Abstract
The antibody repertoire is the fundamental unit that enables development of antigen specific adaptive immune responses against pathogens. Different species have developed diverse genetic and structural strategies to create their respective antibody repertoires. Here we review the shark, chicken, camel, and cow repertoires as unique examples of structural and genetic diversity. Given the enormous importance of antibodies in medicine and biological research, the novel properties of these antibody repertoires may enable discovery or engineering of antibodies from these non-human species against difficult or important epitopes.
Introduction
The adaptive immune system is a crucial adaptation associated with the evolution of vertebrates. The ability to create repertoires of antigen binding molecules and then select those which bind with high affinity to their cognate antigen in order to remove evading pathogens is the key feature of any adaptive immune system. While it may appear that the antibody response is capable of binding an infinite number of epitopes, this is probably not true. The germline repertoires of different species have evolved both for the ability to create diversity, but also in a Darwinian way to select antibody features specific for each organism’s antigenic load. Indeed, knockout of a single VH region in mice can result in impaired ability to fight certain infections associated with a key neutralizing epitope [1]. Thus, while different species may be capable of producing billions of different paratopes in their antibody repertoire, these repertoires are still limited by the scaffold and germline genetic composition of their antibody genes. In this regard, various species have evolved novel structural features that presumably were selected based on their unique struggles with specific foreign invaders. Across all species, therefore, the ‘paratope universe’ is certainly dramatically larger than the individual repertoire of any specific species.
A challenge in the creation of the antibody repertoire at the genetic level is that the number of genes required for millions of antigen binding molecules could theoretically exceed the amount of DNA in the genome. In this regard, strategies to diversify antibodies based on the combinatorial rearrangement of genetic elements to produce single antibodies per cell has been accomplished in several species [2–4,5••]. In the jawless fish (e.g. lamprey and hagfish), the combinatorial association of different variable lymphocyte receptor genes produces an array of antigen receptors based on the leucine rich repeat motif, which is the only known repertoire that does not use the immunoglobulin domain as a structural scaffold [2,6]. In humans and mice V(D)J recombination combinatorially produces genetic diversity by using a multitude of different V, D, and J gene segments, as well as N and P nucleotide junctional diversity, to create the naïve antibody repertoire [7,8]. This recombination event particularly provides diversity within CDR H3 of the paratope which is often a major contact of antigen (Figure 1).
Figure 1.
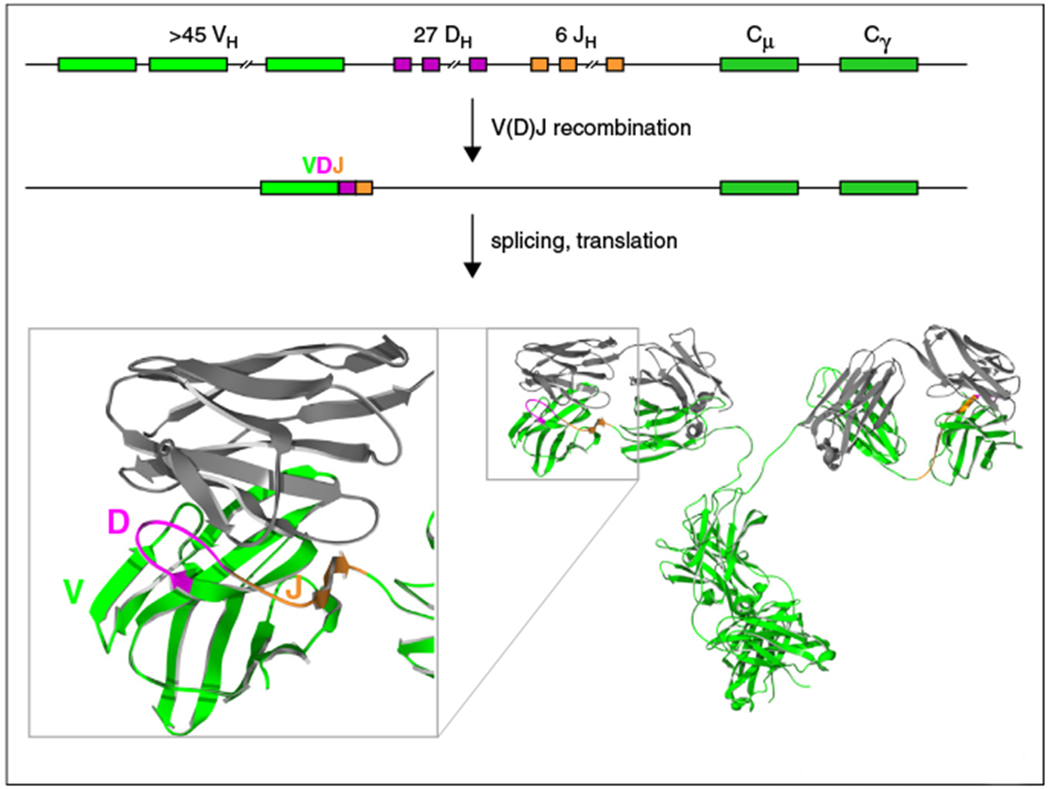
V(D)J recombination and the structure of human and mouse antibodies. A schematic of the human heavy chain locus is shown, where multiple VH, DH, and JH regions can rearrange in any given B-cell to produce a functional VDJ unit that encodes the heavy chain variable region. A close-up of the antigen binding region (bottom left) illustrates the regions encoded by the V, D, and J gene segments. A full length antibody is shown on the bottom right.
Antibodies are major tools in biotechnology; they function as research tools, diagnostic reagents, and are now an important class of drugs for the pharmaceutical industry [9,10]. Historically, the generation of an antibody was accomplished through immunization and hybridoma techniques [11], which resulted in reagents for ELISAs, western blots, flow cytometry, immunoprecipitation, and other important immunochemical techniques. Many therapeutic antibodies were also originally discovered through these techniques [12]. However, as sophistication in drug discovery has increased, along with an explosion in data in genetics and structural biology, it is clear that antibodies with certain binding and functional properties would be ideal for various specific applications. For example, nearly all currently FDA approved antibodies are high affinity antagonists, however antibodies with agonist, antagonist, modulator, or other activities may be mediated by binding to limited epitopes with exquisite specificity and affinity, and may be highly desireable for certain indications and targets [13]. Additionally, binding to enzymatic active sites, allosteric epitopes, or to important regions on multipass membrane proteins may be difficult using standard techniques with the canonical human or mouse antibody scaffolds. Therefore, alternative paratope structures derived from alternate species may allow unique physicochemical binding characteristics not available in traditionally used mouse or human antibodies.
Certain antigens and epitopes can be particularly challenging to develop antibodies against. It is a well appreciated difficulty to generate antibodies with pharmacologic activity against multipass transmembrane proteins like GPCRs and ion channels [14,15]. While this difficulty may in part be due to the challenges associated with producing stable purified protein in functional form, it is also that the typical flat antibody paratope may not be optimal for interacting with grooves, pores, or other concave epitopes in these receptors. Similarly, antibodies that ‘reach’ into enzymatic active sites are relatively rare. Even certain microorganisms present challenges in eliciting antibodies against neutralizing epitopes. In the case of HIV, while antibodies can be raised against the spike protein gp120, these antibodies are typically not neutralizing, and rarely broadly neutralizing [16,17]. However, extremely rare antibodies with long CDR H3s have been identified which pierce the glycan shield and contact key conserved epitopes on the protein and can neutralize multiple viral clades. Given that the human and mouse repertoires appear limited in its ability to target these epitopes, utilization of other species with alternative antibody scaffolds could lend itself to identifying new antibody tools to study the fine aspects of viral neutralization, and potentially to bind more difficult antigens or epitopes.
Here we review strategies for repertoire diversity in shark, camel, chicken, and cow as unique representatives of novel mechanisms at both the genetic and structural levels. Within vertebrates the differences in genetic and structural strategies to create an antibody repertorie are significant (Figure 2), and with relatively few species studied in detail, it seems probable that further variations have yet to be discovered.
Figure 2.
