Abstract
Identifying the genetic mechanisms of adaptation requires the elucidation of links between the evolution of DNA sequence, phenotype, and fitness1. Convergent evolution can be used as a guide to identify candidate mutations that underlie adaptive traits2–4, and new genome editing technology is facilitating functional validation of these mutations in whole organisms1,5. We combined these approaches to study a classic case of convergence in insects from six orders, including the monarch butterfly (Danaus plexippus), that have independently evolved to colonize plants that produce cardiac glycoside toxins6–11. Many of these insects evolved parallel amino acid substitutions in the α-subunit (ATPα) of the sodium pump (Na+/K+-ATPase)7–11, the physiological target of cardiac glycosides12. Here we describe mutational paths involving three repeatedly changing amino acid sites (111, 119 and 122) in ATPα that are associated with cardiac glycoside specialization13,14. We then performed CRISPR–Cas9 base editing on the native Atpα gene in Drosophila melanogaster flies and retraced the mutational path taken across the monarch lineage11,15. We show in vivo, in vitro and in silico that the path conferred resistance and target-site insensitivity to cardiac glycosides16, culminating in triple mutant ‘monarch flies’ that were as insensitive to cardiac glycosides as monarch butterflies. ‘Monarch flies’ retained small amounts of cardiac glycosides through metamorphosis, a trait that has been optimized in monarch butterflies to deter predators17–19. The order in which the substitutions evolved was explained by amelioration of antagonistic pleiotropy through epistasis13,14,20–22. Our study illuminates how the monarch butterfly evolved resistance to a class of plant toxins, eventually becoming unpalatable, and changing the nature of species interactions within ecological communities2,6–11,15,17–19.
Convergently evolved substitutions in ATPα have been hypothesized to contribute to cardiac glycoside resistance in the monarch butterfly and other specialized insects via target-site insensitivity (TSI) in the sodium pump6–11. However, it is unclear whether the changes are sufficient for resistance in whole organisms6–11,15,18,23 or are ‘molecular spandrels’—candidate adaptive alleles that do not confer a fitness advantage when tested more rigorously1,5. In addition, the evolutionary order of substitutions suggests a constrained adaptive walk11,13,14,20–22,24, but an in vivo genetic dissection has not been conducted, so it is not possible to draw conclusions about the adaptive role of these substitutions1–5,15.
We have identified a core set of amino acid substitutions in cardiac glycoside-specialized insects that define potential mutational paths to resistance and TSI. We focused on the first extracellular loop (H1–H2) of ATPα, where most candidate TSI-conferring substitutions occur7–11 (Fig. 1a). We used maximum likelihood to reconstruct ancestral states for cardiac glycoside specialization (feeding and sequestering) and amino acids within the H1–H2 loop of ATPα across a species phylogeny (Fig. 1b, Methods, Supplementary Text, Supplementary Tables 1–3). Sites 111 and 122 underwent frequent parallel substitutions associated with specialization7–11 (Fig. 1b, Extended Data Fig. 1). In addition, site 119 experienced repeated substitutions in specialized insects, and co-evolved with site 111. However, substitutions at site 119 were not associated with specialization, because they also occurred in non-specialized insects (Fig. 1b, Extended Data Fig. 1). To determine whether substitutions at sites 111 or 122 appeared in an ordered fashion relative to substitutions at site 119, we compared the mutational order in 21 specialized lineages to a random permutation null model (Fig. 1c), and found that the ordering was unlikely to have occurred by chance (Fig. 1d). A replacement at site 119 always occurred before or with a replacement at site 122, and repeated substitutions at the three sites evolved concurrently with specialization (Fig. 1b).
Fig. 1. Mutational paths in ATPα associated with insect specialization on cardiac glycoside-producing plants are constrained.
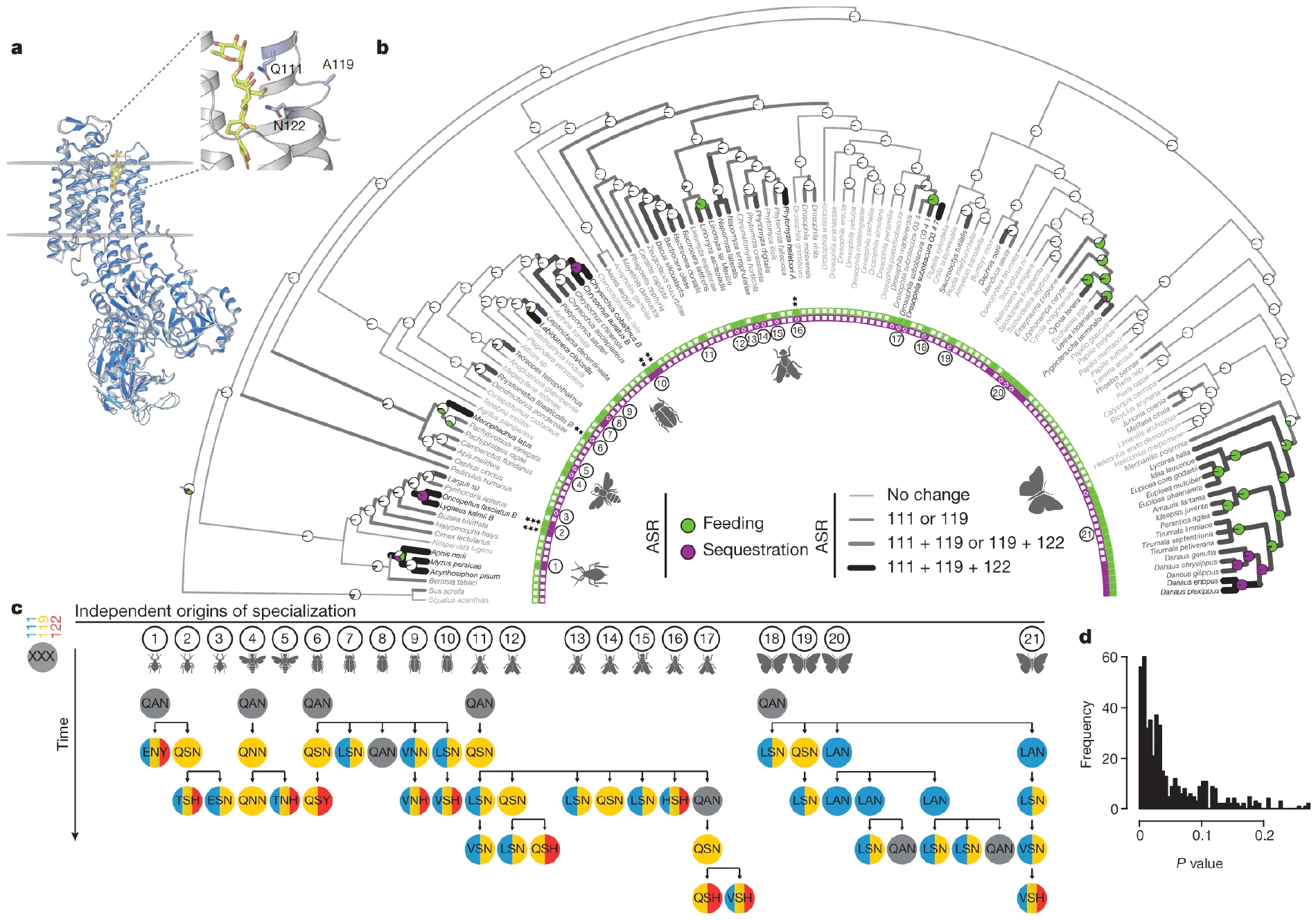
a, Protein homology model of Drosophila melanogaster ATPα (navy) superimposed on a Sus scrofa ATPα crystal structure (light grey) with ouabain (yellow) in the binding pocket. Residues 111, 119 and 122 (sticks) within the H1–H2 extracellular loop are associated with feeding on cardiac glycoside-producing plants and toxin sequestration. b, Maximum likelihood phylogeny based on 4,890 bp from Atpα and coi, with maximum likelihood ancestral state reconstruction (ASR) of feeding and sequestering states, estimated from the states of extant species (inner band of squares). Reconstructions are shown as nodal pie graphs (white, neither feeding nor sequestering; green, feeding; purple, feeding and sequestering), and the number of substituted sites at positions 111, 119 and 122 along branches in grey-scale (light grey 0, medium grey 1, dark grey 2, black 3), based on maximum likelihood ASR of H1–H2 loop amino acid sequences. Black asterisks indicate the Atpα copy number for species with multiple paralogues. c, ATPα substitutions inferred from ASR at positions 111 (blue), 119 (yellow) and 122 (red) in 21 lineages where specialization occurred independently. d, P value distribution from a set of randomized tests to determine the reproducibility of substitutions observed along mutational paths among sub-sampled groups compared to randomly permuted substitutions. On average, 4.9% (considering all mutational steps) of randomly permuted trajectories demonstrate a degree of ordering equal to or greater than observed mutational paths.
The mutational paths lead to three predictions for how substitutions at sites 111, 119 and 122 affect fitness. First, the mutational paths provide stepwise fitness advantages at increasing toxin concentrations. Second, the mutational paths contribute to sequestration of cardiac glycosides through passive toxin accumulation. Third, given the ordered appearance of the substitutions, interactions between substitutions (epistasis) increase fitness and mitigate the pleiotropic fitness costs of adaptive substitutions. We focused on the mutational path taken by the monarch lineage, which includes species that do not feed on cardiac glycoside-producing plants and those that sequester the toxins11 (Fig. 2a).
Fig. 2. Drosophila flies with edited genomes retrace the mutational path of ATPα in the monarch butterfly lineage and show increased survival when fed cardiac glycosides.
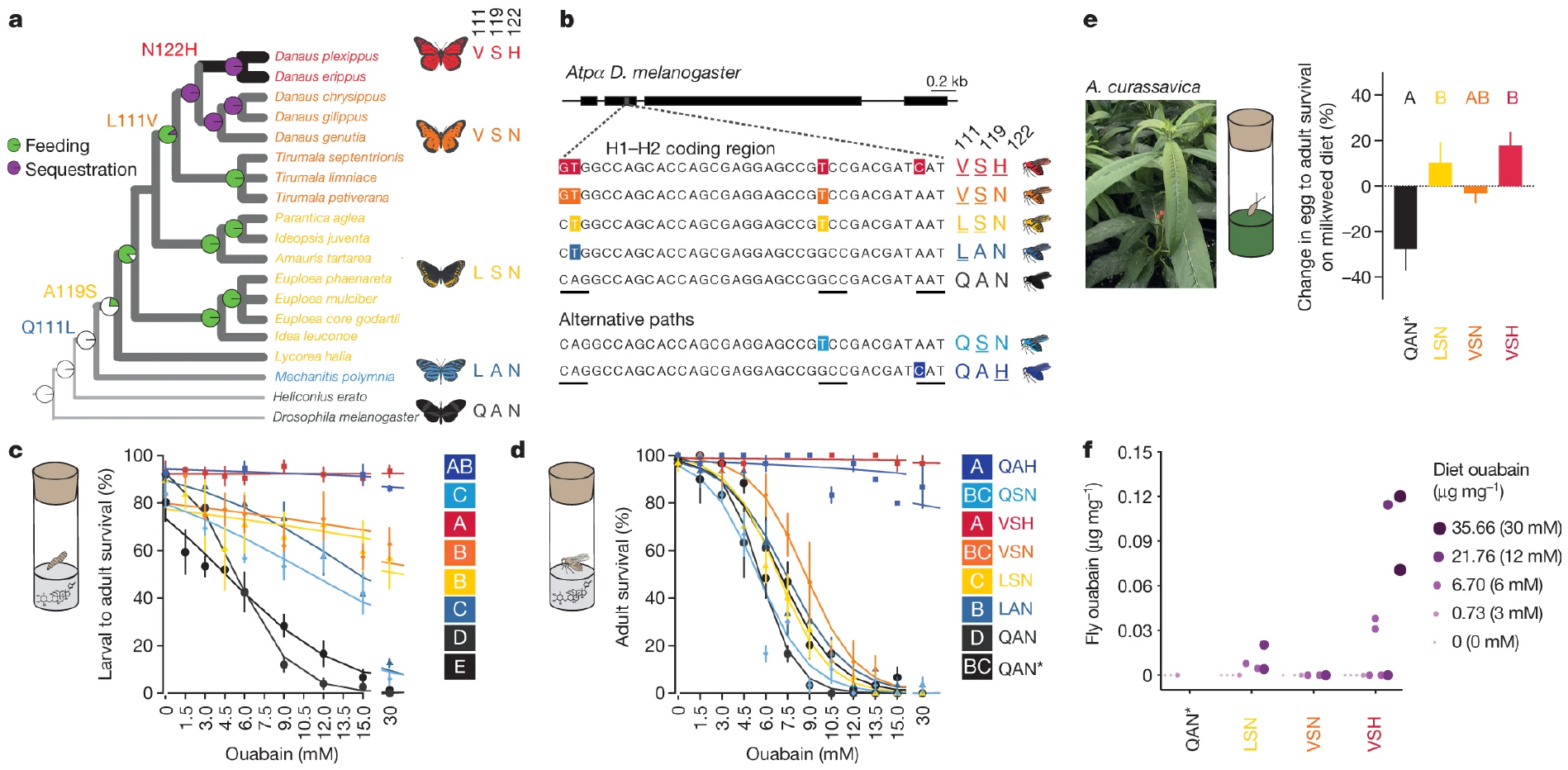
a, The monarch butterfly lineage ratio test on the significance of the interaction term between genotype (line) and ouabain concentration in a logistic regression for each pair of lines (letters). e, Egg–adult survival on diet supplemented with Asclepias with the substitutions observed in the H1–H2 loop of ATPα (adapted from Petschenka et al.)11. b, Non-synonymous point mutations in the edited DNA sequence of the native Atpα in Drosophila knock-in lines code for the substitutions at sites 111, 119 and 122. Codons are underlined. c, d, Larval–adult survival (c) and adult survival (d) of flies reared on diets with ouabain were different between monarch lineage knock-in lines and control lines (QAN = engineered control; QAN* = w1118 wild type). Symbols represent the mean ± s.e.m. of 3–6 biological replicates (50 larvae and 10 females per replicate in c and d, respectively). Curves were fit using a logistic regression model for each line. Pairwise differences in survivorship trajectories between lines were evaluated with a likelihood curassavica leaves relative to control diets (n = 3–4; 100–200 eggs per replicate, see Methods; mean ± s.e.m.) was different between monarch lineage knock-in lines and QAN* (one-way ANOVA, P = 0.0035 followed by post hoc Tukey’s tests (letters)). f, Ouabain concentrations in diet versus adult fly bodies among monarch lineage knock-in lines (n = 2–4 biological replicates per group). Adult flies had not fed since eclosion. Genotype and dietary ouabain concentration influenced the probability of detecting ouabain in post-eclosion flies (logistic regression and likelihood ratio test, genotype two-sided P = 0.024, dietary ouabain concentration two-sided P = 6.344 × 10−5). Further information on experimental design and statistical test results is found in the Source Data.
