Abstract
Staufen1 (STAU1) is a dsRNA binding protein mediating mRNA transport and localization, translational control and STAU1-mediated mRNA decay (SMD). The STAU1 binding site (SBS) within human ADP-ribosylation factor1 (ARF1) 3′UTR binds STAU1 and this downregulates ARF1 cytoplasmic mRNA levels by SMD. However, how STAU1 recognizes specific mRNA targets is still under debate. Our structure of the ARF1 SBS–STAU1 complex uncovers target recognition by STAU1. STAU1 dsRNA binding domain (dsRBD) 4 interacts with two pyrimidines and one purine from the minor groove side via helix α1, the β1–β2 loop anchors the dsRBD at the end of the dsRNA and lysines in helix α2 bind to the phosphodiester backbone from the major groove side. STAU1 dsRBD3 displays the same binding mode with specific recognition of one guanine base. Mutants disrupting minor groove recognition of ARF1 SBS affect in vitro binding and reduce SMD in vivo. Our data thus reveal how STAU1 recognizes minor groove features in dsRNA relevant for target selection.
INTRODUCTION
STAU1 is a dsRNA-binding protein (dsRBP) mediating mRNA transport and localization as well as regulation of mRNA processing, stability and translational efficiency (1). Important functions of the protein were first observed in Drosophila melanogaster where it was shown to be essential for the establishment of the anterior-posterior body pattern (2–5). Two paralogs, STAU1 and STAU2, and isoforms thereof have been described in mammals. STAU1 is found in most tissues, whereas STAU2 is preferentially present in the brain (6–8). The two STAU paralogs differ mainly in the number of dsRBDs. Both contain dsRBD2, 3 and 4, of which dsRBD3 and 4 adopt the canonical α−β−β−β−α dsRBD fold with three principal dsRNA interaction modules (Figure 1A): helix α1 and loop β1–β2 interacting with dsRNA from the minor groove while conserved lysines in helix α2 insert into the major groove (9). STAU1 lacks the dsRBD1, while STAU2 has a truncated dsRBD5 and both proteins contain a tubulin-binding domain (TBD) and a STAU-swapping motif (SSM) for homo- and heterodimerization (6,10–11). STAU proteins are mainly cytoplasmic with enrichment in the periplasmic region and at the rough endoplasmic reticulum where they associate with translating ribosomes depending on protein-protein interactions via dsRBD4 and TBD and on RNA-protein interactions via mRNA and dsRBD3 (6,12). Moreover, a bipartite NLS can target STAU1 to the nucleus where it was linked to the regulation of alternative splicing and nuclear export (13,14). Both STAU paralogs bind different, only partially overlapping subsets of mRNA substrates with protein functions in transport, transcription and cell-cycle control (15–20). The target mRNAs clearly display enrichment in GC-content and secondary structure in their 3′UTRs (Figure 1B) and the structure of Drosophila melanogaster dmSTAU dsRBD3 revealed only RNA phosphodiester backbone interactions with an artificial 12 base pair (bp) stem-loop (9). As a consequence, it is still unclear how STAU proteins recognize many specific mRNA targets in diverse post-transcriptional gene expression pathways.
Figure 1.
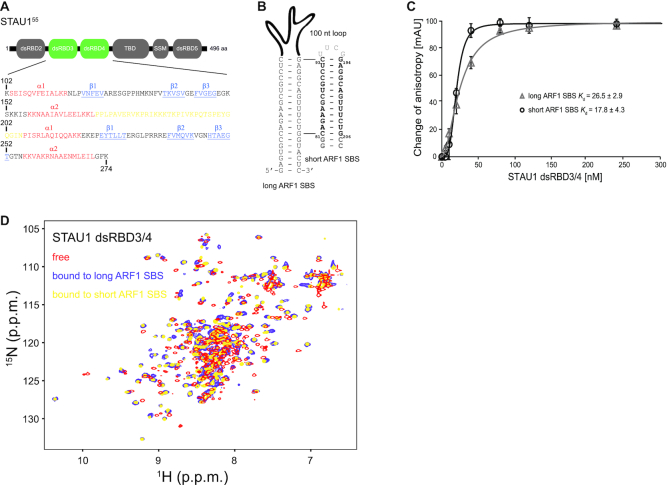
Interaction studies of STAU1 dsRBD3/4 with ARF1 SBS dsRNA. (A) Schematic representation of the human STAU1 domains. STAU1 dsRBD3/4 is located between dsRBD2 and the TBD, the SSM and dsRBD5. Numbering according to the full-length protein sequence (O95793–2). The sequence of recombinant STAU1 dsRBD3/4 used in this study (aa 102–274) is shown. β-strands are shown in blue, α-helices in red and linker amino acids in yellow. (B) The 19 bp SBS dsRNA of human ARF1 3′UTR is shown together with a short construct capped by a UUCG tetraloop which was used for structure determination. Numbering as in (24). (C) Kd values of STAU1 dsRBD3/4 measured by FA using 3′-fluorescein-labeled 19 bp and short ARF1 SBS dsRNA both capped with a UUCG tetraloop. The graph shows the average of three independent measurements with error bars. (D) 1H-15N HSQC of free STAU1 dsRBD3/4 and bound to 19 bp ARF1 SBS dsRNA or the shorter construct. Large chemical shift perturbations occur in both protein domains and are virtually the same for both the 19 bp and the short ARF1 SBS dsRNA.
Beside the well-established roles of STAU in mRNA transport and localization as well as translational control, SMD is the latest addition to the functional repertoire of STAU proteins. In contrast to nonsense-mediated mRNA decay (NMD) (21), a quality control pathway that degrades newly synthesized mRNAs with premature termination codons, SMD regulates mRNA abundance via STAU–3′UTR interactions (22). SMD is a strictly translation-dependent process which requires a double-stranded STAU binding site (SBS) within the 3′UTR downstream of the stop codon and recruitment of the NMD factor Upf1 via STAU dsRBD4 and its TBD (23,24). A bona fide SMD target is characterized by an upregulation of the mRNA with increased mRNA half-life upon depletion of STAU and Upf1 and a SBS downstream of the termination codon (24). SBS can be formed either within the 3′UTR through intramolecular base pairing of sequences up to 1kb apart (20) or by intramolecular base pairing of Alu elements within 3′UTRs with Alu elements of cytoplasmic, polyadenylated long noncoding RNAs (lncRNA) or even Alu elements in other mRNAs (25,26). The best characterized SMD target is ARF1 mRNA which contains a 19 bp stem-loop within the 3′UTR (Figure 1B) required for STAU1 binding and this sole interaction downregulates ARF1 mRNA levels in vivo (23,24).
The recent crystal structure of the complex of human STAU1 RBD3/4 with 19 bp ARF1 SBS at 2.9 Å resolution uncovered that the tandem dsRBDs dimerize upon RNA binding and recognize A-form dsRNA mainly through electrostatic interactions with the RNA backbone (27). The study showed that residues from both dsRBDs also make direct minor groove contacts with G and C bases in Watson–Crick base pairs of the target dsRNA, suggesting some sequence specificity. In the crystal structure, two copies of STAU1 dsRBD3/4 were bound to the ARF1 SBS dsRNA forming an interface between the dsRBDs. However, mutations of residues at the interface did not affect cooperative binding of the dsRBDs (27). Moreover, the crystal structure contained one partially disordered dsRBD4 copy and a second completely disordered copy of dsRBD4. This could suggest steric clashes within the crystal lattice which might influence the positioning of the individual dsRBDs along the target dsRNA (27).
To resolve these issues and gain new insights into dsRNA target recognition by STAU1, we determined the solution structure of the human ARF1 SBS dsRNA - STAU1 dsRBD3/4 complex by high-resolution nuclear magnetic resonance (NMR) spectroscopy. Our structure reveals that STAU1 dsRBD3 and dsRBD4 bind to opposite sides of the ARF1 SBS dsRNA without forming a protein-protein interface. STAU1 dsRBD3/4 directly reads out RNA bases of the ARF1 SBS dsRNA via the minor groove and both dsRBDs participate in the target recognition. STAU1 dsRBD4 binds two pyrimidines and one purine on opposite strands via helix α1 and the β1-β2 loop anchors dsRBD4 at the end of the dsRNA in the minor groove. Lysines in helix α2 bind to the phosphodiester backbone from the major groove. STAU1 dsRBD3 displays the same conserved binding mode and recognizes one guanine base via helix α1 interaction in the minor groove. STAU1 mutations of the amino acids involved in minor groove recognition of ARF1 SBS dsRNA only mildly affect RNA binding affinity in vitro but strongly impact SMD target mRNA binding in vivo and lead to upregulation of mRNA levels.
MATERIALS AND METHODS
The cDNA cloning of human STAU1 dsRBDs and PCR site-directed mutagenesis
The cDNA fragments encoding human STAU1 dsRBD3/4 (102–274) and subdomains (dsRBD3+11aa linker: 102–181, dsRBD4: 205–274) were prepared by polymerase chain reaction (PCR) from Flag-hSTAU1(20) using appropriate primers (Supplementary Table S2) and subcloned into pET28a vector (Novagen) with a TEV instead of the original thrombin cleavage site and a N-terminal His6-Strep-His6-tag, His6-tag or His6-lipo-tag (28) using the NdeI and XhoI restriction sites. Protein mutants were obtained by PCR-based site-directed mutagenesis using pET28a dsRBD3/4 (102–274), dsRBD3+11aa linker (102–181) or dsRBD4 (205–274) plasmid as a DNA template and following the QuickChange protocol (Stratagene) and efficient one-step mutagenesis protocol (29).
Purification of the recombinant STAU1 dsRBD proteins
The dsRBD3/4 (102–274) recombinant protein, subdomains (dsRBD3+11aa linker: 102–181, dsRBD4: 205–274) and mutant proteins were expressed in Escherichia coli BL21-CodonPlus (DE3)-RIL BL21 competent cells (Novagen) in LB rich or M9 minimal media supplemented with 15NH4Cl or 15NH4Cl and 13C-glucose. Cultures were grown to OD600 0.8–1.2 at 37°C, then set to 16°C for 30 min. Protein expression was induced by addition of 1 mM isopropyl β-D-thiogalactoside (IPTG) and further incubation for 14 to 18 h at 16°C. Cells were harvested by centrifugation at 4°C for 10 min at 2600 g and the cell pellet was resuspended in His A buffer (20 mM HEPES pH 7.5, 1 M NaCl, 10% glycerol (w/v), 30 mM imidazole and 3.5 mM 2-mercaptoethanol) with a tablet of protease inhibitor cocktail (cOmplete, ETDA-free, Roche). After lysis with Emulsiflex (Avestin) and centrifugation at 4°C for 30 min at 30 000 g, the supernatant was loaded onto two 5 ml HiTrap chelating columns (HiTrap, GE Healthcare) in series charged with nickel sulfate. All column chromatography was performed with an ÄKTA prime FPLC system equipped with a 50 ml superloop (GE Healthcare) at 4°C. After loading, the column was washed with 50 ml of lysis buffer followed by elution of the protein over a 90 ml gradient with His B buffer (His A buffer with 500 mM NaCl and 0.3–1 M imidazole). The pooled fractions were dialyzed against 2 l of His A buffer (500 mM NaCl and no imidazole) in the presence of TEV protease (1 mg/100 ml) to cleave the affinity-tag at RT overnight. TEV cleavage reaction mixtures were reloaded onto HiTrap chelating columns to remove the His6-TEV protease and the cleaved His6-Strep-His6-tag as well as minor contaminating proteins. The pooled fractions were dialyzed against 2 l of buffer A (50 mM potassium phosphate buffer pH 6.8, 100 mM NaCl, 3.5 mM 2-mercaptoethanol), loaded onto one 5 mL Q-sepharose column (HiTrap Q HP, GE Healthcare) to remove any remaining bound nucleic acids from the proteins. The protein fractions from the flow-through of the column were pooled, and after addition of 10% glycerol (w/v) frozen and stored at −20°C.