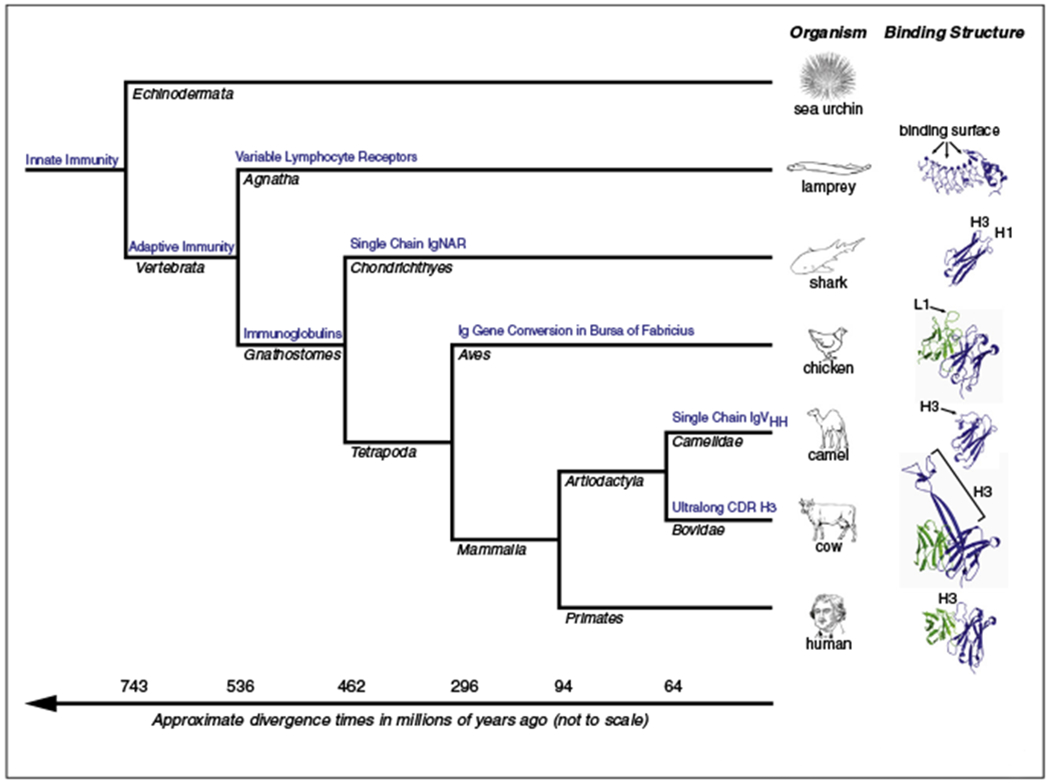
Evolutionary relationships among species: useful antibody innovations evolved several times in vertebrates. Deuterostome phylogeny highlights the divergence times between some lineages that are discussed in this review (in bold). Agnathans such as the lamprey have lymphocyte classes similar to jawed vertebrates, but employ diverse repertoires of variable lymphocyte receptors from somatically rearranged loci employing activation induced cytidine deaminase (AID). Gnathostomes such as the shark began using immunoglobulin superfamily receptors on lymphocytes and first evolved an immunoglobulin heavy chain in the absence of light chains. Birds use AID to gene convert an initially very restricted recombination activating gene (RAG) generated locus via sequence donation from variable segment pseudogenes. Camelids convergently evolved heavy-chain only antibody variants of IgG, and at least some members of the family Bovidae employ heavy chains with an ultralong CDR H3, creating another diverse disulfide stabilized domain. (Right) Structures of antibody binding fragments from diverse species. Ribbon diagrams of lamprey (PDB, 3E6J), shark (4HGK), chicken (4P48), camel (1ZVY), cow (4K3D), and human (1N8Z) antigen binding fragments. Key distinguishing features are that lampreys utilize a leucine rich repeat scaffold, as opposed to other species which use the immunoglobulin domain. Shark and camelid fragments are devoid of light chains, with camelids often using a longer disulfide-bonded CDR H3. Chickens have novel canonical classes of CDR H1, and disulfides in CDR H3. Cows have evolved an ultralong CDR H3 repertoire with a disulfide bonded knob that sits atop a β-ribbon stalk.
Chicken
The chicken has been a model organism for studying B-cell development for over half a century. The role of the bursa of Fabricius in B-cell development was revealed when bursectomized chicks failed to produce antibodies [18,19]. In fact, the name ‘B-cell’ is derived from this discovery regarding the importance of the bursa. Subsequent work revealed B-cell and repertoire developmental pathways in birds. Chickens have serum IgM, IgA, and IgY, the first two of which are homologs of their mammalian counterparts, however they do not have IgE or IgD [20]. The IgY appears related to both mammalian IgG and IgE [3], and may be an evolutionary common ancestor to both [21].
Genetics of chicken repertoire generation
Chickens utilize a diversity creating process that is distinctly unique compared to humans or mice. Chicken antigen receptor genes undergo a single V(D)J recombination event, followed by gene conversion utilizing multiple upstream V-region pseudogenes [3] (Figure 3a). Rearranged variants of the pseudogenes can further diversify the CDR H3 region by inserting sequence into the DH region. Interestingly, the gene conversion process is dependent on the activation-induced cytidine deaminase (AID) enzyme [22], the same factor which is required for performing somatic hypermutation (SH) and class switch recombination in other species [23]. While framework diversity is limited in chicken antibodies [24••], probably due to the need for significant homology at the DNA level for efficient gene conversion, the potential diversity of creating different CDR combinations and content through multiple theoretical gene conversion recombination events is enormous. Although not discussed in detail here, rabbits also use a gene conversion strategy to diversify their antibody repertoire [25].
Figure 3.
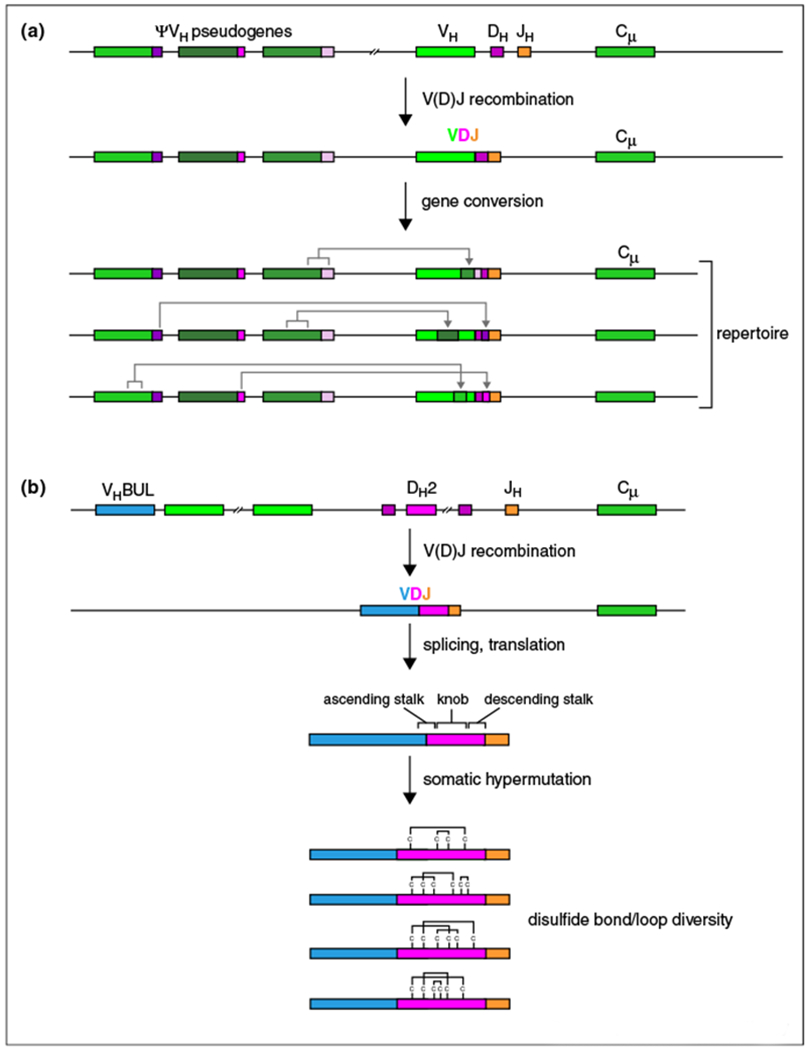
Novel processes for producing genetic diversity in chickens and cows. (a) Unlike humans or mice, chickens use a gene conversion strategy to create a diverse antibody repertoire. A single VDJ event occurs on the heavy chain locus, followed by gene conversion using sequence homology from a number of 5′ pseudogenes (ψVH) which can be in rearranged form and donate diverse genetic fragments into the rearranged V region. (b) Ultralong CDR H3 and repertoire development in the cow. Cow antibodies with ultralong CDR H3s appear to use only one VH region, VHBUL, which recombines with a long D region, DH2 to produce a germline VDJ recombined V-region. The single rearrangement then undergoes somatic hypermutation which may mutate residues to or from cysteine, changing the disulfide patterns of the repertoire, in addition to adding sequence diversity.
Structure of chicken antibodies
While chickens have been a model system for repertoire and lymphocyte development, few detailed structural studies on chicken antibodies have been performed. In repertoire analysis by gene sequencing, Wu et al. analyzed the amino acid content of chicken heavy chains [24••]. Interestingly, cysteine content was substantially higher in chicken CDRs (9.4%) compared to mice (0.25%) or humans (1.6%). They also identified six families of putatively different disulfide patterns, which may include disulfide bonds within CDR H3, or between CDR H3 and CDR H1 or CDR H2. Tyrosine, an amino acid reportedly important and abundant in antibody CDRs [26–28], is found less frequently in chicken CDR H3 (9.2%) compared to humans (16.8%). While on average chicken CDR H3s are not longer than humans or camelids, certain CDR H3s may form longer and unique disulfide stabilized structures. Selection of high affinity antibodies of these unusual structural subtypes could be accomplished in phage-based discovery systems [24••]. Future crystallography of examples of these antibody fragments, and their comparison with typical human CDR H3s or the unusual cow antibody CDR H3s described below will be an interesting comparative study in this regard.
To date only three structures are available for chicken antibody fragments. In structural studies of two chicken scFvs Conroy et al. identified new unique canonical classes of CDR L1, and a disulfide bonded CDR H3 [29•]. The extended loop of L1 along with disulfide bonded CDR H3 could present a unique paratope that is structurally distinct from mouse or human antibodies and, along with the sequence analysis of Wu et al. strongly suggests that chickens may have a novel repertoire of paratopes.
Engineering chicken antibodies
As a model system for immunology, chicken antibodies have been made for multiple research applications [30]. Chicken IgY can be made by several companies using immunization, and phage display of chicken antibody fragments has been accomplished against multiple antigens [31–33]. From a protein engineering standpoint, the DT40 cell line has been manipulated to serve as a host for SH of antigen receptor genes [34], and human pseudogenes have been engineered into DT40 and used to create human repertoires using gene conversion, a process where a donor DNA sequence replaces a homologous sequence in the genome [35]. In this regard, chickens with the antibody loci knocked out have been produced [36•], which may enable further transgenic engineering of human antibody V-regions or pseudogenes to produce transgenic chickens with human antibodies that utilize the novel gene conversion diversity generating systems of birds.
