Abstract
Chronic obstructive pulmonary disease (COPD) and squamous cell lung carcinoma (SCC) are smoking-related diseases. However, the connection between the two is poorly understood. Microarray gene expression profiles in bronchial epithelium from patients with SCC with or without COPD were downloaded from the Gene Expression Omnibus repository. Differentially expressed genes associated with COPD and SCC were identified and visualized using the Advanced Network Merger module in Cytoscape. COPD-associated genes in SCC progression were further identified using the BisoGenet plug-in in Cytoscape. The genetic interaction network was predicted using the Network Analysis function. Heat shock protein 90 α family class A member 1 (HSP90AA1), adrenoceptor β2 (ADRB2), transducin β like 1 X-linked receptor 1 (TBL1XR1) and heat shock protein family B (small) member 1 (HSPB1) were identified to be differentially expressed in SCC and COPD cases. The overall survival rate associated with the gene signatures was investigated using clinical samples from patients with SCC and COPD from the PROGgene database. The results suggest that the pathogenesis of SCC caused by COPD is regulated by HSP90AA1, ADRB2, TBL1XR1 and HSPB1. These genes may serve as potential therapeutic targets for the treatment of patients with COPD-related SCC.
Keywords: chronic obstructive pulmonary disease, squamous cell lung carcinoma, interaction network
Introduction
As the third leading cause of mortality in the United States in 2018, chronic obstructive pulmonary disease (COPD) is a chronic inflammatory lung disease characterized by obstruction of lung airflow (1). In 2017, squamous cell lung cancer, or squamous cell carcinoma of the lung (SCC), which is a type of non-small cell lung cancer (NSCLC), accounted for ~30% of all lung cancer cases worldwide (2). However, the connection between COPD and SCC remains poorly defined (3).
Current literature has extended the idea that inflammation is a basic part of tumorigenesis (3). Inflammatory cells, which interact with the tumor environment, foster proliferation, survival and migration of tumor cells (4). Tumor cells use certain signaling molecules of the innate immune system, such as selectins and chemokines, to foster proliferation, migration and metastasis (4). However, little is known about the role of inflammation in lung cancer progression.
Systems biology has been applied to biological systems for the investigation of cancer (5). A biological system is any system that applies to organic frameworks (6). Systems biology enables the identification of the molecular and cellular networks involved in disease progression (7–9). The present study generated a genetic interaction network using microarray gene expression data from patients with SCC and COPD, and explored the potential molecular mechanisms involved using a gene regulatory networks approach.
Materials and methods
Microarray dataset
The microarray dataset (GSE12472) (6) used in the current study was downloaded from the Gene Expression Omnibus repository (www.ncbi.nlm.nih.gov/geo). All differently expressed genes associated with SCC with or without COPD were screened from this dataset. In the original study, the authors compared gene expression profiles in bronchial epithelia from patients with SCC with and without COPD (6). Bronchial tissues from the larger airways (bronchus diameter >2 mm, surrounded by cartilage) were resected from 10 individuals without COPD and 18 with COPD. Additionally, centrally located primary tumor tissues were collected from 35 patients with SCC, including 17 patients without COPD and 18 with COPD. The most recent post-bronchodilator spirometric results prior to surgery were used to assess the GOLD stage for each patient. Bronchial tissues from 17 patients without COPD were obtained during thoracotomy for NSCLC. Bronchial tissues from 18 patients with COPD were obtained during thoracotomy for NSCLC (n=11) or lung transplantation (n=7). Total RNA was isolated and purified from the samples, and the Agilent Whole Human Genome Oligo microarray (GPL1708; Agilent Technologies, Inc.) was used to determine gene expression levels.
Identification of differentially expressed genes
The raw microarray data was downloaded directly from GSE12472 as the matrix .txt files. Significance analysis of microarrays (SAM; version 2.11; statweb.stanford.edu/~tibs/SAM) was used to identify upregulated and downregulated genes from the comparison between bronchial epithelia of patients with SCC with and without COPD (7). To identify COPD-associated genes that drive SCC progression, genes identified in both the normal bronchial epithelium and COPD tissue were excluded from the list of genes in SCC with COPD using the Advanced Network Merge function in Cytoscape.
Differently expressed gene visualization
Differently expressed genes were visualized using Cluster and TreeView (version 3.0; bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) as previously described (8).
