Abstract
High-grade gliomas are lethal brain cancers whose progression is robustly regulated by neuronal activity. Activity-regulated growth factor release promotes glioma growth, but this alone is insufficient to explain the effect that activity exerts on glioma progression. Here, we use single-cell transcriptomics, electron microscopy, whole-cell patch-clamp electrophysiology and calcium imaging to demonstrate that neuron-glioma interactions include electrochemical communication through bona fide AMPA receptor-dependent neuron-glioma synapses. Neuronal activity also evokes non-synaptic activity-dependent potassium currents that are amplified through gap junction-mediated tumor interconnections forming an electrically-coupled network. Glioma membrane depolarization assessed with in vivo optogenetics promotes proliferation, while pharmacologically or genetically blocking electrochemical signaling inhibits glioma xenograft growth and extends mouse survival. Emphasizing positive feedback mechanisms by which gliomas increase neuronal excitability and thus activity-regulated glioma growth, human intraoperative electrocorticography demonstrates increased cortical excitability in glioma-infiltrated brain. Together, these findings indicate that synaptic and electrical integration in neural circuits promotes glioma progression.
High-grade gliomas are the leading cause of central nervous system (CNS) cancer-related death in both children and adults. This clinical intractability indicates that current understanding of glioma pathophysiology is insufficient. Gliomas infiltrate extensively within the brain and spinal cord, but growth outside the CNS is exceedingly rare. Glioma progression is regulated not only by cell-intrinsic mechanisms, but also by important microenvironmental dependencies. Neurons are a critically important component of the glioma microenvironment and regulate malignant growth in an activity-dependent manner1,2. Activity-regulated release of neuroligin-3 (NLGN3)1,2 is required for glioma progression2, indicating a fundamental role in glioma pathophysiology incompletely explained by stimulation of classical oncogenic signaling pathways alone2. We previously found that neuroligin-3 induces glioma expression of numerous synaptic genes2, raising the intriguing possibility that glioma may engage in synaptic communication. Synapses exist between neurons and normal oligodendroglial precursor cells (OPCs)3,4, and electrochemical signaling can regulate proliferation, differentiation or survival of OPCs and other neural precursor cells (NPCs)5–9. As cellular subpopulations within gliomas closely resemble OPCs10,11, we hypothesized that gliomas may similarly engage in synaptic communication and that this integration into neural circuits may be fundamental to glioma progression.
Synaptic gene expression in glioma
To examine synaptic gene expression in primary human glioma, we analyzed single cell transcriptomic datasets generated from pre-treatment biopsy samples of the major classes of adult and pediatric high-grade gliomas, including adult IDH-mutant glioma11, adult IDH-WT glioma10, and pediatric histone-3 mutant (H3K27M) diffuse midline glioma10. We found broad expression of glutamate receptor genes and post-synaptic structural genes in malignant glioma cells (Fig. 1a, Extended Data Fig. 1a). Unsupervised principal component analysis revealed enrichment of synaptic gene expression within distinct malignant cellular subpopulations (Fig. 1b). Gliomas are comprised of cellular subpopulations that resemble various stages of astrocytic and oligodendrocytic differentiation10,11. Synaptic gene enrichment was chiefly found in glioma cells that resemble OPCs (Fig. 1b, Extended Data Fig. 2c), the only glial cell type that normally functions as a post-synaptic cell3,4. These observations are consistent with the principle that malignant cellular subpopulations assume distinct roles in the heterogeneous cancer ecosystem. Concordant with these findings from primary biopsy tissue (Fig. 1b), single cell transcriptomics of patient-derived H3K27M+ glioma xenografts demonstrated synaptic gene enrichment in the OPC-like subpopulation (Extended Data Fig. 1b, Extended Data Fig. 2a–b).
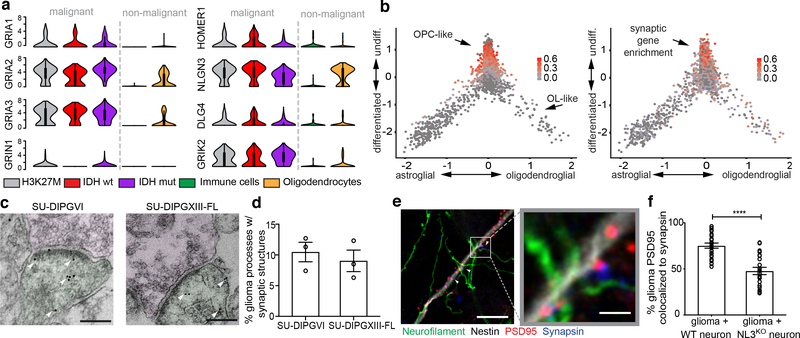
Figure 1: Transcriptomic and structural evidence for glioma synapses.
a, Primary human biopsy single cell transcriptomic data illustrating synapse-associated gene expression levels from H3K27M+ diffuse midline glioma (DMG; grey; n=2,259/6 cells/subjects), IDHwt adult high-grade glioma (red; n=599/3 cells/subjects), IDHmut adult high-grade glioma (purple; n=5,096/10 cells/subjects) malignant cells, and tumor-associated, non-malignant immune cells (green; n=96/5 cells/subjects) and oligodendrocytes (yellow; n=232 cells). For each individual violin plot, y-axis represents log2 TPM (transcripts per million), x-axis represents number of individual cells with indicated expression value, and thick and thin black lines represent interquartile and 1.5x interquartile range, respectively. b, Plot of the lineage (x-axis) and stemness (undifferentiated to differentiated, y-axis) scores for H3K27M+ DMG malignant single cells sorted from primary biopsies (n=2,259 cells). Overlay of grey-red enrichment score indicates relative score for OPC-like genes (left) and synapse-related genes (right; pearson correlation rho=0.47, P<0.0001). c, Immuno-electron microscopy of patient-derived glioma SU-DIPGVI (left) and SU-DIPGXIII-FL (right) xenografts in mouse hippocampus. Immuno-gold particles labeling GFP (white arrowheads). Post-synaptic density in GFP+ tumor cells (pseudo-colored green), synaptic cleft, and clustered synaptic vesicles in apposing presynaptic neuron (pseudo-colored magenta) identify synapses. Scale bar=200μm. g, Quantification of neuron:glioma synaptic structures in SU-DIPGVI and SU-DIPGXIII-FL xenografts expressed as percent of total identified glioma cell processes forming unambiguous synaptic structures (n=3 mice/group; mean±s.e.m). e, Representative confocal image of neurons co-cultured with PSD95-RFP-labeled glioma cells. White box and arrowhead highlight region of synaptic puncta colocalization, zoomed-in view (right). Green=neurofilament (axon); white=nestin (tumor cells); blue=synapsin (pre-synaptic puncta); red=PSD95-RFP (post-synaptic puncta). Scale bars=10μm (left), 2μm (right). f, Quantification of post-synaptic glioma-derived PSD95-RFP colocalized with neuronal pre-synaptic synapsin in co-cultures of glioma cells (SU-DIPGVI) with WT (n=22 cells/10 coverslips) or Nlgn3y/- (NL3KO; n=21 cells/10 coverslips) neurons. Data shown as % colocalization, mean±s.e.m. P-value determined by two-tailed Student’s t-test, ****P<0.0001.
Neuron-to-glioma synapses
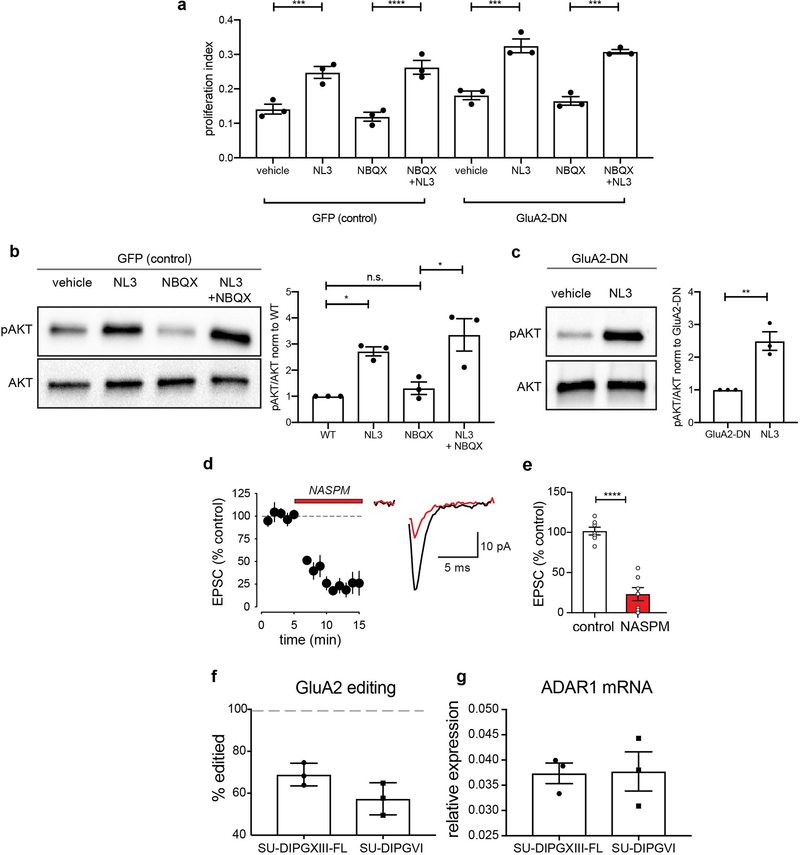
Having established that primary glioma cells express a repertoire of synaptic genes, we next assessed whether structural synapses form between glioma cells and neurons in the tumor microenvironment. Examination of primary glioblastoma tissue ultrastructure using electron microscopy (EM) revealed clear synaptic structures (Extended Data Fig. 2d). To confirm that glioma cells participate in such putative neuron-glioma synapses, we performed immuno-EM in GFP-labeled, patient-derived glioma xenografts. Immuno-EM analyses unambiguously identified GFP+ glioma cells on the post-synaptic side of synaptic structures, with synapses on ~10% of GFP+ glioma processes (Fig. 1c,d and Extended Data Fig. 2e–g). To test the contribution of microenvironmental NLGN3 to neuron-glioma synaptogenesis, we next co-cultured glioma cells expressing fluorescently-tagged PSD95 with WT or Nlgn3y/− neurons. We found marked reduction in the co-localization of neuronal presynaptic puncta (synapsin) with glioma post-synaptic puncta (PSD95-RFP) in co-cultures with Nlgn3y/− neurons compared to WT neurons (Fig.1e–f, Extended Data Fig. 2h). This further implicates NLGN3 in glioma synapse formation, a function distinct from its role stimulating classical oncogenic signaling pathways (Extended Data Fig.3a–c).
Focusing on pediatric gliomas, we next tested if neurons and glioma cells establish electrophysiologically functional synapses using four distinct patient-derived orthotopic xenograft models (Extended Data Table 1). GFP-labeled glioma cells were stereotactically xenografted into the CA1 region of the hippocampal circuit. Following a period of engraftment and growth, acute hippocampal slices were prepared for whole cell patch clamp recordings of GFP+ glioma cells. Stimulation of Schaffer collateral/commissural afferent axons arising from CA3 while patch clamping CA1 region glioma cells enables measurement of the glioma response to axonal activity (Fig. 2a–b). Voltage-clamp recordings revealed stimulation-evoked fast (<5 ms) inward currents consistent with excitatory postsynaptic currents (EPSCs, Fig. 2c). Current-clamp recordings demonstrated that these inward currents were depolarizing (Fig. 2d). Glioma EPSCs were blocked by the voltage-gated sodium channel blocker tetrodotoxin (TTX; Fig. 2e), illustrating dependence on neuronal action potentials. Measuring the current-voltage relationship (I-V curve) illustrated reversal at approximately 0 mV (Fig. 2f) and glioma EPSCs displayed facilitation in response to paired stimuli (Fig. 2g), electrophysiological characteristics suggesting synaptic communication through AMPA receptors (AMPAR, a type of ionotropic glutamate receptor). Concordantly, glioma EPSCs were blocked by NBQX, an AMPAR antagonist (Fig. 2g–i) and decreased by NASPM, an antagonist of calcium-permeable AMPARs (Extended Data Fig. 3d–e). AMPARs lacking GluA2 or containing GluA2 that has not undergone RNA editing of its Q/R site are calcium-permable12,13. GluA2 is broadly expressed in gliomas (Fig. 1a); examination of RNA editing of the Q/R site in pediatric glioma demonstrated GluA2 under-editing (~50–70% edited; Extended Data Fig. 3f–g). Taken together, these results indicate that axon stimulation-evoked, millisecond timescale glioma cell currents require action potentials and are mediated by AMPARs, properties consistent with the conclusion that subpopulations of glioma cells form bona fide synapses with neurons. This conclusion is additionally supported by the evidence for neuron-to-glioma synapses described by Venkataramani and colleagues in this issue (REF).
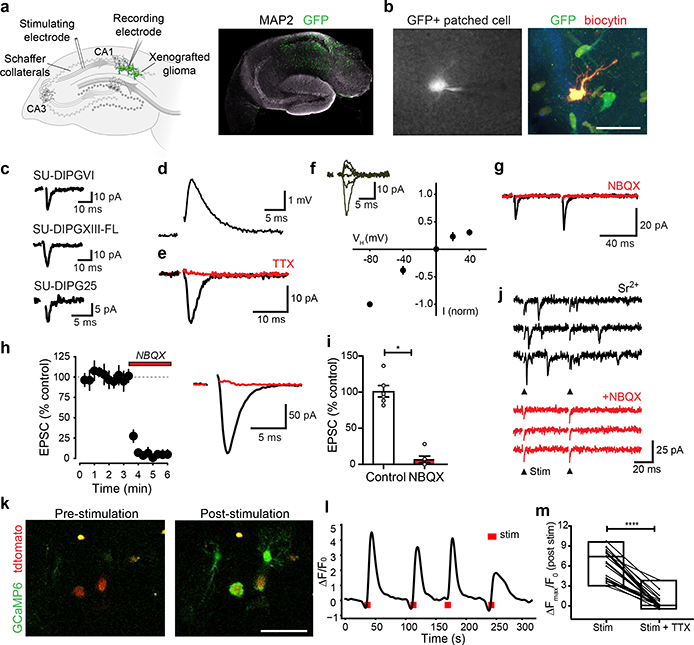
Figure 2: Synaptic AMPAR-mediated EPSCs in glioma.
a, Electrophysiological model. GFP+ glioma cells (green) xenografted in mouse hippocampus CA1 region with Schaffer collateral afferent stimulation (left). Right, representative hippocampal slice micrograph, GFP+ glioma cells=green, MAP2=white, n=8 biological replicates. b, Representative micrograph of patched GFP+ glioma cell with whole-cell pipette containing biocytin (left). Right, biocytin (red)-filled glioma cell co-labeled with GFP (green). Scale bar=50μm; n=21 biological replicates. c, Representative traces of evoked EPSCs in patient-derived glioma xenografts. d, Representative glioma EPSP in current-clamp with Schaffer collateral stimulation. e, Representative evoked glioma EPSC before (black) and after (red) tetrodotoxin (TTX, 0.5μM). f, Current:voltage relationship of evoked EPSCs with representative traces shown as inset (−80mV, n=18/9 cells/mice; −40mV, n=5/2; 0mV, n=18/9; +20mV, n=7/2; +40mV, n=16/7). g, Paired-pulse facilitation of evoked glioma cell EPSC (black) with block by NBQX (red; 10 μM; 50 ms inter-stimulus interval: P2/P1=1.75 ± 0.12; n=8/8 cells/mice). h, Timecourse of evoked glioma cell EPSC block by NBQX (10 μM, duration=red bar; n=6/6 cells/mice; left); Representative trace before (black) and after (red) NBQX (right). i, Quantification of (h). j, Evoked miniature-EPSCs in the presence of strontium (4 mM; top) and block by NBQX (10 μM; bottom; n=4/2 cells/mice). Stimulation timepoint=arrowhead (downward deflection in NBQX is stim artifact). k, Two-photon in situ calcium imaging in SU-DIPGVI xenograft with Schaffer collateral stimulation. Representative frames shown pre- (left) and post-stimulation (right), glioma td-tomato nuclear tag=red, glioma GCaMP6s=green. Scale bar=50μm, n=12/4 slices/mice. l, GCaMP6s intensity trace in representative glioma cell with electrical stimulation (red bar) over time (s). Data plotted as ρF/Fo. Timepoint of stimulation=red mark; n=4 mice. m, Individual cell GCaMP6s response to electrical stimulation +/− TTX (0.5μM). Data plotted as ρFmax/Fo; n=26 cells/3 mice. Data shown as mean±s.e.m (h,i,m). P-values determined by one-tailed Wilcoxon matched-pairs signed rank test (i,m).*P<0.05, ****P<0.0001.