Camels
Camels have conventional hetero-tetrameric antibodies with identical heavy chains paired with identical light chains. These canonical IgG molecules represent approximately 25% of total circulating antibodies [37]. In a significant departure from other mammals, camels also have an unusual antibody type that is similar to a conventional IgG molecule but has identical heavy chains that lack the CH1 domain and does not pair with light chains. These heavy-chain only antibodies represent a significant fraction of the immunoglobulins in the serum, constituting up to 75% of total proteins binding to a protein A column. These molecules have been termed heavy-chain antibodies (HCAbs) and have a dedicated variable domain referred to as the VHH. The VHH domains are structurally and functionally similar to an Fv fragment derived from typical IgG molecules but have only three CDR variable loops to define the antigen binding surface. HCAbs have a molecular weight of about 90 kDa compared to the conventional IgG molecular weight of about 150 kDa [38]. Another differentiating feature of the VHH from canonical IgGs are the long loops that comprise the CDR H3. These long CDR H3 regions may enable VHH’s to interact with and inhibit unusual targets or epitopes not available to the flat binding surface of conventional antibodies such as enzyme active sites or other recessed crevices [39]. Both alpacas and llamas also have HCAbs that are very similar to those found in camels [40].
Camel antibodies have received much interest due to the small size of the VHH, robust biophysical characteristics and unusual antigen binding surface. Currently 5 VHH domains are in clinical development by Ablynx (Ghent, Belgium) and many more are in the early research phase in both academia and the pharmaceutical industry [12].
Genetics of camel repertoire generation
Camel HCAbs share the same gene locus as their conventional IgG tetrameric counterpart [41]. In addition, both HCAbs and IgGs have dedicated variable region genes encoded in germline sequences and undergo classical V(D)J recombination [42]. Repertoire diversity is driven by over 30 unique variable region (VHH) sequences, possible unique splicing events of the mRNA, and promiscuous V genes that can produce either VH (which will also pair with VL molecules) or VHH domains, each of which can undergo SH to produce further diversity [42–44].
Structure of HCAbs
Camel homodimeric HCAbs have an unusual structure compared to conventional IgG molecules. These HCAbs lack the CH1 domain, and the VHH variable region is not compatible with pairing to light chains, resulting in an antibody structure with a molecular weight under 100 kDa. A G to A point mutation that disrupts a consensus splicing sequence may be the cause for HCAbs lacking a CH1 domain. This sequence is located at the 5′ end of the intron between the CH1 hinge-side of the exon and increases the propensity to splice out the CH1 portion of the mRNA [45]. Different isoforms of these HCAbs have been identified and classified by the length of the hinge region sequence between the VHH domain and the CH2 domain. Shorter hinge length isoforms are referred to as IgG3 and the longer hinge regions as IgG2 [38].
Camel HCAbs have unique characteristics that differentiate them from canonical IgGs. Specific mutations in conserved locations are prevalent in HCAbs to possibly compensate for the lack of CH1 and light chains. For example, the mutation Leu11Ser, replaces interactions that classically make contact with the CH1 domain. While protein sequences of VH and VHH framework and hypervariable regions reveal similar structural organization, amino acids at key positions in VHH that define the VL/VH interface were substituted at Val37, Phe42, Gly44, Leu 45, Trp47, Glu49, Arg50 and Gly52 with smaller amino acids in VHH relative to wild-type VH regions that form heterodimers [46,47]. Additionally, a co-crystal structure of a camel VHH complexed with lysozyme highlights an unusually large convex H3 derived paratope that bound inside of the enzyme active site [48••]. The longer H3 loops may provide added surface area to compensate for the loss of the three VL CDR loops that classically contribute to antigen contacts [39] and can achieve binding affinities similar to conventional IgGs [49]. VHH CDR H3 loops are not only longer than canonical IgG H3’s but also have a significantly greater prevalence of intramolecular disulfide bonds that potentially define specific paratope topology [50]. Disulfide bonds form between cysteine residues located at H1 and H3 and between H3 and framework region 2 (Figure 4a). Cysteine is generally limited to positions 30, 32 and 33 in H1, position 50 in FR2 and can occur in several different positions throughout H3 [51]. The constrained disulfide structures may help stabilize the H3 antigen-loop complex, potentially leading to stronger antigen interactions. The additional stability of the H3 region may reduce the entropic cost of restricting the conformation of a flexible loop, lowering the energy barrier for antigen contact and association [52••], and also may help compensate for the lack of electrostatic interaction classically found in IgG H3 loops between Arg or Lys94 and Asp101 (by Kabat numbering) [53,46].
Figure 4.
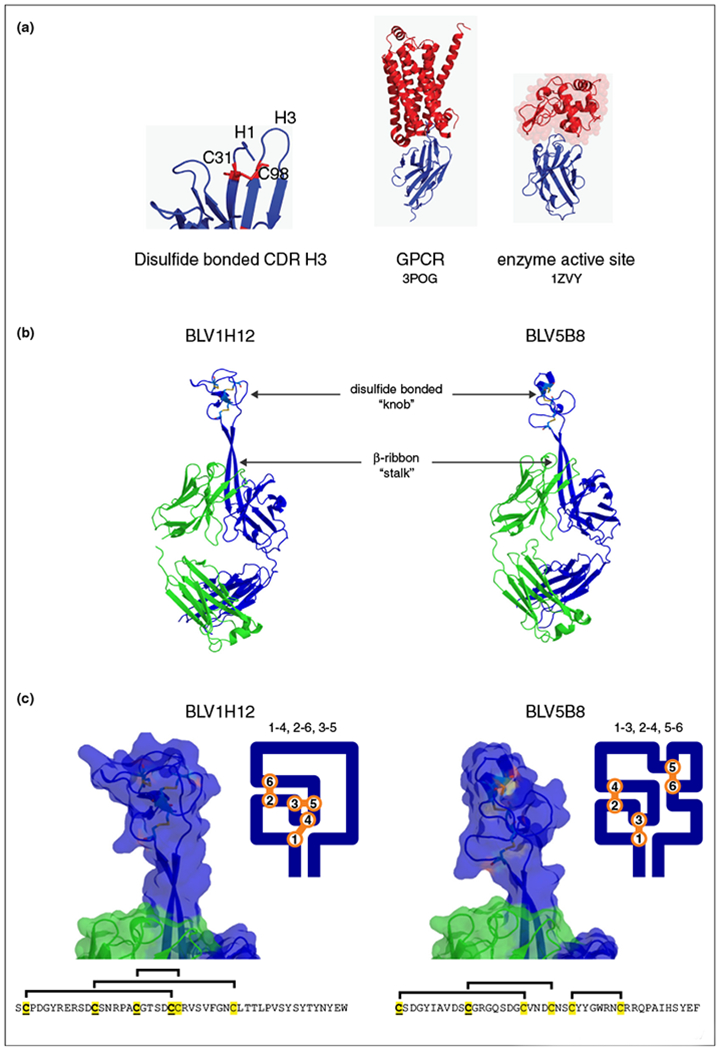
Protruding CDR H3 structures of camelid and cow antibodies. (a) CDR H3 structure and epitope binding of camel antibody fragments. (Left) Closeup of the structure of a camelid Fv, showing the long CDR H3 loop with a disulfide bond between CDR H3 and CDR H1. Unique epitopes bound by camelid Fvs, including the β-adrenergic GPCR (middle) and enzymatic active site of lysozyme (right). Note the protruding CDR H3 which reach into concave epitopes on each of these antigens. (b) Structures of two cow Fab fragments, BLV1H12 and BLV5B8, showing ultralong CDR H3s that are composed of a β-ribbon ‘stalk’ and disulfide bonded ‘knob’ structural minidomains. (c) Closeup views of the surface of the knob domains of BLV1H12 and BLV5B8. The sequences of the knobs are shown below, and a two-dimensional representation of the loops are shown to the right. The disulfide patterns of the two knobs are different, as indicated above the sequences and in the two dimensional loop schematic.
In addition to the unusual structure that potentially enables unique target access, VHH domains are thermodynamically stable and reversibly melt without common complications such as aggregation and loss of binding activity after refolding. While capable of retaining activity after refolding from denaturation, VHH domains do not exceed the thermostability of human subclass VH3 domains [54].
Engineering HCAbs
The small size, stability and ability to tightly bind to antigens have made the industrialization of camel VHH domains, termed ‘nanobodies’, of great interest [55]. Therapeutic applications of nanobodies and most foreign proteins may require some degree of engineering to incorporate as much human-like sequence as possible to minimize potential immunogenicity. Several camel VHH domain sequences have been shown to be homologous with the human VH3 and VH4 germline sequences [39,43]. The high degree of homology between the camel VHH framework sequences with those of the human sequences may aid or even bypass the need to engineer and humanize [44]. Regardless, in an effort to reduce potential immunogenicity, several key conserved residues in the camel FR2 region have been explored by mutating to the human consensus amino acids. Interestingly, biophysical data on several of these engineered molecules such as Glu49Gly and Arg50Leu resulted in more thermodynamically robust constructs while other mutations in FR2 negatively affected antigen binding due to impact of the H3 loop position [55]. H3 loop length is also correlated with the characteristics of surrounding residues on the scaffold and perhaps should be considered when engineering camel VHH domains [56].
There are several examples of camel antibodies with unique characteristics, including many with clinical applications [57•]. Of particular note, the small size of the VHH and its associated longer CDR H3 may allow binding to antigens or epitopes not easily accessible by typical mouse or human antibodies. In this regard, crystallization of the active state β2 adrenoceptor, a multipass transmembrane GPCR, was facilitated by a specific camel antibody fragment, an achievement that produced a key component of the knowledge that ultimately was awarded the Nobel Prize in chemistry in 2012 in the area of GPCR structure and function [58•] (Figure 4a, middle).
