Abstract
Cancer stem cells (CSCs) underpin the resistance of breast cancer (BC) cells to therapy. Dendritic cell (DC)-based treatment is efficacious and safe, but the efficiency of this technique for targeting CSCs in BC treatment requires further investigation. The present study aimed to investigate the ability of DCs pulsed with breast CSC antigens to activate effector lymphocytes for killing BC cells. CD44+/CD24− CSCs were isolated from BCA55-121, an in-house patient-derived BC cell line, and acquisition of stemness properties was confirmed by upregulated expression of OCT4A and a superior proliferative capacity in colony formation assays compared with whole population of BCA55-121 (BCA55-121-WP). DCs were differentiated from monocytes from peripheral blood of healthy donors and pulsed with CSC total RNA. Maturation of the CSC RNA-pulsed DCs was confirmed by increased expression of CD11c, CD40, CD83, CD86 and HLA-DR, as well as reduced CD14 expression compared with monocytes. Total lymphocytes co-cultured with CSC RNA-pulsed DCs were analyzed by flow cytometry for markers including CD3, CD4, CD8, CD16 and CD56. The results revealed that the co-cultures contained mostly cytotoxic CD8+ T lymphocytes followed by CD4+ T lymphocytes and smaller populations of natural killer (NK) and NKT cells. ELISA was used to measure IFN-γ production, and it was revealed that activated CD4+ and CD8+ lymphocytes produced more IFN-γ compared with naïve T cells, suggesting that CD8+ T cells were effector T cells. CSC RNA was a more efficient antigen source compared with RNA from mixed BC cells for activating tumor antigen-specific killing by T cells. These CSC-specific effector T cells significantly induced BC cell apoptosis at a 20:1 effector T cell:tumor cell ratio. Of note, the breast CSCs cultures demonstrated resistance to effector T cell killing, which was in part due to increased expression of programmed death ligand 1 in the CSC population. The present study highlights the potential use of CSC RNA for priming DCs in modulating an anticancer immune response against BC.
Keywords: breast cancer, cancer stem cell, dendritic cell, T lymphocytes, programmed death ligand 1
Introduction
Breast cancer (BC) is the most common cancer in women worldwide with an estimated 2.4 million cases in 2015 (1). Standard treatments of patients with BC include loco-regional therapy (surgery and radiation), hormonal therapy, chemotherapy and molecular targeted therapy. The afore-mentioned treatment regimens have existed for a number of years, however, drug resistance and cancer recurrence still pose a problem for the treatment of patients with BC (2,3). A subpopulation of cells in tumors exhibit stem cell properties, including dormancy, self-renewal, infinite proliferation and multi-lineage differentiation (4,5). Studies in various types of cancer, in particular BC, have demonstrated that cancer stem cells (CSCs) subpopulations play a role in cancer recurrence, therapy resistance, metastasis and aggressive relapse (6–8). Therefore, CSCs are becoming important targets for cancer therapy (8,9). In a number of studies investigating BC, distinct cell subpopulations with high tumorigenicity and stem cell properties have been isolated (10,11). A tumorigenic subpopulation of BC cells that express CD44 with low or negative CD24 expression (CD44+/CD24−/low) has been identified as a major breast CSC population (12).
Dendritic cell (DC)-based treatment has been widely studied over the past two decades (13–15). Most of the clinical trials of DC-based treatment in melanoma, prostate cancer, malignant glioma and renal cell carcinoma demonstrated an increase in median overall survival with minimal toxicity (16). In a previous study, human breast CSCs were injected into NOD/SCID mouse mammary fat pads and used to prime human DCs; after mature DCs were re-injected into mice, they exhibited longer survival times and lower tumor masses compared with mice that had not received DC treatment (17). These studies suggested that tumor antigen-activated DCs may be a promising therapy for BC and that CSC antigens may be used for the production of efficacious DC-based treatment in treating refractory human cancers in general.
To the best of our knowledge, no study has evaluated DC-based therapy using human breast CSC antigens to activate effector T cells for cancer cell killing to date. Thus, the aim of this study was to explore whether CSC antigens may evoke an effective immune response against both CSC and non-CSC cells, which may cause tumor regression and prevent tumor resistance.
Materials and methods
Isolation and characterization of primary BC cell culture
The study protocol was evaluated and approved by the Siriraj Institutional Review Board (approval nos. Si520/2010 and Si321/2016; Bangkok, Thailand). Written informed consent was obtained from all individual participants prior to enrollment in the study between January 2010 and December 2018. A fresh luminal BC tissue sample (0.5×0.5×1.0 cm3) isolated from BC tissues surgically resected from a 48-year old Thai female patient with T2N1aM0 (stage IIB) BC was incubated in 10X antibiotic mixture (1 U/ml penicillin G sodium and 1 mg/ml streptomycin; Thermo Fisher Scientific, Inc.) diluted in DMEM/F12. Following incubation, the cells were washed three times with PBS to remove red blood cells (RBC) and debris. The obtained tissue was chopped into 0.1×0.1×0.1 cm3 sections and plated in a culture dish. Cell trypsinization was performed using 0.25% trypsin and 0.9 mM EDTA (Thermo Fisher Scientific, Inc.) to activate cell proliferation. The cancer cell line obtained using the aforementioned protocol was termed BCA55-121, and karyotype analysis was performed by Giemsa staining (Division of Medical Genetics, Office for Research and Development, Faculty of Medicine Siriraj Hospital). Chromosomes were observed and counted under microscope. Growth curves of BCA55-121 were generated using a Live-Cell imager (IncuCyte® Zoom; Sartorius AG) according to the manufacturer's instructions. HLA typing was performed at the Department of Transfusion Medicine, Faculty of Medicine Siriraj Hospital. BCA55-121 cells were cultured in DMEM/F12 (Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) and placed in a 5% CO2 incubator at 37°C. The presence of protein markers including pan-cytokeratin (pan-CK), programmed death ligand 1 (PD-L1), fibroblast-activation protein (FAP) was confirmed in BCA55-121 cells by immunofluorescence following incubation with anti-panCK (1:100; cat. no. sc-8018; Santa Cruz Biotechnology, Inc.), anti-PD-L1 (1:100; cat. no. ab205921; Abcam), and anti-FAP (1:100; cat. no. ab53066; Abcam) for 2–3 h at room temperature. Cells were then incubated with appropriate secondary antibodies, including anti-mouse horseradish peroxidase (HRP) conjugate Cy3 (1:2,000; cat. no. 115-166-071) and anti-rabbit HRP conjugated FITC (1:400; cat. no. ab6717; Abcam) at room temperature for 1 h in the dark. Nucleus was stained using Hoechst 33342 (1:1,000; Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at room temperature. The proliferation of BCA55-121 cells was analyzed by Incucyte® and analyzed in Incucyte® Zoom System, and was compared with the proliferation of the commercial cell lines MCF-7 and MDA-MB-231.