Gene-gene interaction network construction
The COPD-associated genetic interaction network in SCC was expanded using the BisoGenet plug-in in Cytoscape 3.0 (www.cytoscape.org) as previously reported (8). BisoGenet allows users to customize searches by specifying a target set of genes/proteins to retrieve interaction data, and allows searching for molecular interactions in SysBiomics embedded within Cytoscape (9). SysBiomics integrates data from well-known interaction databases, including the Database of Interacting Proteins (dip.mbi.ucla.edu/dip), BIOGRID (thebiogrid.org), the Human Protein Reference Database (www.hprd.org), the Biomolecular Interaction Network Database (bind.ca), the Molecular INTeraction database (mint.bio.uniroma2.it/mint) and EMBL-EBI INTACT (https://www.ebi.ac.uk/intact/).
Network topological parameters
Topological parameters were calculated using the Network Analysis function of Cytoscape 3.0. Node size and color was mapped based on the degree of connectivity (8).
Survival analysis
The prognostic value of gene signatures in SCC was assessed using PROGgene version 2.0 (watson.compbio.iupui.edu/chirayu/proggene/database/index.php) (10,11). The overall survival rate was compared between patients with high and low mRNA expression of the selected genes using the median gene expression as the cut-off value. For survival analysis, Kaplan-Meier survival curves and the log-rank test were used in this study.
Statistical analysis
For SAM, gene expression was considered significantly different when the false discovery rate (FDR) was ≤5% and the fold change was ≥2.
Results
Differentially expressed genes associated with SCC progression
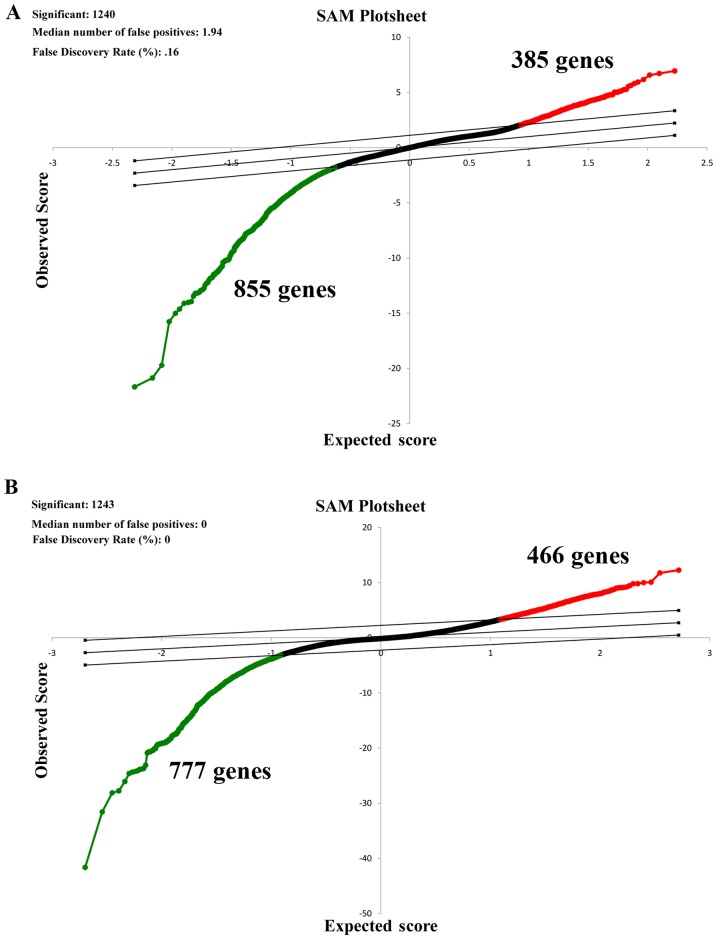
Gene expression in patients with SCC and normal tissues with COPD was compared. As presented in Fig. 1A, a total of 385 upregulated and 855 downregulated genes were identified by SAM when a FDR ≤5% and fold change ≥2 were employed.
Figure 1.
SAM of patients with SCC patients with or without COPD. First, patients with SCC and the normal tissues with COPD were compared. Subsequently, patients with SCC and the normal tissues without COPD were compared. (A) Representative SAM plot for patients with SCC without COPD. (B) Representative SAM plot for patients with SCC and COPD. Red represents upregulated genes and green represents downregulated genes. Black represents gene with no changes in expression. SAM, significance analysis of microarrays; SCC, squamous cell lung carcinoma; COPD, chronic obstructive pulmonary disease.
