The authors review the molecular mechanisms regulating IL-10 production and response and describe classic and novel functions of IL-10 in immune and non-immune cells. They further discuss the therapeutic potential of IL-10 in different diseases and the outstanding questions underlying an effective application of IL-10 in clinical settings.
Abstract
The cytokine IL-10 is a key anti-inflammatory mediator ensuring protection of a host from over-exuberant responses to pathogens and microbiota, while playing important roles in other settings as sterile wound healing, autoimmunity, cancer, and homeostasis. Here we discuss our current understanding of the regulation of IL-10 production and of the molecular pathways associated with IL-10 responses. In addition to IL-10’s classic inhibitory effects on myeloid cells, we also describe the nonclassic roles attributed to this pleiotropic cytokine, including how IL-10 regulates basic processes of neural and adipose cells and how it promotes CD8 T cell activation, as well as epithelial repair. We further discuss its therapeutic potential in the context of different diseases and the outstanding questions that may help develop an effective application of IL-10 in diverse clinical settings.
Introduction
The immune system evolved to fight infection while minimizing host damage. Several regulatory mechanisms are in place to ensure the delicate balance between an effective immune response and the appearance of tissue pathology. An anti-inflammatory response, of which IL-10 is the paradigm, is one such mechanism.
IL-10 (Fiorentino et al., 1989; Moore et al., 1990) is the founding member of a family of cytokines that also includes IL-19, IL-20, IL-22, IL-24, IL-26, IL-28A, IL-28B, and IL-29 (reviewed in Ouyang and O’Garra, 2019). The involvement of IL-10 in many disease states has been demonstrated, both in animal models and in humans with mutations in the IL-10/IL-10R axis (reviewed in Engelhardt and Grimbacher, 2014; Shouval et al., 2014b). However, despite considerable progress in IL-10 biology, many outstanding questions still exist. In this review, we revisit the discovery of IL-10, highlight the latest developments toward understanding the metabolic regulation of IL-10 in various cell types, and discuss the molecular signals downstream of the IL-10R in responding cells. We present an overview of the biological functions of IL-10, including some surprising new findings on nonclassical roles for this cytokine. We finish by summarizing the progress made toward the therapeutic manipulation of IL-10.
The discovery of IL-10: A historic perspective
IL-10 was discovered 30 yr ago as a secreted cytokine synthesis inhibitory factor, produced by T helper (Th) 2 cell clones shown to inhibit cytokine production by Th1 cells (Fiorentino et al., 1989). The mouse and human IL-10–coding genes were subsequently cloned, and the predicted protein sequences were found to be highly homologous to an Epstein–Barr virus–encoded protein, BCRF1 (Moore et al., 1990; Vieira et al., 1991). This was the first suggestion that viruses may exploit the inhibitory properties of IL-10 as a mechanism of immune evasion. Indeed, recombinant BCRF1 protein was shown to mimic the activity of IL-10 (Hsu et al., 1990), namely inhibition of cytokine synthesis by activated human peripheral blood mononuclear cells and by a mouse Th1 cell clone (Vieira et al., 1991). Since then, a number of other viruses have also been shown to encode a homologue of the Il10 gene (Fleming et al., 1997; Kotenko et al., 2000; Jayawardane et al., 2008). Soon after the discovery of IL-10, its pleiotropic action was unveiled initially in the mouse, not only as a cytokine synthesis inhibitory factor, but additionally as a mast cell (Thompson-Snipes et al., 1991) and thymocyte (MacNeil et al., 1990) growth factor, and as an activator of B cells (Go et al., 1990; Rousset et al., 1992).
The mechanisms underlying the ability of IL-10 to inhibit cytokine production by Th1 cells were soon unveiled. IL-10–mediated inhibition of IFN-γ secretion by Th1 cells was demonstrated to occur via its action on the APC function of macrophages (Fiorentino et al., 1991b) and by its inhibition of cytokine production by activated macrophages and dendritic cells (DC; Bogdan et al., 1991; Fiorentino et al., 1991a; Macatonia et al., 1993). Additionally, IL-10 inhibited the killing of intracellular pathogens (Gazzinelli et al., 1992; Frei et al., 1993; Vouldoukis et al., 1997). In complementary studies, IL-10 was shown to prevent antigen-specific proliferation of human T cells by inhibition of the antigen-presenting capacity of monocytes through the down-regulation of class II MHC (de Waal Malefyt et al., 1991b). Collectively, these initial studies placed IL-10 as a key mediator of the anti-inflammatory response.
Genetic ablation of Il10 showed its key role in controlling inflammation in vivo, as IL-10–deficient mice developed colitis (Kühn et al., 1993). These findings prompted many genetic association studies between deficiencies in the IL10 or IL10R genes and susceptibility to colitis, an association that is now well acknowledged (Glocker et al., 2009, 2010, 2011; Jostins et al., 2012; Moran et al., 2013; Engelhardt and Grimbacher, 2014; Ellinghaus et al., 2016). However, the role of IL-10 clearly exceeds the regulation of intestinal inflammation, as a function for this molecule has been also described in several other settings, from inflammatory or neurodegenerative diseases to infection or cancer (reviewed in Ouyang et al., 2011; Lobo-Silva et al., 2016; Ouyang and O’Garra, 2019; Wang et al., 2019).
In summary, IL-10 has emerged as a major suppressor of the immune response and a key player in human disease, and is thus an attractive therapeutic target. However, as discussed below, there is recent evidence that IL-10 may play a previously under-appreciated dual role, in some contexts stimulating the immune response instead of suppressing it. This depends on specific cell types and contexts, thus raising important issues related to its modulation in the clinic and calling for further research into the pleiotropic nature of this cytokine.
Molecular mechanisms regulating IL-10 expression
IL-10–producing cells
Although Th2 cells were the first identified cellular source of IL-10 (Fiorentino et al., 1989), the production of this cytokine by other CD4 and also CD8 T cells was soon demonstrated (Yssel et al., 1992; Tanchot et al., 1998). We now know that cells of both the myeloid and lymphoid lineages secrete IL-10 in response to different stimuli (reviewed in Saraiva and O’Garra, 2010; Ouyang and O’Garra, 2019). This includes macrophages, monocytes, DCs, neutrophils, mast cells, eosinophils, and natural killer cells, in addition to CD4 and CD8 T cells and B cells (reviewed in Moore et al., 2001; Gabryšová et al., 2014). More recently, the group of IL-10–producing cells was expanded to resident macrophages, such as microglia (reviewed in Lobo-Silva et al., 2016) and cardiac macrophages (Hulsmans et al., 2018). Notably, some nonhematopoietic cells, including epithelial cells, are also able to produce IL-10 (Jarry et al., 2008). Finally, tumor cells can also produce IL-10, which has been correlated with their ability to cause immunosuppression (Chen et al., 1994; Itakura et al., 2011).
The regulation of IL-10 production: Global concepts
The expression of IL-10 is tightly regulated to avoid diseases related to its excess or deficiency. Both common and cell-specific regulatory mechanisms including the triggering of specific signaling pathways, the expression and activation of particular transcription factors, and post-transcriptional and epigenetic regulation ensure the appropriate production of IL-10 (reviewed in Saraiva and O’Garra, 2010; Gabryšová et al., 2014; Ouyang and O’Garra, 2019). In myeloid cells, the expression of IL-10 accompanies that of pro-inflammatory cytokines, downstream of the activation of multiple pattern recognition receptors (PRRs; reviewed in Saraiva and O’Garra, 2010; Rutz and Ouyang, 2016; Gabryšová et al., 2018; Ouyang and O’Garra, 2019). MAPK are among the key pathways regulating IL-10. Activation of ERK1/2 downstream of the MAP3 kinase tumor progression locus 2 (Tpl2) regulates the expression of Il10 by TLR-activated macrophages and DCs, as shown by the reduction of IL-10 production observed in Tpl2-deficient or upon chemical inhibition of ERK1/2 in macrophages (Yi et al., 2002; Banerjee et al., 2006; Liu et al., 2006; Kaiser et al., 2009). The MAPK p38 is also activated downstream of TLR signaling and shown to regulate IL-10 production by these cells (Yi et al., 2002; Chi et al., 2006; Kim et al., 2008). Several transcription factors known to regulate the transcription of the Il10 gene in macrophages and DCs are activated downstream of ERK1/2 or p38, as is the case for cAMP response element binding protein (CREB), cyclic AMP-dependent transcription factor 1 (ATF1), cFos, and Sp1 (reviewed in Saraiva and O’Garra, 2010; Rutz and Ouyang, 2016; Gabryšová et al., 2018; Ouyang and O’Garra, 2019). Cooperation between ERK and p38 activates the mitogen- and stress-activated protein kinase 1 (MSK1) and MSK2, which promote IL-10 production in TLR4-stimulated macrophages, through the transcription factors CREB and ATF1 (Ananieva et al., 2008). The regulation of IL-10 by MSK/CREB has been further demonstrated in the context of the activation of TLR2 in intestinal macrophages by a Helicobacter hepaticus–derived polysaccharide (Danne et al., 2017). NF-κB activation downstream of PRR also regulates IL-10 production in myeloid cells. Specifically, binding of several NF-κB family members to the Il10 locus promote IL-10 production in TLR4-stimulated macrophages (Kanters et al., 2003; Saraiva et al., 2005; Chakrabarti et al., 2008). Crosstalk between the NF-κB p105 subunit and Tpl2 has also been reported (Banerjee et al., 2006). Finally, the phosphoinositide-3-kinase (PI3K)/serine/threonine protein kinase B (Akt) cascade also regulates the expression of Il10 in macrophages by blocking the kinase glycogen synthase kinase 3β, which inhibits IL-10 production (Ohtani et al., 2008; Nandan et al., 2012), and conversely potentiating IL-10 production through ERK1/2 (Martin et al., 2003) and mechanistic target of rapamycin (mTOR; Ohtani et al., 2008; Weichhart et al., 2008) activation. Thus, activation of ERK1/2 is at the crossroads of several other signaling cascades, demonstrating its importance as a central pathway for Il10 gene regulation in myeloid cells. In addition to direct PRR signaling, coreceptors and other cytokines regulate IL-10 production by macrophages and DCs. CD40, dectin-1, and DC-specific intercellular adhesion molecule-3–grabbing non-integrin enhance PRR-induced IL-10 (reviewed in Saraiva and O’Garra, 2010; Rutz and Ouyang, 2016; Gabryšová et al., 2018; Ouyang and O’Garra, 2019). Similarly, type I IFN has been shown to potentiate IL-10 production by LPS-stimulated (Chang et al., 2007a; Howes et al., 2016) and Mycobacterium tuberculosis–infected (McNab et al., 2014) bone marrow–derived macrophages. Mechanistically, type I IFN induces transcription of the Il10 gene and stabilizes the resulting mRNA, via STAT1 and ERK activation (Howes et al., 2016).
IL-10 is produced by all T cell subsets. In CD4 Th cells, IL-10 production accompanies that of the hallmark cytokines IFN-γ, IL-4/IL-5/IL-13, and IL-17 (reviewed in Saraiva and O’Garra, 2010; Rutz and Ouyang, 2016; Gabryšová et al., 2018; Ouyang and O’Garra, 2019). IL-10 production occurs downstream of TCR triggering, which activates Ras and subsequently MAPK ERK1/2 and AP1, which enhance Il10 transcription (Jones and Flavell, 2005; Wang et al., 2005; Saraiva et al., 2009). Transcription factors of the NFAT family also promote IL-10 production by T cells (Im et al., 2004; Lee et al., 2009). Additionally, STAT4, 6, and 3 activation during Th1, Th2, or Th17 CD4 T cell differentiation, respectively, have been suggested to contribute to Il10 gene expression (Saraiva et al., 2009), as have the basic leucine zipper ATF-like transcription factor and interferon regulatory factor (IRF) 4, which are critical for Th2 and Th17 differentiation (Ciofani et al., 2012; Glasmacher et al., 2012; Li et al., 2012b). Therefore, the mechanisms required for the differentiation and activation of Th cells seem to be required for regulation of IL-10 production, ensuring a feedback regulatory loop to limit immune pathology. In further support of this, the Th2 and Th17 cell lineage transcription factors GATA binding protein 3 (GATA3) and retinoid-related orphan receptor γt, respectively, have been described to modulate IL-10 production by CD4 T cells. GATA3 binds the Il10 promoter, inducing histone acetylation and remodeling the Il10 locus to facilitate Il10 gene expression (Shoemaker et al., 2006; Chang et al., 2007b), although it is not required for IL-10 production by differentiated Th2 cells (Zhu et al., 2004). In contrast, the Th17-associated transcription factor RORγt has been described to repress IL-10 production by Th17 cells (Ciofani et al., 2012). Several reports attest to the importance of PR domain zinc finger protein 1 (Blimp-1) in the regulation of IL-10 production by T cells (reviewed in Rutz and Ouyang, 2016; Ouyang and O’Garra, 2019). Blimp-1 is induced by STAT4 downstream of the IL-12R and shown to be absolutely required for IL-10 expression by IL-12–driven effector Th1 cells both in vitro and in vivo (Neumann et al., 2014). Furthermore, the induction of IL-10 by IL-27 in CD4 T cells requires Blimp-1 (Neumann et al., 2014), likely downstream of STAT1/STAT3 signaling (Stumhofer et al., 2007). IL-27 together with TCR triggering induced the differentiation of naive T cells into IL-10–producing type 1 regulatory T cells (Tr1; Pot et al., 2009). The induction of IL-10 in Tr1 cells is dependent on the cooperation between the transcription factors Eomes and Blimp-1 (Zhang et al., 2017). Of note, in vitro generation of Tr1 cells in the presence of TGF-β, in addition to IL-27, shifts the transcriptional requirements from Blimp-1 to cMaf and aryl hydrocarbon receptor (AhR; Apetoh et al., 2010; Neumann et al., 2014). Blimp-1 is also required for IL-10 expression by CD8 T cells (Sun et al., 2011).
Recent data place the leucine zipper transcription factor cMaf as a master regulator of IL-10 production, common to both myeloid and T cells (reviewed in Saraiva and O’Garra, 2010; Gabryšová et al., 2014; Rutz and Ouyang, 2016; Ouyang and O’Garra, 2019). cMaf was initially shown to bind to the Il10 promoter and enhance gene transcription in TLR4-stimulated macrophages (Cao et al., 2005). More recently, several studies support a role of cMaf in also regulating T cell–derived IL-10 in vitro (Xu et al., 2009; Apetoh et al., 2010; Ciofani et al., 2012; Gabryšová et al., 2018). IL-10 production in vivo by Th1, Th2, and Th17 cells in experimental disease models requires cMaf (Gabryšová et al., 2018). A link between cMaf and IL-10 expression by human Tr1 and Th17 cells has also been proposed (Gandhi et al., 2010; Aschenbrenner et al., 2018). cMaf cooperates with other transcription factors to regulate IL-10 expression. cMaf may require Blimp-1 to enhance IL-10 production by Th1 cells (Neumann et al., 2014). Although not absolutely required, the transcription factor AhR allows maximal IL-10 production by LPS-stimulated macrophages (Kimura et al., 2009) and T cells, where it directly binds to cMaf and synergistically transactivates the Il10 gene promoter (Apetoh et al., 2010). The transcription factor Bhlhe40 has been recently revealed as a repressor of IL-10 production in T cells. Mice deficient in Bhlhe40 are resistant to the development of experimental autoimmune encephalomyelitis, presumably by increasing the IL-10 production by Th1 and Th17 pathogenic cells (Lin et al., 2014). Targeted deletion of Bhlhe40 in either CD4 T cells or CD11c+ cells correlated with increased IL-10 and decreased IFN-γ production and resulted in increased bacterial burdens during infection with M. tuberculosis, rendering Bhlhe40-deficient mice more susceptible to infection (Huynh et al., 2018). Additionally, Bhlhe40 is a molecular switch for IL-10 production in Th1 cells and as a result, mice with conditional deletion of Bhlhe40 in T cells succumbed to Toxoplasma gondii infection (Yu et al., 2018). In both T cells and bone marrow–derived DCs, Bhlhe40 binds to a characterized enhancing region of the Il10 locus (Huynh et al., 2018), where the activating transcription factors AP1 (Jones and Flavell, 2005) and IRF4 (Ahyi et al., 2009) have been shown to bind. The opposing effects of Bhlhe40 and cMaf on the regulation of IL-10 may result from the interaction between these two transcription factors. In the absence of cMaf, up-regulation of Bhlhe40 in T cells has been described (Gabryšová et al., 2018); conversely, deficiency of Bhlhe40 resulted in the up-regulation of Maf (Yu et al., 2018). Very recently, in macrophages, Bhlhe40 has been shown to bind to promoter elements of the Maf gene, repressing its transcription (Jarjour et al., 2019).