Breast CSC isolation and analysis
BCA55-121 cells were incubated with Allophycocyanin (APC)-labeled anti-CD44 (1:5; cat. no. 21270446; ImmunoTools GmbH) and FITC-labeled anti-CD24 (1:5; cat. no. 21270443; ImmunoTools GmbH) antibodies for 30 min at 4°C. A CytoFLEX flow cytometer (Beckman Coulter, Inc.) was used for flow cytometric analysis using CytExpert software version 2.1 (Beckman Coulter, Inc.). For CSC isolation, 1×107 BCA55-121 cells labeled with the fluorescent antibodies were suspended in 2 ml 2% FBS/PBS, and CD44+CD24- cells were collected using a FACSAria II cell sorter (BD Biosciences). The purity of the obtained cells was assessed using the CytoFLEX flow cytometer.
Soft agar colony formation assay
A 0.5% base agar layer was prepared by mixing 1% agarose with 2X DMEM (or DMEM/F12) supplemented with 20% FBS in a 1:1 ratio. An over-layer of 0.35% agar cell suspension was prepared by mixing 0.7% agarose with 2X DMEM (or DMEM/F12) in a ratio of 1:1. BCA55-121 cells and their CSCs were suspended (4×104 cells/ml), in this mixture, which was incubated at 37°C in 5% CO2 for 14–21 days. Subsequently, cells were fixed with 5% glutaraldehyde and stained with 0.5% crystal violet at room temperature for 15 and 30 min, respectively. Colonies with a diameter >500 µm were counted and surface areas of the formed colonies were measured using an inverted microscope IX71 (original magnification, ×100) at 1 h (baseline) and at the end of the experiment. The data obtained from CSCs are presented compared with the whole population of cultured BC cells.
Western blotting of CSC-related proteins and programmed death ligand-1 (PD-L1)
Proteins (20 µg) extracted from BCA55-121 cells using extraction buffer [0.25 M Tris-HCl pH 6.8, 5% glycerol, 4% SDS, 10% β-mercaptoethanol, 0.001% (w/v) bromophenol blue] were separated using 12% polyacrylamide gels (Sigma-Aldrich; Merck KGaA). Proteins were then transferred to 0.45-µm nitrocellulose membranes. The membranes were blocked for 1 h with 5% BSA in TBST (25 mM Tris-HCl, pH 7.5; 125 mM NaCl; 0.05% Tween 20) at room temperature and incubated overnight at 4°C with anti-human SOX2 rabbit monoclonal antibody (1:1,000; cat. no. 3579; Cell Signaling Technology, Inc.), anti-human OCT4A rabbit monoclonal antibody (1:1,000; cat. no. 2840; Cell Signaling Technology, Inc.), anti-human homeobox protein Nanog (NANOG) rabbit monoclonal antibody (1:1,000; cat. no. 4903, Cell Signaling Technology, Inc.) or anti-human PD-L1 rabbit monoclonal antibody (1:500; cat. no. ab205921; Abcam). The membranes were then washed three times with TBST and incubated with a HRP-conjugated goat anti-rabbit IgG antibody (1:1,000; cat. no. ab6721; Abcam) for 2 h at room temperature. The proteins were visualized using SuperSignal West Pico Chemi-luminescent Substrate (Thermo Fisher Scientific, Inc.) under a Gel Documentation System (G:Box; version EF; Syngene). Anti-β-actin antibody (1:5,000; Santa Cruz Biotechnology, Inc.; cat. no. sc-47778) was incubated for 1 h at room temperature to detect β-actin as a loading control.