We used CRISPR–Cas9 genome editing coupled with homology-directed repair (HDR) to generate viable, homozygous Atpα knock-in Drosophila lines carrying the precise substitutions at sites 111, 119 and 122 of 4 consecutive Atpα genotypes in the monarch lineage: LAN, LSN, VSN, and VSH11,25,26 (substitutions Q111L, A119S, L111V, and N122H, respectively; Fig. 2b, Methods and Supplementary Tables 4–7). We also generated control lines in the same genetic background, and viable, homozygous lines carrying the key single substitutions A119S (QSN) and N122H (QAH), which evolved along the monarch lineage mutational path, but not alone (Fig. 2b, Extended Data Fig. 2). These lines did not carry exogenous DNA sequences or other non-synonymous mutations in the edited region (Supplementary Table 6). We found substantial variation for egg–adult survival among the lines (Extended Data Fig. 3), but Atpα mRNA levels and baseline sodium pump activity were not detectably altered (Extended Data Fig. 3, Supplementary Table 8). A congener of Drosophila melanogaster, D. subobscura, is naturally segregating at positions 111, 119 and 122 for the monarch butterfly genotype (VSH) and was reared from cardiac glycoside-producing plants, indicating that Drosophila is a reasonable model for studying cardiac glycoside resistance and TSI27,28.
We obtained in vivo evidence for adaptation in monarch lineage Atpα through larval–adult and adult survival experiments. Knock-in fly lines were reared on yeast medium with increasing concentrations of ouabain, a hydrophilic cardiac glycoside6 (Fig. 2c, d, Extended Data Figs. 4, 5). LAN, the first genotype to evolve, increased larval–adult survival at lower ouabain concentrations, but survival declined sharply as concentrations increased. LAN also increased adult survival at lower ouabain concentrations. LSN, the second genotype to evolve, increased larval–adult survival at the highest ouabain concentrations. The next step, VSN, provided the same larval–adult and adult survival benefit as LSN. Finally, the survival of ‘monarch flies’ carrying the monarch butterfly genotype (VSH) was unaffected by even the highest levels of ouabain in larvae and adults6,9,11,18 (Fig. 2c, d), which was not due to reductions in feeding rate or toxin ingestion (Extended Data Fig. 6).
When knock-in line eggs were placed on medium containing the suite of cardiac glycosides found in the leaves of the milkweed species Asclepias curassavica and A. fascicularis6, monarch lineage fly genotypes generally showed increased egg–pupal and egg–adult survival rates (Fig. 2e, Extended Data Fig. 7), although not always for VSN (Extended Data Figs. 3, 7). The LSN, VSN and VSH genotypes may enable insects to cope with the complex milieu of cardiac glycosides encountered during host shifts to these plants.
The monarch butterfly ATPα substitutions at positions 111, 119 and 122 may unlock a passive evolutionary route to cardiac glycoside sequestration, as we found small amounts of ouabain in newly emerged adult ‘monarch flies’ reared as larvae on a diet containing ouabain (Fig. 2f). However, toxin concentrations were far lower than in monarch butterflies, and the location of ouabain in flies is unclear6,17,18.
At the physiological level, sodium pump enzymatic assays using head extracts from knock-in fly lines showed that each sequential monarch lineage genotype had a neutral-to-positive effect on TSI to ouabain (Fig. 3a). LAN provided a small increase in TSI, while the next genotypes to evolve, LSN and VSN, increased TSI by about ten times (Fig. 3a). Remarkably, TSI rose about a thousand times in mutant flies carrying VSH, the monarch butterfly genotype. ‘Monarch fly’ sodium pumps were as insensitive to ouabain as those of monarch butterflies (Fig. 3a). Sodium pump enzymatic assays using extracts of Spodoptera frugiperda Sf9 cells transiently expressing Drosophila ATPα proteins with key monarch lineage mutations at sites 111, 119, and 122 mirrored these patterns24 (Extended Data Fig. 8). VSH is sufficient for ‘monarch flies’ to achieve the same degree of TSI to ouabain as the monarch butterfly butterfly, suggesting that TSI is the predominant biological mechanism for the in vivo toxin resistance observed above.
Fig. 3. The mutational path of ATPα in the monarch butterfly lineage sequentially increases TSI to ouabain and is shaped by epistasis.
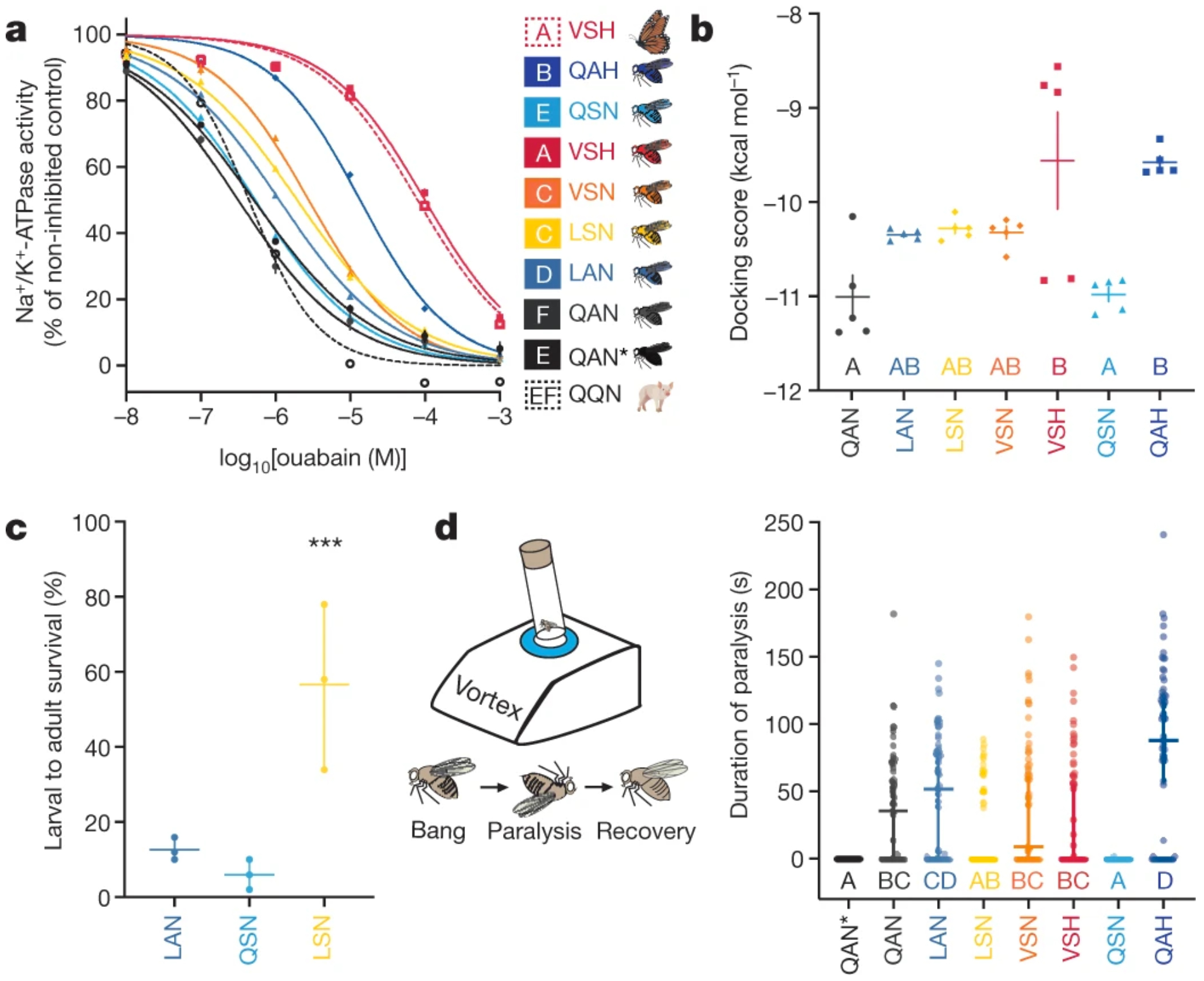
a, In vitro ouabain sensitivity of Na+/K+-ATPase activity in extracts of monarch lineage knock-in and control line fly heads (solid lines; QAN, engineered control; QAN*, w1118 wild type), against activity in extracts of monarch butterfly and pig nervous tissue (positive and negative control, dashed red and black line, respectively). Symbols represent the mean ± s.e.m. of 3–7 biological replicates. log10[IC50] (half-maximum inhibitory concentration) values for the Na+/K+-ATPases were estimated after fitting four-parameter logistic regression curves, and were different between genotypes (one-way ANOVA (P < 0.0001) with post hoc Tukey’s tests (letters)). b, Mean docking scores (± s.e.m. of five replicate calculations) from molecular simulations of ouabain binding to the Na+/K+-ATPases found along the monarch lineage showed differences between genotypes (one-way ANOVA (P = 0.0001) with post hoc Tukey’s tests (letters)). c, Effects of the substitutions Q111L, A119S and their combination on larval–adult survival on diets with 30 mM ouabain. Symbols represent the mean ± s.e.m. of three biological replicates (50 larvae each). The effect of mutations A119S and Q111L together was nearly threefold greater than the combined individual effects on survivorship (logistic regression, interaction effect between mutations: ***P = 2.36 × 10−15), indicating positive epistasis. d, Duration of paralysis following mechanical shocks (that is, bang sensitivity; n = 60 five-to-six-day-old adult flies). Bang sensitivity was affected by genotype (Kruskal–Wallis test (P < 0.0001) with post hoc Dunn’s multiple comparisons tests (letters); medians with 95% confidence intervals), and was higher for QAH than for all other genotypes (P < 0.05), except for LAN, which showed higher bang sensitivity than LSN (P = 0.0134). Further information on experimental design and statistical test results can be found in the Source Data.
Our results provide a starting point for identifying a biochemical mechanism for TSI through molecular docking simulations of ouabain bound to homology-modelled Drosophila Na+/K+-ATPase. In keeping with in vivo and enzymatic results (Figs. 2c–f, 3a), docking scores predicted that the first three Atpα genotypes to evolve resulted in slight increases in TSI to ouabain, followed by a larger increase for VSH, consistent with ouabain binding affinity measurements (Fig. 3b, Extended Data Fig. 9). Elucidation of the full biochemical mechanism for TSI will require additional research.
Site 119 co-evolves with site 111 (Extended Data Fig. 1c), and substitutions at site 119 always occurred before or with TSI-conferring substitutions at site 122 (Fig. 1c), suggesting that antagonistic pleiotropy and epistasis may have shaped mutational paths to resistance and TSI. We further addressed this hypothesis using knock-in lines for N122H (QAH), which is often the last substitution to evolve, and A119S (QSN), a substitution found in both specialists and non-specialists (Fig. 1b, c). QSN did not increase larval–adult survival at lower ouabain concentrations, but provided a survival benefit as ouabain concentrations rose, before survival dropped sharply at the highest concentration, as in LAN (Fig. 2c). In adults, QSN provided a slight survival benefit against ouabain (Fig. 2d), and low TSI to ouabain in sodium pump enzymatic assays (Fig. 3a). At lower ouabain concentrations, LSN provided an increase in larval–adult survival similar to that of LAN, but survival increased by about 40% over LAN and QSN at the highest concentration (Fig. 3c). This points to a role for synergistic, beneficial epistasis in dietary ouabain resistance between Q111L and A119S. QAH provided nearly as much survival benefit as VSH in larval–adult and adult feeding experiments (Fig. 2c, d), and in sodium pump enzymatic assays, TSI to ouabain in QAH was second only to VSH (Fig. 3a). Finally, the binding affinity of ouabain to Na+/K+-ATPase is high for QSN, and low for QAH, according to docking scores (Fig. 3b, Extended Data Fig. 9). N122H conferred the highest TSI of any substitution, yet appeared last in the adaptive walk, and was contingent on a substitution at site 119 (Fig. 1c, d), which suggests that N122H imposes high fitness costs that are mitigated by A119S. To investigate this, we phenotyped monarch lineage knock-in flies for neurological seizures upon shaking (bang sensitivity), a common phenotype in hypomorphic Na+/K+-ATPase mutants29,30. Bang sensitivity varied widely within and among knock-in fly lines; this variation could be due to the intentionally introduced mutations or to unidentified (epi)mutations that arose from base editing and resulted in cryptic decanalization effects on the function of the nervous system. Despite this uncertainty, QSN flies were the least bang sensitive, and QAH flies were the most (Fig. 3d). Furthermore, the first step in the adaptive walk, LAN, resulted in higher bang sensitivity than the second step, LSN. Some costs of Q111L are therefore mitigated by A119S. When N122H was added to VSN, resulting in VSH (the monarch butterfly genotype), the cost of N122H was also reduced, in part, by A119S (Fig. 3d). A119S ameliorates the pleiotropic costs of resistance substitutions at sites 111 and 122 throughout the adaptive walk.