Differentially expressed genes associated with COPD-related SCC progression
Patients with SCC and normal tissues without COPD were compared. According to the SAM plot (Fig. 1B), a total of 1,243 differently expressed genes (466 upregulated and 777 downregulated) were identified.
Aberrant COPD-related genes facilitate SCC progression
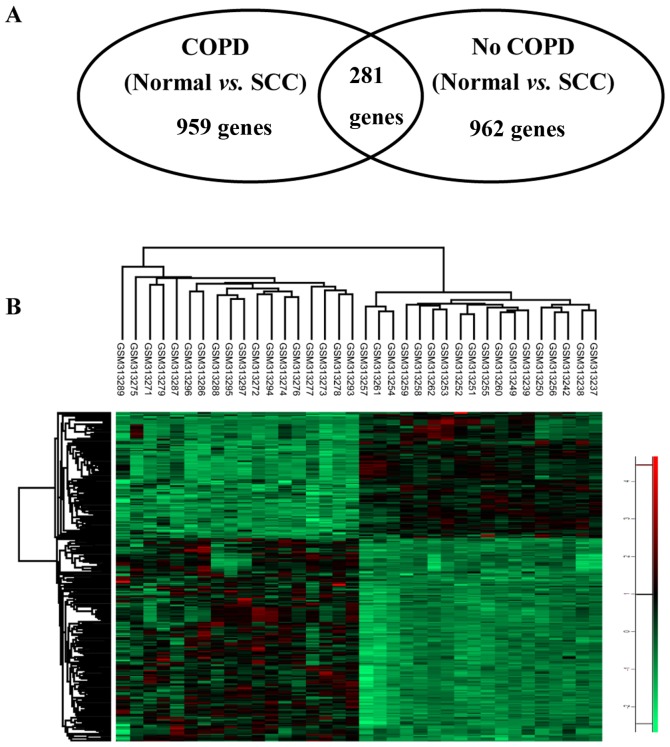
To identify COPD-associated genes that drive SCC progression, genes identified in both the normal bronchial epithelium and COPD tissue were excluded from the list of genes in SCC with COPD. A total of 281 differentially expressed genes for SCC with COPD remained (Fig. 2).
Figure 2.
Hierarchical heatmap clustering visualization of genes specifically expressed in patients with SCC and COPD. (A) Venn diagram of the differentially expressed COPD-associated SCC genes. (B) Heatmap of differentially expressed genes in COPD-associated SCC tissues. Red represents upregulated genes and green represents downregulated genes. SCC, squamous cell lung carcinoma; COPD, chronic obstructive pulmonary disease.
Genetic interaction network of genes in SCC and COPD
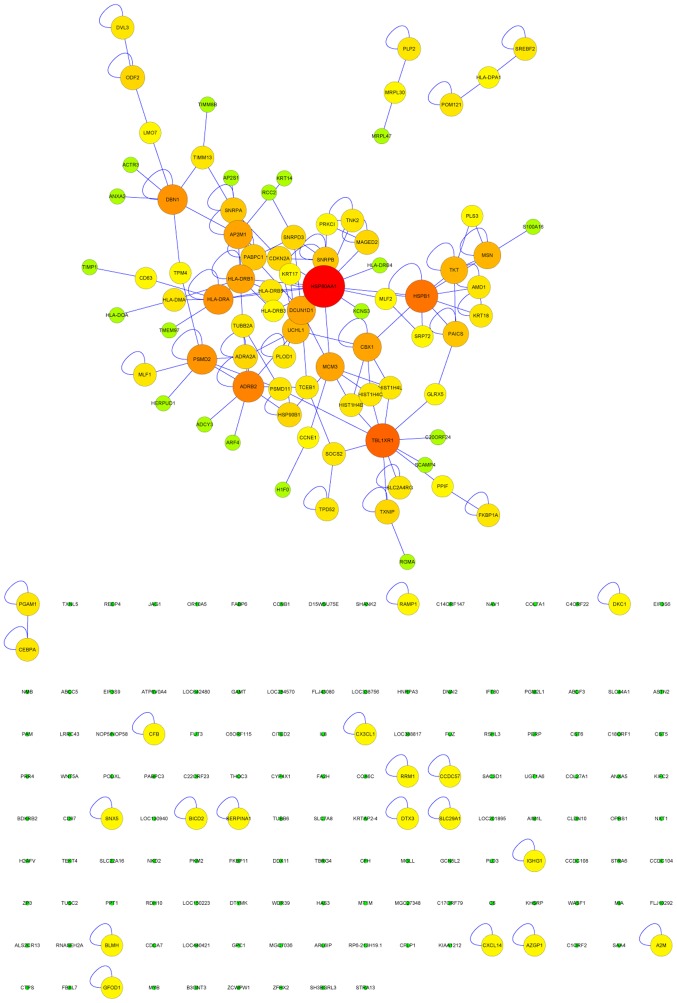
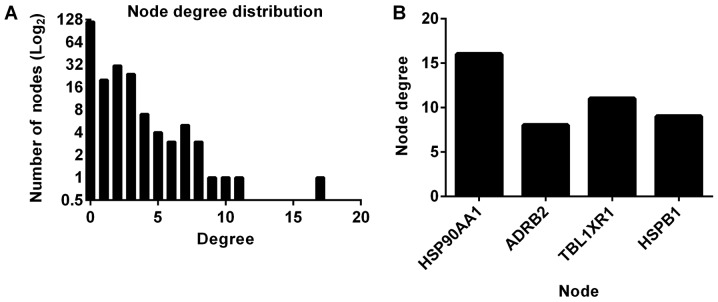
A genetic interaction network investigating the interactions between genes in SCC with COPD was constructed and expanded using BisoGenet. The interaction network consisted of 220 nodes and 163 edges (Fig. 3). The clustering coefficient and