Dendritic cell activation and immunophenotyping assay
Venous blood (50 ml) from three healthy donors with aged between 25 and 30 years with no underlying diseases was collected in 0.1% heparin solution and peripheral blood mononuclear cells (PBMCS) were isolated using Lymphosep® lymphocyte separation media (Biowest) according to the manufacturer's instructions. Briefly, heparinized blood was overlaid on Lymphosep® media and centrifuged at 400 × g at 20°C for 30 min. PBMCs were collected and washed three times at room temperature using 200 × g centrifugations for 10 min with PBS or RBC lysis buffer (150 mM NH4Cl, 10 mM NaHCO3 and 1.26 mM EDTA), and then resuspended in AIM-V® medium (Thermo Fisher Scientific, Inc.) and plated onto 6-well plates at 5×106 cells/well. After 2 h of incubation at 37°C and pH 6.8–7.3, non-adherenT cells were gently removed and suspended in 900 µl human type AB serum (Merck KGaA) with 100 µl DMSO and cryopreserved at −80°C as a source of lymphocytes. The adherent monocyte-enriched cells were cultured at 37°C in 5% CO2 incubator in AIM-V® medium containing granulocyte-macrophage colony stimulating factor (50 ng/ml) and interleukin (IL)-4 (25 ng/ml) (ImmunoTools GmbH) for 6 days to produce immature dendritic cells (iDCs). The iDCs were then matured to mature dendritic cells (mDCs) after incubation at 37°C in 5% CO2 with tumor necrosis factor-α (50 ng/ml) and interferon (IFN)-γ (50 ng/ml) (ImmunoTools GmbH) in AIM-V® medium for 2 days. Subsequently, wells containing ~5×105 mDCs were pulsed with 10 µg cancer cell-derived RNA. For phenotypic analysis of DCs, 5×105 DCs were detached with 5 mM EDTA, washed in 2% FBS/PBS and incubated with fluorescence-conjugated monoclonal antibodies (1:50) at 4°C for 30 min. Following washing in 2% FBS/PBS twice, cells were analyzed using a CytoFLEX flow cytometer. To determine the phenotype of monocytes and DCs, anti-CD11c APC (1:50; cat. no. 21487116; ImmunoTools GmbH), anti-CD14 APC (1:50; 21620146; ImmunoTools GmbH), anti-CD14 FITC (1:50; cat. no. 21620143; ImmunoTools GmbH), anti-CD40 FITC (1:50, cat. no. 21270403; ImmunoTools GmbH), anti-CD83 phycoerythrin (PE; 1:20; cat. no. 12-0839-42; Thermo Fisher Scientific, Inc.), anti-CD86 PE (1:20; cat. no. 12-0869-42; Thermo Fisher Scientific, Inc.) and anti-HLA-DR FITC (1:50; cat. no. 21278993; ImmunoTools GmbH) monoclonal antibodies were used. To determine human leukocyte antigen-A2 (HLA-A2) expression, PBMCs from healthy donors were collected using the method described above and analyzed using anti-HLA-A2 APC antibody (1:50; cat. no. 17-9876-42; ImmunoTools GmbH) and the CytoFLEX Flow cytometer.
Total effector lymphocytes and CD8+ T cell preparation
Cryopreserved lymphocytes were thawed and resuspended in RPMI medium Thermo Fisher Scientific, Inc.) supplemented with 5% human AB serum (Merck KGaA). The lymphocytes were activated by co-culture with RNA-pulsed mDCs at a ratio of 10:1 for 1 day. Subsequently, lymphocytes were cultured at 37°C in RPMI medium supplemented with 5% human AB serum, IL-2 (20 ng/ml), IL-7 (10 ng/ml) and IL-15 (20 ng/ml) (ImmunoTools GmbH) for 9 days. The culture medium was replaced every other day.
Cytotoxic T cells were isolated using a CD8+ T cell Isolation kit (Miltenyi Biotec, Inc.) according to the manufacturer's protocol. In brief, 1×107 lymphocytes were centrifuged at 300 × g for 10 min at room temperature and resuspended in 40 µl buffer containing PBS, 0.5% bovine serum albumin (Capricorn Scientific) and 2 mM EDTA. The cell suspension was mixed and incubated with 10 µl of a CD8+ T cell biotin-antibody cocktail (Miltenyi Biotec, Inc.) for 5 min, followed by 20 µl of a CD8+ T cell micro-bead cocktail (Miltenyi Biotec, Inc.) for 10 min at room temperature. The cell mixture was then magnetically separated using a magnetic-activated cell sorting (MACS) column and MACS separator (Miltenyi Biotec, Inc.), and the flow-through enriched in CD8+ T cells were collected. To determine the phenotype of effector immune cells, anti-CD3 FITC (cat. no. 21850033; ImmunoTools GmbH), anti-CD4 APC (cat. no. 21850046; ImmunoTools GmbH), anti-CD8 APC (cat. no. 21620086; ImmunoTools GmbH), anti-CD16 APC (cat. no. 21278166; ImmunoTools GmbH) and anti-CD56 PE (BioLegend, Inc.) monoclonal antibodies were used. Phenotype markers of lymphocytes, including CD3, CD4, CD8, CD16 and CD56 were analyzed using a FACSCalibur flow cytometer (BD Biosciences) using Flow Jo (Treestar) software version X.
IFN-γ measurement by ELISA
Amounts of IFN-γ secreted by CSC RNA-pulsed DC-activated T cells in comparison with naïve T cells was measured using an IFN-γ ELISA kit (cat. no. DIF50, R&D Systems, Ltd.) according to the manufacturer's protocol. Cytokine concentration was determined by measuring optical density using a microplate reader at 450/570 nm.
Tumor cell killing by effector T cells
BC target cells (T), either the whole culture population (BCA55-121-WP) or the enriched BCA55-121-CSCs, were plated into separate wells of 96-well plates (~3,000 cells/well) for 24 h for the assessment of immune cell killing activity. Subsequently, 50 µl Caspase-3/7 green reagent (Sartorius AG) at a 20 µM concentration was added to detect apoptosis. Total lymphocytes (E) from healthy donors with HLA matched to that of BCA55-121 cells (HLA-A2) were added as effectors and co-cultured with the tumor target cells (T) at effector to target (E:T) ratios of 5:1, 10:1 and 20:1 for 24 h. Cancer cells with apoptotic activity were detected using an IncuCyte® live-cell analysis system.
