Multidrug efflux systems belonging to the resistance-nodulation-division (RND) superfamily are ubiquitous in Gram-negative bacteria. RND efflux systems are often associated with multiple antimicrobial resistance and also contribute to the expression of diverse bacterial phenotypes including virulence, as documented in the intestinal pathogen Vibrio cholerae, the causative agent of the severe diarrheal disease cholera.
KEYWORDS: cholera, OmpR, two-component regulatory systems, virulence regulation
ABSTRACT
Multidrug efflux systems belonging to the resistance-nodulation-division (RND) superfamily are ubiquitous in Gram-negative bacteria. RND efflux systems are often associated with multiple antimicrobial resistance and also contribute to the expression of diverse bacterial phenotypes including virulence, as documented in the intestinal pathogen Vibrio cholerae, the causative agent of the severe diarrheal disease cholera. Transcriptomic studies with RND efflux-negative V. cholerae suggested that RND-mediated efflux was required for homeostasis, as loss of RND efflux resulted in the activation of transcriptional regulators, including multiple environmental sensing systems. In this report, we investigated six RND efflux-responsive regulatory genes for contributions to V. cholerae virulence factor production. Our data showed that the V. cholerae gene VC2714, encoding a homolog of Escherichia coli OmpR, was a virulence repressor. The expression of ompR was elevated in an RND-null mutant, and ompR deletion partially restored virulence factor production in the RND-negative background. Virulence inhibitory activity in the RND-negative background resulted from OmpR repression of the key ToxR regulon virulence activator aphB, and ompR overexpression in wild-type cells also repressed virulence through aphB. We further show that ompR expression was not altered by changes in osmolarity but instead was induced by membrane-intercalating agents that are prevalent in the host gastrointestinal tract and which are substrates of the V. cholerae RND efflux systems. Our collective results indicate that V. cholerae ompR is an aphB repressor and regulates the expression of the ToxR virulence regulon in response to novel environmental cues.
INTRODUCTION
The Gram-negative bacterium Vibrio cholerae is the causative agent of the life-threatening diarrheal disease cholera. V. cholerae is an aquatic organism that infects humans following the consumption of V. cholerae-contaminated food or water. After ingestion, V. cholerae colonizes the epithelium of the small intestine to cause disease by a process that is dependent upon virulence factor production. The two most important V. cholerae virulence factors are the toxin-coregulated pilus (TCP), which mediates intestinal colonization, and cholera toxin (CT), an enterotoxin that is responsible for the secretory diarrhea that is the hallmark of the disease cholera. CT and TCP production is under the control of a hierarchical regulatory system known as the ToxR regulon (1, 2). Activation of the ToxR regulon begins with the expression of two cytoplasmic transcriptional regulators, aphA and aphB (3, 4). AphA and AphB function synergistically to activate tcpP expression. TcpP then binds along with ToxR to the toxT promoter to activate toxT expression. ToxT directly activates the expression of the genes that encode CT and TCP production (2).
The expression of adaptive responses is important for the success of V. cholerae as a pathogen. This includes tight regulation of the ToxR regulon, which is known to limit virulence factor production to the host gastrointestinal tract. Thus, the ToxR regulon has evolved to respond to specific environmental signals within the host (5). Other genes which are important for survival and persistence in aquatic ecosystems must be repressed during host entry for successful colonization (6–8). Late in infection, in preparation for host exit, V. cholerae downregulates virulence genes while activating genes required for dissemination and transmission (9–12). Although the genetic mechanisms involved in ToxR regulon activation have been extensively studied, less is known about how environmental signals influence ToxR regulon expression in vivo.
V. cholerae is exposed to disparate environments within both the aquatic ecosystem and the human gastrointestinal tract. V. cholerae survival and growth in these niches require rapid adaptation to environmental conditions. V. cholerae enters humans from aquatic ecosystems that are typically aerobic and alkaline. The bacterium must then pass through the gastric acid barrier of the stomach before entering the duodenum and migrating to the epithelial surface, where it colonizes the crypts of the small intestine. Successful transition between these dissimilar environments requires that V. cholerae modulates its transcriptional responses so that specific genes are only expressed during colonization of appropriate niches. In V. cholerae, like most bacteria, this is achieved by environmentally responsive regulatory systems that monitor the extracellular environment by use of a range of membrane-bound sensors such as ToxR and two-component signal transduction systems (TCSs) (13).
TCSs are widespread phospho-relay systems that modulate gene expression in response to environmental cues. They consist of a membrane-bound histidine kinase sensor protein coupled with a cytosolic response regulator. In the presence of appropriate stimuli, the sensor autophosphorylates a conserved histidine residue before transferring the phosphate to a conserved aspartate residue on the response regulator, activating the response regulator. Activated response regulators function to modulate adaptive responses by effecting the expression of target genes. Response regulators are typically transcription factors but can also function by other mechanisms (14). The adaptive responses mediated by TCSs are broad and include virulence, motility, metabolism, and stress responses.
One of the better characterized TCSs is the EnvZ-OmpR system that is ubiquitous in Gram-negative bacteria (15). EnvZ is the membrane-associated sensor kinase, and OmpR the response regulator that functions as a transcription factor. EnvZ-OmpR was first discovered in Escherichia coli and was shown to regulate the expression of its two major outer membrane porin proteins (OMP), ompC and ompF, in response to environmental osmolarity (16–18). The function of OmpR as an osmoregulator has been extended to a number of other bacterial genera (19–21). OmpR has also been linked to other phenotypes in Gram-negative bacteria, including virulence (19, 20, 22–26) and acidic tolerance (21, 27–31). The V. cholerae OmpR homolog, open reading frame (ORF) VC2714, has been little studied, and its role in V. cholerae biology is unknown.
The RND efflux systems are ubiquitous tripartite transporters in Gram-negative bacteria that play critical roles in antimicrobial resistance. Many RND efflux systems exhibit broad substrate specificity and have the capacity to efflux multiple substrates that are both structurally and functionally unrelated (32, 33). The RND systems play critical roles in antimicrobial resistance by exporting toxic compounds from the cytosol and periplasm into the extracellular environment. Although RND efflux pumps have been widely studied for their role in multiple antibiotic resistance, they also impact many other physiological phenotypes in bacteria (34). This was recently documented in V. cholerae, where the RND systems were shown to be required for cell homeostasis (35, 36). The absence of RND efflux in V. cholerae resulted in downregulation of the ToxR regulon and altered expression of genes involved in metabolic and environmental adaptation (37, 38), including several TCSs. The results of these studies suggested that RND-mediated efflux modulated homeostasis by effluxing cell metabolites, which served as concentration-dependent environmental cues to initiate transcriptional responses via periplasmic sensing systems. This observation suggested the possibility that these TCSs may have contributed to the virulence attenuation observed in RND efflux-impaired V. cholerae.
In this work, we investigated six regulatory genes that were induced in the absence of RND-mediated efflux for their contribution to virulence factor production in V. cholerae. This revealed that VC2714, encoding a homolog of E. coli OmpR, functioned as a virulence repressor in V. cholerae. We documented that VC2714 repressed the expression of the key virulence regulator aphB. We further showed that ompR expression was regulated in response to detergent-like compounds, which are prevalent in the host gastrointestinal tract and are substrates of the RND transporters. Our collective results suggest that the V. cholerae EnvZ-OmpR TCS has evolved to regulate virulence in response to novel environmental stimuli.