Standard molecular library techniques, particularly phage display, have been utilized to discover and engineer camelid antibodies [57•]. Immunization of an animal followed by construction of a nanobody phage display library result in discovery of antigen specific binders [59]. However, removal of large animal immunization can allow rapid molecular evolution techniques in the laboratory if large ‘naïve’ libraries are constructed. Monegal et al., 2009 constructed a large naïve llama phage display library that enabled identification of constituents with high affinity binding to FGFR1 [60]. In a related approach, a synthetic phage display nanobody library was built based on a conserved camel single-domain antibody fragment (VHH) framework and diversity was introduced into the CDR H3 by randomization using synthetic oligonucleotides [61]. While immunization and phage display approaches have met with success in identifying specific binders, Eschrichia coli display with camel VHH’s has also resulted in the identification of high affinity binders [62].
Several camel VHH domain antibodies have been discovered and are in early preclinical development in areas including oncology, infectious, inflammatory and neurodegenerative diseases. Ablynx currently has five humanized camel VHH domains in clinical development. Clinical targets include von Willibrand’s Factor, TNF-α, IL-6, RANKL and Respiratory Syncytial Virus [12,49,60,63]. Camel VHH’s have also been identified that can neutralize toxins such as the botulinum neurotoxin serotype E [64].
Shark
Nearly half a billion years ago the adaptive immune system based on immunoglobulins, T cell receptors and the major histocompatibility complex evolved in cartilaginous fish (reviewed in [65]). Thus, sharks represent man’s most distant cousins that share the fundamental components of ‘our’ immune system. There are, however, some interesting differences between the lymphocyte antigen receptors of shark and man.
Sharks have three IgH chain isotypes. IgM (μ) appears developmentally and ontologically ancestral and is found in sharks and nearly all jawed vertebrates, with the coelacanth being the only known exception [66]. IgW (ω) is also extant in sharks and orthologous to IgD of other vertebrates, and there is good evidence for a form of class switch recombination between IgM and IgW [67]. Lastly, IgNAR is a cartilaginous fish lineage-specific isotype that does not associate with light chains [68]. Unlike humans or mice which have two light chain classes, in cartilaginous fish IgM and IgW have four IgL with which to heterodimerize: λ, κ, σ, and σ-cart [69]. Sharks have the α, β, γ and δ T cell receptor chains found in other vertebrates [70], and employ the doubly rearranging NARTCR form of TCRd that includes a variable domain very similar to that of the IgL-less Ig-NAR antibody [71]. As a unique antigen receptor, we will focus on the IgL-less IgNAR immunoglobulin isotype, as it has generated much interest beyond comparative immunologists.
Genetics of IgNAR repertoire generation
Although humoral and cellular lymphocyte lineages with diverse antigen receptor repertoires exist in the older, jawless lamprey and hagfish [6], V(D)J recombination of immunoglobulin and T cell receptor variable domain genes is a shared characteristic of the jawed vertebrates (reviewed in [72]).
Shark TCR are in the typical translocon arrangement [73], but their Ig loci exist in a multiple cluster organization throughout the genome [74] making class switch and successful haplotype exclusion [75] especially intriguing. Some of the Ig loci of cartilaginous fish are partially (VDJ) or completely (VDJ) germline joined [76], that is, the animal inherited the locus in a rearranged state from its parents. The fusion of V, D, and J elements in the germline (as opposed to a somatically developing lymphocyte) is a product of gonadal RAG expression in sharks [77]. Whether these preloaded paratopes bind antigens of particularly common or mortal pathogens of elasmobranchs still needs to be determined, but one germline-joined IgM is preferentially used in young sharks [78]. Although shark IgH arise from simpler loci with fewer elements, sequence differences between clusters and junctional diversification by N and P additions produce a repertoire as diverse as that of other vertebrates [79,80].
The variable domains of IgNAR share as much homology with other TCR as Ig (thus the moniker new antigen receptor). It has been suggested that IgNAR may have arisen evolutionarily via the invasion of an IgW cluster by a V gene of NARTCR [81]. More clusters of Ig often exist than IgNAR, yet upon antigen exposure IgNAR is heavily mutated [82]. Low IgNAR levels in young sharks, the slow rise of serum levels of IgNAR in the first year of shark development, and the clear affinity maturation of the molecule have suggested that this isotype serves a role in the shark immune battery analogous to mammalian IgG and that it may be T dependent [83,84]. A memory response characterized by specific antigen production (with secondary response kinetics after the primary response has resolved upon boost without adjuvant) is clearly capable with the IgNAR isotype in nurse shark [85]. SH in sharks may proceed through novel mechanisms as tandem non-template mutations of 2–5 contiguous bases are common [86,87].
Structure of IgNAR
Surface IgNAR has one amino-terminal V domain and either three or five constant domains. The first and third constant domains homodimerize and lend to the antibody stability, and the resolution of the constant region structure showed stabilizing motifs that also stabilize mammalian antibodies when engineered into them [88]. IgNAR can exist as a monomer but at least in the spiny dogfish has been found to multimerize [89]. The variable domains of IgNAR are unusual in that CDR H2 can form a belt around the side of the domain for a structure similar to C1-type immunoglobulin superfamily domains [90]. This peripheral orientation of CDR H2 to the antigen binding site is probably not always the case though, as IgNAR CDR H2 can have selected hypermutations and can be important for antigen binding [91•].
The single V domain antibody general quaternary structure has independently evolved at least twice in vertebrate natural history, in cartilaginous fish and camelids [92•]. There are genetic differences in these structural and analogic convergences, however. IgNAR is encoded at dedicated loci, whereas camelids use an IgG variant that has evolved to encode structural modifications to eschew the IgL [38]. It has been suggested that comparative immunology studies in more diverse clades will discover more vertebrate groups that utilize the single V antibody strategy [93].
Whereas the mobility and tissue-penetrance of canonical hetero-tetrameric antibodies are constrained by their large size, this is less of a problem for the 12 kDa IgNAR homodimer and less still for the IgNAR V domain by itself. This IgNARV is to date the smallest antigen binding domain known in the animal kingdom [94]. An increased frequency of polar and charged amino acids at the solvent exposed regions corresponding to the traditional VH–VL interface makes IgNAR particularly soluble.
Libraries and engineering
The relative stability of the IgNAR IgH homodimer and the somewhat simpler genetics encoding the single variable domain have fueled applied research to adapt this molecule to biotechnology and immunotherapy. Much is known of the optimal immunization protocols necessary for affinity matured humoral IgNAR responses in several elasmobranch species [95]. Phage display technologies have been easily adapted to analyze IgNAR antibodies from immunized sharks [96]. These technologies have been used to create recombinant shark antibodies to many pathogens of human health relevance, including Ebola [97]. The immunotherapeutics arms of some pharmaceutical companies are now exploiting the unique properties of shark IgNAR to make target-specific single V domain antibodies [98]. Despite only (at most) four hypervariable regions to bind antigen (and CDR H2 sometimes does not contribute to the paratope) IgNAR can bind antigen with very high affinities [90,96]. Diverse intradomain disulfide binding patterns divide IgNAR V domains into four types [99,100], only one of which (type III) has not yet been shown to give rise to high affinity binders.
Many different species of shark have now been used to develop libraries and screened with different display technologies, including phage display [96,101], ribosome display [102] and yeast surface display [103]. These libraries can be created from naïve sharks, immunized sharks, or synthetic vNAR libraries, but immunized sharks have been the most successful source till date, possibly due to the large phylogenetic distance between shark and man, reducing the chance of target-specific tolerance of conserved antigens [94].
The amino acid identity between IgNAR and human IgHV domains can be as low as 25%, thus the immunogenic potential is great [96], and progress has recently been made towards IgNARV humanization. A shark vNAR domain against human serum albumin (HAS) was engineered by converting more than half of the framework amino acids to those of the human germline IgL kappa variable sequence DPK9 [91•]. This humanized molecule lost very little affinity over the product of the parental construct, and the humanized IgNARV displayed negligible immunogenicity in dendritic cell assays [104].
The smaller paratope of the shark IgNARV has contributed to its appeal for creating therapeutic binders for target antigen conformations that have been impervious to development of effective blocking or neutralizing antibodies by traditional paratopes. Discriminating binders of cholera toxin were identified from spiny and smooth dogfish naïve libraries, and these monoclonals were found to be more heat resistant than traditional mammalian cholera toxin reagents [105]. A shark IgNAR was also shown to be an effective ‘intrabody’ against the hepatitis B virus precore protein in the endoplasmic reticulum [106]. A list of notable IgNARV binders to relevant targets is shown in Supplementary Table 1 (adapted from [94,104]).
Cow
While sharks, camels, and chickens provide unique examples of alternate antibody structures both in terms of subunit composition (sharks, camels) and diversification mechanisms (chicken), recent discoveries in cow antibody structure and genetics reveal an unusual paradigm for creating both genetic and structural diversity. It has been known for quite some time, largely through the work of Kaushik and colleagues [107–113], that cow antibodies can have unusually long CDR H3 regions that can reach lengths of over sixty amino acids long. In fact, it appears that cows actually generate an ultralong CDR H3 repertoire alongside a shorter repertoire; but even this shorter repertoire is quite long compared to mouse or human antibodies, with cow ‘short’ H3 sequences often being over forty amino acids in length. In a departure from mouse and human antibodies on both a genetic and structural level, cows appear to use SH to create both amino acid content as well as disulfide bonded loop pattern diversity within the ‘stalk’ and ‘knob’ minidomains of their ultralong CDR H3.