Statistical analysis
All data are presented as the mean ± standard deviation (SD). The data were tested for normal distribution by the Shapiro-Wilk test. The data from two groups were analyzed by paired Student's t-test and from multiple groups by one-way repeated-measure analysis of variance (ANOVA) followed by Tukey's post-hoc test using GraphPad Prism software version 7.04 (GraphPad Software, Inc.) or SigmaPlot 16.0 (Systat Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Characterization of the primary BC cell line BCA55-121
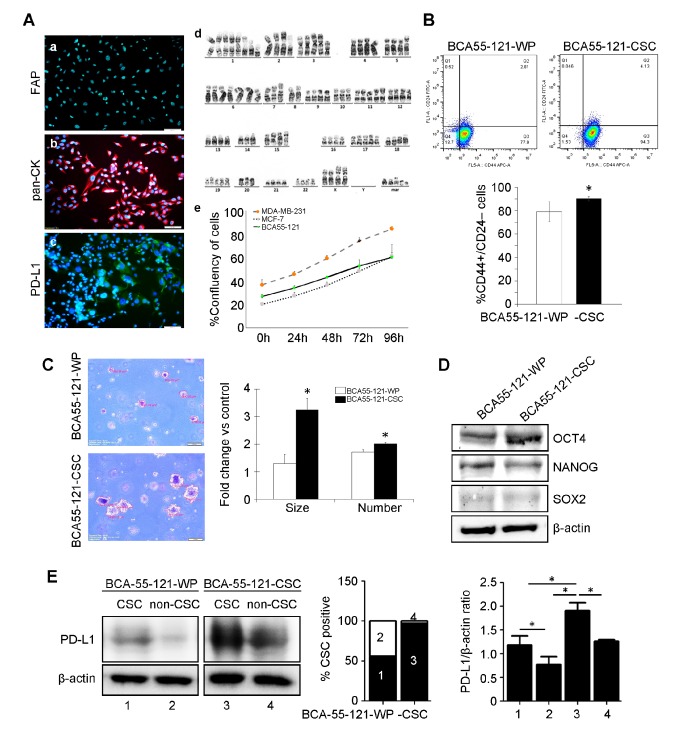
The tumor size measured 4.5×4×2 cm3, and was diagnosed as an invasive ductal carcinoma (moderately-differentiated) positive for estrogen receptor (>75%) and progesterone receptor (<10%) and negative for HER2/neu. Angiolymphatic invasion was absent, however, one lymph node was positive for macro-metastatic cancer cells. The cancer cells were visualized as cobblestone shaped under phase-contrast microscopy (data not shown). The primary cell line was positive for pan-cytokeratin, confirming that the cells were of epithelial origin (18) and without stromal fibroblast contamination in the culture (Fig. 1A). The cell line also tested negative for fibroblast activation protein (FAP), and PD-L1 was expressed at a very low level (Fig. 1A). Chromosomal analysis revealed an aneuploid aberration of chromosome copies. BCA55-121 cells exhibited a higher proliferation rate compared with the MCF-7 cell line, but lower than that of MDA-MB-231 cells (Fig. 1A). Breast CSCs, defined as CD44+/CD24− cells, were separated from the BCA55-121-WP cells by fluorescence-activated cell sorting. These CSC markers were expressed in ~75.2±0.45% of the BCA55-121-WP population (Fig. 1B). CSCs were subsequently sorted to enrich the cells with this phenotype to 92.9±5.83% purity (Fig. 1B). The tumorigenic ability of the BCA55-121-CSCs were tested, and the results were compared with BCA55-121-WP cells. Soft agar colony formation assay, a 3D spheroid cell culture assay that measures cell proliferation in semi-solid matrices (11) was conducted. The assay selectively enriches cells that exhibit a malignant transformation, anchorage-independent proliferation and progenitor cell-like tumorigenic properties. Significantly greater proliferative and tumorigenic abilities (2–3 folds) of the BCA55-121-CSC population were indicated by colony count and colony size compared with unsorted BCA55-121-WP cells (Fig. 1C). The level of OCT4 was increased in BCA55-121-CSCs compared with the unsorted cells (Fig. 1D). NANOG expression did not change in BCA55-121-CSC compared with BCA55-121 WP, and the level of SOX2 was very low. These results confirmed the greater tumorigenic and stemness properties of the CSC-enriched population. In addition, the expression of PD-L1 was significantly increased in the CSC population of the CSC culture compared with the WP culture (Fig. 1E).
Figure 1.
Characterization of the BCA55-121-WP and BCA55-121-CSC BC cell lines. (A) Immunofluorescent staining of (a) pan-CK, (b) PD-L1 and (c) FAP. (d) Chromosomal analysis of BCA55-121 cells displaying aneuploid chromosomes. (e) Growth curve of BCA55-121-WP cells compared with reference BC cell lines, MDA-MB-231 and MCF-7. (B) Flow cytometric analysis of CD44+/CD24− cells in BCA55-121 before and after stem-cell enrichment. (C) Representative fields of cancer cell colonies from the soft agar colony formation assay. Colony size and colony count are presented as fold-changes between BCA55-121-CSC and BCA55-121-WP cells. (D) Protein expression levels of stem cell-associated proteins OCT4, NANOG and SOX2. (E) PD-L1 expression in the CSC and non-CSC populations in BCA55-121-WP and BCA55-121-CSC culture demonstrated using western blot analysis with β-actin as a loading control. Bars represent mean ± SD of two independent experiments. *P<0.05. WP, whole population; BC, breast cancer; CD, cluster of differentiation; CSC, cancer stem cell; pan-CK, pan-cytokeratin; PD-L1, programmed death ligand 1; FAP, fibroblast-activation protein.