network diameter were 0.04 and 10, respectively. The node degree distribution across the entire network was calculated (Fig. 4A). The four nodes with the greatest degree of connectivity [Heat shock protein 90 α family class A member 1 (HSP90AA1); adrenoceptor β2 (ADRB2); transducin β like 1 X-linked receptor 1 (TBL1XR1) and heat shock protein family B (small) member 1 (HSPB1)] were identified (Fig. 4B), and were selected for evaluation of their effect on survival.
Figure 3.
Genetic interaction network in chronic obstructive pulmonary disease-related squamous cell lung carcinoma using the BisoGenet plug-in in Cytoscape. Circles represent genes and the connections represent interactions. Colors represent the node degree of connectivity; red represents genes high connectivity, green represents low connectivity. Only the largest clusters were considered as the genetic interaction network in chronic obstructive pulmonary disease-related squamous cell lung carcinoma. The nodes without connections and/or single nodes (bottom) were not included in the network analysis.
Figure 4.
Network parameter calculation and hub identification. (A) Node degree distribution result for the genetic network. (B) Top nodes that may be involved in chronic obstructive pulmonary disease-associated squamous cell lung carcinoma pathogenesis. ADRB2, adrenoceptor β2; TBL1XR1, transducin β like 1 X-linked receptor 1; HSPB1, heat shock protein family B (small) member 1.
