Abstract
Lung cancer is the most common cancer in males and females and ~40% of lung cancer cases are adenocarcinomas. Previous studies have demonstrated that myristoylated alanine rich protein kinase C substrate (MARCKS) is upregulated in several types of cancer and is associated with poor prognosis in patients with breast cancer. However, its expression level and role in lung adenocarcinoma remain unknown. Therefore, the aim of the present study was to investigate the expression level and biological functions of MARCKS like 1 (MARCKSL1), a member of the MARCKS family, in lung adenocarcinoma. The expression level of MARCKSL1 was examined in human lung adenocarcinoma tissues and cell lines. MARCKSL1-specific small interfering RNAs effectively suppressed its expression level and significantly inhibited the proliferation, migration and invasion of lung adenocarcinoma cells. Additionally, the role of MARCKSLI in the regulation of metastasis was examined. Silencing MARCKSL1 decreased the expression of the epithelial-mesenchymal transition (EMT)-associated proteins E-cadherin, N-cadherin, vimentin and snail family transcriptional repressor 2, and decreased the phosphorylation level of AKT. The results obtained in the current study suggested that MARCKSL1 promoted the progression of lung adenocarcinoma by regulating EMT. MARCKSLI may have prognostic value and serve as a novel therapeutic target in lung adenocarcinoma.
Keywords: myristoylated alanine rich protein kinase C substrate like 1, lung adenocarcinoma, metastasis, epithelial-mesenchymal transition, protein kinase B
Introduction
Lung cancer was the leading cause of cancer-associated mortalities in males and females worldwide in 2018 (1). It was estimated that 222,500 new cases of lung cancer were diagnosed, and 155,870 mortalities due to this disease were recorded in the United States in 2017, accounting for 25% of all cancer-associated mortalities (2). The main reason for the high mortality associated with lung cancer is the high metastatic potential of the disease (3). Therefore, the investigation of the proteins and signaling pathways that promote the progression and metastasis of lung cancer may contribute to the development of novel therapies for the disease.
Myristoylated alanine-rich C kinase substrate (MARCKS) is an important protein kinase C (PKC) substrate. MARCKS reversely binds to several structural and regulatory molecules including actin, calmodulin and phospholipid phosphatidylinositol 4,5-bisphosphate (4). MARCKS is implicated in a number of biological processes such as phagocytosis, membrane trafficking and secretion of mucin (5,6). Furthermore, the role of MARCKS in regulating mucin secretion in airway epithelial cells has been extensively studied (5). The actin-binding property of MARCKS suggests that it may be involved in the regulation of cell adhesion and mobility. In fact, MARCKS was implicated in the migration of neutrophils (7), vascular endothelial cells (4) and smooth muscle cells (8). MARCKS serves an important role during embryo development. Blockade of MARCKS protein expression in Xenopus and zebrafish embryogenesis resulted in defective morphogenetic movements of gastrulation by affecting cortical actin formation and cell adhesion, protrusive activity and polarity (9,10). Therefore, MARCKS may serve as a novel biomarker and therapeutic target for cancer, as metastasis is associated with changes in cell morphology and cell migration. Upregulation of MARCKS has been shown to promote the progression of several types of cancer, such as prostate cancer (11), osteosarcoma (12), breast cancer (13), ovarian cancer (14) and hepatocellular carcinoma (15).
MARCKS like 1 (MARCKSL1) is another member of the MARCKS family (16). It is an important cellular substrate for PKC and serves as an actin binding protein (16). The effector domain, which allows MARCKSL1 to bind to actin, shares 87% homology with the corresponding domain in MARCKS (17). Both MARCKS and MARCKS1 have been associated with migration of breast cancer cells (17,18). Deletion of MARCKSL1 prevents neural tube closure in the developing brain in mice, an event dependent on actin binding, cell elongation and migration (19). MARCKSL1 is upregulated in breast cancer tissues compared with normal tissues and is associated with poor prognosis (20). Jonsdottir et al (21) reported that the level of MARCKSL1 protein expression is a strong prognostic indicator in lymph node-negative breast cancer. Patients with high MARCKSL1 expression exhibit a 50% lower survival rate compared with patients with low expression. Furthermore, knockdown of MARCKSL1 in vitro using siRNA decreases the migratory potential of breast cancer cells (22). In addition to breast cancer, significant upregulation of MARCKSL1 has been reported in esophageal squamous cell carcinoma (23), muscle-derived cancer (17) and uterine cancer (17). However, the expression and the exact role of MARCKSL1 in lung cancer have not been extensively studied. The present study revealed the therapeutic potential of MARCKSL1 in lung adenocarcinoma and determined its contribution to the progression of the disease.
Materials and methods
Cell culture
The human lung adenocarcinoma cell lines H2122, H23, A549, H1975 and H820 and the normal human lung epithelial cell line BEAS-2B were purchased from Jennio Bioech Co., Ltd. Cells were cultured in RPMI 1640 medium (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone; GE Healthcare Life Sciences), 2 mM glutamine (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin (Sigma-Aldrich; Merck KGaA) in an incubator containing 5% CO2 at 37°C.
Immunohistochemistry (IHC)
A total of five formalin-fixed, paraffin-embedded human lung adenocarcinoma tissues and one healthy lung tissue were purchased from Shanghai Outdo Biotech Co., Ltd. Tissues were incubated in 10% formalin at room temperature for 72 h. The tissue sections (6-µm thick) were deparaffinized using xylene at room temperature and antigen retrieval was subsequently performed by incubating the sections in 1X citrate buffer (cat. no. C999; Sigma-Aldrich, Merck KGaA) at 100°C for 10 min. Tissue sections were then blocked with the 2.5% normal horse serum (cat. no. S-2012; Vector Laboratories, Inc.) at room temperature for 30 min and incubated with primary antibodies directed against MARCKSL1 (cat. no. PA5-56495; 1:2,000; Thermo Fisher Scientific, Inc.) and biotin (cat. no. SP-2001; 1:10; Vector Laboratories, Inc.) overnight at 4°C. After that, tissue sections were incubated with a ready-to-use biotinylated pan-specific antibody (cat. no. BP-1400; Vector Laboratories, Inc.) for 30 min at room temperature. The slides were subsequently stained with 3,3′-diaminobenzidine (cat. no. SK-4100; Vector Laboratories, Inc.) for 3 min at room temperature and counterstained with hematoxylin (cat. no. H-3404; Vector Laboratories, Inc.) for 1 min at room temperature. The stained slides were scanned with a PathScope pathology slide scanner (Gene Tech Biotechnology Co., Ltd.).