Design of STAU1 dsRBD3 and dsRBD4 for protein ligation with Sortase A
For protein ligation by Sortase A enzyme (30,31), the cDNA fragments were prepared by PCR from dsRBD3/4 (102–274) using appropriate primers (Supplementary Table S2) and subcloned into pET28a vector (Novagen) with a TEV cleavage site and a N-terminal His6-Strep-His6-tag or His6-lipo tag. We engineered the C-terminus of dsRBD3 to end with the sequence Leu174-Pro175-Ala176-Thr-Gly-His6 (His6-lipo-TEV-dsRBD3_linker_LPATG-His6) and the N-terminus of dsRBD4 to start with Gly-Val177-Glu178-Arg179 (His6-Strep-His6-TEV-G_linker_dsRBD4). This inserts a threonine and a glycine between Ala176 and Val177 of the linker and results in the LPTXG sequence required for ligation by Sortase A (30,31).
Preparation of segmentally isotope-labeled STAU1 dsRBD3/4
The His6-lipo-TEV-dsRBD3_linker_LPATG-His6 fragment was expressed in M9 minimal medium supplemented with 15NH4Cl and 13C-glucose and the His6-Strep-His6-TEV-G_linker_dsRBD4 fragment was expressed LB rich medium as described above for STAU1 dsRBD3/4. The His6-Strep-His6-TEV-G_linker_dsRBD4 fragment was purified as described above. To facilitate the purification of the ligated product as described before (31), the His6-lipo-TEV-dsRBD3_linker_LPATG-His6 fragment was not cleaved with TEV protease and thus only purified by one run of HiTrap chelating column chromatography. Purified uncleaved His6-lipo-TEV-dsRBD3_linker_LPATG-His6 was subjected to anion exchange chromatography to remove remaining bound nucleic acids and protein impurities as described above.
To determine optimal conditions for the ligation reaction, a series of small scale test ligations was performed varying the concentration of fragments and Sortase A by using a dialysis approach as previously described (31). The final ligation reaction was setup in ligation buffer (50 mM Tris pH 8.0, 300 mM NaCl, 30 mM CaCl2, 10% glycerol (w/v), 3.5 mM 2-mercaptoethanol) and the reaction mixture was dialyzed against ligation buffer for 17–20 h. The ligation process was monitored by denaturing polyacrylamide gel electrophoresis (PAGE) and the product purified by two consecutive runs of HiTrap chelating chromatography with TEV protease cleavage between the runs as described above. Purified, ligated, segmentally isotope-labeled dsRBD3/4 was incubated with SUPERase In RNase inhibitor (Ambion) and then further purified by size-exclusion chromatography with a Sephadex S-75 gel filtration column (GE Healthcare) using gel filtration buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 3.5 mM 2-mercaptoethanol) at 4°C. Fractions corresponding to the ligated protein product were pooled, concentrated and dialyzed three times against 1 l NMR buffer (50 mM potassium phosphate pH 6.5, 250 µM ethylenediaminetetraacetic acid (EDTA)) using Slide-A-Lyzer 0.5 mL dialysis cassettes with 3500 MWCO (Thermo Scientific). The mass of the final product was confirmed by MALDI-TOF.
Transcription and purification of ARF1 SBS dsRNA
For unlabeled and isotope-labeled ARF1 SBS dsRNA a cDNA fragment was prepared by PCR using three primers (Supplementary Table S2) and subcloned into pUC18 as previously described (32–34). Unlabeled and 13C-labeled RNA oligonucleotides were transcribed in vitro from plasmid DNA linearized with BbsI/BstV2 I (SibEnzyme) using T7 RNA polymerase as previously described (32–34). Optimal magnesium chloride concentration for efficient transcription was determined by 25 μl trial reactions. After 2 to 4 h of incubation at 37°C, the crude transcription reaction was stopped upon addition of EDTA to a final concentration of 50 mM. Purification of the ARF1 SBS dsRNA was performed with three 5 ml HiTrap DEAE-sepharose FastFlow columns (GE Healthcare) connected in series as previously described (33). Fractions corresponding to pure, monomeric ARF1 SBS dsRNA were pooled, dialyzed three times against 2 l 10 mM sodium phosphate buffer (pH 6.0), frozen and stored at −20°C. If required, fractions containing pure ARF1 SBS dsRNA were concentrated using Vivaspin 20 ml centrifugal devices with 5000 MWCO (Sartorius) and passed over a Sephadex S-75 gel filtration column (GE Healthcare) with gel filtration buffer at 4°C. Before the experiments stored RNA was thawed, concentrated as before and dialyzed three times against 1 l NMR buffer using Slide-A-Lyzer 0.5 or 3 ml dialysis cassettes with 3500 MWCO (Thermo Scientific). The precise concentration was then determined based on the base composition of the hydrolyzed RNA as previously described (35).
Preparation of RNA–protein complexes
Stored, defrosted STAU1 proteins were incubated with SUPERase In RNase inhibitor (Ambion) for 30 min at RT and subjected to size-exclusion chromatography with a Sephadex S-75 gel filtration column (GE Healthcare) using gel filtration buffer at 4°C to remove aggregates. Pure, monomeric protein fractions were pooled, concentrated to up to 500 µM and dialyzed three times against 1 l NMR buffer using Slide-A-Lyzer 0.5 ml dialysis cassettes with 3000 MWCO (Thermo Scientific). Final concentrations of proteins were determined using optical absorbance at 280 nm. Assembly of RNA–protein complexes was monitored by 2D 1H-15N HSQC experiments titrating STAU1 proteins in a stepwise manner with ARF1 SBS dsRNA until the spectra remained unchanged (ratio 1:1). All further complexes were assembled in lab in same way by mixing RNA and protein.
NMR measurement
All NMR measurements for the RNA–protein complexes were performed in NMR buffer at 308K using Bruker Avance III HD 700, 850 and 950 MHz spectrometers all equipped with cryoprobes. Data were processed using Topspin 3.2/3.5 (Bruker) and analyzed with CARA (http://cara.nmr.ch) or Sparky (http://www.cgl.ucsf.edu/home/sparky/).
Protein sequence-specific backbone and side chain assignments were achieved using unlabeled ARF1 SBS dsRNA in complex with 13C,15N-labeled dsRBD4 or dsRBD3+11aa linker in 5 to 10% D2O. The following experiments were performed: 2D 1H-15N HSQC, 2D 1H-13C HSQC, 3D HNCA, 3D HNCO, 3D CBCACONH, 3D HNCACB, 3D HBHACONH, 3D HC(C)H TOCSY, 3D (H)CCH TOCSY and 3D NOESY 1H-13C HSQC aliphatic (36). Aromatic proton assignments were performed using 3D NOESY 1H-13C HSQC aromatic experiments. 3D NOESY 1H-15N HSQC spectra were collected on complexes of unlabeled RNA with 15N-labeled protein. 3D backbone and side chain experiments were performed with non-uniform sampling (NUS) schemes and processed with NMRpipe (37). For NUS data collection and processing, Poisson disk sampling scheme and the modified CLEAN processing scheme with the conservative sparsity of about 20% were used (38,39).
Initial ARF1 SBS dsRNA resonance assignments were performed in the free form of the RNA with 2D 1H-1H TOCSY, 2D 1H-1H NOESY in 5–10% or 100% D2O using unlabeled RNA. Complete ribose and aromatic proton assignments for ARF1 SBS dsRNA were achieved using 13C-labeled RNA to collect 2D 1H-13C HSQC, 3D HC(C)H TOCSY, 3D NOESY 1H-13C HSQC ribose and 3D NOESY 1H-13C HSQC aromatic experiments in 100% D2O optimized for RNA (40). ARF1 SBS dsRNA resonance (H1′-H6/H8 walk) assignments in the ARF1 SBS dsRNA–dsRBD3/4 complex were performed with unlabeled ARF1 SBS dsRNA in complex with segmentally isotope-labeled dsRBD3/4 or dsRBD3+11aa linker by using filtered experiments as described above for in the ARF1 SBS dsRNA–dsRBD4 complex.
ARF1 SBS dsRNA resonance (H1′-H6/H8 walk) assignments in the ARF1 SBS dsRNA–dsRBD4 complex were performed with unlabeled ARF1 SBS dsRNA in complex with 13C,15N-labeled protein using 2D 13C 1F-filtered 2F-filtered NOESY experiments (41) in 100% D2O and 13C-labeled ARF1 SBS dsRNA in complex with 15N-labeled dsRBD4 using 2D 1H-13C HSQC and 3D NOESY 1H-13C HSQC ribose experiments in 100% D2O optimized for RNA (40).
Intermolecular NOEs were obtained by recording 2D 1H-1H, 13C 2F-filtered NOESY and 3D 13C F1-filtered F2-edited HSQC-NOESY (42) in 100% D2O using unlabeled ARF1 SBS dsRNA in complex with 13C,15N-labeled dsRBD4 or segmentally isotope-labeled dsRBD3/4. Unambiguous assignments of intermolecular NOEs from protein side chains to RNA ribose and aromatic protons were also obtained from 3D NOESY 1H-13C HSQC ribose experiments recorded on 13C-labeled ARF1 SBS dsRNA in complex with 15N-labeled dsRBD4 in 100% D2O.
NOESY spectra were recorded with a mixing time of 80 or 150 ms, the 3D (H)CCH- and HC(C)H TOCSY spectra with a mixing time of 23 ms and the 2D 1H-1H TOCSY spectra with mixing times of 20 or 50 ms.
Structure calculation and refinement
The AtnosCandid software (43,44) integrated into the Unio software package was used to generate preliminary structures and a list of automatically assigned NOE distance constraints for ARF1 SBS dsRNA–protein complexes. For ARF1 SBS dsRNA in complex with dsRBD4, peak peaking and NOE assignments were performed on 3D NOESY (15N- and 13C-edited) spectra recorded on 15N-labeled or 13C,15N-labeled dsRBD4 in complex with unlabeled ARF1 SBS dsRNA. Seven iterations were performed and 200 independent structures were calculated at each iteration step. Structures were calculated with CYANA by adding the manually assigned intramolecular RNA and intermolecular RNA–protein distance restraints obtained from dsRBD4–ARF1 SBS dsRNA complex. For ARF1 SBS dsRNA in complex with dsRBD3/4, peak peaking and NOE assignments were performed on 3D NOESY (15N- and 13C-edited) spectra recorded on 15N-labeled or 13C,15N-labeled dsRBD3+11aa linker in complex with unlabeled ARF1 SBS dsRNA as above. In this case, structures were calculated with CYANA by adding the manually assigned intramolecular RNA, intermolecular RNA–protein distance restraints obtained from segmentally isotope-labeled dsRBD3/4–ARF1 SBS dsRNA and dsRBD4–ARF1 SBS dsRNA complexes, respectively, and intramolecular dsRBD4 protein restraints obtained from the structure calculation of the dsRBD4–ARF1 SBS dsRNA complex. For each CYANA run, 200 independent structures were calculated. In both cases, the 50 lowest energy structures were refined with the SANDER module of AMBER 7.0 using a simulated annealing protocol (45). The 20 best structures based on energy and restraint violation energies were analyzed with PROCHECK (46). Chemical shifts for dsRBD3/4–ARF1 SBS dsRNA complex were combined from dsRBD4–ARF1 SBS dsRNA (205–274) and dsRBD3+11aa linker—ARF1 SBS dsRNA (102–181) complexes and submitted to PDB OneDep deposition.