Association of HSP90AA1, ADRB2, TBL1XR1 and HSPB1 with the overall survival rate in COPD-related SCC
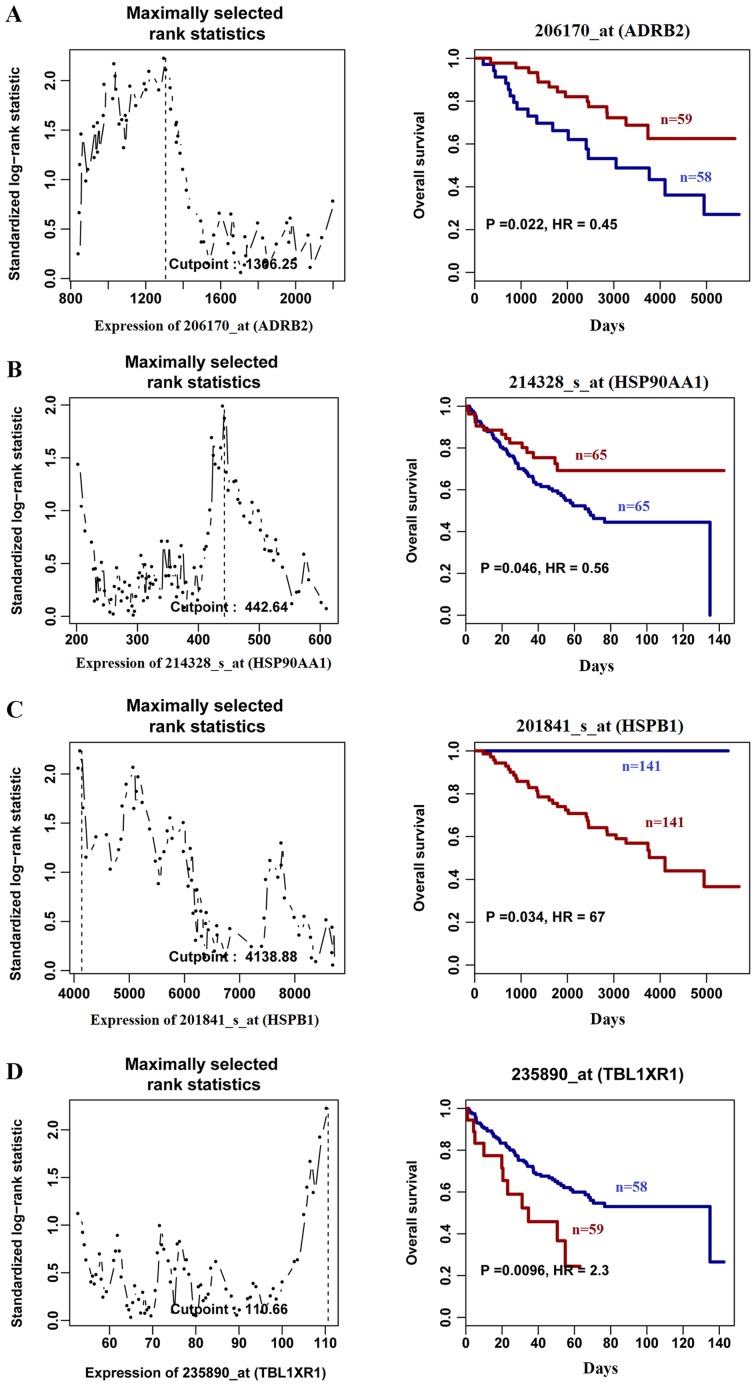
The association between the expression of the aforementioned genes and overall survival rate was evaluated using PROGgene. Compared with patients with SCC with high gene expression, patients with lower gene expression of ADRB2 (Fig. 5A) and HSP90AA1 (Fig. 5B) had a decreased overall survival rate. However, high expression of HSPB1 (Fig. 5C) and TBL1XR1 (Fig. 5D), which were highly expressed in SCC compared with COPD tissues, was associated with decreased overall survival rate compared with low expression. The survival analysis also revealed that patients with low expression of HSPB1 in SCC were still alive at the end of the follow-up period (13.6 years), suggesting that HSPB1 may be a potential therapeutic target in SCC. The results obtained in the present study demonstrate the clinical significance of HSP90AA1, ADRB2, TBL1XR1 and HSPB1 in COPD-related SCC.
Figure 5.
Overall survival analysis according to (A) Gene ID 206170_at (ADRB2), (B) Gene ID 214328_at_(HSP90AA1), (C) Gene ID 201841_s_at (HSPB1) and (D) Gene ID 235890_s_(TBL1XR1) expression, based on a clinical microarray dataset using PROGgene. Left panel represents gene expression distribution in SCC samples. Right panel represents overall survival time. Red and blue represent high and low expression levels, respectively. ADRB2, adrenoceptor β2; HSP90AA1, heat shock protein 90 α family class A member 1; TBL1XR1, transducin β like 1 X-linked receptor 1; HSPB1, heat shock protein family B (small) member 1.
Discussion
COPD is a chronic lung disease that results in decreased airflow in the lungs. Symptoms include trouble with breathing, coughing, mucus (sputum) generation and wheezing (12). This is caused by exposure to irritant compounds or particulate matter, predominantly from tobacco smoke (13). Individuals with COPD are at an increased risk of developing lung tumors and other conditions, such as emphysema and chronic bronchitis (13). COPD is a risk factor for SCC and the association between the two remains unclear (13). Therefore, the association between COPD and SCC needed to be elucidated the present study identified marker genes uniquely associated with SCC with COPD. The results of the present study indicated that high levels of ADRB2 and HSP90AA1 were associated with a longer survival time compared with the respective low expression groups. However, high expression of TBL1XR1 and HSPB1 predicted poor overall survival in patient with SCC.