Activation of PBMC-derived DCs and lymphocytes
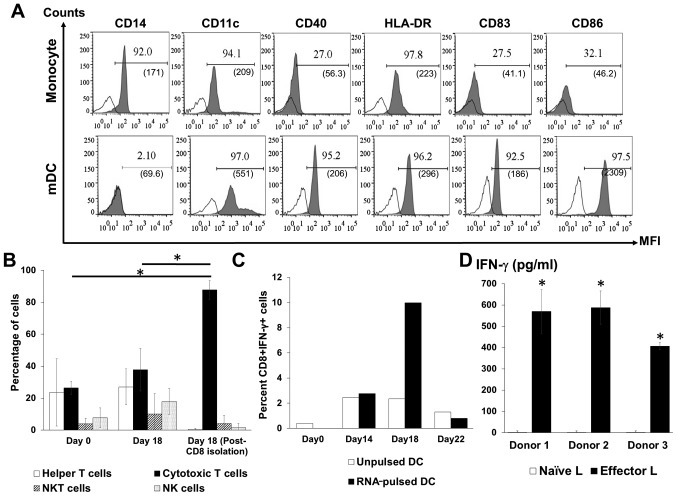
DC phenotype was analyzed on days 1 (monocytes), 6 (iDCs) and 9 (mDCs). Monocytes are the only cell type that expresses CD14 (19), and this was diminished during subsequent maturation and replaced by a DC marker (CD11c), a DC maturation marker (CD83), an antigen-presenting molecule (HLA-DR) and co-stimulatory molecules (CD40 and CD86; Fig. 2A) demonstrating DC maturation in vitro. The subsets of cells in the activated lymphocyte population following co-culture with DCs were analyzed by flow cytometry. The highest proportion of cytotoxic T lymphocytes in the total lymphocytes was observed after co-culture with RNA-pulsed DCs followed by CD4+ T cells (Fig. 2B). NK cell and NKT cell populations were also identified among the activated lymphocytes (Fig. 2B). In addition, a significantly increased number of IFN-γ-positive CD8+ T cells were observed on day 18 of the lymphocyte activation protocol (day 9 after co-culture with RNA-pulsed DC; Fig. 2C). For each of the three healthy donors, higher levels of secreted IFN-γ were present in the supernatants of activated T cells compared with the supernatants of naïve T cells (Fig. 2D).
Figure 2.
Analysis of activated DCs and lymphocyte phenotypes. (A) Profiles of the specific markers CD14, CD11c, CD40, HLA-DR, CD83 and CD86 on monocytes and mDCs. (B) Percentage distribution of lymphocyte subpopulations before and after activation and following the isolation of CD8+ T cells. (C) The percentage of IFN-γ-positive CD8+ T lymphocytes after co-culture with unpulsed or RNA-pulsed DCs from one donor. (D) Level of IFN-γ in the culture media of effector lymphocytes from three donors. Bars represent mean ± SD of three independent experiments. *P<0.05. CD, cluster of differentiation; DCs, dendritic cells; mDC, mature dendritic cell; HLA-DR, human leukocyte antigen DR; IFN, interferon; L, lymphocyte; MFI, mean fluorescence intensity; NK, natural killer.
Cancer cell killing assay
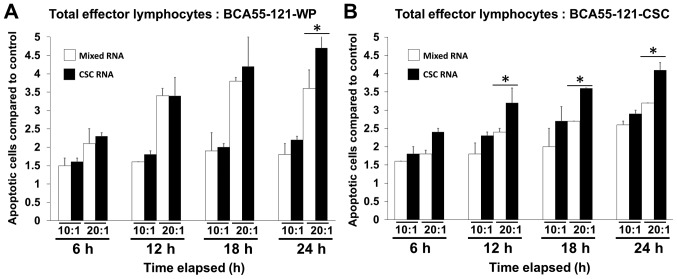
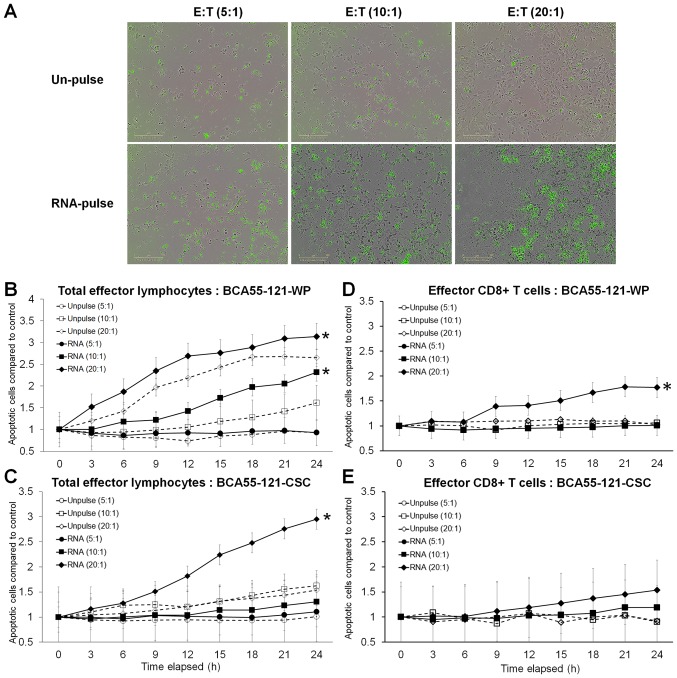
Total RNA from BCA55-121-CSC was loaded onto DCs, and the efficacy in activating effector lymphocytes to kill cancer cells was measured using Caspase-3/7 reagents for apoptosis with the Live-cell imager. To prevent the killing of cancer cells by other immune cells, not effector CD8+ T cells, HLA-A2 matched lymphocytes and BCA55-121 cells were used. The efficacy of tumor cell killing between lymphocytes activated by BCA55-121-WP and CSC RNA-pulsed DCs was compared (Fig. 3). The killing activity in the CSC RNA-activated lymphocytes was greater compared with that of BCA55-121-WP or CSCs, and this was at either 10:1 or 20:1 E:T tumor cells with significance reached at 20:1 E:T ratio (Fig. 3). Green fluorescence, representing cells with activated Caspase-3/7 at different E:T ratios, is displayed in Fig. 4A. Although increasing apoptotic activity over time was observed in both experiments, a 10:1 ratio of total activated lymphocytes did not result in enhanced killing of CSC targets, although BCA55-121-WP cells were killed (Fig. 4B and C). Additionally, at a 20:1 ratio, more BCA55-121-WP cells were killed compared with CSCs at all time points (Fig. 4B and C). These results demonstrated that apoptotic resistance may be a property of CSCs. In addition, CD8+ effector T cells isolated from the CSC RNA-activated lymphocyte population were less effective compared with the whole lymphocyte population against either BCA55-121-WP or BCA55-121-CSC targets (Fig. 4D and E). Limited killing activity was observed by CD8+ T cells that were activated by DCs without antigen exposure, which suggested that cancer cell killing was antigen specific.
