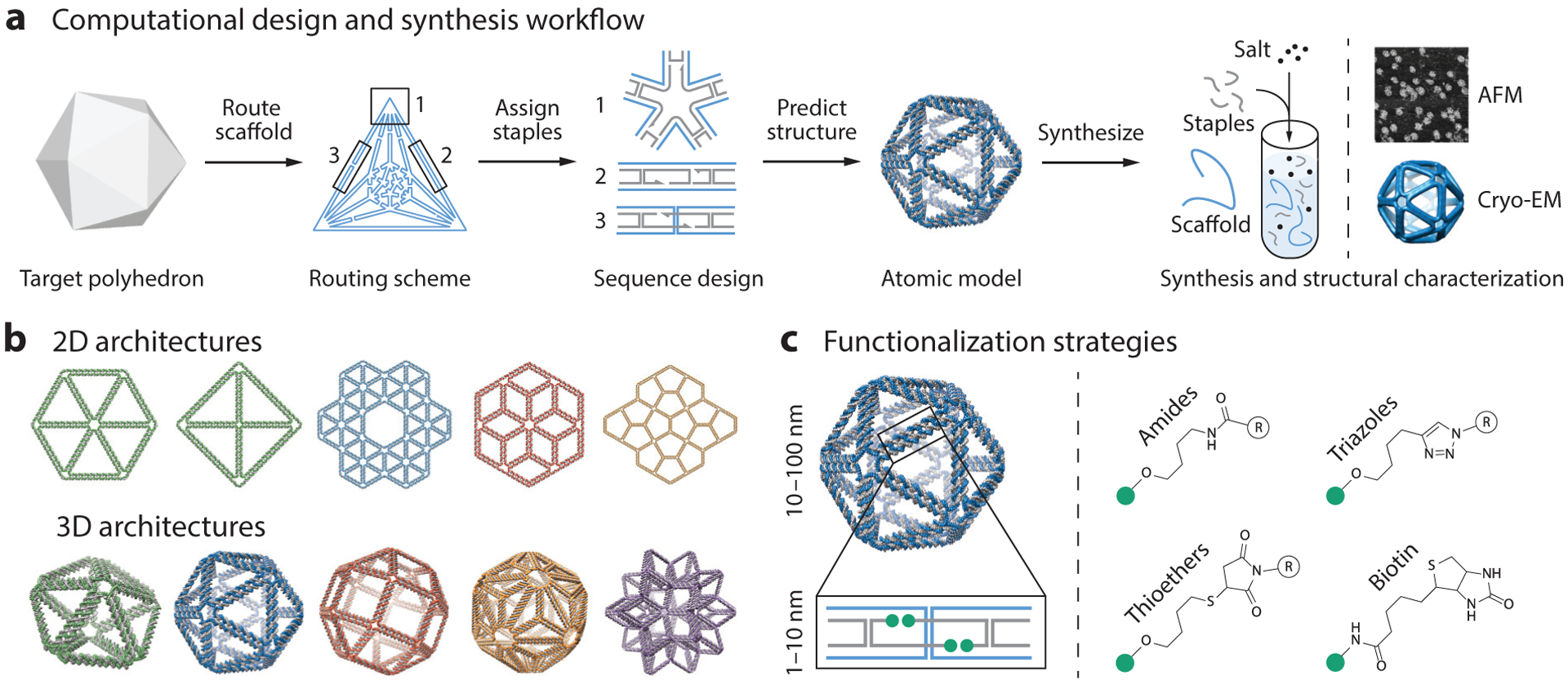
Design and synthesis of wireframe scaffolded DNA origami nanostructures for biophysical research. (a) This panel shows the workflow for designing DNA origami objects using the software DAEDALUS (186). First, a target polyhedron is used as input to the algorithm. DAEDALUS then automatically routes the scaffold through every edge and assigns the staples needed to fold the target object. An atomic model is also generated to allow researchers to visualize the object and design molecular functionalization patterns. Using the computationally designed set of oligonucleotide sequences, the DNA nanoparticle is self-assembled via thermal annealing with an excess of staples over scaffold, which is typically based on the M13 phage or a synthetic sequence. Folding is characterized using agarose gel electrophoresis and structural analysis using atomic force microscopy (AFM), transmission electron microscopy (TEM), or 3D cryo-electron microscopy (cryo-EM). Adapted with permission from Reference 186. (b) A vast number of different wireframe architectures of arbitrary shape can now be designed from the top down using algorithms such as DAEDALUS (186; http://daedalus-dna-origami.org/) and PERDIX (86; http://perdix-dna-origami.org/). Adapted with permission from References 86 and 186. (c) Biomolecular functionalization on wireframe objects can be controlled with single-molecule precision. Strategies for the functionalization of staples at 3′ or 5′ locations include hybridization of single-stranded DNA overhangs with complementary locked nucleic acid or peptide nucleic acid sequences and direct, covalent chemical modifications.
Figure 2.
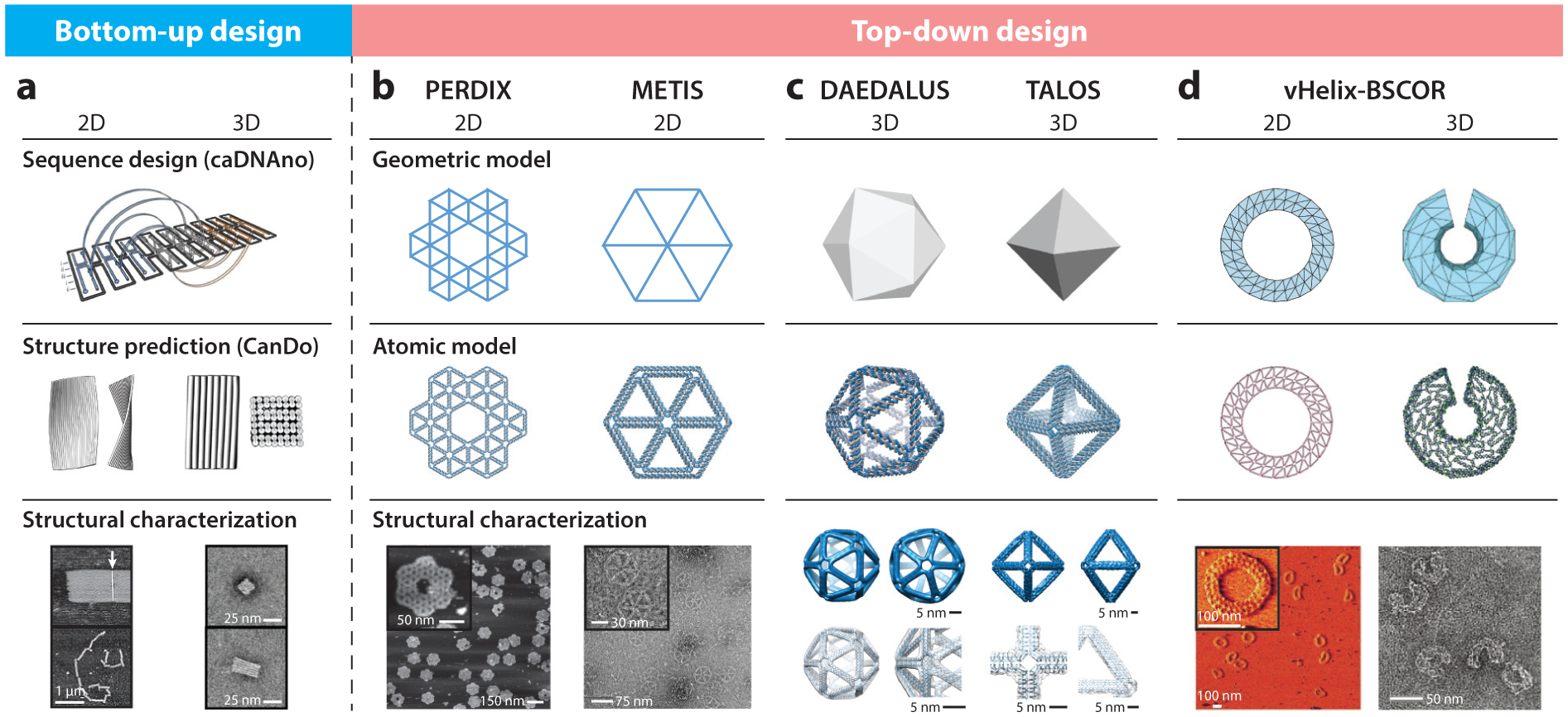
Computational design strategies for scaffolded DNA origami nanostructures. Following the early development of the bottom-up sequence design program caDNAno for rectilinear, brick-like scaffolded DNA origami assemblies, several approaches for the top-down, fully automated design of wireframe scaffolded DNA origami objects were developed, including PERDIX, METIS, DAEDALUS, TALOS, and vHelix-BSCOR. (a) Sequence designs from caDNAno can be imported into the online software CanDo to predict three-dimensional (3D) solution structures. Adapted with permission from References 42, 43, 92, and 143. (b) Both PERDIX and METIS enable the design of any free-form 2D geometry using either DX- or honeycomb-based edges, respectively. Adapted with permission from Reference 86. (c) DAEDALUS and TALOS program arbitrary 3D polyhedral geometries using DX- or honeycomb-based edges, respectively. Adapted with permission from References 186 and 85. (d) vHelix-BSCOR enables the semiautomated top-down design of both 2D and 3D wireframe objects using predominantly single-helix edge architectures (right). Adapted with permission from References 17 and 18. Structural characterization of the target objects is typically achieved using atomic force microscopy for 2D assemblies (a, bottom left, b, bottom left, and d, bottom left, where the white arrow indicates blunt-end stacking of Rothemund rectangles in panel a), cryo-electron microscopy with 3D reconstruction (c, bottom left and right) or transmission electron microscopy (a, bottom right; b, bottom right; d, bottom right)
These preceding advances from the past decade, namely scaffolded DNA origami, wireframe design, and full automation of sequence design, together with recently developed strategies for sequence-specific scaffold production using either templated polymerase chain reaction (PCR) (95, 186, 187) or phage production (120, 134, 163a), have enabled the facile fabrication of nearly any 2D or 3D target nucleic acid assembly imaginable on the 10–100 nm scale.
The general workflow employed for wireframe DNA origami fabrication is illustrated with the top-down design software DAEDALUS (Figure 1a) (186). First, the user generates as input to the software a target 3D polyhedron of arbitrary shape. This target polyhedron is then converted automatically into the staple sequences needed to fold the desired scaffold into the target shape through unsupervised computational steps that include scaffold routing and staple design to ensure robust, high-yield folding. Additionally, an atomic model is generated for 3D visualization for custom design applications that include chemical modifications. Once staple sequences have been determined, wireframe origami synthesis proceeds as conventionally performed (28, 143) by thermal annealing with a tenfold molar excess of staples over scaffold, using a folding buffer (40 mM Tris, 20 mM acetic acid, 2 mM EDTA, and 12 mM MgCl2 at pH 8.0), which can be exchanged postfolding with phosphate buffered saline or other physiological buffers for applications in molecular and cellular biophysics (186). Purification is then typically performed using agarose gel electrophoresis, centrifugal concentrators, density gradients, polyethylene glycol precipitation, or chromatography (28, 100, 134, 146, 161).
Following DNA origami synthesis, the predicted atomic model may be validated experimentally using atomic force microscopy or transmission electron microscopy. Although technically more challenging, cryo–electron microscopy (cryo-EM) offers the possibility of 3D reconstruction to elucidate 3D structure with near single-duplex resolution (12, 86, 186). Advanced single-molecule fluorescence imaging using points accumulation for imaging in nanoscale topography (PAINT) (84, 87, 175) or single-molecule Förster resonance energy transfer (FRET) (145, 173) can also be applied to characterize nanoscale topology and dynamic intramolecular distances, whereas quantitative PCR (170, 186), ultraviolet-visible spectroscopy, and solution-based FRET (146) can be used to characterize thermodynamic stability.
Physics-based computational models complement experimental data and target geometric models to offer insight into conformational flexibility, mechanical properties, and potential structural deviations from target designs. Although molecular dynamics simulations represent the gold standard for atomic-level structural modeling, they are limited to a few select groups due to the intensive computational resources needed to simulate DNA origami even on relatively short timescales of nanoseconds. Hence, the coarse-grained modeling approaches CanDo, which uses finite elements (28, 92, 129, 130), and OxDNA, which uses a bead-based approach (169, 176), have played leading roles in the practical design and simulation of DNA origami assemblies. They are commonly used to elucidate subtle structural details such as twist and bend in rectilinear origami objects (28, 92), as well as multiway junction conformations and conformational flexibility.