Fluorescence anisotropy
Fluorescence anisotropy (FA) was measured on a Tecan Microplate Reader Infinite F500 (Tecan, Austria) equipped with a plate reader using 96-well plates. All measurements were performed in NMR buffer at 35°C. Measurements were performed in 10 µl reaction volume in which 10 nM 3′end fluorescein labeled ARF1 SBS dsRNA was titrated with STAU1 dsRBD WT or mutant proteins. The data points represent the average of three measurements. The fitting was performed with Origin software (OriginLab, Northampton, MA, USA) (47). All data were normalized for adequate visualization.
Cell culture and manipulation
Mammalian cells HEK293T-REx Flip-In (Invitrogen™) and T-REx-HeLa Flip-In (Invitrogen™) were maintained in Dulbecco's-modified Eagle's medium supplemented with 10% fetal calf serum at 37°C in the presence of 5% CO2.
Preparation of constructs and stable cell lines with inducible expression of STAU1 variants
The coding sequence of human STAU1 (55 kDa) was PCR amplified using appropriate primers (Supplementary Table S2) and cloned into pcDNA5 FRT/TO-N-terminal-3×FLAG using NotI and XhoI sites. We used site-directed mutagenesis and appropriate primers (Supplementary Table S2) to obtain mutants E135A/L138A, Q212A/Q215A, E135A/L138A/Q212A/Q215A. The cloning and mutagenesis were subsequently verified by sequencing analysis with primers (Supplementary Table S2). The individual constructs for STAU1 variants were transfected together with plasmid pOG44 (Invitrogen) to HEK293T-REx Flip-In cells and T-REx-HeLa Flip-In, respectively by using Lipofectamine 3000 (Invitrogen™) reagent. Cells with stably integrated constructs were selected with Hygromycin B according to the manufacturer's protocol.
RNA immunoprecipitation (RIP)
Cells grown at 15 cm plate were induced at 70% confluency with 200 ng/ml Doxycycline for 24 h. Cells were washed with ice cold phosphate-buffered saline (PBS) and collected into 1.5 ml tube and lysed for 10 min on ice in 750 μl lysis buffer (LB) containing 150 mM NaCl, 50 mM Tris pH 8,1% Triton X-100, EDTA-free Complete Protease Inhibitor Cocktail (Roche), 1 mM DTT. Each tube contained 120 U RNAse Inhibitor (Biotechrabbit) before the lysis. Lysates were incubated with 4.3 U Turbo DNase (Fermentas) for 15 min at 37°C and insoluble fraction was removed by 10 min centrifugation 11 000 g at 4°C. Supernatants were pre-cleared with 50 μl magnetic beads without antibodies (Dynabeads Protein G, Invitrogen) for 1 h at 4°C rotating. 1/20 volume (37.5 μl) of the lysate was mixed with TriPure reagent (Roche) for the ‘input’ sample RNA extraction. 50 μl of FLAG-M2 Magnetic beads (Sigma) per sample were used for immunoprecipitation. FLAG-M2 magnetic beads were first washed three times with 500 μl LB, then pre-blocked with 1 μg of yeast tRNA resuspended in 100 μl of LB for 1 h at 4°C and again washed three times with 500 μl of LB. The pre-cleared lysates were gently mixed with the pre-blocked and washed FLAG-M2 beads and incubated for 2 h at 4°C rotating. Beads were washed once with 500 μl LB for 1 min and twice with 500 μl wash buffer (350 mM NaCl, 50 mM Tris pH 8, 1% Triton X-100, EDTA-free Complete Protease Inhibitor Cocktail - Roche, 1 mM DTT) for 1 min at 4°C. In the last step, the beads were resuspended in 500 μl LB and an aliquot of 20 μl was mixed with 4× sodium dodecyl sulphate (SDS)-PAGE loading buffer to monitor immunoprecipitation efficiency by western blot. The LB was removed and the rest of the beads were mixed with 1 ml of the TriPure reagent (Roche) to extract the coprecipitated RNA. The ‘elution’ RNA was isolated according to manufacturer's instructions with minor changes: 1 μl GlycoBlue coprecipitant (Thermofisher) was added for isopropanol-based precipitation and RNA was precipitated at least one hour at −80°C. The precipitated RNA was centrifuged for 1h 12 000 g at 4°C, the pellet was washed with 85% ethanol and centrifuged for 20 min 12 000 g at 4°C. RNA was then resuspended in 10 μl of H2O. The ‘input’ RNA was isolated according to manufacturer's instructions, resuspended in 16 μl of H2O, treated with 1.2 U Turbo DNase (Fermentas) and frozen in liquid nitrogen to inactivate the Turbo DNase. Input RNA (2 μg) and 1/2 volume of RNA isolated from RIP eluate (5 μl from the total volume of 10 μl) was used as a template for cDNA synthesis by using Superscript III reverse transcriptase (Invitrogen) and random hexamers (Thermo Scientific) according to manufacturer's instructions. The obtained cDNA was subsequently analyzed by real-time quantitative PCR (RT-qPCR).
Western blot analysis
The protein lysates from whole cell lysate (input), unbound fraction (flow-through) and purified proteins (elution) samples from the RIP experiment were mixed with SDS-PAGE sample buffer, separated by SDS-PAGE and transferred to nitrocellulose membrane by semi-dry blotting (BioRad). Proteins were detected with protein and tag-specific antibodies, respectively. The FLAG antibody (Sigma) was used at dilution 1:3000, STAU1 antibody (Proteintech) at dilution 1:1500 and α-Tubulin antibody (Sigma) at dilution 1:5000.
Real-time PCR analysis
Quantitative PCR (qPCR) was performed with the LightCycler 480 SYBR Green I Master Mix (Roche) and gene-specific primers on LightCycler 480 (Roche). Three to four biological replicates were analyzed. Transcript abundances were calculated by the ΔΔCt method (48). The input data were normalized to an internal control of the housekeeping gene GAPDH mRNA, elution data were normalized to the corresponding Input samples normalized to GAPDH. P-values were calculated by two-tailed paired t-test; P-values < 0.1 were considered significant.
RNA steady-state levels analysis
HEK293T-REx and T-REx-HeLa control and stable-cell lines expressing WT and mutant forms of STAU1 were grown at 6-well plates to 70% confluency and induced for 24 h with doxycycline, at the concentration 200 ng/ml for HEKs and 1000 ng/ml for HeLas. Cells were washed with cold PBS and collected in 500 μl of TriPure reagent (Roche) per one well. The total RNA was isolated according to manufacturer's instructions. Two μg of RNA was used as a template for cDNA synthesis by using Superscript III reverse transcriptase (Invitrogen™) and random hexamers (Thermo Scientific). The cDNA was subsequently analyzed by RT-qPCR. Three to six biological replicates were analyzed. Transcript abundances were calculated by the ΔΔCt method (48). The data were normalized to an internal control of the housekeeping gene GAPDH mRNA and to the control cell line. P-values were calculated by two-tailed paired t-test; P-values < 0.1 were considered significant.
SAXS measurements of the STAU1–ARF1 SBS dsRNA complexes
Small angle X-ray scattering (SAXS) data acquisition is summarized in Supplementary Table S1. Data were collected using BioSAXS-1000, Rigaku (CEITEC, Czech Republic) and at the B21 instrument (Diamond Light Source, UK). BioSAXS-1000 datasets were collected at 308 K, six separate two-dimensional images were collected for buffer and sample with an exposure time of 10 min per image (Pilatus 100K, Dectris). Radial averaging, data reduction and buffer subtractions were performed using SAXSLab3.0.0r1, Rigaku. B21 datasets were collected in the size-exclusion mode (SEC-SAXS) at 288 K, where two-dimensional images with an exposure time of 3 s per image (Pilatus 2M, Dectris) were continuously collected during size exclusion chromatography of the sample. Five subsequent 3 s exposures have been averaged to reduce the noise level of the data. SEC-SAXS data were analyzed using Chromixs (49). All datasets were cropped from the first data point of the Guinier region determined by PRIMUS/qt ATSAS v.2.8.3 (50) to maximum of q = 0.3 Å−1 for further analysis. Integral structural parameters and MW estimate (Supplementary Table S1) were determined using PRIMUS/qt. Evaluation of the solution scattering and fitting to experimental scattering curves was performed using CRYSOL ATSAS v.2.8.3, where automatic constant subtraction was allowed, while other parameters were kept at default.
RESULTS
Characterization of STAU1 dsRBD3/4–ARF1 SBS dsRNA complexes
The principal RNA binding module of STAU1 comprises dsRBD3/4 with canonical α−β−β−β−α topology separated by a basic 34 amino acid linker (Figure 1A) (6,8). The long ARF1 SBS dsRNA required for STAU1 binding in vivo comprises a continuous 19 bp stem with canonical Watson–Crick and two GU base pairs formed between nt 75–93 and 194–212 of the ARF1 3′UTR (Figure 1B) (23,24). Monitoring complex formation using FA measurements reveals high affinity binding (Kd = 26.5 ± 2.9 nM) of recombinant STAU1 dsRBD3/4 to the long ARF1 SBS dsRNA capped by a UUCG tetraloop (Figure 1C). Complex formation was also assessed by NMR titration experiments monitored by recording 1H-15N HSQC spectra. Upon step-wise addition of long ARF1 SBS dsRNA to recombinant 15N-labeled STAU1 dsRBD3/4, many protein resonances broaden beyond detection indicative of binding kinetics in the intermediate to slow exchange regime (Supplementary Figure S1A). When a 1:1 protein to dsRNA ratio is reached, protein resonances sharpen again displaying numerous chemical shift changes indicative of stable complex formation (Figure 1D and Supplementary Figure S1A). To further characterize the complex in solution, we performed SAXS measurements on the STAU1 dsRBD3/4 bound to long ARF1 SBS dsRNA. The SAXS measurements reveal a 1:1 stoichiometry of the complex as determined by a Porod volume of 52800 Å3 (Supplementary Table S1).
A single dsRBD usually displays a register variability of 8–10 bp for sequence-specific minor groove contacts with a dsRNA (51). The long ARF1 SBS dsRNA could therefore bind the STAU1 dsRBD3/4 either in a staggered manner as previously observed for the adenosine deaminase acting on RNA 2 (ADAR 2) bound to dsRNA (52) or in a side-by-side manner on opposite sides of the long ARF1 SBS dsRNA. To test these possibilities, we designed shorter ARF1 SBS dsRNA comprising either the top or bottom two-thirds of the long, 19 bp ARF1 stem (Supplementary Figure S1B). Complex formation monitored by recording 1H-15N HSQC and 1H,1H-TOCSY spectra shows the same spectra with minor differences only when STAU1 dsRBD3/4 is bound to the long and the top two-thirds of ARF1 SBS dsRNA but the complex of dsRBD3/4 with the bottom two-thirds of the long ARF1 SBS dsRNA displays different spectra indicative of an altered binding mode (Figure 1D, Supplementary Figure S1C-D and Table 1). FA measurements using either the top or the bottom two-thirds of ARF1 SBS dsRNA shows a higher affinity interaction for the top two-thirds (Kd = 17.8 ± 4.3 nM) as compared to the bottom two-thirds (Kd = 31.0 ± 0.1 nM) of the ARF1 SBS dsRNA (Figure 1C, Supplementary Figure S1E and Table 1). SAXS measurements of STAU1 dsRBD3/4-bound to the top two-thirds of ARF1 SBS dsRNA reveal a 1:1 stoichiometry of the complex as determined by a Porod volume of 38900 Å3 (Supplementary Table S1). These data suggest that both dsRBDs bind in the same 15 bp region formed between nt 81–93 and 194–206 of the ARF1 3′UTR (Figure 1B).