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Total RNA was reverse transcribed into cDNA using a SuperScript™ III First-Strand Synthesis system (cat. no. 18080-051; Invitrogen, Thermo Fisher Scientific, Inc.) containing first strand Moloney murine leukemia virus reverse transcriptase (cat. no. 18080-051; Invitrogen, Thermo Fisher Scientific, Inc.), random hexamers, dNTPs and RT buffer as previously described (24). The qPCR MARCKSL1 primer set was purchased from Thermo Fisher Scientific, Inc. (cat. no. 4331182) The sequence of the forward primer was 5′-CAGGCTACAGAGCCATCCACTC-3′ and the sequence of the reverse primer was 5′-GCAGCTTAGAGATCACCCACCT-3′. qPCR was performed on a CFX96™ real-time PCR detection system (Bio-Rad Laboratories, Inc.) using the DyNamo ColorFlash Probe qPCR Kit (cat. no. F465S; Thermo Fisher Scientific, Inc.). The thermocycling conditions used were as follows: 7 min at 95°C followed by 3 sec at 95°C and 30 sec at 60°C for 40 cycles with data collection at 60°C. MARCKSL1 mRNA levels were quantified using the 2−ΔΔCq method (25) and normalized to the internal reference gene GAPDH (forward primer, 5′-CATCACTGCCACCCAGAAGACTG-3′ and reverse, 5′-ATGCCAGTGAGCTTCCCGTTCAG-3′).
Cell transfection
Two small interfering (si) RNAs targeting human MARCKSL1 (cat. nos. J-018697-06-0002, sequence, 5′-CCAAGAAGAAGAAGAAAUU-3′; J-018697-09-0002, sequence, 5′-GGAGAAUGGCCACGUGAAAUU-3′) and a non-targeting siRNA (cat. no. D-001810-01-05, sequence, 5′-UGGUUUACAUGUCGACUAA-3′) were obtained from GE Healthcare Dharmacon, Inc. A549 and H1975 cells were seeded at a density of 2.5×105 cells/well in a 6-well plate 24 h prior to transfection. 50 pM siRNA and 10 µl DharmaFECT transfection reagent (cat. no. T-2001-01; Dharmacon, Inc.) were added to 190 µl Opti-MEM media (cat. no. 31985062; Thermo Fisher Scientific, Inc.) in two separate tubes and allowed to incubate for 5 min at room temperature. The contents of the two tubes were subsequently combined, gently stirred and incubated for 20 min at room temperature. The cell culture medium RPMI 1640 (Thermo Fisher Scientific, Inc.) was replaced with 400 µl transfection medium in 2 ml RPMI 1640 medium (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone; GE Healthcare Life Sciences) and 2 mM glutamine (Gibco; Thermo Fisher Scientific, Inc.), without penicillin and streptomycin. RT-qPCR and western blotting were performed to assess the knockdown efficiency 48 h after transfection.
Western blot analysis
BEAS-2B, H2122, H23, A549, H1975 and H820 cells were lysed using RIPA buffer (150 mM sodium chloride, 1% NP-40, 0.5% SDS, 0.1% sodium deoxycholate and 50 mM Tris; pH 7.4) supplemented with protease and phosphatase inhibitor cocktail. Total protein was quantified using a bicinchoninic acid assay and 20 µg protein per lane were separated via SDS-PAGE on 4–20% gels (cat. no. 4561096; Bio-Rad Laboratories, Inc.). The separated proteins were subsequently transferred onto PVDF membranes and blocked in 1% casein in TBS at room temperature for 1 h. The membrane was incubated with primary antibodies against MARCKSL1 (cat. no. PA5-56495; 1:2,000; Thermo Fisher Scientific, Inc.), E-cadherin (cat. no. 610181; 1:2,500; BD Biosciences), N-cadherin (cat. no. 610920; 1:2,500; BD Biosciences), vimentin (cat. no. 5741; 1:1,000; Cell Signaling Technology), snail family transcriptional repressor 2 (SNAI2; cat. no. 9585; 1:1,000; Cell Signaling Technology), phosphorylated-AKT (p-AKT; cat. no. 4060; 1:1,000; Cell Signaling Technology), AKT (cat. no. 4691; 1:1,000; Cell Signaling Technology) and GAPDH (cat. no. 5174; 1:5,000; Cell Signaling Technology) overnight at 4°C. Membranes were then washed with TBST buffer and probed with secondary peroxidase-conjugated antibodies (cat. nos. 7074 and 7076; both 1:2,000; Cell Signaling Technology) for 1 h at room temperature. The proteins were visualized using Immobilon Western Chemiluminescent HRP substrate (cat. no. WBKLS0500; EMD Millipore).
Cell proliferation analysis
A total of 50,000 A549 and H1975 cells per well were plated in RPMI 1640 medium supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin in 12-well plates and incubated at 37°C. A total of 24, 48 and 72 h after plating, the cells were harvested and resuspended in 500 µl PBS. A total of 500 µl 0.4% trypan blue solution (cat. no. 15250061; Thermo Fisher Scientific, Inc.) was added to each sample and the cells were counted using a Bio-Rad TC20 automated cell counter (Bio-Rad Laboratories, Inc.).
Cell viability assay
A total of 2,000 A549 and H1975 cells were plated per well in a 96-well plate and incubated for 72 h at 37°C. The cells were subsequently allowed to equilibrate to room temperature for ~30 min and 100 µl CellTiter-Glo reagent (Promega Corporation) was added per well. Luminescence was recorded using a microplate luminometer.
Wound-healing assay
A total of 2×105 A549 and H1975 cells per well were plated in RPMI 1640 medium supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin in a 12-well plate. When the cells reached 95–100% confluence, the medium was replaced with RPMI 1640 supplemented with 0.5% FBS, 2 mM glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin. After overnight incubation, a linear wound was made with a 200 µl pipette tip. Cell migration into the wound area was monitored at 0 and 28 h using a light microscope (magnification, ×5).