RESULTS
V. cholerae OmpR represses virulence factor production.
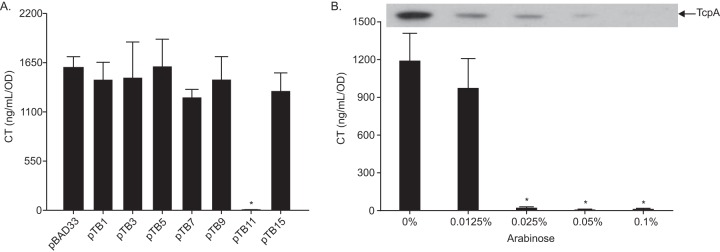
The loss of RND-mediated efflux resulted in downregulation of the ToxR regulon and diminished CT and TCP production (37), suggesting that there are one or more factors linking efflux to virulence factor production. Transcriptional profiling of an RND-negative (ΔvexB ΔvexD ΔvexF ΔvexH ΔvexK ΔvexM) V. cholerae mutant during growth under AKI conditions (i.e., virulence-inducing conditions) showed that the expression of a number of regulatory genes, including several TCSs, was increased in the absence of RND efflux (38). We hypothesized that one or more of these regulatory genes may have contributed to RND efflux-dependent virulence repression. To test this, we expressed six regulators (i.e., VC0486, VC1320 and VC1319, VC1081, VC1638, VC1825, VC1320, and VC2714) from the arabinose-regulated promoter in pBAD33 in wild-type (WT) V. cholerae during growth under AKI conditions in the presence of 0.05% arabinose and quantified CT production (39). VC0486 encodes an uncharacterized DeoR family regulator. VC1320 (carR) and VC1319 (carS) encode the CarRS TCS that is involved in regulating lipopolysaccharide (LPS) remodeling and vps production (40–42); carR (pTB15) and carRS (pTB3) were independently expressed. VC1081 encodes an uncharacterized response regulator. VC1638 was recently shown to regulate the expression of VCA0732 in response to polymyxin B (43). VC1825 is an AraC family regulator that regulates a phosphotransferase system (PTS) transporter (44). VC2714 encodes an uncharacterized response regulator. The results showed that only pTB11, expressing VC2714, repressed CT production (Fig. 1A). VC2714 encodes a homolog of the E. coli osmotic stress regulator OmpR with 92.1% amino acid sequence similarity and hereafter will be referred to as ompR.
FIG 1.
Overexpression of ompR represses virulence factor production. (A) WT V. cholerae harboring pBAD33 or the indicated expression plasmids was cultured under AKI conditions with 0.05% arabinose for 24 h when culture supernatants were used for CT quantitation by GM1 enzyme-linked immunosorbent assay (GM1-ELISA). The pBAD33-based expression plasmids carry the following genes: pTB1, VC0486; pTB3, VC1320 and VC1319; pTB5, VC1081; pTB7, VC1638; pTB9, VC1825; pTB11, VC2714; and pTB15, VC1320. The data represent the mean ± standard deviation (SD) of three independent experiments. *, P < 0.0001 relative to pBAD33 determined by Dunnett’s multiple-comparison test. (B) WT V. cholerae harboring pTB11 (pBAD33-ompR) was cultured under AKI conditions with indicated arabinose concentrations for 24 h when culture supernatants were used for CT quantitation by GM1 ELISA and the cell pellets were used for TcpA immunoblotting. CT data represent the mean ± SD of a minimum of three independent experiments. *, P < 0.0001 relative to 0%, determined by Dunnett’s multiple-comparison test. The TcpA immunoblot is representative of a minimum of three independent experiments.
To further verify that V. cholerae OmpR was a virulence repressor, we repeated the above experiment in WT V. cholerae harboring plasmid pTB11 during growth under AKI conditions in the presence of increasing arabinose concentrations and quantified CT and TcpA production. The results showed an arabinose-dependent inhibition of both CT and TcpA production (Fig. 1B). Based on these results, we conclude that OmpR functions as a virulence repressor in V. cholerae.
OmpR contributes to virulence repression in RND efflux-deficient V. cholerae.
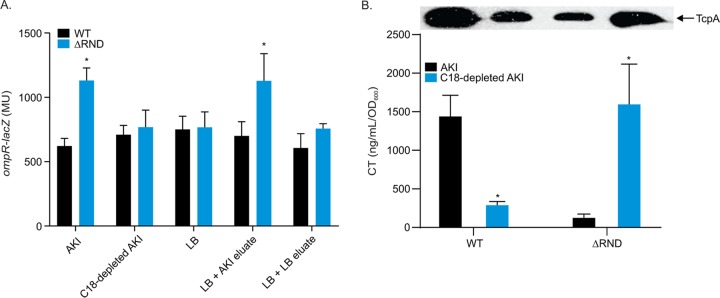
To verify that ompR was upregulated in RND-deficient V. cholerae as previously indicated in a transcriptomics data set (38), we introduced the ompR-lacZ transcriptional reporter plasmid pKD9 into WT and isogenic RND efflux-negative (ΔRND) V. cholerae strains and quantified ompR expression in both strains following growth in LB broth, in minimal T-medium, and under AKI conditions. The results showed significantly increased ompR expression in ΔRND relative to WT during growth under AKI conditions but no significant difference in LB broth or minimal T-medium (Fig. 2A). These findings confirm the previous study and suggest that the RND efflux-dependent induction of ompR is dependent on growth under AKI growth conditions.
FIG 2.
OmpR represses virulence factor production in RND efflux-negative V. cholerae. (A) WT and ΔRND V. cholerae strains harboring an ompR-lacZ reporter plasmid were cultured under the indicated conditions for 5 h when β-galactosidase activity was quantified. Data represent the mean ± SD of three independent experiments performed in triplicate. *, P < 0.0001 relative to WT determined by a t test. (B) The indicated V. cholerae strains were cultured under AKI conditions with the ΔRNDΔompR::pBAD33-ompR strain cultured in the presence of 0.05% arabinose. At 24 h, culture samples were collected, and CT and TcpA production was assessed by GM1 ELISA and TcpA immunoblotting, respectively. The CT data represent the mean ± SD of a minimum of three independent experiments. ND, nondetectable levels of CT. *, P < 0.05; **, P < 0.01 relative to the parental strain, determined by Tukey’s multiple-comparison test. TcpA immunoblot is representative of a minimum of three independent experiments.
We next tested if ompR contributed to the virulence repression observed in the RND-negative strain. To address this, we created ompR deletion strains in WT and RND-negative V. cholerae and quantified CT and TcpA production in WT, ΔRND, and their respective isogenic ΔompR mutants. Confirming previous studies (37), the RND-negative strain produced significantly reduced amounts of CT and TcpA relative to WT (Fig. 2B). Consistent with the finding that OmpR is a V. cholerae virulence repressor, deletion of ompR in WT resulted in a slight but not statistically significant increase in both CT and TcpA production (Fig. 2B). Deletion of ompR in the ΔRND background partially restored CT and TcpA production in the ΔRNDΔompR strain relative to the parental ΔRND strain, but the magnitude of the increase did not reach WT levels (Fig. 2B). To determine if deletion of ompR was sufficient for the increased virulence factor production found between the ΔRNDΔompR and ΔRND strains, ompR was complemented in the ΔRNDΔompR strain by use of plasmid pTB11. Complementation of ompR in the ΔRNDΔompR strain resulted in repression of both CT and TcpA production, indicating that deletion of ompR was in fact responsible for increased virulence factor production in the ΔRNDΔompR strain. Together, these data suggest that ompR contributes to virulence attenuation in the RND-negative background but that other factors are also involved in virulence repression.