To test this hypothesis further, we replaced extracellular calcium with strontium, a manipulation that facilitates asynchronous presynaptic vesicle release14 (Fig. 2j). In the presence of strontium we detected small, fast inward currents consistent with miniature EPSCs (mEPSCs), indicating quantal responses to synaptic vesicles14 in glioma cells. Quantal glioma mEPSCs were similarly blocked by NBQX (Fig. 2j). No fast, large currents reminiscent of action potentials were observed in any of >640 glioma cell recordings. Taken together, these results indicate that synaptic transmission occurs between neurons and a subset of xenografted human glioma cells, exhibiting properties similar to synapses formed with normal OPCs3,4.
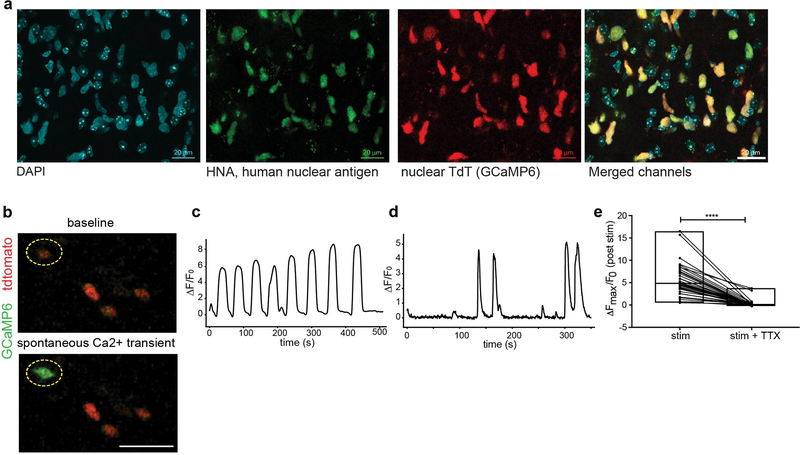
Further exploring the consequences of activity-dependent currents in glioma, we performed in situ two-photon calcium imaging of xenografted glioma expressing the genetically encoded calcium indicator GCaMP6s. Glioma-specific expression of the calcium indicator was validated by co-staining for human nuclear antigen (Extended Data Fig. 4a). Spontaneous calcium transients were consistently observed (Extended Data Fig. 4b–d). Stimulation of Schaffer collateral/commissural afferents elicited calcium transients in glioma cells located in the CA1 target area of the stimulated axons (Fig. 2k–l), providing additional evidence that endogenous circuit activity may exert functionally relevant effects on glioma cells. These evoked calcium transients were blocked by TTX (Fig. 2m, Extended Data Fig. 4e).
Activity-dependent potassium currents
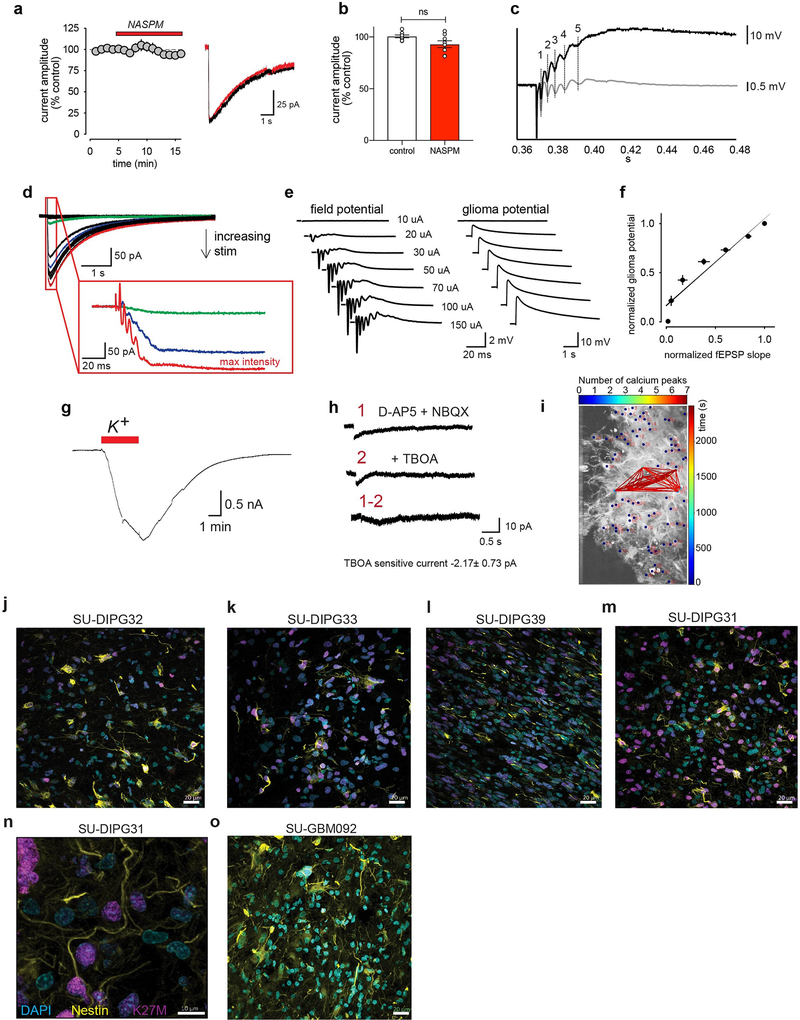
A longer duration electrophysiological response to neuronal activity was found in a subset of glioma cells (Fig. 3a). Distinct from the classical EPSCs (<5ms) described above, these prolonged currents (>1 sec) exhibited kinetics inconsistent with a synaptic response and are instead reminiscent of the neuronal activity-evoked currents observed in normal astrocytes. Supporting the idea that these prolonged currents are distinct from the synaptic responses described above, the calcium-permeable AMPAR inhibitor NASPM had no effect (Extended data Fig. 5a–b). These prolonged glioma currents were blocked by TTX (Fig. 3b–c). Further illustrating a response coupled to neuronal population firing, the morphology of the prolonged currents revealed spike-like waveforms phase-locked to neuronal field potential waveforms that scaled with increased axonal stimulation intensity (Extended Data Fig. 5c–d). Simultaneous whole cell current clamp and field potential recordings reveal that the prolonged current amplitude scaled directly with field potential, meaning that prolonged glioma current amplitude increases with increasing neuronal activity (Extended Data Fig. 5e–f). In normal astrocytes, activity-dependent currents are attributable to glutamate transporter currents and inward potassium currents due to a rise in extracellular potassium from neurons15–17. Consistent with a direct role for increases in extracellular potassium in generating these prolonged currents, large prolonged glioma currents were elicited by application of potassium alone with neuronal activity pharmacologically blocked (Fig. 3d, Extended Data Fig. 5g). Further, activity-dependent prolonged currents were largely diminished by barium, an ion that blocks inwardly-rectifying potassium channels (Fig.3e–f). In contrast, the glutamate transporter antagonist TBOA had a negligible effect (Extended Data Fig. 5h). Taken together, these results support the interpretation that non-synaptic, prolonged glioma currents chiefly reflect potassium flux attributable to a rise in extracellular potassium with neuronal activity.
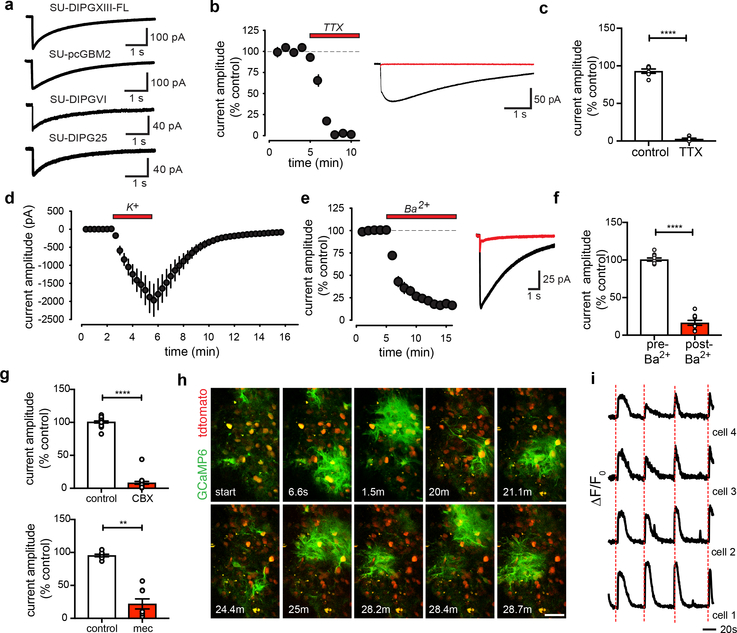
Figure 3: Neuronal activity-dependent potassium currents in glioma.
a, Representative voltage-clamp traces of evoked prolonged current in multiple patient-derived glioma xenograft models. b, Timecourse of evoked current blocked by TTX (duration=red bar; n=6/6 cells/mice; left). Representative trace before (black) and after (red) TTX (right). c, Quantification of (b). d, Timecourse of glioma cell current induced by addition of extracellular potassium (K+, 15mM, duration=red bar; n=9/2 cells/mice) with concurrent neuronal activation blockade. e, As in (b), but with barium (200μM); n=10/3 cells/mice. f, Quantification of (e). g, Quantification of current amplitude decrease with carbenoxolone (CBX; 100μM; n=19/11 cells/mice; top), or meclofenamate (100μM; n=8/3 cells/mice; bottom). h, Two-photon in situ calcium imaging of hippocampal slice xenografted with GCaMP6s-expressing glioma (SU-DIPGXIII-FL); 30-min timecourse; Red=glioma td-Tomato nuclear tag, green=glioma GCaMP6s. Scale bar, 50μm; n=14 mice. i, Phase-locked traces of GCaMP6s intensity over time in four synchronous glioma cells with axonal stimulation (red line). Data plotted as ρF/Fo; n=40/4 cells/mice. Data shown as mean±s.e.m (b,c,d,e,f,g). P-values determined by two-tailed paired Student’s t-test (c,f), or one-tailed Wilcoxon matched-pairs signed rank test (g). **P<0.01, ****P<0.0001.
Gap-junction coupling amplifies currents
Glioma cells with prolonged currents exhibit strikingly low input resistance (Extended Data Fig. 6a), reminiscent of astrocytes. Extensive gap-junctional coupling is partially responsible for low membrane resistance in astrocytes18. Gap junctions couple adult glioma cells through long processes called tumor microtubes19, which we demonstrated in primary pediatric glioma tissue (Extended Data Fig. 5j–o). Following biocytin dye-filling of single cells exhibiting prolonged currents, biocytin diffused to a network of glioma cells (Extended Data Fig. 6b), supporting the existence of a gap junction-coupled network. To test this conclusion, we applied the gap junction blockers carbenoxolone (CBX) or meclofenamate, which reduced the amplitude of prolonged glioma currents (Fig. 3g, Extended Data Fig. 6c–e, 6h–i) while simultaneously increasing glioma input resistance (Extended Data Fig. 6f–g, 6j–k). Together, these observations strongly suggest that activity-regulated increases in extracellular potassium concentrations cause glioma depolarization and that a gap junction-coupled glioma network amplifies the consequences of activity-induced changes in the extracellular ionic environment.
Two-photon calcium imaging further revealed distinct synchronous network calcium transients that both occur spontaneously (Fig. 3h, Extended Data Fig. 5i, Supplementary Video 1) and are elicited by afferent stimulation (Fig. 3i, Supplementary Video 2). This synchronicity could be explained by gap-junction coupling, and accordingly was blocked by application of CBX (Extended Data Fig. 6l–o, Supplementary Video 3), further indicating a functional glioma network through which depolarizing currents propagate. Demonstration of glutamatergic chemical synapses (Fig. 2) and activity-dependent, non-synaptic potassium currents (Fig. 3) build upon early work illustrating glutamate-dependent currents in glioblastoma20 to underscore the surprising observation that this cancer is an electrically active tissue (see Supplementary Videos 1, 2, and 3).
Gliomas exhibit intratumoral and intertumoral cellular heterogeneity, with subpopulations of cancer cells assuming particular roles and even very small cellular fractions proving essential for cancer progression21. Considering all pediatric glioma cells examined (n=643), we find that ~5–10% of glioma cells exhibit synaptic EPSCs, ~40% exhibit prolonged currents in response to neuronal activity. While all four patient-derived xenograft models exhibited neuronal activity-evoked inward currents, the proportion of cells displaying fast EPSCs or prolonged currents varies between patient-derived models. This intertumoral heterogeneity is evident even within a molecularly-defined subtype such as H3K27M+ glioma and predicted by the varied composition of OPC-like and astrocyte-like compartments in individual tumors (Extended Data Fig. 2b, 7a).