Table 1.
Binding affinities of wild-type and mutant STAU1 dsRBDs and ARF1 SBS dsRNA
| STAU1 proteina | dsRNAb | K d (nM)c | Affinity factord |
|---|---|---|---|
| dsRBD3/4 | ARF1 SBS (5′-GGCGAGUGCCAGAAGCUGUCUCCUUCGGGAGGCAGUUUCUGGUACUCGCC-3′) | 26.5 ± 2.9 | 1.5 |
| dsRBD3/4 | top 2/3 ARF1 SBS (5′-GGCAGAAGCUGCCUCUUCGGAGGCAGUUUCUGCC-3′) = sARF1 SBS | 17.8 ± 4.3 | 1.0 |
| dsRBD3/4 | bottom 2/3 ARF1 SBS (5′-GGAGUGCCAGAAGCUACGGUUUCUGGUACUCC-3′) | 31.0 ± 0.1 | 1.7 |
| dsRBD3 | sARF1 SBS | 41.4 ± 1.5 | 2.3 |
| dsRBD4 | sARF1 SBS | 74.8 ± 17.6 | 4.3 |
| dsRBD3 I105A S106A | sARF1 SBS | 147.1 ± 35.6 | 3.6 |
| dsRBD3 E110A L113A | sARF1 SBS | 31.5 ± 1.7 | 0.8 |
| dsRBD3 F109A | sARF1 SBS | 98.2 ± 8.7 | 2.4 |
| dsRBD4 Q212A Q215A | sARF1 SBS | 121.2 ± 15.0 | 1.6 |
| dsRBD4 S208A Q212A Q215A | sARF1 SBS | 389.6 ± 38.4 | 5.2 |
| dsRBD3/4 I105A S106A | sARF1 SBS | 28.8 ± 2.7 | 1.6 |
| dsRBD3/4 E110A L113A | sARF1 SBS | 18.5 ± 4.3 | 1.0 |
| dsRBD3/4 I105A S106A E110A L113A | sARF1 SBS | 21.8 ± 6.4 | 1.2 |
| dsRBD3/4 Q212A Q215A | sARF1 SBS | 15.3 ± 0.7 | 0.9 |
| dsRBD3/4 E110A L113A Q212A Q215A | sARF1 SBS | 18.5 ± 1.1 | 1.0 |
| dsRBD3/4 S208A | sARF1 SBS | 28.0 ± 4.4 | 1.6 |
| dsRBD3/4 S208A Q212A Q215A | sARF1 SBS | 87.2 ± 3.0 | 4.9 |
| dsRBD3/4 I105A S106A S208A Q212A Q215A | sARF1 SBS | 43.7 ± 5.2 | 2.5 |
| dsRBD3/4 K133A | sARF1 SBS | 23.2 ± 9.7 | 1.3 |
| dsRBD3/4 R234A R236A | sARF1 SBS | 26.0 ± 9.1 | 1.5 |
| dsRBD3/4 F109A | sARF1 SBS | 29.7 ± 5.8 | 1.7 |
| dsRBD3/4 R209A | sARF1 SBS | 17.7 ± 4.1 | 1.0 |
| dsRBD3/4 F109A R209A | sARF1 SBS | 27.7 ± 5.5 | 1.6 |
aSTAU1 protein construct and mutations are indicated.
bARF1 SBS dsRNA used for FA measurements are indicated.
cAll FA experiments were performed in technical triplicates and the reported Kd values are the average of three experiments with error bars. dAffinity factor for variant RNA or protein relative to top 2/3 ARF1 (sARF1) SBS dsRNA bound to wild-type STAU1 dsRBD3, dsRBD4 or dsRBD3/4, respectively.
To assess the contributions of the individual STAU1 dsRBDs to the binding affinity, we designed two smaller protein constructs comprising dsRBD3 with 11 amino acids of the linker sequence (102–181) and dsRBD4 (205–274) and compared their binding affinity (Supplementary Figure S2A). FA measurements show that dsRBD3 binds with 2-fold higher affinity (Kd = 41.4 ± 1.5 nM) to top two-thirds ARF1 SBS dsRNA as compared to dsRBD4 (Kd = 74.8 ± 17.6 nM). In both cases, the affinity is lower than observed for the STAU1 dsRBD3/4 construct indicating weak cooperative binding of the tandem dsRBDs driven by the higher affinity of dsRBD3.
Design of STAU1 dsRBD3/4–ARF1 SBS dsRNA complexes for structure determination
The complexity of the 1H-15N HSQC spectrum with highly overlapping resonances from the unstructured linker prompted us to simplify the spectra by employing a ‘divide-and-conquer’ approach working with the individual dsRBDs bound to the top two-thirds of ARF1 SBS dsRNA, termed short ARF1 (sARF1) SBS dsRNA hereafter, separately. We assessed complex formation of the individual STAU1 dsRBDs with sARF1 SBS dsRNA by recording 1H-15N HSQC spectra of the individual subcomplexes and compared them to the 1H-15N HSQC spectrum obtained for the STAU1 dsRBD3/4 complex. While the 1H-15N HSQC spectrum of dsRBD4 displays an almost identical fingerprint of the protein amide resonances as compared to the tandem dsRBD3/4, dsRBD3 shows deviations and broadening of resonances indicating that the latter has different binding characteristics when taken out of the context of the tandem dsRBD3/4 protein construct (Supplementary Figure S2B). The difference is also apparent from 1H,1H-TOCSY spectra recorded on sARF1 SBS dsRNA bound to the tandem or individual dsRBDs. The spectra show that sARF1 SBS dsRNA adopts the same conformation when bound to dsRBD3/4 and dsRBD4 while peak doubling for several pyrimidines upon dsRBD3 binding indicates multiple binding registers or alternate binding orientations along the dsRNA or both (Supplementary Figure S2C).
To reveal the reason for this difference, we recorded 3D 13C F1-filtered F2-edited HSQC-NOESY(42) spectra on the dsRBD3–and dsRBD4–ARF1 SBS dsRNA complexes separately and assigned intermolecular nuclear Overhauser effect (NOE)-derived contacts. While dsRBD4 binds sARF1 SBS dsRNA in one preferred orientation placing the helix α1 at the lower part of the sARF1 SBS dsRNA, dsRBD3 showed multiple binding registers with helix α1 binding the 5′end of the sARF1 SBS dsRNA and the middle part of the stem (Supplementary Figure S3A and B). This promiscuous, non-specific binding mode has also been observed for the isolated Drosophila dsRBD3 bound to an artificial 12 bp stem-loop (9). The data suggest that the dsRBD4–sARF1 SBS dsRNA can be determined in isolation while structure determination of the dsRBD3 interaction with sARF1 SBS dsRNA requires the context of the tandem dsRBD3/4 protein. To alleviate spectral overlap, we prepared segmentally labeled STAU1 dsRBD3/4 protein, where only dsRBD3 and part of the linker is 13C,15N-labeled while the rest of the linker and dsRBD4 is unlabeled and therefore does not contribute to NMR signals in heteronuclear NMR experiments (Supplementary Figure S3C). When this segmentally labeled protein was used for complex formation with sARF1 SBS dsRNA, we observed no strong deviations and broadening of resonances in the 1H-15N HSQC spectrum (Supplementary Figure S3D). Likewise, the intermolecular NOEs indicated one preferred orientation also placing the helix α1 of dsRBD3 at the middle part of the sARF1 SBS dsRNA (Supplementary Figure S3E). With these optimized protein–dsRNA complexes, we determined the NMR solution structure of the STAU1 dsRBD4–sARF1 SBS dsRNA subcomplex and of the STAU1 dsRBD3/4–sARF1 SBS dsRNA complex using standard NMR techniques (53,54).
Structure of STAU1 dsRBD4–sARF1 SBS dsRNA complex reveals sequence readout from the minor groove
The final ensemble of the STAU1 dsRBD4–sARF1 SBS dsRNA complex is well-defined and is represented by twenty conformers with a root-mean-square (r.m.s.) deviation of 0.91 Å for all heavy atoms (Table 2 and Figure 2A). Resonance assignment of the sARF1 SBS dsRNA as well as unambiguous assignment of intermolecular contacts was aided by using 13C,15N-labeled sARF1 SBS dsRNA in complex with 15N-labeled STAU1 dsRBD4 in addition to 13C,15N-labeled STAU1 dsRBD4–unlabeled sARF1 SBS dsRNA complex commonly used for extracting intermolecular NOEs and RNA assignments (53,54). This combined approach yielded a sufficient number of intermolecular (178) and intra-RNA (717) NOEs for high-resolution structure determination (Table 2).
Table 2.
Structural statistics of Staufen1 dsRBD4 and dsRBD3/4 in complex with sARF1 SBS dsRNA
| dsRBD4–sARF1 SBS dsRNA | dsRBD3/4–sARF1 SBS dsRNA | ||
|---|---|---|---|
| dsRBD4 | sARF1 SBS dsRNA | dsRBD3/4 | |
| NMR distance and dihedral restraints | |||
| Distance restraints | |||
| Total NOE | 1359 | 717 | 2472 |
| intraresidual | 390 | 358 | 719 |
| interresidual | |||
| sequential (|i - j| = 1) | 350 | 258 | 664 |
| medium range (1 < | i-j | < 5) | 272 | 35 | 469 |
| long range (| i-j | > = 5) | 347 | 66 | 620 |
| RNA–Protein intermolecular | 178 | 224 | |
| RNA-dsRBD3 | 46 | ||
| RNA-dsRBD4 | 178 | ||
| hydrogen bondsa | 0 | 37 | 0 |
| RNA–Protein hydrogen bondsa | 13 | 19 | |
| Dihedral restraintsb | 0 | 168 | 0 |
| Structure statistics c | |||
| Mean Violations | |||
| Number of distance constraints > 0.1–0.2Å | 5.2 ± 1.0 | 14.8 ± 2.4 | |
| Number of distance constraints > 0.2–0.3Å | 0.0 | 1.1 ± 0.8 | |
| Number of distance constraints > 0.3–0.4Å | 0.0 | 0.1 ± 0.3 | |
| Number of distance constraints > 0.4Å | 0.0 | 0.0 | |
| Number of dihedral constraints < 5° | 4.2 ± 0.8 | 7.1 ± 1.8 | |
| Number of dihedral constraints > 5° | 0.0 | 0.0 | |
| Max. distance constraint violation (Å) | 0.18 | 0.33 | |
| Max. dihedral constraint violation (°) | 0.43 | 0.65 | |
| Mean Deviation from ideal covalent geometry | |||
| Bond Length (Å) | 0.0109 ± 0.0001 | 0.0108 ± 0.0001 | |
| Bond Angle (°) | 2.373 ± 0.016 | 2.369 ± 0.016 | |
| Ramachandran plot statistics c,d,e | |||
| Residues in most favored regions (%) | 77.7 ± 4.2 | 71.7 ± 3.5 | |
| Residues in additionally allowed regions (%) | 21.7 ± 4.2 | 26.0 ± 3.5 | |
| Residues in generously allowed regions (%) | 0.2 ± 0.6 | 1.4 ± 1.3 | |
| Residues in disallowed regions (%) | 0.3 ± 0.6 | 0.9 ± 0.9 | |
| Average pairwise r.m.s. deviation from mean structure d | |||
| Protein | |||
| Backbone atoms | 0.80 ± 0.13 | 1.79 ± 0.36 | |
| Heavy atoms | 1.24 ± 0.11 | 2.21 ± 0.36 | |
| RNA | |||
| Heavy atoms | 0.76 ± 0.23 | 1.23 ± 0.46 | |
| RNA–protein complex | |||
| Heavy atoms | 0.91 ± 0.15 | 1.95 ± 0.31 | |
aHydrogen bond constraints were identified in the course of structure refinement and for imino resonances protected from exchange with H2O.
bDihedral angle constraints for sARF1 SBS dsRNA are based on regular A-form geometry and previous structures of the UUCG tetraloop (60).
cStatistics are computed for the deposited bundle of 20 structures which had lowest violation energy and were selected out of 30 structures with lowest total amber energy from a total of 50 calculated structures. With the exception of maximum constraint violations which give the largest violation among the 20 selected structures, the statistics give the mean and standard deviation.
dBased on structured residue range as defined: Asn205–Lys 274 (dsRBD4) Lys102–Pro173,Asn205–Lys274 (dsRBD3/4) for the proteins and 1–34 for the sARF1 SBS dsRNA.
eRamachandran plot, as defined by the program Procheck (46).