Metabolic regulation of IL-10 production
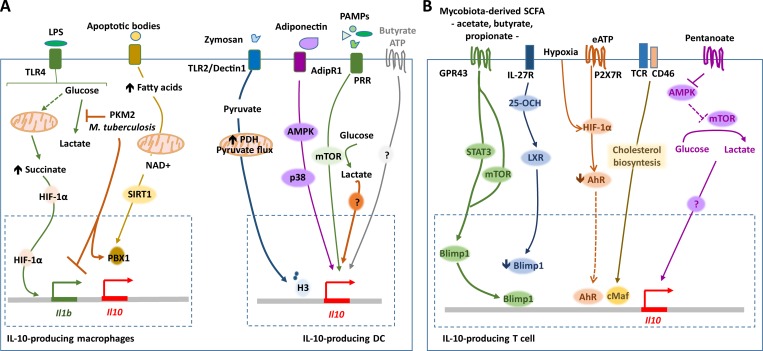
Recent findings show a metabolic reprogramming of immune cells in response to environmental cues (O’Neill and Pearce, 2016). In response to LPS and other stimuli, the metabolic profile of macrophages and DCs shifts toward glycolysis, with the accumulation of metabolites such as citrate and succinate that in turn regulate cytokine gene expression (O’Neill and Pearce, 2016). IL-10 is no exception, its production being regulated by alterations in the cell metabolic networks, as well as by specific metabolites (Fig. 1 A). The LPS-induced metabolic regulator pyruvate kinase isozyme M2 (PKM2) was shown to inhibit the cellular glycolytic reprogramming, succinate accumulation, and IL-1β production, while favoring IL-10 production by macrophages (Palsson-McDermott et al., 2015). Similarly, inhibition of the glycolytic shift promoted by M. tuberculosis infection of macrophages was shown to increase IL-10 production, while decreasing that of IL-1β (Gleeson et al., 2016). In a different setting, macrophages activated by efferocytosis during wound injury up-regulated their production of IL-10 via the SIRTUIN1 signaling cascade and the transcription factor PBX1, driven by the respiratory chain (Zhang et al., 2019). In an experimental model of colitis, chemical inhibition of the nicotinamide phosphoribosyltransferase enzyme reduced mucosal nicotinamide adenine dinucleotide levels and the activities of nicotinamide adenine dinucleotide–dependent enzymes, while favoring the polarization of monocyte/macrophages toward IL-10 production (Gerner et al., 2018). IL-10 production by DCs is also modulated by metabolic pathways. Several metabolites, such as butyrate, adenosine, ATP, or lactic acid, have been proposed to favor IL-10 production by DCs (Li et al., 2012a; Nasi et al., 2013; Singh et al., 2014). Furthermore, activation of mTOR, a central integrator of cellular metabolism, in DCs has been shown to enhance IL-10 (Cao et al., 2008; Amiel et al., 2012; Boor et al., 2013; Hussaarts et al., 2013). Adiponectin also enhances IL-10 production by DCs through a pathway that depends on the cellular energy sensor AMP-activated protein kinase (AMPK; Tan et al., 2014). Moreover, a recent study showed that in DCs stimulated with zymozan, the production of IL-10 depended on the maintenance of pyruvate flux, as it reinforces the presence of acetylated histone 3 molecules bound to the Il10 gene promoter (Márquez et al., 2019).
Figure 1.
IL-10 production is metabolically regulated. (A) Macrophages stimulated via TLR4 undergo a metabolic reprogramming toward glycolysis, which leads to the production of lactate and also to a break of the TCA cycle. As a result, succinate accumulates and activates HIF-1α, and the transcription of the Il1b gene is enhanced. Inhibition of this reprogramming, via the metabolic regulator PKM2 or during infection with M. tuberculosis, inhibits Il1b gene transcription, while promoting that of Il10. An increase of IL-10 production as a result of enhanced fatty acid metabolism has also been described, and is mediated by the transcription factor PBX1. In DCs, the maintenance of the pyruvate flux downstream of zymosan stimulation has been shown to reinforce the presence of acetylated histone 3 at the Il10 gene promoter, thus favoring IL-10 production. Other metabolic regulators, such as AMPK and mTOR, and metabolites, such as ATP, lactate, or butyrate, also contribute to IL-10 production by DCs, in different settings, but the molecular mechanisms in place remain largely unknown. (B) Gut microbiota derived short chain fatty acids signal through GPR43 and activate STAT3 and mTOR in differentiated Th1 cells, up-regulating the expression of Blimp-1 and consequently that of IL-10. In IL-27–induced Tr1 cells, IL-27R signaling induces the oxysterol 25-hydroxycholesterol (25-OHC). 25-OHC activates liver X receptor (LXR) and presumably decreases the expression of the transcription factor Blimp-1, thus limiting the expression of the Il10 gene. In IL-27–differentiated Tr1 cells, extracellular ATP (eATP) has been shown to increase the amounts of HIF-1α and consequently reduce those of AhR. This stabilization of HIF-1α also occurs under hypoxic conditions. Given the role of AhR in enhancing IL-10 expression in Tr1 cells, HIF-1α stabilization ultimately decreases IL-10 production. The Th1 switching from IFN-γ single-producing cells, to IFN-γ/IL-10 double-producing ones, and eventually IL-10 single-producing cells, is coupled to the biosynthesis of cholesterol, via cMaf. In Th17 cells, a short chain fatty acid, pentanoate, has been suggested to inhibit AMPK activation, thus releasing the mTOR pathway, increasing the glycolytic flux, and leading to the production of IL-10. In parallel, through an epigenetic-mediated mechanism, pentanoate reduces IL-17A expression.
T cell immunity also relies on metabolic reprogramming (Buck et al., 2015), but how this affects IL-10 production is only now starting to be understood (Fig. 1 B). Initial evidence for this came from the finding that extracellular ATP or hypoxia induces the transcription factor HIF-1α in Tr1 cells, which in turn down-regulates AhR and IL-10 production (Apetoh et al., 2010; Mascanfroni et al., 2015). In addition, oxysterols, oxidized forms of cholesterol produced downstream of the IL-27R, negatively regulate IL-10 secretion in Tr1 cells via activation of the liver X receptor signaling and possibly through inhibition of the IL-10–inducing transcription factor Blimp-1 (Vigne et al., 2017). Furthermore, inhibition of cholesterol biosynthesis blocked the induction of cMaf and consequently IL-10 production by IFN-γ–producing Th1 cells (Perucha et al., 2019). Finally, the short-chain fatty acid pentanoate has been shown to induce IL-10 production in lymphocytes by reprogramming their metabolic-epigenetic crosstalk toward elevated glucose oxidation, while in parallel suppressing Th17 cells (Luu et al., 2019). Of note, in the same study, pentanoate has been shown to enhance Il10 gene expression in regulatory B cells (Luu et al., 2019). Interestingly, gut microbiota–derived short-chain fatty acids have also been shown to induce Th1 cells to produce IL-10 to maintain intestinal homeostasis, through the activation of STAT3 and mTOR, upstream of Blimp-1 up-regulation (Sun et al., 2018). Despite these studies, the molecular mechanisms concerning IL-10 regulation by metabolic changes or metabolites across different subsets of T cells remain elusive. However, careful interpretation of experiments should be exercised. Indeed, findings from a recent study show that the widely used glucose analogue 2-deoxyglucose inhibited IL-10 production by Th1, Th2, and Th17 cells not through the inhibition of glycolysis, but instead through the modulation of glycosylation on cell surface proteins (Gabryšová et al., 2018).
The regulation of IL-10 in other cell types
In contrast to macrophages/DCs and T cells, the mechanisms regulating IL-10 in other cells are less explored and understood. IL-10 production by PRR-stimulated microglia follows the general basic mechanisms seen in peripheral myeloid cells (reviewed in Lobo-Silva et al., 2016). Molecules present in the microenvironment, such as adenosine, ATP, or glutamate, have been described to potentiate IL-10 production by microglia, but the precise mechanisms underlying these effects are mostly unknown (Seo et al., 2004, 2008; Werry et al., 2011; Koscsó et al., 2012; and reviewed in Lobo-Silva et al., 2016). Furthermore, other cytokines may impact IL-10 secretion by microglia. Although the effect of IFN-γ in regulating microglia-derived IL-10 is still controversial (Veroni et al., 2010; Ishii et al., 2013), type I IFN has been shown to up-regulate IL-10 in PRR-induced microglia (Lobo-Silva et al., 2017), similar to what has been described for bone marrow–derived macrophages (reviewed in Ouyang and O’Garra, 2019).
The regulation of IL-10 production by epithelial cells is also not fully understood. Engagement of CD1d on intestinal epithelial cells has been shown to induce IL-10 (Colgan et al., 1999), via a mechanism that involves the activation of STAT3 and STAT3-dependent transcription of IL-10 (Olszak et al., 2014). Crosstalk between TLR4-activated macrophages and intestinal epithelial cells has also been shown to sustain the expression of IL-10 in the latter, by the action of nuclear receptor peroxisome proliferator-activated receptor γ (Hyun et al., 2015). Furthermore, microbial and/or environmental oxazoles have been shown to induce tryptophan-derived metabolites and activate AhR in intestinal epithelial cells. The activated AhR in turn limits CD1d-restricted production of IL-10 by these cells (Iyer et al., 2018). Thus, metabolites in the intestine regulate IL-10 production. Given the importance of intestinal epithelial cells in maintaining the intestinal barrier and of IL-10 in controlling intestinal inflammation, more research in this area is needed.
The effects of IL-10 in responding cells
The IL-10R signaling cascade in brief
The cellular response to IL-10 depends on engagement of the IL-10R and intracellular signaling cascades (Fig. 2). IL-10, as the other family members, signals through a heterodimeric receptor (Pestka et al., 2004). In the case of IL-10, the receptor is composed two subunits: the high-affinity IL-10Rα, mainly expressed in leukocytes, and the ubiquitously expressed IL-10Rβ, which is shared by the receptor complex of other IL-10 family members (reviewed in Zdanov, 2010; Ouyang and O’Garra, 2019). The intracellular signaling cascades downstream of the IL-10R have been described in detail in previous reviews (Donnelly et al., 1999; Murray, 2006b; Ouyang et al., 2011). Building upon the extensive literature in this topic, an IL-10/IL-10R signaling map has been assembled (Verma et al., 2016).
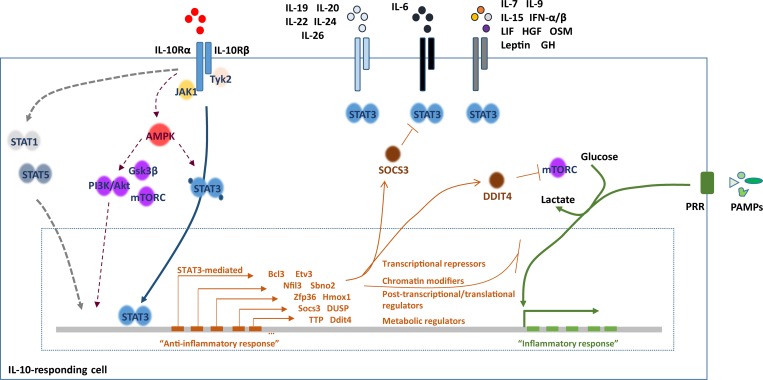
Figure 2.
The anti-inflammatory response initiated downstream of the IL-10R. Binding of IL-10 to its receptor activates a major JAK1-TYK2-STAT3 signaling cascade, which culminates with the induction of the STAT3-mediated anti-inflammatory response. The STAT3 transcriptional reprogramming results in a number of molecules that include transcriptional repressors, chromatin modifiers, and post-transcriptional or post-translational regulators. Together, these molecules will limit the inflammatory response induced for example downstream of PRR activation. Recently, IL-10 has been shown to modulate the transcription of several metabolic regulators, including DDIT4, in a STAT3-dependent way. As DDIT4 is a regulator of mTORC, the IL-10R signaling limits the glycolytic flux downstream of TLR4 activation, which in turn controls the inflammatory response of the macrophage. In addition to the JAK-STAT3 pathway, STAT1, STAT5, and the PI3K-Akt cascade have also been reported to mediate IL-10 responses. The metabolic regulator AMPK has been reported to activate the PI3K-Akt-mTORC cascade, but also STAT3, in response to IL-10. STAT3 is moreover activated downstream of receptors for other IL-10 family cytokines, IL-6, and a number of other cytokines and growth factors. How the cell distinguishes the anti- from the pro-inflammatory reprogramming downstream of STAT3 activation is not fully understood, but the IL-10–induced SOCS3 has been reported to play an important role by blocking STAT3 at least downstream of the IL-6R, but not of the IL-10R. DUSP, dual-specifity phosphatase; Gsk3β, glycogen synthase kinase 3β; Sbno2, protein strawberry notch 2; TTP, tristetraprolin.
Engagement of the IL-10Rα leads to its oligomerization with the IL-10Rβ subunit (Yoon et al., 2006). This in turn allows the phosphorylation of JAK1 associated with the IL-10Rα subunit and of tyrosine kinase 2 (TYK2) associated with the IL-10Rβ subunit (Rodig et al., 1998; Riley et al., 1999). These kinases further phosphorylate two functional tyrosine residues on the intracellular domain of the IL-10Rα, needed for the recruitment of STAT3 (Weber-Nordt et al., 1996). Activation of STAT3 by its phosphorylation allows its translocation to the nucleus, with the initiation of a specific transcriptional program that largely defines the IL-10–mediated anti-inflammatory response (Takeda et al., 1999; Lang et al., 2002; Williams et al., 2004a; reviewed in Murray, 2006b; Hutchins et al., 2013). In addition to the JAK/STAT3 pathway, the signaling cascade PI3K/Akt/GSK3β has also been shown to mediate IL-10–induced transcription in macrophages (Crawley et al., 1996; Antoniv and Ivashkiv, 2011). Furthermore, IL-10 stimulation of primary monocytes results in PI3K-mediated mTORC1 activity (Crawley et al., 1996). Interestingly, IL-10 activation of the both STAT3 and PI3K/Akt/mTORC1 pathways in macrophages was shown to require AMPK (Zhu et al., 2015). Of note, activation of STAT1 and STAT5 downstream of the IL-10R has also been reported (Lehmann et al., 1994; Wehinger et al., 1996; Rahimi et al., 2005; Emmerich et al., 2012).
Interestingly, STAT3 activation has also been described for other cytokines in addition to IL-10, most notably IL-6 (Fig. 2). It is surprising that both pro- and anti-inflammatory cytokines may signal through the same molecule even in the same cell type, despite inducing profoundly different responses, and resulting in a different gene activation program (reviewed in Murray, 2006a, 2007; Hutchins et al., 2013). A likely explanation for this apparent paradox relies on the cooperation of STAT3 with selective cofactors to deliver a different gene expression program. One such cofactor is the suppressor of cytokine signaling 3 (SOCS3), which, despite being induced by both IL-10 and IL-6 in macrophages, only suppresses the activity of the IL-6R (gp130; Croker et al., 2003; Lang et al., 2003). Indeed, the IL-10R appears refractory to the effects of all members of the SOCS family (Williams et al., 2004b). Additionally, it is possible that in response to different stimuli, the epigenetic landscape of the STAT3-regulated genes changes, thus allowing for selective transcription of anti-inflammatory or inflammatory mediators (reviewed in Murray, 2006a, 2007; Hutchins et al., 2013).
Molecular basis for IL-10/STAT3 anti-inflammatory activities
As discussed above, a wealth of data positions the IL-10/STAT3 axis as a major transcriptional inhibitor of genes encoding cytokines, chemokines, cell-surface molecules, and other molecules required for a full immune response (Murray, 2005; reviewed in Murray, 2006b). The requirement for STAT3 as a mediator of IL-10 responses is well-established (Takeda et al., 1999; Lang et al., 2002; Williams et al., 2004a). Macrophage/neutrophil-restricted deletion of STAT3 mimics IL-10 deficiency itself, resulting in the development of enterocolitis and high susceptibility to LPS-mediated shock and septic peritonitis (Takeda et al., 1999; Kobayashi et al., 2003; Matsukawa et al., 2003, 2005). How STAT3 activation results in the anti-inflammatory activity of IL-10 is not fully understood (Fig. 2).
Several independent reports performed over the years globally show that STAT3 acts indirectly by inducing the expression of a series of genes that execute the inhibitory effects associated with an anti-inflammatory response (reviewed in Hutchins et al., 2013). Among the STAT3-dependent anti-inflammatory mediators are several transcription factors, such as the E26 transformation–specific family transcriptional repressor, ETV3, and a helicase family corepressor, Strawberry notch homologue 2, that inhibit NF-κB–mediated transcription in LPS-stimulated macrophages (El Kasmi et al., 2007); and Nfil3, which negatively regulates the production of IL-12p40 by LPS-stimulated myeloid cells (Smith et al., 2011). Other molecular targets of IL-10–activated STAT3 have been described, including the up-regulation of the dual-specificity phosphatase 1, which impairs the inflammatory response of LPS-induced macrophages by down-regulating the activity of the MAPK p38 (Hammer et al., 2005). Also, in LPS-stimulated macrophages, a link between IL-10/STAT3 and the induction of the RNA destabilizing factor tristetraprolin has been proposed, and shown to be mediated by a delayed activation of p38 MAPK, resulting in diminished mRNA and protein levels of proinflammatory cytokines (Schaljo et al., 2009). Finally, in LPS-activated macrophages, IL-10 was shown to inhibit the transcription of the microRNA miR-155 in a STAT3-dependent manner, leading to increased expression of SH2 domain-containing inositol 5′-phosphatase 1 (SHIP1; McCoy et al., 2010), a negative regulator of TLR signaling (An et al., 2005). Subsequently, SHIP1 was shown to inhibit TNF translation in LPS-activated macrophages through an IL-10–dependent, STAT3-independent mechanism (Chan et al., 2012). Of note, IL-10 induction of miR-155 was also shown to regulate the expression of Aicda, a critical component of antibody class-switching in B cells (Fairfax et al., 2015).
Two genome-wide studies were performed to reveal the universe of STAT3-regulated genes. The obtained data support a model of indirect STAT3 action in response to IL-10. The first study integrated STAT3 chromatin immunoprecipitation, RNA-sequencing, and computational approaches performed in IL-10–treated resting or LPS-stimulated macrophages (Hutchins et al., 2012). The second study used chromatin immunoprecipitation sequencing to analyze the binding of STAT3 at the genomic level in DCs following histone deacetylases inhibition (Sun et al., 2017). Regardless of the critical role of STAT3 in mediating the response downstream of the IL-10R, it is possible that the STAT3 pathway may not be sufficient to mediate the full action of IL-10. Other anti-inflammatory mediators have been described downstream of the IL-10R, but their dependency on STAT3 is as yet unclear. Such is the case for Bcl3, which inhibits TNF production in LPS-treated macrophages via impairment of NF-κB (Kuwata et al., 2003) and A20-binding inhibitor of NF- κB activation 3, which attenuates NF-κB signaling in human but not mouse macrophages (Weaver et al., 2007). IL-10–induced heme-oxygenase 1 has also been shown to contribute to the anti-inflammatory response triggered in macrophages (Lee and Chau, 2002).