V. cholerae OmpR represses aphB expression.
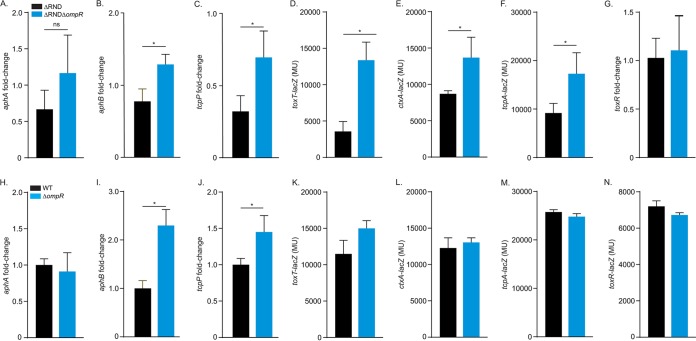
The above results suggested that OmpR was a virulence repressor, but the mechanism by which it attenuated virulence factor production was unclear. As CT and TCP production is positively regulated by the ToxR regulon, we hypothesized that OmpR repressed components of the ToxR regulon. If this was true, then ompR deletion in ΔRND should increase the expression of the affected ToxR regulon genes relative to the parental strain ΔRND. We therefore compared ToxR regulon gene expression in ΔRND and its isogenic ΔompR mutant during growth under AKI conditions. The results showed that ompR deletion in the RND-negative strain ΔRND did not significantly affect aphA expression (Fig. 3A) but did result in increased expression of aphB and the ToxR regulon genes downstream from aphB (i.e., tcpP, toxT, ctxA, and tcpA) (Fig. 3B through F). ΔRND and ΔRNDΔompR had comparable levels of toxR expression, indicating that virulence repression by OmpR was not due to reduced toxR expression (Fig. 3G). As aphB is one of the most upstream regulators in the ToxR regulon, these results suggested that OmpR attenuated virulence factor production by repressing aphB in ΔRND.
FIG 3.
OmpR represses the ToxR regulon. V. cholerae strains were cultured under AKI conditions when gene expression was assessed using lacZ promoter reporters (panels D to F and K to N) or reverse transcription-quantitative PCR (qRT-PCR) (panels A to C and G to J) as described in Materials and Methods. (A to G) Reporter gene expression in ΔRND and ΔRNDΔompR V. cholerae strains. (H to N) Reporter gene expression in WT and ΔompR V. cholerae strains. The results presented in panels A to C and H to J were generated at 3.5 h postinoculation; results from the remaining assays were generated at 5 h postinoculation. Data represent mean and SD of at least three independent experiments performed in triplicate. *, P < 0.05 relative to parental strain determined by a t test.
To test if OmpR affected ToxR regulon expression in efflux-sufficient cells, we repeated the above experiments in the WT during growth under AKI conditions. The results showed that ompR deletion in the WT did not affect aphA expression but resulted in increased expression of aphB and its downstream target tcpP (Fig. 3I and J) but not the other ToxR regulon genes (Fig. 3H, J, K, L, M, and N). This is consistent with the observation that deletion of ompR did not significantly affect CT or TcpA production in the WT (Fig. 2B). Collectively, these results support the conclusion that OmpR is an aphB repressor and that OmpR regulation of aphB is relevant in WT cells during growth under AKI conditions.
Ectopic ompR expression represses aphB transcription in E. coli.
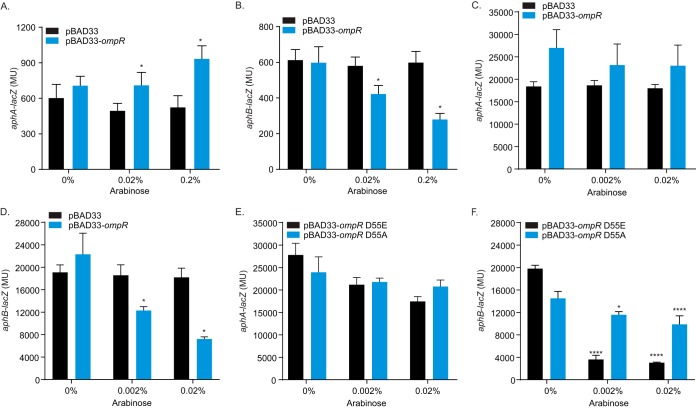
To further confirm that OmpR can repress aphB, we tested if ectopic ompR expression altered aphB expression in V. cholerae and the heterologous host E. coli. In the first set of experiments, we expressed ompR from pTB11 in WT V. cholerae bearing lacZ transcriptional reporters for aphA and aphB during growth under AKI conditions in the presence of varying arabinose concentrations to induce ompR expression. The results showed a small arabinose dose-dependent increase in aphA expression (Fig. 4A); the biological significance of this finding is unclear. By contrast, we observed an arabinose dose-dependent decrease in aphB expression (Fig. 4B), confirming that OmpR is an aphB repressor. Although OmpR may have a weak ability to induce aphA expression, its ability to repress aphB appears to be dominant, as the net consequence of ompR regulation of aphA and aphB is repression of tcpP (Fig. 3C and J).
FIG 4.
V. cholerae OmpR represses aphB expression. (A and B) WT V. cholerae harboring either pBAD33 or pBAD33-ompR (pTB11) with either aphA-lacZ (pXB202) or aphB-lacZ (pXB203) reporter plasmids was cultured under AKI conditions for 5 h with the indicated arabinose concentrations when β-galactosidase activity was quantified. Data represent the mean ± SD of three independent experiments performed in triplicate. (C and D) E. coli strain EC100 harboring either pBAD33 or pBAD33-ompR with either aphA-lacZ or aphB-lacZ reporter plasmids was cultured in LB broth for 5 h with the indicated arabinose concentrations when β-galactosidase activity was quantified. Data represent the mean ± SD of three independent experiments performed in triplicate. (E and F) E. coli strain EC100 harboring indicated pBAD33-ompR aspartate 55 mutant constructs (i.e., D55E or D55A) with either aphA-lacZ or aphB-lacZ reporter plasmids was cultured in LB broth with the indicated arabinose concentrations for 5 h when β-galactosidase activity was quantified. Data represent the mean ± SD of three independent experiments performed in triplicate. *, P < 0.01; ****, P < 0.001 relative to 0% arabinose determined by Sidak’s multiple-comparison test.
In the second set of experiments, we expressed V. cholerae ompR from pTB11 in E. coli bearing aphA-lacZ or aphB-lacZ transcriptional reporters to address whether OmpR acted directly at the respective promoters. The E. coli strains were cultured to mid-log phase in the presence of varying arabinose concentrations when aphA-lacZ or aphB-lacZ expression was quantified. The results showed that arabinose addition had little effect on aphA expression (Fig. 4C). By contrast, there was an arabinose dose-dependent decrease in aphB expression (Fig. 4D), consistent with OmpR being an aphB repressor. These results suggest that OmpR may act directly at the aphB promoter; however, we cannot exclude the possibility that OmpR could be acting through an intermediate that is present in both E. coli and V. cholerae.
As OmpR has been shown to function as an active transcription factor in both the unphosphorylated and phosphorylated forms, we investigated if the phosphorylation of V. cholerae OmpR plays a role in its repression of aphB. To this end, OmpR point mutants mimicking constitutive phosphorylation (D55E) and constitutive dephosphorylation (D55A) of the conserved aspartate were cloned into pTB11. The mutant ompR constructs were then expressed in E. coli, and aphA and aphB expression was quantified as before. Consistent with the above findings, expression of the phosphomimic ompR mutants did not significantly affect aphA expression (Fig. 4E). By contrast, expression of the ompRD55E allele strongly repressed aphB expression, whereas expression of the ompRD55A allele only marginally repressed aphB, and to a far lesser extent than the ompRD55E allele (Fig. 4F). Taken together, these results indicate that both phosphoforms of V. cholerae OmpR have the capacity to repress aphB transcription, but the phosphorylated form (i.e., ompRD55E) is predominately responsible for aphB repression. Collectively, these results support the conclusion that OmpR negatively regulates the ToxR regulon via directly repressing aphB transcription, and they suggest that the phosphorylated form of OmpR is responsible for this repression.
V. cholerae ompR is induced by bile salts and detergents.
While the above data showed that OmpR functions as a virulence repressor through repression of aphB, we wished to address the environmental factors that modulate ompR expression in V. cholerae. OmpR has been extensively studied in Enterobacteriaceae, where it has been shown to function as an osmoregulator that mediates adaptive responses to osmotic stress (18, 22, 45). We therefore tested if V. cholerae ompR responded to changes in medium osmolarity by quantifying ompR-lacZ expression during growth under AKI conditions in standard AKI broth (86 mM NaCl), AKI broth with low NaCl (21.5 mM), and AKI with excess NaCl (250 mM). As shown in Fig. 5A, the NaCl concentration did not significantly affect ompR expression, suggesting that ompR was not regulated in response to osmolarity. Consistent with this, growth analysis showed that ompR was dispensable for growth in high-osmolarity broth up to 500 mM NaCl in both WT and ΔRND backgrounds (Fig. 5B and C). From these results, we conclude that V. cholerae OmpR is not regulated in response to medium osmolarity and therefore likely responds to different environmental stimuli than what is observed in Enterobacteriaceae.