Boyden chamber invasion assay
A total of 15,000 cells in serum-free RMPI 1640 medium were seeded in Matrigel-coated chambers (8-µm pore size; cat. no. 354480; BD Biosciences). RPMI 1640 medium supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin was added to each well outside the chambers. The cells were incubated at 37°C for 24 h. The invading cells were subsequently fixed in 100% methanol for 10 min at room temperature, stained with 0.5% crystal violet for 20 min at room temperature and visualized using a light microscope in eight randomly selected fields (magnification, ×10).
Spheroid invasion assay
Spheroids were formed by adding 100 µl media containing 5,000 cells to each well in a 96-well round bottom low attachment plate (Corning, Inc.). Spheroids were incubated at 37°C for 3 days. Spheroids were then plated in 8-well LabTek chambered slide with 250 µl PureCol® collagen solution (2 mg/ml; PureCol). After 24 h, invading cells were recorded using a light microscope (magnification, ×5).
Oncomine analysis
The Oncomine database (www.oncomine.org), a bioinformatics tool containing >18,000 cancer gene expression microarrays (26), was researched for MARCKSL1 gene. ‘Cancer versus normal analysis’ was chosen for analysis type and ‘lung cancer’ was chosen for cancer type so that the data sets containing MARCKSL1 expression data for lung cancer versus normal lung tissue was analyzed and displayed.
Statistical analysis
All data are derived from at least three independent experiments and are presented as the mean ± standard deviation. One-way ANOVA and the Bonferroni post hoc test were used to compare replicate means where each column in a row was compared with all other columns using GraphPad Prism software (version 7; GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference. Survival curves were plotted using the Kaplan-Meier method as described previously (27), and compared using the hazard ratio (HR), 95% CI and log-rank P-values.
Results
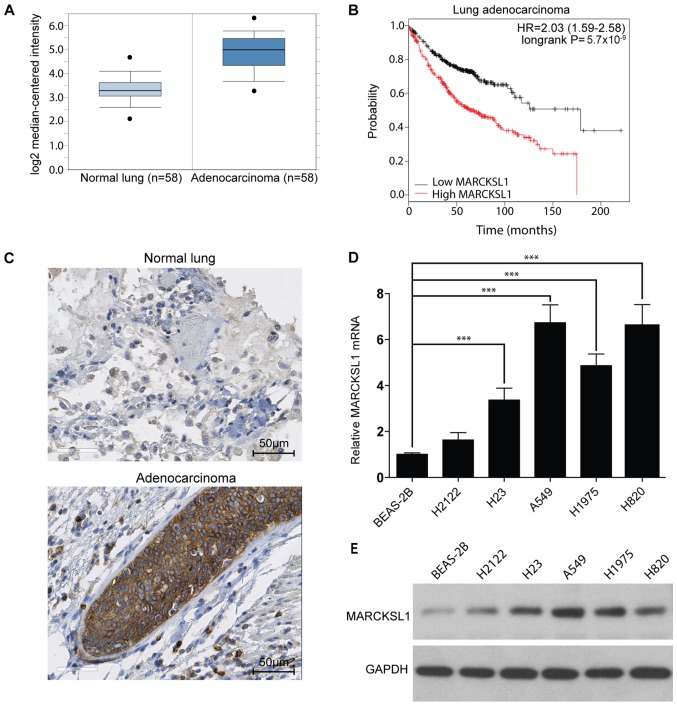
MARCKSL1 expression is upregulated in lung adenocarcinoma
The MARCKSL1 gene expression level in lung cancer and normal tissues was investigated. The Oncomine database (www.oncomine.org), which is a bioinformatics tool containing >18,000 cancer gene expression microarrays (26), was used. A total of six datasets (28–33) containing gene expression profiles for tumor and normal tissues downloaded from the Oncomine database. MARCKSL1 mRNA levels were significantly increased in tumor tissues compared with normal tissues in the six datasets downloaded from the Oncomine database. Data from one database are shown in Fig. 1A (fold change, 2.93; P<0.001) and all six results are presented in Table SI. In addition, the Kaplan-Meier plotter online database (kmplot.com/analysis) was used to investigate the effect of MARCKSL1 expression on the survival rate of patients with lung adenocarcinoma. A total of 720 patients were included in the analysis. The expression range was from 280 to 14,567 and the cut-off value was 2,763. High expression of MARCKSL1 was associated with poor survival rate in lung adenocarcinoma patients (Fig. 1B). To further investigate MARCKSL1 expression in tumor tissues, the expression pattern of MARCKSL1 in five human lung adenocarcinoma and one normal lung samples (Table SII) was investigated using IHC. The level of MARCKSL1 protein expression was increased in lung adenocarcinoma tissue compared with normal lung tissues (Fig. 1C). For further validation, the MARCKSL1 mRNA expression level in five lung adenocarcinoma cell lines (H2122, H23, A549, H1975 and H820) and one normal human lung epithelial cell line (BEAS-2B) was assessed using RT-qPCR. The present results suggested that MARCKSL1 mRNA level was significantly increased in four lung adenocarcinoma cell lines (H23, A549, H1975 and H820) compared with the BEAS-2B cell line (Fig. 1D). Furthermore, the protein expression level of MARCKSL1 was increased in the aforementioned four lung adenocarcinoma cells compared with the BEAS-2B cell line (Fig. 1E), in line with the RT-qPCR results. Taken together, the present data indicated that MARCKSL1 was upregulated in lung adenocarcinoma, suggesting that it may serve a role in tumor progression.
Figure 1.
MARCKSL1 is dysregulated in human lung adenocarcinoma and cell lines. (A) MARCKSL1 mRNA levels were analyzed in six lung adenocarcinoma and normal lung tissue microarray datasets downloaded from the Oncomine database. The results of a representative dataset are presented. (B) KM plots of patients with lung adenocarcinoma with different MARCKSL1 mRNA expression levels (high vs. low) as found in the KM-Plotter database. (C) Representative normal lung and lung adenocarcinoma specimens stained with MARCKSL1. The MARCKSL1 (D) mRNA and (E) protein levels in five lung adenocarcinoma cell lines and a normal lung epithelial cell line were determined by reverse transcription-quantitative PCR and western blotting, respectively. Data are presented as the mean ± standard error of the mean of three independent experiments. ***P<0.001. MARCKSL1, MARCKS like 1; KM, Kaplan-Meier; HR, hazard ratio.