Some lines of evidence suggest that chromatin accessibility might also be a mechanism of regulation of gene expression by the IL-10/STAT3 axis. For example, HDAC4, which acts as a repressor of gene expression by deacetylating histones, was identified as a STAT3 target (Hutchins et al., 2012). Furthermore, in adipose tissue, IL-10 has been shown to repress transcription of thermogenic genes by altering chromatin accessibility in a STAT3-dependent manner (Rajbhandari et al., 2018).
Classic roles of IL-10
In addition to being produced by many different cell types, IL-10 functionally targets diverse cells, resulting in a broad anti-inflammatory activity. As compared with most other cell types, macrophages express higher levels of the IL-10R (Moore et al., 2001). Compelling evidence provided by the study of cell-restricted IL-10R deficiency has pointed to macrophages being the main targets of the IL-10 inhibitory effects (Shouval et al., 2014a; Zigmond et al., 2014). Indeed, soon after its discovery, IL-10 was shown to trigger a robust immune suppressive response in macrophages and other APCs, mainly via the transcriptional inhibition of cytokines and chemokines, as well as of MHCII, and costimulatory and adhesion molecules (Bogdan et al., 1991; de Waal Malefyt et al., 1991a,b; Fiorentino et al., 1991a,b; Ding and Shevach, 1992; D’Andrea et al., 1993; Willems et al., 1994; Creery et al., 1996). Overall, IL-10 both modulates the local cytokine micro-environment and limits antigen presentation, thus preventing the efficient development of T cell responses. Additionally, IL-10 also limits basic microbicidal mechanisms, namely the induction of nitric oxide synthase and the production of nitric oxide, as well as the release of reactive oxygen intermediates (Bogdan et al., 1991) by murine macrophages stimulated through PRR in the context of IFN-γ (Cunha et al., 1992).
The suppressive effect of IL-10 appears to be targeted to specific genes. This is well illustrated by a gene expression screen for IL-10–induced genes performed in LPS-stimulated macrophages, which identified a 15–20% inhibition of the LPS-induced genes (Lang et al., 2002). A more recent study further elucidated the mechanisms underlying the suppressive activity of IL-10 in LPS-stimulated macrophages, through genome-wide approaches. In line with previous reports, inhibition of transcription was found to be the primary mechanism for IL-10–mediated suppression (Conaway et al., 2017). Novel targets of IL-10 in macrophages have been recently revealed, including the modulation of the macrophage metabolic reprogramming downstream of TLR4 activation (Ip et al., 2017). Specifically, IL-10 inhibited LPS-induced glucose uptake and glycolysis, promoting a shift toward oxidative phosphorylation (Ip et al., 2017).
Inhibition of cytokine and chemokine production by IL-10 is also documented for neutrophils (Cassatella et al., 1993; Kasama et al., 1994) and for resident macrophages, such as microglia (Balasingam and Yong, 1996; and reviewed in Lobo-Silva et al., 2016). However, and most interestingly, the comparison of the anti-inflammatory response induced by IL-10 across different LPS-stimulated myeloid cells showed a divergent pattern. Specifically, in macrophages, IL-10 mostly inhibited NF-κB target genes, whereas in DCs and mast cells, an indirect disruption of IRF was proposed, and in neutrophils, IL-10 appeared to induce both IRF disruption and indirect NF-κB inhibition (Hutchins et al., 2015).
Classically, IL-10 is known to inhibit T cell responses via APCs (Macatonia et al., 1993). However, IL-10 can also limit T cell responses by acting directly on CD4 T cells to induce nonresponsiveness or anergy (Groux et al., 1996). More recently, IL-10 has been shown to act directly on memory/effector T cells (Kamanaka et al., 2011) in experimental colitis, in Th17 cells during intestinal inflammation (Huber et al., 2011), and in Th2 cells in a murine model of house dust mite allergy (Coomes et al., 2017). Surprisingly, IL-10–stimulatory effects on T cells have also been described. IL-10 enhances CD8 T cell proliferation, cytotoxic activity, and IFN-γ production (Groux et al., 1998; Mumm et al., 2011; Naing et al., 2018). IL-10 promotes the survival and action of Foxp3 regulatory T cells (Murai et al., 2009; Chaudhry et al., 2011). Stimulatory activities of IL-10 on mast cells and B cells have also been well documented (Thompson-Snipes et al., 1991; Rousset et al., 1992; Itoh and Hirohata, 1995).
Nonclassic roles of IL-10
Although the effects of IL-10 have been classically studied in immune cells, the range of IL-10–responding cells is expanding (Fig. 3). It is now evident that IL-10 also acts in nonhematopoietic cells, where it plays important homeostatic roles. An example of this comes from central nervous system (CNS) and peripheral nervous system cells, which respond to IL-10 and other IL-10 family members (reviewed in Lobo-Silva et al., 2016; Burmeister and Marriott, 2018). In this context, IL-10 plays important roles in limiting neuronal damage during infection or other inflammatory processes, as well as in regulating homeostatic processes. IL-10 has been associated with increased neuronal survival (Zhou et al., 2009a,b) and the regulation of adult neurogenesis (Perez-Asensio et al., 2013; Pereira et al., 2015). Administration of IL-10 in different settings proved beneficial for axon regeneration, through mechanisms that implicate STAT3 and the regulation of NF-κB signaling (reviewed in Vidal et al., 2013). Finally, IL-10 was shown to reduce vulnerability of neurons to CNS ischemia and trauma (Knoblach and Faden, 1998; Grilli et al., 2000). Importantly, whether the effects of IL-10 on neurons and oligodendrocytes are direct or indirect is still controversial. To support a direct effect on cells of the CNS, resting astrocytes express the IL-10R and respond to IL-10 (Molina-Holgado et al., 2001; Ledeboer et al., 2002; Xin et al., 2011; Perriard et al., 2015). IL-10 limits reactive astrogliosis in response to pathogens and promotes the expression of TGF-β by astrocytes (Balasingam and Yong, 1996; Norden et al., 2014; and reviewed in Burmeister and Marriott, 2018). At the organismal level, variations in IL-10 expression have been associated with altered depressive-like behavior. IL-10–deficient female mice displayed increased depressive-like behavior, whereas IL-10–overexpressing mice displayed a decreased depressive-like behavior (Mesquita et al., 2008). More research is clearly needed to fully elucidate how IL-10 regulates the CNS in health and disease.
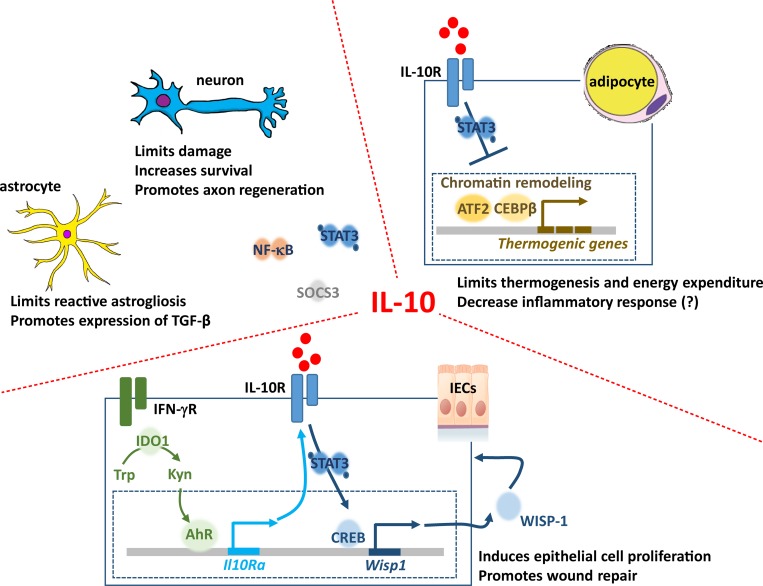
Figure 3.
Nonclassic roles of IL-10. The functions of IL-10 surpass its effects in immune cells. IL-10 plays an important role in the CNS, limiting the damage effects of neuroinflammation, but also contributing to several homeostatic processes. The molecular mechanisms operating in neurons or astrocytes in response to IL-10 remain elusive, but STAT3 and SOCS3 activation, as well as NF-κB, regulation have been proposed. Bone marrow–derived IL-10 activates the IL-10R/STAT3 signaling cascade, altering the chromatin landscape and the transcription factor occupancy at regulatory regions of thermogenic genes. The thermogenic gene expression is thus repressed and the energy expenditure limited. As a result, mice lacking IL-10 are protected against diet-induced obesity. In epithelial cells, IL-10 has been recently associated with wound healing. This is particularly important in intestinal epithelial cells (IECs), where the underlying mechanisms are starting to be unveiled. In response to IFN-γ, the enzyme 2,3-dioxygenase 1 (IDO1) catalyses the conversion of tryptophan (Trp) to kynurenine (Kyn), which activates the AhR, thus inducing the transcription of the IL-10Rα subunit. Activation of the IL-10R in response to macrophage-derived IL-10 leads to the STAT3-dependent CREB activation and the transcription of WSP-1, a promoter of epithelial cell proliferation and repair.
Another nonclassic function ascribed to IL-10 relates to its regulation of adipose tissue (Fig. 3). Epidemiological studies link a low production of IL-10 with metabolic syndrome and type 2 diabetes (van Exel et al., 2002), but in animal models, ablation of IL-10 did not support an anti-obesity function for this cytokine (den Boer et al., 2006; Miller et al., 2011; Mauer et al., 2014). However, previous studies proposed that IL-10 might promote the activity of M2 macrophages in adipose tissue (Lumeng et al., 2007; Hong et al., 2009; Gao et al., 2013; Xie et al., 2014), or act directly on adipocytes to decrease their inflammatory response (Lira et al., 2012). Very recently, and contrary to the previously established idea that IL-10 might stimulate brown tissue activity by limiting inflammation, direct IL-10 signaling in adipocytes was strikingly shown to limit thermogenesis and energy expenditure (Rajbhandari et al., 2018). Mechanistically this effect involved remodeling the chromatin landscape of adipocytes and specific transcriptional down-regulation of thermogenic genes (Rajbhandari et al., 2018). As a result, mice lacking IL-10 had increased energy expenditure and were protected against diet-induced obesity (Rajbhandari et al., 2018). It will be interesting in future to evaluate the therapeutic value of IL-10 manipulation in obesity, taking all these findings into consideration.
The last “nonclassic” role of IL-10 that we will discuss relates to its function as a promoter of epithelial wound repair (Fig. 3). Several studies highlight the IL-10–mediated pro-repair activities, which have been classically attributed to its anti-inflammatory activities (reviewed in Sziksz et al., 2015). However, this concept is changing, and an effect of IL-10 as a regulator of the extracellular matrix and fibroblast function is emerging (Balaji et al., 2017; and reviewed in King et al., 2014). Similarly, the critical role of IL-10 in gut homeostasis is well documented, with the majority of the studies focusing on its anti-inflammatory activities (reviewed in Kole and Maloy, 2014). Recent evidence reveals an additional role for IL-10 as an orchestrator of epithelial wound repair (Lorén et al., 2015); however, the molecular bases for this are only starting to be unveiled. Macrophage-derived IL-10 was shown to activate CREB signaling in intestinal epithelial cells, leading to the secretion of the WNT1-inducible signaling protein 1, which in turn induced epithelial cell proliferation and wound closure (Quiros et al., 2017). Furthermore, up-regulation of the IL-10R in intestinal epithelial cells has been described (Kominsky et al., 2014), and recent studies not only confirm the importance of the IL-10R in wound healing but also implicate tryptophan metabolites and AhR in its up-regulation (Lanis et al., 2017).
Therapeutic manipulation of IL-10
Given the wide spectrum of the anti-inflammatory properties attributed to IL-10, therapeutic manipulation of this cytokine has attracted a great deal of interest (reviewed in O’Garra et al., 2008; Ouyang and O’Garra, 2019; Wang et al., 2019). We next discuss the therapeutic opportunities for IL-10 in various pathologies, and the strategies used to manipulate IL-10 levels in the organism (Fig. 4).
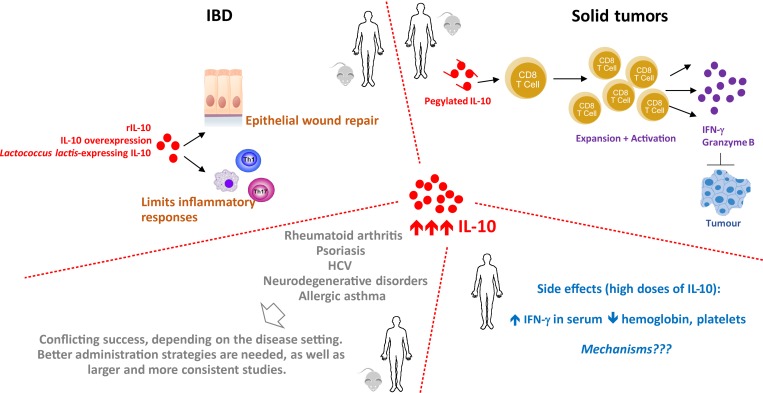
Figure 4.
The potential of enhancing IL-10 to treat disease. Several IL-10 enhancing strategies have been tried in a number of diseases and models of disease. The best understood effects of IL-10 at the mechanistic level have been described in the case of IBD and cancer. Side effects of IL-10 administration have been reported in both patients and healthy volunteers, but the underlying mechanisms remain unknown. The human figure represents clinical data, and the mouse figure preclinical findings. HCV, hepatitis C virus.
Enhancing IL-10 in inflammatory bowel disease (IBD)
IBD is perhaps the best studied disease in the context of IL-10, owing to the initial findings that IL-10–deficient mice develop colitis (Kühn et al., 1993), together with the profound genetic association of IBD with deficient IL-10 responses (Glocker et al., 2009, 2010, 2011; Jostins et al., 2012; Moran et al., 2013; Engelhardt and Grimbacher, 2014; Ellinghaus et al., 2016). Administration of IL-10 or IL-10 overexpression in different animal models of colitis has proved consistently beneficial (Hagenbaugh et al., 1997; Steidler et al., 2000; Cardoso et al., 2018). In humans, systemic (intravenous or subcutaneous) administration of recombinant IL-10 has been tested to treat IBD in different trials, but in general it did not significantly improve the clinical outcome of the patients (van Deventer et al., 1997; Fedorak et al., 2000; Schreiber et al., 2000; Colombel et al., 2001). A likely explanation for this relies on the possible low concentration of IL-10 achieved locally. Thus, efforts have been placed in designing strategies to specifically deliver IL-10 to the intestine. An interesting approach has been the development of bacteria engineered for intestinal IL-10 delivery. Specifically, the dairy microbe Lactococcus lactis has been modified to express the Il10 gene and intragastrically administered to mice (Steidler et al., 2000). This strategy has proved efficient in dextran sulfate sodium–induced and spontaneous (IL-10–deficient) colitis models (Steidler et al., 2000). Subsequent improvements of the engineered L. lactis have been devised, all with positive effects in ameliorating experimental colitis (reviewed in Shigemori and Shimosato, 2017; Wang et al., 2019). Genetic modification of Bifidobacterium bifidum for IL-10 expression has also been described (Mauras et al., 2018), thus expanding the species of bacteria amenable for IL-10 intestinal delivery. Although only tested in a small cohort of Crohn’s disease patients, therapeutic L. lactis expressing human IL-10 ameliorated the clinical score of disease (Braat et al., 2006), which, together with the promising findings obtained in the mouse model, suggests that further improving these approaches may render them applicable for human therapy.
IL-10 as an immunotherapy for cancer
The immunosuppressive role of IL-10 has led to the general view that its presence during cancer would facilitate tumor immune escape (Dunn et al., 2004). This hypothesis is validated in several animal models. Overexpression of IL-10 resulted in failure to control an immunogenic tumor (Hagenbaugh et al., 1997), and blockade of IL-10Rα increased the therapeutic benefit of chemotherapy in a breast cancer model (Ruffell et al., 2014) and to other transplantable tumors in the context of CpG immunostimulatory oligonucleotide by reversal of tumor-induced DC paralysis (Vicari et al., 2002). Likewise, IL-10 deficiency conferred resistance to UV-induced skin carcinogenesis (Loser et al., 2007). In patients, the presence of IL-10 has been described in the tumor-microenvironment of different cancers and overall correlated with poor prognosis (Nemunaitis et al., 2001; Mannino et al., 2015; reviewed in O’Garra et al., 2008). However, early lines of evidence suggest an unanticipated protective role for IL-10 in cancer. Both IL-10 overexpression and IL-10 administration have been reported to associate with tumor shrinkage and rejection (Berman et al., 1996; Zheng et al., 1996; Groux et al., 1999). More recently, administration of pegylated IL-10 to mouse models of breast cancer or squamous cell carcinoma provided compelling preclinical evidence on the efficacy of IL-10 as an immunotherapy for cancer (Mumm et al., 2011). Mechanistically, IL-10 increased CD8 T cell infiltration in tissue, induced IFN-γ production, and favored effective T cell memory responses (Mumm et al., 2011; Emmerich et al., 2012). Prompted by these studies, a pegylated human IL-10 was developed (Naing et al., 2016) and shown to induce hallmarks of CD8 T cell immunity in patients with solid tumors (Naing et al., 2018). Thus, IL-10 may in some types of cancer be useful as an immunotherapy by potentiating the activity of antitumor CD8 T cells (reviewed in Autio and Oft, 2019; Ouyang and O’Garra, 2019; Wang et al., 2019). However, a recent study has shown that tumor-infiltrating regulatory T cells through their expression of IL-10 and IL-35 may contribute to the exhaustion of intratumoral CD8 T cells, via the up-regulation of the transcription factor Blimp-1 (Sawant et al., 2019). Several factors may explain the discrepancies between these different studies, including the cell targeted by IL-10 in the different tumors (for example, myeloid versus T cell), or differences in the T cell response to IL-10 at different effector stages. Thus, it is critical to determine the context whereby IL-10 may provide a protective or detrimental role in the treatment of tumors, in order to appropriately tailor cancer therapy.