FIG 5.
V. cholerae ompR does not respond to osmolarity but is induced by membrane-intercalating agents. (A) WT V. cholerae harboring an ompR-lacZ reporter plasmid (pDK9) was cultured under AKI conditions with the indicated NaCl concentrations for 5 h when β-galactosidase activity was quantified. (B and C) Growth analysis of V. cholerae ΔompR mutants. Overnight LB cultures of V. cholerae WT, ΔompR, ΔRND, and ΔRNDΔompR were diluted 1:10,000 in either fresh LB medium (0.172 M NaCl) (B) or LB medium containing a total concentration of 0.5 M NaCl (C) and cultured at 37°C with constant shaking in a microtiter plate reader. Growth was recorded as the OD600 every 30 min. Data indicate the average of at least three independent experiments performed in triplicate. (D and E) WT V. cholerae harboring an ompR-lacZ reporter plasmid was cultured in LB broth for 4 h when the indicated RND efflux substrates (D) or ethanol (E) was added to the culture medium. The cultures were then incubated with shaking for an additional hour when β-galactosidase activity was quantified. Data indicate the average ± SD of three independent experiments performed in triplicate. UT, untreated samples. *, P < 0.01; **, P < 0.001 relative to untreated determined by Dunnett’s multiple-comparison test.
The finding that ompR was induced in the absence of RND-mediated efflux (Fig. 2A) suggested that small molecules that accumulate intracellularly in the absence of RND efflux may play a role in ompR expression. Previous studies showed that a major function of the V. cholerae RND efflux systems was in resistance to hydrophobic and detergent-like molecules, including bile salts, fatty acids, and detergents (37, 46). We therefore tested if bile salts or detergents affected ompR expression as described above. The results showed that the addition of deoxycholate, bile salts, oxgall, and SDS to the growth media increased ompR expression (Fig. 5D). We also tested another small molecule, indole. Indole is a V. cholerae metabolite that is an RND efflux substrate and virulence repressor (46, 47). The data showed that indole did not affect ompR expression, suggesting that altered ompR expression was specific for compounds with detergent-like properties. As detergents are associated with envelope stress due to their membrane-intercalating properties, we hypothesized that ompR may be induced in response to envelope stress. To test this, we quantified ompR expression following the induction of membrane stress by ethanol treatment (48). The results of these experiments showed that there was an ethanol dose-dependent increase in ompR expression (Fig. 5E). Taken together, these findings suggest that V. cholerae ompR is likely regulated in response to membrane perturbations resulting from exposure to membrane-intercalating agents.
C18-depleted AKI medium nullifies ompR induction in an RND-negative V. cholerae strain.
Based on the results above, we hypothesized that hydrophobic and/or nonpolar compounds present in the AKI medium were accumulating in the RND efflux-deficient strain ΔRND and activating ompR transcription. To test this, we generated a depleted AKI medium by passing AKI broth through a Sep Pak C18 cartridge to deplete nonpolar and hydrophobic compounds from the medium. We then quantified ompR expression in WT and RND-negative strains harboring pDK9 (ompR-lacZ) following growth under AKI conditions in AKI broth and in the C18-depleted AKI medium. The results showed increased ompR expression in ΔRND during growth in AKI broth as expected (Fig. 6). Growth of the WT in the C18-depleted AKI medium did not affect ompR expression when compared to expression in the standard AKI medium. However, growth of ΔRND in the C18-depleted AKI medium alleviated the increase in ompR transcription observed in the standard AKI medium. To determine if the hydrophobic compounds from the AKI medium that were retained on the C18 column were responsible for ompR induction in ΔRND, we eluted the retained compounds from the C18 cartridges used to extract AKI and LB broth. We then determined if the respective eluates contained ompR-inducing compounds by adding them to LB broth cultures of ΔRND and WT and quantifying ompR-lacZ expression. The results showed that the addition of the AKI medium C18 column eluate, but not LB C18 column eluate, activated ompR expression in ΔRND, while neither eluate had an effect on ompR expression in the WT (Fig. 6). Collectively, these data suggest that hydrophobic and/or nonpolar compounds present in the AKI medium are responsible for increased ompR expression in the RND-negative strain. The fact that the C18-depleted AKI medium did not affect ompR expression in the WT indicated that this phenotype was RND dependent. Significantly, we also observed that the increase in ompR expression in ΔRND was not dependent on growth under AKI conditions (i.e., static growth followed by shaken growth), as ompR expression was also enhanced in cultures grown in AKI broth under noninducing conditions (not shown). This observation, combined with the finding that ompR was not induced in ΔRND during growth in LB broth or T-medium (Fig. 2A), suggested that the ompR-inducing molecules were only present in the AKI medium. From these experiments, we conclude that hydrophobic and/or nonpolar compounds that are present in AKI medium, but not the LB medium, are responsible for ompR activation in ΔRND. Further, because this phenotype was RND efflux dependent, we infer that the inducing compounds are substrates for the V. cholerae RND efflux systems.
FIG 6.
C18-depleted AKI medium abolishes the RND efflux-dependent induction of ompR expression in V. cholerae. (A) WT and ΔRND V. cholerae strains harboring an ompR-lacZ reporter plasmid (pDK9) were cultured in the indicated media for 5 h when ompR-lacZ expression was quantified. *, P < 0.01 relative to WT determined by a t test. (B) WT and ΔRND V. cholerae strains were cultured under AKI conditions in either an AKI medium or a C18-depleted AKI medium for 24 h when CT and TcpA production was assessed by GM1 ELISA and TcpA immunoblotting, respectively. Data represent the mean ± SD of three independent experiments. *, P < 0.01 relative to AKI, determined by t test. C18-depleted AKI medium and C18 column AKI medium eluates were prepared as described in Materials and Methods.
The observation that growth in C18-depleted AKI medium resulted in an RND efflux-dependent repression of ompR expression, coupled with the finding that OmpR is a virulence regulator, led us to explore if C18 depletion of the AKI medium also affected virulence factor production. To explore this, CT and TcpA production was quantified in WT and ΔRND strains cultured under AKI conditions in either standard AKI medium or a C18-depleted AKI medium. As previously illustrated, the ΔRND strain had reduced CT and TcpA production compared to WT when cultured in the standard AKI medium. Interestingly, growth of WT in the C18-depleted AKI medium resulted in a reduction in both CT and TcpA production (Fig. 6B). Inversely, growth of a ΔRND strain in the C18-depleted AKI medium resulted in an increase in both CT and TcpA production to levels relative to standard AKI-cultured WT. Collectively, these data suggest that the nonpolar and hydrophobic compounds that are extracted from the AKI medium by the Sep Pak C18 column play a role in both ompR expression and virulence factor production. The nature of these molecules will require further investigation.
DISCUSSION
V. cholerae is an inhabitant of the aquatic ecosystem that can colonize the human gastrointestinal tract to cause disease. The ability of V. cholerae to replicate in these two disparate ecosystems is dependent upon its ability to rapidly adapt to the changing environments it encounters. For example, upon host entry, V. cholerae must adjust to dramatic changes in temperature, pH, salinity, oxygen tension, and the presence of antimicrobial compounds. At the same time, colonization of the intestinal tract requires the expression of niche-specific genes (e.g., virulence factors). Prior to exiting the host, V. cholerae must also regulate the expression of genes that are important for transmission and dissemination (9–12). How all of these responses are integrated within the dynamic environment in the host is poorly understood. What is clear is that periplasmic sensing systems play a critical role in the process. This includes ToxR, which regulates host entry; the Cad system, which contributes to acid tolerance; the CarRS TCS, which mediates antimicrobial peptide resistance; OscR, which regulates response to osmolality; and stress responses like the Cpx system that alleviate stress due to the presence of antimicrobial compounds in this host (1, 40–42, 49–51).