Genetics of cow ultralong CDR H3 repertoire generation
Cows have limited combinatorial diversity potential with only 12 VH regions that comprise one highly homologous heavy chain family (compared to seven in humans) which is related to the human VH4 family [114–116,117•]. Cows perform V(D)J recombination similarly to other species, however the ultralong subset of cow antibodies appears to preferentially use a single VH (termed VHBUL) and an ultralong DH2 [5••]. While the cow genome project has been published [118], the difficulty in assembly and annotation of the heavy chain locus leaves open the possibility that alternative VH or DH regions could still be discovered. Even if this were the case, deep sequencing analysis reveals that any undiscovered VH or DH regions would be highly similar to VHBUL and DH2 [5••]. Unlike mice or humans, several species like cows and sheep are unusual in activating SH during development of the naïve primary repertoire, with AID induced mutations compensating for the limited V(D)J combinatorial diversity [117•,119,120]. An unusual feature of cow germline DH regions is that they encode multiple cysteines. In the ultralong DH2 region a repeat of Gly-Tyr-Gly or Gly-Tyr-Ser sequences use the relatively uncommon codons GGT (for Gly) and AGT (for Ser) and TAT (for Tyr), each of which can be mutated to cysteine with one nucleotide change [5••]. Furthermore, there are numerous RGYW SH ‘hotspots’ throughout the DH region, making it potentially highly mutagenic. Thus, this cow DH region appears to be primed to mutate to cysteine through AID induced SH. The DH2 encodes four cysteines, which could potentially form two disulfide bonds. Cow antibody sequences, however, contain multiple cysteines that do not appear to align with those in DH2. This lack of clear conservation to the germline was resolved when deep sequencing analysis revealed mutations both to and from cysteine in antibody sequences undergoing SH [5••]. Alignment of CDR H3 sequences with the germline DH2 revealed almost complete conservation of the first cysteine with progressively less conservation of the three remaining germline cysteines moving towards the C-terminus. Additionally, multiple ‘new’ cysteines that were generated somatically could be identified. Importantly these results suggest a novel mechanism for structural diversity generation; mutating to and from cysteines can alter the disulfide patterns in CDR H3, resulting in wholesale changes in loop structures and compositions [5••] (Figure 3b).
Structure of cow antibodies
Wang et al. solved the structures of two bovine antibody Fab fragments which contained ultralong CDR H3s [5••]. These two fragments, BLV1H12 and BLV5B8, were originally cloned from B-cells transformed with Bovine Leukemia Virus and their cognate antigens are not known [110]. While both had ultralong CDR H3s, their sequences in the CDR H3 regions were highly dissimilar. Remarkably, however, both structures had two unusual motifs never seen before in antibody CDR H3 structures: a β-ribbon ‘stalk’ that protruded far from the traditional antibody paratope, and a distal disulfide bonded ‘knob’ which rested upon the stalk (Figure 4b and c). This ‘stalk and knob’ feature is reminiscent of a mushroom protruding out from the surface of the antibody (Figure 4b). Both CDR H3s had six cysteines, however their positions were not conserved. When the first cysteine was ‘fixed’ and aligned with the germline DH2 region, three of the cysteines could be aligned positionally, however only the first two were conserved with the DH2 germline. The structures clearly revealed different disulfide bonding patterns in the knobs of BLV1H12 and BLV5B8 where the former had a 1–4, 2–6, 3–5 pattern and the latter had 1–3, 2–4, 5–6 (Figure 4c). Despite the unique ‘stalk and knob’ features, the sequences, disulfide bonding pattern, surface shape and charge, and loop lengths within the knob were highly dissimilar between the two antibody fragments. Structurally, the remaining five CDRs and variable region framework regions were nearly superimposable and had little sequence variation. Thus the diversity of these cow antibodies appears to all reside within the ultralong CDR H3 region. Deep sequencing also revealed that most sequences contained an even number of cysteines at diverse positions, further strengthening the hypothesis that disulfide bonds, and their associated loops, are an important component of the structural repertoire [5••]. The unique ability to use a single germline genetic construct as a template to mutationally derive different disulfide patterns, and thus minifolds, is a novel way to create structural diversity in an antigen binding repertoire (Figure 3b).
Engineering cow antibodies
Compared to other species discussed above, little work has been done on discovery and engineering bovine antibodies. Phage display has been successful using cow antibody repertoires [121], however it is unclear whether the multiple disulfide bonds found in the knob of ultralong CDR H3s can be successfully selected using prokaryotic based systems which may lack the machinery to efficiently secrete and fold these complex molecules. An antigen specific ultralong CDR H3 antibody targeting the NS2-3 protein of Bovine Diarrheal Virus was expressed and its binding properties analyzed by site directed mutagenesis [5••]. All of the binding was found to reside within the ultralong CDR H3 ‘knob’ domain, with the other CDRs being irrelevant for antigen binding. Given the unusual paratope of ultralong CDR H3s, engineering efforts in replacing the ‘knob’ with known bioactive peptides have been undertaken. Interestingly, the knob could be replaced by G-CSF (granulocyte colony stimulating factor) [122] and erythropoietin [123], with biological function of the engineered protein being maintained. Liu et al. replaced the knob domain of BLV1H12 with a β-hairpin peptide known to bind CXCR4, producing a molecule with high affinity and activity [124]. This ‘knob replacement’ engineering could endow small peptides with long half-life and effector function of traditional antibodies, and may be a way to improve dosing regimens for some recombinant therapeutic proteins. Humanization of the bovine scaffold has not yet been reported, however the homology with human VH4 suggests this may be feasible, albeit with the caveat that the other cow CDRs besides H3 may play an important structural and stability role through their contacts with the ultralong ‘stalk’. The structure of the ultralong CDR H3, whose knob is of similar size and shape to other small disulfide bonded peptides like knottins, chemokines, toxins, protease inhibitors, and defensins, certainly suggests that unique epitopes could be bound by these unusual antibodies.
Conclusions
Different species have taken unique approaches to create diverse antigen receptor repertoires. At the genetic level, while V(D)J recombination is a key molecular mechanism amongst all jawed vertebrates, its role in the actual creation of diversity ranges from providing a template for gene conversion with a single V(D)J event in chickens, to being a major source of combinatorial diversity in mice and humans through unique V(D)J events in each B-cell. Similarly, the AID enzyme is primarily utilized to create the secondary repertoire by SH in mice and humans, but appears to play a major role in creating the primary repertoire in cows and perhaps other species [117•].
From a structural standpoint, mice and humans use a heterotetrameric IgG to bind antigens, with diversity occurring in all six CDRs, and combinatorial and junctional diversity focused on the CDR H3 and L3. These paratopes typically form a flat or undulating binding surface for contacting antigen. By contrast, camels and sharks have convergently developed heavy chain antibodies devoid of light chains. In an additional twist, the cow ultralong CDR H3 repertoire appears to contain all of its diversity within CDR H3, and none in the other five CDRs which provide a structural role for the CDR H3 ‘stalk’. Although cows use a light chain, the ultralong repertoire is limited in using a single light chain VL which pairs with the single VHBUL V-region. Chickens appear to have unique CDR L1 structures, and similarly to camelids, have disulfide bonds within CDR H3 or between H3 and H1 or H2. While the limited structural work done thus far shows major differences in antibodies from different species, further study on these different species should shed light on just how different the paratope repertoires are from one another as well as human and mouse antibodies.
Why significantly different genetic and structural strategies have evolved in different species is an interesting question. It appears as though both shark and camelids have evolved heavy chain-only antibodies in a convergent manner. Similarly, the strategy of using gene conversion appears to also have convergently evolved in both chickens and rabbits. Thus, both the genetic mechanism of gene conversion and the structural paradigm of heavy chain only antibodies must have certain evolutionary advantages, however what these may be are currently unclear. The structural differences between antibodies of different species suggest that different antigen structures (or structural classes) may have applied evolutionary pressure on individual species to develop alternative paratopes to enable an effective immune response. It has been suggested that the smaller Fv binding unit of camelid HCAbs may enable binding to unique concave epitopes, and camelid antibodies with protruding CDR H3s have been raised against difficult GPCR and enzyme active site epitopes (Figure 4a). The ‘knob’ regions of ultralong CDR H3 cow antibodies, which are even smaller than a nanobody, might be able to similarly ‘reach’ into such concave epitopes. Of note, both camelids and cows are ruminants which have substantial antigen load in their rumen stomach compartment that is made up of high titers of both prokaryotic and eukaryotic microorganisms [125–128]. Whether these microorganisms provided evolutionary pressure to develop novel antibody repertoires in cows and camels will require significant further study.
Given the enormous importance of antibodies in research and medicine as reagents, diagnostics, and therapeutics, the potential use of the immune system of alternative species could provide unique molecules that may have biochemical properties not present in antibodies derived from classic species like humans or mice. The paratopes provided by unique L1s, disulfide bonded or ultralong CDR H3s, or antibody Fv regions devoid of light chains certainly will allow different modes of binding to antigens, or even enable binding to certain epitope structures. Additionally, distant species may allow a more robust immune response to human antigens, due to their evolutionary distance and potential ability to overcome tolerance. Given these unique advantages, future efforts in discovery of antibodies and study of alternative species’ immune systems is sure to continue to uncover novel mechanisms for antibody diversity generation as well as further variations on the structural theme of how to design an antibody repertoire. With the recent progress in deep sequencing antibody repertoires and whole genomes, detection of novel features of antibody repertoires in diverse species could progress rapidly in the near future [129,130].
Supplementary Material
Acknowledgement
This work was supported by NIH Grant 1R01GM105826-01A1 to VS.
Footnotes
Conflict of interest statement
MdlR and VS have equity interests in Sevion Therapeutics which is developing cow antibodies.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.sbi.2015.06.002.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Mi QS, Zhou L, Schulze DH, Fischer RT, Lustig A, Rezanka LJ, Donovan DM, Longo DL, Kenny JJ: Highly reduced protection against streptococcus pneumoniae after deletion of a single heavy chain gene in mouse. Proc Natl Acad Sci U S A 2000, 97:6031–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z: Diversity and function of adaptive immune receptors in a jawless vertebrate. Science 2005, 310:1970–1973. [DOI] [PubMed] [Google Scholar]
- 3.Parvari R, Avivi A, Lentner F, Ziv E, Tel-Or S, Burstein Y, Schechter I: Chicken immunoglobulin gamma-heavy chains: limited VH gene repertoire, combinatorial diversification by D gene segments and evolution of the heavy chain locus. EMBO J 1988, 7:739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tonegawa S: Somatic generation of antibody diversity. Nature 1983, 302:575–581. [DOI] [PubMed] [Google Scholar]
- 5.••.Wang F, Ekiert DC, Ahmad I, Yu W, Zhang Y, Bazirgan O, Torkamani A, Raudsepp T, Mwangi W, Criscitiello MF, Wilson IA et al. : Reshaping antibody diversity. Cell 2013, 153:1379–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]; First structural description of ultralong CDR H3 antibodies in cattle, as well as the genetics and deep sequencing of the ultralong repertoire.