Toward functional biophysical applications of DNA origami, conjugation of bioactive molecules or fluorophores at the 3′ and 5′ ends of staples is routinely performed using either covalent chemistries or simple hybridization to single-stranded 3′ or 5′ overhangs (Figure 1c). Internal oligonucleotide modifications can also be utilized, although they are generally costlier unless in-house DNA synthesis is performed. More sophisticated DNA origami functionalization strategies have recently been reviewed in detail elsewhere (62). Because each DNA base in a given scaffolded DNA origami object is known a priori from the target design with single-molecule precision and addressability, applications in which asymmetric positioning of bioactive molecules is utilized are straightforward to realize. This is particularly powerful in the case of wireframe assemblies that offer both internal and external presentation of 3′ and 5′ staple ends, which are easily modified to adjust their inward versus outward facing angles within approximately 34° and 0.34-nm accuracy in the B-form DNA duplex. This powerful spatial control of molecular presentation offered by DNA origami, combined with the versatility in geometric control offered by wireframe design, now offers full control over the ability to mimic and interrogate biophysical processes using structured presentation of nucleic acids, proteins, carbohydrates, and small molecules. Integration of external signals to perform logical operations offers yet another entirely distinct arena of temporal control over structured DNA assemblies, which is reviewed comprehensively elsewhere (15, 167, 200).
These foregoing properties render structured DNA assemblies unique biophysical research tools compared with other nanomaterials including liposomes, dendrimers, polymers, and viral protein mimics (32, 36, 104), which offer low-cost, large-scale production for in vitro applications, but lack the ease and versatility of geometric design, together with the capability of asymmetric, orthogonal internal and external molecular functionalization. To highlight the unique power and versatility of this synthetic structural biology approach, we feature three select research applications of wireframe DNA origami that we anticipate will have transformative impact on molecular and cellular biophysics in the coming decade: light harvesting and nanoscale energy transport, RNA structural biology, and immune receptor signaling.
BIOLOGICALLY INSPIRED LIGHT HARVESTING
Natural photosynthetic complexes absorb sunlight and transport energy to their reaction centers with remarkable efficiencies. These efficiencies are due to several design principles, including directional energy transport and a high cross-sectional area for light absorption and transfer, such as in the chlorosome found in the light-harvesting complex of green sulfur bacteria (Figure 3a) and the phycobilisome antenna found in cyanobacteria (Figure 3b). Strong electronic interaction of chromophores because of dense packing in these light-harvesting complexes leads to coherent sharing of light excitation or excited-state delocalization among several chromophores, which in turn alters the absorption cross section and energy-transfer rates in photosynthetic antennae (149). These design principles are achieved by embedding photosynthetic chromophores within protein complexes (Figure 3c,d). Considering that this leads to highly efficient energy absorption, transfer across nanometer to micrometer length scales, and energy conversion, a fundamental question is whether natural photosynthetic complexes can be mimicked using synthetic, biologically inspired light-harvesting systems. DNA nanotechnology offers the ability to implement the hierarchical, dense organization of chromophores that is typically observed in natural photosynthetic complexes. Specifically, the bottom-up synthesis of biologically inspired light-harvesting systems based on DNA, with unprecedented single-molecule addressability in the position and orientation of chromophores, offers the unique opportunity to test long-standing hypotheses in the field of light harvesting regarding nanoscale energy transport.
Figure 3.
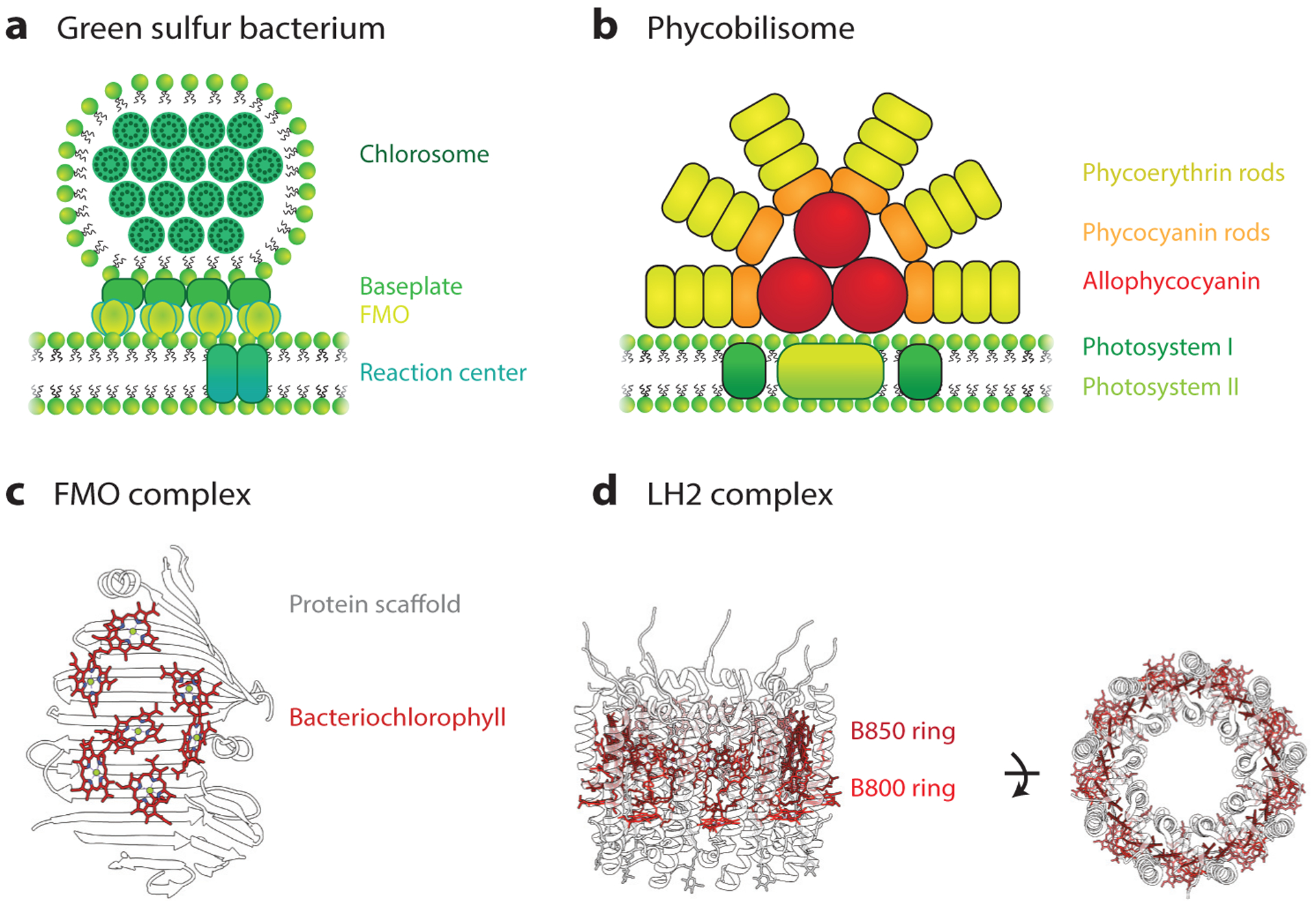
Natural photosynthetic light-harvesting systems. (a) Schematic representation of a light-harvesting complex in the green sulfur bacterium. Chlorosomes have high-absorption cross sections that harvest light and transfer energy to the Fenna-Matthews-Olson (FMO) complex through the baseplate. Absorbed energy is then transferred to reaction centers. (b) Illustrative representation of the phycobilisome light-harvesting antennae. Bilin-containing proteins, phycoerythrin, phycocyanin, and allophycocyanin, absorb light in the green, orange, and red regions of the visible light spectrum. The energy-transfer cascade from the phycoerythrin to allophycocyanin via phycocyanin funnels the energy to the photosystem I and II reaction centers (59, 60, 119). (c) Protein scaffolds in the FMO control the spatial organization and influence the site energies of bacteriochlorophyll pigments (184) [Protein Data Bank identification (PDB ID) 3EOJ]. (d) The light-harvesting complex (LH2) in purple bacteria has a characteristic circular structure, which contains the B800 and B850 rings (PDB ID 2FKW) (31). Proteins control the spatial organization of bacteriochlorophyll pigments in the B800 and B850 rings. These different organizations lead to exciton delocalization (118) and fast energy transfer (105).
Toward this end, several synthetic DNA-dye systems have been fabricated to operate in the weak-coupling regime where incoherent or hopping transport is prevalent and can be described using Förster theory. One-dimensional energy transfer along DNA duplexes is the most common approach to investigating energy transfer. For this purpose, dyes can be incorporated onto the DNA duplex scaffold through intercalation (35, 68), groove binding (8, 88, 156, 171, 196), or covalent attachment (4, 47). Energy transfer between similar dyes, or energy migration along the DNA duplex, can be mediated by these incorporated dyes. Energy migration functions as a bridge between primary donors and acceptors in donor-bridge-acceptor systems. Another approach to bridge primary donors to acceptors is to design a downhill energy-transfer pathway. This can be achieved by covalently attaching dyes with cascading excited-state energies on the DNA structural scaffold (70, 112). In contrast to an energy-migration approach in which energy transport proceeds through a random walk, cascaded energy transfer can be directed toward the primary acceptor. This approach can be used to realize downhill energy-transfer distances of up to several tens of nanometers with relatively high efficiencies (70).
Scaffolded DNA origami enables the synthesis of larger, more modular and rigid light-harvesting structures with denser dye functionalization patterns compared with earlier approaches, such as DNA duplexes and branched DNA structures (22). Because of the versatility of DNA origami, it is feasible to modify energy-transfer pathways of donor-acceptor pairs and control nanoscale energy transport (71, 123). Perhaps even more importantly, DNA origami allows for the investigation of energy transfer in larger 3D geometries, emulating energy-transfer pathways in natural light-harvesting systems (Figure 4a) (44, 128).
Figure 4.
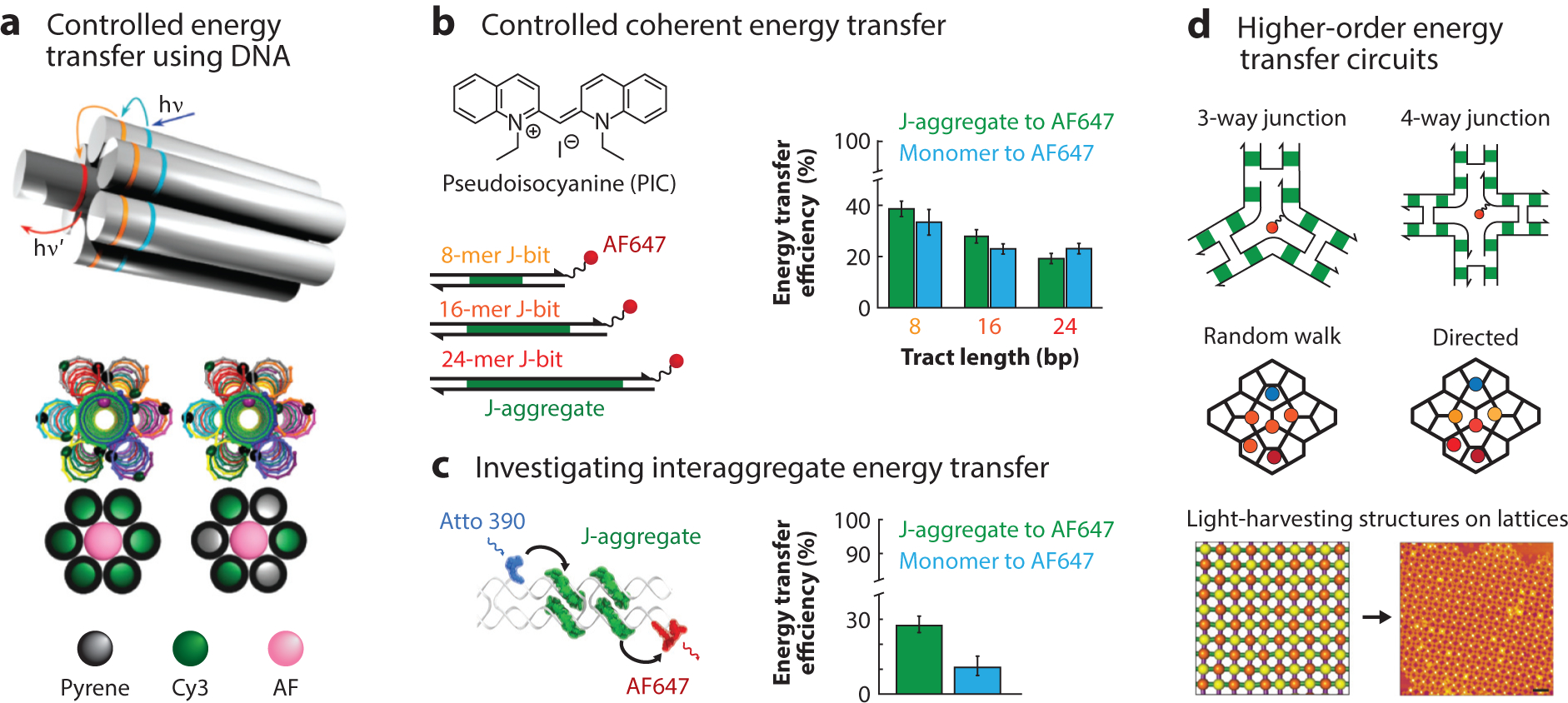
Biologically inspired artificial light-harvesting systems. (a) Three-dimensional energy transfer on DNA duplex bundles. DNA nanostructures enable the controlled variation of the distances and numbers of dyes scaffolded to investigate directed energy transfer. Adapted with permission from Reference 44. (b) The energy-transfer efficiency of DNA-templated PIC J-aggregates to Alexa Fluor® 647 decreases with the length of the DNA template due to static disorder. Adapted with permission from Reference 14. (c) The sequence-selectivity of PIC aggregation presents an opportunity to create higher-order excitonic circuits to understand the dynamics of interaggregate energy transfer. Adapted with permission from Reference 19. (d) DNA nanostructures can be leveraged as designer nanoscale scaffolds to understand energy funneling and directed energy transport mechanisms that are typically found in natural light-harvesting systems. The ability to program light-harvesting structures across different length scales using DNA, from the nanoscale distance of dyes to the microscale organization of light-harvesting DNA constructs (102), provides a path toward mimicking photosynthesis. Adapted with permission from Reference 102. Abbreviations: AF, Alexa Fluor® AF647, Alexa Fluor® 647; bp, base pair; Cy3, C3-indocyanine; PIC, pseudoisocyanine; hv, energy of an incoming photon; hv′, energy of an outgoing photon; J-bit, specific, noncovalent complex of aggregated PIC monomers templated by an A-tract of duplex DNA.