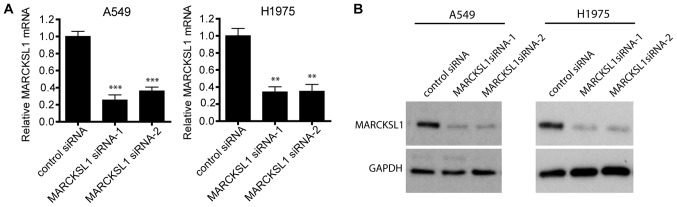
siRNA effectively suppresses MARCKSL1 expression in lung adenocarcinoma cells
To investigate the functional role of MARCKSL1 in lung adenocarcinoma, two lung adenocarcinoma cell lines, A549 and H1975, which exhibit high levels of MARCKSL1 expression, were selected for further study. A549 and H1975 cells were transfected with two MARCKSL1-specific siRNAs and RT-qPCR revealed that both siRNAs resulted in MARCKSL1 downregulation compared with control cells transfected with scrambled siRNA (Fig. 2A). Western blotting produced similar results, suggesting that MARCKSL1 protein expression was significantly suppressed following transfection with MARCKSL1 siRNAs in A549 and H1975 cell lines (Fig. 2B).
Figure 2.
MARCKS expression in the lung adenocarcinoma cell lines A549 and H1975 is suppressed following siRNA knockdown. A549 and H1975 cells were transfected with either non-targeting siRNA or MARCKSL1-directed siRNAs, and the (A) mRNA and (B) protein levels of MARCSKL1 in these cells were determined by reverse transcription-quantitative PCR and western blotting, respectively. **P<0.01 and ***P<0.001 vs. control siRNA. MARCKSL1, MARCKS like 1; si, small interfering.
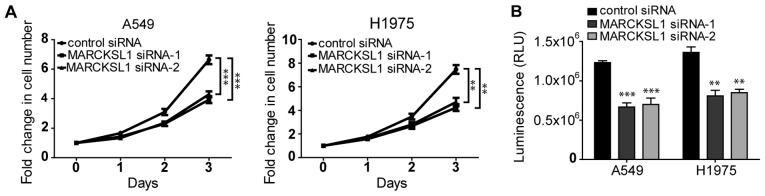
Silencing of MARCKSL1 suppresses the proliferation and viability of lung adenocarcinoma cells
It has been shown that MARCKS increases cell proliferation in renal cell carcinoma (34). As a MARCKS family member that shares ~50% amino acid homology with MARCKS, MARCKSL1 was demonstrated to promote the proliferation of certain cells, including retinal progenitor cells (35). The effect of MARCKSL1 on the proliferation of lung adenocarcinoma cells was investigated. As shown in Fig. 3A, the proliferation of A549 and H1975 cells transfected with MARCSKL1-specific siRNAs was significantly decreased compared cells transfected with control siRNA. Similar results were obtained using the CellTiter-Glo assay, which is based on ATP levels in the cells. The luminescence in cells transfected with MARCKSL1 siRNAs was significantly decreased compared with cells transfected with control siRNA (Fig. 2B), suggesting that the cells were less viable. Therefore, downregulation of MARCKSL1 expression inhibited the proliferation and viability of lung adenocarcinoma cells.
Figure 3.
MARCKL1 knockdown inhibits cell proliferation and decreased cell viability. (A) The proliferation of transfected A549 and H1975 cells was analyzed using trypan blue and an automated cell counter. (B) The effect of MARCKSL1 knockdown on cell viability was evaluated by CellTiter-Glo assay. Data are presented as the mean ± standard error of the mean of three independent measurements. **P<0.01 and ***P<0.001 vs. control siRNA. MARCKSL1, MARCKS like 1; si, small interfering; RLU, relative luminescence unit.
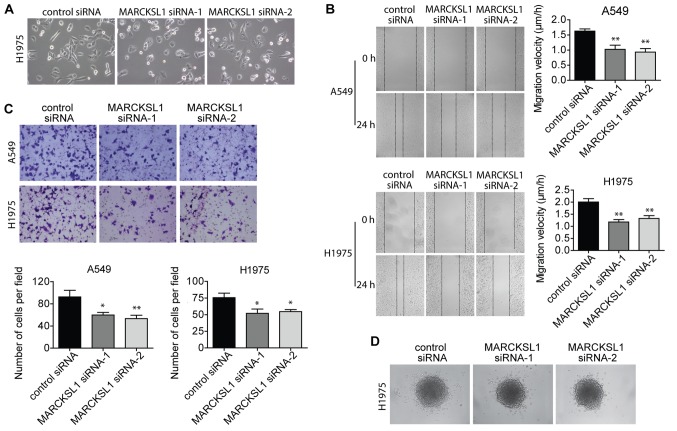
Suppression of MARCKSL1 inhibits migration and invasion of lung adenocarcinoma cells
When culturing the cells, the extent of cell spreading and surface protrusions was reduced in A549 and H1975 cells transfected with MARCKSL1 siRNAs compared with control siRNA (Fig. 4A). This suggested an inhibition of filopodia and lamellopodia formation, and a reduction of cell motility. Therefore, wound healing and Boyden chamber invasion assays were performed to evaluate cell migration and invasion, respectively. The migratory ability of A549 and H1975 cells was significantly impaired in the MARCKSL1 siRNA group compared with the control siRNA group (Fig. 4B). A similar decrease in cell invasion ability was observed (Fig. 4C). The effect of MARCKSL1 on H1975 cell invasion was further investigated using a 3D model. H1975 cells transfected with MARCKSL1 siRNAs or control siRNA were used to form spheroids, which were subsequently embedded in collagen gel. After 24 h, fewer cells transfected with MARCKSL1 siRNAs had moved out of the spheroids and into the surrounding collagen compared with cells transfected with the siRNA control (Fig. 4D). These results suggested that MARCKSL1 increased the migration and invasion abilities of lung adenocarcinoma cells.
Figure 4.