- 6.Pancer Z, Amemiya CT, Ehrhardt GR, Ceitlin J, Gartland GL, Cooper MD: Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 2004, 430:174–180. [DOI] [PubMed] [Google Scholar]
- 7.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG: The rag proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol 2000, 18:495–527. [DOI] [PubMed] [Google Scholar]
- 8.Smider V, Chu G: The end-joining reaction in V(D)J recombination. Semin Immunol 1997, 9:189–197. [DOI] [PubMed] [Google Scholar]
- 9.Nelson AL, Dhimolea E, Reichert JM: Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov 2010, 9:767–774. [DOI] [PubMed] [Google Scholar]
- 10.Reichert JM, Rosensweig CJ, Faden LB, Dewitz MC: Monoclonal antibody successes in the clinic. Nat Biotechnol 2005, 23:1073–1078. [DOI] [PubMed] [Google Scholar]
- 11.Kohler G, Milstein C: Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256:495–497. [DOI] [PubMed] [Google Scholar]
- 12.Strohl WR: Antibody discovery: sourcing of monoclonal antibody variable domains. Curr Drug Discov Technol 2014, 11:3–19. [DOI] [PubMed] [Google Scholar]
- 13.Mao H, Graziano JJ, Chase TM, Bentley CA, Bazirgan OA, Reddy NP, Song BD, Smider VV: Spatially addressed combinatorial protein libraries for recombinant antibody discovery and optimization. Nat Biotechnol 2010, 28:1195–1202. [DOI] [PubMed] [Google Scholar]
- 14.Hutchings CJ, Koglin M, Marshall FH: Therapeutic antibodies directed at G protein-coupled receptors. MAbs 2010, 2:594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J-H, Park C-K, Chen G, Han Q, Xie R-G, Liu T, Ji R-R, Lee S-Y: A monoclonal antibody that targets a NaV1.7 channel voltage sensor for pain and itch relief. Cell 2014, 157:1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Kwong PD, Wilson IA: HIV-1 and influenza antibodies: seeing antigens in new ways. Nat Immunol 2009, 10:573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R et al. : A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 2011, 334:1097–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper MD, Peterson RDA, South MA, Good RA: The functions of the thymus system and the bursa system in the chicken. J Exp Med 1966, 123:75–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glick B, Chang TS, Jaap RG: The bursa of fabricius and antibody production. Poultry Sci 1956, 35:224–225. [Google Scholar]
- 20.Ratcliffe MJ: Antibodies, immunoglobulin genes and the bursa of fabricius in chicken B cell development. Dev Comp Immunol 2006, 30:101–118. [DOI] [PubMed] [Google Scholar]
- 21.Warr GW, Magor KE, Higgins DA: IgY: clues to the origins of modern antibodies. Immunol Today 1995, 16:392–398. [DOI] [PubMed] [Google Scholar]
- 22.Arakawa H, Hauschild J, Buerstedde JM: Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 2002, 295:1301–1306. [DOI] [PubMed] [Google Scholar]
- 23.Fugmann SD, Schatz DG: One aid to unite them all. Science 2002, 295:1244–1245. [DOI] [PubMed] [Google Scholar]
- 24.••.Wu L, Oficjalska K, Lambert M, Fennell BJ, Darmanin-Sheehan A, Ní Shúilleabháin D, Autin B, Cummins E, Tchistiakova L, Bloom L, Paulsen J et al. : Fundamental characteristics of the immunoglobulin VH repertoire of chickens in comparison with those of humans, mice, and camelids. J Immunol 2012, 188:322–333. [DOI] [PubMed] [Google Scholar]; Detailed amino acid analysis of chicken CDR H3s including groups based on potential disulfide bond patterns.
- 25.Becker RS, Knight KL: Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell 1990, 63:987–997. [DOI] [PubMed] [Google Scholar]
- 26.Fellouse FA, Wiesmann C, Sidhu SS: Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc Natl Acad Sci U S A 2004, 101:12467–12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanov II, Schelonka RL, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW: Development of the expressed ig CDR-H3 repertoire is marked by focusing of constraints in length, amino acid use, and charge that are first established in early B cell progenitors. J Immunol 2005, 174:7773–7780. [DOI] [PubMed] [Google Scholar]
- 28.Nikula TK, Bocchia M, Curcio MJ, Sgouros G, Ma Y, Finn RD, Scheinberg DA: Impact of the high tyrosine fraction in complementarity determining regions: measured and predicted effects of radioiodination on IgG immunoreactivity. Mol Immunol 1995, 32:865–872. [DOI] [PubMed] [Google Scholar]
- 29.•.Conroy PJ, Law RHP, Gilgunn S, Hearty S, Caradoc-Davies TT, Lloyd G, O’Kennedy RJ, Whisstock JC: Reconciling the structural attributes of avian antibodies. J Biol Chem 2014, 289:15384–15392. [DOI] [PMC free article] [PubMed] [Google Scholar]; Structures of chicken scFv antibody fragments showing novel CDR L1 canonical structures.
- 30.Spillner E, Braren I, Greunke K, Seismann H, Blank S, du Plessis D: Avian IgY antibodies and their recombinant equivalents in research, diagnostics and therapy. Biologicals 2012, 40:313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies EL, Smith JS, Birkett CR, Manser JM, Anderson-Dear DV, Young JR: Selection of specific phage-display antibodies using libraries derived from chicken immunoglobulin genes. J Immunol Methods 1995, 186:125–135. [DOI] [PubMed] [Google Scholar]
- 32.van Wyngaardt W, Malatji T, Mashau C, Fehrsen J, Jordaan F, Miltiadou D, du Plessis DH: A large semi-synthetic single-chain Fv phage display library based on chicken immunoglobulin genes. BMC Biotechnol 2004, 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamanaka HI, Inoue T, Ikeda-Tanaka O: Chicken monoclonal antibody isolated by a phage display system. J Immunol 1996, 157:1156–1162. [PubMed] [Google Scholar]
- 34.Magari M, Kanehiro Y, Todo K, Ikeda M, Kanayama N, Ohmori H: Enhancement of hypermutation frequency in the chicken B cell line DT40 for efficient diversification of the antibody repertoire. Biochem Biophys Res Commun 2010, 396:353–358. [DOI] [PubMed] [Google Scholar]
- 35.Schusser B, Yi H, Collarini EJ, Izquierdo SM, Harriman WD, Etches RJ, Leighton PA: Harnessing gene conversion in chicken B cells to create a human antibody sequence repertoire. PLoS One 2013, 8:e80108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.•.Schusser B, Collarini EJ, Yi H, Izquierdo SM, Fesler J, Pedersen D, Klasing KC, Kaspers B, Harriman WD, van de Lavoir M-C, Etches RJ et al. : Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc Natl Acad Sci U S A 2013, 110:20170–20175. [DOI] [PMC free article] [PubMed] [Google Scholar]; Illustrates early attempts at a transgenic approach that may ultimately harness the chicken diversity system for recombinant human antibodies.
- 37.Griffin LM, Snowden JR, Lawson AD, Wernery U, Kinne J, Baker TS: Analysis of heavy and light chain sequences of conventional camelid antibodies from camelus dromedarius and camelus bactrianus species. J Immunol Methods 2014, 405:35–46. [DOI] [PubMed] [Google Scholar]
- 38.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R: Naturally occurring antibodies devoid of light chains. Nature 1993, 363:446–448. [DOI] [PubMed] [Google Scholar]
- 39.Riechmann L, Muyldermans S: Single domain antibodies: comparison of camel VH and camelised human VH domains. J Immunol Methods 1999, 231:25–38. [DOI] [PubMed] [Google Scholar]
- 40.Harmsen MM, Ruuls RC, Nijman IJ, Niewold TA, Frenken LG, de Geus B: Llama heavy-chain V regions consist of at least four distinct subfamilies revealing novel sequence features. Mol Immunol 2000, 37:579–590. [DOI] [PubMed] [Google Scholar]
- 41.Achour I, Cavelier P, Tichit M, Bouchier C, Lafaye P, Rougeon F: Tetrameric and homodimeric camelid IgGs originate from the same IgH locus. J Immunol 2008, 181:2001–2009. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen VK, Hamers R, Wyns L, Muyldermans S: Camel heavy-chain antibodies: diverse germline V(H)H and specific mechanisms enlarge the antigen-binding repertoire. EMBO J 2000, 19:921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen VK, Muyldermans S, Hamers R: The specific variable domain of camel heavy-chain antibodies is encoded in the germline. J Mol Biol 1998, 275:413–418. [DOI] [PubMed] [Google Scholar]
- 44.Deschacht N, De Groeve K, Vincke C, Raes G, De Baetselier P, Muyldermans S: A novel promiscuous class of camelid single-domain antibody contributes to the antigen-binding repertoire. J Immunol 2010, 184:5696–5704. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen VK, Hamers R, Wyns L, Muyldermans S: Loss of splice consensus signal is responsible for the removal of the entire C(H)1 domain of the functional camel IGG2A heavy-chain antibodies. Mol Immunol 1999, 36:515–524. [DOI] [PubMed] [Google Scholar]
- 46.Muyldermans S, Atarhouch T, Saldanha J, Barbosa JA, Hamers R: Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng 1994, 7:1129–1135. [DOI] [PubMed] [Google Scholar]
- 47.Chothia C, Novotny J, Bruccoleri R, Karplus M: Domain association in immunoglobulin molecules. The packing of variable domains. J Mol Biol 1985, 186:651–663. [DOI] [PubMed] [Google Scholar]
- 48.••.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L: Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci U S A 2006, 103:4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]; Describes the crystal structure of a camel VHH domain that highlights a large convex paratope of the H3 that binds inside the active site of lysozyme.
