Mammalian complex-type N-glycan synthesis is a multi-step process that results in heterogeneous glycosylation of proteins. Heterogeneity in therapeutic glycoproteins causes difficulties for protein purification and process reproducibility and can lead to variable therapeutic efficacy. Here we report engineered mammalian cell lines that have a shortened Golgi N-glycosylation pathway, which express proteins with small, sialylated trisaccharide N-glycans. This glycoengineering strategy, which we call GlycoDelete, results in proteins with substantially reduced glycan heterogeneity.
Each step in mammalian N-glycan biosynthesis (Fig. 1a, upper part) is <100% efficient and some enzymes compete for substrates, resulting in many different glycoforms. Heterogenous glycosylation presents problems in the production of therapeutic proteins. For example, glycans can affect pharmacokinetics and biological activities1,2. However, N-glycans are often crucial for protein folding and so these difficulties cannot be overcome by removing N-glycosylation sites. The yeast Pichia pastoris has been engineered3,4 to produce proteins with relatively homogenous, humanized complex-type N-glycans. Several mammalian cell glycoengineering strategies have been developed. For example, the antibody-dependent cell-mediated cytotoxicity of IgG antibodies can be enhanced by modifying the Fc-linked N-glycan. This is a desirable feature for antibodies used in tumor treatment5,6. However, no generally applicable mammalian cell glycoengineering technology is available to produce glycoproteins in which N-glycosylation is reduced to small structures that simplify protein manufacturing, and this is the goal of this study.
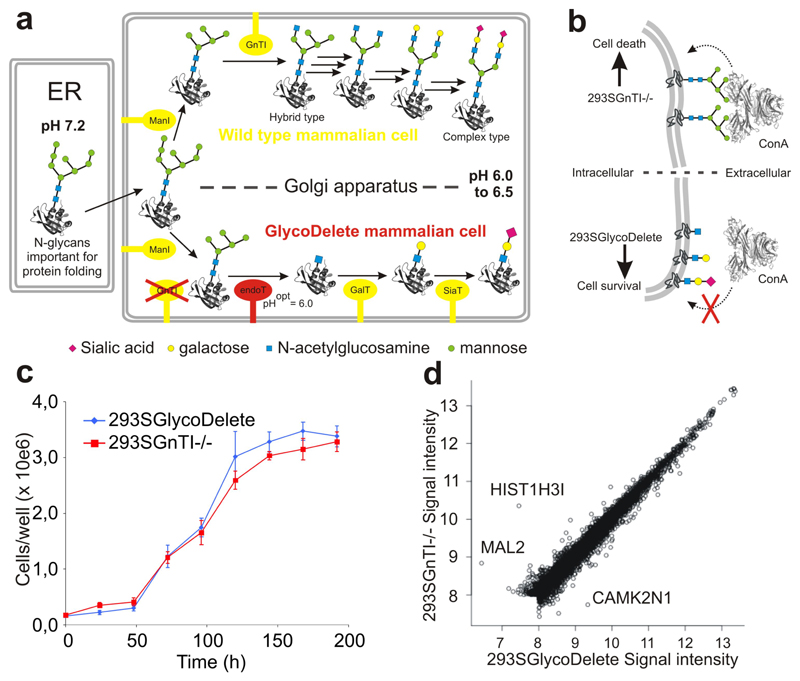
Fig. 1. The GlycoDelete strategy & cell line characterization.
(a) In mammalian cells with intact glycosylation machinery (a; upper part), oligomannose glycans entering the Golgi are trimmed by class I mannosidases (ManI) to Man5GlcNAc2. They are committed to hybrid or complex type N-glycans after modification by N-acetylglucosaminyltransferase 1 (GnTI) with a β-1,2-N-acetylglucosamine on the α-1,3-mannose. Several glycosylhydrolases and glycosyltransferases further model complex type N-glycans through many biosynthetic steps (black arrows; enzymes not shown), leading to substantial heterogeneity. In GlycoDelete cells, (a; lower part) GnTI is deleted and endoT is targeted to the Golgi, resulting in hydrolysis of oligomannose N-glycans by endoT. The resulting single GlcNAc stumps can be elongated by Golgi-resident galactosyl- and sialyltransferases. (b) Concanavalin A selection directly selects for the desired GlycoDelete glycan phenotype, as deglycosylation of cell surface glycoproteins by endoT results in the absence of ConA ligands, rendering cells resistant to ConA. The parental 293SGnTI-/- cells die when treated with ConA. (c) Growth curve for 293SGnTI-/- and 293SGlycoDelete cells in 6-well culture plates, counted every 24 hours. Error bars are standard deviations (S.D.), n=3. Numerical data for this graph are in Supplementary Table 2. (d) Scatterplot of average (n=3) gene expression values of 7344 genes for 293SGlycoDelete versus 293SGnTI-/- cells. The correlation coefficient is 0.9865. Significantly differentially expressed genes (P<0.01) are labeled with their names. Microarray signal intensities lower than 8 on the represented scale were too low for reliable detection.
N-glycans are required for protein folding in the endoplasmic reticulum; in the Golgi some mannose groups are removed and the glycans are then elaborated by several enzymes, starting with N-acetylglucosaminyltransferase I (GnTI). Previously, human embryonic kidney 293S cells have been engineered to produce glycoproteins bearing Man5GlcNAc2 N-glycans by deleting GnTI (293SGnTI-/- cells)7. Several endo-β-N-acetylglucosaminidases are known that can hydrolyze such glycans to leave a single asparagine-linked N-acetylglucosamine (GlcNAc) residue. Here we show that Golgi-targeted expression of a endo-β-N-acetylglucosaminidase in 293SGnTI-/- cells produces single GlcNAc N-glycan ‘stumps’ that can be recognized and modified by galactosyltransferases and sialyltransferases, resulting in glycoproteins with a sialylated type II branch (Fig. 1a (lower panel)); a common feature of N- and O-glycans. This three-step pathway results in substantially reduced heterogeneity of N-glycans.
We chose endoT8 from the fungus Hypocrea jecorina as the endo-β-N-acetylglucosaminidase to target to the Golgi, because optimum pH for endoT function (pH 6.0) is close that in the mammalian trans-Golgi apparatus. In order to target endoT to the trans-Golgi of 293SGnTI-/- cells, we fused the endoT coding sequence without its predicted signal sequence to Golgi targeting domains from two human enzymes normally present in the Golgi (Supplementary Fig.1). When the endoT catalytic domain was fused to the targeting domain of the human β-galactoside-α-2,6-sialyltransferase 1 (ST6GAL1)9 (referred to as the ST-endoT fusion protein), it was retained intact in the cells. Transient expression of ST-endoT in 293SGnTI-/- cells resulted in in vivo deglycosylation of a stably expressed and secreted Flt3 receptor extracellular domain10 (Supplementary Fig. 2).