MARCKL1 knockdown inhibits the migration and invasion of the lung adenocarcinoma cell lines H1975 and A549. (A) The morphology of H1975 cells transfected with MARCKSL1 siRNA and control siRNA was evaluated using a light microscope (magnification, ×10). (B) Wound-healing assay was performed to evaluate the cell migratory ability of transfected A549 and H1975 cells using a light microscope (magnification, ×5). (C) The invasion ability of A549 and H1975 cells was evaluated by a Boyden chamber invasion assay using a light microscope (magnification, ×10). (D) The 3D spheroid invasion assay was performed to analyze the effect of MARCKSL1 knockdown on cell invasion using a light microscope (magnification, ×5). *P<0.05 and **P<0.01 vs. control siRNA. MARCKSL1, MARCKS like 1; si, small interfering.
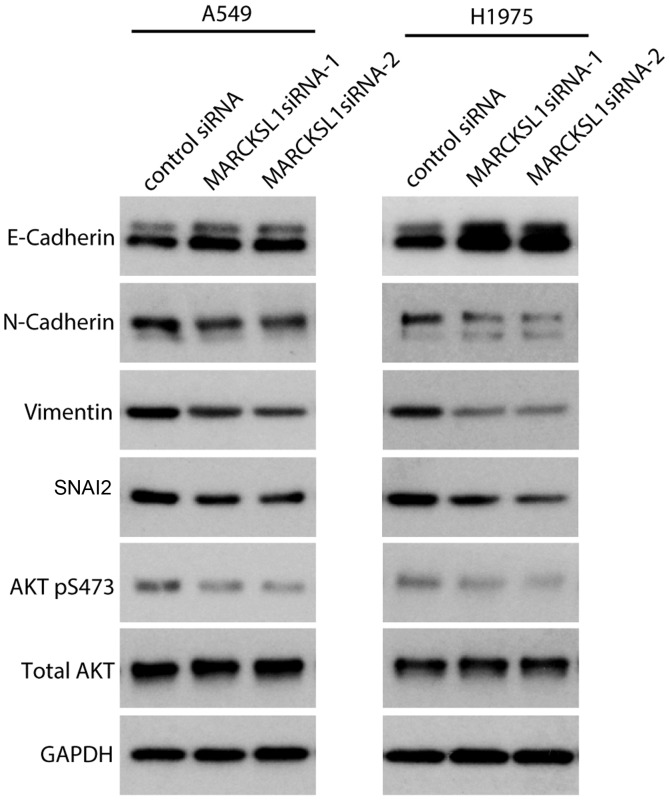
MARCKSL1 promotes epithelial-mesenchymal transition (EMT) in lung adenocarcinoma cells
EMT plays an important role in normal development as well as tumor progression, and EMT causes epithelial cells to acquire mesenchymal and fibroblast-like properties and to show reduced intercellular adhesion and increased mobility. The expression of EMT-associated genes in siRNA-transfected A549 and H1975 cells was investigated. Western blot analysis revealed that the expression of the epithelial marker E-cadherin was upregulated, whereas the expression of the mesenchymal markers N-cadherin and vimentin were downregulated in A549 and H1975 cells transfected with MARCKSL1 siRNAs compared with control siRNA (Fig. 5). SNAI2, a key regulator of EMT that suppresses the transcription of E-cadherin (36), was also suppressed following MARCKSL1 knockdown. It has been shown that the phosphoinositide 3-kinase (PI3K)/AKT signaling pathway plays an important role in promoting EMT in various types of cancer including breast, lung and colorectal cancer (37–39). Therefore, the effect of MARCKSL1 expression on the PI3K/AKT signaling pathway was investigated in A549 and H1975 cells. AKT is activated by phosphorylation of the carboxy terminus at S473 (40). Compared with A549 and H1975 cells transfected with control siRNA, cells transfected with MARCKSL1 exhibited decreased AKT phosphorylation at S473, while the total AKT remained unchanged (Fig. 5). The results obtained in the current study revealed that MARCKSL1 activated the AKT signaling pathway, facilitated EMT and promoted the migration and invasion of lung adenocarcinoma cells in vitro.
Figure 5.
Knockdown of MARCKSL1 suppresses epithelial-mesenchymal transition-associated proteins and AKT activation. Expression of E-cadherin, N-cadherin, vimentin, SNAI2, p-AKT and AKT was evaluated by western blotting. GAPDH served as loading control. Multiple bands were observed for E-cadherin and N-cadherin due to post-translational modifications as previously described (67,68). MARCKSL1, MARCKS like 1; AKT, protein kinase B; si, small interfering; SNAI2, snail family transcriptional repressor 2; p, phosphorylated.
Discussion
Since the MARCKS protein was identified in the early 1980s, its role in regulating cell movement has been extensively studied. MARCKS plays important roles during embryonic development (9,10) and in tumor progression (14,34). However, the expression levels and function of MARCKSL1 in lung cancer remain unclear.
In the present study, IHC revealed that MARCKSL1 protein levels were increased in human lung adenocarcinoma specimens compared with adjacent noncancerous and normal lung tissues. Furthermore, MARCKSL1 mRNA and protein levels were increased in a number of adenocarcinoma cell lines compared with the normal human lung epithelial cell line BEAS-2B. In six sets of microarray data downloaded from the Oncomine database, which included MARCKSL1 expression levels in lung adenocarcinoma and normal lung tissues, the expression levels of MARCKSL1 were significantly increased in lung adenocarcinoma tissues compared normal tissues. The results in the present study were similar to previous results investigating the Human Protein Atlas, where MARCKSL1 expression was detected in six lung adenocarcinoma specimens using IHC, with high-intensity staining observed in 4/6 samples and medium-intensity staining observed in 2/6 samples (41). Therefore, MARCKSL1 may serve as a potential therapeutic target for the diagnosis and prognosis prediction of lung adenocarcinoma. Interestingly, MARCKL1 was detected in 3/5 squamous cell carcinoma specimens in the Human Protein Atlas, with medium- and low-intensity staining observed in two and one samples, respectively. These previous results suggested that the expression level of MARCKSL1 may vary among different types of lung tumors (42).