Substitutions at three amino acid sites in ATPα are sufficient together, but not alone, to explain the evolution of resistance and TSI to cardiac glycosides achieved by the monarch butterfly at organismal, physiological and biochemical levels. The adaptive walk follows theoretical predictions on the length of such walks2–4,13,14, involves epistasis13,14,20,22, and minimizes pleiotropic fitness costs3,4,13,14,21, and variations of it convergently re-appeared across lineages that diverged more than three hundred million years ago7–11. Genome editing technology facilitates functional tests of adaptation across levels of biological organization5,25,26. Although mutational paths to adaptive peaks have been identified in microorganisms2–4,13,14,22, this is, to our knowledge, the first in vivo validation of a multi-step adaptive walk in a multicellular organism, and illustrates how complex organismal traits can evolve by following simple rules.
Methods
Data reporting.
We performed preliminary experiments to assess variance and estimate sample sizes before conducting all bioassays. We used the online tool random.org (https://www.random.org/sequences/) to randomize all bioassays, except for the feeding experiments with milkweed and the sequestration assay. We were blind to the allocation of individuals to treatment groups. We were also blind to the treatment groups when taking measurements during all bioassays, except for the feeding experiments with milkweed and the sequestration assay.
Species phylogeny and ancestral state reconstructions.
To identify amino acid changes associated with transitions to specialization on cardiac glycoside-producing host plants and cardiac glycoside sequestration, we first constructed a species maximum likelihood (ML) phylogeny. Phylogenetic relationships between all surveyed species were taken from previous studies (see Supplementary Table 1 for references). The phylogeny included insects adapted to feeding on such plants, their non-adapted relatives, and two vertebrate outgroup species. The species sampling method is described in the Supplementary Text. Because we constructed this supertree from disparate studies that used overlapping but non-identical molecular markers, we estimated branch lengths under this fixed topology using the full-length coding sequences of Atpα and the widely used gene coding for the mitochondrial cytochrome c oxidase subunit 1 (coi) for a combined total of 4,890 bp of sequence (see Supplementary Table 2 for ID numbers, source databases and references). Branch length estimation was implemented in IQ-TREE version 1.6.631. The amino acid residues were numbered after the mature Sus scrofa protein residue numbering as in previous phylogenetic studies of Atpα in insects7–11,24. For each of the species with Atpα duplications, we included only the paralogue that is mostly expressed in the insect’s body rather than copies that show expression bias towards the nervous tissue, so as to be conservative in downstream analyses10. Our reasoning is that the body-expressed copies experience stronger selective pressure from exposure to dietary cardiac glycosides than nervous tissue-expressed copies because the nervous tissue is protected from cardiac glycosides by the haemolymph–brain barrier (perineurium)6,23,32. Thus, we focused on the origin of substitutions that fixed as a result of interactions with cardiac glycosides, rather than adaptation to the nervous system after release from this agent of selection.
We performed ASR of feeding on cardiac glycoside-producing plants and cardiac glycoside sequestration by ML using the ace function of the ape package in R33. We also performed ASR of amino acid sequences of the H1–H2 extracellular loop using joint ML methods34, implemented in HyPhy version 2.3.1435. Amino acid sites that were (1) identified as coevolving (see description of Bayesian graphical model (BGM) methods below), (2) significantly associated with cardiac glycoside feeding and sequestration (see TraitRateProp analysis below), and (3) highly variable and evolving convergently (see section Reproducibility of mutational order)—sites 111, 119, and 122—were chosen as the focus of this study. Using the ASR of amino acid sequences, the number of substitutions at these three sites was mapped onto the species tree, along with ancestral states of cardiac glycoside feeding and sequestration. The tree was visualized using the Interactive Tree Of Life (iTOL)36, and can be found here: https://itol.embl.de/tree/1361521431149621561136381.
Using the joint likelihood framework of TraitRateProp37, we identified amino acid residues within the H1–H2 loop of ATPα for which the rate of sequence evolution was associated with changes in the cardiac glycoside feeding and sequestering character state described above. More details of this analysis can be found in the Supplementary Text.
To detect epistatic or co-evolving interactions between amino acid sites within the H1–H2 loop that might constrain or enable the evolution of ATPα, we used the Spidermonkey/BGM method38 in HyPhy version 2.3.14. Sites in this analysis were filtered with a default minimum number of five substitutions across the phylogeny.
Interactions between sites with a marginal posterior probability (PP) > 0.5 were considered biologically meaningful.
Convergent substitutions in the amino acid residues of the H1–H2 loop of Atpα.
We aimed to identify the most variable and convergently evolving amino acid residues within the H1–H2 loop in extant insects of 21 lineages in which specialization on cardiac glycoside-producing host plants and toxin sequestration evolved independently according to ASR analyses. At each position in the H1–H2 loop, we quantified the number of times a particular substitution occurred across these independent lineages (for example, the number of times Q111L occurred). We used the results from the ASR for feeding on cardiac glycoside-producing plants and sequestration to define these 21 independent lineages that evolved these traits, and we used the ASR for the H1–H2 loop to identify independent amino acid changes along these lineages (see above). We used the following set of rules to estimate the number of substitution occurrences across these lineages: (1) we considered substitutions only in extant species that were different from the ancestral sequence of all insect orders, (2) ancestrally shared substitutions in the H1–H2 loop were counted once to avoid overrepresentation, and (3) we did not score for reversion substitutions to the ancestral state. For further information on the estimation of independent substitution occurrences see the Source Data for Extended Data Fig. 1.
Reproducibility of mutational order.
We identified amino acid residues 111, 119 and 122 of the H1–H2 loop of cardiac glycoside-adapted insects as the most variable and convergently evolving (to the same amino acid states). Next, we used a statistical method that compared the observed evolutionary pattern of mutations at ATPα positions 111, 119 and 122 in specialized insects to a random permutation null model to test whether the ordered appearance of these mutations could have occurred by chance14. The reproducibility of mutational order (RMO) score was defined as previously described14. The RMO score of two ordered sequences of ATPα mutations along insect lineages is defined by the number of shared mutation pairs that occurred in the same order, from which we subtract the number of shared mutation pairs that occurred in reverse order—for example, two sequences that experienced mutations among six sites, [1, 2, 3, 4] and [1, 6, 4, 3], would have an RMO score of 1 (two shared mutation pairs occurred in the same order [1 and 3; 1 and 4], but one shared mutation pair occurred in the opposite order [3 and 4]). We used the results from the ASR of feeding and sequestering in conjunction with the ASR results of the H1–H2 loop sequence to define the mutational path in each lineage examined, starting from an ancestor that was estimated to be neither feeding on cardiac glycoside-producing plants nor sequestering the toxins and ending at extant species that feed and/or sequester.
The RMO score for a set of two paths requires two assumptions that are violated in this study: that the exact order in which mutations arose can be discerned, and that each mutational path is independent14. Multiple mutations that co-occurred along the same branch leave the order in which each mutation occurred ambiguous. In addition, different paths can be dependent on one another as mutations arising on internal branches in the phylogeny can appear in multiple different paths towards branch tips that derived from these internal branches. Under this dependency, we find 13 groups of independent paths. To evaluate the probability of seeing these paths by chance, conditional on these two complicating factors, we used a two-step randomization process. First, we randomly sampled one path from each of the 13 groups of paths, for a total of 13 independent paths (see Source Data for Fig. 1d). Second, for any co-occurring mutations, we randomly chose a sequential ordering of them. We then calculated the RMO score for this set of sampled paths. Next, we randomly permuted the ordering of the mutations within each path, and calculated the RMO score of this random order. This random permutation was done 100,000 times to get 100,000 different RMO scores. We calculated the fraction of these RMO scores that were greater than or equal to the RMO score of the 13 paths originally sampled. The resulting P value represents the probability that the observed level of ordering in the 13 paths could have been produced by random chance, with a smaller P value indicating stronger evidence for non-random ordering of the observed mutations. This two-step randomization process was repeated 500 times, to generate a distribution of 500 unique P values. Through this process, we found a mean P value of 0.0495, indicating that on average less than 5% of the random 100,000 permutations induced an ordering of the same magnitude as our observed data. We also observed that 66.6% of the 500 P values generated were lower than 0.05. The heavy tail in large P values (the other 33.3%) can be attributed to the single path in which mutations at sites 111, 119, and 122 all occur on a single branch, and the ordering is left ambiguous. When we randomized the ordering of these three mutations, we found a mean P value of 0.0149 when the mutation at position 119 is more ancient than that at 122 on this specific path. Conversely, when the randomization chooses 122 to be more ancient than 119, we find a much larger mean P value of 0.073.
Generation of knock-in fly lines.
Genomic engineering strategy.
We used a two-step genome editing approach using CRISPR–Cas9 coupled with HDR to introduce the different non-synonymous point mutation combinations of interest into the native Atpα of D. melanogaster in the region coding for the enzyme subunit’s H1–H2 extracellular loop (Extended Data Fig. 2). In the first step, the Atpα region encoding the H1–H2 loop was replaced with a 3×P3::GFP marker through CRISPR–Cas9-mediated HDR. This generated the deletion allele AtpαDeletion (GFP+). This line served as a common stock for the second step, in which the 3×P3::GFP marker was replaced with each of the point mutation alleles in Atpα through an additional round of CRISPR–Cas9-mediated HDR. This generated a first set of knock-in lines with the non-synonymous point mutation alleles AtpαQ111L/A119S (LSN), AtpαQ111V/A119S (VSN), and AtpαQ111V/A119S/N122H (VSH), and later, a second set of knock-in lines with the non-synonymous point mutation alleles AtpαQ111L (LAN), AtpαA119S (QSN), and AtpαN122H (QAH). With this second set, we also generated a CRISPR–Cas9-engineered control line following the same two-step strategy but without introducing any non-synonymous changes (QAN; Supplementary Table 6).
Construction of guide RNAs and donor plasmids.
All guide RNAs (gRNAs) used in the first and second rounds of CRISPR–Cas9-mediated HDR were cloned into the pCFD3-dU6:3gRNA vector26, a gift from S. Bullock (Addgene plasmid 49410). All gRNAs were tested for potential off-target effects using the flyCRISPR website39 (http://flycrispr.molbio.wisc.edu/). No potential off-target sites elsewhere in the genome were predicted, but they cannot be ruled out. generation of the deletion line was created through Golden Gate assembly via BsaI digestion and subsequent ligation of four DNA fragments: a selection cassette containing the 3×P3::GFP marker (GenetiVision), two ~1-kb-sized, PCR-amplified homologous arms (by Phusion polymerase) and a pUC57-Kan vector backbone. Each of the donor plasmids for the generation of the mutant alleles (replacement vectors) was also created through Golden Gate assembly via BsaI digestion and subsequent ligation of four DNA fragments: the donor DNA containing the mutations of interest, two ~1-kb-sized, PCR-amplified homologous arms (by Phusion polymerase) and a pUC57-Kan vector backbone. For the replacement vectors, an initial donor plasmid was built that contained two point mutations at the gRNA PAM sequences to prevent CRISPR–Cas9 retargeting. Because the mutation in the region of the downstream gRNA codes for amino acid, a silent mutation at E176 (GAG to GAA) was included in the design as a control. This initial donor plasmid served as a template for introducing the point mutations in the donor DNAs for all alleles through template extension PCR. The sequences of the gRNAs and recovered Atpα in all homozygous knock-in lines are provided in Supplementary Tables 4 and 6, respectively.
Embryo injection, crosses and mutant screening.
For the generation of the Atpα deletion line, a mixture of two gRNAs (at 50 ng/μl each) and the donor plasmid carrying the 3×P3::GFP marker (at 150 ng/μl) was injected into embryos of BL#54591 carrying the nos-Cas9 gene26. The adult flies that survived microinjection (G0) were crossed with w1118 and G1s were screened for the presence of green fluorescence in the compound eyes. Positive male heterozygous G1s were selected and genotyped by PCR with cassette-specific primers and Sanger sequencing (in both directions) to confirm that the cassette insertion was in the expected location. For this, two sets of PCR primers were used that covered both the left homologous arm, plus its junction upstream of the GFP cassette, and the right arm, plus its junction downstream of the cassette. The positive G1s were then crossed to the double-balanced TM3, Sb/TM6, ebony fly line and screened for the absence of the ebony phenotype. Positive G2 ATPαDeletion (GFP+)/TM3, Sb flies were obtained and the Atpα deletion line was established.
For the generation of each Atpα knock-in line, a mixture of two gRNAs (at 50 ng/μl each) and each donor plasmid carrying the respective point mutation(s) (at 150 ng/μl) was injected into embryos of the Atpα deletion line carrying nos-Cas9. To achieve this, the Atpα deletion line was crossed with BL#54591 again to introduce nos-Cas9 into the X chromosome of the deletion line. The injected G0 flies were crossed with the double-balanced TM3, Sb/TM6, ebony fly line and screened for the absence of green fluorescence in the compound eyes. Positive male heterozygous G1s were selected, genotyped by PCR with primers covering the region with the knock-in mutations, and balanced with TM3, Sb. Positive heterozygous G2 flies were intercrossed and G3 homozygous flies were obtained carrying the desired knock-in mutations. These flies were genotyped with primers flanking the area affected by the genome engineering process at both steps of HDR. The overall crossing scheme for the generation of the Atpα knock-in lines is presented in Extended Data Fig. 2.
