Abstract
The transferrin receptor (TfR) of the bloodstream form (BSF) of Trypanosoma brucei is a heterodimer comprising glycosylphosphatidylinositol (GPI)-anchored expression site–associated gene 6 (ESAG6 or E6) and soluble ESAG7. Mature E6 has five N-glycans, consisting of three oligomannose and two unprocessed paucimannose structures. Its GPI anchor is modified by the addition of 4–6 α-galactose residues. TfR binds tomato lectin (TL), specific for N-acetyllactosamine (LacNAc) repeats, and previous studies have shown transport-dependent increases in E6 size consistent with post-glycan processing in the endoplasmic reticulum. Using pulse-chase radiolabeling, peptide-N-glycosidase F treatment, lectin pulldowns, and exoglycosidase treatment, we have now investigated TfR N-glycan and GPI processing. E6 increased ∼5 kDa during maturation, becoming reactive with both TL and Erythrina cristagalli lectin (ECL, terminal LacNAc), indicating synthesis of poly-LacNAc on paucimannose N-glycans. This processing was lost after exoglycosidase treatment and after RNAi-based silencing of TbSTT3A, the oligosaccharyltransferase that transfers paucimannose structures to nascent secretory polypeptides. These results contradict previous structural studies. Minor GPI processing was also observed, consistent with α-galactose addition. However, increasing the spacing between E6 protein and the GPI ω-site (aa 4–7) resulted in extensive post-translational processing of the GPI anchor to a form that was TL/ECL-reactive, suggesting the addition of LacNAc structures, confirmed by identical assays with BiPNHP, a non-N-glycosylated GPI-anchored reporter. We conclude that BSF trypanosomes can modify GPIs by generating structures reminiscent of those present in insect-stage trypanosomes and that steric constraints, not stage-specific expression of glycosyltransferases, regulate GPI processing.
Keywords: glycobiology, transferrin, glycosylphosphatidylinositol (GPI anchor), trypanosome, N-linked glycosylation, glycosylphosphatidylinositol processing, kinetoplastid protozoa, N-glycan processing, transferrin receptor, trypanosome
Introduction
African trypanosomes of the Trypanosoma brucei ssp. (referred to hereafter as “trypanosomes”) are kinetoplastid protozoa that are the causative agents of human and veterinary trypanosomiasis throughout sub-Saharan Africa, wherever the insect vector (tsetse flies, Genus Glossina) is found (1). Trypanosomes represent an ancient branching group within the Excavata (2), the Kinetoplastida, and whereas they obey the letter of the “eukaryotic law,” they bend the spirit in many remarkable ways. For instance, nuclear gene expression involves concerted polycistronic transcription and trans-splicing to generate typical mature mRNAs with a 5′ cap and 3′ poly-A tail (3). Even more “otherworldly” is mitochondrial RNA editing in which as much as 50% of mitochondrially encoded mRNAs are formed by post-transcriptional insertion/deletion of uridine residues (4). Another example is the compartmentalization of glycolytic enzymes in a peroxisome-like organelle called the glycosome (5), presumably for enhanced enzymatic efficiency.
Although perhaps not as flashy as RNA editing, trypanosomes have also offered some remarkable insights into the full range of eukaryotic glycobiology. Most striking are glycosylphosphatidylinositol (GPI)4 membrane anchors, which were first fully characterized in bloodstream form (BSF) T. brucei, not surprisingly, as the GPI-anchored variant surface glycoprotein (VSG) comprises 10% of total cellular protein. Rapid attachment of preformed GPIs to nascent GPI-anchored proteins in the ER (6, 7), the core GPI structure (8), and the GPI biosynthetic pathway (reviewed in Ref. 9) were all first determined in trypanosomes and subsequently shown to be largely the same throughout eukaryotic phyla. All GPIs, with minor variations, have the same core structure (ethanolamine-P-6Manα1–2Manα1–6Manα1–4GlcNα1–6PI), and these can be decorated with various modifications (reviewed in Ref. 10). In yeast and mammals, a set configuration of ethanolamine moieties are in phosphodiester linkage to the trimannosyl core, and these regulate the exit of nascent GPI anchor precursor from the ER. Limited and variable attachment of hexoses to the core can also occur. In trypanosomes, the common core GPI structure, albeit with life cycle stage–specific lipid configurations, is attached to nascent polypeptides in the ER, and subsequent processing leads to distinct glycan structures in BSF and procyclic insect stage (PCF) parasites (Fig. 1A). In BSFs, a spectrum of side-chain galactose residues are added during post-ER trafficking (11, 12). On procyclin, the major surface protein of PCF trypanosomes, a complex branched poly-N-acetyllactosamine (Galβ1–4GlcNAc) and lacto-N-biose (Galβ1-3GlcNAc) structure is elaborated, presumably also in post-ER compartments, although this has not been formally shown. Upon arrival at the cell surface, this GPI structure serves as the acceptor for transfer of terminal sialic acid residues from host serum sialoglycoproteins by an endogenous cell surface trans-sialidase (13).
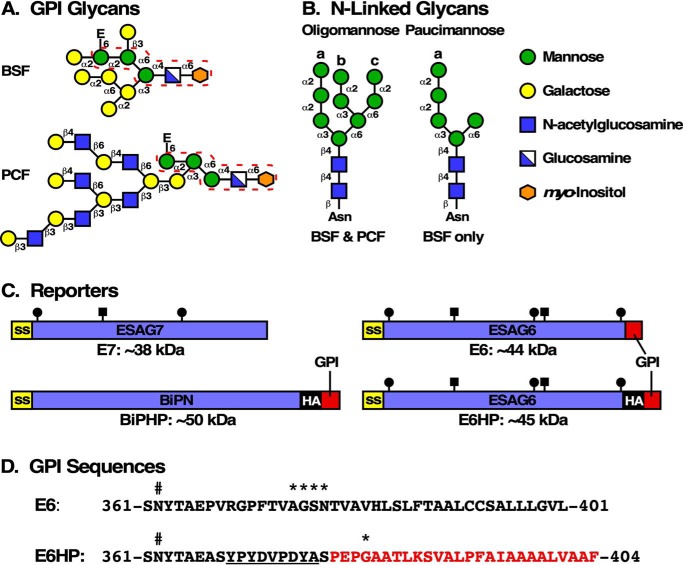
Figure 1.
Trypanosome stage-specific glycan and reporter structures. A, stage-specific GPI structures. The common core GPI glycan structure is found in both BSF and PCF cells (dashed red line). In BSF trypanosomes, the core is decorated with variable galactose residues (known maximum in VSG shown). In PCF trypanosomes, an extensive branched poly-N-acetyllactosamine structure is synthesized. Terminal sialic acid residues that are transferred by trans-sialidase at the cell surface are not shown. In each case, the phosphoethanolamine bridge to the C terminus of the polypeptide is indicated (E). B, dolichol-linked and N-linked glycans that are synthesized and transferred to nascent secretory polypeptides in trypanosomes. The full triantennary Man9GlcNAc2 oligomannose structure is transferred by the TbSTT3B oligosaccharyltransferase in both BSF and PCF trypanosomes. The intermediate biantennary Man5GlcNAc2 paucimannose structure is transferred to acidic sequons by TbSTT3A in BSF cells only. A and B, diagrams adapted from Ref. 10. C, diagrams of TfR subunits (E6 and E7) and the E6HP and BiPNHP reporters. N-terminal signal sequences (ss), mature coding regions (blue boxes), HA epitopes (black, HA), GPI attachment sequences (red, GPI), and N-glycosylation sites (lollipops, oligomannose; squares, paucimannose) are indicated. The calculated polypeptide molecular masses (not including signal sequences and GPI attachment signals) are shown below each diagram. The GPI attachment peptide of BiPNHP and E6HP derives from the EP1 procyclin coding region (33); the signal sequence of E6HP derives from EP1. Replacement of the native E6 C terminus with the HP sequence results in a net gain of 4–7 residues. Diagrams of the central coding regions (blue, E7, E6, and BiPN) are to scale. D, the C-terminal sequences of E6 and E6HP are aligned. The C-terminal N-glycan site (Asn-362, hashtag), the HA epitope (underlined), and the EP procyclin GPI attachment peptide (red) are all indicated. The known procyclin ω-site (53), and the potential TfR ω-sites (see “Experimental procedures”), are indicated by asterisks.
Trypanosomes also differ markedly from the standard eukaryotic model systems in regard to N-glycosylation. Both yeast and mammals (and most other eukaryotes) transfer glucosylated triantennary Glc3Man9GlcNac2 structures from lipid-linked oligosaccharide (LLO) donors to Asn-X-Ser/Thr acceptor sites (sequons) in nascent secretory polypeptides. This reaction is carried out in the ER by a multicomponent oligosaccharyltransferase complex (OST) (14, 15), in which the catalytic subunit is STT3. Although these OSTs can utilize abbreviated LLOs, in vivo there is an overwhelming preference for the full-sized glycan structure. In contrast, the kinetoplastids typically transfer nonglucosylated structures (16) and have stand-alone single-subunit TbSTT3 OSTs (14). In T. brucei, there are three isoforms, TbSTT3A–C, with distinct LLO and sequon substrate specificities (17). TbSTT3B, which is constitutively expressed in BSF and PCF stages, has a strong preference for Man9GlcNAc2 oligomannose LLOs (Fig. 1B) and is nondiscriminatory for sequon acceptors. TbSTT3A is up-regulated in BSF trypanosomes, is highly specific for biantennary Man5GlcNAc2 paucimannose LLOs (Fig. 1B), and has strong preference for acidic sequons. TbSTT3C is not expressed in BSF or PCF stages, and nothing is known about its in vivo substrate specificities. All three genes are found as a tandem repeat in a single genetic locus (17).