The present study investigated the function of MARCKSL1 in two lung adenocarcinoma cell lines, A549 and H1975. Silencing of MARCKSL1 by siRNA transfection significantly decreased the proliferation, migration and invasion of these cells compared with cells transfected with control siRNA. Metastases are responsible for the majority of cancer-associated mortalities, including lung cancer (3,43,44). EMT promotes metastasis in epithelial-derived carcinoma as it allows cells to acquire mesenchymal, fibroblast-like properties, consequently increasing motility and invasiveness (37). An important molecular feature of EMT is the downregulation of E-cadherin, a cell adhesion molecule located on the surfaces of normal epithelial and carcinoma cells (45,46). Increased E-cadherin levels are often associated with reduced invasion and metastasis of human breast epithelial cells (47). N-cadherin, another member of the cadherin family, promotes invasion in breast, prostate, pancreatic and squamous cell carcinoma (48). Vimentin is an intermediate filament protein expressed in mesenchymal cells and is a widely used marker of EMT (49–51). SNAI2, a zinc finger transcriptional factor, suppresses the expression of E-cadherin and serves as an important inducer of EMT (52). The present study revealed the upregulation of E-cadherin and downregulation of N-cadherin, vimentin and SNAI2 in A549 and H1975 cells following MARCKSL1 knockdown, indicating that EMT was suppressed. The present data support the hypothesis that MARCKSL1 promotes EMT and increases the invasion and migration abilities of lung adenocarcinoma cells.
Accumulating evidence has demonstrated that the activation of the AKT signaling pathway is a key feature of EMT (53–55). AKT signaling inhibits SNAIL1 phosphorylation while its inhibition decreases the level of SNAIL1 expression (54). SNAIL1 downregulates E-cadherin and stimulates EMT in human colon adenocarcinoma HT-29-M6 cells (56). Furthermore, AKT phosphorylates and activates hypoxia inducible factor 1 subunit α without a physical heat shock, leading to the upregulation of SNAI2 and promoting EMT (57). The present study revealed that AKT phosphorylation was decreased following MARCKSL1 knockdown, suggesting that the AKT/SNAI2 signaling pathway may serve a role in MARCSKL1-induced EMT. As AKT serves important roles in several signaling pathways and is involved in a number of cellular processes, including glucose metabolism and apoptosis (58), MARCKSL1 may serve as a novel therapeutic target that may be used in combination with other agents in cancer treatment (59–63).
MARCSKL1 is an important regulatory molecule that mediates mucin granule release by human bronchial epithelial cells (64). A peptide identical to the N-terminal 24-amino acid fragment of MARCKS, the MARCKS N-terminus sequence (MANS) peptide, competitively inhibits the binding of MARCKS to the membranes of mucin-secreting granules and attenuates mucus release from goblet cells (65). Interestingly, MANS peptide treatment inhibited MARCKS function during fibroblast and lung cancer migration and impaired the metastatic potential of invasive lung cancer cells in vivo (33,66). Since MARCKSL1 is a MARCKS homolog, and both proteins have highly conserved regions and similar functions in regulating cell migration (16), it is likely that peptides identical to the N-terminus of MARCKSL1 may inhibit its function and suppress lung adenocarcinoma cell proliferation, migration and invasion. Further studies are required to evaluate the efficacy of such peptides.
In summary, the results obtained in the current study suggested that MARCKSL1 expression was significantly increased in human lung adenocarcinoma, suggesting that MARCKSL1 may be a negative prognostic factor. Furthermore, MARCKSL1 promoted cell proliferation, migration and invasion of lung adenocarcinoma cells by regulating the AKT/SNAI2 pathway-induced EMT. MARCKSL1 may therefore serve as a potential therapeutic target for lung adenocarcinoma and the potential application of MANS-similar peptides to inhibit MARCKSL1 should be further investigated.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- MARCKS
myristoylated alanine rich protein kinase C substrate
- MARCKSL1
MARCKS like 1
- PKC
protein kinase C
- EMT
epithelial-mesenchymal transition
- siRNA
small interfering RNA
Funding
The present study was supported by the Applied Basic Research Foundation of Changzhou (grant no. CJ20160057).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XY conceived and designed the experiments. RG and MY provided experimental guidance and analyzed and interpreted the data. XW, KC, XS, CH, YL, YW, LS and JC performed the experiments. WL analyzed the data and wrote the paper.
Ethics approval and consent to participate
As the human tissues used in the current study were obtained from a commercial source (Shanghai Outdo Biotech Co., Ltd.), the requirement for ethical approval was waived by the Ethics Committee of The Affiliated Changzhou Second Hospital of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Min TR, Park HJ, Park MN, Kim B, Park SH. The root bark of morus alba L. suppressed the migration of human non-small-cell lung cancer cells through inhibition of epithelial-mesenchymal transition mediated by STAT3 and Src. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20092244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalwa H, Michel T. The MARCKS protein plays a critical role in phosphatidylinositol 4,5-bisphosphate metabolism and directed cell movement in vascular endothelial cells. J Biol Chem. 2011;286:2320–2330. doi: 10.1074/jbc.M110.196022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green TD, Crews AL, Park J, Fang S, Adler KB. Regulation of mucin secretion and inflammation in asthma: A role for MARCKS protein? Biochim Biophys Acta. 2011;1810:1110–1113. doi: 10.1016/j.bbagen.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arbuzova A, Schmitz AA, Vergères G. Cross-talk unfolded: MARCKS proteins. Biochem J. 2002;362:1–12. doi: 10.1042/bj3620001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheats MK, Sung EJ, Adler KB, Jones SL. In vitro neutrophil migration requires protein kinase C-delta (δ-PKC)-mediated myristoylated alanine-rich C-kinase substrate (MARCKS) phosphorylation. Inflammation. 