The sequences of all primers used for screening and validating the mutant lines are provided in Supplementary Table 5. The validated sequences of the knock-in and control lines are provided in Supplementary Table 6. The number of embryos injected, the number of independent lines established, and the ability of mutant lines to give rise to homozygous progeny are provided in Supplementary Table 7. GenetiVision performed all injections as well as the initial mutant screening.
Egg lethality assay.
To compare egg–adult survival among the knock-in lines and the w1118 (QAN*) line, we introduced 100 eggs of each line into narrow vials containing ~10 ml of 22% Instant Drosophila Medium in Millipore water (w/v). Each of the groups was tested in 7 or 8 replicate vials. Pupariation, adult eclosion, and survival were monitored over a period of ~21 days until all adult flies had eclosed. Real-time quantitative PCR. Total RNA was extracted from five- or six-day-old females using the ReliaPrep RNA Tissue MiniPrep System including the DNase treatment step. From this, 900 ng total RNA was converted to cDNA using the ProtoScript II cDNA synthesis kit (NEB). Real-time quantitative PCR (qPCR) reactions were run on the StepOne Real-Time PCR System (ThermoFisher Scientific). Reaction volumes were as follows: 10 μl 2× DyNAmo HS SYBR Green qPCR Kit, 0.15 μl ROX Passive Reference Dye, 0.5 μl 60 μM forward and reverse primers, and 20 ng cDNA to a total reaction volume of 20 μl. qPCR was performed for Atpα using rpl32 as an internal control. The primer sequences are provided in Supplementary Table 8. The rpl32 primer sequences were as previously reported40. All runs included an initial 10-min denaturation step at 95 °C, followed by 40 cycles of 95 °C for 15 s and 58 °C for 1 min. The runs were finished with a melt curve ramp from 60 °C to 95 °C during which data were collected every +3 °C. The expression of Atpα for each genotype was assayed in two or three biological replicates and two technical replicates.
In vitro sodium pump assays using extracts of heads and nervous tissue.
Sodium pump preparations.
Sodium pump preparations were performed as described11. For each genotype, 100 frozen flies (50 females and 50 males that were eight days old) were decapitated under a dissecting microscope, on dry ice, using a clean scalpel. Heads were then suspended in 2.2 ml Millipore water, homogenized (Wheaton glass grinder) to yield a single stock preparation, and pipetted into 250-μl aliquots (each aliquot contained the equivalent of 12.5 fly heads). Similarly, a pooled stock of monarch butterfly (Danaus plexippus) nervous tissue homogenate was prepared from adult butterflies that had been frozen alive at −80 °C and thawed on ice for dissection. In brief, 16 butterfly brains (with thoracic ganglia) were homogenized as above, pooled in 8 ml Millipore water, and pipetted into 375-μl aliquots (each aliquot contained the equivalent of three-quarters of a brain). All aliquots were frozen at −80 °C, freeze-dried overnight, and then stored at −80 °C until use. Purified porcine sodium pump (A7510, Sigma) was stored at −80 °C, at a concentration of 1 unit/ml in Millipore water, in 50-μl aliquots.
Monarch butterfly and fly sodium pump preparations were thawed on ice, resuspended in cold Millipore water (fly preparations: 425 μl, monarch butterfly preparation: 2 ml; this equates to 12.5 fly heads per 425 μl and 75 butterfly heads per 2 ml), vortexed, sonicated (ultra-sonic bath), and centrifuged for 5 min at 5,000 r.p.m. to remove pelleted material. Porcine sodium pump preparation was thawed on ice and diluted with deionized water to yield a concentration of 0.05 units/ml. All enzyme preparations were kept on ice during experiments.
Plate assays.
For each enzyme type, we analysed activity against a serial dilution of ouabain (ouabain octahydrate, O3125, Sigma) ranging from 10−3 M to 10−8 M, in 10% DMSO. Three or four technical replicates of each fly preparation were assayed across three microplates, such that each plate included at least one replicate of each fly line, as well as monarch and pig sodium pump. Reactions were performed as described11, except for the following change: instead of pre-incubation without ATP, we prepared a ‘mastermix’ for each enzyme type, on ice. This mastermix contained all reagents except for the inhibitor, and was dispensed directly onto the ouabain solution in the microplate just before incubation. After 20 min, reactions were stopped and inorganic phosphate was stained.
Curve fitting and estimation of IC50.
The absorbance values of reactions were corrected for the background value (full inhibition) for each enzyme type and calculated as per cent residual activity using reference points of the non-inhibited reaction as 100% and the completely inhibited reaction as 0% residual activity. The results for each enzyme type were fitted to the four-parameter logistic equation log(inhibitor) versus response–variable slope: Y = Bottom + (Top – Bottom)/1 + 10((log10[IC50]−X)×Hillslope) where Y is the percentage of non-inhibited control; Bottom is the percentage of non-inhibited control (constrained to 0 as 0% residual activity of the reaction is expected in high doses of ouabain); Top is the percentage of non-inhibited control (constrained to 100 as 100% residual activity of reaction is expected in the absence of ouabain); IC50 is the concentration of ouabain at which 50% of sodium pump activity is inhibited; X is the concentration of ouabain; and Hillslope is the steepness of the curve.
Finally, to calculate the absolute activity of the Na+/K+-ATPase, a phosphate calibration curve ranging from 0.2 to 1.2 mM KH2PO4 was run on each plate, ouabain inhibition of the samples was analysed, and phosphate released (nmol/μl) was determined at 700 nm according to a previously described photometric method41. The activity of the Na+/K+-ATPase of each sample can be determined as the difference in the amount of phosphate released between the non-inhibited control (all ATPases active) and a reaction in which only the contaminating ATPases are active (a buffer system without KCl42). As the reactions were performed per fly head, the final Na+/K+-ATPase activity is expressed as nmol ATP hydrolysed per fly head per min.
Feeding experiments with knock-in fly lines.
We performed all feeding experiments in a growth chamber at ~21–23 °C and ~50–60% humidity, on a 12 h:12 h light:dark cycle. In all feeding experiments, vials for independent trials were coded and placed in a randomized order in rows on cardboard trays.
Feeding experiment with knock-in adults and larvae reared on medium containing ouabain.
For the immatures, we introduced fifty second-instar larvae in wide vials containing 10 ml of 27.5% Ward’s Instant Drosophila Medium (470024–740, Ward’s Science) in Millipore water (w/v) supplemented with a series of ouabain concentrations. Ouabain (≥95% purity) was obtained from Sigma-Aldrich (11018-89-6). We used second-instar larvae for the experiment because of the occurrence of variably penetrant embryonic lethality in the knock-in lines (Extended Data Fig. 3a). We selected the Instant Drosophila Medium diet because our knock-in lines and the w1118 flies (QAN*) performed well on it. We assessed larval performance for all genotypes on diets that ranged in ouabain concentration from 0 mM to 30 mM. A concentration higher than 30 mM could not be prepared because ouabain crystallized in water above this concentration. Each of the treatment and control groups was tested in triplicate vials. Pupariation, adult eclosion, and survival were monitored over a period of ~21 days until all adult flies had eclosed.
For the adults, we introduced ten females (4–7 days old) in wide vials contain-ing 5 ml of 5% sucrose/1% agar in Millipore water supplemented with a series of ouabain concentrations. We performed experiments with females because they have been reported to be more sensitive to ouabain than males when ouabain is ingested orally at (near-)millimolar concentrations23,43. We reared flies on medium with ouabain concentrations ranging from 0 to 30 mM for all genotypes. Each treatment and control group was tested in triplicate vials. Survival of females was monitored over a 3–6-day period in each vial.
CAFE assays with knock-in adults.
We measured the effects of different dietary ouabain concentrations on fly survival and food consumption rate using the CApillary FEeder (CAFE) assay as described23, with small modifications. We also estimated the lethal ouabain dose necessary to kill 50% of the flies in each treatment group (LD50). We introduced three females (four to seven days old) in each vial. Flies were supplied daily with two capillary tubes (5-μl calibrated pipets, 53432–706, VWR) containing 5% sucrose in Millipore water (w/v) supplemented with concentrations of ouabain that ranged from 0 to 30 mM for all genotypes. The daily replacement of the capillaries ensured the ad libitum availability of food. Each genotype was tested with five replicate vials for all treatment and control groups. The experiment lasted for seven days. Consumption was measured as described44.
To estimate LD50 we measured the cumulative amount of food consumed (in μl per fly) over the average number of days it took for the second out of three flies to die in each replicate vial. The average number of days was rounded to the nearest whole number of days. As no more than half of the flies died in some of the treatment groups, we proportionally extrapolated the amount of food it took to cause the proportion of deaths we observed in such a treatment group to the amount of food it would then take to cause the death of half of the flies in that treatment group. The daily consumption rate per fly (in μl per fly per day) was calculated over the number of days for which at least one fly was alive in each replicate vial. Consumption data from the day(s) leading up to the death of the last fly in a vial were not considered for rate calculation if the last living fly was no longer mobile. Survival was expressed as the treatment group average of the median number of days of fly survival per vial during the experiment.
Fly survival on medium supplemented with milkweed leaf powder.
We performed two experiments in which we raised three knock-in (LSN, VSN and VSH) and w1118 control (QAN*) line flies on Drosophila medium with and without pulverized leaves of two milkweed species (A. fascicularis or A. curassavica). The purpose of these experiments was twofold: (1) to compare the survival of the knock-in lines to the control line in fly medium; and (2) to compare the effects of milkweed leaves on survival of the knock-in and control lines. The experimental procedure was the same for both experiments. We introduced eggs of each line in narrow vials containing ~10 ml of 22% Instant Drosophila Medium in Millipore water (w/v) with or without 8% dried milkweed leaf powder. We added milkweed leaf powder to the fly diet at an amount similar to that added in the semi-artificial monarch butterfly diet45. We dried the milkweed leaves of each species in a drying oven at ~45 °C with airflow for ~48 h and then ground them to powder with a mortar and pestle. We collected leaves of A. fascicularis growing naturally outside the Berkeley Oxford Track Greenhouses (Berkeley, CA) in January 2018. We grew A. curassavica plants in the greenhouse of the VLSB building at UC Berkeley and collected leaves in April 2018.
We introduced 100 eggs into each vial for knock-in lines LSN and VSH, and the w1118 control (QAN*) line. We introduced 200 eggs for knock-in line VSN into each vial because of the higher embryonic lethality in this line (Extended Data Fig. 3a). Each of the treatment and control groups was tested in quadruplicate vials.
Pupariation, adult eclosion, and survival were monitored over a period of ~24 days until all adult flies had eclosed.
Sequestration assays.
Collection of fly samples.
To assess sequestration of ouabain in flies, we examined extracts of freshly eclosed adult females and males of each knock-in (LSN, VSN, VSH) and w1118 control (QAN*) line raised as larvae in white fly medium (Ward’s Instant Drosophila Medium) supplemented with a series of ouabain concentrations (0 mM, 3 mM, 6 mM, 12 mM and 30 mM). We let 25 female flies (~five to eight days old) lay eggs for four days in wide vials containing a diet with each respective concentration of ouabain in white medium on which larvae fed after eclosion. When larvae completed development, we transferred puparia into fresh, empty food vials on humidified filter paper for each condition to prevent freshly emerging adult flies from feeding and subsequently excreting ouabain before collection. We then collected newly eclosed adult flies within 2–3 h of emergence. The flies were stored immediately in groups of 25 in 1.5-ml collection tubes at −20 °C.
Preparation of fly samples for high-performance liquid chromatography (HPLC).
Frozen flies were lyophilized and each set of 25 individuals was weighed on a microbalance. Samples were pooled, within fly line and diet type, into sets of 50–100 flies and transferred to 2-ml screw-cap tubes containing 0.9 g zirconia/silica beads (2.3 mm; Biospec). Samples were then extracted in 1 ml methanol using a FastPrep homogenizer (MP Biomedicals; three 45-s sessions at 6.5 m/s), and centrifuged for 12 min at 14,000 r.p.m. Then, 750 μl of cleared supernatant was transferred to a fresh tube and dried to completion in a vacuum concentrator (Labconco). Samples were then filtered (0.2 μm; Millipore) and 15 μl extract was injected on an Agilent 1100 series HPLC unit (Agilent). Compounds were separated on a Gemini C18 reversed-phase column (3 μm, 150 × 4.6 mm, Phenomenex). Cardiac glycosides were eluted on a constant flow of 0.7 ml/min with an acetonitrile–water gradient as follows: 0–8 min: 16% acetonitrile; 25 min: 70% acetonitrile; 30–40 min: 95% acetonitrile. Peaks were detected by diode array at 218 nm, and absorbance spectra were recorded from 200 to 300 nm. The ouabain peak was identified in sample extracts by comparing its retention time to an external ouabain standard (O3125, Sigma), and confirmed by observing a symmetrical spectral absorbance peaking at 218 nm46. Sample ouabain concentrations were then estimated relative to the known concentration of the external standard.
Fitting HPLC data to a multiple logistic regression model.
The HPLC data for the knock-in lines were fitted to a multiple logistic regression model in R (dependent variable = ouabain detected or not): Y = intercept + X where Y is the presence of ouabain in the fly; intercept is fixed to X = 0 and Y = 0 (because in the absence of ouabain in the food there is no ouabain in the fly); and X is the mean amount of ouabain in the diet (μg/mg). The w1118 control (QAN*) line was omitted from this analysis because this line did not survive well enough on diets with millimolar levels of ouabain. In all cases, the diet’s intended ouabain concentrations were used as a predictor variable rather than the measured concentrations in the diet. A likelihood ratio test in R was used to test for significance of each model term (genotype and diet).
In vitro sodium pump assays using extracts of transiently transfected Sf9 cells.