These peculiarities of N-glycan addition have significant consequences for subsequent stage-specific N-glycan processing. In PCF trypanosomes, because TbSTT3B is the dominant OST, all sequons are occupied by oligomannose structures. These can be trimmed down to triantennary Man5GlcNAc2 structures, but no further because the Golgi α-mannosidase II that removes the inner α1–3- and α1–6-linked mannoses of the “b” and “c” arms (Fig. 1B) is not present in trypanosomes (18). In contrast, because the biantennary Man5GlcNAc2 glycans attached by TbSTT3A to acidic sequons in BSF trypanosomes are already missing these mannose residues, they can be elaborated in the Golgi into complex type N-glycans, following removal of the α1–2-linked mannoses of the “a” arm. Typically, this involves the addition of a limited number of N-acetyllactosamine (LacNAc) moieties, sometimes capped with terminal α1–3 galactose residues (19). However, in some cases, hypermodification occurs, leading to the production of giant poly-N-acetyllactosamine (pNAL) glycans averaging 54 LacNAc units (20, 21)—the largest known N-glycans in all of glycobiology. All of these features are illustrated by p67, a highly N-glycosylated (14 sequons) lysosomal type I transmembrane protein (22, 23). In PCF trypanosomes, p67 is synthesized as a 100-kDa glycoform (gp100) that contains only oligomannose N-glycans. In BSF trypanosomes, p67 is likewise synthesized as a gp100 precursor but contains a mixture of oligo- and paucimannose structures. During subsequent transit of the Golgi, at least a few of the latter are processed to hypermodified pNAL-containing N-glycans, thereby generating a mature gp150 glycoform.
Another trypanosomal protein that displays a wide range of glyco-modifications is the BSF-specific transferrin receptor (TfR). TfR is a heterodimer of the expression site–associated genes 6 and 7 (ESAG6 and ESAG7) (24–26), both of which are in the larger VSG family (27). The two subunits are highly similar (Fig. 1C), but whereas ESAG6 has a GPI anchor that is structurally similar to the galactosylated GPIs found on VSGs, with up to six galactose residues (28), ESAG7 is truncated at the C terminus and has no such modification. Structural analyses using lectin reactivity and exoglycosidase treatment of purified TfR and high-performance TLC of released TfR N-glycans indicated that each is synthesized with a mixture of oligo- and paucimannose N-glycans (29). ESAG6 (E6) has three oligomannose (N28, N237, and N362) and two paucimannose (N112 and N252) structures; ESAG7 (E7) has two oligomannose (N28 and N236) structures and one paucimannose (N112) structure (numbering relative to start codon). All of these have been trimmed by the removal of some or all of the external α1–2-linked mannose residues during intracellular trafficking, but no evidence for processing of the paucimannose glycans to complex type LacNAc-containing oligosaccharides on either subunit was observed. Overall then, this paints a picture of modest post-ER processing—trimming of oligo- and paucimannose N-glycans and attachment of limited GPI side-chain galactose residues.
However, other data sets suggest a more complex situation. The existence of pNAL-containing N-glycans was first inferred from binding studies with tomato lectin (TL) (20), which has a strong specificity for linear LacNAc repeats (>3) (30). It also has weaker reactivity with the chitobiose core on trimmed oligomannose structures (31, 32). TfR was shown to be part of the total set of TL-binding proteins in BSF trypanosomes, which includes p67, suggesting some degree of conversion of paucimannose oligosaccharides to complex LacNAc-containing N-glycans on TfR (20). Whether this rises to the level of “giant pNAL” is not clear. Second, in our experience, pulse-chase radiolabeling experiments consistently show a time- and transport-dependent increase in the size of E6 (∼5 kDa), but not E7 (33–35) (see below). Such an increase in size cannot be accounted for by minor trimming of N-glycans and limited GPI galactosylation.
In this work, we use in vivo biosynthetic assays in conjunction with sequential lectin pulldowns to investigate the relative contributions of N-glycan and GPI processing to the maturation of native TfR, specifically the GPI-anchored E6 subunit. Our results both confirm (GPI processing) and contradict (N-glycan processing) the previous structural studies of these glycoconjugates (see Refs. 28 and 29, respectively). However, manipulation (loosening) of the steric environment immediately upstream of the GPI anchor results in significant GPI glycan processing reminiscent of the branched poly-LacNAc side chains seen on the GPI anchors of procyclic trypanosomes. These results suggest that stage-specific differences in GPI structure are controlled by steric access of substrates in the Golgi to constitutively expressed glycosyltransferases responsible for side-chain attachment, rather than by stage-specific expression of the glycosyltransferases themselves.
Results
Glycosylation of E6 and E7
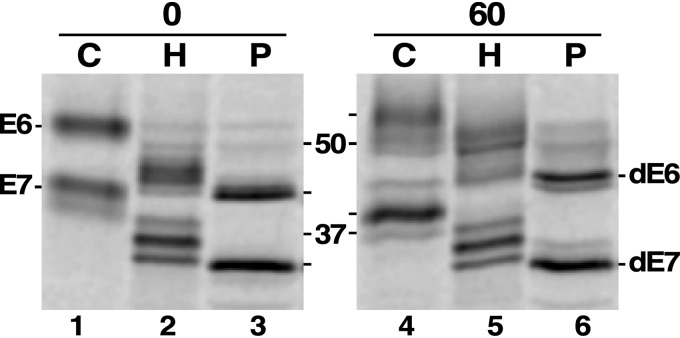
To initiate our studies, we first reinvestigated the N-glycosylation status of newly synthesized E6 and E7 and the corresponding mature glycoforms by pulse-chase radiolabeling (Fig. 2; shown in annotated format in Fig. S1). In each case, radiolabeled polypeptides were specifically immunoprecipitated and then treated with either endo-β-N-acetylglycosidase H (Endo H), which removes oligomannose N-glycans, or peptide-N-glycanase F) (PNGase F), which removes all N-glycans. Note that Endo H will not cleave paucimannose Man5GlcNAc2 structures (11, 17). At T0, both newly synthesized E6 and E7 are detected as single prominent species, which PNGase F treatment reduces to fully deglycosylated forms consistent with the removal of five and three N-glycans, respectively (Fig. 2, lane 1 versus lane 3). Endo H treatment generates ladders of partially de-N-glycosylated species, indicating the presence of variable numbers of resistant paucimannose glycans (Fig. 2, lane 2), but in each case, the major remaining glycoform is consistent with the assignments of Mehlert et al. (28) (E6, 2 paucimannose; E7, 1 paucimannose). Equivalent analyses of mature E6 and E7 from the end of the chase period (T60) indicates that, whereas the overall patterns of Endo H sensitivity are unaltered (Fig. 2, lane 2 versus lane 5), a number of changes indicative of glycan processing have occurred. First is a decrease in the size of mature E7 (Fig. 2, lane 1 versus lane 4), which we ascribe to trimming of terminal α1–2-linked mannose residues on all three N-glycans (Fig. S1, shift 1). Second is an increase in the size of mature E6 (lane 1 versus lane 4) that is mirrored in the partially de-N-glycosylated species (Fig. 2, lane 2 versus lane 5). These shifts could be due to processing of paucimannose N-glycans to complex glycoforms and/or processing of the GPI anchor (Fig. S1, shifts 2 and 3, respectively). However, there is also a clear increase in size of the fully de-N-glycosylated species (Fig. 2, lane 3 versus lane 6) that, absent some heretofore unknown post-translational modification, must be attributed to processing of the GPI anchor (Fig. S1, shift 4). As this processing is not large enough to account for the full increase (∼5 kDa) seen in intact E6, we conclude that both N-glycan and GPI modification occur during intracellular transport of TfR.
Figure 2.
Enzymatic de-N-glycosylation of TfR. Cultured BSF trypanosomes were pulse-chase (15/60 min) radiolabeled with [35S]Met/Cys in the presence of FMK024 (20 μm) to prevent degradation in the lysosome. After immunoprecipitation with anti-TfR, the samples were mock-treated (C) or treated with Endo H (H) or PNGase F (P). Samples were analyzed by SDS-PAGE (107 cell equivalents/lane) and visualized by phosphorimaging. Chase times (min) and molecular mass markers (kDa) are indicated. Mobilities of intact E6 and E7 and de-N-glycosylated E6 and E7 (dE6 and dE7), are shown on the left and right, respectively. Panels were digitally separated after image processing. Detailed identification of individual glycoforms and mobility shifts is presented in Fig. S1.
Processing of E6 and E7 N-glycans
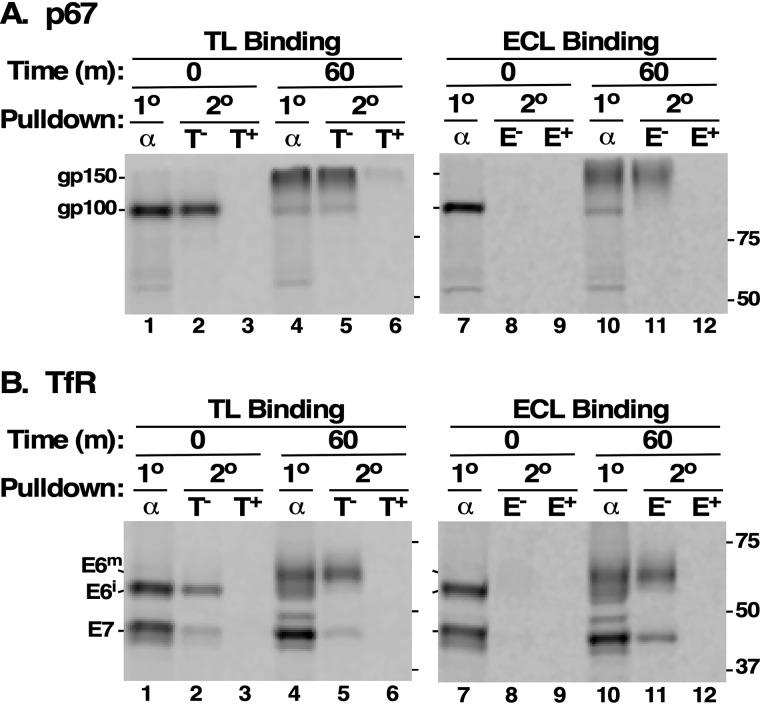
We have previously defined a rubric for assessing the post-ER addition of N-acetyllactosamine to N-glycans on trypanosome secretory proteins using TL and Erythrina cristagalli lectin (ECL) (32). TL binds linear repeats of three or more LacNAc, but also binds to paucimannose glycans (30, 31). ECL is specific for terminal LacNAc units (36). This rubric is illustrated with endogenous p67. Cells were pulse-chase radiolabeled, and p67 polypeptides were immunoprecipitated. Precipitates were then solubilized, and sequential pulldown was performed with either TL or ECL. Free ligands (chitin hydrolysate or lactose, respectively) were included as competitors to confirm specificity. Initially, p67 is detected as the newly synthesized gp100 glycoform (Fig. 3A, lanes 1 and 7). This species is readily seen in sequential pulldown with TL due to reactivity with paucimannose glycans (Fig. 3A, lane 2), but not with ECL, because LacNAc addition cannot occur at this early time point (Fig. 3A, lane 8). At the end of the chase, p67 has been converted to the larger Golgi gp150 glycoform by pNAL addition (Fig. 3A, lanes 4 and 10), as borne out by reactivity with both TL and ECL (Fig. 3A, lanes 5 and 11, respectively).