Depolarization promotes glioma proliferation
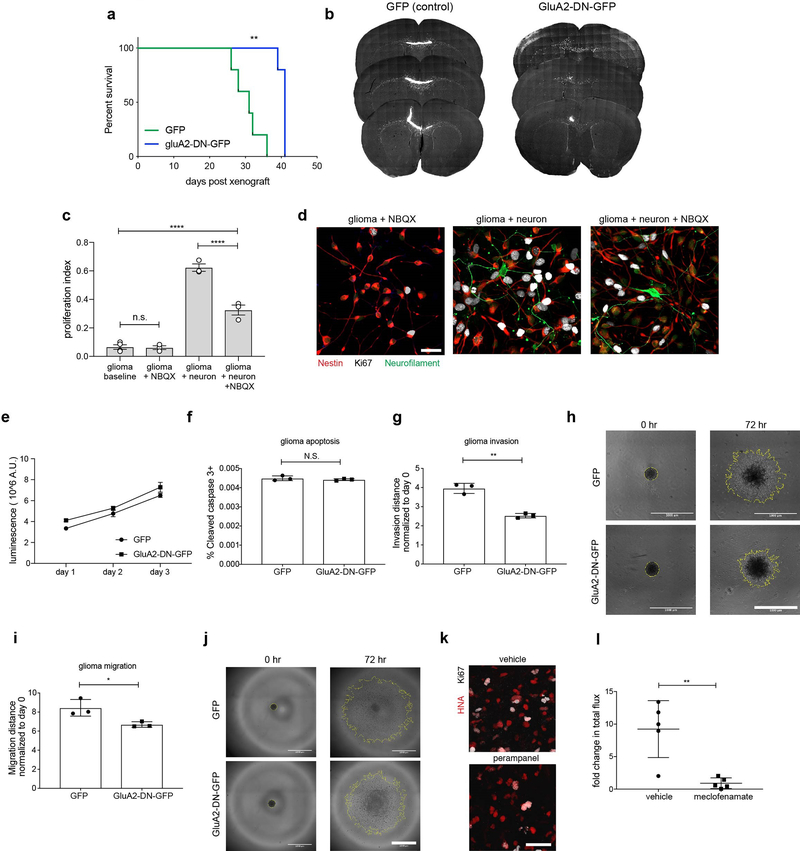
Depolarization can profoundly affect cellular behavior5–9, and we have found two distinct mechanisms by which neuronal activity induces glioma cell membrane depolarization. To test if glioma cell membrane depolarization promotes proliferation, we used in vivo optogenetic techniques to depolarize xenografted glioma cells expressing the blue light-sensitive cation channel channelrhodopsin-2 (ChR2; Extended Data Fig. 7b). Glioma cells expressing ChR2-YFP were xenografted to the cortex, and after a period of engraftment and growth blue light was delivered to depolarize the glioma xenograft. Compared to mock-stimulated control groups, we found that glioma depolarization robustly promoted glioma xenograft proliferation (Fig. 4a–d). Blue light exposure alone had no effect on proliferation nor apoptosis in control glioma xenografts (Extended Data Fig. 7c–e).
Figure 4: Glioma membrane depolarization promotes glioma progression.
a, Optogenetic paradigm for glioma depolarization. ChR2-expressing glioma (blue), region of analysis (light blue). b, Proliferation index of SU-DIPGXIII-FL-ChR2 xenograft after mock stimulation (mock stim) or blue light stimulation (stim) measured as percent of GFP+/HNA+ cells expressing Ki67 (mock stim, n=8; stim, n=9 mice). c, As in (b), but SU-DIPGVI-ChR2 xenografts (n=6 mice/group). d, Representative confocal micrographs from (c), illustrating proliferating SU-DIPGVI-ChR2. Red=human nuclei; white=Ki67. Scale bar=50μm. e-f, Kaplan-Meier survival curves of SU-DIPGXIII-P* xenografts overexpressing e, GFP-only (green) or GluA2-WT-GFP (red) and f, GFP-only (in 80% of cells, green) or GluA2-DN-GFP (in 80% of cells, blue); n=5 mice/group. g, Competitive outgrowth of non-GluA2-DN-GFP-expressing cells in (f), determined by GFP/total human nuclei pixel intensity; (n=3 mice/group). h, Representative confocal micrographs of (f-g). White=human nuclei; green=GFP. Scale bar=50μm. i, Representative confocal images of SU-DIPGXIII-FL xenografts expressing GFP-only control (top) or GluA2-DN-GFP (bottom). Gray=MBP; White=glioma-GFP. Scale bar=500μm. j, Quantification of (i) (n=8 mice/group). k, Proliferation index of SU-DIPGVI xenografts treated with perampanel (AMPAR blocker) or vehicle control; (n=8 mice/group). l, Proliferation index of SU-DIPGXIII-FL in mice treated with meclofenamate (gap junction blocker) or vehicle control; (n=9 vehicle, n=8 treated mice). Data shown as mean±s.e.m (b,c,g,j,k,l). **P<0.01. ***P<0.001, ****P<0.0001. P-values determined by two-tailed unpaired Student’s t-test (b,c,g,k,l); two-tailed log rank analyses (e,f); two-sided Mann-Whitney test (j).
Targeting electrochemical communication
As membrane depolarization promotes glioma proliferation, we next tested the relative functional contributions of each mechanism of electrochemical communication, beginning with AMPAR-mediated EPSCs. We over-expressed either WT-GluA2 subunit fused to GFP, GFP alone, or a dominant-negative GluA2 (GluA2-DN-GFP) in glioma and confirmed decreased conductance in GluA2-DN-GFP-expressing glioma cells (Extended Data Fig. 7f–g). Mice bearing xenografts overexpressing WT-GluA2-GFP survived a shorter time than GFP-only xenografts, while mice bearing GluA2-DN-GFP xenografts exhibited improved survival and decreased tumor burden compared to GFP-only controls (Fig. 4e–j, Extended Data Fig. 8a–b). To test for a possible in vivo growth advantage of GluA2-expression, we xenografted a mixture of 80% GluA2-DN-GFP construct-expressing and 20% non-expressing cells and found that tumors were composed almost entirely of non-GluA2-DN-GFP expressing cells at the survival endpoint (Fig. 4g–h).
Similar to in vivo experiments, co-culture of glioma cells with neurons markedly increased proliferation. NBQX partially reduced this effect (Extended Data Fig. 8c–d); neuronal secreted factors such as NLGN3 accounts for residual elevated proliferation1. Given the stark effect of AMPAR function on glioma growth in vivo and in co-culture, we next tested the relative contributions of cell-intrinsic glutamate signaling mechanisms. While paracrine/autocrine AMPA signaling may promote adult glioblastoma growth22,23, NBQX had no effect on pediatric glioma proliferation in the absence of neurons (Extended Data Fig. 8c–d). Testing cell-intrinsic effects of GluA2-DN expression, we similarly found no difference in growth rate or apoptosis in pediatric glioma cells outside of the neuronal microenvironment (Extended Data Fig. 8e–f). In contrast, pediatric glioma cell migration and invasion were influenced by GluA2-DN expression in a cell-intrinsic manner (Extended Data Fig. 8g–j), consistent with previous reports in adult gliomas24. Taken together, these findings indicate that glioma AMPAR activation promotes pediatric glioma growth chiefly through microenvironmental interactions such as neuron-to-glioma synaptic transmission.
Next, we sought to therapeutically target glioma currents using existing drugs. Using an AMPAR-blocking anti-epileptic drug (perampanel), we found a ~50% decrease in pediatric glioma proliferation in perampanel-treated mice compared to vehicle-treated controls (Fig. 4k, Extended Data Fig. 8k). We then targeted gap junction-mediated amplification of neuronal activity-dependent potassium currents using the brain-penetrant gap-junction blocker meclofenamate25. Meclofenamate treatment similarly decreased pediatric glioma xenograft proliferation and growth (Fig. 4l, Extended Data Fig. 8l).
Gliomas increase neuronal excitability
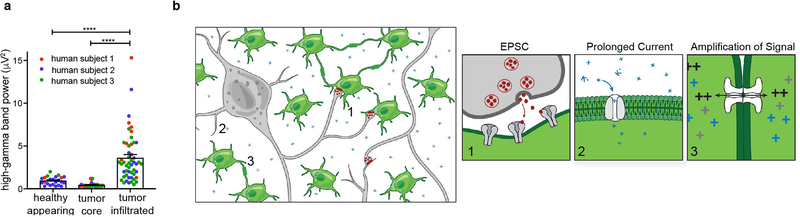
Neuron-glioma interactions are bidirectional; neuronal activity increases glioma growth1, and gliomas are thought to increase neuronal activity. In preclinical adult glioblastoma models, glioma cells induce neuronal hyperexcitability and seizures26,27 through non-synaptic glutamate secretion26,28, and through secretion of synaptogenic factors27. To assess neuronal hyperexcitability in primary human glioblastoma, we performed intraoperative electrocorticography in three awake adult human subjects with cortical high-grade gliomas (IDH WT) prior to surgical resection (Fig. 5). High gamma frequency (70–110 Hz) power, a measure that correlates with neuronal firing rate and local field potential29 and that is elevated by cortical hyperexcitability30, was sampled over a 3-minute period while the subjects were in a resting state. Outside of the necrotic nodular core of the tumor, we found markedly increased high gamma power in tumor-infiltrated brain compared to normal-appearing brain (Fig. 5a–b, Extended Data Fig. 9a). Concordantly, we found neuronal hyperexcitability in the microenvironment of pediatric glioma xenografts (Fig. 5c). These findings support the concept26,27 of hyperexcitable neurons in the glioma microenvironment, which would potentiate mechanisms of activity-regulated glioma progression. As neuronal action potentials result in extracellular potassium rise31,32, neuronal hyperexcitability in the glioma microenvironment would promote non-synaptic prolonged glioma potassium currents, as well as synaptic neuron-to-glioma EPSCs (Extended Data Fig. 9b).
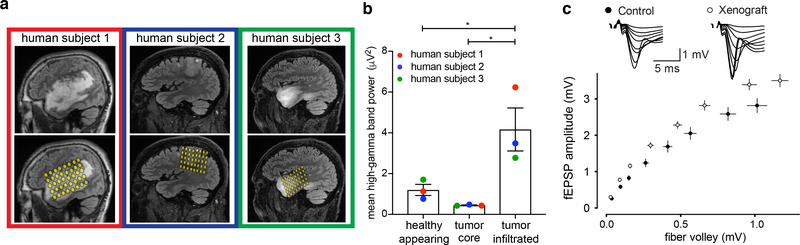
Figure 5: Increased neuronal excitability in glioma-infiltrated brain.
a-b, Human neuronal hyperexcitability in glioma: a, top, sagittal brain MRI FLAIR sequences of adult IDH WT cortical glioblastoma in three individuals (human subject 1,2,3 outlined in red, blue, green, respectively). Bottom, intraoperative cortical electrode placement. b, Electrocorticography signals (mean high-gamma frequency-filtered power (μV2)) in each of healthy-appearing, tumor core, and tumor-infiltrated brain (n=3 patients with n=23, n=29, n=51 total channels, respectively). Data points colored by patient as in (a). c, Mouse neuronal hyperexcitability in pediatric glioma xenograft (SU-DIPG-XIIIFL): plot of presynaptic fiber volley vs. amplitude of field EPSP at varying axonal stimulation intensities (10,20,30,50,75,100,150,200 μA) in glioma-bearing or control hippocampus (n=17/3 control and n=18/3 glioma-bearing slices/mice at each datapoint). Data fit to a non-linear regression and compared using the extra-sum-of-squares F test; F=61.61, P<0.0001). Representative traces of field responses to varying intensities shown above. Data shown as mean±s.e.m (b,c). P-values determined by one-way ANOVA with Tukey’s post-hoc analysis. *P<0.05, ****P<0.0001.
Discussion
Membrane depolarization and depolarization-induced calcium transients promote normal NPC development, in part through voltage-gated calcium channel signaling5–9. Non-synaptic neurotransmitter release mediates synchronous calcium transients in gap-junction-coupled NPCs in many contexts33–35, but synapses between presynaptic neurons and post-synaptic NPCs are also well-described, including transient synapses onto migrating neuroblasts during corticogenesis36 and synapses onto OPCs in the developing and adult brain3,4. High-grade gliomas integrate into electrical networks and depolarizing current promotes glioma progression similarly to the well-established effect in normal NPCs. In glioma, we have demonstrated bona fide neuron-to-glioma synapses, reminiscent of the synapses found on normal OPCs. As well, we have shown neuronal activity-evoked potassium currents in glioma cells, reminiscent of activity-dependent currents in normal astrocytes37. Neuronal activity-induced glioma membrane depolarization by either mechanism promotes glioma proliferation and growth, through voltage-sensitive signaling pathway(s) to be fully elucidated in future work.
Neuronal activity is emerging as a critical regulator of glioma progression. The electrochemical communication described here joins activity-regulated secretion of growth factors1,2 as mechanisms that mediate this important microenvironmental interaction. NLGN3, an activity-regulated secreted factor fundamental to glioma progression2, functions both by stimulating classical oncogenic signaling pathways1,2 and by promoting neuron-to-glioma synaptogenesis. Appreciating the crosstalk between neurons and the glioma cells invading and integrating into neural circuitry elucidates promising therapeutic targets, including activity-regulated secreted growth factors1,2, neuron-to-glioma neurotransmission, ion channels and gap-junction coupling. Modulating the influence of glioma on neuronal excitability represents an important opportunity to dampen the magnitude of activity-regulated glioma growth. Taken together, the findings presented here identify synaptic neurotransmission and activity-dependent potassium currents as mechanisms driving glioma growth and elucidate the previously unexplored potential to target glioma circuit dynamics for therapy of these lethal cancers.
Methods
Single Cell Sequencing Analysis
We combined publicly-available single cell datasets processed and annotated by Filbin et al.10 and by Venteicher et al.11 all sequenced using smart-seq2 protocol. Following the quality control measures taken in these studies we filtered the data to keep cells with more than 400 detected genes, and genes which were expressed in more than 3 cells. We assessed the single cell transcriptome from 5,096 adult IDH-mutant glioma cells derived from 10 subjects, 599 adult IDH-WT glioma cells derived from 3 subjects, and 2,259 pediatric histone-3 mutant (H3K27M) diffuse midline glioma (DMG) cells derived from 6 subjects. Malignant cells were inferred by expression programs and detection of tumor-specific genetic alterations. For each sample we performed first cell-level normalization, and then centered the gene expression around 0 to allow Principal Component Analysis (PCA) computation. Following the PCA reduction we clustered the cells using shared nearest neighbor clustering. We examined the synapse signature of each of the cells in each cluster by taking the mean expression of genes listed below, and subtracting the average expression of a control set of genes chosen from the same expression distribution (as explained in Venteicher et. al.11 supplementary methods).
Xenograft dissociation
NSG mice were sacrificed six weeks following xenograft of patient-derived DIPG cell lines (SU-DIPGVI and SU-DIPGXIII-FL) into the right hippocampus. The brain was isolated and sliced on a Leica vibratome VT1200S at a thickness of 350 microns. Acute brain slices were placed in oxygenated artificial cerebrospinal fluid before being visually inspected under a fluorescent microscope for presence and proper placement of GFP+ DIPG cells. Slices containing DIPG cells were then minced with a scalpel and enzymatically dissociated using the Miltenyi brain tumor dissociation kit P (130–095-942). The resultant cell suspension was passed through a 70-micron SmartStrainer from Miltenyi (130–098-462). Myelin debris was then removed using the Miltenyi myelin removal beads II (130–096-733).
Fluorescence-activated cell sorting (FACS)
Cell suspensions from xenograft dissociation were stained with APC/Cy7 anti-human HLA-A, B, C from Biolegend (311426) and Calcein Violet 450 AM Viability Dye from eBioscience (65–0854-39). Gates were first drawn to exclude calcein negative (dead) cells. Double-positive, viable DIPG cells (HLA+ and GFP+) were index sorted on a FACSAria Fusion Special Order Instrument (Becton Dickinson) as singlets by adhering to a strict forward scatter by area gating scheme. Cells were sorted into skirted 96-well plates, each well of which contained 4 microliters of lysis buffer as described in Picelli et al.38
Whole-transcriptome amplification, library construction, sequencing, and processing
96-well plates of single sorted cells were processed using the smart-seq2 protocol as described in Picelli et al.38 Next-generation sequencing libraries were then generated using the Nextera XT kit from Illumina (FC-131–1096) and cells were multiplexed in batches of 96 using the IDT for Illumina dual-unique indexing scheme (20027213). All pools were sequenced in paired-end mode on the Illumina Next500 to an average target depth of ~1 million reads per cell with mid-output kits (~130 million reads). Paired-end, 75-basepair reads were mapped to the UCSC hg19 human transcriptome using Bowtie 0.12.7 with parameters “-q–phred33-quals -n 1 -e 99999999 -l 25 -I 1 -X 2000 -a -m 15 -S -p 6”, which allows alignment of sequences with single base changes, such as point mutations in the H3F3A gene. Expression values were calculated from SAM files using RSEM 1.2.19 in paired-end mode using parameters “–estimate-rspd–paired end -sam -p 6”, from which TPM values for each gene were extracted. Matrices of TPM (transcripts per million) values were then processed and analyzed using the Seurat (V2.0) package. Individual cells were scored for their expression of a curated housekeeping gene set and then TPM values were normalized, analyzed for variable gene expression and scaled for downstream dimensionality reduction. PCA was run and then clusters defined from significant PCs.