Genetically engineered D. melanogaster sodium pumps expressed in Sf9 cells were obtained as described24. The enzymes combined the β-subunit Nrv2.2 (accession no. NM_001273235) with the α-subunit ATPα (accession no. HE962487) into which the substitutions Q111L and A119S had been introduced either singly or in combination by site-directed mutagenesis (QuickChange II XL Kit; Agilent Technologies). A triple-mutated construct coding for the Q111V, A119S, and N122H substitutions was also generated. The constructs were expressed by baculovirus infection of Sf9 cells (Bac-to-Bac Baculovirus Expression System, Thermo Fisher Scientific), and membranes of infected Sf9 cells were collected according to previously established protocols24. Extracted membrane proteins were subjected to sodium pump enzyme assays as in the assays with extracts of heads and nervous tissue above, the only differences being that assays were started by the addition of ATP as described11, and that absolute activities of the Na+/K+-ATPase were expressed as nmol ATP hydrolysed per mg protein per min after calculation24. Curve fitting and estimation of IC50. Curve fitting was performed as for the data on sodium pump activity from the assays with extracts of heads and nervous tissue. Protein homology modelling and in silico mutagenesis. The structure of the D. melanogaster Na+/K+-ATPase is not available in the protein data bank (PDB) and had to be obtained via multi-template homology modelling47. We started by selecting the crystal structures of high-affinity Na+/K+-ATPase from Sus scrofa (PDB ID: 4HYT)48 and Bos taurus (PDB ID: 4XE5)49 kidney as templates. The Na+/K+-ATPase α-subunits of these species share 75.9%, and 76.1% sequence identities with the D. melanogaster ATPase α-subunit, respectively. We then performed homology modelling of the structure via Modeller47 and PyMod 250, using a PyMOL51 plugin to prepare, align and perform the modelling calculations. Homologous structures for the query sequence were identified through the BLAST search tool, after which the template and target sequences were aligned using ClustalW. For more accurate modelling the existing ligands were removed from the templates, and the N-terminal excess amino acids were taken out of the target sequence. The modelled structure was minimized using 80 steps of Steepest Descent, 20 steps of Conjugate Gradients, 20 steps of Quasi Newton, and 100 steps of Molecular Dynamics at room temperature.
Molecular docking.
Protein and ligand preparation for molecular docking.
We extracted the ouabain structure from the co-crystal structure of high-affinity Na+/K+-ATPase from S. scrofa (PDB ID: 4HYT)48, and prepared the ligand (ouabain) and protein files for docking simulations using the AutoDock Tools (ADT) package52 of MGLTools 1.5.653. During docking calculations the protein structure was kept rigid and the ligand flexible, while all water molecules were removed and only polar hydrogen atoms were added to the modelled protein structure used. We assigned Gasteiger charges to the ligand, and saved the ligand and protein structures in PDBQT format for docking simulations.
Molecular docking.
For docking calculations of the ouabain ligand into the modelled ATPase structure we used Autodock4 (version 4.2)52. Grid maps were prepared with the AutoGrid tool of the ADT package, resulting in a grid box of 60 × 60 × 80 Å, centred around the coordinates of ouabain in the co-crystal structure with 0.375 Å of spacing between grid points. We ran a Lamarckian genetic algorithm (LGA) to search for the best conformers during the calculations, keeping the docked ouabain coordinates closest to the coordinates of the ouabain ligand in the co-crystal structure as best conformer for each set of docking calculations. Autodock4 was run at default parameter settings for all docking simulations with the exception of the number of GA runs, which was set to 100. Repeating docking calculations five times for each system allowed for statistical analysis of the results. Each docked structure was inspected visually and all of the structure representations were prepared in PyMOL. A lower binding affinity in the output of the docking simulations corresponds to a better docking of the ligand to the receptor.
Bang sensitivity assays.
We performed paralysis assays with five- or six-day-old females. Females were kept with males in standard molasses fly medium (UC Berkeley Biosciences Divisional Services, Fly Food Facility) until the day of the experiment. In the paralysis assay trials, vials were coded and placed in a randomized order in rows on cardboard trays. Independent experiments were performed across each cohort as described54, but with small modifications. Individual, female adults of each genotype were transferred to empty, clean, culture vials 10 min before the start of the assay. Each vial with one female was vortexed at maximum speed (3,000 r.p.m.) for 10 s using a standard laboratory vortexer (Analogue vortex mixer, VWR), after which the time until recovery from paralysis was measured. The time until recovery is defined as the time after vortexing that is required for the fly to regain the ability to stand upright. Data from three independent runs of the experiment were pooled for each line and medians were compared using the Kruskal–Wallis tests with Dunn’s post hoc tests corrected for multiple comparisons.
Statistical analysis.
For all bioassays, we performed pilot experiments to estimate sample sizes. For the feeding experiments, the concentration ranges for ouabain were determined using pilot experiments. We used the Prism software package (GraphPad Prism 8) to plot and statistically analyse all data, except for the data described in Figs. 1d, 2c, d, f, 3c. For the RMO analysis in Fig. 1d we used a package in R that was based on code developed originally for MATLAB14. For Fig. 2c, d, to determine whether the genotype of fly lines had an effect on survival across ouabain concentrations, we tested for each pair of knock-in lines whether the two predicted survival curves were significantly different from one another. We first performed a logistic regression on the probability of survival against model terms for genotype and ouabain concentration, as well as an interaction term for the effects of genotype and ouabain concentration combined. For two lines, L1 and L2, the logistic regression is of the form log(P(survival)/1 − P(survival)) = B0 + B1ouabain + B2I(L2) + B3(ouabain × I(L2)), where I(L2) is the indicator function for the genotype of line L2, and ouabain represents the concentration in mM. We then used a likelihood ratio test (LRT, d.f. = 1), to evaluate the hypothesis that the coefficient of the interaction term is different from 0, with a non-zero coefficient indicating that the genotypes of the two lines have significantly different effects on survival at different ouabain concentrations (B2 captures the general effect on survival, independent of ouabain concentration). This LRT provided a P value which we compared to a Bonferroni-corrected significance level of α = 0.05/28 = 0.0018, where 28 was the number of pairwise tests performed between all knock-in and control lines.
For the ouabain sequestration experiment in Fig. 2f we used an LRT to evaluate the significance of each model term in the logistic regressions. For Fig. 3c, we tested for potential epistasis between the Q111L and A119S mutations for larval–adult survivorship at the maximum ouabain concentration of 30 mM by performing a logistic regression with interaction term: log(P(survival)/1 − P(survival)) = B0 + BSI(S) + BLI(L) + BSLI(S)I(L), where I(S) and I(L) are the indicator functions for the presence of the A119S and Q111L mutations, respectively. We tested the hypothesis that the effect of mutations A119S and Q111L together, BSL, was equal to the sum of each individual effect on survivorship (if BSL = BS + BL). In all other experiments, we tested for differences between genotypes with either one-way or two-way ANOVA with Tukey’s post hoc tests corrected for multiple comparisons, or the non-parametric Kruskal–Wallis test with Dunn’s post hoc tests corrected for multiple comparisons, as appropriate. In experiments where four-parameter logistic equations were fitted to the data (Fig. 3a, Extended Data Fig. 8), we used global fitting followed by the extra sum-of-squared F test as a method to test whether the model parameters differed between data sets. We provide the full statistical test results in the Source Data files for each figure.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Extended Data
Extended Data Fig. 1. Substitutions at ATPα amino acid residues 111, 119 and 122 are directly or indirectly associated with insect specialization on plants that produce cardiac glycosides.
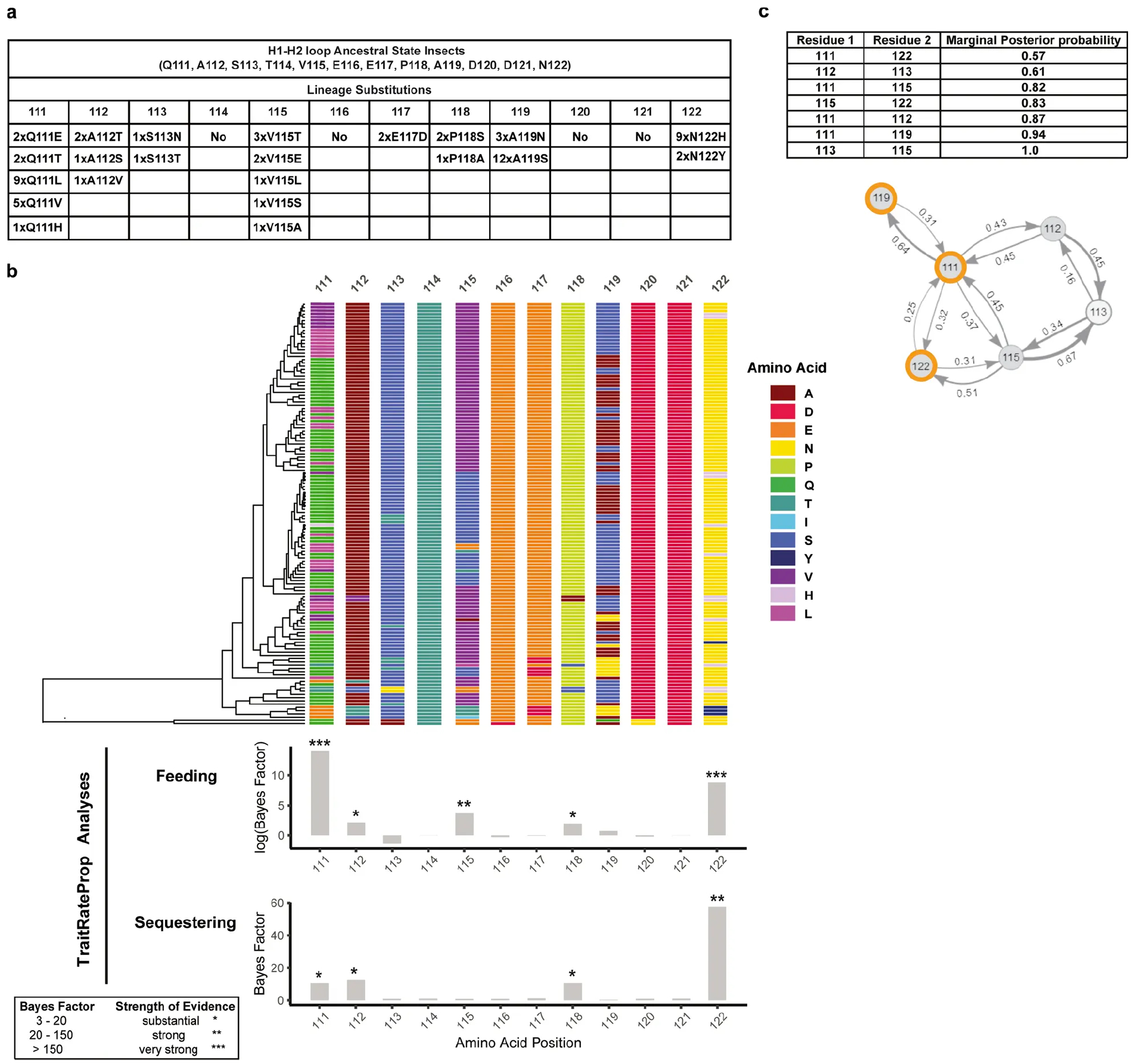
a, The number of occurrences of each substitution across the 21 lineages in which specialization evolved independently. b, TraitRateProp analysis of the H1–H2 loop of ATPα across insects shows amino acid residues that are strongly associated with feeding on cardiac glycoside-producing plants and toxin sequestration. Bayes factor values in the top histogram indicate per-site associations between feeding and sequence rate evolution, values in the bottom histogram indicate per-site associations between sequestration and sequence rate evolution. Values over 10 were considered different (asterisks). For information on the species included in the analysis please see Supplementary Text. Colours in the multi-sequence alignment represent individual amino acids. c, BGM shows the correlated evolution of amino acid sites within the H1–H2 loop of ATPα. The table shows the marginal posterior probabilities (PP) between amino acid interactions, where the PP exceeds a default cut-off of 0.5. The residue interactions are depicted graphically, with amino acid sites represented by the nodes and the PP associated with a given epistatic or co-evolutionary interaction indicated by the values at the arrows. Nodes circled in orange indicate amino acid sites that are the focus of experiments in this study. Sites 111 and 122 are very strongly associated with feeding and sequestering, and site 119 co-evolves with site 111.
Extended Data Fig. 2. A two-step genome editing approach using CRISPR–Cas9 and HDR to generate knock-in Atpα lines of Drosophila melanogaster.

In the first step, the region encoding the H1–H2 extracellular domain was replaced with a 3×P3::GFP marker through CRISPR–Cas9-mediated HDR. This generated a common parent line with the deletion allele AtpαDeletion (GFP+). In the second step, the 3×P3::GFP marker was replaced with each of the synonymous and non-synonymous point mutation alleles through an additional round of CRISPR–Cas9-mediated HDR to generate the knock-in lines. The crossing schemes to establish the deletion line and the knock-in lines following the first and second rounds of HDR, respectively, are also shown. See also Methods and Supplementary Tables 4–7 for further details on the genome engineering strategy and crosses behind the establishment of the knock-in lines.
Extended Data Fig. 3. Point mutations have some effect on adult emergence, but do not lead to major changes in baseline Atpα expression or Na+/K+-ATPase activity.