Figure 3.
Sequential pulldown of TfR with lectins. A and B cultured trypanosomes were pulse-chase (15/60 min) radiolabeled with [35S]Met/Cys in the presence of FMK024 (20 μm) to prevent degradation in the lysosome, and cell extracts were prepared under native conditions. Labeled polypeptides were immunoprecipitated with either anti-p67 (A) or anti-TfR (B) antibodies (α). These primary (1°) immunoprecipitates were solubilized under denaturing conditions and affinity-selected (2°) with TL:Bio (left, T) or ECL:Bio (right, E) in the absence (−) or presence (+) of competing chitin hydrolysate (1:1000) or lactose (200 mm), respectively. Primary immunoprecipitates and subsequent lectin:streptavidin pulldowns were fractionated by SDS-PAGE (107 cell equivalents/lane), and radiolabeled polypeptides were visualized by phosphorimaging. The mobilities of p67 glycoforms (gp150 and gp100) and TfR subunits (E6 and E7) are indicated on the left. Mobilities of molecular mass standards are indicated on the right (kDa). All data shown are representative of multiple experiments (n = 4). The measured conversion of immature (E6i) to mature (E6m) E6 glycoform is ∼5 kDa.
An almost identical pattern of glycosylation was observed with TfR subunits. Newly synthesized E6 and E7 of the expected sizes were observed at T0 (Fig. 3B, lanes 1 and 7). Both were weakly reactive with TL, but not with ECL, in concordance with the presence of one (E7) or two (E6) paucimannose glycans (Fig. 3B, lanes 2 and 8, respectively). At the end of the chase period, E7 had decreased slightly in size, whereas E6 had increased ∼5 kDa, as was seen in Fig. 2 (Fig. 3B, lanes 4 and 10). The mature E7 was weakly reactive with both TL and ECL, suggesting limited addition of LacNAc to the single paucimannose glycan (Fig. 3B, lanes 5 and 11). In contrast, mature E6 was strongly reactive with both TL and ECL, indicating significant addition of LacNAc to the two paucimannose glycans. These results strongly suggest that the paucimannose glycans on E6 are subject to post-ER conversion to complex type structures containing LacNAc repeats. However, these results do not formally rule out the addition of LacNAc to the GPI anchor, although this modification is typically found only in PCF trypanosomes.
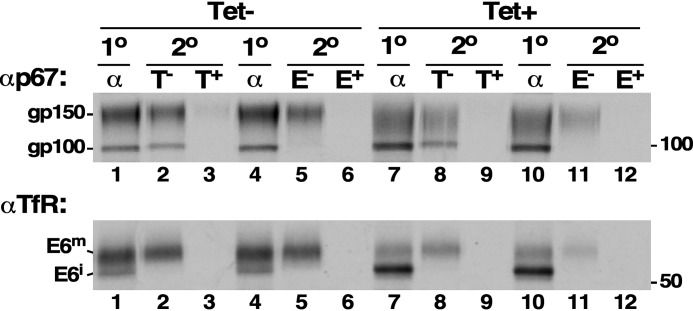
We took a genetic approach to confirm that processing of E6 paucimannose N-glycans was indeed occurring, as such modifications have been previously discounted (29). Using a heterozygous cell line in which a single allele of the TbSTT3 locus was knocked out, we introduced an inducible RNAi construct specifically targeting TbSTT3A, the OST responsible for transfer of paucimannose oligosaccharides. This strategy was used to originally characterize the sequon and N-glycan specificities of the TbSTT3A and TbSTT3B (17). Because complex glycan processing in T. brucei can only occur on paucimannose precursors, we reasoned that ablation of TbSTT3A would reduce the size and lectin reactivity profile of mature E6 if such structures are normally present. In agreement with the work of Izquierdo et al. (17), knockdown of the remaining TbSTT3A allele had no effect on cell viability (Fig. S2A), despite a ∼50% reduction in TbSTT3A mRNA (Fig. S2B). Presumably, this represents a cumulative reduction of ∼75% because one allele has been eliminated by knockout. Our standard lectin profiling was then performed on mature E6 labeled from pulse-chase reactions. Again, mature p67 was used as a positive control for the presence of paucimannose oligosaccharides. As seen above, p67 polypeptides recovered from unsilenced cells were mostly processed gp150 (TL- and ECL-reactive) with residual amounts of precursor gp100 (TL-reactive only) (Fig. 4, top, lanes 1–6). In contrast, ablation of TbSTT3A resulted in a marked decrease in the amount and size of processed gp150 (Fig. 4, top, lanes 7–12), with a clear compensatory increase in the amount of unprocessed gp100 (Fig. S2C, 2.2 ± 0.48-fold, mean ± S.D., n = 3). These results confirm the replacement of paucimannose with oligomannose structures, with subsequent reduction of glycan processing, as expected for ablation of TbSTT3A (17). Essentially the same result was seen with E6 (Fig. 4, bottom). TbSTT3A knockdown resulted in a dramatic decrease in mature E6 with a compensatory increase in immature E6 (Fig. S2C, 2.8 ± 0.35-fold, n = 3) and loss of lectin reactivity. These results again strongly indicate that processing of E6 involves paucimannose oligosaccharides, likely by the addition of LacNAc.
Figure 4.
TbSTT3A knockdown. The TbSTT3A RNAi cell line was cultured without (tet−) or with (tet+) tetracycline to initiate specific dsRNA synthesis. On day 3, cells were pulse-chase radiolabeled exactly as in Fig. 3, and cell extracts were prepared under native conditions from the final (60-min) chase fractions. Labeled polypeptides were immunoprecipitated with either anti-p67 (top) or anti-TfR (bottom) antibodies (α). Secondary pulldowns were then performed with lectins and specific inhibitors as in Fig. 3. All pulldowns were fractionated by SDS-PAGE (107 cell equivalents/lane) and visualized by phosphorimaging. The mobilities of p67 glycoforms (gp150 and gp100) and TfR (E6 species only) are indicated on the left. Mobilities of molecular mass standards are indicated on the right (kDa). All data shown are representative of multiple experiments (n = 3).
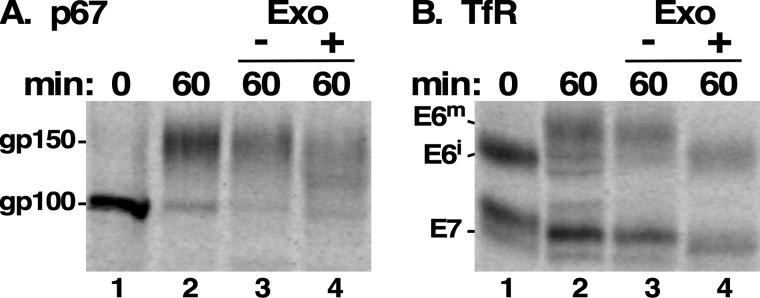
Finally, to confirm that this processing does involve the addition of LacNAc, we treated immunoprecipitates of mature p67 and TfR with combined exo-β1–4-galactosidase and exo-β1–2,3,4,6-N-acetylglucosaminidase, resulting in a partial reduction in the size of mature gp150 p67, which is known to have hypermodified pNAL-containing N-glycans (Fig. 5A, lane 3 versus lane 4). The failure to completely reduce the size of p67 may be due to the highly branched nature of pNAL glycans (21), but this result nevertheless validates the assay. The same treatment reduced mature E6 to a size essentially equivalent to that of the immature precursor (Fig. 5B, lane 4 versus lane 1). A slight, but quantitative, decrease in the size of E7 was also seen (Fig. 5B, lane 3 versus lane 4). These results corroborate the lectin reactivities and provide compelling evidence that paucimannose N-glycans on E6 and E7 are processed by the addition of LacNAc repeats.
Figure 5.
Exoglycosidase treatment. Trypanosomes were pulse-chase (15/60 min) radiolabeled with [35S]Met/Cys in the presence of FMK024 (20 μm) to prevent degradation in the lysosome, and cell extracts were prepared at the beginning (lane 1) and end (lane 2) of the chase period for immunoprecipitation with anti-p67 (A) or anti-TfR (B). Matched end point immunoprecipitates were solubilized and either mock-treated (lane 3) or treated with exo-β-gal and exo-β-N-acetylglucosaminidase (lane 4) as described under “Experimental procedures.” A representative experiment is presented (107 cell equivalents/lane). Both panels are from the same gel and phosphorimage and were digitally separated for presentation after image processing. Mobilities of glycoforms are indicated.
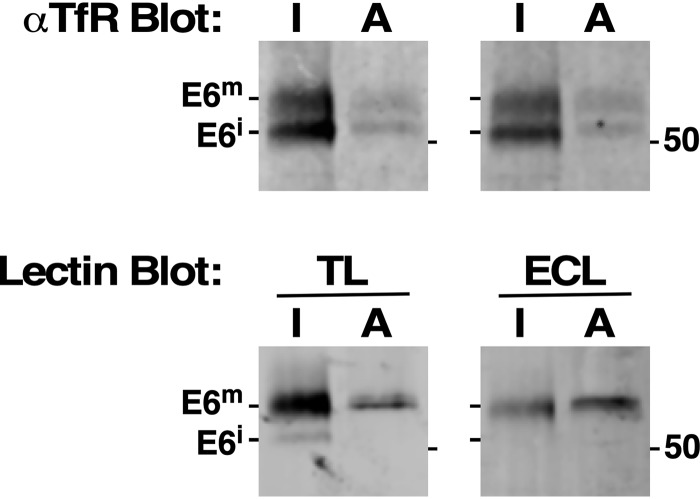
These results contrast with those of Mehlert et al. (29), who found no evidence for LacNAc modification of paucimannose N-glycans on affinity-purified TfR. In an attempt to reconcile these differing results, we collected TfR by parallel pulldown with anti-TfR and Tf-beads and subjected the bound materials to blotting with either TL or ECL, followed by immunoblotting with anti-TfR. The immunoblot revealed roughly equal amounts of both immature and mature TfR in the steady state pool of both pulldowns (Fig. 6, top). The relative inefficiency of the affinity pulldown may be because TfR is rapidly ligated with serum transferrin upon arrival in the flagellar pocket, rendering it unavailable for binding to Tf-beads (34). Thus, the affinity-purified material likely represents newly synthesized TfR en route to the cell surface. Importantly, the mature species is reactive with both TL and ECL, indicating the presence of LacNAc on steady-state TfR (Fig. 6, bottom). Thus, in our hands, affinity-purified TfR does contain processed paucimannose glycans. Factors contributing to the discrepancy between our results and those of Mehlert et al. (29) are discussed below.