The potential of IL-10 to treat other diseases
In addition to IBD and cancer, the therapeutic manipulation of IL-10 has been envisaged in the context of several other pathologies (reviewed in O’Garra et al., 2008; Ouyang and O’Garra, 2019; Wang et al., 2019). In both rheumatoid arthritis and psoriasis, IL-10 administration has yielded some promising results at the preclinical and clinical levels (McInnes et al., 2001; Trachsel et al., 2007; Galeazzi et al., 2014). In allergic asthma, where pathological responses to inhaled allergens develop due to a failure of immune tolerance, successful therapeutic strategies are linked to an increase of IL-10. This is the case for the glucocorticoid dexamethasone, which favors IL-10 production by human CD4 and CD8 T cells (Richards et al., 2000; Barrat et al., 2002). Furthermore, severe steroid-resistant asthma associates with failure of patient cells to enhance IL-10 in response to dexamethasone (Xystrakis et al., 2006; Gupta et al., 2014). A recent study showed that the dexamethasone-driven IL-10 production by human memory CD4 T cells is accompanied by that of IL-17A, IL-17F, and IL-22. However, the presence of high doses of IL-2 skewed the response to dexamethasone toward single IL-10–producing cells (Mann et al., 2019). Therefore, combined therapies that enhance glucocorticoid-driven IL-10 production in the absence of IL-17 are of potential interest in asthma. Several strategies to enhance IL-10 in the context of neurological disorders with an immune component have also been developed (reviewed in Lobo-Silva et al., 2016; Burmeister and Marriott, 2018). These included administration of recombinant IL-10, enhancement of IL-10 production through agonists, or delivery of IL-10 through viral vectors (reviewed in Kwilasz et al., 2015). However, the preclinical success of IL-10 in this setting has been conflicting, and targeting IL-10 to treat neuroimmune disorders has not been tested in the clinic. An opposing strategy consists of IL-10 blockade, in pathologies where its excess is detrimental. This is the case of systemic lupus erythematous. A small clinical trial performed in systemic lupus erythematous patients showed an improvement of symptoms following IL-10 blockade (Llorente et al., 2000).
Given the anti-inflammatory action of IL-10, its therapeutic manipulation in infection is appealing. However, the effects of IL-10 in infection significantly depend on the infecting agent (reviewed in Couper et al., 2008; Redford et al., 2011). In a number of experimental models of infection including T. gondii (Gazzinelli et al., 1996), malaria (Li et al., 1999), and Trypanosoma cruzi (Hunter et al., 1997), ablation of IL-10 led to minimal pathogen clearance, while allowing fatal tissue damage. Interestingly, administration of IL-10 to a small cohort of chronic hepatitis C patients who had not previously responded to IFN-based therapies did not impact antiviral activity and resulted in improved liver histology and function, as well as in reduced liver fibrosis in a large proportion of patients receiving treatment (Nelson et al., 2000). In studies using experimental models of Listeria monocytogenes (Dai et al., 1997), lymphocytic choriomeningitis virus (Brooks et al., 2006), Leishmania major (Belkaid et al., 2001), Leishmania donovani (Murray et al., 2002), and M. tuberculosis (Redford et al., 2010), IL-10 deficiency resulted in increased pathogen clearance, beneficial to the host. In the case of tuberculosis, both human and mouse studies highlight a role for IL-10 in limiting the host protective immune response (reviewed in Redford et al., 2011). Interestingly, a recent study has shown that the delivery of aerosolized peptide inhibitors targeting the IL-10/STAT3 pathway to mice chronically infected with M. tuberculosis led to a reduction of bacterial burden accompanied by the up-regulation of antimicrobial effector molecules and of apoptotic/autophagy mediators (Upadhyay et al., 2018).
Despite the large number of disease settings where IL-10 modulation has been tried, of which we only discussed a few, the overall data are conflicting, and a clear argument in favor of IL-10 as a therapy is still nonexistent. Several variables need to be fine-tuned, including the anatomical location where IL-10 is needed for treatment and the context of diseases where IL-10 has a stimulatory or anti-inflammatory effect.
Secondary effects associated with increased doses of IL-10
Most data assessing the side effects of IL-10 administration have been obtained in the context of systemic administration of recombinant IL-10 to Crohn’s disease patients. The overall results show that systemic administration of recombinant IL-10 is well-tolerated, particularly at low doses (van Deventer et al., 1997). However, when higher doses of IL-10 were administered, some unexpected side effects have been observed (Fig. 4), including fatigue and headache, accompanied by a reduction of red blood cells and thrombocytes (Buruiana et al., 2010). Additionally, the induction of the pro-inflammatory cytokine IFN-γ and decreased hemoglobin and platelet counts have been reported (Tilg et al., 2002b). Similar side effects have been described during the administration of recombinant IL-10 to treat psoriatic arthritis (McInnes et al., 2001). Importantly, these effects seemed to be reversible (Tilg et al., 2002a) and did not appear to be disease-related, as even in healthy volunteers, an up-regulation of IFN-γ in the serum has been observed (Lauw et al., 2000). The mechanisms underlying this immune-stimulatory role of IL-10 remain unknown. It has been suggested that the observed anemia might be caused by an IL-10–driven deregulation of iron metabolism (Tilg et al., 2002a). Whether the up-regulation of IFN-γ and granzyme B result from the activation of CD8 T cells, as shown in the case of pegylated IL-10, or from alternative mechanisms remains unknown. More research is needed to elucidate the aforementioned effects of IL-10, as they may provide key insights on combinatory therapies to increase the efficacy of IL-10 in inflammatory conditions.
Concluding remarks
The wide range of IL-10–producing and IL-10–responding cells offers the immune system the possibility of regulating the immune response in different situations and anatomical sites. However, its opposing context-specific anti-inflammatory and stimulatory effects make the therapeutic manipulation of IL-10 challenging. Progress in this area will require a deeper understanding of IL-10 biology, specifically of the molecular mechanisms that govern IL-10 production and action, as well as of the contrasting roles this cytokine may play in different contexts. Also important will be obtaining information on nonclassic functions of IL-10, which are only now being unveiled. Given the history of IL-10 research, exciting new findings on the biology of this cytokine are expected in the future.
Acknowledgments
M. Saraiva is financed by Fundo Europeu de Desenvolvimento Regional funds through the COMPETE 2020 Operational Program for Competitiveness and Internationalisation, Portugal 2020, and by Portuguese funds through Fundação para a Ciência e Tecnologia in the framework of the project ‘‘Institute for Research and Innovation in Health Sciences’’ (POCI-01-0145-FEDER-007274), and by Fundação para a Ciência e Tecnologia through Estimulo Individual ao Emprego Científico. P. Vieira is funded by Agence National de la Recherche, through the project MYELOTEN (ANR-13-ISV1-0003-01), and by the Institut Pasteur, France. A. O’Garra is funded by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001126), the UK Medical Research Council (FC001126), and the Wellcome Trust (FC001126).
The authors declare no competing financial interests.
References
- Ahyi A.N., Chang H.C., Dent A.L., Nutt S.L., and Kaplan M.H.. 2009. IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. J. Immunol. 183:1598–1606. 10.4049/jimmunol.0803302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel E., Everts B., Freitas T.C., King I.L., Curtis J.D., Pearce E.L., and Pearce E.J.. 2012. Inhibition of mechanistic target of rapamycin promotes dendritic cell activation and enhances therapeutic autologous vaccination in mice. J. Immunol. 189:2151–2158. 10.4049/jimmunol.1103741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H., Xu H., Zhang M., Zhou J., Feng T., Qian C., Qi R., and Cao X.. 2005. Src homology 2 domain-containing inositol-5-phosphatase 1 (SHIP1) negatively regulates TLR4-mediated LPS response primarily through a phosphatase activity- and PI-3K-independent mechanism. Blood. 105:4685–4692. 10.1182/blood-2005-01-0191 [DOI] [PubMed] [Google Scholar]
- Ananieva O., Darragh J., Johansen C., Carr J.M., McIlrath J., Park J.M., Wingate A., Monk C.E., Toth R., Santos S.G., et al. . 2008. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat. Immunol. 9:1028–1036. 10.1038/ni.1644 [DOI] [PubMed] [Google Scholar]
- Antoniv T.T., and Ivashkiv L.B.. 2011. Interleukin-10-induced gene expression and suppressive function are selectively modulated by the PI3K-Akt-GSK3 pathway. Immunology. 132:567–577. 10.1111/j.1365-2567.2010.03402.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetoh L., Quintana F.J., Pot C., Joller N., Xiao S., Kumar D., Burns E.J., Sherr D.H., Weiner H.L., and Kuchroo V.K.. 2010. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 11:854–861. 10.1038/ni.1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschenbrenner D., Foglierini M., Jarrossay D., Hu D., Weiner H.L., Kuchroo V.K., Lanzavecchia A., Notarbartolo S., and Sallusto F.. 2018. An immunoregulatory and tissue-residency program modulated by c-MAF in human TH17 cells. Nat. Immunol. 19:1126–1136. 10.1038/s41590-018-0200-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autio K., and Oft M.. 2019. Pegylated Interleukin-10: Clinical Development of an Immunoregulatory Cytokine for Use in Cancer Therapeutics. Curr. Oncol. Rep. 21:19 10.1007/s11912-019-0760-z [DOI] [PubMed] [Google Scholar]
- Balaji S., Wang X., King A., Le L.D., Bhattacharya S.S., Moles C.M., Butte M.J., de Jesus Perez V.A., Liechty K.W., Wight T.N., et al. . 2017. Interleukin-10-mediated regenerative postnatal tissue repair is dependent on regulation of hyaluronan metabolism via fibroblast-specific STAT3 signaling. FASEB J. 31:868–881. 10.1096/fj.201600856R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasingam V., and Yong V.W.. 1996. Attenuation of astroglial reactivity by interleukin-10. J. Neurosci. 16:2945–2955. 10.1523/JNEUROSCI.16-09-02945.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Gugasyan R., McMahon M., and Gerondakis S.. 2006. Diverse Toll-like receptors utilize Tpl2 to activate extracellular signal-regulated kinase (ERK) in hemopoietic cells. Proc. Natl. Acad. Sci. USA. 103:3274–3279. 10.1073/pnas.0511113103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat F.J., Cua D.J., Boonstra A., Richards D.F., Crain C., Savelkoul H.F., de Waal-Malefyt R., Coffman R.L., Hawrylowicz C.M., and O’Garra A.. 2002. In vitro generation of interleukin 10-producing regulatory CD4(+) T cells is induced by immunosuppressive drugs and inhibited by T helper type 1 (Th1)- and Th2-inducing cytokines. J. Exp. Med. 195:603–616. 10.1084/jem.20011629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y., Hoffmann K.F., Mendez S., Kamhawi S., Udey M.C., Wynn T.A., and Sacks D.L.. 2001. The role of interleukin (IL)-10 in the persistence of Leishmania major in the skin after healing and the therapeutic potential of anti-IL-10 receptor antibody for sterile cure. J. Exp. Med. 194:1497–1506. 10.1084/jem.194.10.1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman R.M., Suzuki T., Tahara H., Robbins P.D., Narula S.K., and Lotze M.T.. 1996. Systemic administration of cellular IL-10 induces an effective, specific, and long-lived immune response against established tumors in mice. J. Immunol. 157:231–238. [PubMed] [Google Scholar]
- Bogdan C., Vodovotz Y., and Nathan C.. 1991. Macrophage deactivation by interleukin 10. J. Exp. Med. 174:1549–1555. 10.1084/jem.174.6.1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boor P.P., Metselaar H.J., Mancham S., van der Laan L.J., and Kwekkeboom J.. 2013. Rapamycin has suppressive and stimulatory effects on human plasmacytoid dendritic cell functions. Clin. Exp. Immunol. 174:389–401. 10.1111/cei.12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braat H., Rottiers P., Hommes D.W., Huyghebaert N., Remaut E., Remon J.P., van Deventer S.J., Neirynck S., Peppelenbosch M.P., and Steidler L.. 2006. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin. Gastroenterol. Hepatol. 4:754–759. 10.1016/j.cgh.2006.03.028 [DOI] [PubMed] [Google Scholar]
- Brooks D.G., Trifilo M.J., Edelmann K.H., Teyton L., McGavern D.B., and Oldstone M.B.. 2006. Interleukin-10 determines viral clearance or persistence in vivo. Nat. Med. 12:1301–1309. 10.1038/nm1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M.D., O’Sullivan D., and Pearce E.L.. 2015. T cell metabolism drives mmunity. J. Exp. Med. 212:1345–1360. 10.1084/jem.20151159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister A.R., and Marriott I.. 2018. The Interleukin-10 Family of Cytokines and Their Role in the CNS. Front. Cell. Neurosci. 12:458 10.3389/fncel.2018.00458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buruiana F.E., Solà I., and Alonso-Coello P.. 2010. Recombinant human interleukin 10 for induction of remission in Crohn’s disease. Cochrane Database Syst. Rev. (11):CD005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Liu J., Song L., and Ma X.. 2005. The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J. Immunol. 174:3484–3492. 10.4049/jimmunol.174.6.3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Manicassamy S., Tang H., Kasturi S.P., Pirani A., Murthy N., and Pulendran B.. 2008. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat. Immunol. 9:1157–1164. 10.1038/ni.1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso A., Gil Castro A., Martins A.C., Carriche G.M., Murigneux V., Castro I., Cumano A., Vieira P., and Saraiva M.. 2018. The Dynamics of Interleukin-10-Afforded Protection during Dextran Sulfate Sodium-Induced Colitis. Front. Immunol. 9:400 10.3389/fimmu.2018.00400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassatella M.A., Meda L., Bonora S., Ceska M., and Constantin G.. 1993. Interleukin 10 (IL-10) inhibits the release of proinflammatory cytokines from human polymorphonuclear leukocytes. Evidence for an autocrine role of tumor necrosis factor and IL-1 beta in mediating the production of IL-8 triggered by lipopolysaccharide. J. Exp. Med. 178:2207–2211. 10.1084/jem.178.6.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A., Sadler A.J., Kar N., Young H.A., Silverman R.H., and Williams B.R.. 2008. Protein kinase R-dependent regulation of interleukin-10 in response to double-stranded RNA. J. Biol. Chem. 283:25132–25139. 10.1074/jbc.M804770200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.S., Ming-Lum A., Golds G.B., Lee S.J., Anderson R.J., and Mui A.L.. 2012. Interleukin-10 inhibits lipopolysaccharide-induced tumor necrosis factor-α translation through a SHIP1-dependent pathway. J. Biol. Chem. 287:38020–38027. 10.1074/jbc.M112.348599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E.Y., Guo B., Doyle S.E., and Cheng G.. 2007a Cutting edge: involvement of the type I IFN production and signaling pathway in lipopolysaccharide-induced IL-10 production. J. Immunol. 178:6705–6709. 10.4049/jimmunol.178.11.6705 [DOI] [PubMed] [Google Scholar]
- Chang H.D., Helbig C., Tykocinski L., Kreher S., Koeck J., Niesner U., and Radbruch A.. 2007b Expression of IL-10 in Th memory lymphocytes is conditional on IL-12 or IL-4, unless the IL-10 gene is imprinted by GATA-3. Eur. J. Immunol. 37:807–817. 10.1002/eji.200636385 [DOI] [PubMed] [Google Scholar]
- Chaudhry A., Samstein R.M., Treuting P., Liang Y., Pils M.C., Heinrich J.M., Jack R.S., Wunderlich F.T., Brüning J.C., Müller W., and Rudensky A.Y.. 2011. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 34:566–578. 10.1016/j.immuni.2011.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Daniel V., Maher D.W., and Hersey P.. 1994. Production of IL-10 by melanoma cells: examination of its role in immunosuppression mediated by melanoma. Int. J. Cancer. 56:755–760. 10.1002/ijc.2910560524 [DOI] [PubMed] [Google Scholar]