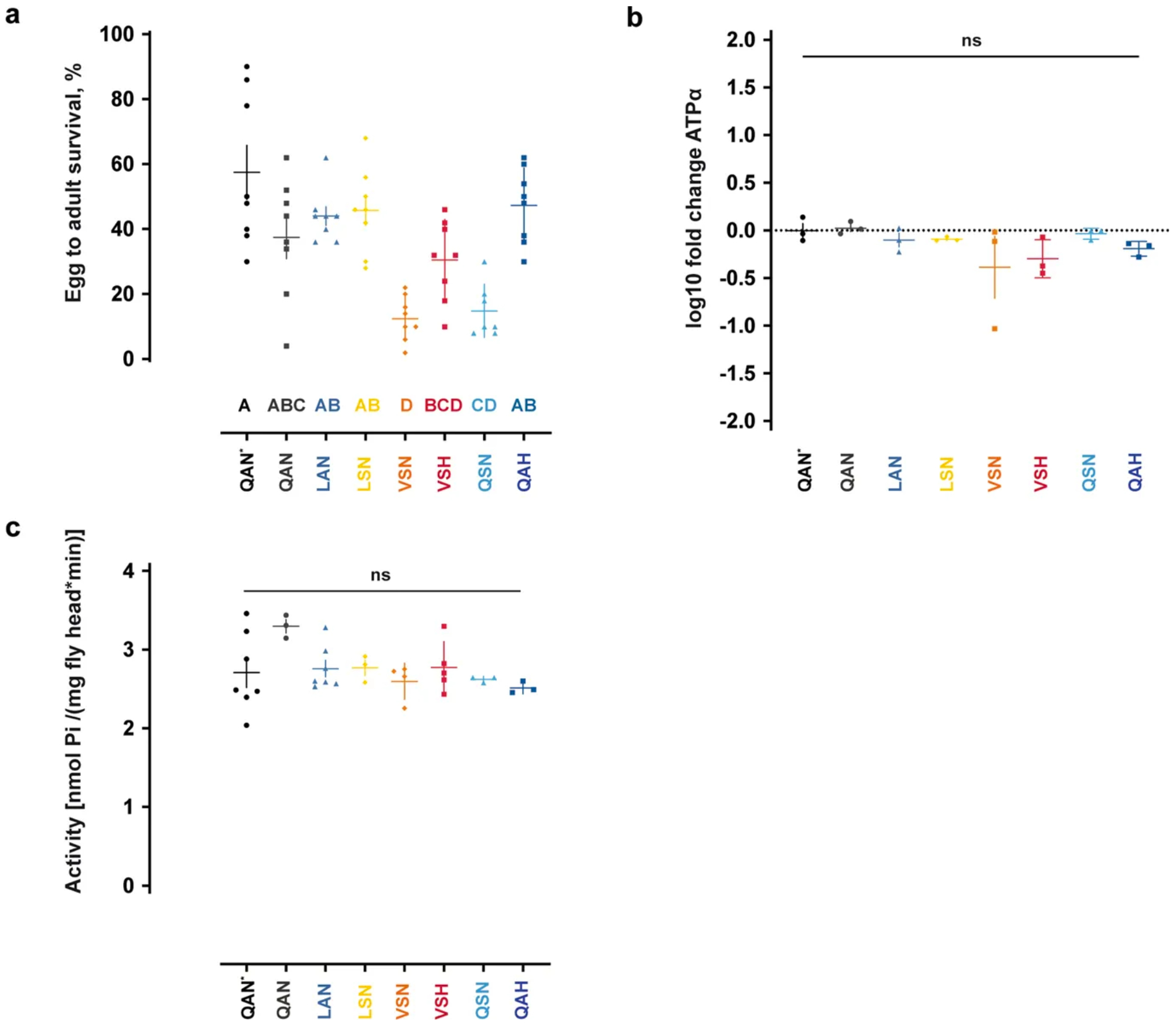
a, Percentages of emerging adults of the knock-in and control lines on standard Drosophila medium (n = 7–8 vials, 100 eggs per vial, mean ± s.e.m.). Survival of the knock-in lines and control lines QAN (engineered control) and QAN* (w1118 wild type) was compared using one-way ANOVA (P < 0.001). Survival differed between QAN* and some of the knock-in lines, but not between the engineered control line QAN and any of the knock-in lines except VSN (post hoc Tukey’s tests (letters)). b, Atpα expression was not different among the engineered Drosophila knock-in lines or w1118 wild-type flies (QAN*). Atpα transcript level differences were assayed by qPCR. Expression was assayed in three biological replicates (symbols represent the mean ± s.e.m.), with two technical replicates per biological replicate (averaged for each biological replicate), of five- to six-day-old females as fold change standardized against rpl32 expression in QAN* flies. The expression fold change between genotypes was compared using one-way ANOVA (P = 0.3197). c, None of the sequential Atpα genotypes found along the monarch lineage affected base-line levels of pump activity in a sodium pump enzymatic assay using extracts of fly heads (one-way ANOVA, P = 0.1377; symbols represent the mean ± s.e.m. of 3–7 biological replicates). Further information on experimental design and statistical test results is in the Source Data. ns, not significant.
Extended Data Fig. 4. Drosophila flies with edited genomes show increased larval–adult survival when fed cardiac glycosides.
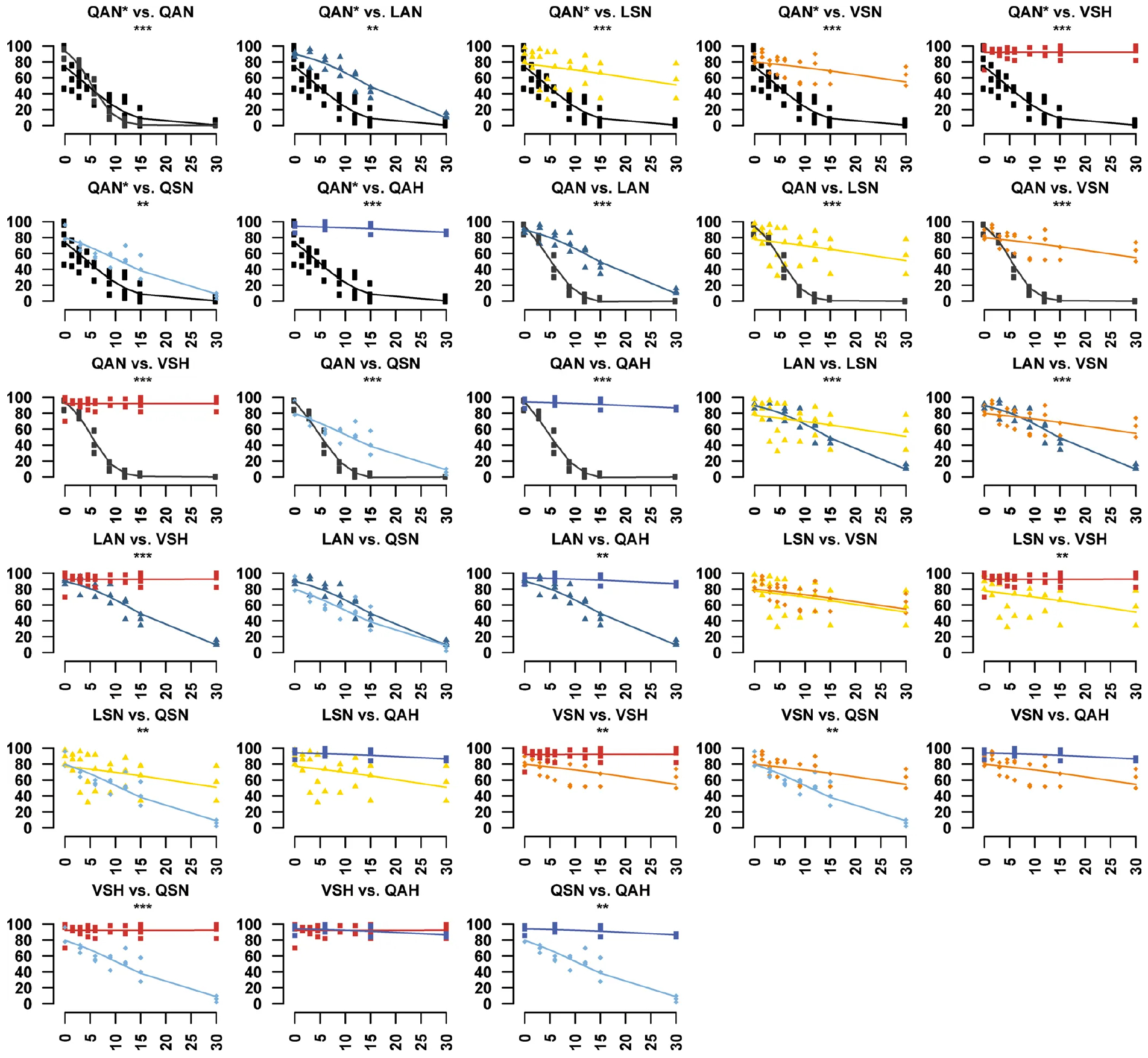
These panels accompany Fig. 2c. Larval–adult survival when reared on diets with a range of ouabain concentrations was different between monarch lineage knock-in lines relative to control lines (QAN, engineered control; QAN*, w1118 wild type). Symbols represent the mean ± s.e.m. of 3–6 biological replicates (50 larvae per replicate). Curves were fit through a univariate logistic regression (effect of ouabain concentration on survival), and the difference in survivorship trajectories between each pair of fly lines (genotypes) was evaluated by performing an LRT to assess the significance of the inclusion of an interaction term between genotype and ouabain concentration in the logistic regression for a pair of lines (**P < 0.01, ***P < 0.001). Further information on experiment design and statistical test results is in the Source Data.
Extended Data Fig. 5. Drosophila flies with edited genomes show increased adult survival when fed cardiac glycosides.
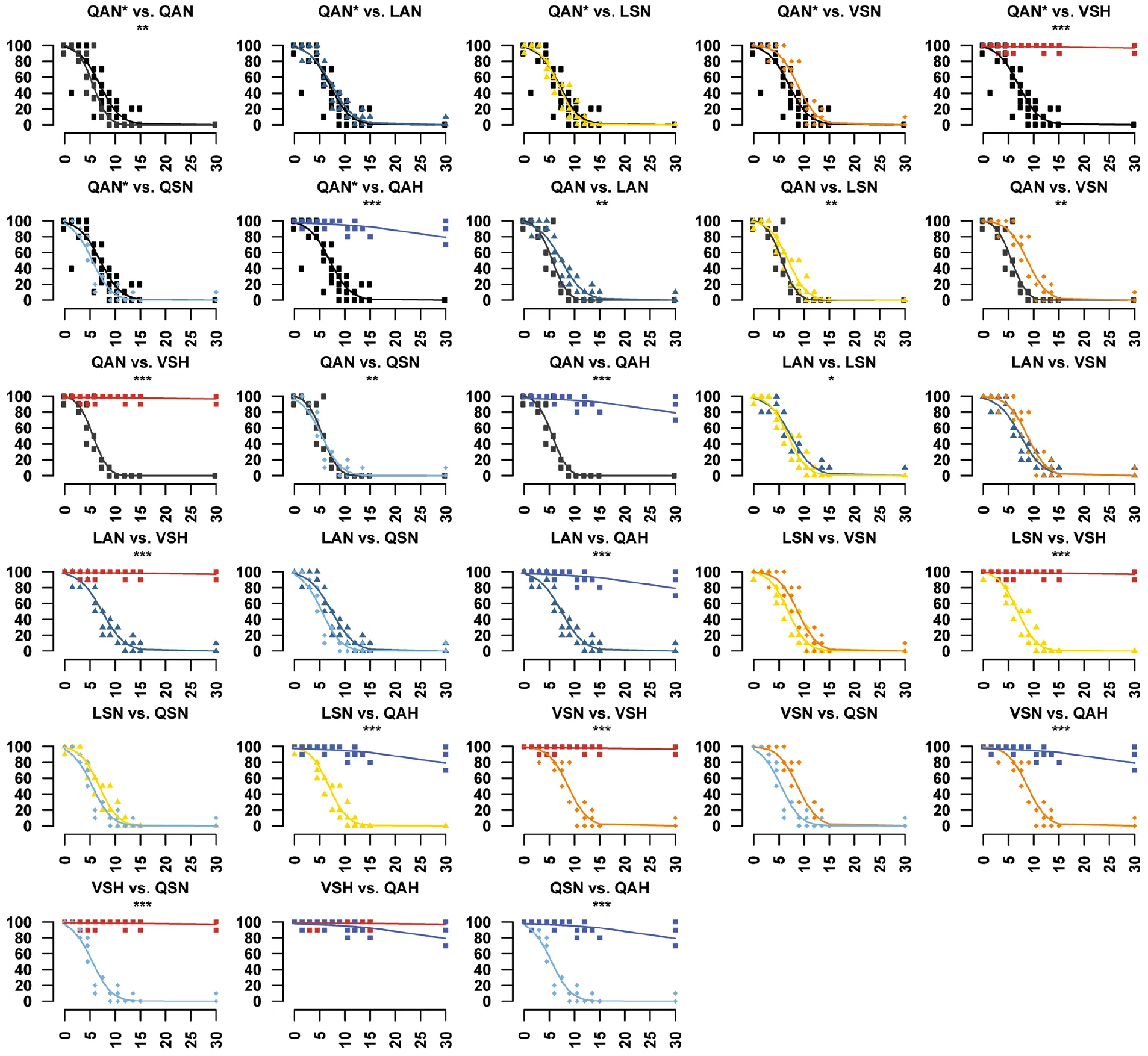
These panels accompany Fig. 2d. Adult survival when reared on diets with a range of ouabain concentrations was different between monarch lineage knock-in lines and control lines (QAN, engineered control; QAN*, w1118 wild type). Symbols represent the mean ± s.e.m. of 3–6 biological replicates. Curves were fit through a univariate logistic regression (the effect of ouabain concentration on survival), and a difference in survivorship trajectories between each pair of fly lines (genotypes) was evaluated by performing an LRT to assess the significance of the inclusion of an interaction term between genotype and ouabain concentration in the logistic regression for a pair of lines (*P < 0.05, **P < 0.01, ***P < 0.001). Further information on experimental design and statistical test results is in the Source Data.