2-D plots
(Generated by methods described in Tirosh et al.39) For the DIPG biopsies and xenografts plots, the y-axis value for each cell was calculated as the difference between a sK27M-specific OPC-like stem cell signature and the sum of astroglial and oligodendroglial-differentiation scores defined in Filbin et al.10 In other words, increasing y-values correspond to stronger stem-like characteristics. Conversely, negative y-values indicate a differentiated cell state. The x-axis was plotted by assessing each cell for astroglial and oligodendroglial module scores and plotting the larger value of the two. If a cell scored negatively for both differentiation scores, it was assigned a value of zero and jitter was applied to the ggplot function in R to facilitate visualization of these cells (height=0.1, width=0.1). Lastly, if a cell scored more highly for the astroglial than oligodendroglial module, then the sign of the astroglial score was switched to allow for plotting both differentiation paths along one axis. Plots were generated using ggplot in RStudio 1.0.136.
Derivation of synapse gene set from DIPG biopsies
In the case of DIPG biopsies, the OPC-score is the OPC-shared score defined in Filbin et al.10 The DIPG synapse score was derived from a single biopsy (BCH869) by a method described below and is distinct from the OPC-shared score. Following removal of non-malignant cells, DIPG biopsy BCH869 was isolated for analysis via the Seurat (V2.0) pipeline. TPM expression values were normalized and variable genes were identified. Data were then scaled for dimensionality reduction via PCA and tSNE. Shared nearest neighbor clustering of DIPG cells revealed a cluster statistically enriched for genes involved in synapse formation and function. The resulting synaptic gene set used is as follows: PTPRS, ARHGEF2, GRIK2, DNM3, LRRTM2, GRIK5, NLGN4X, NRCAM, MAP2, INA, TMPRSS9.
Signature correlations
To compute a gene signature for a set of genes, we use the function AddModuleScore by Seurat40 package, which calculates the average expression levels of the gene set subtracted by the aggregated expression of 100 randomly chosen control gene sets, where the control gene sets are chosen from matching 25 expression bins corresponding to the tested gene set expression. To calculate the correlation between two gene expression scores we used pearson correlation and report the correlation coefficient (rho) and the pearson p-value.
Cell Culture
For all human tissue studies, informed consent was obtained and tissue was used in accordance with protocols approved by the Stanford University Institutional Review Board (IRB). For all patient-derived cultures, mycoplasma testing is routinely performed and short tandem repeat (STR) DNA fingerprinting was performed every three months to verify authenticity.
The STR fingerprints and clinical characteristics for the patient-derived cultures and xenograft models used have been previously reported 1,2, 41, 42, with the exception of SU-DIPG25 which is X/X (AMEL), 12/12 (CSF1PO1), 8/11 (D13S317), 12/13 (D16S539), 30/35 (D21S11), 11/13 (D5S818), 10/12 (D7S820), 9/9 (TH01), 7/8 (TPOX), 14/18 (vWA). SU-DIPG25 is a H3.3K27M mutant tumor that was derived from the brainstem at the time of autopsy from a female who was 5 years of age, treated with radiotherapy, and survived 11 months (clinical characteristics are also reported in Extended Data Table 1).
Regarding primary tissue samples SU-DIPG-39 and SU-GBM092: SU-DIPG39 is a H3.3K27M mutant tumor that was derived from the brainstem at the time of autopsy from a male who was 5 years of age at diagnosis, treated with radiotherapy, and survived 6 months. SU-GBM092 is an adult hemispheric glioma, sampled at the time of autopsy from a male who was 47 years of age at diagnosis, treated with surgical resection, radiotherapy and temozolomide, and survived 6 months.
All high-grade glioma cultures were generated as previously described1. In brief, tissue was obtained from high-grade glioma (WHO grade III or IV) tumors at the time of biopsy or from early post-mortem donations. Tissue was dissociated both mechanically and enzymatically and grown in a defined, serum-free medium designated ‘tumor stem media’ (TSM), consisting of Neurobasal(-A) (Invitrogen), B27(-A) (Invitrogen), human-bFGF (20 ng ml−1; Shenandoah), human- EGF (20 ng ml−1; Shenandoah), human PDGF-AA (10 ng ml−1) and PDGF-BB (10 ng ml−1; Shenandoah) and heparin (2 ng ml−1; Stem Cell Technologies). All cell cultures were routinely tested for mycoplasma.
PSD-95-RFP cloning
PSD-95-pTagRFP (Plasmid #52671) was purchased from Addgene. Primers were used to excise and amplify the PSD-95-pTagRFP ORF from the original vector. This amplicon was inserted by Gibson cloning (New England Biolabs #E2611) downstream of an EF1α-promoter in a pCDH vector (System Biosciences #CD811A-1) linearized by XbaI and NotI. Forward primer for Gibson assembly: 5’-AAA TCG GAT CCG CGG CCG CGG CCA CCA TGG ACT GTC TC-3’ and reverse primer for Gibson assembly: 5’- ATC CAG AGG TTG ATT GTC GAT CAA TTA AGT TTG TGC CCC AG-3’. A PGK promoter in the same plasmid drives GFP expression. Once cloned, the plasmid was packaged together with helper plasmids (pΔ8.9 and VSV-g) to generate replication-deficient lentivirus from adherent 293T cells. One million target SU-DIPGVI or SU-DIPGXIII-FL cells were infected with Lenti-X (Takara) concentrated viral supernatant and allowed to recover for 1 week. RFP-positive cells were isolated for purity by FACS (BD FACS Aria) and returned to culture.
Neuron-glioma co-culture experiments
Neurons were isolated from the brains of P1 NSG; Nlgn3WT or NSG;Nlgn3y/− animals using the ‘Neural Tissue Dissociation Kit - Postnatal Neurons’ (Miltenyi), and followed by the ‘Neuron Isolation Kit, Mouse’ (Miltenyi) per manufacturer’s instructions. After isolation, 300,000 neurons were plated onto circular glass coverslips (Electron Microscopy Services) pre-treated for 1-hour RT with poly-L-lysine (Sigma) and then 3 hours at 37°C with 5 μg/mL mouse laminin (Thermo Fisher). Neurons are cultured in BrainPhys neuronal medium (Stemcell Technologies) supplemented with 1x glutamax (Invitrogen), pen/strep (Invitrogen), B27 supplement (Invitrogen), BDNF (10ng/mL; Shenandoah), and GDNF (5ng/mL; Shenandoah), TRO19622 (5μM; Tocris), beta-mercaptoethanol (Gibco), and 2% fetal bovine serum. Half of the medium was replenished on DIV 1 and UFDU was added at 1μM. This was repeated at DIV 3. On DIV 5, half of the medium was replaced with serum-free in the morning. In the afternoon, the medium was again replaced with half serum-free medium containing 75,000 glioma cells expressing PSD-95-RFP. Glioma cells were cultured with neurons for 72 hours and then fixed with 4% PFA for 20 minutes at room temperature and stained for immunofluorescence analysis as described below.
Synaptic Puncta Staining and Visualization
For immunohistochemistry, fixed coverslips were incubated in blocking solution (3% normal donkey serum, 0.3% Triton X-100 in TBS) at room temperature for 1 hour. Primary antibodies, mouse anti-nestin (1:500, Abcam), guinea pig anti-synapsin1/2 (1:500; Synaptic Systems), rabbit anti-RFP (1:500; Rockland), or chicken anti-neurofilament (M+H; 1:1000; Aves Labs) were diluted in antibody diluent solution (1% normal donkey serum in 0.3% Triton X-100 in TBS) and incubated overnight at 4°C. Samples were then rinsed three times in TBS and incubated in secondary antibody solution (Alexa 488 donkey anti-chicken IgG; Alexa 594 donkey anti-rabbit IgG, Alexa 647 donkey anti-mouse IgG, and Alexa 405 donkey anti-guinea pig IgG all used at 1:500 (Jackson Immuno Research)) in antibody diluent solution at 4°C overnight. Coverslips were rinsed three times in TBS and mounted with ProLong Gold Mounting medium (Life Technologies). Images were collected using a 63x oil-immersion objective on a Zeiss LSM800 confocal microscope and processed with airyscan. Coverslips were used to quantify colocalization as described below.
Confocal puncta quantification
Colocalization of all synaptic puncta images from neuron:glioma co-cultures described above were analyzed using a custom ImageJ processing script written at the Stanford Shriram Cell Science Imaging Facility to define each pre and post synaptic puncta and determine colocalization within a defined proximity of 1.5μM. In order to partially subtract local background, we used the ImageJ rolling ball background subtraction (https://imagej.net/Rolling_Ball_Background_Subtraction). The peaks were found using the imglib2 DogDetection plugin (https://github.com/imglib/imglib2algorithm/blob/master/src/main/java/net/imglib2/algorithm/dog/DogDetection.java). In this plugin, the difference of Gaussians is used to enhance the signal of interest using two different sigma: a “smaller” sigma, which defines the smallest object to be found and a “larger” sigma, for the largest object. The plugin then identifies the objects that are above the min peak value, and assigns ROIs to each channel. The number of neuron and glioma ROIs are counted and the script extracts the number of glioma ROIs within 1.5μm of the neuron ROIs. This script was implemented in Fiji/ImageJ43,44,45 using the ImgLib246 and ImageJ Ops (https://imagej.net/ImageJ_Ops) libraries.
Mice and housing conditions
All in vivo experiments were conducted in accordance with protocols approved by the Stanford University Institutional Animal Care and Use Committee (IACUC) and performed in accordance with institutional guidelines. Animals were housed according to standard guidelines with free access to food and water in a 12 h light/12 h dark cycle. For brain tumor xenograft experiments, the IACUC does not set a limit on maximal tumor volume but rather on indications of morbidity. In no experiments were these limits exceeded as mice were euthanized if they exhibited signs of neurological morbidity or if they lost 15% or more of their body weight.
Orthotopic Xenografting
For all immuno-electron microscopy, electrophysiology, single cell sorting, or calcium imaging experiments, a single-cell suspension from cultured SU-pcGBM2-GFP, SU-DIPGVI-GFP, SU-DIPGXIII-FL-GFP, SU-DIPGVI-GCaMP6s, or SU-DIPGXIII-FL-GCaMP6s neurospheres were prepared in sterile PBS immediately prior to the xenograft procedure. Animals at P28–30 were anesthetized with 1%–4% isoflurane and placed in a stereotactic apparatus. The cranium was exposed via midline incision under aseptic conditions. 600,000 cells in 3μl sterile PBS were stereotactically implanted into the CA1 region of the hippocampus through a 31-gauge burr hole, using a digital pump at infusion rate of 0.4 μL/min and 31-gauge Hamilton syringe. Stereotactic coordinates used were as follows: 1.5 mm lateral to midline, 1.8 mm posterior to bregma, –1.4 mm deep to cranial surface. At the completion of infusion, the syringe needle was allowed to remain in place for a minimum of 2 minutes, then manually withdrawn at a rate of 0.875 mm/min to minimize backflow of the injected cell suspension.
All other xenografts were done using similar techniques with variations in the location of the injection site based on the needs of the experiment. SU-DIPGXIII-FL-mockGFP and SU-DIPGXIII-FL-GluA2DN for tumor burden analysis, and SU-DIPGXIII-FL for meclofenamate treatment, were xenografted into the premotor cortex; stereotactic coordinates used were as follows: 0.5 mm lateral to midline, 1.0 mm anterior to bregma, − 1.75 mm deep to cranial surface. SU-DIPGVI-ChR2, SU-DIPGXIII-FL-YFP, and SU-DIPGXIII-FL-ChR2 were xenografted more superficially into the premotor cortex to optimize blue light penetration; stereotactic coordinates used were as follows: 0.5 mm lateral to midline, 1.0 mm anterior to bregma, − 1.0 mm deep to cranial surface. SU-DIPGXIII-P*-mockGFP, SU-DIPGXIII-P*-GluA2WT and SU-DIPGXIII-P*-GluA2DN for survival studies, and SU-DIPGVI for perampanel treatment were xenografted into the pons; stereotactic coordinates used were as follows: 1.0 mm lateral to midline, − 0.8 mm posterior to lambda, − 5.0 mm deep to cranial surface.
Sample preparation and image acquisition for electron microscopy
8–12 weeks after xenografting, mice were sacrificed by transcardial perfusion with Karnovsky’s fixative: 2% glutaraldehyde (EMS 16000) and 4% paraformaldehyde (EMS 15700) in 0.1M sodium cacodylate (EMS 12300), pH 7.4. Primary adult glioma tissue taken at surgical resection was fixed in the same solution. Transmission electron microscopy was performed in the tumor mass within the CA1 region of the hippocampus for all xenograft analysis. The samples were then post-fixed in 1% osmium tetroxide (EMS #19100) for 1 hour at room temperature, washed 3 times with ultrafiltered water, then en bloc stained for 2 hours at room temperature. Samples were dehydrated in graded ethanol (50%, 75%, and 95%) for 15 minutes each at 4°C; the samples were then allowed to equilibrate to room temperature and were rinsed in 100% ethanol 2 times, followed by acetonitrile for 15 minutes. Samples were infiltrated with EMbed-812 resin (EMS #14120) mixed 1:1 with acetonitrile for 2 hours followed by 2:1 EMbed-812:acetonitrile for 2 hours. The samples were then placed into EMbed-812 for 2 hours, then placed into TAAB capsules filled with fresh resin, which were then placed into a 65°C oven overnight. Sections were taken between 40 and 60 nm on a Leica Ultracut S (Leica) and mounted on 100 mesh Ni grids (EMS #FCF100-Ni). For immunohistochemistry, microetching was done with 10% periodic acid and eluting of osmium with 10% sodium metaperiodate for 15 minutes at room temperature on parafilm. Grids were rinsed with water three times in between and followed by 0.5M Glycine quench. Grids were incubated in blocking solution (0.5% BSA, 0.5% Ovalbumin in PBST) at room temperature for 20 minutes. Primary rabbit anti-GFP (1:300; MBL International) was diluted in the same blocking solution and incubated overnight at 4°C. The following day, grids were rinsed in PBS three times, and incubated in secondary antibody (1:10 10nm Gold conjugated IgG TED Pella 15732) for one hour at room temperature and rinsed with PBST followed by water. For each staining set, samples that did not contain any GFP-expressing cells were stained simultaneously to control for any non-specific binding. Grids were contrast stained for 30 seconds in 3.5% uranyl acetate in 50% acetone followed by staining in 0.2% lead citrate for 90 seconds. Samples were imaged using a JEOL JEM-1400 TEM at 120kV and images were collected using a Gatan Orius digital camera.