As synthetic light-harvesting scaffolds, DNA nanostructures have provided numerous experimental demonstrations of programmed energy-transfer pathways that are designed through precise control over the positions and orientations of chromophores. Extending these designer characteristics of DNA scaffolds to also control the electronic coupling strength of chromophores may provide mechanistic insights into the role of strongly interacting chromophores, a molecular feature observed in photosynthetic chromophore assemblies in natural light harvesting. Indeed, several examples of strongly interacting dye systems scaffolded by DNA have emerged recently. Cyanine (25, 109) and squaraine dyes (109) that are covalently attached to DNA as dimers across the DNA duplex show spectroscopic features suggestive of Davydov excited-state splitting, characteristic of strongly coupled dye aggregates. Similarly, porphyrins, a class of molecular heterocycles involved in photosynthetic light harvesting, show strong electronic interactions when covalently attached to DNA (188). Recent studies of the sequence-selective formation of pseudoisocyanine J-aggregates on DNA duplexes (Figure 4b,c) (14, 19) by our group observed excited-state delocalization lengths that are comparable to measured excited-state delocalization lengths in natural light-harvesting systems (118, 135). Scaffolding these J-aggregates on DNA constructs allowed for the investigation of the effect of inhomogeneity of excited-state energies on the overall energy-transfer process (14) and interaggregate energy transfer (19), photophysical processes that are challenging to probe in other self-assembled aggregates (Figure 4c) (20, 65).
The preceding applications provide a broad overview of the potential for using DNA as a scaffolding material for distinct chromophores, including the programming of both long-range and short-range energy transfer. Remaining challenges include integrating these systems into photosynthetic antennae or energy storage systems, with energy-transfer efficiencies that approach those of natural systems. Specifically, in a DNA-dye complex, the following spatial design principles for light harvesting are necessary to compete with the efficiencies found in nature: (a) designing unidirectional energy-transfer pathways to funnel absorbed photon energies to artificial reaction centers, (b) positioning and orienting chromophores precisely on DNA scaffolds, and (c) integrating the hierarchical organization of different light-harvesting and energy conversion elements (Figure 4d).
In addition to long-range energy-transfer mechanisms in natural photosynthesis, the importance of short-range coherent effects between electronic and vibrational states on energy-transfer efficiency remains a point of debate. Electronic coherence occurs when electronic states of closely packed chromophores strongly interact with one another, creating a delocalized manifold of excited states. Dynamic motion of these excited states along the chromophores can occur on femtosecond timescales. In contrast, electronic-vibrational coherence occurs when the strength of electronic interaction between chromophores is similar in magnitude to a vibrational mode. Although the existence of these types of coherences has been observed in photosynthetic systems (46), the function of coherence in the context of natural light harvesting remains elusive. For example, persistent amplitude oscillations observed in the multidimensional spectra of the Fenna-Matthew-Olson complex, a photosynthetic apparatus (Figure 3c), revealed the complex mixing of electronic and vibrational states that can affect photosynthetic energy transfer (106, 179). DNA origami has the potential to develop tractable model systems that have diverse dye geometries, orientations, and electronic structures to reveal coherence effects present in natural light harvesting (141).
Due to their ubiquity in photosynthetic complexes, closely packed chromophores offer an important technological capability for synthetic DNA-dye scaffolded complexes. These DNA-dye complexes may be used to elucidate the role of quantum coherent states in natural and synthetic systems, as well as to investigate the interplay between long- and short-range energy-transfer effects. Single-molecule control over chromophores templated by DNA origami scaffolds offers a versatile molecular toolbox to investigate distinct energy-transfer pathways and mimic key structural elements found in natural light-harvesting systems. Exploration of the convergence of synthetic light-harvesting technologies with natural light-harvesting systems will likely be of major interest in the next decade.
RNA STRUCTURAL BIOLOGY
In contrast to the high fidelity of Watson-Crick base-pairing that leads to highly predictable DNA secondary structure formation, which has enabled the field of structural DNA nanotechnology, the prediction of secondary and tertiary structures of long RNAs remains challenging (116, 160). This is largely due to the relatively subtle chemical differences between DNA and RNA, which also lead to diverse RNA catalytic activities that have been exploited in synthetic biology applications, expertly reviewed elsewhere (80). Two examples of chemical differences are the additional 2′ hydroxyl group on the ribose and the uracil instead of the thymine bases found in DNA, complicating the prediction of RNA secondary and tertiary structure due to noncanonical base-pairing through sugar edge and guanine-uracil wobble base-pairing interactions, respectively. Taken together, these complexities motivate the need for new tools to determine the 3D structural and dynamical properties of long coding as well as noncoding RNAs.
In the cell, the dynamics and heterogeneity of secondary and tertiary RNA structures are leveraged to regulate translational and catalytic activity (23, 138, 157). Furthermore, viral RNA genomes, including Ebola and human immunodeficiency virus (HIV), contain numerous unique structural motifs due to their large mutation space and selective pressure toward maintaining small genomic sizes (76). Elucidating these preceding genomic structures and architectures may aid in identifying new targets for drug discovery (108), analogous to the discovery of novel structure-activity relationships garnered from understanding mechanisms of antibiotic binding to the ribosome (192). New insights into RNA tertiary structure formation should also improve the rational design of RNA aptamers (45, 185) and ribozymes for biological engineering applications (83, 89, 127, 159).
In structural RNA nanotechnology, mechanistic sequence-based knowledge of secondary and tertiary RNA structure formation has already enabled substantial advances. Specifically, instrumental work pioneered the generation of nanoparticles using the tectonics approach that leverages the combination of structural subunits such as tRNAs with naturally occurring junctions (2, 37, 158, 164, 190). In addition, employing native RNA interactions such as kissing loops or paranemic crossovers has programmed RNA-based structures to fold predictably from a single long strand (1, 58, 66, 93), with applications ranging from molecular capture and sensing in cells to the development of therapeutics (74).
Notwithstanding, little remains known about the native tertiary structure of long RNAs, including their stability, dynamic structural interconversions, and structural stabilization by RNA-binding proteins. And while the number of known RNA tertiary structures is growing, most studies report on small fragments or rigid ribonucleoprotein complexes like the ribosome. In contrast, a paucity of 3D structural data is available for viral RNA genomes. For example, only 3% of the HIV genome has known tertiary structure according to an analysis of the Protein Data Bank. This is largely due to the lack of 3D structural characterization tools for longer RNA molecules that adopt variable conformations, which are not amenable to nuclear magnetic resonance or crystallization for X-ray crystallography. Consequently, methods to probe local secondary structures and higher-order tertiary interactions in large RNAs are still actively being explored, as previously reviewed (116, 160). Notable successes for secondary structure prediction have been realized using physics-and homology-based modeling approaches together with chemical probing, and, to a lesser degree, in tertiary structure prediction (9, 38, 50, 73, 114–116, 139, 165, 205). In contrast with long, flexible RNAs, compact RNAs are often structurally well-ordered, making them amenable to X-ray crystallography to resolve their high resolution tertiary structure (13, 64, 90, 148, 183, 193, 203). In addition, their small size makes them suitable for characterization using nuclear magnetic resonance (91, 202) and small angle X-ray scattering (34, 48) in solution, as well as single-molecule FRET (37, 132, 182) to resolve their conformational dynamics. Cryo-EM offers an alternative for high-resolution RNA tertiary structure determination (57), yet to date the number of available high-resolution structures is limited, likely due to conformational heterogeneity that limits the accuracy of class averages needed to reconstruct 3D architecture.
A major exception, and indeed one of the greatest achievements of RNA structural biology, is the high-resolution tertiary structure of the ribosome (Figure 5a). Through decades of research, we now know with near atomic resolution the organization and dynamic motions of the ribosome, including the peptidyl-transferase center (PTC) with its coordination of tRNAs at the P and A sites. This center facilitates catalytic transfer of the growing polypeptide chain to the next amino acid while ratcheting along mRNA in a cyclical, coordinated dance (174). Atomic-resolution structures of the entire ribosome and its assembly intermediates have allowed for a considerably deeper understanding of the folding and reactivity of the PTC (Figure 5b) (124). The ability to obtain these high-resolution structures, however, was in part due to the conformational stabilization of ribosomal RNA by protein-RNA interactions, a principle that might be viewed as nature’s RNA origami, and indeed may also inform strategies for controlling other RNA tertiary structures for structural and synthetic biology.
Figure 5.