Figure 3.
Tumor cell apoptosis caused by effector T cells activated by BCA55-121-WP-RNA-pulsed DCs and those activated by BCA55-121-CSC-RNA-pulsed DCs. Cancer cell apoptosis was assessed by an immune cell killing assay using the IncuCyte® live cell analysis system. Time-dependent cancer cell apoptotic count induced by total lymphocytes against (A and B) BCA55-121-WP and BCA55-121-CSC compared with the apoptotic cell count of each condition at 0 h. The y-axis represents the numbers of apoptotic cells compared with those in the control group at the starting point of each treatment condition. The data are from triplicate wells of one experiment. *P<0.05 vs. T cells from the unpulsed DCs. CSC, cancer stem cell; DCs, dendritic cells; WP, whole population.
Figure 4.
Tumor cell apoptosis caused by CSC RNA-pulsed DC-activated T cells. Cancer cell apoptosis was assessed by an immune cell killing assay using the IncuCyte® live cell analysis system. (A) Representative field images of cell-mediated cancer cell apoptosis at effector T cells at E:T ratios of 5:1, 10:1, and 20:1. (B and C) Time course of apoptotic cell counts of cancer cells exposed to different E:T ratios of total effector T lymphocytes to targets (B) BCA55-121-WP (C) BCA55-121-CSC and (D and E) Time course of apoptotic cell counts of cancer cells exposed to different ratios of CD8+ T cells in (D) BCA55-121-WP and (E) BCA55-121-CSC. The y-axis represents the numbers of apoptotic cells compared with those in the control at the starting point of each treatment condition. Results shown are from triplicate wells from two independent experiments. *P<0.05 vs. T cells from the unpulsed DCs. CSC, cancer stem cell; DCs, dendritic cells; WP, whole population; E, effector T cells; T, target cells.
Discussion
The present study explored the potential of DC-primed T cells to alleviate treatment-refractory BC. CSCs were selected as a major target to alleviate cancer recurrence and aggressiveness. CD44+/CD24− was used as a CSC marker, and subsequent analyses of this subpopulation revealed its superior proliferative and tumorigenic capability in an anchorage-independent 3D culture assay compared with BCA55-121-WP. This ability was associated with high expression of stem cell-associated proteins, confirming the stem cell properties of a new in-house BC cell line, BCA55-121-CSC. In the present study, higher expression of PD-L1 was observed in the CSC population of the CSC culture compared with the CSC population of the WP culture. These results were supported by previous studies, in which PD-L1 expression was positively associated with the expression of stemness markers (20,21), and supported the notion that BCA55-121-CSC cultures possessed CSC properties. In the present study, CSC RNA could be presented as tumor antigens by DCs and activated T cells in the whole T cell population and enriched CD8+ T cells, to stimulate an enhanced ability to kill cancer cells superior to that obtained using RNA from the total BCA55-121 cell population. BCA55-121-CSCs were more resistant to activated effector T cells compared with the whole BCA55-121 cell population. In the current study, total lymphocytes yielded an improved killing effect on both BCA55-121-WP and BCA55-121-CSC populations compared with effector CD8+ T cells as the total lymphocyte population contains CD4+, NK and NKT cells, which may have an effect on cancer cell killing.
Tumor-associated antigens (TAAs), in the form of peptides, protein lysates or total RNA, were capable of priming DCs for effector lymphocyte activation (22,23). Peptide antigens can be loaded directly onto MHC molecules; however, it is mandatory to match the immunogenic epitopes relevant to a particular cancer cell with the complementary specific HLA haplotype (15). The single best immunogenic TAA epitope in BC is yet to be defined, cancer-derived RNA has been used to present a broad range of TAA epitopes on various MHC haplotypes present on DCs (24,25). The RNA-based antigen approach has been most commonly used in clinical studies, offering HLA class I and II cross-presentation and in turn inducing humoral and cellular immune responses in CD4+ and CD8+ T cells (26–28). The present study compared the cancer cell killing ability of T cells activated by DCs pulsed with total RNA with those activated by protein lysate-pulsed DCs and demonstrated that the ability to induce cancer cell apoptosis was greater in the former (data not shown). Moreover, the use of RNA avoids the effects of the immunosuppressive factors present in the tumor protein lysate; therefore, pulsing DCs with RNA may result in improved function (24,25). Taken together, this evidence supports the selection of total RNA as the antigen source in the present study. The present study demonstrated that CSC-RNA-pulsed DCs induced effector lymphocytes to kill both BCA55-121-WP and BCA55-121-CSCs, and the killing activity was superior compared with that of DCs that received RNA from whole cancer cells, which contained a mixture of both non-CSCs and CSCs. These results emphasized the potential of DC-based protocols using CSC RNA to prime DCs for eradication of tumor cells via tumor antigen-specific effector T cells.
In the present study, the T cell population activated by co-culture with RNA-primed DCs consisted of mainly cytotoxic and helper T cells together with some NKT and NK cells. These activated T cells produced high levels of IFN-γ, which is characteristic of effector T cells (29). These findings were consistent with another report that confirmed the potential of ex vivo activation of effector T cells with an enhanced specificity for cancer antigens appropriate for use in adoptive transfer (30). In the present study, cancer cell killing by the effector T cells generated with CSC RNA-pulsed DCs was significantly greater compared with T cells activated with unpulsed DCs, and the effect was dose-dependent. Substantial killing was observed in cultures of unpulsed DC-activated T cells, which may be due to two main factors. The first is the presence of non-antigen specific cancer killing cells in the total lymphocyte population, comprising NK and NKT cells, which are also activated by DCs. In the present study, non-specific killing by other immune cells was addressed by isolating CD8+ T cells from the activated total lymphocyte population. The tumor-killing effect of the activated CD8+ T cells demonstrated a greater specificity of killing, potentially by antigen-dependent cytotoxic T cells, since little or no killing was observed among unpulsed-DC-activated CTLs. The efficacy of tumor killing between lymphocytes activated by whole culture RNA-pulsed DCs and CSC RNA-pulsed DCs was further compared. The results of the present study revealed superior tumor killing activity with the CSC RNA-activated cells, especially at 20:1 and 10:1 E:T ratios. Inferior apoptotic activity was observed towards BCA55-121-CSC target cells. The second potential mechanism of killing in unpulsed cultures maybe the mismatch of HLA molecules between effector cells derived from healthy donors and the target cancer cells. HLA-A2 matched donors were used in the present study as the BCA55-121 cancer cell line is HLA-A2 positive; however, residual HLA haplotype mismatch may have stimulated a small amount of non-specific killing and prevented optimal antigen presentation and effector cell activation leading to lower specific cytotoxicity in RNA-pulsed cultures. However, in the clinical application of DC-based vaccines, autologous DCs are activated by autologous tumor antigens and thus eliminate this confounder (14,16). The limitation of this study was the lack of investigation of HLA class I expression in DC. In addition, using only one donor was a limitation of the experiment presented in this study.