To establish a 293SGnTI-/- derived cell line stably expressing ST-endoT fusion protein, we selected for cells with the desired glycan phenotype using concanavalin A (ConA). ConA is a tetrameric cytotoxic lectin that binds to oligomannose and hybrid-type N-glycans. Full deglycosylation of cell surface glycoproteins by endoT would result in the absence of ConA ligands, thus rendering the cells resistant to this lectin (Figure 1b). Four weeks after transfection, we obtained clones resistant to ConA (used at the lowest concentration that killed all of the parental 293SGnTI-/- cells). Two clones were selected for robust growth, and that with the highest ConA-resistance was named 293SGlycoDelete (Supplementary Fig. 3). Genomic integration and expression of ST-endoT were validated by PCR and immunoblotting, respectively (Supplementary Fig. 4). 293SGlycoDelete cells have similar morphology to 293SGnTI-/- cells and have an indistinguishable growth rate (Fig. 1c). However, we noticed that 293SGlycoDelete cells are less adherent than 293SGnTI-/- cells; this is a desirable feature for suspension cultivation, as used in biopharmaceutical production.
We profiled the transcriptomes of 293SGnTI-/- and 293SGlycoDelete cells using exon microarrays and found that only 3 of the 7344 genes that had detectable expression were more than 2-fold differentially expressed (P < 0.01) between the two cell lines (Fig. 1d). Transcriptional comparison of 293SGnTI-/- line and the 293S parent showed differential transcription of about 70 genes (Supplementary Fig. 5), without clear enrichment for particular pathways. We have observed substantial genomic rearrangement in the 293SGnTI-/- line (L.M., M.B., N.C., unpublished observations), which may account for these differences. Therefore, GlycoDelete engineering does not substantially alter the transcriptional profile of the cells. The absence of a transcriptional signature of the unfolded protein response11 in the 293S GlycoDelete cells demonstrates that the GlycoDelete strategy does not noticeably interfere with the role of N-glycans in ER-quality control.
To assess the efficiency of N-glycan remodelling in the 293SGlycoDelete line, granulocyte-macrophage colony-stimulating factor (GM-CSF)12 was transiently expressed in 293S, 293SGnTI-/- and 293SGlycoDelete cells and purified from the culture medium. GM-CSF produced in 293S or 293SGnTI-/- cells runs as three main bands (corresponding to occupancy of none, one or two of the N-glycosylation sites13), which are converted to a lower molecular weight (MW) form of the protein by treatment with PNGaseF (which cleaves the N-glycosidic bond between the Asn side chain and N-glycans that contain at least the chitobiose core) (Fig. 2a). O-glycosylation13 was responsible for at least some of the remaining heterogeneity in GM-CSF MW, as indicated by its partial disappearance upon sialidase digestion. GM-CSF purified from 293SGlycoDelete cells ran at a lower MW (Fig. 2a) and PNGaseF treatment did not cause any change in the observed pattern, demonstrating the absence of chitobiose core-containing N-glycans. Treatment with sialidase caused a more substantial shift in the MW of GlycoDelete GM-CSF than for GM-CSF from 293S or 293SGnTI-/- cells, indicating the presence of more sialic acid residues on GlycoDelete GM-CSF (Fig. 2a). This conclusion is also supported by the glycan analytics described below (Fig. 2b, Supplementary Fig. 6 and 7). After digestion with both PNGaseF and sialidase, GM-CSF from all three cell lines ran as single bands with indistinguishable mobility (note that these gels do not resolve non-glycosylated proteins from those modified with the small GlycoDelete N-glycan stumps), supporting the conclusion that the differences between GM-CSF from 293S, 293SGnTI-/- and 293SGlycoDelete cells were due to glycosylation differences; this was confirmed by mass spectrometrical analysis of the intact proteins (Supplementary Fig. 8).
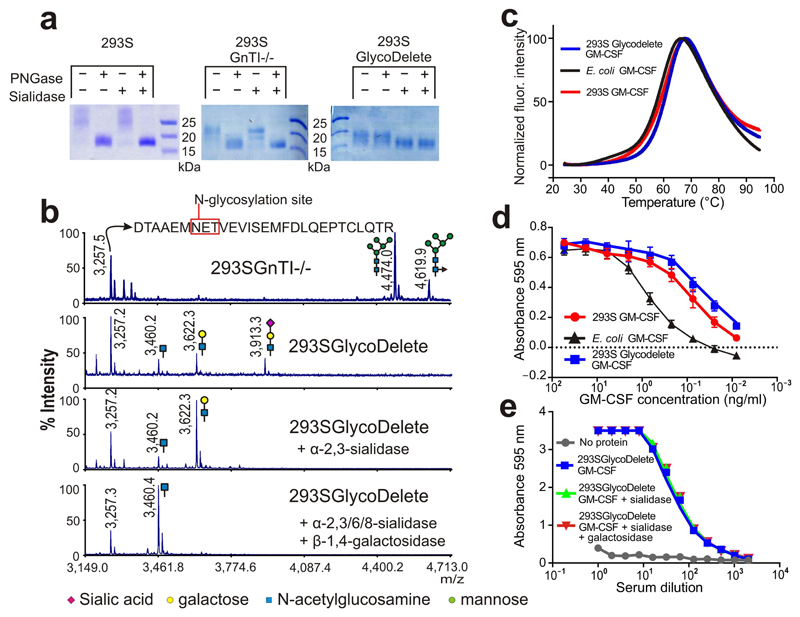
Fig. 2. GlycoDelete glycan characterization.
(a) SDS-PAGE of 293S, 293SGnTI-/- and 293SGlycoDelete GM-CSF samples. Each sample was treated with PNGaseF, sialidase or both enzymes, analysed on an SDS-PAGE gel and stained with coomassie brilliant blue. The non-cropped can be found in Supplementary Fig. 15. (b) MALDI-TOF-MS spectra of GM-CSF samples. Peaks are labeled with their m/z values. The spectrum of the 293SGnTI-/- GM-CSF reveals the presence of Man5GlcNAc2 and fucosylated Man5GlcNAc2 on the glycopeptide containing N37 (top spectrum; left and right glycans, respectively). These glycoforms are absent in GlycoDelete GM-CSF (2nd spectrum). New peaks at m/z values corresponding to HexNAc, Hex-HexNAc and Sia-Hex-HexNAc modified glycopeptides are detected. Spectra of exoglycosidase-digested GlycoDelete GM-CSF N-glycans with α-2,3-sialidase or both a broad spectrum sialidase and β-1,4-galactosidase are shown. These spectra show that N-glycans on GlycoDelete GM-CSF N37 are Neu5Ac-α-2,3-Gal-β-1,4-GlcNAc and Gal-β-1,4-GlcNAc. (c) Thermofluor assay of 293S, 293SGlycoDelete and E. coli produced GM-CSF. We observed similar average (n=3) melting curves for all GM-CSF glycoforms (Tm is approximately 60°C). (d) Bioactivity of 293S and 293SGlycoDelete produced GM-CSF as measured in a TF1 erythroleukemia cell proliferation assay (n=3). E. coli produced GM-CSF serves as a non-glycosylated control sample. The error bars are S.D. Numerical data for this graph are in Supplementary Table 3. (e) ELISA analysis of anti-glycan antibody titers in GlycoDelete GM-CSF immunized rabbit serum. Removal of sialic acid and galactose monosaccharides from the GlycoDelete glycan does not reduce serum antibody recognition. Numerical data for this graph are in Supplementary Table 4.