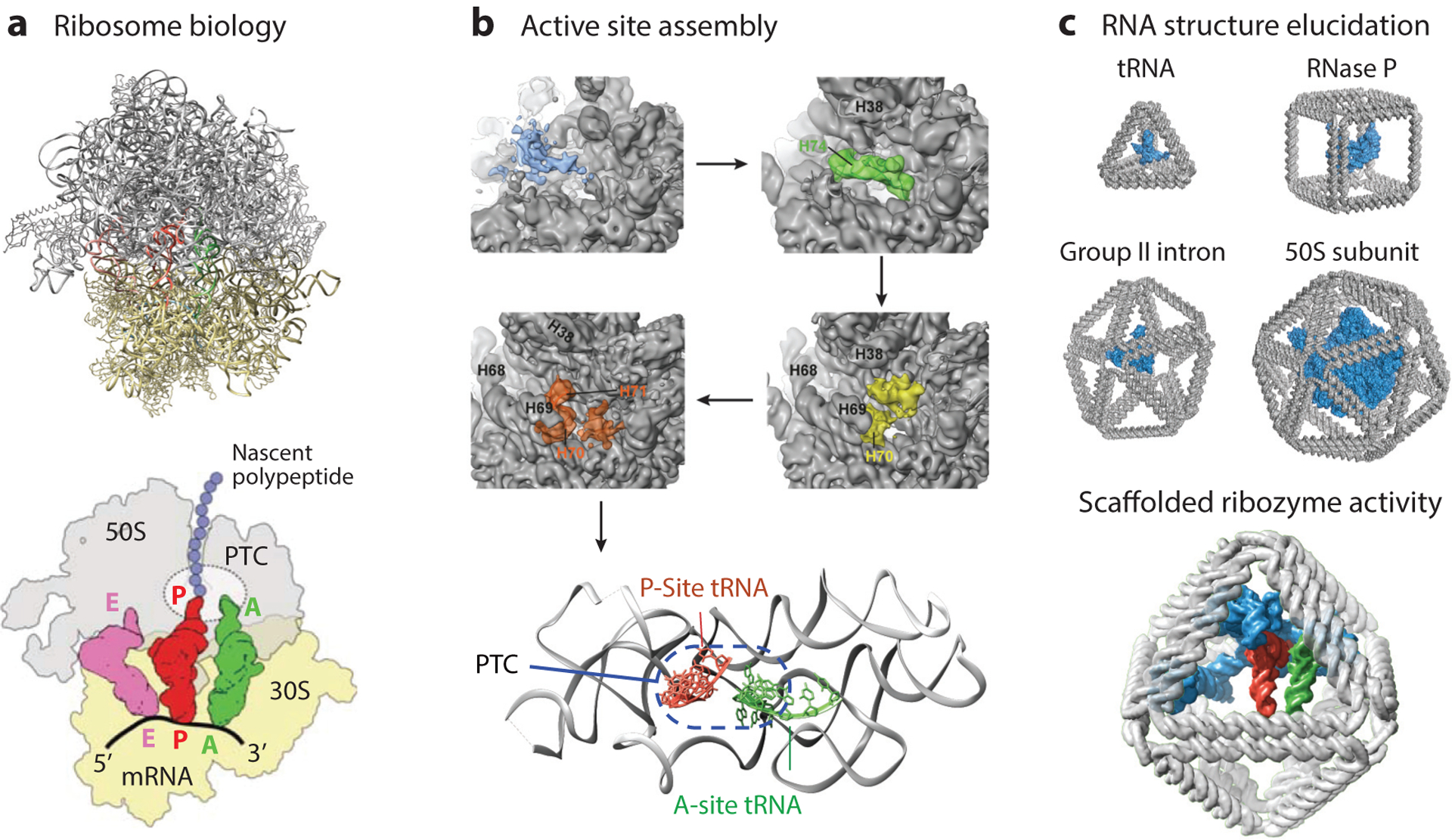
RNA tertiary structure and function in natural and synthetic systems. (a) The structure of the ribosome has been solved to atomic resolution (top) [Protein Data Bank identification (PDB ID) 1GIY and 1JGO; References 198, 199], which has yielded biological insight into ribosome activity (bottom; adapted with permission from Reference 7), including coordination of tRNAs in the peptidyl-transferase center (PTC). (b) Folding intermediates of the ribosome show late assembly of an active PTC (adapted with permission from Reference 124), with the active conformation in the bottom panel (PDB ID 4V6F; Reference 82). (c) DNA origami objects of different sizes and shapes may be used to coordinate RNA structures, modeled for size comparison here with a tRNA (PDB ID 1WZ2), RNase P (PDB ID 3Q1R; Reference 140), group II intron (PDB ID 4DS6; Reference 29), and the 50S subunit of a ribosome (PDB ID 2WWQ; Reference 155), for applications in RNA structural biology (top) and synthetic catalysts (bottom) with a hypothetical minimized PTC, shown in blue (modified from PDB ID 1GIY; Reference 198), modeled into an octahedral origami. The P-site tRNA is shown in red, the A-site tRNA in green, and the E-site tRNA in purple.
One path toward engineering such conformational stability might employ wireframe DNA origami. Specifically, wireframe DNA origami may be used to coordinate RNAs in 3D space to elucidate their tertiary structure, probe conformational population dynamics, interrogate protein-RNA interactions, and investigate RNA domain folding and maturation (Figure 5c). Indeed, furthering structural biology has been a longstanding goal of DNA nanotechnology, as originally proposed by Seeman (151), with recent advances including the coordination of proteins using DNA origami assemblies (172, 180) and the formation of crystalline DNA lattices (168, 204). The coordination of long RNAs using engineered loops as part of wireframe DNA origami staple or scaffold sequences might allow for the structural stabilization of an RNA molecule in a native conformation within the origami. In an ideal case, restricting RNA tertiary conformation using such tertiary interactions could subsequently help cryo-EM elucidate biologically relevant tertiary structures of long RNAs. Integrating such structural studies with single-molecule FRET may further reveal conformational heterogeneity and substates that are otherwise impossible to resolve using crystallography and solution- or surface-based studies that interrogate RNAs in their isolated, native state.
Alternatively, the ability to complex RNAs to wireframe DNA origami in precise spatial positions and orientations may offer the opportunity to engineer synthetic PTC activity outside of the ribosome, which otherwise requires coordination by ribosomal proteins (Figure 5c) (5). Successful applications of structured DNA nanoparticles in this regard may even enable engineering orthogonal translation systems (121, 125) and provide new tools to investigate RNA modification biochemistry (63, 136, 137), and ribosome assembly and folding (178), while also contributing to the development of minimal synthetic translation systems (51, 163).
IMMUNE RECEPTOR SIGNALING
Cell membrane receptors facilitate the sensing of biochemical information and the transport of material across the plasma membrane, enabling cells to interact with, and respond to, their environment. Among them, immune receptors represent an important class of molecules that includes the innate immune pathogen recognition receptors as well as highly specific antigen receptors of the adaptive immune system on the surface of T and B cells (Figure 6a). These receptors have evolved to differentially sense a variety of antigens and pathogen-associated molecular patterns (PAMPs) including carbohydrates and lipids, as well as protein or peptide epitopes (53). Important parameters of immune recognition include the spatial organization of antigens and PAMPs, specifically the number and stoichiometry of ligands (72) and their 3D organization (24, 33). They govern the coordination of downstream signaling and thereby help to distinguish self from foreign pathogens (113), and also differentiate pathogens from commensal microbes (3). Understanding underlying mechanisms of immune receptor function promises new insights into cellular responses that drive allergic reactions (56, 147), autoimmune diseases (111), and cancers (131, 150). In addition, this improved understanding may impact the design and development of more efficient vaccines and cancer immunotherapies.
Figure 6.
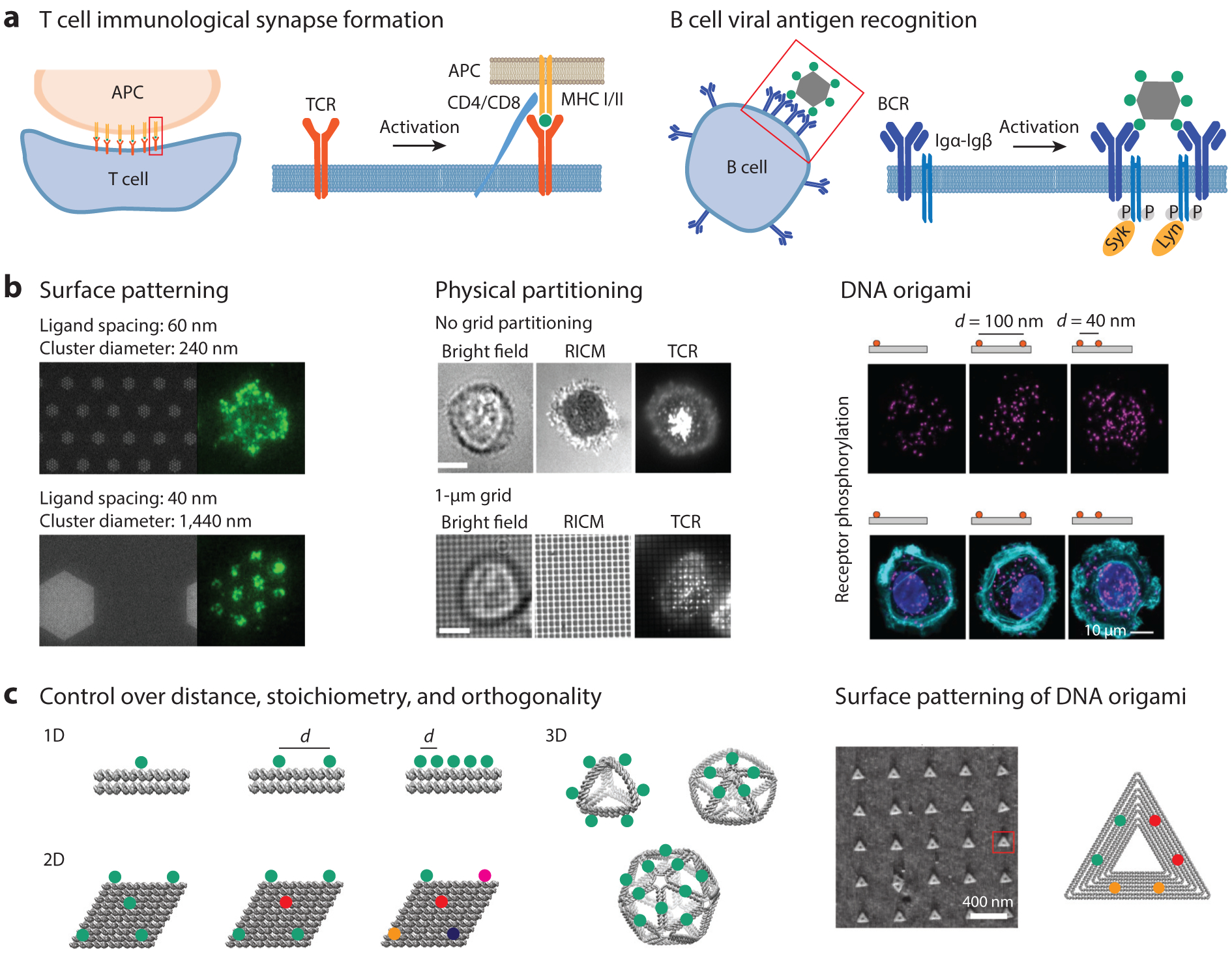
Templated antigen and ligand systems to probe immune cell surface receptor signaling. (a) Schematic of TCR and BCR activation showing the importance of the nanoscale organization of receptors on triggering immune responses. (b) Effects of TCR ligand density and cluster size on TCR activation (left). Adapted with permission from Reference 24. A minimum number of TCR ligands is needed to induce T cell synapse formation (middle). Adapted with permission from Reference 107. DNA origami has been used to present ligands at specific distances to study receptor activation (right). Adapted with permission from Reference 162. (c) DNA origami nanoparticles can be designed to investigate the effects of antigen distance, stoichiometry, dimensionality, and multiplexing through site-specific control of functionalization. Additionally, DNA origami can be used for surface patterning to study the effect of antigen precluster distance on immune receptor activation. Atomic force microscopy data adapted with permission from Reference 61. Abbreviations: APC, antigen-presenting cell; BCR, B cell receptor; CD4/8, cluster of differentiation 4/8; Igα/β, immunoglobulin-α/β; MHC, major histocompatibility complex; RICM, reflection interference contrast microscopy; TCR, T cell receptor; Syk, spleen tyrosine kinase; Lyn, Lck/Yes novel tyrosine kinase.
Immune receptors are expressed on a variety of cell types, including nonimmune cells, and perform their functions both on the surface of the cell and within internal compartments. Often, cellular sensing of PAMPs or antigens occurs in the context of a specialized tissue compartment such as the lymph node, involving either a portion of a, or an entire, pathogen surface. Sensing may be coordinated with a fixed complement, and it may occur in the context of cell–cell synaptic adhesions. Immune receptor signaling is commonly regulated by coreceptors and cross talk between transmembrane receptors that sense environmental cues in a spatially dependent manner (27, 77, 81). Coordination between immune cell surface receptors has been shown to be involved in HIV entry into T cells and other viral infections (101). Although these complex factors can influence the behavior and fate of the cell downstream from receptor signaling events, understanding mechanistically how receptor clustering of, and coordination between, adaptive immune receptors (Figure 6a) influences signaling and cellular fate remains challenging (69).
Materials that mimic the nanoscale organization of antigens or PAMPs allow for certain features of immune receptor activation to be reproduced in vitro, facilitating a mechanistic understanding of their relative contributions (Figure 6b). These materials include supported lipid bilayers with tethered antigens (24), which facilitate the study of surface-tethered antigens, and can be extended to systems mimicking antigen-presenting cells. Lipid vesicles (26) as well as patterned surfaces (40, 107) have been used to control clustering and partitioning of T cell receptor (TCR) ligands to study immunological synapse formation, including the critical number of ligands needed for TCR activation (Figure 6b). In addition, multivalent chemical polymers (16), synthetic antigen peptides (10), engineered phages (94), and viral (11) systems have helped shed light on the effects of affinity and multivalency on receptor activation.
Importantly, co-ligation of receptors and coreceptors has highlighted the cooperativity between immune receptors (27, 78), and trivalent presentation of Toll-like receptor 9 (TLR9), TLR7, and TLR4 agonists has demonstrated synergistic behavior between distinct TLRs (181). Many of these systems display limitations, however, with regard to controlling rigidity, stoichiometry, orthogonality, and placement of ligands and antigens. By contrast, DNA-based materials, and in particular scaffolded DNA origami, are able to meet these demands because they offer independent, high degrees of control over antigen placement, stoichiometry, type, dimensionality, and mechanical compliance (42, 85, 86, 143, 186) (Figure 6c), as well as biocompatibility (177, 186).