The microarray analysis employed in the current study revealed that ADRB2 and HSP90AA1 are downregulated in SCC tissues compared with healthy adjacent non-cancerous tissue, suggesting that they may serve important roles in the progression of SCC. Previous studies reported that ADRB2 is an important regulator of airway smooth muscle tone (14–16). High levels of ADRB2 are associated with reduced lung function, asthma and COPD (14). Additionally, ADRB2 is an anti-inflammatory gene and was previously reported to be involved in hyperinflammation (17) and autoimmune disorders such as rheumatoid arthritis (18). Furthermore, ADRB2 has been demonstrated to be downregulated in prostate and breast cancer, and functional gain of ADRB2 decreases cancer progression (19,20). However, the association between ADRB2 and COPD-associated SCC remains unknown. In the present study, TBL1XR1 mRNA was highly expressed in samples from patients with SCC with COPD, but not SCC without COPD. TBL1XR1 is a member of the WD40 repeat-containing gene family. The protein encoded by this gene composes an integral subunit of both nuclear receptor corepressor and histone deacetylase 3 complexes, and serves an essential role in regulating the activation of transcriptional and intracellular signaling pathways, such as JNK and ribonuclease pathways (21,22). Upregulation of TBL1XR1 gene expression has been observed in various cancer cell lines and solid tumors from patients (23,24), lymphoma (25) and acute leukemia (26), and are associated with advanced tumor stage, metastasis and poor prognosis. In SCC, elevated mRNA and protein expression of TBL1XR1 was confirmed in cells lines and patient samples (27). While a direct association between COPD and TBL1XR1 has not been revealed, this gene has been reported to regulate several important signaling pathways that modulate the growth of lung α-smooth muscle actin-positive cells and airway reconstruction (28). Previously, Zou et al (29) reported that HSP90AA1 affects the response of patients with systemic lupus erythematosus to glucocorticoid treatment and the lipopolysaccharide-induced inflammatory response, including tumor necrosis factor secretion by monocytes. Furthermore, the role of HSP90AA1 in cancer pathogenesis is currently an area of active investigation. A study suggested that high expression of HSP90AA1 was an independent factor associated with mortality in patients with breast cancer (triple-negative and human epidermal growth factor receptor 2-negative/estrogen receptor-positive subtypes) (30).
HSPB1 encodes HSP20, a member of the heat shock protein family (31,32). The association between increased expression levels of HSPA1A and HSPB1 in plasma and lymphocytes and the risk of COPD has been reported and verified by lung tissue proteomics (31). Additionally, upregulated expression of HSPB1 was observed in SCC and the endothelial-to-mesenchymal transition, and was associated with poor survival (33). Upregulation of HSPB1 has also been observed in leukemia, cervical cancer, breast cancer and NSCLC. In the present study, the possible clinical importance of HSPB1 in COPD-related SCC are in agreement with previous studies, which suggests that it requires further research (34).
In the present study, patients with SCC who exhibited low expression of HSPB1 were still alive after 13.6 years, suggesting that HSPB1 may be a potential therapeutic target in SCC. The efficacy of HSPB1 as a therapeutic target requires further investigation and validation by in vivo experiments.
In conclusion, ADRB2, HSP90AA1, TBL1XR1 and HSPB1 may serve important roles in COPD-associated SCC and may provide novel therapeutic targets for the treatment of lung cancer. The limited number of studies existing on these four genes in COPD-related SCC progression is a limitation that impacts the interpretation of the findings from the current in silico study and limits the applications of these signatures within clinical practice.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Hunan Provincial Science and Technology Department Project (grant no. 2015SK20402).
Availability of data and materials
The datasets analyzed during the current study are available in the GEO repository with no. GSE12472.