In this study, we interrogated the function of six regulatory genes on virulence factor production in V. cholerae. All of the tested regulatory genes were identified as being upregulated in an RND efflux-negative V. cholerae mutant (38). As RND-mediated efflux is required for virulence factor production, these induced regulatory genes represented potential efflux-dependent virulence repressors. We found that ompR contributed to repression of virulence factor production in the RND-null strain by repressing aphB expression. AphB is a key regulator in the ToxR virulence regulon (3). Previous studies have shown that AphB activity is modulated by low oxygen and acidic pH, but it was unknown whether expression of aphB was itself regulated (52). To our knowledge, OmpR is the first regulator shown to modulate aphB expression in V. cholerae. We further demonstrated that ompR was activated in response to membrane-intercalating compounds that are abundant in the host, suggesting that this regulatory circuit may be relevant in vivo.
Although the function of OmpR has been widely explored in Enterobacteriaceae, the function of the V. cholerae OmpR homolog has not been investigated previously. OmpR is known as an osmoregulator in Enterobacteriaceae. Expression of ompR is induced at high salt concentrations, and OmpR regulates transcriptional responses to alleviate osmotic stress (16, 53, 54). Herein we report that V. cholerae ompR was not induced in response to osmolality and that ompR was dispensable for growth at high salt concentrations. These findings were consistent with two previous studies on V. cholerae responses to osmolarity (50, 55), neither of which identified ompR as one of the genes to respond to increased osmolarity. In the latter study, OscR was identified as an osmoregulator which regulated motility and biofilm formation (50). We did not observe any effect of ompR on either of these two phenotypes (not shown), suggesting that OscR and OmpR function independently. Taken together, these results suggest that V. cholerae OmpR has evolved to respond to different environmental stimuli and fulfill new functions.
Bacterial regulatory networks evolve in response to evolutionary pressures placed on individual species, as they inhabit specific niches (56, 57). TCSs have been suggested to evolve under such selective pressures to respond to novel stimuli and regulate diverse target genes to meet the needs of specific bacterial species (58). EnvZ-OmpR is an example of this. While EnvZ-OmpR is ubiquitous in Gammaproteobacteria, its function appears to have evolved divergently in several bacterial species (20, 23, 24, 31, 59). Our results suggest that this divergent evolution has also occurred in V. cholerae. We speculate that the lifestyle of V. cholerae, which involves growth in murine environments and the human host gastrointestinal tract, has selected for OmpR to respond to novel stimuli and to fulfill a novel physiological role in V. cholerae. Sequence comparison of the V. cholerae ompR locus to the E. coli ompR locus supports this hypothesis. While V. cholerae OmpR is 83% identical in amino acid sequence to its E. coli homolog, the V. cholerae EnvZ sensor kinase is only 47% identical to its E. coli counterpart.
OmpR functioned as a repressor of virulence factor production, and its expression was induced by the addition of membrane-intercalating compounds such as detergents and ethanol. Further, our OmpR phosphomimic studies suggested that it was the phosphorylated form of OmpR that was responsible for virulence repression. This likely explains the upregulation of ompR in the RND-negative background, as cells lacking RND-mediated efflux are hypersensitive to membrane-intercalating compounds due to the RND mutant's diminished ability to actively efflux these compounds from within the cell (37, 46, 60). We speculate that virulence repression in the RND-null mutant resulted from the intracellular accumulation of nonpolar and hydrophobic molecules that are present in AKI medium (e.g., fatty acids and detergent-like molecules). This hypothesis is supported by the finding that WT and RND-negative V. cholerae strains have comparable ompR expression when cultured in a C18 cartridge-depleted AKI medium. These molecules are likely the substrates of the RND transporters and thus accumulated in the RND-negative mutant, resulting in ompR induction and subsequent virulence repression. This is supported by the observations that OmpR contributes to virulence repression in an RND-negative strain and that growth of the RND-negative strain in the C18-extracted AKI medium attenuated ompR expression.
Bile salts are found at high concentrations in the lumen of the small intestine. The finding that ompR was activated in response to bile salts suggests the possibility that OmpR could contribute to spatial and temporal virulence regulation that has been observed in vivo (61). Following ingestion, V. cholerae enters the host small intestine, migrates to the intestinal epithelium, and activates virulence factor production while colonizing the intestinal epithelium. This suggests the possibility that high bile salt concentrations within the intestinal lumen may activate ompR to repress virulence factor production. Once V. cholerae traverses the mucus layer to colonize the epithelial surface, where bile salt concentrations are reduced, OmpR-mediated virulence repression would be alleviated. This tight regulation of virulence factor production is paramount to the pathogenic success of V. cholerae. It is interesting to speculate that V. cholerae OmpR is one of multiple factors that converge on the ToxR regulon to ensure that it is only expressed in the appropriate in vivo niche. It is noteworthy that bile salts and fatty acids have pleiotropic effects on the ToxR regulon. Fatty acids have been shown to negatively affect ToxT activity (62). Bile salts have been implicated in intermolecular disulfide bond formation in TcpP (63). Bile acids, fatty acids, and other detergent-like compounds also signal through ToxR to repress aphA (38, 64). Thus, there seems to be a coordinated response by V. cholerae to these environmental cues to effect virulence factor production.
The induction of V. cholerae ompR in response to nonspecific membrane-intercalating agents suggests that OmpR could also function as part of a generalized membrane stress response. Consistent with this, there is evidence that OmpR in Enterobacteriaceae may be a component of other stress response systems (65, 66). A conserved response to membrane stress in bacteria includes suppressing membrane protein production as a mechanism to alleviate envelope stress. Thus, OmpR-dependent virulence repression in V. cholerae could conceivably contribute to a membrane stress response because the ToxR regulon controls the expression of many membrane-bound and secreted proteins, including the two major outer membrane porins OmpU and OmpT (67). However, analysis of WT and ΔompR whole-cell lysates by SDS-PAGE staining did not reveal any effect of OmpR on the production of OmpU and OmpT (not shown), which is consistent with the finding that OmpR did not affect toxR expression or protein production (Fig. 3 and data not shown). This contrasts what is observed in other bacterial species where OmpR regulates the expression of outer membrane porins (16–18, 21, 68).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains EC100Dpir+ and SM10λpir were used for cloning and plasmid conjugation, respectively. V. cholerae strain JB58 was used as the WT in all experiments. Bacterial strains were routinely grown at 37°C in lysogeny broth or on LB agar. AKI growth conditions were used to induce V. cholerae virulence gene expression as previously described (69). The modified T medium was prepared as previously described (70). Antibiotics were used at the following concentrations: streptomycin (58), 100 μg/ml; carbenicillin (Cb), 100 μg/ml; and chloramphenicol (Cm), 20 μg/ml for E. coli and 2 μg/ml for V. cholerae. The C18-conditioned AKI medium was prepared as follows. Sep-Pak C18 cartridges (Waters) were preconditioned with 10 ml of 100% methanol followed by 10 ml of sterile double-distilled water (ddH2O) before 50 ml of AKI broth was passed through the cartridge and the flowthrough collected and used as a conditioned AKI broth. Molecules that were retained on the C18 columns following the passage of LB or AKI broth medium were eluted from the column with 10 ml of 100% methanol. The eluates were concentrated by evaporation. The resulting residue was resuspended in a volume of LB broth that was identical to the volume of the extracted AKI broth and filter sterilized prior to use.