Figure 2.
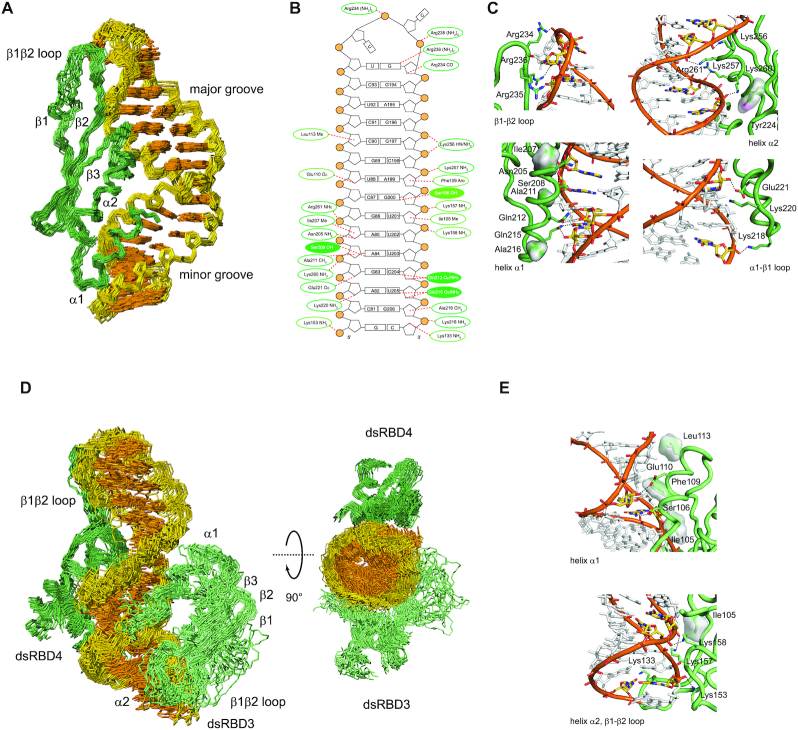
Structure of STAU1 dsRBD4 and dsRBD3/4 in complex with sARF1 SBS dsRNA. (A) Structural ensemble of the STAU1 dsRBD4–sARF1 SBS dsRNA complex. Heavy-atom superposition of the ensemble of the 20 lowest-energy structures (Table 2). The protein backbone is shown in dark green and the RNA heavy atoms of the bases in orange and those of the ribose-phosphodiester backbone are shown in gold (omitting phosphate and 2′-OH oxygens). (B) Schematic representation of sARF1 SBS dsRNA showing interactions of dsRBD3 as well as dsRBD4 as dotted lines. Interactions with the ribose-phosphodiester backbone are circled in dark green for dsRBD4 and light green for dsRBD3 while base interactions are shown as filled circles. (C) Details of the interactions of dsRBD4 β1–β2 loop, helix α2, helix α1 and α1–β2 loop with sARF1 SBS dsRNA. Amino acid side chains mediating interactions with the dsRNA are shown as sticks, interacting dsRNA residues are shown in yellow with nitrogen atoms in blue and oxygen atoms in red and the phosphodiester backbone is in orange with the phosphate oxygens in red. Protein residues making hydrophobic contacts are shown with their Van der Waals surface and hydrogen bonds are indicated by black dotted lines. (D) Structural ensemble of the STAU1 dsRBD3/4–sARF1 SBS dsRNA complex. Heavy-atom superposition of the ensemble of the 20 lowest-energy structures (Table 2). The protein backbone of dsRBD4 is shown in dark green, the one of dsRBD3 in light green and the RNA heavy atoms of the bases in orange and those of the ribose-phosphodiester backbone are shown in gold (omitting phosphate and 2′-OH oxygens). A second view rotated by 90° along the horizontal axis shows that dsRBD3 and dsRBD4 interact with sARF1 SBS dsRNA on opposite sides of the helix. The linker residues (172–204) are omitted. (E) Details of the interactions of dsRBD3 helix α1, helix α2 and β1–β2 loop with sARF1 SBS dsRNA. Color scheme as in C.
The STAU1 dsRBD4 adopts the canonical α−β−β−β−α dsRBD fold and interacts with the sARF1 SBS dsRNA in the expected three regions (Figure 2A). Region 3, the N-terminal end of helix α2 of STAU1 dsRBD4 contains the conserved KKNAK-motif which makes contacts with the phosphate oxygens from the major groove side via its lysine side chains (Figure 2B and C) (51). In the case of STAU1 dsRBD4, these electrostatic interactions with the phosphodiester backbone occur in pairs with Lys256 hydrogen bonding the phosphate oxygens of G197 and C198 via the backbone amide and side chain amino groups, respectively, and Lys257 hydrogen bonding the phosphate oxygens of A199 via the side chain amino group. On the other side of the major groove of the sARF1 SBS dsRNA, a second pair with Lys260 and Arg261 hydrogen bonds the phosphate oxygens of A84 and G86 via the side chain amino and guanidinium groups, respectively (Figure 2B and C). The interaction of Lys260 is also stabilized by a hydrophobic interaction of its side chain with Tyr224 from the β1 strand. These interactions aid to recognize A-form dsRNA and are crucial for dsRNA binding affinity since replacement of basic amino acids in the KKNAK-motif by acidic amino acids as found in STAU1 dsRBD5 is incompatible with dsRNA binding (11).
In region 2, interactions of Arg234, Arg235 and Arg236 place the β1-β2 loop in the minor groove of the UUCG tetraloop which we used to cap the end of sARF1 SBS dsRNA. Arg234 hydrogen bonds to the phosphodiester backbone via its guanidinium group as well as to a ribose 2′ hydroxyl group via its backbone carbonyl oxygen (Figure 2B and C). The guanidinium group of Arg235 hydrogen bonds to the phosphodiester backbone and Arg236 hydrogen bonds to the O6 carbonyl oxygen of the guanosine of the UUCG tetraloop which adopts a syn-conformation. The latter contact is not sequence-specific and could also occur with the O2 carbonyl oxygen of pyrimidines which is consistent with a uracil in this position of the 100 nt loop of ARF1 SBS (23,24). Overall, the β1–β2 loop interactions read out minor groove features and shape of an RNA hairpin loop in a non-sequence-specific manner thereby anchoring the STAU1 dsRBD4 at the end of sARF1 SBS dsRNA.
The last canonical interaction point of a dsRBD with dsRNA is region 1 which usually comprises residues 3, 4, 7, 8 and 11 of helix α1 making partially sequence-specific contacts to the minor groove of dsRNA (51). In the case of STAU1 dsRBD4 spacing of residues is maintained but shifted by one amino acid involving Ile207, Ser208, Ala211, Gln212 and Gln215 (Figure 2B). Ile207 makes hydrophobic contacts with the ribose moiety of A85 assisting placement of helix α1 in the minor groove of the sARF1 SBS dsRNA (Figure 2B and C). The side chain hydroxyl of Ser208 forms a hydrogen bond with the N3 of A84 which is considered non-sequence-specific since it could also form with the N3 of guanosine or the O2 carbonyl oxygen of pyrimidines (Figure 2B and C). The methyl groups of Ala211 and also Ala216 are involved in stacking interactions with the ribose moieties of A84 and G206, respectively, further anchoring helix α1 in the minor groove (Figure 2B and C). This interaction is supported by numerous NOEs between the Ala211 and Ala216 methyl group protons and the riboses of G83, A84 and G206, respectively. The base paired partner of G83, C204 shows canonical interactions with the amide side chain of Gln212 contacting the O2 carbonyl oxygen and the 2′ hydroxyl group of the ribose. This type of contact could be maintained by the N3 of a purine and is therefore not considered purely sequence-specific (55). The last position also involves side chain contacts of Gln215 with O2 carbonyl oxygen and the 2′ hydroxyl group of U205 (Figure 2B and C). Beside the contacts via helix α1, also one linker residue, namely Asn205, contacts the minor groove side of the dsRNA by forming a hydrogen bond of the side chain amide to the 2′ hydroxyl group of A85 (Figure 2C).
Beside the canonical contacts from regions 1–3 of the STAU1 dsRBD4, we also observed three electrostatic contacts from Lys218, Lys220 and Glu221 of the α1–β1 loop. The side chain amino group of Lys218 hydrogen bonds to the phosphodiester backbone and the side chain carboxyl of Glu221 hydrogen bonds the 2′ hydroxyl group of G83. Lys220 also approaches the phosphodiester backbone of A82, suggesting probably a water-mediated interaction (Figure 2C).
Thus STAU1 dsRBD4 interacts with two consecutive pyrimidines on one strand and a purine on the opposite strand in an A-form dsRNA helix and additional hydrogen bonding and hydrophobic contacts to the minor and major grooves assist these interactions. The conservation of the interacting residues among STAU1 dsRBD4 from different species suggests that this binding mode is conserved (Supplementary Figure S4A).
Structure of STAU1 dsRBD3/4–sARF1 SBS dsRNA complex
The structured domains of the final ensemble of the STAU1 tandem dsRBD3/4–sARF1 SBS dsRNA complex are well-defined and the twenty conformers display a r.m.s. deviation of 1.95 Å for all heavy atoms (Table 2 and Figure 2D). The flexible linker between the dsRBDs could only be partially assigned and is therefore unstructured in the final ensemble of structures (Supplementary Figure S4B). Using the segmentally labeled STAU1 dsRBD3/4 protein with only dsRBD3 and part of the linker being 13C,15N-labeled allowed us to extract 46 intermolecular NOEs from a 3D 13C F1-filtered F2-edited HSQC-NOESY spectrum and to define the interaction of STAU1 dsRBD3 with sARF1 SBS dsRNA (Supplementary Figure S3C-E). The lower number of intermolecular NOEs for the dsRBD3–sARF1 SBS dsRNA interaction as compared to dsRBD4 is reflected in a lower r.m.s. deviation of 2.68 Å for all protein heavy atoms as compared to 1.37 Å for dsRBD4 (Supplementary Figure S4C). The structure also rationalizes the observed minor differences in the 1H-15N HSQC spectra of dsRBD3/4 bound to long ARF1 or sARF1 SBS dsRNA (Figure 1D and Supplementary Figure S1C). Upon shortening of the ARF1 SBS dsRNA signal, differences occur in both helices of dsRBD3 and in the β1–β2 loop and β3 strand of dsRBD4 (Supplementary Figure S5A). All the differences in the amide chemical shifts occur in residues not involved in interaction with the dsRNA and the chemical shift pattern in 3D 15N NOESY-HSQC spectra is preserved (Supplementary Figure S5B and C). Thus, these differences in amide chemical shifts suggest stabilization of secondary structure elements when bound to the sARF1 SBS dsRNA rather than differences in the interaction with the dsRNA. In dsRBD3 which is more dynamic in the free form as compared to dsRBD4, the shift differences might indicate stabilization of the C-terminus of helix α1 (K114) and the N-terminus of helix α2 (S156) upon sARF1 SBS dsRNA binding, and in dsRBD4 the differences cluster in one region of the protein indicating a stabilization of the β1–β2 loop and adjacent β3 strand upon binding to the tetraloop of sARF1 SBS dsRNA (Supplementary Figure S5C).