- Chi H., Barry S.P., Roth R.J., Wu J.J., Jones E.A., Bennett A.M., and Flavell R.A.. 2006. Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. Proc. Natl. Acad. Sci. USA. 103:2274–2279. 10.1073/pnas.0510965103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkhurst C.N., Muratet M., et al. . 2012. A validated regulatory network for Th17 cell specification. Cell. 151:289–303. 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan S.P., Hershberg R.M., Furuta G.T., and Blumberg R.S.. 1999. Ligation of intestinal epithelial CD1d induces bioactive IL-10: critical role of the cytoplasmic tail in autocrine signaling. Proc. Natl. Acad. Sci. USA. 96:13938–13943. 10.1073/pnas.96.24.13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombel J.F., Rutgeerts P., Malchow H., Jacyna M., Nielsen O.H., Rask-Madsen J., Van Deventer S., Ferguson A., Desreumaux P., Forbes A., et al. . 2001. Interleukin 10 (Tenovil) in the prevention of postoperative recurrence of Crohn’s disease. Gut. 49:42–46. 10.1136/gut.49.1.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway E.A., de Oliveira D.C., McInnis C.M., Snapper S.B., and Horwitz B.H.. 2017. Inhibition of Inflammatory Gene Transcription by IL-10 Is Associated with Rapid Suppression of Lipopolysaccharide-Induced Enhancer Activation. J. Immunol. 198:2906–2915. 10.4049/jimmunol.1601781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomes S.M., Kannan Y., Pelly V.S., Entwistle L.J., Guidi R., Perez-Lloret J., Nikolov N., Müller W., and Wilson M.S.. 2017. CD4+ Th2 cells are directly regulated by IL-10 during allergic airway inflammation. Mucosal Immunol. 10:150–161. 10.1038/mi.2016.47 [DOI] [PubMed] [Google Scholar]
- Couper K.N., Blount D.G., and Riley E.M.. 2008. IL-10: the master regulator of immunity to infection. J. Immunol. 180:5771–5777. 10.4049/jimmunol.180.9.5771 [DOI] [PubMed] [Google Scholar]
- Crawley J.B., Williams L.M., Mander T., Brennan F.M., and Foxwell B.M.. 1996. Interleukin-10 stimulation of phosphatidylinositol 3-kinase and p70 S6 kinase is required for the proliferative but not the antiinflammatory effects of the cytokine. J. Biol. Chem. 271:16357–16362. 10.1074/jbc.271.27.16357 [DOI] [PubMed] [Google Scholar]
- Creery W.D., Diaz-Mitoma F., Filion L., and Kumar A.. 1996. Differential modulation of B7-1 and B7-2 isoform expression on human monocytes by cytokines which influence the development of T helper cell phenotype. Eur. J. Immunol. 26:1273–1277. 10.1002/eji.1830260614 [DOI] [PubMed] [Google Scholar]
- Croker B.A., Krebs D.L., Zhang J.G., Wormald S., Willson T.A., Stanley E.G., Robb L., Greenhalgh C.J., Förster I., Clausen B.E., et al. . 2003. SOCS3 negatively regulates IL-6 signaling in vivo. Nat. Immunol. 4:540–545. 10.1038/ni931 [DOI] [PubMed] [Google Scholar]
- Cunha F.Q., Moncada S., and Liew F.Y.. 1992. Interleukin-10 (IL-10) inhibits the induction of nitric oxide synthase by interferon-gamma in murine macrophages. Biochem. Biophys. Res. Commun. 182:1155–1159. 10.1016/0006-291X(92)91852-H [DOI] [PubMed] [Google Scholar]
- D’Andrea A., Aste-Amezaga M., Valiante N.M., Ma X., Kubin M., and Trinchieri G.. 1993. Interleukin 10 (IL-10) inhibits human lymphocyte interferon gamma-production by suppressing natural killer cell stimulatory factor/IL-12 synthesis in accessory cells. J. Exp. Med. 178:1041–1048. 10.1084/jem.178.3.1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W.J., Köhler G., and Brombacher F.. 1997. Both innate and acquired immunity to Listeria monocytogenes infection are increased in IL-10-deficient mice. J. Immunol. 158:2259–2267. [PubMed] [Google Scholar]
- Danne C., Ryzhakov G., Martínez-López M., Ilott N.E., Franchini F., Cuskin F., Lowe E.C., Bullers S.J., Arthur J.S.C., and Powrie F.. 2017. A Large Polysaccharide Produced by Helicobacter hepaticus Induces an Anti-inflammatory Gene Signature in Macrophages. Cell Host Microbe. 22:733–745.e5. 10.1016/j.chom.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boer M.A., Voshol P.J., Schröder-van der Elst J.P., Korsheninnikova E., Ouwens D.M., Kuipers F., Havekes L.M., and Romijn J.A.. 2006. Endogenous interleukin-10 protects against hepatic steatosis but does not improve insulin sensitivity during high-fat feeding in mice. Endocrinology. 147:4553–4558. 10.1210/en.2006-0417 [DOI] [PubMed] [Google Scholar]
- de Waal Malefyt R., Abrams J., Bennett B., Figdor C.G., and de Vries J.E.. 1991a Interleukin 10(IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J. Exp. Med. 174:1209–1220. 10.1084/jem.174.5.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waal Malefyt R., Haanen J., Spits H., Roncarolo M.G., te Velde A., Figdor C., Johnson K., Kastelein R., Yssel H., and de Vries J.E.. 1991b Interleukin 10 (IL-10) and viral IL-10 strongly reduce antigen-specific human T cell proliferation by diminishing the antigen-presenting capacity of monocytes via downregulation of class II major histocompatibility complex expression. J. Exp. Med. 174:915–924. 10.1084/jem.174.4.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., and Shevach E.M.. 1992. IL-10 inhibits mitogen-induced T cell proliferation by selectively inhibiting macrophage costimulatory function. J. Immunol. 148:3133–3139. [PubMed] [Google Scholar]
- Donnelly R.P., Dickensheets H., and Finbloom D.S.. 1999. The interleukin-10 signal transduction pathway and regulation of gene expression in mononuclear phagocytes. J. Interferon Cytokine Res. 19:563–573. 10.1089/107999099313695 [DOI] [PubMed] [Google Scholar]
- Dunn G.P., Old L.J., and Schreiber R.D.. 2004. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 21:137–148. 10.1016/j.immuni.2004.07.017 [DOI] [PubMed] [Google Scholar]
- El Kasmi K.C., Smith A.M., Williams L., Neale G., Panopoulos A.D., Watowich S.S., Häcker H., Foxwell B.M., and Murray P.J.. 2007. Cutting edge: A transcriptional repressor and corepressor induced by the STAT3-regulated anti-inflammatory signaling pathway. J. Immunol. 179:7215–7219. 10.4049/jimmunol.179.11.7215 [DOI] [PubMed] [Google Scholar]
- Ellinghaus D., Jostins L., Spain S.L., Cortes A., Bethune J., Han B., Park Y.R., Raychaudhuri S., Pouget J.G., Hübenthal M., et al. Psoriasis Association Genetics Extension (PAGE) . 2016. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 48:510–518. 10.1038/ng.3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich J., Mumm J.B., Chan I.H., LaFace D., Truong H., McClanahan T., Gorman D.M., and Oft M.. 2012. IL-10 directly activates and expands tumor-resident CD8(+) T cells without de novo infiltration from secondary lymphoid organs. Cancer Res. 72:3570–3581. 10.1158/0008-5472.CAN-12-0721 [DOI] [PubMed] [Google Scholar]
- Engelhardt K.R., and Grimbacher B.. 2014. IL-10 in humans: lessons from the gut, IL-10/IL-10 receptor deficiencies, and IL-10 polymorphisms. Curr. Top. Microbiol. Immunol. 380:1–18. [DOI] [PubMed] [Google Scholar]
- Fairfax K.A., Gantier M.P., Mackay F., Williams B.R., and McCoy C.E.. 2015. IL-10 regulates Aicda expression through miR-155. J. Leukoc. Biol. 97:71–78. 10.1189/jlb.2A0314-178R [DOI] [PubMed] [Google Scholar]
- Fedorak R.N., Gangl A., Elson C.O., Rutgeerts P., Schreiber S., Wild G., Hanauer S.B., Kilian A., Cohard M., LeBeaut A., and Feagan B.. 2000. Recombinant human interleukin 10 in the treatment of patients with mild to moderately active Crohn’s disease. The Interleukin 10 Inflammatory Bowel Disease Cooperative Study Group. Gastroenterology. 119:1473–1482. 10.1053/gast.2000.20229 [DOI] [PubMed] [Google Scholar]
- Fiorentino D.F., Bond M.W., and Mosmann T.R.. 1989. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J. Exp. Med. 170:2081–2095. 10.1084/jem.170.6.2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentino D.F., Zlotnik A., Mosmann T.R., Howard M., and O’Garra A.. 1991a IL-10 inhibits cytokine production by activated macrophages. J. Immunol. 147:3815–3822. [PubMed] [Google Scholar]
- Fiorentino D.F., Zlotnik A., Vieira P., Mosmann T.R., Howard M., Moore K.W., and O’Garra A.. 1991b IL-10 acts on the antigen-presenting cell to inhibit cytokine production by Th1 cells. J. Immunol. 146:3444–3451. [PubMed] [Google Scholar]
- Fleming S.B., McCaughan C.A., Andrews A.E., Nash A.D., and Mercer A.A.. 1997. A homolog of interleukin-10 is encoded by the poxvirus orf virus. J. Virol. 71:4857–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei K., Nadal D., Pfister H.W., and Fontana A.. 1993. Listeria meningitis: identification of a cerebrospinal fluid inhibitor of macrophage listericidal function as interleukin 10. J. Exp. Med. 178:1255–1261. 10.1084/jem.178.4.1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabryšová L., Howes A., Saraiva M., and O’Garra A.. 2014. The regulation of IL-10 expression. Curr. Top. Microbiol. Immunol. 380:157–190. [DOI] [PubMed] [Google Scholar]
- Gabryšová L., Alvarez-Martinez M., Luisier R., Cox L.S., Sodenkamp J., Hosking C., Pérez-Mazliah D., Whicher C., Kannan Y., Potempa K., et al. . 2018. c-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4+ T cells. Nat. Immunol. 19:497–507. 10.1038/s41590-018-0083-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeazzi M., Bazzichi L., Sebastiani G.D., Neri D., Garcia E., Ravenni N., Giovannoni L., Wilton J., Bardelli M., Baldi C., et al. . 2014. A phase IB clinical trial with Dekavil (F8-IL10), an immunoregulatory ‘armed antibody’ for the treatment of rheumatoid arthritis, used in combination wiIh methotrexate. Isr. Med. Assoc. J. 16:666. [PubMed] [Google Scholar]
- Gandhi R., Kumar D., Burns E.J., Nadeau M., Dake B., Laroni A., Kozoriz D., Weiner H.L., and Quintana F.J.. 2010. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nat. Immunol. 11:846–853. 10.1038/ni.1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Zhang C., Ma Y., Bu L., Yan L., and Liu D.. 2013. Hydrodynamic delivery of mIL10 gene protects mice from high-fat diet-induced obesity and glucose intolerance. Mol. Ther. 21:1852–1861. 10.1038/mt.2013.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzinelli R.T., Oswald I.P., James S.L., and Sher A.. 1992. IL-10 inhibits parasite killing and nitrogen oxide production by IFN-gamma-activated macrophages. J. Immunol. 148:1792–1796. [PubMed] [Google Scholar]
- Gazzinelli R.T., Wysocka M., Hieny S., Scharton-Kersten T., Cheever A., Kühn R., Müller W., Trinchieri G., and Sher A.. 1996. In the absence of endogenous IL-10, mice acutely infected with Toxoplasma gondii succumb to a lethal immune response dependent on CD4+ T cells and accompanied by overproduction of IL-12, IFN-gamma and TNF-alpha. J. Immunol. 157:798–805. [PubMed] [Google Scholar]
- Gerner R.R., Klepsch V., Macheiner S., Arnhard K., Adolph T.E., Grander C., Wieser V., Pfister A., Moser P., Hermann-Kleiter N., et al. . 2018. NAD metabolism fuels human and mouse intestinal inflammation. Gut. 67:1813–1823. 10.1136/gutjnl-2017-314241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasmacher E., Agrawal S., Chang A.B., Murphy T.L., Zeng W., Vander Lugt B., Khan A.A., Ciofani M., Spooner C.J., Rutz S., et al. . 2012. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 338:975–980. 10.1126/science.1228309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson L.E., Sheedy F.J., Palsson-McDermott E.M., Triglia D., O’Leary S.M., O’Sullivan M.P., O’Neill L.A., and Keane J.. 2016. Cutting Edge: Mycobacterium tuberculosis Induces Aerobic Glycolysis in Human Alveolar Macrophages That Is Required for Control of Intracellular Bacillary Replication. J. Immunol. 196:2444–2449. 10.4049/jimmunol.1501612 [DOI] [PubMed] [Google Scholar]
- Glocker E.O., Kotlarz D., Boztug K., Gertz E.M., Schäffer A.A., Noyan F., Perro M., Diestelhorst J., Allroth A., Murugan D., et al. . 2009. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361:2033–2045. 10.1056/NEJMoa0907206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker E.O., Frede N., Perro M., Sebire N., Elawad M., Shah N., and Grimbacher B.. 2010. Infant colitis--it’s in the genes. Lancet. 376:1272 10.1016/S0140-6736(10)61008-2 [DOI] [PubMed] [Google Scholar]
- Glocker E.O., Kotlarz D., Klein C., Shah N., and Grimbacher B.. 2011. IL-10 and IL-10 receptor defects in humans. Ann. N. Y. Acad. Sci. 1246:102–107. 10.1111/j.1749-6632.2011.06339.x [DOI] [PubMed] [Google Scholar]
- Go N.F., Castle B.E., Barrett R., Kastelein R., Dang W., Mosmann T.R., Moore K.W., and Howard M.. 1990. Interleukin 10, a novel B cell stimulatory factor: unresponsiveness of X chromosome-linked immunodeficiency B cells. J. Exp. Med. 172:1625–1631. 10.1084/jem.172.6.1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grilli M., Barbieri I., Basudev H., Brusa R., Casati C., Lozza G., and Ongini E.. 2000. Interleukin-10 modulates neuronal threshold of vulnerability to ischaemic damage. Eur. J. Neurosci. 12:2265–2272. 10.1046/j.1460-9568.2000.00090.x [DOI] [PubMed] [Google Scholar]
- Groux H., Bigler M., de Vries J.E., and Roncarolo M.G.. 1996. Interleukin-10 induces a long-term antigen-specific anergic state in human CD4+ T cells. J. Exp. Med. 184:19–29. 10.1084/jem.184.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groux H., Bigler M., de Vries J.E., and Roncarolo M.G.. 1998. Inhibitory and stimulatory effects of IL-10 on human CD8+ T cells. J. Immunol. 160:3188–3193. [PubMed] [Google Scholar]
- Groux H., Cottrez F., Rouleau M., Mauze S., Antonenko S., Hurst S., McNeil T., Bigler M., Roncarolo M.G., and Coffman R.L.. 1999. A transgenic model to analyze the immunoregulatory role of IL-10 secreted by antigen-presenting cells. J. Immunol. 162:1723–1729. [PubMed] [Google Scholar]
- Gupta A., Dimeloe S., Richards D.F., Chambers E.S., Black C., Urry Z., Ryanna K., Xystrakis E., Bush A., Saglani S., and Hawrylowicz C.M.. 2014. Defective IL-10 expression and in vitro steroid-induced IL-17A in paediatric severe therapy-resistant asthma. Thorax. 69:508–515. 10.1136/thoraxjnl-2013-203421 [DOI] [PubMed] [Google Scholar]
- Hagenbaugh A., Sharma S., Dubinett S.M., Wei S.H., Aranda R., Cheroutre H., Fowell D.J., Binder S., Tsao B., Locksley R.M., et al. . 1997. Altered immune responses in interleukin 10 transgenic mice. J. Exp. Med. 185:2101–2110. 10.1084/jem.185.12.2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M., Mages J., Dietrich H., Schmitz F., Striebel F., Murray P.J., Wagner H., and Lang R.. 2005. Control of dual-specificity phosphatase-1 expression in activated macrophages by IL-10. Eur. J. Immunol. 35:2991–3001. 10.1002/eji.200526192 [DOI] [PubMed] [Google Scholar]
- Hong E.G., Ko H.J., Cho Y.R., Kim H.J., Ma Z., Yu T.Y., Friedline R.H., Kurt-Jones E., Finberg R., Fischer M.A., et al. . 2009. Interleukin-10 prevents diet-induced insulin resistance by attenuating macrophage and cytokine response in skeletal muscle. Diabetes. 58:2525–2535. 10.2337/db08-1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes A., Taubert C., Blankley S., Spink N., Wu X., Graham C.M., Zhao J., Saraiva M., Ricciardi-Castagnoli P., Bancroft G.J., and O’Garra A.. 2016. Differential Production of Type I IFN Determines the Reciprocal Levels of IL-10 and Proinflammatory Cytokines Produced by C57BL/6 and BALB/c Macrophages. J. Immunol. 197:2838–2853. 10.4049/jimmunol.1501923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu D.H., de Waal Malefyt R., Fiorentino D.F., Dang M.N., Vieira P., de Vries J., Spits H., Mosmann T.R., and Moore K.W.. 1990. Expression of interleukin-10 activity by Epstein-Barr virus protein BCRF1. Science. 250:830–832. 10.1126/science.2173142 [DOI] [PubMed] [Google Scholar]
- Huber S., Gagliani N., Esplugues E., O’Connor W. Jr., Huber F.J., Chaudhry A., Kamanaka M., Kobayashi Y., Booth C.J., Rudensky A.Y., et al. . 2011. Th17 cells express interleukin-10 receptor and are controlled by Foxp3− and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity. 34:554–565. 10.1016/j.immuni.2011.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsmans M., Sager H.B., Roh J.D., Valero-Muñoz M., Houstis N.E., Iwamoto Y., Sun Y., Wilson R.M., Wojtkiewicz G., Tricot B., et al. . 2018. Cardiac macrophages promote diastolic dysfunction. J. Exp. Med. 215:423–440. 10.1084/jem.20171274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C.A., Ellis-Neyes L.A., Slifer T., Kanaly S., Grünig G., Fort M., Rennick D., and Araujo F.G.. 1997. IL-10 is required to prevent immune hyperactivity during infection with Trypanosoma cruzi. J. Immunol. 158:3311–3316. [PubMed] [Google Scholar]