The present study revealed that CSCs were more resistant compared with the whole cancer cell population to effector T cells. This may be explained by the finding that the CSC population in CSC cultures expressed high levels of PD-L1 (20), which may induce apoptosis of effector T cells (31). These in vitro findings need to be investigated in an ex vivo system and in clinical trials for the development of DC-based activation of T cells against breast CSCs. A combination of drugs targeting CSCs and activated effector T cells may be a potential approach for resolving the issue of the resistant CSC population. This treatment needs to be developed alone or in conjuction with other treatments to optimize antitumor immune responses and overcome the relatively immunosuppressive stage, during which DCs in the tumor and the surrounding microenvironment develop tolerance (32,33). Personalized cancer therapy may be achieved by the activation of patient-derived DCs by using autologous tumor antigens from surgical tissue-derived primary cell culture. Several studies performed using the personalized DC-based vaccine have confirmed potent anti-tumor immunity activity mediated by a broad range of TAAs relevant to each patient (34–36). This approach has the potential to improve the efficacy of treatment and quality of life for patients with BC.
Acknowledgments
Not applicable.
Funding
This research project was supported by the Faculty of Medicine, Siriraj Hospital, Mahidol University (grant no. R015933006) and by the Thailand Science Research and Innovation (grant no. RSA6280091), Ministry of Higher Education, Science, Research and Innovation, Thailand awarded to CT.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
NS, NJ, PJ and ST performed the experiments and acquired the data. CT conceived and designed the study, acquired, analyzed and interpreted the data. TC, PY and PT analyzed the data. MW and PO collected the breast cancer tissue samples and verified clinicopathological data. NS, NJ and CT drafted the article. CT revised the manuscript for important intellectual content. All authors have read and approved the manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all individual participants prior to enrollment in the study. The present study was approved by the Siriraj Institutional Review Board (approval nos. Si520/2010 and Si321/2016).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colleoni M, Sun Z, Price KN, Karlsson P, Forbes JF, Thürlimann B, Gianni L, Castiglione M, Gelber RD, Coates AS, Goldhirsch A. Annual hazard rates of recurrence for breast cancer during 24 years of follow-up: Results from the international breast cancer study group trials I to V. J Clin Oncol. 2016;34:927–935. doi: 10.1200/JCO.2015.62.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerber B, Freund M, Reimer T. Recurrent breast cancer: Treatment strategies for maintaining and prolonging good quality of life. Dtsch Arztebl Int. 2010;107:85–91. doi: 10.3238/arztebl.2010.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawood S, Austin L, Cristofanilli M. Cancer stem cells: Implications for cancer therapy. Oncology (Williston Park) 2014;28:1101–1107, 1110. [PubMed] [Google Scholar]
- 5.Palucka K, Banchereau J. Dendritic-cell-based therapeutic cancer vaccines. Immunity. 2013;39:38–48. doi: 10.1016/j.immuni.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdullah LN, Chow EK. Mechanisms of chemoresistance in cancer stem cells. Clin Transl Med. 2013;2:3. doi: 10.1186/2001-1326-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrasco E, Alvarez PJ, Prados J, Melguizo C, Rama AR, Aránega A, Rodríguez-Serrano F. Cancer stem cells and their implication in breast cancer. Eur J Clin Invest. 2014;44:678–687. doi: 10.1111/eci.12276. [DOI] [PubMed] [Google Scholar]
- 8.Dey P, Rathod M, De A. Targeting stem cells in the realm of drug-resistant breast cancer. Breast Cancer (Dove Med Press) 2019;11:115–135. doi: 10.2147/BCTT.S189224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Islam F, Gopalan V, Smith RA, Lam AK. Translational potential of cancer stem cells: A review of the detection of cancer stem cells and their roles in cancer recurrence and cancer treatment. Exp Cell Res. 2015;335:135–147. doi: 10.1016/j.yexcr.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 10.Britton KM, Kirby JA, Lennard TW, Meeson AP. Cancer stem cells and side population cells in breast cancer and metastasis. Cancers (Basel) 2011;3:2106–2130. doi: 10.3390/cancers3022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akrap N, Andersson D, Bom E, Gregersson P, Ståhlberg A, Landberg G. Identification of distinct breast cancer stem cell populations based on single-cell analyses of functionally enriched stem and progenitor pools. Stem Cell Reports. 2016;6:121–136. doi: 10.1016/j.stemcr.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao J, Fan W, Ma B, Wu Y. Breast cancer stem cells expressing different stem cell markers exhibit distinct biological characteristics. Mol Med Rep. 2016;14:4991–4998. doi: 10.3892/mmr.2016.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelao L, Criscitiello C, Esposito A, De Laurentiis M, Fumagalli L, Locatelli MA, Minchella I, Santangelo M, De Placido S, Goldhirsch A, Curigliano G. Dendritic cell-based vaccines: Clinical applications in breast cancer. Immunotherapy. 2014;6:349–360. doi: 10.2217/imt.13.169. [DOI] [PubMed] [Google Scholar]