STAU1 dsRBD3 and dsRBD4 bind on the opposite sides of the sARF1 SBS dsRNA and thus do not form a protein-protein interface consistent with the absence of interdomain NOEs in the 3D 13C F1-filtered F2-edited HSQC-NOESY spectrum recorded on the segmentally labeled STAU1 dsRBD3/4–sARF1 SBS dsRNA complex (Figure 2D). As seen for dsRBD4, dsRBD3 also interacts with sARF1 SBS dsRNA in the canonical 3 regions. Helix α1 is placed in the minor groove through stacking interactions of Ile105, Phe109 and Leu113 with the ribose moieties of U201 and A199 on one strand and C90 on the other strand. This placement results in an interaction of the side chain carboxyl group of Glu110 with the 2′ hydroxyl of the ribose moiety of U88 (Figure 2E). Furthermore, the side chain hydroxyl group of Ser106 interacts with the amino group of G200 which is considered a specific interaction for G-C or C-G Watson–Crick base pairs (Figure 2E) (51). In region 3, Lys157 and Lys158 interact with the phosphodiester backbone of one RNA strand and Lys153 on the opposite strand. The positioning of Lys158 is additionally stabilized by a hydrophobic contact with Ile105 from helix α1. Finally, in the ß1–ß2 loop, the side chain amino group of Lys133 contacts the ribose O4′ or O2′ oxygens probably in a water-mediated manner (Figure 2E).
Comparison of the NMR and crystal structures of the STAU1 dsRBD3/4–sARF1 SBS dsRNA complex
The recently published crystal structure of STAU1 dsRBD3/4 bound to the long, 19 bp ARF1 SBS duplex RNA identifies the same set of interacting protein residues with minor deviations but differs both in stoichiometry and arrangement of the dsRBDs along the dsRNA (27). In the crystal structure, long ARF1 SBS dsRNA binds two copies of dsRBD3 on opposite sides of the RNA helix and one copy of dsRBD4 interacting with one copy of dsRBD3 and the dsRNA while the second copy of dsRBD4 is disordered (Supplementary Figure S6A). In our hands, STAU1 dsRBD3/4 did not form a stable 2:1 complex with the long ARF1 SBS dsRNA as shown by our NMR titration experiments which are indicative of stable complex formation only at a protein to RNA ratio of 1:1 (Figure 1D and Supplementary Figure S1A). Similarly, the molecular weight derived from SAXS measurements of STAU1 dsRBD4 and dsRBD3/4–sARF1 SBS dsRNA complexes and overall good fit of solution scattering from atomic models to experimental SAXS data (CRYSOL χ2 = 1.30 and 1.07, respectively) also supports a 1:1 complex formation in solution (Supplementary Table S1 and Figure S6B–E).
Furthermore, based on our initial NMR experiments, we further shortened the ARF1 SBS dsRNA and showed that complex formation is the same as observed with the longer dsRNA (Supplementary Figure S1B–E), again with a 1:1 stoichiometry. The difference in stoichiometry for the longer ARF1 SBS dsRNA might arise from the fact that our long ARF1 SBS is capped by a stable tetraloop while in the crystal structure a plain dsRNA was used allowing one copy of the two dsRBD3 to interact with a neighboring dsRNA in the crystal lattice via the ß1-ß2 loop (27). As a possible consequence, one copy of the dsRBD3 binds in the same region of the ARF1 SBS dsRNA as observed in the NMR structure while the second copy of dsRBD3 partly occupies the dsRBD4 binding side observed in the NMR structure (Supplementary Figure S6A). For one dsRBD3 copy, we detect very similar interactions to the same RNA bases and phosphodiester backbone oxygens as observed in the crystal structure, only the guanine-specific interaction of the ß1-ß2 loop is lost due to our shortened sARF1 SBS dsRNA construct and some interactions of the N-terminal amino acids (K102, S103) are not detected either. For dsRBD4, we obtained 178 intermolecular NOEs and we detect more contacts than in the crystal structure where one copy of dsRBD4 binds in the opposite direction as compared to the NMR structure and displays high B-factors while the other one is completely disordered. In region 3 of dsRBD4, we observe one additional interaction of an arginine with the phosphodiester backbone and in region 2 we observe contacts from 3 arginines with the phosphodiester backbone and riboses which are not present in the disordered ß1–ß2 loop of the crystal structure. The helix α1 interactions of dsRBD4 occur through the same amino acid side chains (Q212 and Q215) but with other bases (C204 and U205 instead of C90 in the crystal structure) and we observe a minor groove base contact between A84 and Ser208 which is absent in the crystal structure. Moreover, additional contacts with riboses and the phosphodiester backbone from two lysines and a glutamate in the adjacent α1–ß1 loop are observed. In summary, one dsRBD3 copy shows the same binding characteristics both in the solution and crystal structure while in solution, dsRBD4 binds in the opposite direction along the dsRNA and partly occupies the position of the second dsRBD3 copy observed in the crystal. Regardless of this difference in positioning and orientation of the dsRBDs, the same amino acid side chains interact with the same type of bases thereby preserving shape and sequence read-out from the minor groove.
Minor groove contacts of STAU1 dsRBD3/4 play little role for binding affinity in vitro but are important for target selection in vivo
To test the importance of the minor groove base contacts, we mutated key interacting residues of dsRBD3 and dsRBD4 to alanine and tested the effect of these substitutions on sARF1 SBS dsRNA binding affinity by FA measurements. In dsRBD4, mutation of Gln212 and Gln215 to alanine showed only a 1.6-fold reduction in binding affinity, while combination of these mutations with the third minor groove interacting amino acid, Ser208 showed a 5.2-fold drop in affinity (Table 1 and Supplementary Figure S7A). Likewise, mutation of the guanine-interacting Ser106 in dsRBD3 together with the neighboring hydrophobic sugar contact of Ile105 displayed a 3.6-fold drop in affinity (Table 1 and Supplementary Figure S7B). Interestingly, another hydrophobic sugar contact (Phe109) showed a similarly strong 2.4-fold reduction while alanine mutation of the 2′ hydroxyl (Glu110) and a hydrophobic sugar contact (Leu113) in helix α1 of dsRBD3 did not affect the affinity (Table 1 and Supplementary Figure S7B).
We next examined the effect of these mutations in the context of the STAU1 dsRBD3/4 protein construct. Alanine mutations in dsRBD3 helix α1 (Ile105/Ser106–1.6-fold, Glu110/Leu113–1.0-fold, and Ile105/Ser106/Glu110/Leu113–1.2-fold) of STAU1 dsRBD3/4 showed only minor or no loss in affinity (Table 1 and Supplementary Figure S7C). Similarly, alanine mutations in dsRBD4 helix α1 (Gln212/Gln215–0.9-fold) or in combination with dsRBD3 helix α1 mutations (Glu110/Leu113/Gln212/Gln215–1.0-fold) also showed wild-type (WT) binding affinities confirming that these residues are not major determinants of binding affinity as already seen with the isolated dsRBDs (see above, Table 1 and Supplementary Figure S7C). On the other hand, alanine mutations that had larger effects in the isolated dsRBDs also showed consistently stronger loss in the STAU1 dsRBD3/4 construct (Ser208–1.6-fold, Ser208/Gln212/Gln215–4.9-fold, and Ile105/Ser106/Ser208/Gln212/Gln215–2.5-fold) showing that these minor groove interactions are important for dsRNA target binding. In addition, we also tested alanine mutants in β1–β2 loop in STAU1 dsRBD3/4 (Lys133–1.3-fold, Arg234/Arg236–1.5-fold) which showed only minor loss in affinity. The alanine mutant of Phe109 which showed a 2.4-fold loss in affinity in the isolated dsRBD3 also showed a 1.7-fold loss in affinity in the STAU1 dsRBD3/4, while the alanine mutant of Arg209 which is not in contact with the dsRNA in showed no loss in affinity. Combination of the latter two mutations in STAU1 dsRBD3/4 showed a 1.6-fold loss in affinity confirming that only Phe109 but not Arg209 contribute to binding affinity (Table 1 and Supplementary Figure S7D). Overall, the binding experiments suggest that minor groove interactions are not the major determinants for binding affinity in vitro and that the interaction between STAU1 dsRBD3/4 and sARF1 SBS dsRNA is mainly driven by electrostatic interactions along the phosphodiester backbone as also described for the crystal structure of the complex (27).
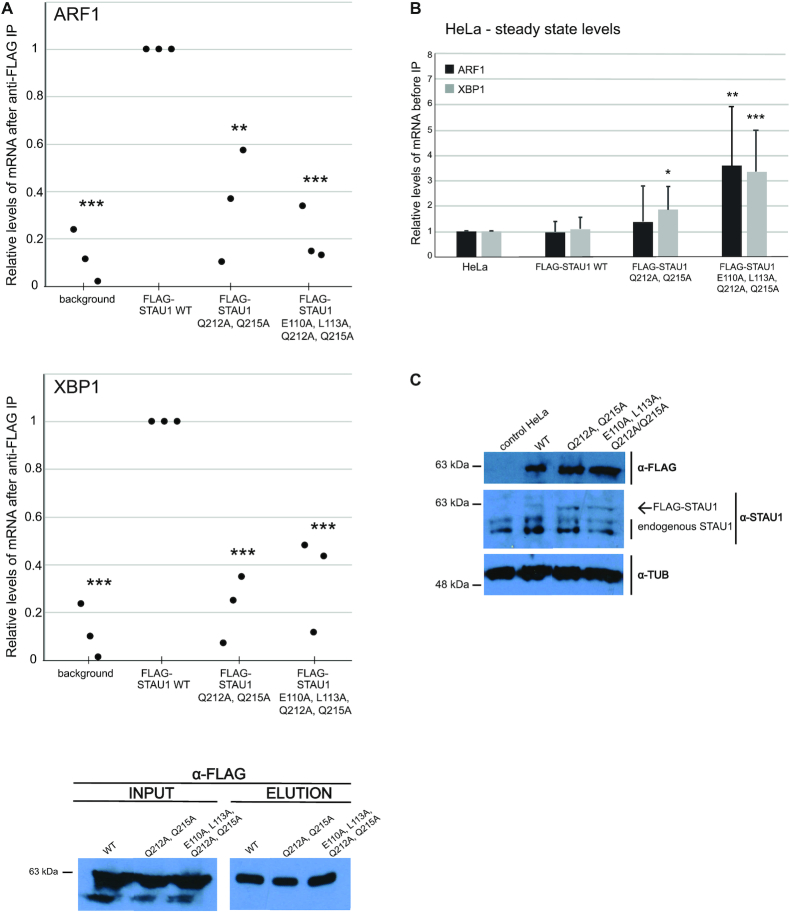
To test the relevance of minor groove interactions on STAU1 in vivo function, we prepared stable HEK293T-REx and T-REx-HeLa cell lines with inducible expression of FLAG-tagged WT and mutant versions of STAU1, respectively. We aimed to evaluate the effect of these mutation on mRNA binding and steady state levels of ARF1 and X-box binding protein 1 (XBP1), two bona-fide targets of SMD(20,23). To assess mRNA binding, we performed RNA-immunoprecipitations (RIP) of STAU1 using the FLAG antibody and quantified the co-precipitated RNAs by RT-qPCR with gene-specific primers directed to either the stem-loop region in the 3′UTR or the coding sequence (Supplementary Figure S8A). Interestingly, the dsRBD4 mutant in Q212A/Q215A that displayed only minor effect on the RNA binding in vitro showed decreased RNA binding in vivo as evaluated based on the level of co-precipitated RNAs (Figure 3A, Supplementary Figure S8B). The overexpression of this dsRBD4 mutant (Q212A/Q215A) was accompanied by a significant, 2-fold upregulation of XBP1 mRNA and insignificant increase in ARF1 mRNA in HeLa cells (Figure 3B). The overexpression of STAU1 containing mutations in both dsRBD3 and 4 (E110A/L113A/Q212A/Q215A) lead to significant (3- to 4-fold) accumulation of both mRNAs in HeLa cells (Figure 3B) and a similar trend was observed in HEK293T cells (Supplementary Figure S8C). Similar results were obtained when we normalized mRNA levels to their corresponding pre-mRNAs in both cell types upon expression of WT and mutant FLAG-tagged forms of STAU1 (Supplementary Figure S8D). This further supported the conclusion that the dsRBD mutations affect STAU1 function in SMD.