- Hussaarts L., Smits H.H., Schramm G., van der Ham A.J., van der Zon G.C., Haas H., Guigas B., and Yazdanbakhsh M.. 2013. Rapamycin and omega-1: mTOR-dependent and -independent Th2 skewing by human dendritic cells. Immunol. Cell Biol. 91:486–489. 10.1038/icb.2013.31 [DOI] [PubMed] [Google Scholar]
- Hutchins A.P., Poulain S., and Miranda-Saavedra D.. 2012. Genome-wide analysis of STAT3 binding in vivo predicts effectors of the anti-inflammatory response in macrophages. Blood. 119:e110–e119. 10.1182/blood-2011-09-381483 [DOI] [PubMed] [Google Scholar]
- Hutchins A.P., Diez D., and Miranda-Saavedra D.. 2013. The IL-10/STAT3-mediated anti-inflammatory response: recent developments and future challenges. Brief. Funct. Genomics. 12:489–498. 10.1093/bfgp/elt028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins A.P., Takahashi Y., and Miranda-Saavedra D.. 2015. Genomic analysis of LPS-stimulated myeloid cells identifies a common pro-inflammatory response but divergent IL-10 anti-inflammatory responses. Sci. Rep. 5:9100 10.1038/srep09100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J.P., Lin C.C., Kimmey J.M., Jarjour N.N., Schwarzkopf E.A., Bradstreet T.R., Shchukina I., Shpynov O., Weaver C.T., Taneja R., et al. . 2018. Bhlhe40 is an essential repressor of IL-10 during Mycobacterium tuberculosis infection. J. Exp. Med. 215:1823–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun J., Romero L., Riveron R., Flores C., Kanagavelu S., Chung K.D., Alonso A., Sotolongo J., Ruiz J., Manukyan A., et al. . 2015. Human intestinal epithelial cells express interleukin-10 through Toll-like receptor 4-mediated epithelial-macrophage crosstalk. J. Innate Immun. 7:87–101. 10.1159/000365417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im S.H., Hueber A., Monticelli S., Kang K.H., and Rao A.. 2004. Chromatin-level regulation of the IL10 gene in T cells. J. Biol. Chem. 279:46818–46825. 10.1074/jbc.M401722200 [DOI] [PubMed] [Google Scholar]
- Ip W.K.E., Hoshi N., Shouval D.S., Snapper S., and Medzhitov R.. 2017. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science. 356:513–519. 10.1126/science.aal3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H., Tanabe S., Ueno M., Kubo T., Kayama H., Serada S., Fujimoto M., Takeda K., Naka T., and Yamashita T.. 2013. ifn-γ-dependent secretion of IL-10 from Th1 cells and microglia/macrophages contributes to functional recovery after spinal cord injury. Cell Death Dis. 4:e710 10.1038/cddis.2013.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Huang R.R., Wen D.R., Paul E., Wünsch P.H., and Cochran A.J.. 2011. IL-10 expression by primary tumor cells correlates with melanoma progression from radial to vertical growth phase and development of metastatic competence. Mod. Pathol. 24:801–809. 10.1038/modpathol.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., and Hirohata S.. 1995. The role of IL-10 in human B cell activation, proliferation, and differentiation. J. Immunol. 154:4341–4350. [PubMed] [Google Scholar]
- Iyer S.S., Gensollen T., Gandhi A., Oh S.F., Neves J.F., Collin F., Lavin R., Serra C., Glickman J., de Silva P.S.A., et al. . 2018. Dietary and Microbial Oxazoles Induce Intestinal Inflammation by Modulating Aryl Hydrocarbon Receptor Responses. Cell. 173:1123–1134.e11. 10.1016/j.cell.2018.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarjour N.N., Schwarzkopf E.A., Bradstreet T.R., Shchukina I., Lin C.C., Huang S.C., Lai C.W., Cook M.E., Taneja R., Stappenbeck T.S., et al. . 2019. Bhlhe40 mediates tissue-specific control of macrophage proliferation in homeostasis and type 2 immunity. Nat. Immunol. 20:687–700. 10.1038/s41590-019-0382-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarry A., Bossard C., Bou-Hanna C., Masson D., Espaze E., Denis M.G., and Laboisse C.L.. 2008. Mucosal IL-10 and TGF-beta play crucial roles in preventing LPS-driven, IFN-gamma-mediated epithelial damage in human colon explants. J. Clin. Invest. 118:1132–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardane G., Russell G.C., Thomson J., Deane D., Cox H., Gatherer D., Ackermann M., Haig D.M., and Stewart J.P.. 2008. A captured viral interleukin 10 gene with cellular exon structure. J. Gen. Virol. 89:2447–2455. 10.1099/vir.0.2008/001743-0 [DOI] [PubMed] [Google Scholar]
- Jones E.A., and Flavell R.A.. 2005. Distal enhancer elements transcribe intergenic RNA in the IL-10 family gene cluster. J. Immunol. 175:7437–7446. 10.4049/jimmunol.175.11.7437 [DOI] [PubMed] [Google Scholar]
- Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., et al. International IBD Genetics Consortium (IIBDGC) . 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 491:119–124. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser F., Cook D., Papoutsopoulou S., Rajsbaum R., Wu X., Yang H.T., Grant S., Ricciardi-Castagnoli P., Tsichlis P.N., Ley S.C., and O’Garra A.. 2009. TPL-2 negatively regulates interferon-beta production in macrophages and myeloid dendritic cells. J. Exp. Med. 206:1863–1871. 10.1084/jem.20091059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamanaka M., Huber S., Zenewicz L.A., Gagliani N., Rathinam C., O’Connor W. Jr., Wan Y.Y., Nakae S., Iwakura Y., Hao L., and Flavell R.A.. 2011. Memory/effector (CD45RB(lo)) CD4 T cells are controlled directly by IL-10 and cause IL-22-dependent intestinal pathology. J. Exp. Med. 208:1027–1040. 10.1084/jem.20102149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanters E., Pasparakis M., Gijbels M.J., Vergouwe M.N., Partouns-Hendriks I., Fijneman R.J., Clausen B.E., Förster I., Kockx M.M., Rajewsky K., et al. . 2003. Inhibition of NF-kappaB activation in macrophages increases atherosclerosis in LDL receptor-deficient mice. J. Clin. Invest. 112:1176–1185. 10.1172/JCI200318580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasama T., Strieter R.M., Lukacs N.W., Burdick M.D., and Kunkel S.L.. 1994. Regulation of neutrophil-derived chemokine expression by IL-10. J. Immunol. 152:3559–3569. [PubMed] [Google Scholar]
- Kim C., Sano Y., Todorova K., Carlson B.A., Arpa L., Celada A., Lawrence T., Otsu K., Brissette J.L., Arthur J.S., and Park J.M.. 2008. The kinase p38 alpha serves cell type-specific inflammatory functions in skin injury and coordinates pro- and anti-inflammatory gene expression. Nat. Immunol. 9:1019–1027. 10.1038/ni.1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Naka T., Nakahama T., Chinen I., Masuda K., Nohara K., Fujii-Kuriyama Y., and Kishimoto T.. 2009. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J. Exp. Med. 206:2027–2035. 10.1084/jem.20090560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A., Balaji S., Le L.D., Crombleholme T.M., and Keswani S.G.. 2014. Regenerative Wound Healing: The Role of Interleukin-10. Adv. Wound Care (New Rochelle). 3:315–323. 10.1089/wound.2013.0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach S.M., and Faden A.I.. 1998. Interleukin-10 improves outcome and alters proinflammatory cytokine expression after experimental traumatic brain injury. Exp. Neurol. 153:143–151. 10.1006/exnr.1998.6877 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Kweon M.N., Kuwata H., Schreiber R.D., Kiyono H., Takeda K., and Akira S.. 2003. Toll-like receptor-dependent production of IL-12p40 causes chronic enterocolitis in myeloid cell-specific Stat3-deficient mice. J. Clin. Invest. 111:1297–1308. 10.1172/JCI17085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole A., and Maloy K.J.. 2014. Control of intestinal inflammation by interleukin-10. Curr. Top. Microbiol. Immunol. 380:19–38. [DOI] [PubMed] [Google Scholar]
- Kominsky D.J., Campbell E.L., Ehrentraut S.F., Wilson K.E., Kelly C.J., Glover L.E., Collins C.B., Bayless A.J., Saeedi B., Dobrinskikh E., et al. . 2014. IFN-γ-mediated induction of an apical IL-10 receptor on polarized intestinal epithelia. J. Immunol. 192:1267–1276. 10.4049/jimmunol.1301757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscsó B., Csóka B., Selmeczy Z., Himer L., Pacher P., Virág L., and Haskó G.. 2012. Adenosine augments IL-10 production by microglial cells through an A2B adenosine receptor-mediated process. J. Immunol. 188:445–453. 10.4049/jimmunol.1101224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S.V., Saccani S., Izotova L.S., Mirochnitchenko O.V., and Pestka S.. 2000. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. USA. 97:1695–1700. 10.1073/pnas.97.4.1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn R., Löhler J., Rennick D., Rajewsky K., and Müller W.. 1993. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 75:263–274. 10.1016/0092-8674(93)80068-P [DOI] [PubMed] [Google Scholar]
- Kuwata H., Watanabe Y., Miyoshi H., Yamamoto M., Kaisho T., Takeda K., and Akira S.. 2003. IL-10-inducible Bcl-3 negatively regulates LPS-induced TNF-alpha production in macrophages. Blood. 102:4123–4129. 10.1182/blood-2003-04-1228 [DOI] [PubMed] [Google Scholar]
- Kwilasz A.J., Grace P.M., Serbedzija P., Maier S.F., and Watkins L.R.. 2015. The therapeutic potential of interleukin-10 in neuroimmune diseases. Neuropharmacology. 96(Pt A):55–69. 10.1016/j.neuropharm.2014.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R., Patel D., Morris J.J., Rutschman R.L., and Murray P.J.. 2002. Shaping gene expression in activated and resting primary macrophages by IL-10. J. Immunol. 169:2253–2263. 10.4049/jimmunol.169.5.2253 [DOI] [PubMed] [Google Scholar]
- Lang R., Pauleau A.L., Parganas E., Takahashi Y., Mages J., Ihle J.N., Rutschman R., and Murray P.J.. 2003. SOCS3 regulates the plasticity of gp130 signaling. Nat. Immunol. 4:546–550. 10.1038/ni932 [DOI] [PubMed] [Google Scholar]
- Lanis J.M., Alexeev E.E., Curtis V.F., Kitzenberg D.A., Kao D.J., Battista K.D., Gerich M.E., Glover L.E., Kominsky D.J., and Colgan S.P.. 2017. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol. 10:1133–1144. 10.1038/mi.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauw F.N., Pajkrt D., Hack C.E., Kurimoto M., van Deventer S.J., and van der Poll T.. 2000. Proinflammatory effects of IL-10 during human endotoxemia. J. Immunol. 165:2783–2789. 10.4049/jimmunol.165.5.2783 [DOI] [PubMed] [Google Scholar]
- Ledeboer A., Brevé J.J., Wierinckx A., van der Jagt S., Bristow A.F., Leysen J.E., Tilders F.J., and Van Dam A.M.. 2002. Expression and regulation of interleukin-10 and interleukin-10 receptor in rat astroglial and microglial cells. Eur. J. Neurosci. 16:1175–1185. 10.1046/j.1460-9568.2002.02200.x [DOI] [PubMed] [Google Scholar]
- Lee T.S., and Chau L.Y.. 2002. Heme oxygenase-1 mediates the anti-inflammatory effect of interleukin-10 in mice. Nat. Med. 8:240–246. 10.1038/nm0302-240 [DOI] [PubMed] [Google Scholar]
- Lee C.G., Kang K.H., So J.S., Kwon H.K., Son J.S., Song M.K., Sahoo A., Yi H.J., Hwang K.C., Matsuyama T., et al. . 2009. A distal cis-regulatory element, CNS-9, controls NFAT1 and IRF4-mediated IL-10 gene activation in T helper cells. Mol. Immunol. 46:613–621. 10.1016/j.molimm.2008.07.037 [DOI] [PubMed] [Google Scholar]
- Lehmann J., Seegert D., Strehlow I., Schindler C., Lohmann-Matthes M.L., and Decker T.. 1994. IL-10-induced factors belonging to the p91 family of proteins bind to IFN-gamma-responsive promoter elements. J. Immunol. 153:165–172. [PubMed] [Google Scholar]
- Li C., Corraliza I., and Langhorne J.. 1999. A defect in interleukin-10 leads to enhanced malarial disease in Plasmodium chabaudi chabaudi infection in mice. Infect. Immun. 67:4435–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Huang L., Ye H., Song S.P., Bajwa A., Lee S.J., Moser E.K., Jaworska K., Kinsey G.R., Day Y.J., et al. . 2012a Dendritic cells tolerized with adenosine A2AR agonist attenuate acute kidney injury. J. Clin. Invest. 122:3931–3942. 10.1172/JCI63170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Spolski R., Liao W., Wang L., Murphy T.L., Murphy K.M., and Leonard W.J.. 2012b BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 490:543–546. 10.1038/nature11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.C., Bradstreet T.R., Schwarzkopf E.A., Sim J., Carrero J.A., Chou C., Cook L.E., Egawa T., Taneja R., Murphy T.L., et al. . 2014. Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat. Commun. 5:3551 10.1038/ncomms4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira F.S., Rosa J.C., Pimentel G.D., Seelaender M., Damaso A.R., Oyama L.M., and do Nascimento C.O.. 2012. Both adiponectin and interleukin-10 inhibit LPS-induced activation of the NF-κB pathway in 3T3-L1 adipocytes. Cytokine. 57:98–106. 10.1016/j.cyto.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Liu Y.W., Chen C.C., Tseng H.P., and Chang W.C.. 2006. Lipopolysaccharide-induced transcriptional activation of interleukin-10 is mediated by MAPK- and NF-kappaB-induced CCAAT/enhancer-binding protein delta in mouse macrophages. Cell. Signal. 18:1492–1500. 10.1016/j.cellsig.2005.12.001 [DOI] [PubMed] [Google Scholar]
- Llorente L., Richaud-Patin Y., García-Padilla C., Claret E., Jakez-Ocampo J., Cardiel M.H., Alcocer-Varela J., Grangeot-Keros L., Alarcón-Segovia D., Wijdenes J., et al. . 2000. Clinical and biologic effects of anti-interleukin-10 monoclonal antibody administration in systemic lupus erythematosus. Arthritis Rheum. 43:1790–1800. [DOI] [PubMed] [Google Scholar]
- Lobo-Silva D., Carriche G.M., Castro A.G., Roque S., and Saraiva M.. 2016. Balancing the immune response in the brain: IL-10 and its regulation. J. Neuroinflammation. 13:297 10.1186/s12974-016-0763-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo-Silva D., Carriche G.M., Castro A.G., Roque S., and Saraiva M.. 2017. Interferon-β regulates the production of IL-10 by toll-like receptor-activated microglia. Glia. 65:1439–1451. 10.1002/glia.23172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorén V., Cabré E., Ojanguren I., Domènech E., Pedrosa E., García-Jaraquemada A., Mañosa M., and Manyé J.. 2015. Interleukin-10 Enhances the Intestinal Epithelial Barrier in the Presence of Corticosteroids through p38 MAPK Activity in Caco-2 Monolayers: A Possible Mechanism for Steroid Responsiveness in Ulcerative Colitis. PLoS One. 10:e0130921 10.1371/journal.pone.0130921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loser K., Apelt J., Voskort M., Mohaupt M., Balkow S., Schwarz T., Grabbe S., and Beissert S.. 2007. IL-10 controls ultraviolet-induced carcinogenesis in mice. J. Immunol. 179:365–371. 10.4049/jimmunol.179.1.365 [DOI] [PubMed] [Google Scholar]
- Lumeng C.N., Bodzin J.L., and Saltiel A.R.. 2007. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 117:175–184. 10.1172/JCI29881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu M., Pautz S., Kohl V., Singh R., Romero R., Lucas S., Hofmann J., Raifer H., Vachharajani N., Carrascosa L.C., et al. . 2019. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat. Commun. 10:760 10.1038/s41467-019-08711-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macatonia S.E., Doherty T.M., Knight S.C., and O’Garra A.. 1993. Differential effect of IL-10 on dendritic cell-induced T cell proliferation and IFN-gamma production. J. Immunol. 150:3755–3765. [PubMed] [Google Scholar]
- MacNeil I.A., Suda T., Moore K.W., Mosmann T.R., and Zlotnik A.. 1990. IL-10, a novel growth cofactor for mature and immature T cells. J. Immunol. 145:4167–4173. [PubMed] [Google Scholar]
- Mann E.H., Gabryšová L., Pfeffer P.E., O’Garra A., and Hawrylowicz C.M.. 2019. High-Dose IL-2 Skews a Glucocorticoid-Driven IL-17+IL-10+ Memory CD4+ T Cell Response towards a Single IL-10-Producing Phenotype. J. Immunol. 202:684–693. 10.4049/jimmunol.1800697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannino M.H., Zhu Z., Xiao H., Bai Q., Wakefield M.R., and Fang Y.. 2015. The paradoxical role of IL-10 in immunity and cancer. Cancer Lett. 367:103–107. 10.1016/j.canlet.2015.07.009 [DOI] [PubMed] [Google Scholar]
- Márquez S., Fernández J.J., Mancebo C., Herrero-Sánchez C., Alonso S., Sandoval T.A., Rodríguez Prados M., Cubillos-Ruiz J.R., Montero O., Fernández N., and Sánchez Crespo M.. 2019. Tricarboxylic Acid Cycle Activity and Remodeling of Glycerophosphocholine Lipids Support Cytokine Induction in Response to Fungal Patterns. Cell Reports. 27:525–536.e4. 10.1016/j.celrep.2019.03.033 [DOI] [PubMed] [Google Scholar]
- Martin M., Schifferle R.E., Cuesta N., Vogel S.N., Katz J., and Michalek S.M.. 2003. Role of the phosphatidylinositol 3 kinase-Akt pathway in the regulation of IL-10 and IL-12 by Porphyromonas gingivalis lipopolysaccharide. J. Immunol. 171:717–725. 10.4049/jimmunol.171.2.717 [DOI] [PubMed] [Google Scholar]