- 14.Constantino J, Gomes C, Falcão A, Cruz MT, Neves BM. Antitumor dendritic cell-based vaccines: Lessons from 20 years of clinical trials and future perspectives. Transl Res. 2016;168:74–95. doi: 10.1016/j.trsl.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Sabado RL, Balan S, Bhardwaj N. Dendritic cell-based immunotherapy. Cell Res. 2017;27:74–95. doi: 10.1038/cr.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anguille S, Smits EL, Lion E, van Tendeloo VF, Berneman ZN. Clinical use of dendritic cells for cancer therapy. Lancet Oncol. 2014;15:e257–e267. doi: 10.1016/S1470-2045(13)70585-0. [DOI] [PubMed] [Google Scholar]
- 17.Pham PV, Le HT, Vu BT, Pham VQ, Le PM, Phan NL, Trinh NV, Nguyen HT, Nguyen ST, Nguyen TL, Phan NK. Targeting breast cancer stem cells by dendritic cell vaccination in humanized mice with breast tumor: Preliminary results. Onco Targets Ther. 2016;9:4441–4451. doi: 10.2147/OTT.S105239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andergassen U, Vogl A, Mumm JN, Kölbl AC, Hutter S, Rack B, Friese K, Jeschke U. Immunocytochemical characterization of disseminated tumour cells from bone marrow of breast cancer patients. Anticancer Res. 2016;36:3217–3222. [PubMed] [Google Scholar]
- 19.Zarif JC, Hernandez JR, Verdone JE, Campbell SP, Drake CG, Pienta KJ. A phased strategy to differentiate human CD14+monocytes into classically and alternatively activated macrophages and dendritic cells. Biotechniques. 2016;61:33–41. doi: 10.2144/000114435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darvin P, Sasidharan Nair V, Elkord E. PD-L1 Expression in human breast cancer stem cells is epigenetically regulated through posttranslational histone modifications. J Oncol. 2019;2019:3958908. doi: 10.1155/2019/3958908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao L, Guo Q, Li X, Yang X, Ni H, Wang T, Zhao Q, Liu H, Xing Y, Xi T, Zheng L. MiR-873/PD-L1 axis regulates the stemness of breast cancer cells. EBioMedicine. 2019;41:395–407. doi: 10.1016/j.ebiom.2019.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin T, Shi P, Gou S, Shen Q, Wang C. Dendritic cells loaded with pancreatic cancer stem cells (CSCs) lysates induce antitumor immune killing effect in vitro. PLoS One. 2014;9:e114581. doi: 10.1371/journal.pone.0114581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie BH, Yang JY, Li HP, Zhang B, Chen W, Zhou B, Peng BG, Liang LJ, He Q. Dendritic cells transfected with hepatocellular carcinoma (HCC) total RNA induce specific immune responses against HCC in vitro and in vivo. Clin Transl Oncol. 2014;16:753–760. doi: 10.1007/s12094-013-1145-7. [DOI] [PubMed] [Google Scholar]
- 24.Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, Mellman I, Prindiville SA, Viner JL, Weiner LM, Matrisian LM. The prioritization of cancer antigens: A national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15:5323–5337. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Criscitiello C. Tumor-associated antigens in breast cancer. Breast Care (Basel) 2012;7:262–266. doi: 10.1159/000342164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garg NK, Dwivedi P, Prabha P, Tyagi RK. RNA pulsed dendritic cells: An approach for cancer immunotherapy. Vaccine. 2013;31:1141–1156. doi: 10.1016/j.vaccine.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 27.McNamara MA, Nair SK, Holl EK. RNA-based vaccines in cancer immunotherapy. J Immunol Res. 2015;2015:794528. doi: 10.1155/2015/794528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen DS, Mellman I. Oncology meets immunology: The cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Bhat P, Leggatt G, Waterhouse N, Frazer IH. Interferon-γ derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis. 2017;8:e2836. doi: 10.1038/cddis.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen ST, Nguyen HL, Pham VQ, Nguyen GT, Tran CD, Phan NK, Pham PV. Targeting specificity of dendritic cells on breast cancer stem cells: In vitro and in vivo evaluations. Onco Targets Ther. 2015;8:323–334. doi: 10.2147/OTT.S77554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duraiswamy J, Kaluza KM, Freeman GJ, Coukos G. Dual blockade of PD-1 and CTLA-4 combined with tumor vaccine effectively restores T-cell rejection function in tumors. Cancer Res. 2013;73:3591–3603. doi: 10.1158/0008-5472.CAN-12-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zong J, Keskinov AA, Shurin GV, Shurin MR. Tumor-derived factors modulating dendritic cell function. Cancer Immunol Immunother. 2016;65:821–833. doi: 10.1007/s00262-016-1820-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimodaira S, Sano K, Hirabayashi K, Koya T, Higuchi Y, Mizuno Y, Yamaoka N, Yuzawa M, Kobayashi T, Ito K, Koizumi T. Dendritic cell-based adjuvant vaccination targeting Wilms' tumor 1 in patients with advanced colorectal cancer. Vaccines (Basel) 2015;3:1004–1018. doi: 10.3390/vaccines3041004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Yao K, Wang B, Qing J, Liu G. Potent dendritic cell vaccine loaded with latent membrane protein 2A (LMP2A) Cell Mol Immunol. 2008;5:365–372. doi: 10.1038/cmi.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hobo W, Strobbe L, Maas F, Fredrix H, Greupink-Draaisma A, Esendam B, de Witte T, Preijers F, Levenga H, van Rees B, et al. Immunogenicity of dendritic cells pulsed with MAGE3, survivin and B-cell maturation antigen mRNA for vaccination of multiple myeloma patients. Cancer Immunol Immunother. 2013;62:1381–1392. doi: 10.1007/s00262-013-1438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.