While only several examples have been published to date, the potential for structured DNA assemblies to offer mechanistic insight into the geometric requirements of immune receptor signaling and function has been demonstrated. For example, trimeric DNA duplexes were used to template dinitrophenyl antigens in a mast cell–IgE system. By increasing the spacer distance between the dinitrophenyl monomers, the authors showed a dependence of IgE receptor signaling on interantigen distance, suggesting cooperativity between the receptors (166).
Several studies on nonimmune receptors have also been reported and highlight the value of DNA nanostructures for controlled ligand presentation. Recently, a large scaffolded DNA origami nanostructure was used to present dimers of ephrin ligands to MDA-MB-231 cells expressing the ephrin receptor EphA2 (Figure 6b) (162). Here, the authors compared monovalent rod-like DNA nanoparticles with dimers displaying either 40-nm or 100-nm interligand distances to demonstrate the impact of ligand organization on receptor signaling, and established the maximum distance allowed for EphA2 activation. In a separate study, Niemeyer and colleagues (6) used a surface-based DNA origami system to pattern the ligand epidermal growth factor (EGF), exploring the effects of interligand distance and stoichiometry on EGF receptor activation. Their results suggested that placing EGF 5 nm apart yielded lower cellular responses than spacings greater than 30 nm.
Taken together, these studies indicate that nanoscale control over ligand organization may be used to further our understanding of the roles played by multivalent binding, cooperativity, and receptor clustering on immune recognition. Future work might pursue reading out engaged ligand-receptor binding interactions while controlling ligand presentation using DNA origami. DNA-PAINT has already proven partially useful for this aim, having been used to confirm the incorporation and accessibility of a single-stranded target DNA present on DNA origami (175). Structured DNA has also been used to understand how stoichiometry impacts TLR signaling. Indeed, immune cells are particularly sensitive to CpG DNA sequences, which are abundant in bacterial genomes, and are recognized by the pattern recognition receptor TLR9, as well as other pattern recognition receptors (21, 117). Comparing the immunostimulatory response of DNA nanoribbons functionalized with CpG to monomeric presentation of CpG showed a marked increase in immune stimulation of the highly repetitive nanoribbon (126). Moreover, similar structures have already been used in vivo as vaccine platforms (103) and in modulating immune responses (79).
DNA-based materials have only just begun to shed light on the molecular requirements of immune cell surface receptor function. To date, studies have focused largely on CpG-modified DNA nanomaterials because of the facile attachment of oligonucleotides using base-pairing interactions. Functionalization of DNA origami with antigens can facilitate the study of a broader range of immune receptors, where functionalization relies on a variety of chemical techniques to attach peptide and protein ligands to preserve antigen and ligand function following attachment to scaffolded DNA origami objects (Figure 6c). These features can add to the growing body of knowledge on the cooperativity between distinct cell surface receptors, serving as tools to study complex signaling networks and pathogen discrimination by immune cells. Improved tools for the design and synthesis of DNA origami may aid efforts to understand the dependence of receptor responses on inter-antigen spacing and other geometric features, where the facile design of customizable DNA origami allows for full site addressability of ligands and antigens in arbitrary nanoscale geometries.
CONCLUSIONS AND PERSPECTIVES
Controlling the spatial organization of biomolecules using structured DNA assemblies offers tremendous potential for molecular and cellular biophysical research. Unprecedented control over the stoichiometric loading, geometric configuration, and mechanical properties of scaffolded DNA origami offers researchers the ability to mimic and manipulate biomolecular organization on the 1–100-nm scale to advance our mechanistic understanding of biological structure and function in areas ranging from light harvesting to RNA structural biology to immune receptor signaling.
In biologically inspired light harvesting, early approaches employed linear and branched DNA duplexes to organize chromophores to control directional energy transport and enhance absorption efficiency. In comparison, DNA origami has recently enabled the synthesis of considerably more elaborate, modular light-harvesting structures that mimic important aspects of natural photosynthetic systems (44, 71, 195). The continued exploration of these biologically inspired systems will be further enabled by advances in structural DNA nanotechnology that leverage 2D and 3D wireframe designs. These advances include the ability to construct hierarchical chromophore assemblies that integrate long-range and short-range transfer mechanisms to reveal fundamental insight into natural mechanisms of nanoscale energy transport and light harvesting (14, 19).
Similarly, wireframe DNA origami offers a uniquely powerful tool to address challenging questions in RNA structural biology. Structured DNA nanoparticles with convex exteriors and concave hollow interiors can be employed to capture flexible viral RNAs by presenting selective single-stranded overhang motifs that are complementary to single-stranded RNA regions revealed by chemical footprinting. This approach may result in the stabilization of specific tertiary structure conformations that can subsequently be elucidated using single-particle cryo-EM. Incorporation of RNA-binding proteins and chemical modifications can be used to test their impact on structural heterogeneity. Alternatively, the precisely controlled 3D geometry of structured DNA assemblies can be leveraged to scaffold RNA motifs to test hypotheses surrounding the structural origins of their catalytic activity, such as in scaffolding catalytic RNAs to synthesize artificial PTCs, with the potential to provide fundamental insights into ribosome evolution and function.
Finally, controlling the 1D, 2D, and 3D spatial organizations of immunogens on the 1–100-nm scale offers the important potential to reveal mechanisms of immune recognition and signaling. Unlike natural or synthetic viral constructs for immunogen presentation, structured DNA assemblies offer full control over copy number, spacing, and the dimensionality of presentation, providing room to study how these independent structural features of pathogens have evolved to interact with immune cells. Basic questions surrounding immune receptor clustering or kinetic segregation of phosphatases and kinases that are thought to depend on these structural features can now be tested directly using DNA origami.
The emergence of fully automated top-down design strategies for 2D and 3D wireframe structures over the last few years, together with established protocols for molecular functionalization and scaffold synthesis, should help to disseminate more broadly the application of scaffolded DNA origami to diverse research areas in molecular and cellular biophysics. In addition to the several highlighted areas of focus in this article, namely light harvesting, RNA structural biology, and immunology, we anticipate significant impact in related areas as DNA origami continues to become more accessible and its unique power to address fundamental biological questions continues to be demonstrated.
Although this article focused largely on static structural assemblies, DNA nanotechnology offers equal if not more room for impact when leveraging its ability to sense external signals and respond dynamically using logical operations both in vivo (41, 98) and in vitro (110). These dynamic operations offer additional spatial-temporal control, for example, in the induction of immune signaling or ligand binding. Further, the mechanical properties of origami can be controlled passively as well as dynamically to sense and transduce mechanical stresses in situ, offering additional opportunity for studying membrane biophysics and signaling, as well as other areas of molecular biophysics (55, 97, 122).
Given the very recent rise of computational tools to perform fully automated sequence design, as well as predict 2D and 3D structural and mechanical properties, substantial research is now needed to further our understanding of these properties experimentally. Although Seeman’s (151) landmark conception of structural DNA nanotechnology took place over 30 years ago, and Rothemund’s (143) conception of scaffolded DNA origami took place over 10 years ago, major advances in fully automated top-down sequence design and scaffold production have only occurred recently, and we therefore believe that structural DNA nanotechnology remains in its infancy as this still nascent field expands its reach to biophysicists worldwide, with plenty of room at the bottom (49).
SUMMARY POINTS.
DNA nanotechnology offers the ability to spatially organize or interface with biomolecules in one, two, and three dimensions. Unprecedented geometric control at the 1–100-nm scale in combination with single-molecule addressability and programmability render these nanostructures powerful tools for biophysical research.
Recent advances in the automated top-down design of 2D and 3D wireframe architectures are democratizing scaffolded DNA origami. Nonexperts working in biophysical research and related fields can now leverage DNA origami to manipulate or mimic macromolecular biological assemblies and their related cellular processes.
We highlight intriguing applications of wireframe DNA origami to biologically inspired light harvesting, RNA structural biology, and immunology, for which we anticipate substantial impact in the coming decade.
Photosynthetic systems leverage the hierarchical and dense organization of dyes to enable the absorption of sunlight and directional energy transfer at remarkable efficiency. Both long-range and short-range energy-transfer mechanisms contribute to this efficiency. By controlling the organization of chromophores on the 1–100-nm scale, DNA origami offers unique opportunities to integrate both mechanisms to investigate or mimic fundamental aspects of light harvesting.
The elucidation of RNA tertiary structures remains challenging, particularly for long RNAs, predominantly owing to their conformational heterogeneity. DNA origami-based wireframe assemblies might be used to stabilize conformational substates and thereby facilitate structure elucidation using single-particle cryo-EM. Moreover, we envision these assemblies might enable the organization of catalytic RNAs to generate artificial ribozymes, potentially mimicking the PTC of the ribosome.
The clustering of immune receptors, the recruitment of coreceptors, and the kinetic segregation of phosphatases and kinases represent fundamental processes governing immune recognition. The organization of antigens or PAMPs at the 1–100-nm scale using DNA origami is ideally suited to systematically evaluate these processes and to infer their impact on the induction of endocytosis and immune cell signaling.
FUTURE ISSUES.
It will be important to further decrease the synthetic costs associated with DNA nanotechnology.
Strategies for the combinatorial synthesis of functionalized DNA origami libraries may enable high-throughput functional screening and their associated downstream applications.
Mimicking the local chemical environments of dyes and biomolecules typically embedded in protein or lipid systems may be essential for native biophysical studies.
Incorporating physiologically relevant conditions into RNA structural studies will be essential to understanding their native folds and functionally relevant conformational substates.
Immune cell responses to diverse structured DNA motifs and sequences present in DNA origami will be important to understand and control.
BOTTOM-UP COMPUTATIONAL DESIGN OF SCAFFOLDED DNA ORIGAMI.
A major bottleneck that has limited the widespread application of scaffolded DNA origami objects to biophysical studies is the complexity of designing the hundreds of staple strands needed to hybridize and fold the long scaffold strand such that it adopts the target 2D or 3D structure of interest. The computational design tool caDNAno has played a major role in overcoming this barrier for rectilinear, brick-like origami by enabling semiautomated staple design (43), which is typically combined with the 3D structure-prediction software CanDo to optimize staple crossovers prior to synthesis (Figure 2) (28, 92, 130). However, scaffold routing and staple design for wireframe assemblies are prohibitively complex using caDNAno, even for experts, and therefore fully automated sequence design algorithms are needed.
TOP-DOWN COMPUTATIONAL DESIGN OF WIREFRAME SCAFFOLDED DNA ORIGAMI.
Manual scaffold routing and staple design for wireframe assemblies are prohibitively complex. The top-down computational design tools PERDIX, METIS, DAEDALUS, TALOS, and vHelix-BSCOR have therefore been developed to perform automated scaffold routing and staple sequence design based only on a target geometry or shape (Figure 2). PERDIX performs fully automated scaffold routing and staple sequence design for any free-form 2D geometry using exclusively double-crossover (DX)-based edges (86; http://perdix-dna-origami.org), whereas METIS designs any 2D geometry using mechanically stiffer honeycomb edges (H. Jun, X. Wang, & M. Bathe, unpublished information; http://metis-dna-origami.org). DAEDALUS solves the scaffold routing and staple design problem fully automatically for any 3D polyhedral surface using solely DX-based edges (186; http://daedalus-dna-origami.org), whereas TALOS renders any 3D polyhedral surface using mechanically stiffer honeycomb edges, thereby also requiring greater scaffold length for the same particle geometry and size (85; http://talos-dna-origami.org). vHelix-BSCOR is an alternative computational approach in which BSCOR performs the necessary scaffold routing and vHelix performs the staple design for 2D and 3D wireframe objects (17, 18). This algorithm utilizes hybrid single-duplex and DX edges together with physics-based modeling to solve the scaffold routing and staple design problems by iteratively adjusting wireframe edge lengths empirically, whereas PERDIX, METIS, DAEDALUS, and TALOS all solve the top-down sequence design problem fully automatically using uniformly DX or honeycomb edges, without any iterative adjustments to edge lengths.