- Mascanfroni I.D., Takenaka M.C., Yeste A., Patel B., Wu Y., Kenison J.E., Siddiqui S., Basso A.S., Otterbein L.E., Pardoll D.M., et al. . 2015. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nat. Med. 21:638–646. 10.1038/nm.3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukawa A., Takeda K., Kudo S., Maeda T., Kagayama M., and Akira S.. 2003. Aberrant inflammation and lethality to septic peritonitis in mice lacking STAT3 in macrophages and neutrophils. J. Immunol. 171:6198–6205. 10.4049/jimmunol.171.11.6198 [DOI] [PubMed] [Google Scholar]
- Matsukawa A., Kudo S., Maeda T., Numata K., Watanabe H., Takeda K., Akira S., and Ito T.. 2005. Stat3 in resident macrophages as a repressor protein of inflammatory response. J. Immunol. 175:3354–3359. 10.4049/jimmunol.175.5.3354 [DOI] [PubMed] [Google Scholar]
- Mauer J., Chaurasia B., Goldau J., Vogt M.C., Ruud J., Nguyen K.D., Theurich S., Hausen A.C., Schmitz J., Brönneke H.S., et al. . 2014. Signaling by IL-6 promotes alternative activation of macrophages to limit endotoxemia and obesity-associated resistance to insulin. Nat. Immunol. 15:423–430. 10.1038/ni.2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauras A., Chain F., Faucheux A., Ruffié P., Gontier S., Ryffel B., Butel M.J., Langella P., Bermúdez-Humarán L.G., and Waligora-Dupriet A.J.. 2018. A New Bifidobacteria Expression SysTem (BEST) to Produce and Deliver Interleukin-10 in Bifidobacterium bifidum. Front. Microbiol. 9:3075 10.3389/fmicb.2018.03075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy C.E., Sheedy F.J., Qualls J.E., Doyle S.L., Quinn S.R., Murray P.J., and O’Neill L.A.. 2010. IL-10 inhibits miR-155 induction by toll-like receptors. J. Biol. Chem. 285:20492–20498. 10.1074/jbc.M110.102111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes I.B., Illei G.G., Danning C.L., Yarboro C.H., Crane M., Kuroiwa T., Schlimgen R., Lee E., Foster B., Flemming D., et al. . 2001. IL-10 improves skin disease and modulates endothelial activation and leukocyte effector function in patients with psoriatic arthritis. J. Immunol. 167:4075–4082. 10.4049/jimmunol.167.7.4075 [DOI] [PubMed] [Google Scholar]
- McNab F.W., Ewbank J., Howes A., Moreira-Teixeira L., Martirosyan A., Ghilardi N., Saraiva M., and O’Garra A.. 2014. Type I IFN induces IL-10 production in an IL-27-independent manner and blocks responsiveness to IFN-γ for production of IL-12 and bacterial killing in Mycobacterium tuberculosis-infected macrophages. J. Immunol. 193:3600–3612. 10.4049/jimmunol.1401088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita A.R., Correia-Neves M., Roque S., Castro A.G., Vieira P., Pedrosa J., Palha J.A., and Sousa N.. 2008. IL-10 modulates depressive-like behavior. J. Psychiatr. Res. 43:89–97. 10.1016/j.jpsychires.2008.02.004 [DOI] [PubMed] [Google Scholar]
- Miller A.M., Wang H., Bertola A., Park O., Horiguchi N., Ki S.H., Yin S., Lafdil F., and Gao B.. 2011. Inflammation-associated interleukin-6/signal transducer and activator of transcription 3 activation ameliorates alcoholic and nonalcoholic fatty liver diseases in interleukin-10-deficient mice. Hepatology. 54:846–856. 10.1002/hep.24517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Holgado E., Vela J.M., Arévalo-Martín A., and Guaza C.. 2001. LPS/IFN-gamma cytotoxicity in oligodendroglial cells: role of nitric oxide and protection by the anti-inflammatory cytokine IL-10. Eur. J. Neurosci. 13:493–502. 10.1046/j.0953-816x.2000.01412.x [DOI] [PubMed] [Google Scholar]
- Moore K.W., Vieira P., Fiorentino D.F., Trounstine M.L., Khan T.A., and Mosmann T.R.. 1990. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science. 248:1230–1234. 10.1126/science.2161559 [DOI] [PubMed] [Google Scholar]
- Moore K.W., de Waal Malefyt R., Coffman R.L., and O’Garra A.. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19:683–765. 10.1146/annurev.immunol.19.1.683 [DOI] [PubMed] [Google Scholar]
- Moran C.J., Walters T.D., Guo C.H., Kugathasan S., Klein C., Turner D., Wolters V.M., Bandsma R.H., Mouzaki M., Zachos M., et al. . 2013. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm. Bowel Dis. 19:115–123. 10.1002/ibd.22974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm J.B., Emmerich J., Zhang X., Chan I., Wu L., Mauze S., Blaisdell S., Basham B., Dai J., Grein J., et al. . 2011. IL-10 elicits IFNγ-dependent tumor immune surveillance. Cancer Cell. 20:781–796. 10.1016/j.ccr.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Murai M., Turovskaya O., Kim G., Madan R., Karp C.L., Cheroutre H., and Kronenberg M.. 2009. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat. Immunol. 10:1178–1184. 10.1038/ni.1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P.J. 2005. The primary mechanism of the IL-10-regulated antiinflammatory response is to selectively inhibit transcription. Proc. Natl. Acad. Sci. USA. 102:8686–8691. 10.1073/pnas.0500419102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P.J. 2006a STAT3-mediated anti-inflammatory signalling. Biochem. Soc. Trans. 34:1028–1031. 10.1042/BST0341028 [DOI] [PubMed] [Google Scholar]
- Murray P.J. 2006b Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr. Opin. Pharmacol. 6:379–386. 10.1016/j.coph.2006.01.010 [DOI] [PubMed] [Google Scholar]
- Murray P.J. 2007. The JAK-STAT signaling pathway: input and output integration. J. Immunol. 178:2623–2629. 10.4049/jimmunol.178.5.2623 [DOI] [PubMed] [Google Scholar]
- Murray H.W., Lu C.M., Mauze S., Freeman S., Moreira A.L., Kaplan G., and Coffman R.L.. 2002. Interleukin-10 (IL-10) in experimental visceral leishmaniasis and IL-10 receptor blockade as immunotherapy. Infect. Immun. 70:6284–6293. 10.1128/IAI.70.11.6284-6293.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naing A., Papadopoulos K.P., Autio K.A., Ott P.A., Patel M.R., Wong D.J., Falchook G.S., Pant S., Whiteside M., Rasco D.R., et al. . 2016. Safety, Antitumor Activity, and Immune Activation of Pegylated Recombinant Human Interleukin-10 (AM0010) in Patients With Advanced Solid Tumors. J. Clin. Oncol. 34:3562–3569. 10.1200/JCO.2016.68.1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naing A., Infante J.R., Papadopoulos K.P., Chan I.H., Shen C., Ratti N.P., Rojo B., Autio K.A., Wong D.J., Patel M.R., et al. . 2018. PEGylated IL-10 (Pegilodecakin) Induces Systemic Immune Activation, CD8+ T Cell Invigoration and Polyclonal T Cell Expansion in Cancer Patients. Cancer Cell. 34:775–791.e3. 10.1016/j.ccell.2018.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandan D., Camargo de Oliveira C., Moeenrezakhanlou A., Lopez M., Silverman J.M., Subek J., and Reiner N.E.. 2012. Myeloid cell IL-10 production in response to leishmania involves inactivation of glycogen synthase kinase-3β downstream of phosphatidylinositol-3 kinase. J. Immunol. 188:367–378. 10.4049/jimmunol.1100076 [DOI] [PubMed] [Google Scholar]
- Nasi A., Fekete T., Krishnamurthy A., Snowden S., Rajnavölgyi E., Catrina A.I., Wheelock C.E., Vivar N., and Rethi B.. 2013. Dendritic cell reprogramming by endogenously produced lactic acid. J. Immunol. 191:3090–3099. 10.4049/jimmunol.1300772 [DOI] [PubMed] [Google Scholar]
- Nelson D.R., Lauwers G.Y., Lau J.Y., and Davis G.L.. 2000. Interleukin 10 treatment reduces fibrosis in patients with chronic hepatitis C: a pilot trial of interferon nonresponders. Gastroenterology. 118:655–660. 10.1016/S0016-5085(00)70134-X [DOI] [PubMed] [Google Scholar]
- Nemunaitis J., Fong T., Shabe P., Martineau D., and Ando D.. 2001. Comparison of serum interleukin-10 (IL-10) levels between normal volunteers and patients with advanced melanoma. Cancer Invest. 19:239–247. 10.1081/CNV-100102550 [DOI] [PubMed] [Google Scholar]
- Neumann C., Heinrich F., Neumann K., Junghans V., Mashreghi M.F., Ahlers J., Janke M., Rudolph C., Mockel-Tenbrinck N., Kühl A.A., et al. . 2014. Role of Blimp-1 in programing Th effector cells into IL-10 producers. J. Exp. Med. 211:1807–1819. 10.1084/jem.20131548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden D.M., Fenn A.M., Dugan A., and Godbout J.P.. 2014. TGFβ produced by IL-10 redirected astrocytes attenuates microglial activation. Glia. 62:881–895. 10.1002/glia.22647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Garra A., Barrat F.J., Castro A.G., Vicari A., and Hawrylowicz C.. 2008. Strategies for use of IL-10 or its antagonists in human disease. Immunol. Rev. 223:114–131. 10.1111/j.1600-065X.2008.00635.x [DOI] [PubMed] [Google Scholar]
- O’Neill L.A., and Pearce E.J.. 2016. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213:15–23. 10.1084/jem.20151570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani M., Nagai S., Kondo S., Mizuno S., Nakamura K., Tanabe M., Takeuchi T., Matsuda S., and Koyasu S.. 2008. Mammalian target of rapamycin and glycogen synthase kinase 3 differentially regulate lipopolysaccharide-induced interleukin-12 production in dendritic cells. Blood. 112:635–643. 10.1182/blood-2008-02-137430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T., Neves J.F., Dowds C.M., Baker K., Glickman J., Davidson N.O., Lin C.S., Jobin C., Brand S., Sotlar K., et al. . 2014. Protective mucosal immunity mediated by epithelial CD1d and IL-10. Nature. 509:497–502. 10.1038/nature13150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W., and O’Garra A.. 2019. IL-10 Family Cytokines IL-10 and IL-22: from Basic Science to Clinical Translation. Immunity. 50:871–891. 10.1016/j.immuni.2019.03.020 [DOI] [PubMed] [Google Scholar]
- Ouyang W., Rutz S., Crellin N.K., Valdez P.A., and Hymowitz S.G.. 2011. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 29:71–109. 10.1146/annurev-immunol-031210-101312 [DOI] [PubMed] [Google Scholar]
- Palsson-McDermott E.M., Curtis A.M., Goel G., Lauterbach M.A.R., Sheedy F.J., Gleeson L.E., van den Bosch M.W.M., Quinn S.R., Domingo-Fernandez R., Johnston D.G.W., et al. . 2015. Pyruvate Kinase M2 Regulates Hif-1α Activity and IL-1β Induction and Is a Critical Determinant of the Warburg Effect in LPS-Activated Macrophages. Cell Metab. 21:347 10.1016/j.cmet.2015.01.017 [DOI] [PubMed] [Google Scholar]
- Pereira L., Font-Nieves M., Van den Haute C., Baekelandt V., Planas A.M., and Pozas E.. 2015. IL-10 regulates adult neurogenesis by modulating ERK and STAT3 activity. Front. Cell. Neurosci. 9:57 10.3389/fncel.2015.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Asensio F.J., Perpiñá U., Planas A.M., and Pozas E.. 2013. Interleukin-10 regulates progenitor differentiation and modulates neurogenesis in adult brain. J. Cell Sci. 126:4208–4219. 10.1242/jcs.127803 [DOI] [PubMed] [Google Scholar]
- Perriard G., Mathias A., Enz L., Canales M., Schluep M., Gentner M., Schaeren-Wiemers N., and Du Pasquier R.A.. 2015. Interleukin-22 is increased in multiple sclerosis patients and targets astrocytes. J. Neuroinflammation. 12:119 10.1186/s12974-015-0335-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perucha E., Melchiotti R., Bibby J.A., Wu W., Frederiksen K.S., Roberts C.A., Hall Z., LeFriec G., Robertson K.A., Lavender P., et al. . 2019. The cholesterol biosynthesis pathway regulates IL-10 expression in human Th1 cells. Nat. Commun. 10:498 10.1038/s41467-019-08332-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S., Krause C.D., Sarkar D., Walter M.R., Shi Y., and Fisher P.B.. 2004. Interleukin-10 and related cytokines and receptors. Annu. Rev. Immunol. 22:929–979. 10.1146/annurev.immunol.22.012703.104622 [DOI] [PubMed] [Google Scholar]
- Pot C., Jin H., Awasthi A., Liu S.M., Lai C.Y., Madan R., Sharpe A.H., Karp C.L., Miaw S.C., Ho I.C., and Kuchroo V.K.. 2009. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J. Immunol. 183:797–801. 10.4049/jimmunol.0901233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros M., Nishio H., Neumann P.A., Siuda D., Brazil J.C., Azcutia V., Hilgarth R., O’Leary M.N., Garcia-Hernandez V., Leoni G., et al. . 2017. Macrophage-derived IL-10 mediates mucosal repair by epithelial WISP-1 signaling. J. Clin. Invest. 127:3510–3520. 10.1172/JCI90229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi A.A., Gee K., Mishra S., Lim W., and Kumar A.. 2005. STAT-1 mediates the stimulatory effect of IL-10 on CD14 expression in human monocytic cells. J. Immunol. 174:7823–7832. 10.4049/jimmunol.174.12.7823 [DOI] [PubMed] [Google Scholar]
- Rajbhandari P., Thomas B.J., Feng A.C., Hong C., Wang J., Vergnes L., Sallam T., Wang B., Sandhu J., Seldin M.M., et al. . 2018. IL-10 Signaling Remodels Adipose Chromatin Architecture to Limit Thermogenesis and Energy Expenditure. Cell. 172:218–233.e17. 10.1016/j.cell.2017.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redford P.S., Boonstra A., Read S., Pitt J., Graham C., Stavropoulos E., Bancroft G.J., and O’Garra A.. 2010. Enhanced protection to Mycobacterium tuberculosis infection in IL-10-deficient mice is accompanied by early and enhanced Th1 responses in the lung. Eur. J. Immunol. 40:2200–2210. 10.1002/eji.201040433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redford P.S., Murray P.J., and O’Garra A.. 2011. The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol. 4:261–270. 10.1038/mi.2011.7 [DOI] [PubMed] [Google Scholar]
- Richards D.F., Fernandez M., Caulfield J., and Hawrylowicz C.M.. 2000. Glucocorticoids drive human CD8(+) T cell differentiation towards a phenotype with high IL-10 and reduced IL-4, IL-5 and IL-13 production. Eur. J. Immunol. 30:2344–2354. [DOI] [PubMed] [Google Scholar]
- Riley J.K., Takeda K., Akira S., and Schreiber R.D.. 1999. Interleukin-10 receptor signaling through the JAK-STAT pathway. Requirement for two distinct receptor-derived signals for anti-inflammatory action. J. Biol. Chem. 274:16513–16521. 10.1074/jbc.274.23.16513 [DOI] [PubMed] [Google Scholar]
- Rodig S.J., Meraz M.A., White J.M., Lampe P.A., Riley J.K., Arthur C.D., King K.L., Sheehan K.C., Yin L., Pennica D., et al. . 1998. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 93:373–383. 10.1016/S0092-8674(00)81166-6 [DOI] [PubMed] [Google Scholar]
- Rousset F., Garcia E., Defrance T., Péronne C., Vezzio N., Hsu D.H., Kastelein R., Moore K.W., and Banchereau J.. 1992. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc. Natl. Acad. Sci. USA. 89:1890–1893. 10.1073/pnas.89.5.1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell B., Chang-Strachan D., Chan V., Rosenbusch A., Ho C.M., Pryer N., Daniel D., Hwang E.S., Rugo H.S., and Coussens L.M.. 2014. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 26:623–637. 10.1016/j.ccell.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz S., and Ouyang W.. 2016. Regulation of Interleukin-10 Expression. Adv. Exp. Med. Biol. 941:89–116. 10.1007/978-94-024-0921-5_5 [DOI] [PubMed] [Google Scholar]
- Saraiva M., and O’Garra A.. 2010. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10:170–181. 10.1038/nri2711 [DOI] [PubMed] [Google Scholar]
- Saraiva M., Christensen J.R., Tsytsykova A.V., Goldfeld A.E., Ley S.C., Kioussis D., and O’Garra A.. 2005. Identification of a macrophage-specific chromatin signature in the IL-10 locus. J. Immunol. 175:1041–1046. 10.4049/jimmunol.175.2.1041 [DOI] [PubMed] [Google Scholar]
- Saraiva M., Christensen J.R., Veldhoen M., Murphy T.L., Murphy K.M., and O’Garra A.. 2009. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity. 31:209–219. 10.1016/j.immuni.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawant D.V., Yano H., Chikina M., Zhang Q., Liao M., Liu C., Callahan D.J., Sun Z., Sun T., Tabib T., et al. . 2019. Adaptive plasticity of IL-10+ and IL-35+ Treg cells cooperatively promotes tumor T cell exhaustion. Nat. Immunol. 20:724–735. 10.1038/s41590-019-0346-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaljo B., Kratochvill F., Gratz N., Sadzak I., Sauer I., Hammer M., Vogl C., Strobl B., Müller M., Blackshear P.J., et al. . 2009. Tristetraprolin is required for full anti-inflammatory response of murine macrophages to IL-10. J. Immunol. 183:1197–1206. 10.4049/jimmunol.0803883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S., Fedorak R.N., Nielsen O.H., Wild G., Williams C.N., Nikolaus S., Jacyna M., Lashner B.A., Gangl A., Rutgeerts P., et al. . 2000. Safety and efficacy of recombinant human interleukin 10 in chronic active Crohn’s disease. Crohn’s Disease IL-10 Cooperative Study Group. Gastroenterology. 119:1461–1472. 10.1053/gast.2000.20196 [DOI] [PubMed] [Google Scholar]