EM data analysis
Sections from the xenografted hippocampi of mice were imaged as above using TEM imaging. Here 101 sections of SU-DIPGVI across 3 mice and 104 sections form SU-DIPGXIII-FL across 3 mice were analyzed. EM images were taken at 6000x with a field of view of 15.75 μm2. Synapses were inspected by two distinct investigators, including board-certified pathologist and head of Stanford University School of Medicine Department of Neuropathology, Dr. Hannes Vogel. Glioma cells were counted and analyzed after unequivocal identification of immunogold particle labeling with 3 or more particles. Further, in order to determine synaptic structures all 3 of the following criteria had to be clearly met: (1) presence of synaptic vesicle clusters; (2) visually apparent synaptic cleft; (3) identification of clear post-synaptic density in glioma cell. In order to quantify the percent of glioma cells forming synaptic structures, number of glioma:neuron synapses identified was divided over total number of glioma cells analyzed.
Slice preparation for electrophysiology and calcium imaging experiments
Hippocampal slices (225 μm thick) were prepared from mice (4–8 weeks after xenografting) in accordance with a protocol approved by Stanford University APLAC as previously described47 and maintained for 30 minutes in warm (30 °C) oxygenated (95%O2/5%CO2) ACSF containing (in mM): 119 NaCl, 26 NaHCO3, 10 glucose, 2.5 KCl, 1 NaH2PO4, 1.3 MgSO4, and 2.5 CaCl2 before being allowed to equilibrate at room temperature for an additional hour.
Electrophysiology
Slices were transferred to a recording chamber and perfused with oxygenated, warmed (28–30 °C) ACSF containing picrotoxin (50 μM). In some experiments CaCl2 was replaced with SrCl2 (4 mM). GFP-labeled glioma cells were identified with an epifluorescent microscope equipped with DIC optics (Olympus BX50WI). Whole-cell recording pipettes (3–5 MΩ) were filled with a solution containing (in mM): 135 CsMeSO4, 12 HEPES, 8 NaCl, 0.25 EGTA, 2 MgCl2, 1 Mg2ATP, 0.3 Na3GTP and 5 phosphocreatine for voltage-clamp recordings (CsMeSO4 was replaced with equimolar KMeSO4 for current-clamp recordings; pH adjusted to 7.4, 292–296 mOsM). In some experiments biocytin (0.5%) was added to the pipette solution. Glioma and CA1 pyramidal cells were voltage-clamped at −80 mV and −70 mV, respectively. Synaptic responses were evoked with a bipolar electrode placed near the DIPG cells in s. radiatum or, in some cases, s. oriens to maximize the chance of activating a synaptic input onto the cells. Population field EPSPs (fEPSPs) were recorded with a pipette filled with 1 M NaCl solution containing 10 mM HEPES (pH adjusted to 7.4) and placed in proximal s. radiatum of CA1. fEPSPs for input-output experiments were recorded in picrotoxin-free ACSF. Field recordings from infected slices were interleaved with control slices from the uninfected contralateral hippocampus of the same mouse. The sensitivity of glioma cells to extracellular K+ was performed in the presence of D-AP5 (100 μM), NBQX (10 μM) and TTX (0.5 μM) to block neuronal activity. Optogenetic currents were evoked with a 470 nm collimated LED connected to the microscope and controlled by a t-cube LED driver (Thorlabs). Stimulation duration, frequency and intensity were set by a Master 8 programmable pulse generator (A.M.P.I.) connected to an Iso-flex stimulus isolator (A.M.P.I.) for synaptic stimulation or the LED driver for optogenetic stimulation or a Picospritzer II (Parker Hannifin Corp.) for local application of S-AMPA (6 psi, 1s). Synaptic, optogenetic and agonist-induced signals were acquired with a MultiClamp 700B amplifier (Molecular Devices) and digitized at 10 kHz with a National Instruments data acquisition device (BNC-2090). Data were recorded and analyzed using a custom program written for Igor Pro software (Wavemetrics). For representative traces shown in Fig. 2c, d, e, f, g, h; Fig. 3a, b, e; Fig. 5c; Extended Data Fig. 3d; Extended Data Fig. 5a, d, e, h; Extended Data Fig. 6c and i, stimulus artifacts preceding the EPSC/P or field potential have been removed for clarity.
Inhibitors
Drugs and toxins used for electrophysiology were picrotoxin (50μM; Sigma), TTX (0.5μM; Tocris), NBQX (10μM; Tocris), NASPM (100 μM; Tocris), D-AP5 (100μM; Tocris), carbenoxolone (100μM; Alfa Aesar), Meclofenamate (100 μM; Selleckchem), S-AMPA (500 μM; Tocris), Cyclothiazide (100 μM; Tocris), TBOA (200 μM; Tocris), and biocytin (Invitrogen). When used for in vitro slice application drugs were made up as a 1000x stock in dH20 or DMSO and dissolved to their final concentrations in ACSF before exposure to slices.
Calcium Imaging
For calcium imaging, the genetically encoded calcium indicator GCaMP6s was lentivirally transduced into SU-DIPGVI and SU-DIPGXIII-FL (pLV-ef1-GCaMP6s-P2A-nls-tdTomato). In this case, glioma cells containing the GCaMP6s reporter can be identified using the tdTomato nuclear tag. These cells were isolated and xenografted into the CA1 region of the hippocampus as described above. Xenografts were post-fixed and stained for tdTomato and human nuclei markers to ensure only glioma cells expressed the construct (Extended Data Fig.4).
Two photon calcium imaging experiments were performed using Prairie Ultima XY upright two-photon microscope for tissue slices equipped with an Olympus LUM Plan FI W/IR-2 40x water immersion objective. The temperature of the perfusion media, ACSF as described above, was kept at 30°C, and perfused through the system at rate of 2ml/min. Excitation light was provided at a wavelength of 920 nm through a tunable Ti:Sapphire laser (Spectra Physics Mai Tai DeepSee) to allow for excitation of both tdTomato and GCaMP6s. The actual laser power reaching the scanhead for each scope is dynamically controlled by Pockels cells via software interface. Pockels cell were set at 10 for all experiments, and PMTs were set at 800 for each channel. For these settings, power at back aperture of the objective was about ~30 mW at 920 nm. The wavelength ranges for the emission filters were PMT1: 607nm center wavelength with 45nm bandpass (full width at half-max) and PMT2: 525nm center wavelength with 70nm bandpass (full width at half-max). Recordings were made at 0.65 frames per second (~1.5Hz) for about 30 minutes in the case of spontaneous activity and 10 minutes in the case of response to periodic electrical stimulation. Cells were identified via the expression for the nuclear tdTomato tag and were only imaged in the area of interest, specifically in the CA1 region of the hippocampus. Similar to the electrophysiology paradigm, for neuronal stimulation experiments, the electrode was placed in the hippocampus to stimulate the neuronal inputs originating in CA3. For electrical stimulation, ~20μA over 200μs was delivered to local axons using a stimulating bipolar microelectrode. For all inhibitor experiments, inhibitors were directly diluted into the ACSF perfusion media at desired concentration, oxygenated, and delivered to the slices through the perfusion system.
Calcium Imaging Analysis
Two photon live image sequences were analyzed using a custom image processing pipeline at the Stanford Shriram Cell Science Imaging Facility. In order to correct for any motion artifact we identify the relative shift between adjacent images in the “nucleus” (tdTomato) channel using cross-correlation48 and applied the correction to both the nucleus and calcium channels. To segment the image into regions representing individual cells we first divide the image into superpixels using the simple linear iterative clustering (SLIC)49. We then manually adjust a threshold to separate background superpixels from foreground (cells) superpixels using their median intensity. The ΔF/F traces of each cell were computed by subtracting the mean intensity of the cell at every time point with the 20th percentile of an adjustable time window preceding the time point and dividing it by the same number. To find the local maxima of calcium signals, the data are first smoothed using a 20-frame window and peaks are found using an algorithm written by Marco Duarte (http://nbviewer.jupyter.org/github/demotu/BMC/blob/master/notebooks/DetectPeaks.ipynb). To measure synchronicity between cells, we used the definition proposed by Osswald et al.19. Synchronous cells, the number of synchronous communications, and the time point of the synchronous firing were determined using a window of 6 frames (~4sec) around each peak. The correlation image for each pixel is computed by averaging the correlation coefficients (taken over time) of each pixel with its four immediate neighbors50. This algorithm was implemented in Python using the scikit-image library51. Full 30-minute spontaneous activity was assessed for synchronicity in Fig. 3h, however for clarity, only 10 minutes of analysis is shown in Fig. 5i. For gap junction blocker, CBX experiments, synchronicity was calculated for spontaneous transients over the course of 15 minutes in multiple xenografts. CBX was then perfused through the media, and calcium transients were recorded for another 15 minutes. Transients were analyzed for synchronicity as above before and after addition of inhibitor.
Quantitative fluorescence intensity analysis was done on calcium transients which were reliably evoked by axonal stimulation. In order to determine the effect of tetrodotoxin on the calcium transients in response to electrical stimulation of the CA3 Schaffer collaterals, the field of cells were stimulated 3 times in 1-minute intervals to ensure synaptic connectivity. 500μM tetrodotoxin was then perfused into the slices and the stim repeated on the same field of cells to gauge direct effect of TTX on stimulation response. For analysis, ROIs of each responding nucleus were set and ΔFmax/F0 (maximum difference in fluorescence intensity normalized to background fluorescence) measurements were determined as above before and after TTX treatment.
Fiber Optic Placement and In Vivo Optogenetic Stimulation
Fiber optic placement was performed as previously described1 a minimum of 7 days prior to optogenetic stimulation. Animals were anesthetized with 1%–4% isoflurane and placed in a stereotactic apparatus. The cranium was exposed using a midline incision under aseptic conditions. A fiber optic ferrule (Doric Lenses) was placed at the premotor cortex (M2) of the right hemisphere using the following coordinates: 0.5 mm lateral to midline, 1.0 mm anterior to bregma, 0.65 mm deep to cranial surface in the right hemisphere. At 6 weeks post-xenograft (allowing a minimum of 7 days of recovery following ferrule placement procedure), all animals were connected to a 100-mW 473-nm DPSS laser system with a mono fiber patch cord, which freely permits wakeful behavior of the animal. Pulses of light with ~5 mW measured output at tip of the patch cord were administered at a frequency of 20 Hz for periods of 30 s, followed by 90 s recovery periods, for a total session duration of 30 minutes per day for 5 consecutive days. This power represents ~30 mW/cm2 light density at the tip of the patch cord; with the optical ferrule placed just below the pial surface this would deliver ~3 mW/cm2 approximately midway through the cortex52. Mice were sacrificed 24 hours following the final (5th) stimulation session.
Perfusion and Immunohistochemistry
Animals were anesthetized with intraperitoneal Avertin (tribromoethanol), then transcardially perfused with 20 ml of PBS. Brains were fixed in 4% paraformaldehyde overnight at 4°C, then transferred to 30% sucrose for cryoprotection. Brains were then embedded in Tissue-Tek O.C.T. (Sakura) and sectioned in the coronal plane at 40 mm using a sliding microtome (Microm HM450; Thermo Scientific).
For immunohistochemistry, coronal sections were incubated in blocking solution (3% normal donkey serum, 0.3% Triton X-100 in TBS) at room temperature for 30 minutes. Chicken anti-GFP (1:500, Abcam), mouse anti-human nuclei clone 235–1(1:100; Millipore), rabbit anti-Ki67 (1:500; Abcam), rabbit anti-cleaved caspase 3 (1:200, Cell Signaling), mouse anti-MAP2 (1:1000; Millipore), mouse anti-vimentin, (1:500; Dako), rabbit anti-histone H3.3 K27M mutant (1:500; Millipore), mouse anti-nestin (1:500, Abcam), or Streptavidin Alex Fluor 594 conjugate (1:500, Thermo Fisher), were diluted in antibody diluent solution (1% normal donkey serum in 0.3% Triton X-100 in TBS) and incubated overnight at 4°C. Sections were then rinsed three times in TBS and incubated in secondary antibody solution (Alexa 488 donkey anti-chicken IgG; Alexa 594 donkey anti-rabbit IgG, Alexa 647 donkey anti-mouse IgG, Alexa 405 donkey anti-guinea pig IgG, Alexa 647 donkey anti-rabbit IgG, or Alexa 594 donkey anti-mouse IgG all used at 1:500 (Jackson Immuno Research) in antibody diluent at 4°C overnight. Sections were rinsed three times in TBS and mounted with ProLong Gold Mounting medium (Life Technologies).
Confocal imaging and quantification of cell proliferation and tumor burden
Cell quantification within xenografts was performed by a blinded investigator using live counting on a 40x oil immersion objective of a Zeiss LSM700 scanning confocal microscope and Zen 2011 imaging software (Carl Zeiss). For Ki67 analysis, the area for quantification was selected as follows: of a 1 in 6 series of 40-μm coronal sections, 4 consecutive sections were selected. For optogenetic studies, since ~10% of the light reaches 0.5mm through the cortex (roughly midway)52 in each of the four consecutive sections, four 160 × 160-μm field area for quantification were selected in superficial cortical layer 2/3 through layer 5A/B of M2.
For overall tumor burden analysis of SU-DIPGXIII-FL-GluA2DN mice (Fig. 4i–j), the area for quantification was selected as follows: of a 1 in 2 series of 40-μm coronal sections, 6 consecutive sections were selected at approximately 1.1–0.86 mm anterior to bregma; using our stereotactic coordinates for tumor xenograft, these sections are expected to include the tissue most proximal to the site of tumor cell implantation in the coronal plane. Whole tissue slices were imaged simultaneously and imaged for total human nuclei and glioma derived GFP. Confocal microscopy images were analyzed using a custom image processing script written at the Stanford Shriram Cell Science Imaging Facility. To isolate the brain section we use the ImageJ IJ_IsoData threshold algorithm. First, we did a partial local background subtraction using the ImageJ rolling ball background subtraction followed by a Gaussian blur using a sigma of 7. Then, to isolate the foreground pixels, we used the ImageJ Triangle threshold algorithm. Data is presented as mean intensity (total intensity/area of slice).
For overall GFP+ cell outgrowth burden analysis of SU-DIPGXIII-P*-GluA2DN mice (Fig. 4g), both overall human nuclei and GFP were imaged for analysis. %GFP was noted based upon the ratio of overall GFP/HNA signal as a measure of the percentage of glioma cells co-labeled with GFP.
For drug treatment studies, a 1 in 6 series of coronal brain sections were selected with 4 consecutive slices analyzed with respect to overall tumor burden. Within each field, all human nuclear antigen (HNA) and GFP-positive tumor cells were quantified to determine tumor burden within the areas quantified. HNA and GFP-positive tumor cells were then assessed for co-labeling with Ki67. To calculate the proliferation index (the percentage of proliferating tumor cells for each animal), the total number of HNA and GFP-positive cells co-labeled with Ki67 across all areas quantified was divided by the total number of cells counted across all areas quantified (Ki67+/GFP+/HNA+).
To measure cell apoptosis triggered by optogenetic stimulation, cleaved caspase-3 was colocalized with HNA in stimulated and mock-stimulated brain tissue. Tissue was stained in a 1 in 12 series, and three tumor-containing slices were selected for analysis. Tiled confocal images were taken of the entire tumor region in each slice at using a 20x objective, and total counts of caspase-3+/HNA+ cells were counted.