ACKNOWLEDGMENTS
The authors gratefully acknowledge funding from the Office of Naval Research (N00014-12-1-0621, N00014-14-1-0609, N00014-13-1-0664, N00014-16-1-2506, N00014-16-1-2181, N00014-16-1-2953, N00014-17-1-2609, and N00014-18-1-2290), the National Science Foundation (CCF-1547999, CCF-1564025, CMMI-1334109, CBET-1729397, CHE-1839155, PHY-1305537, and PHY-1707999), the Army Research Office (W911NF1210420), the Human Frontier Science Program (RGP0029/2014), the Department of Energy (DE-SC0016353 and DE-SC0001088), and the National Institutes of Health (U01-MH106011, R01-MH112694, and R21-EB026008). Support for this research was provided by a core center grant (P30-ES002109) from the National Institute of Environmental Health Sciences, National Institutes of Health.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Afonin KA, Cieply DJ, Leontis NB. 2008. Specific RNA self-assembly with minimal paranemic motifs. J. Am. Chem. Soc 130:93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonin KA, Viard M, Kagiampakis I, Case CL, Dobrovolskaia MA, et al. 2015. Triggering of RNA interference with RNA-RNA, RNA-DNA, and DNA-RNA nanoparticles. ACS Nano 9:251–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801 [DOI] [PubMed] [Google Scholar]
- 4.Albinsson B, Hannestad JK, Börjesson K. 2012. Functionalized DNA nanostructures for light harvesting and charge separation. Coord. Chem. Rev 256:2399–413 [Google Scholar]
- 5.Anderson RM, Kwon M, Strobel SA. 2007. Toward ribosomal RNA catalytic activity in the absence of protein. J. Mol. Evol 64:472–83 [DOI] [PubMed] [Google Scholar]
- 6.Angelin A, Weigel S, Garrecht R, Meyer R, Bauer J, et al. 2015. Multiscale origami structures as interface for cells. Angew. Chem. Int. Ed. Engl 54:15813–17 [DOI] [PubMed] [Google Scholar]
- 7.Arenz S, Wilson DN. 2016. Bacterial protein synthesis as a target for antibiotic inhibition. Cold Spring Harb. Perspect. Med 6:a025361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armitage BA. 2005. Cyanine dye–DNA interactions: intercalation, groove binding, and aggregation In DNA Binders and Related Subjects, ed. Waring MJ, Chaires JB, pp. 55–76. Berlin/Heidelberg: Springer [Google Scholar]
- 9.Auffinger P, Bielecki L, Westhof E. 2004. Symmetric K+ and Mg2+ ion-binding sites in the 5S rRNA loop E inferred from molecular dynamics simulations. J. Mol. Biol 335:555–71 [DOI] [PubMed] [Google Scholar]
- 10.Avalos AM, Bilate AM, Witte MD, Tai AK, He J, et al. 2014. Monovalent engagement of the BCR activates ovalbumin-specific transnuclear B cells. J. Exp. Med 211:365–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachmann M, Rohrer U, Kundig T, Burki K, Hengartner H, Zinkernagel R. 1993. The influence of antigen organization on B cell responsiveness. Science 262:1448–51 [DOI] [PubMed] [Google Scholar]
- 12.Bai XC, Martin TG, Scheres SH, Dietz H. 2012. Cryo-EM structure of a 3D DNA-origami object. PNAS 109:20012–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905–20 [DOI] [PubMed] [Google Scholar]
- 14.Banal JL, Kondo T, Veneziano R, Bathe M, Schlau-Cohen GS. 2017. Photophysics of J-aggregate-mediated energy transfer on DNA. J. Phys. Chem. Lett 8:5827–33 [DOI] [PubMed] [Google Scholar]
- 15.Bath J, Turberfield AJ. 2007. DNA nanomachines. Nat. Nanotechnol 2:275–84 [DOI] [PubMed] [Google Scholar]
- 16.Bennett NR, Zwick DB, Courtney AH, Kiessling LL. 2015. Multivalent antigens for promoting B and T cell activation. ACS Chem. Biol 10:1817–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benson E, Mohammed A, Bosco A, Teixeira AI, Orponen P, Hogberg B. 2016. Computer-aided production of scaffolded DNA nanostructures from flat sheet meshes. Angew. Chem. Int. Ed. Engl 55:8869–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benson E, Mohammed A, Gardell J, Masich S, Czeizler E, et al. 2015. DNA rendering of polyhedral meshes at the nanoscale. Nature 523:441–44 [DOI] [PubMed] [Google Scholar]
- 19.Boulais É, Sawaya NPD, Veneziano R, Andreoni A, Banal JL, et al. 2017. Programmed coherent coupling in a synthetic DNA-based excitonic circuit. Nat. Mater 17:159–66 [DOI] [PubMed] [Google Scholar]
- 20.Bricker WP, Banal JL, Stone MB, Bathe M. 2018. Molecular model of J-aggregated pseudoisocyanine fibers. J. Chem. Phys 149:024905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. 2015. Innate immune pattern recognition: a cell biological perspective. Ann. Rev. Immunol 33:257–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckhout-White S, Spillmann CM, Algar WR, Khachatrian A, Melinger JS, et al. 2014. Assembling programmable FRET-based photonic networks using designer DNA scaffolds. Nat. Commun 5:5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burkhardt DH, Rouskin S, Zhang Y, Li GW, Weissman JS, Gross CA. 2017. Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. eLife 6:e22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai H, Muller J, Depoil D, Mayya V, Sheetz MP, et al. 2018. Full control of ligand positioning reveals spatial thresholds for T cell receptor triggering. Nat. Nanotechnol 13:610–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cannon BL, Patten LK, Kellis DL, Davis PH, Lee J, et al. 2018. Large Davydov splitting and strong fluorescence suppression: an investigation of exciton delocalization in DNA-templated Holliday junction dye aggregates. J. Phys. Chem 122:2086–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carbone CB, Kern N, Fernandes RA, Hui E, Su X, et al. 2017. In vitro reconstitution of T cell receptor-mediated segregation of the CD45 phosphatase. PNAS 114:E9338–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter R, Fearon D. 1992. CD19: lowering the threshold for antigen receptor stimulation of B lymphocytes. Science 256:105–7 [DOI] [PubMed] [Google Scholar]
- 28.Castro CE, Kilchherr F, Kim DN, Shiao EL, Wauer T, et al. 2011. A primer to scaffolded DNA origami. Nat. Methods 8:221–29 [DOI] [PubMed] [Google Scholar]
- 29.Chan RT, Robart AR, Rajashankar KR, Pyle AM, Toor N. 2012. Crystal structure of a group II intron in the pre-catalytic state. Nat. Struct. Mol. Biol 19:555–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen JH, Seeman NC. 1991. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350:631–33 [DOI] [PubMed] [Google Scholar]
- 31.Cherezov V, Clogston J, Papiz MZ, Caffrey M. 2006. Room to move: Crystallizing membrane proteins in swollen lipidic mesophases. J. Mol. Biol 357:1605–18 [DOI] [PubMed] [Google Scholar]
- 32.Chevalier A, Silva DA, Rocklin GJ, Hicks DR, Vergara R, et al. 2017. Massively parallel de novo protein design for targeted therapeutics. Nature 550:74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cochran JR, Cameron TO, Stone JD, Lubetsky JB, Stern LJ. 2001. Receptor proximity, not intermolecular orientation, is critical for triggering T cell activation. J. Biol. Chem 276:28068–74 [DOI] [PubMed] [Google Scholar]
- 34.Comandur R, Olson ED, Musier-Forsyth K. 2017. Conservation of tRNA mimicry in the 5′-untranslated region of distinct HIV-1 subtypes. RNA 23:1850–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunningham PD, Bricker WP, Díaz SA, Medintz IL, Bathe M, Melinger JS. 2017. Optical determination of the electronic coupling and intercalation geometry of thiazole orange homodimer in DNA. J. Chem. Phys 147:055101. [DOI] [PubMed] [Google Scholar]
- 36.Czapar AE, Tiu BDB, Veliz FA, Pokorski JK, Steinmetz NF. 2018. Slow-release formulation of cowpea mosaic virus for in situ vaccine delivery to treat ovarian cancer. Adv. Sci 5:1700991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daher M, Mustoe AM, Morriss-Andrews A, Brooks CL III, Walter NG. 2017. Tuning RNA folding and function through rational design of junction topology. Nucleic Acids Res. 45:9706–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das R, Baker D. 2007. Automated de novo prediction of native-like RNA tertiary structures. PNAS 104:14664–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dietz H, Douglas SM, Shih WM. 2009. Folding DNA into twisted and curved nanoscale shapes. Science 325:725–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doh J, Irvine DJ. 2006. Immunological synapse arrays: patterned protein surfaces that modulate immunological synapse structure formation in T cells. PNAS 103:5700–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Douglas SM, Bachelet I, Church GM. 2012. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335:831–34 [DOI] [PubMed] [Google Scholar]
- 42.Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM. 2009. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459:414–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Douglas SM, Marblestone AH, Teerapittayanon S, Vazquez A, Church GM, Shih WM. 2009. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37:5001–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dutta PK, Varghese R, Nangreave J, Lin S, Yan H, Liu Y. 2011. DNA-directed artificial light-harvesting antenna. J. Am. Chem. Soc 133:11985–93 [DOI] [PubMed] [Google Scholar]
- 45.Ellington AD, Szostak JW. 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–22 [DOI] [PubMed] [Google Scholar]
- 46.Engel GS, Calhoun TR, Read EL, Ahn TK, Mancal T, et al. 2007. Evidence for wavelike energy transfer through quantum coherence in photosynthetic systems. Nature 446:782–86 [DOI] [PubMed] [Google Scholar]
- 47.Ensslen P, Wagenknecht H-A. 2015. One-dimensional multichromophor arrays based on DNA: from self-assembly to light-harvesting. Acc. Chem. Res 48:2724–33 [DOI] [PubMed] [Google Scholar]
- 48.Fang X, Wang J, O’Carroll IP, Mitchell M, Zuo X, et al. 2013. An unusual topological structure of the HIV-1 Rev response element. Cell 155:594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feynman RP. 1959. There’s plenty of room at the bottom. Eng. Sci 23(5):22–36 [Google Scholar]
- 50.Flores SC, Altman RB. 2010. Turning limited experimental information into 3D models of RNA. RNA 16:1769–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forster AC, Church GM. 2006. Towards synthesis of a minimal cell. Mol. Syst. Biol 2:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franklin RE, Gosling RG. 1953. Molecular configuration in sodium thymonucleate. Nature 171:740–41 [DOI] [PubMed] [Google Scholar]
- 53.Franz KM, Kagan JC. 2017. Innate immune receptors as competitive determinants of cell fate. Mol. Cell 66:750–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu TJ, Seeman NC. 1993. DNA double-crossover molecules. Biochemistry 32:3211–20 [DOI] [PubMed] [Google Scholar]
- 55.Funke JJ, Ketterer P, Lieleg C, Schunter S, Korber P, Dietz H. 2016. Uncovering the forces between nucleosomes using DNA origami. Sci. Adv 2:e1600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gangloff SC, Guenounou M. 2004. Toll-like receptors and immune response in allergic disease. Clin. Rev. Allergy Immunol. 26:115–25 [DOI] [PubMed] [Google Scholar]
- 57.Garmann RF, Gopal A, Athavale SS, Knobler CM, Gelbart WM, Harvey SC. 2015. Visualizing the global secondary structure of a viral RNA genome with cryo-electron microscopy. RNA 21:877–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geary C, Rothemund PW, Andersen ES. 2014. RNA nanostructures. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science 345:799–804 [DOI] [PubMed] [Google Scholar]
- 59.Glazer AN. 1984. Phycobilisome a macromolecular complex optimized for light energy transfer. Biochim. Biophys. Acta Rev. Bioenerg 768:29–51 [Google Scholar]
- 60.Glazer AN. 1985. Light harvesting by phycobilisomes. Annu. Rev. Biophys. Biophys. Chem 14:47–77 [DOI] [PubMed] [Google Scholar]
- 61.Gopinath A, Rothemund PW. 2014. Optimized assembly and covalent coupling of single-molecule DNA origami nanoarrays. ACS Nano 8:12030–40 [DOI] [PubMed] [Google Scholar]
- 62.Gothelf KV. 2017. Chemical modifications and reactions in DNA nanostructures. MRS Bull. 42:897–903 [Google Scholar]
- 63.Green R, Noller HF. 1996. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA 2:1011–21 [PMC free article] [PubMed] [Google Scholar]
- 64.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35:849–57 [DOI] [PubMed] [Google Scholar]
- 65.Haedler AT, Kreger K, Issac A, Wittmann B, Kivala M, et al. 2015. Long-range energy transport in single supramolecular nanofibres at room temperature. Nature 523:196–99 [DOI] [PubMed] [Google Scholar]