To further characterize the N-glycans on GM-CSF from 293SGlycoDelete and 293SGnTI-/- cells, we analysed the samples by MALDI mass spectrometry (Fig. 2b and Supplementary Fig. 6). 293S GM-CSF glycans were analysed by capillary electrophoresis (Supplementary Fig. 7) and revealed a typical heterogenous mixture of multibranched complex-type glycans. The level of sialylation was low, likely due to the rapid transfer of the cells to serum-free medium during protein production. The glycopeptide containing N37 of 293SGnTI-/- GM-CSF was detected as Man5GlcNAc2(Fuc) N-glycosylated peptide (Fig. 2b, upper trace), in agreement with previous findings7,14. These ions are absent from the spectrum of GM-CSF produced in 293SGlycoDelete cells, in which we detected three new glycopeptide masses. These masses are consistent with HexNAc-glycopeptide, Hex-HexNAc-glycopeptide and Neu5Ac-Hex-HexNAc-glycopeptide (Fig. 2b). Similar observations were made for the glycopeptide containing N27 (Supplementary Fig. 6).
To confirm the identity and linkage of the hexose and Neu5Ac units on GlycoDelete GM-CSF glycopeptides, we performed exoglycosidase digests with an α-2,3-sialidase or α-2,3/6/8-sialidase and β-1,4-galactosidase (Fig. 2b). This allowed us to establish the Gal-β-1,4-GlcNAc and Neu5Ac-α-2,3-Gal-β-1,4-GlcNAc identity of the di-and trisaccharide modified peptides. The presence of these glycans - not just the single GlcNAc endoT digestion product - on proteins produced in the GlycoDelete cells shows that galactosyl- and sialyltransferases in the Golgi act on the GlcNAc stumps generated by endoT. This confirms that endoT deglycosylation of GM-CSF must have occurred intracellularly and not post-secretionally. Quantification of the relative peak intensities of the pre- and post-sialidase treated spectra indicated that approximately 75% of glycans on GM-CSF from GlycoDelete cells were sialylated.
We then investigated the influence of the GlycoDelete glycan alteration on properties of GM-CSF. A thermofluor assay15 showed that the melting temperatures of GM-CSF from E. coli (non-glycosylated, Tm = 58.9±0.6°C), 293S cells (complex type N-glycosylation, Tm = 61.2±3.2°C) and 293SGlycoDelete cells (Tm = 61.5±0.2°C) were not significantly different (Kruskal-Wallis test, n=4, p>0.05) (Fig. 2c). Furthermore, in a TF1 human erythroleukemia cell proliferation assay (Fig. 2d)16, the bioactivity of GM-CSF from 293S and 293SGlycoDelete cells was indistinguishable.
To assess whether GlycoDelete glycans contribute to the antigenicity of GM-CSF, we immunized rabbits with GM-CSF from 293SGlycoDelete cells. Binding of serum antibodies to undigested, sialidase treated or sialidase and galactosidase treated 293SGlycoDelete GM-CSF was determined by ELISA. GM-CSF from which the GlycoDelete glycan structures had been removed and that with the GlycoDelete glycans present were recognised equally well, indicating that the GlycoDelete glycans did not form new immunogenic epitopes on GM-CSF in rabbits (Fig. 2e).
To further explore the scope of the GlycoDelete technology, the monoclonal anti-CD20 antibody GA101 (ref. 17) was transiently expressed and purified from 293S and 293SGlycoDelete cells. The cell lines produced similar amounts of the molecule (Supplementary Fig. 9). Anti-CD20 produced by 293S cells carries core-fucosylated biantennary N-glycans, typical of IgG antibodies18 on the single N-glycosylation site (N297) in the heavy chain Cγ2-domain (Supplementary Fig. 10). As expected, treatment with PNGaseF reduced the MW (Fig. 3a). By contrast, the antibody produced in 293SGlycoDelete cells ran at approximately the same MW as the PNGaseF treated antibody from 293S cells, and the MW was not further reduced by PNGaseF treatment (Fig. 3a). This result is consistent with the N-glycans on this hIgG having been cleaved by endoT.
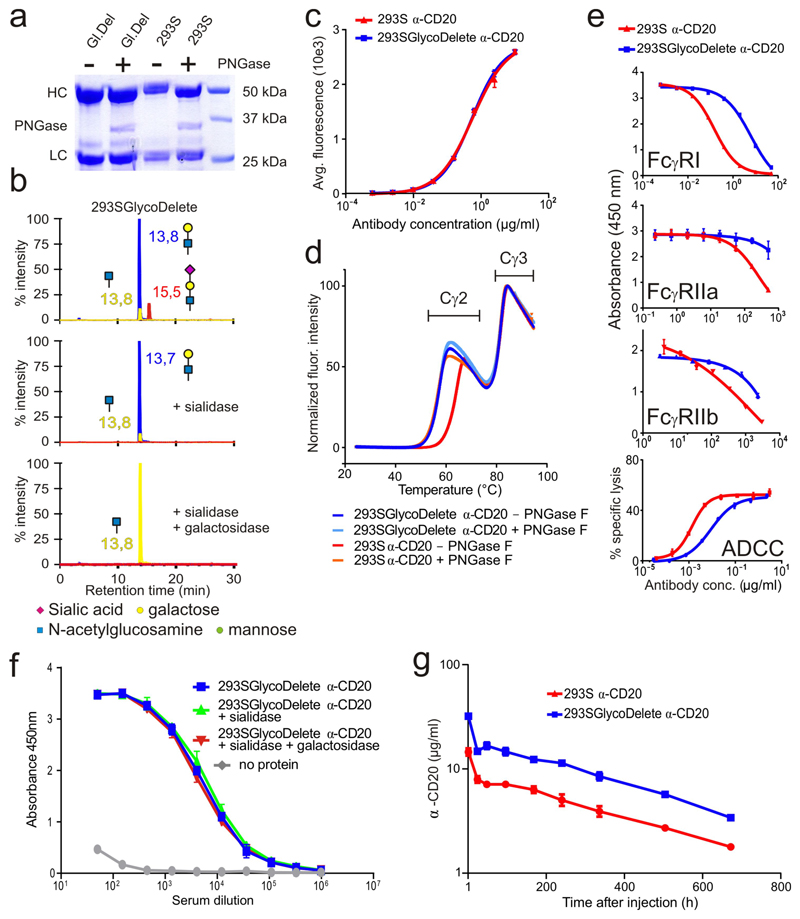
Fig. 3. Functional and immunological characterization of GlycoDelete anti-CD20.