- 49.Harmsen MM, De Haard HJ: Properties, production, and applications of camelid single-domain antibody fragments. Appl Microbiol Biotechnol 2007, 77:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sircar A, Sanni KA, Shi J, Gray JJ: Analysis and modeling of the variable region of camelid single-domain antibodies. J Immunol 2011, 186:6357–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conrath KE, Wernery U, Muyldermans S, Nguyen VK: Emergence and evolution of functional heavy-chain antibodies in camelidae. Dev Comp Immunol 2003, 27:87–103. [DOI] [PubMed] [Google Scholar]
- 52.••.Govaert J, Pellis M, Deschacht N, Vincke C, Conrath K, Muyldermans S, Saerens D: Dual beneficial effect of interloop disulfide bond for single domain antibody fragments. J Biol Chem 2012, 287:1970–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]; Camelid disulfide bonds stabilize the CDR H3 antigen binding loops and lead to stronger antigen interactions.
- 53.Kabat EA, Wu TT: Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J Immunol 1991, 147:1709–1719. [PubMed] [Google Scholar]
- 54.Ewert S, Cambillau C, Conrath K, Pluckthun A: Biophysical properties of camelid V(HH) domains compared to those of human V(H)3 domains. Biochemistry 2002, 41:3628–3636. [DOI] [PubMed] [Google Scholar]
- 55.Vincke C, Loris R, Saerens D, Martinez-Rodriguez S, Muyldermans S, Conrath K: General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem 2009, 284:3273–3284. [DOI] [PubMed] [Google Scholar]
- 56.Kastelic D, Soler N, Komel R, Pompon D: The global sequence signature algorithm unveils a structural network surrounding heavy chain CDR3 loop in camelidae variable domains. Biochim Biophys Acta 2013, 1830:3373–3381. [DOI] [PubMed] [Google Scholar]
- 57.•.Desmyter A, Spinelli S, Roussel A, Cambillau C: Camelid nanobodies: killing two birds with one stone. Curr Opin Struct Biol 2015, 32:1–8. [DOI] [PubMed] [Google Scholar]; Updated review of camelid nanobodies.
- 58.•.Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A et al. : Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 2011, 469:175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]; Application of a camelid nanobody to achieve a GPCR crystal structure.
- 59.Abbady AQ, Al-Mariri A, Zarkawi M, Al-Assad A, Muyldermans S: Evaluation of a nanobody phage display library constructed from a brucella-immunised camel. Vet Immunol Immunopathol 2011, 142:49–56. [DOI] [PubMed] [Google Scholar]
- 60.Monegal A, Ami D, Martinelli C, Huang H, Aliprandi M, Capasso P, Francavilla C, Ossolengo G, de Marco A: Immunological applications of single-domain llama recombinant antibodies isolated from a naive library. Protein Eng Des Sel 2009, 22:273–280. [DOI] [PubMed] [Google Scholar]
- 61.Yan J, Li G, Hu Y, Ou W, Wan Y: Construction of a synthetic phage-displayed nanobody library with CDR3 regions randomized by trinucleotide cassettes for diagnostic applications. J Translat Med 2014, 12:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salema V, Marin E, Martinez-Arteaga R, Ruano-Gallego D, Fraile S, Margolles Y, Teira X, Gutierrez C, Bodelon G, Fernandez LA: Selection of single domain antibodies from immune libraries displayed on the surface of E. Coli cells with two beta-domains of opposite topologies. PLoS One 2013, 8:e75126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei G, Meng W, Guo H, Pan W, Liu J, Peng T, Chen L, Chen CY: Potent neutralization of influenza A virus by a single-domain antibody blocking M2 ion channel protein. PLoS One 2011, 6:e28309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bakherad H, Mousavi Gargari SL, Rasooli I, Rajabibazl M, Mohammadi M, Ebrahimizadeh W, Safaee Ardakani L, Zare H: In vivo neutralization of botulinum neurotoxins serotype e with heavy-chain camelid antibodies (VHH). Mol Biotechnol 2013, 55:159–167. [DOI] [PubMed] [Google Scholar]
- 65.Flajnik MF, Rumfelt LL: The immune system of cartilaginous fish. Curr Top Microbiol Immunol 2000, 248:249–270. [DOI] [PubMed] [Google Scholar]
- 66.Amemiya CT, Alfoldi J, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, Manousaki T, Schneider I, Rohner N, Organ C et al. : The African coelacanth genome provides insights into tetrapod evolution. Nature 2013, 496:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu C, Lee V, Finn A, Senger K, Zarrin AA, Du Pasquier L, Hsu E: Origin of immunoglobulin isotype switching. Curr Biol 2012, 22:872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF: A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 1995, 374:168–173. [DOI] [PubMed] [Google Scholar]
- 69.Criscitiello MF, Flajnik MF: Four primordial immunoglobulin light chain isotypes, including lambda and kappa, identified in the most primitive living jawed vertebrates. Eur J Immunol 2007, 37:2683–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Criscitiello MF, Ohta Y, Saltis M, McKinney EC, Flajnik MF: Evolutionarily conserved tcr binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J Immunol 2010, 184:6950–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Criscitiello MF, Saltis M, Flajnik MF: An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc Natl Acad Sci U S A 2006, 103:5036–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Litman GW, Rast JP, Fugmann SD: The origins of vertebrate adaptive immunity. Nat Rev Immunol 2010, 10:543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen H, Kshirsagar S, Jensen I, Lau K, Covarrubias R, Schluter SF, Marchalonis JJ: Characterization of arrangement and expression of the T cell receptor gamma locus in the sandbar shark. Proc Natl Acad Sci U S A 2009, 106:8591–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hinds KR, Litman GW: Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature 1986, 320:546–549. [DOI] [PubMed] [Google Scholar]
- 75.Malecek K, Lee V, Feng W, Huang JL, Flajnik MF, Ohta Y, Hsu E: Immunoglobulin heavy chain exclusion in the shark. PLoS Biol 2008, 6:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kokubu F, Litman R, Shamblott MJ, Hinds K, Litman GW: Diverse organization of immunoglobulin VH gene loci in a primitive vertebrate. EMBO J 1988, 7:3413–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee SS, Fitch D, Flajnik MF, Hsu E: Rearrangement of immunoglobulin genes in shark germ cells. J Exp Med 2000, 191:1637–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rumfelt LL, Avila D, Diaz M, Bartl S, McKinney EC, Flajnik MF: A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc Natl Acad Sci U S A 2001, 98:1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kokubu F, Hinds K, Litman R, Shamblott MJ, Litman GW: Extensive families of constant region genes in a phylogenetically primitive vertebrate indicate an additional level of immunoglobulin complexity. Proc Natl Acad Sci U S A 1987, 84:5868–5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kokubu F, Hinds K, Litman R, Shamblott MJ, Litman GW: Complete structure and organization of immunoglobulin heavy chain constant region genes in a phylogenetically primitive vertebrate. EMBO J 1988, 7:1979–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pettinello R, Dooley H: The immunoglobulins of cold-blooded vertebrates. Biomolecules 2014, 4:1045–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Diaz M, Flajnik MF: Evolution of somatic hypermutation and gene conversion in adaptive immunity. Immunol Rev 1998, 162:13–24. [DOI] [PubMed] [Google Scholar]
- 83.Castro CD, Ohta Y, Dooley H, Flajnik MF: Noncoordinate expression of J-chain and Blimp-1 define nurse shark plasma cell populations during ontogeny. Eur J Immunol 2013, 43:3061–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rumfelt LL, McKinney EC, Taylor E, Flajnik MF: The development of primary and secondary lymphoid tissues in the nurse shark ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand J Immunol 2002, 56:130–148. [DOI] [PubMed] [Google Scholar]
- 85.Dooley H, Flajnik MF: Shark immunity bites back: affinity maturation and memory response in the nurse shark, ginglymostoma cirratum. Eur J Immunol 2005, 35:936–945. [DOI] [PubMed] [Google Scholar]
- 86.Diaz M, Velez J, Singh M, Cerny J, Flajnik MF: Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian ig genes and to spontaneous mutations in evolution: the translesion synthesis model of somatic hypermutation. Int Immunol 1999, 11:825–833. [DOI] [PubMed] [Google Scholar]
- 87.Lee SS, Tranchina D, Ohta Y, Flajnik MF, Hsu E: Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity 2002, 16:571–582. [DOI] [PubMed] [Google Scholar]
- 88.Feige MJ, Grawert MA, Marcinowski M, Hennig J, Behnke J, Auslander D, Herold EM, Peschek J, Castro CD, Flajnik M, Hendershot LM et al. : The structural analysis of shark ignar antibodies reveals evolutionary principles of immunoglobulins. Proc Natl Acad Sci U S A 2014, 111:8155–8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith LE, Crouch K, Cao W, Muller MR, Wu L, Steven J, Lee M, Liang M, Flajnik MF, Shih HH, Barelle CJ et al. : Characterization of the immunoglobulin repertoire of the spiny dogfish (Squalus acanthias). Dev Comp Immunol 2012, 36:665–679. [DOI] [PubMed] [Google Scholar]
- 90.Dooley H, Stanfield RL, Brady RA, Flajnik MF: First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. Proc Natl Acad Sci U S A 2006, 103:1846–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.•.Kovalenko OV, Olland A, Piche-Nicholas N, Godbole A, King D, Svenson K, Calabro V, Muller MR, Barelle CJ, Somers W, Gill DS et al. : Atypical antigen recognition mode of a shark immunoglobulin new antigen receptor (IgNAR) variable domain characterized by humanization and structural analysis. J Biol Chem 2013, 288:17408–17419. [DOI] [PMC free article] [PubMed] [Google Scholar]; Work illustrating progress towards humanized shark single chain therapeutics.