2015;38:1126–1141. doi: 10.1007/s10753-014-0078-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu D, Makkar G, Strickland DK, Blanpied TA, Stumpo DJ, Blackshear PJ, Sarkar R, Monahan TS. Myristoylated alanine-rich protein kinase substrate (MARCKS) regulates small GTPase Rac1 and Cdc42 activity and is a critical mediator of vascular smooth muscle cell migration in intimal hyperplasia formation. J Am Heart Assoc. 2015;4:e002255. doi: 10.1161/JAHA.115.002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iioka H, Ueno N, Kinoshita N. Essential role of MARCKS in cortical actin dynamics during gastrulation movements. J Cell Biol. 2004;164:169–174. doi: 10.1083/jcb.200310027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang YW, Wei CY, Dai HP, Zhu ZY, Sun YH. Subtractive phage display technology identifies zebrafish marcksb that is required for gastrulation. Gene. 2013;521:69–77. doi: 10.1016/j.gene.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 11.Dorris E, O'Neill A, Hanrahan K, Treacy A, Watson RW. MARCKS promotes invasion and is associated with biochemical recurrence in prostate cancer. Oncotarget. 2017;8:72021–72030. doi: 10.18632/oncotarget.18894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Su P, Zhi L, Zhao K. miR-34c-3p acts as a tumor suppressor gene in osteosarcoma by targeting MARCKS. Mol Med Rep. 2017;15:1204–1210. doi: 10.3892/mmr.2017.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manai M, Thomassin-Piana J, Gamoudi A, Finetti P, Lopez M, Eghozzi R, Ayadi S, Lamine OB, Manai M, Rahal K, et al. MARCKS protein overexpression in inflammatory breast cancer. Oncotarget. 2017;8:6246–6257. doi: 10.18632/oncotarget.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, Xu S, Jin P, Yang X, Li X, Wan D, Zhang T, Long S, Wei X, Chen G, et al. MARCKS contributes to stromal cancer-associated fibroblast activation and facilitates ovarian cancer metastasis. Oncotarget. 2016;7:37649–37663. doi: 10.18632/oncotarget.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song J, Wang Q, Luo Y, Yuan P, Tang C, Hui Y, Wang Z. miR-34c-3p inhibits cell proliferation, migration and invasion of hepatocellular carcinoma by targeting MARCKS. Int J Clin Exp Pathol. 2015;8:12728–12737. [PMC free article] [PubMed] [Google Scholar]
- 16.El Amri M, Fitzgerald U, Schlosser G. MARCKS and MARCKS-like proteins in development and regeneration. J Biomed Sci. 2018;25:43. doi: 10.1186/s12929-018-0445-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Björkblom B, Padzik A, Mohammad H, Westerlund N, Komulainen E, Hollos P, Parviainen L, Papageorgiou AC, Iljin K, Kallioniemi O, et al. c-Jun N-terminal kinase phosphorylation of MARCKSL1 determines actin stability and migration in neurons and in cancer cells. Mol Cell Biol. 2012;32:3513–3526. doi: 10.1128/MCB.00713-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong LWR, Yang DC, Chen CH. Myristoylated alanine-rich C kinase substrate (MARCKS): A multirole signaling protein in cancers. Cancer Metastasis Rev. 2017;36:737–747. doi: 10.1007/s10555-017-9709-6. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Chang S, Duncan SA, Okano HJ, Fishell G, Aderem A. Disruption of the MacMARCKS gene prevents cranial neural tube closure and results in anencephaly. Proc Natl Acad Sci USA. 1996;93:6275–6279. doi: 10.1073/pnas.93.13.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Jarrett J, Huang CC, Satcher RL, Jr, Levenson AS. Identification of estrogen-responsive genes involved in breast cancer metastases to the bone. Clin Exp Metastasis. 2007;24:411–422. doi: 10.1007/s10585-007-9078-6. [DOI] [PubMed] [Google Scholar]
- 21.Jonsdottir K, Zhang H, Jhagroe D, Skaland I, Slewa A, Björkblom B, Coffey ET, Gudlaugsson E, Smaaland R, Janssen EA, Baak JP. The prognostic value of MARCKS-like 1 in lymph node-negative breast cancer. Breast Cancer Res Treat. 2012;135:381–390. doi: 10.1007/s10549-012-2155-9. [DOI] [PubMed] [Google Scholar]
- 22.Salem O, Erdem N, Jung J, Münstermann E, Wörner A, Wilhelm H, Wiemann S, Körner C. The highly expressed 5′isomiR of hsa-miR-140-3p contributes to the tumor-suppressive effects of miR-140 by reducing breast cancer proliferation and migration. BMC Genomics. 2016;17:566. doi: 10.1186/s12864-016-2869-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karagoz K, Lehman HL, Stairs DB, Sinha R, Arga KY. Proteomic and metabolic signatures of esophageal squamous cell carcinoma. Curr Cancer Drug Targets. 2016 doi: 10.2174/1568009616666160203113721. [DOI] [PubMed] [Google Scholar]
- 24.Wei CY, Wang HP, Zhu ZY, Sun YH. Transcriptional factors smad1 and smad9 act redundantly to mediate zebrafish ventral specification downstream of smad5. J Biol Chem. 2014;289:6604–6618. doi: 10.1074/jbc.M114.549758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lánczky A, Nagy Á, Bottai G, Munkácsy G, Szabó A, Santarpia L, Győrffy B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 28.Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, Lin CH, Whang-Peng J, Hsu SL, Chen CH, Huang CY. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA, Jr, Johnson GL, Hirsch FR, Merrick DT, Franklin WA, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167:1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen CH, Thai P, Yoneda K, Adler KB, Yang PC, Wu R. A peptide that inhibits function of Myristoylated Alanine-Rich C Kinase Substrate (MARCKS) reduces lung cancer metastasis. Oncogene. 2014;33:3696–3706. doi: 10.1038/onc.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen CH, Fong LWR, Yu E, Wu R, Trott JF, Weiss RH. Upregulation of MARCKS in kidney cancer and its potential as a therapeutic target. Oncogene. 2017;36:3588–3598. doi: 10.1038/onc.2016.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J, Izumi T, Nunomura K, Satoh S, Watanabe S. MARCKS-like protein, a membrane protein identified for its expression in developing neural retina, plays a role in regulating retinal cell proliferation. Biochem J. 2007;408:51–59. doi: 10.1042/BJ20070826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naber HP, Drabsch Y, Snaar-Jagalska BE, ten Dijke P, van Laar T. Snail and Slug, key regulators of TGF-β-induced EMT, are sufficient for the induction of single-cell invasion. Biochem Biophys Res Commun. 2013;435:58–63. doi: 10.1016/j.bbrc.2013.04.037. [DOI] [PubMed] [Google Scholar]