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or primer | Genotype, characteristics, or sequence (5′–3′) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| EC100Dpir+ | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ- rpsL (StrR) nupG pir+ | Epicenter |
| SM10λpir | thi-1 thr leu tonA lacY supE recA::RP4-2-4-Tc::Mu Kmr (λ pirR6K) | 67 |
| V. cholerae | ||
| JB58 | 01 El Tor strain N16961 ΔlacZ Smr | Lab collection |
| ΔRND | JB58 ΔvexB ΔvexD ΔvexF ΔvexH ΔvexK ΔvexM | 37 |
| DK243 | JB58 ΔompR | This work |
| DK246 | ΔRND ΔompR | This work |
| Plasmids | ||
| pBAD33 | Arabinose-regulated expression vector | 39 |
| pTB1 | pBAD33 expressing VC0486 | This work |
| pTB3 | pBAD33 expressing VC1320-VC1319 (carRS) | This work |
| pTB5 | pBAD33 expressing VC1081 | This work |
| pTB7 | pBAD33 expressing VC1638 | This work |
| pTB9 | pBAD33 expressing VC1825 | This work |
| pTB11 | pBAD33 expressing VC2714 (ompR) | This work |
| pDK14 | pBAD33 expressing VC2714-D55E | This work |
| pDK15 | pBAD33 expressing VC2714-D55A | This work |
| pTB15 | pBAD33 expressing VC1320 (carR) | This work |
| pTL61T | Reporter plasmid for making transcriptional fusions to lacZ, Ampr | 72 |
| pDK9 | pTL61T containing the ompR promoter | This work |
| pXB192 | pTL61T containing the toxT promoter | 37 |
| pXB193 | pTL61T containing the ctxAB promoter | 73 |
| pXB194 | pTL61T containing the tcpA promoter | 73 |
| pXB201 | pTL61T containing the toxRS promoter | 73 |
| pXB202 | pTL61T containing the aphA promoter | 73 |
| pXB203 | pTL61T containing the aphB promoter | 73 |
| pXB266 | pTL61T containing the leuO promoter | 64 |
| pWM91 | Suicide vector used for allelic exchange, Ampr | 67 |
| pWM91-ΔompR | Suicide vector used for deletion of ompR | This work |
| Primers | ||
| P-VC2714-F-XhoI | GGCTCGAGAACTCGATTGAGTATGAGAAAGG | |
| P-VC2714-R-XbaI | AATCTAGACCATGATCCCACCTAACTGTTGTTC | |
| VC2714-F1-XhoI | TTCTCGAGTGCGGCTTTGCTGTCGGCGAC | |
| VC2714-R1-BamHI | GCGGATCCCACCTTGGCTGCGATTGCTAAC | |
| VC2714-F2 | ATGCGCGCTTGCGTTCCTGATGGTAAAGCCGCCAAC | |
| VC2714-R2 | ACCATCAGGAACGCAAGCGCGCATCATCATCTACCAC | |
| VC2714-F-SacI | AAGAGCTCAACAGTTAGGTGGGATCATG | |
| VC2714-R-SmaI | TTCCCGGGCTAAAAGAAGTTAGTTGGCGGC | |
| aphA-F | GCAGAACCTTACCGTCTGCAA | |
| aphA-R | GCGTAATAAGCGGCTTCGATT | |
| aphB-F | ATCGGTGAAGTGAAAGACATTTTGG | |
| aphB-R | GATGTTGATGCAACTCTTCAGCAT | |
| ToxRS-F | CGTCAAAACGGTTCCGAAACG | |
| ToxRS-R | CGCGAGCCATCTCTTCTTCAA | |
| tcpP-F | TAGCCGGCATTACTCATGATCTAC | |
| tcpP-R | TTGTTATCCCCGGTAACCTTGC | |
| gyrA-F | CAATGCCGGTACACTGGTACG | |
| gyrA-R | AAGTACGGATCAGGGTCACG |
Plasmid and mutant construction.
Oligonucleotides used in this study are listed in Table 1. Chromosomal DNA from the WT was used as the PCR template for cloning experiments. The ompR-lacZ reporter plasmid pDK9 was generated as follows. The ompR promoter region was amplified by PCR using the P-VC2714-F-XhoI and P-VC2714-R-XbaI oligonucleotide primers. The resulting amplicon was digested with XhoI and XbaI restriction endonucleases and ligated into a similarly digested pTL61T vector to generate the plasmid pDK9. The ompR expression vector pTB11 was created by amplifying ompR using the VC2714-F-SacI and VC2714-R-SmaI oligonucleotide primers. The resulting 766-bp fragment was digested with SacI and SmaI restriction endonucleases and ligated into similarly digested pBAD33 to generate pTB11. The other expression plasmids (pTB3, pTB5, pTB7, pTB9, and pTB15) were made in a similar manner. The OmpR aspartate 55 mutant expression plasmids were constructed as follows. Internal fragments (364 bp) of the ompR gene containing D55E and D55A mutations were custom synthesized by Integrated DNA Technologies. The resulting fragments were digested with PmlI and EcoRI before being ligated into similarly digested pTB11 to generate plasmids pDK14 and pDK15. The ompR (VC2714) deletion construct was constructed as follows. Primer pairs ompR-F1/ompR-R2 and ompR-F2/ompR-R1 were used in separate PCRs with N16961 genomic DNA. The two resulting amplicons (∼1.5 kb each) were collected and used as the template for the second-round PCR amplification with the flanking ompR-F1 and ompR-R1 PCR primers. The resulting ∼3-kb amplicon was then digested with the SpeI and SmaI restriction endonucleases before being ligated into similarly digested pWM91 vector to generate pWM91-ΔompR. pWM91-ΔompR was then used to delete ompR through allelic exchange as previously described (37). All plasmids were validated via DNA sequencing.
Transcriptional reporter assays.
V. cholerae and E. coli strains containing the indicated lacZ reporters were cultured under AKI conditions, in LB broth, or in a modified T medium. At the indicated times, aliquots were collected in triplicate, and β-galactosidase activity was quantified as previously described (71). The experiment quantifying ompR expression during growth under varying NaCl concentrations was performed as follows. WT strains harboring pDK9 were cultured under virulence factor-inducing conditions in AKI medium containing the indicated NaCl concentrations for 5 h. Culture aliquots were then collected in triplicate, and β-galactosidase production was assessed. The experiments quantifying gene expression responses to bile salts, deoxycholate, SDS, oxgall, indole, and ethanol were performed as follows. The indicated strains were grown in LB broth at 37°C with shaking for 4 h when the indicated compounds were added to the cultures. Thereafter, the cultures were then incubated with shaking for an additional hour before culture aliquots were collected in triplicate and β-galactosidase production was assessed. All of the transcriptional reporter experiments were performed independently at least three times.
Determination of CT and TcpA production.
CT production was determined by ganglioside GM1 enzyme-linked immunosorbent CT assays as previously described by use of purified CT (Sigma) as a standard (37). The production of TcpA was determined by Western immunoblotting as previously described (10).
Growth curve experiments.
Growth curves were generated in microtiter plates. Overnight cultures of WT, ΔRND, ΔompR, and ΔRNDΔompR strains grown in LB broth were washed in phosphate-buffered saline (PBS) and then diluted 1:10,000 in fresh LB broth containing either 0.172 M NaCl or 0.5 M NaCl. Two hundred microliters of the diluted cultures was then aliquoted in triplicate wells of a 96-well microtiter plate. The microtiter plates were then incubated at 37°C with constant shaking, and the optical density at 600 nm (OD600) was measured every 30 min using a BioTek Synergy microplate reader.
Quantitative real-time PCR.
V. cholerae strains were grown under AKI conditions for 3.5 h when total RNA was isolated from the cultures using TRIzol (Invitrogen) per the manufacturer’s directions. cDNA was generated from the purified RNA using the Maxima First Strand cDNA synthesis kit (Thermo). The expression levels of specific genes were quantified by amplifying 25 ng of cDNA with 0.3 μM primers using the SYBR green PCR mix (Thermo) on a StepOnePlus real-time PCR System (Applied Biosystems). The relative expression levels of genes in the mutant and WT cultures were calculated using the 2−ΔΔCT method. The presented results are the means ± standard deviations from three biological replicates, with each biological replicate being generated from three technical replicates. DNA gyrase (gyrA) was used as the internal control.