Figure 6.
Affinity purification of TfR. Native extracts were prepared from log-phase cells, and TfR was selected by immunoprecipitation with αTfR (I, 107 cell equivalents) or affinity pulldown with Tf-beads (A, 108 equivalents). Matched samples were then sequentially blotted with either TL or ECL (bottom), followed by immunoblotting with αTfR (top) to reveal steady-state E6. A representative data set is presented. Mobilities of immature and mature forms of TfR and molecular mass markers are indicated.
Processing of the E6 GPI anchor
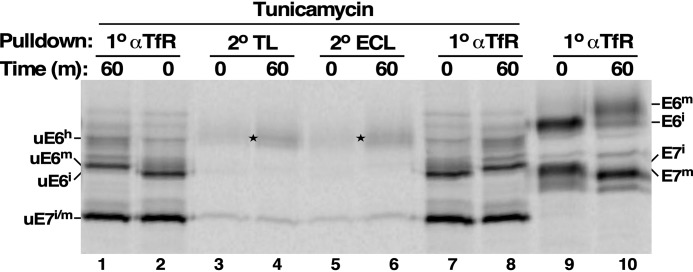
To investigate the role of the GPI glycan in the overall processing of E6, we pretreated cells with tunicamycin to flush out all LLO precursors. Pulse-chase radiolabeling followed by sequential lectin pulldowns was then performed (Fig. 7; shown in annotated format in Fig. S3). At T0, un-N-glycosylated E6 and E7 species of the expected size based on the prior PNGase F treatment (Fig. 2) were observed (Fig. 7, lanes 2 and 7 versus lane 9). During the subsequent chase, there was no change in the mobility of E7, as expected, because there are no glycans of any kind. However, there was a clear but modest increase in the size of E6 (Fig. 7, lanes 2 and 7 versus lanes 1 and 8; Fig. S3, shift 1). This mature un-N-glycosylated species was nonreactive with either TL or ECL, confirming the absence of LacNAc (Fig. 7, lanes 3–6). We therefore ascribe this processing to the post-ER addition of ∼6 galactose residues, consistent with the TfR GPI structural analyses of Mehlert et al. (28). Interestingly, there was a minor amount of time-dependent hypermodification of E6 generating a larger species that was reactive with both TL and ECL (Fig. 7, lanes 4 and 6; discussed below). These results indicate that GPI processing makes a minor contribution to the overall ∼5-kDa increase in mass of E6 during post-ER trafficking, the bulk of which is due to conversion of paucimannose N-glycans to LacNAc-containing complex glycans.
Figure 7.
TfR GPI anchor binds TL and ECL. Cultured trypanosomes were pretreated with tunicamycin (1 h, 300 ng/ml) and then pulse-chase (15/60 min) radiolabeled in the continued presence of tunicamycin to inhibit N-glycan synthesis and FMK024 (20 μm) to block turnover in the lysosome. Lysates were prepared under native conditions at the beginning and end of the chase and subjected to primary immunoprecipitation with anti-TfR (1° αTfR, lanes 1 and 2 and lanes 7 and 8). Precipitates were solubilized, and secondary pulldowns were performed with TL:Bio (2° TL, lanes 3 and 4) or ECL:Bio (2° ECL, lanes 5 and 6) as indicated. Samples were analyzed by SDS-PAGE (107 cell equivalents/lane) and visualized by phosphorimaging. The mobilities of the un-N-glycosylated species of immature E6 (uE6i), mature E6 (uE6m), hypermodified mature E6 (uE6h), and E7 (uE7) are indicated on the left. As a control for fully glycosylated TfR subunits, primary immunoprecipitates from cultures without tunicamycin are presented (lanes 9 and 10). The mobilities of these species are indicated on the right. Stars indicate TL- and ECL-reactive hypermodified uE6h that appears during the chase period. Detailed identification of individual glycoforms and mobility shifts are presented in Fig. S3.
Processing of alternate GPI substrates (E6HP/BiPNHP)
The appearance of a small amount of hypermodified E6 when N-glycosylation is blocked is reminiscent of the processing we previously observed when an procyclin reporter (EPMH) was expressed in BSF cells (Fig. 2 of Ref. 33). EPMH was synthesized as a 40-kDa precursor that was quantitatively converted to an ∼55 kDa smear during intracellular transport. EPMH has a single N-glycosylation site (sequon pI 3.28) that most certainly is occupied by a paucimannose structure in BSF trypanosomes. Furthermore, the mature glycoform, but not the precursor, was strongly reactive with TL, consistent with the addition of pNAL. However, when N-glycosylation was blocked with tunicamycin, EPMH was still subject to substantial modification, increasing in size ∼7–8 kDa (TL reactivity was not determined). We attributed this residual processing to modification of the GPI anchor because it was not present in a matched GPI-minus reporter, and a second reporter used in the same study supported that conclusion (Fig. 7 of Ref. 33; BiPNHP). BiPNHP is the globular N-terminal ATPase domain of BiP, a resident ER Hsp70 chaperone, fused at the C terminus to an HA tag (H) and the EP procyclin GPI attachment peptide (P) (Fig. 1C). BiPNHP was also subject to marked processing during transport, increasing from a sharp ∼55-kDa precursor to a heterogeneous ∼65-kDa mature glycoform. Because BiPNHP has no N-glycosylation sites, and the matched soluble BiPN reporter is not modified, we concluded that it too was subject to GPI processing.
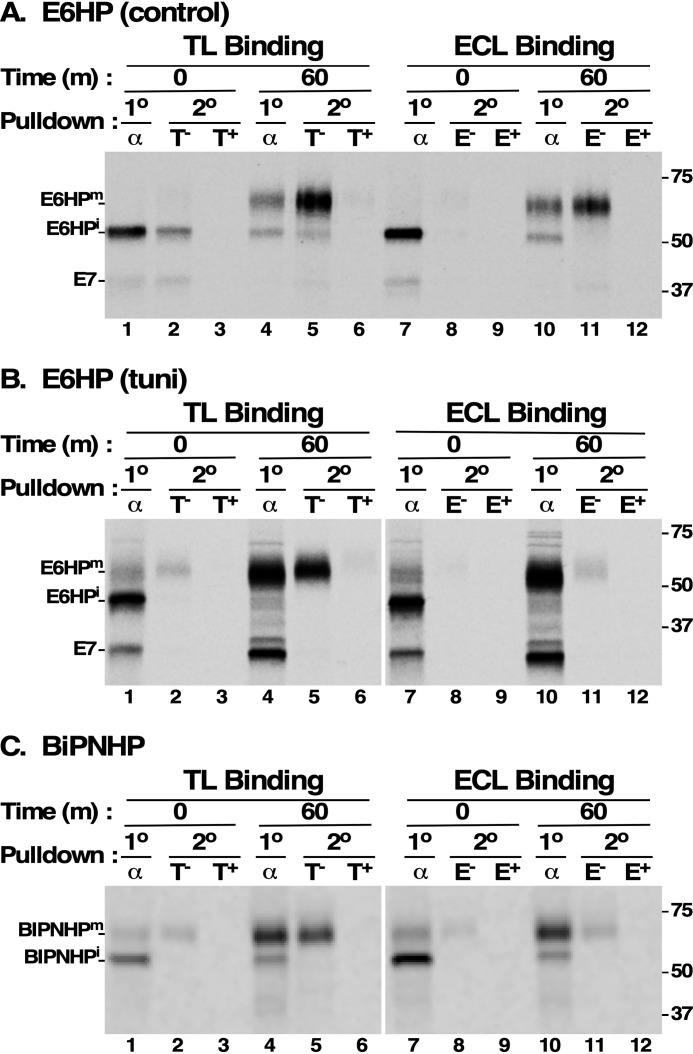
To determine whether the hypermodification of E6 in the absence of N-glycans was due to GPI processing, and if so whether this might be influenced by steric considerations, we replaced the native C terminus with the fused HA:EP GPI attachment peptide (E6HP), resulting in a net gain of 4–7 residues upstream of the predicted GPI attachment site (Fig. 1D; the exact ω-site of E6 is not known). This construct was used to replace the native E6 ORF in the active expression site, and we then repeated the sequential lectin pulldowns (Fig. 8A). The overall pattern of E6HP synthesis and processing was essentially the same as native E6 (compare Fig. 8A (lanes 1–12) with Fig. 3B (lanes 1–12)), the only difference being a larger quantitative increase in size of the immature precursor (Fig. 8A, lanes 1 and 7) to the mature TL+/ECL+ glycoform (Fig. 8A, lanes 4 and 5 and lanes 10 and 11) (∼12 kDa for E6HP versus ∼5 kDa for E6). This size increase is roughly equivalent to the minor amount of hypermodification seen with native E6.
Figure 8.
Sequential pulldown of E6HP and BiPNHP with lectins. Cultured BSF trypanosomes (107 cell equivalents/lane) expressing E6HP (A and B) or BiPNHP (C) were [35S]Met/Cys pulse-chase (15/60 min) radiolabeled in the presence of FMK024 (20 μm). As indicated, tunicamycin was included to block N-glycosylation (B, tuni). Cell extracts were prepared as described in Fig. 3. E6HP and BiPNHP polypeptides were immunoprecipitated with anti-HA antibodies (α), and then solubilized lysates were affinity-selected (2°) with TL:Bio (T, left) or ECL:Bio (E, right) in the absence (−) or presence (+) of competing chitin hydrolysate (1:1000) or lactose (200 mm), respectively. The mobilities of mature (m) and immature (i) E6HP and BiPNHP species are indicated on the left, and molecular mass standards are indicated on the right (kDa). Measured conversion of normal E6HPi to E6HPm is ∼12 kDa (A, lane 1 versus lane 4); measured conversion of non-N-glycosylated E6HP is ∼7 kDa (B, lane 1 versus lane 4); and measured conversion of BiPNHPi to BiPNHPm is ∼10 kDa (C, lane 1 versus lane 4).