Figure 3.
Mutations in dsRBD3 and dsRBD4 impair binding to target mRNAs in vivo. (A) RT-qPCR analysis of the amount of ARF1 and XBP1 mRNAs coprecipitated with WT and mutant STAU1 from HEK293T-REx cells. The Y-axis represents the enrichment of mRNAs coprecipitated with STAU1 variants relative to the level in the whole cell lysate (input). Input ARF1 and XBP1 mRNA levels were normalized to GAPDH as an internal control. The qPCR results were analyzed by the ΔΔCt method. Scatter plot represents relative enrichments of precipitated mRNAs with FLAG-tagged STAU1 mutants relative to the FLAG-WT STAU1. Background is the level of mRNAs unspecifically bound to FLAG-beads from control HEK293T-REx where no FLAG-tagged protein was expressed. Error bars SD (n = 3–4 biological replicates), *P-value < 0.1, **P-value < 0.01, ***P-value < 0.001; P-values were calculated by two-tailed paired t-test. The western blot below the graphs shows the efficiency of FLAG immunoprecipitation. Input is the whole cell lysate before IP. (B) RT-qPCR analysis of steady-state levels of ARF1 and XBP1 mRNA in T-REx-HeLa cells upon overexpression of stably integrated STAU1 variants. Data were analyzed by ΔΔCt calculation method and normalized to GAPDH as an internal control. Bar plot represents fold enrichments relatively to control T-REx-HeLa cells. Error bars SD (n = 3 biological replicates), *P-value < 0.1; P-values were calculated by two-tailed paired t-test. (C) Western blot analysis of STAU1 protein levels upon doxycyclin induction using antibodies as indicated on the right. Tubulin was used as a loading control. Control HeLa is a cell line without any integration and expression of a FLAG-tagged protein.
Accordingly, the dsRBD3 and dsRBD4 mutations led to significantly decreased binding to both mRNAs in vivo (Figure 3A and Supplementary Figure S8B). In summary, combined mutations in minor groove binding residues in STAU1 dsRBD3/4 show decreased dsRNA binding and mRNA accumulation when expressed in the context of the full-length STAU1 in vivo. Thus minor groove contacts are the determinants for target selection in vivo.
DISCUSSION
Our solution structure of STAU1 dsRBD3/4 in complex with sARF1 SBS dsRNA reveals how STAU1 selects target mRNAs for SMD. Both dsRBDs show the canonical binding and while dsRBD4 in isolation binds sARF SBS dsRNA in one preferred orientation, dsRBD3 shows promiscuous binding in different regions of the dsRNA (Supplementary Figure S3). Only in the context of the tandem dsRBD3/4, dsRBD3 selects a single binding site on the dsRNA. To extract this structural information, we used a segmentally labeled STAU1 protein where only dsRBD3 is isotope labeled and dsRBD4 is not and thus invisible in heteronuclear NMR experiments. With this approach, we could perform isotope-filtered NMR experiments which allowed us to extract two types of information: first, NOE-distance information required for definition of the dsRBD3 binding site on the sARF1 SBS dsRNA on the opposite side of dsRBD4 and secondly that the two dsRBD modules are not involved in protein–protein contacts as seen in the crystal structure (27).
In isolation, dsRBD3 binds with 2-fold higher affinity to sARF1 SBS dsRNA as compared to isolated dsRBD4. This might suggest that the dsRBD3 module binds A-form dsRNA first in a promiscuous manner scanning the A-form dsRNA and this aids in recruiting dsRBD4 for dsRNA binding. The dsRBD4 module then selects a defined binding site on the dsRNA and as a consequence, dsRBD3 also binds in a defined location on the dsRNA. In addition, β1–β2 loop interaction of both dsRBDs with opposite ends of the dsRNA might further assist final binding site selection on the dsRNA.
Binding affinity of dsRBDs is mainly driven by electrostatic interactions with the phosphodiester backbone and 2′ hydroxyl groups, interactions which select for dsRNA over dsDNA and thus for regular A-form geometry (51). As such, mutations of charged, basic residues in regions 2 and 3 of STAU1 dsRBD3/4 lead to a 4- to 8-fold reduction in binding affinity as compared to WT protein (27). While these interactions constitute the basis for binding affinity for dsRNA, only very few minor groove base contacts in the right register length are providing sequence specificity and thus determine target selection (51). As a result, altering minor groove readout by mutating key protein residues often has only very little effect on affinity (Supplementary Figure S7). This contrasts protein interactions with ssRNA which are often characterized by an intricate network of hydrogen bonds and Van der Waals interactions and thus very sensitive to protein mutations affecting sequence readout.
We observed differential in vitro RNA binding affinities of STAU1 dsRBD3/4 mutations when tested in individual dsRBDs or in the context of tandem dsRBD3/4 (Table 1). Interestingly, these minor groove contacting mutations showed much stronger effect in the full-length protein in vivo. The combined mutant in dsRBD3/4 (E110A/L113A/Q212A/Q215A) appeared to significantly affect SMD of ARF1 and XBP1 mRNAs. The differences between the in vitro and in vivo dsRNA binding can be a consequence of presenting the mutant STAU1 in the context of the full-length protein and testing endogenous full-length mRNA targets. In HeLa cells, the level of expression of FLAG-tagged STAU1 forms was comparable to the level of endogenous STAU1 (Figure 3C). Yet, we observed more than 3-fold upregulation of both mRNA targets in cells expressing mutations in dsRBD3 and 4 (Figure 3B). It is possible that the introduced STAU1 mutations affected yet additional STAU1 activities in vivo, such as effects on STAU1 localization or its role in mRNA export or translation.
STAU1 recognizes many different dsRNA targets ranging from mRNA transport and localization elements (1), 5′UTR signals to regulate translation (56) to SMD signals in 3′UTRs to regulate mRNA levels (23) and expansion segments in 18S and 28S rRNA (20). Beside ARF1 SBS, the vast number of STAU1 dsRNA targets, especially transport and localization elements, has not been characterized in more detail (15–20). Many target RNAs form long double-stranded helices, such as duplexes formed by long non-coding RNAs and 3′UTRs which also activate SMD (25) and also transport signals are often complex such as the one of Drosophila bicoid mRNA which dimerizes to provide an extended interaction surface for Drosophila dmSTAU binding (2,57). The recognition of such large signals by STAU1 might not only require minor groove interactions by dsRBD3/4 but also in vivo dimerization (11) and other domains, such as dsRBD2. STAU2, for instance, contains an additional dsRBD1 beside dsRBD2/3/4 and both tandem dsRBD1/2 and dsRBD3/4 modules bind dsRNA with similar affinities and kinetics suggesting cooperative binding of all four domains for stable complex formation and target dsRNA selection (58).
The question remains how STAU1 and STAU2 proteins exhibit their diverse functions in post-transcriptional regulation of gene expression when only a few minor groove interactions determine dsRNA target selection. It is feasible that depending on the complexity of the dsRNA signal, the overall shape of STAU1 or STAU2 complexes with target RNAs could differ. This would then expose different STAU1 or STAU2 protein of surfaces for interaction with other proteins mediating RNA transport, translational control, or SMD and thus determine the cellular function of this interaction. Whether this additional layer of complexity confers the diverse functions of STAU1 and STAU2 proteins in post-transcriptional regulation of gene expression needs to be explored with future studies of different full-length STAU1 or STAU2–dsRNA complexes functioning in diverse gene expression pathways.
DATA AVAILABILITY
The coordinates and chemical shifts of the structural ensembles of STAU1 dsRBD4–sARF1 SBS dsRNA and the STAU1 dsRBD3/4–sARF1 SBS dsRNA complexes have been deposited in the OneDep Protein Data Bank under PDB accession number 6SDY and 6SDW, and BMRB accession number 34422 and 34421, respectively. SAXS datasets, fits and experimental details have been deposited in Small Angle Scattering Biological Data Bank (www.sasbdb.org) (59) as entry SASDF29 (STAU1 dsRBD4–sARF1 SBS dsRNA), SASDF39 (STAU1 dsRBD3/4–sARF1 SBS dsRNA), and SASDF49 (STAU1 dsRBD3/4-long ARF1 SBS dsRNA).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Prof. M. Sattler for providing plasmid of WT and mutant Sortase A, Dr J. Ule for providing the Flag-hSTAU1 plasmid, helpful staff from all CEITEC core facilities and previous as well as current lab members for their useful discussion and technical help during this study.
Authors contributions’: P.J.L. and D.K.Y. designed the project. D.K.Y. expressed and purified proteins and performed in vitro RNA transcription and purification. D.K.Y. optimized the STAU1-ARF1 complexes and prepared all samples for structural studies. M.Z., M.K., C.B. and D.K.Y. prepared samples for FA measurements. D.K.Y. performed all NMR experiments with help of P.J.L. D.K.Y. and P.J.L. analyzed NMR data and performed structure calculations. M.Z., M.K., C.B. and D.K.Y performed FA measurements and analysis. T.K., performed and analyzed SAXS measurements. D.Z. performed all the experiments in mammalian cells, constructs, stable cell lines, mRNA quantifications by RT-qPCR, and western blotting, D.Z. and S.V analyzed the data. P.J.L. wrote the manuscript, D.Z. and S.V. wrote method and results part on the in vivo experiments in mammalian cells. All authors discussed the results and approved the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EU Seventh Framework Programme under the ‘Capacities’ specific programme [286154–SYLICA]; INBIOR [CZ.1.07/2.3.00/20.0042]; CEITEC 2020 (LQ1601) project with financial contribution made by the Ministry of Education, Youths and Sports of the Czech Republic (MEYS CR) within special support paid from the National Programme for Sustainability II funds; CIISB Research Infrastructure project LM2015043 funded by MEYS CR; Diamond Light Source; B21 Beamline Scientists; Marie Curie Action-Career Integration Grant [PCIG14-GA-2013–630758]; EMBO Installation Grant [3014]; Czech Science Foundation Grant [P305/18/08153S]; Human Frontier Science Program Grant [RGP0024/2008]; Czech Science Foundation Grant [16–21341S to S.V., D.Z.]. Funding for open access charge: Grantová Agentura České Republiky [P305/18/08153S].