(a) Anti-CD20 SDS-PAGE. Gl. Del: 293SGlycoDelete anti-CD20. HC: antibody heavy chain. LC: antibody light chain. The non-cropped gel is in Supplementary Fig. 15. (b) LC-MS/MS in SRM mode of GlycoDelete anti-CD20 glycopeptides. Peak labels state LC elution times. Peak color-codes: trisaccharide- (red), disaccharide- (blue) or monosaccharide-modified (yellow) glycopeptides. Exoglycosidase digests with sialidase and β-1,4-galactosidase illustrate identical glycans as observed for GM-CSF. (c) CD20-binding by anti-CD20 as assessed by flow cytometry. Numerical data for this graph are in Supplementary Table 5. (d) Average melting curves (n=3) as determined in a thermofluor assay for untreated or PNGase-digested 293S and 293SGlycoDelete anti-CD20. (e) Competition ELISA (top three graphs) and ADCC assay to assess effector function of the anti-CD20 Fc. Concentration series of 293S and 293SGlycoDelete anti-CD20 were compared in their competition with a coated anti-Fc antibody. The error bars indicate standard error of the mean (S.E.M.) with n=3. The 4th graph shows specific lysis in an ADCC assay. The error bars indicate S.D. and n=3. Numerical data for this graph are in Supplementary Table 6. (f) Anti-glycan antibody ELISA analysis of 293SGlycoDelete anti-CD20 immunized rabbit serum. Anti-CD20 recognition by antibodies in the serum of rabbits immunized with GlycoDelete GM-CSF was analysed. Anti-CD20 samples were treated with sialidase, sialidase and galactosidase or no enzyme. Error bars show S.D. and n=3. Numerical data for this graph are in Supplementary Table 7. (g) Anti-CD20 pharmacokinetics in mice. Blood concentrations of anti-CD20 were measured over time after intravenous injection of 293S or 293SGlycoDelete anti-CD20. Error bars are S.E.M. and n=4. Numerical data for this graph are in Supplementary Table 8.
To further characterize the glycans on 293SGlycoDelete anti-CD20, the different glycoforms of the tryptic IgG peptide containing the N-glycosylation site were quantified using liquid chromatography–electrospray ionisation mass spectrometry (LC-MS/MS) in selected reaction monitoring (SRM) mode (Fig. 3b). Furthermore, we performed LC-MS analysis of the intact antibody with and without dissociation of the chains through reduction (Supplementary Fig. 11). The LC-MS/MS analysis revealed that the GlycoDelete protein was modified with HexNAc, Gal-HexNAc and NeuNAc-Gal-HexNAc N-glycans, as we had also observed for GM-CSF. Quantification of the relative glycopeptide peak areas of pre- and post-sialidase treated samples allowed to establish that 19% of the anti-CD20 carries the sialylated trisaccharide and 72% carries the Gal-GlcNAc disaccharide, the remainder being the GlcNAc-modified peptide. In SRM mode LC-MS/MS peptide analysis, the Hex5-HexNAc2-glycopeptide that is dominant in 293SGnTI-/-IgG was below the detection limit for 293SGlycoDelete IgG. Intact protein LC-MS analysis revealed a very small remaining fraction of the Hex5-HexNAc2 glycoform, both in 293S and 293SGlycoDelete antibodies. The amount of Hex5HexNAc2 in both preparations was quantified at 2.5% of the total glycan pool by DNA-sequencer carbohydrate electrophoresis of a 1:1 mixture of both antibodies (data not shown).
In addition, flow cytometric analysis of binding to CD20+-cells revealed that GlycoDelete anti-CD20 antigen binding was identical to that of 293S anti-CD20 (Fig. 3c), demonstrating that the antigen-binding fold is unaffected. As N-glycans make up part of the fold packing contacts in the Cγ2 domain, size reduction of these glycans is expected to lead to a Tm drop. Accordingly, the Tm for Cγ2 melting is approximately 64°C for complex-type N-glycosylated 293S anti-CD20 and it drops to 57°C for 293S GlycoDelete anti-CD20, similar to PNGaseF-digested WT anti-CD20 (Fig. 3d). We found no evidence of aggregation of anti-CD20 expressed by 293S or 293SGlycoDelete cells by gel filtration chromatography (Supplementary Fig. 12).
Glycosylation on heavy chain N297 has a major influence on the affinity of binding of antibodies to Fc gamma receptors (FcγRs)19, so we assessed the binding of 293S and 293SGlycoDelete anti-CD20 to different human Fcγ receptors. Surface plasmon resonance experiments (Supplementary Table 1) showed that the human and mouse neonatal Fc receptors (FcRn) have a similar affinities for both anti-CD20 glycoforms. This is expected because the FcRn binding site is not located near the Cγ2 N-glycan site20. We set up a competition ELISA for FcγRI, FcγRIIa and FcγRIIb in which the anti-CD20 antibodies compete in solution for FcγR binding with a precoated IgG. In all three cases we detected >10x reduced binding competition by 293SGlycoDelete anti-CD20 compared to the 293S anti-CD20 (Fig. 3e). FcγRIIIa binding affinity, as assessed by Bio-Layer Interferometry (Supplementary Table 1), was 5.8 times lower for 293SGlycoDelete anti-CD20 than for 293S anti-CD20. Similarly, in an antibody-dependent cell-cytotoxicity (ADCC) assay using natural killer (NK) cells as effectors (Fig. 3e) we found that the EC50 of the specific lysis with 293SGlycoDelete anti-CD20 is 6.6 times higher than with 293S anti-CD20. Overall, the GlycoDelete glycosylation of hIgG1 Fc leads to reduced binding to FcγRs; in the context of producing neutralizing antibodies, this might be desirable to improve safety21.
To assess whether GlycoDelete glycans on the IgG are immunogenic, we performed a similar immunization experiment as for GM-CSF (Fig. 3f) and again concluded that GlycoDelete glycans do not substantially contribute to antigenicity of the anti-CD20 molecule.
Remarkably, pharmacokinetic analysis in mice showed that a higher serum level of 293SGlycoDelete antibody is attained relative to the 293S variant. Beyond the peak concentration, both glycoforms were cleared at a very similarly slow rate (Fig. 3g, Supplementary Fig. 13), as was anticipated from the identical FcRn affinity. Consequently, it takes 10-12 days longer for the concentration of the GlycoDelete antibody to drop below any required therapeutic threshold concentration. Further studies are needed to address the mechanism for this immediate reduced clearance of the GlycoDelete antibody. A possibility is that higher sialylation would lead to lower clearance through reduced binding to liver and macrophage lectin receptors. Together with a clear scope for further enhancing the sialylation levels of GlycoDelete IgG, this offers exciting prospects for reduced frequency of dosing for neutralizing therapeutic IgGs that often require long circulation periods in the blood.
To establish that GlycoDelete is compatible with stable transfection-based protein production and that it can process membrane proteins, we stably expressed the 5HT1D22 G-protein coupled receptor (GPCR) in 293SGlycoDelete cells. We again observed complete deglycosylation of the receptor (Supplementary Fig. 14).