GluA2 editing efficiency
RNA was extracted using TRIZOL based on the manufacturer’s instructions from 3 sets of cell pellets obtained from the same flask. 600 ng of RNA was incubated with 1μl of DNaseI (Maxima First Strand cDNA Synthesis Kit for RT-qPCR) and 1μl of DNaseI buffer brought to a total volume of 10μl at 37°C for 2 minutes and cooled down in ice for 2 minutes. 4μl of 5X Reaction mix and 1μl of Maxima Enzyme mix were added to the DNase-I treated reaction and brought to a total volume of 20μl with addition of nuclease-free water. The tubes were incubated at 25°C for 10 minutes followed by 65°C for 15 minutes and the reaction was terminated by heating at 85°C for 5 minutes. For each reaction, we also ran a reverse transcriptase minus control.
The GluA2 PCR nested primers were based on Kawahara et al.53 The first PCR was done using accuprime Taq HiFi (ThermoFisher Scientific) on 2μl cDNA and 10 μM primers (GluA2-F1: 5’-TTC CTG GTC AGC AGA TTT AGC C-3’; GluA2-R1: 5’- TTC CCT TTG GAC TTC CGC AC-3’) in 50μl total volume. Initial denaturation was done for 2 minutes at 94°C; followed by 37 cycles of denaturation for 30 seconds at 94°C, annealing for 30 seconds at 60°C, and extension for 30 seconds at 68°C. Final extension was done for 2 minutes at 68°C. The nested PCR was done using 2 μl of the first PCR product and primers (GluA2-F1 and GluA2-R2: 5’-TGG GAG ACA CCA TCC TCT CTA CAG-3’) at similar PCR conditions, except annealing was done at 65°C. The correct size bands were cut and gel extraction was done using a Qiagen kit (catalog# 28706). 800 ng of eluted DNA was digested with 2U of BbvI enzyme and 10X Cutsmart buffer in a total volume of 40 μl at 37°C for 12 hours and the reaction was stopped at 65°C for 2 minutes. The digested volume was cleaned up using Zymo DNA cleanup kit and submitted to Agilent Bioanalyzer QC. For GluA2, the editing efficiency was calculated by dividing molarity of 228-bp band (edited) to molarity of 49-bp band (both edited and unedited).
Quantitative polymerase chain reaction
The primers were synthesized from Elim Biopharm based on Kawahara et al.53 RNA was extracted using TRIZOL based on manufacturer’s instructions from 500,000 cells in a 6-well plate with 1ml growth media 24 hours after passaging. The cDNA synthesis was performed exactly as mentioned in GluA2 editing efficiency methods. Both no template control and reverse transcriptase minus control were included. qPCR was performed in a BIO-RAD thermocycler as triplicate. The cDNA was diluted 1:10 with water and 2μl of it was used in the PCR reaction along with 5μl of Maxima SYBR Green/ROX qPCR Master Mix (2X) (Thermo Fisher Scientific, K0222) and 3μM qPCR primers, all brought to a total volume of 10μl. β-actin was used as an endogenous control and data was normalized based on the ΔCt method. Oligos used were as follows: ADAR1 qPCR-F1, GAT GCC TTT GCA GAA CAC CA; ADAR1 qPCR-R1, TGC CTT CTG ATG CTG AGA ACC; β-actin qPCR-F, CGA GGA CTT TGA TTG CAC AT; β-actin qPCR-R, ACA CGA AAG CAA TGC TAT CA.
GluA2-DN cloning
To generate dominant negative GluA2 subunits of AMPA receptor we carried out two amino acid substitutions in the pore region: Methionine to Arginine (M/R: 606), and Glutamine to Arginine (Q/R: 607); AMPA receptors that incorporate this subunit are non-conducting54. We obtained the CAG-EGFP-GluA2DN construct generated by Amit Agarwal and Dwight Bergles and cloned the EGFP-GluA2DN region from the original CAG-EGFP-GluA2DN (pAA018) into a lentiviral system. We introduced Mlu-I with Kozak sequence (5’TAA GCA ACG CGT GCC ACC ATG GTG AGC AAG GGC GAG-3’) and Xma-I restriction sites (5’-TGC TTA CCC GGG CTA AAT TTT AAC ACT CTC-3’) on 5’ EGFP end and 3’ GluA2DN respectively by PCR. 1μg of pLV-EF1a-IRES-Hygro (addgene#85134) was digested with MluI-HF & XmaI in NEB cutsmart buffer overnight at 37°C to remove the IRES-HYGRO region. The digested product was run in 1% agarose gel and the correct digested band was gel purified using Qiagen gel extraction kit. At a 20ng/μl concentration, the digested vector was treated with FAST Alkaline Phosphatase (FERMENTAS) at 37°C for 10 minutes and deactivated at 75°C for 5 minutes. 300 ng of the PCR product with restriction sites were digested with MluI-HF and XmaI for 30 minutes and column purified using Zymo DNA Clean & Concentrator™−5 D4013. The vector and insert were ligated at 5:1 insert DNA: vector DNA using T4 DNA ligase (EL0011; Fisher Scientific) for 1 hour at room temperature. The cells were transformed in homemade Stbl3 cells overnight and the colonies were picked the next day and sent for Sanger sequencing to ElimBio.
GluA2-WT cloning
We introduced SpeI (5’-TAAGCAactagtATGCAAAAGATTATGCAT-3’) and XmaI (5’-TGCTTAcccgggCTAAATTTTAACACTTTCGAT-3’) restriction sites in full length human GRIA2 clone (GluA2 WT) obtained from Origene (RC212599) by PCR. 1μg of pLV-EF1a-eGFP-GluA2DN was digested with SpeI-HF & XmaI in NEB cutsmart buffer overnight at 37°C to remove the GluA2DN region. The digested product was run in 1% agarose gel and the correct digested band was gel purified using Qiagen gel extraction kit. At a 20ng/μl concentration, the digested vector was treated with FAST Alkaline Phosphatase (FERMENTAS) at 37°C for 10 minutes and deactivated at 75°C for 5 minutes. 300 ng of the PCR product with restriction sites were digested with SpeI-HF and XmaI for 30 minutes and column purified using Zymo DNA Clean & Concentrator™−5 D4013.The vector and insert were ligated and cells were transformed as described above.
CellTiter-Glo assay
To assess overall cell number, 5000 SU-DIPGXIII-FL-mockGFP or SU-DIPGXIII-FL-GluA2DN glioma cells were seeded in growth media in a 96-well plate. After 24, 48, or 72 hours, CellTiter-Glo reagent (Promega) was added at a 1:1 ratio. Luminescence was measured after a 10-minute incubation at room temperature to stabilize signal.
Three-dimensional (3D) invasion and migration assays
3D invasion assays were performed as previously described55 with some modifications. Briefly, in ULA 96-well round-bottomed plates (Costar, Corning Inc.), 50μl of Matrigel Basement Membrane Matrix (Corning Life Sciences) was added to each well containing neurospheres of ~200μm in diameter in 50μl of media. The plates were incubated at 37°C, 5% CO2, 95% humidity for 1hr before addition of 50μl/well of culture medium. Images were obtained using the Cytation 5 Cell Imaging Multi-Mode reader (Biotek) at time zero and 72 hours post Matrigel encapsulation. Image analysis was carried out using ImageJ by measuring the diameter of the invasive area. The degree of cell invasion into the Matrigel was measured for 6 replicate wells normalizing to each initial spheroid diameter at time-zero and the data plotted as a mean ratio for 3 biological repeats. 3D migration assays were similarly performed as previously described56 with some modifications. Briefly, flat-bottomed 96-well plates (Falcon) were coated with 2.5μg/50μl laminin/well (Thermo Fisher) in sterile H2O. Once coating was complete, a total of 200 μl/well of culture medium was added to each well. A total of 100μl medium was removed from ULA 96-well round-bottomed plates containing neurospheres of ~200μm in diameter and the remaining medium including the neurosphere were transferred into the pre-coated plates. Images were obtained using the Cytation 5 Cell Imaging Multi-Mode reader (Biotek) at time zero and 72 hours post matrigel encapsulation. Image analysis was carried out using ImageJ by measuring the diameter of the invasive area. The degree of cell migration on the laminin was measured for 6 replicate wells normalizing to each spheroid time zero diameter and the data plotted as a mean ratio for 3 biological repeats.
Neuron:glioma co-culture EdU incorporation assay
For EdU incorporation assays, coverslips were prepared as described above. Again, at DIV 5, glioma cells were added to the neuron cultures. 48 hours after addition of glioma cells, slides were treated with 10μM EdU with or without NBQX (10μM; Tocris). Cells were fixed after and additional 24 hours using 4% PFA and stained using the Click-iT EdU kit and protocol (Invitrogen). Proliferation index was then determined by quantifying percentage of EdU labelled glioma cells (identified by EdU+/nestin+) over total number of glioma cells using confocal microscopy.
Bioluminescence imaging
For in vivo monitoring of tumor growth, bioluminescence imaging was performed using an IVIS imaging system (Xenogen). Mice orthotopically xenografted with luciferase-expressing glioma cells were placed under isofluorane anaesthesia and injected with luciferin substrate. Animals were imaged at baseline and randomized based on tumor size by a blinded investigator so that experimental groups contained an equivalent range of tumor sizes. All total flux values were then normalized to baseline values to determine fold change of tumor growth.
Mouse drug treatment studies
For all drug studies, NSG mice were xenografted as above with either SU-DIPGVI or SU-DIPGXIII-FL cells. Four weeks post-xenograft, SU-DIPGXIII-FL-bearing animals were treated with systemic administration of meclofenamate sodium (20mg/kg; Selleck Chemicals; formulated in 10% DMSO in PBS) via intraperitoneal injection for two weeks (5 days/week). Similarly, four weeks post-xenograft, SU-DIPGVI-bearing animals were treated with oral administration of AMPA receptor blocker, perampanel (0.75 mg/kg; Adooq Biosciences; formulated in 10% DMSO, 60% PEG300, 30% water) via oral gavage for three weeks (5 days/week). For both studies, controls were treated with an identical volume of the relevant vehicle. Drug treatment began four weeks after xenografting and continued through week six or seven. Bioluminescence imaging was performed before treatment and every 7 days thereafter using an IVIS imaging system (Xenogen) under isoflurane anaesthesia. Tumor burden was assessed as fold change in total flux from the beginning to end of treatment.
Survival Studies
For survival studies, morbidity criteria used were either: reduction of weight by 15% initial weight, or severe neurological motor deficits consistent with brainstem dysfunction (i.e. hemiplegia or an incessant stereotyped circling behavior seen with ventral midbrain dysfunction). Kaplan-Meier survival analysis using log rank testing was performed to determine statistical significance.
ECoG Electrode Localization and Data Analyses
Intra-operative photographs with and without subdural electrodes present were used to localize each electrode contact. Images were registered using landmarks from gyral anatomy and vascular arrangement to preoperative T1- and T2-weighted MRI scans. Tumor boundaries were localized on MRI scans and electrodes within 10 mm of necrotic tumor core tissue were identified as ‘tumor’ contacts. Electrodes overlying the hypointense core of the tumor were identified as “core”, electrodes extending from the contrast enhancing rim extending to the edge of FLAIR were considered “infiltrative margin”, and electrodes completely outside of any T1 post gadolinium or FLAIR signal were considered “healthy”.
Electrocorticographic (ECoG) signals were acquired during a three-minute resting-state period after stopping the administration of anesthetics and the patient was judged to be alert and awake. Post-operative videos were re-analyzed to ensure all data was collected in true resting state. Recordings were acquired at 4800 Hz and down-sampled to 1200 Hz during the initial stages of processing. Channels with excessive noise artifacts were visually identified and removed. Following the rejection of artifactual channels, data were referenced to a common average, high-pass filtered at 0.1 Hz to remove slow drift artifacts, and band-pass filtered between 70–110 Hz using a 300-Order FIR filter to focus the analyses on the high-gamma band range, which is strongly related to local mean population spiking rates. High-gamma band power (HGp) was then calculated using the square of the Hilbert transform on the filtered data. HGp was then averaged across the resting-state time-series, yielding a single measure of neural responsivity for each electrode contact. HGp levels were then compared between tumor and non-tumor channels using independent samples t-tests separately for each patient.
Statistical analyses
Statistical tests were conducted using Prism (GraphPad) software unless otherwise indicated. Gaussian distribution was confirmed by the Shapiro–Wilk normality test. For parametric data, unpaired two-tailed Student’s t-tests or one-way ANOVA with Tukey’s post hoc tests to further examine pairwise differences were used as indicated. Paired two-tailed Student’s t-tests were used in the case of same cell experiments (as in electrophysiological recordings). For non-parametric data, a two-sided unpaired Mann–Whitney test was used as indicated or one-tailed Wilcoxon matched-pairs signed rank test was used in the case of same-cell experiments. Two-tailed log rank analyses were used to analyze statistical significance of Kaplan Meier survival curves. For I/O curve comparison, non-linear regression fits were compared using the extra-sum-of-squares F-test to compare separate vs. individual best-fit models between datasets. A level of P < 0.05 was used to designate significant differences. On the basis of the variance of xenograft growth in control mice, we used at least 3 mice per genotype to give 80% power to detect an effect size of 20% with a significance level of 0.05. For all animal experiments, the number of independent mice used is listed in figure legend.
Data Availability
All data is available in the article, source data, or from the corresponding author upon reasonable request. Custom code created is available on GitHub. RNA sequencing of single-cell patient derived xenografts is available on GEO (GSE134269).
Extended Data
Extended Data Figure 1: Synaptic gene expression in single cell primary glioma and patient-derived xenografts.
a, Primary human biopsy single cell transcriptomic data illustrating synapse associated and ion-channel gene expression in H3K27M+ diffuse midline glioma (DMG; grey, n=2,259 cells/6 subjects), IDHwt adult high-grade glioma (red, n=599 cells/3 subjects), IDHmut adult high-grade glioma (purple, n=5,096 cells/10 subjects) malignant cells, and tumor-associated, non-malignant immune cells (green; n=96 cells/5 subjects) and oligodendrocytes (yellow; n=232 cells). b, As in (a) for single cell transcriptomic analysis of H3K27M+ DMG xenograft models (SU-DIPVI, blue and SU-DIPGXIII-FL, yellow) illustrating broad synaptic gene expression similar to that found in primary DMG tissue samples as shown in Fig. 1a. For each individual violin plot, y-axis represents log2 TPM (transcripts per million), and x-axis represents number of individual cells with indicated expression value.