Conflict of interest statement. None declared.
REFERENCES
- 1. Heraud-Farlow J.E., Kiebler M.A.. The multifunctional Staufen proteins: conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci. 2014; 37:470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. St Johnston D., Beuchle D., Nusslein-Volhard C.. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991; 66:51–63. [DOI] [PubMed] [Google Scholar]
- 3. Ephrussi A., Dickinson L.K., Lehmann R.. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991; 66:37–50. [DOI] [PubMed] [Google Scholar]
- 4. Ferrandon D., Elphick L., Nusslein-Volhard C., St Johnston D.. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994; 79:1221–1232. [DOI] [PubMed] [Google Scholar]
- 5. St Johnston D. The intracellular localization of messenger RNAs. Cell. 1995; 81:161–170. [DOI] [PubMed] [Google Scholar]
- 6. Wickham L., Duchaine T., Luo M., Nabi I.R., DesGroseillers L.. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell Biol. 1999; 19:2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Monshausen M., Putz U., Rehbein M., Schweizer M., DesGroseillers L., Kuhl D., Richter D., Kindler S.. Two rat brain staufen isoforms differentially bind RNA. J. Neurochem. 2001; 76:155–165. [DOI] [PubMed] [Google Scholar]
- 8. Duchaine T.F., Hemraj I., Furic L., Deitinghoff A., Kiebler M.A., DesGroseillers L.. Staufen2 isoforms localize to the somatodendritic domain of neurons and interact with different organelles. J. Cell Sci. 2002; 115:3285–3295. [DOI] [PubMed] [Google Scholar]
- 9. Ramos A., Grunert S., Adams J., Micklem D.R., Proctor M.R., Freund S., Bycroft M., St Johnston D., Varani G.. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000; 19:997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martel C., Dugre-Brisson S., Boulay K., Breton B., Lapointe G., Armando S., Trepanier V., Duchaine T., Bouvier M., Desgroseillers L.. Multimerization of Staufen1 in live cells. RNA. 2010; 16:585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gleghorn M.L., Gong C., Kielkopf C.L., Maquat L.E.. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nat. Struct. Mol. Biol. 2013; 20:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luo M., Duchaine T.F., DesGroseillers L.. Molecular mapping of the determinants involved in human Staufen-ribosome association. Biochem. J. 2002; 365:817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ravel-Chapuis A., Belanger G., Yadava R.S., Mahadevan M.S., DesGroseillers L., Cote J., Jasmin B.J.. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J. Cell Biol. 2012; 196:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elbarbary R.A., Li W., Tian B., Maquat L.E.. STAU1 binding 3′UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev. 2013; 27:1495–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Furic L., Maher-Laporte M., DesGroseillers L.. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA. 2008; 14:324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heraud-Farlow J.E., Sharangdhar T., Li X., Pfeifer P., Tauber S., Orozco D., Hormann A., Thomas S., Bakosova A., Farlow A.R. et al.. Staufen2 regulates neuronal target RNAs. Cell Rep. 2013; 5:1511–1518. [DOI] [PubMed] [Google Scholar]
- 17. Laver J.D., Li X., Ancevicius K., Westwood J.T., Smibert C.A., Morris Q.D., Lipshitz H.D.. Genome-wide analysis of Staufen-associated mRNAs identifies secondary structures that confer target specificity. Nucleic Acids Res. 2013; 41:9438–9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Lucas S., Oliveros J.C., Chagoyen M., Ortin J.. Functional signature for the recognition of specific target mRNAs by human Staufen1 protein. Nucleic Acids Res. 2014; 42:4516–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ricci E.P., Kucukural A., Cenik C., Mercier B.C., Singh G., Heyer E.E., Ashar-Patel A., Peng L., Moore M.J.. Staufen1 senses overall transcript secondary structure to regulate translation. Nat. Struct. Mol. Biol. 2014; 21:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sugimoto Y., Vigilante A., Darbo E., Zirra A., Militti C., D’Ambrogio A., Luscombe N.M., Ule J.. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015; 519:491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Popp M.W., Maquat L.E.. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu. Rev. Genet. 2013; 47:139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park E., Maquat L.E.. Staufen-mediated mRNA decay. Wiley Interdiscip. Rev. RNA. 2013; 4:423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim Y.K., Furic L., Desgroseillers L., Maquat L.E.. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005; 120:195–208. [DOI] [PubMed] [Google Scholar]
- 24. Kim Y.K., Furic L., Parisien M., Major F., DesGroseillers L., Maquat L.E.. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007; 26:2670–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gong C., Maquat L.E.. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′UTRs via Alu elements. Nature. 2011; 470:284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gong C., Tang Y., Maquat L.E.. mRNA-mRNA duplexes that autoelicit Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 2013; 20:1214–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lazzaretti D., Bandholz-Cajamarca L., Emmerich C., Schaaf K., Basquin C., Irion U., Bono F.. The crystal structure of Staufen1 in complex with a physiological RNA sheds light on substrate selectivity. Life Sci. Alliance. 2018; 1:e201800187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elantak L., Wagner S., Herrmannova A., Karaskova M., Rutkai E., Lukavsky P.J., Valasek L.. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J. Mol. Biol. 2010; 396:1097–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu H.T., Naismith J.H.. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008; 8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen I., Dorr B.M., Liu D.R.. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:11399–11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Freiburger L., Sonntag M., Hennig J., Li J., Zou P., Sattler M.. Efficient segmental isotope labeling of multi-domain proteins using Sortase A. J. Biomol. NMR. 2015; 63:1–8. [DOI] [PubMed] [Google Scholar]
- 32. Lukavsky P.J., Puglisi J.D.. Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides. RNA. 2004; 10:889–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Easton L.E., Shibata Y., Lukavsky P.J.. Rapid, nondenaturing RNA purification using weak anion-exchange fast performance liquid chromatography. RNA. 2010; 16:647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zlobina M., Sedo O., Chou M.Y., Slepankova L., Lukavsky P.J.. Efficient large-scale preparation and purification of short single-stranded RNA oligonucleotides. Biotechniques. 2016; 60:75–83. [DOI] [PubMed] [Google Scholar]
- 35. Ryder S.P., Recht M.I., Williamson J.R.. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 2008; 488:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sattler M., Schleucher J., Griesinger C.. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Mag. Res. Spectrosc. 1999; 34:93–158. [Google Scholar]
- 37. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 38. Kazimierczuk K., Zawadzka A., Kozminski W.. Optimization of random time domain sampling in multidimensional NMR. J. Magn. Reson. 2008; 192:123–130. [DOI] [PubMed] [Google Scholar]
- 39. Stanek J., Kozminski W.. Iterative algorithm of discrete Fourier transform for processing randomly sampled NMR data sets. J. Biomol. NMR. 2010; 47:65–77. [DOI] [PubMed] [Google Scholar]
- 40. Lukavsky P.J., Puglisi J.D.. RNAPack: an integrated NMR approach to RNA structure determination. Methods. 2001; 25:316–332. [DOI] [PubMed] [Google Scholar]
- 41. Peterson R.D., Theimer C.A., Wu H., Feigon J.. New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA-protein complexes. J. Biomol. NMR. 2004; 28:59–67. [DOI] [PubMed] [Google Scholar]
- 42. Lee W., Revington M.J., Arrowsmith C., Kay L.E.. A pulsed field gradient isotope-filtered 3D 13C HMQC-NOESY experiment for extracting intermolecular NOE contacts in molecular complexes. FEBS Lett. 1994; 350:87–90. [DOI] [PubMed] [Google Scholar]
- 43. Herrmann T., Guntert P., Wuthrich K.. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR. 2002; 24:171–189. [DOI] [PubMed] [Google Scholar]
- 44. Herrmann T., Guntert P., Wuthrich K.. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002; 319:209–227. [DOI] [PubMed] [Google Scholar]
- 45. Case D.A., Cheatham T.E. 3rd, Darden T., Gohlke H., Luo R., Merz K.M. Jr, Onufriev A., Simmerling C., Wang B., Woods R.J.. The Amber biomolecular simulation programs. J. Comput. Chem. 2005; 26:1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M.. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996; 8:477–486. [DOI] [PubMed] [Google Scholar]
- 47. Pollard T.D. A guide to simple and informative binding assays. Mol. Biol. Cell. 2010; 21:4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 49. Panjkovich A., Svergun D.I.. CHROMIXS: automatic and interactive analysis of chromatography-coupled small-angle X-ray scattering data. Bioinformatics. 2018; 34:1944–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Franke D., Petoukhov M.V., Konarev P.V., Panjkovich A., Tuukkanen A., Mertens H.D.T., Kikhney A.G., Hajizadeh N.R., Franklin J.M., Jeffries C.M. et al.. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017; 50:1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Masliah G., Barraud P., Allain F.H.. RNA recognition by double-stranded RNA binding domains: a matter of shape and sequence. Cell Mol. Life Sci. 2013; 70:1875–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stefl R., Oberstrass F.C., Hood J.L., Jourdan M., Zimmermann M., Skrisovska L., Maris C., Peng L., Hofr C., Emeson R.B. et al.. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010; 143:225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dominguez C., Schubert M., Duss O., Ravindranathan S., Allain F.H.. Structure determination and dynamics of protein-RNA complexes by NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2011; 58:1–61. [DOI] [PubMed] [Google Scholar]
- 54. Yadav D.K., Lukavsky P.J.. NMR solution structure determination of large RNA-protein complexes. Prog. Nucl. Magn. Reson. Spectrosc. 2016; 97:57–81. [DOI] [PubMed] [Google Scholar]
- 55. Gan J.H., Tropea J.E., Austin B.P., Court D.L., Waugh D.S., Ji X.H.. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006; 124:355–366. [DOI] [PubMed] [Google Scholar]
- 56. Dugre-Brisson S., Elvira G., Boulay K., Chatel-Chaix L., Mouland A.J., DesGroseillers L.. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005; 33:4797–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ferrandon D., Koch I., Westhof E., Nusslein-Volhard C.. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997; 16:1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Heber S., Gaspar I., Tants J.N., Gunther J., Moya S.M.F., Janowski R., Ephrussi A., Sattler M., Niessing D.. Staufen2-mediated RNA recognition and localization requires combinatorial action of multiple domains. Nat. Commun. 2019; 10:1659–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Valentini E., Kikhney A.G., Previtali G., Jeffries C.M., Svergun D.I.. SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res. 2015; 43:D357–D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lukavsky P.J., Kim I., Otto G.A., Puglisi J.D.. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003; 10:1033–1038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and chemical shifts of the structural ensembles of STAU1 dsRBD4–sARF1 SBS dsRNA and the STAU1 dsRBD3/4–sARF1 SBS dsRNA complexes have been deposited in the OneDep Protein Data Bank under PDB accession number 6SDY and 6SDW, and BMRB accession number 34422 and 34421, respectively. SAXS datasets, fits and experimental details have been deposited in Small Angle Scattering Biological Data Bank (www.sasbdb.org) (59) as entry SASDF29 (STAU1 dsRBD4–sARF1 SBS dsRNA), SASDF39 (STAU1 dsRBD3/4–sARF1 SBS dsRNA), and SASDF49 (STAU1 dsRBD3/4-long ARF1 SBS dsRNA).