ACKNOWLEDGMENTS
This work was supported by the NIH under award numbers R01AI132460 and R21AI141934. D.E.K. was supported in part by training grant AI049820.
The content is solely the responsibility of the authors.
REFERENCES
- 1.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A 84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Childers BM, Klose KE. 2007. Regulation of virulence in Vibrio cholerae: the ToxR regulon. Future Microbiol 2:335–344. doi: 10.2217/17460913.2.3.335. [DOI] [PubMed] [Google Scholar]
- 3.Kovacikova G, Skorupski K. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J Bacteriol 181:4250–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skorupski K, Taylor RK. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol 31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 5.Peterson KM, Gellings PS. 2018. Multiple intraintestinal signals coordinate the regulation of Vibrio cholerae virulence determinants. Pathog Dis 76:ftx126. doi: 10.1093/femspd/ftx126. [DOI] [PubMed] [Google Scholar]
- 6.Cakar F, Zingl FG, Moisi M, Reidl J, Schild S. 2018. In vivo repressed genes of Vibrio cholerae reveal inverse requirements of an H+/Cl- transporter along the gastrointestinal passage. Proc Natl Acad Sci U S A 115:E2376–E2385. doi: 10.1073/pnas.1716973115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsiao A, Liu Z, Joelsson A, Zhu J. 2006. Vibrio cholerae virulence regulator-coordinated evasion of host immunity. Proc Natl Acad Sci U S A 103:14542–14547. doi: 10.1073/pnas.0604650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cakar F, Zingl FG, Schild S. 2019. Silence is golden: gene silencing of V. cholerae during intestinal colonization delivers new aspects to the acid tolerance response. Gut Microbes 10:228–234. doi: 10.1080/19490976.2018.1502538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bina J, Zhu J, Dziejman M, Faruque S, Calderwood S, Mekalanos J. 2003. ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc Natl Acad Sci U S A 100:2801–2806. doi: 10.1073/pnas.2628026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bina XR, Taylor DL, Vikram A, Ante VM, Bina JE. 2013. Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo(Phe-Pro). mBio 4:e00366-13. doi: 10.1128/mBio.00366-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larocque RC, Harris JB, Dziejman M, Li X, Khan AI, Faruque AS, Faruque SM, Nair GB, Ryan ET, Qadri F, Mekalanos JJ, Calderwood SB. 2005. Transcriptional profiling of Vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect Immun 73:4488–4493. doi: 10.1128/IAI.73.8.4488-4493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schild S, Tamayo R, Nelson EJ, Qadri F, Calderwood SB, Camilli A. 2007. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2:264–277. doi: 10.1016/j.chom.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng AT, Ottemann KM, Yildiz FH. 2015. Vibrio cholerae response regulator VxrB controls colonization and regulates the type VI secretion system. PLoS Pathog 11:e1004933. doi: 10.1371/journal.ppat.1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galperin MY. 2010. Diversity of structure and function of response regulator output domains. Curr Opin Microbiol 13:150–159. doi: 10.1016/j.mib.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forst S, Delgado J, Inouye M. 1989. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci U S A 86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarma V, Reeves P. 1977. Genetic locus (ompB) affecting a major outer-membrane protein in Escherichia coli K-12. J Bacteriol 132:23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall MN, Silhavy TJ. 1981. Genetic analysis of the major outer membrane proteins of Escherichia coli. Annu Rev Genet 15:91–142. doi: 10.1146/annurev.ge.15.120181.000515. [DOI] [PubMed] [Google Scholar]
- 18.Slauch JM, Garrett S, Jackson DE, Silhavy TJ. 1988. EnvZ functions through OmpR to control porin gene expression in Escherichia coli K-12. J Bacteriol 170:439–441. doi: 10.1128/jb.170.1.439-441.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin TH, Chen Y, Kuo JT, Lai YC, Wu CC, Huang CF, Lin CT. 2018. Phosphorylated OmpR is required for type 3 fimbriae expression in Klebsiella pneumoniae under hypertonic conditions. Front Microbiol 9:2405. doi: 10.3389/fmicb.2018.02405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tipton KA, Rather PN. 2017. An ompR-envZ two-component system ortholog regulates phase variation, osmotic tolerance, motility, and virulence in Acinetobacter baumannii strain AB5075. J Bacteriol 199:e00705-16. doi: 10.1128/JB.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao H, Zhang Y, Han Y, Yang L, Liu X, Guo Z, Tan Y, Huang X, Zhou D, Yang R. 2011. Phenotypic and transcriptional analysis of the osmotic regulator OmpR in Yersinia pestis. BMC Microbiol 11:39. doi: 10.1186/1471-2180-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernardini ML, Fontaine A, Sansonetti PJ. 1990. The two-component regulatory system ompR-envZ controls the virulence of Shigella flexneri. J Bacteriol 172:6274–6281. doi: 10.1128/jb.172.11.6274-6281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee AK, Detweiler CS, Falkow S. 2000. OmpR regulates the two-component system SsrA-ssrB in Salmonella pathogenicity island 2. J Bacteriol 182:771–781. doi: 10.1128/jb.182.3.771-781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva IN, Pessoa FD, Ramires MJ, Santos MR, Becker JD, Cooper VS, Moreira LM, Silva IN, Pessoa FD, Ramires MJ, Santos MR, Becker JD, Cooper VS, Moreira LM. 2018. The OmpR regulator of Burkholderia multivorans controls mucoid-to-nonmucoid transition and other cell envelope properties associated with persistence in the cystic fibrosis lung. J Bacteriol 200:e00216-18. doi: 10.1128/JB.00216-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwan WR. 2009. Survival of uropathogenic Escherichia coli in the murine urinary tract is dependent on OmpR. Microbiology 155:1832–1839. doi: 10.1099/mic.0.026187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reboul A, Lemaitre N, Titecat M, Merchez M, Deloison G, Ricard I, Pradel E, Marceau M, Sebbane F. 2014. Yersinia pestis requires the 2-component regulatory system OmpR-EnvZ to resist innate immunity during the early and late stages of plague. J Infect Dis 210:1367–1375. doi: 10.1093/infdis/jiu274. [DOI] [PubMed] [Google Scholar]
- 27.Bang IS, Kim BH, Foster JW, Park YK. 2000. OmpR regulates the stationary-phase acid tolerance response of Salmonella enterica serovar typhimurium. J Bacteriol 182:2245–2252. doi: 10.1128/jb.182.8.2245-2252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stincone A, Daudi N, Rahman AS, Antczak P, Henderson I, Cole J, Johnson MD, Lund P, Falciani F. 2011. A systems biology approach sheds new light on Escherichia coli acid resistance. Nucleic Acids Res 39:7512–7528. doi: 10.1093/nar/gkr338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heyde M, Portalier R. 1987. Regulation of major outer membrane porin proteins of Escherichia coli K 12 by pH. Mol Gen Genet 208:511–517. doi: 10.1007/bf00328148. [DOI] [PubMed] [Google Scholar]
- 30.Hu Y, Lu P, Wang Y, Ding L, Atkinson S, Chen S. 2009. OmpR positively regulates urease expression to enhance acid survival of Yersinia pseudotuberculosis. Microbiology 155:2522–2531. doi: 10.1099/mic.0.028381-0. [DOI] [PubMed] [Google Scholar]
- 31.Quinn HJ, Cameron AD, Dorman CJ. 2014. Bacterial regulon evolution: distinct responses and roles for the identical OmpR proteins of Salmonella Typhimurium and Escherichia coli in the acid stress response. PLoS Genet 10:e1004215. doi: 10.1371/journal.pgen.1004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li XZ, Plesiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Bambeke F, Glupczynski Y, Plesiat P, Pechere JC, Tulkens PM. 2003. Antibiotic efflux pumps in prokaryotic cells: occurrence, impact on resistance and strategies for the future of antimicrobial therapy. J Antimicrob Chemother 51:1055–1065. doi: 10.1093/jac/dkg224. [DOI] [PubMed] [Google Scholar]