To determine whether the large increase in size of E6HP could be attributed solely to GPI glycan processing, we repeated this experiment with tunicamycin treatment to eliminate the contribution of N-glycans (Fig. 8B). An identical pattern of processing was observed, albeit with global size reductions due to the absence of N-glycans. Importantly, the immature precursor was still quantitatively converted to a larger (∼7 kDa) mature form during intracellular transport (Fig. 8B, compare lane 1 with lane 4 and lane 7 with lane 10; see also Fig. 9 for a direct comparison). This increase can only be due to GPI glycan processing, and the TL and ECL reactivity of the mature species (Fig. 8B, lanes 5 and 11, respectively) suggests that it represents the addition of poly-LacNAc, as in the native procyclin GPI structure (see Fig. 1A). The relatively weaker reactivity of the mature E6HP GPI anchor with ECL may be due to the presence of terminal GlcNAcβ1–6Gal or lacto-N-biose (Galβ1–3GlcNAc) disaccharides. The small amount of mature TL reactive E6HP at the T0 time point is likely due to rapid transport of this reporter from the ER to the Golgi during the 15-min labeling period. Finally, to unequivocally establish that the TL/ECL reactivity of mature un-N-glycosylated E6HP was due to GPI glycan processing, we repeated these sequential pulldowns with the BiPNHP reporter, which has no N-glycans (Fig. 8C). Newly synthesized BiPNHP of the expected size (∼55 kDa) was detected, and this species was completely nonreactive with either TL or ECL (Fig. 8C, lane 1 versus lane 2 and lane 7 versus lane 8). During the chase, this was converted overwhelmingly to an ∼65-kDa mature glycoform that was robustly reactive with TL and less so with ECL (Fig. 8C, lane 4 versus lane 5 and lane 10 versus lane 11). As seen with E6HP, the small amount of mature TL/ECL-reactive BiPNHP at the early time point is due to rapid transport, and the weak reactivity with ECL may be due to terminal non-LacNAc disaccharides. These results confirm our previous findings (33) and further establish that facile addition of LacNAc to the GPI core can occur in BSF trypanosome if the proper reporter is used.
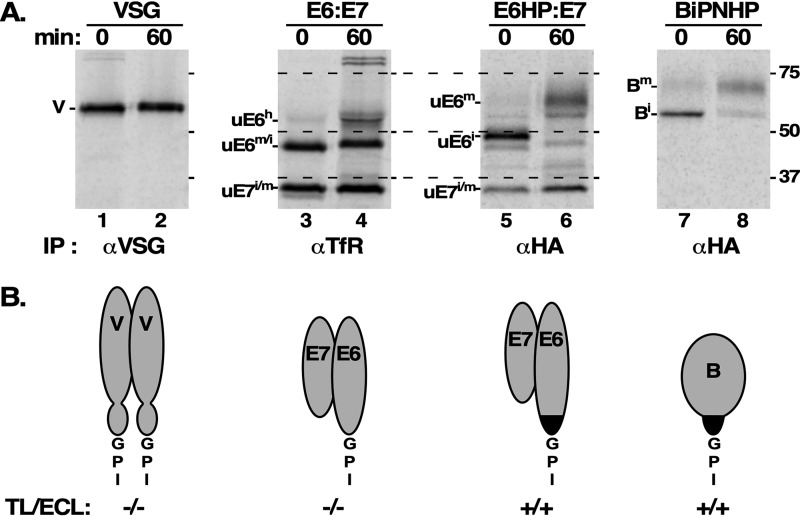
Figure 9.
GPI processing of VSG, E6, E6HP, and BiPNHP. A, all cultured BSF trypanosomes were pretreated with tunicamycin (300 ng/ml, 1 h) to inhibit N-glycan synthesis (BiPNHP cells were untreated). Cells were then [35S]Met/Cys pulse-chase (15/60 min) radiolabeled in the presence of FMK024 (20 μm) to prevent degradation in the lysosome. Lysates were prepared under native conditions, and immunoprecipitation (IP) was performed with anti-VSG, anti-TfR, or anti-HA as indicated. Precipitates were analyzed by SDS-PAGE and phosphorimaging: VSG, 106 cell equivalents/lane; all others, 107 cell equivalents/lane. The mobilities of un-N-glycosylated glycoforms of VSG (V); E6, E7, and E6HP subunits (E6 and E7); and BiPNHP (B) are indicated on the left of the appropriate panels. Immature, mature, and hypermodified species are indicated as superscripts (i, m, and h). Mobilities of molecular mass markers are on the right (kDa). All samples were run and imaged in consecutive lanes of the same gel and then digitally separated for clearer labeling/presentation. All mobilities are directly comparable between panels; dashed lines allow easy comparison of E6 and E6HP glycoforms. B, corresponding cartoons of the various reporters to highlight the differences between each. VSG is a homodimer with large N-terminal domains and smaller membrane-proximal domains. TfR is a heterodimer of E6 and E7, both of which have similar folds to the VSG N-terminal domain. E6HP and BiPNHP have the same C-terminal fusion of the HA tag and the EP procyclin GPI attachment sequence (HP, black). The net difference between E6 and E6HP is 4–7 residues upstream of the GPI anchor. Reactivity of the mature GPI structures with TL and ECL are indicated (+/−). VSG221 reactivity is based on published structural data (37), and all others are from presented experimental data.
These findings strongly suggest that polypeptide steric constraints at the extreme C terminus govern processing of the GPI glycan. To demonstrate this in a directly comparative manner, we pulse-chase radiolabeled tunicamycin-treated cells and analyzed immunoprecipitates of the various reporters in relation to native VSG221. VSG221 had a barely perceptible increase in size during intracellular transport (Fig. 9, lanes 1 and 2), consistent with the addition of 2–4 GPI galactose residues (37). Likewise, as seen in Fig. 2, native E6 increased in size consistent with the addition of 4–6 GPI galactose moieties (28) (Fig. 9, lanes 3 and 4). And as before, a limited amount of GPI hypermodification was also seen. In contrast, E6HP, which is slightly larger that E6 due to an additional 4–7 C-terminal amino acid residues (Fig. 9, lane 3 versus lane 5), increases dramatically in size during the chase (Fig. 9, lanes 5 and 6). Likewise, BiPNHP shows a dramatic size increase (Fig. 9, lanes 7 and 8). The size increase of E6HP and BiPNHP can only be due to significant processing of the GPI glycan, and the lectin reactivity of each (Fig. 8) indicates this is due to the addition of poly-LacNAc, as seen with the procyclin GPI anchor in PCF trypanosome. The structural differences between these reporters (Fig. 9, cartoons) and their effects on GPI processing are discussed below.
Discussion
Structural studies of purified glycoconjugates from T. brucei transferrin receptor indicate the presence of trimmed oligo- and paucimannose N-glycans with no evidence of further processing (e.g. no addition of LacNAc to paucimannose oligosaccharides as seen in other BSF glycoproteins) (29). However, our own biosynthesis assays have consistently indicated that the E6 subunit of TfR is subject to significant processing during intracellular transport (33–35). Furthermore, TfR was one of the original BSF trypanosome proteins identified as containing TL-reactive N-glycans (20). Finally, all published works that have assessed steady-state populations of TfR by immunoblotting, including Mehlert et al. (29) typically see a heterogenous “smear” of E6 polypeptides, consistent with N-glycan processing (25–27, 29, 38–40). To resolve this issue, we have now performed detailed biosynthesis studies in conjunction with sequential lectin pulldowns, reverse genetics, and exoglycosidase treatments.
In our biosynthesis assays, the E6 subunit of TfR is processed with a net gain of ∼5 kDa. A minor component of this is processing of the GPI anchor consistent with the addition of limited galactose residues as per Ref. 28 (discussed below). However, five observations indicate that the larger part of this processing is the addition of LacNAc repeats to paucimannose oligosaccharides in the Golgi. First, the time-dependent increase in size is blocked by tunicamycin, indicating processing of N-glycans. Second, radiolabeled mature E6 is strongly reactive with both TL and ECL, indicating the presence of multiple LacNAc units. Third, silencing of TbSTT3A, the OST responsible for attachment of paucimannose N-glycans, limits conversion of immature to mature E6. Fourth, exoglycosidase digestion reduces the size of mature E6 to that of immature. Finally, lectin blotting of affinity-purified steady-state TfR confirms reactivity of mature E6 with both TL and ECL. Collectively, these results provide compelling evidence that paucimannose N-glycans on E6, and to a lesser extent E7, are modified during intracellular transport by the addition of multiple LacNAc residues.
Our results conflict with the prior structural studies of Mehlert et al. (29), but we feel that several factors may contribute to this seeming contradiction. First, careful examination of the blotting data (Figs. 2 and S1 of Ref. 29) actually reveals a similar pattern of lectin reactivity. Using affinity-purified TfR, anti-TfR detected both small (immature) and large (mature) steady-state E6 species in an approximately equal ratio, concanavalin A also strongly detected both species, and both ECL and ricin detected the large species. The latter results were admittedly weak and might have been discounted by the authors. However, TL reactivity was completely negative. Overall then, excepting TL, these results are actually quite consistent with our own. A second factor may be in the glycan structural methodology, which relied on high performance TLC and Dionex ion-exchange chromatography of [3H]borohydride-labeled N-glycans (Figs. 3 and S3 of Ref. 29). In the first case, glycans with multiple LacNAc units will not leave the origin and thus will be excluded from consideration. Likewise in the second case, such structures are unlikely to elute from the column under the conditions used. Unfortunately, we can offer no further insight into this discrepancy.
In contrast to N-glycans, our findings with GPI processing of native E6 (a slight increase in size) are fully consistent with Mehlert et al. (4–6 α-galactose sidechain residues) (28). However, we did observe a small amount of GPI hypermodification that is reminiscent of the processing we have previously seen with several GPI-anchored reporters in BSF trypanosomes (33). In that work, two different monomeric GPI reporters, EPMH and BiPNHP, were subject to 7–10-kDa increases in mass during intracellular trafficking that could only be ascribed to GPI processing. BiPNHP was particularly telling, as it has no post-translational modifications other than a GPI anchor. This led us to ask whether more extensive GPI processing of the TfR GPI anchor can occur, and if so, whether steric constraints at the C terminus might influence access of GPI substrates to glycosyltransferases during transit of the Golgi. Indeed, replacing the native E6 GPI attachment signal with the HP peptide resulted in quantitative GPI processing of ∼7 kDa, in keeping with that seen originally with BiPNHP. The processed E6HP and BiPNHP GPI anchors were reactive with both TL and ECL, indicating the addition of a poly-LacNAc side-chain structure. We made repeated attempts to degrade this structure with combined exo-β-N-acetylglucosaminidase/exo-β-galactosidase treatment, with and without neuraminidase or α-galactosidase, without success. Nevertheless, the lectin results are compelling. Thus, the addition of just 4–7 amino acids upstream of the ω site for GPI addition allows quantitative synthesis of a side chain that is likely similar to that found on the native GPI anchor of procyclin in procyclic insect stage trypanosomes (Fig. 1A).