Extended Data Fig. 6. The mutational path of ATPα in the monarch butterfly lineage increases dietary tolerance to ouabain in vivo in engineered Drosophila without affecting feeding rate.
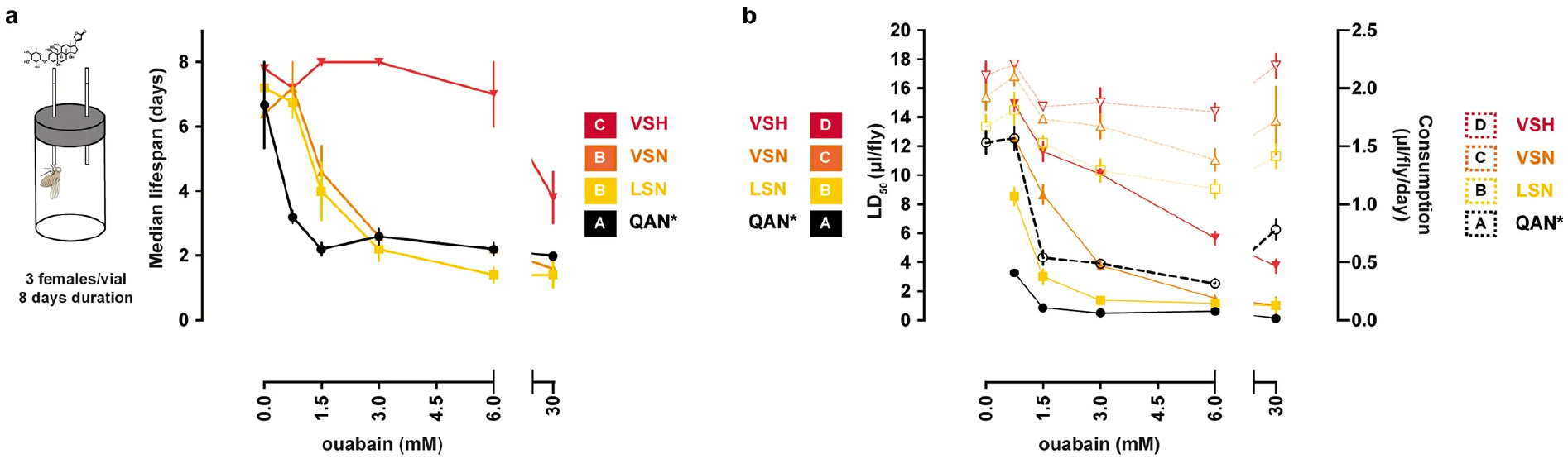
a, Estimation of mean lifespan (days) in adult females (four to seven days old at the start of the experiment) of the knock-in and control lines in CAFE assays across a range of ouabain concentrations. Each data point represents the mean ± s.e.m. of five biological replicates. Both ouabain concentration and genotype affect the survival time (two-way ANOVA (P < 0.0001) with post hoc Tukey’s tests (letters indicate pairwise differences between genotypes)). b, Estimation of LD50 (μl per fly; solid lines) and feeding rates (μl per fly per day; dashed lines) in the same individuals as in a. Each data point represents the mean ± s.e.m. of five biological replicates. Both ouabain concentration and genotype affect LD50 (two-way ANOVA (P < 0.0001) with post hoc Tukey’s tests (letters indicate pairwise differences between genotypes)). Further information on experimental design and statistical test results is in the Source Data.
Extended Data Fig. 7. Survival of knock-in lines on fly diets supplemented with dried, pulverized leaves of two milkweed species that host monarch butterflies in nature (A. curassavica and A. fascicularis).
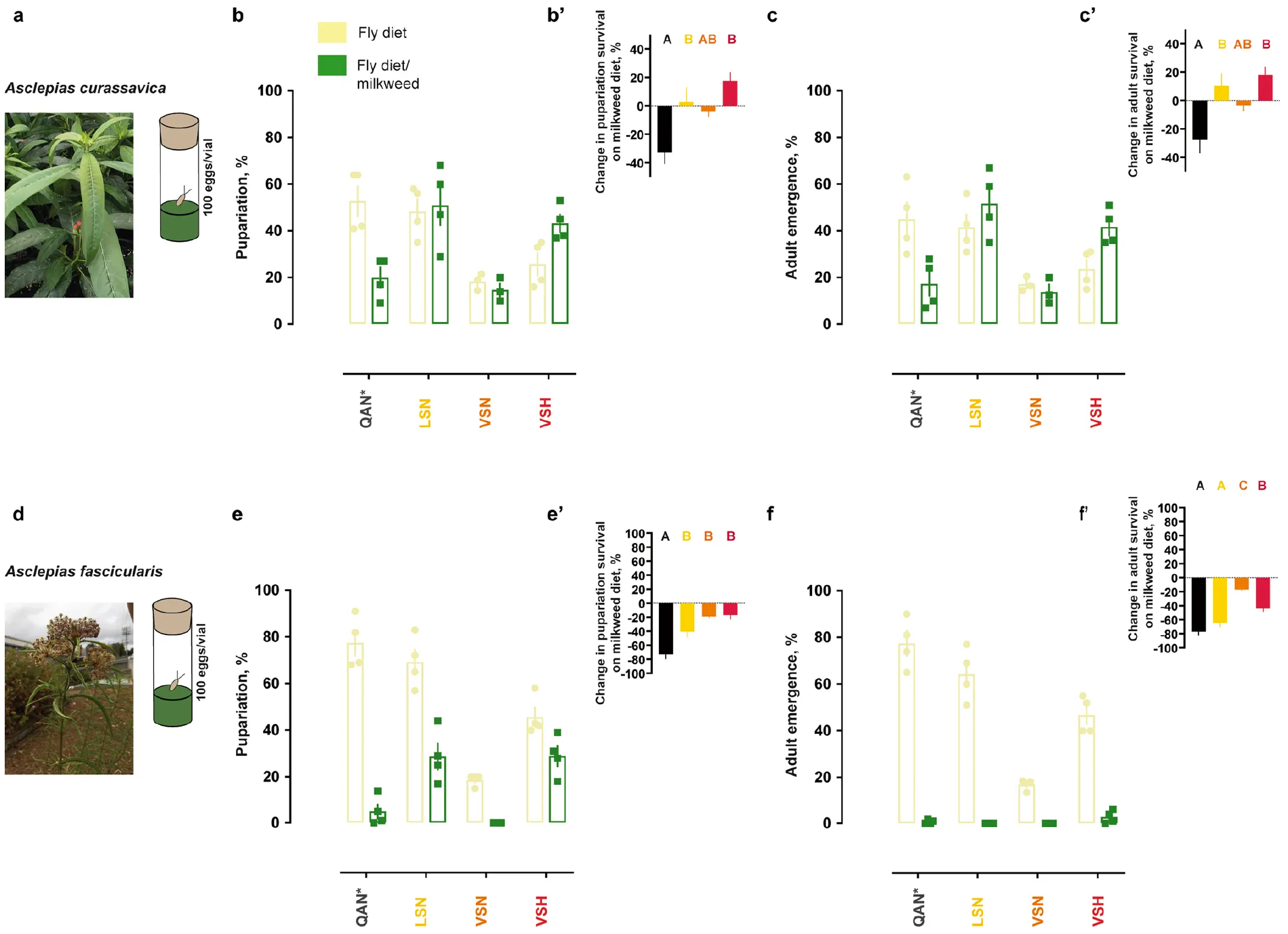
a, Photograph of A. curassavica plant used in this study. b, c, Percentages of pupariating larvae and emerging adults of the knock-in and control (wild-type w1118: QAN*) lines on fly diet with and without A. curassavica leaf material (n = 3–4, mean ± s.e.m.). b′, c′, Differences in pupariation and emergence percentages on a fly diet with milkweed relative to percentages on a control diet (n = 3–4, mean ± s.e.m.). Mean differences between percentages in b′ and c′ were tested with one-way ANOVA (P < 0.01) followed by post hoc Tukey’s tests (letters). These panels accompany Fig. 2e. d, Photograph of A. fascicularis plant used in this study. e, f, Percentages of pupariating larvae and emerging adults of the knock-in and control lines on fly diet with and without A. fascicularis leaf material (n = 4, mean ± s.e.m.). e′, f′, Differences in pupariation and emergence percentages on a fly diet with milkweed relative to percentages on a control diet (n = 4, mean ± s.e.m.). Mean differences between percentages in e′ and f′ were tested with one-way ANOVA (P < 0.001) followed by post hoc Tukey’s tests (letters). Experiments were performed once, and adding leaf material of either of the two milkweed species to the fly diet had largely consistent effects on survival of the monarch lineage knock-in and control fly lines.
Extended Data Fig. 8. The Atpα genotypes found along the monarch lineage sequentially increase TSI to ouabain without affecting baseline levels of sodium pump activity.
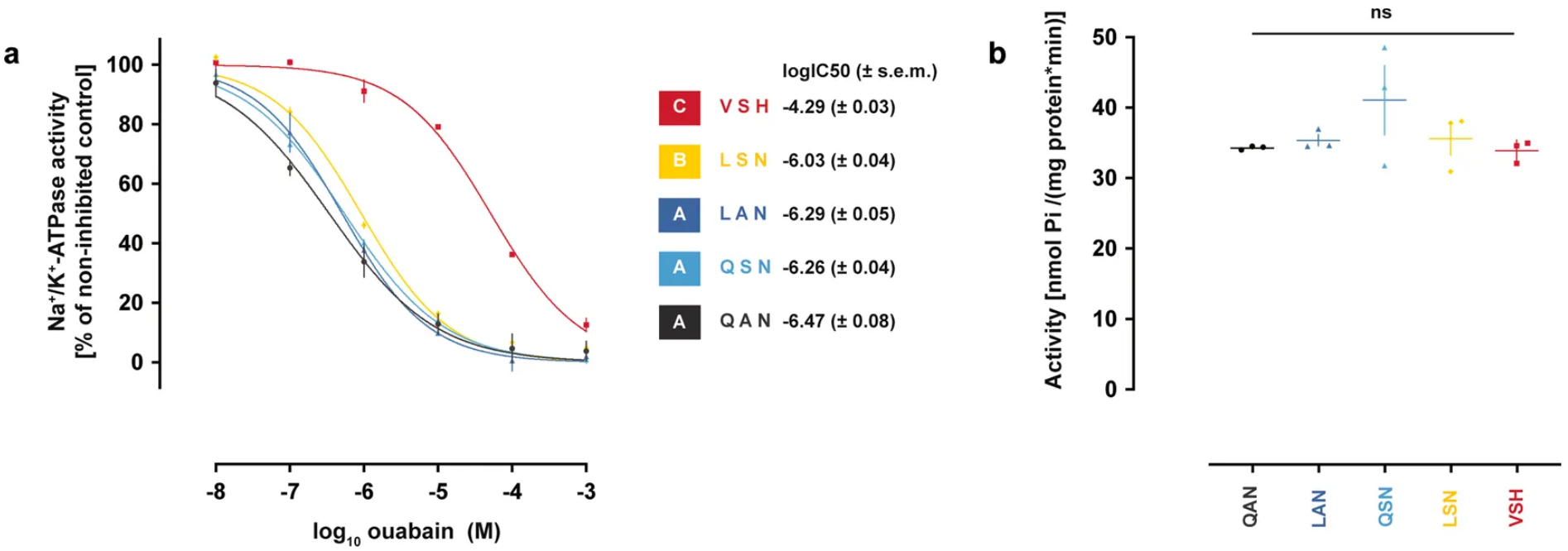
a, In vitro ouabain sensitivity of engineered Drosophila Na+/K+-ATPases transiently expressed in Sf9 cell lines. Each data point represents the mean ± s.e.m. of three biological replicates. log10[IC50] for each type of Na+/K+-ATPase was estimated after four-parameter logistic curve fitting, and statistical differences between log10[IC50] values were tested with one-way ANOVA (P < 0.0001) followed by post hoc Tukey’s tests (letters). b, None of the sequential Atpα genotypes found along the monarch lineage affected baseline levels of pump activity in the enzymatic assay with extracts of transiently transfected Sf9 cells (one-way ANOVA, P = 0.3197). Each data point represents the mean ± s.e.m. of three biological replicates.
Extended Data Fig. 9. Molecular docking simulations show stepwise reductions in ouabain binding to Na+/K+-ATPases with monarch lineage substitutions in ATPα.