- Seo D.R., Kim K.Y., and Lee Y.B.. 2004. Interleukin-10 expression in lipopolysaccharide-activated microglia is mediated by extracellular ATP in an autocrine fashion. Neuroreport. 15:1157–1161. 10.1097/00001756-200405190-00015 [DOI] [PubMed] [Google Scholar]
- Seo D.R., Kim S.Y., Kim K.Y., Lee H.G., Moon J.H., Lee J.S., Lee S.H., Kim S.U., and Lee Y.B.. 2008. Cross talk between P2 purinergic receptors modulates extracellular ATP-mediated interleukin-10 production in rat microglial cells. Exp. Mol. Med. 40:19–26. 10.3858/emm.2008.40.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemori S., and Shimosato T.. 2017. Applications of Genetically Modified Immunobiotics with High Immunoregulatory Capacity for Treatment of Inflammatory Bowel Diseases. Front. Immunol. 8:22 10.3389/fimmu.2017.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker J., Saraiva M., and O’Garra A.. 2006. GATA-3 directly remodels the IL-10 locus independently of IL-4 in CD4+ T cells. J. Immunol. 176:3470–3479. 10.4049/jimmunol.176.6.3470 [DOI] [PubMed] [Google Scholar]
- Shouval D.S., Biswas A., Goettel J.A., McCann K., Conaway E., Redhu N.S., Mascanfroni I.D., Al Adham Z., Lavoie S., Ibourk M., et al. . 2014a Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. 40:706–719. 10.1016/j.immuni.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shouval D.S., Ouahed J., Biswas A., Goettel J.A., Horwitz B.H., Klein C., Muise A.M., and Snapper S.B.. 2014b Interleukin 10 receptor signaling: master regulator of intestinal mucosal homeostasis in mice and humans. Adv. Immunol. 122:177–210. 10.1016/B978-0-12-800267-4.00005-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Gurav A., Sivaprakasam S., Brady E., Padia R., Shi H., Thangaraju M., Prasad P.D., Manicassamy S., Munn D.H., et al. . 2014. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 40:128–139. 10.1016/j.immuni.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.M., Qualls J.E., O'Brien K., Balouzian L., Johnson P.F., Schultz-Cherry S., Smale S.T., and Murray P.J.. 2011. A distal enhancer in Il12b is the target of transcriptional repression by the STAT3 pathway and requires the basic leucine zipper (B-ZIP) protein NFIL3. J. Biol. Chem. 286:23582 10.1074/jbc.M111.249235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidler L., Hans W., Schotte L., Neirynck S., Obermeier F., Falk W., Fiers W., and Remaut E.. 2000. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 289:1352–1355. 10.1126/science.289.5483.1352 [DOI] [PubMed] [Google Scholar]
- Stumhofer J.S., Silver J.S., Laurence A., Porrett P.M., Harris T.H., Turka L.A., Ernst M., Saris C.J., O’Shea J.J., and Hunter C.A.. 2007. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol. 8:1363–1371. 10.1038/ni1537 [DOI] [PubMed] [Google Scholar]
- Sun J., Dodd H., Moser E.K., Sharma R., and Braciale T.J.. 2011. CD4+ T cell help and innate-derived IL-27 induce Blimp-1-dependent IL-10 production by antiviral CTLs. Nat. Immunol. 12:327–334. 10.1038/ni.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Wu W., Chen L., Yang W., Huang X., Ma C., Chen F., Xiao Y., Zhao Y., Ma C., et al. . 2018. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun. 9:3555 10.1038/s41467-018-05901-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Iyer M., McEachin R., Zhao M., Wu Y.M., Cao X., Oravecz-Wilson K., Zajac C., Mathewson N., Wu S.J., et al. . 2017. Genome-Wide STAT3 Binding Analysis after Histone Deacetylase Inhibition Reveals Novel Target Genes in Dendritic Cells. J. Innate Immun. 9:126–144. 10.1159/000450681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sziksz E., Pap D., Lippai R., Béres N.J., Fekete A., Szabó A.J., and Vannay Á.. 2015. Fibrosis Related Inflammatory Mediators: Role of the IL-10 Cytokine Family. Mediators Inflamm. 2015:764641 10.1155/2015/764641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Clausen B.E., Kaisho T., Tsujimura T., Terada N., Förster I., and Akira S.. 1999. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 10:39–49. 10.1016/S1074-7613(00)80005-9 [DOI] [PubMed] [Google Scholar]
- Tan P.H., Tyrrell H.E., Gao L., Xu D., Quan J., Gill D., Rai L., Ding Y., Plant G., Chen Y., et al. . 2014. Adiponectin receptor signaling on dendritic cells blunts antitumor immunity. Cancer Res. 74:5711–5722. 10.1158/0008-5472.CAN-13-1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanchot C., Guillaume S., Delon J., Bourgeois C., Franzke A., Sarukhan A., Trautmann A., and Rocha B.. 1998. Modifications of CD8+ T cell function during in vivo memory or tolerance induction. Immunity. 8:581–590. 10.1016/S1074-7613(00)80563-4 [DOI] [PubMed] [Google Scholar]
- Thompson-Snipes L., Dhar V., Bond M.W., Mosmann T.R., Moore K.W., and Rennick D.M.. 1991. Interleukin 10: a novel stimulatory factor for mast cells and their progenitors. J. Exp. Med. 173:507–510. 10.1084/jem.173.2.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg H., Ulmer H., Kaser A., and Weiss G.. 2002a Role of IL-10 for induction of anemia during inflammation. J. Immunol. 169:2204–2209. 10.4049/jimmunol.169.4.2204 [DOI] [PubMed] [Google Scholar]
- Tilg H., van Montfrans C., van den Ende A., Kaser A., van Deventer S.J., Schreiber S., Gregor M., Ludwiczek O., Rutgeerts P., Gasche C., et al. . 2002b Treatment of Crohn’s disease with recombinant human interleukin 10 induces the proinflammatory cytokine interferon gamma. Gut. 50:191–195. 10.1136/gut.50.2.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel E., Bootz F., Silacci M., Kaspar M., Kosmehl H., and Neri D.. 2007. Antibody-mediated delivery of IL-10 inhibits the progression of established collagen-induced arthritis. Arthritis Res. Ther. 9:R9 10.1186/ar2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay R., Sanchez-Hidalgo A., Wilusz C.J., Lenaerts A.J., Arab J., Yeh J., Stefanisko K., Tarasova N.I., and Gonzalez-Juarrero M.. 2018. Host Directed Therapy for Chronic Tuberculosis via Intrapulmonary Delivery of Aerosolized Peptide Inhibitors Targeting the IL-10-STAT3 Pathway. Sci. Rep. 8:16610 10.1038/s41598-018-35023-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deventer S.J., Elson C.O., and Fedorak R.N.. Crohn’s Disease Study Group . 1997. Multiple doses of intravenous interleukin 10 in steroid-refractory Crohn’s disease. Gastroenterology. 113:383–389. 10.1053/gast.1997.v113.pm9247454 [DOI] [PubMed] [Google Scholar]
- van Exel E., Gussekloo J., de Craen A.J., Frölich M., Bootsma-Van Der Wiel A., and Westendorp R.G.. Leiden 85 Plus Study . 2002. Low production capacity of interleukin-10 associates with the metabolic syndrome and type 2 diabetes: the Leiden 85-Plus Study. Diabetes. 51:1088–1092. 10.2337/diabetes.51.4.1088 [DOI] [PubMed] [Google Scholar]
- Verma R., Balakrishnan L., Sharma K., Khan A.A., Advani J., Gowda H., Tripathy S.P., Suar M., Pandey A., Gandotra S., et al. . 2016. A network map of Interleukin-10 signaling pathway. J. Cell Commun. Signal. 10:61–67. 10.1007/s12079-015-0302-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veroni C., Gabriele L., Canini I., Castiello L., Coccia E., Remoli M.E., Columba-Cabezas S., Aricò E., Aloisi F., and Agresti C.. 2010. Activation of TNF receptor 2 in microglia promotes induction of anti-inflammatory pathways. Mol. Cell. Neurosci. 45:234–244. 10.1016/j.mcn.2010.06.014 [DOI] [PubMed] [Google Scholar]
- Vicari A.P., Chiodoni C., Vaure C., Aït-Yahia S., Dercamp C., Matsos F., Reynard O., Taverne C., Merle P., Colombo M.P., et al. . 2002. Reversal of tumor-induced dendritic cell paralysis by CpG immunostimulatory oligonucleotide and anti-interleukin 10 receptor antibody. J. Exp. Med. 196:541–549. 10.1084/jem.20020732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal P.M., Lemmens E., Dooley D., and Hendrix S.. 2013. The role of “anti-inflammatory” cytokines in axon regeneration. Cytokine Growth Factor Rev. 24:1–12. 10.1016/j.cytogfr.2012.08.008 [DOI] [PubMed] [Google Scholar]
- Vieira P., de Waal-Malefyt R., Dang M.N., Johnson K.E., Kastelein R., Fiorentino D.F., deVries J.E., Roncarolo M.G., Mosmann T.R., and Moore K.W.. 1991. Isolation and expression of human cytokine synthesis inhibitory factor cDNA clones: homology to Epstein-Barr virus open reading frame BCRFI. Proc. Natl. Acad. Sci. USA. 88:1172–1176. 10.1073/pnas.88.4.1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigne S., Chalmin F., Duc D., Clottu A.S., Apetoh L., Lobaccaro J.A., Christen I., Zhang J., and Pot C.. 2017. IL-27-Induced Type 1 Regulatory T-Cells Produce Oxysterols that Constrain IL-10 Production. Front. Immunol. 8:1184 10.3389/fimmu.2017.01184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vouldoukis I., Bécherel P.A., Riveros-Moreno V., Arock M., da Silva O., Debré P., Mazier D., and Mossalayi M.D.. 1997. Interleukin-10 and interleukin-4 inhibit intracellular killing of Leishmania infantum and Leishmania major by human macrophages by decreasing nitric oxide generation. Eur. J. Immunol. 27:860–865. 10.1002/eji.1830270409 [DOI] [PubMed] [Google Scholar]
- Wang X., Wong K., Ouyang W., and Rutz S.. 2019. Targeting IL-10 Family Cytokines for the Treatment of Human Diseases. Cold Spring Harb. Perspect. Biol. 11:a028548 10.1101/cshperspect.a028548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Sato H., Kusam S., Sehra S., Toney L.M., and Dent A.L.. 2005. Regulation of IL-10 gene expression in Th2 cells by Jun proteins. J. Immunol. 174:2098–2105. 10.4049/jimmunol.174.4.2098 [DOI] [PubMed] [Google Scholar]
- Weaver B.K., Bohn E., Judd B.A., Gil M.P., and Schreiber R.D.. 2007. ABIN-3: a molecular basis for species divergence in interleukin-10-induced anti-inflammatory actions. Mol. Cell. Biol. 27:4603–4616. 10.1128/MCB.00223-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Nordt R.M., Riley J.K., Greenlund A.C., Moore K.W., Darnell J.E., and Schreiber R.D.. 1996. Stat3 recruitment by two distinct ligand-induced, tyrosine-phosphorylated docking sites in the interleukin-10 receptor intracellular domain. J. Biol. Chem. 271:27954–27961. 10.1074/jbc.271.44.27954 [DOI] [PubMed] [Google Scholar]
- Wehinger J., Gouilleux F., Groner B., Finke J., Mertelsmann R., and Weber-Nordt R.M.. 1996. IL-10 induces DNA binding activity of three STAT proteins (Stat1, Stat3, and Stat5) and their distinct combinatorial assembly in the promoters of selected genes. FEBS Lett. 394:365–370. 10.1016/0014-5793(96)00990-8 [DOI] [PubMed] [Google Scholar]
- Weichhart T., Costantino G., Poglitsch M., Rosner M., Zeyda M., Stuhlmeier K.M., Kolbe T., Stulnig T.M., Hörl W.H., Hengstschläger M., et al. . 2008. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 29:565–577. 10.1016/j.immuni.2008.08.012 [DOI] [PubMed] [Google Scholar]
- Werry E.L., Liu G.J., Lovelace M.D., Nagarajah R., Hickie I.B., and Bennett M.R.. 2011. Lipopolysaccharide-stimulated interleukin-10 release from neonatal spinal cord microglia is potentiated by glutamate. Neuroscience. 175:93–103. 10.1016/j.neuroscience.2010.10.080 [DOI] [PubMed] [Google Scholar]
- Willems F., Marchant A., Delville J.P., Gérard C., Delvaux A., Velu T., de Boer M., and Goldman M.. 1994. Interleukin-10 inhibits B7 and intercellular adhesion molecule-1 expression on human monocytes. Eur. J. Immunol. 24:1007–1009. 10.1002/eji.1830240435 [DOI] [PubMed] [Google Scholar]
- Williams L., Bradley L., Smith A., and Foxwell B.. 2004a Signal transducer and activator of transcription 3 is the dominant mediator of the anti-inflammatory effects of IL-10 in human macrophages. J. Immunol. 172:567–576. 10.4049/jimmunol.172.1.567 [DOI] [PubMed] [Google Scholar]
- Williams L.M., Ricchetti G., Sarma U., Smallie T., and Foxwell B.M.. 2004b Interleukin-10 suppression of myeloid cell activation--a continuing puzzle. Immunology. 113:281–292. 10.1111/j.1365-2567.2004.01988.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Fu Q., Ortega T.M., Zhou L., Rasmussen D., O’Keefe J., Zhang K.K., and Chapes S.K.. 2014. Overexpression of IL-10 in C2D macrophages promotes a macrophage phenotypic switch in adipose tissue environments. PLoS One. 9:e86541 10.1371/journal.pone.0086541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin J., Wainwright D.A., Mesnard N.A., Serpe C.J., Sanders V.M., and Jones K.J.. 2011. IL-10 within the CNS is necessary for CD4+ T cells to mediate neuroprotection. Brain Behav. Immun. 25:820–829. 10.1016/j.bbi.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yang Y., Qiu G., Lal G., Wu Z., Levy D.E., Ochando J.C., Bromberg J.S., and Ding Y.. 2009. c-Maf regulates IL-10 expression during Th17 polarization. J. Immunol. 182:6226–6236. 10.4049/jimmunol.0900123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xystrakis E., Kusumakar S., Boswell S., Peek E., Urry Z., Richards D.F., Adikibi T., Pridgeon C., Dallman M., Loke T.K., et al. . 2006. Reversing the defective induction of IL-10-secreting regulatory T cells in glucocorticoid-resistant asthma patients. J. Clin. Invest. 116:146–155. 10.1172/JCI21759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi A.K., Yoon J.G., Yeo S.J., Hong S.C., English B.K., and Krieg A.M.. 2002. Role of mitogen-activated protein kinases in CpG DNA-mediated IL-10 and IL-12 production: central role of extracellular signal-regulated kinase in the negative feedback loop of the CpG DNA-mediated Th1 response. J. Immunol. 168:4711–4720. 10.4049/jimmunol.168.9.4711 [DOI] [PubMed] [Google Scholar]
- Yoon S.I., Logsdon N.J., Sheikh F., Donnelly R.P., and Walter M.R.. 2006. Conformational changes mediate interleukin-10 receptor 2 (IL-10R2) binding to IL-10 and assembly of the signaling complex. J. Biol. Chem. 281:35088–35096. 10.1074/jbc.M606791200 [DOI] [PubMed] [Google Scholar]
- Yssel H., De Waal Malefyt R., Roncarolo M.G., Abrams J.S., Lahesmaa R., Spits H., and de Vries J.E.. 1992. IL-10 is produced by subsets of human CD4+ T cell clones and peripheral blood T cells. J. Immunol. 149:2378–2384. [PubMed] [Google Scholar]
- Yu F., Sharma S., Jankovic D., Gurram R.K., Su P., Hu G., Li R., Rieder S., Zhao K., Sun B., and Zhu J.. 2018. The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J. Exp. Med. 215:1813–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdanov A. 2010. Structural analysis of cytokines comprising the IL-10 family. Cytokine Growth Factor Rev. 21:325–330. 10.1016/j.cytogfr.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Lee J.S., Gartlan K.H., Schuster I.S., Comerford I., Varelias A., Ullah M.A., Vuckovic S., Koyama M., Kuns R.D., et al. . 2017. Eomesodermin promotes the development of type 1 regulatory T (TR1) cells. Sci. Immunol. 2:eaah7152 10.1126/sciimmunol.aah7152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Weinberg S., DeBerge M., Gainullina A., Schipma M., Kinchen J.M., Ben-Sahra I., Gius D.R., Yvan-Charvet L., Chandel N.S., et al. . 2019. Efferocytosis Fuels Requirements of Fatty Acid Oxidation and the Electron Transport Chain to Polarize Macrophages for Tissue Repair. Cell Metab. 29:443–456.e5. 10.1016/j.cmet.2018.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L.M., Ojcius D.M., Garaud F., Roth C., Maxwell E., Li Z., Rong H., Chen J., Wang X.Y., Catino J.J., and King I.. 1996. Interleukin-10 inhibits tumor metastasis through an NK cell-dependent mechanism. J. Exp. Med. 184:579–584. 10.1084/jem.184.2.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Peng X., Insolera R., Fink D.J., and Mata M.. 2009a IL-10 promotes neuronal survival following spinal cord injury. Exp. Neurol. 220:183–190. 10.1016/j.expneurol.2009.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Peng X., Insolera R., Fink D.J., and Mata M.. 2009b Interleukin-10 provides direct trophic support to neurons. J. Neurochem. 110:1617–1627. 10.1111/j.1471-4159.2009.06263.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Min B., Hu-Li J., Watson C.J., Grinberg A., Wang Q., Killeen N., Urban J.F. Jr., Guo L., and Paul W.E.. 2004. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat. Immunol. 5:1157–1165. 10.1038/ni1128 [DOI] [PubMed] [Google Scholar]
- Zhu Y.P., Brown J.R., Sag D., Zhang L., and Suttles J.. 2015. Adenosine 5′-monophosphate-activated protein kinase regulates IL-10-mediated anti-inflammatory signaling pathways in macrophages. J. Immunol. 194:584–594. 10.4049/jimmunol.1401024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E., Bernshtein B., Friedlander G., Walker C.R., Yona S., Kim K.W., Brenner O., Krauthgamer R., Varol C., Müller W., and Jung S.. 2014. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. 40:720–733. 10.1016/j.immuni.2014.03.012 [DOI] [PubMed] [Google Scholar]