These results strongly suggest that steric constraints do affect GPI processing, presumably by hindering (less processing) or allowing (more processing) access to glycosyltransferases during transit of the Golgi. That this occurs is supported by direct comparison of the degree of processing seen in the reporters used in this work, relative to the processing seen with endogenous VSG (Fig. 9). VSG is a homodimer and presumably has the most constrained “GPI environment” in proximity to the membrane—it is processed by the addition of side-chain galactose residues. The amount of modification (0–6 hexoses) varies, and this has been ascribed to C-terminal structural differences between different VSG classes (i.e. steric constraint) (37, 41). Native TfR is a VSG-like heterodimer in which the E7 subunit is truncated. This presumably creates a more open environment around the single GPI anchor on E6, but still processing is largely restricted to the addition of variable galactose residues, albeit on the upper end of what is seen in VSG (4–6 hexose) (28). As noted above, the addition of just 4–7 residues at the GPI addition site apparently releases steric constraints enough to allow quantitative production of a procyclin-like poly-LacNAc type side chain. Finally, the attachment of the HP GPI anchor peptide to BiPN, the globular N-terminal ATPase domain of the ubiquitous ER chaperone, also results in extensive GPI poly-LacNAc modification.
It is probable that the effect of steric constraint on GPI processing is also at play in procyclic trypanosomes. Ectopically expressed MITat1.4 VSG (VSG117) in procyclic parasites received a procyclic-type GPI anchor in regard to lipid arrangement (42), but GPI processing was not evident. However, when the native GPI peptide was replaced with the HP peptide, for a net gain of 15 amino acids, processing occurred consistent with the addition of a poly-LacNAc side chain (note: no lectin binding or exoglycosidase assays were performed). In another instance, a totally different VSG (AnTat11.17) was expressed in procyclics (43). It too received a procyclin-type GPI anchor and, although biosynthesis assays were not performed, exoglycosidase treatment suggested that it was processed by LacNAc addition. Presumably then, the GPI environment in AnTat11.17 is more relaxed than that in MITat1.4, leading to processing of the native protein.
Collectively, these findings suggest that the modification of glycans on secretory glycoproteins in trypanosomes is not extensively regulated by differential expression of the glycosyltransferases involved. Rather, it is other factors that take precedence. In the case of N-glycans, it is the stage-specific expression of OST isoforms with different substrate preferences (LLO and sequon), coupled with the lack of Golgi α-mannosidase II, that limits such processing to the bloodstream stage (17, 18). TbSTT3A is expressed only in BSF trypanosomes, and it has a preference for biantennary Man5GlcNAc2 structures that can be extended by LacNAc addition in the Golgi without prior mannose trimming. However, in the absence of TbSTT3A in procyclic trypanosomes, TbSTT3B transfers triantennary Man9GlcNAc2 structures that cannot be trimmed to a form that can then be modified in the Golgi. In the case of GPI anchors, it is apparently protein steric constraint on access to constitutively expressed glycosyltransferases that regulates the extent of processing, although the rate of transit of individual reporters through the Golgi may also contribute. Steric constraint is a concept that has already been invoked in relation to the variable amount of side-chain galactose residues attached to VSGs of differing C-terminal classes expressed in BSF trypanosomes (37, 41). Our work broadens this concept to include stage-specific side-chain modifications found on GPI anchors across the trypanosome life cycle. This, however, does not rule out a contribution of stage-specific expression of glycosyltransferases to the diversity of glycoconjugates found in trypanosomes. Indeed, expression of at least one transferase critical for complex N-glycan formation, GlcNAc transferase II, is 15-fold higher in BSF than PCF trypanosomes (44). It has been estimated that there are a minimum of 38 distinct glycosidic linkages found in T. brucei (19), each requiring a unique glycosyltransferase, and it seems likely that more instances of stage-specific regulation will be found. Altogether then, this paints a rich tapestry of how diversity of glycoconjugate structure is generated in trypanosomes.
Experimental procedures
Cell lines and culture
All experiments were carried out with the bloodstream form Lister 427 strain of T. brucei brucei (MITat1.2 expressing VSG221) or the derivative SM221 cell line (45), which expresses both T7 RNA polymerase and tetracycline repressor for conditional expression. Cells were grown at 37 °C in HMI9 medium (46) supplemented with 10% fetal bovine serum (tetracycline-free). For all experiments, cells were harvested at mid-to-late log phase (0.5–1 × 106).
Construction of reporter cell lines
All GPI-anchored reporter proteins used in this study are shown in Fig. 1C. Generation of the BiPNHP reporter cell line has been described (33). This construct has the globular N-terminal ATPase domain from the ER molecular chaperone BiP joined in-frame with a C-terminal HA-tag:EP1 procyclin GPI signal (codons 120–145) fusion. The sequence of this “HP” segment following GPI attachment is ASYPYDVPDYASPEPG (where 5′-AS is the NheI cloning site; HA tag underlined).
The E6HP construct was assembled in pXS6 (47) as follows (5′–3′): 5′-UTR–targeting region (nt −484 to +1; relative to the E6 ORF); hygromycin resistance cassette; βα-tubulin intergenic region; EP1 procyclin signal sequence (Tb927.10.20160; nt 1–81); RNAi-resistant E6 ORF minus the native signal and GPI sequences (nt 58–1098, codons 20–366) fused in frame with the HP sequence derived from BiPNHP; and 3′-UTR–targeting region (nt 1–601; relative to E6 stop codon). All E6 segments were derived from the cloned BES1 expression site (clone H25N7 (48), gift of Professor Gloria Rudenko, Imperial College). All segments were confirmed by sequencing. The resultant construct was excised with ClaI/FseI for homologous replacement of the endogenous E6 gene in the active ES1 of a TfR RNAi cell line (described previously in Refs. 34 and 49). Note that this construct was originally generated for other reasons and has been repurposed for the experiments described herein. Consequently, silencing of native TfR was never employed. Because of the replacement of the native E6 C terminus with the C-terminal HP segment, the expressed E6HP protein following GPI addition is expected to have a longer C-terminal amino acid sequence. The TfR GPI ω-site has not been mapped experimentally, but is predicted to be Ala-375 by the big-PI algorithm (50) (http://mendel.imp.ac.at/gpi/gpi_server.html).5 However, visual inspection of the E6 C-terminal sequence suggests other potential ω-sites (Gly-376, Ser-377, and Asn-378). Thus, a net increased spacing between the GPI anchor and the bulk of the E6 polypeptide of 4–7 residues is predicted in the E6HP reporter (Fig. 1D).
The TbSTT3A RNAi cell line used in this work was generated by a similar approach as in Ref. 17. An TbSTT3A dsRNAi construct was generated in the pLEW100v5X:Pex11 stem loop vector (47). A 498-bp TbSTT3A target sequence (Tb927.5.890, nt 1236–1734 relative to the start codon) was amplified from T. brucei gDNA by PCR. The amplicon was sequentially inserted upstream of the Pex11 stuffer using HindIII/XhoI and then downstream in the opposite orientation using NdeI/XbaI. All cloning steps were confirmed by sequencing, and the resultant plasmid was linearized with NotI for transfection into the TbSTT3A,B,C+/− cell line (single TbSTT3 locus knockout, a generous gift of Professor Mike Ferguson, University of Dundee). All transfections and clonal selections were as described (34).
Antibodies and blotting reagents
The following antibodies have been described in our prior publications (47, 49, 51): rabbit anti-VSG221, mouse mAb anti-HA (HA7, Sigma), mouse monoclonal anti-p67. Monoclonal anti-HA1.1 (HA) was from BioLegend (San Diego, CA; formerly Covance). Rabbit anti-TfR (BES1-specific) was a generous gift of Drs. Piet Borst and Henri Luenen (Netherlands Cancer Institute, Amsterdam). Tomato lectin-biotin (TL:Bio), E. cristagalli lectin:biotin (ECL:Bio), lactose, and chitin hydrolysate were from Vector Laboratories (Burlingame, CA). Protein A–Sepharose (GE Healthcare) and NeutraAvidin beads (Thermo Fisher Scientific) were used for primary immunoprecipitations and secondary pulldowns, respectively. Secondary reagents for lectin and immunoblotting blotting were IRDye800CW streptavidin and IRDye680-conjugated goat anti-rabbit IgG (LI-COR Biosciences, Lincoln, NE). Quantitative fluorescent signals were scanned on an Odyssey CLx Imager (LI-COR Biosciences).
Radiolabeling and immunoprecipitation
Pulse-chase metabolic radiolabeling with [35S]Met/Cys (PerkinElmer Life Sciences) was performed as described previously (33, 34), with and without the lysosomal thiol protease inhibitor FMK024 (morpholinourea-phenylalanine-homophenylalanine-fluoromethylketone, 20 μm; MP Biomedicals, Aurora, OH) as indicated in the figure legends. Also as indicated, cells were pretreated (1 h) with tunicamycin (300 ng/ml; Sigma) and then radiolabeled in the continued presence of FMK024 and tunicamycin. Subsequent immunoprecipitations of specific radiolabeled proteins from cell lysates were performed as described previously (33, 34). Solubilization of primary immunoprecipitations and subsequent reconstitution for secondary pulldowns with TL:Bio or ECL:Bio were performed as described (32). Specific pulse and chase times are indicated in the figure legends. Immunoprecipitates and pulldowns were fractionated by SDS-PAGE and analyzed by phosphorimaging using a Typhoon FLA 9000 with ImageJ (National Institutes of Health).