Extended Data Figure 2: Synaptic gene expression and structural synapses in glioma.
a, Plot of the lineage (x-axis) and stemness (y-axis) scores for H3K27M+ DMG malignant single cells (dots) sorted from patient-derived glioma xenograft models (SU-DIPG-VI and SU-DIPG-XIII-FL). Overlay of grey-red enrichment score indicates relative score for OPC-like genes (left) and synapse-related genes (right; n=335/4 cells/mice; pearson correlation rho=0.57, P<0.0001). b, Plot of the lineage vs. stemness scores for H3K27M+ DMG malignant single cells in (a), colored by xenograft model. Red=SU-DIPGXIII-FL, blue=SU-DIPGVI. Though each xenograft model clearly contains cells across all lineages, SU-DIPGXIII-FL has a prominent astrocyte-like cell population, while SU-DIPGVI has a prominent OPC-like population. c, Unbiased principal component analysis of single cell gene expression from an individual patient primary biopsy sample of H3K27M+ glioma (BCH869) reveals cellular clusters that resemble proliferating cells (cell-cycle), OPCs (OPC-like), astrocytes (AC-like) and mixed astroglial/oligodendroglial cells (AC/OC). Synaptic gene enrichment for individual cells shown below and co-localizes in the OPC-like cluster. d, Electron microscopy of primary adult glioblastoma tumor sample illustrating clear synaptic structure. Scale bar, 500μm. e, Original (non-pseudo-colored) immuno-EM images shown in Fig.1c. f, Additional examples of neuron:glioma synapses identified by immuno-electron microscopy in patient-derived xenografts of SU-DIPGVI and SU-DIPGXIII-FL (n=3 mice/group). Arrowheads indicate immuno-gold particle labeling of GFP. Scale bar, 200μm. g, Quantification of neuron:glioma synaptic structures in SU-DIPGVI (n=101 sections) and SU-DIPGXIII-FL (n=104 sections) xenografts. For each individual violin plot, y-axis represents number of identified unambiguous neuron:glioma synapses in each section, x-axis represents number of individual cells with indicated value. h, Quantification of colocalized post-synaptic glioma-derived PSD95-RFP with pre-synaptic synapsin in co-cultures of SU-DIPGXIII-FL glioma cells with WT or Nlgn3y/− neurons (NL3KO; n=21 cells in 10 coverslips/group). Data shown as mean ± s.e.m. P-values determined by two-sided Mann-Whitney test. **** P < 0.0001.
Extended Data Figure 3: Mitogenic effects of NLGN3 are independent from AMPAR signaling and properties of glioma AMPARs.
a, Proliferative response of GFP (control) and GluA2-dominant negative subunit expressing glioma cells (GluA2-DN) after 24-hour exposure to soluble extracellular neuroligin-3 (NLGN3; 100nm) in the presence and absence of AMPA-receptor blocker, NBQX (10μM). b, Western blot analysis of phospho-AKT (Ser473) and total AKT in GFP (control) glioma cells in response to 5-minute exposure of soluble extracellular neuroligin-3 (NLGN3; 100nm) in the presence and absence of AMPA-receptor blocker, NBQX (10μM); left. Quantitative analysis of the ratio of pAKT/AKT normalized to vehicle (right). c, Western blot analysis of phospho-AKT (Ser473) and total AKT in GluA2-DN expressing glioma cells in response to 5-minute exposure of soluble extracellular neuroligin-3 (NLGN3; 100nm; left). Quantitative analysis of the ratio of pAKT/AKT normalized to vehicle (right). d, Time course of evoked glioma cell EPSC block by NASPM (100 μM, duration=red bar (n=7/5 cells/mice; left); Representative trace before (black) and after (red) addition of NASPM (right). e, Quantification of (d). f, GluA2 subunit Q/R editing efficiency in SU-DIPGXIII-FL and SU-DIPGVI cells as measured by PCR and expressed as % edited. g, Expression of ADAR1, the enzyme responsible for Q/R editing of GluA2 mRNA. Plot illustrates ADAR1 enzyme mRNA expression relative to beta-actin as measured by qPCR. Analyses in a,b,c,f,g were calculated from three independent sets of cells. Data shown as mean ± s.e.m. P values determined by one-way ANOVA with Tukey’s post-hoc analysis (a,b), by two-tailed Student’s t-test (c), by two-tailed paired Student’s t-test (e). All data shown as mean ± s.e.m. *P<0.01, **P<0.001, ***P<0.001, ****P<0.0001, NS = not significant.
Extended Data Figure 4: Glioma xenograft calcium imaging with GCaMP6.
a, Confocal micrographs of xenografted SU-DIPGXIII-FL cells expressing GCaMP6s-tdTomato into the hippocampus stained with DAPI (blue), human nuclear antigen (HNA, green), and TdTomato nuclear tag (red). Merged image shown on the right illustrates specificity of Tdtomato tag to HNA+ cells. Scale bar = 20μm for all images. Immunostaining independently replicated in three mice. b, Spontaneous calcium transients in SU-DIPGVI xenograft visualized by two-photon in situ calcium imaging. Representative frames shown with red = glioma td-Tomato nuclear tag; green = GCaMP6s in glioma cells. Scale bar = 50μm; n=5 mice. See also Supplementary Video 1. c, Trace of normalized GCaMP6s intensity over time in an individual xenografted glioma (SU-DIPGVI) cell exhibiting an oscillatory spontaneous transient. Data plotted as ρF/Fo. Results were replicated across n=3 mice. d, As in (c), trace of normalized GCaMP6s intensity over time in an individual xenografted glioma (SU-DIPGXIII-FL) cell exhibiting a less regular spontaneous transient. This type of transient is more frequently observed in glioma xenografts. Data plotted as ρF/Fo. Results were replicated across n=3 mice. e, Individual xenografted glioma (SU-DIPGXIII-FL) cellular responses to axonal stimulation before and after application of tetrodotoxin (0.5μM) as measured by GCaMP6s intensity. Data plotted as ρFmax/Fo; n=40/4 cells/mice. P-values determined by one-tailed Wilcoxon matched-pairs signed rank test. **** P < 0.0001.
Extended Data Figure 5: Prolonged glioma currents and pediatric glioma tumor microtubes.
a, Time course of prolonged current block by NASPM (100μM; duration=red bar; n=8/5 cells/mice. Data are means ± s.e.m. (left); Representative traces of evoked prolonged current (block) unaffected by NASPM (red). b, Quantification of (a). P values determined by two-tailed paired Student’s t-test. NS = not significant. c, Alignment of phase-locked simultaneous recording of glioma prolonged potential with the field potential of firing neuronal population. d, Representative prolonged current traces with increasing stimulation intensity. Maximum intensity, red; intermediate intensities, blue and green (inset). Zoomed-in view illustrates distinct spike-like waveforms consistent with response to neuronal population firing. e, Relationship of extracellular field potential to magnitude of prolonged current (SU-DIPGXIII-FL xenograft) illustrated by simultaneous field potential (fEPSP) and whole-cell glioma current-clamp recordings. f, Prolonged glioma potential amplitudes vs. slope of fEPSPs elicited by electrical stimulation (10, 20, 30, 50, 70, 100 and 150 μA; R2= 0.92; n=14/4 cells:fields/mice for each except n=11/3 cells:fields/mice for 30 μA). g, Representative trace of potassium (K+)-induced prolonged current in SU-DIPGXIII-FL xenografts (n=9/2 cells/mice). h, Effect of TBOA on prolonged current in glioma (SU-DIPGXIII-FL). (1) Representative trace of residual current left after application with D-AP5 + NBQX in prolonged response to stimulation (top). D-AP5 + NBQX likely reduces the prolonged current due to the effect on CA1 pyramidal neurons, not through direct effect on the glioma cells themselves; (2) Representative trace of residual current after trace 1 was then treated with glutamate transporter blocker, 200μM TBOA (middle); (1 minus 2) Subtraction of trace 2 from trace 1 reveals 2pA current, that can be accounted for by TBOA. It should be noted that a small residual current remains (bottom; n=5/2 cells/mice). i, Synchronicity analysis of calcium peaks in glioma cells in Figure 3 shown over the course of 10 minutes. Red lines indicate cells synchronized with one another at various timepoints during indicated period. j-o, Confocal micrographs of primary human glioma tissue samples illustrate density and length of nestin immunopositive tumor microtubes. j-n, primary human tissue samples of pediatric H3K27M+ diffuse midline gliomas of the pons (also known as diffuse intrinsic pontine glioma, DIPG), sampled at the time of autopsy. o, Analysis in a primary tissue sample from an adult glioblastoma (SU-GBM092) illustrates similarity to adult glioma. For all images, blue, DAPI; yellow, nestin; magenta, H3K27M (tumor-specific antigen). Scale bar=20μm, except in (n), scale bar is 10μm. Data independently replicated in three sections per sample.
Extended Data Figure 6: Prolonged currents in pediatric glioma amplified by gap-junction coupling.
a, Input resistance of non-responding and depolarizing cells (n=29 fast EPSC, n=247 prolonged current, n=319 no response). b, Biocytin dye filling illustrates coupling of xenografted glioma cells that exhibit prolonged currents (n=7/2 slices/mice); Red = streptavidin-biocytin; green = GFP. Scale bar, 100μm. c, Prolonged currents in glioma largely blocked by carbenoxolone (CBX, 100μM). Red box highlights zoomed in view of representative trace to illustrate residual slow current after application of CBX. d, Time course of prolonged current in glioma cells (black) and pyramidal cell EPSC (blue) responses to addition of CBX with subsequent washout of inhibitor (duration=red bar; n=6/6 glioma cells/mice; n=5/2, neurons/mice). e, Bar graph with additional data quantifies the drop in current amplitude after addition of CBX (n=19/11 cells/mice; data also shown in Fig.3l). f, Input resistance of cells in (d) in response to CBX. g, Quantification of (f) (n=19/11 cells/mice). h, Time course of prolonged current in glioma in response to addition of meclofenamate (100μM; duration=red bar; n=8/3 cells/mice). i, Quantification of (h); data also shown in Fig.3l; left. Representative traces illustrating block of prolonged current amplitude after addition of meclofenamate (red, right). j, Input resistance of cells in (h) in response to addition of meclofenamate (n=8/3 cells/mice). k, Quantification of (j). l, Representative frames from two photon calcium imaging of in situ SU-DIPGXIII-FL glioma xenografts illustrating spontaneous transients before (top) or after addition of CBX (bottom). Red = tdTomato nuclear tag in glioma cells; green = GCaMP6s. Scale bar, 50μm; Results replicated across 3 mice. m, Synchronicity analyses of spontaneous calcium transients before (top) or after addition of CBX (bottom). n, Representative trace of GCaMP6s peak intensity over time (s) in a single cell before (black) and after addition of CBX (red). Data plotted as ρF/Fo; n=3 mice. o, Synchronicity scores of individual cells within glioma xenograft before and after addition of CBX (n=164 cells across 3 mice). y-axis represents synchronicity score, x-axis represents number of cells with specific score. For a, d, e, f, g, h, i, j, and k, data are shown as means ± s.e.m. P-values determined by one-way ANOVA for (a), and by one-tailed Wilcoxon matched-pairs signed rank test. for (e,g,i,k,o). **P<0.01. ****P<0.0001. NS indicates P > 0.05.
Extended Data Figure 7: Heterogeneity of glioma cell electrophysiological response and validation of ChR2 and DN-GluA2 function in glioma.
a, Electrophysiological responses by model. Number of whole cell patch clamp recordings from cells in xenografted hippocampal slices separated by electrophysiological response to local electrical stimulation. b, Demonstration of depolarizing inward current in SU-DIPXIII-FL-ChR2 cells in response to single stimulation and 20Hz pulses of blue light as measured in current clamp (top) and voltage clamp (bottom). c, Proliferation index of xenografted SU-DIPGXIII-FL-YFP control glioma cells (no opsin expressed) in response to blue light stimulation or mock stimulation as measured by the proportion of GFP+/HNA+ cells expressing Ki67 24-hours after five optogenetic stimulation sessions (n=3 mice, mock stim; n=4 mice, stim). d, Quantification of cleaved caspase-3 in xenografted SU-DIPGXIII-FL-YFP control glioma cells in response to blue light stimulation or mock stimulation as measured by total number of HNA+ cells co-labeled with cleaved caspase-3 (n=3 mice/group). e, As in (d), quantification of cleaved caspase-3 in xenografted SU-DIPXIII-FL-ChR2 glioma cells (n=3 mice, mock stim; n=4 mice, stim). f, Validation of GluA2-dominative negative AMPA receptor subunit expressing construct. Representative traces of whole-cell voltage-clamp recording of WT (black) and GluA2-DN expressing (grey) SU-DIPGVI cells in response to 500μM (S)-AMPA (n=6 cells). g, Representative traces of whole-cell voltage-clamp recording in WT (black) and GluA2-DN expressing (grey) SU-DIPGXIII-FL cells in response to 500μM (S)-AMPA (n=6 cells). SU-DIPGXIII-FL cells are unable to homogeneously express the dominant construct, and therefore may be connected to WT GluA2 expressing cells, which accounts for the remaining current in the illustrated trace. Incorporation of the GluA2-DN construct thus results in a significantly abrogated AMPAR-dependent depolarization. Data shown as mean ± s.e.m for (c,d,e). All P-values determined by two-tailed Student’s t-test. NS = not significant.
Extended Data Figure 8: Glioma AMPAR function in vitro, in co-culture and in vivo.
a, Kaplan-Meier survival curves of second cohort of mice orthotopically xenografted with control GFP-only or GluA2-DN-GFP over-expressing cells (SU-DIPGXIII-P* xenograft model; n=5 mice per group). b, Representative coronal sections of mouse brains bearing SU-DIPGXIII-FL xenografts either expressing control GFP construct (left) or GluA2-DN-GFP construct; right). Gray, MBP; White, glioma-GFP. c, Proliferation indices of SU-DIPGXIII-FL cells at baseline in neuronal medium, in response to 10μM NBQX, in co-culture with neurons, or in co-culture with neurons in the presence of 10μM NBQX (n=3 biological replicates/group, except n=4 for baseline). d, Representative images of neuron-glioma co-cultures in the presence and absence of NBQX. Green = neurofilament (neuronal processes); Red = nestin (glioma cell processes); White = Ki67. Scale bar = 50μm. e, in vitro growth analysis of control GFP or GluA2-DN-GFP cells monitored over 3 days. f, in vitro apoptosis analysis of control GFP or GluA2-DN-GFP as measured by % of total cells co-stained with cleaved-caspase. g, 3D Matrigel invasion assay in WT (GFP) and GluA2-DN (GluA2-DN-GFP) expressing SU-DIPGXIII-FL cells 72 hours after seeding. h, Representative images of (g) at time 0 hr (left) and 72 hr (right) in control GFP-expressing (top) and GluA2-DN-GFP expressing cells (bottom). Scale bar = 1000μm. i, 3D migration assay in WT (GFP) and GluA2-DN (GluA2-DN-GFP) expressing SU-DIPGXIII-FL cells 72 hours after seeding. j, Representative images of (i) at time 0 hr (left) and 72 hr (right) in control GFP-expressing (top) and GluA2-DN-GFP expressing cells (bottom). Scale bar =1000μm. k, Representative confocal micrographs illustrating proliferating SU-DIPGVI cells in vehicle or perampanel-treated mice (n=8 mice/group). Red = human nuclei; white = Ki67. Scale bar = 50μm. l, IVIS bioluminescence analysis of overall tumor growth in SU-DIPGXIII-FL xenografts treated with vehicle or meclofenamate over a two-week period. Data represented as fold change in total flux; n=5 mice/group. Data shown as mean ± s.e.m. for (c,e,f,g,i,l). For analyses in (d-j), n=3 biological replicates. P-values determined by two-tailed log rank analyses (a), by one-way ANOVA with post-hoc analysis (c), by two-tailed unpaired Student’s t-test (f,g,i,l). *P<0.05, **P< 0.01,****P<0.0001. NS = not significant.