In conclusion, this study introduces the GlycoDelete glycoengineering strategy, which represents a radical approach to solving the issue of N-glycosylation heterogeneity in mammalian cell-based glycoprotein production. It consists of the inactivation of a single glycosyltransferase (GnTI, encoded by the MGAT1 gene) and simple overexpression of a deglycosylating enzyme followed by lectin selection. GlycoDelete cells produce proteins with the Gal-GlcNAc disaccharide or its α-2,3-sialylated trisaccharide derivative, and some of the monosaccharide intermediate. This is in contrast to the dozens of glycan structures in WT mammalian cells. The GlycoDelete strategy allows striking an optimal balance between retaining the folding-enhancing functions of N-glycans while avoiding the extensive heterogeneity introduced through mammalian Golgi N-glycan processing. Apart from the advantages of GlycoDelete’s drastically reduced N-glycan complexity in biopharmaceutical manufacturing, examples of the therapeutic benefit of similar in vitro generated short, simple N-glycans have been reported23–25. Furthermore, we have shown that GlycoDelete engineering favorably alters the characteristics of antibodies of which the therapeutic goal is antigen neutralization without the need for additional effector function. The technology is likely to lead to ‘biobetters’, a development of active interest in the biopharmaceutical industry26. Future work will explore which other modulations of biopharmaceutical function can be achieved by the GlycoDelete structures.
Online Methods
General cell culture and transfection
We maintained 293SGnTI-/- cells in a humidified incubator at 37°C and 5% CO2 in DMEM/F12 (Gibco) with 10% fetal bovine serum, 292 μg/mL L-glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin (all Sigma-Aldrich).
For small-scale transfections, the cells were plated 48 hours prior to transfection at ~150,000 cells per 6-well. They were transfected using the TransIT®-293 Transfection Reagent (Mirus Bio LLC) according to the manufacturer’s instructions. For transient or large-scale transfections, cells were transfected with the calcium phosphate transfection method. Raji cells were cultured in RPMI 1640 + 10% FCS + 2 mM L-Glutamine.
All cell lines were routinely tested for mycoplasma contamination with the Plasmotest kit (InvivoGen).
Transient endoT expression
The endoT fusion constructs (pCAGGS-GM2S-endoT and pCAGGS-ST-endoT) and the secreted endoT construct (pCAGGS-s-endoT) were transiently transfected to 293SGnTI-/- cells as described above. Supernatant and cell lysate samples were analysed to assess targeting domain performance (Supplementary Fig. 1).
In vivo de-N-glycosylation by transient transfection of endoT-fusions
De-N-glycosylation by endoT was evaluated by transfecting all endoT constructs to 293SGnTI-/- cells stably and inducibly expressing the Flt3 receptor extracellular domain (Supplementary Fig. 2).
Construction of pcDNA3.1(-)Zeo-ST-endoT
We cloned the ST-endoT PCR fragment (primers: Supplementary Note 1) into pcDNA3.1/zeo(-) using the XhoI and KpnI restriction sites.
Stable cell line generation
We transfected 293SGnTI-/- cells in a small-scale transfection with pcDNA3.1(-)Zeo-ST-endoT. We initiated selection with 15 μg/mL concanavalin A at 48 hours post-transfection. After 14 days, the cells were trypsinized and replated in conditioned medium (medium of 2 days old 293SGnTI-/- cultures, sterile filtered and mixed with 50% (v/v) fresh DMEM/F12) containing 10 μg/mL ConA. After 14 days, five large and nicely separated colonies were picked and expanded in the presence of 10 μg/mL ConA. The two fastest growing clones were further analysed.
293SGnTI-/- and 293SGlycoDelete growth curve
Cells from a 70 – 80% confluent culture were first diluted to ~60,000 cells/mL, counted again (time point 0 hours) and transferred to a 6-well plate (180,000 cells/well). At each time point, 3 wells were detached by pipetting up and down the medium and the viable cells were counted for each well using trypan blue exclusion and a hemocytometer. The result shown in Figure 1c represents one of two replicate experiments.
Gene expression analysis
RNA isolation and sample preparation for analysis on GeneChip Human Exon 1.0 ST Arrays (Affymetrix) are described in Supplementary Note 2.
GM-CSF production and purification
pORF-hGM-CSF-6xHis was transiently transfected to both 293SGnTI-/- and 293SGlycoDelete cell lines. The secreted GM-CSF was purified from the medium as described in Supplementary Note 3.
Anti-CD20 production and purification
Anti-CD20 was transiently expressed in both 293S and 293SGlycoDelete cell lines as described above and purified as described in Supplementary Note 3.
Sialidase, galactosidase and PNGaseF digests and SDS-PAGE
We diluted the glycoproteins in 50 mM of phosphate buffer pH 7.0 containing 40 mM of β-mercapto-ethanol and 0.5% SDS. Samples were incubated for 10 minutes at 98°C. After cooling, 1% Igepal CA630 and the appropriate enzymes were added: 100 U of PNGaseF (in house production), 200 mU of Arthrobacter ureafaciens sialidase (in house production), 2mU of Streptococcus pneumoniae β-1,4-galactosidase (Prozyme) or combinations. The samples were incubated overnight at 37°C and analysed the following day on a tricine SDS-PAGE gel.
Thermofluor assays
Assays were performed as described15. Briefly, purified protein was diluted to 20 μl in a buffer (PBS for GM-CSF and 25 mM histidine, 125 mM NaCl, pH 6.00 for anti-CD20) and Sypro orange dye (Life technologies) was added. Each experiment was run as a technical triplicate, with a triplicate blank measurement without test protein. Fluorescence in function of the temperature was recorded in a 348-well Lightcycler 480 (Roche) from 25°C to 95°C at 0.01°C/s.
Melting temperatures were calculated as the V50 value of a Boltzmann sigmoidal curve fitted to the averaged data points of the three replicates in each experiment. For graphing, the raw datasets were averaged, (averaged) blank corrected and then normalized (minimal value = 0%, maximal value = 100%).
For the GM-CSF samples, an average Tm was calculated from a set of independent experiments (E. coli: n=4, 293S: n= 3, 293SGlycoDelete: n=3). We tested whether the average Tm’s were statistically significantly different in a Kruskal-Wallis (P=0.05) and Dunn test for multiple comparisons (α=0.05).
MALDI glycopeptide analysis
GM-CSF of the different cell lines (1-4 μg of protein in 20 μL) was supplemented with 10 μL of 3x Tricine gel loading buffer (1.5 M Tris-HCl, pH 8.45, 35% glycerol, 10% SDS, 0.01% coomassie and 30 mM of DTT) and incubated for 10 minutes at 98°C. 3 μL of a 500 mM iodoacetamide stock was added and the samples were incubated for 1 hour in the dark. We separated the samples on a 12% tricine SDS-PAGE gel and cut out the bands. Detailed methods for in-gel tryptic digestion are described in Supplementary Note 4. We treated the tryptic peptides with either no enzyme, 50 mU of α-2,3-sialidase (Takara Bio Inc.), or both 200 of mU A. ureafaciens sialidase and 2 mU of Streptococcus pneumoniae β-1,4-galactosidase (Prozyme). All digests were incubated for 24 hours at 37°C, dried in a speedvac, reconstituted with 10 μL of 0.2% trifluoroacetic acid (TFA) (Sigma-Aldrich) and cleaned up with C18 ZipTip® pipette tips (Millipore) according to the manufacturer’s instructions. Samples were analysed with 6-aza-2-thiothymine (ATT) matrix saturated in 50% acetonitrile containing 0.1% TFA, on a 4800 MALDI TOF/TOF™ Analyzer (Applied Biosystems) in the positive ion mode. The reported m/z values were observed in several iterations of technical optimizations and the results of the fully optimized experiments are shown.