Authors' contributions
LW and HZ conceived and designed the study, and analyzed and interpreted the data. LW and HZ analyzed the microarray data. LZ, HL, QC and XZ performed the survival analysis. LW and HZ wrote the manuscript. All the authors approved the manuscript.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board Committee of the Xiangya Hospital of Central South University. The data were obtained from a publicly available dataset; therefore, no written informed consent was required.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Balkissoon R, Lommatzsch S, Carolan B, Make B. Chronic obstructive pulmonary disease: A concise review. Med Clin North Am. 2011;95:1125–1141. doi: 10.1016/j.mcna.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Harjes U. Non-small cell lung cancer: Where there's smoke. Nat Rev Cancer. 2017;17:634–635. doi: 10.1038/nrc.2017.95. [DOI] [PubMed] [Google Scholar]
- 3.Houghton AM. Mechanistic links between COPD and lung cancer. Nat Rev Cancer. 2013;13:233–245. doi: 10.1038/nrc3477. [DOI] [PubMed] [Google Scholar]
- 4.Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, Simithy J, Lan Y, Lin Y, Zhou Z, et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature. 2017;550:402–406. doi: 10.1038/nature24050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: A network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boelens MC, Gustafson AM, Postma DS, Kok K, van der Vries G, van der Vlies P, Spira A, Lenburg ME, Geerlings M, Sietsma H, et al. A chronic obstructive pulmonary disease related signature in squamous cell lung cancer. Lung Cancer. 2011;72:177–183. doi: 10.1016/j.lungcan.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan S, Li X, Tie L, Pan Y, Li X. KIAA0101 is associated with human renal cell carcinoma proliferation and migration induced by erythropoietin. Oncotarget. 2016;7:13520–13537. doi: 10.18632/oncotarget.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin A, Ochagavia ME, Rabasa LC, Miranda J, Fernandez-de-Cossio J, Bringas R. BisoGenet: A new tool for gene network building, visualization and analysis. BMC Bioinformatics. 2010;11:91. doi: 10.1186/1471-2105-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goswami CP, Nakshatri H. PROGgene: Gene expression based survival analysis web application for multiple cancers. J Clin Bioinforma. 2013;3:22. doi: 10.1186/2043-9113-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goswami CP, Nakshatri H. PROGgeneV2: Enhancements on the existing database. BMC Cancer. 2014;14:970. doi: 10.1186/1471-2407-14-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng Z, Yang D, Huang X, Xiao Z. Effect of carbocisteine on patients with COPD: A systematic review and meta-analysis. Int J Chron Obstruct Pulmon Dis. 2017;12:2277–2283. doi: 10.2147/COPD.S140603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho T, Cusack RP, Chaudhary N, Satia I, Kurmi OP. Under- and over-diagnosis of COPD: A global perspective. Breathe (Sheff) 2019;15:24–35. doi: 10.1183/20734735.0346-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen AO, Jensen CS, Arredouani MS, Dahl R, Dahl M. Variants of the ADRB2 gene in COPD: Systematic review and meta-analyses of disease risk and treatment response. COPD. 2017;14:451–460. doi: 10.1080/15412555.2017.1320370. [DOI] [PubMed] [Google Scholar]
- 15.Zhao H, Wu X, Dong CL, Wang BY, Zhao J, Cao XE. Association between ADRB2 genetic polymorphisms and the risk of chronic obstructive pulmonary disease: A case-control study in a chinese population. Genet Test Mol Biomarkers. 2017;21:491–496. doi: 10.1089/gtmb.2017.0030. [DOI] [PubMed] [Google Scholar]
- 16.Hussein MH, Sobhy KE, Sabry IM, El Serafi AT, Toraih EA. Beta2-adrenergic receptor gene haplotypes and bronchodilator response in Egyptian patients with chronic obstructive pulmonary disease. Adv Med Sci. 2017;62:193–201. doi: 10.1016/j.advms.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Agac D, Estrada L, Farrar D. Beta-2 adrenergic receptor signaling prevents hyperinflammation through early release of IL-10. J Immunology. 2017;198:221–224. [Google Scholar]