Quantitative RT-PCR
TbSTT3A mRNA levels were determined using quantitative RT-PCR. Total RNA was isolated from log-phase cultures using an RNeasy Mini kit (Qiagen). RNA was treated with DNase I on-column using an RNase-Free DNase Set (Qiagen), and corresponding cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad), both according to the manufacturer's instructions. Quantitative RT-PCR was performed using diluted cDNAs and Power SYBR Green PCR Master Mix (Life Technologies, Inc.) with oligonucleotide pairs targeting the transcripts: TbSTT3A from nt 1668 to 1764 and TbZFP3 (Tb927.3.720) from nt 241 to 301. Amplification was performed using an Applied Biosystems StepOne Real-Time PCR System (Life Technologies). For each transcript, post-amplification melting curves indicated a single dominant product. TbSTT3A RNA transcripts were normalized to the internal reference gene TbZFP3 (52). All reactions were performed in technical triplicate, and means ± S.D. for three biological replicates are presented.
Deglycosylation and exoglycosidase treatment
Enzymatic deglycosylation of TfR using Endo H and PNGase F was performed as described previously (32). Briefly, trypanosomes were pulse-chase radiolabeled with [35S]Met/Cys, and polypeptides were immunoprecipitated with anti-TfR. Precipitates were solubilized in 1% SDS, collected by ethanol precipitation, and then treated with the glycosidases mentioned above according to the manufacturer's specifications (New England Biolabs, Ipswich MA). Samples were then fractionated by SDS-PAGE and visualized by phosphorimaging.
Exoglycosidase treatment of p67 and TfR N-glycans by exo-β1–4-galactosidase and exo- β1–2,3,4,6-N-acetylglucosaminidase (New England Biolabs) was done using a modified version of the protocol described above. After precipitation with ethanol, the pellets were solubilized in 10 μl of 0.5% SDS, 40 mm DTT and heated at 95 °C for 10 min. The denatured glycoproteins were chilled on ice and microcentrifuged for 10 s. The supernatants were used as substrates for the exoglycosidase treatment. The substrates were mock-treated or not in a 50-μl final volume of 5 mm CaCl2, 50 mm sodium acetate, pH 5.5, 8 units of exo-β1–2,3,4,6-N-acetylglucosaminidase, and 16 units of exo-β1–4-galactosidase at 37 °C for 16 h. Samples were then fractionated by SDS-PAGE and visualized by phosphorimaging.
Affinity purification of TfR
TfR was affinity-purified with transferrin-beads using a methodology similar to that of Mehlert et al. (29). BSF cells were harvested, washed, and lysed at 5 × 108 cells/ml in TEN buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA) containing 1% Nonidet P-40 and protease inhibitor mixture (33). Lysates were incubated at (37 °C, 10 min) to allow generation of soluble form TfR by GPI hydrolysis and then clarified by centrifugation. Holo-transferrin (Sigma-Aldrich) was coupled to Amino-LinkTM beads according to the manufacturer's directions (Thermo Fisher Scientific). The lysate was mixed with Tf-beads (1 ml of lysate, 600 μl of 75% slurry, 4 °C, overnight). After washing, bound TfR was eluted twice using 300 μl of 100 mm glycine, pH 2.5, and rapidly neutralized with 40 μl of 1 m Tris-HCl, pH 7.5. The pooled eluates were concentrated to 100 μl by spin filtration, and 20 μl (108 cell equivalents) was analyzed by lectin and immunoblotting. As a positive control, TfR was immunoprecipitated using anti-TfR as described above.
Author contributions
C. M. K., C. T., K. J. S., and J. D. B. conceptualization; C. M. K., C. T., and J. D. B. data curation; C. M. K., C. T., and J. D. B. formal analysis; C. M. K., C. T., and K. J. S. investigation; J. D. B. funding acquisition; J. D. B. project administration.
Supplementary Material
Acknowledgments
We are grateful to Professors Piet Borst and Henri Luenen (Netherlands Cancer Institute, Amsterdam) for generously providing anti-TfR antibodies.
This work was supported by NIAID, National Institutes of Health, Grant R01 AI035739 and funds from the Jacobs School of Medicine and Biomedical Sciences (to J. D. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- GPI
- glycosylphosphatidylinositol
- BSF
- bloodstream form
- VSG
- variant surface glycoprotein
- PCF
- procyclic form
- LLO
- lipid-linked oligosaccharides
- OST
- oligosaccharyltransferase
- LacNac
- N-acetyllactosamine
- pNAL
- poly-N-acetyllactosamine
- Tf
- transferrin
- TfR
- transferrin receptor
- ESAG
- expression site–associated gene
- E6 and E7
- ESAG6 and ESAG7, respectively
- TL
- tomato lectin
- Endo H
- endo-β-N-acetylglycosidase H
- PNGase F
- peptide-N-glycanase F
- ECL
- Erythrina cristagalli lectin
- TL:Bio
- tomato lectin-biotin
- ECL:Bio
- E. cristagalli lectin:biotin
- FMK024
- morpholinourea-phenylalanine-homophenylalanine-fluoromethylketone
- ER
- endoplasmic reticulum
- nt
- nucleotide(s)
- Man
- mannose.
References
- 1. Welburn S. C., Molyneux D. H., and Maudlin I. (2016) Beyond tsetse: implications for research and control of human African trypansomiasis epidemics. Trends Parasitol. 32, 230–241 10.1016/j.pt.2015.11.008 [DOI] [PubMed] [Google Scholar]
- 2. Walker G., Dorrell R. G., Schlacht A., and Dacks J. B. (2011) Eukaryotic systematics: a user's guide for cell biologists and parasitologists. Parasitology 138, 1638–1663 10.1017/S0031182010001708 [DOI] [PubMed] [Google Scholar]
- 3. Clayton C. E. (2016) Gene expression in kinetoplastids. Curr. Opin. Microbiol. 32, 46–51 10.1016/j.mib.2016.04.018 [DOI] [PubMed] [Google Scholar]
- 4. Read L. K., Lukeš J., and Hashimi H. (2016) Trypanosome RNA editing: the complexity of geting U in and taking U out. Wiley Interdiscip. Rev. RNA 7, 33–51 10.1002/wrna.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haanstra J. R., González-Marcano E. B., Gualdrón-López M., and Michels P. A. M. (2016) Biogenesis, maintenance, and dynamics of glycosomes in trypanosomatid parasites. Biochim. Biophys. Acta 1863, 1038–1048 10.1016/j.bbamcr.2015.09.015 [DOI] [PubMed] [Google Scholar]
- 6. Bangs J. D., Hereld D., Krakow J. L., Hart G. W., and Englund P. T. (1985) Rapid processing of the carboxyl terminus of a trypanosome variant surface glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 82, 3207–3211 10.1073/pnas.82.10.3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferguson M. A. J., Duszenko M., Lamont G. S., Overath P., and Cross G. A. M. (1986) Biosynthesis of Trypanosoma brucei variant surface glycoprotein: N-glycosylation and addition of a phosphatidylinositol membrane anchor. J. Biol. Chem. 261, 356–362 [PubMed] [Google Scholar]
- 8. Ferguson M. A. J., Homans S. W., Dwek R. A., and Rademacher T. W. (1988) Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science 239, 753–759 10.1126/science.3340856 [DOI] [PubMed] [Google Scholar]
- 9. Ferguson M. A. J. (1999) The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 112, 2799–2809 [DOI] [PubMed] [Google Scholar]
- 10. Ferguson M. A. J., Hart G. W., and Kinoshita T. (2017) Glycosylphosphatidylinositol anchors. in Essentials of Glycobiology, 3rd Ed. (Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L., and Seeberger P. H., eds) pp. 2015–2017, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 11. Bangs J. D., Doering T. L., Englund P. T., and Hart G. W. (1988) Biosynthesis of a variant surface glycoprotein of Trypanosoma brucei: processing of the glycolipid membrane anchor and N-linked oligosaccharides. J. Biol. Chem. 263, 17697–17705 [PubMed] [Google Scholar]
- 12. Mayor S., Menon A. K., and Cross G. A. M. (1992) Galactose-containing glycosylphosphatidylinositols in Trypanosoma brucei. J. Biol. Chem. 267, 754–761 [PubMed] [Google Scholar]
- 13. Pontes de Carvalho L. C., Tomlinson S., Vandekerckhove F., Bienen E. J., Clarkson A. B., Jiang M.-S., Hart G. W., and Nussenzweig V. (1993) Characterization of a novel trans-sialidase of Trypanosoma brucei procyclic trypomastigotes and identification of procyclin as the main sialic acid acceptor. J. Exp. Med. 177, 465–474 10.1084/jem.177.2.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kelleher D. J., and Gilmore R. (2006) An evolving view of eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R 10.1093/glycob/cwj066 [DOI] [PubMed] [Google Scholar]
- 15. Mohorko E., Glockshuber R., and Aebi M. (2011) Oligosaccharyltransferase: the central enzyme of N-linked glycosylation. J. Inherit. Metab. Dis. 34, 869–878 10.1007/s10545-011-9337-1 [DOI] [PubMed] [Google Scholar]
- 16. Parodi A. J. (1993) N-Glycosylation in trypanosomatid protozoa. Glycobiology 3, 193–199 10.1093/glycob/3.3.193 [DOI] [PubMed] [Google Scholar]
- 17. Izquierdo L., Schulz B. L., Rodriques J. A., Güther M. L., Procter J. B., Barton G. J., Aebi M., and Ferguson M. A. J. (2009) Distinct donor and acceptor specificities of Trypanosoma brucei oligosaccharyltransferases. EMBO J. 28, 2650–2661 10.1038/emboj.2009.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manthri S., Güther M. L., Izquierdo L., Acosta-Serrano A., and Ferguson M. A. J. (2008) Deletion of the TbALG3 gene demonstrates site-specific N-glycosylation and N-glycan processing in Trypanosoma brucei. Glycobiology 18, 367–383 10.1093/glycob/cwn014 [DOI] [PubMed] [Google Scholar]
- 19. Izquierdo L., Nakanishi M., Mehlert A., Machray G., Barton G. J., and Ferguson M. A. J. (2009) Identification of a glycosylphosphatidylinositol anchor-modifying β1–3N-acetylglucosaminyl transferase in Trypanosoma brucei. Mol. Microbiol. 71, 478–491 10.1111/j.1365-2958.2008.06542.x [DOI] [PubMed] [Google Scholar]