LC-ESI-MS glycopeptide analysis
We diluted 9 μg of anti-CD20 in 20 μL of 50 mM phospate buffer pH 7.0. Either no enzyme, 100 mU of Arthrobacter ureafaciens sialidase (in house production) or 2mU of β-1,4-galactosidase (Streptococcus pneumoniae) and 100mU of sialidase were added and the mixture was incubated for 4 hours at 37°C. The samples were denatured in a 2 M ureum, 10 mM DTT, 50 mM ammonium bicarbonate buffer for 30 minutes at 60°C. Iodoacetamide was added to a concentration of 20 mM and incubated in the dark for 30 minutes. Next, the samples were digested with 1/50 (w/w) trypsin (Promega) and incubated overnight at 37°C.
The samples were loaded directly on an Acclaim PepMap 100 analytical column (L x ID 15 cm x 75 μm, C18, 3 μm, 100 Å) (Thermo) at a flow rate of 300 nL/min, on a U3000-RSLC system (Thermo). Mobile phases were 0.1% HCOOH in H2O (solvent A) and 0.1% HCOOH in acetonitrile (ACN) (solvent B). The samples were separated with a 30 minute gradient, ranging from 2% to 40% solvent B, and the eluting peptides were sprayed directly into a 4000 QTRAP mass spectrometer (AB Sciex) with the NanoSpray II ESI source (AB Sciex). A selected reaction monitoring (SRM) method was used to target the glycosylated peptide EEQYNSTYR, where the triple quadrupole cycled through the following SRM transition list with a dwell time of 250 ms: Pep-GlcNAc: 696.8 (2+)/526.3 (+) and 696.8 (2+)/1189.5 (+) (DP 81.9 V, CE 39.8 eV), Pep-GlcNAc-Gal: 777.8 (2+)/526.3 (+) and 777.8 (2+)/1189.5 (+) (DP 87.8 V, CE 43.9 eV), Pep-GlcNAc-Gal-Sial: 923.4 (2+)/526.3 (+) and 923.4 (2+)/1189.5 (+) (DP 98.4 V, CE 51.2 eV). The 526.3 Da fragment ion (y4-ion, STYR) was used as quantifier, and the 1189.5 Da fragment ion (loss of sugar-modification group) was used as qualifier. The analysis and processing of the data was done with Skyline27. This experiment was performed two times. One of the experiments was conducted as a technical duplicate, the other one as a technical triplicate.
Ratio of sialylated and galactosylated glycans
See Supplementary Note 5.
GM-CSF bioactivity experiments, TF-1 proliferation assay
TF1 cells (ATCC n° CRL-2003) were maintained in RPMI 1640, 10% (v/v) fetal bovine serum, 2 mM of L-Gln and 2 ng/mL of recombinant human GM-CSF at 37°C, 5% CO2. Before starting the assay, cells were washed three times with medium without cytokines. The cells were subsequently put back in medium (200,000 cells/mL) without cytokines and left for 2 hours at 37°C.
Upon initiation of the assay, cells were plated in a 96-well plate (20,000 cells/100μL/well) and serial dilutions (54 ng/mL – 8 pg/mL) of the different glycoforms of GM-CSF were added. Cells were incubated for 48h, 72h and 96h before performing the MTT assay (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) as described28. Briefly, 20 μl of MTT (5 mg/mL stock) was added per well and incubated. After 4h at 37°C, 80 μL of stop solution (10% SDS in 0.01 M HCl) was added and the plate was further incubated overnight at 37°C. Finally, optical density was measured at 595 nm. The data points plotted in Figure 2d represent mean values from 3 technical replicates. The error bars are S.D. The reported differences between the GM-CSF glycoforms were observed in several iterations of technical optimization of these experiments. The results of the fully optimized bioactivity experiment are shown.
Rabbit immunizations
New Zealand White male or female rabbits, aged 13-16 weeks (2 rabbits for each antigen, only one rabbit shown in Figures 2 and 3) were injected with 293S GM-CSF, Glycodelete GM-CSF, 293S anti-CD20 or Glycodelete anti-CD20. 50 μg of antigen in 500 μL of antigen solution (50 μg of protein diluted in 0.9% NaCl solution up to 500 μL) + 500 μL of complete Freund’s adjuvant was injected subcutaneously at days 0, 14, 28 and 56. Rabbits were bled on day 0 (preimmune bleeding), day 38, day 66 and day 80 (final bleeding). The immunization was performed by CER Groupe and approved by the CER Groupe ethical committee.
Serum ELISAs with GlycoDelete proteins
Glycosidase digestions were performed as described above. Wells of Maxisorp microtiter plates were coated (ON, 4°C) with 0.25 μg/mL of GM-CSF or 0.15 μg/mL of anti-CD20 in 50 μl of coating buffer (0.05 M Na2CO3, 0.05 M NaHCO3, pH 9.6) washed three times with PBS + 0.1% Tween, and blocked with 1% BSA in PBS with 250 mM glycine for 2h at room temperature. Blocking buffer was removed and the plates were dried overnight.
Detection antibodies (anti-GM-CSF rabbit serum, final bleeding; anti- (anti-CD20) rabbit serum, final bleeding) were added in PBS + 0.1% Tween20 + 0.1% goat serum and incubated for 2h at room temperature.
Plates were washed 4 times with wash buffer before adding donkey anti-rabbit HRP (1:2000) (Catalog number NA934, GE healthcare) in PBS + 1% BSA and incubating for 1h at room temperature.
We washed the plates again 3 times with wash buffer, upon which the TMB (3,3',5,5'-Tetramethylbenzidine, BD OptEIA) substrate (1:1, 100 μL/well) was added and the plate was incubated at room temperature for 30 minutes. Finally, we added 50 μL of stop solution (2 N H2SO4) and the absorbance was measured at 450 nm.
The ELISA with GM-CSF (Figure 2e) was performed once with two biological replicates (2 rabbits immunized, only one shown). The ELISA with anti-CD20 was performed once with two biological replicates (2 rabbits immunized) and one of the biological replicates was then repeated with three technical replicates. The result of the latter experiment is shown in Figure 3f. The data points plotted in this figure represent mean values from the 3 technical replicates. The error bars are S.D.
CD20 binding by anti-CD20
Fc receptors on the Raji cells were blocked with anti-CD32 antibodies IV.329 (in house production) and AT10 (Catalog number MCA1075, AbD serotec) at 10 μg/mL, incubated with the cells for one hour on ice. Next, the cells were plated into a 96-well plate (105 cells/well) and the 293S or 293SGlycoDelete anti-CD20 was added in a dilution series starting from 10 μg/mL. The cells were incubated for 1 hour at 4°C and then washed twice with PBS + 2% BSA. To detect the anti-CD20, an anti-F(Ab)2 secondary antibody conjugated to dylight 649 (Catalog number: 109-496-097, Jackson laboratories) was added at a 1:200 dilution. The cells were again incubated for 30 minutes at 4°C and washed twice with PBS + 2% BSA. To fix the cells, 150 μL of fixative (CellFIX, Becton Dickinson) was added in each well and incubated for 1 hour at 4°C. The secondary antibody was detected through flow cytometry (FACSCalibur, Becton Dickinson). The data points plotted in Figure 3c represent mean values from 3 technical replicates. The error bars are S.D. This experiment was conducted twice.