- 66.Han D, Qi X, Myhrvold C, Wang B, Dai M, et al. 2017. Single-stranded DNA and RNA origami. Science 358:6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han DR, Pal S, Nangreave J, Deng ZT, Liu Y, Yan H. 2011. DNA origami with complex curvatures in three-dimensional space. Science 332:342–46 [DOI] [PubMed] [Google Scholar]
- 68.Hannestad JK, Sandin P, Albinsson B. 2008. Self-assembled DNA photonic wire for long-range energy transfer. J. Am. Chem. Soc 130:15889–95 [DOI] [PubMed] [Google Scholar]
- 69.Hartman NC, Groves JT. 2011. Signaling clusters in the cell membrane. Curr. Opin. Cell Biol. 23:370–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heilemann M, Tinnefeld P, Sanchez Mosteiro G, Garcia Parajo M, Van Hulst NF, Sauer M. 2004. Multistep energy transfer in single molecular photonic wires. J. Am. Chem. Soc 126:6514–15 [DOI] [PubMed] [Google Scholar]
- 71.Hemmig EA, Creatore C, Wünsch B, Hecker L, Mair P, et al. 2016. Programming light-harvesting efficiency using DNA origami. Nano Lett. 16:2369–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Henrickson SE, Mempel TR, Mazo IB, Liu B, Artyomov MN, et al. 2008. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat. Immunol 9:282–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hofacker IL. 2003. Vienna RNA secondary structure server. Nucleic Acids Res. 31:3429–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoiberg HC, Sparvath SM, Andersen VL, Kjems J, Andersen ES. 2018. An RNA origami octahedron with intrinsic siRNAs for potent gene knockdown. Biotechnol. J 26:e1700634. [DOI] [PubMed] [Google Scholar]
- 75.Holliday R 1964. Mechanism for gene conversion in fungi. Genet. Res 5:282–307 [DOI] [PubMed] [Google Scholar]
- 76.Holmes EC. 2009. The evolutionary genetics of emerging viruses. Annu. Rev. Ecol. Evol. Syst 40:353–72 [Google Scholar]
- 77.Horton HM, Bernett MJ, Pong E, Peipp M, Karki S, et al. 2008. Potent in vitro and in vivo activity of an Fc-engineered anti-CD19 monoclonal antibody against lymphoma and leukemia. Cancer Res. 68:8049–57 [DOI] [PubMed] [Google Scholar]
- 78.Horton HM, Chu SY, Ortiz EC, Pong E, Cemerski S, et al. 2011. Antibody-mediated coengagement of FcγRIIb and B cell receptor complex suppresses humoral immunity in systemic lupus erythematosus. J. Immunol 186:4223–33 [DOI] [PubMed] [Google Scholar]
- 79.Huang L, Lemos HP, Li L, Li M, Chandler PR, et al. 2012. Engineering DNA nanoparticles as immunomodulatory reagents that activate regulatory T cells. J. Immunol 188:4913–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Isaacs FJ, Dwyer DJ, Collins JJ. 2006. RNA synthetic biology. Nat. Biotechnol 24:545–54 [DOI] [PubMed] [Google Scholar]
- 81.Janeway CA. 1992. The T cell receptor as a multicomponent signalling machine: CD4/CD8 coreceptors and CD45 in T cell activation. Annu. Rev. Immunol 10:645–74 [DOI] [PubMed] [Google Scholar]
- 82.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. 2010. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol 17:555–60 [DOI] [PubMed] [Google Scholar]
- 83.Johansson HE, Liljas L, Uhlenbeck OC. 1997. RNA recognition by the MS2 phage coat protein. Semin. Virol 8:176–85 [Google Scholar]
- 84.Johnson-Buck A, Nangreave J, Kim DN, Bathe M, Yan H, Walter NG. 2013. Super-resolution fingerprinting detects chemical reactions and idiosyncrasies of single DNA pegboards. Nano Lett. 13:728–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jun H, Shepherd TR, Zhang K, Bricker WP, Li S, et al. 2019. Automated sequence design of 3D polyhedral wireframe DNA origami with honeycomb edges. ACS Nano 13:2083–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jun H, Zhang F, Shepherd TR, Ratanalert S, Qi X, et al. 2019. Autonomously designed free-form 2D DNA Origami. Sci. Adv 5(1):eaav0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jungmann R, Steinhauer C, Scheible M, Kuzyk A, Tinnefeld P, Simmel FC. 2010. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 10:4756–61 [DOI] [PubMed] [Google Scholar]
- 88.Karlsson HJ, Eriksson M, Perzon E, Åkerman B, Lincoln P, Westman G. 2003. Groove-binding unsymmetrical cyanine dyes for staining of DNA: syntheses and characterization of the DNA-binding. Nucleic Acids Res. 31:6227–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Katz ZB, English BP, Lionnet T, Yoon YJ, Monnier N, et al. 2016. Mapping translation ‘hot-spots’ in live cells by tracking single molecules of mRNA and ribosomes. eLife 5:e10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. 2005. Crystal structure of a bacterial ribonuclease P RNA. PNAS 102:13392–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, et al. 2015. RNA structure. Structure of the HIV-1 RNA packaging signal. Science 348:917–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim DN, Kilchherr F, Dietz H, Bathe M. 2012. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 40:2862–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ko SH, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. 2010. Synergistic self-assembly of RNA and DNA molecules. Nat. Chem 2:1050–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kouskoff V, Famiglietti S, Lacaud G, Lang P, Rider JE, et al. 1998. Antigens varying in affinity for the B cell receptor induce differential B lymphocyte responses. J. Exp. Med 188:1453–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krieg E, Shih WM. 2018. Selective nascent polymer catch-and-release enables scalable isolation of multi-kilobase single-stranded DNA. Angew. Chem. Int. Ed. Engl 57:714–18 [DOI] [PubMed] [Google Scholar]
- 96.Krishnan Y, Bathe M. 2012. Designer nucleic acids to probe and program the cell. Trends Cell Biol. 22:624–33 [DOI] [PubMed] [Google Scholar]
- 97.Le JV, Luo Y, Darcy MA, Lucas CR, Goodwin MF, et al. 2016. Probing nucleosome stability with a DNA origami nanocaliper. ACS Nano 10:7073–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li S, Jiang Q, Liu S, Zhang Y, Tian Y, et al. 2018. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol 36:258–64 [DOI] [PubMed] [Google Scholar]
- 99.Li X, Yang X, Qi J, Seeman NC. 1996. Antiparallel DNA double crossover molecules as components for nanoconstruction. J. Am. Chem. Soc 118:6131–40 [Google Scholar]
- 100.Lin C, Perrault SD, Kwak M, Graf F, Shih WM. 2013. Purification of DNA-origami nanostructures by rate-zonal centrifugation. Nucleic Acids Res. 41:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, et al. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367–77 [DOI] [PubMed] [Google Scholar]
- 102.Liu W, Halverson J, Tian Y, Tkachenko AV, Gang O. 2016. Self-organized architectures from assorted DNA-framed nanoparticles. Nat. Chem 8:867–73 [DOI] [PubMed] [Google Scholar]
- 103.Liu X, Xu Y, Yu T, Clifford C, Liu Y, et al. 2012. A DNA nanostructure platform for directed assembly of synthetic vaccines. Nano Lett. 12:4254–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu Y, Gonen S, Gonen T, Yeates TO. 2018. Near-atomic cryo-EM imaging of a small protein displayed on a designed scaffolding system. PNAS 115:3362–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma Y-Z, Cogdell RJ, Gillbro T. 1997. Energy transfer and exciton annihilation in the B800–850 antenna complex of the photosynthetic purple bacterium Rhodopseudomonas acidophila (strain 10050). A femtosecond transient absorption study. J. Phys. Chem 101:1087–95 [Google Scholar]
- 106.Maiuri M, Ostroumov EE, Saer RG, Blankenship RE, Scholes GD. 2018. Coherent wavepackets in the Fenna-Matthews-Olson complex are robust to excitonic-structure perturbations caused by mutagenesis. Nat. Chem 10:177–83 [DOI] [PubMed] [Google Scholar]
- 107.Manz BN, Jackson BL, Petit RS, Dustin ML, Groves J. 2011. T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. PNAS 108:9089–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Marcheschi RJ, Tonelli M, Kumar A, Butcher SE. 2011. Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication. ACS Chem. Biol 6:857–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Markova LI, Malinovskii VL, Patsenker LD, Haner R. 2013. J- vs. H-type assembly: pentamethine cyanine (Cy5) as a near-IR chiroptical reporter. Chem. Commun 49:5298–300 [DOI] [PubMed] [Google Scholar]
- 110.Marras AE, Zhou L, Su HJ, Castro CE. 2015. Programmable motion of DNA origami mechanisms. PNAS 112:713–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marshak-Rothstein A 2006. Toll-like receptors in systemic autoimmune disease. Nat. Rev. Immunol 6:823–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Massey M, Ancona MG, Medintz IL, Algar WR. 2015. Time-gated DNA photonic wires with Förster resonance energy transfer cascades initiated by a luminescent terbium donor. ACS Photonics 2:639–52 [Google Scholar]
- 113.Medzhitov R 2002. Decoding the patterns of self and non-self by the innate immune system. Science 296:298–300 [DOI] [PubMed] [Google Scholar]
- 114.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc 127:4223–31 [DOI] [PubMed] [Google Scholar]
- 115.Miao Z, Adamiak RW, Antczak M, Batey RT, Becka AJ, et al. 2017. RNA-puzzles round III: 3D RNA structure prediction of five riboswitches and one ribozyme. RNA 23:655–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Miao Z, Westhof E. 2017. RNA structure: advances and assessment of 3D structure prediction. Annu. Rev. Biophys 46:483–503 [DOI] [PubMed] [Google Scholar]
- 117.Mogensen TH. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev 22:240–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Monshouwer R, Abrahamsson M, van Mourik F, van Grondelle R. 1997. Superradiance and exciton delocalization in bacterial photosynthetic light-harvesting systems. J. Phys. Chem 101:7241–48 [Google Scholar]
- 119.Mullineaux CW. 2007. Phycobilisome-reaction centre interaction in cyanobacteria. Photosynth. Res 95:175–82 [DOI] [PubMed] [Google Scholar]
- 120.Nafisi PM, Aksel T, Douglas SM. 2018. Construction of a novel phagemid to produce custom DNA origami scaffolds. Synth. Biol 3:ysy015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. 2010. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464:441–44 [DOI] [PubMed] [Google Scholar]
- 122.Nickels PC, Wunsch B, Holzmeister P, Bae W, Kneer LM, et al. 2016. Molecular force spectroscopy with a DNA origami-based nanoscopic force clamp. Science 354:305–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nicoli F, Barth A, Bae W, Neukirchinger F, Crevenna AH, et al. 2017. Directional photonic wire mediated by homo-Förster resonance energy transfer on a DNA origami platform. ACS Nano 11:11264–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nikolay R, Hilal T, Qin B, Mielke T, Burger J, et al. 2018. Structural visualization of the formation and activation of the 50S ribosomal subunit during in vitro reconstitution. Mol. Cell 70:881–93.e3 [DOI] [PubMed] [Google Scholar]
- 125.Orelle C, Carlson ED, Szal T, Florin T, Jewett MC, Mankin AS. 2015. Protein synthesis by ribosomes with tethered subunits. Nature 524:119–24 [DOI] [PubMed] [Google Scholar]
- 126.Ouyang X, Li J, Liu H, Zhao B, Yan J, et al. 2013. Rolling circle amplification-based DNA Origami nanostructures for intracellular delivery of immunostimulatory drugs. Small 9:3082–87 [DOI] [PubMed] [Google Scholar]
- 127.Paige JS, Wu KY, Jaffrey SR. 2011. RNA mimics of green fluorescent protein. Science 333:642–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pan K, Boulais E, Yang L, Bathe M. 2014. Structure-based model for light-harvesting properties of nucleic acid nanostructures. Nucleic Acids Res. 42:2159–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pan K, Bricker WP, Ratanalert S, Bathe M. 2017. Structure and conformational dynamics of scaffolded DNA origami nanoparticles. Nucleic Acids Res. 45:6284–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pan K, Kim DN, Zhang F, Adendorff MR, Yan H, Bathe M. 2014. Lattice-free prediction of three-dimensional structure of programmed DNA assemblies. Nat. Commun 5:5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pardoll DM. 2012. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12:252–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Parks JW, Kappel K, Das R, Stone MD. 2017. Single-molecule FRET-Rosetta reveals RNA structural rearrangements during human telomerase catalysis. RNA 23:175–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pinheiro AV, Han D, Shih WM, Yan H. 2011. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol 6:763–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Praetorius F, Kick B, Behler KL, Honemann MN, Weuster-Botz D, Dietz H. 2017. Biotechnological mass production of DNA origami. Nature 552:84–87 [DOI] [PubMed] [Google Scholar]