- 18.Pont-Kingdon G, Bohnsack J, Sumner K, Whiting A, Clifford B, Guthery SS, Jorde LB, Lyon E, Prahalad S. Lack of association between beta 2-adrenergic receptor polymorphisms and juvenile idiopathic arthritis. Scand J Rheumatol. 2009;38:91–95. doi: 10.1080/03009740802541488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braadland PR, Ramberg H, Grytli HH, Tasken KA. β-Adrenergic receptor signaling in prostate cancer. Front Oncol. 2015;4:375. doi: 10.3389/fonc.2014.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pérez-Sayáns M, Somoza-Martín JM, Barros-Angueira F, Diz PG, Gándara Rey JM, García-García A. Beta-adrenergic receptors in cancer: Therapeutic implications. Oncol Res. 2010;19:45–54. doi: 10.3727/096504010X12828372551867. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Dormady SP, Basch RS. Identification of four human cDNAs that are differentially expressed by early hematopoietic progenitors. Exp Hematol. 2000;28:1286–1296. doi: 10.1016/S0301-472X(00)00539-7. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/S1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 23.Ma M, Yu N. Over-expression of TBL1XR1 indicates poor prognosis of serous epithelial ovarian cancer. Tohoku J Exp Med. 2017;241:239–247. doi: 10.1620/tjem.241.239. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, He Y, Cao Q, Liu N, Zhang W. TBL1XR1 is highly expressed in gastric cancer and predicts poor prognosis. Dis Markers. 2016;2016:2436518. doi: 10.1155/2016/2436518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott DW, Mungall KL, Ben-Neriah S, Rogic S, Morin RD, Slack GW, Tan KL, Chan FC, Lim RS, Connors JM, et al. TBL1XR1/TP63: A novel recurrent gene fusion in B-cell non-Hodgkin lymphoma. Blood. 2012;119:4949–4952. doi: 10.1182/blood-2012-02-414441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campregher PV, Halley ND, Vieira GA, Fernandes JF, Velloso ED, Ali S, Mughal T, Miller V, Mangueira CL, Odone V, Hamerschlak N. Identification of a novel fusion TBL1XR1-PDGFRB in a patient with acute myeloid leukemia harboring the DEK-NUP214 fusion and clinical response to dasatinib. Leuk Lymphoma. 2017;58:2969–2972. doi: 10.1080/10428194.2017.1318437. [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Han H, Hu Y, Yang W, Lv Y, Wang L, Zhang L, Ji J. MicroRNA-130a-3p suppresses cell migration and invasion by inhibition of TBL1XR1-mediated EMT in human gastric carcinoma. Mol Carcinog. 2018;57:383–392. doi: 10.1002/mc.22762. [DOI] [PubMed] [Google Scholar]
- 28.Moiseenko A, Kheirollahi V, Chao CM, Ahmadvand N, Quantius J, Wilhelm J, Herold S, Ahlbrecht K, Morty RE, Rizvanov AA, et al. Origin and characterization of alpha smooth muscle actin-positive cells during murine lung development. Stem Cells. 2017;35:1566–1578. doi: 10.1002/stem.2615. [DOI] [PubMed] [Google Scholar]
- 29.Zou YF, Xu JH, Gu YY, Pan FM, Tao JH, Wang DG, Xu SQ, Xiao H, Chen PL, Liu S, et al. Single nucleotide polymorphisms of HSP90AA1 gene influence response of SLE patients to glucocorticoids treatment. Springerplus. 2016;5:222. doi: 10.1186/s40064-016-1911-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng Q, Chang JT, Geradts J, Neckers LM, Haystead T, Spector NL, Lyerly HK. Amplification and high-level expression of heat shock protein 90 marks aggressive phenotypes of human epidermal growth factor receptor 2 negative breast cancer. Breast Cancer Res. 2012;14:R62. doi: 10.1186/bcr3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui X, Xing J, Liu Y, Zhou Y, Luo X, Zhang Z, Han W, Wu T, Chen W. COPD and levels of Hsp70 (HSPA1A) and Hsp27 (HSPB1) in plasma and lymphocytes among coal workers: A case-control study. Cell Stress Chaperones. 2015;20:473–481. doi: 10.1007/s12192-015-0572-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15:e493–e503. doi: 10.1016/S1470-2045(14)70263-3. [DOI] [PubMed] [Google Scholar]
- 33.Choi SH, Nam JK, Kim BY, Jang J, Jin YB, Lee HJ, Park S, Ji YH, Cho J, Lee YJ. HSPB1 inhibits the endothelial-to-mesenchymal transition to suppress pulmonary fibrosis and lung tumorigenesis. Cancer Res. 2016;76:1019–1030. doi: 10.1158/0008-5472.CAN-15-0952. [DOI] [PubMed] [Google Scholar]
- 34.Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d'Assignies MS, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available in the GEO repository with no. GSE12472.