- 92.•.Flajnik MF, Deschacht N, Muyldermans S: A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol 2011, 9:e1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]; Review of heavy chain only antibodies from sharks and camelids.
- 93.Criscitiello MF: What the shark immune system can and cannot provide for the expanding design landscape of immunotherapy. Expert Opin Drug Dis 2014, 9:725–739. [DOI] [PubMed] [Google Scholar]
- 94.Zielonka S, Empting M, Grzeschik J, Konning D, Barelle CJ, Kolmar H: Structural insights and biomedical potential of IgNAR scaffolds from sharks. mAbs 2015, 7:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Flajnik MF, Dooley H: The generation and selection of single-domain, V region libraries from nurse sharks. Methods Mol Biol 2009, 562:71–82. [DOI] [PubMed] [Google Scholar]
- 96.Dooley H, Flajnik MF, Porter AJ: Selection and characterization of naturally occurring single-domain (IgNAR) antibody fragments from immunized sharks by phage display. Mol Immunol 2003, 40:25–33. [DOI] [PubMed] [Google Scholar]
- 97.Goodchild SA, Dooley H, Schoepp RJ, Flajnik M, Lonsdale SG: Isolation and characterisation of ebolavirus-specific recombinant antibody fragments from murine and shark immune libraries. Mol Immunol 2011, 48:2027–2037. [DOI] [PubMed] [Google Scholar]
- 98.Muller MR, O’Dwyer R, Kovaleva M, Rudkin F, Dooley H, Barelle CJ: Generation and isolation of target-specific single-domain antibodies from shark immune repertoires. Methods Mol Biol 2012, 907:177–194. [DOI] [PubMed] [Google Scholar]
- 99.Stanfield RL, Dooley H, Flajnik MF, Wilson IA: Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 2004, 305:1770–1773. [DOI] [PubMed] [Google Scholar]
- 100.Streltsov VA, Varghese JN, Carmichael JA, Irving RA, Hudson PJ, Nuttall SD: Structural evidence for evolution of shark Ig new antigen receptor variable domain antibodies from a cell-surface receptor. Proc Natl Acad Sci U S A 2004, 101:12444–12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nuttall SD, Krishnan UV, Hattarki M, De Gori R, Irving RA, Hudson PJ: Isolation of the new antigen receptor from wobbegong sharks, and use as a scaffold for the display of protein loop libraries. Mol Immunol 2001, 38:313–326. [DOI] [PubMed] [Google Scholar]
- 102.Kopsidas G, Roberts AS, Coia G, Streltsov VA, Nuttall SD: In vitro improvement of a shark IgNAR antibody by qbeta replicase mutation and ribosome display mimics in vivo affinity maturation. Immunol Lett 2006, 107:163–168. [DOI] [PubMed] [Google Scholar]
- 103.Zielonka S, Weber N, Becker S, Doerner A, Christmann A, Christmann C, Uth C, Fritz J, Schafer E, Steinmann B, Empting M et al. : Shark attack: high affinity binding proteins derived from shark vNAR domains by stepwise in vitro affinity maturation. J Biotechnol 2014, 191:236–245. [DOI] [PubMed] [Google Scholar]
- 104.Kovaleva M, Ferguson L, Steven J, Porter A, Barelle C: Shark variable new antigen receptor biologics — a novel technology platform for therapeutic drug development. Expert Opin Biol Ther 2014, 14:1527–1539. [DOI] [PubMed] [Google Scholar]
- 105.Liu JL, Anderson GP, Delehanty JB, Baumann R, Hayhurst A, Goldman ER: Selection of cholera toxin specific IgNAR single-domain antibodies from a naive shark library. Mol Immunol 2007, 44:1775–1783. [DOI] [PubMed] [Google Scholar]
- 106.Walsh R, Nuttall S, Revill P, Colledge D, Cabuang L, Soppe S, Dolezal O, Griffiths K, Bartholomeusz A, Locarnini S: Targeting the hepatitis B virus precore antigen with a novel IgNAR single variable domain intrabody. Virology 2011, 411:132–141. [DOI] [PubMed] [Google Scholar]
- 107.Kaushik AK, Kehrli ME Jr, Kurtz A, Ng S, Koti M, Shojaei F, Saini SS: Somatic hypermutations and isotype restricted exceptionally long CDR3H contribute to antibody diversification in cattle. Vet Immunol Immunopathol 2009, 127:106–113. [DOI] [PubMed] [Google Scholar]
- 108.Koti M, Kataeva G, Kaushik A: Organization of DH-gene locus is distinct in cattle. Dev Biol 2008, 132:307–313. [DOI] [PubMed] [Google Scholar]
- 109.Ramsland PA, Kaushik A, Marchalonis JJ, Edmundson AB: Incorporation of long CDR3S into V domains: implications for the structural evolution of the antibody-combining site. Exp Clin Immunogenet 2001, 18:176–198. [DOI] [PubMed] [Google Scholar]
- 110.Saini SS, Allore B, Jacobs RM, Kaushik A: Exceptionally long CDR3H region with multiple cysteine residues in functional bovine igm antibodies. Eur J Immunol 1999, 29:2420–2426. [DOI] [PubMed] [Google Scholar]
- 111.Saini SS, Farrugia W, Ramsland PA, Kaushik AK: Bovine igm antibodies with exceptionally long complementarity-determining region 3 of the heavy chain share unique structural properties conferring restricted VH + Vlambda pairings. Int Immunol 2003, 15:845–853. [DOI] [PubMed] [Google Scholar]
- 112.Saini SS, Kaushik A: Extensive CDR3H length heterogeneity exists in bovine foetal VDJ rearrangements. Scand J Immunol 2002, 55:140–148. [DOI] [PubMed] [Google Scholar]
- 113.Shojaei F, Saini SS, Kaushik AK: Unusually long germline DH genes contribute to large sized CDR3H in bovine antibodies. Mol Immunol 2003, 40:61–67. [DOI] [PubMed] [Google Scholar]
- 114.Berens SJ, Wylie DE, Lopez OJ: Use of a single VH family and long CDR3S in the variable region of cattle Ig heavy chains. Int Immunol 1997, 9:189–199. [DOI] [PubMed] [Google Scholar]
- 115.Hosseini A, Campbell G, Prorocic M, Aitken R: Duplicated copies of the bovine JH locus contribute to the Ig repertoire. Int Immunol 2004, 16:843–852. [DOI] [PubMed] [Google Scholar]
- 116.Lopez O, Perez C, Wylie D: A single VH family and long CDR3S are the targets for hypermutation in bovine immunoglobulin heavy chains. Immunol Rev 1998, 162:55–66. [DOI] [PubMed] [Google Scholar]
- 117.•.Zhao Y, Jackson SM, Aitken R: The bovine antibody repertoire. Dev Comp Immunol 2006, 30:175–186. [DOI] [PubMed] [Google Scholar]; Review of cow antibody development.
- 118.Consortium TBGSA, Elsik CG, Tellam RL, Worley KC: The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science 2009, 324:522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liljavirta J, Ekman A, Knight JS, Pernthaner A, Iivanainen A, Niku M: Activation-induced cytidine deaminase (AID) is strongly expressed in the fetal bovine ileal peyer’s patch and spleen and is associated with expansion of the primary antibody repertoire in the absence of exogenous antigens. Mucosal Immunol 2013, 6:942–949. [DOI] [PubMed] [Google Scholar]
- 120.Liljavirta J, Niku M, Pessa-Morikawa T, Ekman A, Iivanainen A: Expansion of the preimmune antibody repertoire by junctional diversity in Bos taurus. PLoS One 2014, 9:e99808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.O’Brien PM, Aitken R, O’Neil BW, Campo MS: Generation of native bovine mabs by phage display. Proc Natl Acad Sci U S A 1999, 96:640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang Y, Wang D, de Lichtervelde L, Sun SB, Smider VV, Schultz PG, Wang F: Functional antibody CDR3 fusion proteins with enhanced pharmacological properties. Angew Chem Int Ed 2013, 52:8295–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang Y, Wang D, Welzel G, Wang Y, Schultz PG, Wang F: An antibody CDR3-erythropoietin fusion protein. ACS Chem Biol 2013, 8:2117–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu T, Liu Y, Wang Y, Hull M, Schultz PG, Wang F: Rational design of CXCR4 specific antibodies with elongated CDRS. J Am Chem Soc 2014, 136:10557–10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hungate RE: The rumen microbial ecosystem. Annu Rev Ecol Syst 1975, 6:39–66. [Google Scholar]
- 126.Hungate RE, Bryant MP, Mah RA: The rumen bacteria and protozoa. Annu Rev Microbiol 1964, 18:131–166. [DOI] [PubMed] [Google Scholar]
- 127.Russell JB, Rychlik JL: Factors that alter rumen microbial ecology. Science 2001, 292:1119–1122. [DOI] [PubMed] [Google Scholar]
- 128.Sharpe ME, Latham MJ, Reiter B: The occurrence of natural antibodies to rumen bacteria. J Gen Microbiol 1969, 56:353–364. [DOI] [PubMed] [Google Scholar]
- 129.Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR: The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotechnol 2014, 32:158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lavinder JJ, Hoi KH, Reddy ST, Wine Y, Georgiou G: Systematic characterization and comparative analysis of the rabbit immunoglobulin repertoire. PLoS One 2014, 9:e101322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