FcγR Surface plasmon resonance (SPR) experiments
A Biacore 2000 SPR biosensor (GE Healthcare) was used to assay the interaction of FcRn with the different anti-CD20 glycoforms. All experiments were performed at 25°C. A CM5 chip was activated for crosslinking for 7 minutes with an EDC/NHS (1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide/N-hydroxysuccinimide) solution at a flow rate of 10 μL/min. Next, 10 μg/mL of streptavidin (Roche) in a 10 mM acetate buffer, pH 5.0, was immobilized at the same flow rate for 7 minutes, resulting in densities ranging from 1180 to 1280 resonance units (RU). After immobilization, the chip was blocked by injecting 1 M of ethanolamine for 7 min. To finalize the immobilization, the chip was washed three times with 20 μL of a 40 mM NaOH/1 M NaCl buffer.
To immobilize the hFcRn on the streptavidin sensor surface, the pH was brought to 8.0 by priming with HBS-EP buffer pH 8.0 (GE Healthcare). Biotinylated hFcRn (produced at NovImmune30) was diluted in HBS-EP buffer and immobilized on the chip. Then, the system was primed with HBS-EP buffer at pH 6.0.
IgG was injected at different concentrations ranging from 67 nM to 2 nM, and diluted in HBS-EP buffer at pH 6.0. Each injection was performed for 3 min at a flow rate of 30 μL/min and every time in duplicate. The dissociation was monitored for 12 min. HBS-EP buffer at pH 8.0 was used for regeneration. Results were double referenced and analysed using a Langmuir 1:1 fitting model (BIAeval software version 4.1).
Competition ELISAs
The wells of Maxisorp microtiter plates were coated overnight at 4°C with coating antibody (8 μg/mL of an anti-idiotype antibody for the FcγRI ELISA; for FcγRIIA and FcγRIIB respectively 16 and 10 μg/mL of HZ 15C1, a humanized anti-TLR4 IgG1 (NovImmune)), in 50 μl of PBS and were then washed five times with washing buffer (PBS + 0.05% Tween) and blocked with 250 μL of 3% BSA in PBS per well for 1h at 37°C. After blocking, the plates were washed 5 times with washing buffer.
50 μL of anti-CD20 was added to the wells in a serial dilution in PBS together with 50 μL of the His-tagged FcγR (FcγRI: 0,030 μg/mL, FcγRIIaR: 0,056 μg/mL, FcγRIIB: 1 μg/mL (all from R&D Systems). The plates were incubated for 1.5h at 37°C, followed by five washes with washing buffer. HRP labeled anti-His antibody (Catalog number 34660, Qiagen) was added at a 1:2000 dilution in PBS and the plates were incubated for 1 hour at 37°C. The plates were washed five times with washing buffer before adding 50 μL of TMB super slow (Diarect) substrate. The plates were then incubated in the dark for 30 minutes. Finally, 50 μL of stop solution (2 N H2SO4) was added. Absorbance at 450 nm was measured with a Synergy HT plate reader (Biotek).
The data points plotted in Figure 3e, top three panels, represent mean values from 3 technical replicates. The error bars are S.E.M. The reported differences between the 293S and 293SGlycoDelete-produced antibody were observed in several iterations of technical optimization of these experiments and the results of the fully optimized ELISAs are shown.
Biolayer interferometry assay
Real-time binding of purified IgG to FcγRIIIaV was evaluated using biolayer interferometry (BLI) on an Octet RED96 system (Fortebio). For more details see Supplementary note 6.
ADCC assay
Peripheral blood mononuclear cells (PBMCs) were isolated from fresh blood after centrifugation in a ficoll tube (Vacutainer tube CPT, Becton Dickinson). Natural killer (NK) cells were isolated from the PBMC pool using a negative NK Cell Isolation Kit (Miltenyi Biotec). These cells were activated overnight in growth medium (RPMI 1640 + 10% FCS + 2 mM glutamine) + 10 ng/mL of IL-2.
Raji cells were seeded in a 96-well plate at 20,000 cells per well. 25 μL samples of anti-CD20 antibodies were added in a 1:5 dilution series (in ADCC medium: RPMI 1640 + 1% BSA + 2 mM of glutamine + 25 μg/mL of Gentamicin), starting 5 μg/mL. The plates were then incubated for 30 min at 37°C and 5% CO2. NK cells were added to the Raji cells in a ratio of 1:5 (Raji:NK) and the plate was incubated at 37°C and 5% CO2 for 4 hours. Finally we determined the specific lysis by measuring the lactate dehydrogenase (LDH) levels for each well (Cytotoxicity Detection Kit PLUS, Roche).
The data points in Figure 3e, bottom panel, represent mean values from 3 technical replicates. The error bars are S.D. The reported profiles were observed in several iterations of technical optimization of these experiments and the results of the fully optimized experiment are shown.
Pharmacokinetics
Two groups of 36 female, 8 weeks old C57BL/6J mice (Charles River) were randomly assigned to be intravenously injected with 18,5 μg (1 mg/kg) of either 293S or 293SGlycoDelete anti-CD20. At each time point (1h, 24h, 48h, 4d, 7d, 10d, 14d, 21d and 28d), 4 mice per treatment group were sacrificed for a final bleeding and the concentration of anti-CD20 was determined with the FastELYSA human IgG kit (RD-Biotech) according to the manufacturer’s instructions. The data points shown in Figure 3g are the mean values (4 mice) for each time point. The error bars are S.E.M. This experiment was repeated with bleedings at earlier time points post injection (see Supplementary Fig 13). For practical reasons, the investigators were not blinded to the treatment group assignment of the mice. This experiment was approved by the local ethical committees.
Supplementary Material
Acknowledgments
We thank Guillemette Pontini, Yves Poitevin, Lorène Bernasconi, Delphine Schrag and Sylvain Raimondi (NovImmune) for their help with anti-CD20 generation and characterization, Prof. Savvas Savvides (UGent) for providing the 293SGnTI-/- clone expressing the Flt3 receptor extracellular domain, Prof. Els Van Damme (UGent) for providing ConA, Dr. Eef Dirksen and Karin Nooijen (Merck) for the LC-MS analysis of anti-CD20. L. M. and M. B. are supported by predoctoral fellowships and N. F. by a post-doctoral fellowship of the Fund for Scientific Research-Flanders (FWO). F.S. and S.D. are supported by predoctoral fellowships of IWT Flanders (Strategic Basic Research fellowships no. 101456 and 111252). This research was supported by VIB, UGent-IOF Advanced Grant nr. 041 and FWO research project grant no. G.0.541.08.N.10.