- 20. Nolan D. P., Geuskens M., and Pays E. (1999) N-Linked glycans containing linear poly-N-acetyllactosamine as sorting signals in endocytosis in Trypanosoma brucei. Curr. Biol. 9, 1169–1172 10.1016/S0960-9822(00)80018-4 [DOI] [PubMed] [Google Scholar]
- 21. Atrih A., Richardson J. M., Prescott A. R., and Ferguson M. A. J. (2005) Trypanosoma brucei glycoproteins contain novel giant poly-N-acyetyllactosamine carbohydrate chains. J. Biol. Chem. 280, 865–871 10.1074/jbc.M411061200 [DOI] [PubMed] [Google Scholar]
- 22. Kelley R. J., Brickman M. J., and Balber A. E. (1995) Processing and transport of a lysosomal membrane glycoprotein is developmentally regulated in African trypanosomes. Mol. Biochem. Parasitol. 74, 167–178 10.1016/0166-6851(95)02493-X [DOI] [PubMed] [Google Scholar]
- 23. Alexander D. L., Schwartz K. J., Balber A. E., and Bangs J. D. (2002) Developmentally regulated trafficking of the lysosomal membrane protein p67 in Trypanosoma brucei. J. Cell Sci. 115, 3253–3263 [DOI] [PubMed] [Google Scholar]
- 24. Salmon D., Geuskens M., Hanocq F., Hanocq-Quertier J., Nolan D., Ruben L., and Pays E. (1994) A novel heterodimeric transferrin receptor encoded by a pair of VSG expression site-associated genes in T. brucei. Cell 78, 75–86 10.1016/0092-8674(94)90574-6 [DOI] [PubMed] [Google Scholar]
- 25. Ligtenberg M. J. L., Bitter W., Kieft R., Steverding D., Janssen H., Calafat J., and Borst P. (1994) Reconstitution of a surface transferrin binding complex in insect form Trypanosoma brucei. EMBO J. 13, 2565–2573 10.1002/j.1460-2075.1994.tb06546.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Steverding D., Stierhof Y.-D., Fuchs H., Tauber R., and Overath P. (1995) Transferrin-binding protein complex is the receptor for transferrin uptake in Trypanosoma brucei. J. Cell Biol. 131, 1173–1182 10.1083/jcb.131.5.1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salmon D., Hanocq-Quertier J., Paturiaux-Hanocq F., Pays A., Tebabi P., Nolan D. P., Michel A., and Pays E. (1997) Characterization of the ligand-binding site of the transferrin receptor in Trypanosoma brucei demonstrates a structural relationship with the N-terminal domain of the variant surface glycoprotein. EMBO J. 16, 7272–7278 10.1093/emboj/16.24.7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mehlert A., and Ferguson M. A. J. (2007) Structure of the glycosylphosphatidylinositol anchor of the Trypanosoma brucei transferrin receptor. Mol. Biochem. Parasitol. 151, 220–223 10.1016/j.molbiopara.2006.11.001 [DOI] [PubMed] [Google Scholar]
- 29. Mehlert A., Wormald M. R., and Ferguson M. A. J. (2012) Modeling of the N-glycosylated transferrin receptor suggests how transferrin binding can occur within the surface coat of Trypanosoma brucei. PLoS Pathog. 8, e1002618 10.1371/journal.ppat.1002618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Merkle R. K., and Cummings R. D. (1987) Relationship of terminal sequences to the length of poly-N-acetyllactosamine chains in asparagine-linked oligosaccharides from the mouse lymphoma cell line BW5147. J. Biol. Chem. 262, 8179–8189 [PubMed] [Google Scholar]
- 31. Oguri S. (2005) Analysis of sugar chain-binding specificity of tomato lectin using lectin blot: recognition of high mannose-type N-glycans produced by plants and yeast. Glycoconj. J. 22, 453–461 10.1007/s10719-005-5329-4 [DOI] [PubMed] [Google Scholar]
- 32. Schwartz K. J., Peck R. F., and Bangs J. D. (2013) Intracellular trafficking and glycobiology of TbPDI2, a stage-specific protein disulfide isomerase in Trypanosoma brucei. Eukaryot. Cell 12, 132–141 10.1128/EC.00293-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schwartz K. J., Peck R. F., Tazeh N. N., and Bangs J. D. (2005) GPI valence and the fate of secretory membrane proteins in African trypanosomes. J. Cell Sci. 118, 5499–5511 10.1242/jcs.02667 [DOI] [PubMed] [Google Scholar]
- 34. Tiengwe C., Bush P. J., and Bangs J. D. (2017) Controlling transferrin receptor trafficking with GPI-valence in bloodstream stage African trypanosomes. PLoS Pathog. 13, e1006366 10.1371/journal.ppat.1006366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Umaer K., Bush P. J., and Bangs J. D. (2018) Rab11 mediates selective recycliing and endocytic trafficking in Trypanosoma brucei. Traffic 19, 406–420 10.1111/tra.12565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Itakura Y., Nakamura-Tsuruta S., Kominami J., Sharon N., Kasai K., and Hirabayashi J. (2007) Systematic comparison of oligosaccharide specificity of Ricinus communis agglutinin I and Erythrina lectins: a search by frontal affinity chromatography. J. Biochem. 142, 459–469 10.1093/jb/mvm153 [DOI] [PubMed] [Google Scholar]
- 37. Mehlert A., Richardson J. M., and Ferguson M. A. J. (1998) Structure of the glycosylphosphatidylinositol membrane anchor of a class-2 variant surface glycoprotein from Trypanosoma brucei. J. Mol. Biol. 277, 379–392 10.1006/jmbi.1997.1600 [DOI] [PubMed] [Google Scholar]
- 38. Steverding D., Stierhof Y.-D., Chaudhri M., Ligtenberg M., Schell D., Beck-Sickinger A. G., and Overath P. (1994) ESAG 6 and 7 products of Trypanosoma brucei form a transferrin binding protein complex. Eur. J. Cell Biol. 64, 78–87 [PubMed] [Google Scholar]
- 39. Maier A., and Steverding D. (1996) Low affinity of Trypanosoma brucei transferrin receptor to apotransferrin at pH 5 explains the fate of the ligand during endocytosis. FEBS Lett. 396, 87–89 10.1016/0014-5793(96)01073-3 [DOI] [PubMed] [Google Scholar]
- 40. Mussmann R., Engstler M., Gerrits H., Kieft R., Toaldo C. B., Onderwater J., Koerten H., van Luenen H. G. A. M., and Borst P. (2004) Factors affecting the level and localization of the transferrin receptor in Trypanosoma brucei. J. Biol. Chem. 279, 40690–40698 10.1074/jbc.M404697200 [DOI] [PubMed] [Google Scholar]
- 41. Zitzmann N., Mehlert A., Carroué S., Rudd P. M., and Ferguson M. A. J. (2000) Protein structure controls the processing of the N-linked oligosaccharides and glycosylphosphatidylinositol glycans of variant surface glycoproteins expressed in bloodstream form Trypanosoma brucei. Glycobiology 10, 243–249 10.1093/glycob/10.3.243 [DOI] [PubMed] [Google Scholar]
- 42. Bangs J. D., Ransom D. M., McDowell M. A., and Brouch E. M. (1997) Expression of bloodstream variant surface glycoproteins in procyclic stage Trypanosoma brucei: role of GPI anchors in secretion. EMBO J. 16, 4285–4294 10.1093/emboj/16.14.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paturiaux-Hanocq F., Zitzmann N., Hanocq-Quertier J., VanHamme L., Rolin S., Geuskens M., Ferguson M. A. J., and Pays E. (1997) Expression of a variant surface glycoprotein of Trypanosoma gambiense in procyclic forms of Trypanosoma brucei shows that cell type dictates the nature of the glycosylphosphatidylinositol membrane anchor attached to the glycoprotein. Biochem. J. 324, 885–895 10.1042/bj3240885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Damerow M., Graalfs F., Güther M. L., Mehlert A., Izquierdo L., and Ferguson M. A. J. (2016) A gene of the β3-glycosyltransferase family encodes N-acetylglucosaminyltransferase II function in Trypanosoma brucei. J. Biol. Chem. 291, 13834–13845 10.1074/jbc.M116.733246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wirtz E., Leal S., Ochatt C., and Cross G. (1999) A tightly regulated inducible expression system for conditional gene knockouts and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 10.1016/S0166-6851(99)00002-X [DOI] [PubMed] [Google Scholar]
- 46. Hirumi H., and Hirumi K. (1994) Axenic culture of African trypanosome bloodstream forms. Parasitol. Today 10, 80–84 10.1016/0169-4758(94)90402-2 [DOI] [PubMed] [Google Scholar]
- 47. Silverman J. S., Schwartz K. J., Hajduk S. L., and Bangs J. D. (2011) Late endosomal Rab7 regulates lysosomal trafficking of endocytic but not biosynthetic cargo in Trypanosoma brucei. Mol. Microbiol. 82, 664–678 10.1111/j.1365-2958.2011.07842.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Berriman M., Hall N., Sheader K., Bringaud F., Tiwari B., Isobe T., Bowman S., Corton C., Clark L., Cross G. A. M., Hoek M., Zanders T., Berberof M., Borst P., and Rudenko G. (2002) The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 122, 131–140 10.1016/S0166-6851(02)00092-0 [DOI] [PubMed] [Google Scholar]
- 49. Tiengwe C., Muratore K. A., and Bangs J. D. (2016) Surface proteins, ERAD and antigenic variation in Trypanosoma brucei. Cell Microbiol. 18, 1673–1688 10.1111/cmi.12605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eisenhaber B., Bork P., and Eisenhaber F. (1999) Prediction of potential GPI-modification sites in proprotein sequences. J. Mol. Biol. 292, 741–758 10.1006/jmbi.1999.3069 [DOI] [PubMed] [Google Scholar]
- 51. Peck R. F., Shiflett A. M., Schwartz K. J., McCann A., Hajduk S. L., and Bangs J. D. (2008) The LAMP-like protein p67 plays an essential role in the lysosome of African trypanosomes. Mol. Microbiol. 68, 933–946 10.1111/j.1365-2958.2008.06195.x [DOI] [PubMed] [Google Scholar]
- 52. MacGregor P., Savill N. J., Hall D., and Matthews K. R. (2011) Transmission stages dominate trypanosome within-host dynamics during chronic infections. Cell Host Microbe 9, 310–318 10.1016/j.chom.2011.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Clayton C. E., and Mowatt M. R. (1989) The procyclic acidic repetitive proteins of Trypanosoma brucei: purification and post-translational modifications. J. Biol. Chem. 264, 15088–15093 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.