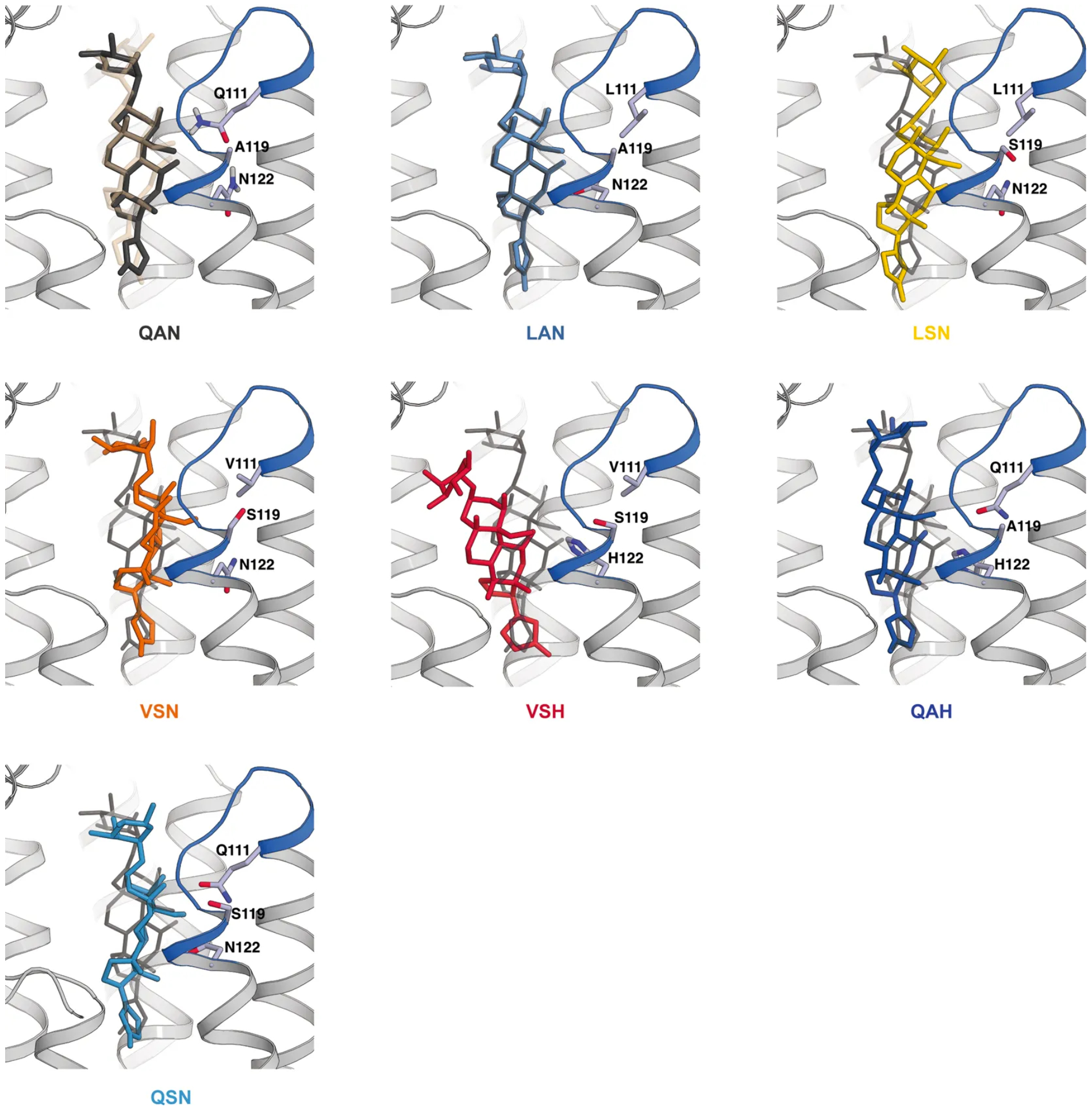
The ouabain binding pocket structure obtained from molecular docking simulations for each Na+/K+-ATPase with mutated ATPα. The mutated residues are shown in sticks and are labelled. The H1–H2 loop of ATPα is shown in blue. The extracellular region of the α-subunit is removed for simplicity. For the wild-type (QAN) ATPase, ouabain is shown in its co-crystal structure coordinates (white, transparent) together with its best-docked position. For all other ATPases only the best-docked positions (closest to the co-crystal structure) are shown together with ouabain’s docking position for the wild-type ATPase (dark grey). The triple-mutated VSH ATPase has two distinct docking scores: one is similar to the docking energy for the wild-type ATPase and the other has the lowest binding energy compared to all other mutated ATPases. The potential existence of both states might be related to a trend of reduced bang sensitivity for this genotype compared to some of the single-mutant genotypes. A119 is not directly part of the ouabain binding pocket, and therefore, A119S alone does not change ouabain binding. Although the consequences of A119S are relatively subtle, the mutation may disrupt the local hydrogen bonding network and cause structural or dynamic changes in the loop or in its vicinity.
Supplementary Material
Acknowledgements
We thank V. Wagschal, who helped with the construction and testing of in vitro cell lines; E. Toprak, who provided MATLAB code; T. O’Connor, who aided in the sequestration analyses; the Essig Museum of Entomology for photographs of milkweed butterfly specimens; M. Fa and K. O’Connor-Giles for advice on the development of the fly lines; and E. LaPlante for assistance with feeding assays. D. Bachtrog, K. Mooney, P. Moorjani, M. Nachman, R. Nielsen, P. Sudmant, R. Tarvin and B. Walsh provided feedback that improved the manuscript. Access to the HPC resources of Aix-Marseille Université was supported by the Equip@Meso (ANR-10-EQPX-29-01) project of the Investissements d’Avenir supervised by the Agence Nationale de la Recherche. This project was supported by grants from the Gordon and Betty Moore Foundation (Life Sciences Research Foundation Postdoctoral Fellowship Grant GBMF2550.06 to S.C.G.), the German Research Foundation (DFG, grant Do527/5-1 to S.D.), the Agence National de la Recherche (grant no. BioHSFS ANR-15-CE11-0007 to F.S. and F.R.), the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 772257 to F.S. and F.R.), the National Geographic Society (grant 9097-12 to N.K.W.), the National Science Foundation (grant DEB-1256758 to N.K.W. and IOS-1907491 to A.A.A.), the John Templeton Foundation (grant ID 41855 to A.A.A., S.D., and N.K.W.), and the National Institute of General Medical Sciences of the National Institutes of Health (award no. R35GM119816 to N.K.W.).
Footnotes
Data availability
The data supporting the findings of this study are available within the paper and its Supplementary Information.
Code availability
The code to compute RMO (reproducibility of mutational order) scores in the presence of unordered mutations and correlated pathways can be accessed in Github (https://github.com/gaguerra/ModifiedRMO). The set of R scripts implements the RMO score, first proposed by Toprak and co-workers14, with the new additions of accounting for non-independent mutational pathways (in the presence of shared ancestry) and partially unresolved mutational pathways.
Competing interests The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41586-019-1610-8.
Peer review information Nature thanks Joseph W. Thornton and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-019-1610-8.
References
- 1.Barrett RD & Hoekstra HE Molecular spandrels: tests of adaptation at the genetic level. Nat. Rev. Genet 12, 767–780 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Agrawal AA Toward a predictive framework for convergent evolution: integrating natural history, genetic mechanisms, and consequences for the diversity of life. Am. Nat 190, S1–S12 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Stern DL The genetic causes of convergent evolution. Nat. Rev. Genet 14, 751–764 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Storz JF Causes of molecular convergence and parallelism in protein evolution. Nat. Rev. Genet 17, 239–250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siddiq MA, Loehlin DW, Montooth KL & Thornton JW Experimental test and refutation of a classic case of molecular adaptation in Drosophila melanogaster. Nat. Ecol. Evol 1, 0025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agrawal AA, Petschenka G, Bingham RA, Weber MG & Rasmann S Toxic cardenolides: chemical ecology and coevolution of specialized plant–herbivore interactions. New Phytol. 194, 28–45 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Petschenka G, Wagschal V, von Tschirnhaus M, Donath A & Dobler S Convergently evolved toxic secondary metabolites in plants drive the parallel molecular evolution of insect resistance. Am. Nat 190, S29–S43 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Holzinger F, Frick C & Wink M Molecular basis for the insensitivity of the Monarch (Danaus plexippus) to cardiac glycosides. FEBS Lett. 314, 477–480 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Dobler S, Dalla S, Wagschal V & Agrawal AA Community-wide convergent evolution in insect adaptation to toxic cardenolides by substitutions in the Na,K-ATPase. Proc. Natl Acad. Sci. USA 109, 13040–13045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhen Y, Aardema ML, Medina EM, Schumer M & Andolfatto P Parallel molecular evolution in an herbivore community. Science 337, 1634–1637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petschenka G et al. Stepwise evolution of resistance to toxic cardenolides via genetic substitutions in the Na+/K+-ATPase of milkweed butterflies (lepidoptera: Danaini). Evolution 67, 2753–2761 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Horisberger JD Recent insights into the structure and mechanism of the sodium pump. Physiology (Bethesda) 19, 377–387 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Weinreich DM, Delaney NF, Depristo MA & Hartl DL Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Toprak E et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet 44, 101–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whiteman NK & Mooney KA Evolutionary biology: Insects converge on resistance. Nature 489, 376–377 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Berenbaum MR in Molecular Aspects of Insect–Plant Associations (eds Brattsten LB & Ahmad S) 257–272 (Springer, 1986). [Google Scholar]
- 17.Brower LP, Ryerson WN, Coppinger LL & Glazier SC Ecological chemistry and the palatability spectrum. Science 161, 1349–1350 (1968). [DOI] [PubMed] [Google Scholar]
- 18.Petschenka G & Agrawal AA Milkweed butterfly resistance to plant toxins is linked to sequestration, not coping with a toxic diet. Proc. R. Soc. B 282, 20151865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhan S et al. The genetics of monarch butterfly migration and warning colouration. Nature 514, 317–321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarvin RD et al. Interacting amino acid replacements allow poison frogs to evolve epibatidine resistance. Science 357, 1261–1266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hague MTJ et al. Large-effect mutations generate trade-off between predatory and locomotor ability during arms race coevolution with deadly prey. Evol. Lett 2, 406–416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blount ZD, Barrick JE, Davidson CJ & Lenski RE Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489, 513–518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groen SC et al. Multidrug transporters and organic anion transporting polypeptides protect insects against the toxic effects of cardenolides. Insect Biochem. Mol. Biol 81, 51–61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalla S & Dobler S Gene duplications circumvent trade-offs in enzyme function: Insect adaptation to toxic host plants. Evolution 70, 2767–2777 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Lin S, Staahl BT, Alla RK & Doudna JA Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Port F, Chen H-M, Lee T & Bullock SL Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl Acad. Sci. USA 111, E2967–E2976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pegueroles C et al. Inversions and adaptation to the plant toxin ouabain shape DNA sequence variation within and between chromosomal inversions of Drosophila subobscura. Sci. Rep 6, 23754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shorrocks B in The Genetics and Biology of Drosophila (eds Ashburner M et al. ) 385–428 (Academic, 1982). [Google Scholar]
- 29.Ashmore LJ et al. Novel mutations affecting the Na, K ATPase alpha model complex neurological diseases and implicate the sodium pump in increased longevity. Hum. Genet 126, 431–447 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubiger M, Feng Y, Fambrough DM & Palka J A mutation of the Drosophila sodium pump α-subunit gene results in bang-sensitive paralysis. Neuron 12, 373–381 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Trifinopoulos J, Nguyen L-T, von Haeseler A & Minh BQ W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petschenka G, Pick C, Wagschal V & Dobler S Functional evidence for physiological mechanisms to circumvent neurotoxicity of cardenolides in an adapted and a non-adapted hawk-moth species. Proc. R. Soc. B 280, 20123089 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paradis E, Claude J & Strimmer K APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Pupko T, Pe’er I, Shamir R & Graur D A fast algorithm for joint reconstruction of ancestral amino acid sequences. Mol. Biol. Evol 17, 890–896 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Pond SLK, Frost SDW & Muse SV HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Letunic I & Bork P Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy Karin E, Ashkenazy H, Wicke S, Pupko T & Mayrose I TraitRateProp: a web server for the detection of trait-dependent evolutionary rate shifts in sequence sites. Nucleic Acids Res. 45, W260–W264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poon AF, Lewis FI, Pond SL & Frost SD An evolutionary-network model reveals stratified interactions in the V3 loop of the HIV-1 envelope. PLOS Comput. Biol 3, e231 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gratz SJ et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponton F, Chapuis MP, Pernice M, Sword GA & Simpson SJ Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol 57, 840–850 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Taussky HH & Shorr E A microcolorimetric method for the determination of inorganic phosphorus. J. Biol. Chem 202, 675–685 (1953). [PubMed] [Google Scholar]
- 42.Petschenka G & Dobler S Target-site sensitivity in a specialized herbivore towards major toxic compounds of its host plant: the Na+K+-ATPase of the oleander hawk moth (Daphnis nerii) is highly susceptible to cardenolides. Chemoecology 19, 235–239 (2009). [Google Scholar]
- 43.Beikirch H Toxicity of ouabain on Drosophila melanogaster. Experientia 33, 494–495 (1977). [DOI] [PubMed] [Google Scholar]
- 44.Ja WW et al. Prandiology of Drosophila and the CAFE assay. Proc. Natl Acad. Sci. USA 104, 8253–8256 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glass HW Jr & Pan ML Laboratory rearing of monarch butterflies (Lepidoptera: Danaiidae), using an artificial diet. Ann. Entomol. Soc. Am 76, 475–476 (1983). [Google Scholar]
- 46.Malcolm SB & Zalucki MP Milkweed latex and cardenolide induction may resolve the lethal plant defence paradox. Entomol. Exp. Appl 80, 193–196 (1996). [Google Scholar]
- 47.Šali A, Potterton L, Yuan F, van Vlijmen H & Karplus M Evaluation of comparative protein modeling by MODELLER. Proteins 23, 318–326 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Laursen M, Yatime L, Nissen P & Fedosova NU Crystal structure of the high-affinity Na+K+-ATPase-ouabain complex with Mg2+ bound in the cation binding site. Proc. Natl Acad. Sci. USA 110, 10958–10963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregersen JL, Mattle D, Fedosova NU, Nissen P & Reinhard L Isolation, crystallization and crystal structure determination of bovine kidney Na+,K+- ATPase. Acta Crystallogr. F 72, 282–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janson G, Zhang C, Prado MG & Paiardini A PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics 33, 444–446 (2017). [DOI] [PubMed] [Google Scholar]
- 51.The PyMOL molecular graphics system v.1.8 (Schrödinger LLC, 2015). Morris GM et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility J. Comput. Chem 30, 2785–2791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanner MF Python: a programming language for software integration and development. J. Mol. Graph. Model 17, 57–61 (1999). [PubMed] [Google Scholar]
- 53.Ganetzky B & Wu CF Indirect suppression involving behavioral mutants with altered nerve excitability in Drosophila melanogaster. Genetics 100, 597–614 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