Footnotes
Author contributions
L.M.: Cell engineering, cell characterization, GM-CSF analytics, thermofluor assays, co-wrote the manuscript. F.S.: GM-CSF analytics, anti-CD20 analytics, pharmacokinetics experiment, co-wrote the manuscript. G.E.: Experimental design, supervision and interpretation of anti-CD20 bioanalytics experiments, manuscript correction. N.F.: Bioactivity, immunogenicity ELISA and pharmacokinetics experiments, contributed to the manuscript. M.B.: Gene expression experiments and dataprocessing. A.D.S.: ADCC, ELISA FcγR binding and CD20-binding assays. S.D.: LC-ESI-MS experiments and analysis. F.R.: anti-CD20 expression construct. E.P.: Assisted with immunogenicity ELISA experiments. E.H.: pharmocokinetics experiment. P.M. and G.M.: SPR and BLI experiments. L. Cons and L. Chatel: pharmacokinetics experiment. B.D.: Supervision of LC-ESI-MS experiments. N.C.: Conceived the GlycoDelete technology, initiated the project, assisted in experimental design and interpretation and co-wrote the manuscript.
Competing financial interests statement
The authors declare not to have competing financial interests beyond their inventorship (L.M. and N.C.) on patent applications covering the GlycoDelete technology (WO/2010/015722).
References
- 1.Ferrara C, et al. Modulation of therapeutic antibody effector functions by glycosylation engineering: Influence of Golgi enzyme localization domain and co-expression of heterologous β1, 4-N-acetylglucosaminyltransferase III and Golgi α-mannosidase II. Biotechnol Bioeng. 2006;93:851–861. doi: 10.1002/bit.20777. [DOI] [PubMed] [Google Scholar]
- 2.Elliott S, et al. Control of rHuEPO biological activity: The role of carbohydrate. Exp Hematol. 2004;32:1146–1155. doi: 10.1016/j.exphem.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs PP, Geysens S, Vervecken W, Contreras R, Callewaert N. Engineering complex-type N-glycosylation in Pichia pastoris using GlycoSwitch technology. Nat Protoc. 2009;4:58–70. doi: 10.1038/nprot.2008.213. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton SR, et al. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science. 2006;313:1441–1443. doi: 10.1126/science.1130256. [DOI] [PubMed] [Google Scholar]
- 5.Herter S, et al. Preclinical Activity of the Type II CD20 Antibody GA101 (Obinutuzumab) Compared with Rituximab and Ofatumumab In Vitro and in Xenograft Models. Mol Cancer Ther. 2013;12:2031–2042. doi: 10.1158/1535-7163.MCT-12-1182. [DOI] [PubMed] [Google Scholar]
- 6.Kanda Y, et al. Comparison of cell lines for stable production of fucose-negative antibodies with enhanced ADCC. Biotechnol Bioeng. 2006;94:680–688. doi: 10.1002/bit.20880. [DOI] [PubMed] [Google Scholar]
- 7.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stals I, et al. Identification of a gene coding for a deglycosylating enzyme in Hypocrea jecorina. FEMS Microbiol Lett. 2010;303:9–17. doi: 10.1111/j.1574-6968.2009.01849.x. [DOI] [PubMed] [Google Scholar]
- 9.Grundmann U, Nerlich C, Rein T, Zettlmeissl G. Complete cDNA sequence encoding human beta-galactoside alpha-2,6-sialyltransferase. Nucleic Acids Res. 1990;18:667. doi: 10.1093/nar/18.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verstraete K, et al. Structural insights into the extracellular assembly of the hematopoietic Flt3 signaling complex. Blood. 2011;118:60–68. doi: 10.1182/blood-2011-01-329532. [DOI] [PubMed] [Google Scholar]
- 11.Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 12.Lee F, et al. Isolation of cDNA for a human granulocyte-macrophage colony-stimulating factor by functional expression in mammalian cells. Proc Natl Acad Sci. 1985;82:4360–4364. doi: 10.1073/pnas.82.13.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forno G, et al. N- and O-linked carbohydrates and glycosylation site occupancy in recombinant human granulocyte-macrophage colony-stimulating factor secreted by a Chinese hamster ovary cell line. Eur J Biochem. 2004;271:907–919. doi: 10.1111/j.1432-1033.2004.03993.x. [DOI] [PubMed] [Google Scholar]
- 14.Crispin M, et al. Inhibition of hybrid- and complex-type glycosylation reveals the presence of the GlcNAc transferase I-independent fucosylation pathway. Glycobiology. 2006;16:748–756. doi: 10.1093/glycob/cwj119. [DOI] [PubMed] [Google Scholar]
- 15.Ericsson UB, Hallberg BM, DeTitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 16.Kitamura T, et al. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol. 1989;140:323–334. doi: 10.1002/jcp.1041400219. [DOI] [PubMed] [Google Scholar]
- 17.Mössner E, et al. Increasing the efficacy of CD20 antibody therapy through the engineering of a new type II anti-CD20 antibody with enhanced direct and immune effector cell–mediated B-cell cytotoxicity. Blood. 2010;115:4393–4402. doi: 10.1182/blood-2009-06-225979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nallet S, et al. Glycan variability on a recombinant IgG antibody transiently produced in HEK-293E cells. New Biotechnol. 2012;29:471–476. doi: 10.1016/j.nbt.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov. 2009;8:226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 20.Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat Rev Immunol. 2007;7:715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 21.Lux A, Yu X, Scanlan CN, Nimmerjahn F. Impact of Immune Complex Size and Glycosylation on IgG Binding to Human FcγRs. J Immunol. 2013;190:4315–4323. doi: 10.4049/jimmunol.1200501. [DOI] [PubMed] [Google Scholar]
- 22.Hamblin MW, Metcalf MA. Primary structure and functional characterization of a human 5-HT1D-type serotonin receptor. Mol Pharmacol. 1991;40:143–148. [PubMed] [Google Scholar]
- 23.Tradtrantip L, Ratelade J, Zhang H, Verkman AS. Enzymatic deglycosylation converts pathogenic neuromyelitis optica anti–aquaporin-4 immunoglobulin G into therapeutic antibody. Ann Neurol. 2013;73:77–85. doi: 10.1002/ana.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nandakumar KS, et al. Dominant suppression of inflammation by glycan-hydrolyzed IgG. Proc Natl Acad Sci. 2013;110:10252–10257. doi: 10.1073/pnas.1301480110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Allhorn M, Collin M. Sugar-free Antibodies—The Bacterial Solution to Autoimmunity? Ann NY Acad Sci. 2009;1173:664–669. doi: 10.1111/j.1749-6632.2009.04739.x. [DOI] [PubMed] [Google Scholar]
- 26.Biosimilar, biobetter and next generation therapeutic antibodies. MAbs. 2011;3:107–110. doi: 10.4161/mabs.3.2.14785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tada H, Shiho O, Kuroshima K, Koyama M, Tsukamoto K. An improved colorimetric assay for interleukin 2. J Immunol Methods. 1986;93:157–165. doi: 10.1016/0022-1759(86)90183-3. [DOI] [PubMed] [Google Scholar]
- 29.Ramsland PA, et al. Structural basis for Fc gammaRIIa recognition of human IgG and formation of inflammatory signaling complexes. J Immunol. 2011;187:3208–3217. doi: 10.4049/jimmunol.1101467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magistrelli G, et al. Robust recombinant FcRn production in mammalian cells enabling oriented immobilization for IgG binding studies. J Immunol Methods. 2012;375:20–29. doi: 10.1016/j.jim.2011.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.