- 34.Piddock LJ. 2006. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol 4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 35.Taylor DL, Bina XR, Slamti L, Waldor MK, Bina JE. 2014. Reciprocal regulation of resistance-nodulation-division efflux systems and the Cpx two-component system in Vibrio cholerae. Infect Immun 82:2980–2991. doi: 10.1128/IAI.00025-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunkle DE, Bina XR, Bina JE, Kunkle DE, Bina XR, Bina JE. 2017. The Vibrio cholerae VexGH RND efflux system maintains cellular homeostasis by effluxing Vibriobactin. mBio 8:e00126-17. doi: 10.1128/mBio.00126-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bina XR, Provenzano D, Nguyen N, Bina JE. 2008. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect Immun 76:3595–3605. doi: 10.1128/IAI.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bina XR, Howard MF, Taylor-Mulneix DL, Ante VM, Kunkle DE, Bina JE. 2018. The Vibrio cholerae RND efflux systems impact virulence factor production and adaptive responses via periplasmic sensor proteins. PLoS Pathog 14:e1006804. doi: 10.1371/journal.ppat.1006804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bilecen K, Fong JCN, Cheng A, Jones CJ, Zamorano-Sánchez D, Yildiz FH. 2015. Polymyxin B resistance and biofilm formation in Vibrio cholerae are controlled by the response regulator CarR. Infect Immun 83:1199–1209. doi: 10.1128/IAI.02700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bilecen K, Yildiz FH. 2009. Identification of a calcium-controlled negative regulatory system affecting Vibrio cholerae biofilm formation. Environ Microbiol 11:2015–2029. doi: 10.1111/j.1462-2920.2009.01923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herrera CM, Crofts AA, Henderson JC, Pingali SC, Davies BW, Trent MS. 2014. The Vibrio cholerae VprA-VprB two-component system controls virulence through endotoxin modification. mBio 5:e02283-14. doi: 10.1128/mBio.02283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matson JS, Livny J, DiRita VJ. 2017. A putative Vibrio cholerae two-component system controls a conserved periplasmic protein in response to the antimicrobial peptide polymyxin B. PLoS One 12:e0186199. doi: 10.1371/journal.pone.0186199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayes CA, Dalia TN, Dalia AB. 2017. Systematic genetic dissection of PTS in Vibrio cholerae uncovers a novel glucose transporter and a limited role for PTS during infection of a mammalian host. Mol Microbiol 104:568–579. doi: 10.1111/mmi.13646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brzostek K, Raczkowska A, Zasada A. 2003. The osmotic regulator OmpR is involved in the response of Yersinia enterocolitica O:9 to environmental stresses and survival within macrophages. FEMS Microbiol Lett 228:265–271. doi: 10.1016/S0378-1097(03)00779-1. [DOI] [PubMed] [Google Scholar]
- 46.Taylor DL, Ante VM, Bina XR, Howard MF, Bina JE. 2015. Substrate-dependent activation of the Vibrio cholerae vexAB RND efflux system requires vexR. PLoS One 10:e0117890. doi: 10.1371/journal.pone.0117890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howard MF, Bina XR, Bina JE, Howard MF, Bina XR, Bina JE. 2019. Indole inhibits ToxR regulon expression in Vibrio cholerae. Infect Immun 87:e00776-18. doi: 10.1128/IAI.00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dombek KM, Ingram LO. 1984. Effects of ethanol on the Escherichia coli plasma membrane. J Bacteriol 157:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merrell DS, Camilli A. 1999. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol Microbiol 34:836–849. doi: 10.1046/j.1365-2958.1999.01650.x. [DOI] [PubMed] [Google Scholar]
- 50.Shikuma NJ, Yildiz FH. 2009. Identification and characterization of OscR, a transcriptional regulator involved in osmolarity adaptation in Vibrio cholerae. J Bacteriol 191:4082–4096. doi: 10.1128/JB.01540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogt SL, Raivio TL. 2012. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol Lett 326:2–11. doi: 10.1111/j.1574-6968.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- 52.Kovacikova G, Lin W, Skorupski K. 2010. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J Bacteriol 192:4181–4191. doi: 10.1128/JB.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alphen WV, Lugtenberg B. 1977. Influence of osmolarity of the growth medium on the outer membrane protein pattern of Escherichia coli. J Bacteriol 131:623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chakraborty S, Kenney LJ. 2018. A new role of OmpR in acid and osmotic stress in Salmonella and E. coli. Front Microbiol 9:2656. doi: 10.3389/fmicb.2018.02656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu X, Liang W, Du P, Yan M, Kan B. 2014. Transcript changes in Vibrio cholerae in response to salt stress. Gut Pathog 6:47. doi: 10.1186/s13099-014-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lozada-Chavez I, Janga SC, Collado-Vides J. 2006. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res 34:3434–3445. doi: 10.1093/nar/gkl423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor TB, Mulley G, Dills AH, Alsohim AS, McGuffin LJ, Studholme DJ, Silby MW, Brockhurst MA, Johnson LJ, Jackson RW. 2015. Evolutionary resurrection of flagellar motility via rewiring of the nitrogen regulation system. Science 347:1014–1017. doi: 10.1126/science.1259145. [DOI] [PubMed] [Google Scholar]
- 58.Perez JC, Shin D, Zwir I, Latifi T, Hadley TJ, Groisman EA. 2009. Evolution of a bacterial regulon controlling virulence and Mg2+ homeostasis. PLoS Genet 5:e1000428. doi: 10.1371/journal.pgen.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cameron AD, Dorman CJ. 2012. A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet 8:e1002615. doi: 10.1371/journal.pgen.1002615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor DL, Bina XR, Bina JE. 2012. Vibrio cholerae VexH encodes a multiple drug efflux pump that contributes to the production of cholera toxin and the toxin co-regulated pilus. PLoS One 7:e38208. doi: 10.1371/journal.pone.0038208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee SH, Hava DL, Waldor MK, Camilli A. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 62.Lowden MJ, Skorupski K, Pellegrini M, Chiorazzo MG, Taylor RK, Kull FJ. 2010. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc Natl Acad Sci U S A 107:2860–2865. doi: 10.1073/pnas.0915021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang M, Liu Z, Hughes C, Stern AM, Wang H, Zhong Z, Kan B, Fenical W, Zhu J. 2013. Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc Natl Acad Sci U S A 110:2348–2353. doi: 10.1073/pnas.1218039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ante VM, Bina XR, Bina JE. 2015. The LysR-type regulator LeuO regulates the acid tolerance response in Vibrio cholerae. Microbiology 161:2434–2443. doi: 10.1099/mic.0.000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerken H, Misra R. 2010. MzrA-EnvZ interactions in the periplasm influence the EnvZ/OmpR two-component regulon. J Bacteriol 192:6271–6278. doi: 10.1128/JB.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jubelin G, Vianney A, Beloin C, Ghigo JM, Lazzaroni JC, Lejeune P, Dorel C. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol 187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gibson MM, Ellis EM, Graeme-Cook KA, Higgins CF. 1987. OmpR and EnvZ are pleiotropic regulatory proteins: positive regulation of the tripeptide permease (tppB) of Salmonella typhimurium. Mol Gen Genet 207:120–129. doi: 10.1007/bf00331499. [DOI] [PubMed] [Google Scholar]
- 69.Iwanaga M, Kuyyakanond T. 1987. Large production of cholera toxin by Vibrio cholerae O1 in yeast extract peptone water. J Clin Microbiol 25:2314–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mey AR, Craig SA, Payne SM. 2012. Effects of amino acid supplementation on porin expression and ToxR levels in Vibrio cholerae. Infect Immun 80:518–528. doi: 10.1128/IAI.05851-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 72.Linn T, St Pierre R. 1990. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol 172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bina XR, Bina JE. 2010. The cyclic dipeptide cyclo(Phe-Pro) inhibits cholera toxin and toxin-coregulated pilus production in O1 El Tor Vibrio cholerae. J Bacteriol 192:3829–3832. doi: 10.1128/JB.00191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]