Extended Data Figure 9: Hyperexcitability in the glioma microenvironment and working model of neuron-glioma interactions in the tumor microenvironment.
a, Individual channel electrocorticography signals (mean high-gamma frequency-filtered power (μV2)) in each of healthy-appearing, tumor core, and tumor-infiltrated brain across three patients (n=23, n=29, n=51 total channels, respectively). b, Working model of glioma integration in neural circuity, with hyperexcitability of neurons (grey) exacerbating activity-dependent mechanisms of glioma (green) growth. 1, Neuron-to-glioma synapses, synaptic vesicles = red; AMPA receptors = grey. 2, Inward potassium (K+) current = blue, potassium channel = grey 3, Gap junction (white) coupling in glioma amplifies current.
Extended Data Table 1.
Characteristics of patient-derived high-grade glioma cell cultures / xenografts
| Culture ID | Tumor type, location and grade | Age at diagnosis (years) | Patient sex | Histone-3 mutational status | Other genomic characteristics | Timepoint tissue obtained | Prior therapy | Survival (months) |
|---|---|---|---|---|---|---|---|---|
| SU-pcGBM2 | Pediatric cortical glioblastoma; frontal lobe; WHO grade IV | 15 | M | WT | P53 mutated EGFR amplified | Biopsy at diagnosis | none | N/A |
| SU-DIPGVI | DIPG; pons; WHO grade IV | 7 | F | H3.3K27M | N/A | Early postmortem autopsy | XRT; vorinostat | 6 |
| SU-DIPGXIII-P* | DIPG; pons; WHO grade IV | 6 | F | H3.3K27M | N/A | Early postmortem autopsy | XRT | 4 |
| SU-DIPGXIII-FL | DIPG; frontal lobe met; WHO grade IV | 6 | F | H3.3K27M | N/A | Early postmortem autopsy | untreated | 4 |
| SU-DIPG25 | DIPG; pons; WHO grade IV | 5 (at death) | F | H3.3K27M | N/A | Early postmortem autopsy | XRT | 11 |
| SU-DIPG31 | DIPG; pons; WHO grade IV | 18 | M | H3.3K27M | N/A | Early postmortem autopsy | XRT; etoposide | N/A |
| SU-DIPG32 | DIPG; pons; WHO grade IV | 5 | M | H3.1K27M | N/A | Early postmortem autopsy | XRT; avastin | N/A |
| SU-DIPG33 | DIPG; pons; WHO grade IV | 8 | M | H3.1K27M | N/A | Early postmortem autopsy | XRT | N/A |
| SU-DIPG39 | DIPG; pons; WHO grade IV | 5 (at death) | M | H3.3K27M | N/A | Early postmortem autopsy | XRT | 6 |
| SU-GBM092 | GBM; hemispheric; WHO grade IV | 47 | M | N/A | N/A | Early postmortem autopsy | Resection; XRT; TMZ | 6 |
| SF0047 (human subject 1) | GBM; left temporal lobe; WHO grade IV | 57 | M | N/A | Neg IDH1 R132H; MGMT unmethylate d | Resection at diagnosis | none | N/A |
| SF0109 (human subject 2) | GBM; left parietal lobe; WHO grade IV | 63 | M | N/A | Neg IDH1 R132H | Resection at diagnosis | none | N/A |
| SF0144 (human subject 3) | HGG; left temporal lobe; WHO grade III | 74 | M | N/A | Neg IDH1 R132H; ATRX WT | Resection at diagnosis | none | N/A |
WHO = World Health Organization; DIPG = diffuse intrinsic pontine glioma; XRT = radiotherapy; TMZ = temozolomide. Note that SU-DIPGXIII-P* and SU-DIPGXIII-FL were tissue samples acquired from the same patient at autopsy; in this case the pontine tumor (P) received focal radiotherapy, whereas the frontal lobe (FL) tumor was an untreated metastasis from the primary pontine tumor to the frontal lobe.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge support from the National Institutes of Health Director’s Common Fund (DP1 NS111132 to M.M.) National Institute of Neurological Disorders and Stroke (R01 NS092597 to M.M., K08 NS110919 to S.H.J.), National Cancer Institute (F31 CA200273 to H.S.V.), National Institutes of Mental Health (P50 MH086403 to R.C.M.), Michael Mosier Defeat DIPG Foundation (to M.M.), ChadTough Foundation (to M.M.), V Foundation (to M.M.), Department of Defense (NF140075 to M.M.), McKenna Claire Foundation (to M.M.), Alex’s Lemonade Stand Foundation (to M.M.), The Cure Starts Now Foundation and DIPG Collaborative (to M.M.), Unravel Pediatric Cancer (to M.M.), N8 Foundation (to M.M.), Abbie’s Army Foundation (to M.M.), Brantley’s Project supported by Ian’s Friends Foundation (to M.M.), Waxman Family Research Fund (to M.M.), Joey Fabus Childhood Cancer Foundation (to M.M.), Virginia and D.K. Ludwig Fund for Cancer Research (to M.M.), Bio-X Institute (to L.T.T. and A.C.G.), Maternal and Child Health Research Institute at Stanford (to M.M., A.C.G and H.S.V.), Anne T. and Robert M. Bass Endowed Faculty Scholarship in Pediatric Cancer and Blood Diseases (to M.M.), Dr. Mildred Scheel Cancer Foundation (57406718 to M.A.), Damon Runyan Foundation (to K.R.T.), Sontag Foundation Distinguished Scientist Award (to M.L.S.), Howard Hughes Medical Institute (to A.R.), Klarman Cell Observatory (to A.R.), Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to D.E.B.), Chica and Heinz Schaller Research Foundation (to A.A.), Deutsche Forschungsgemeinschaft (AG 287/1-1 to A.A.), The Robert Wood Johnson Foundation (74259 RWJF to S.H.J.)
We thank Sigrid Knemeyer at SciStories for illustrations, Andrew Olsen at the Stanford Neuroscience Imaging Core (supported by National Institutes of Neurological Disorders and Stroke grant NS069375), Sofia Kakaizada from the Hervey-Jumper research group and Marni Shore from the Suva laboratory.
Footnotes
The authors declare the following competing interests: MM is an SAB member of Cygnal Therapeutics. AR is a founder and equity holder of Celsius Therapeutics and an SAB member of ThermoFisher Scientific and Syros Pharmaceuticals.
Supplementary Information, including Supplementary Videos 1–3 and Extended Data Table 1 is available in the online version of the paper
References
- 1.Venkatesh HS et al. Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion. Cell 161, 803–816, doi: 10.1016/j.cell.2015.04.012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkatesh HS et al. Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature 549, 533–537, doi: 10.1038/nature24014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergles DE, Roberts JD, Somogyi P & Jahr CE Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature 405, 187–191, doi: 10.1038/35012083 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Karadottir R, Cavelier P, Bergersen L & Attwell D NMDA receptors are expressed in oligodendrocytes and activated in ischaemia. Nature 438, 1162–1166, doi: 10.1038/nature04302 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LoTurco JJ, Owens DF, Heath MJ, Davis MB & Kriegstein AR GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron 15, 1287–1298 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Luk KC & Sadikot AF Glutamate and regulation of proliferation in the developing mammalian telencephalon. Dev Neurosci 26, 218–228, doi: 10.1159/000082139 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Wang Q, Haydar TF & Bordey A Nonsynaptic GABA signaling in postnatal subventricular zone controls proliferation of GFAP-expressing progenitors. Nat Neurosci 8, 1179–1187, doi: 10.1038/nn1522 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deisseroth K et al. Excitation-neurogenesis coupling in adult neural stem/progenitor cells. Neuron 42, 535–552, doi:S0896627304002661 [pii] (2004). [DOI] [PubMed] [Google Scholar]
- 9.Kougioumtzidou E et al. Signalling through AMPA receptors on oligodendrocyte precursors promotes myelination by enhancing oligodendrocyte survival. Elife 6, doi: 10.7554/eLife.28080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filbin MG et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360, 331–335, doi: 10.1126/science.aao4750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venteicher AS et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, doi: 10.1126/science.aai8478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sommer B, Kohler M, Sprengel R & Seeburg PH RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67, 11–19 (1991). [DOI] [PubMed] [Google Scholar]
- 13.Hollmann M, Hartley M & Heinemann S Ca2+ permeability of KA-AMPA--gated glutamate receptor channels depends on subunit composition. Science 252, 851–853 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Oliet SH, Malenka RC & Nicoll RA Bidirectional control of quantal size by synaptic activity in the hippocampus. Science 271, 1294–1297 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Bergles DE & Jahr CE Synaptic activation of glutamate transporters in hippocampal astrocytes. Neuron 19, 1297–1308 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Luscher C, Malenka RC & Nicoll RA Monitoring glutamate release during LTP with glial transporter currents. Neuron 21, 435–441 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Sibille J, Pannasch U & Rouach N Astroglial potassium clearance contributes to short-term plasticity of synaptically evoked currents at the tripartite synapse. J Physiol 592, 87–102, doi: 10.1113/jphysiol.2013.261735 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKhann GM, 2nd, D’Ambrosio, R. & Janigro, D. Heterogeneity of astrocyte resting membrane potentials and intercellular coupling revealed by whole-cell and gramicidin-perforated patch recordings from cultured neocortical and hippocampal slice astrocytes. J Neurosci 17, 6850–6863 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osswald M et al. Brain tumour cells interconnect to a functional and resistant network. Nature 528, 93–98, doi: 10.1038/nature16071 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Labrakakis C, Patt S, Hartmann J & Kettenmann H Glutamate receptor activation can trigger electrical activity in human glioma cells. Eur J Neurosci 10, 2153–2162 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Singh SK et al. Identification of human brain tumour initiating cells. Nature 432, 396–401, doi:nature03128 [pii] 10.1038/nature03128 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Ishiuchi S et al. Ca2+-permeable AMPA receptors regulate growth of human glioblastoma via Akt activation. J Neurosci 27, 7987–8001, doi: 10.1523/JNEUROSCI.2180-07.2007 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sontheimer H A role for glutamate in growth and invasion of primary brain tumors. J Neurochem 105, 287–295, doi: 10.1111/j.1471-4159.2008.05301.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyons SA, Chung WJ, Weaver AK, Ogunrinu T & Sontheimer H Autocrine glutamate signaling promotes glioma cell invasion. Cancer Res 67, 9463–9471, doi: 10.1158/0008-5472.CAN-07-2034 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Q et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 533, 493–498, doi: 10.1038/nature18268 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell SL, Buckingham SC & Sontheimer H Human glioma cells induce hyperexcitability in cortical networks. Epilepsia 53, 1360–1370, doi: 10.1111/j.1528-1167.2012.03557.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.John Lin CC et al. Identification of diverse astrocyte populations and their malignant analogs. Nat Neurosci 20, 396–405, doi: 10.1038/nn.4493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckingham SC et al. Glutamate release by primary brain tumors induces epileptic activity. Nat Med 17, 1269–1274, doi: 10.1038/nm.2453 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ray S, Crone NE, Niebur E, Franaszczuk PJ & Hsiao SS Neural correlates of high-gamma oscillations (60–200 Hz) in macaque local field potentials and their potential implications in electrocorticography. J Neurosci 28, 11526–11536, doi: 10.1523/JNEUROSCI.2848-08.2008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yizhar O et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477, 171–178, doi: 10.1038/nature10360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodgkin AL & Katz B The effect of sodium ions on the electrical activity of giant axon of the squid. J Physiol 108, 37–77 (1949). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ransom BR & Goldring S Ionic determinants of membrane potential of cells presumed to be glia in cerebral cortex of cat. J Neurophysiol 36, 855–868, doi: 10.1152/jn.1973.36.5.855 (1973). [DOI] [PubMed] [Google Scholar]
- 33.Bittman KS & LoTurco JJ Differential regulation of connexin 26 and 43 in murine neocortical precursors. Cereb Cortex 9, 188–195 (1999). [DOI] [PubMed] [Google Scholar]
- 34.LoTurco JJ, Blanton MG & Kriegstein AR Initial expression and endogenous activation of NMDA channels in early neocortical development. J Neurosci 11, 792–799 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marins M et al. Gap junctions are involved in cell migration in the early postnatal subventricular zone. Dev Neurobiol 69, 715–730, doi: 10.1002/dneu.20737 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Ohtaka-Maruyama C et al. Synaptic transmission from subplate neurons controls radial migration of neocortical neurons. Science 360, 313–317, doi: 10.1126/science.aar2866 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Kuffler SW Neuroglial cells: physiological properties and a potassium mediated effect of neuronal activity on the glial membrane potential. Proc R Soc Lond B Biol Sci 168, 1–21, doi: 10.1098/rspb.1967.0047 (1967). [DOI] [PubMed] [Google Scholar]
References
- 38.Picelli S et al. Full-length RNA-seq from single cells using Smart-seq2. Nature protocols 9, 171–181, doi: 10.1038/nprot.2014.006 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Tirosh I et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313, doi: 10.1038/nature20123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 36, 411–420, doi: 10.1038/nbt.4096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin GL et al. Non-inflammatory tumor microenvironment of diffuse intrinsic pontine glioma. Acta Neuropathol Commun 6, 51, doi: 10.1186/s40478-018-0553-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin EY et al. Neural Precursor-Derived Pleiotrophin Mediates Subventricular Zone Invasion by Glioma. Cell 170, 845–859 e819, doi: 10.1016/j.cell.2017.07.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rueden CT et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC bioinformatics 18, 529, doi: 10.1186/s12859-017-1934-z (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nature methods 9, 676–682, doi: 10.1038/nmeth.2019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nature methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pietzsch T, Preibisch S, Tomancak P & Saalfeld S ImgLib2--generic image processing in Java. Bioinformatics 28, 3009–3011, doi: 10.1093/bioinformatics/bts543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu D et al. Postsynaptic synaptotagmins mediate AMPA receptor exocytosis during LTP. Nature 544, 316–321, doi: 10.1038/nature21720 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guizar-Sicairos M, Thurman ST & Fienup JR Efficient subpixel image registration algorithms. Opt Lett 33, 156–158 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Achanta R et al. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell 34, 2274–2282, doi: 10.1109/TPAMI.2012.120 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Pnevmatikakis EA et al. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89, 285–299, doi: 10.1016/j.neuron.2015.11.037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Walt S et al. scikit-image: image processing in Python. PeerJ 2, e453, doi: 10.7717/peerj.453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yizhar O, Fenno LE, Davidson TJ, Mogri M & Deisseroth K Optogenetics in neural systems. Neuron 71, 9–34, doi:S0896–6273(11)00504–6 [pii] 10.1016/j.neuron.2011.06.004 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Kawahara Y, Ito K, Sun H, Kanazawa I & Kwak S Low editing efficiency of GluR2 mRNA is associated with a low relative abundance of ADAR2 mRNA in white matter of normal human brain. Eur J Neurosci 18, 23–33 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Sobolevsky AI, Rosconi MP & Gouaux E X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756, doi: 10.1038/nature08624 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vinci M, Box C & Eccles SA Three-dimensional (3D) tumor spheroid invasion assay. J Vis Exp, e52686, doi: 10.3791/52686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vinci M, Box C, Zimmermann M & Eccles SA Tumor spheroid-based migration assays for evaluation of therapeutic agents. Methods Mol Biol 986, 253–266, doi: 10.1007/978-1-62703-311-4_16 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the article, source data, or from the corresponding author upon reasonable request. Custom code created is available on GitHub. RNA sequencing of single-cell patient derived xenografts is available on GEO (GSE134269).