- 135.Pullerits T, Chachisvilis M, Sundström V. 1996. Exciton delocalization length in the B850 antenna of Rhodobacter sphaeroides. J. Phys. Chem 100:10787–92 [Google Scholar]
- 136.Punekar AS, Liljeruhm J, Shepherd TR, Forster AC, Selmer M. 2013. Structural and functional insights into the molecular mechanism of rRNA m6 A methyltransferase RlmJ. Nucleic Acids Res. 41:9537–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Punekar AS, Shepherd TR, Liljeruhm J, Forster AC, Selmer M. 2012. Crystal structure of RlmM, the 2′O-ribose methyltransferase for C2498 of Escherichia coli 23S rRNA. Nucleic Acids Res. 40:10507–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pyle AM. 2016. Group II intron self-splicing. Annu. Rev. Biophys 45:183–205 [DOI] [PubMed] [Google Scholar]
- 139.Ramani V, Qiu R, Shendure J. 2015. High-throughput determination of RNA structure by proximity ligation. Nat. Biotechnol 33:980–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A. 2010. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468:784–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Romero E, Novoderezhkin VI, van Grondelle R. 2017. Quantum design of photosynthesis for bioinspired solar-energy conversion. Nature 543:355–65 [DOI] [PubMed] [Google Scholar]
- 142.Rosenberg JM, Seeman NC, Kim JJP, Suddath FL, Nicholas HB, Rich A. 1973. Double helix at atomic resolution. Nature 243:150–54 [DOI] [PubMed] [Google Scholar]
- 143.Rothemund PWK. 2006. Folding DNA to create nanoscale shapes and patterns. Nature 440:297–302 [DOI] [PubMed] [Google Scholar]
- 144.Sa-Ardyen P, Vologodskii AV, Seeman NC. 2003. The flexibility of DNA double crossover molecules. Biophys. J 84:3829–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sacca B, Ishitsuka Y, Meyer R, Sprengel A, Schoneweiss EC, et al. 2015. Reversible reconfiguration of DNA origami nanochambers monitored by single-molecule FRET. Angew. Chem. Int. Ed. Engl 54:3592–97 [DOI] [PubMed] [Google Scholar]
- 146.Sacca B, Meyer R, Niemeyer CM. 2009. Temperature-dependent FRET spectroscopy for the high-throughput analysis of self-assembled DNA nanostructures in real time. Nat. Protoc 4:271–85 [DOI] [PubMed] [Google Scholar]
- 147.Salazar F, Ghaemmaghami AM. 2013. Allergen recognition by innate immune cells: critical role of dendritic and epithelial cells. Front. Immunol 4:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102:615–23 [DOI] [PubMed] [Google Scholar]
- 149.Scholes GD, Fleming GR, Olaya-Castro A, van Grondelle R. 2011. Lessons from nature about solar light harvesting. Nat. Chem 3:763–74 [DOI] [PubMed] [Google Scholar]
- 150.Schreiber RD, Old LJ, Smyth MJ. 2011. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331:1565–70 [DOI] [PubMed] [Google Scholar]
- 151.Seeman NC. 1982. Nucleic acid junctions and lattices. J. Theor. Biol 99:237–47 [DOI] [PubMed] [Google Scholar]
- 152.Seeman NC. 2010. Nanomaterials based on DNA. Annu. Rev. Biochem 79:65–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Seeman NC, Rosenberg JM, Suddath FL, Kim JJ, Rich A. 1976. RNA double-helical fragments at atomic resolution. I. The crystal and molecular structure of sodium adenylyl-3′,5′-uridine hexahydrate. J. Mol. Biol 104:109–44 [DOI] [PubMed] [Google Scholar]
- 154.Seeman NC, Sleiman HF. 2017. DNA nanotechnology. Nat. Rev. Mater 3:17068 [Google Scholar]
- 155.Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache JP, et al. 2009. Structural insight into nascent polypeptide chain-mediated translational stalling. Science 326:1412–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Seifert JL, Connor RE, Kushon SA, Wang M, Armitage BA. 1999. Spontaneous assembly of helical cyanine dye aggregates on DNA nanotemplates. J. Am. Chem. Soc 121:2987–95 [Google Scholar]
- 157.Serganov A, Patel DJ. 2007. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet 8:776–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. 2010. A polyhedron made of tRNAs. Nat. Chem 2:772–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, et al. 2006. Aptamers evolved from live cells as effective molecular probes for cancer study. PNAS 103:11838–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shapiro BA, Le Grice SF. 2016. Advances in RNA structure determination. Methods 103:1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shaw A, Benson E, Hogberg B. 2015. Purification of functionalized DNA origami nanostructures. ACS Nano 9:4968–75 [DOI] [PubMed] [Google Scholar]
- 162.Shaw A, Lundin V, Petrova E, Fordos F, Benson E, et al. 2014. Spatial control of membrane receptor function using ligand nanocalipers. Nat. Methods 11:841–46 [DOI] [PubMed] [Google Scholar]
- 163.Shepherd TR, Du L, Liljeruhm J, Samudyata, Wang J, et al. 2017. De novo design and synthesis of a 30-cistron translation-factor module. Nucleic Acids Res. 45:10895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163a.Shepherd TR, Du RR, Huang H, Wamhoff E-C, Bathe M. 2019. Bioproduction of pure, kilobase-scale single-stranded DNA. Sci. Rep 9:6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. 2011. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol 6:658–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11:959–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sil D, Lee JB, Luo D, Holowka D, Baird B. 2007. Trivalent ligands with rigid DNA spacers reveal structural requirements for IgE receptor signaling in RBL mast cells. ACS Chem. Biol 2:674–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Simmel FC, Schulman R. 2017. Self-organizing materials built with DNA. MRS Bull. 42:913–19 [Google Scholar]
- 168.Simmons CR, Zhang F, Birktoft JJ, Qi X, Han D, et al. 2016. Construction and structure determination of a three-dimensional DNA crystal. J. Am. Chem. Soc 138:10047–54 [DOI] [PubMed] [Google Scholar]
- 169.Snodin BE, Randisi F, Mosayebi M, Sulc P, Schreck JS, et al. 2015. Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. J. Chem. Phys 142:234901. [DOI] [PubMed] [Google Scholar]
- 170.Sobczak JPJ, Martin TG, Gerling T, Dietz H. 2012. Rapid folding of DNA into nanoscale shapes at constant temperature. Science 338:1458–61 [DOI] [PubMed] [Google Scholar]
- 171.Sovenyhazy KM, Bordelon JA, Petty JT. 2003. Spectroscopic studies of the multiple binding modes of a trimethine-bridged cyanine dye with DNA. Nucleic Acids Res. 31:2561–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sprengel A, Lill P, Stegemann P, Bravo-Rodriguez K, Schoneweiss EC, et al. 2017. Tailored protein encapsulation into a DNA host using geometrically organized supramolecular interactions. Nat. Commun 8:14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Stein IH, Schuller V, Bohm P, Tinnefeld P, Liedl T. 2011. Single-molecule FRET ruler based on rigid DNA origami blocks. ChemPhysChem 12:689–95 [DOI] [PubMed] [Google Scholar]
- 174.Steitz TA. 2008. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 9:242–53 [DOI] [PubMed] [Google Scholar]
- 175.Strauss MT, Schueder F, Haas D, Nickels PC, Jungmann R. 2018. Quantifying absolute addressability in DNA origami with molecular resolution. Nat. Commun 9:1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sulc P, Romano F, Ouldridge TE, Rovigatti L, Doye JP, Louis AA. 2012. Sequence-dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys 137:135101. [DOI] [PubMed] [Google Scholar]
- 177.Surana S, Shenoy AR, Krishnan Y. 2015. Designing DNA nanodevices for compatibility with the immune system of higher organisms. Nat. Nanotechnol 10:741–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Talkington MW, Siuzdak G, Williamson JR. 2005. An assembly landscape for the 30S ribosomal subunit. Nature 438:628–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Thyrhaug E, Tempelaar R, Alcocer MJP, Žídek K, Bína D, et al. 2018. Identification and characterization of diverse coherences in the Fenna-Matthews-Olson complex. Nat. Chem 10:780–86 [DOI] [PubMed] [Google Scholar]
- 180.Timm C, Niemeyer CM. 2015. Assembly and purification of enzyme-functionalized DNA origami structures. Angew. Chem. Int. Ed. Engl 54:6745–50 [DOI] [PubMed] [Google Scholar]
- 181.Tom JK, Dotsey EY, Wong HY, Stutts L, Moore T, et al. 2015. Modulation of innate immune responses via covalently linked TLR agonists. ACS Cent. Sci 1:439–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tomescu AI, Robb NC, Hengrung N, Fodor E, Kapanidis AN. 2014. Single-molecule FRET reveals a corkscrew RNA structure for the polymerase-bound influenza virus promoter. PNAS 111:E3335–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. 2005. Crystal structure of the RNA component of bacterial ribonuclease P. Nature 437:584–87 [DOI] [PubMed] [Google Scholar]
- 184.Tronrud DE, Wen J, Gay L, Blankenship RE. 2009. The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria. Photosynth. Res 100:79–87 [DOI] [PubMed] [Google Scholar]
- 185.Tuerk C, Gold L. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–10 [DOI] [PubMed] [Google Scholar]
- 186.Veneziano R, Ratanalert S, Zhang K, Zhang F, Yan H, et al. 2016. Designer nanoscale DNA assemblies programmed from the top down. Science 352:1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Veneziano R, Shepherd TR, Ratanalert S, Bellou L, Tao C, Bathe M. 2018. In vitro synthesis of gene-length single-stranded DNA. Sci. Rep 8:6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Vybornyi M, Nussbaumer AL, Langenegger SM, Häner R. 2014. Assembling multiporphyrin stacks inside the DNA double helix. Bioconjug. Chem 25:1785–93 [DOI] [PubMed] [Google Scholar]
- 189.Watson JD, Crick FHC. 1953. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171:737–38 [DOI] [PubMed] [Google Scholar]
- 190.Westhof E, Masquida B, Jaeger L. 1996. RNA tectonics: towards RNA design. Fold Des. 1:R78–88 [DOI] [PubMed] [Google Scholar]
- 191.Wilkins MHF, Stokes AR, Wilson HR. 1953. Molecular structure of deoxypentose nucleic acids. Nature 171:738–40 [DOI] [PubMed] [Google Scholar]
- 192.Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol 12:35–48 [DOI] [PubMed] [Google Scholar]
- 193.Wimberly BT, Brodersen DE, Clemons WM Jr., Morgan-Warren RJ, Carter AP, et al. 2000. Structure of the 30S ribosomal subunit. Nature 407:327–39 [DOI] [PubMed] [Google Scholar]
- 194.Winfree E, Liu F, Wenzler LA, Seeman NC. 1998. Design and self-assembly of two-dimensional DNA crystals. Nature 394:539–44 [DOI] [PubMed] [Google Scholar]
- 195.Woehrstein JB, Strauss MT, Ong LL, Wei B, Zhang DY, et al. 2017. Sub-100-nm metafluorophores with digitally tunable optical properties self-assembled from DNA. Sci. Adv 3:e1602128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wu S, Schuster M, Bagshaw CR, Rant U, Burley GA. 2011. Site-specific assembly of DNA-based photonic wires by using programmable polyamides. Angew. Chem. Int. Ed. Engl 50:2712–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. 2003. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 301:1882–84 [DOI] [PubMed] [Google Scholar]
- 198.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, et al. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883–96 [DOI] [PubMed] [Google Scholar]
- 199.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. 2001. The path of messenger RNA through the ribosome. Cell 106:233–41 [DOI] [PubMed] [Google Scholar]
- 200.Zhang DY, Seelig G. 2011. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem 3:103–13 [DOI] [PubMed] [Google Scholar]
- 201.Zhang F, Jiang S, Wu S, Li Y, Mao C, et al. 2015. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotechnol 10:779–84 [DOI] [PubMed] [Google Scholar]
- 202.Zhang K, Keane SC, Su Z, Irobalieva RN, Chen M, et al. 2018. Structure of the 30 kDa HIV-1 RNA dimerization signal by a hybrid cryo-EM, NMR, and molecular dynamics approach. Structure 26:490–98.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhao C, Rajashankar KR, Marcia M, Pyle AM. 2015. Crystal structure of group II intron domain 1 reveals a template for RNA assembly. Nat. Chem. Biol 11:967–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zheng J, Birktoft JJ, Chen Y, Wang T, Sha R, et al. 2009. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461:74–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–15 [DOI] [PMC free article] [PubMed] [Google Scholar]