- 37.Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: Role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene. 2005;24:7443–7454. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 38.Ning J, Liu W, Zhang J, Lang Y, Xu S. Ran GTPase induces EMT and enhances invasion in non-small cell lung cancer cells through activation of PI3K-AKT pathway. Oncol Res. 2013;21:67–72. doi: 10.3727/096504013X13747716581417. [DOI] [PubMed] [Google Scholar]
- 39.Wei R, Xiao Y, Song Y, Yuan H, Luo J, Xu W. FAT4 regulates the EMT and autophagy in colorectal cancer cells in part via the PI3K-AKT signaling axis. J Exp Clin Cancer Res. 2019;38:112. doi: 10.1186/s13046-019-1043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manning BD, Toker A. AKT/PKB signaling: Navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, et al. A subcellular map of the human proteome. Science. 2017;356 doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 42.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 43.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 44.Seyfried TN, Huysentruyt LC. On the origin of cancer metastasis. Crit Rev Oncog. 2013;18:43–73. doi: 10.1615/CritRevOncog.v18.i1-2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65:3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 48.Cavallaro U. N-cadherin as an invasion promoter: A novel target for antitumor therapy? Curr Opin Investig Drugs. 2004;5:1274–1278. [PubMed] [Google Scholar]
- 49.Mendez MG, Kojima S, Goldman RD. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010;24:1838–1851. doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vuoriluoto K, Haugen H, Kiviluoto S, Mpindi JP, Nevo J, Gjerdrum C, Tiron C, Lorens JB, Ivaska J. Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene. 2011;30:1436–1448. doi: 10.1038/onc.2010.509. [DOI] [PubMed] [Google Scholar]
- 51.Kidd ME, Shumaker DK, Ridge KM. The role of vimentin intermediate filaments in the progression of lung cancer. Am J Respir Cell Mol Biol. 2014;50:1–6. doi: 10.1165/rcmb.2013-0314TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medici D, Hay ED, Olsen BR. Snail and Slug promote epithelial-mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3. Mol Biol Cell. 2008;19:4875–4887. doi: 10.1091/mbc.e08-05-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adh Migr. 2015;9:317–324. doi: 10.1080/19336918.2015.1016686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grille SJ, Bellacosa A, Upson J, Klein-Szanto AJ, van Roy F, Lee-Kwon W, Donowitz M, Tsichlis PN, Larue L. The protein kinase Akt induces epithelial mesenchymal transition and promotes enhanced motility and invasiveness of squamous cell carcinoma lines. Cancer Res. 2003;63:2172–2178. [PubMed] [Google Scholar]
- 56.Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, García De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 57.Carpenter RL, Paw I, Dewhirst MW, Lo HW. Akt phosphorylates and activates HSF-1 independent of heat shock, leading to Slug overexpression and epithelial-mesenchymal transition (EMT) of HER2-overexpressing breast cancer cells. Oncogene. 2015;34:546–557. doi: 10.1038/onc.2013.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–7464. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 59.Dalva-Aydemir S, Bajpai R, Martinez M, Adekola KU, Kandela I, Wei C, Singhal S, Koblinski JE, Raje NS, Rosen ST, Shanmugam M. Targeting the metabolic plasticity of multiple myeloma with FDA-approved ritonavir and metformin. Clin Cancer Res. 2015;21:1161–1171. doi: 10.1158/1078-0432.CCR-14-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bajpai R, Matulis SM, Wei C, Nooka AK, Von Hollen HE, Lonial S, Boise LH, Shanmugam M. Targeting glutamine metabolism in multiple myeloma enhances BIM binding to BCL-2 eliciting synthetic lethality to venetoclax. Oncogene. 2016;35:3955–3964. doi: 10.1038/onc.2015.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mishra RK, Wei C, Hresko RC, Bajpai R, Heitmeier M, Matulis SM, Nooka AK, Rosen ST, Hruz PW, Schiltz GE, Shanmugam M. In silico modeling-based identification of glucose transporter 4 (GLUT4)-selective inhibitors for cancer therapy. J Biol Chem. 2015;290:14441–14453. doi: 10.1074/jbc.M114.628826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei C, Bajpai R, Sharma H, Heitmeier M, Jain AD, Matulis SM, Nooka AK, Mishra RK, Hruz PW, Schiltz GE, Shanmugam M. Development of GLUT4-selective antagonists for multiple myeloma therapy. Eur J Med Chem. 2017;139:573–586. doi: 10.1016/j.ejmech.2017.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei C, Heitmeier M, Hruz PW, Shanmugam M. Evaluating the efficacy of GLUT inhibitors using a seahorse extracellular flux analyzer. Methods Mol Biol. 2018;1713:69–75. doi: 10.1007/978-1-4939-7507-5_6. [DOI] [PubMed] [Google Scholar]
- 64.Li Y, Martin LD, Spizz G, Adler KB. MARCKS protein is a key molecule regulating mucin secretion by human airway epithelial cells in vitro. J Biol Chem. 2001;276:40982–40990. doi: 10.1074/jbc.M105614200. [DOI] [PubMed] [Google Scholar]
- 65.Singer M, Martin LD, Vargaftig BB, Park J, Gruber AD, Li Y, Adler KB. A MARCKS-related peptide blocks mucus hypersecretion in a mouse model of asthma. Nat Med. 2004;10:193–196. doi: 10.1038/nm983. [DOI] [PubMed] [Google Scholar]
- 66.Ott LE, Sung EJ, Melvin AT, Sheats MK, Haugh JM, Adler KB, Jones SL. Fibroblast migration is regulated by myristoylated alanine-rich C-kinase substrate (MARCKS) protein. PLoS One. 2013;8:e66512. doi: 10.1371/journal.pone.0066512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Geng F, Zhu W, Anderson RA, Leber B, Andrews DW. Multiple post-translational modifications regulate E-cadherin transport during apoptosis. J Cell Sci. 2012;125:2615–2625. doi: 10.1242/jcs.096735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wahl JK, III, Kim YJ, Cullen JM, Johnson KR, Wheelock MJ. N-cadherin-catenin complexes form prior to cleavage of the proregion and transport to the plasma membrane. J Biol Chem. 2003;278:17269–17276. doi: 10.1074/jbc.M211452200. [DOI] [PubMed] [Google Scholar]
- 69.Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The eighth edition lung cancer stage classification. Chest. 2017;151:193–203. doi: 10.1016/j.chest.2016.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.